INTRODUCTION
Phosphorus (P) is an essential macronutrient for all living organisms. It serves various basic biological functions as a structural element in nucleic acids and phospholipids, in energy metabolism, in the activation of metabolic intermediates, as a component in signal transduction cascades, and the regulation of enzymes.
Of the major nutrients, P is the most dilute and the least mobile in soil. High sorbing capacity for P in the soil (e.g. sorbtion to metal oxides), P mineralization (e.g. calcium phosphates such as apatite), and/or fixation of P in organic soil matter (by converting soluble P into organic molecules) result in low availability of this macronutrient for uptake into plants (Marschner, 1995). P is absorbed by plants as orthophosphate (Pi, inorganic phosphate). Pi concentration in the soil solution hardly reaches 10 µM and may even drop to submicromolar levels at the root/soil interface, where Pi uptake by plants and root surface-colonizing microorganisms leads to the generation of a zone of Pi depletion around the root cylinder that is maintained due to slow diffusion of Pi from regions distant to the root surface (Figure 1).
Fig. 1.

A transverse section through the tip of a primary root. The dotted line indicates the outer border of the P depletion zone. The arrow indicates the direction of growth.
In industrialized countries, low P availability in agricultural soils is compensated by a high input of P fertilizer to guarantee high crop productivity and yield. Water run-off, soil erosion and leakage in highly fertilized agricultural soils may cause environmental problems such as eutrophication of lakes and rivers. As forecasted by Tilman et al. (2001), during the next 50 years, which is likely to be the final period of rapid agricultural expansion, demand for food by global population will be a major driver of global environmental change. Conversion of natural ecosystems to agriculture by 2050 will be accompanied by an approximate 2.5-fold increase in nitrogen- and P- driven eutrophication of terrestrial, freshwater, and near-shore marine ecosystems. Modern agricultural soils are almost universally maintained at high fertilization. Selection of new cultivars is usually made under such conditions and will not normally distinguish between plants varying in nutrient efficiency (Stevens and Rick, 1986). To alleviate the forecasted adverse negative effects of agricultural expansion, scientists have started to use classical breeding strategies and biotechnology to improve crop plants, based on the current knowledge and aiming at an improved crop yield with a lower input of fertilizer, thus protecting the environment.
In contrast, in many developing tropical countries, subsistence farmers can not buy enough fertilizer due to limited financial capacities or poor infrastructure (Sanchez et al., 1997). As a consequence, P deprivation dramatically limits crop yield and is one of the reasons for poverty and malnutrition. In the future, agriculture from both developed as well as developing countries could thus benefit from modern crop varieties with enhanced P efficiency, thus leading to improved fertilizer management and increased crop yield on low-P soils. Thorough knowledge of the plant's response to P deprivation stress will contribute to the rational and targeted breeding of P efficient crop plants. Therefore, the authors of this chapter focus on summarizing the current state of research covering physiology, biochemistry, and molecular genetics of P acquisition and allocation, and P homeostasis within the plant. Although this review will mainly focus on knowledge acquired on Arabidopsis thaliana, some specific results obtained with other plant species will also be included in this work. For example, formation of mycorrhizae, which is observed in most vascular plants and strongly contributes to plant P nutrition, does not occur in Brassicaceae and therefore Arabidopsis is not a suitable model for mycorrhizae studies.
PHOSPHATE TRANSPORT ACROSS MEMBRANES
The textbook tells us that the development of a barrier such as the cell membrane was a prerequisite for the development of life, and enabled single cells to support metabolic, reproductive, and developmental activities under stable physico-chemical conditions (Buchanan et al., 2000). Much later during the evolution of life, the development of an endomembrane system in eukaryotic cells allowed the formation of organelles, each with its own chemical ‘inner world’ leading to a compartmentalization of solutes within the cell. It is the hydrophobic nature of the lipid bilayer that allows sequestration of hydrophilic compounds, such as most nutrients and metabolites, on either side of the membrane. This generation of different (bio)chemical environments enabled cells to establish a division and separation of biosynthetic, catabolic and storage functions, allowing metabolic flexibility and efficiency. Integration of these functions in the network of the cell and of the entire organism needs a selective transport of compounds across the membrane barriers. This selective permeability is mediated by membrane-spanning proteins within the lipid bilayer. These so-called transport or carrier systems are often highly specific for a certain compound (e.g. a carbohydrate such as sucrose) or a group of similar substances (e.g. divalent cations).
It was the pioneering work of Emmanuel Epstein (Epstein and Hagen, 1952; Epstein et al., 1963) which demonstrated that ion uptake processes across the plasma membrane follow Michaelis-Menten kinetics comparable to those of enzymatic processes known from biochemistry, thus allowing calculations of functional parameters such as pH optimum, Michaelis constant Km, uptake velocity Vmax, and minimal concentration of the ion at which transport occurs, i.e. Cmin, Analysis of nutrient uptake kinetics into plant roots using a radiotracer medium-depletion method (Drew et al., 1984) revealed that Pi uptake kinetics in plants are generally hyperbolic and monophasic at low Pi concentrations (µM range) in the medium and biphasic when extended to high Pi concentrations (mM range), thus suggesting Pi uptake being mediated by high- and low-affinity transport mechanisms, respectively (Schachtman et al., 1998). However, in suspension-cultured tobacco cells, only one kind of Michaelis-Menten-type Pi transport system exhibiting a high-affinity for Pi has been described with no evidence for a low-affinity Pi transport (Shimogawara and Usuda, 1995). Moreover, concentration-dependent Pi influx of barley mesophyll protoplasts shows a combination of one Michaelis-Menten-type kinetics at low Pi concentrations and linear increase at higher Pi concentrations (Mimura et al., 1990). At Pi concentrations in the µM range, which corresponds to the natural conditions in cultivated soils, high-affinity transport occurs (Cogliatti and Clarkson, 1983; Drew et al., 1984). As there are micro- or even submicromolar concentrations of available Pi at the root-soil interface, but millimolar concentrations of intracellular Pi, Pi uptake across the cell boundary must be effected against a steep concentration gradient. This is accomplished by transport of the anion across the membrane coupled to the transport of protons (H+-symport). Plants and fungi use a proton P-type ATPase pump to generate an electrochemical gradient across the plasma membrane at the expense of ATP (Figure 2). This leads to the formation of a large membrane potential with a negative potential of the cytoplasm (−150 to −200 mV). Consequently, the transport of Pi and other anions is usually coupled to protons in a secondary transport process. Thus, the driving force for Pi influx is the proton gradient generated by the P-type H+-ATPase (Schachtman et al., 1998; Sze et al., 1999; Thibaud et al., 1988; Ullrich-Eberius et al., 1984).
Fig. 2.
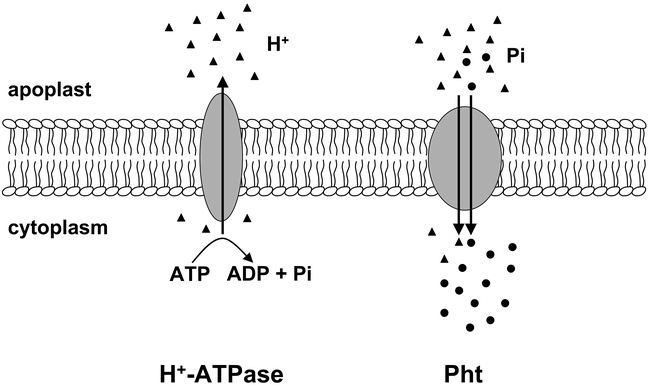
A model for secondary Pi transport across the plasma membrane. The H+ (triangles) gradient across the lipid bilayer is generated by the activity of the H+-ATPase at the expense of ATP. H+/Pi (circles) co-transport is mediated by the Pi transporter protein (Pht). The orientation of the transport is marked by arrows. The orientation of the plasma membrane is as indicated (apoplast, cytoplasm)
Pi is transported from the root to the shoot via the xylem, which consists of dead cells and thus reflects the extracellular space. During leaf senescence or Pi starvation conditions, Pi is redistributed from old source leaves to young sink organs or to the starved root. Transport steps involved in Pi acquisition and distribution within the plant include: Pi uptake at the root periphery, secretion from xylem parenchyma cells prior to xylem loading, loading of shoot tissues with Pi released from the xylem. Subsequent processes, such as Pi uptake into the sieve element/companion cell complex of the vascular tissue during Pi redistribution from source leaves, or Pi release from phloem cells in sink organs and subsequent Pi transfer to surrounding cells, are still poorly understood. Once inside the cell, Pi passes the membranes of cell organelles, such as plastids (e.g. chloroplasts and amyloplasts), mitochondria, and vacuoles, often in exchange with other solutes or protons. Metabolites that are exchanged between the plastids and the cytosol have to be transported across the two envelope membranes. Whereas the outer envelope allows the passage of small molecules, the inner envelope membrane is the actual permeability barrier and the site of Pi and metabolite transport (Flügge, 2001). The solutes to be transported, in exchange with inorganic Pi, are triose phosphates, pentose-phosphates, hexose-phosphates, or phosphoenolpyruvate (PEP). The stoichiometry of these exchanges is important, because it ensures that every molecule of phosphoester transported across either side of the plastid membrane is counterbalanced by the translocation of a Pi molecule to the opposite site (cytosol or plastid).
Mitochondria are the major sites of energy transduction in eukaryotes and uptake of Pi into the mitochondrial matrix is essential for the oxidative phosphorylation of ADP to ATP. Despite this central importance in metabolism, only very little is known about plant Pi transport across the mitochondrial inner membrane (see Kiiskinen et al., 1997 and references therein). Plant mitochondrial Pi transport systems probably catalyze Pi influx as a homodimer either via a Pi/H+ symport, a Pi/OH− antiport mechanism, or via Pi/Pi exchange.
Pi homeostasis (from the Greek words for “same” and “steady”), that is the processes used to actively maintain fairly stable Pi conditions in the cell, is essential for cell integrity and includes the tight regulation of Pi concentrations and transport in cellular compartments, i.e. the cytosol, plastids, mitochondria, and vacuoles (Mimura, 1999). In vacuolated cells of higher plants, the vacuole acts as a storage pool, or ‘non-metabolic pool’, of Pi, and at adequate Pi supply of the plants about 85 – 95% of the total Pi is located in the vacuole (Bieleski, 1973). In contrast, in leaves of Pi-deficient plants, virtually all Pi is localized to the cytosol and chloroplasts, thus representing the ‘metabolic pool’ of Pi in the plant (Bieleski, 1973; Marschner, 1995). Therefore, vacuolar Pi is used to buffer the cytoplasmic Pi level against fluctuations caused by variable external Pi supply or metabolic activities. To date, only a few reports investigating Pi transport across the tonoplast have been published. Pi uptake measurements with intact isolated vacuoles and in situ using 31P-nuclear magnetic resonance (NMR) analysis suggest the existence of Pi transport systems in the tonoplast with ATP being important for Pi influx (Massonneau et al., 2000; Mimura, 1999).
Generally, the understanding of the molecular and biochemical mechanisms involved in Pi uptake at root interfaces (including mycorrhizal interfaces) and subsequent allocation of Pi to different plant tissues and organelles is central for the understanding of Pi homeostasis and is just beginning to be better understood on the molecular physiological level.
PHOSPHATE TRANSPORTERS
The model plant Arabidopsis performs the same functions and contains essentially the same genes as other flowering plants. With completion of its genome sequence (The Arabidopsis Initiative, 2000), Arabidopsis is the main model system for laboratory studies in basic plant biology. Understanding Arabidopsis nutrient transport will assist in understanding and improving nutrient acquisition efficiency in more familiar crop plants which could finally lead to the development of an environmentally sound agriculture with low input of fertilizer. However, it has to be noted that some biological phenomena can not be studied in this model plant, such as e.g. some species-specific plant-pathogen interactions, mycorrhizal symbiosis or nitrogen fixation.
Comparison of membrane transport processes between Arabidopsis, animals, fungi and prokaryotes has identified over 600 predicted membrane transport systems in Arabidopsis(http://www.biology.ucsd.edu/~ipaul-sen/transport/), a similar number to that of the worm Caenorhabditis elegans (∼700 transporters) and over twofold greater than either baker's yeast (Saccharomyces cerevisiae) or the bacterium Escherichia coli (∼300 transporters) (The Arabidopsis Initiative, 2000). Recently, rapid progress has been made in the molecular biology of Pi uptake. Using sequence information of Arabidopsis, expressed sequence tag (EST) clones (accession numbers 134M11T7, 178H14T7, and ATTS2854) (Mitsukawa et al., 1997; Muchhal et al., 1996; Newman et al., 1994; Smith et al., 1997), which exhibited sequence similarity to the yeast high-affinity Pi transporter PHO84 (Bun-Ya et al., 1991), full-length cDNAs and their genomic clones were isolated from DNA libraries of Arabidopsis. An Arabidopsis Information Resource (TAIR) Gene Search at http://arabidopsis.org/info/genefamily/genefamily.html reveals the existence of nine genes in the haploid Arabidopsis genome coding for inorganic Pi transporters that exhibit high sequence similarity to each other. According to the rules recommended by the Commission on Plant Gene Nomenclature (http://mbclserver.rutgers.edu/CPGN/) this plant gene family was named the Pht1 family (Bucher et al., 2001; table 1). The plant Pht1 family together with the yeast and fungal homologs belong to the Pi:H+ symporter (PHS) family which is a member of the large major facilitator superfamily (MFS)(Pao et al., 1998). These nine Pht1 genes are located on chromosomes 1, 2, 3, and 5, respectively (Figure 3a). Interestingly, Pht1;1, Pht1;2, Pht1;3, and Pht1;6 are clustered within 24,200 bp on chromosome 5, suggesting the occurrence of several gene duplication events during the molecular evolution of the Pht1 family in the respective region of the Arabidopsis genome to give rise to four individual Pht1 genes on chromosome 5. To give a visual impression of the similarities between the deduced amino acid sequences, an unrooted phylogenetic tree diagram of the Arabidopsis Pht1 family members is presented in Figure 4. The close relatedness of Pht1;1, Pht1;2, and Pht1;3 sequences furthermore suggests that the three genes may share a common ancestral gene and are evolutionary younger than Pht1;6. This hypothesis is supported by the presence of an intron in the first three Pht1 members whereas no intron is contained in the Pht1;6 genomic region (Figure 3b). The Arabidopsis Pht1 genes encode proteins of approximately 520 amino acids with comparable predicted secondary structures derived from computational analysis, each is characterized by twelve hydrophobic domains presumably spanning the plasma membrane, a hydrophilic N and C terminus localized in the cytoplasm and a large hydrophilic loop between transmembrane spanning domains 6 and 7 (Figure 5). Comparison of Arabidopsis and other plant species Pht1 polypeptide sequences with those of Pi transporters from distantly related species, such as yeast, Neurospora crassa and Glomus versiforme, reveals a high degree of conservation at three positions which could be the target for posttranscriptional processing, such as N-glycosylation and protein phosphorylation (Rausch et al., 2001). Such protein modifications may be of structural significance or of functional importance for transport activity. To date, functionality of Pht1 Pi transporters can only be analyzed in a hydrophobic plasma membrane environment of either yeast or plant cells. Yeast mutants which are defective in either one or two genes encoding high-affinity Pi transporters, respectively, were used for functional complementation to analyze the biochemistry of Pht1 protein-mediated Pi uptake into yeast cells (Daram et al., 1998; Leggewie et al., 1997; Liu et al., 1998b; Muchhal et al., 1996; Rausch et al., 2001). The cDNAs encoding the transporters were cloned into a yeast expression vector behind a strong yeast promoter. Growth of complemented yeast strains and net uptake rates of Pi into the cells indicated that Pi transport of the yeast mutants had been restored by the expression of the plant genes. Expression of Arabidopsis Pht1;1 gene at high levels in tobacco-cultured cells increased the rate of Pi uptake and cell growth under Pi-limited conditions (Mitsukawa et al., 1997). The Km values determined from the Pi absorption curves of either complemented yeast cells or transformed tobacco cells were in the micromolar range. Maximal uptake velocity was at around pH 5 and was reduced by inhibitors of H+-ATPase activity and by the action of protonophores. This is consistent with a proton symport mechanism operating in the plasma membrane. It can therefore be concluded that the Pht1 gene family encodes high-affinity proton/Pi cotransporters that mediate Pi uptake across the plasma membrane. Analysis of Pi transport kinetics of more Pht1 family members will reveal whether this holds true for all Pht1 proteins or whether there are some members, for example, displaying low-affinity Pi uptake. RNA gel blot analysis was used to show that Arabidopsis Pht1 genes are predominantly expressed in roots (Smith et al., 1997; Mucchal et al., 1996). Pht1;4 is, although at low levels, also expressed in Arabidopsis leaves (M. Bucher, unpublished data). More detailled investigations of gene expression in tomato and the legume plant Medicago truncatula using mRNA in situ hybridization and immunolocalization have shown that at least two members of the Pht1 family are predominantly expressed in cells of the root epidermis (Figure 6) including root hairs and in the root cap cells (Chiou et al., 2001; Daram et al., 1998; Liu et al., 1998a). During Pi deprivation conditions, tomato Pht1:1 (LePT1) gene transcripts accumulated in the stelar tissue suggesting a role for the respective protein as a Pi-retrieval system in the stele, reloading apoplastic Pi, which did not readily enter the xylem, in xylem parenchyma cells (Daram et al., 1998). It was shown that Pht1 proteins localize to the plasma membrane and that their abundance is predominantly regulated at the transcriptional level (Muchhal and Raghothama, 1999). Therefore, a primary role in Pi uptake at the root-soil interface can be assigned to this gene family. Moreover, expression analyses in mycorrhized plant species has shown that Pht1 members and homologous proteins are expressed in cells and tissues at three different interfaces involved in Pi acquisition, namely in the hyphal network of mycorrhizal fungi at the soil-fungus interface and in the rhizodermal cells at the soil-root interface, both to allow Pi uptake from the soil solution into the fungal hyphae or the root, respectively, and in root cortex cells colonized with mycorrhizal fungi at the fungus-root interface to allow Pi transfer from the fungus to the host plant (see Rausch and Bucher, 2002 for a review and references therein). The recent completion of the Arabidopsis thaliana genome has allowed expression analysis of all Pht1 members in this species. Promoter-reporter gene fusions for all nine Pht1 genes have been generated using either the green fluorescent protein (GFP) or the β-glucuronidase (GUS) reporter genes (Mudge et al., 2002). Similarly, Karthikeyan et al. (2002) have fused the Pht1;1 and Pht1;4 promoters to the GUS or the luciferase reporter genes, respectively. Four of these promoters were shown to direct reporter gene expression to the root epidermis, outer cortex cells, endodermis and stele, and were induced under low Pi conditions in a manner similar to the previously described Pht1 genes. Other promoters directed reporter gene expression to non-root tissues, such as flowers, leaf vascular tissue, hydathodes of cotyledons, shoot buds, and pollen. Although promoter-reporter gene studies do not necessarily always reflect the expression profile of a gene in vivo, the presented work clearly suggests that Pht1 proteins do not only mediate Pi uptake from the soil solution, but play important roles in Pi translocation within the plant, thus significantly expanding our view of the role of Pht1 Pi transporters in plant nutrition.
