Abstract
Over the past few years, a vast amount of research has illuminated the workings of the secretory system of eukaryotic cells. The bulk of this work has been focused on the yeast Saccharomyces cerevisiae, or on mammalian cells. At a superficial level, plants are typical eukaryotes with respect to the operation of the secretory system; however, important differences emerge in the function and appearance of endomembrane organelles. In particular, the plant secretory system has specialized in several ways to support the synthesis of many components of the complex cell wall, and specialized kinds of vacuole have taken on a protein storage role—a role that is intended to support the growing seedling, but has been co-opted to support human life in the seeds of many crop plants. In the past, most research on the plant secretory system has been guided by results in mammalian or fungal systems but recently plants have begun to stand on their own as models for understanding complex trafficking events within the eukaryotic endomembrane system.
I. INTRODUCTION
While many different plants have been used in studies of the plant secretory system, many labs have focused their attention on the model plant Arabidopsis thaliana. Because of the amount of sequence information, both prior to and (especially) after the recent completion of sequencing of the genome, Arabidopsis was an excellent subject for identifying the molecular machinery required for the operation of the secretory system. With the availability of sequence data for other models, as well as crop plants, other plants may serve as useful comparisons to the Arabidopsis model that has been developed. In the following work, we will attempt to provide an updated review of the secretory system as found in Arabidopsis (using work from other plants as needed). We will report the details of the molecular mechanisms for secretory system function, highlighting the tools available from the many labs working on Arabidopsis. We will also attempt to provide a road map for future research that will help us to better understand the function of the system in Arabidopsis and other plants.
II. THE VESICLE TRAFFICKING MACHINERY
The endomembrane system is composed of many organelles, each of which must maintain a unique composition of membrane and cargo proteins (Figure 1). Traffic flows in two main “default” pathways through the secretory system: secretory and endocytic (Figure 2). The secretory pathway represents the major biosynthetic route (the “anterograde” direction) that starts with newly synthesized proteins that have been targeted to the endoplasmic reticulum (ER) membrane, then flows first to the Golgi, and subsequently to the plasma membrane (PM)/extracellular matrix (ECM). Conversely, the endocytic route represents a counter flow of cargo endocytosed from the PM/ECM, which travels by way of endosomes to the vacuole. Aside from these default pathways, there are several “signal-mediated” pathways that redirect specific proteins from either default pathway (Figure 2). So-called “retrograde” pathways serve as recycling mechanisms to retrieve material back from later steps in either pathway, or serve to direct newly synthesized cargo destined for the vacuole from the default secretory pathway. In addition to these bulk-trafficking methods, recent evidence has indicated that peroxisomes may also be a semi-autonomous part of the endomembrane system (Titorenko and Mullen, 2006), and evidence has even suggested that some plastid proteins may travel through the endomembrane system as well (Nanjo et al., 2006).
Figure 1.
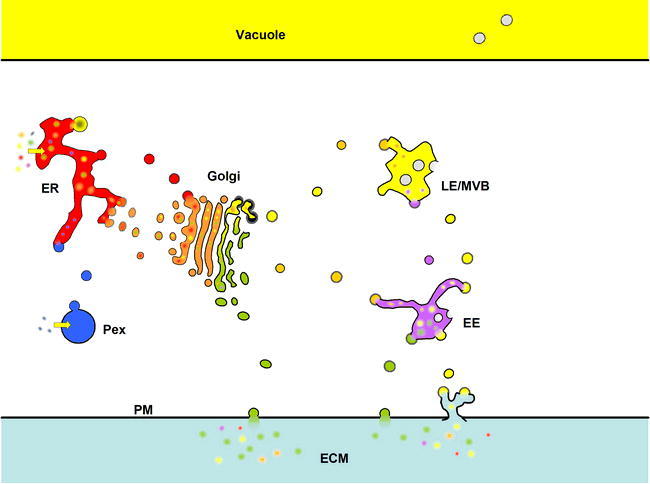
The Secretory/Endomembrane System of Arabidopsis
The endomembrane system of all eukaryotes consists of those membrane-bound organelles that exchange lipid and cargo by vesicle trafficking. Despite the constant exchange of vesicles, each organelle/compartment has a relatively constant array of resident membrane and lumenal proteins. In the figure, resident proteins of the endoplasmic reticulum (ER) are red, those of the Golgi are orange, those of the vacuole are yellow, those of the endosomes (EE) are purple. Secreted proteins (green) end up either in the plasma membrane (PM) or are released into the extracellular matrix (ECM; equivalent to the “cell wall” or “apoplast”). Peroxisomal (Pex) proteins, some of which are first trafficked through the ER, are indicated in blue.
Figure 2.
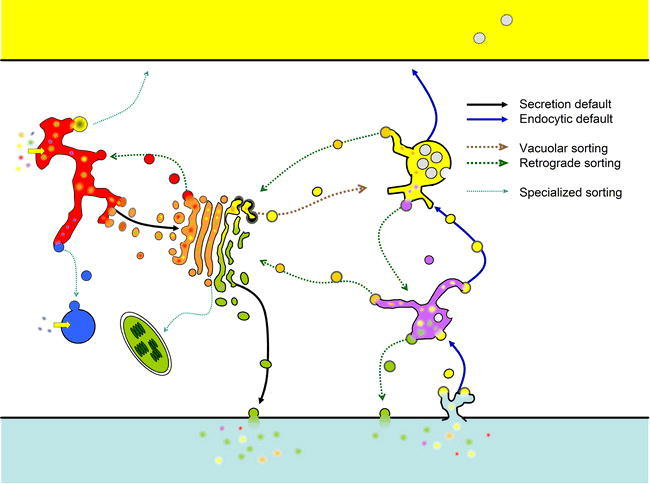
The Secretory Pathway, Endocytic Pathway and Retrograde/Recycling Pathways.
The traditional secretory pathways begins by translocation of proteins (yellow arrow) into the endoplasmic reticulum (ER), followed by transport in vesicles to the Golgi by bulk flow (default secretion, solid black line). From the Golgi, secretory proteins are transported to the Plasma Membrane (PM) or extra-cellular matrix (ECM, “cell wall”). Also in the Golgi, vacuolar proteins are re-directed to the vacuole (by way of the late endosome) due to specific sorting signals (vacuolar sorting, dashed brown line). Cargo that is endocytosed from the PM/ECM is transported through the endosomes by bulk flow (default endocytosis, solid blue line), first to the early endosomes, then to the late endosomes where endocytic cargo meets vacuolar cargo, and finally the cargo arrives in the vacuole. Many signal-mediated retrograde pathways operate to recycle specific cargo at most compartments (dashed green lines). Finally, several specialized sorting pathways serve to transport peroxisomal proteins from the ER to peroxisomes or to transport some glycosylated proteins from the Golgi to the plastid (dashed cyan lines; see text).
It is widely believed that all of these organelles exchange information through the use of small membrane-enclosed transport vesicles. These vesicles bud from a donor compartment using a complex and ordered system of coat proteins that nucleate on the site of vesicle formation to collect cargo and deform the local membrane until a vesicle is freed by scission (Figure 3). Once free, these vesicles then travel through the cytosol by association with cytoskeletal motors or other docking factors. Finally, through the action of docking and tethering factors as well as members of the SNARE family of proteins, these vesicles must identify their target compartment among all the other possibilities and then fuse their lipid bilayer with the target to deliver their cargo (Figure 4).
Figure 3.
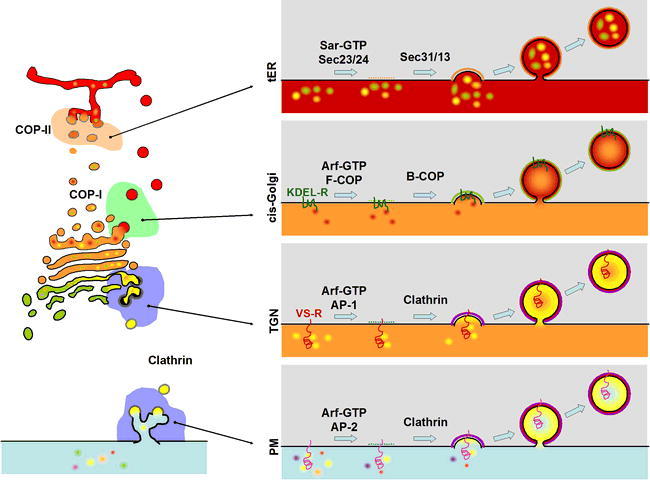
Vesicle Formation at the Donor Compartment
Vesicle formation is accomplished by a similar process at each compartment, though the individual protein machinery varies at the different compartments. The basic process is the same at each compartment: a coat-GTPase cooperates with a cargo-selective coat subunit to collect cargo molecules at the bud site, next a “cage” subunit is recruited from the cytosol to physically deform the lipid bilayer into a vesicle. Vesicle formation at the transitional ER (tER) is accomplished by the COP-II coat system (top panel at right). At the cis-Golgi, vesicle formation is accomplished by the COP-I coat system (second panel at right). Recovery of ER-resident proteins is accomplished by a cargo receptor called the KDEL-Receptor (KDEL-R, encoded by ERD2) which is a membrane protein that binds to ER-retrieval signals (KDEL-COOH) on the lumenal side, and recruits coat components on the cytosolic side. At the TGN, vacuolar cargo is selected by a cargo receptor (VS-R; see text) which recognizes vacuolar sorting signals on the lumenal side and recruits the clathrin adaptor complex (AP-1) on the cytosolic side (third panel at right). Clathrin coats (in this case using AP-2 adaptor) also operate at the PM to accomplish the first step of endocytosis (lower panel at right). In other organisms (i.e., mammals) there are many cargo receptors which serve to internalize extracellular cargo but little is known about such proteins in plants.
Figure 4.
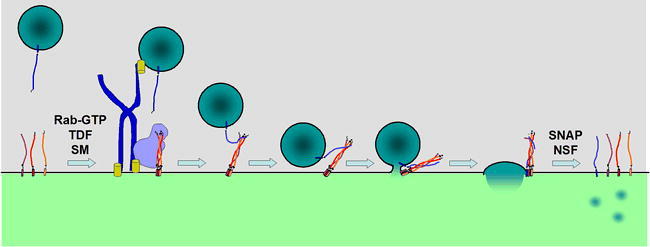
Docking and Fusion of a Vesicle at the Target Compartment.
Docking and fusion of a vesicle is also accomplished by a similar process at each trafficking step, again the individual protein machinery varies. Identification of the target compartment for a particular vesicle is mediated by a member of the Rab family of small GTPases which acts on both the vesicle and target membrane to recruit tethering and docking factors (TDF) and other effectors that serve to attach the vesicle to the target membrane. Once a vesicle is tethered, a member of the SM family of proteins mediates assembly of the target-(t)-SNAREs into a three SNARE-helix bundle that serves as a binding site for the vesicle-(v)-SNARE helix. Mutual twisting of the SNARE helices pulls the membranes into close proximity and drives fusion of the bilayers. (Only one set of SNAREs is shown, fusion is likely mediated by multiple SNARE complexes surrounding the fusion site.) Following fusion of the vesicle, lumenal cargo is delivered into the target compartment lumen and vesicle membrane proteins and lipids are now part of the target membrane. The final four-helix SNARE complex is resolved by the action of a complex of proteins (SNAP and NSF) that use ATP energy to unwind the helices of the individual SNARE proteins for subsequent rounds of fusion (t-SNAREs) or for recycling back to the donor compartment (v-SNARE).
Clearly, some manner of regulation is required at each of these steps to maintain order. One would expect each donor compartment to have a peculiar array of coat proteins regulated by specific coat GTPases that would create a particular transport vesicle (or equivalent membrane-bound carrier). In turn, each target compartment should be uniquely labeled by Rab GTPases, arrays of docking and tethering factors and a distinct collection of SNAREs. In the following sections, we will discuss these particular parts of the vesicle traffic machinery.
II.A. Coat GTPases and Coats: Making a Vesicle
Formation of a transport vesicle is not a spontaneous event. First, a mechanism for cargo selection and concentration is required to decide what belongs in the vesicle. Secondly, some signal must be sent from the lumen to the cytoplasm (where a coat must form) to indicate a site for vesicle formation. Most importantly, some method of membrane deformation and scission from the donor compartment is needed to actually produce the vesicle. This is the job of a large collection of proteins that are referred to as coat proteins. Each type of coat is distinct, though some are related, and a particular coat is responsible for vesicle formation at a particular type of organelle. An overall similarity in the coating mechanism is found (See Figure 3), even though the protein content of the coats may vary. The first step requires recruitment of the cargo to a site on the donor membrane. The details of such a step are still a matter of debate, however it most likely involves the cytoplasmic tails of integral membrane proteins. These proteins may be themselves cargo, or may serve as cargo receptors, or may serve simply in signaling the site of budding. Coincident with this step is the GTP-cycle of a coat-GTPase which helps to coordinate the coating process. This coat-GTPase differs depending on the particular coat proteins, and may be of the ARF-family or of the SAR1 family of small G-proteins. The coat-GTPase for the COP-II coat is called SAR1, and is a single copy gene in yeast, though mammals and plants have multiple SAR1-homologues (TABLE 1). The ARF family of G-proteins is required for both COP-I and clathrin-type coat formation. Consistent with these expanded roles, the ARF-family is large in most eukaryotes, and may have as many as 21 members in Arabidopsis (TABLE 1).
Table 1.
Arabidopsis Coat Complexes and Coat-GTPase
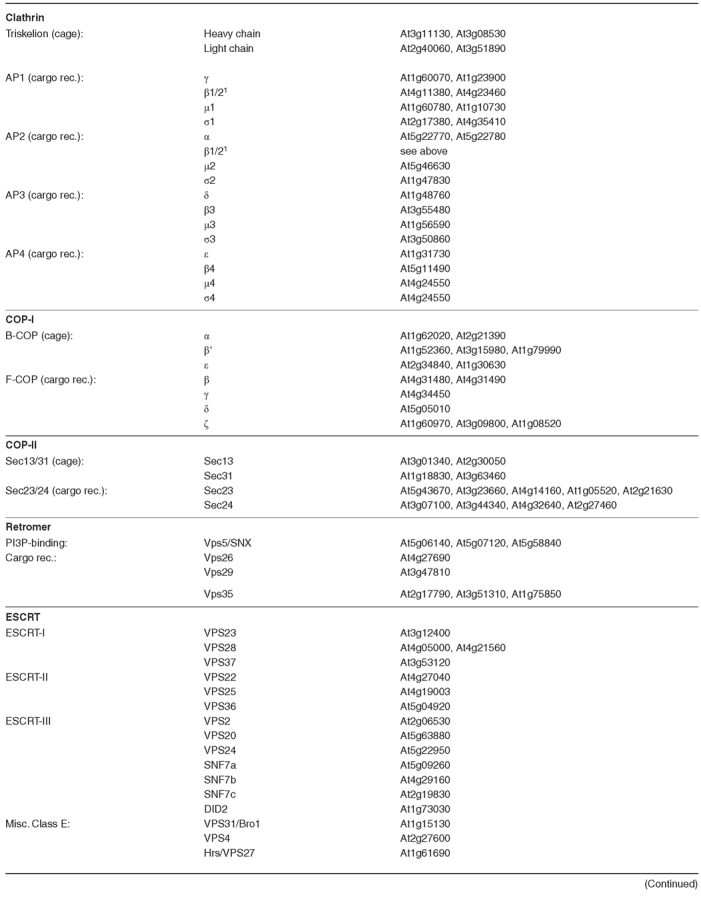
Table 1.
(continued)
The coat-GTPases typically exist in a soluble GDP-bound state while inactive. A GTP-exchange factor (GEF) localized to the coating site serves to recruit the coat-GTPase to the donor membrane and then triggers nucleotide exchange, such that the coat-GTPase assumes its active GTP-bound state. The GEF for the Sar1p-type G-protein is a member of the SEC12-family, a group of ER-integral membrane proteins found in all eukaryotes. The Arabidopsis SEC12 is somewhat atypical in that both SEC12-like genes are smaller than the orthologues found in other eukaryotes, though each seems to function as normal. The GEF for the ARF-type G-protein varies somewhat, since the ARF-type coat-GTPase is involved in many different coating steps. Typically, the ARF-GEF will contain a Sec7-like domain, which is named for the Sec7p protein that is an ARF-GEF used for budding vesicles at the yeast Golgi (reviewed in Jackson and Casanova, 2000). The Sec7-domain proteins fall into 3 main classes. The Gea/GNOM/GBF class contains the Arabidopsis protein GNOM (as well as other proteins related to GNOM; Geldner et al., 2003), mutants of which are embryo lethal due to defects in the polar transport of auxin transporters, and other developmental and physiological defects (Busch et al., 1996; Steinmann et al., 1999). The Sec7/BIG class contains a family of Arabidopsis proteins that show high homology to the metazoan members, though their function has yet to be examined in Arabidopsis. Finally, the ARNO class of proteins is found only in mammals. The interface between the Sec7-domain and the ARF is the target for brefeldin A, a drug used commonly in the study of Golgi function in eukaryotes (Peyroche et al., 1999; Robineau et al., 2000). Treatment of eukaryotic cells with brefeldin generally leads to a breakdown of the Golgi stacks and a complete block in most vesicle transport, illustrating the importance of the ARF-cycle to vesicle transport.
