Abstract
In the past decade the first Arabidopsis genes encoding cytoskeletal proteins were identified. A few dozen genes in the actin and tubulin cytoskeletal systems have been characterized thoroughly, including gene families encoding actins, profilins, actin depolymerizing factors, α-tubulins, and β-tubulins. Conventional molecular genetics have shown these family members to be differentially expressed at the temporal and spatial levels with an ancient split separating those genes expressed in vegetative tissues from those expressed in reproductive tissues. A few members of other cytoskeletal gene families have also been partially characterized, including an actin-related protein, annexins, fimbrins, kinesins, myosins, and villins. In the year 2001 the Arabidopsis genome sequence was completed. Based on sequence homology with well-characterized animal, fungal, and protist sequences, we find candidate cytoskeletal genes in the Arabidopsis database: more than 150 actin-binding proteins (ABPs), including monomer binding, capping, cross-linking, attachment, and motor proteins; more than 200 microtubule-associated proteins (MAPs); and, surprisingly, 10 to 40 potential intermediate filament (IF) proteins. Most of these sequences are uncharacterized and were not identified as related to cytoskeletal proteins. Several Arabidopsis ABPs, MAPs, and IF proteins are represented by individual genes and most were represented as as small gene families. However, several classes of cytoskeletal genes including myosin, eEF1α, CLIP, tea1, and kinesin are part of large gene families with 20 to 70 potential gene members each. This treasure trove of data provides an unprecedented opportunity to make rapid advances in understanding the complex plant cytoskeletal proteome. However, the functional analysis of these proposed cytoskeletal proteins and their mutants will require detailed analysis at the cell biological, molecular genetic, and biochemical levels. New approaches will be needed to move more efficiently and rapidly from this mass of DNA sequence to functional studies on cytoskeletal proteins.
Keywords: actin-binding proteins, microfilaments, tubulin, microtubule-binding proteins, intermediate filaments, myosin, kinesin, plant proteome
Introduction
This review is focused on the actin and tubulin based systems of structural genes and proteins that make up the Arabidopsis cytoskeleton. The Arabidopsis actins, tubulins, actin-binding proteins (ABP), microtubule-associated proteins (MAPs), and intermediate filament (IF) proteins will all be explored. For the sake of simplicity the motor proteins, because they bind filamentous actin (F-actin) or mictotubules (MTs) directly, are included as ABPs and MAPs and are not defined as separate classes. No attempt was made to discuss the hundreds of signal transduction proteins (e.g., families of small GTP binding proteins) affecting cytoskeletal organization, regulation, and dynamic rearrangement (Valster et al., 2000; Wu et al., 2000). Nor was any attempt made to examine the nuclear encoded structural proteins of bacterial origin that might function within mitochondria and chloroplasts, such as the tubulin homologue FtsZ (Erickson and Stoffler, 1996) and actin homologue MreB (van den Ent et al., 2001).
Several cytoskeletal gene families including the actins, profilins, actin depolymerizing factors, α-tubulin, and β-tubulin have been well characterized in Arabidopsis. Each family has been as large or larger than their animal counterparts. In the case of the actin family, it is almost as ancient as the vertebrate actin family, tracing its origin to a single common plant-like gene 400 million years ago. All five of these gene families appear to have an ancient split separating them into constitutive and reproductive classes based on organ-specific expression patterns (Meagher et al., 1999a; Meagher et al., 1999b). This trend for cytoskeletal gene families to be divided into constitutive and reproductive classes of isovariants appears to represent a landmark genomic duplication event in the origin of some cytoskeletal genes and their regulation.
Based on the animal, protist, and yeast literature we can expect to find hundreds of different genes encoding ABP, MAP, and possibly IF proteins in plant genomes, particularly if plants continue to have larger than average gene family sizes. The Arabidopsis genome sequence was recently completed and is extremely well annotated, making its analysis more straightforward than that of the human genome. Therefore, in order to survey the plant cytoskeletal proteome we searched the Arabidopsis genome for actins and tubulins and for more than one hundred different ABP, MAP, and intermediate filament coding sequences. Several hundred genes encoding potential ABP, MAP, and intermediate filament proteins were identified as likely candidates to encode proteins that function in the plant cytoskeleton. These results are presented in Tables 1, 2, and 3, respectively. The majority of these potential ABP, MAPs, and IF proteins were identified by amino acid (a.a.) sequence similarity with well-characterized proteins from the other three eukaryotic kingdoms and a preliminary analysis of the significance of possible protein homologies. In addition to the isoforms of actin, actin-related proteins, and tubulins themselves, these cytoskeletal sequences include those homologous or similar to actin monomer binding, F-actin capping, F-actin severing, F-actin crosslinking, F-actin binding motor proteins, several classes of MAPs and microtubule motor proteins, and possible relatives of the lamins and other IF proteins.
Table 1.
Actins and potential ABPs in the Arabidopsis thaliana database
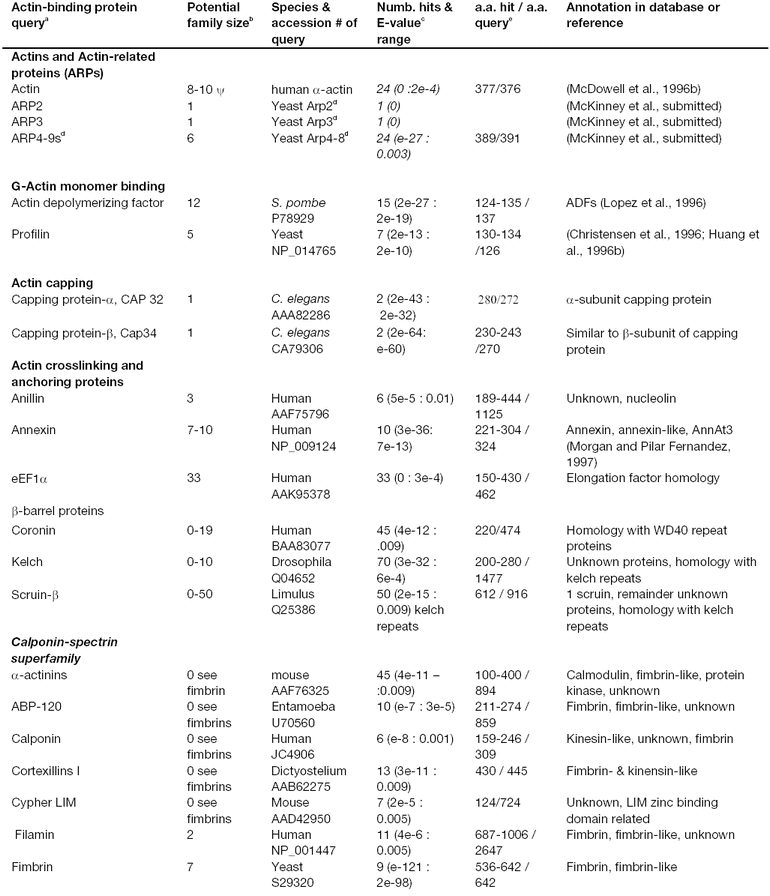
Table 2.
Tubulin and microtubule-associated proteins (MAPs) predicted from the Arabidopsis thaliana sequence database
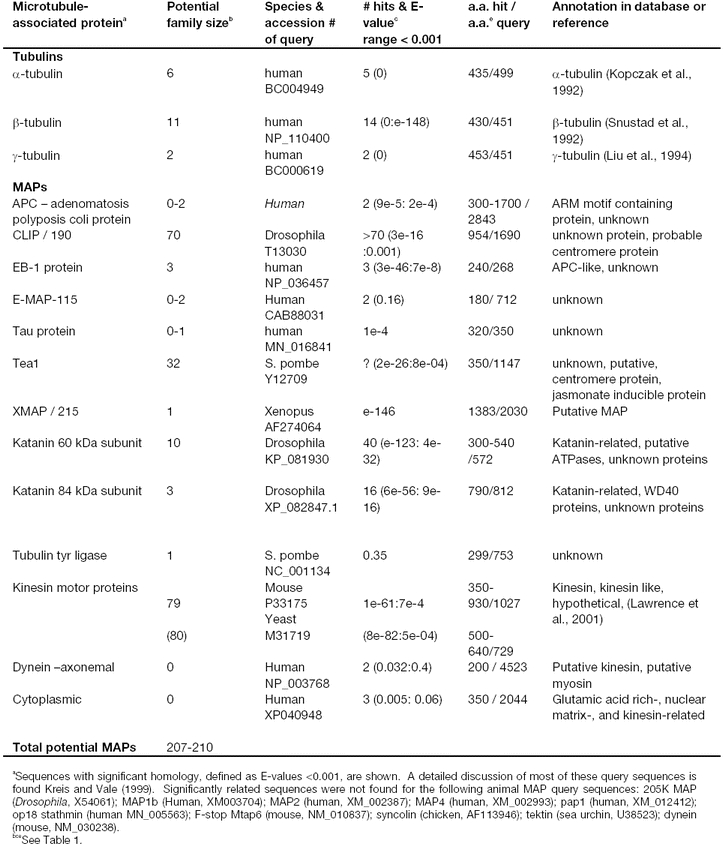
Table 3.
Potential intermediate filament coding sequences in the Arabidopsis thaliana database
The Arabidopsis Cytoskeletal Genome and Proteome
Arabidopsis is now widely accepted as a model plant for studying fundamental aspects of of the biology of higher plants or examining applications of biotechnology to forest and crop species. Arabidopsis has many useful features including simple genetics, small size, rapid life cycle, and a small genome relative to most other plant species. The sequence of the 125-megabase Arabidopsis genome has just been completed, revealing at first approximation 25,500 genes in 11,000 gene families (Arabidopsis Genome Initiative, 2000). Thus, because Arabidopsis contains one of the simplest plant genomes, we can deduce that all plants have a minimum complexity approaching 85% of that of the human genome with its 29,000 genes. This plant genome sequence also serves a model for the larger and more difficult genomes of important crop species (Dennis and Surridge, 2000). Considering the vast potential for alternate splicing and protein modification, the actual size of the Arabidopsis proteome should be expected to be a few fold larger still (Garrels et al., 1997; Brett et al., 2000). The Arabidopsis cytoskeletal proteome is of unknown complexity, but considering the large numbers of ABPs, MAPs, and IFs identified in animals, protists, and yeast it is not surprising that several hundred Arabidopsis genes encoding cytoskeletal proteins were found in this current study. Initial publications on the Arabidopsis genome did not focus on cytoskeletal protein sequences, and only a small fraction (∼40%) of the candidate cytoskeletal genes identified in this study was annotated as such. No list or overview of cytoskeletal genes has been reported. We have taken advantage of this new database to make an initial focused survey of Arabidopsis cytoskeletal proteins. These data will provide an overview of the potential complexity of the plant cytoskeleton.
Expectation-values place confidence limits on identification of proteins in sequence searches
The Arabidopsis cytoskeletal proteins described in this study were identified using BLAST searches. During BLAST searches for related sequences, a method was developed to directly approximate alignments that optimize local similarity. This measure of the statistical significance of alignments is given as an E-value (Allen and Kropf, 1993). Simply stated, E-values can be defined as the expectation of finding two sequences with a given amount of similarity by chance. The lower the E-value, the lower the chance that such a match could have been found at random. For example, an E-value of 0.0001 (e x 10−4, written as e-4 in Tables 1, 2, and 3) suggests that you should not find this level of sequence relatedness to the query by chance more than once in 10,000 searches of a database of this complexity. Potential Arabidopsis homologues to animal, protist, or yeast ABPs, MAPs, and IF are listed in Tables 1, 2, and 3, respectively. Most Arabidopsis sequences included in Tables 1, 2, and 3 had expectation ratios (E-values) less than 0.001 (Gerstein, 1998; Pearson, 2000), a value considered statistically significant in large databases. In many cases E-values less than 0.001 will not be stringent enough to decide relationships of homology (see below), but this low ratio does identify statistically meaningful relatedness. However, in most cases decisions on which potential ABPs, MAPs, or IFs to score as likely homologues was not based on E-values alone. Because proteins and protein subdomains evolve at different rates, any single statistical value will be too inclusive for some complex sequences in “extended” gene families. A few examples how subdomain homologies, extremely short or long query sequences, and orthologues in complex extended gene families effect sequence relatedness are discussed for particular Arabidopsis ABPs, MAPs, and IFs described below. Those query sequences that failed to detect significantly related sequences in the Arabidopsis database are listed in a footnote at the bottom of the appropriate table. Several classes of Arabidopsis sequences passed criteria we set for statistical significance (E < 0.001), but we feel particularly uncertain of a relationship of evolutionary homology to the query sequence. In these cases, the number of sequences identified in the search and their E-values are included in each table (fourth column), but the gene family size is listed as zero (second column of each table).
The majority of cytoskeletal gene sequences discussed herein are likely homologues of their named animal, protist, and fungal counterparts. The rewards for delineating the functional roles and significance of these plant sequences to plant growth and development should be a greater understanding of how each cell both organizes its own structural foundation and contributes to building an organism. In addition, studies of the roles of the plant cytoskeleton in response to stress and pathogens will lead to advances in agricultural biotechnology. However, there will be problems starting with sequence information alone and trying to proceed with full confidence to functional characterization. Perhaps as many as 5 to 20% of the sequences identified as potential ABPs, MAPs, or IFs will be artifacts resulting from an examination of such a large database. Other sequences may be placed in the wrong class of ABPs, MAPs, or IFs due to accidents of selective sequence convergence and neutral drift. Undoubtedly, even larger numbers of cytoskeletal-associated genes have missed detection in our study due to rapid rates of plant sequence divergence from their counterparts in other eukaryotes or evolution of novel motifs and sequences within plants. Some of our gene family size estimates are likely too high due to duplicate listing of sequences in the database that were not resolved (see legend to Table I). In summary, we caution that it is impossible to guarantee that this compilation is 100% accurate. Nonetheless, this collection of sequences should serve to focus future experimental studies of the plant cytoskeleton.
