Background: Caenorhabditis elegans CeFAT-2 was previously characterized as a fatty acid Δ12-desaturase.
Results: CeFAT-2 can introduce second double bond in a range of fatty acid substrates that have a Δ9 double bond.
Conclusion: CeFAT-2 is a bifunctional Δ12/Δ15-desaturase.
Significance: This is the first evidence that the nematode Δ12-desaturase is a bifunctional desaturase that can produce highly unusual desaturated fatty acids.
Keywords: Caenorhabditis elegans, FAD, FAT, Fatty Acid, GC-MS, Genetics, Lipid Metabolism, FAT-2, Bifunctional, Unusual ω1 or ω2 Fatty Acids
Abstract
Caenorhabditis elegans FAT-2 has been characterized as fatty acid Δ12-desaturase able to desaturate C16 and C18 fatty acids. However, in this report we show that when expressed in yeast cells this enzyme can also catalyze Δ15 desaturation. This results in the production of both linoleic acid (ω6 C18:2Δ9,12) and linolenic acid (ω3 C18:3Δ9,12,15) from oleic acid (C18:1Δ9) substrate, and hexadecadienoic acid (ω4 C16:2Δ9,12) and hexadecatrienoic acid (ω1 C16:3Δ9,12,15) from palmitoleic acid (C16:1Δ9) substrate. In addition, this enzyme can also produce C14:2Δ9,12, C15:2Δ9,12, C17:2Δ9,12, and C18:4Δ6,9,12,15 when C14:1Δ9, C15:1Δ9, C17:1Δ9, and C18:3Δ6,9,12 substrates are available in yeast cells. Mass spectrometry analysis of 2,4-dimethyloxazoline modification of fatty acid methyl esters confirms the positions of all newly formed double bonds. These results indicate that when expressed in yeast the C. elegans Δ12-desaturase CeFAT-2 shows a characteristic of a bifunctional Δ12/Δ15-desaturase and has a great deal of elasticity with respect to fatty acid chain length in being able to accept fatty acids ranging from C14 to C18. Interestingly, despite possessing a bifunctional Δ12/Δ15 desaturation activity, phylogenetic analysis suggests that C. elegans Δ12-desaturase CeFAT-2 might have arisen independently from other reported dual Δ12/Δ15-desaturases from fungi and protozoa.
Introduction
In recent years, fatty acid synthesis pathways in many species have been genetically dissected, leading to the identification of fatty acid desaturation and chain elongation genes. For example, the Caenorhabditis elegans polyunsaturated fatty acid synthesis pathway has been shown to contain at least seven desaturases and two elongases (1, 2). These include Δ9-desaturases (FAT-5, FAT-6, FAT-7), Δ12-desaturase (FAT-2), Δ15-desaturase or ω3-desaturase (FAT-1), Δ5-desaturase (FAT-4), and Δ6-desaturase (FAT-3). Among the desaturases, FAT-2 was earlier confirmed as a Δ12-desaturase acting on oleic acid (C18:1Δ9) to produce linoleic acid (LA,2 ω6, C18:2Δ9,12) (3) whereas FAT-1 was shown to be a Δ15-desaturase acting on ω6 fatty acid LA, for example, to form α-linolenic acid (ω3, C18:3Δ9,12,15) (4–6).
Fatty acid desaturases possess at least five specificities: chemoselectivity (desaturation versus hydroxylation), regioselectivity (double bond position), stereoselectivity (cis versus trans), chain length specificity, and acyl carrier specificity (acyl-CoA versus acyl-lipid substrate). Historically, three main types of regioselectivities have been distinguished (6). Desaturases with Δx regioselectivity introduce a first double bond x carbon atoms away from the carboxyl end. A second group comprises ωy-desaturases, and these insert a double bond at position y referenced from the methyl end (for example, ω3-desaturase). Finally, there are the v+z-desaturases which require a preexisting double bond as reference point (v) and catalyze the formation of a new double bond z carbon atoms further along the acyl chain, either toward methyl end (i.e. Δ12-desaturase) or toward carboxyl end (i.e. front end Δ4- or Δ5-desaturase). These modes of regioselectivity are not mutually exclusive as illustrated by several bifunctional fungal Δ12/ω3-desaturases. For example, in addition to the Δ12-fatty acid desaturase members, separate bifunctional Δ12/ω3-fatty acid desaturases have been identified from fungi Fusarium moniliforme (7) and Aspergillus nidulans (8). These bifunctional enzymes desaturate oleic acid C18:1Δ9 at Δ12 position to form C18:2Δ9,12 and can also carry out a further desaturation at the Δ15 position to form ω3-fatty acid C18:3Δ9,12,15. Additionally, these fungal desaturases also convert ω6 substrates, including LA, γ-linolenic acid (C18:3Δ6,9,12), dihomo-γ-linolenic acid (C20:3Δ8,11,14), and arachidonic acid (C20:4Δ5,8,11,14) to their ω3 products and show an obvious preference for the C18 substrates, especially LA (7, 8). A bifunctional Δ12/Δ15-desaturase that directs the synthesis of highly unusual hexadecatrienoic acid (C16:3Δ6,9,12, ω1) has also been isolated from the free-living soil protozoon Acanthamoeba castellanii (9) and phytopathogenic fungus, Claviceps purpurea (10).