Fig. 3.

A Chromosomal location of the members of the Pht1 and Pht2 gene families of Pi transporters in Arabidopsis thaliana. Chromosomes 1, 2, 3, and 5 are shown in scale. The length of the chromosomes is given in megabase pairs on top. The location of the individual genes is marked by diamonds as indicated at bottom. B Structure of the transcription unit of the Pht1 and Pht2 gene family members of Arabidopsis thaliana. The diagram depicts introns (thin lines) and exons (black boxes). The 5′ and 3′ untranslated sequences are omitted. Sizes are given in base pairs.
Fig. 4.

Phylogenetic analysis of Arabidopsis phosphate transporter protein sequences. Protein names are given as listed in table 1. Individual subfamilies as based on clustering of similar sequences are encircled.
Fig. 5.

Predicted topology of the tomato Pht1;1 (LePT1) Pi transporter (Daram et al., 1998) typical for Pht1 proteins, with 12 transmembrane helices, a cytoplasmic N and C terminus and a long cytoplasmic loop between transmembrane helices 6 and 7. Numbers indicate amino acids with the start methionine marked as number 1.
Fig. 6.

Localization of tomato Pht1;1 (LePT1) transcripts in tomato primary roots. Bright-field microscopy of sections of tomato primary roots from seedlings germinated under Pi sufficient conditions. The sections were hybridized with DIG-labeled LePT1 antisense RNA probes. A Longitudinal section of an emerging seedling root, B cross section from early elongation zone with cytoplasmically dense cells, and C from the top of the elongation zone, where cells begin to acquire their final differentiated attributes. Bar size is 500 µm in A, 100 mm in B and C. Reprinted from Daram et al. (1998), with permission by Springer-Verlag.
Recently, the Arabidopsis cDNA ARAth;Pht2;1 (Pht2;1), which encodes a novel Pi transporter, the first member of the Pht2 Pi transporter family in vascular plants, has been cloned (Table 1; Daram et al., 1999). The Pht2;1 cDNA encodes a 61-kD protein which is structurally similar but distinct from the proteins of the Pht1 family in having a large hydrophilic loop between the transmembrane helices 8 and 9 and a long hydrophilic N terminus. Computational modeling suggested extracellular localization of both the N and C termini. Two boxes of eight and nine amino acids, respectively, which are located in the N- and C-terminal domains of the Pht2;1 protein are highly conserved among species across all kingdoms (eubacteria, archeae, fungi, plants, and animals), including sodium-dependent Pi transporters from yeast and mammals. These conserved domains may be of importance for the Pi transport activity or for the structure of the protein. The Pht2;1 gene is predominantly expressed in green tissue. Although the protein sequence is highly similar to that of eukaryotic sodium-dependent Pi transporters, functional analysis of the Pht2;1 protein in mutant yeast cells indicated that it is a H+/Pi symporter. Its fairly high apparent Km for Pi (0.4 mM) and high mRNA abundance around the vascular tissue in leaves, based on RNA in situ hybridization studies, suggested a role for Pi loading of shoot organs (Daram et al., 1999). The independent efforts of three groups subsequently showed that Pht2;1 is localized to the chloroplast. Fusions of the entire Arabidopsis Pht2;1 protein or the N-terminal part of the transporter, respectively, to GFP indicated transport of the fusion proteins to the chloroplast (Versaw and Harrison, 2002; Bucher et al., unpublished). A subcellular proteomics approach, which was developed to identify the most hydrophobic chloroplast membrane proteins in spinach, in combination with immunolocalization and GFP fusion studies allowed Ferro et al. (2002) to identify the spinach Pht2;1 protein as an integral protein of the inner envelope membrane of chloroplasts. Pht2;1 thus differs from members of the Pht1 Pi transporter family in primary structure, affinity for Pi, subcellular localization and hence presumed function.
Arabidopsis Pht3 genes encode a small family of mitochondrial Pi transporters (Table 1; see http://mbclser-ver.rutgers.edu/CPGN/IonXWeb/Pht.group.html and http://arabidopsis.org/info/genefamily/Chloroplast.html). Pht3;1 (NCBI accession number BAB08283; Nakamura et al., 1997), Pht3;2 (PIR Entry T49281), and Pht3;3 (PIR Entry B84550) encode proteins of 309 to 375 amino acids in length which contain 4 to 6 possible transmembrane helices as is suggested by TMpred, a software for the prediction of transmembrane regions (accessible at http://www.ch.embnet.org/software/TMPRED_form.html; Hofmann and Stoffel, 1993). The protein sequences are similar and are about 50% identical in their amino acid sequence. Pht3 genes show homology to the first plant mitochondrial Pi translocator cDNA (Mpt1) cloned from birch (Betula pendula Roth) in a screening for ozone-inducible genes (Kiiskinen et al., 1997b). The Mpt1 cDNA encodes a 364 amino acid polypeptide which is 66% similar to bovine mitochondrial Pi translocator protein isoform B. Birch Mpt1 transcript abundance remains low during leaf development and is lower in roots and leaves when compared to young shoots undergoing wood formation and lignification.
The envelope membrane of plastids contains specific translocators that are involved in transport processes of photosynthetic intermediates. Different classes of inner membrane plastidic Pi translocators that mediate the transport of phosphorylated compounds in exchange with inorganic Pi have recently been characterized (Table 1; Flügge, 1999). The members of the Pi translocator family exhibit partially overlapping substrate specificities. This allows the efficient uptake of individual phosphorylated substrates even in the presence of high concentrations of other phosphorylated metabolites. Moreover, this multiplicity of transporters relates to specific plastid types and differences are seen even within the same plastid population, reflecting the flexibility of plastid metabolism (Bowsher and Tobin, 2001). Arabidopsis TPT is a nuclear gene encoding the chloroplast triose-phosphate/Pi translocator precursor. The mature protein is involved in the export of carbon fixed during the day from the chloroplasts into the cytosol in the form of triose phosphates (Flügge, 1999). The need for phosphoenolpyruvate, as an immediate precursor for the synthesis of secondary products via the shikimic acid pathway or as a precursor for fatty acid or aromatic amino acid biosynthesis, necessitates a plastidic phosphoenolpyruvate/Pi translocator encoded by the PPT gene (Fischer et al., 1997). The PPT protein exhibits approximately 33% identity to the TPT. The corresponding gene is expressed in both photosynthetically active tissues and in non-green tissues, although transcripts were more abundant in non-green tissues. Expression of the coding region in transformed yeast cells and subsequent transport measurements of the purified recombinant translocator showed that the protein mediates transport of Pi in exchange with C3 compounds phosphorylated at C-atom 2, particularly phosphoenolpyruvate. Forward genetics analysis of the previously characterized cue1 Arabidopsis mutant (Li et al., 1995) revealed that PPT is required for synthesis of aromatic amino acids from the shikimic acid pathway, as well as palisade cell development (Streatfield et al., 1999). Non-photosynthetic plastids must import carbon in the form of hexose phosphates via the glucose 6-phosphate Pi translocator, named GPT. The Arabidopsis GPT exhibits 38 and 36% identity with the TPT and PPT proteins, respectively. Thus, GPT proteins represent a third group of plastidic Pi antiporters. Transport experiments with Arabidopsis GPT purified from transformed yeast cultures demonstrated that the GPT protein mediates a 1:1 exchange of glucose 6-phosphate mainly with inorganic Pi, released during starch biosynthesis, and triose phosphates, generated from the oxidative pentose pathway (Kammerer et al., 1998). The fourth type of plastidic Pi translocator is encoded by the pentose phosphate/Pi translocator gene from Arabidopsis, named XPT due to its preference for the substrate xylulose 5-phosphate (Xul-5-P) that is transported in exchange with inorganic Pi or triose phosphates, respectively (Eicks et al., 2002). The XPT protein shares 35, 34, and 47% identity with TPT, PPT, and GPT proteins, respectively. XPT transcripts are present in all organs. In the Arabidopsis genome, a total of eight genes are grouped into the Arabidopsis TPT translocator family containing the four types described above (http://arabidopsis.org/info/genefamily/Antiporters.html). Characterization of the other four family members is needed to unequivocally assign a biological function to each gene.
An essential step in Pi uptake into the shoot is the loading of the xylem with Pi absorbed by the roots. The PHO1 gene, isolated by positional cloning from a mutant deficient in loading Pi to the xylem, is likely to encode the Pi efflux carrier involved in this process (Hamburger et al., 2002). The pho1 mutant and its corresponding gene will be described in further detail in a later section.
Although multiple cDNA clones encoding Pi transporters have been isolated and characterized from several plants, a vacuolar Pi transporter has not been identified yet.
RESPONSES TO PI DEFICIENCY
Plants have evolved a series of morphological and metabolic adaptations that are triggered by phosphate deficiency. These adaptations are aimed at increasing the acquisition of this vital but poorly available nutrient from the soil as well as sustain plant growth and survival under low P availability. At the morphological level, the architecture of the root system is modified, with an increase in root/shoot ratio, increase in the length and density of root hairs, as well as proliferation of lateral roots. In some plants, there is also a reduction in the gravitropism of roots. All these changes improve the capacity of the plant root system to better explore and mine the soil for phosphate. Under Pi deprivation stress, the roots enhance secretion of protons or organic acids in order to enhance the solubilization of insoluble inorganic phosphate complexes. Roots also release phosphatases and nucleases to acquire phosphate from organic sources. The kinetics of Pi transport into the root and across the plant is modified through changes in transporter abundance and affinity for Pi. Biochemical pathways less dependant on intracellular Pi level are activated, such as the replacement of phospholipids by sulfolipids. Although several of these responses to Pi deprivation stress have been documented for a number of agriculturally important plants, the use of Arabidopsis offers the opportunity to get a better understanding of the genetic and biochemical basis of Pi-stress responses.
Morphology of the root system
Growth of plant in Pi-deficient media has a marked effect on root hair elongation and density (Figure 7) (Bates and Lynch 1996, Ma et al., 2001a). At 16 days, root hairs of low Pi plants were three times longer compared to root hairs of high Pi plants (Bates and Lynch, 1996). Increase in root hair length was found to be a result of both increased growth duration and increased growth rate (Bates and Lynch, 1996). Root hair density is about five-time greater in roots grown in low Pi media compared to high Pi media. Both root hair length and density were found to decrease logarithmically in response to increasing Pi media concentration. The anatomy of the Arabidopsis root is significantly changed under low Pi. The root diameter is slightly increased and there are 45% more cortical cells. The number of epidermal cells is also increased while their size is decreased (Figure 8). In Arabidopsis, epidermal cells lying over the anticlinal wall separating two cortical cells differentiate into cells that can bear root hairs (trichoblast), whereas those located over the outer periclinal cortical cell walls differentiate into hairless cells (atrichoblast) (Dolan et al., 1994) The larger number of cortical cells increased the number of trichoblast files from 8 to 12. The increase in root hair density can be explained by a combination of factors including an increase in the number of trichoblasts, an increase in the likelihood that a trichoblast will form a root hair, and a greater stacking of hairs from smaller trichoblast (Ma et al., 2001a). An increase in the frequency of recruitment of atrichoblasts to form root hair also occurs in Pi-deficient roots (Schikora and Schmidt, 2001). These results clearly show that Pi deficiency can affect root cell differentiation and patterning.
Fig. 7.

Changes in root hairs in plants grown under Pi deficiency. Arabidopsis was grown for 10 days in agar medium containing either 5 mM (+Pi) or 5 µM (−Pi) inorganic phosphate.
Fig. 8.

Changes in root anatomy in plants grown under Pi deficiency. Cross sections of Arabidopsis roots grown in agar medium containing either 1 mM (high P) or 1 µM (low P) inorganic phosphate. Asterisks indicate trichoblasts. Reprinted from Ma et al. (2001), with permission by Blackwell Science Ltd.
The increase in both root hair length and density in plants grown under low Pi allows exploration of a larger volume of soil by the root system for Pi acquisition. This increased surface of contact is thought to be a particularly important adaptation for the acquisition of ions which have a limited potential for diffusion in soil, such as for Pi and Fe, in contrast to nutrient that move mainly through diffusion, such as nitrate. In agreement with this, it is found that both Fe and Pi deprivation stress induce a similar increase in root hair length and density, while deficiency in other ions has little effect on root hairs (Bates and Lynch, 1996; Ma et al., 2001a; Schmidt and Schikora, 2001). The importance of root hairs in the acquisition of Pi in Arabidopsis has been demonstrated by Bates and Lynch (2000a, b, 2001) using mutants of Arabidopsis having either a deficiency in root hair elongation (rdh2) or reduced root hair density (rhd6). In plants grown in media with 1.5 µM Pi, wild type plants had greater shoot biomass, total root surface area, absolute growth rate, total P, Pi content per unit length and specific Pi absorption than the two root hair mutants (Bates and Lynch 2000a, b). In contrast, in plants grown in high Pi (54 µM) media, no differences between wild type and mutants were measured. A cost-benefit analysis revealed that under low Pi, wild type roots acquired more P per unit of carbon respired or unit of P invested into the roots than either of the mutants. The competitive advantage of plants with root hairs was shown in an experiment where both mutants and wild type plants were grown together at constant density in media containing either high or low Pi (Bates and Lynch, 2001). The relative competitive advantage of one genotype was estimated using the Relative Crowding Coefficient (RCC) measured as (yield of wild type/yield of rdh2 mutant, in a mixed culture)/ (yield of wild type/yield of rdh2 mutant, in a monoculture). While at high Pi availability, both wild type and mutant had a RCC equal to one, under low Pi nutrition, the wild type had an RCC value greater than one while the hairless mutant had a RCC value less than one. These results demonstrated that root hairs increase the competitiveness of plants under low Pi availability but do not affect growth or competitiveness under high Pi availability (Bates and Lynch, 2001). Geometric models of root growth pattern under Pi deprivation stress substantiated the hypothesis that increased root hair length and density act synergistically on Pi acquisition (Ma et al., 2001b).
