Abstract
Antisense oligonucleotides (ASOs) and small interfering RNA (siRNA) promise specific correction of disease-causing gene expression. Therapeutic implementation, however, has been forestalled by poor delivery to the appropriate tissue, cell type, and subcellular compartment. Topical administration is considered to circumvent these issues. The availability of inhalation devices and unmet medical need in lung disease has focused efforts in this tissue. We report the development of a novel cell sorting method for quantitative, cell type-specific analysis of siRNA, and locked nucleic acid (LNA) ASO uptake and efficacy after intratracheal (i.t.) administration in mice. Through fluorescent dye labeling, we compare the utility of this approach to whole animal and whole tissue analysis, and examine the extent of tissue distribution. We detail rapid systemic access and renal clearance for both therapeutic classes and lack of efficacy at the protein level in lung macrophages, epithelia, or other cell types. We nevertheless observe efficient redirection of i.t. administered phosphorothioate (PS) LNA ASO to the liver and kidney leading to targeted gene knockdown. These data suggest delivery remains a key obstacle to topically administered, naked oligonucleotide efficacy in the lung and introduce inhalation as a potentially viable alternative to injection for antisense administration to the liver and kidneys.
Introduction
Throughout their turbulent, 26-year history, oligonucleotide therapeutics have been faced with two major problems: lack of tissue targeting and inefficient cellular uptake.1 Presently, these are believed to be insignificant in lung disease applications: lung epithelia and macrophages are directly accessible through inhalation. Moreover, they constitute the primary entry point for respiratory viruses and contribute substantially to the underlying inflammation of asthma and chronic obstructive pulmonary disease.2 Thus, the clinical utility of both classical antisense oligonucleotides (ASOs) and small interfering RNA (siRNA) as inhaled therapeutics is under intense investigation. Notably, whilst the most advanced ASO candidates utilize chemical modifications that impart stability such as phosphorothioate (PS) backbones and 2′-OMethyl nucleosides,3 siRNA equivalents are 1st generation, unmodified molecules with phosphodiester backbones (PO).4,5 Perplexingly, preclinical reports of efficacy have been contradictory: tissue distribution, cell type-specific uptake, extent, and duration of activity as well as immune-stimulatory potential are all matters that have been addressed in a number of different animal models of disease and across rodent and primate species. While the results appear mixed and have been generally attributed to differences in experimental design, it is generally accepted that an efficacious delivery system might be advantageous.1,6,7,8,9
Recently, locked nucleic acid (LNA)-containing ASOs have been shown to deliver to the liver of nonhuman primates with remarkable efficacy following parenteral administration.10 Use of LNA results in substantial increases in the ASO-binding efficiency to its' target, improving efficacy.11 Consequently, considerably shorter ASO (12-16-mer) can be used with equivalent specificity to classical, >25 nucleotide long ASO.12 Moreover, their potency in vivo has been reported to be comparable, if not better than that of siRNA,10,13 without the requirement of delivery modification or formulation. Studies in multiple cell lines14,15 have indicated that at least in vitro, the phenomenon can be replicated in the absence of cationic delivery systems (e.g., lipofectamine), in a process deemed “gymnosis, ” correlating in vitro to in vivo efficacy.14 On the basis of these promising early findings we set out to explore the utility of this novel class of LNA ASO in the lung.
Results
Oligonucleotide activity in the lung has so far been reported at the whole tissue level. However, as a number of studies have indicated variable uptake of labeled oligos, efficacy is measured as the average achieved across differentially transfected lung cell types.1 To directly address this, we developed a tissue disruption and cell sorting method (TDCS) for isolation of disease-relevant cells for downstream analysis. We focused on epithelial cells and macrophages, two of the most important target cells in lung disease. Fresh mouse lung tissues were disrupted using a mechanical/enzymatic approach yielding single-cell suspensions. Suspensions were then treated by magnetic bead separation and/or fluorescent-activated cell sorting to obtain populations of live epithelial cells, macrophages or the remaining cell types (other) (Supplementary Figures S1 and S2).
