Background: Arginine is a key amino acid in cellular metabolism in bacteria.
Results: ArgR1 and AhrC mediate arginine-dependent expression of arginine acquisition and virulence genes in the human pathogen Streptococcus pneumoniae.
Conclusion: Arginine regulation in S. pneumoniae significantly differs from that in other model bacteria.
Significance: Understanding metabolic regulation increases insights into the molecular pathogenesis of S. pneumoniae.
Keywords: Bacteria, Gene Regulation, Nitrogen Metabolism, Streptococcus, Transcriptomics, Streptococcus pneumoniae, Arginine, Human Pathogen
Abstract
In this study, we investigated for the first time the transcriptional response of the human pathogen Streptococcus pneumoniae to fluctuating concentrations of arginine, an essential amino acid for this bacterium. By means of DNA microarray analyses, several operons and genes were found, the expression of which was affected by the concentration of arginine in the medium. Five of the identified operons were demonstrated to be directly repressed in the presence of high arginine concentrations via the concerted action of the ArgR-type regulators ArgR1 and AhrC. These ArgR1/AhrC targets encompass the putative amino acid transport genes artPQ, abpA, abpB, and aapA; the arginine biosynthetic genes argGH; and the virulence genes aliB and lmB/adcAII-phtD encoding an oligopeptide-binding lipoprotein and cell surface Zn2+-scavenging units, respectively. In addition, the data indicate that three of the amino acid transport genes encode an arginine ATP-binding cassette transporter unit required for efficient growth during arginine limitation. Instead of regulating arginine biosynthetic and catabolic genes as has been reported for other Gram-positive bacteria, our findings suggest that the physiological function of ArgR1/AhrC in S. pneumoniae is to ensure optimal uptake of arginine from the surrounding milieu.
Introduction
The human pathogen Streptococcus pneumoniae is responsible for infections such as pneumonia, sepsis, otitis media, and meningitis, especially in children and the elderly (1, 2). Several virulence factors that contribute to efficient colonization and invasion of its host have been identified and characterized (1, 3, 4). Ongoing efforts to improve understanding of the molecular biology of pneumococcus will aid in the development of strategies to combat infection by this bacterium.
Proper acquisition and metabolism of nutrients are important for the lifestyle of S. pneumoniae. Several studies have investigated pneumococcal nitrogen metabolism and regulation. It has been shown that the nutritional regulator CodY, which regulates mainly genes involved in nitrogen metabolism, as well as virulence genes like pcpA and ami/aliA in response to the concentration of branched-chain amino acids contributes to pneumococcal colonization (5). In addition, the regulon of the glutamine-dependent repressor GlnR contributes to pneumococcal adhesion to nasopharyngeal epithelial cells and fitness in mice (6, 7), as do glutamine uptake systems (7, 8).
Another important amino acid, the metabolism of which is closely linked to that of glutamine, is arginine (see Fig. 1A). This amino acid is present in the human plasma at a concentration of around 45 μm (9), although in other body sites, different concentrations are found, as in muscle (>1000 μm (9)) and in cerebrospinal fluid ((13 μm (10)). At sites of inflammation, the concentration of free arginine might decrease to very low levels (11, 12), which could lead to decreased T- and B-cell activity (13). In the human host, arginine is also an important donor of NO, which is used by macrophages to kill invasive microbes (14).
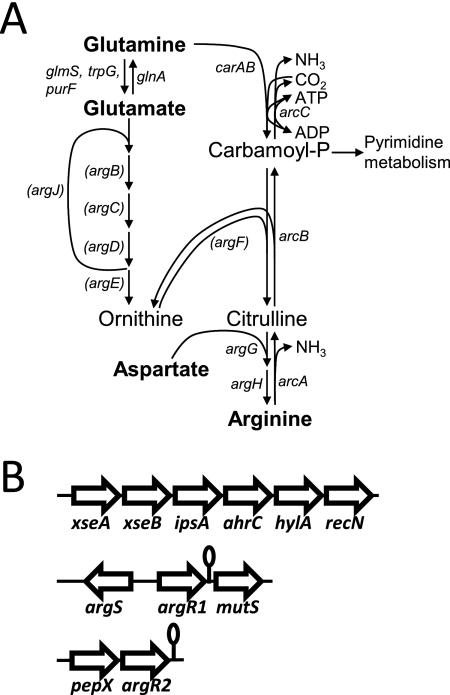
FIGURE 1.
Overview of genes (putatively) involved in arginine metabolism in S. pneumoniae. A, schematic representation of arginine metabolic pathways in S. pneumoniae D39. The arginine biosynthetic pathway encompasses the arg and car genes; the catabolic pathway consists of the arc genes. For completion, the full arginine biosynthesis pathway is depicted. However, genes in parentheses are not present in S. pneumoniae D39 and in the strains present in the Sybil database. Only the most important reaction products and substrates are indicated. Genes encode enzymes as follows: glnA, glutamine synthetase; glmS, glucosamine-fructose-6-phosphate aminotransferase; trpG, anthranilate synthase component II/glutamine amidotransferase; purF, amidophosphoribosyltransferase; argJ, ornithine acetyltransferase; argB, N-acetylglutamate 5-phosphotransferase; argC, N-acetylglutamate 5-semialdehyde dehydrogenase; argD, N2-acetylornithine 5-aminotransferase; argE, acetylornithine acetyltransferase; argF, anabolic ornithine carbamoyltransferase; argG, argininosuccinate synthetase; argH, argininosuccinase; carA/carB, carbamoylphosphate synthetase; arcA, arginine deiminase; arcB, catabolic ornithine carbamoyltransferase; arcC, carbamate kinase. B, genetic neighborhood of the three ArgR-type transcriptional regulators encoded by the D39 genome (not drawn to scale). Lollipops, predicted terminator structures. For further details, see the text.
Previous studies indicate that arginine metabolism and regulation have a link with the virulence of several pathogenic bacteria such as Mycobacterium tuberculosis, Listeria monocytogenes, Legionella pneumophila, and Mycobacterium bovis (15–19). In Streptococcus pyogenes, cells with a deletion in the arcA homologue, the first gene of the arginine catabolic pathway, show decreased eukaryotic cell invasion and have a reduced capacity to grow intracellularly compared with wild-type cells (20). In addition, arginine deiminase (ArcA) activity in S. pyogenes is associated with the inhibition of immune cell proliferation (21). In group A streptococci, growth in amniotic fluid affects the expression of many genes involved in arginine uptake, breakdown, and synthesis (22).
Studies on the regulation of arginine metabolism by ArgR-type regulators have been performed in among others Lactococcus lactis (23–25), Lactobacillus plantarum (26), Enterococcus faecalis (27), Bacillus subtilis (28, 29) and Escherichia coli (30), in which these regulators control transcription of arginine biosynthesis and catabolism. In Streptococcus suis, ArgR is a transcriptional activator of the arginine catabolic arc operon, and both are required for acid resistance (31). In addition, the S. suis arc operon is induced by arginine, reduced O2, temperature, and iron starvation and is subject to carbon catabolite repression (32–34). In Streptococcus gordonii, a similar way of regulation was reported (35, 36). In Streptococcus agalactiae, the expression of two transcriptional operons involved in arginine metabolism, the putative arginine transport genes artPQ (homologous to artPQ investigated in this study in S. pneumoniae D39) and arginine biosynthetic genes argGH, was down-regulated in a mutant of the LysR-type virulence regulator MtaR (37). In S. pyogenes, the transcriptional regulator of virulence factors Rgg also regulates arginine catabolism (38, 39).
Although organisms like B. subtilis and E. coli have only one ArgR-type regulator, several organisms contain two or even three paralogues (40). Also S. pneumoniae contains three ArgR-type regulators. Given the apparent paradox between the high number of ArgR-type regulators and the absence of a complete set of arginine biosynthetic genes (41), as well as the importance of arginine metabolism in other pathogenic bacteria, we were impelled to investigate the response of S. pneumoniae to fluctuating arginine levels by transcriptome analyses. In subsequent experiments, we found that two of three ArgR-type regulators present in this bacterium, namely ArgR1 and AhrC, directly mediate the response to arginine in a cooperative manner. Instead of being dedicated to the regulation of arginine biosynthesis and breakdown, as is the case in other bacteria, pneumococcal ArgR1 and AhrC control the expression of genes involved in arginine and peptide uptake (abpA, abpB, artPQ, aapA, and aliB), as well as the Zn2+-scavenger and virulence genes phtD and lmB/adcAII.
EXPERIMENTAL PROCEDURES
DNA Techniques, β-Galactosidase Assays, Bacterial Strains, and Growth Conditions
All DNA manipulation techniques, growth conditions, and media were the same as described previously (6, 42) unless indicated otherwise. β-Galactosidase assays were performed as described previously (6). Strains and plasmids used or constructed in this study are listed in supplemental Table S1. Primers are listed in supplemental Table S2.