Once activated at the budding site, the active coat-GTPase then functions in the recruitment of “effector”-proteins as well as “cargo-selective” subunits of the coat complexes from the cytoplasm (Figure 3). Effectors can function as enzymes to alter the lipid-characteristics of the budding site, or simply recruit other factors (i.e.: v-SNAREs, Rab-type GTPases, etc.) into the budding vesicle. Finally, the “cage” subunit of the coat complex is nucleated onto the bud site (Figure 3), and functions as a mechanical device to drive membrane deformation (Stagg et al., 2007). Scission of the membrane to free the vesicle is either driven by the coats themselves, by the peculiar lipid composition of the vesicle “neck” region, by other molecular machines such as the dynamin family of large GTPases, or by a combination of all three. Subsequently, a GTPase Activation Protein (GAP) triggers the intrinsic GTPase activity of the coat-GTPase. This step occurs either directly after vesicle formation (COP-II), or can be triggered by some other event (COP-I or clathrin). The GDP-bound G-protein now triggers uncoating of the vesicle, releasing the coatomers back to the cytoplasm for future coating steps. The uncoated vesicle is now free to travel to its target membrane where the SNARE-mediated fusion process occurs.
COP-II Coats
The COP-II coat is conserved in all eukaryotes and is involved in export of secretory cargo from the ER in vesicles (See Figures 3 and 5). In most organisms, ER-exit sites (ERES) are discrete points along the ER membrane, although in some organisms (like budding yeast) the entire ER membrane seems capable of acting as an ER-exit site (reviewed in Glick, 2001). The exit sites are points at which the COP-II machinery is concentrated, and also where anterograde cargo is collected. How cargo becomes concentrated remains unclear, with some arguing for a “bulk-flow” model, whereas others invoke specific cargo receptors. In a bulk-flow model, all cargo not specifically retained in the ER is subject to COP-II-directed transport to the Golgi. On the other hand, some propose that a large class of conserved proteins is involved in clustering cargo for packaging at exit sites. Evidence for both models has accumulated, and it is possible that both occur depending on the cell-type or developmental state or particular cargo molecule.
Figure 5.
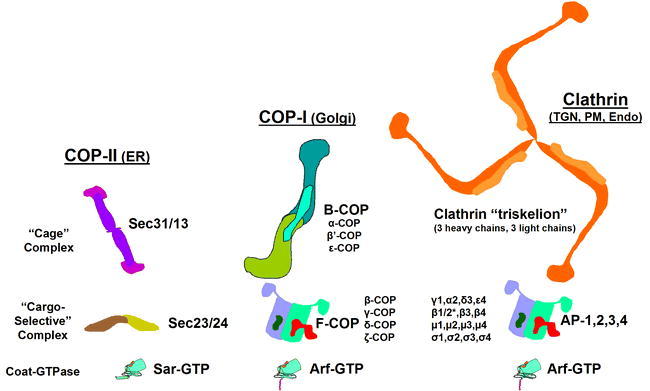
The Sub-Complexes of the Three Major Coat Systems.
Each coat system can be considered to represent three major sub-complexes: a coat-GTPase, a “cargo-selective” complex and a “cage” complex. In some cases, these individual subunits have a clear evolutionary relationship: The Sar and Arf GTPases share a common ancestor among the small GTPases, and the subunits of the four clathrin adaptors are clear examples of gene duplication and also share a common ancestor with the components of F-COP. The three cage subunits also share many common protein folds like the α-solenoid and WD-40 motifs, but this state may be convergent rather than ancestral (see Stag et al., 2007).
The coat-GTPase for COP-II is SAR1, which is found in all eukaryotes. SAR1 serves to recruit the “cargo selective” subunit which is made of two highly-conserved proteins (SEC23 and SEC24; Figure 5). The “cage” sub-unit of COP-II is made from two proteins (SEC13 and SEC31; Figure 5) and assembles onto the nucleation site as directed by the Sec23/24 subunit. The Arabidopsis COPII coat subunits are each encoded by multiple genes (TABLE 1), though whether this indicates redundancy has yet to be investigated. The COP-II coated vesicles are quite unstable as the GAP for SAR1 turns out to be a component of the coat itself (SEC23), thus the vesicles are uncoated soon after budding. This may be significant since in most eukaryotes, the COP-II-derived vesicles are short lived. These vesicle quickly fuse with each other into larger vesicles which (according to the predominant “cisternal maturation” model for Golgi transport) forms a de novo cis-Golgi stack upon docking with the Golgi matrix. Upon uncoating, Sar1p and the coat proteins are released into the cytoplasm for further rounds of vesicle budding.
COP-II mediated sorting has been recently been examined in plants, with all indications that it is similar to that found in other eukaryotes. Takeuchi et al. (2000) showed that dominant negative mutants of the SAR1-GTPase block ER-to-Golgi trafficking. Philipson et al (2001) later showed that transient overexpression of SEC12 (which would presumably prolong the GTP-bound state of SAR1, mimicking the dominant negative) had a similar effect. Interestingly, this SEC12-overexpression effect was not noted previously by Bar-Peled and Raikhel (1997) when they overexpressed SEC12 in Arabidopsis. Since Bar-Peled and Raikhel used stably-transformed plants, some compensation mechanism during the growth of the plants may have occurred and it would not be noted in the transient assays. Some interesting hints concerning the role of vacuolar targeting signals in export from the ER has been reported by Törmäkangas et al. (2001), and surely future research will reveal more interesting observations about the ER in plants.
Plant COP-II proteins have a unique subcellular distribution. Fluorescent protein fusions of a tobacco Sar1 and most of the Arabidopsis COPII coat proteins have been found restricted to mobile ER exit sites localized in the near vicinity of Golgi stacks (Stefano et al., 2006; Hanton et al., 2007; Hanton et al., 2008). However, similarly to mammalian Sar1 and Sec12, in addition to a distribution to ER exit sites, two isoforms of Arabidopsis SAR1 have been found to be associated extensively with ER membranes (Hanton et al., 2008) where SEC12 is also localized (daSilva et al., 2004; Bar-Peled and Raikhel, 1997).
It has been shown that increased synthesis of membrane cargo destined to the plant Golgi induces a signal-mediated recruitment to ER exit sites of a tobacco SAR1 isoform (daSilva et al., 2004) and SEC24 (Hanton et al., 2007). However, soluble proteins that are secreted by bulk-flow do not appear to up-regulate membrane recruitment of COPII, showing that the availability of membrane cargo influences the level of COPII recruitment to ER exit sites. Furthermore, the assembly of COPII coat proteins, such as SEC23 and SEC24, at the plant ER exit sites depends on retrograde protein transport from the Golgi (Stefano et al., 2006). In tobacco leaves, disruption in the activity of Arf1, the GTPase that regulates the retrograde protein route from the Golgi apparatus to the ER (see below), leads to redistribution of SEC23 and SEC24 COPII coat to the cytosol (Stefano et al., 2006). Disruption of plant ERESs due to blockage of the retrograde pathway is reversible as recruitment of COPII coat proteins at the ER exit sites can be re-established by release from Arf1 inhibition (Stefano et al., 2006). This effect suggests that the plant ER is capable of accommodating reversible changes in COPII assembly at ERESs. Gene depletion experiments have demonstrated that ER exit site-associated proteins such as Sec16 contribute to differentiation of ERESs and to efficient ER protein export in mammals and yeast (Connerly et al., 2005; Watson et al., 2006). A role of plant proteins with analogous functions to Sec16 has yet to be established.
COP-I Coats
COP-I coats are typically associated with Golgi trafficking (See Figures 3 and 5), mainly in the retrograde sorting pathway (though in the past it has been argued that anterograde COP-I vesicles act to move cargo forward in Golgi stacks). COP-I coats may also be involved in forming coated vesicles at the ER and endosomal compartments (Kirchausen, 2000). The coat GTPase that drives COP-I coat formation is ARF1, which is typically represented by a large gene family (e.g., 6 ARFA genes in Arabidopsis, see TABLE 1) that also function in other coating processes (See clathrin below). ARF1 appears to have multiple roles in plant cells. It has been shown that ARF1 is distributed to the Golgi and to post-Golgi compartments that bud from the Golgi apparatus and may be involved in endocytosis (Xu and Scheres, 2005; Stefano et al., 2006; Matheson et al., 2007). ARF1 appears to be involved in the recruitment of a golgin, GDAP1, on these Golgi-derived compartments (Matheson et al., 2007), but the functional role of this protein is yet to be determined.
The COP-I coatomer is made up of two multi-subunit complexes called F-COP and B-COP that are conserved among all eukaryotes (Figure 5). F-COP serves as the cargo-selective subunit and is made from four proteins (γ-COP, β-COP, δ-COP, and ζ-COP), while the B-COP forms the “cage” complex and consists of (α-COP, β'-COP and ϵ-COP) As with COP-II coat proteins, the COP-I- proteins are encoded by multiple genes (except for γ and δ) in Arabidopsis (TABLE 1); whether this represents redundancy, or may help to explain the morphologically distinct types of COPI vesicles observed on plant and algal Golgi by Donohoe et al. (2007) remains to be investigated.
Clathrin
Clathrin coated vesicles (CCVs) were the first to be described, and are found in all eukaryotes (Reviewed in Kirchausen, 2000). Coats containing clathrin are found on two major compartments in most organisms, the TGN and the PM, and are involved in sorting cargo into various endosomes from both organelles (See Figure 3). Aside from a role in vesicle formation, clathrin-coated pits (membranes coated with clathrin coatomers but not forming a vesicle) also are essential for signal transduction pathways, and for defining distinct membrane patches both at the PM and the TGN. The clathrin coatomer, made of two protein chains (heavy and light), forms a microscopically visible structure called a triskelion named for its three-legged shape (Fotin et al., 2004). Some organisms, such as yeast, have only a single type of light chain, whereas mammals can have two different light chains (type A and B) in a single triskelion. Evidence from purified plant CCVs suggests that plants may also have more than one type of light-chain peptide (see Robinson et al., 1998), and the fact that two genes are found to encode light-chain like proteins (TABLE 1) supports this hypothesis. Clathrin triskelions are the cage subunit for CCVs, and they require a cargo-selective subunit which is typically a member of the adaptor protein (AP)-family, the GGA-family, or another type of protein (Figure 5). In addition, CCVs require the action of a coat-GTPase from the ARF family.
The AP-complexes were the first clathrin-coordinating complex described, and come in four different types (reviewed in Boehm et al., 2001). The AP-complexes function as the cargo-selective subunit for CCVs, but in some cases these adaptors can act to form coated vesicles without clathrin. Interestingly, the individual subunits of the AP-complexes are evolutionarily related not only to each other, but also to the subunits of F-COP, indicating a “duplication and specialization” mechanism for creating new coatomer specificities (Boehm et al., 2001). In mammals, there are four distinct AP-complexes (AP-1, to -4), each made of 4 subunits: two large subunits (one of α, γ, δ, or ϵ and one of β1- β4; note that these subunits are distinct from the similarly named COP-I components), one medium subunit (μ1- μ4), and one small subunit (σ1- σ4). While mammals have distinct β1 and β2 proteins, many other organisms, including plants, have only a single “β1/2” subunit that probably functions in both the AP-1 and AP-2 complexes (Boehm et al., 2001; Dacks et al., 2008). The AP-complexes recognize specific peptide motifs in the cytoplasmic tails of cargo proteins. These motifs fall into two classes, tyrosine motifs and dileucine motifs. The tyrosine motif consists of a Yxxϕ sequence (where Y is tyrosine, x is any amino acid, and ϕ is any bulky hydrophobic residue). Different AP-complexes have different preferences for variants of the tyrosine motif depending on the nature of the x residues, the context of the signal, and even the phosphorylation state of nearby residues (Ohno et al., 1998). The dileucine motif contains some variant of two adjacent bulky hydrophobic residues (i.e.: LL or LV, etc.), and different AP-complexes respond differently to the context of these residues (e.g.: Rodionov and Bakke, 1998; Honing et al., 1998; Rapoport et al., 1998). These motifs are recognized mainly by either the μ or the β subunit of the complexes, though other subunits may also contribute to binding. Upon binding cargo, some of the AP-complexes are capable of recruiting the clathrin triskelion through a clathrin-binding domain in the large subunits; however, some AP-complexes seem able to form coated vesicles in the absence of clathrin.
The AP-1 complex (γ, β1, μ1, and σ1) is a clathrin-interacting complex that functions in coated vesicle formation at the TGN and endosomal compartments in many eukaryotes (reviewed in Hirst and Robinson, 1998). Arabidopsis encodes two γ, μ1, and σ1 subunits (and a single β1/2-orthologue that probably also acts in the AP-2 complex; Dacks et al., 2008; TABLE 1), suggesting that there may be different AP-1-isoforms present, though this is speculative. Mammalian cells have multiple isoforms of some AP-1 subunits, with some tissues having specific subunits (Meyer et al., 2000). The AP-1/clathrin complex was generally believed to be involved in packaging vacuolar-bound cargo into CCVs at the TGN in most eukaryotes. In Arabidopsis, it has been reported that the AP-1 complex interacts with the cytoplasmic tail of the vacuolar sorting receptor ELP (Sanderfoot et al., 1998), and may thus play a role in sorting of vacuolar cargo. Recent research has indicated that this step may actually be mediated by the GGA-type adaptors in yeast, and perhaps mammals, although a role for AP-1 has not been ruled out. Other roles for AP-1 in recycling back from endosomes to the TGN have also been proposed.
The AP-2 complex (subunits α, β2, μ2, and σ2) is intimately involved in the endocytosis of many (though not all) cargo proteins from the PM (reviewed in Hirst and Robinson, 1998). Arabidopsis encodes single genes for most of these subunits (except for the β2, which is probably substituted for by the β1/2-type gene; Dacks et al., 2008), though genes for the α-type are found as a tandem repeat on chromosome 5 (TABLE VI.A). The AP-2 complex, like the AP-1, requires clathrin for coat formation. To direct endocytosis of a vesicle, many other proteins are important either for recruiting AP-2, or for recruiting clathrin directly to the site of vesicle formation. The AP-2-associated mammalian proteins such as AP180, epsin, auxilin, and perhaps synaptojanin have homologues in the Arabidopsis genome (see table), whereas amphiphysin, endophilin, intersectin, stonin, and synaptotagmin do not. The mammalian AP-2 complex appears to be essential, though the homologous complex in yeast is not essential for endocytosis in these cells (Yeung et al., 1999). The importance of the AP-2 complex in plants, and of the potential orthologues of some of its associated proteins, has yet to be investigated.
The AP-3 complex (δ, β3, μ3, and σ3) is found in most eukaryotes (reviewed in Ordoizzi et al., 1998), though the ability to interact with clathrin has only been shown for the mammalian complex (and even there, this is controversial; Drake et al., 2000; Le Borgne et al., 1998; Dell'Angelica et al., 1998). It is possible that the AP-3 complex forms vesicles in the absence of clathrin. The role of the AP-3 complex is restricted to the TGN and endosomes, and seems to be essential for the targeting of membrane proteins directly to the vacuole/lysosomal membrane in a step that bypasses the late endosome/MVB (Stepp et al., 1997). So, for example yeast enzymes like alkaline phosphatase, or mammalian enzymes like tyrosinase are trafficked directly from the Golgi to the vacuolar/lysosomal membrane. Mutations in these subunits lead to inheritable defects in mammalian cells, and have been associated with known diseases in humans (Dell'Angelica et al., 2000a), indicating the importance of this pathway. Plants encode single genes for each of the subunits (TABLE 1). This is different than mammals, which again have brain-specific isoforms of many AP-3 subunits. The AP-3 complex seems to have a preference for a particular kind of dileucine signal, a so-called acidic dileucine, where 3-5 residues upstream are found either aspartic or glutamic acid residues. In addition, the AP-3 complex can recognize many kinds of tyrosine motifs, at least in vitro.
The AP-4 complex (ϵ, β4, μ4, and σ4) has been described in mammalian cells (Dell'Angelica et al., 1999; Hirst et al., 1999), though it is not found in all fungi. Arabidopsis encodes a single gene homologous to each subunit (TABLE 1). In mammals, AP-4 is found on the TGN, and may or may not be associated with clathrin. A clear role has not been assigned for the AP-4-mediated trafficking, though it seems likely it may be responsible for a sub-set of Golgi-to-endosomal transport.
Other Coats
Another novel coat complex has been described in yeast as being essential for recycling of membrane proteins from the late endosomes to the Golgi, and is called the “Retromer complex” (Seaman et al., 1998; Figure 6). This complex consists of five subunits, Vps5p, Vps17p, Vps26p, Vps29p, and Vps35p. These coatomers interact with the cytoplasmic tails of membrane proteins (i.e.: Vps35p interacts with the tail of Vps10p, the vacuolar sorting receptor) and lead to their packaging into vesicles bound for the Golgi complex (Notwehr et al., 2000). In the absence of this complex, membrane proteins are not recycled, and instead accumulate in the LE before eventual degradation in the vacuole. Orthologues of most of these coat components (except Vps17p) are found in mammals, and it is likely that they function in a similar manner. In fact, the mammalian Vps5p-homologues are members of the sorting nexin group of proteins, a group that is involved in sorting of membrane proteins in the endosomal system. Arabidopsis also contain orthologues of all but Vps17p, in fact having 3 each for Vps5p and Vps35p (TABLE 1). The Arabidopsis retromer has been implicated in recycling of the VSR vacuolar cargo receptors and also in the localization of the auxin efflux carriers PIN1 and PIN2 (Jaillais et al., 2006; Oliviusson et al., 2006; Shimada et al., 2006; Jaillais et al., 2007; Yamazaki et al., 2008) (See III.D).