Actin-based Cytoskeleton
The plant actin cytoskeleton directs numerous subcellular functions and the development of complex organ structures. The different plant processes in which actin has demonstrated or proposed roles include: 1) establishing cell polarity (Bouget et al., 1996); 2) determining the location of the division plane (by positioning the preprophase band); 3) preprogramming of cell wall development and deposition; 4) cell elongation; 5) tip growth (pollen tubes) (Vidali et al., 2001); 6) positioning receptors and transmembrane transport; 7) transporting mRNAs within the cell; 8) streaming cytoplasm; and 9) orienting chloroplasts to light and positioning the nucleus (Staiger and Lloyd, 1991; Meagher and Williamson, 1994). In addition, plant responses to stress involve rapid cytoskeletal responses (Roman and Ecker, 1995; Fischer and Schopfer, 1998; Jin et al., 1999; Aon et al., 2000). Because cell death and migration do not play a significant role in plant development, plant organ structure depends primarily upon control of periclinal and anticlinal cell divisions in the apical meristem (Meyerowitz, 1996; Meyerowitz, 1997a; Meyerowitz, 1997b) and the subsequent expansion and maturation of these cells in the developing organ. Thus, the first four of these actin functions (establishing polarity, division plane determination, cell elongation, directing cell wall deposition) are critical to all of plant development and morphology. Polarity provides the positional information and spatial cues essential for morphogenesis of multicellular structures (Gallagher and Smith, 1997; Cleary and Smith, 1998).
The Arabidopsis actin genes
The actin gene family of the crucifer Arabidopsis thaliana has been established as a valuable model system to study plant actins. There are only ten actin genes in Arabidopsis; all have been cloned, sequenced, and characterized in detail (McDowell et al., 1996c). Eight of the ten actin genes appear to be strongly expressed at some time and place during plant development (An et al., 1996a; An et al., 1996b; Huang et al., 1996a; McDowell et al., 1996a; Y-Q. et al., 1996; Huang et al., 1997). The remaining two members of the Arabidopsis family appear to be pseudogenes (McDowell et al., 1996c), because no mRNA has been detected, but neither contains stop codons that would prevent protein expression. The eight functional Arabidopsis actin genes are dispersed in the genome (McKinney and Meagher, 1998).
A query of the complete Arabidopsis genome with any conventional actin from other kingdoms identifies about 24 homologous sequences (Table 1). Among these, only the ten conventional actins previously described in the literature are present. The remaining 14 sequences include eight actin-related protein (ARP) sequences discussed below, making a total of 18 actins and ARPs. Six sequences identified as actins are redundant sequences in the database that were inadvertently grouped and named separately from these 18. Among the ten genes in the actin family, eight are known to be functional expressed genes. These eight actins can be divided into two ancient classes that are expressed predominantly in vegetative (ACT2, 7, 8) or reproductive (ACT1, 3, 4, 11, 12) organs and tissues. An analysis of steady-state RNA levels and the expression of actin translational fusions to a β-glucuronidase (GUS) reporter in transgenic plants has been performed on all eight functional actin genes (An et al., 1996a; An et al., 1996b; Huang et al., 1996a; McDowell et al., 1996a; Y-Q. et al., 1996; Huang et al., 1997).
Arabidopsis actin gene and mRNA regulatory patterns can be clearly categorized as being one of two complementary types: either vegetative/constitutive or reproductive. For example, all three vegetative actins, ACT2, ACT7, and ACT8, are strongly expressed in roots, stems, and leaves of germinating seedlings, young plants, and mature plants (Meagher et al., 1999b). These three actins represent two ancient actin subclasses long diverged from the rest of the actin family. Vegetative gene expression includes some organs of the floral organ complex (e.g., sepals, petals, filaments, stigma, style). Strong ACT2 expression is maintained even in older tissues, suggesting an important role in maintaining the plant cytoskeleton in mature vegetative tissues. ACT7 is the primary actin gene responding to phytohormones, has exceptionally high expression levels in young rapidly growing tissues, and in contrast to ACT2, is low in mature tissues (McDowell et al., 1996a; Kandasamy et al., 2001). Little or no expression of the vegetative genes was observed in mature pollen sacs, ovules, embryos, or seeds.
In contrast, reproductive actins are characterized by expression in pollen, ovules, and/or seeds. For example, all five reproductive actins, ACT1, ACT3, ACT4, ACT11, and ACT12, are strongly expressed in mature pollen (Huang et al., 1996a; Y-Q. et al., 1996; Huang et al., 1997). ACT1, ACT3, and ACT11 are expressed in developing floral meristems and the emerging carpel and ovules (Meagher et al., 1999b). ACT11 predominates during embryo and seed development. Little or no reproductive gene expression is detected in vegetative organs, such as root, stems, leaves, sepals, and petals. Each actin gene shows significant organ variability in steady-state RNA levels, ranging from eight-fold for ACT2 to 5000-fold for ACT1. There are a few exceptions to the distinct separation into vegetative and reproductive patterns. For example, some of the vegetative actins are expressed early in pollen and ovule development and all five reproductive actins are expressed at low levels in vascular tissue development.
Using actin evolution rates as a measure, these vegetative and reproductive gene classes appear to have diverged from a single common ancestral plant actin sequence some 400 MY ago (Meagher et al., 1999b). Gene and protein trees suggest that over the past 150 – 350 MY both classes of actin have further subdivided, resulting in a total of five actin subclasses. Additional support for the topological and age relationships of subclasses in the tree comes from a recent immunochemical characterization of an ancient, pollen-specific epitope, centered on Asn79, which is found in all angiosperms and the most recently evolved gymnosperms (Kandasamy et al., 1999). This epitope is shared by the two most recently derived reproductive actin subclasses, but is not found in any other plant or animal actins. It is reasonable to assume that this amino acid change arose in a common ancestral gene. Thus, the branch of the actin tree containing the pollen-specific actins (ACT1, 3, 4, 12) traces its origin to late gymnosperms about 220 MY ago. Similarly, the phytohormonally controlled Arabidopsis actin ACT7 is the only member of ancient actin subclass 2 in Arabidopsis (McDowell et al., 1996a; McDowell et al., 1996c). This actin is required for normal wound and auxin responses in Arabidopsis (Kandasamy et al., 2001). A nearly identical wound-responsive actin gene sequence is found in mallow (Jin et al., 1999). Mallow (Malva pusilla) is in the cotton family and as such is a close relative of Arabidopsis, having shared a common ancestor in the last 60 MY (Cronquist, 1981; Taylor, 1981).
Actin-related proteins (ARPs)
In the last decade a series of actin-related proteins have been found in eukaryotes (Schafer and Schroer, 1999). They exhibit 10–60% amino acid sequence identity to true actins, and vary more widely in length than bona fide actins. Arp2 and Arp3 are nearly 60% identical with bona fide actins and are involved in nucleation and branching of F-actin polymers. A recent genomic analysis revealed a small family of Arabidopsis ARPs (McKinney et al., 2002) and a detailed examination was made of the expression of an Arabidopsis Arp2 homologue (Klahre and Chua, 1999). Animal Arp2 and Arp3 each detected one corresponding Arabidopsis sequence of the correct length that were more related than any of the true actins (Table 1). These two sequences are undoubtedly Arp2 and Arp3 orthologues, respectively. Other less-well-conserved ARP query sequences such as yeast Arp4-9 detected Arabidopsis actin-related sequences with higher, but still quite convincing E-values (e-102/e-06) and homology over the full length of the query. These six other ARPs are not actins, Arp2s, or Arp3s and are included in the total of eight ARPs listed in Table 1. McKinnney et al. (2002) detected expressed cDNAs for most of these ARPs suggesting that each is expressed at the RNA level and that they are probably not pseudogenes.
In contrast to the success with most ARP query sequences no Arp1 homologue is found in Arabidopsis. When animal or yeast Arp1 is used to search the Arabidopsis database, Arp1 sequences were more similar to the known bona fide Arabidopsis actins than to any potential ARP sequence. Among complex extended gene families like the actins it can be difficult to identify true orthologues. Arp1, 2, and 3 in animals, protists, and fungi are all about 60% identical in amino acid sequence to the true actins. Yeast Arp1 has significant homology and aligns over the full 376 a.a. length with all the true actins. However, Arp1 homologues among animals, protists, and fungi are much closer to each other than any true actin. Thus, when we query the Arabidopsis genome with Arp1, the true actins serve as negative controls. With these built in controls, it is absolutely clear that there are no true Arp1 orthologues in Arabidopsis. The lack of Arp1 orthologues in plants served as one piece of evidence for the lack of the entire dynactin complex of proteins in higher plants (Lawrence et al., 2001b) (also see dynein section below).
Actin-binding Protein Sequences (ABPs) in Arabidopsis
Constructing a working list of Arabidopsis ABPs:
Starting with prototypical a.a. sequences for more than 70 actin-binding proteins (ABPs) identified in animals, protists, and fungi (Kreis and Vale, 1999), we searched the Arabidopsis database for homologous sequences. While some of the 70 ABP query sequences share common functional domains, most represent distinct protein sequence classes and contain multiple identifiable domains. Our data on potential Arabidopsis ABPs are summarized in Table 1. At least 36 of the 70 query sequences turned up statistically significant sequence matches in the Arabidopsis genome. Some of the ABP query sequences belonged to families with relatively complex domain and repeat structures. Therefore, we cartooned their structures in Figures 1–3 as a guide. These figures will be used to illustrate the potential for finding Arabidopsis sequences overlapping in their relatedness to different query sequences. By identifying the Arabidopsis sequences most closely related to animal, protist, or fungal sequences and using existing nomenclature for better characterized animal genes, only 20 of the 34 classes of Arabidopsis sequences should be termed separate classes. In other words, the Arabidopsis ABPs identified can all be traced to approximately 20 common ancestral sequences in other eukaryotes. In most cases, the Arabidopsis ABP sequences were in small (1–3 genes) to moderate (5–10 genes) sized gene families, although a few were found in very large families.
Figure 1.
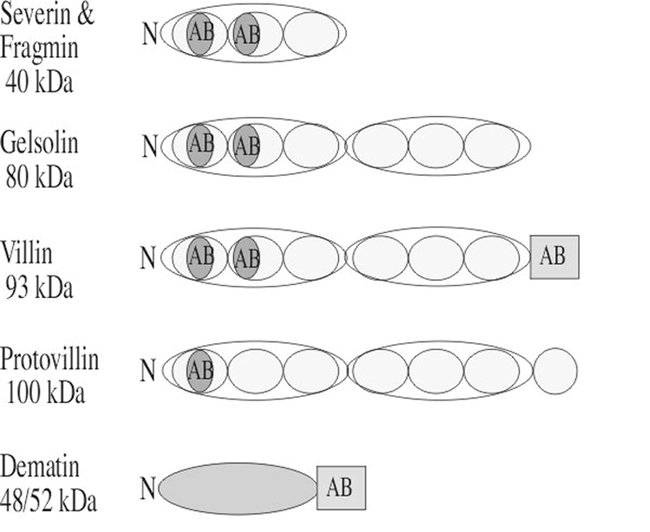
Domain structures of the animal and protist villin- and gelsolin-related actin-binding proteins used as query sequences. Most members of this superfamily including severin, fragmin gelsolin, villin, and protovillin share a common actin-binding domain (AB; oval drawn in blue). Members of this family also share protein repeat units (yellow ovals) with three repeats in severin and fragmin and to groups of these three repeats for a total of six repeats in gelsolin, villin, and protovilin. Dematin is related through its common actin-binding domain with villin (AB; rectangle drawn in blue). The approximate molecular weights of example proteins in kilo Daltons (kDa) are shown on the left.
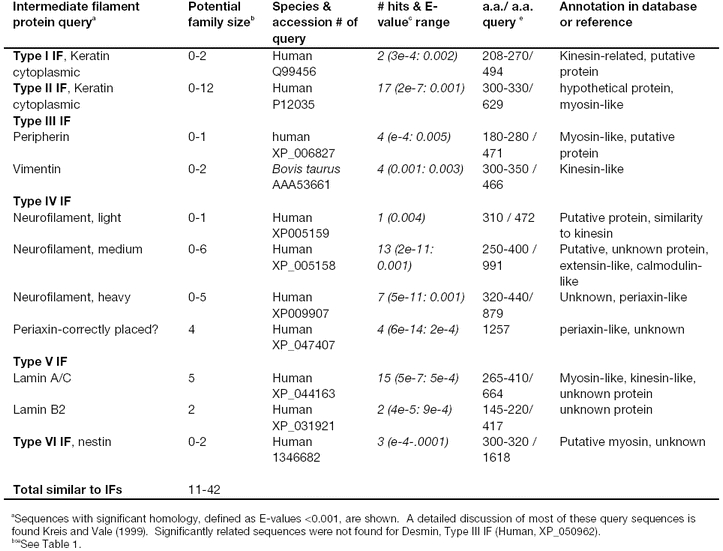
Figure 3.
