Background: Dysfunction of the peroxisomal ABC transporter ALDP and elevated very long chain fatty acids are the hallmark of X-ALD.
Results: Peroxisomal ABC transporters interact with proteins functioning in fatty acid synthesis and activation.
Conclusion: We identified a new fatty acid synthesis-transport machinery adapted to different chain lengths and metabolic conditions.
Significance: This machinery extends the knowledge on peroxisomal β-oxidation and X-ALD pathogenesis.
Keywords: ABC Transporter, Bioluminescence Resonance Energy Transfer (BRET), Fatty Acid Metabolism, Neurodegenerative Diseases, Peroxisomes, ACLY, ALDP, FASN, PMP70, X-ALD
Abstract
The neurodegenerative disease X-linked adrenoleukodystrophy (X-ALD) is characterized by the abnormal accumulation of very long chain fatty acids. Mutations in the gene encoding the peroxisomal ATP-binding cassette half-transporter, adrenoleukodystrophy protein (ALDP), are the primary cause of X-ALD. To gain a better understanding of ALDP dysfunction, we searched for interaction partners of ALDP and identified binary interactions to proteins with functions in fatty acid synthesis (ACLY, FASN, and ACC) and activation (FATP4), constituting a thus far unknown fatty acid synthesis-transport machinery at the cytoplasmic side of the peroxisomal membrane. This machinery adds to the knowledge of the complex mechanisms of peroxisomal fatty acid metabolism at a molecular level and elucidates potential epigenetic mechanisms as regulatory processes in the pathogenesis and thus the clinical course of X-ALD.
Introduction
The adrenoleukodystrophy protein (ALDP; ABCD1)3 belongs to the ATP-binding cassette (ABC) transporter protein family. In addition to PMP70 (ABCD3) and ALDR (ABCD2), ALDP is one of three ABC half-transporters residing in the peroxisomal membrane of mammals (1–4). ALDP has a pivotal role in peroxisome function, as mutations in the ABCD1 gene encoding ALDP are responsible for the most common inherited peroxisomal disorder, X-linked adrenoleukodystrophy (X-ALD). This complex and devastating neurodegenerative disorder is characterized by the abnormal accumulation of saturated very long chain fatty acids (VLCFAs), especially in brain white matter, adrenal cortex, and testis (5–7). To date, 567 pathogenic ABCD1 mutations are known, of which 357 are missense (human gene mutation database (HGMD)). They are distributed throughout the gene without a mutational hotspot. 138 of the ABCD1 mutations are associated with a considerable reduction or total lack of the ALDP protein, whereas in the presence of 31 mutations the ALDP protein is detectable. Phenotypic variability in X-ALD is extremely pronounced. One and the same mutation can lead to different disease patterns in patients, ranging from the severe childhood cerebral form characterized by fatal progressive demyelination, over-adrenomyeloneuropathy, and adrenal insufficiency without neurological symptoms to asymptomatic individuals. The pathogenesis of X-ALD, as well as the mechanism of function of the peroxisomal ABC half-transporters and their assembly in the peroxisomal membrane, is still unsolved, and hence a defect in ALDP alone is not satisfactory to explain the broad range of clinical X-ALD phenotypes.
ABC transporters generally consist of two homologous halves, each containing a membrane-spanning domain with multiple hydrophobic transmembrane helices and a hydrophilic cytosolic nucleotide-binding fold comprising the highly conserved motifs, Walker A, Walker B, and 19-mer, which are important for substrate specificity and nucleotide binding (8–10). ABC transporters can exist either as full-size transporters comprising 12 transmembrane helices and two nucleotide-binding folds, e.g. the multiple drug resistance transporter (11) and the cystic fibrosis transmembrane regulator (12), or as half-transporters with only six transmembrane helices and one nucleotide-binding fold. ALDP, PMP70, and ALDR belong to the group of half-transporters, which implies that their functionality depends on dimerization. It was demonstrated previously that ALDP and PMP70 can form homodimers as well as heterodimers with other peroxisomal ABC half-transporters (13–16). ALDP as a homodimer is able to transport VLCFAs, most probably VLCFA-CoAs, from the cytosol into the peroxisomal lumen for β-oxidation, as demonstrated in yeast (17). PMP70 seems to be responsible for the import of long chain fatty acids and branched chain acyl-CoAs, as well as intermediates of bile acid synthesis, into the peroxisomal lumen (18, 19). In Drosophila the uptake of precursors for the synthesis of red and brown pigments that determines eye color is controlled by various combinations of the ABC half-transporters, scarlet, brown, and white, respectively (20). It remains to be elucidated whether the formation of different ABC full-size transporter subspecies in Homo sapiens has any influence on the transported substrates.