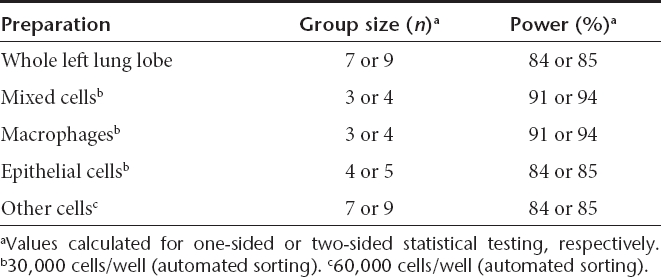
We next carried out pilot studies in luciferase-expressing transgenic mice to establish statistical power for successfully measuring 50% knockdown. Luminometry was carried out at the whole body, whole tissue, mixed cell, or cell type-specific level (n = 10). Whereas whole body bioluminescence was considerably variable, lung and liver tissue bioluminescence was surprisingly consistent (Supplementary Figure S1). Nevertheless, a minimum of seven animals per group was found necessary to achieve 84–85% power at the tissue level (one- or two-sided t-test). TDCS luminometry measurements, however, enabled up to 66% reduction in animal use (Table 1): as few as three animals per experimental group were calculated as sufficient to reveal a 50% knockdown at a power of 91–94%.
Table 1. Statistical power calculations for luminometry.
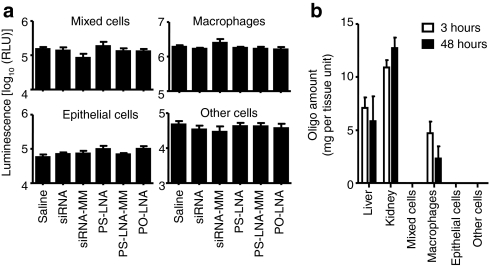
To compare the utility of topically administered LNA ASO and siRNA, animals were dosed intratracheally (i.t.) with sequences against luciferase and lungs were collected 48 hours post-administration. Disappointingly, neither class of oligonucleotide achieved knockdown of luciferase across any lung cell type (Figure 1a). Furthermore, mass spectrometry analysis of TDCS preparations revealed that PS-LNA was the only oligo retained in lung cells and only in macrophages. Unexpectedly, a substantial proportion of the PS-LNA dose was detected in the liver at both 3 and 48 hours after dosing [26.4% (±6.2) and 15.3% (±13.7), respectively]. Interestingly, kidneys also accumulated substantial quantities (7.4% (±0.8) and 5.5 (±3.0) at 3 and 48 hours, respectively), loading more oligo per gram tissue (Figure 1b). The fate of the PO-LNA and siRNA remained elusive.
Figure 1.
Direct administration of small interfering RNA (siRNA) and locked nucleic acid (LNA) antisense oligonucleotide (ASO) to mouse lung results in no efficacy and accumulation in liver, kidney, and lung macrophages. (a) Luciferase activity expressed in relative light units (RLU) in specific lung cell types isolated by tissue disruption and cell sorting method (TDCS) from luciferase-expressing, transgenic mouse lungs 48 hours after intratracheal administration of siRNA, PS-LNA, PO-LNA or their mismatch controls (MM). (b) Oligo accumulation in tissues (per gram of tissue) or TDCS pneumonocyte preparation (per 30,000 cells) at 3 and 48 hours post-intratracheal administration of PS-LNA ASO. All measurements are presented as means + SD.
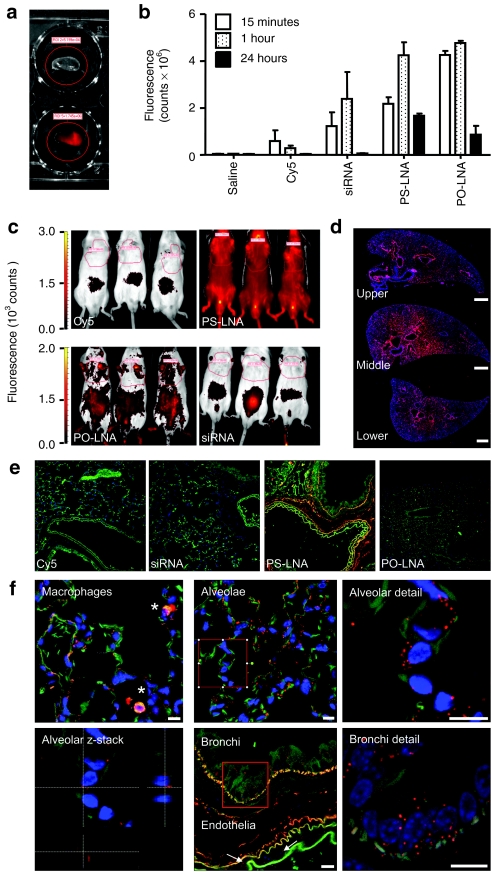
Our findings put into question a number of theories on the lung utility and activity of siRNA. We therefore decided to further examine the fate of the different classes of oligos after i.t. administration using sulphonated Cy5 dye as a molecular beacon. Curious over the role of lung surfactant in lung cell uptake of siRNA16 and the impact of its' cationic charge on aggregation with anionic oligonucleotides toward hypothesized in situ particle formation in the lung lumen, we decided to examine the lung tissue across