Phytohormones have a considerable impact on root hair growth. Externally applied auxin increases both root hair length and density in the wildtype (Bates and Lynch, 1996, 2000a), while application of auxin restores root hair density in the rdh6 mutants (Masucci and Schiefelbein, 1994). Similarly, treatment of roots with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) results in development of more root hairs in wild type and restoration of normal root hair density in the rdh6 mutant (Masucci and Schiefelbein, 1994). Furthermore, the application of ethylene synthesis inhibitor or ethylene perception agonist reduces root hair number (Masucci and Schiefelbein, 1994). The implication of auxin, ABA and ethylene on the morphological changes induced by Pi deprivation stress has been studied. Comparison of the responses of wild type and ABA mutants aba1 and abi2 to Pi deprivation stress revealed no significant differences in the developmental responses of roots (Trull et al., 1997), implying that ABA does not have a major role in coordinating the Pi deprivation stress response. Although application of IAA increases root hair length and density in plants grown on high Pi, it has little effects on root hairs in plants grown on low Pi (Bates and Lynch, 1996; Ma et al. 2001a). Furthermore, low Pi stimulated root hair elongation in the hairless auxin-resistant mutant axr1 and axr2, as well as in the auxin-insensitive mutant aux1, in a manner similar to wild type (Figure 9) (Ma et al., 2001a; Schikora and Schmidt 2001; Schmidt and Schikora, 2001). Thus, although auxin does affect root hair development, it appears that low Pi can increase root hair length and density in a manner that is at least partially independent of auxin. Similar conclusions were also obtained regarding the role of ethylene, since ethylene-resistant etr1 and ethylene-insensitive ein2 grown under Pi limitation show an increase in root hair length and density similar to wild type plants (Schmidt 2001; Schmidt and Schikora, 2001) (Figure 9). It is interesting to note, however, that despite the similarity in the response of Arabidopsis root hair growth to low Pi and Fe, the use of the same mutants revealed that both auxin and ethylene are indeed required to mediate changes in root hair growth under Fe stress response (Figure 9) (Schmidt 2001; Schmidt and Schikora, 2001).
Fig. 9.

Root tips of Arabidopsis wild type and of various hormone-related mutants grown in nutrient-sufficient media (control) or in the absence of Pi or Fe. A, Col-0 control; B, Col-0 –Fe; C, Col-0 –Pi; D, axr1 control; E, axr1 –Fe; F, axr1 –Pi; G, axr2 control; H, axr2 –Fe; I, axr2 –Pi; J, ein2 control; K, ein2 –Fe; L, ein2 –Pi; M, etr1 control, N, etr1 –Fe; O, etr1 –Pi; P, eto3 control; Q, eto3 –Fe; R, eto3 –Pi. Bar = 0.25 mm. Reprinted from Schmidt and Schikora (2001), with permission by the American Society of Plant Biologists.
In addition to changes in the development of root hairs, Pi deficiency affects the development of lateral roots. In beans, Pi deprivation stress increases the density of lateral roots, reduces the length of the primary root, as well as reduces the gravitropism of lateral roots so that the root system becomes shallower. This response is thought to promote Pi acquisition since shallower root systems are more efficient because their roots are more dispersed and suffer less inter-root competition (Lynch and Brown, 1998). In Arabidopsis, growth on low Pi media results in a reduction in primary root length and internode length, as well as in an increase in lateral root length. There is, thus, a redistribution of root growth from the primary root to the lateral roots, the later becoming more dense (López-Bucio et al., 2002; Williamson et al., 2001) (Figure 10). Local supply of high phosphate in a patch results in a decrease of primary root growth after the root left the patch, while lateral root growth is enhanced but lateral root density remains unaffected (Linkohr et al., 2002). No effects of Pi deficiency on the gravitropic responses of lateral roots have been described.
Fig. 10.
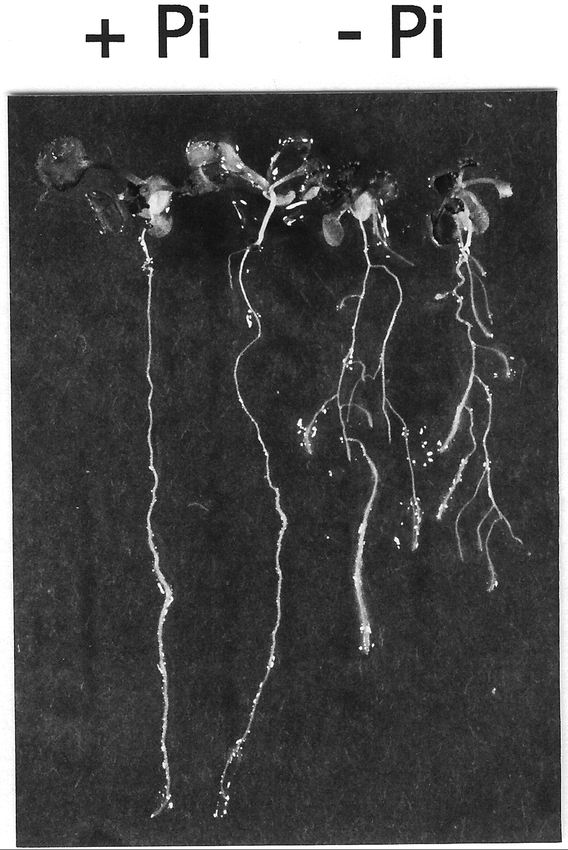
Changes in root architecture in plants grown under Pi deficiency. Arabidopsis was grown for 12 days in agar medium containing either 5 mM (+Pi) or 5 µM (−Pi) inorganic phosphate.
Since addition of exogenous auxin or ethylene inhibits primary root elongation and alters lateral root formation, the role of auxin and ethylene in the changes of the root system under Pi deficiency was addressed using the auxin transport mutants aux1 and eir1, the auxin response mutants axr1, axr2, axr4 and iaa28, the ethylene-insensitive mutants etr1, ein2, ein3, and hls, the ethylene-overproducing mutant eto1 and the ethylene constitutive response mutant ctr1 (López-Bucio et al., 2002; Williamson et al., 2001). Under Pi deprivation stress, all auxin-related mutants showed a decrease in primary root length as well as an increase in lateral root number and density that were similar to wild type plants. The only exception is the iaa28 mutant which is severely defective in lateral root formation in plants grown in either Pi-sufficient or Pi-deficient conditions (López-Bucio et al., 2002). The root system of all ethylene mutants respond to Pi deprivation by a decrease in primary root growth and increase in lateral root formation, although the response of the ctr1 and eto1 mutants was reduced compared to wild type (López-Bucio et al., 2002). Together, these experiments indicate that changes in root architecture induced by Pi deficiency is largely independent of the auxin and ethylene pathways.
Phosphate mobilization
Numerous plants have been found to respond to Pi starvation by an increase in the production of nucleases and phosphatases. Considering that the organic phosphorus content can reach up to 80% in some soils, extracellular nucleases and phosphatases are thought to play an important role in scavenging the soil for Pi from organic sources. Similarly, intracellular phosphatases and nucleases may be involved in scavenging and regulating the supply of Pi from intracellular organic sources.
Enhanced production of phosphatase under Pi deficiency is a response found among a wide spectrum of organisms, including bacteria, yeast and plants (Duff et al., 1994; Golstein et al., 1988; Lenburg and O'Shea, 1996; Torriani, 1990). Phosphatases have been traditionally classified as being either alkaline or acid phosphatase according to their optimal pH for catalysis (Duff et al., 1994). Plant alkaline phosphatases generally display rather strict substrate specificity and play defined roles in metabolism. These include fructose 1,6-bisphosphatase and sucrose 6-phosphate phosphatase. In contrast, acid phosphatases generally have rather unspecific activities, with the exception of the intracellular phosphoenolpyruvate phosphatase (Duff et al. 1989). While the role of some of these unspecific intracellular acid phosphatases can be inferred from physiological studies, such as the role of phytase in the release of Pi from phytate, considerable uncertainty remains on the role of others, such as the vegetative storage acid phosphatases found in soybean and Arabidopsis (Berger et al., 1995). Most studies on the relationship between Pi deprivation stress and phosphatases in Arabidopsis have been done with extracellular acid phosphatases.
Acid phosphatase activity associated with Arabidopsis roots can be easily revealed by staining the roots with the chromogenic substrate 5-bromo-4-chloro-3-indolyl-phosphate (BCIP). Under Pi deprivation stress, roots show a strong increase in phosphatase activity (Figure 11) (Llyod et al., 2001; Trull et al., 1997; Trull and Deikman, 1998). Similar activities have been found in roots of lupin, rice, wheat and tomato (Tadano and Sakai, 1991). In Arabidopsis, protein extracts from shoots and roots revealed the presence of several proteins having acid phosphatase activity, ranging from approximately 30 to 120 kD, which increase following Pi deprivation stress (del Pozo et al., 1999; Trull and Deikman, 1998). Purification of a 34 kD acid phosphatase from Arabidopsis led to the isolation of the corresponding AtACP5 gene (del Pozo et al., 1999). The AtACP5 protein contained an N-terminal extension of 31 amino acids relative to the mature purified protein, with the characteristics of a signal peptide. This indicates that AtACP5 is most likely an extracellular phosphatase. The Arabidopsis protein showed high similarity to mammalian type 5 purple acid phosphatase and displayed peroxidation activity. Northern analysis and promoter-GUS fusions showed that the gene was expressed in shoots and roots and was strongly up-regulated under Pi deficiency as well as under salt stress, hydrogen peroxide and ABA treatment, and in senescing tissues (del Pozo et al., 1999). Together, these data indicate that AtACP5 is involved in both phosphate mobilization and in the metabolism of reactive oxygen species in stressed or senescent parts of the plant.
Fig. 11.

Secretion of acid phosphatase from Arabidopsis roots grown in Pi-deficient media. Plants were grown in Pi-sufficient (left petri) or Pi-deficient (right petri) media and overlaid with an agar solution containing the substrate BCIP. The blue color reveals the presence of acid phosphatase. Photograph kindly provided by Ann Lloyd (Exeter University, United Kingdom).
A distinct gene has been identified in Arabidopsis roots which would encode a 42–46 kD acid phosphatase (Haran et al., 2000). Promoter-GUS studies revealed a strong regulation of the promoter activity by Pi availability. Recombinant GFP containing the N-terminal sequence of the cloned acid phosphatase was secreted by the roots into the medium, indicating that the enzyme was extracellular.
A mutant of Arabidopsis has been isolated which showed reduced staining for root acid phosphatase in roots of plants grown under Pi starvation (Trull and Deikman, 1998). This mutant, named pup1 (phosphatase-underproducer) was shown to be missing only one (160 kD) of the multiple acid phosphatase found in Arabidopsis. The mutant was otherwise normal in most of the classical responses to Pi deprivation stress, including up-regulation of other acid phosphatases, accumulation of anthocyanins and changes in root growth and morphology (Trull and Deikman, 1998).
Recent analysis of the Arabidopsis genome has revealed the presence 29 genes encoding proteins belonging to the purple acid phosphatase family (Li et al., 2002). Expression studies of a subset of seven genes revealed that only two genes, distinct from AtACP5, were upregulated under Pi deprivation stress. These data indicate that only some members of the purple acid phosphatase gene family are regulated by Pi nutrition.
Numerous studies have shown that Pi deprivation induces the production of ribonucleases in plants. Pi starvation of tomato cell cultures leads to the co-regulated induction of several extracellular and intracellular (vacuolar and extravacuolar) ribonucleases (Dodds et al., 1996; Köck et al., 1995; Löffler et al., 1992; Nürnberger et al., 1990). Arabidopsis was shown to contain at least 16 proteins with ribonuclease activity (Yen and Green, 1991). The ribonucleases known as S-RNases are mainly expressed in the style and are involved in gametophytic self-incompatibility (Bariola and Green, 1997). In contrast, the role of S-like RNases is less well defined. S-like RNases are expressed in a variety of tissues and do not participate in the control of self-incompatibility (Bariola and Green, 1997). Three genes encoding for S-like RNases have been cloned and studied in some detail in Arabidopsis. RNS1 is an extracellular ribonuclease. The RNS1 gene is expressed almost exclusively in flowers in Pi sufficient plants (Bariola et al., 1994). Under Pi deprivation stress, there is a strong up-regulation of gene expression as well as protein accumulation in whole plants, including leaves (Bariola et al., 1999). RNS2 has a broader expression pattern, being expressed in most tissues in Pi sufficient plants. Like RNS1, RNS2 gene expression and protein accumulation is up-regulated by Pi starvation (Bariola et al., 1994, 1999; Taylor et al., 1993). The protein is intracellular and contains a C-terminal extension that has features similar to vacuolar-targeting sequences, suggesting that it may be vacuolar, although ER localization is also possible. The RNS3 gene is expressed in root, leaves and stems and is not regulated by Pi availability (Bariola et al., 1994). All three RNS genes are up-regulated by tissue senescence. Antisense down-regulation of RNS1 and RNS2 expression was correlated with elevated levels of anthocyanin accumulation in both Pi-sufficient and Pi-deficient plants (Bariola et al., 1999). Since anthocyanin production is triggered by a variety of stress other than Pi limitation, such as wounding, low temperature, high light intensity and ozone exposure, it is unclear whether the increase in anthocyanin observed in these antisense plants is directly related to Pi metabolism or not. Since Arabidopsis is known to be able to grow in media containing DNA as the main Pi source (Chen et al., 2000), it would be interesting to determine whether plants with a down-regulation of RNS1 and RNS2 are less able to utilize extracellular organic phosphorus as a Pi source.
Tomato cells have been shown to produce an extracellular cyclic nucleotide phosphodiesterase as an accessory nucleolytic activity during phosphate starvation (Abel et al., 2000). The combination of ribonucleases and cyclic nucleotide phosphodiesterase would enable the complete utilization of Pi found in nucleotides (Figure 12). It is presently unknown if a similar cyclic nucleotide phosphodiesterase is produced in Arabidopsis plants grown in Pi-deficient media.
Fig. 12.
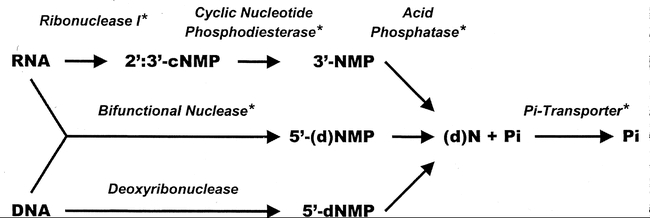
Model of extracellular nucleic acid degradation and Pi recycling by secretory nucleolytic enzymes. Asterisks indicate proteins known to be inducible by Pi starvation in some plants. Reprinted from Abel et al. (2000), with permission by the American Society of Plant Biologists.
Enhanced mobilization of extracellular organic phosphorus has also been achieved in transgenic plants overexpressing an extracellular phytase (Richardson et al., 2001). Phytate (myo-inositol-hexakisphosphate) is the main form of storage of P in the seeds (Brinch-Pedersen et al., 2002). Phytate is also found in soils where it can account between 20 to 50% of the total soil organic P. Despite its relative abundance, soil phytate appears to be poorly utilized by the root system. While Arabidopsis growing in artificial agar medium is able to obtain Pi from a range of organic phosphorus substrates, such as RNA, DNA, AMP, ATP, 3-phosphoglyceric acid and glucose 1-phosphate, it is unable to use phytate (Richardson et al., 2001). In agreement with this result is the fact that phytase activity in roots is very low and not found as an extracellular enzyme. Furthermore, although a number of acid phosphatases are secreted by the root system under Pi deficiency, the activity of these enzymes towards phytate appears very limited. A large improvement in the acquisition of Pi from extracellular phytate was achieved when Arabidopsis was transformed with a phytase gene from Aspergillus niger that was modified for secretion of the protein by addition of a signal peptide sequence from the carrot extensin gene. Expression of the same phytase intracellularly did not result in an increase in Pi acquisition from extracellular phytate. These results raise the interesting question as to why evolution has not favored the production of extracellular phytase in the roots as an efficient way of acquiring Pi from phytate in soil.