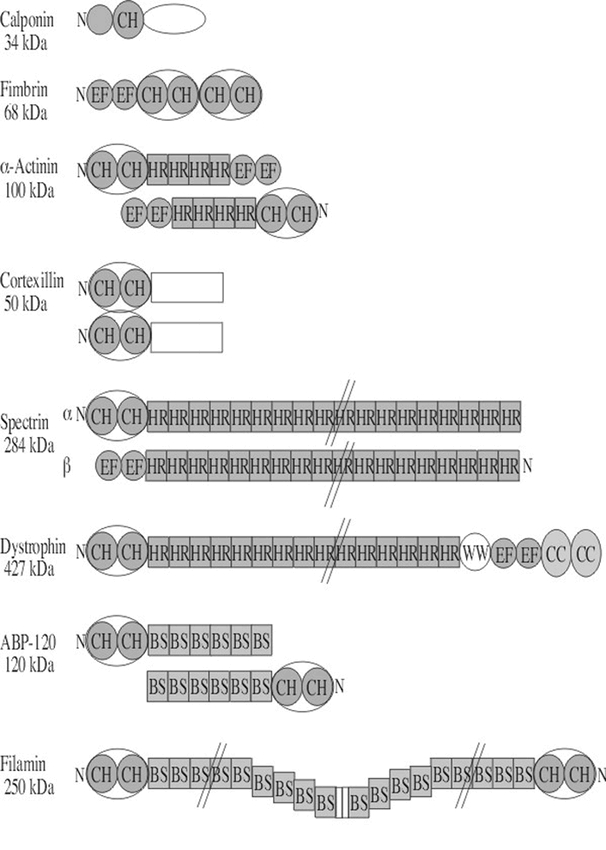
Domain structures of several animal and protist calponin-related actin crosslinking and anchoring proteins used as query sequences. The domain structures of several crosslinking and anchoring proteins are summarized in this diagram. Subdomains are labeled with the following abbreviations: BS, β-sheet repeats also called filamin repeats; CC, α-helical coiled coil domain; CH, actin-binding calponin homology domain of ∼100 a.a. shared by all members of this family drawn in blue; EF, EF hand calcium binding domain; HR, helical repeat common to spectrin also called spectrin repeats; WW, two adjacent tryptophan residues conserved in dystrophins. The N-terminal region of each protein is marked with the letter N. In the cases where the proteins function as dimers or heterodimers (α-actinin, spectrin, ABP120, and filamin), the N- to C-terminal orientation may be flipped to show the second molecule. Calponin is not a crosslinking or anchoring protein, but is included in this figure for comparison to the numerous proteins containing a calponin homology (CH) domain. The approximate molecular weights of single example polypeptide chains in kilo Daltons (kDa) are shown on the left.
We generally used query sequences from other kingdoms in homology searches to avoid becoming too focused on small differences between subclasses of plant proteins within a gene/protein family. In addition, we have attempted to clarify some of the confusion generated with plant sequences previously named and thus classified as being more like one non-plant ABP than another. For example, 10 Arabidopsis sequences have already been named based on their being related to animal villins. The villins are discussed in detail below with the other F-actin capping and severing proteins. We presume the first Arabidopsis homologue was matched to an animal villin, and all other Arabidopsis sequences were matched to this plant “villin” sequence. Our study suggests that half of these sequences may share closer ancestry with the related animal gelsolins.
All total 150–300 potential Arabidopsis ABP sequences were identified, including sequences similar to and in most cases clearly homologous to actin monomer binding, capping, severing, crosslinking, anchoring, and motor proteins. Many of these (>50%) are novel plant sequences previously classified as unknown open reading frames or hypothetical proteins in the database. A critical interpretation of these data is essential if we are to find the majority of novel ABPs without wasting our time on false positives or missing homologues with low levels of homology. We have attempted to adjust for the complex nomenclature among animal, protist, and fungal cytoskeletal protein by identifying the Arabidopsis sequences by the name of the most homologous or best characterized similar ancestral ABP. Table 1 cross-references other related sequences that are homologues to the most closely related class of plant sequences.
Actin Monomer Binding Proteins
Actin depolymerizing factor (ADF):
ADFs or cofilins are small ABPs with homologues in all eukaryotic kingdoms. ADFs accelerate the rate of actin filament turnover, and they have much lower turnover activity in their phosphorylated form. A 137 a.a. query sequence from fission yeast (P78929) detected 11 distinct Arabidopsis sequences with a high level of homology and sequence alignment over at least 124 a.a. residues (McKinney and Meagher, unpublished). Thus, the family appears a little larger than its counterpart in mammals. The three ADFs characterized in maize, whose transcript and protein levels have been assayed, are easily distinguished by strong expression either in reproductive or vegetative patterns, similar to the patterns of the ancient classes of actins and profilins (Lopez et al., 1996; Jiang et al., 1997; Staiger et al., 1997; Staiger, 2000). Detailed in vitro kinetic studies have characterized some of the physical and chemical properties of Arabidopsis ADFs (Carlier et al., 1997; Didry et al., 1998; Ressad et al., 1999; Dong et al., 2001a; Dong et al., 2001b), and they are quite comparable to human ADFs. In particular, there appears to be a synergy between ADF-induced F-actin turnover and the interactions of other ABPs (e.g., profilin, gelsolin, Arp2/3 complex) with actin filaments (Didry et al., 1998; Ressad et al., 1998; Ressad et al., 1999).
Profilin:
Profilins are small (130 a.a.) proteins that sequester actin monomers and have complex effects on actin polymerization. For example, they can inhibit nucleation, but in their active ATP form, profilactin complexes can accelerate addition of actin monomers to the barbed ends of growing F-actin filaments. Vertebrate profilins barely align with the five known Arabidopsis profilin sequences (Christensen et al., 1996; Huang et al., 1996b) due to an increase in the rate of vertebrate profilin sequence divergence relative to the rate for profilins in lower animals and other kingdoms (Huang et al., 1996b). Yeast, protist, and insect profilins are more closely related to each other than to vertebrate profilins. These non-vertebrate profilins can be easily aligned to the plant proteins with more than 30% identity.
Using the yeast profilin sequence as a query, seven Arabidopsis sequences with reasonable E-values (E = 2e-13 - 2e-10) were found (Table 1). If there had been much less conservation, a full alignment of yeast profilin with plant profilin homologues would have been difficult. While the conserved N- and C-terminal domains align easily, the internal a.a. sequences are poorly related. Identifying the profilins by sequence searches of the Arabidopsis database serves as a useful control for the identification of other ABPs. Clearly, E-values in this relatively low range are accurately identifying functional homologues. No new profilins were found that had not been described previously. The five known profilin sequences designated PRF1, 2, 3, 4 and 5 were found by extensive searches of clone libraries with hybridization or by PCR of cDNAs with degenerate primers (Christensen et al., 1996; Huang et al., 1996b). The two new profilin sequences identified appear to be redundantly named sequences with sequencing mistakes or alleles of existing sequences with a few base differences from the previously characterized profilins.
The profilin gene family is nearly as large as and even more diverse in sequence than the Arabidopsis actin family (Christensen et al., 1996; Huang et al., 1996b). Like actins, plant profilins can be placed into either vegetative (constitutive) or reproductive classes based on sequence and expression patterns. PRF1, PRF2, and PRF3 are expressed in most vegetative tissues, but are also expressed in embryo and endosperm, unlike the vegetative actins (Kandasamy et al., 2002b). In contrast, Arabidopsis PRF4 and PRF5 are expressed strongly in pollen. Detailed examination of the tissue-specific expression of promoter-reporter fusions from one vegetative (PRF2) and one reproductive (PRF4) profilin (Christensen et al., 1996) show patterns quite similar to possible actin counterparts, ACT2 and ACT1, respectively. Maize profilins closely related in sequence to the Arabidopsis reproductive sequences have also been reported. The class I maize profilin sequences are also specifically expressed in pollen (Staiger et al., 1993), while the class II sequences are expressed in most tissues, ovules, and endosperm and hence were termed constitutive (Gibbon et al., 1998; Gibbon and Staiger, 2000; Kovar et al., 2000b). These observations have led us to suggest that the evolution of the vegetative (constitutive) and reproductive classes of actins and profilins might have been concordant (Huang et al., 1996b; Meagher et al., 1999b).
Initial studies with Arabidopsis plants defective in profilin production explore the role of profilins in plant development. Suppression of PRF1 and the closely related members of the constitutive class of profilins was achieved by expressing antisense RNA to PRF1 (PFN1) coding sequence (Ramachandran et al., 2000). These plants were dwarfed and most cells were smaller than normal. In particular, cells were shorter than wild-type, appearing to be defective in elongation. Supporting the idea that there is a positive role for profilin in cell elongation is the fact that overexpression of PRF1 produced plants with longer than normal roots, root hairs, and longer root cells. An independent study examined the PRF1 allele prf1-1, which contains a T-DNA insertion in the promoter, 100 bp upstream of the TATA (McKinney et al., 2001). This allele destroys the light repression characteristic of PRF1, increasing its expression in light-grown hypocotyls. When grown in the light these plants had longer than normal hypocotyls and raised cotyledons, defects similar to light and circadian responses. The positive correlation of PRF1 expression with cell length is consistent with the requirement for profilactin complexes to add actin monomers to growing F-actin filaments. In contrast, if the role of profilins as an actin monomer-sequestering protein had dominated, then profilin levels might have negatively correlated with cell length.
Thymosin family:
β-thymosin is a 45 a.a. animal actin-binding and sequestering protein found in high concentrations in vertebrate cells. Actobindin and ciboulot are related proteins that contain two and three thymosin-like repeats, respectively (Vandekerckhove et al., 1990; Boquet et al., 2000). The human β-thymosin query gave very high and unremarkable E-values (1.6–2.8) with two hypothetical ORFs in Arabidopsis that should be taken as statistically insignificant. However, thymosin is not a well-conserved sequence even among animals and it is too short to give good E-values without high levels of homology. Similarly, actobindin gave high E-values suggesting the relationships are insignificant (Table 1 bottom). Thus, Arabidopsis either lacks thymosin-like sequences, or has homologs that are too distantly related to detect. Even if homologous in origin, short amino acid sequences with low to moderate similarity yield high E-values in the insignificant range.
Actin Capping and Severing Proteins
α and β capping protein subunits:
Capping protein is a highly conserved heterodimeric protein (α- and β-subunits) that caps the barbed ends of actin filaments (Cooper and Schafer, 2000; Hart et al., 2000). Capping protein heterodimers also bind G-actin monomers and can help nucleate actin polymerization. Using the 272 a.a. α-subunit of C. elegans capping protein as a query, we found one striking homologue with ∼30% sequence identity that extended over most of the sequence and had very significant E-values (2e-43, Table I). A C. elegans β-subunit query also identifies one highly conserved homologue in Arabidopsis (2e-60, Table I). The presence of highly significant sequences for both the alpha and beta subunits indicates the presence of capping protein as a regulator of actin dynamics in Arabidopsis. Capping protein is effectively inhibited by phosphoinositides. Thus, this highly conserved protein has the potential to play essential roles in signaling changes in plant actin polymerization.
Villin- and gelsolin-related superfamily:
The villin- and gelsolin-related superfamily members share three repeats (severin, fragmin, and CapG) or six repeats (gelsolin, villin, protovillin) of a common subdomain structure as shown in Figure 1. All members of the family have the ability to cap the barbed ends of actin filaments, and some members also sever (gelsolin, villin, severin, fragmin), or bundle (villin, dematin) actin filaments, or nucleate F-actin filament growth (villin, gelsolin, fragmin, severin). The actin capping, severing, and nulceating activities of these proteins are activated by Ca++, and inhibited by phosphoinositides. The bundling activity of villin requires the presence of an additional actin-binding site located in the C-terminal “headpiece” domain. This domain is also present in the actin bundling protein dematin.
There are 11 Arabidopsis genomic sequences that match closely to animal or protist villin- and gelsolin-related query sequences. Ten of these plant sequences are designated in the database as encoding villins, while none are classified as gelsolin-, severin-, or dematin-related. However, only four of the sequences have C-terminal extensions containing the KKEK actin-binding motif that defines animal villins. Furthermore, this part of two of the Arabidopsis proteins has been shown to possess actin-binding activity (Klahre et al., 2000). A plant homologue of villin has been associated with the actin filament network in the streaming cytoplasm of lilly root hairs (Tominaga et al., 2000). Thus, some of the plant sequences detected are clear villin homologues. A sequence-by-sequence comparison suggests that at least four of the remaining sequences listed as Arabidopsis villins are more closely related to known animal gelsolins. In addition, a few of the sequences detected are redundant in the database. Further, none of the Arabidopsis sequences is predicted to encode a putative three domain protein homologous to severin or CapG. Careful biochemical analysis will be required to determine which of these proteins exhibit actin filament capping, severing, nucleating, and cross-linking activities observed for the protist and animal homologues. It will be of significant interest to determine whether their activities are regulated by PIP2 and/or Ca2+.
Actin Crosslinking Proteins
Anillin:
Anillins are multidomain proteins found in animals that are associated with the nucleus in interphase and localizes in the cleavage furrow during mitosis. They bind and bundle actin filaments in vitro. While nothing like furrow formation occurs during plant cell division, a small family of three to six weakly related anillin-like Arabidopsis sequences were identified using a human query sequence. They align within the actin-binding and nuclear localization domains. These plant sequences are previously listed as unknown or nucleolin-like.
eEF1α:
Translational elongation factor 1α (eEF1α is known to co-localize with actin in plant endosperm and other tissues (Sun et al., 1997). This is of general interest to both molecular and cell biology because of evidence demonstrating that polyribosomes and some mRNAs may be linked to the actin cytoskeleton (Ramaekers et al., 1983; Davies et al., 1991; Klyachko et al., 2000; Gramolini et al., 2001). Further, in Dictyostelium eEF1α has been extensively characterized as an actin bundling protein (Edmonds et al., 1998) and a microtubule binding, bundling, and severing protein as reviewed in Furukawa et al. (2001). Using a human sequence as a query, 33 eEF1α homologues were found in the Arabidopsis genome, and 10 of these aligned over more than 430 amino acids. It is difficult to anticipate if this large family of proteins exists to service specialized needs of translational elongation, localization of the translational machinery, or organization of the plant cell cytoskeleton.
Espins:
Espin is an actin cross-linking protein that is present in the ectoplasmic specializations of Sertoli cells in testis. A short isoform of espin; along with villin and fimbrin; is present in the microfilament core bundle in intestinal epithelial cells (Bartles, 2000). Espin contains multiple binding sites for actin. A search of the Arabidopsis database with a human espin query produced 20 putative homologs with convincing E-values ranging from e-20 to 6e-9 and alignment over the N-terminal half of the 745 a.a. query. A search with rat espin, the first espin sequence characterized (Bartles et al., 1998) produced similar results (not shown). It will be interesting to see whether these sequences do encode actin bundling proteins, and whether they promote formation of a subset of bundled actin arrays in specialized cell types, as they do in animal cells.