multiple time points. Strikingly, even 15 minutes after i.t. administration, urinal excretion was observed indicating rapid absorption from the lung: the classical fight or flight response to handling resulted in emiction in most animals, with urine from dye control-, siRNA-, and PO-LNA-treated animals being of blue color, most likely on account of the dark blue color of the sulphonated Cy5 dye. This was confirmed by mass spectrometry analysis of group-pooled urine samples (Supplementary Figure S3). Moreover, the sulphonated Cy5-labeled siRNA antisense sequence and PO-LNA, were present in urine samples at higher concentrations than that measured for PS-LNA indicating greater retention of the PS-LNA within the system (Supplementary Table S1). Fluorescent imaging of excised, fixed lung tissues indicated that by 24 hours PS-LNA followed by PO-LNA were retained in the lung in a protracted fashion, whereas siRNA was undetectable (Figure 2a,b). Imaging the entire animals at 24 hours postdosing identified the urinary bladder as a distinct location of siRNA concentration. Importantly, by stark contrast, substantial quantities of PS-LNA were still in circulation (Figure 2c). These data reaffirmed the functional observations obtained with unlabeled material, indicating that inhaled siRNA, PO, and PS oligos rapidly escape the lung via systemic circulation, with most of the siRNA and PO oligo doses being renally excreted, potentially in a matter of minutes.
Figure 2.
Phosphorothioate backbones abrogate rapid systemic access and renal clearance of intratracheally administered oligos. (a) An example of explanted tissue fluorescence emission 15 minutes after intratracheal administration of saline (upper panel) or sulphonated Cy5-labeled (lower panel) oligo. (b) Time course of left lung lobe fluorescence emission following intratracheal administration of sulphonated Cy5, or dye-labeled small interfering RNA (siRNA), phosphate backbone (PO-LNA) or phosphorothioate backbone (PS-LNA) antisense oligonucleotides. (c) Whole animal bioimaging of sulphonated Cy5, or dye-labeled siRNA, PO-LNA, or PS-LNA 24 hours after intratracheal administration. (d) Collapsed Z-stack imaging of sulphonated Cy5-labeled PS-LNA antisense oligonucleotide (ASO) (red channel) across upper, middle, and lower left lung lobe formalin fixed, paraffin embedded (FFPE) sections counterstained for DNA by DAPI (blue channel) confirms uniform distribution in central and distal parts of the lobe 1 hour after administration. Bar = 0.5 mm. (e) Labeled oligo detection (red channel) in FFPE lung sections (green channel: autofluorescence) counterstained for DNA by DAPI (blue channel). (f) Endosome-like structures are formed in alveolae, bronchi, and endothelia (white arrows) within 15 minutes of PS-LNA ASO administration to the mouse lung. Detailed examination (red squares) and z-stacking analysis indicates perinuclear localization in alveolae. Substantial accummulation in alveolar macrophages (*) is documented. Bars = 0.01 mm. All measurements are presented as means + SD. LNA, locked nucleic acid.