Construction of Transcriptional lacZ Fusions
Ectopic lacZ fusions to the abpA, artP, aapA, abpB, aliB, and arcA promoters were made in pPP2 with primer pairs Pspd_0109_1/Pspd_0109_2, Pspd_0719_1/Pspd_0719_2, Pspd_0887_1/Pspd_0887_2, Pspd_1226_1/Pspd_1226_2, Pspd_1357_1/Pspd_1357_2, and ParcA_ccpA_mut-1/ParcA_ccpA_mut-2, respectively. Mutations in the above promoters were introduced by fusing PCR products generated with the following primers and cloning them in pPP2: Pspd_0109_1/Pspd_0109_mut2 + Pspd_0109_2.2/Pspd_0109_mut1, Pspd_0719_1/Pspd_0719_mut2 + Pspd_0719_2/Pspd_0719_mut2, Pspd_1226_1/Pspd_1226_mut2 + Pspd_1226_2.2/Pspd_1226_mut1, Pspd_1357_1/Pspd_1357_mut2 + Pspd_1357_2.2/Pspd_1357_mut2, and ParcA_ccpA_mut-1/ParcA_ccpA_mut-2 + ParcA_ccpA_mut-3/ParcA_ccpA_mut-4. These constructs are listed in supplemental Table S1 (pAS5–pAS14). In all cases, E. coli EC1000 was used as the cloning host. The lacZ fusion constructs were introduced into D39 wild type and the D39 ΔargR1, D39 ΔahrC, and D39 ΔargR1ΔahrC mutant strains by means of integration via double crossover in the bgaA (spd_0562) gene, yielding strains AS15–AS50. All plasmid constructs were checked by sequencing, and new loci created with these plasmids were verified by PCR.
Construction of Deletion Mutants
In-frame marker-free deletions of argR1, ahrC, abpA, and artP were constructed with plasmid pORI280 essentially as described (42) using primer pairs argR_KO-1/argR_KO-2 and argR_KO-3/argR_KO-4, ahrC_D39_KO1/ahrC_D39_KO2 and ahrC_D39_KO3/ahrC_D39_KO4, SPD_0109_KO1/SPD_0109_KO2 and SPD_0109_KO3/SPD_0109_KO4, and SPD_0719_KO1/SPD_0719_KO2 and SPD_0109_KO3/SPD_0109_KO4. Allelic replacement mutants of aapA, abpB, and aliB were made using primer pairs SPD_0887_KO1/SPD_0887_KO2 and SPD_0887_KO3/SPD_0887_KO4, SPD_1226_KO1/SPD_1226_KO2 and SPD_1226_KO3/SPD_1226_KO4, and SPD_1357_KO-1/SPD_1357_KO-2 and SPD_1357_KO-3/SPD_1357_KO-4 by overlap extension PCR (43). These mutants and combinations thereof are listed in supplemental Table S1 (strains AS1–AS14). Mutations were verified by PCR and/or DNA sequencing.
Microarray Analyses
Microarray analyses were done essentially as described previously (6, 44, 45). The transcriptome data for the ahrC and argR1ahrC mutants were obtained from hybridizations of two biological replicates to two slides containing three spots per gene. The transcriptome data for the argR1 mutant were obtained from hybridizations of three biological replicates to three slides containing two spots per gene. The transcriptome data for the high/low arginine comparison were obtained from hybridizations of two biological replicates to two slides containing two spots per gene. In all cases, genes were considered significantly differentially expressed when the Bayesian p value was <0.001 except for the high/low arginine comparison where the cutoff was set to 0.01 because of the lower number of spots per gene. See Table 1 for the detailed criteria applied for the ratio cutoff. Microarray data have been deposited to the Gene Expression Omnibus (GEO) and have accession number GSE33043.
TABLE 1.
Summary of DNA microarray analyses of D39 ΔargR1, D39 ΔahrC, and D39 ΔargR1ΔahrC compared with D39 wild type grown in CDM + 10 mm arginine and of D39 wild type grown in CDM + 0.05 mm arginine compared with 10 mm arginine
The table shows significantly differentially expressed genes (based on the Bayesian p values) with ratios (mutant/wild type, 0.05 mm Arg/10 mm Arg) ≤0.66 and ≥1.5 but only when at least in one of the four conditions the ratio was ≤0.5 or ≥2.0. In several cases, neighboring genes were also included. PTS, phosphotransferase.
| Locus tag | Annotation | Gene name | Low Arg | ΔargR1 | ΔahrC | ΔargR1 ΔahrC |
|---|---|---|---|---|---|---|
| spd_0053 | Amidophosphoribosyltransferase | purF | 0.5 | |||
| spd_0054 | Phosphoribosylaminoimidazole synthetase | purM | 1.5 | 0.4 | ||
| spd_0055 | Phosphoribosylglycinamide formyltransferase | purN | 0.4 | |||
| spd_0056 | VanZ protein, putative | vanZ | 0.4 | |||
| spd_0057 | Bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | purH | 0.3 | |||
| spd_0109 | Amino acid ABC transporter, periplasmic amino acid-binding protein, putative | apbA | 10.1 | 38.6 | 21.5 | 10.8 |
| spd_0110 | Argininosuccinate synthase | argG | 6.8 | 29.9 | 9.6 | 5.1 |
| spd_0111 | Argininosuccinate lyase | argH | 6.6 | 19.6 | 4.6 | 2.8 |
| spd_0114 | Hypothetical protein SPD_0114 | 0.2 | 0.3 | 0.4 | ||
| spd_0115 | Hypothetical protein SPD_0115 | 0.2 | 0.3 | 0.3 | ||
| spd_0116 | Hypothetical protein SPD_0116 | 0.2 | 0.3 | 0.5 | ||
| spd_0117 | Hypothetical protein SPD_0117 | 0.3 | 0.6 | |||
| spd_0118 | Hypothetical protein SPD_0118 | 0.3 | 0.5 | 0.3 | ||
| spd_0119 | Hypothetical protein SPD_0119 | 0.2 | 0.4 | 0.2 | ||
| spd_0121 | Hypothetical protein SPD_0121 | 0.2 | 0.3 | 0.2 | ||
| spd_0122 | Hypothetical protein SPD_0122 | 0.2 | 0.3 | 0.3 | ||
| spd_0123 | Hypothetical protein SPD_0123 | 0.2 | 0.3 | 0.3 | ||
| spd_0124 | Hypothetical protein SPD_0124 | 0.2 | 0.5 | 0.3 | ||
| spd_0125 | Hypothetical protein SPD_0125 | 0.5 | 1.8 | |||
| spd_0161 | Hypothetical protein SPD_0161 | 0.4 | ||||
| spd_0228 | Transcriptional regulator, AraC family protein | 0.8 | 0.5 | 0.6 | ||
| spd_0311 | Glucan 1,6-α-glucosidase | dexB | 2.0 | |||
| spd_0313 | Hypothetical protein SPD_0313 | 0.4 | 0.5 | 0.6 | ||
| spd_0405 | Acetolactate synthase 3 regulatory subunit | ilvH | 0.6 | 0.5 | ||
| spd_0406 | Ketol-acid reductoisomerase | ilvC | 0.7 | |||
| spd_0407 | Hypothetical protein SPD_0407 | 0.4 | 0.4 | |||
| spd_0408 | Hypothetical protein SPD_0408 | 0.4 | 0.3 | |||
| spd_0409 | Threonine dehydratase | ilvA | 0.5 | 0.5 | ||
| spd_0450 | Type I restriction-modification system, S subunit, putative | 0.2 | ||||
| spd_0451 | Type I restriction-modification system, S subunit, putative | 0.2 | ||||
| spd_0452 | Integrase/recombinase, phage integrase family protein | 3.9 | ||||
| spd_0473 | Immunity protein BlpY | blpY | 0.6 | 4.7 | 1.5 | |
| spd_0559 | PTS system, IIA component, putative | 2.3 | ||||
| spd_0560 | PTS system, IIB component, putative | 2.6 | ||||
| spd_0561 | PTS system, IIC component, putative | 2.6 | ||||
| spd_0562 | β-Galactosidase precursor, putative | bgaA | 2.5 | |||
| spd_0610 | Hypothetical protein SPD_0610 | 0.4 | ||||
| spd_0611 | Hypothetical protein SPD_0611 | 0.5 | ||||
| spd_0719 | Amino acid ABC transporter, permease protein | artP | 1.9 | 2.1 | 1.9 | 1.5 |
| spd_0720 | Amino acid ABC transporter, ATP-binding protein | artQ | 1.9 | 1.8 | 1.7 | 1.4 |
| spd_0721 | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase | foldD | 1.5 | 1.8 | 1.5 | 1.5 |
| spd_0781 | Hypothetical protein SPD_0781 | 0.5 | ||||
| spd_0852 | Dihydroorotate dehydrogenase 1B | pyrD | 0.5 | |||