Figure 6.
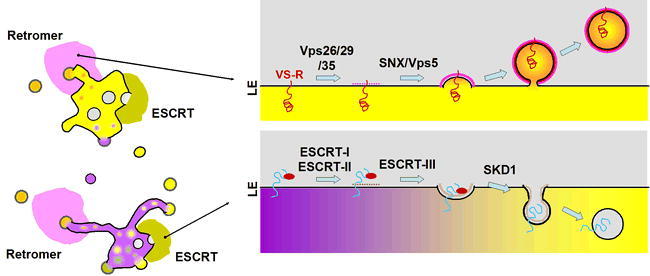
Retromer and ESCRT Coat Machinery
The retromer coat is responsible for recycling of proteins (e.g., the vacuolar cargo receptor) from the endosomal compartments back to the Golgi. Coat formation is superficially similar to that of the other coat complexes (see Figure 3), with the complex of Vps26, Vps29 and Vps35 functioning to bind and collect cargo proteins and the sorting nexin/Vps5 subunit functioning as a cage, but the mechanisms appears to be distinct. Similarly, the ESCRT system has subunits that function in cargo recognition and some variation of a “cage”, but it is topologically reversed to create a vesicle inside the lumen of the endosomal compartments (though evidence of the protein coat having a role in membrane deformation as shown in the figure is controversial). The ESCRT-I, -II and –III sub-complexes act sequentially to identify cargo and form a “cage” that leads to the invagination in the endosomal membranes. A large ATPase (SKD1) then acts to release the ESCRT complexes back to the cytosol prior to enclosure of the lumenal vesicle. These internal vesicles are ultimately delivered to the vacuolar lumen where their contents are degraded.
Though many coat complexes are well conserved among all eukaryotes, this is not universally the case. Yeast and mammals also have a coatomer called the GGA-type that has recently been shown to be essential for Golgi to endosomal traffic. The GGA stands for Golgi-localized, Gamma (γ)-ear-containing, ARF-binding proteins (Hirst et al. 2000; Dell'Angelica et al., 2000b). The name describes their localization, the presence of the clathrin-interacting γ-ear (from the AP-1 γ-subunit) domain found in their carboxy-terminus, and their ability to interact with the ARF-GT-Pases mentioned earlier as coat-forming GTPases. Based upon this description, it should not be surprising that these proteins play an essential role in the binding of the cytoplasmic tails of cargo proteins, and then triggering clathrin coat formation through the ARF and clathrin-interaction domains (reviewed in Tooze, 2001). Yeast contain two partially redundant genes encoding Gga1 and Gga2p, whereas mammals contain three orthologues (GGA1-3) (Hirst et al. 2000; Dell'Angelica et al., 2000b). Arabidopsis and other plants have no obvious homologues to GGA-type coatomers. It is possible that this role has been usurped by other coats (i.e.: AP-1 adaptors?), or that some unknown protein may have a GGA-like function.
A third class of “coat-like” components are represented by the ESCRT complex, which function “in reverse” to make the internal vesicles of the MVB (see Figure 6 and III.D). Arabidopsis (like other plants) encodes all of the expected subunits of the ESCRT complexes (TABLE 1). Other coats are certain to exist in cells. One clear example is the coat that likely envelops the “Dense Vesicles” (DVs) which carry the storage proteins to the PSV in many plant cells. No molecular details of such a coat have been established. Other potential coats have been suggested by researchers working in various organisms, but none are widely studied and such studies are not as well characterized at this point. It is also important to note that the molecular details concerning the type of coat (if there is even such a coat required) that forms secretory vesicles (i.e.: the Golgi-to-PM pathway) have not been reported in any eukaryote. Clearly many experiments remain to be done.
II.B. Rab-GTPases and SM-proteins: Preparing the Membranes for Fusion
Formation of a vesicle through the action of the coat proteins is only the first step of vesicle trafficking. Once freed from the cage of the coat through uncoating, the vesicle now must traverse the cytosol in search of a unique target compartment that must be distinguished from all the other endomembranes of the cell. These steps are the role of the Rab-GTPases and a variety of tethering and docking proteins. Though there is much recent work concerning these tethering and docking complexes in mammals and yeast, very little work on these proteins has been done in plants. A recent study by Koumandou et al. (2007) has examined the evolutionary conservation of these proteins across eukaryotes, and we will not repeat such analysis here. Instead, we will focus on the two classes of proteins that have been examined in plants, the Rab-GTPases and the SM-family of proteins.
Rab-GTPases
Rabs represent a large subgroup of the small G-protein family, and have a standard G-nucleotide binding cycle. In general, Rabs are prenylated at a C-terminal Cys-motif (although one type of plant specific Rab is instead myristoylated at the N-terminus), and through this lipid modification, can cycle on and off of membranes in a G-nucleotide-specific manner. Like the rest of the vesicle trafficking machinery, Rabs are a eukaryotic specific protein family, and are found in all eukaryotes examined thus far (e.g., see Pereira-Leal et al., 2003). Through genomic analysis, there are thought to be eight types of Rab proteins that are conserved among the majority of eukaryotes, and are thought to represent the “minimal set” of Rab proteins (Pereira-Leal and Seabra, 2001; Field et al., 2006). Appropriately enough, plants have members of each of these groups, and the groups have been named A-H (i.e., “RabA”; Pereira-Leal and Seabra, 2001; Vernoud et al., 2003; TABLE 2.). Though this nomenclature suffers in comparison to that of mammalian Rabs, it has the advantage of instantly identifying the different ancestral Rab groups. It is thought that each of these ancestral groups would mediate a particular vesicle trafficking event, and this has tended to be the case based on research in disparate eukaryotes (reviewed in Vernoud et al., 2003; Field et al., 2006). Some organisms have additional types of Rabs, while almost all have multiple members of each type. Plants, for instance, tend to have small gene families of two-to-nine members in addition to sub-groups within each ancestral type.
Table 2.
Docking and Tethering factors: Rab-GTPases and SM-Proteins

Table 2.
(continued)
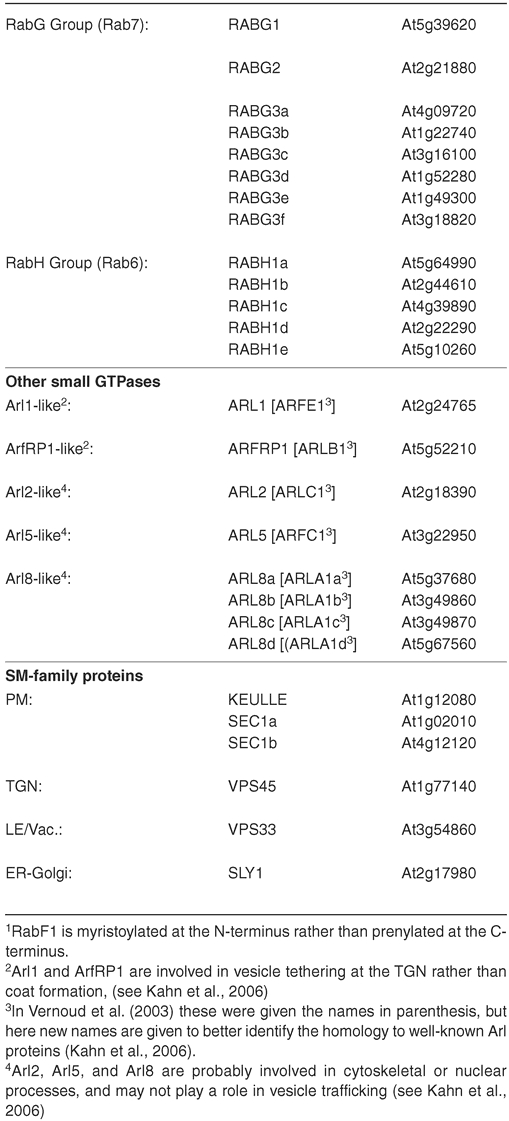
In eukaryotes, it is known that the Rab/Ypt-family is involved in cargo selection, recruitment of molecular motor proteins, and interactions with docking factors (Figure 4). Some work has indicated a similar function in plant cells. Arabidopsis contains 57 members of the Rab-type of small GTPase (reviewed in Vernoud et al, 2002; TABLE 2). Like the coat-GTPases, the Rabs also have an essential G-protein cycle. While in the GDP-bound state, the Rabs are inactive and are typically found in the cytoplasm bound to a protein called RabGDI (GDP disassociation inhibitor). When recruited to a particular membrane by a Rab-GEF (Guanine Exchange Factor), the GDP is exchanged for GTP, and the Rab becomes associated with the membrane. The activated Rab then further recruits other effectors to the bud site, including SNAREs, so called docking factors, as well as molecular motors that will tether the vesicle to the cytoskeleton. Such effectors have been recently reviewed elsewhere for yeast and mammalian cells (Pfeffer, 1999; Lowe, 2000), and since very little is known about plant factors, we will not discuss these further at this time.
Studies in Arabidopsis have generally found the members of the various Rab groups to localize to the compartments expected based on homology, and often Rabs are used as markers for a particular compartment. For example, members of the RabB and RabD groups label the ER and Golgi compartments (Batoko et al., 2000; Cheung et al., 2002) as expected from homology (to the Rab2 and Rab1 type of mammalian/fungal Rabs, respectively; TABLE 2). The Golgi itself is marked by members of the RabH group (Bednarek et al., 1994; Latijnhouwers et al., 2007) as expected by the homology with mammalian Rab6. The members of the RabG family label the vacuolar compartments (e.g., Nahm et al., 2003) as expected from their homology to the lysosomal/vacuolar Rab7 proteins from mammals and fungi. The other groups of Rabs (RabA, RabC, RabE and RabF) all label endocytic compartments of some kind, which is expected from homology to the mammalian orthologues (Rab11, Rab18, Rab8, and Rab5, respectively; see TABLE 2). Importantly, their differential localization among the endocytic compartments may greatly influence the ”maturity” of a particular endosome or of a specific domain within an endosomal membrane (for example, see Ueda et al., 2004; Preuss et al., 2004; Haas et al., 2007; Chow et al., 2008). However, because most Rabs are encoded by large gene families in Arabidopsis, only in a few cases has a particular Rab been functionally implicated in targeting to a given compartment (for example, see Sohn et al., 2003; Zheng et al., 2005; Preuss et al., 2006). “Stacking” of mutants, together with other molecular genetic techniques may eventually reveal individual functions for Rab isoforms.
SM (Sec1-Munc) Family Proteins
The SM-family of proteins are known to interact with the syntaxin (Qa) family of SNAREs (reviewed in Hanson, 2000). This interaction is believed to trigger a change of conformation in the syntaxin (from “closed” to “open”) allowing formation of SNARE complexes (e.g.: Dulubova et al., 1999). Arabidopsis has 6 members of the SM family and other plants have similar numbers (Sanderfoot et al., 2007; TABLE 2). This number is many-fold lower than the number of syntaxins in Arabidopsis, so clearly, the Sec1-family members must interact with more than one syntaxin, or other mechanisms for triggering the conformational change in syntaxins must exist. Three members of the Arabidopsis Sec1- family have been investigated. VPS45 is a Sec1-family member that is localized to the TGN in Arabidopsis (Bassham et al., 2000). Interestingly, while it is able to complement a defect in a homologous gene in yeast (also called VPS45; Bassham and Raikhel, 1998), in Arabidopsis its localization and the syntaxins with which it interacts (Bassham et al., 2000) suggest that it plays a distinct role in plant cells (discussed in Bassham and Raikhel, 2000). VPS33 of Arabidopsis is found on late endosomes and the vacuole, and seems to interact as part of a “Class-C-Complex” similar to the orthologues in yeast and mammals (Rojo et al., 2005). Whereas the other types of SM-family proteins appear to exist in single-copy, the PM-type of SM-protein is represented by three genes in Arabidopsis (KEULLE, SEC1a, and SEC1b). Of these three, only one has been investigated thoroughly: KEULLE interacts with the cytokinesis-specific syntaxin KNOLLE (Assaad et al., 2001), and likely regulates the function of this syntaxin since keule mutants have similar phenotypes to knolle, and double mutants display an additive phenotype (Waizeneggar et al., 2000).
II.C. SNAREs: The Machines of Membrane Fusion
The various members of the SNARE family of proteins are distributed among the organelles in a specific way. Particular SNAREs label unique target membranes, while others are incorporated into vesicles from only certain donor compartments. Of course, since v-SNAREs must travel between compartments, they will be found on more than a single organelle. In fact, due to flexibility in SNARE-SNARE interactions, different combinations of SNAREs can lead to additional levels of organization among the organelles. SNARE proteins have been reviewed many times recently, including for several plants (Sanderfoot, 2007). The basic aspects of the SNARE hypothesis (see above) have been well supported, and all results are consistent with SNARE proteins operating as both target-specificity and membrane-fusion determinants in vesicle trafficking.
Examining the sequences available from many eukaryotes has indicated that most eukaryotes have a common set of SNARE proteins, with the SNARE helices of these proteins falling into 4 main types (Qa-, Qb-, Qc- and R; Table 3). Moreover, through such genomic analysis, one can also identify a “core set” that likely represents the genes from the last common ancestor of the extant eukaryotes (Sanderfoot, 2007). This core set would include five Qa-SNAREs, four Qb-SNAREs, four Qc-SNAREs and three R-SNAREs. In addition to these 16 putatively ancestral SNAREs, a few others are shared across several major clades, but appear to have been lost in others (Sanderfoot, 2007). For example, only animals and fungi seem to have a class of R-SNAREs called “brevins”; while most eukaryotes outside of the Animal + Fungi clade have a type of Qb-SNARE and Qc-SNARE that probably was missing in the opisthokont ancestor. There is also a double-SNARE-helix type (Qb+Qc-SNAREs; e.g., mammalian SNAP25, fungal Sec9p or plant SNAP33) that has a very spotty appearance among eukaryotes, appearing in mainly the multi-cellular lineages, but absent from many of the unicellular organisms (Sanderfoot, 2007). It has previously been noted that land plants have an extreme surplus of SNARE-encoding genes (Sanderfoot et al., 2000). While this surplus can partially be explained by the presence of small gene families, it seems that the plant gene families are not simply examples of redundancy, and may actually represent a large degree of complexity in vesicle trafficking.
Table 3.
Arabidopsis SNARE Proteins (see Sanderfoot, 2007)
Table 3.
(continued)

Qa-SNAREs
Qa-SNAREs (“syntaxins”) are the thought to be the anchor point of the SNARE complexes. Members of each of the other classes of SNARE have been proposed to act as vesicle-(v)-SNAREs, but the Qa-SNAREs seem to always act on the target membranes, and have been considered to be useful markers for particular organelles on which they reside. For example, the various groups of Qa-SNAREs in land plants seem to mark a non-redundant set of organelles (see below).
The Qa-SYP8 family is the likely syntaxin of the plant ER system, represented in Arabidopsis by a single gene, SYP81 (Sanderfoot, 2000). Little work has been done in plants, though fusing a fluorescent protein to SYP81 and expressing the fusion in protoplasts results in localization to the ER (Uemura et al., 2004). Since green plants tend to have a very distributed ER which has many essential functions in plant cells, members of the SYP8 family should be a useful target for research in the future.
Members of the Qa-SYP3 family have been localized to the ER and Golgi in many plants (Rancour et al., 2004; Uemura et al., 2004), and their homology to the mammalian syntaxin 5 and fungal Sed5p support this role. In Arabidopsis, the two members of the gene family (SYP31 and SYP32) may serve distinct functions (Rancour et al., 2004), though this needs further investigation.
Later in the Golgi system, at the TGN, the Qa-SYP4 family takes over. Research has indicated that members of the SYP4 family are found on the trans-face of the Golgi stacks (Bassham et al., 2000; Geldner et al., 2003, Uemura et al., 2004) and participate in several SNARE complexes at that point (Bassham et al., 2000; Sanderfoot et al., 2001b; Chen et al., 2005). Evidence indicates that these proteins are essential in Arabidopsis (Sanderfoot et al., 2001a), indicating the importance of vesicle trafficking at this organelle.
Among the endosomes, the Qa-SNARE is from the Qa-SYP2 family. Proteins in this family have been shown to localize to the TGN, late endosomes and to the vacuolar membrane (Conceição et al., 1997; Sato et al., 1997; Sanderfoot et al., 1999; Rojo et al., 2005), and form several SNARE complexes in this region (Bassham et al., 2000; Sanderfoot et al., 1999). Members of this gene family also seem to have major physiological roles in plants, since mutants in these genes are either lethal, or greatly alter plant morphology (Sanderfoot et al., 2001b; Yano et al., 2003).