The β-barrel repeat proteins:
The β-barrel (βB) repeat family of proteins in animal and fungi is quite diverse, and all are related by the presence of at least four repeats of a ∼ 50 a.a. sequence that assumes a stacked β-sheet structure. Each repeat forms the face of a barrel or propeller (Adams et al., 2000). The members have been further subdivided into the kelch repeat subfamily including kelch and scruin, and the WD repeat family including coronin, and the β subunits of heterotrimeric G proteins. The domain structures of these βB proteins are compared in Figure 2. The β-barrel domains can participate directly in actin-binding as shown for scruin in Figure 2. The β-barrels of other family members bind directly to other partners, such as Gβ. Although only a subset of these proteins are actin-binding proteins, the family is discussed here as a group due to the inherent tendency of BLAST searches to identify both closely related and more distantly related sequences.
Figure 2.
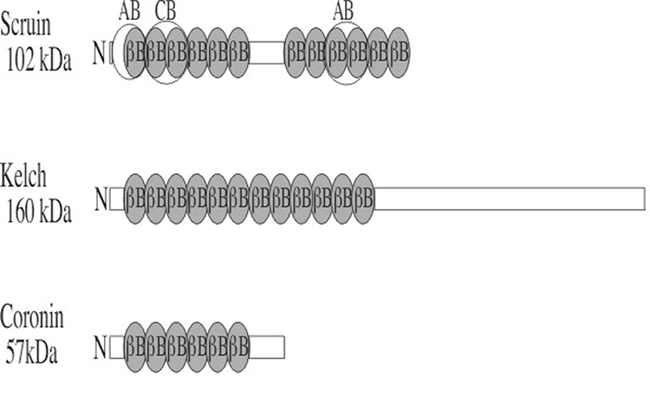
Domain structures of the animal β-barrel actin-binding proteins used as query sequences. Coronin, scruin, and kelch all contain β-barrels repeats (βB, drawn in blue), or kelch repeats. The portions of these repeats that bind actin are not well defined, but their approximate positions are indicated in the structure of scruin (AB). CB is a calmodulin-binding domain. The βB repeat in coronin is also known as the widely distributed WD40 repeat found in G-β proteins that do not bind actin. The approximate molecular weights of example proteins in kilo Daltons (kDa) are shown on the left.
Kelch was first identified as a likely structural protein in Drosophila ring canals. Using Drosophila kelch as a query, a very large family of Arabidopsis proteins was identified with significant E-values as high as 3e-32 (Table 1) and most were aligning with the kelch repeat region.
Scruins are 918 a.a. actin bundling proteins found in horse shoe crab sperm in a 1:1 molar ratio with actin. They are thought to help build the long acrosomal processes in Limulus sperm. Animal scruins contain two sets of six kelch β-barrel repeats (Figure 2). A horseshoe crab scruin query identifies a large family of 50 scruin-related sequences in the Arabidopsis database. With the exception of one sequence, the other potential scruin-like sequences are listed as unknown sequences in Arabidopsis. The degree of sequence similarity is low, but the alignment extends over more than 600 of the 900 a.a. query sequence. Each contains homology with multiple 50 a.a. repeats in the two β-barrel domains separated by a 200 a.a. linker domain. The possible binding of these putative Arabidopsis scruin homologs to actin or other putative partners will require biochemical and cell biological investigation.
Coronin was first identified in the protist Dicytostelium. Coronin null cells show defects in locomotion, endocytosis, and cytokinesis. Coronins are also found in yeast and humans and due to potential endocytotic functions they might be expected to be found in plants. The coronins including the human query we used have five N-terminally located WD40 repeats and this region detects a very large number of plant sequences with low scores, most of which are probably not coronins. Nineteen Arabidopsis sequences show a region of approximately 200 to 270 a.a. that aligns with the 472 a.a. human coronin query and the first 150 a.a. of this region aligns with the WD40 repeat region. The remainder of the Arabidopsis sequence alignment extends downstream of or C-terminal to the WD40 repeat region. The fact that the region of homology extends beyond the WD40 repeat region suggests that some of these sequences may encode coronin-related proteins. Confirming that these Arabidopsis sequences constitute a coronin family is difficult without more sequence comparison and functional data. The existence of hundreds of WD40 repeat proteins creates the possibility that convergent evolution and/or exon shuffling has created some non-coronin WD40 repeat proteins. Careful study will be required to resolve these issues.
Calponin-related superfamily:
Calponin, also called CaP, is an F-actin- and calmodulin-binding protein. The sequence of the calponin F-actin binding domain (CH, in Figure 3) defines the entire calponin and spectrin superfamily of proteins, but calponin is not itself an actin crosslinking protein. When calponin binds to F-actin it blocks the myosin-binding site, but how it functions in vivo to regulate F-actin or F-actin's motor function with myosin is unknown. We found a small family of Arabidopsis sequences with moderate homology to calponin containing a region of 159–246 a.a. aligning with the 309 a.a. human calponin query (Table 1). However, these sequences were more related to kinesins and fimbrins and were previously identified as such. While it is still quite possible that some of the more distant sequences identified by the calponin query with scores greater than 0.001 are homologues, it will require more extensive analysis of these sequences and their proteins products to make the connection.
The calponin-spectrin superfamily is comprised of a diverse group of crosslinking and anchoring proteins including spectrin, fimbrin, filamin, dystrophin, α-actinin, and ABP-120 found in animals, protists, and/or yeast. These ABPs share a common CH (calponin homology) actin binding domain, and proteins with this domain were used as query sequences to identify putative homologous families of Arabidopsis proteins. The actin-binding domain present in spectrin superfamily members contains two calponin homology repeats (Goldsmith et al., 1997; Hanein et al., 1998). The structure of several well-characterized members of this ABP superfamily are shown in Figure 3. A provisional attempt has been made to determine which of these diverse and well-characterized proteins are most related to the Arabidopsis sequences in order to estimate the sizes of the Arabidopsis families. It appears that plants contain direct homologues of both the fimbrins and filamins. The degree of sequence relatedness with calponin, spectrin, α-actinin, and ABP-120 was less significant.
Fimbrins are moderate sized (68kDa), crosslinking/bundling proteins and are in the highest concentration at the leading edge of motile animal cells. Fimbrin sequences begin with an N-terminal, EF-hand calcium binding domain (EF) and end with four copies of the calponin homology (CH), actin-binding domain that comprise the two actin-binding regions as shown in Figure 3. Fimbrin functions as a monomer. When the 642 a.a. yeast fimbrin is used as a query, nine Arabidopsis fimbrin sequences were identified with E-values less than 2e-98 (Table 1). These plant sequences are obviously homologues of animal sequences that contain the CH actin-binding domain. Most of the Arabidopsis sequences align well with the multiple CH domains that make up the majority of the fimbrin sequence. In addition, two of these plant sequences had significant homology that also extended into the N-terminal, EF-hand calcium binding domain (EF, Figure 3). Similar results are found with animal and protist fimbrin query sequences. Arabidopsis fimbrin AtFim1 shares 40% amino acid identity with fimbrins from other kingdoms and has many of the properties of previously characterized fimbrins (McCurdy and Kim, 1998; Kovar et al., 2000a). AtFim1 crosslinks F-actin in a calcium independent manner, and stabilizes actin filaments against depolymerization by profilin.
Surprisingly, fimbrin query sequences do not detect other members of the calponin/spectrin superfamily in plants as they would in animals. The E-values of the next most related sequences after those already identified in Arabidopsis as fimbrins are all greater than 0.36. Either the CH domains of these other members are too divergent to be recognized or they are not present on other Arabidopsis actin-binding proteins.
Filamins are long flexible crosslinking proteins that function as homodimers. Filamin contains two N-terminal, CH actin-binding domains, and these are followed by many repeats of a β-sheet sequence (filamin repeats or BS, Figure 3) making up the C-terminal 80% of the protein. ABP120 sequences have the same structure as filamin, but many fewer BS repeats (Figure 3). Using a human filamin query (NP_001447), many Arabidopsis sequences were identified with homology to the CH region and all with homology to this region were clearly more related to fimbrins. The β-sheet repeat detected three likely homologues of filamins with homology extending over 1000 to 1500 a.a. of the 2647 a.a. query (Table I). All three are homologous to filamin through the BS domain repeats, but none of these had a significantly related N-terminal CH domain. Similarly, sequences identified using the query ABP 120, which also contains BS repeats, did not appear to be likely Arabidopsis homologues of this protein.
The dystrophins (syntrophins) are enormous proteins found in animals (Koenig et al., 1988). Dystrophins help link the actin-based cytoskeleton to the extracellular matrix via a multimeric, membrane-associated glyco-protein complex. Dystrophins contain two N-terminal, CH actin-binding domains; a large number of spectrin helical repeats; two EF hand calcium binding domains; and two C-terminal, α-helical coiled coil (CC) domains (Figure 3). Using the 3685 a.a. human dystrophin as a query, we found a family of nine related sequences with moderately good expectation scores. However, all were aligning with the CH domain, were not long enough to encode dystrophin homologues, and were more closely related to fimbrins.
Cortexillin were first observed to accumulate in the cortex of Dicytostelium cells. Cortexillin crosslinks actin filaments in anti-parallel orientation. Each monomer contains two N-terminal, calponin-related actin-binding domains (CH), a coiled coil region (CC) that effects parallel dimerization, and a C-terminal PIP2 binding domain. Using Dicytostelium cortexillin I as a query, the sequence showed significant similarity to all the Arabidopsis fimbrin-related sequences through the CH domain (Figure 3), but no true homologues of cortexillins were detected. Dictyostelium cortexillin II gave a similar result (not shown).
Other members of this superfamily, α-actinins and spectrin (Figure 3), contain alpha-helical spectrin repeats. Significantly related sequences were detected in the Arabidopsis database through these conserved domains (Figure 3), but none of the plant sequences were clear homologues of the query sequences. For example, the α-actinins contain the N-terminal CH domain, helical spectrin repeats, and a C-terminal calcium binding domain (Figure 3). While each of these domains taken individually from within the mouse α-actinin found Arabidopsis targets that were related with statistical significance (Table 1), the query did not find any convincing true homologues of α-actinin containing all three domains.
Actin Anchoring Proteins
Annexins:
Animal annexins are calcium-dependent phospholipid binding proteins. They form trimers that polymerize into sheets on acidic membrane lipid surfaces, to which the actin cytoskeleton can be anchored. Several excellent homologues of human annexin are found in the Arabidopsis database with 200–300 a.a. aligning with the 324 a.a. query. The plant annexins share significant sequence homology with their animal counterparts (Morgan and Fernandez, 1997; Morgan and Pilar Fernandez, 1997) and can function in liposome aggregation (Hoshino et al., 1995). The amino terminal heme-binding activity of some Arabidopsis annexins has implicated their role in protecting against oxygen stress (Gidrol et al., 1996).
Talin:
Talins and their distant homologs Sla2p are found in animals, Dictyostelium, and yeast. In animals, talin is thought to be a key player in binding the F-actin cytoskeleton to integral membrane receptors, and is thus believed to be essential to cell-cell and cell-substrate adhesion. These proteins have domains that bind G-actin, F-actin, α-actinin, vinculin, integrins, and membranes. Using a 2541 a.a. human talin query we found four Arabidopsis sequences with reasonable sequence relatedness extending no more than 300 a.a. (Table I). Three of these plant sequences aligned with regions of the talin N-terminal actin-binding domain and one with the talin C-terminal actin-binding domain. None of these plant sequences were among the ezrin or Sla2p-related sequences listed in Table 1.
Sla2p is a yeast protein found associated with cortical actin and actin patches. Among several membrane-associated proteins functioning in osmolarity sensing and endocytosis, they are also thought to be involved in cell polarity determination. This function is of great interest to all plant developmental biologists. The two Arabidopsis sequences contain 260 and 380 a.a. regions, respectively, with moderate homology to the 968 a.a. yeast query sequence (Table I). One is listed as clathrin-like; the other as unknown. These Sla2p-related sequences are not among the distantly related ezrin or talin sequences detected in this study. However, the region of homology is not within the putative actin-binding domain, but within the less well-characterized N-terminal domain.
Tensin:
The tensins are large actin capping proteins originally defined in chicken. They bind to the barbed ends of F-actin filaments and are usually localized to the plus ends of actin filaments at membrane junctions. They also have the I/LWEQ actin-binding motif. A 1744 a.a. chicken tensin query identified one 1200 a.a. Arabidopsis homologue (Table I). It contains both the tyrosine phosphatase-like domain and the SH2 domain separated by a large unaligned region similar in spacing to the chicken sequence. Numerous Arabidopsis proteins were identified with homology to either of these subdomains alone, but further effort would be needed to explore their relationship to tensins in greater depth. Thus, tensin-like sequences may play a role in signaling changes in the structure of the plant cytoskeleton.
Vinculin:
Vinculin is an anchor protein responsible for attaching actin filaments to the plasma membrane in animals. Its 1066 a.a. sequence contains binding sites for α-actinin, talin, polyproline binding proteins, and actin, which are distributed respectively from the N-terminal to C-terminal ends of vinculin. Several Arabidopsis sequences with 240 to 640 a.a. stretches of weak homology to the 1066 a.a. mouse vinculin are present in the database. Most are listed as unknown, one as a proton pump interactor, and one as myosin-like.
ERM Family:
The ezrin, moesin, and radixin (ERM) proteins are all quite similar in structure. They contain an N-terminal membrane association domain, C-terminal actin-binding domain, and two ERMADs (ERM-association domains) that mediate formation of homo- and heterotypic oligomers. These approximately 580 a.a. proteins are thought to function as membrane-microfilament linking proteins. Arabidopsis contains a large family of 30 to 40 sequences with 180–280 a.a. homology with mouse or Ciona (a primative cordate) ezrin sequences. They all show significant homology to the highly charged amino acid-rich (e.g., glutamic, lysine, arginine) helical domain of ezrin. A few of the Arabidopsis sequences also contain homology to the proline rich domain (PR) and the C-terminal actin-binding domain (ER) common to some members of the ERM superfamily. The sequences are nearly all listed as hypothetical and unknown proteins in the database. One of the 40 sequences is listed as putative auxilin and another as a putative vicilin. Thus, while 10 to 20 of these plant sequences are possible sequence homologues of ERMs, the lack of clear homology to the N-terminal domains makes it difficult to determine their true relationship. Further investigation is needed to study the possible presence of ERM proteins in Arabidopsis. Arabidopsis contains a large number of proteins with long stretches rich in glutamic, lysine, and arginine, which are likely to be structural proteins, but it is not clear of what class.