| spd_0887 | Amino acid permease family protein | aapA | 1.5 | 1.7 | 1.7 | 1.5 |
| spd_0888 | Adhesion lipoprotein | lmB/adcAII | 2.0 | 1.7 | 1.5 | 1.8 |
| spd_0889 | Pneumococcal histidine triad protein D precursor | phtD | 2.1 | 1.7 | 1.7 | 2.0 |
| spd_1063 | Transcriptional regulator of arginine metabolism expression, putative | ahrC | 0.4 | 0.3 | ||
| spd_1225 | Hypothetical protein SPD_1225 | 2.2 | 3.2 | 3.4 | 2.6 | |
| spd_1226 | Amino acid ABC transporter, amino acid-binding protein | abpB | 2.3 | 3.7 | 4.1 | 3.4 |
| spd_1356 | ABC transporter, ATP-binding protein authentic frameshift | 2.3 | 3.5 | 3.2 | 3.0 | |
| spd_1357 | Oligopeptide ABC transporter, oligopeptide-binding protein AliB | aliB | 2.9 | 4.1 | 2.3 | 2.1 |
| spd_1515 | Hypothetical protein SPD_1515 | 0.5 | 0.6 | 0.6 | ||
| spd_1516 | Hypothetical protein SPD_1516 | 0.5 | 0.6 | 0.6 | ||
| spd_1517 | Hypothetical protein SPD_1517 | 0.5 | 0.5 | 0.3 | ||
| spd_1634 | Galactokinase | galK | 2 | |||
| spd_1731 | Hypothetical protein SPD_1731 | 0.5 | ||||
| spd_1903 | DNA mismatch repair protein HexA, authentic frameshift | 3.0 | 2.6 | |||
| spd_1904 | Arginine repressor | argR1 | 0.1 | 0.2 | ||
| spd_1911 | Phosphate ABC transporter, permease protein PstC | pstC | 1.7 | |||
| spd_1912 | Phosphate ABC transporter, permease protein PstA | pstA | 1.7 | |||
| spd_1913 | Phosphate ABC transporter, ATP-binding protein | 2.0 | ||||
| spr0112 | Hypothetical protein | 0.3 | 0.6 | 0.3 | ||
| spr0116 | Hypothetical protein | 0.2 | 0.3 | 0.2 | ||
| spr0470 | Hypothetical protein | |||||
| spr1134 | Hypothetical protein | 2.1 | ||||
| sp_0594 | Hypothetical protein, fusion | 0.4 |
Overexpression and Purification of Strep-tagged ArgR1 and AhrC
For the overexpression of N-terminally Strep-tagged variants of ArgR1 and AhrC, their respective genes were amplified from D39 chromosomal DNA using primers ArgR1_OX_1_strep/ArgR1_OX_2 and AhrC_OX_1_strep/AhrC_OX_2. The resulting PCR products were digested with RcaI/XbaI and cloned into the NcoI/XbaI sites of pNG8048E, yielding plasmids pAS15 and pAS16. Overexpression in L. lactis NZ9000 was done essentially as described (46). Purification of Strep-ArgR1 and Strep-AhrC from L. lactis was performed using the Strep-Tactin column from IBA according to the supplier's instructions. The purified proteins were stored at a concentration of around 0.1 mg/ml in the elution buffer (100 mm Tris-HCl, pH 8, 150 mm NaCl, 2.5 mm desthiobiotin, 1 mm β-mercaptoethanol, and 1 mm EDTA) with 10% glycerol at −80 °C.
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays (EMSAs) were performed with [γ-33P]ATP-labeled probes in buffer containing 20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 0.1 mm dithiothreitol (DTT), 8.7% (w/v) glycerol, 62.5 mm KCl, 1 mm EDTA, 25 μg/ml bovine serum albumin, 25 μg/ml poly(dI-dC), and 3000 cpm [γ-33P]ATP-labeled PCR product. As probes, the promoter regions of PabpA, PartP, PaapA, PabpB, and PaliB were used that were PCR-amplified from plasmids pAS5–pAS9. In addition, PabpB-mut and PaliB-mut were used that were PCR-amplified from chromosomal DNA from strains A28 and A32 using primer pairs Pspd_1226_1/RNlacZ-fw and Pspd_1357_1/RNlacZ-fw. As a negative control, a PCR fragment of Pspd_1049 amplified from D39 chromosomal DNA with primers Pspd_1049−1/Pspd_1049-2 was used. Reactions were incubated at 37 °C for 10 min before loading on gel. Gels were run in 0.44 m Tris borate buffer, pH 8.3 at 100 V for 90 min.
DNase I footprints were carried our essentially as described before (47, 48). As probes, PCR products of PabpA and PabpB were used that were generated with plasmids pAS5 and pAS8 as a template, respectively. In both cases, the forward primer was labeled with [γ-33P]ATP.
RESULTS
In Silico Analysis of Genes Involved in Arginine Metabolism in S. pneumoniae
In contrast to many organisms like L. lactis, L. plantarum, and B. subtilis, which contain a functional biosynthesis pathway for arginine (26, 49–51), the analysis of the genome sequence of S. pneumoniae D39 showed that it contains only two “arg” genes that are putatively involved in arginine biosynthesis, i.e. argG and argH encoding the enzymes for the conversion of citrulline (and aspartate) to arginine (Fig. 1A, overview of arginine metabolic pathways in S. pneumoniae). S. pneumoniae also possesses carA and carB encoding carbamoylphosphate synthetase that catalyzes the production of carbamoyl phosphate from glutamine, which is an intermediate in both arginine and pyrimidine synthesis. However, the anabolic argF gene that catalyzes citrulline production from carbamoyl phosphate is not encoded within the D39 pneumococcal genome, like the argBCDE genes, which produce ornithine, the other substrate of ArgF, out of glutamate.
In addition, strain D39 contains an arc operon (spd_1975–77) encoding the putative arginine catabolic genes arcA, arcB, and arcC (Fig. 1A), although there is an authentic frameshift in the arcA gene giving rise to two 744- and 450-bp ORFs instead of the single full-length 1230-bp arcA gene as is present in strain TIGR4 for example (41). In TIGR4 as well as in >60% of the 33 sequences of pneumococcal strains present in the Sybil database, argGH are not (intactly) present. In contrast, the arc genes are encoded by almost all pneumococcal strains in Sybil except BS293 and BS397. This indicates that there are strain to strain variations in arginine metabolism.
In the D39 genome, three regulators can be found that have high homology to the ArgR and AhrC regulators of arginine biosynthesis and catabolism of L. lactis (23). SPD_1904 and SPD_0787 have the highest homology to ArgR from L. lactis (the homology with SPD_1904 is slightly higher) and are therefore named ArgR1 and ArgR2, respectively. SPD_1063 has the highest homology to AhrC of L. lactis and is therefore named AhrC. All three regulators are conserved in all 34 S. pneumoniae strains present in the Sybil database, although in a few strains, orthologues of spd_0787 seem truncated at the 5′ part (data not shown). In addition, the genomic context is always the same (Fig. 1B) with spd_0787 (argR2) lying downstream and in tandem with a pepX gene (X-prolyl-dipeptidyl aminopeptidase), spd_1904 (argR1) being flanked by argS (arginyl-tRNA synthetase), and mutS (DNA mismatch repair protein) and spd_1063 (ahrC) being surrounded by genes encoding DNA recombination and repair protein RecN, hemolysin A, geranyltranstransferase IspA, and the exonuclease VII XseAB. Interestingly, the genetic neighborhood of ahrC is also highly conserved in other species such as Bacillus sp., Streptococcus sp., and other gram-positive bacteria, suggesting a functional relationship (40). The conservation of these ArgR-type regulatory genes in all pneumococcal strains indicates that they fulfill important roles in S. pneumoniae.