Here, we report the identification of a novel undescribed catalytic function for the previously characterized C. elegans Δ12-desaturase CeFAT-2 enzyme. When expressed in yeast cells, CeFAT-2 displays characteristics of a bifunctional Δ12/Δ15-desaturase and has a great deal of elasticity with respect to fatty acid chain length in being able to desaturate fatty acids ranging from C14 to C18. Finally, phylogenetic analysis suggests that despite possessing a bifunctional Δ12/Δ15 desaturation activity, CeFAT-2 appears to have arisen independently from other reported dual Δ12/Δ15-desaturases from fungi (7, 8) and protozoa (9).
EXPERIMENTAL PROCEDURES
Fatty Acids and Fatty Acid Methyl Ester (FAME)
Free fatty acids and FAME standards C8–C24 were purchased from Sigma. C18:1Δ12 was chemically synthesized. Other fatty acid substrates were purchased from Sigma or Cayman Chemical Company.
Gene Subcloning
The C. elegans fat-2 gene (GenBank accession number AF240777) was PCR-amplified from cDNA with restriction sites HindIII and XhoI at the ends of PCR primers (forward, 5′-GTAAAGCTTGGCCCCCGACG and reverse, 5′-TGTCTCGAGATTTTGTAGTTTATTG) and cloned in yeast expression vector pYES2 (Invitrogen) with a Gal1 promoter to generate pXZP278. As a control, the Arabidopsis Δ12-desaturase gene AtFAD2 (At3g12120) coding sequence was also PCR-amplified with primers (forward, 5′-TCCGTCGACATGGGTGCAGGTGGAAGA and reverse, 5′-CGGGGTACCTCATAACTTATTGTTGTACCAG). The PCR product was cloned into pGEM-T Easy (Promega) and subcloned into pYES2 as EcoRI blunt-ended SpeI fragment to generate plasmid pXZP279. The gene sequences in these plasmids were confirmed by sequencing the entire amplified regions. The C. elegans fat-1 gene (GenBank accession number L41807) was synthesized by GeneArt and cloned into yeast expression vector with a Gal1 promoter as an EcoRI-XhoI fragment in which restriction sites were built in before and after the start and stop codons, generating plasmid pXZP586.
Expression of Genes in Yeast and Gas Chromatography Analysis
Plasmids were introduced into yeast strain S288C (11) cells by heat shock as described previously (12). Yeast feeding with different exogenous fatty acids was essentially as described previously (12). FAMEs were analyzed with a Agilent N6890 GC on a 60-m BPX70 column (0.25-mm inner diameter, 0.25-μm film thickness, SGE), or Thermo Polaris Q GC-MS on a 30-m BPX70 column. The column temperature was programmed as an initial temperature at 170 °C holding for 1 min, ramping to 200 °C at 5 °C/min and 210 °C at 2.5 °C/min then to 240 °C at 10 °C/min, and finally holding for 3 min on Agilent GC, or 60 °C holding for 1 min, ramping to 120 °C at 9 °C/min then to 250 °C at 50 °C/min, and holding for 1 min on a Thermo GC-MS. Helium was the carrier gas. Identification of peaks was based on comparison of retention time with standard FAME and confirmed with MS acquired and processed with XcaliburTM software from Thermo Electron Corporation.
Double bond positions in the FAME were confirmed with 2,4-dimethyloxazoline (DMOX) adducts by derivatizing the FAMEs with 2-amino-2-methyl-propanol at 190 °C overnight, cooling, and acidifying with 0.1 n HCl before extracting with dichloromethane (13), and analyzed as described previously for FAMEs (12).
RESULTS
C. elegans Δ12-Desaturase CeFAT-2 Also Carries Out Δ15 Desaturation
In this study, CeFAT-2 is expressed under the control of an inducible Gal1 promoter in Saccharomyces cerevisiae strain S288C (MATa, his3-d200 leu2-d1 trp-1-d63 lys2-d202 ura3-52). In the absence of any exogenous fatty acid feeding, it produces two new peaks in addition to the new but expected C16:2Δ9,12 or C18:2Δ9,12 (Fig. 1). As expected, the main fatty acids in yeast cells carrying empty vector are C16:0, C16:1Δ9, C18:0, and C18:1Δ9 (Fig. 1A). Trace amounts of C16:1Δ9trans, C17:0, and C18:1Δ11 are also found. The two abundant new peaks present in the FAMEs of yeast cells expressing both Arabidopsis AtFAD2 and nematode CeFAT-2 (both enzymes were previously demonstrated as Δ12-desaturases) are confirmed as hexadecadienoic acid (C16:2Δ9,12) and LA (C18:2Δ9,12), as shown in Fig. 1, B and C. In addition, trace amounts of C18:2Δ11,14, presumably elongated from C16:2Δ9,12, were also identified. Interestingly, additional new peaks in the FAMEs of yeast expressing CeFAT-2 are also observed (Fig. 1C). One of them has the same retention time as the C18:3Δ9,12,15 standard and the dominant fatty acid present in Arabidopsis leaf (Fig. 1D). The other new peak appeared before C18:0 in Fig. 1C and has a retention time comparable with C16:3Δ7,10,13, an abundant Arabidopsis leaf fatty acid (Fig. 1D). The identities of these new peaks are confirmed by MS analysis (see supplemental Fig. S1). The mass spectra matched to the mass spectra of standards C16:2 (234, 266 m/z), C16:3 (232, 264 m/z), C18:2 (262, 294 m/z) and C18:3 (260, 292 m/z).