All three peroxisomal transporters are synthesized on free ribosomes in the cytosol and are then inserted into the peroxisomal membrane. Localization of the ABC transporters to the peroxisomal membrane is mediated by interaction with PEX19 via a targeting motif at the N-terminal part of the ABC transporters (21, 22). To bring light on the role of these transporters in the peroxisome membrane and thus the pathogenesis of X-ALD, we searched for new interaction partners of ALDP and PMP70. We identified positive protein-protein interactions (PPI) of ALDP, PMP70, ATP-citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty-acid synthase (FASN), and FATP4 by use of GST pulldown experiments followed by mass spectrometry (LC/MS) and co-immunoprecipitation assays, as well as bioluminescence resonance energy transfer (BRET) measurements. A more detailed characterization of these interactions revealed a stratified interaction pattern with varying relative binding affinities among different protein pairs. These findings are in line with potential protein complexes of alternating composition that might be adjusted to different metabolic needs or pathways. As these proteins are involved in sequential steps of lipid metabolism, our findings are in line with a novel, fatty acid synthesis-transport machinery located at the peroxisomal membrane.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
All cDNA fragments were amplified from human cDNA using Pfu polymerase (Promega). Sequences for the oligonucleotides used for plasmid constructions are listed in supplemental Table 5. To amplify GST-C-ALDP (aa 357–745), oligonucleotides “C-term. ALDP for.2” and “ALDPStop-NotI rev” were used. For GST-C-PMP70 (aa 339–659), oligonucleotides “C-term. PMP70 for” and “PMP70NotI” were used. GST-C-ALDP and GST-C-PMP70 were cloned into pGEX-6P-2 (GE Healthcare). EYFP-ALDP (aa 1–745) was amplified using oligonucleotides “ALDP up” and “ALDP down,” and the amplification product was cloned into pEYFP-C1 vector (Clontech). ECFP-PMP70 (aa 1–659) was cloned into pECFP-C1 vector (Clontech) after amplification with primers “PMP70 up” and “PMP70 down.” For FATP4myc oligonucleotides “FATP4myc for” and “FATP4myc rev” were used, and the product was cloned into pcDNATM3.1/myc-His(−)B (Invitrogen) in which a stop codon was inserted between the Myc tag and the His tag.
For the construction of BRET vectors, respective ORFs of ALDP (BC015541), FASN (BC007909), ACLY (NM_001096), PMP70, (BC009712) and FATP4 (NM_005094) were obtained as Gateway entry clones (Invitrogen) based on pDONRTM223 from a copy of the Mammalian Gene Collection or amplified by PCR from p3xFLAG-CMV-7.1 or pcDNA3.1 and introduced into the pDONRTM221 by recombinational cloning. Expression clones coding for N- and C-terminal fusion proteins with Renilla reniformis luciferase (Rluc) and yellow fluorescent protein (YFP), respectively, were generated by a recombination of pDONRTM223 or pDONRTM221 entry clones with the respective BRET destination vectors, resulting in four expression vectors for each protein of interest. All constructs were verified by sequence analysis and test expression following Western blotting.
GST Pulldown Assays
GST fusion proteins and GST were produced in Escherichia coli BL21 cells (Stratagene). At 3 h after induction with 1 mm isopropyl β-d-thiogalactoside at 30 °C, cells were harvested, resuspended in cold PBST (140 mm NaCl, 2.5 mm KCl, 6.5 mm Na2 HPO4, 1.5 mm K2 HPO4, pH 7.25 + 0.1% (v/v) Tween 20) plus protease inhibitors (Roche Applied Science), and lysed by sonication. Lysis was enhanced by freezing and thawing. GST and fusion protein lysates were clarified by centrifugation (18,000 × g, 30 min, 4 °C). Supernatants were incubated with a 50% glutathione-Sepharose 4B slurry (GE Healthcare). Recombinant proteins immobilized to the Sepharose beads were incubated with HeLa cell lysates at 4 °C overnight. HeLa cell lysate was prepared from eight confluent 10-cm plates. 1 ml of lysis buffer (120 mm NaCl, 50 mm Tris pH 7.5, 1 mm EDTA, 0.1% Nonidet P-40, 0.1% Triton X-100, and protease inhibitors) was used per plate. Cell lysates were prepared by homogenization on ice followed by centrifugation (14,000 × g, 10 min, 4 °C). After overnight incubation with HeLa cell lysate, beads were washed three times with lysis buffer. Bound proteins were eluted by heating at 95 °C in SDS sample buffer and visualized in SDS-PAGE by colloidal Coomassie staining.
Protein Identification Using LC/MS
Tryptic in-gel digestion was performed according to Shevchenko et al. (23) with sequencing grade trypsin (Promega). Tryptic peptides were analyzed on an UltiMate 3000 nano-HPLC system (Dionex) operated with Xcalibur (Thermo Scientific) through DCMS Link (Dionex) with μ-PrecolumnTM cartridge Acclaim PepMap100 C18, 5 μm, 100 Å, 300-μm inner diameter × 5 mm (Dionex P/N 160454), analytical capillary column; 75-μm inner diameter × 15 cm, Acclaim PepMap100 C18, 3 μm (Dionex P/N 160321), ion trap mass spectrometer (LCQ DecaXPplus, Thermo Scientific); and a PicoTipTM emitter FS360-20-10 (New Objective). For LC/MS, a 10-μl sample was loaded on a μ-PrecolumnTM cartridge and washed for 8 min at a flow rate of 30 μl/min. The gradient nano-flow (200 nl/min) transferred the peptides to the analytical capillary column. A solvent gradient from 5% B (84% acetonitrile, 16% water, 0.07% formic acid) to 50% B within 40 min was applied. Columns were cleansed with 95% B followed by pre-equilibration (5% B). Peptides eluting during the 40-min gradient were online transferred to MS by electrospray ionization (ESI) through the use of a PicoTipTM emitter and a spray voltage of 1.5 kV. During the LC gradient the mass spectrometer cycled through the acquisition of a full MS scan within the mass range of 300 to 1400 Da followed by four data-dependent, collision-induced, MS/MS spectra of the four most intense ions. The dynamic exclusion was set at exclusion mass width ±3 Da, repeat count 2, repeat duration 0.5 min, exclusion list size 50, and exclusion duration 1 min. Spectra were collected in centroid mode. MS2 spectra analysis was done with the BioworksBrowser 3.3.1 package using the NCBI non-redundant database. Search parameters for TurboSEQUEST were: (i) precursor ion mass tolerance less than 1.4 atomic mass units, (ii) fragment ion mass tolerance less than 1.0 atomic mass unit, (iii) up to three missed tryptic cleavages allowed, and (iv) fixed cysteine modification by carboxyamidomethylation (plus 57.05 atomic mass units) and variable modification by methionine oxidation (plus 15.99 atomic mass units). Matching peptides for protein identification had to pass the following filters: (i) cross-correlation scores (Xcorr) over 2.5, (ii) p value below 5.0E-02, and (iii) primary scores (Sp) of at least 500.