To explore the mechanism of systemic access from the lung lumen, we carried out histological studies with the sulphonated Cy5-labeled oligos. Hematoxylin and eosin staining documented the absence of any appreciable lymphocytic infiltrate (Supplementary Figure S4). Confocal imaging of formalin fixed, paraffin embedded sections further reaffirmed the notion of rapid PO oligo clearance: PS-LNA was uniformly detected in sections across lung lobes (Figure 2d) whereas no siRNA signal was detected, and a very weak PO-LNA signal was only observed in scattered lung macrophages (Figure 2e). Examination of the subcellular localization of PS-LNA at 15 minutes indicated the presence of discreet, particulate structures within macrophages >>> alveolar epithelia > bronchial epithelia >> endothelia (Figure 2f). Moreover, diffuse submucosal fiber staining was also detected in larger airways. Interestingly, no particulate structures were observed in the lung lumen challenging the surfactant::oligo interaction hypothesis. Alternatively, the luminal residence time of such particles might be substantially <15 minutes, or extracellular complexes could be lost during preparation of formalin fixed, paraffin embedded sections. By 1 hour accumulation of oligos was further enhanced in alveolar macrophages, and cells reminiscent of type II alveolar epithelia, suggesting ongoing uptake and accumulation in the deep lung. Interestingly, membrane spreading in type I cells and bronchial distribution remained similar (Supplementary Figure S5a). By 24 hours diffuse cytosolic staining was observed in scattered bronchial epithelia, whereas the majority of oligo appeared to associate with submucosal membranes and elastin fibers (Supplementary Figure S5b). Collectively, these data indicate that only i.t. administered PS-LNA but not siRNA or PO-LNA are able to enter lung cells in an efficient manner to form discreet intracellular structures following internalization, yet are unable to mediate target gene knockdown. The distinct lack of nuclear localization of PS-LNA across all the time points studied is in accordance with the recent, in vitro-derived proposition that this might be a requirement for efficacy.15 Curiously, the A549 type II alveolar epithelium cell line has been indicated to exhibit similar PS-LNA ASO uptake patterns in gymnotic experiments in vitro and to be equally refractory to target-specific knockdown.15 These observations and the data in our study further support the early report of in vitro–in vivo correlation of efficacy for gymnotic experiments.14 However, the extent to which nuclear15 or cytosolic GW182-body accumulation14 is a universally necessary phenomenon or a cell-type specific requirement remains to be determined. The nature of the intracellular, punctuate structures formed with PS-LNA in our in vivo studies are the subject of ongoing investigations, though our early attempts have failed to find colocalization with well-established markers of early endosomes (EEA1) or caveosomes (caveolin 1).17,18,19,20
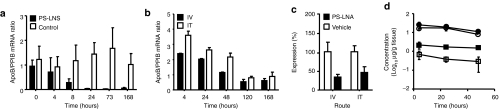
Previous reports of systemic pharmacokinetics of i.t. administered PS ASO,21 our own observations of liver loading for PS-LNA ASO, and the disappointing performance of inhaled oligos in the lung encouraged us to investigate the utility of this noninvasive route for PS-LNA ASO delivery to the liver. Initially, mice were dosed intravenously (i.v.) with a known oligo against apolipoprotein B (ApoB)10 and necropsy was performed at time points to provide a kinetic profile of knockdown in the liver. Significant knockdown (P = 0.0079) was achieved after a single dose, with maximal effect being achieved by 24 hours, maintained for up to 7 days (168 hours) (Figure 3a). Variability within the negative control groups was consistent with normal behavior of gene expression, possibly driven by circadian rhythms and feeding patterns. Due to the high potency exhibited in vivo, a comparison between i.v. and i.t. administration was performed at a 10× lower dose. Again, animals were sacrificed at various time points to provide a kinetic insight into knockdown of the target (Figure 3b). Over 50% knockdown was achieved using either administration route at 168 hours compared to vehicle treatment (Figure 3c). Overall, i.t. dosing of the ApoB PS-LNA attained 25% and 80% of the exposure observed in the liver and kidney, respectively when compared to i.v. dosing (Figure 3d). This demonstrates that an appreciable amount of PS-LNA ASO enters the liver after i.t. dosing, which in this case is sufficient to drive significant knockdown of the target mRNA. Notably, the time to maximal effect was delayed in the i.t. dosed group (Figure 3b). This is most likely due to protracted absorption from the lung into the systemic circulation slowing the time to maximal liver concentration. However, upon chronic weekly administration, as would be expected in the clinic, such small impedance would be expected to be insignificant.
Figure 3.
Intratracheal administration results in comparable efficacy in the liver to that achieved by intravenous injection. (a) Time course of apolipoprotein B (ApoB) mRNA knockdown in louse livers after intravenous administration of a 30 mg/kg dose of a highly potent PS-LNA antisense oligonucleotide (ASO) or a control ASO. (b) Kinetics of ApoB mRNA expression in the mouse liver following intravenous (i.v.) or intratracheal (i.t.) administration of a 3 mg/kg dose of. (c) Levels of ApoB mRNA in the livers of mice 7 days after i.v. or i.t. administration expressed relative to the mRNA levels in time-matched, vehicle-treated animals. (d) Concentrations of ApoB PS-LNA determined in both liver (squares) and kidney (circles) for i.v. (filled symbols) and i.t. (empty symbols) dose routes. All measurements are presented as means + SD. LNA, locked nucleic acid.