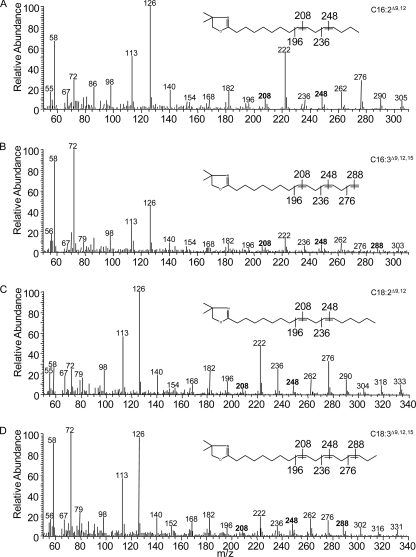
FIGURE 1.
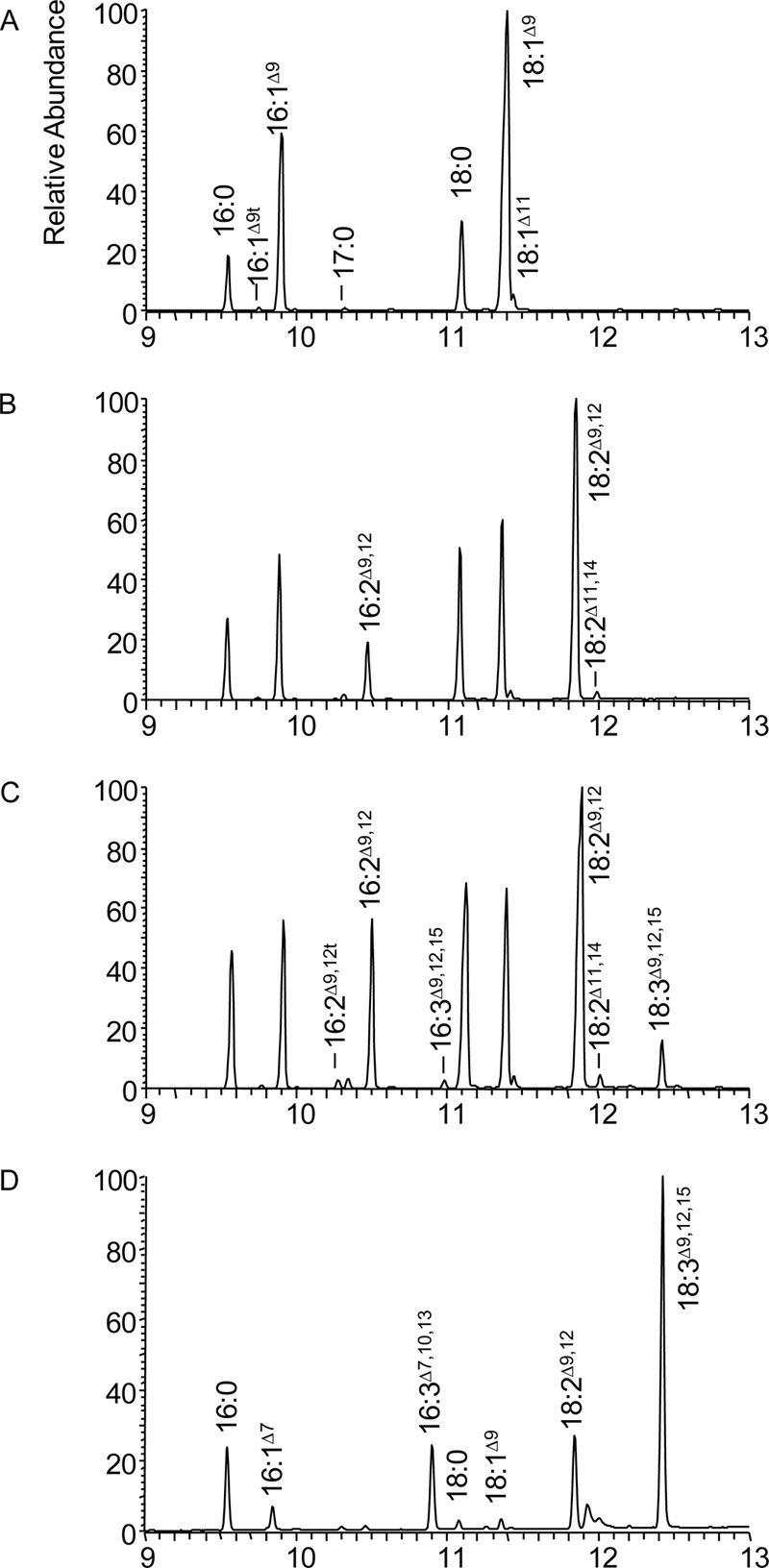
Partial gas chromatogram of FAMEs from yeast cells or Arabidopsis leaf. A–C, yeast cells expressing vector pYES2 only, AtFAD2, and CeFAT-2. D, Arabidopsis leaf.