Cell Culture and Transient Transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; PAA Laboratories) plus 10% fetal calf serum and 1% l-glutamine at 37 °C and 5% CO2. The day before transfection, 1 × 106 cells were seeded. Cells were transiently transfected with DNA coding for the indicated proteins using Effectene transfection reagent (Qiagen). HEK293 cells were cultured in Hyperflask cell culture vessels (Corning) in DMEM high glucose (4.5 g/liter; PAA Laboratories) supplemented with 10% fetal calf serum, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 0.25 μg/ml amphotecerin B. For BRET measurement, transfection based on electroporation was performed in a 96-well format using the Amaxa 96-well shuttle system (Lonza).
Co-immunoprecipitation and Immunoblotting (Western Blotting)
HeLa cells were grown in 10-cm plates. Cells from 6 plates/condition were harvested and transiently transfected with the indicated DNA constructs. Twenty-four hours post-transfection, cells were lysed in 500 μl of co-immunoprecipitation lysis buffer/plate (150 mm NaCl, 20 mm Tris pH 7.5, 1 mm EDTA, 1% Triton X-100, and protease inhibitors) by homogenization followed by a freeze-thaw cycle. Lysates were clarified by centrifugation (18,000 × g, 10 min, 4 °C). Half of the supernatant was rotary incubated overnight at 4 °C with antibodies as indicated, i.e. anti-GFP (JL8, Clontech, 1:50), anti-FASN (Cell Signaling, 1:50), or anti-Myc (Cell Signaling, 1:1000). The remaining half served as a control and was incubated without antibody. The next day 50 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology) was added for 2 h. After five washing steps with co-immunoprecipitation lysis buffer, the precipitated proteins were eluted by heating at 95 °C in SDS sample buffer and analyzed by Western blotting. Proteins separated by SDS-PAGE were transferred to nitrocellulose membranes using blotting buffer (48 mm Tris-HCl, 39 mm glycine, 0.04% w/v SDS, and 20% methanol). Membranes were blocked in TBST plus 5% defatted milk followed by overnight incubation at 4 °C with primary antibodies as indicated, i.e. anti-GFP (JL8, Clontech, 1:1000), anti-ACLY (Cell Signaling, 1:1000), anti-FASN (Cell Signaling, 1:1000), anti-Myc (Cell Signaling, 1:1000), and anti-ALDP (2B4, Euromedex, 1:1000). Incubation with the appropriate secondary HRP-labeled antibody was followed by detection with Lumi-Light Western blotting substrate (Roche Applied Science).
Immunofluorescence Staining
HeLa cells were seeded on glass coverslips. For immunofluorescence, cells were washed with PBS and fixed with 4% paraformaldehyde solution for 15 min. After three washing steps with PBS, cells were permeabilized with PBS supplemented with 1% (v/v) Triton X-100 for 5 min. After three washing steps with PBS, primary antibodies were added for 1 h (polyclonal anti-FASN, Cell Signaling, 1:50; monoclonal anti-ALDP, 2B4, Euromedex, 1:1000). After washing three times with PBS, secondary antibodies were added for an additional hour (anti-mouse-488, Molecular Probes, 1:1000; anti-rabbit-Cy3, Jackson ImmunoResearch, 1:1000). After three final washing steps with PBS, cells were mounted using ProLong Gold (Invitrogen) and examined using a confocal microscope.
BRET Measurements
The binary interaction of proteins in living cells was analyzed using bioluminescence resonance energy transfer (BRET) as described previously (24). Interactions were tested in all eight possible combinations of the proteins either fused to Rluc (energy donor) or YFP (energy acceptor). All combinations were tested in duplicates as well as in two independent experiments. HEK293 cells were co-transfected with a total of 0.8 μg of DNA at an acceptor to donor ratio of 3:1 and incubated at 30 °C. BRET saturation experiments to determine the maximal BRET ratio (BRETmax) and relative binding affinity (BRET50) were performed as described previously (24). Cells were co-transfected with the indicated respective DNAs with increasing acceptor to donor ratios, keeping the total DNA amount at a constant level. After 48 h, 30 μm coelenterazine was added as the Rluc substrate, and light emission was collected in a 96-well microplate luminometer (LUMIstar OPTIMA, BMG Labtech) over 10 s at 475 nm (Rluc signal) and 535 nm (BRET signal). The signals were analyzed by calculating the BRET ratio based on equation 1,
 |
where R is the BRET ratio, IA is the intensity of light emission at 535 nm, ID is the intensity of light emission at 475 nm, and cf is correction factor = (BRETcontrol/Rluccontrol) with the negative control being the co-transfection of the donor fusion protein with YFP in the absence of the protein of interest. A positive interaction of two investigated protein pairs was assumed, if at least one of eight tested tag combinations resulted in a BRET ratio above an experimentally derived method-specific threshold of 0.094. A positive control interaction (bJun-bFos) and the expression of Rluc-YFP as a positive control construct were included in every individual experiment. In addition, negative controls analyzing the interaction of ALDP with putatively non-interacting proteins (IL10RB and ACADS) were performed (supplemental Table 2).