Discussion
Collectively, these data demonstrate that after direct administration to the lung siRNA and PO-LNA are not readily taken up by lung cells but are instead rapidly absorbed and renally cleared. Thus, the accuracy of early reports of efficacious topical siRNA administration to the lung, challenged in studies examining the utility of both liposomal22 and polymeric23 delivery systems in improving topical administration efficacy, are put further into question. Indeed, increasing evidence has amassed that innate immunity activation might have accounted for what numerous groups interpreted as RNA interference.1 The most striking demonstration of the power of these responses was delivered in a study indicating that siRNA are mild Toll-like receptor 3 agonists: intraperitoneal dosing and Toll-like receptor 3 activation was shown to suppress choroid neovascularization in the rodent eye.24 Elsewhere, siRNA-mediated Toll-like receptor 7/8 activation was shown to be responsible for curtailing viral infection, including some instances previously reported as on-target RNA interference in pulmonary viral infection models.25 Taken together, these data suggest that siRNA administered topically to the lung either intranasally or i.t. and without any form of delivery enhancement, do not deliver to lung cells and cannot enact RNA interference. We nonetheless caution extrapolation of our findings to primate species, or larger PS ASO molecules claimed to be efficacious following topical administration in the lung lung.26,27 It is rather our view that this body of evidence makes a strong case for examining more closely inhaled oligonucleotide fate in more clinically relevant species or in man, to robustly underpin proof of mechanism conclusions.
Importantly, in stark contrast to PO-LNA or siRNA, PS-LNA ASO appeared to be retained in lung epithelia, though appreciable amounts of oligo were detected by mass spectrometry only in macrophages. To our knowledge this is the first report on the subcellular localization of inhaled oligos in the lung across multiple time points and as early as 15 minutes after dosing. Understanding the entry mechanisms and trafficking of PS-LNA in the various cell types of the lung will undoubtedly be important in addressing the lack of efficacy documented in this study. Crucially, despite the observed retention of PS-LNA ASO in the lung, these oligos were also inactive in disease-relevant cell types, suggesting that efficacy in the lung is not an issue of stability, but an issue of delivery. However, PS-LNA ASO also access systemic circulation for an appreciable duration, and are taken up by the liver to exhibit knockdown of a liver mRNA target. These results indicate that inhalation approaches might be of utility to systemically targeted oligonucleotide therapeutics such as aptamers against circulatory or endothelial targets, or liver- and kidney-targeted antisense therapies. Further clinical relevance evaluation, such as the long-term impact of lung administration is, however, necessary: whereas alveolar macrophages were observed to perform their well-documented role of detritus clearance from the deep lung, accumulation in type II alveolar epithelial precursors, the resident lung stem cells, was indicated. If the intracellular bodies observed in our studies are the outcome of serendipitous interaction with membrane proteins, this phenomenon might prove sequence-specific and ultimately inconsequential.
In conclusion, we report a novel TDCS technique that has enabled conclusive demonstration of lack of siRNA or LNA ASO-mediated activity in either macrophages or epithelia of the mouse lung after topical administration. Nonphosphorothioated oligos partitioned rapidly to systemic circulation and were eliminated via the renal route within minutes. Use of a PS backbone in LNA ASO resulted in appreciable lung macrophage accumulation but with no demonstrable activity. Our histological examinations demonstrate that whilst tissue penetration of the lung is ubiquitous, rapid transport mechanisms might account for systemic access. Crucially, as PS-LNA ASO are relatively refractory to rapid renal clearance, sufficient amounts of lung-absorbed oligo remain in circulation and accumulate in substantial quantities in the liver and kidney. In the case of an ApoB-targeted oligonucleotide, the amount of drug absorbed from the lung was sufficient to accumulate in the liver at a pharmacologically active level, leading to knockdown of ApoB mRNA. Thus, this report pioneers the prospect of noninvasive administration of oligonucleotide therapeutics for circulatory, liver, and kidney diseases.
Materials and Methods
Oligonucleotides. We screened in vitro (Supplementary Materials and Methods) a collection of custom designed siRNA and LNA ASO (Eurogentec, Southampton, UK) targeting luciferase and identified sequences (Table 2) with IC50 values of 0.23 nmol/l and 97.58 nmol/l post-transfection or gymnotic uptake respectively at 48 hours. No significant cytotoxicity was observed (<10%). Mismatch control siRNA (MM-siRNA) and PS-LNA (MM-PS-LNA) were designed which yielded no appreciable activity (no detectable IC50). A previously described PS-LNA ASO against ApoB10 with a gymnotic IC50 of 490 nmol/l was used for the liver efficacy studies. Quantification of oligonucleotide uptake in cells and tissues was performed by mass spectrometry (Supplementary Materials and Methods).