The positions of the double bonds in the new products are confirmed as C16:2Δ9,12, C16:3Δ9,12,15, C18:2Δ9,12, and C18:3Δ9,12,15 by MS analysis of DMOX adducts of the FAMEs, as shown in Fig. 2. Fig. 2A shows the structure of C16:2 (molecular ion of m/z 305), with the double bond present in positions Δ9 and Δ12 located by the gap of 12 atomic mass units between m/z 196 and 208, 236 and 248, respectively. Fig. 2B shows the structure of C16:3 with molecular ion of m/z 303 and indicates that this C16 fatty acid contains three double bonds. These double bonds are confirmed to be present at positions 9, 12, and 15 and located by the gap of 12 atomic mass units between m/z 196 and 208, 236 and 248, 276 and 288, respectively. Similar structures of DMOX adducts of C18:2 and C18:3 are shown in Fig. 2, C and D. These results suggest that CeFAT-2 can also function as a Δ15-desaturase and produce a ω3 C18:3Δ9,12,15 product from C18:1Δ9 as well as a highly unusual ω1 C16:3Δ9,12,15 fatty acid from C16:1Δ9. It is noteworthy that Δ12 desaturation activity of CeFAT-2 is 8–10 times higher than its Δ15-desaturase activity (Table 1). Finally, other tiny peaks can also be observed in FAMEs from yeast expressing CeFAT-2. These are confirmed either as trans-isomers of corresponding cis-unsaturated fatty acids (data not shown) or as possible elongation products produced by yeast cells (see below).
FIGURE 2.
Mass spectrometry of DMOX derivatives of major CeFAT-2 products. A–D, DMOX adducts of C16:2Δ9,12, C16:3Δ9,12,15, C18:2Δ9,12, and C18:3Δ9,12,15, respectively.
TABLE 1.
Fatty acid compostion (weight %) in yeast cells expressing empty vector only or Δ12-desaturases
Trace amounts of C15, C17, and trans-C16 or C18 are not included.
| Fatty acid | Vector | AtFAD2 | CeFAT-2 |
|---|---|---|---|
| C16:0 | 6.55 | 8.68 | 9.64 |
| C16:1Δ9 | 25.60 | 15.38 | 11.57 |
| C16:1Δ11 | <0.01 | <0.01 | <0.01 |
| C16:2Δ9,12 | 0.00 | 6.62 | 10.39 |
| C16:3Δ9,12,15 | 0.00 | 0.00 | 0.46 |
| C18:0 | 10.59 | 14.88 | 18.34 |
| C18:1Δ9 | 55.60 | 16.97 | 13.17 |
| C18:1Δ11 | 1.02 | 0.46 | 0.47 |
| C18:2Δ9,12 | 0.00 | 35.30 | 31.79 |
| C18:2Δ11,14 | 0.00 | 0.42 | 0.62 |
| C18:3Δ9,12,15 | 0.00 | 0.00 | 3.26 |
| Δ12 Desaturation on C16:1 (%) | 0.0 | 30.1 | 47.3 |
| Δ12 Desaturation on C18:1 (%) | 0.0 | 67.5 | 72.7 |
| Δ15 Desaturation on C16:2 (%) | 0.0 | 0.0 | 4.3 |
| Δ15 Desaturation on C18:2 (%) | 0.0 | 0.0 | 9.3 |
Divergent Fatty Acid Substrates Desaturated by CeFAT-2
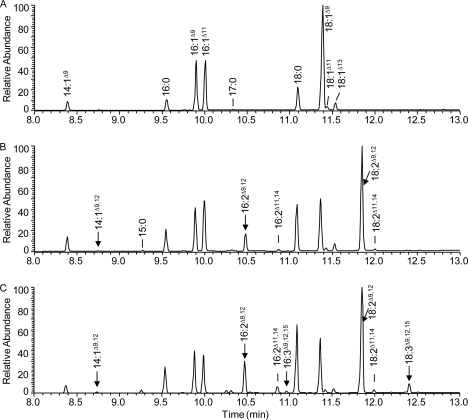
Further extensive yeast feeding with exogenous fatty acids shows that CeFAT-2 can desaturate C14:1Δ9, C15:1Δ9, and C17:1Δ9 at the Δ12 position to produce C14:2Δ9,12, C15:2Δ9,12, and C17:2Δ9,12. Fig. 3 shows the GC analysis of yeast FAMEs expressing vector only, AtFAD2, and CeFAT-2. When fed with C14:1Δ9, in addition to the C16:2 and C18:2 products, a new peak with mass spectra of C14:2 can be found only in cells expressing either of the Δ12-desaturases (Fig. 3 and supplemental Fig. S2). The new products are confirmed by DMOX adducts of the FAMEs as C14:2Δ9,12, C16:2Δ9,12, and C18:2Δ9,12 (supplemental Fig. S2 and data not shown). In addition, low amounts of C16:3 and C18:3 are also detected in FAMEs of yeast expressing CeFAT-2 but not in yeast expressing AtFAD2, again confirming the additional Δ15 desaturation activity of CeFAT-2. Also, in cells expressing both AtFAD2 and CeFAT-2, trace amounts of additional dienoic peaks are detected eluting after the C16:2Δ9,12 and C18:2Δ9,12 peaks. These peaks display the typical mass spectra of C16:2 and C18:2 (234, 266 m/z and 262, 294 m/z, respectively). These are confirmed by DMOX adducts of the FAMEs to be C16:2Δ11,14, C18:2Δ11,14, possibly elongated from C14:2Δ9,12 and C16:2Δ9,12. When provided with C14:1Δ9, yeast cells can elongate this fatty acid to C16:1Δ11 substantially, as shown by the new peak eluting after C16:1Δ9 in FAMEs of all yeast samples (Fig. 3). Trace amounts of C18:1Δ11 and C18:1Δ13 are also detected, presumably the elongation products of C16:1Δ9 and C16:1Δ11, respectively. The mass spectra of DMOX adducts C16:1Δ11, C16:2Δ11,14, C18:1Δ11, C18:1Δ13, and C18:2Δ11,14 are shown in supplemental Fig. S2.