PPI parameters were calculated by: Y = BRETmax × (X/BRET50 + X). BRETmax is the maximal BRET ratio, and BRET50 is the acceptor to donor ratio required to reach half-maximal BRET, indicating the relative binding affinity of protein-protein interaction partners.
RESULTS
Identification of Novel Interaction Partners of ALDP and PMP70
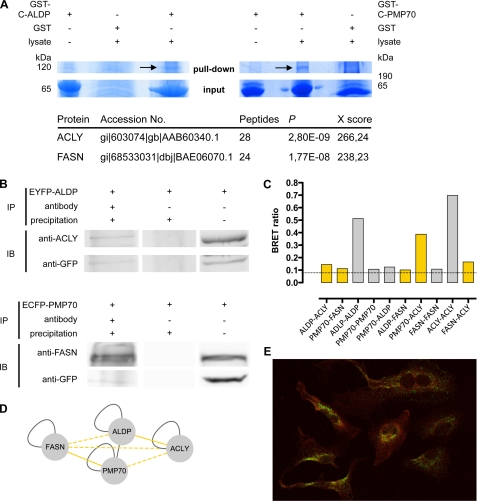
To identify novel interaction partners, we performed GST pulldown experiments with recombinant ALDP and PMP70. Because the N-terminal halves of both proteins were inserted into the peroxisomal membrane and therefore were highly hydrophobic, we expressed the hydrophilic cytoplasmic parts of both transporters in E. coli (aa 357–745 of ALDP and aa 339–659 of PMP70) as GST fusion proteins. Purified GST-C-ALDP and GST-C-PMP70 or GST as a control were coupled to glutathione-Sepharose and incubated with lysates of HeLa cells (Fig. 1A). To ensure specific binding of proteins from the cell lysate to C-ALDP or C-PMP70, pulldown experiments were compared by using GST alone or the corresponding GST fusion proteins, which were not incubated with cell lysate. Protein bands at 67 and 62 kDa represent GST-C-ALDP and GST-C-PMP70 fusion proteins, respectively. Protein bands not present in control lanes were excised, trypsin-digested, and analyzed by LC/MS. For the most prominent band of the C-ALDP pulldown, at around 120 kDa, many high scoring peptides belonging to ACLY were found, giving a total SEQUEST protein score of 266. In addition, a C-PMP70-specific band of more than 200 kDa that was not present in control lanes could be identified as FASN, with a SEQUEST protein score of 238 (see supplemental Table 1 for peptide scores of ACLY and FASN).
FIGURE 1.
The ABC half-transporters ALDP and PMP70 bind ACLY and FASN. A, in two independent GST pulldown experiments, recombinant fusion proteins of the C-terminal part of ALDP (GST-C-ALDP, 67 kDa), the C-terminal part of PMP70 (GST-C-PMP70, 62 kDa) or GST alone were incubated with HeLa cell lysates and subjected to Coomassie-stained SDS-PAGE. Prominent bands (arrows) present in the GST-C-ALDP and the GST-C-PMP70 pulldown but not in the control (GST alone) were analyzed by LC/MS. For the identified proteins, the SEQUEST probability values and the SEQUEST protein scores are given (for peptide Xscores see also supplemental Table 1). In GST-C-ALDP pulldown, ACLY (120 kDa) was identified as the binding partner, and in GST-C-PMP70 pulldown FASN (270 kDa) was found. B, co-immunoprecipitation experiments of HeLa cells overexpressing EYFP-ALDP (upper panel) or ECFP-PMP70 (lower panel). Lane 1 shows the co-immunoprecipitation experiments (IP) using a GFP antibody. Immunoblotting (IB) demonstrates that endogenous ACLY co-precipitates with EYFP-ALDP (upper panel) and that endogenous FASN co-precipitates with ECFP-PMP70 (lower panel). Lane 2 shows the control that was incubated with A/G PLUS-agarose but not with antibody. In lane 3, 50 μl of cell lysates as input was analyzed. C, BRET experiments analyzing binary PPI of ALDP, PMP70, ACLY, and FASN. One of eight tag combinations tested that resulted in the highest BRET ratio (supplemental Table 2) is shown. Data are given as the means of n = 2 independent experiments. The dashed line highlights the method-specific threshold for positive PPI of 0.094. Bars representing PPI reported previously in the literature are colored in gray; bars representing novel interactions are highlighted in yellow. D, results of PPI experiments are depicted in a network, with each line connecting two nodes representing an interaction of the proteins reported previously in the literature (gray line), identified by GST pulldown or co-immunoprecipitation and BRET (solid yellow line), or identified by BRET only (dashed yellow line). E, confocal microscopy to examine the co-localization of FASN and peroxisomes. HeLa cells were co-stained with a monoclonal ALDP antibody and anti-mouse-488 secondary antibody to visualize peroxisomes (green) and a polyclonal FASN-antibody and anti-rabbit-Cy3 secondary antibody (red). The yellow color in the overlay image indicates co-localization of FASN and the peroxisome.