S. pneumoniae D39 Is Auxotrophic for Arginine
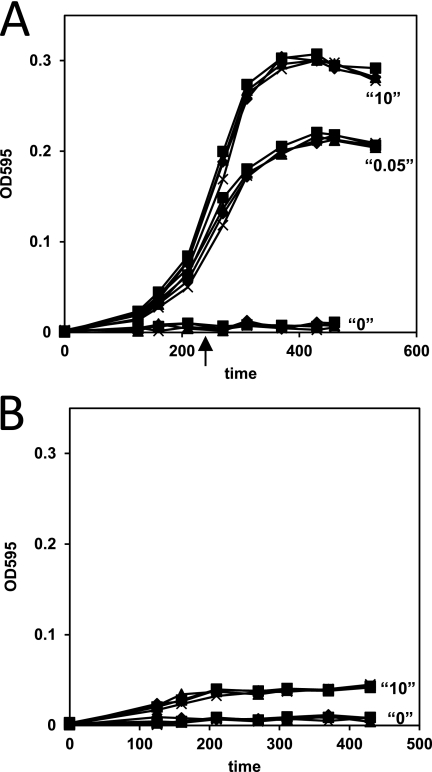
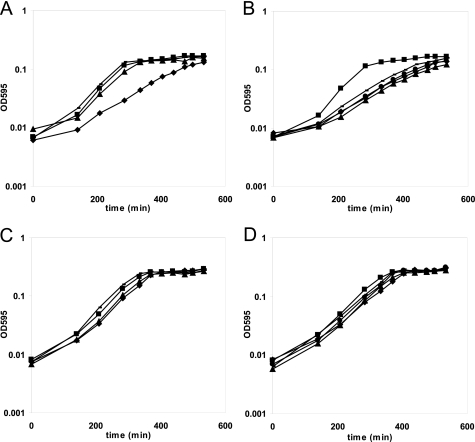
Based on the in silico analysis, it is likely that D39 is auxotrophic for arginine. To test this hypothesis, D39 was grown in the presence of 0, 0.05, and 10 mm arginine (Fig. 2A). As expected, no growth was seen in the absence of arginine (Fig. 2A), confirming previous observations (52). With 0.05 mm arginine, a concentration present in the human blood (9), growth was still slower than with 10 mm, indicating that this is an arginine-limited condition. Regarding the presence of ArgG and ArgH in D39, growth was tested in medium with citrulline, which is a substance that is present in the human tissues and in a concentration similar to that of arginine (9, 53). In the absence of arginine, S. pneumoniae D39 was indeed able to grow on citrulline (Fig. 2B), indicating the functionality of the ArgGH enzymes. However, growth was very poor, suggesting that for optimal growth other ways of arginine acquisition from external sources have to be utilized by S. pneumoniae as well.
FIGURE 2.
Growth of D39 wild type (■), D39 ΔargR1 (▴), D39 ΔahrC (♦), and D39 ΔargR1ΔahrC (×) in CDM medium with 0, 0.05, and 10 mm arginine and no citrulline (A) and in CDM with 0 or 10 mm citrulline and no arginine (B). Aspartate, which is together with citrulline a substrate for ArgG, was present in a concentration of 0.9 mm in the CDM. However, S. pneumoniae D39 is not auxotrophic for aspartate (52). The arrow in A indicates the point of harvesting of the cells for the DNA microarray experiments.
Identification of Genes Regulated by Arginine Concentration
Because (i) arginine is an essential amino acid for S. pneumoniae, (ii) there are three putative ArgR-type regulators encoded by the pneumococcal genome, and (iii) arginine concentrations in the habitat of S. pneumoniae may fluctuate, we hypothesized that arginine is an important regulatory cue for this bacterial pathogen. To investigate the response of D39 to arginine, we performed a transcriptome comparison of S. pneumoniae D39 wild type grown in low arginine chemically defined medium (CDM)3 compared with high arginine CDM. D39 wild type grew slightly slower with a concentration of 0.05 mm arginine as compared with 10 mm arginine and to a lower optical density (Fig. 2A). Therefore, cells were harvested in a comparable stage of growth, i.e. in the midexponential phase (Fig. 2A). 13 genes were up-regulated in arginine-limited growth conditions (Table 1). Interestingly, these included several operons encoding amino acid ABC transporter components to which we gave the tentative names ArtPQ (based on homology to S. pyogenes and E. coli ArtPQ), AbpA (arginine-binding protein A) and the paralogous AbpB, and AapA (amino acid permease A). All of these proteins are conserved in the 33 strains in the Sybil database except AbpA, which seems absent from ∼10% of these strains. Furthermore, the aliB gene encoding an oligopeptide-binding protein that contributes to the uptake of arginine-containing peptides (54) and the arginine biosynthetic genes argGH were up-regulated as well. Five genes were down-regulated (Table 1) among which was pyrD (spd_0852) that is involved in pyrimidine synthesis, which is closely connected to arginine metabolism.
Identification of ArgR1/AhrC Targets by DNA Microarrays
As in L. lactis, the ArgR and AhrC proteins are responsible for the transcriptional regulation in response to arginine (25). Chromosomal deletions were constructed of both argR1 and ahrC in S. pneumoniae D39. To avoid the possibility of polar effects, this was done using a marker-free system (42). Subsequently, these mutants were used to determine the effect of ArgR1 and AhrC on the pneumococcal transcriptome as well as their role in mediating the transcriptional response to arginine as investigated above. The argR1 and ahrC deletion strains grew similarly as the wild-type D39 strain in both high arginine CDM and low arginine CDM (Fig. 2, A and B). Because in L. lactis the effect of deletion of argR and ahrC was most pronounced in medium with a high arginine concentration (25), we compared the transcriptome of D39 wild type with that of the isogenic argR1 and ahrC mutants in CDM with 10 mm arginine (Table 1). Many genes were differentially expressed in both mutants compared with the wild type; the majority of were also affected by varying arginine concentrations. These include the abpA-argGH operon, two additional operons that encode amino acid ABC transporter components (artPQ-folD and apbB), an operon containing an amino acid permease gene (aapA) together with the Zn2+-scavenging genes lmb/adcAII (55–57) and phtD (58–60), and lastly the gene encoding oligopeptide-binding lipoprotein, aliB. Also in the argR1ahrC double mutant these five operons were up-regulated compared with the wild type (Table 1). Additionally, some other effects were observed in the argR1ahrC mutant as well, such as down-regulation of the ilv genes (spd_0405–9) compared with the wild type; this was also the case in the ahrC mutant. There was down-regulation of the spd_0114–0124 and spd_1515–17 hypothetical genes in all three mutants compared with the wild type, but expression of these genes was not affected by arginine (Table 1).
Thus, the microarray analyses indicate that ArgR1 and AhrC affect the expression of a number of genes among which are five operons (abpA-argGH, artPQ-folD, lmB/adcAII-phtD, apbB, and aliB) containing genes putatively involved in arginine metabolism and uptake. These five operons were also up-regulated in medium with limiting arginine. Quantitative RT-PCRs for the first gene in each of these five operons confirmed this expression behavior (supplemental Table S3). We next investigated the regulation of these operons in more detail.
Regulation by ArgR1 and AhrC Is Arginine-dependent
Based on sequence analysis, abpA and aliB seem to be monocistronic transcriptional units, although for abpA, based on the microarray results, there is apparently read-through to the downstream gene spd_1225 (Table 1), which is oppositely orientated; however, the promoter of this gene was not regulated by ArgR1 or AhrC (data not shown). Also for aliB there is downstream read-through, as the frameshifted gene spd_1356 was affected in the same way (Table 1). Based on the DNA microarray data and sequence analyses for promoters and terminators, PabpA, PartP, and PaapA all appear to drive expression of three downstream genes. To investigate in more detail the regulation of these operons, ectopic transcriptional lacZ fusions to the predicted promoter regions of abpA, artP, aapA, abpB, and aliB (see Fig. 3 for sequence analyses of these promoters) were introduced in D39 wild type and the argR1, ahrC, and arg1RahrC isogenic mutants. In the wild type, expression of the five promoters was higher in CDM with a low arginine concentration than in CDM with a high arginine concentration and also higher than in the nitrogen-rich complex medium GM17 (Fig. 4A and supplemental Table S4). This shows that the promoters predicted and cloned are functional and confirms the microarray data on the transcriptional response to arginine. In the arginine regulator mutants, expression was highly derepressed compared with the wild type and not dependent on the concentration of arginine in the medium (Fig. 4A and supplemental Table S4). Thus, ArgR1 and AhrC control the expression from the predicted promoters of these five operons in an arginine-dependent way. In addition, the data show that both regulators are required for the arginine-dependent regulation in agreement with the microarray data on these mutants.
FIGURE 3.
Nucleotide sequences of promoter regions as indicated in figure. Putative −35/−10 sequences are in bold. Translational starts are in italic. Predicted ArgR operators are underlined. Bases that were mutated as described in the text are indicted with asterisks above the sequence. Regions in PabpA and PabpB where binding was detected in the DNase I footprint analysis are in gray shading.
FIGURE 4.
A, specific β-galactosidase activity of D39 WT (wt), D39 ΔargR1 (R), D39 ΔahrC (C), and D39 ΔargR1ΔahrC (RC) containing PabpA-lacZ (abpA), PartP-lacZ (artP), PaapA-lacZ (aapA), PabpB-lacZ (abpB), and PaliB-lacZ (aliB) transcriptional fusions. Cells were grown in GM17 medium (black bars) and in CDM with 10 (gray bars) or 0.025 (white bars) mm arginine and harvested at the midexponential phase of growth. Data are the averages of at least three measurements. p values were <0.005 for the comparisons between 10 and 0.025 mm arginine in the wild type for each of the lacZ fusions as calculated with the Student's t test. B, specific β-galactosidase activity of D39 WT (wt) and D39 ΔargR1 (R) containing PabpA-mut-lacZ (abpA-mut), PartP-mut-lacZ (artP-mut), PabpB-mut-lacZ (abpB-mut), and PaliB-mut-lacZ (aliB-mut) transcriptional fusions. Cells were grown in CDM with 10 (gray bars) or 0.025 (white bars) mm arginine and harvested at the midexponential phase of growth. Data are the averages of at least three measurements, and error bars represent the standard deviation.