FIGURE 3.
Partial gas chromatogram of yeast feeding for CeFAT-2 with C14:1Δ9. A–C, FAMEs of yeast S288C expressing pYES2 vector only, AtFAD2, and CeFAT-2, respectively, fed with C14:1Δ9. New desaturated products from AtFAD2 or CeFAT-2 are indicated by arrows.
When fed with C15:0, yeast cells can elongate part of this fatty acid into C17:0, as shown in FAMEs of yeast cells expressing either pYES2 vector only or Δ12-desaturases (see supplemental Fig. S3). Both C15:0 and C17:0 are desaturated into monounsaturated fatty acid C15:1 and C17:1, by the host cell Δ9-desaturase, as these peaks are also present in cells expressing vector only. These peaks are confirmed as C15:1Δ9 and C17:1Δ9 by DMOX adducts rather than C15:1Δ10 and C17:1Δ10 (see supplemental Fig. S3). There is another C17:1 peak eluting after C17:1Δ9 in all three samples, presumably C17:1Δ11, an elongation product of C15:1Δ9. Compared with FAMEs from yeast cells expressing vector only, both AtFAD2 and CeFAT-2 produce several new peaks, including C15:2Δ9,12 (co-eluting with trace amounts of trans-C16:1Δ9), C16:2Δ9,12, C17:2Δ9,12, and C18:2Δ9,12. In addition, trace amounts of C17:2Δ11,14 and C18:2Δ11,14 are also detectable in yeast cells expressing both AtFAD2 and CeFAT-2, presumably elongated from the desaturation product C15:2Δ9,12 and C16:2Δ9,12, respectively. Again, cells expressing CeFAT-2 also produce C16:3Δ9,12,15 and C18:3Δ9,12,15, evidence of Δ15 desaturation activity of CeFAT-2. Because C17:2Δ9,12 is only produced in low amounts, we cannot confirm whether this product can be further desaturated to C17:3Δ9,12,15 by CeFAT-2.
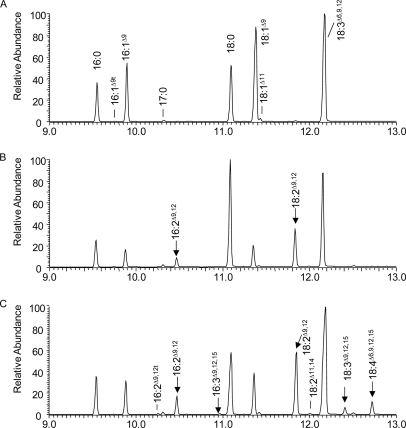
When fed with C18:3Δ6,9,12, there is no detectable product produced from the fed substrate in yeast cells expressing vector only (Fig. 4A) or AtFAD2 (Fig. 4B), although the cells incorporate an abundant amount of the substrate. In contrast, CeFAT-2-expressing cells produce C18:4Δ6,9,12,15 via a desaturation at the Δ15 position (Fig. 4C). The characteristic ion for the n-3 structure, at m/z 107/108 (14), was apparent (see supplemental Fig. S4) as well as the ion for Δ6,9 double bonds at m/z 194. The double bond positions are also confirmed at Δ6,9,12,15 by DMOX adducts, by the gap of 12 atomic mass units with characteristic molecular ion at m/z 166, 206, 246 and 286, respectively.
FIGURE 4.
Partial gas chromatogram of yeast feeding for CeFAT-2 with C18:3Δ6,9,12. A–C, FAMEs of yeast S288C expressing pYES2 vector only, AtFAD2, and CeFAT-2, respectively, fed with C18:3Δ6,9,12.
Other yeast feeding experiments also suggest that C18:1Δ6, C18:1Δ9trans, C18:1Δ11, C18:1Δ12, C18:1Δ13, C18:3Δ9,12,15, C20:1Δ11, and C20:2Δ11,14, C20:3Δ8,11,14, C20:4Δ8,11,14,17 are not accepted as substrates by CeFAT-2 for further desaturation (data not shown). This suggests that the preexistence of a cis-Δ9 double bond is required for second or third double bond formation, and C18:3Δ9,12,15 is not used for further desaturation at other positions (see supplemental Fig. S5).