Because the GST pulldowns were performed with the cytosolic parts of ALDP and PMP70, we sought to verify the interaction of full-length ALDP and PMP70 with endogenous ACLY and FASN by means of immunoprecipitation. The proper peroxisomal localization of ECFP/EYFP-tagged ALDP and PMP70 has been demonstrated previously. Kashiwayama et al. (25) and Hillebrand et al. (16) show that these tags, either at the N-terminal or the C-terminal side of the transporters, do not hamper import into the peroxisomal membrane. ACLY and FASN were detected by specific antibodies in the respective immunoblots after overexpression of EYFP-ALDP and ECFP-PMP70 in HeLa cells and precipitation with anti-GFP (Fig. 1B). These data substantiated the physical interaction of ALDP with ACLY, as well as PMP70 and FASN, and showed that full-length ABC transporters properly sorted to the peroxisomal membrane can form complexes with their endogenously expressed binding partners. Immunofluorescence co-staining of ALDP and FASN was performed in HeLa cells to determine co-localization of the endogenous proteins (Fig. 1E); ALDP (green) shows a punctate peroxisomal staining pattern, whereas FASN (red) shows a ubiquitous staining throughout the cell. Co-localization of FASN with peroxisomes (Fig. 1E, yellow) supported the physical interaction of FASN with peroxisomal ABC transporters.
To confirm PPI found by GST pulldown and co-immunoprecipitation studies, BRET experiments were performed testing binary interactions in vivo. We conducted a PPI mini-screen, probing all possible heteromeric and homomeric interacting pairs of ALDP, PMP70, FASN, and ACLY including previously described homo- or hetero-oligomerizations and potential further interactions. HEK293 cells were co-transfected with constructs of the respective genes carrying N- or C-terminal tags of Rluc as the donor or YFP as the acceptor of energy transfer (supplemental Table 2). For all heteromeric and homomeric protein pairs tested, at least one of eight possible tag combinations resulted in BRET ratios above the method-specific threshold for a positive PPI (0.094) (Fig. 1C). Thus, these data confirm the interactions of ALDP-ACLY (BRET ratio 0.14) and PMP70-FASN (BRET ratio 0.11) identified by GST pulldown and co-immunoprecipitation. Furthermore, they support the previously reported homo- and hetero-oligomerization of ALDP and PMP70 (ALDP-ALDP, 0.51; PMP70-PMP70, 0.11; and ALDP-PMP70, 0.12) as well as for the known homo-oligomerization of ACLY (0.42) and FASN (0.11). Beyond that, BRET experiments identified positive interactions for ALDP with FASN (0.10) and for PMP70 with ACLY (0.39). These data point to the formation of a heteromeric complex of ABC transporters with FASN and ACLY, which was further substantiated by a positive interaction of FASN with ACLY (0.17).
In conclusion, our data have revealed novel interactions of the ABC half-transporters, ALDP and PMP70, with ACLY and FASN and confirmed known homo-oligomerization of all proteins tested. The observation of the mutual interactions of ALDP, PMP70, FASN, and ACLY (Fig. 1D) gave rise to the hypothesis that varying ternary or quaternary complexes of different functionally active dimeric ABC full-size transporters (ALDP-ALDP, PMP70-PMP70, and ALDP-PMP70) and FASN-ACLY, respectively, could be formed.
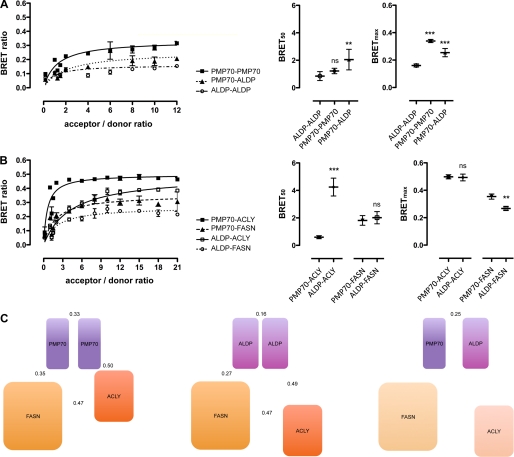
Characterization of Protein Complex Architectures by BRET Saturation Experiments
In a previous study, we demonstrated, by means of live cell FRET microscopy in vivo, that ALDP and PMP70 form homodimers as well as ALDP-PMP70 heterodimers, where ALDP homodimers displayed highest values followed by PMP70-PMP70 and ALDP-PMP70, respectively (16). In this study, the interactions of both half-transporters (ALDP and PMP70) with FASN and ACLY were established. To characterize the architecture of these potential complexes, we first aimed to approximate the likelihood for the formation of different complexes by determining the relative binding affinity. Second, we investigated their structural conformation by BRET-based distance measurements. BRET saturation experiments were performed (Fig. 2 and supplemental Table 3), where the relative binding affinity index could be determined by use of the ratio (acceptor/donor ratio) of proteins carrying the energy acceptor (YFP) to proteins carrying the energy donor (Rluc) at half-maximal BRET (BRET50). For proteins with comparable structure, the maximal BRET ratio (BRETmax) can be taken as a measure of the relative orientation of the tags to each other, hence giving information about differences in protein conformation. The analysis of the relative binding affinity of ALDP and PMP70 to form homo- and hetero-oligomers (Fig. 2A) showed that ALDP had a significant higher affinity (p < 0.01) to form homo-oligomers (BRET50, 0.85) than hetero-oligomers with PMP70 (BRET50, 2.05). These findings are in line with those obtained by FRET (16). In addition, the data may suggest that PMP70 has a comparable affinity to homo-oligomerize (BRET50, 1.22) and is more likely to exist as a homo-oligomeric than a hetero-oligomeric transporter (this difference was not statistically significant). Conformational analyses by BRETmax resulted in higher values for PMP70-PMP70 and ALDP-PMP70 as compared with ALDP-ALDP (Fig. 2A), indicating a different conformation.