Interaction of ArgR1/AhrC with Their Target Promoters Is Dependent on Arginine
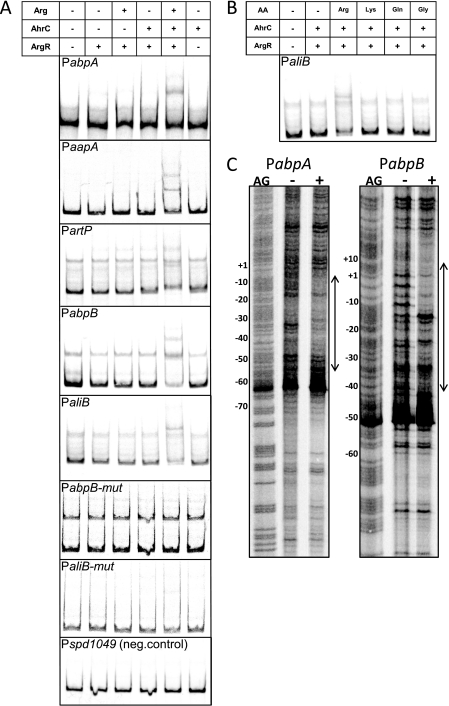
To investigate the mechanism by which ArgR1 and AhrC regulate the expression of the five promoters, EMSAs were performed with ArgR1 and AhrC purified with an N-terminal Strep tag. A high concentration of ArgR1 resulted in binding to all promoters irrespective of the presence of AhrC or arginine, whereas AhrC on its own did not bind any of the tested promoters (data not shown). With a lower concentration of ArgR1, binding only occurred when both AhrC and arginine were present (Fig. 5A). No binding was seen for the negative control (Fig. 5A). Moreover, from several amino acids tested, only arginine was able to induce binding (Fig. 5B). These data show that the ArgR1/AhrC-dependent regulation occurs via direct binding of these regulators to their target promoters. Furthermore, both regulators are required for binding whereby ArgR1 seems to be the main factor in mediating the interaction with the promoter. Lastly, binding is dependent specifically on arginine.
FIGURE 5.
In vitro interaction of ArgR1/AhrC with their target promoters. A and B, binding of N-terminally Strep-tagged ArgR1/AhrC to the indicated promoters as analyzed with EMSAs. Strep-ArgR1 was used in a concentration of 20 nm, and Strep-AhrC was used in a concentration of 50 nm. Amino acids were added in a concentration of 10 mm. The table above the gel pictures indicates which components were added (ArgR, Strep-ArgR1; AhrC, Strep-AhrC; AA, amino acid; Arg, arginine; Lys, lysine; Gln, glutamine; and Gly, glycine). Unshifted bands that migrated slower than the probe probably represent single-stranded DNA due to slightly unequal concentrations of primers used to PCR the probes or due to the high AT content of the DNA (47, 87). C, DNase I footprinting analyses of the binding of Strep-ArgR1 and Strep-AhrC to PabpA and PabpB. Protein was either not added (−) or added in a concentration of 80 nm for Strep-ArgR1 and 200 nm for Strep-AhrC (+). All reactions contained 10 mm arginine. Numbers on the left of the figures indicate the base pair positions relative to the translational starts. AG, Maxam-Gilbert A+G sequence ladder. Regions of protection are also indicated in Fig. 3. neg., negative.
Identification of Binding Site for ArgR1/AhrC
When the E. coli consensus ArgR box (30) (5′-TNTGNATWWWWATNCANA-3′) was used to search the D39 genome using Genome2D (61) allowing one mismatch, 18 hits were scored, but only one was in the promoter of a gene present in the ArgR1/AhrC microarray analyses in this study, namely in PartP (Fig. 3). When the D39 genome was searched in silico with a weight matrix of the L. lactis ArgR box (24) only two of a list of 18 hits were present in promoters of ArgR1/AhrC targets identified in this study, namely in PaliB and PabpB (Fig. 3). By close inspection of PabpA, a similar sequence was found that could function as an Arg box for this promoter (Fig. 3).
To prove that the predicted operators mediate the repression by ArgR/AhrC, point mutations were made in all four (Fig. 3), and the mutant promoters were fused to lacZ. In wild-type D39 grown in high arginine CDM, derepression of expression of the mutated abpB and aliB promoters to a level similar to that seen in the argR1 mutant was observed compared with the wild-type promoters (Fig. 4B). For PabpA-mut and PartP-mut, derepression was very clear as compared with the wild-type promoters albeit not fully to the level of the argR1 mutant, showing that regulation via ArgR1/AhrC was not completely abolished (Fig. 4B). In the wild-type background, the expression from all four mutated promoters was hardly influenced anymore by the concentration of arginine (Fig. 4B). When PCR fragments comprising PabpB-mut and PaliB-mut, which were both completely derepressed to the level of expression in the argR1 mutant (Fig. 4B), were used in EMSAs, no binding of ArgR1/AhrC in the presence of arginine was observed (Fig. 5A). In addition, DNase I footprinting analyses with PabpA and PabpB showed that ArgR1/AhrC bind to a DNA region that comprises the entire predicted operator sequence as well as the sequence downstream of it (Figs. 5C and 3). Taken together, these results show that the positions of the ArgR1/AhrC operators in PabpA, PartP, PabpB, and PaliB are indeed at the locations that were predicted and that the operators mediate interaction of ArgR1/AhrC with the target promoters.
ArgR1/AhrC Do Not Regulate arcABC Operon
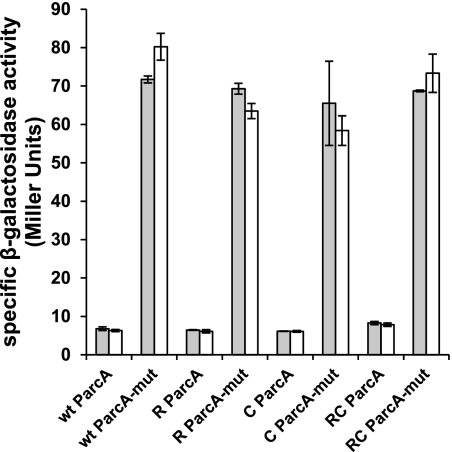
In L. lactis and many other organisms, the arginine catabolic genes are regulated by ArgR (23, 29, 31, 62). In this study, however, no effect of ArgR1 and AhrC was observed on the arginine catabolic arcABC operon of S. pneumoniae. In many gram-positive bacteria, the arc operon is subject to carbon catabolite control (27, 36, 63). By screening the S. pneumoniae D39 genome (64), a catabolite recognition element (cre) was found in the promoter of arcA (5′-TGTAAGCGGTACCC-3′). cre sites are recognized by the carbon catabolite control protein A CcpA (65, 66). To test whether the cre site in ParcA is functional and could mask a possible role of ArgR1/AhrC in S. pneumoniae, it was mutated to 5′-TGTAATTTGTACCC-3′. Subsequently, both the wild-type ParcA and the mutant ParcA (ParcA-mut) were fused to lacZ and introduced into the regulator mutants. Expression of the mutated promoter was indeed highly derepressed compared with the wild type, confirming the functionality of the predicted cre site (Fig. 6). However, expression of both versions of ParcA was not affected by the argR1/ahrC mutations, showing that ArgR1/AhrC have no role in regulating this promoter.
FIGURE 6.
Specific β-galactosidase activity of D39 WT (wt), D39 ΔargR1 (R), D39 ΔahrC (C), and D39 ΔargR1ΔahrC (RC) containing ParcA-lacZ (ParcA) or ParcA-mut transcriptional fusions. Cells were grown in CDM with 10 (gray bars) or 0.025 (white bars) mm arginine and harvested at the midexponential phase of growth. Data are the averages of at least three measurements, and error bars represent the standard deviation.