The numerous members of the Qa-SYP1 family represent the PM-syntaxin in the plant kingdom. This group is made from four smaller clades which likely represent distinct aspects of PM-vesicle trafficking (Sanderfoot, 2007). KNOLLE (SYP111) is expressed in a cell-cycle-specific manner and localizes to the cell plate during cytokinesis, a role that is essential for seedling survival (Lukowitz et al., 1996; Lauber et al, 1997). The ROR2/PEN1-group (PEN1/SYR1/SYP121 and SYP122 in Arabidopsis) seems to specialize in “point defense” against fungal invaders (Collins et al., 2003; Assaad et al., 2004), though it likely also plays roles in general secretion (Kargul et al., 2001). The SYP124-group (SYP123, 124 and 125 in Arabidopsis) plays an unknown role, though their expression is very high in the male gametophyte (Sanderfoot, 2007). The SYP13-group (SYP131 and 132 in Arabidopsis) probably represents the more ancient clade (Sanderfoot, 2007) with roles in general secretion (Catalano et al., 2007; Kalde et al., 2007).
Qb-SNAREs
Qb-SNAREs generally operate as t-SNAREs at many essential parts of the endomembrane system. Though some have been used as markers, they are not as restricted to individual compartments as the Qa-SNAREs.
The Qb-SNARE of the ER-complex is a member of the SEC20 group in animals and fungi (see Dilcher et al., 2003), and this type of SNARE is also found in plants (Sanderfoot, 2007). However, no study of this protein has been reported from plants.
Two different Qb-SNAREs operate in the Golgi, similar to animals and fungi. Qb-MEMB1 (MEMB11 and 12 in Arabidopsis), which is homologous to mammalian Membrin or fungal Bos1p, has been shown to localize to the ER-to-Golgi in plants (Uemura et al., 2004; Chatre et al., 2005; Latijnhouwers et al., 2007). Later in the Golgi, the Membrin/Bos1p-SNARE is replaced by a Qb-GS29/Gos1p-SNARE in mammals and fungi (Hong, 2005), and the equivalent protein in plants is Qb-GOS1. In Arabidopsis, this group of SNAREs is represented by a two member gene family (GOS11 and GOS12), both of which reside in the Golgi as expected (Uemura et al., 2004).
The fourth Qb-SNARE (VTI11-to-14 in Arabidopsis) seems to function in the endosomes and vacuole. Arabidopsis and other plants have multiple paralogues that have been found to take on several overlapping roles in the Golgi/endosomal/vacuole SNARE complexes (Bassham et al., 2000; Sanderfoot et al., 2001b; Zheng et al., 1999; Kato et al., 2002; Surpin et al., 2003). Like the Qa-SYP22 group, members of this group seem to play larger roles in plant physiology, including response to gravitropism (Kato et al., 2002). They also represent an example of partial redundancy among gene family members: individual mutants of VTI1 gene family members are viable yet have distinct physiological phenotypes and SNARE-partner specificity (Kato et al., 2002; Niihama et al., 2005; Surpin et al, 2003).
The fifth kind of Qb-SNARE, NPSN1, was first identified among the green plants (Sanderfoot et al., 2000), though it is now clear that many other eukaryotes (outside of animals or fungi) also have a member of this family (Sanderfoot, 2007). This SNARE seems to operate as part of the endosomal/secretory pathway, and likely plays a role in cytokinesis, at least in Arabidopsis (Uemura et al., 2004; Zheng et al., 2003).
Qc-SNAREs
Qc-SNAREs operate as v-SNAREs in the early endosomal system, and as likely t-SNAREs in the later secretory pathway (Hong, 2005). Among eukaryotes, they are found to form four main groups, with some additional groups seen in some lineages.
A Qc-SNARE that operates in the ER is the Qc-USE1-like SNAREs recently identified by several labs in fungi (Burri et al., 2004; Dilcher et al., 2003). Due to an annotation error early in the Arabidopsis genome annotation, this gene was misidentified with a cation exchange transporter that was adjacent to the Arabidopsis USE11 gene, and a second family member is also found elsewhere in the genome (Sanderfoot, 2007).
The Qc-BET1-homologue of the green plants probably operates as the v-SNARE for ER-derived vesicles in transit to the cis-Golgi (Uemura et al., 2004) and is represented by two genes in Arabidopsis (BET11/Bs14a and BET12/Bs14b). Like both mammals and fungi, green plants have a second BET1-like SNARE that presumably operates in the later stacks of the Golgi. Called the plant-SFT1 clade (Sanderfoot et al., 2000; Sutter et al., 2006), it is represented by two genes (SFT11 and SFT12) in Arabidopsis (Sanderfoot, 2007). Outside of the green lineage, no obvious equivalents to the plant SFT1 clade are found, similar to the fungal Sft1p and the mammalian GS15 being lineage specific expansions of the BET1-group (Hong, 2005).
Three other related groups of Qc-SNAREs are found in the endosomal system. Qc-SYP6 (SYP61/OSM1 in Arabidopsis) has been localized to the TGN and endosomes in Arabidopsis (Uemura et al., 2004; Bassham et al., 2000). Mutations in the Arabidopsis SYP61 gene lead to defects in resistance to changes in osmotic pressure (Zhu et al., 2002), suggesting that this protein may have roles in aspects of secretion similar to mammalian syntaxin 6 (reviewed in Hong, 2005). The Qc-SYP5 group (SYP51 and 52 in Arabidopsis), is probably the equivalent to mammalian syntaxin 8 and fungal Syn8p (Sanderfoot et al., 2000; Sanderfoot, 2007). As with SYP6, SYP5 is found on the TGN, the endosomes and on the vacuole (Uemura et al., 2004; Bassham et al., 2000; Carter et al., 2004). It has also been shown to interact with the SNARE complexes of the late endosome and vacuole (Bassham et al., 2000). Qc-SYP7 (SYP71 and 72 in Arabidopsis) was first identified in the green plants (Sanderfoot et al., 2000), but has since been identified in many other eukaryotes, except in animals and fungi (Sanderfoot, 2007). Members of the Qc-SYP7 family have been found on the PM in several plants (Marmagne et al., 2004; Mongrand et al., 2004; Morel et al., 2006). Arabidopsis SYP71 interacts with Qb-NPSN11 and Qa-KNOLLE as a part of cell plate formation during cell division, and is also found specifically at the basal domain of the PM in non-dividing root cells (L.C. Conner and A.A. Sanderfoot, unpublished data), suggesting a role in specialized secretory processes.
Qb+Qc-SNAREs
The SNAP33-type proteins are an example of a Qb+Qc-SNARE, and have been shown to be essential in such roles as general secretion, pathogen defense, and for cell plate formation during cytokinesis (Sanderfoot et al., 2000; Collins et al., 2003; Heese et al., 2001; Kargul et al., 2001). Compared to animal/fungal Qb+c-SNAP25, the land plant SNAP33 proteins have an N-terminal extension (which does not bear any strong resemblance to the extensions of the mammalian SNAP29, SNAP47 or the fungal Sec9p N-termini), and lack the palmitoylation site of the animal SNAP25-like proteins (Sanderfoot, 2007). Interestingly, this type of SNARE is missing from many unicellular eukaryotes, and instead only seems to be found in multicellular organisms (Sanderfoot, 2007).
R-SNAREs
The classic R-SNAREs are the so-called “Brevins” that are found in both mammals and fungi (for example, Synaptobrevin/VAMP1 in mammals and Snc1p in yeast; Hong, 2005), but the more prevalent type of R-SNARE are the “longin” type (Rossi et al., 2004). Green plants do not appear to have any brevin-type proteins; like most other eukaryotes, they only have the “longin” types of R-SNARE (Sutter et al., 2006; Sanderfoot, 2007). R-SNAREs are most often considered to be v-SNAREs, and thus reside on multiple compartments.
The R-SEC22 type of SNARE (SEC221 and 222 in Arabidopsis) seems to operate in several ER/Golgi SNARE complexes in many eukaryotes, including plants (Chatre et al., 2005; Sanderfoot, 2007). A second type of longin-R-SNARE is the YKT6-type of SNARE, which is somewhat unique in being attached to membranes by lipid modification rather than through a transmembrane domain like other SNAREs (Hong, 2005). YKT6-group members have been found on several membranes in plant cells, and have been reported to form several SNARE complexes with Golgi and endosomal SNAREs (Chen et al., 2005).
A third group of longin-type R-SNAREs similar to mammalian VAMP7 are found in all eukaryotes (Sanderfoot, 2007). Plants have two major clades of VAMP7-like R-SNAREs: One clade, the VAMP71 clade (VAMP711-to-714 in Arabidopsis) is found in all green plants and is phylogenetically more similar to the VAMP7-SNAREs found in other eukaryotes (Sanderfoot, 2007). The members of this group are found on organelles in the late endosomal and vacuolar system (Uemura et al., 2004; Carter et al., 2004), similar to the yeast and mammalian VAMP7 SNAREs (Pryor et al., 2004; Wen et al., 2006). The VAMP72 group (VAMP721-to-728 in Arabidopsis) is only found in green plants (Sanderfoot, 2007), and is involved in secretion (Kwon et al., 2008).
In addition to those SNAREs that operate as fusion machinery, there are also some SNARE proteins that seem to operate in regulation. For example, an evolutionarily conserved family of proteins related to the mammalian protein Tomosyn have a C-terminal R-SNARE domain, but lack a transmembrane span (or any other mechanism for direct attachment to the membrane) and have a large N-terminal domain with several WD-40 repeats (Gerst et al., 2003; Sutter et al., 2006; Sanderfoot, 2007). In Arabidopsis, this is represented by two genes (TYN11 and TYN12; Sutter et al., 2006), but whether this protein regulates SNARE complex formation in plants like it does in animals and fungi has not yet been examined.
III. THE ORGANELLES AND COMPARTMENTS
III.A. ER-to-Golgi Trafficking
The endoplasmic reticulum (ER) is continuous with the nuclear membrane as well as ramifying throughout the cytoplasm as a reticulated sheet (Figure 7). It is likely that some portion of the ER is in close contact with every organelle in the cell, performing various roles with each contact. The ER also has many biochemical and metabolic roles, though its role in initiating the bulk of the secretory system is of most relevance to this chapter. How the ER maintains its reticulate appearance is not known. Novel studies suggest that reticulons may be involved in this aspect of the ER, similarly to other systems (Tolley et al., 2008).
Figure 7.
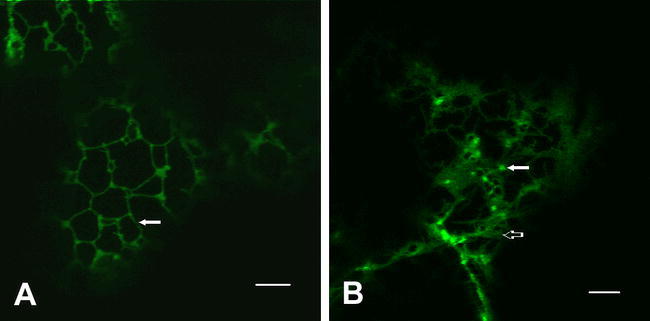
The Early Endomembrane System: ER and Golgi.
(A) The ER is typically observed as a reticulate pattern that ramifies throughout the cytosol, here visualized by confocal microscopy in a leaf epidermal cell expressing GFP-Calnexin (see: Irons et al., 2003). Early Golgi markers, such as GFP-ERD2 (the KDEL-receptor; see Boevink et al., 1998) show bright spots corresponding to the Golgi stacks (arrow), but also have a small percentage of protein that is also found in the ER (open arrow).
Proteins intended for the endomembrane system typically contain N-terminal signal peptides, or have analogous transmembrane domains at other places within the protein that engage the ER-translocation machinery in a similar way (reviewed in Martoglio and Dobberstein, 1998). The overall mechanism and the proteinaceous machinery is remarkably conserved across eukaryotes. Most eukaryotes are capable of both co- and post-translational translocation, and often similar proteins may use either method depending on the species in which they are expressed (reviewed in Kalies and Hartmann, 1998). Regardless, signal peptides can be successfully exchanged between different organisms (Gierasch, 1989), indicating the overall conservation of the process.
Once engaged by the translocation machinery, proteins that lack membrane-spanning domains are released into the lumen of the ER, whereas the membrane proteins are threaded into the membrane (often multiple times) depending upon the folding information within the peptide sequence of the membrane protein. In either case, chaperones and other factors assist in the folding, disulfide bond formation, core-glycosylation, and oligomerization of the proteins. During this process, misfolded proteins are selectively retained by a quality control process which either completes the folding and releases the protein, or marks it for destruction (see review by Vitale and Denecke, 1999).
It has been suggested that the ER has many distinct domains based upon morphology and other criteria (Staehelin, 1997). The domain of most relevance to this review is the transitional ER (tER). The tER (also called ERESs) is the domain where secretory cargo proteins become concentrated for packaging into the COP-II transport vesicles which will carry them to the Golgi stacks. The tER is best studied in mammals and fission yeast, where clear morphological and cytochemical evidence for tER “exit sites” is found (Hammond and Glick, 2000, Rossanese et al., 1999). Structures with similar morphology have been reported in plant cells (Staehelin, 1997). The tER-type domain may not be strictly necessary, as budding yeast does not appear to produce specific tER sites, and instead, it is believed that cargo may exit from any point in this yeast (Rossanese et al., 1999). Packaging of cargo in the tER still remains somewhat controversial. While some of the proteinaceous components of the vesicle forming machinery have been characterized (see COP-II, section II.A), whether there is a selective packaging or simple “bulk-flow” of lumenal contents towards the Golgi in many cell types is unclear. Many so-called ER-to-Golgi “cargo receptors” have been identified in yeast and mammals suggesting specific packaging (e.g.: Lavoie et al., 1999, Denzel et al., 1999, etc.). Whether a bulk-flow or specific mechanism exists has been investigated in plant cells (Phillipson et al., 2001; Törmäkangas et al., 2001), and further work will be required to implicate one over the other. Despite some level of specificity in packaging, ER-residents are often reported to escape as far as the trans-Golgi network, and an efficient Golgi-to-ER recycling system (KDEL-receptor) is found for their retrieval (reviewed in Vitale and Denecke, 1999). Thus, it is likely that some selective process is functioning for particular proteins, whereas some non-selective sampling of the lumenal contents also occurs. Selection of membrane proteins for packaging is likely more selective and is probably dependant on peptide signals within the membrane protein sequence (see COP-II, section II.A).
The Golgi is not the only destination for cargo exiting from the ER. Many plant species create unique Protein Bodies (also called “Precursor Accumulating”, PAC vesicle; Hara-Nishimura et al., 1998) of seed storage proteins by selective distention of an ER subdomain (reviewed in Herman and Larkins, 1999). Seed storage mRNAs are translated upon this portion of the ER membrane leading to quick delivery of the proteins into the forming protein body. When “full”, the protein body buds away from the ER, and may eventually fuse with a Protein Storage Vacuole. Some evidence of a direct (non-Protein Body) pathway from the ER to the Vacuole has also been reported (Jiang and Rogers, 1998), though mechanistically, little is known about this type of trafficking. It has recently become clear that peroxisomes may also be formed (at least partially) by budding from a sub-domain of the ER (reviewed in Mullen et al., 2001).
Since the ER is also a major biosynthetic source of lipids, the ER also must exchange material with other organelles such as plastids, mitochondria, and the plasma membrane, though the mechanics of these exchanges are unclear. Storage of lipids (as an energy source) during seed development occurs in a similar process (reviewed in Galili et al., 1998). Triacylglycerols are funneled between the leaflets of the ER membrane, creating an oil body that is surrounded by a single monolayer. Eventually, this oil body recruits a particular set of proteins, and subsequently buds off the ER and exists free in the cytoplasm. Further studies into these areas may lead to a new understanding of how the ER functions in intracellular traffic apart from the traditional vesicle-mediated secretory pathway.
III.B. The Golgi: Central Sorting Station for the Secretory Pathway
The Golgi apparatus is the common destination for most secretory proteins after departing the ER. It is typically formed of 5–20 individual cisternae which are stacked in a stereotypic manner producing a polarized organelle of cis-to-trans orientation. In Arabidopsis (as in most plants), many individual Golgi bodies (called Dictysomes in some older literature) are found dispersed throughout the cytoplasm, moving quickly throughout the cell by cytoplasmic streaming (for example, see Nebenführ et al., 1999; Boevink et al., 1999; Nebenführ et al., 2000; Figure 7). This type of Golgi distribution is found in many eukaryotes, though is in contrast to the mammalian pattern of a single “super” Golgi stack in the perinuclear region, or to the budding yeast pattern of unstacked Golgi cisternae. The cisternae are distinguished not only by morphological criteria, but also by diagnostic metabolic and enzymatic activities, as well as by the order in which secretory cargo passes. Cargo fresh from the ER first arrives in the cis-Golgi network, passing sequentially through cis-, medial-, and trans-cisterna before arriving at the trans-Golgi network (TGN).