FIGURE 2.
Characterization of the protein complex architecture of ALDP-PMP70-ACLY-FASN. BRET saturation experiments were performed to determine relative binding affinities (BRET50, acceptor to donor ratio at half-maximal BRET ratio) and maximal BRET ratios as a measure of the distance of BRET tags (BRETmax). Binary interactions of ALDP-ALDP, PMP70-PMP70, or PMP70-ALDP (A) and ALDP-FASN, PMP70-FASN, ALDP-ACLY, or PMP70-ACLY (B) are shown. BRET ratios are given as a function of acceptor to donor ratios, and BRET50 and BRETmax were calculated by nonlinear regression analysis (supplemental Table 3). Data are given as means ± S.D. of n = 3 independent experiments. The statistical significance of differences between individual BRET50 and BRETmax values was determined by analysis of variance and refers to the first protein-pair of each individual subgraph (***, p < 0.001; **, p < 0.01; ns, not significant with p > 0.05). C, illustration of the architecture of potential protein complexes comprising ABC full-transporters (PMP70-PMP70, ALDP-ALDP, and ALDP-PMP70), ACLY, and FASN. Symbol sizes are correlated relative to the molecular weights. Symbol distances depict the relative binding affinity of individual protein pairs. Numbers are given for BRETmax values representing the distance of the tags from each other, providing information on complex conformation. The protein complex based on the ALDP-PMP70 full-transporter is shaded to illustrate the unknown binding affinities of ACLY and FASN to the heteromeric ABC transporter.
Both PMP70 (BRET50, 1.81) and ALDP (BRET50, 2.02) bind FASN with a similar relative affinity (p > 0.05), indicating that the likelihood of FASN constituting a complex with ALDP or PMP70 is almost the same (Fig. 2B). A significantly higher relative binding affinity was obtained for ACLY interacting with PMP70 (BRET50, 0.59) than with ALDP (BRET50, 4.25), suggesting that ACLY is far more likely to attach to the complex with PMP70 than to the complex with ALDP. Yet, in contrast to the likelihood of complex formation, the evaluation of maximal BRET ratios may suggest a comparable structural conformation of both ternary complexes (ALDP/ALDP-FASN-ACLY and PMP70/PMP70-FASN-ACLY). The interaction of ALDP-ACLY and PMP70-ACLY occurred with similar BRETmax values (0.49 and 0.50), and the values obtained for the interaction of FASN with ALDP (BRETmax, 0.27) and PMP70 (BRETmax, 0.35), although significantly different (p < 0.01), pointed to a comparable structural conformation (Fig. 2B).
The additional interaction of ACLY and FASN inside the potential complex displayed a low relative binding affinity (BRET50, 3.83) in the same range as ACLY-ALDP. Although the BRET50 values obtained from different protein pairs do not provide absolute values, it was shown that the relative binding affinities (BRET50) correlated with dissociation constants determined by quantitative methods (26).
In summary, BRET experiments substantiated the differences in the formation of homomeric as compared with heteromeric ABC transporters composed of ALDP and PMP70. On the basis of our data, we suggest a model of varying protein complexes involved in the synthesis and transport of fatty acids at the peroxisomal membrane (Fig. 2C). In theory, all combinations of ALDP, PMP70, FASN, and ACLY are possible. However, the likelihood for the formation of a quaternary complex composed of a heteromeric ABC transporter and FASN-ACLY is rather low. Ternary complexes formed by homomeric ABC transporters and FASN-ACLY would be of a similar structure, but the abundance of ACLY in the ALDP-ALDP-based complex is of lower likelihood than for the PMP70-based complex.
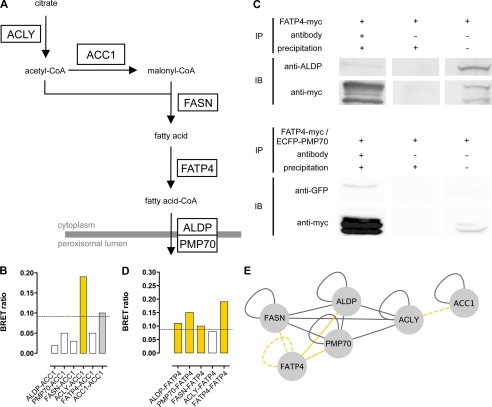
Association of Acetyl-CoA Carboxylase 1 (ACC1) with Newly Identified Protein Complex
Because ACC1 as the malonyl-CoA provider is the enzyme that functionally lies between ACLY and FASN in de novo fatty acid synthesis (Fig. 3A), we hypothesized that ACC1 might also associate with the newly identified complex. Thus, we investigated the binary interaction of ACC1 with ALDP, PMP70, ACLY, FASN, and FATP4 by BRET analysis. Co-transfection of ACC1 with ACLY resulted in BRET ratios above the method-specific threshold for a positive PPI (0.094) in two of eight possible tag combinations, with the highest BRET ratio (0.19) detected in cells co-expressing ACC1-Rluc and ACLY-YFP. However, co-expression of ACC1 with ALDP, PMP70, FASN, or FATP4 did not result in positive BRET ratios (Fig. 3B). These data indicate that ACC1 interacts with ACLY but not with ALDP, PMP70, FASN, or FATP4. Moreover, cells co-expressing ACC1 with N-terminal tags of Rluc and YFP resulted in strong BRET signals with a BRET ratio of 0.1. This finding supports previous reports for the homo-oligomerization of ACC1.