A feeding experiment with C16:1Δ7 results in a low amount (1.3%) of C16:2Δ7,10 at a conversion rate of 3%, close to 12 times lower than converting C16:1Δ9 to C16:2Δ9,12. However, this low activity is not specific for CeFAT-2 because AtFAD2 also produces a trace amount (0.3%) of C16:2Δ7,10 at a conversion rate of 0.7% (supplemental Table S1). The identity of C16:1Δ7 and C16:2Δ7,10 peaks is confirmed by MS analysis on both FAMEs and DMOX adducts of the FAMEs which display mass spectra similar to the standard C16:1Δ7 and C16:2Δ7,10 (data not shown).
C20:5Δ5,8,11,14,17 (ω3) rather than C18:3Δ9,12,15 is a major polyunsaturated fatty acid in C. elegans, thus it is of interest to know whether CeFAT-2 contributes to the C20:5Δ5,8,11,14,17 production. CeFAT-1 is the ω3-desaturase converting ω6 fatty acid C20:4Δ5,8,11,14 to ω3 fatty acid C20:5Δ5,8,11,14,17. We show in yeast cells that CeFAT-1 is able to produce C20:5Δ5,8,11,14,17 from fed C20:4Δ5,8,11,14 at a conversion rate of 18%, but only a trace amount (0.44%) of C20:5Δ5,8,11,14,17 can be produced by CeFAT-2 at conversion rate of 2% (supplemental Table S2). This low desaturation activity is more a ω3 desaturation, rather than a Δ15 desaturation.
In addition, we have fed the yeast cells expressing these genes with ω3 fatty acid C20:4Δ8,11,14,17 to check whether CeFAT-2 can produce ω3 fatty acid C20:5Δ5,8,11,14,17 from this substrate. This Δ5 desaturation step is catalyzed by FAT-4 in C. elegans (4). As expected, neither CeFAT-2 nor CeFAT-1 can use this substrate to produce C20:5Δ5,8,11,14,17.
Relationship of CeFAT-2 to Δ12- and Δ15-Desaturases
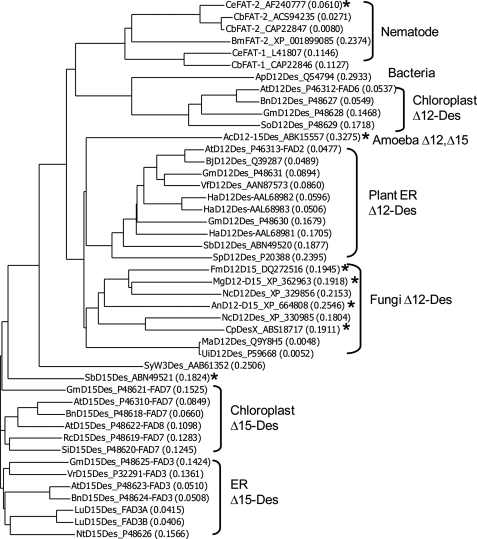
As expected, a BLAST search with the CeFAT-2 sequence identifies a diverse range of desturases, including Δ12- and Δ15-desaturases, in the NCBI data base. Alignment of the desaturase sequences shows that the nematode CeFAT-2 and CeFAT-1 form a separate cluster with bacteria or chloroplast Δ12-desaturases (FAD6), whereas microsomal Δ12-desaturases (FAD2) and Δ15-desaturases (FAD3) form the other two major clusters (Fig. 5). However, CeFAT-2 shares a slightly higher sequence identity to AtFAD3 (29%) than AtFAD2 (27%).
FIGURE 5.
Sequence comparison of CeFAT-2 and related desaturases. Each sequence is followed by an accession number. * indicates bifunctional desaturase activity.
We have also examined whether C. elegans Δ15-desaturase CeFAT-1, when expressed in yeast under the same experimental conditions as CeFAT-2, has any ability to carry out a Δ12 desaturation. CeFAT-1 was confirmed as ω3-desaturase when expressed in yeast cells (6) and in plant (5) and is able to act on C16-C20 substrates. When we express CeFAT-1 under Gal1 promoter in yeast S288C without the addition of exogenous substrate, no Δ12-desaturase activity on C18:1Δ9 is detected (see supplemental Table S3). Additionally, when fed with C18:2Δ9,12, C18:3Δ6,9,12, or C20:4Δ5,8,11,14 substrates, CeFAT-1 clearly shows ω3-desaturase activity resulting in the production of C18:3Δ9,12,15, C18:4Δ6,9,12,15, or C20:5Δ5,8,11,14,17. In addition, CeFAT-1 showed no strong preference for C20 over C18 fatty acid substrate as speculated elsewhere (1) because their observed ω3 desaturation rates were 7.1% and 9.5%, respectively (see supplemental Table S3).