Deletion of artP, abpA, and abpB Leads to Impaired Growth in Low Arginine Medium
All five operons that were shown to be direct targets of ArgR/AhrC contain genes putatively involved in amino acid uptake. To test whether these genes contribute to arginine acquisition, mutants in these genes were constructed and grown in CDM with either a low or a high concentration of arginine. Single mutants of abpA and abpB encoding putative amino acid-binding proteins did not display decreased growth in low arginine CDM (Fig. 7A). Marker-free deletion of artP encoding a putative amino acid ABC transporter permease component led to a growth defect only in CDM with 0.05 mm arginine and not in 10 mm arginine (Fig. 7, A and C). Double and triple mutants displayed the impaired growth behavior as well (Fig. 7, B and D). These results suggest that these three genes (abpA, artP, and abpB) constitute an arginine uptake ABC transporter in which the arginine binding domains can substitute for each other. The data also indicate that S. pneumoniae D39 contains other systems for arginine uptake as the observed growth reduction was modest. A candidate gene involved in arginine uptake is the ArgR1/AhrC target aapA encoding an amino acid permease family protein with homology to RocE and RocC in B. subtilis, two proposed arginine permease genes (67, 68). In Mycobacterium bovis, a protein homologous to RocE and RocC was shown to mediate arginine and γ-aminobutyric acid (GABA) uptake (69). However, mutation of the aapA gene did not lead to decreased growth compared with the wild type in medium containing a low concentration of arginine (data not shown). Also, it did not aggravate the phenotype of the artP mutant in CDM with 0.05 mm arginine (data not shown). Although aliB has been shown to be involved in the utilization by S. pneumoniae of arginine-containing oligopeptides (54), it could be that it has some affinity for arginine in amino acid form as well. However, like the aapA mutant, mutation of aliB did not lead to a phenotype in medium with a low concentration of arginine (data not shown). Therefore, aliB seems not to be involved in the acquisition of arginine in amino acid form.
FIGURE 7.
Mutants in ArgR1/AhrC targets affect growth in low arginine CDM. D39 wild type (■), D39 ΔabpA (—), D39 ΔartP (♦), and D39 ΔabpB (▴) were grown in CDM containing either 0.05 mm arginine (A) or 10 mm arginine (B). Similarly, D39 wild type (■), D39 ΔabpAΔartP (—), D39 ΔabpAΔabpB (●), D39 ΔartPΔabpB (▴), and D39 ΔabpAΔartPΔabpB (♦) were grown in CDM containing 0.05 mm arginine (C) or 10 mm arginine (D). Graphs are representative for the phenotypes of the mutants.
DISCUSSION
In this study, the role of two ArgR-type regulators, ArgR1 and AhrC, was studied in the human pathogen S. pneumoniae strain D39. These regulatory proteins were demonstrated to directly regulate five amino acid transport operons in a cooperative way (see Fig. 8 for a schematic overview of the function of ArgR1 and AhrC). The data suggest that the abpA, abpB, and artPQ genes located in three of the regulated operons encode an arginine ABC uptake unit. Furthermore, the aliB gene, which is directly controlled by ArgR1/AhrC as well, has been shown previously to be necessary for growth in medium with peptides (in particular Arg-Pro-Pro and Arg-Pro-Pro-Gly-Phe) as the sole source of arginine (54). Lastly, ArgR1 and AhrC direct expression of a promoter that drives transcription of the amino acid permease gene aapA and the gene pair lmB/adcAII-phtD encoding a Zn2+-binding surface lipoprotein (70) and the surface-exposed Zn2+/Mn2+-binding histidine triad protein PhtD (58), a highly conserved protein among pneumococcal strains and a promising vaccine candidate (60). ArgR1 and AhrC as well as most of their target genes are highly conserved among different pneumococcal strains, implicating an important role in the lifestyle of S. pneumoniae.
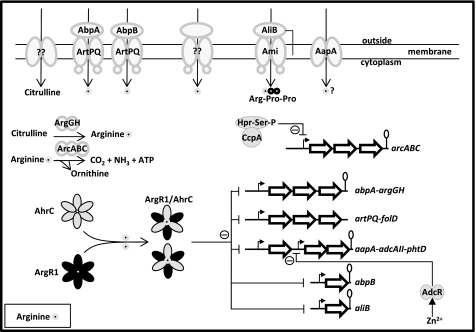
FIGURE 8.
Schematic overview of (regulation of) arginine metabolism in S. pneumoniae D39 based on results in this study and previous studies. In conditions with abundant arginine, ArgR1 and AhrC form a heterohexameric complex bound to the effector molecule arginine. This complex binds to the promoters of the five target operons, thereby blocking transcription of the downstream genes. In conditions of arginine limitation, repression of these operons is relieved, leading to increased arginine uptake via ArtPQ-AbpA/AbpB and Ami/AliB (arginine-containing peptides such as Arg-Pro-Pro). In addition, uptake of arginine proceeds via an unknown import route. When citrulline is available, intracellular arginine levels are also increased by the action of the ArgG and ArgH enzymes, which catalyze the conversion of citrulline to arginine via argininosuccinate. When preferred carbon sources are scarce, CcpA stops repressing the arginine deiminase operon arcABC, allowing arginine to be used as an alternative energy source. In addition, both low arginine conditions and low Zn2+ conditions lead to increased expression of the genes encoding the Zn2+ scavengers AdcAII and PhtD due to derepression of PaapA via ArgR1/AhrC and derepression of PadcAII via the Zn2+-dependent transcriptional regulator AdcR. A citrulline uptake system, an alternative arginine uptake system, and a tentative role of the AapA cationic amino acid permease in arginine uptake are indicated/depicted.
This is the first study on arginine-mediated gene regulation in S. pneumoniae. In recent years, the functioning of the L. lactis arginine-regulatory proteins was unraveled (23–25). In this bacterium, three operons, namely the argCJDBF, gltS-argE, and argGH arginine biosynthetic operons, were repressed by a high concentration of arginine via ArgR/AhrC, whereas the arcABD1C1C2TD2-yvaD operon was activated by ArgR/AhrC in the presence of arginine (23, 24). Also in other organisms like E. coli (30), Pseudomonas aeruginosa (62, 71), B. subtilis (28, 29), L. plantarum (26), and E. faecalis (27), arginine biosynthetic and catabolic genes have been shown to be regulated by ArgR-type regulators. Except for the argGH genes, which we showed to belong to the ArgR1/AhrC regulon in S. pneumoniae, the pneumococcal genome does not contain arginine biosynthetic genes. Instead, we found that ArgR1/AhrC are mainly dedicated to the regulation of arginine acquisition in this bacterium. Notably, although the arc operon is a target of ArgR in many organisms (16, 23, 27, 31, 35, 62), in our study, no effect of arginine and ArgR1/AhrC on expression of the arcA promoter was observed. However, the results suggest an involvement of CcpA. This is different from other streptococcal species such as S. suis (31), Streptococcus rattus, (63), and S. gordonii (35, 36) in which expression of the arc operon is (besides its regulation via CcpA) also arginine/ArgR-dependent. A recent study in S. suis found an effect of one of the three ArgR-type regulators present in this organism exclusively on the expression of the arc operon (31). The reason no transcriptional effects similar to that in our study were observed might be that the S. suis ArgR orthologue subject of that study is most homologous to ArgR2 in S. pneumoniae. However, although so far we have not identified a phenotype for the argR2 mutant, it does not seem to be involved in the regulation of ParcA in S. pneumonia D39,4 indicating that the ArgR-type regulators have different specific functions in different streptococci.
In L. lactis, it was proposed that ArgR and AhrC form a heterohexameric complex in the presence of arginine that has high affinity for the promoters of the arginine biosynthetic genes. At the same time, AhrC sequesters ArgR, preventing it from repressing the promoter of the arginine catabolic arc operon. In this model, AhrC seems to be more important for arginine sensing than ArgR; this is supported by the functional significance of Asp-124 (which is not present in ArgR) in AhrC. This Asp residue is conserved in the pneumococcal AhrC as well but not in ArgR1. Thus, the results in our study indicate that despite the low similarity between the ArgR regulons of L. lactis and S. pneumoniae (the physiological function is arginine biosynthesis and breakdown in L. lactis but mainly arginine acquisition/uptake in S. pneumoniae) the way in which ArgR1/AhrC control their target genes strongly resembles that of the counterparts in L. lactis.
Obviously, an intriguing question is why two regulatory proteins are necessary for arginine-dependent regulation. The first explanation could be that besides reacting to arginine ArgR1 and AhrC respond to other stimuli as well. Second, they could interact with other regulators as has been shown for AhrC in B. subtilis (72) and ArgR in P. aeruginosa (73). Third, the expression and synthesis of ArgR1 and AhrC could also be an important control point in the formation of a functional complex for regulation of the target genes whereby that of ArgR1 could be regulated in a different way than that of AhrC. Indeed, recent studies in B. subtilis found that translation of ahrC mRNA is inhibited by base-pairing with the small RNA SR1, the expression of which is dependent on arginine and ornithine (74, 75). Lastly, given the fact that ahrC is located in a locus with DNA modification genes encoding the DNA recombination and repair protein RecN (76) and the exonuclease VII XseAB, a functional interaction with these genes might be possible. Interestingly, the S. pneumoniae argR1 gene is located upstream of hexA encoding a DNA mismatch repair protein (77). In E. coli, ArgR has a secondary function in recombination at the cer locus during monomerization of ColE1 plasmids (78). Therefore, it is tempting to speculate that the ArgR-type regulators have a role related to DNA modification in S. pneumoniae and other bacteria as well (40). Interestingly, a number of hypothetical genes were down-regulated in the argR1 and ahrC mutants. Expression of these genes was not influenced by the concentration of arginine. However, the consistency of these effects in the argR1, ahrC, and argR1ahrC mutants also argues for a role of ArgR1 and AhrC that lies outside arginine-dependent control of gene expression.