How the cargo passes through the cisternae is still a matter of some controversy (see Pelham and Rothman, 2000). Some evidence suggests that the cisternae are static organelles, and that all traffic between the stacks is vesicle-mediated by COP-I coated vesicles. Other evidence indicates that the stacks may simply be transient structures (i.e.: cisternal maturation models), that are formed new from ER-derived vesicles (or collections of fused vesicles called VTCs in mammalian cells) at the cis-side, then sequentially mature into medial- and trans-cisternae. New cis-stacks form behind the maturing cisternae creating an “assembly-line”-like formation of stacks. Proteins which are specific to an earlier cisterna are collected into COP-I vesicles which remove these proteins back to the earlier stacks, while the maturing stack receives its own specific proteins by collecting COP-I vesicles from a later stack. Finally, at the TGN, the stack is partitioned into secretory vesicles and endosomal compartments. As you may see, the major difference between these two models is the role of the COPI coated vesicles: In the first case, the COP-I coat caries cargo both forward (trans-wise) and backward (cis-wise), whereas in a cisternal maturation-type model, COP-I coats only move in a retrograde manner. Most recent evidence has supported cisternal maturation, especially in plants (e.g. see Otegui et al., 2006; Donohoe et al., 2008), but these models are not necessarily mutually exclusive, and may vary between different organisms at different times of development.
The cis-Golgi is the first cisterna encountered following the ER. The ER-derived vesicles fuse with the cis-Golgi delivering their contents to the lumen or limiting membrane. Within the cis-stack, modifications to the core N-glycosylation of proteins begin. Enzymes such as α-mannosidase remove the terminal mannose residues, creating substrates for other glycosyltransferases which act in the later stacks to produce the unique glycosylation patterns found on many proteins. One of these enzymes, a -mannosidase, has been fused to green fluorescent protein (GFP) by the Staehelin group, and has been used as an in vivo marker for the Golgi stacks, revealing some interesting behaviors in dividing cells (Nebenführ et al., 2000).
The cis-cisternae are also the site of recapture of escaped ER residents. The KDEL-receptor is a transmembrane protein whose lumenal domain specifically recognizes a C-terminal K/HDEL motif found at the C-terminus of ER-resident soluble proteins (Pelham, 1996). Upon binding of proteins with these signals, the KDEL-receptor recruits the COP-I machinery and mediates the return of these proteins to the ER. Recent work has indicated that the plant KDEL receptor appears to exceptionally efficient, and rarely allows any ER residents to escape past the early Golgi stacks (Phillipson et al., 2001). Experiments in mammalian cells have indicated that ER residents may sometimes be allowed passage as far as the TGN before recapture. The passage of some ER residents into the Golgi stacks may be beneficial for the acquisition of Golgi-specific modifications that may be needed in some cases, however the functional significance of this difference is unclear.
Since the number of cisternae in a Golgi apparatus can vary significantly, definition of a “medial” stack is equally variable. Enzymatically, these stacks can often have activities that differ from that of the earlier and later cisternae. Certainly, the spectrum of glycosyltransferases must be somehow distinct, but how this distinction is set-up and maintained remains unclear. Some evidence suggests that synthesis of some cell wall glycans (rhamnogalac-turans and pectins) is initiated in the medial stacks, whereas the trans-cisterna is the typical site of xyloglucan assembly (reviewed in Dupree and Sherrier, 1998). Some of the molecular details of the glycosyltransferases involved in these processes have begun to be worked out, though much remains to be done.
At some point within the trans-cisterna (perhaps earlier), particular cargo proteins have been selected based upon their final destinations. At some level, the cisternae at the TGN are segregated into domains. This has been observed directly in only a few cases; for example, when the localization of two related TGN-localized syntaxins SYP41 and 42 (see SNAREs) were examined they were found to be at distinct locales at the TGN (Bassham et al., 2000). Since that time, using these markers, other proteins have been found to segregate in similar manners: The vacuolar cargo receptor ELP was found to preferentially localize with SYP42 at the TGN (Bassham et al., 2000), the syntaxin SYP51 was also found with only SYP42 (Sanderfoot et al., 2001b), and CTPP-type vacuolar cargo was found to segregate away from ELP at the TGN (Ahmed et al., 2000). Similar results showing a partition of some cargo away from the vacuolar cargo receptor has also been reported by Robinson and co-workers using expanding cotyledon cells of pea as a model system (Hohl et al., 1996; Hinz et al., 1999). Vacuolar cargo (of all types) must also be segregated away from secreted cargo by the Golgi-resident vacuolar sorting receptors (reviewed in Vitale and Raikhel, 1999). These receptors lead to a concentration of the vacuolar cargo into at least one domain of the cisterna. In doing so, secretory cargo is excluded and ends up in distinct regions of the stack. Clearly, the work of various coat proteins and Golgi matrix components are essential for this process, though we are still early in the identification of these factors.
The ELP/BP-80 class of vacuolar sorting receptors (VSR) are found concentrated in the trans-cisternae and the TGN (Paris et al., 1997; Sanderfoot et al., 1998). The VSR proteins are typically represented by large protein families that may exhibit distinct signal specificity. Evidence has indicated that members of this family can specifically recognize both N-terminal and C-terminal vacuolar sorting signals (Kirsch et al., 1994; Cao et al., 2000; Ahmed et al., 2000; Shimada et al., 2003; Otegui et al., 2006), though it remains unclear whether this can be accomplished by the same kind of VSR. It does appear that at least one member of the VSR family is capable of recruiting a clathrin-type coat to the membrane (perhaps through the AP-1 type of coatomer), and directs the packaging of the NTPP-type cargo into CCVs (Ahmed et al., 2000). These CCVs travel onto the late endosomes/prevacuolar compartment (see III.E). Other VSR family members also seem to be able to recognize CTPP-type vacuolar cargo and direct this cargo to a protein storage vacuole (Shimada et al., 2003). In cells that lack a storage vacuole, the specificity of the cargo receptors must be regulated in some way such that NTPP and CTPP cargo can become segregated at the TGN (Ahmed et al., 2000; Otegui et al., 2006). Other potential cargo receptors (i.e., AtRMR; Jiang et al., 2000) have been suggested for targeting of various vacuolar cargo from within the Golgi, but the mechanisms remain elusive and controversial (Otegui et al., 2006; Hunter et al., 2007; Olbrich et al., 2007).
In the absence of other sorting signals, soluble cargo proteins are directed into secretory vesicles and targeted to the plasma membrane. It remains unclear in any eukaryote as to the nature of the coat proteins involved in packaging secretory vesicles. Regardless, some measure of specificity must be involved, especially in polarized cells. Certain proteins are restricted to particular portions of the cell membrane (see III.C), indicating that secretion need not be a non-specific process. However, very little is known at this time about how a plant cell produces or maintains the polarization of its plasma membrane.
Another aspect of Golgi mediated targeting occurs during cytokinesis (reviewed by Jürgens, 2005). Plants have a unique method of cell division whereby a novel membrane structure (the cell plate) is synthesized by Golgi-derived vesicles at the point of cytokinesis. The cell plate is a site of furious vesicle fusion and formation, and eventually, a new plasma membrane is formed by the fusion of the phragmoplast with the maternal plasma membranes (Seguí-Simarro et al., 2007). It seems likely that this organelle is derived from a novel form of regulated secretion, since the components of the vesicle fusion machinery involved in this step (see II.C) are related to the factors involved in general secretion.
III.C. The PM is the Default Destination, but why are the Different Sides Different?
The limiting membrane of the cell is the target for cargo both intended for secretion and for incorporation into the membrane. Thus, secreted proteins and the components of the extracellular matrix are delivered to the apoplast where they can become incorporated or diffuse away. Membrane proteins become incorporated into the plasma membrane where they act as ion channels or membrane transporters, ligand receptors and signaling complexes, or even as physical contact points for both the intracellular cytoskeleton network or for the extracellular matrix. A useful example of a receptor complex is the Clavata system that controls cell fate in the Arabidopsis shoot apical meristem (reviewed in Clark, 2001). In this system, a multi-member transmembrane receptor kinase complex (CLV1/2) is assembled across the plasma membrane and coordinates binding to the extracellular peptide lig-and CLV3 with signal transduction through a cytoplasmic kinase cascade. Disruptions of any of the genes in this pathway prevent signal transduction, resulting in overproliferation of the shoot apical meristem. The secretory system is essential to functioning of this system as well, both in positioning the receptor complex at the PM, and in delivery of the secreted CLV3 ligand. Redirection of the ligand to the vacuole (Rojo et al., 2002) blocks signal transduction, indicating the importance of proper targeting of proteins in the secretory system.
The lipid of the plasma membrane is quite different from that of the other membranes, being enriched in specific lipids and sterols. This lipid composition is unlikely to homogeneous, rather, similar to what is found in other eukaryotes, there are likely to be distinct patches which are highly enriched in particular lipids. These patches serve to concentrate particular types of proteins like receptor/signaling complexes, cytoskeleton attachment points, and endocytic machinery. Most work on the compartmentalization of lipid domains has been done in mammals, where proteins like caveolin are essential for stability of these domains. Although plants lack caveolin (no homologous gene is found in the Arabidopsis genome), it is still likely that similar domains exist, though little work has been reported on such things in plant cells.
The limiting membrane itself also is likely to show sidedness in the sense that the upper (or apical) portion of the cell may contain proteins not found in the lateral or basal (bottom) portion of the cell. This polarity is accomplished both by specific targeting to a particular domain, by specific retrieval though endocytosis, and by peculiar characteristics of the lipid patches found in the domains. This polarity is essential to cell function and to signaling. For example, transport of auxin through the root is accomplished by specific influx transporters in the apical membrane which transport the neutralized auxin into the cytoplasm. In the cytoplasm, the auxin ionizes due to the near neutral pH. At the basal membrane, an active efflux transporter carries the auxin ion back to the apoplasm (reviewed in Jones, 1998). Without the specific localization of the transporter to particular regions, polarity of the auxin transport could not be accomplished. The probable auxin efflux carrier PIN1 is localized specifically to the basal part of root cells (Galweiler et al., 1998; see Figure 8). This specific polarized localization of PIN1 is accomplished through Golgi-mediated trafficking and requires the action of the product of the GNOM gene, a Sec7-domain-containing protein that likely regulates vesicle budding through interaction with an ARF-GTPase at the trans-Golgi (Steinmann et al., 1999; Geldner et al., 2003). Other members of the PIN1-like family are found on the apical or lateral faces rather than basal portion of the PM, suggesting further polarized domains exist in plants (Müller et al., 1998; Friml et al., 2002; Friml et al., 2003; Klein-Vehn et al., 2006). How these domains are established and maintained remains unknown in plants, and continues to be an area of intensive investigation.
Figure 8.
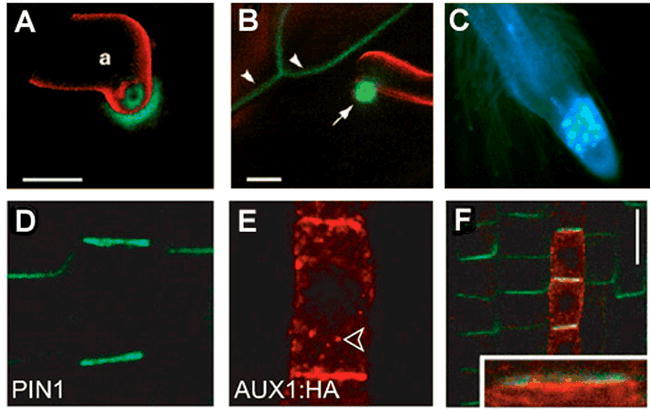
The Plasma Membrane in Polarized Cells
In addition to simply forming the limiting membrane of the plant cell, the PM has recently been found to have multiple kinds of specialized sub-domains in cell that are polarized or that are responding to particular stresses or developmental signals. For example, in studies reported by Assaad et al (2004), an epidermal cell responds to infection by powdery mildew (Erysiphe cichoracearum; red) by establishment of a particular PM structure called papillae. Two PM-SNAREs, Qa-PEN1 (A) and Qa-SYP122 (B), visualized by confocal microscopy in cell expressing GFP- or CFP-fusions (respectively), become enriched in these domains and are essential for resisting the fungal infection. In a similar way, the Qa-SNARE KNOLLE is also important for establishment of the cell plate in dividing cells (C),here visualized in the root tips of plants expressing KNOLLE::CFP-KNOLLE. The best studied examples of polarization in plant cells has been shown by the auxin efflux (PINs) and influx (AUX1s) transporters. For example, in work reported in Kleine-Vehn et al. (2006) the authors performed whole-mount immunofluorescent microscopy to show how the protophloem cells of the root have independent apical and basal domains enriched in AUX1 and PIN1 (respectively (D-F)). Images (A) and (B) are from Assaad et al., 2004; images (D-F) are from Kleine-Vehn et al., 2006.
III.D. Endosomes as a Central Sorting Station for the Late Endomembrane System
Endosomes are organelles that transport and sort cargo from both the biosynthetic pathway (for example proteins coming from the Golgi) and the endocytic pathway (endocytosed plasma membrane molecules). There are several types of endosomes in eu-karyotic cells and they differ in function, structure, and biochemical composition (Figure 9). Endocytosed plasma membrane proteins are transported in vesicles to the “early endosomes.” Early endosomes may have recycling functions and send back to the plasma membrane some of the endocytosed cargo. In mammalian cells, there is a distinct set of endosomes called recycling endosomes that play this recycling role. Other endocytosed proteins that have been targeted for degradation remain in the endosomal system where they are recognized by the ESCRT machinery and sorted into internal vesicles (reviewed by Hurley 2008 and see Coat Proteins). These endosomes containing luminal vesicles are called late endosomes or multivesicular bodies. Multivesicular bodies, frequently called prevacuolar compartments in plants, finally fuse with the vacuolar membrane, releasing the internal vesicles into the vacuolar lumen where they are degraded by hydrolyases.
Figure 9.
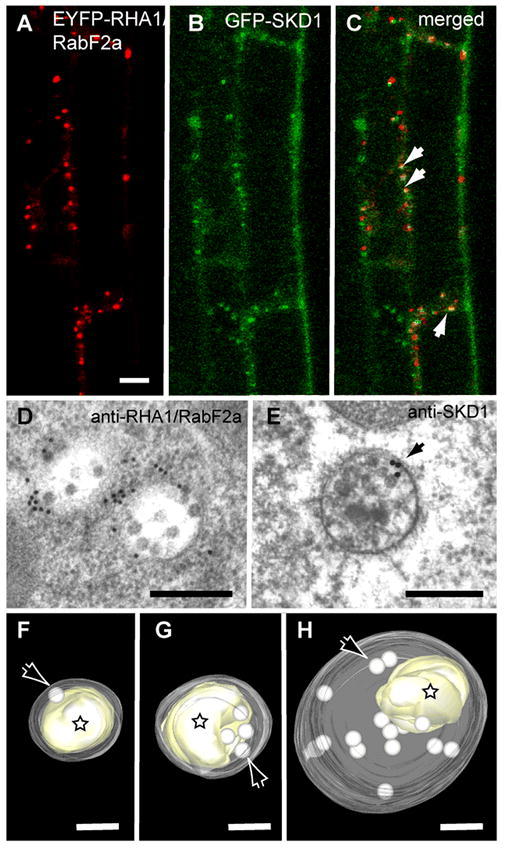
Endosomal Compartments in Plants
Endosomal compartments in plants have long been under examined due to a lack of suitable marker proteins, however, new marker proteins have recently been developed allowing observation of these compartments through both fluorescent and electron microscopy. (A)-(C) Partial co-localization of GFP-SKD1 and a fluorescent fusion protein of the endosomal Rab GTPase RHA1/RabF2a in Arabidopsis roots cells. Arrows indicate areas of colocalization. Bars = 10 m. (D)-(E) Immunogold localization of endosomal proteins in high-pressure frozen/freeze-substituted wild type Arabidopsis roots. Gold labeling shows localization of RHA1/RabF2a (D) and SKD1 (E) on multivesicular bodies or MVBs. Bars = 200 nm. (F)-(H) Tomographic models of prevacuolar compartments/MVBs during formation of protein storage vacuoles in the Arabidopsis embryo reconstructed from electron tomograms. The limiting membranes of MVBs have been made translucent in these tomographic models to visualize the internal vesicles (arrows) and electron dense aggregates of vacuolar storage proteins (stars). Images (A) to (E) are from Haas et al. (2007); images (F)-(H) are from Otegui et al. (2006). Bars= 100 nm
Endosomes also receive Golgi-derived vesicles carrying newly synthesized vacuolar proteins (Kotzer et al. 2004, Otegui, et al. 2006, Tse, et al. 2004) that are delivered to the vacuole. In particular, multivesicular endosomes act as prevacuolar compartments in developing seeds, where they carry storage proteins and processing proteases and deposit them in protein storage vacuoles (Figure 9).
The recycling of vacuolar cargo receptors back to the TGN also happens at the endosomes (Figure 6). The endosomal protein complex called Retromer (see Coat Proteins), that in mammals and yeast is responsible for the recycling of vacuolar cargo receptors, has been identified in Arabidopsis (Jaillais et al. 2006, 2007, Oliviusson et al. 2006, Shimada, et al. 2006). The Arabidopsis retromer subunits VPS35, VPS26 and VPS29 have been shown to localize to Rab5- positive multivesicular endosomes, together with the vacuolar sorting receptor VSR1 (Oliviusson et al., 2006; Jaillais et al, 2006). vps35 mutants exhibit dwarfism, early leaf senescence, mis-sorting of vacuolar seed storage proteins to the extracellular space, and increased accumulation of VSR receptors (Yamazaki et al., 2008). In fact, VPS35 co-immunoprecipitates with VSR1 (Oliviusson et al., 2006). Similar missorting of seed storage proteins has also been reported in the vps29/mag1 retromer mutant (Shimada et al. 2006). In addition, vps29 mutants show abnormal embryo development, seedlings with multiple cotyledons, and reduced primary root growth with fewer lateral roots and altered gravitropic response (Jaillais et al., 2007). Some of these phenotypic alterations seem to be related to a role of the retromer in the subcellular localization of PIN auxin efflux carriers and the subsequent alteration of auxin transport in retromer mutants (Jaillais et al., 2007).