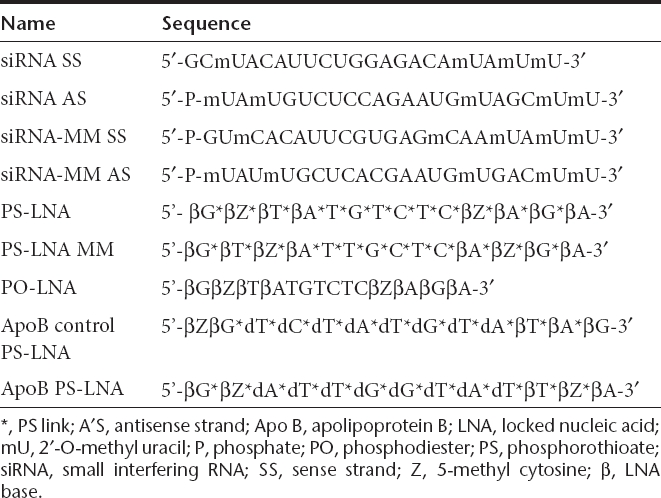
Table 2. Oligonucleotides used in this study.
Animals. Most animal models of asthma and chronic obstructive pulmonary disease focus on innate immunity activation pathways and related biomarker measurements of efficacy. siRNA carry a risk of TLR3, 7 and 8 activation, whereas LNA ASO are currently believed to be refractory to immunostimulation. To minimize any confounding impact of potential oligonucleotide-mediated immune activation we performed our studies in reporter mice expressing luciferase constitutively under the control of a β-actin promoter mice [FVB/N-Tg (β-actin-luc)-Xen mice; Caliper LS, Runcorn, UK ]. Histological studies (see Supplementary Materials and Methods) were performed on male BALB/c mice (20–25 g, Harlan UK, Wyton, UK) (n = 10) and ApoB knockdown studies were performed in CD21 mice (Harlan UK) (Supplementary Materials and Methods). Animals were dosed i.t. under mild inhaled isofluorane anaesthesia (4% vol/vol in O2) with 10 nmol of the designated oligonucleotide or vehicle (saline, 0.02 ml). i.v. dosing was carried out in 0.05 ml volumes. All in vivo procedures were carried out under local ethics approval and in strict accordance with the 1986 Animals (Scientific Procedures) Act.
TDCS. Lungs were dissociated using a collagenase/DNAse I enzyme mix and a GentleMACS tissue dissociator (Miltenyi Biotec, Woking, UK), and remaining red blood cells were removed by hemolysis as described in the Supplementary Materials and Methods section. Mouse lung cell suspensions were prepared for sorting and separated into macrophages (CD11b+/F4/80+), or epithelial cells (CD45-/CD326+) as described in the Supplementary Materials and Methods section.
Ex vivo luminometry. Luciferase activity is proportional to enzyme concentration, and the protein has a reported <4-hour half-life in vivo;28,29 this enables luminometry measurements as a marker of knockdown efficiency. Thus, assays were performed on TDCS preps in white 96-well microtitre plates (Matrix, Loughborough, UK). Cells were centrifuged at 400g for 10 minutes at 4 °C and the supernatant was discarded. 0.1 ml of room temperature DPBS with Ca2+ and Mg2+ (Sigma, Dorset, UK) were added and 0.1 ml of room-temperature Brite Lite Plus reagent (Perkin Elmer, Waltham, MA) was added and the plates were immediately read on an Envision Plate reader using the standard ultrasensitive luminescence protocol (0.1 sec read per well, probe distance from well = 0). Luminescence was expressed as relative light units following background substraction. A threshold of 40,000 relative light units was determined in maintaining linear consistency of detection (Supplementary Figure S6), thus setting the required cell number to 30,000 for mixed cells, macrophages, and epithelia and 60,000 for other cell types. No appreciable loss of viability or bioluminescence activity was observed when TDCS was performed up to 48 hours after fresh tissue excision and 4 °C storage in 2.5% fetal bovine serum -supplemented DPBS.