Miscellaneous ABPs
Tropomyosin:
Tropomyosin forms head-to-tail dimers of its coiled coil domain that bind along actin filaments and modulate interactions of F-actin with myosin and other ABPs. The 248 a.a. yeast query sequence identified more than a dozen weakly related Arabidopsis sequences with a 115 to 240 a.a. region aligning. The coiled coil region is very glutamic rich and accounts for a significant number of the matching residues, making the functional relationship of the plant sequences identified suspect. The best 12 hits were identified either as hypothetical proteins or in three cases as myosin heavy chain-like. The latter similarity may be due to a propensity for formation of coiled coil structures suggesting that additional analyses of these putative Arabidopsis tropomyosins is warranted.
Sucrose synthase:
Sucrose synthase (SuSy) is a proposed actin-binding protein associated with the actin cytoskeleton, although direct binding of SuSy to F-actin has not been demonstrated (Winter et al., 1998; Winter and Huber, 2000). One model for the interaction of SuSy with the cytoskeleton suggests that as the cell wall is being constructed on the outside of the cell, enzyme complexes making cell wall precursors like SySy's synthesis of sucrose are moved below the membrane by actin-myosin motors. Another possible link between SuSy and the F-actin cytoskeleton that may require this protein interaction is the massive transport of sucrose from leaves into the phloem and down to the stems and roots of plants. Using a distant 816 a.a. maize SuSy sequence as a query, 17 SuSy-related sequences are found in the Arabidopsis genome that are not duplicates. If indeed SuSy interacts with actin, the diverse family of SuSy proteins could interact differentially with different enzyme-cytoskeletal complexes in different organs to alter the rates of sucrose transport and the quality of cell wall synthesis.
Myosin Motor Proteins
The myosins are large motor proteins that produce force along F-actin filaments at the expense of ATP hydrolysis. They are involved in a diverse array of movement functions in plant cells, from cytoplasmic streaming and organelle movement to determining polarity and directing changes in plant cell growth and development. In animals, protists, and fungi there are several diverse classes of myosins such as skeletal and smooth muscle, cytoplasmic, and as many as 17 named myosin classes (Mermall et al., 1998; Kreis and Vale, 1999). Myosin V is involved in cytoplasmic organelle movement in animal cells and is a good candidate for a homologous progenitor of plant myosins. Initial studies suggested that plant myosins most resemble animal myosin V proteins, forming dimers with two actin heads and a globular tail of variable length that often contains a membrane interaction domain (Kinkema and Schiefelbein, 1994; Kinkema et al., 1994; Yamamoto et al., 1999). A detailed survey of myosins from several model organisms suggests that the 17 Arabidopsis myosins examined were all slightly more related to animal myosin V sequences than to any other animal, protist, or fungal myosin subclass (Berg et al., 2001). Of the several animal myosin query sequences tested, a human cytoplasmic myosin V query of 1855 a.a. yields the best alignments with the greatest statistical significance in searches the Arabidopsis database. Of the 48 plant sequences detected, 12 sequences showed a 1500–1770 a.a. residue region of high homology, while the worst matches aligned only with the approximately 300 a.a. motor domain. A 1526 a.a. S. pombe myosin II heavy chain query that, among myosins, is not an immediate homologue of myosin V, still identifies more than 50 Arabidopsis sequences with significant myosin homology. While most sequences have less significant scores than for myosin V, a few sequences appeared more highly related to myosin II, but this may not be statistically significant (McKinney and Meagher, unpublished). For 26 sequences, the alignment extends from the motor domain and into the tail region with 800–1250 a.a. residues aligning. The remainder of the sequences align either in a portion of the motor or tail domains (200–700 aa). A third query with D. melanogaster intestinal brush border type 1B myosin sequence of 1026 residues detected 48 Arabidopsis sequences with still lower significance, but 26 of these sequence showed 750–860 residues aligning from the head into the tail regions. An alignment of all these Arabidopsis sequences revealed that most of the 48 sequences were duplicates or truncated versions of longer sequences in the databases. Only about 20 distinct sequences with clear myosin homology were found. The enormous length of the myosin coding regions and large number of splicing events that must be predicted from DNA sequence probably accounts for such a large number of misannotated and truncated sequences. However, it is clear that Arabidopsis has a very large family of diverse myosins.
The question remains as to ancestry of plant myosin subclasses. Shortly after completion of the Arabidopsis database, Reddy and Day (2001a) analyzed the Arabidopsis myosin-related sequences. They concluded that there were only 17 bona fide myosin-related sequences in Arabidopsis. Sequence tree building separated the plant myosins into two groups, those still closely related to algal myosins (myosin class VIII) and a group of higher plant myosins (myosin class XI). None of the plant sequences were more related to animal myosins than they were to each other, although animal myosin V sequences remain one of the closest outgroups consistent with the findings of Berg et al. (2001). In other words, the plant myosins are all related to one common ancestral plant or algal myosin sequence, and the myosin family divergence all occurred after plants and their protist green algal ancestors diverged from other eukaryotes. Thus, all of the diversity in the plant myosin family appears to have a more recent origin that postdates their common ancestry with myosins in other eukaryotic kingdoms. We can conclude that a single progenitor myosin gene may have given rise to all the diversity in the plant kingdom. The only apparent alternative and much less likely explanation would be that convergent evolution in a diverse family of myosin genes had homogenized the plant myosin motor sequences to be more similar to each other. We have left our estimate of the myosin family (Table I) as somewhat larger than that in Reddy (2001a) to include a few more distantly related sequences of uncertain function (McKinney and Meagher, unpublished). The rates of myosin sequence divergence in plants have not yet been estimated, but the significance of plant myosin diversity has been discussed further (Reddy, 2001).
Tubulin-based Cytoskeleton
The plant microtubule (MT) based cytoskeletal system plays many distinct roles in the formation of subcellular structures and in directing plant cell and organ development. Plant cell MTs play essential roles in mitosis, meiosis, cell division, cellulose microfibril alignment, cell wall deposition, and morphogenesis (Amos, 2000; Wick, 2000; Schroer, 2001). While MTs are less elastic than actin filaments, they participate in many but not all of the same cellular processes.
Microtubules (MTs) are relatively rigid tubular structures 24 nm in diameter. Microtubules are assembled from a helical array of α/β tubulin heterodimers that form 13 parallel protofilaments within the microtubule. Microtubule structures are quite dynamic with the minus end anchored at the microtubule organizing center (MTOC). Rapid addition and loss of tubulin heterodimers occurs from the plus end. This capacity of single microtubules to exist in alternate phases of growth or shortening is termed “dynamic instability”, and arises from the presence of terminal subunits with bound GTP or GDP (Mitchison, 1992). Rapid turnover rates of MTs with half-lives measured in minutes are observed during mitosis and cell division, and relatively slow turnover-rates are observed for other structures such as interphase arrays. The γ-tubulins have more specialized functions in MTOC structures, and as such are expressed at much lower levels.
Plant Tubulin Proteins
There are three ancient classes of tubulins, α-, β-, and γ-tubulins, found in all eukaryotes examined so far. Each α−, β-, and γ-tubulin monomer is approximately 50 kDa. Thorough searching of genomic and cDNA libraries and examination of Southern blots of restriction endonuclease digested DNA suggests there are a total of 17 tubulin genes in Arabidopsis. There are six α-tubulin (Carpenter et al., 1992; Kopczak et al., 1992), nine β-tubulin (Snustad et al., 1992), and two γ-tubulin (Liu et al., 1994) gene sequences characterized with these classical molecular techniques. Using human α-, β-, and γ-tubulins as query sequences, 4, 14, and 2 tubulin sequences, respectively, were found in the Arabidopsis data base as shown in Table 2. There is discrepancy in the number of α- and β-tubulins. Two pairs of α-tubulins encode identical protein isovariants (Kopczak et al., 1992) and are listed as one sequence each in the database (TUA2 & TUA4; TUA3 & TUA5). Further, two slightly different TUA1 sequences are listed, perhaps representing two alleles. Two of the β-tubulins are listed more than once in the database, while at least two others appear to represent novel gene and protein sequences, bringing the total to 11 β-tubulins. Thus, the tubulin gene superfamily in Arabidopsis contains at least 19 genes. Most but not all members of the Arabidopsis α- and β-tubulin gene families each can be divided into vegetative (or constitutive) and reproductive classes based both on expression patterns and to some extent on sequence relatedness, and thus the gene family structure has some similarity to the actins and profilins (Meagher et al., 1999b).
The variety of Arabidopsis tubulin proteins are thought to form a diversity of microtubules in different organs, cells, and subcellular locations. The functions of the diverse MTs are controlled by binding of MAPs in different tissues, in different subcellular locations, and at various stages in plant development. Potential Arabidopsis MAPs with significant homology to MAP query sequences are listed in Table 2. Those MAPs that failed to detect homologous sequences in the Arabidopsis database are listed in a note at the bottom of the table.
Microtubule-Associated Protein Sequences (MAPs) in Arabidopsis
We began a working list of putative Arabidopsis microtubule-associated proteins (MAPs) by selecting the prototypical a.a. sequences for approximately 30 MAPs identified in animals, protists, and fungi (Kreis and Vale, 1999) and searching for similar and homologous sequences in the Arabidopsis database. The MAP query sequences represent approximately 20 distinct protein sequence classes and many of these contained several distinct sequence motifs. Our data on potential Arabidopsis MAPs are summarized in Table 2. About 25% of the query sequences turned up significant sequence matches in the Arabidopsis genome. Four were contained in small gene families, while three (i.e., sequences similar to CLIP, Tea1, and kinesin) were found in very large families.
APC:
Adenomatosis polyposis coli (APC) protein was first characterized as a human tumor suppressor gene. Mutations in APC contribute to various cancers, particularly to colon cancer and familial adenomatosis polyposis. APC is a very large and complex 290 kDa protein with homologues in distant animal species such as Drosophila and C. elegans. APC has multiple 42 amino acid armadillo repeats that mediate protein-protein interactions; a number of 15 – 20 a.a. β-catenin binding repeats; a basic MT binding sequence that can stabilize MTs; and a region that can bind the EB1 protein (Kreis and Vale, 1999). The tumor-promoting mutations in APC remove the ability to bind and sequester β-catenin. More recent findings reveal effects of APC in cell adhesion and motility. APC has emerged as a protein that provides links between the microtubule and actin cytoskeletal networks (Dikovskaya et al., 2001). A 2843 a.a. human APC query detected a few similar sequences in Arabidopsis spanning the MT binding domain with moderate to poor similarity and alignment extending over several hundred amino acids as shown in Table 2.
EB1:
EB1 is a MAP that regulates MT orientation, dynamics, and cell polarity (Korinek et al., 2000; Tirnauer and Bierer, 2000) and binds to the APC protein. EB1 localized specifically to the plus ends of growing microtubules. The 268 a.a. human EB1 query identified three clear Arabidopsis homologues with regions aligning over 240 a.a. EB1 also contains an N-terminal CH (calponin homology) domain that may allow for actin-binding. Thus EB1, like APC, has the potential to link the F-actin and microtubule-based cytoskeletal systems.
CLIP:
CLIP is a microtubule binding protein that is localized along short subsections of MT structures near their plus ends in animal cells (Perez et al., 1999). CLIP may link endosomes to microtubules and has also been observed in the mitotic spindle. From the N- to C-terminal region CLIP homologues may contain one or two MT binding sites, serine rich domains, coiled coil repeats, and a metal binding domain. The Drosophila CLIP query sequence that contains all of these sequence motifs identified an extremely large family of more than 70 moderately similar sequences in the Arabidopsis genome. Many of these predicted Arabidopsis protein sequences had regions of 900–1300 amino acids aligning with the distal coil-coil regions of the 1690 a.a. Drosophila query. Only one Arabidopsis sequence showed sequence similarity extending into one of the two MT binding sites on the N-terminal portion of CLIP. CLIP proteins share homology in the MT binding domain with diverse proteins such as glued (Drosophila), kinesin (Drosophila), and cofactors A and B (humans). The Arabidopsis sequences identified were for the most part unknown proteins in the database, while some were previously identified as related to chromosome condensation, MAR binding, myosin-like, kinesin-like, and probable centromere proteins. Most of these were aligning with the coiled coil repeats of the CLIP query. The true identity of all the members of the Arabidopsis CLIP-like family awaits more detailed sequence analysis and functional studies.
E-MAP-115:
The 115 kDa microtubule associated protein E-MAP-115 is bound to subsets of microtubules in epithelial cells of humans. It is thought to be involved in the stabilization and reorganization of microtubules in polarized cells (Masson and Kreis, 1995). E-MAP-115 contains N-terminal domains for MT binding and more distal charged helical segments. Arabidopsis contains a small family of very poorly related sequences with E values below the limit of statistical significance (Table 2). However, the 180 a.a. plant sequence aligning with the 712 a.a. human query spanned most of the MT binding domain, supporting the possible relationship between the animal and putative plant MAP-115-like sequences.
Tau:
Tau is a MAP identified in animals and yeast that stabilizes microtubules and has been shown to promote nucleation and growth. In animals it is thought to assist in the outgrowth of neurites and determine neuronal polarity. Abnormal aggregations of Tau termed “neurofibrillary tangles” are associated with neurodegenerative diseases. The 352 a.a. human query detected one moderately related Arabidopsis sequence within the ∼95 a.a. region aligning (Table 2). The alignment was in proline rich region of Tau, and not in the repeated MT binding domain, suggesting that the Arabidopsis sequence may not encode a MAP.