A number of important plant proteins continuously cycle between the plasma membrane and endosomes in a mechanism called constitutive cycling (reviewed by Murphy et al. 2005). Examples of constitutively recycled proteins include the PIN proteins, (putative auxin efflux regulators), PIP2 (a plasma membrane AT-Pase thought to be a water channel), BRI1 (brassinosteroid receptor), and KAT1 (a K+-channel). This mechanism appears to allow dynamic changes in the distribution of plasma membrane proteins. A key player in constitutive cycling of PIN1 is GNOM, a GDP/GTP exchange factor for the ARF GTPases (Steinmann et al. 1999).
Although it is clear that the endosomal system is well developed in plant cells, the identity of the plant early endosomes has remained elusive despite having been a target of study for decades (see Joachim and Robinson, 1984; Tanchak et al, 1984). Recent studies suggest that a TGN-related compartment received endocytosed material from the plasma membrane, acting as an early endosome (reviewed by Lam et al. 2007).
The sorting of plasma membrane proteins into endocytic vesicles and subsequently into the internal vesicles of multivesicular bodies are crucial steps in the down-regulation of receptors involved in development, signaling, and cell differentiation as well as in general turnover of plasma membrane proteins in eukaryotes. Ubiquitination of membrane-bound receptors is now well established as a signal for both endocytosis at the plasma membrane and sorting into internal vesicles of multivesicular bodies (reviewed by Mukhopadhyay and Riezman 2007). When the ubiquitinated receptors reach the late endosome, flat clathrin coats highly enriched in HRS (hepatocyte growth factor-regulated tyrosine kinase substrate) form on the endosomal membranes. Three multi-subunit complexes called ESCRT-I, -II, -III (endosomal sorting complexes required for transport) are also required for protein sorting into MVB internal vesicles in yeast and mammalian cells.
Once the three ESCRTs are assembled onto the endosomal membrane, the AAA ATPase Vps4p (yeast)/SKD1 (mammals) is required for endosomal membrane invagination, presumably by releasing the ESCRTs from the membrane (Figure 6). The binding of Vps4p/SKD1 to the membrane is regulated by its ATPase cycle, being membrane-associated in its ATP-bound form and cytoplasmic in its ADP-bound form. Vps4p/SKD1 is a crucial player in the MVB pathway: when Vps4p/SKD1 is knocked out or a dominant negative (ATPase deficient) form of Vps4p/SKD1 is expressed, aberrant endosomes form, causing disruptions in the secretory and endocytic pathways (Babst et al. 1998). Whereas there is some variation in the organization of the ESCRT complexes in different organisms, the function of Vps4p/SKD1 appears to be highly conserved in all eukaryotic cells. However, whereas yeast and mammalian cells are able to survive expressing ATPase deficient versions of Vps4p/SKD1, the expression of such a form of AtSKD1 in plant cells causes dramatic alterations in the endomembrane system and cell death, suggesting an essential role of the endo-somal sorting machinery in plants (Haas et al. 2007). Fluorescent fusion proteins of AtSKD1 and ELC, a Vps23p/TSG101 homolog that is part of the ESCRT machinery, have been shown to localize to endosomes in Arabidopsis (Figure 9) (Haas et al., 2007; Spitzer et al., 2006). In addition, AtSKD1 has been shown to partially localize with Rab5-like endosomal GTPases (Figure 9).
III.E. Vacuoles: More than Just a Lysosome
The most conspicuous organelle in a mature plant cell is the large central vacuole, which can occupy as much as 90% of the cell volume (Figure 10). Plant vacuoles play many essential roles for the cell. A major role is storage: of water, ions, sugars, hormones, secondary metabolites, metals, pigments, toxins, and even proteins in specialized types of vacuoles. The vacuole is also the equivalent of the lysosome of mammals, in that it contains the various hydrolyases which are used to degrade and recycle proteins, lipids, carbohydrates, etc (reviewed by Muntz 2007). In concert with the plasma membrane, the vacuole also has the essential role of maintaining turgor pressure in the cell. This turgor is essential for homeostasis and also for growth and development. It is generally thought by expansion of a large water filled vacuole held tight with turgor pressure, a plant cell can grow quickly and become quite large without an equivalent increase in the volume of energy-intensive cytoplasm. The essential importance of the vacuole to plant growth and development was shown by Rojo et al. (2001) with the vacuoleless mutant of Arabidopsis. Plants where the VACUOLELESS gene is disrupted by a T-DNA were found to completely lack a central vacuole, and die as embryos.
Figure 10.
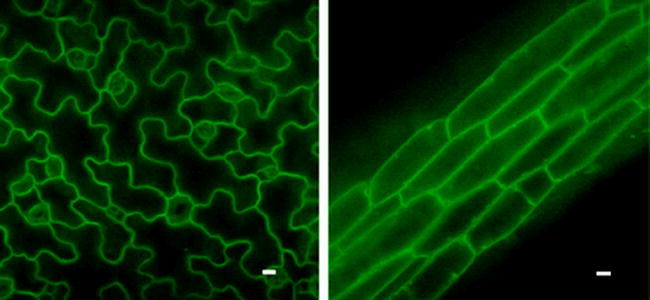
Vacuolar Compartments in Plants
In most mature vegetative cells, the majority of the cell volume is occupied by the large central lytic vacuole. This can be visualized by confocal microscopy of cells expressing 35S::GFP-δ-TIP (J. Zouhar, E. Avila, and N.V.R., unpublished) in either epidermal cells of the cotyledon (left) or hypocotyl cells (right).
Vacuoles differ in their functions and contents and some cell types may contain more than one type of vacuole depending on the developmental stage and environmental conditions (Hoh et al., 1995; Paris et al., 1996; Di Sansebastiano et al., 1998; Swanson et al., 1998; Otegui et al. 2005). Protein storage vacuoles (PSVs) in seed tissues accumulate large amounts of storage proteins, processing proteases, and phytic acid crystals called globoids. In Arabidopsis and legumes, the seed storage proteins destined for the PSV begin to condense in the early stacks of the Golgi and form “dense vesicles” (DVs; Hohl et al., 1996; Hinz et al., 1999; Hillmer et al., 2001, Otegui et al. 2006). The DV expands as additional cargo is packaged within, until finally budding free from the TGN. The DVs are subsequently trafficked to multivesicular bodies where their initial proteolytic processing starts until they are released into the PSV upon multivesicular bodies-PSV fusion. In other species (cucurbits), some storage proteins are packaged into a DV-like organelle (called a PAC vesicle) at the ER (Hara-Nishimura et al., 1998). The ER distends until a PAC buds free into the cytoplasm. A single PAC vesicle may fuse with other PAC vesicles, receive additional cargo delivered from the Golgi, eventually form an multi-vesicular body-like organelle, and then fuse with a PSV.
The lytic functions of vacuoles serve multiple purposes: degradation of storage proteins for amino acid recycling during seed germination, degradation of plasma membrane proteins (see III.D), degradation of cytoplasmic material sent to the vacuole via autophagy (see III.F), defense against pathogens (reviewed in van der Hoorn and Jones, 2004), senescence, and programmed cell death (reviewed by Jones 2001).
The tonoplast or vacuolar membrane contains numerous transporters and channels that mediate the transport of different compounds and ions between the vacuolar lumen and the cytoplasm (Carter et al, 2004; Jaquinod et al., 2007, Schmidt et al., 2007). Most vacuoles seem to contain at least vacuolar H+-ATPases, vacuolar H+-translocating pyrophosphatases and TIP (tonoplast intrinsic protein) aquaporins. The proton gradient generated by both types of proton pumps provides the force that allows other transporters to translocate ions across the tonoplast against their concentration or electrochemical gradient. For example, Na+, Ca2+, NO3−, Zn2+ and several other ions enter the vacuoles through proton antiporters (Hirschi et al. 2004; de Angeli, 2006; Kawachi et al., 2008). In fact, transgenic Arabidopsis plants overexpressing the vacuolar H+ pyrophosphatase AVP1 show higher salt tolerance, likely due to an increased pH gradient across the tonoplast which could facilitate accumulation of ions inside the vacuole (Gaxiola et al. 2001). The vacuole is a major site for Ca2+ storage in the form of free Ca2+, calcium oxalate, or chelated with organic acids. In addition to the Ca2+/H+ antiporter, at least one vacuolar Ca2+ pump and several Ca2+ efflux channels have been identified in the vacuole (Geisler et al, 2000).
As mentioned above, the pressure exerted by the vacuole is important for keeping cell turgidity and allowing changes in cell volume during growth. Rapid transient changes in cell volume, such as in guard cells during stomatal opening/closure or during closure of Venus fly traps to capture insects and pulvinus-mediated leaf movement in Mimosa, also depend on water fluxes through both the plasma membrane and the tonoplast. These water fluxes are facilitated by water channels called aquaporins, which are found in both the plasma membrane and the tonoplast. Arabidopsis contains approximately 35 genes encoding for aqua-porins, from which 10 genes seem to have vacuolar functions. The mechanism by which aquaporins are regulated is not completely clear but it seems to depend on phosphorylation, pH, and osmotic gradients (reviewed by Maurel 2007 and Katsuhara et al. 2008). The water fluxes observed during rapid transient changes in cell volume are induced by changes in osmotic pressure associated with ion fluxes through the plasma membrane and the tonoplast.
In addition, vacuoles play an essential role in plants with CAM (crassulacean acid metabolism)-type of photosynthesis. These plants open their stomata during the night to minimize water loss. The CO2 is incorporated into organic compounds and stored in the vacuole as malic acid. During the day, the CO2 is released from the organic acid to enter the Calvin cycle in the chloroplast. It has been shown that an equivalent of 17% of the total dry mass of leaf cells is transported through the tonoplast in CAM species in a day (Pucher et al., 1949; Holtum et al., 2005).
III.F. Autophagy: Last Desperate Step or just a Normal Part of Life?
Macroautophagy, termed simply autophagy herein, meaning ‘self-eating’, is a pathway for the degradation of proteins and other macromolecules by transport into the vacuole (Gupta, 1988; Aubert et al., 1996; Moriyasu and Ohsumi, 1996). Autophagy is a somewhat unique vesicle trafficking pathway in that double-membrane transport vesicles form de novo in the cytoplasm, engulfing and enclosing cytoplasmic components before being transported to the vacuole (Bassham, 2007). After fusion between the outer autophagosome membrane and the tonoplast, the inner membrane-enclosed contents are deposited in the vacuolar lumen and degraded by resident hydrolases. Unlike most trafficking pathways in plants, autophagy is inducible, with a barely detectable level of transport under normal growth conditions, presumably performing housekeeping functions (Inoue et al., 2006; Xiong et al., 2007a), and a massive induction under stress or senescence conditions (Yoshimoto et al., 2004; Contento et al., 2005).
A number of genes have been identified as required for autophagy, primarily in yeast through genetic screens (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995). Most of these genes are conserved throughout eukaryotes (Meijer et al., 2007), and can be classified based on their function at different stages of the autophagy process. Proteins have been classified as involved in (1) ATG9 cycling, which may be required for membrane recruitment (2) phosphatidylinositol 3-kinase complex, functioning in formation of PI3P at the autophagosome initiation site and (3) ubiquitin-like conjugation systems, required for autophagosome formation and expansion (Xie and Klionsky, 2007).
Plant genes involved in autophagy have been identified by sequence similarity to known yeast autophagy genes (Bassham et al., 2006; Xie and Klionsky, 2007). Arabidopsis knockout mutants have been characterized for representative genes of each of the functional classes described above. The first knockout mutations in predicted Arabidopsis autophagy components, from classes (1) and (3) above, revealed requirements for autophagy in carbon and nitrogen starvation responses (Doelling et al., 2002; Hanaoka et al., 2002). The mutants also showed early senescence, suggesting, as predicted, a role for autophagy in nutrient recycling both under nutrient deficiency and during the senescence program. Knockouts in additional genes of these functional classes show similar phenotypes (Yoshimoto et al., 2004; Suzuki et al., 2005; Thompson et al., 2005; Xiong et al., 2005). Further phenotypic analyses of these mutants may reveal roles for autophagy in additional physiological processes; a function for autophagy in degradation of oxidized proteins during oxidative stress has already been demonstrated in this manner (Xiong et al., 2007b).
Analysis of an Arabidopsis homolog of a class (2) gene, ATG6, has produced very different results. Arabidopsis knockout mutants in ATG6 are gametophyte lethal, with defects in pollen function that preclude the generation of homozygous knockouts (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008). Yeast Atg6 has a dual function in autophagy and vacuolar trafficking (Seaman et al., 1997; Kametaka et al., 1998); the Arabidopsis atg6 mutant phenotype may therefore be due to defects in vacuolar trafficking rather than an autophagy defect.
It has long been suggested that, in addition to autophagy being a survival response during environmental stress conditions, programmed cell death in plants may also occur via activation of autophagy (Bozhkov and Jansson, 2007). Cell death certainly involves the vacuole, and vacuolar proteases have been implicated in activation of cell death pathways in Arabidopsis (Hatsugai et al., 2004; Rojo et al., 2004; Kuroyanagi et al., 2005). However, most of the characterized Arabidopsis autophagy mutants show no developmental abnormalities and are indistinguishable from wild-type plants under normal conditions, suggesting that at least some types of cell death are independent of the autophagy machinery. Research in tobacco provided intriguing evidence for a role for autophagy in regulation of programmed cell death during the hypersensitive response to pathogen attack. Upon exposure to tobacco mosaic virus, autophagy was induced at the infection site, and also systemically in non-infected leaves. Silencing of a tobacco ATG6 homolog prevented this induction and also led to unrestricted spread of the cell death to uninfected regions of the leaf (Liu et al., 2005). This suggests that autophagy is required for restricting programmed cell death, rather than functioning in the cell death process itself.
Recent evidence indicates that this function of autophagy is also conserved in Arabidopsis. To circumvent the lethality of Arabidopsis atg6 mutants (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008), ATG6 antisense lines with reduced expression were generated. In addition to the typical starvation and senescence phenotypes of autophagy mutants (Doelling et al., 2002; Hanaoka et al., 2002), these plants showed unrestricted cell death upon pathogen infection during both a resistance response and during disease (Patel and Dinesh-Kumar, 2008), consistent with results in the tobacco system. While the precise mechanism behind the regulation of programmed cell death by autophagy remains unknown, this is expected to be a fruitful area of future research.
IV. CONCLUSIONS
In the past few years, Arabidopsis has been a major model system for the study of many aspects of plant growth and development, as well as the intricacies of cell biology. In this chapter, we have described the recent results concerning the operation of the secretory system of Arabidopsis. As noted many times in the text, what we know is dwarfed by what is left to be discovered. This is, of course, a good thing, and continuing research using Arabidopsis as an essential model will help to greatly expand our knowledge–both of the secretory system of plants, but also of all eukaryotes.
Acknowledgments
The authors would like to thank Natasha Raikhel for her contributions to the initial version of this chapter (published on March 22, 2003, doi:10.1199/tab.0098).
Footnotes
Citation: Bassham D.C., Brandizzi F., Otegui M.S., and Sanderfoot A.A. (2008) The Secretory System of Arabidopsis. The Arabidopsis Book 6:e0116. doi:10.1199/tab.0116
elocation-id: e0116
This chapter was updated on September 30, 2008. The original version has been archived and is available in PDF format at http://dx.doi.org/10.1199/tab.0098.