Apyrases are enzymes that are able to hydrolyze the γ- and ß-phosphate on ATP or ADP. Expression of a pea apyrase in Arabidopsis was shown to lead to enhanced growth and Pi transport when P is supplied as ATP (Thomas et al., 1999). Although apyrase could lead to the release of Pi from extracellular ATP, the role of apyrase in the overall improvement in Pi acquisition is unclear since similar improvements were also observed when P was supplied as free inorganic Pi. It appears that expression of apyrase may indirectly affect the activity of Pi transporters in yeast and plants (Thomas et al., 1999).
Enhanced secretion of organic acids, such as malate and citrate, from the root system of plants growing in Pi deficient media has been described for several plants, including white lupin (Gardner et al., 1981). These organic acids help to solubilize Pi found in insoluble inorganic complexes, such as aluminium phosphate complexes. Enhanced secretion of organic acids is also linked to aluminium resistance in several plants, since chelated aluminium is less deleterious to plant cells than free Al+3 ions. Increased secretion of malate and citrate in roots of plants grown in medium with low Pi has been reported for a number of Arabidopsis accessions (Narang et al., 2000). Overexpression of plant mitochondrial citrate synthase in either carrots or Arabidopsis has been shown to lead to enhanced levels of citrate excretion from roots and improved growth of plants growing in either media containing poorly soluble aluminium phosphate or in phosphorus-limited acid soil (Koyama et al., 1999, 2000). Similarly, tobacco plants overexpressing a bacterial citrate synthase also showed improvement in root citrate excretion as well as acquisition of Pi from acid soils (López-Bucio et al., 2000), although the results of this work have recently been challenged by Delhaize et al. (2001).
Modification of Pi transport
It was shown in physiological studies that during P limitation in the medium, Pi uptake velocity (Vmax) is enhanced probably through an increased synthesis of Pi transport systems and thus an increased abundance of Pi uptake sites in the plasma membrane (Drew et al., 1984; Lee, 1982; Shimogawara and Usuda, 1995; Ullrich-Eberius et al., 1984). Alternatively, existing uptake systems might be activated by posttranslational modification, thus leading to a higher Vmax of existing transport systems (Cogliatti and Clarkson, 1983). In Arabidopsis plants subject to Pi deprivation, Vmax is drastically increased whereas the apparent Km value remains largely unchanged. This is paralleled by an increased Pi uptake rate and a decrease in internal Pi in shoots and roots (Dong et al., 1999). Accumulation of Pht1;1, Pht1;2, and Pht1;4 mRNA, respectively, during Pi deprivation stress imposed by cultivation of Arabidopsis plants in a hydroponic culture medium lacking Pi is detectable within 3 to 5 days following the initiation of Pi starvation (Muchhal et al., 1996; Smith et al., 1997). Similar to the situation in tomato and potato (Bucher et al., 2001; Daram et al., 1998; Leggewie et al., 1997; Liu et al., 1998a), Arabidopsis Pht1 transporters fall into two subfamilies with five Pht1 genes being predominantly expressed in roots (Muchhal et al., 1996; Mudge et al., 2002; Okumura et al., 1998; Karthikeyan et al., 2002; Smith et al., 1997), while transcripts of four genes were detectable partially in roots but also in shoot organs (Mudge et al., 2002) and suspension-cultured cells (Okumura et al., 1998). Expression of the majority of genes of both subfamilies is enhanced during Pi starvation. To date, the affinity for Pi (Km) of individual Arabidopsis Pht1 transporters has not been published, except that for Pht1;1 which indicated that Pht1;1 is a high-affinity Pi transporter (Mitsukawa et al., 1997). The similarity between estimated Km values of Arabidopsis plants in hydroponic culture and Pht1 Pi transporters from Arabidopsis and other plant species, analysed following heterologous expression of the plant genes in yeast, suggests that a key function of this gene family is Pi uptake from the soil, especially in root hairs (Daram et al., 1998; Dong et al., 1999; Karthikeyan et al., 2002; Leggewie et al., 1997; Liu et al., 1998b; Mudge et al., 2002; Rausch et al., 2001). Thus, Vmax of Pi transport in Arabidopsis roots is likely to be the result of the expression of multiple Pi transporter genes exhibiting similar affinities for Pi. However, allosteric mechanisms in Pi transporter regulation can not be excluded. This hypothesis is based on sequence similarities between Pht1 transporters and their orthologs from non-plant species showing conserved regions for post-translational modification such as protein phosphorylation or glycosylation (Bucher et al., 2001). Transgenic tobacco cells overexpressing the Arabidopsis Pht1;1 gene exhibited increased biomass production under certain Pi limited conditions, establishing gene engineering of Pi transport as one approach towards enhancing plant cell growth (Mitsukawa et al., 1997).
In the field of Pi acquisition in plants, the impact of the symbiosis between fungi and most plants can not be ignored. Arbuscular mycorrhizae are the most common soil-based symbioses formed in the roots of ≥80% of the terrestrial plants. Unfortunately, in Arabidopsis, like Brassicacae species in general, roots are not colonized by arbuscular mycorrhizal fungi and this species is thus not a suitable host to investigate these ecologically and agriculturally important associations (Smith and Read, 1997). Under low P conditions, which is the case in almost all natural ecosystems, the host plant derives mainly P from the fungus, which in turn benefits from plant-based photosynthetic assimilates. In studying the molecular basis of symbiotic P uptake into the plant, a Pi transporter gene StPT3 in potato has recently been identified (Rausch et al., 2001). In a split-root experiment it was shown that StPT3 is expressed exclusively in root parts that are “infected” with a mycorrhizal fungus (Figure 13). RNA localization and reporter gene expression indicated a specific induction of StPT3 expression in root cortex cells containing mycorrhizal structures. Interestingly, StPT3 shares high similarity with Pht1 Pi transporters from Arabidopsis and other plant species, suggesting that high-affinity Pi transport in plants is mediated by similar proteins at two different interfaces, the soil-root interface in all terrestrial plants including non-mycorrhized Arabidopsis and the fungus-root interface in mycorrhized plants. Interestingly, mycorrhiza-specific Pi transporters similar to StPT3 have recently been identified from the two model plants rice (Oryza sativa) (Paszkowski et al., 2002) and Medicago truncatula, a close relative of the forage legume alfalfa (Harrison et al., 2002). Immunohistochemistry localized the Medicago MtPT4 protein to the periarbuscular plant membrane, which surrounds the fungal arbuscule. Future experiments will show how important these peculiar membrane proteins are in the functioning of the symbiosis.
Fig. 13.
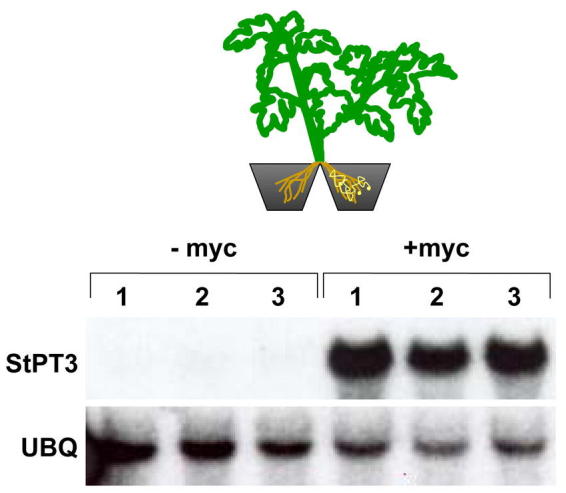
Potato Pht1;3 (StPT3) transcript abundance in mycorrhizas. Schematic view of the split-root system at top with the non-mycorrhized root part at left and the mycorrhized root part at right. AM fungal spores and hyphae are marked in yellow. Pi transporter transcript levels in roots cultured in a split-root system as detected via RNA gel blot analysis. Randomly labeled cDNA probes, as indicated at left, hybridized with RNA on the blot from parts of roots of three individual plants per split-root system (1, 2, and 3) that were cultured without (−myc) or with (+myc) Glomus intraradices, respectively. Both compartments of one system were irrigated with 5 mM Pi. Ubiquitin (UBQ) served as a marker for constitutive gene expression (see also Rausch et al., 2001). Courtesy of C. Rausch, ETH Zürich, Switzerland.
Metabolic adjustments
Inorganic phosphate plays a central role in plant metabolism both as a substrate for photophosphorylation and ATP generation through respiration as well as a key component of the export of carbon from the chloroplasts during photosynthesis. Pi and nucleoside phosphates participate as substrates or products in numerous enzymatic reactions and also act as allosteric modulators. Phosphorylation plays also an important role in the control of several signal transduction pathways. It is thus not surprising that Pi deficiency has a profound effect on various aspects of plant metabolism, including photosynthesis, carbon fixation, glycolysis, and respiration (reviewed in Plaxton and Carswell, 1999). Although numerous studies have focussed on the effects of Pi deprivation on various aspects of plant metabolism, few of them have been done directly in Arabidopsis. It is nevertheless likely that the general conclusions reached from these studies, and in particular for studies done with suspension cells of Brassica nigra, will also generally apply to Arabidopsis.
Phosphate deficiency is known to affect photosynthesis and carbon fixation through several mechanisms, including direct effects on the structural assembly of the photosynthetic apparatus, the energy-transducing systems in the thylakoids, inhibition of several key enzymes of the Calvin cycle and feedback inhibition by the pH gradient across the thylakoids or by the redox state of electron carriers (reviewed in Natr, 1992). Analysis of the effects of Pi deprivation on photosynthesis in Arabidopsis plants grown either in low Pi media or in the pho1 mutant deficient in leaf Pi accumulation revealed little effects on photosynthetic electron transfer, O2 evolution and excitation energy allocation to the photosynthetic reaction centers, despite of considerable structural changes in the photosynthetic apparatus, as indicated by pigment and thylakoid ultrastructure data (Härtel et al., 1998). However, light-saturated photosynthetic electron transport in pho1 plants compared to wild type was reduced by 30% when applying another protocol of plant cultivation (Hurry et al. 2000). This effect was found to be associated with significant changes in both the expression levels of photosynthetic genes and the activities of key enzymes involved in photosynthesis.
In common with many other plants, there is a considerable increase in starch biosynthesis in Pi deficient shoots of Arabidopsis (Cieresko et al., 2001; Zakhleniuk et al., 2001). This increase in starch is at least mediated by both the release of inhibition of the starch biosynthetic enzyme ADP-glucose pyrophosphorylase by the allosteric inhibitor Pi, as well as an increase in the expression of the genes encoding the subunits of the same enzyme (Cieresko et al., 2001). Reduction in the cytoplasmic level of Pi following severe Pi deprivation stress will also reduce the exchange of triose phosphate derived from photosynthesis with Pi across the chloroplast envelope. Thus, one effect of the conversion of triose phosphate to starch is the liberation of Pi within the chloroplast that can then be used to maintain photosynthesis.
An interesting relationship has been uncovered between Pi supply in leaves and the acclimatization of photosynthesis to low temperature in Arabidopsis (Hurry et al., 2000). Transfer of plants from 20 °C to 5-10 °C rapidly inhibits photosynthesis because low temperature inhibits sucrose synthesis, which itself leads to a perturbation of the recycling of inorganic Pi, detected as an increase in phosphorylated intermediates as well as a decrease in ATP/ADP and triose phosphate /3-phosphoglycerate ratios. Low temperature is thus linked to phosphate-limitation of photosynthesis. Prolonged exposure to low temperature leads to a decline of transcripts encoding proteins involved in photosynthesis. Acclimatization of new leaves that develop at low temperature depends on the re-establishment of photosynthesis and the production of sucrose, the later being itself a cryoprotectant and also involved in synthesis of other cryoprotectants, such as proline. Acclimatization is mediated by an increase in the proteins and transcripts for a number of enzymes involved in the Calvin cycle and in sucrose synthesis, as well as by a redistribution of vacuolar Pi towards the cytoplasm for its incorporation into phosphorylated metabolites. Hurry et al. (2000) have examined whether low shoot phosphate can trigger or improve cold acclimatization of photosynthetic carbon metabolism. This was done by comparing wild type to the pho1 and pho2 mutants, which have a decreased and increased shoot Pi level, respectively, while root Pi levels are unchanged (see section mutants affected in phosphate acquisition and homeostasis for a detailed description of the mutants). Cold acclimatization was improved in the pho1 mutant and weakened in pho2. This was mediated, at least in part, by an increased level of the sucrose biosynthetic enzymes sucrose phosphate synthase and fructose 1,6-biphosphatase, as well as an increase of the expression of several genes coding for enzymes of the Calvin cycle in the pho1 mutant. These increases in protein and gene expression were detected in pho1 grown at 23?C and were maintained after the shift at 5°C. Pi deprivation stress, thus, appears to pre-condition plant metabolism in a way that is beneficial to cold acclimatization while high Pi hinders this process.
Although cytoplasmic level of Pi remains generally constant at the expense of vacuolar Pi in plants in the early stage of Pi deficiency, severe Pi deprivation stress will eventually decrease levels of cytoplasmic Pi as well as of nucleoside phosphates, such as ATP and ADP (Plaxton and Carswell, 1999). In contrast, levels of pyrophosphate appear to remain more constant and may thus serve as a substitute energy donor. Reduction in cytoplasmic Pi, ATP, and ADP directly affects glycolysis since several enzymes involved in this pathway uses Pi/ATP/ADP as co-substrate, such as the ATP-dependent phosphofructokinase, NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (NAD-3GPDH), 3-phosphoglycerate kinase, and pyruvate kinase (Figure 14) (Duff et al., 1989, Theodorou et al., 1993). Furthermore, Pi is known to stimulate phosphofructokinase (PFK) activity and inhibit pyrophosphate-dependent phosphofructokinase. Several enzymes are activated under Pi deprivation that contribute to maintenance of the carbon flux through glycolysis despite reduced concentration of cytosolic Pi and nucleoside phosphates. Both the level of the enzyme and of the transcript coding for the UDP-glucose phosphorylase has been shown to be induced under Pi deprivation in Arabidopsis (Cieresko et al., 2001). This enzyme utilizes pyrophosphate to generate glucose 1-phosphate and ultimately glucose-6-phosphate, thus bypassing the ATP-requiring hexose kinase (Figure 14). Similarly, the activity of a pyrophosphate-dependent phosphofructokinase is induced at least 20-fold in Pi deficient Brassica nigra suspension cells (Duff et al., 1989). This enzyme would bypass the ATP-requiring phosphofructokinase. Similarly, a non-phosphorylating NADP-dependent G3PDH is induced under Pi deprivation stress, bypassing the Pi-requiring NAD-G3PDH and ADP-requiring 3-phosphoglycerate kinase (Figure 14). Still in B. nigra cells, Pi stress was shown to result in the induction of a phosphoenolpyruvate (PEP) phosphatase, which would replace the conversion of PEP to pyruvate via the cytoplasmic ADP-requiring pyruvate kinase. Since the PEP phosphatase is located in the vacuole, tonoplast transport mechanisms involved in shuttling PEP and pyruvate are hypothesized (Theodorou et al., 1993). An alternative pathway for the ADP-independent conversion of PEP to pyruvate is via the PEP carboxylase, which is found to be induced 5-fold in Pi-depleted B. nigra cells. Collectively, the activation of these phosphate starvation inducible “bypasses” ensures adequate maintenance of carbon flux through glycolysis while recycling phosphate esters to inorganic Pi (Plaxton and Carswell, 1999).
Fig. 14.

Model indicating several changes in plant metabolism occuring in response to Pi deficiency. The modified pathways are indicated with bold arrows. The enzymes that catalyze the numbered reactions are: 1, invertase; 2, sucrose synthase; 3, hexokinase; 4, fructokinase; 5, UDP-glucose pyrophosphorylase; 6, nucleoside diphosphate kinase; 7, phosphoglucomutase; 8, phosphoglucose isomerase; 9, ATP-dependent phosphofructokinase; 10, pyrophosphate-dependent phosphofructokinase; 11, NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (phosphorylating); 12, 3-phosphoglycerate kinase; 13, NADP-dependent glyceraldehyde 3-phosphate dehydrogenase (non-phosphorylating); 14, pyruvate kinase; 15, phosphoenolpyruvate phosphatase; 16, phosphoenolpyruvate carboxylase; 17, malate dehydrogenase; 18, malic enzyme; 19, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase; 20, 3-dehydroquinate dehydratase; 21, 5-enolpyruvylshikimate 3-phosphate (EPSP) synthase; 22, chorismate synthase; 23, tonoplast H+-ATPase; 24, tonoplast H+-pyrophosphate. Abbreviations are as follow: CHA, chorismate; DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; DHQ, dehydroquinate; 1,3-DPGA, 1,3-diphosphoglycerate; E4P, erythrose 4-phosphate; EPSP, 5-enolpyruvylshikimate 3-phosphate; Fru-6-P, fructose 6-phosphate; Fru-1,6-P2, fructose 1,6-bisphosphate; Glu-1-P, glucose 1-phosphate; Glu-6-P, glucose 6-phosphate; G3P, glyceraldehyde 3-phosphate; G3PDH; glyceraldehyde 3-phosphate dehydrogenase; OAA, oxaloacetate; PEP, phosphoenolpyruvate; 3-PGA, 3-phosphoglycerate; PPi, pyrophosphate; S3P, shikimate 3-phosphate. Reproduced from Plaxton and Carswell (1999) with permission by Marcel Dekker Inc.