FIGURE 3.
ALDP, PMP70, FASN, and ACLY partially interact with further enzymes involved in fatty acid synthesis (ACC1) and metabolism (FATP4). A, scheme of the fatty acid synthesis pathway. ACLY converts citrate to acetyl-coA, which is then carboxylated to malonyl-CoA by ACC1, both serving as substrates for FASN. The multifunctional enzyme FASN performs seven sequential reactions and converts acetyl-CoA and malonyl-CoA to the long chain fatty acid palmitate, which can be subjected to elongation. Following transesterification to acyl-CoA by FATP4, activated fatty acids are transported across the peroxisomal membrane by ALDP and PMP70. B, BRET experiments analyzing binary interactions of ACC1 with ALDP, PMP70, ACLY, and FASN, as well as ACC1 homo-oligomerization. One of eight tag combinations tested that resulted in the highest BRET ratio (supplemental Table 2) is shown. Data are given as the means of n = 2 independent experiments. The dashed line highlights the method-specific threshold for positive PPI of 0.094. Bars representing PPI reported previously in the literature are colored gray; bars representing novel interactions are highlighted in yellow. C, for co-immunoprecipitation, HeLa cells were transfected with FATP4myc (upper panel) or co-transfected with FATP4myc and ECFP-PMP70 (lower panel), respectively. After immunoprecipitation (IP) of FATP4myc with anti-Myc, ALDP (anti-ALDP) and ECFP-PMP70 (anti-GFP) were detected by immunoblotting (IB) (lane 1). In lane 2 the controls without antibody incubation are depicted, and in lane 3, 50 μl of cell lysates as input was analyzed. D, BRET experiments analyzing binary interactions of FATP4 with ALDP, PMP70, and FASN, respectively, as well as FATP4 homo-oligomerization. One of eight tag combinations tested that resulted in the highest BRET ratio (supplemental Table 2) is shown. Data are given as the means of n = 2 independent experiments. The dashed line highlights the method-specific threshold for positive PPI of 0.094. Bars showing positive PPI are colored yellow. E, representation of the PPI network comprising ALDP, PMP70, ACC1, ACLY, FASN, and FATP4. Interactions of FATP4 and ACC1 with ALDP, PMP70, and FASN identified by BRET and co-immunoprecipitation (solid yellow line) or BRET only (dashed yellow line) are depicted, with each line connecting two nodes representing a positive interaction. All other PPI identified in this study or reported previously in the literature are depicted by gray lines.
Identification of FATP4 as a New Interaction Partner for the ABCD-ACLY-FASN Complex
The members of the identified PPI network provide acetyl-CoA as the building block for fatty acids (ACLY), malonyl-CoA (ACC1), long chain fatty acid synthesis (FASN), and transport of CoA-activated fatty acids across the peroxisomal membrane (ALDP or PMP70). Therefore, we searched for the missing link in the complex that might carry out this activation step (Fig. 3A). Fatty acid transport protein 4 (FATP4) was identified as the main VLCFA acyl-CoA synthetase that localizes to the peroxisome and to other organelles (27). To analyze the association of FATP4 with components of the ABCD-ACLY-FASN complex, we performed co-immunoprecipitation and BRET experiments. FATP4myc, overexpressed in HeLa cells, and endogenous ALDP or co-overexpressed ECFP-PMP70 were tested for co-precipitation (Fig. 3C). In immunoblots three bands ranging from ∼58 to 65 kDa were detected for FATP4myc. Whether this was the result of posttranslational modifications or protein degradation was not analyzed, but repeated experiments showed the same pattern. Besides an efficient precipitation of FATP4myc, corresponding bands for ALDP (80 kDa) and ECFP-PMP70 (100 kDa) were detected.
Next, we performed BRET experiments in living cells to verify the interaction of ALDP and PMP70 with FATP4, to analyze whether FATP4 also interacts with the other members of the complex, FASN and ACLY, and to identify possible homo-oligomerization of FATP4. For all but one of the five protein pairs tested, the BRET ratios indicated a positive PPI, confirming the interaction of FATP4 with ALDP (mean 0.11) and PMP70 (mean 0.15), respectively (Fig. 3D and supplemental Table 2). Furthermore, we showed that FATP4 binds to FASN (mean 0.10) and that FATP4 has the capacity to form homo-oligomers (mean 0.19). However, FATP4 did not show a positive PPI with ACLY (mean 0.08). Taken together, these findings show that FATP4 is part of a PPI network (Fig. 3E) of proteins involved in the synthesis, activation, and transport of fatty acids at the peroxisomal membrane. The interaction pattern that we found indicates that FATP4 forms a complex with an ABC full-transporter and FASN. In addition, ACLY is associated via an interaction with PMP70 or, to a lesser extent, ALDP.