REFERENCES
- Ahmed S. U., Bar-Peled M., Raikhel N. V. Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 1997;1146(1):325–336. doi: 10.1104/pp.114.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. U., Rojo E., Kovaleva V., Venkataraman S., Dombrowski J. E., Matsuoka K., Raikhel N. V. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 2000;1496(1):1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F. F., Huet Y., Mayer U., Jürgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J. Cell Biol. 2001;1526(1):531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F. F., Qiu J. L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S. C., Edwards H., Ramonell K., Somerville C. R., Thordal-Christensen H. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell. 2004;156(1):5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S., Gout E., Bligny R., MartyMazars D., Barrieu F., Al-abouvette J., Marty F., Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: Control by the supply of mitochondria with respiratory substrates. J. Cell Biol. 1996;1336(1):1251–1263. doi: 10.1083/jcb.133.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;176(1):2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M., Raikhel N. V. Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 1997;1146(1):315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D. C., Raikhel N. V. An Arabidopsis VPS45p homolog implicated in protein transport to the vacuole. Plant Physiol. 1998;1176(1):407–415. doi: 10.1104/pp.117.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D. C., Raikhel N. V. Unique features of the plant vacuolar sorting machinery. Curr. Opin. Cell Biol. 2000;126(1):491–495. doi: 10.1016/s0955-0674(00)00121-6. [DOI] [PubMed] [Google Scholar]
- Bassham D. C., Sanderfoot A. A., Kovaleva V., Zheng H., Raikhel N. V. AtVPS45 complex formation at the trans-Golgi network. Mol. Biol. Cell. 2000;116(1):2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D. C. Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 2007;106(1):587–593. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Bassham D. C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L. J., Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy. 2006;26(1):2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H. Q., Hawes C., Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal Golgi movement in plants. Plant Cell. 2000;126(1):2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek S. Y., Reynolds T. L., Schroeder M., Grabowski R., Hengst L., Gallwitz D., Raikhel N. V. A small GTP-binding protein from Arabidopsis thaliana functionally complements the yeast YPT6 null mutant. Plant Physiol. 1994;1046(1):591–596. doi: 10.1104/pp.104.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Wasteneys G. O., Jones D. A. The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell. 2000;126(1):1179–1201. doi: 10.1105/tpc.12.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Moore I. Plant cytokinesis: KNOLLE joins the club. Curr. Biol. 2001;116(1):R423–426. doi: 10.1016/s0960-9822(01)00251-2. [DOI] [PubMed] [Google Scholar]
- Bock J. B., Matern H. T., Peden A. A., Scheller R. H. A genomic perspective on membrane compartment organization. Nature. 2001;4096(1):839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Boehm M., Aguilar R. C., Bonifacino J. S. Adaptins: The final recount. Mol. Biol. Cell. 2001;126(1):2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;156(1):441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Bozhkov P., Jansson C. Autophagy and cell-death pro-teases in plants: two wheels of a funeral cart. Autophagy. 2007;36(1):136–138. doi: 10.4161/auto.3600. [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Piper R. C., Weisman L. S., Stevens T. H. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol. 1998;1426(1):651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Sollner T. H., Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2003;1006(1):9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch M., Mayer U., Jürgens G. Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 1996;2506(1):681–691. doi: 10.1007/BF02172979. [DOI] [PubMed] [Google Scholar]
- Carter C., Pan S., Zouhar J., Avila E. L., Girke T., Raikhel N. V. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;166(1):3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Rogers S. W., Butler J., Beevers L., Rogers J. C. Structural requirements for ligand binding by a probable plant vacuolar sorting receptor. Plant Cell. 2000;126(1):493–506. doi: 10.1105/tpc.12.4.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C. M., Czymmek K. J., Gann J. G., Sherrier D. J. Medicago truncatula syntaxin SYP132 defines the symbio-some membrane and infection droplet membrane in root nodules. Planta. 2007;2256(1):541–550. doi: 10.1007/s00425-006-0369-y. [DOI] [PubMed] [Google Scholar]
- Chatre L., Brandizzi F., Hocquellet A., Hawes C., Moreau P. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 2005;1396(1):1244–1254. doi: 10.1104/pp.105.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shin Y. K., Bassham D. C. YKT6 is a core constituent of membrane fusion machineries at the Arabidopsis trans-Golgi network. J. Mol. Biol. 2005;3506(1):92–101. doi: 10.1016/j.jmb.2005.04.061. [DOI] [PubMed] [Google Scholar]
- Cheung A. Y., Chen C. Y., Glaven R. H., de Graaf B. H., Vidali L., Hepler P. K., Wu H. M. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell. 2002;146(1):945–962. doi: 10.1105/tpc.000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. M., Neto H., Foucart C., Moore I. Rab-A2 and Rab-A3 GTPases Define a trans-Golgi Endosomal Membrane Domain in Arabidopsis That Contributes Substantially to the Cell Plate. Plant Cell. 2008;206(1):101–123. doi: 10.1105/tpc.107.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. E. Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2001;26(1):276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- Collins N. C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J. L., Huckelhoven R., Stein M., Freialdenhoven A., Somerville S. C., Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;4256(1):973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Conceição AdS., Marty-Mazars D., Bassham D. C., Sanderfoot A. A., Marty F., Raikhel N. V. The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;96(1):571–582. [PMC free article] [PubMed] [Google Scholar]
- Connerly P. L., Esaki M., Montegna E. A., Strongin D. E., Levi S., Soderholm J., Glick B. S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005;156(1):1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Contento A. L., Xiong Y., Bassham D. C. Visualization of autophagy in Arabidopsis using the fluorescent dye monodan-sylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;426(1):598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Poon P. P., Field M. C. Phylogeny of endo-cytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. USA. 2008;1056(1):588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva L. L., Snapp E. L., Denecke J., Lippincott-Schwartz J., Hawes C., Brandizzi F. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell. 2004;166(1):1753–1771. doi: 10.1105/tpc.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J. M., Thomine S., Gambale F., Barbier-Brygoo H. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature. 2006;4426(1):939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Klumperman J., Stoorvogel W., Bonifacino J. S. Association of the AP-3 adaptor complex with clathrin. Science. 1998;2806(1):431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Mullins C., Bonifacino J. S. AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem. 1999;2746(1):7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Aguilar R. C., Wolins N., Hazelwood S., Gahl W. A., Bonifacino J. S. Molecular characterization of the protein encoded by the Hermansky-Pudlak syndrome type 1 gene. J. Biol. Chem. 2000a;2756(1):1300–1306. doi: 10.1074/jbc.275.2.1300. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 2000b;1496(1):81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel A., Otto F., Girod A., Pepperkok R., Watson R., Rosewell I., Bergeron J. J., Solari R. C., Owen M. J. The p24 family member p23 is required for early embryonic development. Curr. Biol. 2000;106(1):55–58. doi: 10.1016/s0960-9822(99)00266-3. [DOI] [PubMed] [Google Scholar]
- Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;226(1):3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano G. P., Paris N., Marc-Martin S., Neuhaus J. M. Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 1998;156(1):449–457. doi: 10.1046/j.1365-313x.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- Di Sansebastiano G. P., Paris N., Marc-Martin S., Neuhaus J. M. Regeneration of a lytic central vacuole and of neutral peripheral vacuoles can be visualized by green fluorescent proteins targeted to either type of vacuoles. Plant Physiol. 1999;1266(1):78–86. doi: 10.1104/pp.126.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J. H., Walker J. M., Friedman E. M., Thompson A. R., Vierstra R. D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002;2776(1):33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- Donohoe B. S., Kang B. H., Staehelin L. A. Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc. Natl. Acad. Sci. USA. 2008;1046(1):163–168. doi: 10.1073/pnas.0609818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M. T., Zhu Y., Kornfeld S. The assembly of AP-3 adaptor complex-containing clathrin-coated vesicles on synthetic liposomes. Mol. Biol. Cell. 2000;116(1):3723–3736. doi: 10.1091/mbc.11.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Sugita S., Hill S., Hosaka M., Fernandez I., Sudhof T. C., Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;186(1):4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree P., Sherrier D. J. The plant Golgi apparatus. Biochim. Biophys. Acta. 1998;14046(1):259–270. doi: 10.1016/s0167-4889(98)00061-5. [DOI] [PubMed] [Google Scholar]
- Field M. C., Gabernet-Castello C., Dacks J. B. Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv. Exp. Med. Biol. 2007;6076(1):84–96. doi: 10.1007/978-0-387-74021-8_7. [DOI] [PubMed] [Google Scholar]
- Friml J., Wisniewska J., Benkova E., Mendgen K., Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;4156(1):806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;4266(1):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;4326(1):573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;1436(1):1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Sengupta-Gopalan C., Ceriotti A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol. Biol. 1998;386(1):1–29. [PubMed] [Google Scholar]
- Galweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;2826(1):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gaxiola R. A., Li J., Undurraga S., Dang L. M., Allen G. J., Alper S. L., Fink G. R. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA. 2001;986(1):11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Frangne N., Gomes E., Martinoia E., Palmgren M. G. The ACA4 Gene of Arabidopsis Encodes a Vacuolar Membrane Calcium Pump That Improves Salt Tolerance in Yeast. Plant Physiol. 2000;1246(1):1814–1827. doi: 10.1104/pp.124.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;1126(1):219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Gerst J. E. SNARE regulators: matchmakers and match-breakers. Biochim. Biophys. Acta. 2003;16416(1):99–110. doi: 10.1016/s0167-4889(03)00096-x. [DOI] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989;286(1):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Glick B. S. ER export: more than one way out. Curr. Biol. 2001;116(1):R361–363. doi: 10.1016/s0960-9822(01)00194-4. [DOI] [PubMed] [Google Scholar]
- Gupta H. S. Vacuolar autophagy - apparent dilution and elimination of cytoplasmic organelles in acridine orange-treated plant cells. Caryologia. 1988;416(1):347–359. [Google Scholar]
- Haas T. J., Sliwinski M. K. D. E. M., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M. S. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell. 2007;196(1):1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. T., Glick B. S. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell. 2000;116(1):3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;1296(1):1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I. Sec1 gets a grip on syntaxin. Nat. Struct. Biol. 2000;76(1):347–349. doi: 10.1038/75103. [DOI] [PubMed] [Google Scholar]
- Hanton S. L., Chatre L., Renna L., Matheson L. A., Brandizzi F. De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 2007;1436(1):1640–1650. doi: 10.1104/pp.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S. L., Chatre L., Renna L., Matheson L. A., Brandizzi F. Plant Sar1 isoforms with near-identical protein sequences exhibit different localizations and effects on secretion. Plant Molecular Biology in press. 2008;PMID6(1):18322804. doi: 10.1007/s11103-008-9317-5. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I. I., Shimada T., Hatano K., Takeuchi Y., Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell. 1998;106(1):825–836. doi: 10.1105/tpc.10.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T., Morano K., Scott S., Klionsky D. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;1316(1):591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe N., Olsen L. Autophagy Protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. 2008. Autophagy 4. [DOI] [PubMed]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;3056(1):855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- Herman E. M., Larkins B. A. Protein storage bodies and vacuoles. Plant Cell. 1999;116(1):601–614. doi: 10.1105/tpc.11.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M., Gansel X., Sticher L., Wick P., Grebe M., Granier F., Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 2001;1556(1):239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S., Movafeghi A., Robinson D. G., Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J. Cell Biol. 2001;1526(1):41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G., Hillmer S., Baumer M., Hohl I. I. Vacuolar storage proteins and the putative vacuolar sorting receptor BP-80 exit the Golgi apparatus of developing pea cotyledons in different transport vesicles. Plant Cell. 1999;116(1):1509–1524. doi: 10.1105/tpc.11.8.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K. D. The Calcium Conundrum. Both Versatile Nutrient and Specific Signal. Plant Physiol. 2004;1366(1):2438–2442. doi: 10.1104/pp.104.046490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Bright N. A., Rous B., Robinson M. S. Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell. 1999;106(1):2787–2802. doi: 10.1091/mbc.10.8.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 2000;1496(1):67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Robinson M. S. Clathrin and adaptors. Biochim. Biophys. Acta. 1998;14046(1):173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Hoh B., Hinz G., Jeong B. K., Robinson D. G. Protein storage vacuoles form de novo during pea cotyledon development. J. Cell Sci. 1995;1086(1):299–310. doi: 10.1242/jcs.108.1.299. [DOI] [PubMed] [Google Scholar]
- Hohl I., Robinson D. G., Chrispeels M. J., Hinz G. Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J. Cell Sci. 1996;1096(1):2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- Holtum J. A. M., Smith J., Neuhaus H. E. Intracellular transport and pathway of carbon flow in plants with crassulacean acid metabolism. Functional Plant Biology. 2005;326(1):429–449. doi: 10.1071/FP04189. [DOI] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim. Biophys. Acta. 2005;17446(1):120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Honing S., Sandoval I. V., von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 1998;176(1):1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. R., Craddock C. P., Di Benedetto S., Roberts L. M., Frigerio L. Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 2007;1456(1):1371–1382. doi: 10.1104/pp.107.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. ESCRT complexes and the biogenesis of multi-vesicular bodies. Curr. Opin. Cell Biol. 2008;206(1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant and Cell Physiology. 2006;476(1):1641–1652. doi: 10.1093/pcp/pcl031. [DOI] [PubMed] [Google Scholar]
- Irons S. L., Evans D. E., Brandizzi F. The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J. Exp. Bot. 2003;546(1):943–950. doi: 10.1093/jxb/erg102. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Casanova J. E. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;106(1):60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miege C., Rollin C., Gaude T. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006;4436(1):106–109. doi: 10.1038/nature05046. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Santambrogio M., Rozier F., Fobis-Loisy I., Miege C., Gaude T. The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell. 2007;1306(1):1057–1070. doi: 10.1016/j.cell.2007.08.040. [DOI] [PubMed] [Google Scholar]
- Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieux V., Bruley C., Garin J., Bourguignon J. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics. 2007;66(1):394–412. doi: 10.1074/mcp.M600250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Rogers J. C. Integral membrane protein sorting to vacuoles in plant cells: evidence for two pathways. J. Cell Biol. 1998;1436(1):1183–1199. doi: 10.1083/jcb.143.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Phillips T. E., Rogers S. W., Rogers J. C. Biogenesis of the protein storage vacuole crystalloid. J. Cell Biol. 2000;1506(1):755–770. doi: 10.1083/jcb.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim S., Robinson D. G. Endocytosis of cationic ferritin by bean leaf protoplasts. Eur. J. Cell Biol. 1984;346(1):212–216. [PubMed] [Google Scholar]
- Jones A. M. Auxin transport: down and out and up again. Science. 1998;2826(1):2201–2203. doi: 10.1126/science.282.5397.2201. [DOI] [PubMed] [Google Scholar]
- Jones A. M. Programmed Cell Death in Development and Defense. Plant Physiol. 2001;1256(1):94–97. doi: 10.1104/pp.125.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 2004;206(1):481–504. doi: 10.1146/annurev.cellbio.20.082503.103057. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Plant cytokinesis: fission by fusion. Trends Cell Biol. 2005;156(1):277–283. doi: 10.1016/j.tcb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Kalde M., Nühse T. S., Findlay K., Peck S. C. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. USA. 2007;1046(1):11850–11855. doi: 10.1073/pnas.0701083104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K. U., Hartmann E. Protein translocation into the endoplasmic reticulum (ER)—two similar routes with different modes. Eur. J. Biochem. 1998;2546(1):1–5. doi: 10.1046/j.1432-1327.1998.2540001.x. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;2736(1):22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kargul J., Gansel X., Tyrrell M., Sticher L., Blatt M. R. Protein-binding partners of the tobacco syntaxin NtSyr1. FEBS Lett. 2001;5086(1):253–258. doi: 10.1016/s0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- Katsuhara M., Hanba Y. T., Shiratake K., Maeshima M. Expanding roles of plant aquaporins in plasma membranes and cell organelles. Functional Plant Biology. 2008;356(1):1–14. doi: 10.1071/FP07130. [DOI] [PubMed] [Google Scholar]
- Kato T., Morita M. T., Fukaki H., Yamauchi Y., Uehara M., Niihama M., Tasaka M. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;146(1):33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M., Kobae Y., Mimura T., Maeshima M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008;6(1):M707646200. doi: 10.1074/jbc.M707646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 2000;16(1):187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Kirsch T., Paris N., Butler J. M., Beevers L., Rogers J. C. Purification and initial characterization of a potential plant vacuolar targeting receptor. Proc. Natl. Acad. Sci. USA. 1994;916(1):3403–3407. doi: 10.1073/pnas.91.8.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Swarup R., Bennett M., Friml J. Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell. 2006;186(1):3171–3181. doi: 10.1105/tpc.106.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou V. L., Dacks J. B., Coulson R. M., Field M. C. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 2007;76(1):29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer A. M., Brandizzi F., Neumann U., Paris N., Moore I., Hawes C. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 2004;1176(1):6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi M., Yamada K., Hatsugai N., Kondo M., Nishimura M., Hara-Nishimura I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005;2806(1):32914–32920. doi: 10.1074/jbc.M504476200. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi M., Yamada K., Hatsugai N., Kondo M., Nishimura M., Hara-Nishimura I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005;2806(1):32914–32920. doi: 10.1074/jbc.M504476200. [DOI] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H. S., Lipka U., Humphry M., Bau S., Straus M., Kwaaitaal M., Rampelt H., El Kasmi F., Jürgens G., Parker J., Panstruga R., Lipka V., Schulze-Lefert P. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;4516(1):835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Lam S. K., Tse Y. C., Robinson D. G., Jiang L. Tracking down the elusive early endosome. Trends in Plant Science. 2007;126(1):497–505. doi: 10.1016/j.tplants.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., Gillespie T., Boevink P., Kriechbaumer V., Hawes C., Carvalho C. M. Localization and domain characterization of Arabidopsis golgin candidates. J. Exp. Bot. 2007;586(1):4373–4386. doi: 10.1093/jxb/erm304. [DOI] [PubMed] [Google Scholar]