Levels of ADP and Pi are important regulators of plant mitochondrial respiration. Reduced levels of ADP or Pi leads to a reduction in electron flux to the cytochrome pathway and lower cytochrome oxidase activity. Studies done largely in bean have shown that under Pi-deficiency, two non-phosphorylating pathways of mitochondrial electron transport are activated, namely the cyanide-resistant alternative oxidase pathway and the rotenone-insensitive bypass to complex I (Figure 14). The activity of these non-phosphorylating pathways would allow the functioning of the Krebs cycle and the mitochondrial electron transport chain with limited ATP production, which may contribute to enhanced survival of Pi-deficient plants (Plaxton and Carswell, 1999).
Phosphate deprivation in non-photosynthetic and photosynthetic bacteria has been shown to result in the replacement of phospholipids with non-phosphorus-containing lipids, such as sulfolipids (Benning et al., 1993; Minnikin et al., 1974). In Rhodobacter sphaeroides, mutation blocking synthesis of sulfolipids results in a decrease in growth of cells grown under low Pi conditions (Benning et al., 1993). The pho1 mutant of Arabidopsis, which has low leaf Pi, as well as wild type plants grown in media with low Pi, were shown to have a reduction in all phospholipids, including phosphatidylcholine and phosphatidylglycerol, and a corresponding increase in digalactosyl diacylglycerol and sulfoquinovosyl diacylglycerol (Dörmann and Benning, 2002; Essigmann et al., 1998). Pi deprivation has been found to be associated with an upregulation of the SQD1 and SQD2 genes encoding chloroplast proteins involved in sulfolipid biosynthesis (Essigmann et al., 1998; Yu et al., 2002). Insertional inactivation of the SQD2 gene resulted in a complete absence of sulfolipids (Yu et al. 2002). Although the mutant was largely indistinguishable from wild-type, reduced growth under Pi stress was observed. Pi deprivation was also found to induce a novel pathway for the synthesis of extraplastidic galactolipids (Härtel et al., 2000). Two novel genes coding for monogalactosyl diacylglycerol synthases have recently been shown to be induced under phosphate stress, indicating that the corresponding proteins could be involved in the pathway of extraplastidic galactolipids in Pi-stress plants (Awai et al., 2001). More recently, the DGD2 gene of Arabidopsis, encoding a digalactosyldiacylglycerol synthase, was shown to be strongly upregulated under Pi stress (Kelly and Dörmann, 2002). Together, these studies support the hypothesis that plants have adapted to Pi deprivation by functionally replacing phospholipids with non-phosphorus galactolipids and sulfolipids, thus avoiding severe disruption in photosynthesis and other functions relying on membrane lipids (Dörmann and Benning, 2002; Härtel et al., 2000).
One of the classical features of Pi deprivation stress in crop plants is the development of dark-green shoots due to the accumulation of anthocyanins, a class of red/purple colored flavonoids. Anthocyanins could be involved in the prevention of damage to nucleic acid, due to their absorption of UV light, as well as in the attenuation of photoinhibitory damage to chloroplasts that may arise during Pi-limited photosynthesis. Arabidopsis also responds to Pi deprivation by an increase in anthocyanins. This is easily observed in Arabidopsis mutants deficient in leaf Pi accumulation (Poirier et al. 1991; Zakhleniuk et al., 2001). Furthermore, wild type plants grown for 11 days in media deficient in Pi were shown to accumulate 4-times more flavonols compared to plants grown in Pi-sufficient medium, with an increase in quercetin, kaempferol and isorhamnetin concentrations (Stewart et al., 2001).
Phosphate deficiency and gene expression
A number of genes with often poorly defined functions have been found to be up-regulated by Pi deprivation stress. Several of these genes were identified through differential screening of cDNA libraries made from plants grown in media containing various concentrations of Pi. In tobacco cell culture, analysis by differential display of genes induced following the addition of Pi to Pi-stressed cultures led to the isolation of two genes, named PHI-1 and PHI-2 (Sano et al., 1999; Sano and Nagata, 2002). The function of these genes is unknown. A gene encoding a ß-glucosidase has been found to be up-regulated under Pi deprivation in both Brassica nigra suspension cells and in roots of Arabidopsis (Malboobi and Lefebvre, 1995, 1997). The role of this ß-glucosidase in Pi-stress response is unknown. In tomato, the LCA1 gene encoding a Ca+−ATPase was also found to be expressed differentially in phosphate-starved roots (Muchhal et al., 1997). The ARR6 gene of Arabidopsis, encoding a protein with similarity to bacterial response regulators, was shown to be up-regulated by a number of nutrient stress, including phosphorus, nitrogen and potassium deprivation (Coello and Polacco, 1999).
The AtIPS1 and AtIPS2 (or At4) genes of Arabidopsis were found to belong to a small family of genes which includes the TPSI1 gene of tomato and Mt4 gene of Medicago truncatula (Burleigh and Harrison, 1997; Liu et al., 1997; Martin et al., 2000). These genes are characterized by the presence of short and overlapping open reading frames (ORF). For example, the longest ORF for TPSI1 encodes a 58 amino acid peptide, while AtIPS1 contains one ORF starting with an AUG of 24 amino acid long while a second ORF, not starting with an AUG, is 80 amino acid long (Martin et al., 2000). Most of the ORFs of this gene family are non-conserved with the exception of a 4 amino acid peptide that is found in both Arabidopsis AtIPS1 and AtIPS2 genes. At the nucleotide level, AtIPS1 and AtIPS2 have two regions of sequence similarity of nearly 70% in regions of 251 and 96 nucleotides. However, when compared to the tomato TPSI1 and M. truncatula Mt4 genes, only a small stretch of 22 nucleotides is found to be conserved between all members of this gene family (Martin et al., 2000). All genes show strong transcript up-regulation in roots and shoots of plants undergoing Pi starvation (Burleigh and Harrison, 1997; Liu et al., 1997; Martin et al., 2000). No other nutrient stress was found to up-regulate these genes. One possibility is that peptides encoded by the AtIPS/Mt4/TPSI1 gene family are involved in mediating the Pi-stress response in plants. However, the lack of homology between these peptides suggests that perhaps it is rather the RNA molecules themselves, and in particular the homologous stretch of 22 nucleotides, which may be the active component. Clearly, further work is needed to understand the potential role of this gene family in Pi-stress response.
Split-root experiments in M. truncatula indicated that regulation of Mt4 expression in roots is more influenced by the Pi status of the shoot than by the external or local Pi concentration in the root (Burleigh and Harrison, 1999). A similar conclusion has also been reached concerning the regulation of the expression of the tomato Pi transporters LePT1 and LePT2 in roots (Liu et al., 1998). However, the implication of a shoot signal in the regulation of AtIPS1 in roots of Arabidopsis is less clear. The pho1 mutant of Arabidopsis is deficient in phosphate loading to the xylem and has low shoot Pi and high root Pi (Poirier et al., 1991). Transgenic pho1 plants expressing the GUS gene under the control of the AtIPS1 promoter revealed high GUS expression in the leaves but low GUS level in most cells of the root except for the endodermis and in emerging lateral roots. Application of cytokinin to plants growing in low Pi medium suppresses the induction of the AtIPS1, At4, AtACP5 and Pht1;1 (AtPT1) genes in roots but not in shoots (Martin et al., 2000). These results suggest a potential role for cytokinin in the regulation of several genes responding to Pi nutrition in roots.
Complete sequencing of the Arabidopsis genome and all the genomics tools associated with it, including microarray and AffimetrixTM technology, will undoubtedly allow a more comprehensive view of the genes and networks playing an active role in plant Pi-stress adaptation. A first glance at the range of genes that are up-regulated by phosphate stress has been revealed through an array analysis of genes involved in nitrogen nutrition (Wang et al., 2001). Nitrate supply to roots that had been starved for nitrogen was shown to lead to induction of the phosphate transporter LePT2, of a novel Leu zipper transcription factor, as well as of a non-symbiotic hemoglobin gene. Conversely, Pi deprivation was shown to lead to the up-regulation of the same genes. These data reveal the potential of connections and networks between the regulation of nitrogen and phosphorus nutrition (Wang et al., 2001).
MUTANTS AFFECTED IN PHOSPHATE ACQUISITION AND HOMEOSTASIS
The screening and analysis of Arabidopsis mutants affected in mineral nutrition has been a fruitful approach for the study of genes involved in the uptake and metabolism of ions, such as iron and nitrate (Crawford, 1994). To date, at least 6 mutants have been characterized in Arabidopsis that affect Pi uptake and metabolism. These mutants can be classified in two major classes: mutants primarily affected in components of Pi acquisition and distribution in plants, and mutants affected either in the perception or the response to Pi deprivation stress.
The first genetic screen for mutants affected in Pi acquisition was done by Poirier et al. (1991). Analysis of total P present in leaves of 2000 EMS-mutagenized plants enabled the isolation of a single plant having 3–4 times less total P than wild type. The mutant, named pho1, showed classical phenotypes of Pi deficiency, including reduced growth and accumulation of anthocyanins (Figure 15). The amount of free Pi present in leaves of the mutant grown in soil was 20 times lower than wild type, while the amount of P present in lipids, nucleic acid and other esters were largely unchanged. In plants grown in either a hydroponic culture system or in agar, the amount of Pi in leaves was also strongly reduced in the mutant, while Pi in roots was similar to wild type (Delhaize and Randall, 1995; Poirier et al., 1991). The quantity of other ions was mainly unchanged in the pho1 mutant, with the exception of a relatively small reduction in potassium content in leaves of the mutant. The rate of phosphate uptake into the roots was similar between mutant and wild type over a wide range of external phosphate concentration (0.32 to 1000 µM). In contrast, transfer of the Pi from the roots to the shoot was reduced to 3–10% of the wild type levels in plants grown in media with less than 200 µM external Pi. However, Pi that was delivered to the xylem via the hypocotyl (thus bypassing the root system) was transferred to the shoot at a normal rate. The defect in phosphate transfer from the roots to the shoots could be overcome by providing higher levels of Pi in the medium. Transfer of sulfate from the roots to the shoot was essentially normal in the pho1 mutant. Together, these results indicated that the pho1 mutant was impaired in a protein involved in the loading of Pi to the xylem in the root. This mutant provided a genetic evidence for the involvement of distinct protein(s) involved in the uptake of Pi from the external solution into the root and the subsequent transfer of the Pi to the xylem vessel for its transport to the shoot.
Fig. 15.

Phenotype of pho1 mutant. Arabidopsis wild type (left) and pho1 mutant (right) were grown in soil under constant illumination.
The PHO1 gene has recently been identified by a mapped-based positional cloning strategy (Hamburger et al., 2002). The protein is divided in two domains, the N-terminal half is mainly hydrophilic while the C-terminal half has 6 potential membrane-spanning domains. Although the presence of membrane-spanning domains is a feature expected for an integral membrane protein, such as an ion transporter, the structure of PHO1 appears quite distinct from members of the major facilitator superfamily, which typically have twelve membrane-spanning domains grouped in two clusters of six (Pao et al., 1998). Strikingly, PHO1 shows no homology to characterized solute transporters, including the family of Pht1 H+/Pi co-transporters identified in plants and fungi (see paragraph phosphate transporters). PHO1 shows, however, highest homology to the Rcm1 mammalian receptor for xenotropic murine leukemia retrovirus (Battini et al., 1999). Although the function of Rcm1 is still unknown, it is interesting to note that several receptors for mammalian retroviruses have been found to be solute transporters, such as transporters for amino acids and Na+-Pi cotransporters (Kavanaugh and Kabat, 1996; Rasko et al., 1999). PHO1 also shows significant homology to the Saccharomyces cerevisiae Syg1 protein involved in the mating pheromone signal transduction pathway (Spain et al., 1995). PHO1 was found to be mainly expressed in the roots and is only weakly up-regulated by Pi deprivation stress (Hamburger et al. 2002). Promoter-GUS fusion experiments revealed predominant expression of the PHO1 promoter in the stelar cells of the root and the lower part of the hypocotyl (Figure 16). This pattern is consistent with the role of PHO1 in loading Pi to the xylem (Poirier et al., 1991). Expression of PHO1 in yeast and Xenopus oocytes failed to reveal a Pi transport activity associated with protein. It is possible that the heterologous systems used to express PHO1 do not allow a functional reconstitution of transporter activity. Alternatively, it is possible that PHO1 represents only one subunit of a multi-subunit transporter and that the presence of other unidentified components is essential for Pi transport activity. Finally, it is also possible that PHO1 may not directly be involved in Pi transport but that it indirectly influences the activity of a plasma membrane Pi transporter. Clearly further studies are required to reveal the mode of action of PHO1 in Pi loading to the xylem. Interestingly, the Arabidopsis genome contains 10 additional genes showing homology to PHO1 (Hamburger et al., 2002). It is likely that several members of this family may also play a role in Pi homeostasis.
Fig. 16.
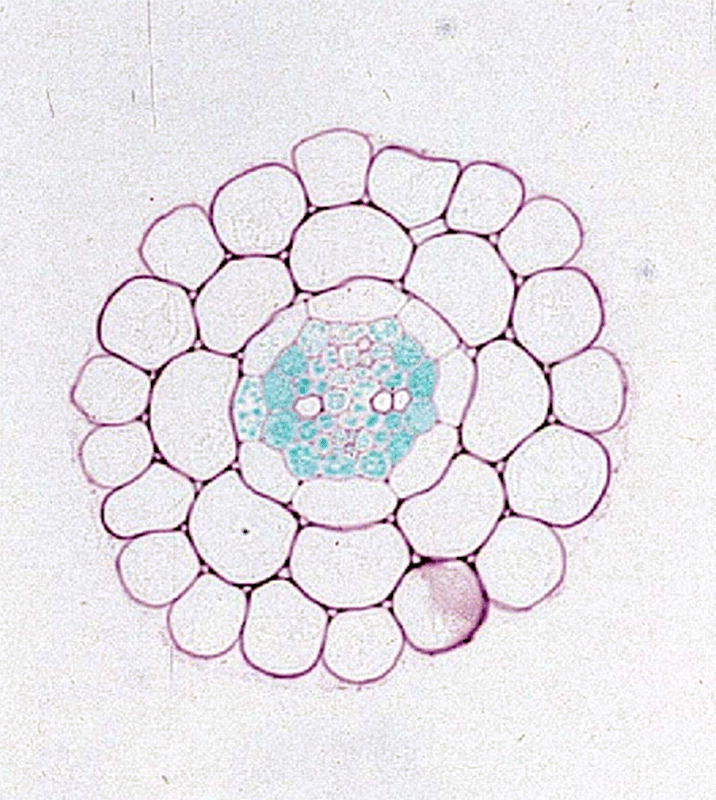
Expression profile of the PHO1 promoter. A 2 kbp promoter region of PHO1 was cloned in front of the GUS reporter gene and transformed in Arabidopsis. GUS expression is detected in the stelar cells of the roots.
Delhaize and Randall (1995) have isolated a second mutant affected in Pi acquisition. Approximately 100 000 EMS mutagenized plants were grown in soil and screened visually for phenotypes consistent with nutrient toxicity or deficiency. Approximately 2000 plants identified in this first screen were subsequently analyzed by X-ray fluorescence spectrometry (Delhaize et al., 1993). From this screen a novel allele of pho1 was identified as well as two alleles of a novel mutant named pho2. This mutant was characterized by a 2- to 4-fold increase in the amount of Pi in leaves compared to wild type while the level of Pi in roots was unchanged (Delhaize and Randall, 1995). There was also an increase of Pi in stems, siliques, and seeds. The Pi toxicity symptoms of the pho2 mutant were apparently affected by the transpiration rate. Under high transpiration conditions in soil-grown plants, the margins of the pho2 leaves became necrotic, and under extreme conditions, the whole leaf senesced. Pho2 mutant plants were approximately 50% smaller (by fresh weight) compared to wild type. Although toxicity symptoms were influenced by transpiration rates, the higher Pi level in leaves of pho2 were observed even in plants grown submerged in liquid solution, indicating that the higher accumulation of Pi in leaves was independent of transpiration.