Tea1p:
The fission yeast protein Tea1p is located at the ends of cells and the ends of MTs suggesting an important role in cell polarity and polar growth. Tea1p is a member of the kelch family of β-barrel proteins (Adams et al., 2000). The S. pombe Tea1p sequence detected a large number of related sequences in Arabidopsis that can be divided into two classes: the most closely related aligned over a region of 300 to 350 residues with the amino terminal kelch repeats of Tea1p, and a less related group aligned with the coiled-coiled region in the C-terminal half. The E values are quite convincing, and the Arabidopsis proteins are very likely kelch family members. However, since β-barrel proteins can participate in various protein-protein interactions, the identity of the Arabidopsis Tea1p homologues as MAPs will require further study.
XMAP215:
XMAP215 is a Xenopus oocyte protein that promotes MT assembly in vitro that is thought to play a role in early animal embryo development. It promotes both elongation and shortening of MTs and undergoes cell-cycle dependent phosphorylation at multiple sites. The 2030 a.a. Xenopus query detected one highly related sequence in the Arabidopsis database with significant homology extending for more than 1400 a.a. The XMAP215 homologue identified is identical to the previously characterized Arabidopsis microtubule organizing center protein, MOR1 (Whittington et al., 2001). Temperature sensitive mutants demonstrate that a functional MOR1 gene is essential for cortical microtubule organization in plant cells.
Katanin:
Katanin is an ATP-dependent microtubule severing protein that breaks MTs down to functional tubulin αβ-heterodimers. Katanin was first characterized from sea urchin eggs as a heterodimer of 60 and 84 kDa. Using a Drosophila katanin query for the 60 kDa subunit we found a moderate sized family of closely related sequences in Arabidopsis with excellent scores. One plant sequence with E = e-123 aligned over nearly the full-length of the 572 a.a. query. Using a Drosophila katanin query for the 84 kDa subunit we found a small family of Arabidopsis sequences related over most of the length of the 812 a.a. query. This latter comparison was confused by another 100 Arabidopsis sequences that shared homology with the WD40 repeat sequence with Katanin, but are probably not katanin homologues.
Tubulin tyrosine ligase:
Tubulin tyrosine ligase catalyzes the reversible detyrosination and tyrosination of the C-terminal tyrosine of α-tubulin. The 753 a.a. S. pombe query sequence detected only one Arabidopsis sequence with statistically weak relatedness over the 340 a.a. region that aligned (Table 2). However, the aligned region in the Arabidopsis sequence showed several short but significant stretches of amino acid identity making it a possible homologue.
Kinesin motors
Kinesins produce force along microtubules at the expense of ATP hydrolysis. The kinesins in animals, protists, and yeast represent a large and extremely diverse group of sequences with great functional diversity. The first kinesins studied were specialized for transport of cargo toward the plus-ends of microtubules, but family members that move toward MT minus ends have also been described. Kinesins are responsible for some changes in and maintenance of cell structure and many are involved in chromosome movement during mitosis and meiosis. Arabidopsis contains a very large superfamily of kinesin homologues. Using a 1027 a.a. mouse conventional “heavy chain” kinesin query with an N-terminal motor domain, 79 distinct sequences with significant levels of sequence homology were identified (Table 2). For 32 of these plant sequences, this homology extended considerably beyond the 300 a.a. motor domain and several aligned over 900 a.a. The longest 34 of these 79 homologous sequences are undoubtedly N-terminal domain kinesins. While the remaining plant sequences clearly encode kinesin motors, their domain organization is not always clear. When a mouse bipolar kinesin with an N-terminal motor domain is used as the query a few Arabidopsis sequences align better overall, but many fewer align over more than just the motor domain. Using a C-terminal motor domain kinesin from yeast as the query, a similarly large family of kinesins was found, with a larger percentage of the alignments extended over most (500–640 aa) of the 729 aa query. In this case, alignment for a few hundred a.a. beyond the motor domain suggested that 30 to 35 of the Arabidopsis kinesins either belong to the C-terminal domain subclass or at least are related kinesins with an internal motor domain. Due to the high degree of conservation of the motor domain, searches using the N-terminal, internal, and C-terminal motor domains as query sequences identify members of all three groups of kinesins. The structural relationship of the motor domain to several plant kinesin sequences is discussed by Reddy (Reddy, 2001; Reddy and Day, 2001b). Thus, the total number of Arabidopsis kinesins is probably as high as 79 or 80. Many of these Arabidopsis kinesins have related maize counterparts, suggesting ancient substructure to the kinesin family phylogeny.
Functional studies have characterized several Arabidopsis kinesin-like proteins. The Arabidopsis mutant zwichel gene ZW1 encodes an N-terminal motor domain kinesin (zw) (Oppenheimer et al., 1997). Mutants in ZW1 have abnormal trichomes with shortened stalks and only two branches instead of the usual three, suggesting an essential role for conventional kinesins in trichome morphogensis. A C-terminal kinesin-motor domain protein from Arabidopsis AtKCBP functions as a motor in vitro showing motor activity directed toward the minus end of glass bound microtubules.
Dynein:
Dynein is a minus-end directed microtubule motor that exists in animals and protists both in axonemes associated with cilia and flagella and in the cytoplasm associated with the dynactin complex. A search of the Arabidopsis database with query sequences for axonemal and cytoplasmic dynein heavy chains produced no close homologs (see Lawrence, et al., (2001b)). This is consistent with the loss of motile sperm and the dynein-containing flagella in the transition from gymnosperms to the more recently evolved angiosperms. Further, the absence of both ARP1 and cytoplasmic dynein indicates that most if not all components of the dynactin complex are absent from Arabidopsis.
Intermediate Filament-related (IF) Protein Sequences in Arabidopsis
Intermediate filament protein (IF) homologues have not been confirmed in the plant kingdom, with a few exceptions like the nuclear lamins (McNulty and Saunders, 1992; Masuda et al., 1997). Indirect evidence for other possible plant IFs has come from detecting proteins that appear antigenically related to animal IFs (Dawson et al., 1985; Shaw et al., 1991). However, convincing verification by widespread duplication of these findings, or by biochemical isolation of plant IF proteins, has been lacking. It is now possible to query the entire Arabidopsis genome with known IF sequences and assess the significance of possible homologues and similar sequences that are identified.
IFs are so named because the sizes of these filaments (10 nm diameter) are between that of F-actin (7–10 nm) and microtubules (25 nm). IFs play structural roles in many animal cells, and can be linked by accessory proteins to both the actin and microtubule cytoskeletal networks. It is possible that the structural role played by IFs in animal cells is rendered moot or significantly modified in plant cells, because they are supported by a cell wall.
Animal IFs have been divided into six classes (Type I–VI) as discussed in Kreis and Vale (Kreis and Vale, 1999) and examples of each were used as query sequences unless noted otherwise. Over three dozen potential homologues of animal IFs were identified as encoded by the Arabidopsis genome, as summarized in Table 3. Most align with the glutamic acid rich α-helical rod (filamentous) domain common among most IF proteins. Because there is scant physical evidence for presence of IFs other than nuclear lamins in plants, these novel plant IF sequences will remain in question until their encoded proteins are shown to be present in filaments in plant cells and/or capable of forming filaments. At the sequence level these potential IFs may be difficult to distinguish from other proteins that form coiled coil structures and/or are rich in acidic charged residues like glutamic acid.
Cytoplasmic Keratins:
The majority of animal IF genes and proteins are classified as Type I or Type II cytoplasmic keratins. These keratins have relatively distinct N- and C-terminal amino acid domains often separated by a repetitious filamentous domain. Using a 494 a.a. human Type I keratin as query we found only two weakly similar sequences in the Arabidopsis genome with 208 to 270 a.a. regions aligning (Table 3). A 629 a.a. human type II keratin query identified several moderately similar sequences in the Arabidopsis genome with the alignment extending over 300 a.a.
Vimentin, desmin, and peripherin:
Type III IF proteins include vimentin, desmin, and peripherin (Table 3). Desmin, once called skeletin, is found in muscle cells and thought to be involved in the structural connection and alignment of the myofibrils within a muscle fiber. Peripherin is so named because it is localized to the peripheral portions of neuronal cells and may play roles in their development and maintenance (Kreis and Vale, 1999). In animal organs and tissues vimentin is expressed in diverse cell types of mesenchymal origin. Vimentin has very resilient viscoelastic properties and is often linked to both MTs and F-actin networks and helps to integrate these systems. Thus, although all Type III IF proteins have specific tasks in animals and they would not be expected to have true functional homologues in plant, they could still have distant sequence homologues performing other tasks in plants. Desmin, vimentin, and peripherin are filamentous proteins of 53, 54, and 58 kDa, respectively.
A human peripherin query identified a few Arabidopsis sequences with potential significance, as shown in Table 3. A bovine vimentin query sequence detected several similar Arabidopsis sequences with E-values at the statistical cutoff for significance (0.001: 0.003). However, the two best sequences showed alignment over 340 a.a. of the 466 a.a. query making the relationship more interesting. Most of the peripherin- and vimentin-related sequences detected are for the most part more related to myosins and kinesins, respectively. These latter homologies may arise from similarities among sequences that form coiled coil regions of protein structure. A human query sequence for desmin did not detect any related sequences in the Arabidopsis genome.
Neurofilaments:
Type IV IF proteins include the light, medium, and heavy neurofilament proteins. Neurofilaments are found primarily in neuronal animal cells. These proteins align in parallel to form long heteropolymeric structures (Kreis and Vale, 1999). The N-terminal half of the neurofilament protein is related in structure to vimentin starting with a short head sequence and a rod domain made from coiled coil sequences. This sequence is followed in all three neurofilament proteins with highly charged glutamic-acid rich sequences. For medium and heavy neurofilament proteins, this glutamic-acid rich region is interrupted by KSP (lys-ser-pro) or KEP (lys-glu-pro) repeats. A human query sequence for a light neurofilament identified only one Arabidopsis sequence with 40% similarity over 320 residues of the 472 a.a. query, but it had a very poor E-value (0.004). A human medium neurofilament of 991 a.a. detected 13 plant sequences with good E-values (2e-11: 0.001), with the most related sequence showing 400 a.a. aligning (Table 3). A human heavy neurofilment query identified several sequences with a higher level of relatedness aligning over 540 a.a. to the 879 a.a. query and having reasonable E-values (5e-11: 0.001). Most of the similarities to the plant proteins are with the potential filamentous coiled coil domain and long glutamic rich sequences of the neurofilaments. None of the Arabidopsis sequences contained the KSP or KEP repeats. Thus, while there may be sequences related to the neurofilaments in plants, further physical proof will be needed to establish their presence and role in the cytoskeleton. As mentioned previously for the ezrin (ERM) family of ABPs, there are large numbers of large unidentified glutamic-acid rich proteins in the Arabidopsis database that are probably structural proteins.
Nuclear lamins:
The nuclear lamins are classified as Type V IFs and have been found in diverse species in the animal kingdom. The lamins are generally found at the nuclear periphery in close association with chromatin, but occasionally in structures within the nucleus. They are thought to play roles in nuclear envelope and chromatin structure. The lamin proteins have an N-terminal, α-helical rod domain flanked by multiple phosphorylation sites. This is followed by a central nuclear localization signal (NLS), a poorly conserved sequence of 100 a.a., and a C-terminal isoprenylation signal. Using human A/C and human B2 lamins as query sequences, several potential Arabidopsis proteins were found with moderate similarity extending for nearly 50% of the 664 a.a. and 417 a.a. query sequences. While some of the sequences of human nuclear lamin A/C are aligning with the helical domains of kinesins and myosins, other plant sequences are listed as unknown or hypothetical proteins. The alignment with both classes of animal lamin is predominantly with the α-helical rod domain, while some of plant sequences also contain a clear homologue of the NLS.
Nestins:
The only known type VI IF protein is nestin. Nestins are approximately 230 kDa proteins found in neuronal cells and thought to have evolved from the neurofilaments. They have an N-terminal α-helical rod domain typical of most IFs, and this is followed by numerous repeats of an 11-amino acid motif. Using a human nestin query, a few significantly related hypothetical proteins are found in Arabidopsis, but most are related through the glutamic rich filamentous region and none have any conservation to the 11-mer nestin protein repeat.
Discussion
The Complexity of the Arabidopsis Cytoskeletal Genome
An initial estimate for the size of the cytoskeletal genome can be made from the minimum of 400 structural genes encoding likely cytoskeletal proteins identified in Tables 1, 2, and 3. The Arabidopsis actin system is comprised of 16 actin and actin-related sequences and approximately another 150 putative ABPs. Another potential 150 ABP were detected that showed weaker homology to query sequences and whose identity is therefore more difficult to certify. The microtubule system is composed of 17 to 19 tubulin sequences and another 220 potential MAPs. Approximately 10 to 40 potential IF protein sequences were identified. A reasonable estimate for the number of genes encoding cytoskeletal structural proteins from these three groups is between 400 and 500. Thus, it can be estimated that approximately 2% of the ∼26,000 genes in Arabidopsis encode cytoskeletal structural proteins or proteins linked directly to them. Considering that many poorly conserved ABPs, MAP, and IF proteins would not be detected by sequence homology, it is possible that the actual number of cytoskeletal genes is even greater. A large percentage of these cytoskeletal genes may encode mRNAs with multiple splice variants, as they do in animals (e.g., fimbrins, ARPs), and many proteins will be secondarily modified by mechanisms such as phosphorylation, methylation, and isoprenylation (Garrels et al., 1997; Brett et al., 2000). Thus, the Arabidopsis cytoskeletal proteome could easily exceed 1000 different proteins.
Future of Plant Cytoskeletal Research
Research on the plant cytoskeleton has been strengthened in the last two decades by combining cell biological approaches with molecular genetic and biochemical tools. Expression of individual cytoskeletal genes is highly regulated both spatially and temporally. The diverse members in a family may distinguish themselves with complementary patterns of expression in reproductive or vegetative tissues, as is the case for most actins, ADFs, profilins, and tubulins.