The operons shown to be directly repressed by ArgR1/AhrC all comprise genes (likely) involved in arginine acquisition. S. pneumoniae contains no ornithine/arginine antiporter (arcD1/2 in L. lactis), which may indicate that ornithine does not accumulate in the cell (given the absence of the arginine biosynthetic genes, the only way could be via the arginine deiminase pathway) or is used as a carbon/nitrogen source. Despite the fact that ArgR1/AhrC in S. pneumoniae regulate three operons containing genes involved in the uptake of arginine, these operons, although improving growth efficiency in low arginine, are not essential for growth in medium with arginine present in amino acid form at low concentration. Thus, it is likely that there is a second, yet unidentified arginine uptake system encoded by the pneumococcal genome. We are currently designing experiments to identify this system. In addition, the role of the predicted amino acid permease AapA remains to be determined.
Notably, there are a couple of virulence genes in the regulon of ArgR1/AhrC. The oligopeptide-binding protein encoded by aliB has been shown to be important for nasopharyngeal colonization together with the paralogous oligopeptide-binding lipoproteins amiA and aliA, which are controlled by the pleiotropic regulator CodY (5, 79). The Ami·AliA-B complex therefore is a nice example of a system regulated by different nutritional stimuli via different regulators, which in this way control the ability of pneumococcus to colonize the nasopharynx. Next to aliB, also the lmB/adcAII gene encoding a Zn2+-binding lipoprotein (70) with a possible role in virulence (55–57, 80–82) and phtD encoding a Zn2+-scavenging surface protein (58, 60) that may also be involved in virulence and has a high potential as vaccine (60, 83, 84) have been shown to belong to the ArgR1/AhrC regulon. The regulon of ArgR1/AhrC intersects at the lmB/adcAII-phtD genes with the regulon of the Zn2+-dependent regulator AdcR, which represses expression of the lmB/adcAII promoter in the presence of Zn2+ (85). Therefore, the expression of these genes is dependent on two totally different environmental cues, which might be advantageous in the lifestyle of S. pneumoniae. As in S. agalactiae, the transcriptional regulator MtaR, which is necessary for virulence, affects the expression of putative arginine ABC transporter genes (37). It could be that the homologous genes (artPQ in D39) are subject to regulation by MtaR (homologous to spd_0588) in S. pneumoniae as well. Previous signature-tagged mutagenesis screens have found that AapA contributes to infection in a lung infection model (86). However, it is not known whether ArtPQ, AbpA, and AbpB contribute to virulence. We are currently exploring this possibility.
Supplementary Material
Acknowledgments
We thank Anne de Jong and Siger Holsappel for help with the production of the DNA microarrays.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4.
T. G. Kloosterman and O. P. Kuipers, unpublished data.
- CDM
- chemically defined medium
- ABC
- ATP-binding cassette.
REFERENCES
- 1. Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008) Nat. Rev. Microbiol. 6, 288–301 [DOI] [PubMed] [Google Scholar]
- 2. Scott J. A., Brooks W. A., Peiris J. S., Holtzman D., Mulholland E. K. (2008) J. Clin. Investig. 118, 1291–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitchell A. M., Mitchell T. J. (2010) Clin. Microbiol. Infect. 16, 411–418 [DOI] [PubMed] [Google Scholar]
- 4. Hammerschmidt S. (2006) Curr. Opin. Microbiol. 9, 12–20 [DOI] [PubMed] [Google Scholar]
- 5. Hendriksen W. T., Bootsma H. J., Estevão S., Hoogenboezem T., de Jong A., de Groot R., Kuipers O. P., Hermans P. W. (2008) J. Bacteriol. 190, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kloosterman T. G., Hendriksen W. T., Bijlsma J. J., Bootsma H. J., van Hijum S. A., Kok J., Hermans P. W., Kuipers O. P. (2006) J. Biol. Chem. 281, 25097–25109 [DOI] [PubMed] [Google Scholar]
- 7. Hendriksen W. T., Kloosterman T. G., Bootsma H. J., Estevão S., de Groot R., Kuipers O. P., Hermans P. W. (2008) Infect. Immun. 76, 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Härtel T., Klein M., Koedel U., Rohde M., Petruschka L., Hammerschmidt S. (2011) Infect. Immun. 79, 44–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canepa A., Filho J. C., Gutierrez A., Carrea A., Forsberg A. M., Nilsson E., Verrina E., Perfumo F., Bergström J. (2002) Nephrol. Dial. Transplant. 17, 413–421 [DOI] [PubMed] [Google Scholar]
- 10. Sethuraman R., Lee T. L., Chui J. W., Tachibana S. (2006) Neurochem. Res. 31, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 11. Currie G. A., Gyure L., Cifuentes L. (1979) Br. J. Cancer 39, 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albina J. E., Mills C. D., Barbul A., Thirkill C. E., Henry W. L., Jr., Mastrofrancesco B., Caldwell M. D. (1988) Am. J. Physiol. Endocrinol. Metab. 254, E459–E467 [DOI] [PubMed] [Google Scholar]
- 13. Wu G. (2009) Amino Acids 37, 1–17 [DOI] [PubMed] [Google Scholar]
- 14. Li P., Yin Y. L., Li D., Kim S. W., Wu G. (2007) Br. J. Nutr. 98, 237–252 [DOI] [PubMed] [Google Scholar]
- 15. Hovel-Miner G., Faucher S. P., Charpentier X., Shuman H. A. (2010) J. Bacteriol. 192, 4504–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ryan S., Begley M., Gahan C. G., Hill C. (2009) Environ. Microbiol. 11, 432–445 [DOI] [PubMed] [Google Scholar]
- 17. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 18. Klarsfeld A. D., Goossens P. L., Cossart P. (1994) Mol. Microbiol. 13, 585–597 [DOI] [PubMed] [Google Scholar]
- 19. Talaue M. T., Venketaraman V., Hazbón M. H., Peteroy-Kelly M., Seth A., Colangeli R., Alland D., Connell N. D. (2006) J. Bacteriol. 188, 4830–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Degnan B. A., Fontaine M. C., Doebereiner A. H., Lee J. J., Mastroeni P., Dougan G., Goodacre J. A., Kehoe M. A. (2000) Infect. Immun. 68, 2441–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Degnan B. A., Palmer J. M., Robson T., Jones C. E., Fischer M., Glanville M., Mellor G. D., Diamond A. G., Kehoe M. A., Goodacre J. A. (1998) Infect. Immun. 66, 3050–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sitkiewicz I., Green N. M., Guo N., Bongiovanni A. M., Witkin S. S., Musser J. M. (2010) PLoS One 5, e9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsen R., Buist G., Kuipers O. P., Kok J. (2004) J. Bacteriol. 186, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen R., Kok J., Kuipers O. P. (2005) J. Biol. Chem. 280, 19319–19330 [DOI] [PubMed] [Google Scholar]
- 25. Larsen R., van Hijum S. A., Martinussen J., Kuipers O. P., Kok J. (2008) Appl. Environ. Microbiol. 74, 4768–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicoloff H., Arsène-Ploetze F., Malandain C., Kleerebezem M., Bringel F. (2004) J. Bacteriol. 186, 6059–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barcelona-Andrés B., Marina A., Rubio V. (2002) J. Bacteriol. 184, 6289–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dennis C. A., Glykos N. M., Parsons M. R., Phillips S. E. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 421–430 [DOI] [PubMed] [Google Scholar]
- 29. Miller C. M., Baumberg S., Stockley P. G. (1997) Mol. Microbiol. 26, 37–48 [DOI] [PubMed] [Google Scholar]
- 30. Maas W. K. (1994) Microbiol. Rev. 58, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fulde M., Willenborg J., de Greeff A., Benga L., Smith H. E., Valentin-Weigand P., Goethe R. (2011) Microbiology 157, 572–582 [DOI] [PubMed] [Google Scholar]
- 32. Gruening P., Fulde M., Valentin-Weigand P., Goethe R. (2006) J. Bacteriol. 188, 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W., Liu L., Chen H., Zhou R. (2009) FEMS Microbiol. Lett. 292, 123–133 [DOI] [PubMed] [Google Scholar]