Analysis of the kinetics of Pi uptake into whole seedlings revealed that the pho2 mutant had about a twofold greater Pi uptake rate than wild type plants under Pi-sufficient conditions. Furthermore, a greater proportion of the acquired Pi was located in the shoot in pho2 (Dong et al., 1998). This difference in the Pi uptake rate between pho2 and wild type seedlings was abolished when shoots were removed, clearly indicating that the mutation primarily affected shoot Pi acquisition. Although pho2 mutants could recycle Pi from the shoots to the roots through the phloem, the proportion of shoot Pi translocated to the roots was less than half of that found for the wild type plants. Two mechanisms have been proposed to explain the phenotype of the pho2 mutant. One model is that pho2 has a specific impairment in a leaf Pi transporter that would influence the phloem transport of Pi between shoots and roots. In this context, it is interesting to note that the pho2 locus does not map close to any of the PHO1 homologues or of the low affinity shoot-specific Pi transporter Pht2;1 gene, which has been hypothesized to play a role in Pi loading of shoot organs (Daram et al., 1999). Alternatively, pho2 could have a defect in a protein involved in sensing leaf Pi concentration, which in turn regulates the activity or expression of Pi transporters in the shoot, leading to uncontrolled accumulation of Pi under Pi-sufficient conditions (Dong et al., 1998). A clearer picture of the role of PHO2 in Pi homeostasis will require the eventual cloning and characterization of the gene.
Four Arabidopsis mutants have been isolated with genetic screens based on the induction of Pi-scavenging proteins and genes, such as phosphatases and nucleases, in response to Pi deprivation stress. For example, the pup1 and pho3 mutants were isolated by screening for an alteration in the induction of root acid phosphatases in plant grown in Pi-deficient media. Acid phosphatases are detected through the conversion of the colorless substrate 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) to a blue compound following cleavage of the phosphate group. Screening of mutagenized plants growing in Pi-deficient agar medium containing BCIP lead to the isolation of three mutants showing reduced blue staining of the roots. Of those mutants, the pup1 (phosphatase underproducer) mutant has been described (Trull and Deikman, 1998). The mutant was shown to be missing only one (160 kD) of the multiple acid phosphatase found in Arabidopsis. The mutant was otherwise normal in most of the classical responses to Pi deprivation stress, including up-regulation of phosphatase activity, accumulation of anthocyanins, change in root growth and morphology, as well as changes in Pi distribution between shoot and roots (Trull and Deikman, 1998). The overall growth pattern of the mutant was largely indistinguishable from wild type. It is likely that the PUP1 gene codes for the 160 kD acid phosphatase.
In a separate screen based on overlaying roots of Pi-deficient plants with an agar layer containing BCIP, a mutant named pho3 was isolated (Zakhleniuk et al., 2001). The mutant had lower level of acid phosphatase activity in roots of Pi-deficient and Pi-sufficient plants compared to wild type, while the shoot acid phosphatase activity was higher in the mutant under the same conditions. The changes in phosphatase activity were not correlated to a particular isoform. The pho3 mutant showed a typical Pi-stress response when grown in soil, including poor growth, production of anthocyanins, and increased accumulation of starch. In the rosette leaves of mature soil-grown plants, the total P amount in the mutant was 50% of wild type levels. In plants grown in agar media containing 0.63 mM Pi, the total P level was reported to be 25% lower than the wild type level in the shoot and about 65% lower in the root. This decrease in total P is, however, not reflected by a decrease in level of free Pi in roots (Lloyd et al., 2001). It is thus unclear at this point which of the various Pi-containing components of the plant cells is reduced in the mutant. The pho3 mutant shares several of the characteristics of the pho1 mutant, including overall reduced growth, lower P level and accumulation of anthocyanins. However, in contrast to pho1, the roots of pho3 mutant appear to show a reduced total P content compared to wild type. As no complementation test has been done between pho1 and pho3, it is still unclear whether the PHO3 gene is distinct from PHO1.
Nucleases are some of the enzymes that are induced under Pi deprivation stress and that are involved in scavenging Pi from the environment. Arabidopsis has been shown to be able to grow in media containing DNA as the main Pi source, indicating that the release of nucleases and phosphatases is sufficient to acquire Pi from extracellular DNA (Chen et al., 2000). A genetic screen has been based on this ability of Arabidopsis to acquire Pi from exogenous DNA. A screen of 50 000 mutagenized seedlings lead to the isolation of 22 mutant lines that showed severe growth defect in media containing DNA as the only Pi source, but which recovered on medium containing soluble inorganic Pi. Further analysis of nine of these lines revealed different levels of growth retardation of roots in media containing either DNA, RNA or inorganic Pi. One line, named psr1 (phosphate starvation response) showed a significant reduction in the level of several isoforms of acid phosphatases as well as of both RNS1 and RNS2 ribonucleases in plants grown in Pi-deficient medium, indicating that the mutant was affected in the production of several enzymes normally induced by Pi deprivation stress (Chen et al., 2000). Genetic analysis indicated that psr1 behaved as a single recessive mutation. These data are consistent with the hypothesis that psr1 may be a mutant in a component of the Pi starvation response pathway in higher plant.
The phr1 (phosphate starvation response) mutant was isolated by screening a mutagenized population containing the GUS reporter gene under the control of the promoter of the AtIPS1 gene (AtIPS1::GUS) (Rubio et al., 2001). In wild type plants transformed with the AtIPS1::GUS construct, GUS expression is strongly induced in Pi-deficient plants (Martin et al., 2000). The phr1 mutant displayed a strong reduction in GUS expression following Pi deprivation stress, as well as a reduction in the expression of several genes normally induced by Pi starvation, such as AtIPS1 and At4 genes, the high affinity Pi transporter Pht1;1 (AtPT1), the acid phosphatase AtACP5, the ribonuclease RNS1, and AtIPS3, a gene encoding a protein of unknown function. Furthermore, the phr1 mutant was deficient in the accumulation of anthocyanins normally induced by Pi deficiency, although anthocyanins accumulation induced by jasmonic acid and abscisic acid was normal. Also normal in the mutant was the increase in density and length of root hairs induced by Pi deprivation stress (Rubio et al., 2001). The PHR1 gene has been cloned and was shown to be a transcription factor (see below for further details). PHR1 thus represents the first identified element of the phosphate starvation signaling in higher plants (Rubio et al., 2001).
In addition to the forward genetic approach of mutant screening and gene isolation, the cloning and characterization of numerous Pi transporters opens the way for the analysis of mutants in these transporters by reverse genetic approaches using T-DNA or transposon tagging. A mutant in the PHT2;1 gene encoding a low affinity Pi transporter located in the chloroplast has recently been described (Versaw and Harrison, 2002). Although only a single copy of the PHT2;1 gene is present in the Arabidopsis genome, the phenotype of the null mutant was subtle. Mutants grown in high Pi media had a slightly reduced rosette size (70–80% of wild type ) and reduced Pi content (74% of wild type). Under low-Pi conditions, no difference were observed in fresh weight of mutants and wild type. However, root Pi content of the mutant was 50–70% that of the wild type, while leaf Pi content of the mutant was 130–150% that of the wild type. Redistribution of Pi from old leaves towards young leaves in plants that were switched from high Pi media to low Pi media was reduced in mutants. Together, these results indicate a deficiency in Pi retranslocation under Pi-deprivation stress in the mutant. Expression analysis of several genes known to be up-regulated by Pi-deprivation stress indicated that the response was stronger in the mutant compared to wild type.
ANALYSIS OF PHOSPHATE ACQUISITION USING ARABIDOPSIS ACCESSIONS
In addition to the uses of mutants in a forward genetic approach using physical, chemical or biological (T-DNA or transposon) mutagenesis, exploitation of the natural variations present among wild type accessions of Arabidopsis can be used to gain insight into the genetic components of Pi acquisition (Alonso-Blanco and Koornneef, 2000). Krannitz et al. (1991a, b) measured a number of parameters associated with Pi acquisition in plants that were grown on Pi-depleted media for 16 days. These parameters included initial Pi uptake rates, the external Pi concentration at which there is no net Pi uptake (referred as Cmin), root length and mass, as well as root/shoot ratio. Highly significant differences were recorded among the accessions for all measured parameters.
Recently, Narang et al. (2000) have used the natural variations found in accessions to analyze the morphological and physiological parameters which have the greatest impact on the ability of Arabidopsis to acquire Pi from a sparingly soluble source. An initial set of 36 accessions were grown on 0.4% agarose-solidified media with Pi supplied either as soluble potassium phosphate (KH2PO4) or as the sparingly soluble hydroxylapatite (Ca5(PO4)3OH). The phosphate acquisition efficiency (PAE) was measured as the mean of the (phosphate content in shoot for plants grown in media containing 0.5 g/L hydroxylapatite / phosphate content in shoot for plants grown in media containing 1 mM potassium phosphate). The accessions C24, Co and Cal exhibited the highest PAE while Col-0 and Te exhibited the lowest PAE. Using these five accessions, significant differences were recorded for root morphology (root length and mass, root hair length and density), phosphate uptake kinetics, rhizosphere acidification, organic acid release, and the ability of the roots to penetrate substrates of different density (agarose concentrations of 0.15% and 0.4%). The high PAE of accessions C24 and Co was mediated by a root system having long and dense root hairs, high Pi uptake per unit root length and high substrate penetration ability. In contrast, the accessions Col-0 and Te having a low PAE produced roots that had a low substrate penetration, short and sparse root hairs, and low uptake efficiency per unit root length. The accession Cal, was intermediate for these parameters, but displayed in addition an enhanced rhizosphere acidification capability. This study highlighted the importance of the architecture of the root system, and in particular of the root hairs, in the acquisition of Pi from sparingly soluble source.
F1 hybrids resulting from a cross between accessions C24 and Col-0 showed superior ability to acquire Pi from hydroxylapatite when compared to either parent, representing a classical case of heterosis or hybrid vigor (Narang and Altmann, 2001). The hybrid inherited the long root hair length and high phosphate transporter expression from C24 while it inherited the long root length trait of Col-0. The root hair density of the hybrid was intermediate between the parents. The data suggests that the superiority of the F1 hybrid is due to the accumulation of favorable dominant genes at numerous loci.
THE PHOSPHATE REGULON IN PLANTS
As described above, plants have evolved a complex series of adaptive responses to Pi deprivation, which includes secretion of Pi-mobilizing enzymes, such as phosphatases, and changes in the Pi transporters. Similar adaptive changes to Pi deprivation also occur in bacteria and fungi. The mechanisms that control the adaptation of Escherichia coli and S. cerevisiae to Pi limitation have been extensively studied and numerous components of the regulatory networks linking Pi levels to gene activation have been described. Genetic screens aimed at understanding how Pi deprivation leads to activation of secreted phosphatases in both E. coli and S. cerevisiae have been central to this research.
In E. coli, the PHO regulon comprises a set of at least 15 genes involved in the acquisition of Pi (Figure 17) (Torriani, 1990). These genes are controlled by a two-component regulatory system encoded by the phoB-phoR operon. PhoR is a histidine protein kinase located in the inner membrane and acts as a Pi sensor. Under Pi deprivation, phoR autophosphorylates itself, which in turn leads to phosphorylation of phoB, the positive regulatory protein. The phosphorylated phoB binds to a specific region (the Pho box) of the promoter of each gene of the Pho regulon and activates transcription.
Fig. 17.

The Pho regulon of E. coli. Abbreviations: OM, outer membrane; IM, inner membrane; AP, alkaline phosphatase; G3P, glycerol 3-phosphate. The Pho regulon is composed of the porin E protein (PhoE), glycerol -3-phosphate binding protein (G-3-PBP), sn-glycerol 3-phosphate uptake system (Ugp A, UgpC, UgpQ), alkaline phophatase (PhoA), phosphate binding protein (PiBP), Pi specific transporter (PstA, PstB, PstC), protein kinase-sensor (PhoR), positive regulator-transcriptional activator (PhoB), and a modulator of PhoR (PhoU). Reproduced from Torriani (1990) with permission by Wiley Publishers.
In S. cerevisiae, the Pho regulon comprises a set of at least 22 genes (Ogawa et al., 2000). The proteins Pho80, Pho81, Pho85 and Pho4 are the key factors controlling expression of the genes under the Pho regulon (Figure 18) (Lenburg and O'Shea, 1996). Pho4 is the transcription factor that binds a consensus nucleotide (Pho4 box) found in the promoters of genes forming the regulon, which includes the Pi transporters Pho84 and Pho 89 as well as the acid phosphatase Pho5. The level of phosphorylation controls activation of Pho4. Whereby the hyperphosphorylated Pho4 is mainly located in the cytoplasm, the hypophosphorylated form is found predominantly in the nucleus where it can act to activate transcription. Phosphorylation of Pho4 is controlled by the Pho80-Pho85 cyclin-cyclin dependent protein kinase complex (CDK), which is itself controlled by the Pho81 CDK inhibitor. Under low Pi conditions, the Pho81 protein inhibits the Pho80-Pho85 complex, preventing phosphorylation of Pho4, and thus activating transcription of the regulon. Alternatively, under high Pi availability, the Pho80-Pho85 kinase phosphorylates Pho4. Pho4 interacts with Pho2 which itself activates transcription of only a subset of Pho4-dependent genes.
Fig. 18.

The Pho regulon of S. cerevisiae. Ovals and boxes represent proteins and genes, respectively. Thick lines mean that the signals are transduced to the downstream component, while dotted lines indicate the absence of an interaction with the downstream component. Open ovals and boxes indicate active states, gray oval and boxes indicate inactive state. Reproduced from Ogawa et al (2000) with permission by the American Society for Cell Biology.
By analogy to E. coli and S. cerevisiae, elucidation of the signaling components linking Pi deprivation to the regulation of genes involved in metabolic and developmental adaptation of plants to nutrient deficiency has been tackled by a genetic approach. As described above, genetic screens have been done in Arabidopsis to isolate mutants that are affected in the response of plant to Pi deficiency. The pup1 and pho3 mutants were isolated based on a reduced activation of extracellular root acid phosphatases following Pi deficiency (Trull and Deikman, 1998; Zakhleniuk et al., 2001). Whereas pup1 is likely to be defective in a gene encoding one particular acid phosphatase, the defect in pho3 is unclear and could involve a Pi transporter. In contrast, the Arabidopsis psr1 mutant, which was found defective in the production of both nucleases and acid phosphatases under Pi deficiency, is likely to encode a gene involved directly in the Pho signal transduction pathway (Chen et al., 2000). The Arabidopsis PSR1 gene has not yet been isolated.
The first gene involved in the Pi-stress signal induction pathway in photosynthetic eukaryotes has been identified in Chlamydomonas reinhardtii (Wykoff et al., 1999). The C. reinhardtii mutant psr1 was isolated from a screen based both on the survival of cells after exposure to high concentration of radioactive 32Pi as well as on the reduced activation of acid phosphatase following Pi deprivation (Shimogawara et al., 1999). The mutant also fails to increase the rate of Pi transport upon exposure to Pi limitation. The C. reinhardtii PSR1 gene was cloned and shown to encode a protein containing at the N-terminal half a region similar to the MYB DNA-binding domain, and at the C-terminal half a glutamine-rich sequence characteristic of transcriptional activators (Wykoff et al., 1999). The level of PSR1 mRNA transiently increases up to 13-fold 8 hours after a shift to Pi-free media. The level of PSR1 protein increases at least 10-fold one day after Pi starvation. Immunolocalization of the protein demonstrated that it was located in the nucleus in both Pi-sufficient and Pi-deficient cells. Together, these data support the conclusion that PSR1 is a transcriptional factor and that under Pi deprivation stress the increased accumulation of PSR1 in the nucleus leads to activation of genes forming the Pho regulon of C. reinhardtii.