Lung delivery studies using sulphonated Cy5-labeled oligos. Fluorescent imaging of sulphonated Cy5-labeled oligos (Supplementary Materials and Methods) in animals and tissues was carried out using an IVIS-Spectrum with excitation set at 640 nm and monitoring emission at 680 nm. Whole animal imaging was carried out 24 hours after dosing under mild inhaled isofluorane anesthesia. Lungs were removed at designated times (0.25, 1, or 24 hours) following dosing. Right lung lobes were flash frozen in liquid nitrogen and left lung lobes inflation fixed with 4% neutral-buffered formalin for 24 hours. After fixation lungs were placed in 6-well plates and imaged as described. Regions of interest were drawn around the lung and total fluorescence measured as photon/second.
Confocal microscopy. DAPI staining was carried out immediately before imaging. Specimens were imaged on an inverted confocal microscope (Leica TCS SP5; Leica; Microsystems, Milton Keynes, UK) using a 63× lens (Leica HCX PL APO CS 63.0 × 1.40 OIL UV). Images for the blue (DAPI), green (autofluorescence) and far red (Cy5) channel were taken in sequential mode using appropriate excitation and emission settings. Images of whole lung sections were assembled from high resolution images covering the whole area of the section and using the Leica tiling algorithm (Leica). Maximum projections of z-stacks (1 µm spacing, covering the height of the entire section) were used for qualitative assessment of label distribution across the section.
Statistics. Ex vivo luminometry studies were performed on either 10 (pilot study) or five animals per group. A nonparametric, two-sided, Mann–Whitney U-test was performed on the liver efficacy studies due to the small sample size (n = 5 per group). Comparisons were considered statistically significant when P < 0.05. All data was analyzed and graphed using GraphPad Prism software (version 5.1).
SUPPLEMENTARY MATERIAL Figure S1 Application of TDCS results in highly consistent luminescence readouts. Figure S2. Characterization of pneumonocyte preparations after TDCS. Figure S3. Extracted ion chromatograms of oligos. Figure S4. Lung histology confirms lack of lymphocytic infiltrate as a consequence of intratracheal oligonucleotide administration. Figure S5. Confocal imagining of intratracheally administered phosphorothioate LNA antisense (PS-LNA) distribution in the mouse lung over time. Figure S6. Linearity of luminescence from β-actin luciferase transgenic mouse lung cells ex vivo. Figure S7. Stability assessment of siRNA using LC-MS/MS. Table S1. Oligo detection by MS in urine samples. Materials and Methods
Acknowledgments
This work presented in this article was funded by Pfizer Inc. This work was conducted in Sandwich, Kent, UK, Dusseldorf, Germany and Boston, Massachusetts, USA. The authors declared no conflict of interest.
Supplementary Material
Application of TDCS results in highly consistent luminescence readouts.
Characterization of pneumonocyte preparations after TDCS.
Extracted ion chromatograms of oligos.
Lung histology confirms lack of lymphocytic infiltrate as a consequence of intratracheal oligonucleotide administration.
Confocal imagining of intratracheally administered phosphorothioate LNA antisense (PS-LNA) distribution in the mouse lung over time.
Linearity of luminescence from β-actin luciferase transgenic mouse lung cells ex vivo.
Stability assessment of siRNA using LC-MS/MS.
Oligo detection by MS in urine samples.
.