New immune reagents, GFP-fusion proteins, improved fixation protocols, and microscope technology allow the plant cytoskeleton to be examined in increasingly beautiful detail and suggest a coming renaissance of our understanding of cytoskeletal structure and function. Results with protein family member-specific antisera demonstrate that protein isovariants are easily distinguished in their expression. For example, isovariant-specific antibodies were used to show that the maize ADF protein family is split between vegetative and reproductive expression (Lopez et al., 1996); that the Arabidopsis actin ACT7 protein responds to auxin stimulation (McDowell et al., 1996a; Kandasamy et al., 2001); and that expression patterns of the late pollen-specific actins can be extrapolated to other distant plant species (Kandasamy et al., 1999). Antibodies have another advantage in that they provide the most detailed resolution of cytoskeletal structures. For example, an unconventional Arabidopisis myosin was shown to be positioned in the post-cytokinetic cell wall (Reichelt et al., 1999) and the ectopic expression of an actin isovariant was shown to create F-actin sheets in place of F-actin bundles in leaf cells (Kandasamy et al., 2002a). The diversity of sequences in cytoskeletal gene families and their great divergence from vertebrate sources favors the isolation of plant isovariant-specific antibodies (Kandasamy et al., 1999). The disadvantage of antibody detection is that chemical fixation protocols often damage the cytoskeleton. The plant actin cytoskeleton is particularly sensitive to proteolytic degradation during prolonged fixation. Transgenic plants expressing fluorescent protein tagged cytoskeletal proteins offer the advantage of easy detection in living tissues. GFP-tagged actin-binding proteins such as GFP-talin can be used to follow F-actin filament rearrangement with excellent precision during cell growth and development (Kost et al., 1998).
Mutational studies are essential to dissecting cytoskeletal gene and protein function and often give surprising results by way of novel phenotype or no observable phenotype for what might be presumed an essential gene (Martienssen and Irish, 1999; Bouche and Bouchez, 2001). Sequence-based screening protocols and the availability of large T-DNA libraries (McKinney et al., 1995)(www.biotech.wisc.edu/Arabidopsis/) and trans-poson libraries with sequence information on flanking regions (www.nadii.com/) have greatly accelerated the acquisition of tagged Arabidopsis mutants. While it is clear that T-DNA mutations in Arabidopsis actins have serious deleterious effects on plant survival (Asmussen et al., 1998; Gilliland et al., 1998), the cell, developmental, and conditional phenotypes of individual mutants need to be described to fully understand actin cytoskeletal function. However, the disruption of single isovariants in a cytosketal gene family often cause subtle phenotypes (McKinney et al., 2001; Gilliland et al., 2002a; Gilliland et al., 2002b), suggesting we have not yet understood the selective pressures that may preserve individual gene family members. Many more mutants and multiple mutant alleles are needed to sort out the functions of the individual isovariants. This demand for disrupting gene and protein function may be more easily met by newer approaches for suppressing RNA and protein expression such as interference RNA, RNAi (Chuang and Meyerowitz, 2000), a method of post-transcriptional gene silencing (PTGS). Stem-loop RNA structures containing sense and antisense RNA for target gene transcripts were used to disrupt four different genes controlling floral or meristem development. RNAi was efficient enough at PTGS to produce a phenotype in greater than 85% of the more than 100 transgenic plants examined for each of four target genes. Resistance to pharmacologic agents that act on the cytoskeleton has not been exploited to its fullest to isolate plant mutants, although natural biotypes of a weedy grass have been identified with resistance to dinitroaniline herbicides encoded by a missense allele of α-tubulin (Yamamoto et al., 1998).
A deeper understanding of the function and dynamics of the various actin cytoskeletal processes should be aided by some additional tools that have recently been developed. Transcriptional profiling of the plant genomes (Schena et al., 1995; Schena, 1996; Xu et al., 2001) should reveal if hundreds of other cytoskeletal genes follow the well-described vegetative and reproductive expression patterns common for the first characterized cytoskeletal gene families (Meagher et al., 1999b). Further, such approaches will provide detailed catalogs of changes in gene expression accompanying cytoskeletal refashioning associated with morphogenesis, response to growth hormones, and stress respones. A fuller understanding of the physical biochemistry of cytoskeletal processes and the distinctions between isovariants depends upon development of cell-free systems for examining plant F-actin and MT polymers and their interacting proteins. These in vitro systems have been commonly used to examine the physical chemical properties of the animal, yeast, and protist cytoskeleton. A few genetic tricks provide a dissection of basic protein functions and can distinguish functions among isovariants. Yeast genetics has been particularly useful in defining minimal protein functions for some cytoskeletal proteins. Arabidopsis profilins complement the loss of viability of yeast profilin mutants (Christensen et al., 1996). Cells of fission the yeast S. pombe have a distinct rod shaped morphology and an easily stained cytoskeleton. Changes in S. pombe morphology have made it an ideal organism in which to screen for the over-expression of Arabidopsis cDNAs encoding cytoskeletal proteins (Xia et al., 1996). Plant genetics should soon be a more powerful tool as the relative availability of Arabidopsis cytoskeletal mutants is put to use. An assay which follows nuclear positioning in the Tradescantia stamen hairs after microinjection of purified proteins (Staiger et al., 1994; Gibbon et al., 1997) can be used to measure association constants and kinetic parameters for individual protein isovariants. The ectopic expression of animal isovariants has been very informative in animal systems (Fyrberg et al., 1998; Hart and Cooper, 1999) and should prove a powerful tool in plants. In the case of Arabidopsis actin the results are equally striking. Ectopic expression of reproductive actin in vegetative Arabidopsis tissues produced extremely dwarf and abnormal plant phenotypes (Kandasamy et al., 2002a), far more extreme than observed by over expressing vegetative actin or observed with any T-DNA insertions inactivating gene expression. Ectopic expression studies in plants will be aided by the relative ease with which Arabidopsis can be transformed with novel gene constructs and regenerated. Defining function for these many newly acquired structural protein sequences will ultimately require the networking of diverse scientific and technical approaches and putting information into an evolutionary context (Meagher, 2002).
Acknowledgments
We would like to thank Elizabeth McKinney for her unpublished data on the ARPs and myosins, Gay Gragson for editing the manuscript, and Chris Somerville for several helpful suggestions. This research was supported by funds from National Institutes of Health (GM 36397-14) and from the National Science Foundation (MCB 9808748).
Footnotes
Citation: Meagher R.B., and Fechheimer M. (2003) The Arabidopsis Cytoskeletal Genome. The Arabidopsis Book 2:e0096. doi:10.1199/tab.0096
elocation-id: e0096
Published on: September 30, 2003
Table 1.
Continued.
REFERENCES
- Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;102(1)(1):17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;2152(3)(1):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amos LA. Focusing-in on microtubules. Curr Opin Struct Biol. 2000;102(2)(1):236–41. doi: 10.1016/s0959-440x(00)00070-1. [DOI] [PubMed] [Google Scholar]
- An Y-Q, Huang S, McDowell JM, McKinney EC, Meagher RB. Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell. 1996;82(1):15–30. doi: 10.1105/tpc.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;102(1):107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Aon MA, Cortassa S, Gomez Casati DF, Iglesias AA. Effects of stress on cellular infrastructure and metabolic organization in plant cells. Int Rev Cytol. 2000;1942(1):239–73. doi: 10.1016/s0074-7696(08)62398-0. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;4082(6814)(1):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Asmussen MA, Gilliland LU, Meagher RB. Detection of deleterious genotypes in multigenerational studies. II. Theoretical and experimental dynamics with selfing and selection. Genetics. 1998;1492(1):727–737. doi: 10.1093/genetics/149.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles JR. Parallel actin bundles and their multiple actin-bundling proteins. Curr Opin Cell Biol. 2000;122(1)(1):72–8. doi: 10.1016/s0955-0674(99)00059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, Wierda A, Chen B. Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol. 1998;1432(1)(1):107–19. doi: 10.1083/jcb.143.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;122(4)(1):780–94. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet I, Boujemaa R, Carlier MF, Preat T. Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell. 2000;1022(6)(1):797–808. doi: 10.1016/s0092-8674(00)00068-4. [DOI] [PubMed] [Google Scholar]
- Bouche N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol. 2001;42(2)(1):111–7. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Bouget F-Y, Gerttula S, Shaw SL, Quatrano RS. Localization of actin mRNA during the establishment of cell polarity and early cell divisions in Fucus embryos. Plant Cell. 1996;82(1):189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Letters. 2000;4742(1):83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;4742(1)(1):83–6. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility [see comments]. J Cell Biol. 1997;1362(6)(1):1307–22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter JL, Ploense S, Snustad DP, Silflow CD. Preferential expression of an a-tubulin gene of Arabidopsis in pollen. Plant Cell. 1992;42(1):557–571. doi: 10.1105/tpc.4.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen HE, Ramachandran S, Tan CT, Surana U, Dong CH, Chua NH. Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 1996;102(2)(1):269–79. doi: 10.1046/j.1365-313x.1996.10020269.x. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;972(9)(1):4985–90. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL, Smith LG. The tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell. 1998;102(11)(1):1875–1888. doi: 10.1105/tpc.10.11.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol. 2000;122(1)(1):97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- Cronquist A. An integrated system of classification of flowering plants., ed., Columbia Unversity Press: New York. 1262. 1981.
- Davies E, Fillingham BD, Oto Y, Abe S. Evidence for the existence of cytoskeleton-bound polysomes in plants. Cell Biol Int Rep. 1991;152(10)(1):973–81. doi: 10.1016/0309-1651(91)90147-b. [DOI] [PubMed] [Google Scholar]
- Dawson PJ, Hulme JS, Lloyd CW. Monoclonal antibody to intermediate filament antigen crossreacts with higher plant cells. J. Cell Biol. 1985;1002(1):1793–1798. doi: 10.1083/jcb.100.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Surridge C. Arabidopsis thaliana genome. Introduction. Nature. 2000;4082(6814)(1):791. doi: 10.1038/35048677. [DOI] [PubMed] [Google Scholar]
- Didry D, Carlier MF, Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem. 1998;2732(40)(1):25602–11. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Dikovskaya D, Zumbrunn J, Penman GA, Nathke IS. The adenomatous polyposis coli protein: in the limelight out at the edge. Trends Cell Biol. 2001;112(9)(1):378–84. doi: 10.1016/s0962-8924(01)02069-4. [DOI] [PubMed] [Google Scholar]
- Dong CH, Kost B, Xia G, Chua NH. Molecular identification and characterization of the Arabidopsis AtADF1, AtADFS and AtADF6 genes. Plant Mol Biol. 2001;452(5)(1):517–27. doi: 10.1023/a:1010687911374. [DOI] [PubMed] [Google Scholar]
- Edmonds BT, Bell A, Wyckoff J, Condeelis J, Leyh TS. The effect of F-actin on the binding and hydrolysis of guanine nucleotide by Dictyostelium elongation factor 1A. J Biol Chem. 1998;2732(17)(1):10288–95. doi: 10.1074/jbc.273.17.10288. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J Cell Biol. 1996;1352(1)(1):5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Schopfer P. Physical strain-mediated microtubule reorientation in the epidermis of gravitropically or phototropically stimulated maize coleoptiles. Plant J. 1998;152(1)(1):119–23. doi: 10.1046/j.1365-313x.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- Furukawa R, Jinks TM, Tishgarten T, Mazzawi M, Morris DR, Fechheimer M. Elongation factor 1beta is an actin-binding protein. Biochim Biophys Acta. 2001;15272(3)(1):130–40. doi: 10.1016/s0304-4165(01)00157-x. [DOI] [PubMed] [Google Scholar]
- Fyrberg EA, Fyrberg CC, Biggs JR, Saville D, Beall CJ, Ketchum A. Functional nonequivalence of Drosophila actin isoforms. Biochem Genet. 1998;362(7–8)(1):271–87. doi: 10.1023/a:1018785127079. [DOI] [PubMed] [Google Scholar]
- Gallagher K, Smith LG. Asymmetric cell division and cell fate in plants. Curr. Opin. Cell. Biol. 1997;92(6)(1):842–848. doi: 10.1016/s0955-0674(97)80086-5. [DOI] [PubMed] [Google Scholar]
- Garrels JI, McLaughlin CS, Warner JR, Futcher B, Latter GI, Kobayashi R, Schwender B, Volpe T, Anderson DS, Mesquita-Fuentes R, Payne WE. Proteome studies of Saccharomyces cerevisiae: identification and characterization of abundant proteins. Electrophoresis. 1997;182(8)(1):1347–60. doi: 10.1002/elps.1150180810. [DOI] [PubMed] [Google Scholar]
- Gerstein M. Measurement of the effectiveness of transitive sequence comparison, through a third ‘intermediate’ sequence. Bioinformatics. 1998;142(8)(1):707–14. doi: 10.1093/bioinformatics/14.8.707. [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Ren H, Staiger CJ. Characterization of maize (Zea mays) pollen profilin function in vitro and in live cells. Biochem J. 1997;3272(1):909–15. doi: 10.1042/bj3270909. (Pt 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Staiger CJ. Chapter 4. 2000;2(1):45–65. Profilin in CJ Staiger, et al., Actin: A dynamic framework for multiple plant cell functions, Klewer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- Gibbon BC, Zonia LE, Kovar DR, Hussey PJ, Staiger CJ. Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell. 1998;102(1):981–993. doi: 10.1105/tpc.10.6.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidrol X, Sabelli PA, Fern YS, Kush AK. Annexin-like protein from Arabidopsis thaliana rescues delta oxyR mutant of Escherichia coli from H2O2 stress. Proc Natl Acad Sci USA. 1996;932(20)(1):11268–73. doi: 10.1073/pnas.93.20.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland LU, Kandasamy MK, Meagher RB. Loss of the predominant Arabidopsis thaliana actin, ACT2, disrupts root hair development. (in prep)