DISCUSSION
The C. elegans CeFAT-2 was isolated and characterized as a nematode fatty acid Δ12-desaturase in an earlier study (3). In the study, the Δ12-desaturase activity of CeFAT-2 was confirmed by heterologous expression with the constitutive ADH1 promoter in S. cerevisiae strain YRP685 (MATa, leu2, lys2, his4, trp1, ura3), where CeFAT-2 was able to desaturate C16:1Δ9 and C18:1Δ9 to produce C16:2Δ9,12 and C18:2Δ9,12. The study concluded that CeFAT-2 was a Δ12-desaturase, and no obvious Δ15 desaturation activity was reported (3). In contrast, we find that when expressed in yeast strain S288C the C. elegans Δ12-desaturase CeFAT-2 can function as a Δ12/Δ15 bifunctional desaturase and behaves as a ν +3′ desaturase (i.e. able to insert double bonds three carbons away from an existing double bond). More specifically, our results show that this enzyme is able to further desaturate a diverse range of fatty acid substrates containing a preexisting Δ9 double bond sequentially at the Δ12 and Δ15 positions. We were unable to find the product C18:2Δ9,15, indicating that the enzyme requires a preexisting double bond at the Δ12 position to carry out desaturation at the Δ15 position. A possible reason for the observed discrepancy between the earlier study and ours could be the different yeast promoters used to drive the expression of the CeFAT-2. For example, the earlier study (3) used a constitutive ADH1 promoter, whereas we have used an inducible promoter, and the failure to detect bifunctional activity in the earlier study might be due to different level of CeFAT-2 protein made. In the earlier study, the accumulation level of C18:2Δ9,12 product was relatively lower, and this might have led to the further desaturated products not being detected. In addition, we also confirm that CeFAT-1, as reported previously (6), is a ω3-desaturase which converts ω6 fatty acids C18:2Δ9,12, C18:3Δ6,9,12, and C20:4Δ5,8,11,14 into ω3 products C18:3Δ9,12,15, C18:4Δ6,9,12,15, and C20:5Δ5,8,11,14,17 and displays no Δ12 desaturation activity. This result indicates that the bifunctional activity of CeFAT-2 observed in our expression system is unlikely to be an artifact of our experimental conditions.
When expressed in yeast, CeFAT-2 can use a range of substrates for desaturation. These include C14:1Δ9, C15:1Δ9, C16:1Δ9, C16:2Δ9,12, C17:1Δ9, C18:1Δ9, C18:2Δ9,12, and C18:3Δ6,9,12 which are converted into C14:2Δ9,12, C15:2Δ9,12, C16:2Δ9,12, C16:3Δ9,12,15, C17:2Δ9,12, C18:2Δ9,12, C18:3Δ9,12,15, and C18:4Δ6,9,12,15 at the ω2, ω3, ω4, ω1, ω5, ω6, ω3, and ω3 positions, respectively. The production of ω1-C16:3, a highly unusual fatty acid containing a terminal double bond, is noteworthy. Production of ω1-C16:3 fatty acid has recently been reported in the yeast by the ectopic expression of an amoeba bifunctional Δ12/Δ15-desaturase (9) or a fungal Δ12,ω3-desaturase (10) and sorghum root hair fatty acid desaturase SbDES3 (15). The CeFAT-2 desaturase hence adds to the growing list of desaturases displaying remarkable inherent elasticity in being able to catalyze the synthesis of fatty acids desaturated the at ω1, ω3, or ω6 position.
The newly discovered Δ12/Δ15 bifunctional desaturation activity of CeFAT-2 partially overlaps with the Δ15 desaturation activity of CeFAT-1. We show in yeast cells that CeFAT-1 is able to desaturate ω6 fatty acids C18:1Δ12, C18:2Δ9,12, C18:3Δ6,9,12, and C20:4Δ5,8,11,14 to ω3 fatty acids C18:2Δ12,15, C18:3Δ9,12,15, C18:4Δ6,9,12,15, and C20:5Δ5,8,11,14,17, but only a trace amount (0.44%) of C20:5Δ5,8,11,14,17 can be produced by CeFAT-2 from C20:4Δ5,8,11,14 (supplemental Tables S2 and S3). From this additional feeding, we can conclude that the CeFAT-2 dual function does not substantially overlap with CeFAT-1, and it has only a minor ω3 desaturation activity. Therefore, it is unlikely that CeFAT-2 makes a substantial contribution to the synthesis of ω3 C20:5Δ5,8,11,14,17 in C. elegans. The fact that the C. elegans fat-1 mutant does not accumulate ω3 polyunsaturated fatty acids prompts the question whether CeFAT-2 is able to carry out any Δ15 desaturation in the nematode (4). Another possibility is that the expression level of CeFAT-2 is lower than CeFAT-1 in nematode, and as it only possesses up to 10% Δ15 desaturation activity compared with Δ12 desaturation activity, the CeFAT-2 Δ15 desaturation activity is not enough for complementing the fat-1 mutation. The bifunctional Δ12/Δ15 characteristic of CeFAT-2 suggests the possible existence of an ancestral progenitor from which both CeFAT-2 and CeFAT-1 might have derived individually. It is thus possibly that CeFAT-2 evolved into a Δ12-desaturase but still maintained some Δ15 desaturation activity, whereas CeFAT-1 evolved into a pure Δ15-desaturase. Alternatively, the nematode ancestor might have evolved the Δ12-desaturase from its Δ15-desaturase after a gene duplication event because the nematode fat-1 and fat-2 genes are transcribed in the same orientation, have similar structures of three exons and two introns, and are separated by ∼5.3 kb of DNA (3). Finally, bifunctional Δ12/Δ15-desaturases have been isolated from fungi (7) and protozoa (9). Interestingly, these bifunctional desaturases (indicated by * in Fig. 5) cluster together with plant microsomal Δ12-desaturases and are separated from CeFAT-2, suggesting that CeFAT-2 has evolved independently from fungi or protozoa dual functional Δ12/Δ15-desaturases.