REFERENCES
- Moschos SA, Spinks K, Williams AE., and, Lindsay MA. Targeting the lung using siRNA and antisense based oligonucleotides. Curr Pharm Des. 2008;14:3620–3627. doi: 10.2174/138161208786898851. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest. 2008;118:3546–3556. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvreau GM, Boulet LP, Cockcroft DW, Baatjes A, Cote J, Deschesnes F.et al. (2008Antisense therapy against CCR3 and the common β chain attenuates allergen-induced eosinophilic responses Am J Respir Crit Care Med 177952–958. [DOI] [PubMed] [Google Scholar]
- Alvarez R, Elbashir S, Borland T, Toudjarska I, Hadwiger P, John M.et al. (2009RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy Antimicrob Agents Chemother 533952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R.et al. (2010A status report on RNAi therapeutics Silence 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weers JG, Bell J, Chan HK, Cipolla D, Dunbar C, Hickey AJ.et al. (2010Pulmonary formulations: what remains to be done J Aerosol Med Pulm Drug Deliv 23 Suppl 2S5–23. [DOI] [PubMed] [Google Scholar]
- Roy I., and, Vij N. Nanodelivery in airway diseases: challenges and therapeutic applications. Nanomedicine. 2010;6:237–244. doi: 10.1016/j.nano.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnappa BC. siRNA for inflammatory diseases. Curr Opin Investig Drugs. 2009;10:418–424. [PubMed] [Google Scholar]
- Lam JK, Liang W., and, Chan HK.2011Pulmonary delivery of therapeutic siRNA Adv Drug Deliv Revepub ahead of print). [DOI] [PMC free article] [PubMed]
- Straarup EM, Fisker N, Hedtjärn M, Lindholm MW, Rosenbohm C, Aarup V.et al. (2010Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates Nucleic Acids Res 387100–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreck J, Wyszko E, Gillen C., and, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C.et al. (2011Silencing of microRNA families by seed-targeting tiny LNAs Nat Genet 43371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M.et al. (2004Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs Nature 432173–178. [DOI] [PubMed] [Google Scholar]
- Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Høg A.et al. (2010Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents Nucleic Acids Res 38e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qu Z, Kim S, Shi V, Liao B, Kraft P.et al. (2011Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection Gene Ther 18326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BJ, Guan Y, Tang Q, Du C, Xie FY, He ML.et al. (2004Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus Antivir Ther (Lond) 9365–374. [PubMed] [Google Scholar]
- Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D.et al. (2010Systems survey of endocytosis by multiparametric image analysis Nature 464243–249. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Porat-Shliom N., and, Cohen LA. Clathrin-independent endocytosis: a unique platform for cell signaling and PM remodeling. Cell Signal. 2009;21:1–6. doi: 10.1016/j.cellsig.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Bürli T, Zerial M., and, Helenius A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Sandvig K, Torgersen ML, Raa HA., and, van Deurs B. Clathrin-independent endocytosis: from nonexisting to an extreme degree of complexity. Histochem Cell Biol. 2008;129:267–276. doi: 10.1007/s00418-007-0376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin PL, Bayley D, Giddings J, Craig SJ, Cummins LL, Hastewell JG.et al. (1998Pulmonary bioavailability of a phosphorothioate oligonucleotide (CGP 64128A): comparison with other delivery routes Pharm Res 15583–591. [DOI] [PubMed] [Google Scholar]
- Griesenbach U, Kitson C, Escudero Garcia S, Farley R, Singh C, Somerton L.et al. (2006Inefficient cationic lipid-mediated siRNA and antisense oligonucleotide transfer to airway epithelial cells in vivo Respir Res 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glud SZ, Bramsen JB, Dagnaes-Hansen F, Wengel J, Howard KA, Nyengaard JR.et al. (2009Naked siLNA-mediated gene silencing of lung bronchoepithelium EGFP expression after intravenous administration Oligonucleotides 19163–168. [DOI] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ.et al. (2008Sequence- and target-independent angiogenesis suppression by siRNA via TLR3 Nature 452591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L.et al. (2008Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation Hum Gene Ther 19991–999. [DOI] [PubMed] [Google Scholar]
- Ripple MJ, You D, Honnegowda S, Giaimo JD, Sewell AB, Becnel DM.et al. (2010Immunomodulation with IL-4Rα antisense oligonucleotide prevents respiratory syncytial virus-mediated pulmonary disease J Immunol 1854804–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XR, Tian XL, Bo JP, Li SG, Liu ZL., and, Niu B. Inhibition of allergic airway inflammation by antisense-induced blockade of STAT6 expression. Chin Med J. 2011;124:26–31. [PubMed] [Google Scholar]
- Ignowski JM., and, Schaffer DV. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol Bioeng. 2004;86:827–834. doi: 10.1002/bit.20059. [DOI] [PubMed] [Google Scholar]
- Brandes C, Plautz JD, Stanewsky R, Jamison CF, Straume M, Wood KV.et al. (1996Novel features of drosophila period Transcription revealed by real-time luciferase reporting Neuron 16687–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Application of TDCS results in highly consistent luminescence readouts.
Characterization of pneumonocyte preparations after TDCS.
Extracted ion chromatograms of oligos.
Lung histology confirms lack of lymphocytic infiltrate as a consequence of intratracheal oligonucleotide administration.
Confocal imagining of intratracheally administered phosphorothioate LNA antisense (PS-LNA) distribution in the mouse lung over time.
Linearity of luminescence from β-actin luciferase transgenic mouse lung cells ex vivo.
Stability assessment of siRNA using LC-MS/MS.
Oligo detection by MS in urine samples.
.