A functional homologue of the C. reinhardtii PSR1 gene has been identified in Arabidopsis (Rubio et al., 2001). The Arabidopsis phr1 mutant was isolated based on the lack of induction of the promoter of the AtIPS1 gene under Pi deprivation stress, as detected by a AtIPS1::GUS construct. The mutant also fails to activate a number of Pi starvation-induced genes and metabolic responses, including accumulation of anthocyanin. The Arabidopsis PHR1 protein shares with the C. reinhardtii PSR1 the presence of a MYB DNA-binding domain and a predicted coiled-coil domain, indicating potential dimerization of the protein with itself or with other proteins. Analysis of the binding of PHR1 to sequences of the AtIPS1 promoter indeed revealed that PHR1 binds as a dimer to an imperfect palindromic sequence. This PHR1-binding sequence is present in the promoter of several genes that are up-regulated following Pi deprivation stress, such as the promoter of the acid phosphatase AtACP5, the ribonuclease RNS1 and the Pi transporter Pht1;1 (AtPT1) (Rubio et al., 2001). PHR1 was found located in the nucleus in both Pi-deficient and Pi-sufficient cells. However, in contrast to the C. reinhardtii PSR1 gene, the PHR1 gene is only weakly up-regulated by Pi deprivation stress. It is concluded that PHR1 is a transcription factor participating in the Pho regulon system of higher plants. Interestingly, the phr1 mutant is not defective in the increase in root hair density and length in response to Pi deprivation stress. Thus, only a subset of Pi starvation responses is mediated via PHR1. At least 15 other proteins are present in Arabidopsis that shares features of the MYB-coiled-coil protein family, raising the possibility that other members of this family may also contribute to the response of plants to Pi deficiency.
Cytokinins have been shown to repress the induction of several genes in roots by Pi deficiency, including AtIPS1 and At4 (Martin et al., 2000). Screening for Arabidopsis plants which have high AtIPS1 gene expression when grown in Pi-deficient media containing kinetin identified several mutants which were allelic to the cre1 mutant (Franco-Zorrilla et al., 2002). CRE1 encodes for a cytokinin receptor that may trigger a cascade of phosphorelay reactions upon perception of the hormone. In addition to AtIPS1, several Pi-starvation response genes showed reduced cytokinin sensitivity in the cre1 mutants, such as AtACP5 and AtPT1. CRE1 was down-regulated by Pi starvation and induced by cytokinins. These results indicate an important contribution and integration of the cytokinin two-component signaling circuitry with the regulation of Pi starvation responses in roots.
Analysis and identification of the promoter sequences and DNA binding proteins that are involved in the regulation of genes induced by Pi deprivation stress is still at an early stage in plants. Proteins binding to the promoter region of the tomato TPSI1 gene and of the Arabidopsis Pi transporter Pht1;4 (AtPT2) were analyzed by DNA mobility-shift assay (Mukatira et al., 2001). These two genes are specifically up-regulated by Pi deprivation. Two regions of these promoters specifically bound nuclear protein factors only from Pi-sufficient plants and not from Pi deprivation-stressed plants. Similar findings have also been found for the B. nigra ß-glucosidase gene induced under Pi deprivation stress (Malboobi et al., 1998). These results indicate that induction of genes by Pi deprivation may be, at least in part, mediated by the decreased interaction of repressor proteins with the promoters of these genes.
Phosphite (also referred as phosphorous acid or phosphonate, HPO32-) is an isotere of the Pi anion in which one oxygen is replaced by a hydrogen. Phosphite is rapidly absorbed by plant cells and competes with Pi, indicating that the same transporters are involved in the entry of Pi and phosphite (MacDonald et al., 2001). However, in contrast to Pi, phosphite cannot be covalently assimilated into organic compounds, as enzymes catalyzing the transfer of Pi do not accept phosphite. Phosphite has been shown to efficiently suppress many of the Pi starvation response observed in Arabidopsis and Brassica nigra. Thus, growth of plant on media without Pi but containing phosphite do not respond to Pi deprivation with changes in root/shoot ratio, root hair formation, anthocyanin accumulation, activities of phosphate-scavenging enzymes such as RNAse, DNAse and phosphatase, as well as changes in the expression of several genes normally induced by Pi-deprivation stress, including AtACP5, AtPT1, AtPT2 and At4 (Ticconi et al., 2001, Varadarajan et al., 2002). These experiments indicate that the cell sensors and/or signal transduction components involved the coordinated response to Pi-deprivation stress actually perceive phosphite as Pi even though the former compound is not metabolized, thus depriving the plants of the mechanisms needed to adapt to this stress. Phosphite may be used as a useful tool for the search of genes involved in the Pi starvation signal transduction pathway (Abel et al., 2002).
CONCLUSIONS
Although progress on the knowledge of the genes and proteins implicated in Pi transport and homeostasis has accelerated in recent years, still much remains to be learned. For example, although the whole genome of Arabidopsis has now been sequenced, we still do not know any gene that is involved in the transport of Pi across the vacuole, an important aspect of Pi homeostasis. Combination of both classical forward genetics along with reverse genetics and all the tools associated with functional genomics should allow to gain new insights into the structure and regulation of Pi transport across all membranes, as well as on the signal transduction pathways and biochemical regulation involved in Pi deprivation stress response and Pi homeostasis. Ultimately, it is hoped that our knowledge on Pi transport and metabolism in Arabidopsis and other plants will permit the development of crops capable of high yield with reduced additional input of Pi in soil.
Acknowledgments
Research on phosphate metabolism in the authors' laboratories has been funded, in part, by grants from the Swiss National Fund (grants 31-41903.94 and 31-61731.00 to YP, grant 31-52934.97 to MB), the Swiss Federal Office for Education and Research (grants 96.0023-2 to YP and 96.0023-1 to MB) which falls within the 1994–1998 Biotechnology Work Programme of the European Union (grant BIO4CT-960-770), the University of Lausanne (YP), the Canton de Vaud (YP), and the ETH Zürich (MB).
Footnotes
Citation: Poirier Y., and Bucher M. (2002) Phosphate Transport and Homeostasis in Arabidopsis. The Arabidopsis Book 1:e0024. doi:10.1199/tab.0024
elocation-id: e0024
Published on: September 30, 2002
Table 1: List of members of Arabidopsis thaliana Pi transporter families. Tissue localization by Northern blots

REFERENCES
- Abel S., Nürnberger T., Ahnert V., Krauss G-J., Glund K. Induction of an extracellular cyclic nucleotide phosphodiesterase as an accessory ribonucleolytic activity during phosphate starvation of cultured tomato cells. Plant Physiol. 2000;1221(1):543–552. doi: 10.1104/pp.122.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S., Ticconi C. A., Delatorre C. A. Phosphate sensing in higher plants. Physiol. Plant. 2002;1151(1):1–8. doi: 10.1034/j.1399-3054.2002.1150101.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 2000;51(1):22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- Awai K., Maréchal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K-I., Ohta H., Joyard J. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2001;981(1):10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola P. A., Green P. J. Plant ribonucleases. 1997;1(1):163–190. In Ribonucleases: Structure and Functions, G. D'Alessio and J.F. Riordan, eds, (New York, USA: Academic Press), pp. [Google Scholar]
- Bariola P. A., Howard C. J., Taylor C. B., Verburg M. T., Jaglan V. D., Green P. J. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;61(1):673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bariola P. A., MacIntosh G. C., Green P. J. Regulation of S-like ribonuclease levels in Arabidopsis: antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol. 1999;1191(1):331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T. R., Lynch J. P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;191(1):529–538. [Google Scholar]
- Bates T. R., Lynch J. P. The efficiency of Arabidopsis thaliana (Brassicaceae) root hairs in phosphorus acquisition. Am. J. Bot. 2000a;871(1):964–970. [PubMed] [Google Scholar]
- Bates T. R., Lynch J. P. Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2000b;871(1):958–963. [PubMed] [Google Scholar]
- Bates T. R., Lynch J. P. Root hairs confer a competitive advantage under low phosphorus availability. Plant Soil. 2001;2361(1):243–250. [Google Scholar]
- Battini J-L., Rasko J. E. J., Miller A. D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA. 1999;961(1):1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C., Beatty J. T., Prince R. C., Somerville C. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl. Acad. Sci. USA. 1993;901(1):1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J. E. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars light and phosphate. Plant Mol. Biol. 1995;271(1):933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Bieleski R. L. Phosphate pools, phosphate transport, and phosphate availability. Ann. Rev. Plant. Physiol. 1973;241(1):225–252. [Google Scholar]
- Brinch-Pedersen H., Sorensen L. D., Holm P. B. Engineering crop plants: getting a handle on phosphate. Trends Plant Sci. 2002;71(1):118–125. doi: 10.1016/s1360-1385(01)02222-1. [DOI] [PubMed] [Google Scholar]
- Bowsher C. G., Tobin A. K. Compartmentation of metabolism within mitochondria and plastids. J. Exp. Bot. 2001;521(1):513–27. [PubMed] [Google Scholar]
- Buchanan B. B., Gruissem W., Russel L. J. Biochemistry and Molecular Biology of Plants. 2000. American Society of Plant Physiologists, Rockville, Maryland.
- Bucher M., Rausch C., Daram P. Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant. Nutr. Soil Sci. 2001;1641(1):209–217. [Google Scholar]
- Bun-Ya M., Nishimura M., Harashima S., Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 1991;111(1):3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh S. H., Harrison M. J. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol. Biol. 1997;341(1):199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- Burleigh S. H., Harrison M. J. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;1191(1):241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. L., Delatorre C. A., Bakker A., Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;2111(1):13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- Chiou T. J., Liu H., Harrison M. J. The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 2001;251(1):281–293. doi: 10.1046/j.1365-313x.2001.00963.x. [DOI] [PubMed] [Google Scholar]
- Cieresko I., Johansson H., Hurry V., Kleczkowski L. A. Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta. 2001;2121(1):598–605. doi: 10.1007/s004250000424. [DOI] [PubMed] [Google Scholar]
- Coello P., Polacco J. C. ARR6, a response regulator from Arabidopsis, is differentially regulated by plant nutritional status. Plant Science. 1999;1431(1):211–220. [Google Scholar]
- Cogliatti D. H., Clarkson D. T. Physiological changes in, and phosphate uptake by potato plants during development of, and recovery from phosphate deficiency. Physiol. Plant. 1983;581(1):287–294. [Google Scholar]
- Crawford N. M. Metabolic and genetic control of nitrate, phosphate, and iron assimilation in plants. 1994;1(1):1119–1146. In Arabidopsis, E. Meyerowitz and C. Somerville, eds (Cold Spring Harbor USA, Cold Spring Harbor Press), pp. [Google Scholar]
- del Pozo J. C., Allona I., Rubio V., Leyva A., de la Peña A., Aragoncillo C., Paz-Ares J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J. 1999;191(1):579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- Daram P., Brunner S., Persson B. L., Amrhein N., Bucher M. Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta. 1998;2061(1):225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- Daram P., Brunner S., Rausch C., Steiner C., Amrhein N., Bucher M. Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell. 1999;111(1):2153–2166. doi: 10.1105/tpc.11.11.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Hebb D. M., Ryan P. R. Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiol. 2001;1251(1):2059–2067. doi: 10.1104/pp.125.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Randall P. J. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;1071(1):207–211. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Randall P. J., Wallace P. A., Pinkerton A. Screening Arabidopsis for mutants in mineral nutrition. Plant Soil. 1993;155/1561(1):131–134. [Google Scholar]
- Dodds P. N., Clarke A. E., Newbigin E. Molecular characterization of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol. Biol. 1996;311(1):227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 1994;1201(1):2465–2474. [Google Scholar]
- Dong B., Ryan P. R., Rengel Z., Delhaize E. Phosphate uptake in Arabidopsis thaliana: dependence of uptake on the expression of transporter genes and internal phosphate concentrations. Plant Cell Environ. 1999;221(1):1455–1461. [Google Scholar]
- Dong B., Rengel Z., Delhaize E. Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta. 1998;2051(1):251–256. doi: 10.1007/s004250050318. [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C. Galactolipids rule in seed plants. Trends Plant Sci. 2002;71(1):112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- Drew M. C., Saker L. R., Barber S. A., Jenkins W. Changes in the kinetics of phosphate and potassium absorption in nutrient-deficient barley roots measured by a solution-depletion technique. Planta. 1984;1601(1):490–499. doi: 10.1007/BF00411136. [DOI] [PubMed] [Google Scholar]
- Duff S. M. G., Moorhead G. B. G., Lefebvre D. D., Plaxton W. C. Phosphate starvation inducible “bypasses” of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;901(1):1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff S. M. G., Sarath G., Plaxton W. C. The role of acid phosphatases in plant phosphorus metabolism. Physiol. Plant. 1994;901(1):791–800. [Google Scholar]
- Eicks M., Maurino V., Knappe S., Flugge U-I., Fischer K. The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in Plants. Plant Physiol. 2002;1281(1):512–522. doi: 10.1104/pp.010576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E., Hagen C. E. A kinetic study of the absorption of alkali cations by barley roots. Plant Physiol. 1952;271(1):457–474. doi: 10.1104/pp.27.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E., Rains D. W., Elzam O. E. Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. U.S.A. 1963;491(1):684–692. doi: 10.1073/pnas.49.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B., Güler S., Narang R. A., Linke D., Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1998;951(1):1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Riviere-Rolland H., Vermat T., Seigneurin-Berny D., Grunwald D., Garin J., Joyard J., Rolland N. Integral membrane proteins of the chloroplast envelope: Identification and subcellular localization of new transporters. Proc Natl Acad Sci USA. 2002;991(1):11487–11492. doi: 10.1073/pnas.172390399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Kammerer B., Gutensohn M., Arbinger B., Weber A., Hausler R. E., Flügge U. I. A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell. 1997;91(1):453–62. doi: 10.1105/tpc.9.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U-I. Plant chloroplasts and other plastids. Encyclopedia of Life Sciences. 2001. Macmillan Publishers Ltd, Nature Publishing Group; www.els.net.