It will be of interest to identify the catalytic residues responsible for the Δ15 desaturation activity of Δ12-desaturase CeFAT-2. Meesapyodsuk et al. (10) identified two critical residues close to the conserved His boxes of C. purpurea Δ12/Δ15 bifunctional desaturase, Val-152 and Ala-206, which are conserved in the bifunctional desaturases but different from that of Δ12-desaturase. These authors confirmed the role of these residues as well as C-terminal amino acid sequence (residues 302–477) in regioselectivity of the enzyme by site-directed mutagenesis or domain swapping. However, sequence alignment analysis suggests that CeFAT-2 is quite divergent from plant, fungi, or protozoa bifunctional Δ12/Δ15-desaturases, and the residues at the equivalent positions for the two key residues from C. purpurea Δ12/Δ15 bifunctional desaturase are different from both Δ12-desaturase or Δ12/Δ15 bifunctional desaturases. Therefore, other residues may also be responsible for the newly confirmed Δ15 desaturation activity of CeFAT-2. For example, residues close to His boxes conserved in either Δ15-desaturases or Δ12/Δ15 bifunctional desaturases can be considered to be such candidates (supplemental Fig. 6S). Their role, if any, in the CeFAT-2 bifunctional activity remains to be studied further.
Supplementary Material
Acknowledgments
We thank Bei Dong, Cheryl Blundell, Geraldine Lester, Luch Hac, and Suzhi Li of Commonwealth Scientific and Industrial Research Organisation (CSIRO) Plant Industry for excellent technical assistance; Dr. Mira Dumancic of CSIRO Ecosystem Sciences for providing C. elegans RNA; and Dr. Florian Graichen of CSIRO Molecular and and Health Technologies for chemical synthesis of C18:1Δ12. We thank Dr. Charles Hocart of Australian National University and Dr. Rosangela Devilla of CSIRO Plant Industry for advice on gas chromatography mass spectrometry, and Dr. Tony Ashton and Dr. Shoko Okada for critical reading. We thank to two anonymous reviewers for valuable comments.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
- LA
- linoleic acid
- AtFAD2
- Arabidopsis Δ12-desaturase
- CeFAT-1
- C. elegans fatty acid desaturase 1
- CeFAT-2
- C. elegans fatty acid desaturase 2
- DMOX
- 2,4-dimethyloxazoline
- FAME
- fatty acid methyl ester.
REFERENCES
- 1. Napier J. A., Michaelson L. V. (2001) Lipids 36, 761–766 [DOI] [PubMed] [Google Scholar]
- 2. Watts J. L. (2009) Trends Endocrinol. Metab. 20, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peyou-Ndi M. M., Watts J. L., Browse J. (2000) Arch. Biochem. Biophys. 376, 399–408 [DOI] [PubMed] [Google Scholar]
- 4. Watts J. L., Browse J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spychalla J. P., Kinney A. J., Browse J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1142–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meesapyodsuk D., Reed D. W., Savile C. K., Buist P. H., Ambrose S. J., Covello P. S. (2000) Biochemistry 39, 11948–11954 [DOI] [PubMed] [Google Scholar]
- 7. Damude H. G., Zhang H., Farrall L., Ripp K. G., Tomb J. F., Hollerbach D., Yadav N. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9446–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M., Hornung E., Busch S., Kassner N., Ternes P., Braus G. H., Feussner I. (2007) J. Biol. Chem. 282, 26666–26674 [DOI] [PubMed] [Google Scholar]
- 9. Sayanova O., Haslam R., Guschina I., Lloyd D., Christie W. W., Harwood J. L., Napier J. A. (2006) J. Biol. Chem. 281, 36533–36541 [DOI] [PubMed] [Google Scholar]
- 10. Meesapyodsuk D., Reed D. W., Covello P. S., Qiu X. (2007) J. Biol. Chem. 282, 20191–20199 [DOI] [PubMed] [Google Scholar]
- 11. Mortimer R. K., Johnston J. R. (1986) Genetics 113, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou X.-R., Robert S., Singh S., Green A. (2006) Plant Sci. 170, 665–673 [Google Scholar]
- 13. Fay L., Richli U. (1991) J. Chromatography A 541, 89–98 [Google Scholar]
- 14. Fellenberg A. J., Johnson D. W., Poulos A., Sharp P. (1987) Biomed. Environ. Mass Spectrom. 14, 127–129 [Google Scholar]
- 15. Pan Z., Rimando A. M., Baerson S. R., Fishbein M., Duke S. O. (2007) J. Biol. Chem. 282, 4326–4335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.