- Lauber M. H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997;1396(1):1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C., Paiement J., Dominguez M., Roy L., Dahan S., Gushue J. N., Bergeron J. J. Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. J. Cell Biol. 1999;1466(1):285–299. doi: 10.1083/jcb.146.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R., Alconada A., Bauer U., Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Biol. Chem. 1998;2736(1):29451–2961. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Talloczy Z., Levine B., Dinesh-Kumar S. P. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;1216(1):567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Lowe M. Membrane transport: tethers and TRAPPs. Curr. Biol. 2000;106(1):R407–409. doi: 10.1016/s0960-9822(00)00505-4. [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Mayer U., Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;846(1):61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Marmagne A., Rouet M. A., Ferro M., Rolland N., Alcon C., Joyard J., Garin J., Barbier-Brygoo H., Ephritikhine G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteomics. 2004;36(1):675–691. doi: 10.1074/mcp.M400001-MCP200. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;86(1):410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Matheson L. A., Hanton S. L., Rossi M., Latijnhouwers M., Stefano G., Renna L., Brandizzi F. Multiple roles of ADP-ribosylation factor 1 in plant cells include spatially regulated recruitment of coatomer and elements of the Golgi matrix. Plant Physiol. 2007;1436(1):1615–1627. doi: 10.1104/pp.106.094953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C. Plant aquaporins: novel functions and regulation properties. FEBS Lett. 2007;5816(1):2227–2236. doi: 10.1016/j.febslet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- McNew J. A., Parlati F., Fukuda R., Johnston R. J., Paz K., Paumet F., Sollner T. H., Rothman J. E. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;4076(1):153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- Meijer W., van der Klei I., Veenhuis M., Kiel J. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;36(1):106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;196(1):2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J. P., Hartmann M. A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J. J. Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 2004;2796(1):36277–3686. doi: 10.1074/jbc.M403440200. [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J. J., Blein J. P., Simon-Plas F. Proteomics of plant detergent-resistant membranes. Mol. Cell Proteomics. 2006;56(1):1396–1411. doi: 10.1074/mcp.M600044-MCP200. [DOI] [PubMed] [Google Scholar]
- Moriyasu Y., Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;1116(1):1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A., Guan C., Galweiler L., Tanzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;176(1):6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R. T., Flynn C. R., Trelease R. N. How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci. 2001;66(1):256–261. doi: 10.1016/s1360-1385(01)01951-3. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;3156(1):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Muntz K. Protein dynamics and proteolysis in plant vacuoles. J. Exp. Bot. 2007;586(1):2391–2407. doi: 10.1093/jxb/erm089. [DOI] [PubMed] [Google Scholar]
- Murphy A. S., Bandyopadhyay A., Holstein S. E., Peer W. A. Endocytotic cycling of PM proteins. Ann. Rev. Plant Biol. 2005;566(1):221–251. doi: 10.1146/annurev.arplant.56.032604.144150. [DOI] [PubMed] [Google Scholar]
- Nahm M. Y., Kim S. W., Yun D., Lee S. Y., Cho M. J., Bahk J. D. Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol. 2003;446(1):1341–1349. doi: 10.1093/pcp/pcg163. [DOI] [PubMed] [Google Scholar]
- Nanjo Y., Oka H., Ikarashi N., Kaneko K., Kitajima A., Mitsui T., Muñoz F. J., Rodríguez-López M., Baroja-Fernández E., Pozueta-Romero J. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;186(1):2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Gallagher L. A., Dunahay T. G., Frohlick J. A., Mazurkiewicza A. M., Meehl J. B., Staehelin L. A. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 1999;1216(1):1127–1142. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Frohlick J. A., Staehelin L. A. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 2000;1246(1):135–151. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M., Uemura T., Saito C., Nakano A., Sato M. H., Tasaka M., Morita M. T. Conversion of functional specificity in Qb-SNARE VTI1 homologues of Arabidopsis. Curr. Biol. 2005;156(1):555–560. doi: 10.1016/j.cub.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Nothwehr S. F., Ha S. A., Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 2000;1516(1):297–310. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Aguilar R. C., Yeh D., Taura D., Saito T., Bonifacino J. S. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 1998;2736(1):25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Cowles C. R., Emr S. D. The AP-3 complex: a coat of many colours. Trends Cell Biol. 1998;86(1):282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Olbrich A., Hillmer S., Hinz G., Oliviusson P., Robinson D. G. Newly formed vacuoles in root meristems of barley and pea seedlings have characteristics of both protein storage and lytic vacuoles. Plant Physiol. 2007;1456(1):1383–1394. doi: 10.1104/pp.107.108985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviusson P., Heinzerling O., Hillmer S., Hinz G., Tse Y. C., Jiang L., Robinson D. G. Plant retromer, localized to the pre-vacuolar compartment and microvesicles in Arabidopsis, may interact with vacuolar sorting receptors. Plant Cell. 2006;186(1):1239–1252. doi: 10.1105/tpc.105.035907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M. S., Herder R., Schulze J. M., Jung R., Staehelin L. A. The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell. 2006;186(1):2567–2581. doi: 10.1105/tpc.106.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M. S., Noh Y. S., Martinez D., Vila Petroff M., Staehelin L. A., Amasino R. M., Guiamet J. J. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 2005;416(1):831–844. doi: 10.1111/j.1365-313X.2005.02346.x. [DOI] [PubMed] [Google Scholar]
- Otegui M. S., Staehelin L. A. Keystone symposium on Golgi apparatus and secretory pathway of eukaryotic cells. 1. Vol. 6. Breckenridge; Colorado: 2004. Analysis of Golgi and multivesicualr body functions during protein storage vacuole formation in Arabidopsis thaliana. p. 106. [Google Scholar]
- Paris N., Rogers S. W., Jiang L., Kirsch T., Beevers L., Phillips T. E., Rogers J. C. Molecular cloning and further characterization of a probable plant vacuolar sorting receptor. Plant Physiol. 1997;1156(1):29–39. doi: 10.1104/pp.115.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris N., Stanley C. M., Jones R. L., Rogers J. C. Plant cells contain two functionally distinct vacuolar compartments. Cell. 1996;856(1):563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;46(1):20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. The dynamic organization of the secretory pathway. Cell Struct. Funct. 1996;216(1):413–419. doi: 10.1247/csf.21.413. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Rothman J. E. The debate about transport in the Golgi—two sides of the same coin. Cell. 2000;1026(1):713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J. B., Seabra M. C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;3136(1):889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C. L. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;36(1):275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Pfeffer S. R. Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1999;16(1):E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Phillipson B. A., Pimpl P., daSilva L. L., Crofts A. J., Taylor J. P., Movafeghi A., Robinson D. G., Denecke J. Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell. 2001;136(1):2005–2020. doi: 10.1105/TPC.010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M. L., Serna J., Falbel T. G., Bednarek S. Y., Nielsen E. The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell. 2004;166(1):1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M. L., Schmitz A. J., Thole J. M., Bonner H. K., Otegui M. S., Nielsen E. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 2006;1726(1):991–998. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor P. R., Mullock B. M., Bright N. A., Lindsay M. R., Gray S. R., Richardson S. C., Stewart A., James D. E., Piper R. C., Luzio J. P. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;56(1):590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucher G., Vickery H., Abrahams M., Leavenworth C. Studies in the metabolism of crassulacean plants: diurna1 variations of organic acids and starch in excised leaves of Bryophyllum calycinum. Plant Physiol. 1949;246(1):610–620. doi: 10.1104/pp.24.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L. J., Gu H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007;176(1):249–263. doi: 10.1038/cr.2007.7. [DOI] [PubMed] [Google Scholar]
- Rancour D. M., Dickey C. E., Park S., Bednarek S. Y. Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 2002;1306(1):1241–1253. doi: 10.1104/pp.011742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I., Chen Y. C., Cupers P., Shoelson S. E., Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;176(1):2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robineau S., Chabre M., Antonny B. Binding site of brefeldin A at the interface between the small G protein ADP-ribo-sylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA. 2000;976(1):9913–9918. doi: 10.1073/pnas.170290597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. G., Hinz G., Holstein S. E. The molecular characterization of transport vesicles. Plant Mol. Biol. 1998;386(1):49–76. [PubMed] [Google Scholar]
- Rodionov D. G., Bakke O. Medium chains of adaptor complexes AP-1 and AP-2 recognize leucine-based sorting signals from the invariant chain. J. Biol. Chem. 1998;2736(1):6005–6008. doi: 10.1074/jbc.273.11.6005. [DOI] [PubMed] [Google Scholar]
- Rojo E., Gillmor C. S., Kovaleva V., Somerville C. R., Raikhel N. V. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell. 2001;16(1):303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- Rojo E., Sharma V. K., Kovaleva V., Raikhel N. V., Fletcher J. C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;146(1):969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Martin R., Carter C., Zouhar J., Pan S., Plotnikova J., Jin H., Paneque M., Sanchez-Serrano J. J., Baker B., Ausubel F. M., Raikhel N. V. VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 2004;146(1):1897–1906. doi: 10.1016/j.cub.2004.09.056. [DOI] [PubMed] [Google Scholar]
- Rojo E., Zouhar J., Kovaleva V., Hong S., Raikhel N. V. The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol. Biol. Cell. 2005;146(1):361–369. doi: 10.1091/mbc.E02-08-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese O. W., Soderholm J., Bevis B. J., Sears I. B., O'Connor J., Williamson E. K., Glick B. S. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 1999;1456(1):69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Banfield D. K., Vacca M., Dietrich L. E., Ungermann C., D'Esposito M., Galli T., Filippini F. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004;296(1):682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. A., Ahmed S. U., Marty-Mazars D., Rapoport I., Kirchhausen T., Marty F., Raikhel N. V. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA. 1998;956(1):9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Raikhel N. V. The specificity of vesicle trafficking: coat proteins and SNAREs. Plant Cell. 1999;116(1):629–642. doi: 10.1105/tpc.11.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Kovaleva V., Zheng H., Raikhel N. V. The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 1999;1216(1):929–938. doi: 10.1104/pp.121.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Assaad F. F., Raikhel N. V. The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;1246(1):1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Pilgrim M., Adam L., Raikhel N. V. Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell. 2001a;136(1):659–666. doi: 10.1105/tpc.13.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Kovaleva V., Bassham D. C., Raikhel N. V. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell. 2001b;126(1):3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A. The expansion of SNARE gene families parallels the rise of multicellularity among the green plants. Plant Physiol. 2007;1446(1):6–17. doi: 10.1104/pp.106.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. H., Nakamura N., Ohsumi Y., Kouchi H., Kondo M., Hara-Nishimura I., Nishimura M., Wada Y. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 1997;2726(1):24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- Schmidt U. G., Endler A., Schelbert S., Brunner A., Schnell M., Neuhaus H. E., Marty-Mazars D., Marty F., Baginsky S., Martinoia E. Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 2007;1456(1):216–229. doi: 10.1104/pp.107.096917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., McCaffery J. M., Emr S. D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 1998;1426(1):665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., Marcusson E. G., Cereghino J. L., Emr S. D. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 1997;1376(1):79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí-Simarro J. M., Otegui M. S., Austin J. R., Staehelin L. A. Plant Cytokinesis – Insights gained from electron tomography studies. In: Verma D. P. S., Hong Z., editors. Plant Cell Monogr:Cell Division Control in Plants. Springer-Verlag; Berlin: 2007. [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2003;1006(1):16095–16100. doi: 10.1073/pnas.2530568100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 2006;476(1):1187–1194. doi: 10.1093/pcp/pcj103. [DOI] [PubMed] [Google Scholar]
- Sohn E. J., Kim E. S., Zhao M., Kim S. J., Kim H., Kim Y. W., Lee Y. J., Hillmer S., Sohn U., Jiang L., Hwang I. Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell. 2003;156(1):1057–1070. doi: 10.1105/tpc.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Muller S., Hanisch F-G., Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;1336(1):4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Staehelin L. A. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;116(1):1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- Stagg S. M., LaPointe P., Balch W. E. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 2007;176(1):221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Chatre L., Hanton S. L., Moreau P., Hawes C., Brandizzi F. In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 2006;466(1):95–110. doi: 10.1111/j.1365-313X.2006.02675.x. [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C. L., Paris S., Galweiler L., Palme K., Jürgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;2866(1):316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Stepp J. D., Huang K., Lemmon S. K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;1396(1):1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M. T., Saito C., Avila E., Blakeslee J. J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;156(1):2885–2899. doi: 10.1105/tpc.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Raikhel N. Traffic jams affect plant development and signal transduction. Nature Rev. Mol. Cell Biol. 2004;56(1):100–109. doi: 10.1038/nrm1311. [DOI] [PubMed] [Google Scholar]
- Sutter J., Campanoni P., Blatt M. R., Paneque M. Setting SNAREs in a different wood. Traffic. 2006;76(1):627–638. doi: 10.1111/j.1600-0854.2006.00414.x. [DOI] [PubMed] [Google Scholar]
- Suzuki N. N., Yoshimoto K., Fujioka Y., Ohsumi Y., Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;16(1):119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Swanson S. J., Bethke P. C., Jones R. L. Barley aleurone cells contain two types of vacuoles. Characterization Of lytic organelles by use of fluorescent probes. Plant Cell. 1998;106(1):685–698. doi: 10.1105/tpc.10.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Ueda T., Sato K., Abe H., Nagata T., Nakano A. A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2000;236(1):517–525. doi: 10.1046/j.1365-313x.2000.00823.x. [DOI] [PubMed] [Google Scholar]
- Tanchak M. A., Griffing L. R., Mersey B. G., Fowke L. C. Endocytosis of cationized ferritin by coated vesicles of soybean protoplasts. Planta. 1984;1626(1):481–486. doi: 10.1007/BF00399912. [DOI] [PubMed] [Google Scholar]
- Thompson A. R., Doelling J. H., Suttangkakul A., Vierstra R. D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;1386(1):2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;3496(1):275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Titorenko V. I., Mullen R. T. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 2006;1746(1):11–17. doi: 10.1083/jcb.200604036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N., Sparkes I. A., Hunter P. R., Craddock C. P., Nuttall J., Roberts L. M., Hawes C., Pedrazzini E., Frigerio L. Over-expression of a Plant Reticulon Remodels the Lumen of the Cortical Endoplasmic Reticulum but Does not Perturb Protein Transport. Traffic. 2008;96(1):94–102. doi: 10.1111/j.1600-0854.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- Tooze S. A. Cell biology. GGAs tie up the loose ends. Science. 2001;2926(1):1663–1665. doi: 10.1126/science.1062239. [DOI] [PubMed] [Google Scholar]
- Törmäkangas K., Hadlington J. L., Pimpl P., Hillmer S., Brandizzi F., Teeri T. H., Denecke J. A vacuolar sorting domain may also influence the way in which proteins leave the endoplasmic reticulum. Plant Cell. 2001;136(1):2021–2032. doi: 10.1105/TPC.000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y. C., Mo B., Hillmer S., Zhao M., Lo S. W., Robinson D. G., Jiang L. Identification of multivesicular bodies as pre-vacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell. 2004;166(1):672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;3336(1):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M. H., Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;406(1):783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 2004;296(1):49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- van der Hoorn R. A. L., Jones J. D. G. The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 2004;76(1):400–407. doi: 10.1016/j.pbi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Vernoud V., Horton A., Yang Z., Nielsen E. Analysis of the Small GTPase Gene Family of Arabidopsis thaliana. Plant Physiol. 2003;1316(1):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Denecke J. The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell. 1999;116(1):615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Raikhel N. V. What do proteins need to reach different vacuoles. Trends Plant Sci. 1999;46(1):149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- Waizenegger I., Lukowitz W., Assaad F., Schwarz H., Jürgens G., Mayer U. The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr. Biol. 2000;106(1):1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- Waters M. G., Hughson F. M. Membrane tethering and fusion in the secretory and endocytic pathways. Traffic. 2000;16(1):588–597. doi: 10.1034/j.1600-0854.2000.010802.x. [DOI] [PubMed] [Google Scholar]
- Watson P., Townley A. K., Koka P., Palmer K. J., Stephens D. J. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;76(1):1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Sollner T. H., Rothman J. E. SNARE-pins: minimal machinery for membrane fusion. Cell. 1998;926(1):759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;96(1):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Bassham D. C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;426(1):535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Bassham D. C. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007a;36(1):257–258. doi: 10.4161/auto.3847. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Nguyen P. Q., Bassham D. C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007b;1436(1):291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. Dissection of Arabidopsis ADP-RI-BOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell. 2005;176(1):525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 2008;496(1):142–156. doi: 10.1093/pcp/pcn006. [DOI] [PubMed] [Google Scholar]
- Yano D., Sato M., Saito C., Sato M. H., Morita M. T., Tasaka M. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA. 2003;1006(1):8589–8594. doi: 10.1073/pnas.1430749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung B. G., Phan H. L., Payne G. S. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell. 1999;106(1):3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;166(1):2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Bednarek S. Y., Sanderfoot A. A., Alonso J., Ecker J. R., Raikhel N. V. NPSN11 is a cell plate associated SNARE protein that interacts with the Syntaxin KNOLLE. Plant Physiol. 2002;1296(1):530–539. doi: 10.1104/pp.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., von Mollard G. F., Kovaleva V., Stevens T. H., Raikhel N. V. The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol. Biol. Cell. 1999;106(1):2251–2264. doi: 10.1091/mbc.10.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Camacho L., Wee E., Batoko H., Legen J., Leaver C. J., Malhó R., Hussey P. J., Moore I. A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell. 2005;176(1):2020–2036. doi: 10.1105/tpc.105.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Gong Z., Zhang C., Song C. P., Damsz B., Inan G., Koiwa H., Zhu J. K., Hasegawa P. M., Bressan R. A. OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell. 2002;146(1):3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]