- Flügge U-I. Phosphate translocators in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;501(1):27–45. doi: 10.1146/annurev.arplant.50.1.27. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Martin A. C., Solano R., Rubio V., Leyva A., Paz-Ares J. Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 2002;321(1):353–360. doi: 10.1046/j.1365-313x.2002.01431.x. [DOI] [PubMed] [Google Scholar]
- Gardner W. K., Parbery D. G., Barber D. A. Proteoid root morphology and function in Linus albus. Plant Soil. 1981;601(1):143–147. [Google Scholar]
- Goldstein A. H., Beartlein D. A., McDaniel R. G. Phosphate starvation inducible metabolism in Lycopersicon esculentum: I. Excretion of acid phosphatase by tomato plants and suspension cultured cells. Plant Physiol. 1988;871(1):711–715. doi: 10.1104/pp.87.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;141(1):889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran S., Logendra S., Seskar M., Bratanova M., Raskin I. Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol. 2000;1241(1):615–626. doi: 10.1104/pp.124.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. J., Dewbre G. R., Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 2002;141(1):2413–2429. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H., Dörmann P., Benning C. DGD1-independant biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2000;971(1):10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H., Essigmann B., Lokstein H., Hoffmann-Benning S., Peters-Kottig M., Benning C. The phospholipid-deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Bioch. Biophys. Acta. 1998;14151(1):205–218. doi: 10.1016/s0005-2736(98)00197-7. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;3471(1):166. [Google Scholar]
- Hurry V., Strand A., Furbank R., Stitt M. The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J. 2000;241(1):383–396. doi: 10.1046/j.1365-313x.2000.00888.x. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., Flügge U-I. Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: The glucose 6-phosphate/phosphate antiporter. Plant Cell. 1998;101(1):105–117. doi: 10.1105/tpc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A. S., Varadarajan D. K., Mukatira U. T., D'Urzo M. P., Damsz B., Raghothama K. G. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002;1301(1):221–233. doi: 10.1104/pp.020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh M. P., Kabat D. Identification and characterization of a widely expressed phosphate transporter/retrovirus receptor family. Kidney Int. 1996;491(1):959–963. doi: 10.1038/ki.1996.135. [DOI] [PubMed] [Google Scholar]
- Kelly A. A., Dörmann P. DGD2, and Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase, is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 2002;2771(1):1166–1173. doi: 10.1074/jbc.M110066200. [DOI] [PubMed] [Google Scholar]
- Kiiskinen M., Korhonen M., Kangasjarvi J. Isolation and characterization of cDNA for a plant mitochondrial phosphate translocator (Mpt1): ozone stress induces Mpt1 mRNA accumulation in birch (Betula pendula Roth). Plant Mol. Biol. 1997;351(1):271–279. doi: 10.1023/a:1005868715571. [DOI] [PubMed] [Google Scholar]
- Köck M., Löffler A., Abel S., Glund K. cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol. Biol. 1995;271(1):45–50. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Koyama H., Kawamura A., Kihara T., Hara T., Takita E., Shibata D. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 2000;411(1):1030–1037. doi: 10.1093/pcp/pcd029. [DOI] [PubMed] [Google Scholar]
- Koyama H., Takita E., Kawamura A., Hara T., Shibata D. Over expression of mitochondrial citrate synthase gene improves the growth of carrot cells in Al-phosphate medium. Plant Cell Physiol. 1999;401(1):482–488. doi: 10.1093/oxfordjournals.pcp.a029568. [DOI] [PubMed] [Google Scholar]
- Krannitz P. G., Aarssen L. W., Lefebvre D. D. Relationships between physiological and morphological attributes related to phosphate uptake in 25 genotypes of Arabidopsis thaliana. Plant Soil. 1991a;1331(1):169–175. [Google Scholar]
- Krannitz P. G., Aarssen L. W., Lefebvre D. D. Correction for non-linear relationships between root size and short term Pi uptake in genotype comparisons. Plant Soil. 1991b;1331(1):157–167. [Google Scholar]
- Lee R. B. Selectivity and kinetics of ion uptake by barley plants following nutrient deficiency. Ann. Bot. 1982;501(1):429–449. [Google Scholar]
- Leggewie G., Willmitzer L., Riesmeier J. W. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: Identification of phosphate transporters from higher plants. Plant Cell. 1997;91(1):381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg M. E., O'Shea E. K. Signaling phosphate starvation. Trends Biochem. Sci. 1996;211(1):383–386. [PubMed] [Google Scholar]
- Li H. M., Culligan K., Dixon R. A., Chory J. Cue1 - a mesophyll cell-specific positive regulator of light-controlled gene-expression in Arabidopsis. Plant Cell. 1995;71(1):1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhu H., Liu K., Liu X., Leggewie G., Udvardi M., Wang D. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002;2771(1):27772–27781. doi: 10.1074/jbc.M204183200. [DOI] [PubMed] [Google Scholar]
- Linkohr B. I., Williamson L. C., Fitter A. H., Leyser H. M. O. Nitrate and phosphate availability and distribution have different effcets on the root system architecture of Arabidopsis. Plant J. 2002;291(1):751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- Liu C. M., Muchhal U. S., Mukatira U., Kononowicz A. K., Raghothama K. G. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998a;1161(1):91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Muchhal U. S., Raghothama K. G. Differential expression of TPSI1, a phosphate starvation-induced gene in tomato. Plant Mol. Biol. 1997;331(1):867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Liu H., Trieu A. T., Blaylock L. A., Harrison M. J. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol. Plant-Microbe Interact. 1998b;111(1):14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Lloyd J. C., Zakhleniuk O. V., Raines C. A. Identification of mutants in phosphorus metabolism. Ann. Appl. Biol. 2001;1381(1):111–115. [Google Scholar]
- Löffler A., Abel S., Jost W., Beintema J. J., Glund K. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiol. 1992;981(1):1472–1478. doi: 10.1104/pp.98.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J., de la Vega O. M., Guevara-García A., Herrera-Estrella L. Enhanced phosphorus uptake in transgenic tobacco plants that over-produce citrate. Nat. Biotechnol. 2000;181(1):450–453. doi: 10.1038/74531. [DOI] [PubMed] [Google Scholar]
- Lu Y-P., Zhen R-G., Rea P. AtPT4: A fourth member of the Arabidopsis phosphate transporter gene family. Plant Physiol. 1997;1141(1):747. [Google Scholar]
- López-Bucio L., Hernández-Abreu E., Sánchez-Calderón L., Nieto-Jacobo M. R., Simpson J., Herrera-Estrella L. Phosphate sensitivity alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;1291(1):244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. P., Brown K. M. Regulation of root architecture by phosphorus availability. 1998;1(1):148–156. In Phosphorus in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes, J.P. Lynch, J. Deikman, eds (Maryland USA: Amer. Soc. Plant Physiol.), pp. [Google Scholar]
- Ma Z., Bielenberg D. G., Brown K. M., Lynch J. P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 2001a;241(1):459–467. [Google Scholar]
- Ma Z., Walk T. C., Marcus A., Lynch J. P. Morphological synergism in root hair length, density, initiation and geometry acquisition in Arabidopsis thaliana: a modeling approach. Plant Soil. 2001b;2361(1):221–235. [Google Scholar]
- MacDonald A. E., Grant B. R., Plaxton W. C. Phosphite (phosphorus acid): its relevance in the environment and agriculture and influence on plant phosphate starvation response. J. Plant Nutr. 2001;241(1):1505–1519. [Google Scholar]
- Malboobi M. A., Lefebvre D. D. Isolation of cDNA clones of genes with altered expression levels in phosphate-starved Brassica nigra suspension cells. Plant Mol. Biol. 1995;281(1):859–870. doi: 10.1007/BF00042071. [DOI] [PubMed] [Google Scholar]
- Malboobi M. A., Hannoufa A., Tremblay L., Lefebvre D. D. Towards an understanding of gene regulation during the phosphate starvation response. 1998;1(1):215–226. In Phosphorus in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes, J.P. Lynch, J. Deikman, eds (Maryland USA: Amer. Soc. Plant Physiol.), pp. [Google Scholar]
- Malboobi M. A., Lefebvre D. D. A phosphate-starvation inducible ß-glucosidase gene (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol. Biol. 1997;341(1):57–68. doi: 10.1023/a:1005865406382. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. 1995. Academic Press, London.
- Martin A. C., del Pozo J. C., Iglesias J., Rubio V., Solano R., de la Pena Leyva A., Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation-responsive genes in Arabidopsis. Plant J. 2000;241(1):559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Massonneau A., Martinoia E., Dietz K. J., Mimura T. Phosphate uptake across the tonoplast of intact vacuoles isolated from suspension-cultured cells of Catharanthus roseus (L.) G. Don. Planta. 2000;2111(1):390–395. doi: 10.1007/s004250000297. [DOI] [PubMed] [Google Scholar]
- Massuci J. D., Schiefelbein J. W. The rdh6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;1061(1):1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin D. E., Abdolrahimzadeh H., Baddily J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature. 1974;2491(1):268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- Mimura T. Regulation of phosphate transport and homeostasis in plant cells. Internat. Rev. Cytol. 1999;1911(1):149–200. [Google Scholar]
- Mimura T., Dietz K., Kaiser W., Schramm M., Kaiser G., Heber U. Phosphate transport across biomembranes and cytosolic phosphate homeostasis in barley leaves. Planta. 1990;1801(1):139–146. doi: 10.1007/BF00193988. [DOI] [PubMed] [Google Scholar]
- Mitsukawa N., Okumura S., Shirano Y., Sato S., Kato T., Harashima S., Shibata D. Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc. Natl. Acad. Sci. U.S.A. 1997;941(1):7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucchal U. S., Liu C., Raghothama K. G. Ca+-ATPase is expressed differentially in phosphate-starved roots of tomato. Physiol. Plant. 1997;1011(1):540–544. [Google Scholar]
- Muchhal U. S., Pardo J. M., Raghothama K. G. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 1996;931(1):10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal U. S., Raghothama K. G. Transcriptional regulation of plant phosphate transporters. Proc. Natl. Acad. Sci. U.S.A. 1999;961(1):5868–5872. doi: 10.1073/pnas.96.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S. R., Rae A. L., Diatloff E., Smith F. W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002;311(1):341–353. doi: 10.1046/j.1365-313x.2002.01356.x. [DOI] [PubMed] [Google Scholar]
- Mukatira U. T., Liu C., Varadarajan D. K., Raghothama K. G. Negative regulation of phosphate starvation-induced genes. Plant Physiol. 2001;1271(1):1854–1862. [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Sato S., Kaneko T., Kotani H., Asamizu E., Miyajima N., Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. III. Sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 1997;41(1):401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Narang R. A., Altmann T. Phosphate acquisition heterosis in Arabidopsis thaliana: a morphological and physiological analysis. Plant Soil. 2001;2341(1):91–97. [Google Scholar]
- Narang R. A., Bruene A., Altmann T. Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol. 2000;1241(1):1786–1799. doi: 10.1104/pp.124.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natr L. Mineral nutrients-a ubiquitous stress factor for photosynthesis. Photosynthetica. 1992;271(1):271–294. [Google Scholar]
- Newman T., de Bruijn F. J., Green P., Keegstra K., Kende H., McIntosh L., Ohlrogge J., Raikhel N., Somerville S., Thomashow M. Genes galore: a summary of methods for accessing results from large- scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;1061(1):1241–55. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T., Abel S., Jost W., Glund K. Induction of an extracellular ribonuclease in cultured cells upon phosphate starvation. Plant Physiol. 1990;921(1):970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N., DeRisi J., O. Brown P. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;111(1):4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura S., Mitsukawa N., Shirano Y., Shibata D. Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 1998;51(1):261–269. doi: 10.1093/dnares/5.5.261. [DOI] [PubMed] [Google Scholar]
- Pao S. S., Paulsen I. T., Saier M. H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;621(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U., Kroken S., Roux C., Briggs S. P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2002;991(1):13324–13329. doi: 10.1073/pnas.202474599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton W. C., Carswell M. C. Metabolic aspects of the phosphate starvation response in plants. 1999;1(1):350–372. In Plant Responses to Environmental Stresses, from Phytohormones to Genome Reorganization, H.R. Lerner, (New York USA: M. Dekker), pp. [Google Scholar]
- Poirier Y., Thoma S., Somerville C., Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;971(1):1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko J. E. J., Battini J-L., Gottschalk R. J., Mazo I., Miller A. D. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA. 1999;961(1):2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C., Bucher M. Molecular mechanisms of phosphate transport in plants. Planta. 2002;216 doi: 10.1007/s00425-002-0921-3. in press. [DOI] [PubMed] [Google Scholar]
- Rausch C., Daram P., Brunner S., Jansa J., Laloi M., Leggewie G., Amrhein N., Bucher M. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature. 2001;4141(1):462–70. doi: 10.1038/35106601. [DOI] [PubMed] [Google Scholar]
- Richardson A. E., Hadobas P. A., Hayes J. E. Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J. 2001;251(1):641–649. doi: 10.1046/j.1365-313x.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martin A. C., Iglesias J., Leyva A., Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Develop. 2001;151(1):2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P., Buresh R., Leakey R. Trees, soils, and food security. Phil. Trans. R. Soc. Lond. B. 1997;3521(1):949–961. [Google Scholar]
- Sano T., Kuraya Y., Amino S., Nagata T. Phosphate as a limiting factor for the cell division of tobacco BY-2 cells. Plant Cell Physiol. 1999;401(1):1–8. doi: 10.1093/oxfordjournals.pcp.a029464. [DOI] [PubMed] [Google Scholar]
- Sano T., Nagata T. The possible involvement of a phosphate induced transcription factor encoded by PHI-2 gene from tobacco in ABA-signaling pathway. Plant Cell Physiol. 2002;431(1):12–20. doi: 10.1093/pcp/pcf002. [DOI] [PubMed] [Google Scholar]
- Schachtman D. P., Reid R. J., Ayling S. M. Phosphorus uptake by plants: from soil to cell. Plant Physiol. 1998;1161(1):447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A., Schmidt W. Acclimative changes in root epidermal cell fate in response to Fe and P deficiency: a specific role for auxins ? Protoplasma. 2001;2181(1):67–75. doi: 10.1007/BF01288362. [DOI] [PubMed] [Google Scholar]
- Schmidt W. From faith to fate: ethylene signaling in morphogenic responses to P and Fe deficiency. J. Plant Nutr. Soil Sci. 2001;1641(1):147–154. [Google Scholar]
- Schmidt W., Schikora A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 2001;1251(1):2078–2084. doi: 10.1104/pp.125.4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K., Usuda H. Uptake of inorganic phosphate by suspension-cultured tobacco cells: Kinetics and regulation by Pi starvation. Plant Cell Physiol. 1995;361(1):341–351. [Google Scholar]
- Shimogawara K., Wykoff D. D., Usuda H., Grossman A. R. Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. 1999;1201(1):685–693. doi: 10.1104/pp.120.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. W., Ealing P. M., Dong B., Delhaize E. The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 1997;111(1):83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. Mycorrhizal Symbiosis. 1997. Academic Press, San Diego, CA.
- Spain B. H., Koo D., Ramakrishnan M., Dzudzor B., Colicelli J. Truncated forms of a novel yeast protein suppress the lethality of a G protein a subunit deficiency by interacting with the b subunit. J. Biol. Chem. 1995;2701(1):25435–25444. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- Stevens M. A., Rick C. M. Chapter 2: Genetics and Breeding. 1986;1(1):35–109. In Atherton, J.G. and Rudich, J. (eds.), The tomato crop. A scientific basis for improvement. Chapman and Hall, London, pp. [Google Scholar]
- Stewart A. J., Chapman W., Jenkins G. I., Graham I., Martin T., Crozier A. The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ. 2001;241(1):1189–1197. [Google Scholar]
- Streatfield S. J., Weber A., Kinsman E. A., Hausler R. E., Li J., Post-Beittenmiller D., Kaiser W. M., Pyke K. A., Flugge U. I., Chory J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;111(1):1609–1622. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H., Li X., Palmgren M. G. Energization of plant cell membranes by H+-pumping ATPases. Regulation and biosynthesis. Plant Cell. 1999;111(1):677–690. doi: 10.1105/tpc.11.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. B., Bariola P. A., del Cardayré S. B., Raines R. T., Green P. J. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc. Natl. Acad. Sci. USA. 1993;901(1):5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;4081(1):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Theodorou M. E., Plaxton W. C. Metabolic adaptation of plant respiration to nutritional phosphate deprivation. Plant Physiol. 1993;1011(1):339–344. doi: 10.1104/pp.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud J-B., Davidian J-C., Sentenac H., Soler A., Grignon C. H+ cotransports in roots as related to the surface pH shift induced by H+ excretion. Plant Physiol. 1988;881(1):1469–1473. doi: 10.1104/pp.88.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Sun Y., Naus K., Lloyd A., Roux S. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol. 1999;1191(1):543–551. doi: 10.1104/pp.119.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi C. A., Delatorre C. A., Abel S. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 2001;1271(1):963–972. [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Fargione J., Wolff B., D'Antonio C., Dobson A., Howarth R., Schindler D., Schlesinger W. H., Simberloff D., Swackhamer D. Forecasting agriculturally driven global environmental change. Science. 2001;2921(1):281–284. doi: 10.1126/science.1057544. [DOI] [PubMed] [Google Scholar]
- Todano T., Sakai H. Secretion of acid phosphatase by the roots of several crop species under phosphorus-deficient conditions. Soil Sci. Plant Nutr. 1991;371(1):129–140. [Google Scholar]
- Torriani A. From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays. 1990;121(1):371–376. doi: 10.1002/bies.950120804. [DOI] [PubMed] [Google Scholar]
- Trull M. C., Deikman J. An Arabidopsis mutant missing one acid phosphatase isoform. Planta. 1998;2061(1):544–550. doi: 10.1007/s004250050431. [DOI] [PubMed] [Google Scholar]
- Trull M. C., Guiltinan M. J., Lynch J. P., Deikman J. The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant Cell Environ. 1997;201(1):85–92. [Google Scholar]
- Ullrich-Eberius C. I., Novacky A., van Bel A. J. E. Phosphate uptake in Lemna gibba G1: energetics and kinetics. Planta. 1984;1611(1):46–52. doi: 10.1007/BF00951459. [DOI] [PubMed] [Google Scholar]
- Varadarajan D. K., Karthikeyan A. S., Matilda P. D., Raghothama K. G. Phosphite, an analogue of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiol. 2002;1291(1):1232–1240. doi: 10.1104/pp.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw W. K., Harrison M. J. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell. 2002;141(1):1751–1766. doi: 10.1105/tpc.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H., Garvin D. F., Kochian L. V. Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol. 2001;1271(1):345–359. doi: 10.1104/pp.127.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson L. C., Ribrioux S. P. C. P., Fitter A. H., Leyser H. M. O. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001;1261(1):875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D. D., Grossman A. R., Weeks D. P., Usuda H., Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 1999;961(1):15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y., Green P. J. Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol. 1991;971(1):1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA. 2002;991(1):5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk O. V., Raines C. A., Lloyd J. C. pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta. 2001;2121(1):529–534. doi: 10.1007/s004250000450. [DOI] [PubMed] [Google Scholar]