- Gilliland LU, McKinney EC, Asmussen MA, Meagher RB. Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics. 1998;1492(2)(1):717–25. doi: 10.1093/genetics/149.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland LU, Pawloski L, Kandasamy MK, Meagher RB. The Arabidopsis actin isovariant, ACT7, plays a vital role in germintion and root growth. (in prep) [DOI] [PubMed]
- Goldsmith SC, Pokala N, Shen W, Fedorov AA, Matsudaira P, Almo SC. The structure of an actin-crosslinking domain from human fimbrin. Nat Struct Biol. 1997;42(9)(1):708–12. doi: 10.1038/nsb0997-708. [DOI] [PubMed] [Google Scholar]
- Gramolini AO, Belanger G, Jasmin BJ. Distinct regions in the 3′ untranslated region are responsible for targeting and stabilizing utrophin transcripts in skeletal muscle cells. J Cell Biol. 2001;102(1):10. doi: 10.1083/jcb.200101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Volkmann N, Goldsmith S, Michon AM, Lehman W, Craig R, DeRosier D, Almo S, Matsudaira P. An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat Struct Biol. 1998;52(9)(1):787–92. doi: 10.1038/1828. [DOI] [PubMed] [Google Scholar]
- Hart MC, Cooper JA. Vertebrate isoforms of actin capping protein b have distinct functions in vivo. J. Cell Biol. 1999;1472(1):1287–1298. doi: 10.1083/jcb.147.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Mizutani A, Chida M, Hidaka H, Mizutani J. Plant annexin form homodimer during Ca(2+)-dependent liposome aggregation. Biochem Mol Biol Int. 1995;352(4)(1):749–55. [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT4/ACT12 actin gene subclass is strongly expressed in post-mitotic pollen. Plant J. 1996;102(1):189–202. doi: 10.1046/j.1365-313x.1996.10020189.x. [DOI] [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen and developing ovules. Plant Mol. Biol. 1997;332(1):125–139. doi: 10.1023/a:1005741514764. [DOI] [PubMed] [Google Scholar]
- Huang S, M J.M., Weise MJ, Meagher RB. The Arabidopsis profilin gene family: Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 1996;1112(1):115–126. doi: 10.1104/pp.111.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CJ, Weeds AG, Hussey PJ. The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 1997;122(5)(1):1035–1043. doi: 10.1046/j.1365-313x.1997.12051035.x. [DOI] [PubMed] [Google Scholar]
- Jin S, Xu R, Wei Y, Goodwin PH. Increased expression of a plant actin gene during a biotrophic interaction between round-leaved mallow, Malva pusilla, and Colletotrichum gloeosporioides f. sp. malvae. Planta. 1999;2092(4)(1):487–494. doi: 10.1007/s004250050752. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Gilliland L, McKinney E, Meagher RB. Expression of phytohormonally regulated actin subclass in Arabidopsis and tobacco tissue culture cells. Plant Cell. 2001. (in press)
- Kandasamy MK, Gilliland LU, McKinney EC, Meagher RB. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell. 2001;132(7)(1):1541–54. doi: 10.1105/TPC.010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney E, Meagher RB. The late pollen specific actins in angiosperms. Plant J. 1999;182(1):681–691. doi: 10.1046/j.1365-313x.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB. Functional non-equivalency of actin isovariants in Arabidopsis. Mol Biol Cell. 2001. in press. [DOI] [PMC free article] [PubMed]
- Kandasamy MK, McKinney EC, Meagher RB. Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil Cytoskelton. submitted. [DOI] [PubMed]
- Kinkema K, Schiefelbein J. A myosin from a higher-plant has structural similarities to class-U myosins. J. Mol. Biol. 1994;239(4) doi: 10.1006/jmbi.1994.1400. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Wang HY, Schiefelbein J. Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant Mol. Biol. 1994;262(4)(1):1139–1153. doi: 10.1007/BF00040695. [DOI] [PubMed] [Google Scholar]
- Klahre U, Chua NH. The Arabidopsis actin-related protein 2 (AtARP2) promoter directs expression in xylem precursor cells and pollen. Plant Mol Biol. 1999;412(1)(1):65–73. doi: 10.1023/a:1006247600932. [DOI] [PubMed] [Google Scholar]
- Klahre U, Friederich E, Kost B, Louvard D, Chua NH. Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 2000;1222(1)(1):35–48. doi: 10.1104/pp.122.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyachko N, Aksenova L, Dunaeva M, Kulikova A. Interaction of plant polysomes with the actin cytoskeleton. Cell Biol Int. 2000;242(6)(1):351–8. doi: 10.1006/cbir.1999.0517. [DOI] [PubMed] [Google Scholar]
- Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;532(2)(1):219–26. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis contains at least six expressed alpha-tubulin genes. Plant Cell. 1992;42(5)(1):539–47. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;2872(5461)(1):2257–9. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;162(3)(1):393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kovar D, Staiger C, Weaver E, McCurdy D. AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 2000;242(5)(1):625–36. doi: 10.1046/j.1365-313x.2000.00907.x. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Drobak BK, Staiger CJ. Maize profilin isoforms are functionally distinct. Plant Cell. 2000;122(4)(1):583–98. doi: 10.1105/tpc.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis T, Vale R. Guidebook to the cytoskeletal and motor proteins, ed. 1999. A Sambrook and Tooze Publicaton at Oxford University Press: New York. 551.
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;22(5)(1):362–3. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. gamma-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. Plant Cell. 1994;62(2)(1):303–14. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez I, Anthony RG, Maciver SK, Jiang CJ, Khan S, Weeds AG, Hussey PJ. Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc. Natl. Acad. Sci., U.S.A. 1996;932(14)(1):7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R, Irish V. Copying out our ABCs: the role of gene redundancy in interpreting genetic hierarchies. Trends Genet. 1999;152(11)(1):435–7. doi: 10.1016/s0168-9525(99)01833-8. [DOI] [PubMed] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res. 1997;2322(1)(1):173–81. doi: 10.1006/excr.1997.3531. [DOI] [PubMed] [Google Scholar]
- McCurdy DW, Kim M. Molecular cloning of a novel fimbrin-like cDNA from Arabidopsis thaliana. Plant Mol Biol. 1998;362(1)(1):23–31. doi: 10.1023/a:1005884112192. [DOI] [PubMed] [Google Scholar]
- McDowell J, An Y-Q, McKinney EC, Huang S, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 1996;1112(1):699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An Y-Q, Meagher RB. Structure and Evolution of the Actin Gene Family inArabidopsis thaliana. Genetics. 1996;1422(1):587–602. doi: 10.1093/genetics/142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2-1 and act4-1. Plant J. 1995;82(4)(1):613–22. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB. Small changes in the regulation of one Arabidopsis profilin isovariant, prf1, alter seedling development. Plant Cell. 2001;132(5)(1):1179–91. doi: 10.1105/tpc.13.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB. The diverse plant actin-related proteins encoded by complex and differentially regulated genes. Genome Research. submitted.
- McKinney EC, Meagher RB. Members of the Arabidopsis actin gene family are widely dispersed in the genome. Genetics. 1998;1492(1):663–675. doi: 10.1093/genetics/149.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty AK, Saunders MJ. Purification and immunological detection of pea nuclear intermediate filaments: evidence for plant nuclear lamins. J Cell Sci. 1992;1032(1):407–414. doi: 10.1242/jcs.103.2.407. [DOI] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Vitale AV. The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet. 1999;152(7)(1):278–84. doi: 10.1016/s0168-9525(99)01759-x. [DOI] [PubMed] [Google Scholar]
- Meagher RB, Williamson RE. 1994. The Plant Cytoskeleton in E Meyerowitz and C Somerville, Arabidopsis, Cold Spring Harbor Laboratory Press, Cold Spring Harbor. 1049–1084.
- Mermall V, Post PL, Mooseker MS. Un-conventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;2792(5350)(1):527–33. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Plant development: local control, global patterning. Curr Opin Genet Dev. 1996;62(4)(1):475–479. doi: 10.1016/s0959-437x(96)80070-0. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;882(3)(1):299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Compare and contrast actin filaments and microtubules. Mol Biol Cell. 1992;32(12)(1):1309–15. doi: 10.1091/mbc.3.12.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RO, Pilar Fernandez M. Distinct annexin subfamilies in plants and protists diverged prior to animal annexins and from a common ancestor. J Mol Evol. 1997;442(2)(1):178–88. doi: 10.1007/pl00006134. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA. 1997;942(12)(1):6261–6. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol. 2000;1322(1):185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;962(4)(1):517–27. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 2000;1242(4)(1):1637–47. doi: 10.1104/pp.124.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaekers FC, Benedetti EL, Dunia I, Vorstenbosch P, Bloemendal H. Polyribosomes associated with microfilaments in cultured lens cells. Biochim Biophys Acta. 1983;7402(4)(1):441–8. doi: 10.1016/0167-4781(83)90093-3. [DOI] [PubMed] [Google Scholar]
- Reddy A. Molecular motors and their functions in plants. Int Rev Cytol. 2001;2042(1):97–178. doi: 10.1016/s0074-7696(01)04004-9. [DOI] [PubMed] [Google Scholar]
- Reddy A, Day I. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001;2(7) doi: 10.1186/gb-2001-2-7-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Day I. Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genomics. 2001;22(1)(1):2. doi: 10.1186/1471-2164-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J. Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 1999;192(5)(1):555–567. doi: 10.1046/j.1365-313x.1999.00553.x. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Egile C, Pantaloni D, Carlier MF. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J Biol Chem. 1999;2742(30)(1):20970–6. doi: 10.1074/jbc.274.30.20970. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;2732(33)(1):20894–902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- Roman G, Ecker JR. Genetic analysis of a seedling stress response to ethylene in Arabidopsis. Philos Trans R Soc Lond B Biol Sci. 1995;3502(1331)(1):75–81. doi: 10.1098/rstb.1995.0140. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Schroer TA. Actin-related proteins. Annu Rev Cell Dev Biol. 1999;152(1):341–363. doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- Schena M. Genome analysis with gene expression microarrays. BioEssays. 1996;182(5)(1):427–431. doi: 10.1002/bies.950180513. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quatitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;2702(1):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr Opin Cell Biol. 2001;132(1)(1):92–6. doi: 10.1016/s0955-0674(00)00179-4. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Fairbairn DJ, Lloyd CW. Cytoplasmic and nuclear intermediate filament antigens in higher plants in CW Lloyd. 1991. The cytoskeletal basis of plant growth and form, Academic, London. 69–81.
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. Plant Cell. 1992;42(5)(1):549–56. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy ASN, Endow SA. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA. 1997;942(1)(1):322–327. doi: 10.1073/pnas.94.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ. Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51 doi: 10.1146/annurev.arplant.51.1.257. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Gibbon BC, Kovar DR, Zonia LE. Profilin and actin-depolymerizing factor: Modulators of actin organization in plants. Trends Plant Sci. 1997;22(1):275–281. [Google Scholar]
- Staiger CJ, Goodbody KC, Hussey PJ, Valenta R, Drøbak BK, Lloyd CW. The profilin multigene family of maize: Differential expression of three isoforms. Plant J. 1993;42(1):631–641. doi: 10.1046/j.1365-313x.1993.04040631.x. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Lloyd CW. The plant cytoskeleton. Curr. Opin. Cell Biol. 1991;32(1):33–42. doi: 10.1016/0955-0674(91)90163-s. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Yuan M, Valenta R, Shaw PJ, Warn RM, Lloyd CW. Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr Biol. 1994;42(3)(1):215–9. doi: 10.1016/s0960-9822(00)00050-6. [DOI] [PubMed] [Google Scholar]
- Sun Y, Carneiro N, Clore AM, Moro GL, Habben JE, Larkins BA. Characterization of maize elongation factor 1A and its relationship to protein quality in the endosperm. Plant Physiol. 1997;1152(3)(1):1101–7. doi: 10.1104/pp.115.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;1492(4)(1):761–6. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Yokota E, Vidali L, Sonobe S, Hepler PK, Shimmen T. The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta. 2000;2102(5)(1):836–43. doi: 10.1007/s004250050687. [DOI] [PubMed] [Google Scholar]
- Valster AH, Hepler PK, Chernoff J. Plant GTPases: the Rhos in bloom. Trends Cell Biol. 2000;102(4)(1):141–6. doi: 10.1016/s0962-8924(00)01728-1. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos L, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;4132(1):39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J, Van Damme J, Vancompernolle K, Bubb MR, Lambooy PK, Korn ED. The covalent structure of Acanthamoeba actobindin. J Biol Chem. 1990;2652(22)(1):12801–5. [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK. Actin polymerization is essential for pollen tube growth. Mol Biol Cell. 2001;122(8)(1):2534–45. doi: 10.1091/mbc.12.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick S. Plant microtubules meet their MAPs and mimics. Nat Cell Biol. 2000;22(11)(1):E204–6. doi: 10.1038/35041129. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC. Identification of sucrose synthase as an actin-binding protein. FEBS Lett. 1998;4302(3)(1):205–8. doi: 10.1016/s0014-5793(98)00659-0. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol. 2000;352(4)(1):253–89. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Wu G, Li H, Yang Z. Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol. 2000;1242(4)(1):1625–36. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Ramachandran S, Hong Y, Chan YS, Simanis V, Chua NH. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 1996;102(4)(1):761–9. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Bak S, Decker A, Paquette SM, Feyereisen R, Galbraith DW. Microarray-based analysis of gene expression in very large gene families: the cytochrome P-450 gene superfamily of Arabidopsis thaliana. Gene. 2001;2722(1–2)(1):61–74. doi: 10.1016/s0378-1119(01)00516-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Zeng L, Baird WV. Alpha-tubulin missense mutations correlate with antimicrotubule drug resistance in Eleusine indica. Plant Cell. 1998;102(2)(1):297–308. doi: 10.1105/tpc.10.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hamada S, Kashiyama T. Myosins from plants. CMLS. 1999;562(1):227–232. doi: 10.1007/s000180050424. [DOI] [PMC free article] [PubMed] [Google Scholar]