- 34. Winterhoff N., Goethe R., Gruening P., Rohde M., Kalisz H., Smith H. E., Valentin-Weigand P. (2002) J. Bacteriol. 184, 6768–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y., Dong Y., Chen Y. Y., Burne R. A. (2008) Appl. Environ. Microbiol. 74, 5023–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong Y., Chen Y. Y., Burne R. A. (2004) J. Bacteriol. 186, 2511–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bryan J. D., Liles R., Cvek U., Trutschl M., Shelver D. (2008) BMC Genomics 9, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaussee M. A., Callegari E. A., Chaussee M. S. (2004) J. Bacteriol. 186, 7091–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chaussee M. S., Somerville G. A., Reitzer L., Musser J. M. (2003) J. Bacteriol. 185, 6016–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larsen R. (2005) Transcriptional Regulation of Central Amino Acid Metabolism in Lactococcus lactis. Ph.D. thesis, University of Groningen, Groningen, The Netherlands [Google Scholar]
- 41. Lanie J. A., Ng W. L., Kazmierczak K. M., Andrzejewski T. M., Davidsen T. M., Wayne K. J., Tettelin H., Glass J. I., Winkler M. E. (2007) J. Bacteriol. 189, 38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kloosterman T. G., Bijlsma J. J., Kok J., Kuipers O. P. (2006) Microbiology 152, 351–359 [DOI] [PubMed] [Google Scholar]
- 43. Song J. H., Ko K. S., Lee J. Y., Baek J. Y., Oh W. S., Yoon H. S., Jeong J. Y., Chun J. (2005) Mol. Cells 19, 365–374 [PubMed] [Google Scholar]
- 44. van Hijum S. A., García de la Nava J., Trelles O., Kok J., Kuipers O. P. (2003) Appl. Bioinformatics 2, 241–244 [PubMed] [Google Scholar]
- 45. van Hijum S. A., de Jong A., Baerends R. J., Karsens H. A., Kramer N. E., Larsen R., den Hengst C. D., Albers C. J., Kok J., Kuipers O. P. (2005) BMC Genomics 6, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuipers O. P., de Ruyter P. G., Kleerebezem M., de Vos W. M. (1998) J. Biotechnol. 64, 15–21 [Google Scholar]
- 47. den Hengst C. D., Curley P., Larsen R., Buist G., Nauta A., van Sinderen D., Kuipers O. P., Kok J. (2005) J. Bacteriol. 187, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kloosterman T. G., van der Kooi-Pol MM, Bijlsma J. J., Kuipers O. P. (2007) Mol. Microbiol. 65, 1049–1063 [DOI] [PubMed] [Google Scholar]
- 49. Belitsky B. R. (2002) in Bacillus subtilis and Its Closest Relatives: from Genes to Cells (Sonenshein A. L., Hoch J. A., Losick R., eds) pp. 203–232, ASM Press, Washington, D. C [Google Scholar]
- 50. Bolotin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S. D., Sorokin A. (2001) Genome Res. 11, 731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bringel F., Frey L., Boivin S., Hubert J. C. (1997) J. Bacteriol. 179, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kazmierczak K. M., Wayne K. J., Rechtsteiner A., Winkler M. E. (2009) Mol. Microbiol. 72, 590–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maarsingh H., Zaagsma J., Meurs H. (2008) Eur. J. Pharmacol. 585, 375–384 [DOI] [PubMed] [Google Scholar]
- 54. Alloing G., de Philip P., Claverys J. P. (1994) J. Mol. Biol. 241, 44–58 [DOI] [PubMed] [Google Scholar]
- 55. Spellerberg B., Rozdzinski E., Martin S., Weber-Heynemann J., Schnitzler N., Lütticken R., Podbielski A. (1999) Infect. Immun. 67, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tenenbaum T., Spellerberg B., Adam R., Vogel M., Kim K. S., Schroten H. (2007) Microbes Infect. 9, 714–720 [DOI] [PubMed] [Google Scholar]
- 57. Wahid R. M., Yoshinaga M., Nishi J., Maeno N., Sarantuya J., Ohkawa T., Jalil A. M., Kobayashi K., Miyata K. (2005) Pediatr. Int. 47, 196–202 [DOI] [PubMed] [Google Scholar]
- 58. Loisel E., Chimalapati S., Bougault C., Imberty A., Gallet B., Di Guilmi A. M., Brown J., Vernet T., Durmort C. (2011) Biochemistry 50, 3551–3558 [DOI] [PubMed] [Google Scholar]
- 59. Adamou J. E., Heinrichs J. H., Erwin A. L., Walsh W., Gayle T., Dormitzer M., Dagan R., Brewah Y. A., Barren P., Lathigra R., Langermann S., Koenig S., Johnson S. (2001) Infect. Immun. 69, 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rioux S., Neyt C., Di Paolo E., Turpin L., Charland N., Labbé S., Mortier M. C., Mitchell T. J., Feron C., Martin D., Poolman J. T. (2011) Microbiology 157, 336–348 [DOI] [PubMed] [Google Scholar]
- 61. Baerends R. J., Smits W. K., de Jong A., Hamoen L. W., Kok J., Kuipers O. P. (2004) Genome Biol. 5, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park S. M., Lu C. D., Abdelal A. T. (1997) J. Bacteriol. 179, 5300–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Griswold A., Chen Y. Y., Snyder J. A., Burne R. A. (2004) Appl. Environ. Microbiol. 70, 1321–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carvalho S. M., Kloosterman T. G., Kuipers O. P., Neves A. R. (2011) PLoS One 6, e26707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zomer A. L., Buist G., Larsen R., Kok J., Kuipers O. P. (2007) J. Bacteriol. 189, 1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lulko A. T., Buist G., Kok J., Kuipers O. P. (2007) J. Mol. Microbiol. Biotechnol. 12, 82–95 [DOI] [PubMed] [Google Scholar]
- 67. Gardan R., Rapoport G., Débarbouillé M. (1995) J. Mol. Biol. 249, 843–856 [DOI] [PubMed] [Google Scholar]
- 68. Calogero S., Gardan R., Glaser P., Schweizer J., Rapoport G., Debarbouille M. (1994) J. Bacteriol. 176, 1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seth A., Connell N. D. (2000) J. Bacteriol. 182, 919–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Loisel E., Jacquamet L., Serre L., Bauvois C., Ferrer J. L., Vernet T., Di Guilmi A. M., Durmort C. (2008) J. Mol. Biol. 381, 594–606 [DOI] [PubMed] [Google Scholar]
- 71. Lu C. D., Yang Z., Li W. (2004) J. Bacteriol. 186, 3855–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gardan R., Rapoport G., Débarbouillé M. (1997) Mol. Microbiol. 24, 825–837 [DOI] [PubMed] [Google Scholar]
- 73. Lu C. D., Winteler H., Abdelal A., Haas D. (1999) J. Bacteriol. 181, 2459–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Heidrich N., Moll I., Brantl S. (2007) Nucleic Acids Res. 35, 4331–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heidrich N., Chinali A., Gerth U., Brantl S. (2006) Mol. Microbiol. 62, 520–536 [DOI] [PubMed] [Google Scholar]
- 76. Reyes E. D., Patidar P. L., Uranga L. A., Bortoletto A. S., Lusetti S. L. (2010) J. Biol. Chem. 285, 16521–16529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Claverys J. P., Lacks S. A. (1986) Microbiol. Rev. 50, 133–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sénéchal H., Delesques J., Szatmari G. (2010) FEMS Microbiol. Lett. 305, 162–169 [DOI] [PubMed] [Google Scholar]
- 79. Kerr A. R., Adrian P. V., Estevão S., de Groot R., Alloing G., Claverys J. P., Mitchell T. J., Hermans P. W. (2004) Infect. Immun. 72, 3902–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Terao Y., Kawabata S., Kunitomo E., Nakagawa I., Hamada S. (2002) Infect. Immun. 70, 993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weston B. F., Brenot A., Caparon M. G. (2009) Infect. Immun. 77, 2840–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Linke C., Caradoc-Davies T. T., Young P. G., Proft T., Baker E. N. (2009) J. Bacteriol. 191, 5814–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Melin M., Di Paolo E., Tikkanen L., Jarva H., Neyt C., Käyhty H., Meri S., Poolman J., Väkeväinen M. (2010) Infect. Immun. 78, 2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ogunniyi A. D., Grabowicz M., Mahdi L. K., Cook J., Gordon D. L., Sadlon T. A., Paton J. C. (2009) FASEB J. 23, 731–738 [DOI] [PubMed] [Google Scholar]
- 85. Shafeeq S., Kloosterman T. G., Kuipers O. P. (2011) Metallomics 3, 609–618 [DOI] [PubMed] [Google Scholar]
- 86. Hava D. L., Camilli A. (2002) Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 87. Albano M., Smits W. K., Ho L. T., Kraigher B., Mandic-Mulec I., Kuipers O. P., Dubnau D. (2005) J. Bacteriol. 187, 2010–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.