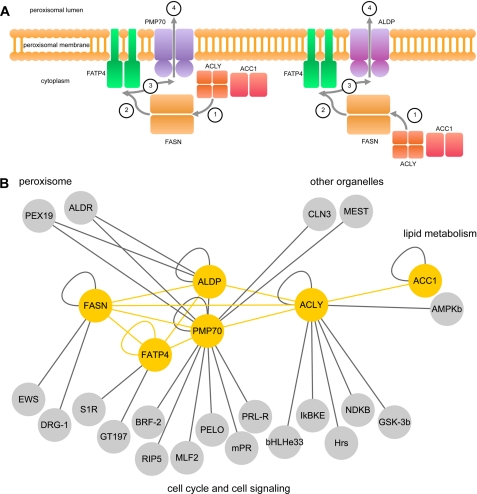
Identification of a New Fatty Acid Synthesis-Transport Machinery at the Peroxisomal Membrane
The network of ABC transporters, ACC1, ACLY, FASN, and FATP4, supports the existence of a fatty acid synthesis-transport machinery providing activated fatty acids for peroxisomal β-oxidation (Fig. 4A). At the functional level, there is a coherent connection between the ABC transporters ACLY, FASN, and FATP4. Thus, a possible functional sequence of the machinery consists of four steps 1) provision of acetyl-CoA by ACLY, 2) synthesis of fatty acids of FASN, 3) activation of fatty acids to their CoA esters by FATP4, and 4) transport of activated fatty acids by ALDP and/or PMP70 across the peroxisomal membrane for consecutive β-oxidation in the peroxisomal matrix. In addition, ACC1 is associated with ACLY and provides malonyl-CoA as a second building block for de novo fatty acid synthesis. ACC1 does not cross-interact with the other members of the complex and thus rather is associated with, rather than being an integral part of, the protein complex described. Based on our data derived from BRET saturation experiments, we propose a model for the newly identified complex that comprises at least three different arrangements with the ALDP homo-oligomer, the PMP70 homo-oligomer, and the ALDP-PMP70 hetero-oligomer in combination with ACLY and FASN, respectively.
FIGURE 4.
Model of a fatty acid synthesis-transport machinery composed of ABC full-transporters, ACC1, ACLY, FASN, and FATP4, showing its relation to the cellular interactome. A, alternative compositions of the fatty acid synthesis-transport machinery and the functional sequence are illustrated schematicically. The protein complexes face the cytoplasmic side of the peroxisome and are anchored to the organellar membrane by the ABC transporters and FATP4. All proteins are depicted as homo-oligomers. ACLY is less likely to be associated to the ALDP-based ABC full-transporter as compared with the PMP70-based complex. ACLY and ACC1 provide acetyl-CoA and malonyl-CoA (step 1) as building blocks for fatty acid de novo synthesis by FASN (step 2), and FATP4 activates LCFA and VLCFA by esterification to CoA (step 3), which are transported across the peroxisomal membrane (step 4) by ABC full-transporters for subsequent β-oxidation. B, PPI network of ALDP, PMP70, ACC1, ACLY, FASN, and FATP4 (shown in yellow, this study) and first-order interactions with the cellular interactome (yellow, this study; gray, databases; see also supplemental Table 4). Interacting proteins are grouped with respect to cellular components (peroxisome and other organelles) and biological processes (lipid metabolism, cell cycle, and cell signaling).
In an attempt to link the fatty acid synthesis-transport machinery to the cellular interactome, we performed a database analysis (HPRD and BioGRID databases) with subsequent manual recuration (supplemental Table 4) and identified 20 primary PPIs (Fig. 4B). The interacting proteins can be assigned to four different functional entities, i.e. peroxisomal, other organelles, fatty acid metabolism, and cell cycle and signaling pathways.
Impact of Mutations in the ABCD1 Gene on Protein-Protein Interactions between ALDP and Members of the Fatty Acid Synthesis-Transport Machinery
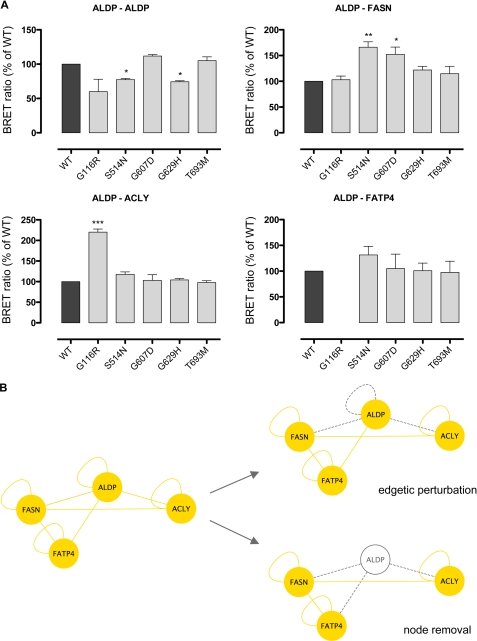
If a mutation leads to total lack of the corresponding protein, all interactions of this protein will be erased (node removal). In the presence of residual protein, a mutation-induced modification or disruption of a single PPI can occur (edgetic perturbation). We analyzed, whether five missense mutations in the ABCD1 gene known to be associated with near normal ALDP protein amounts (G116R, S514N, G607D, G629H, and T693M) would induce edgetic perturbations by altering ALDP homo-oligomerization or the interaction of ALDP with FASN, ACLY, or FATP4 (Fig. 5). Two mutations, S514N and G629H, led to a reduction of the BRET ratio related to ALDP homo-oligomerization. Conversely, S514N and G607D increased the BRET ratio for the interaction of ALDP with FASN, whereas G116R had the same effect on the interaction between ALDP and ACLY. The interaction between ALDP and FATP4 was not affected by any of the mutations.
FIGURE 5.
Disease-causing mutations in the ABCD1 gene lead to edgetic perturbations and are prone to node removal. A, BRET experiments analyzing the impact of ABCD1 gene mutations on binary interactions of ALDP. WT ALDP was compared with ALDP constructs representing disease-causing missense mutations (G116R, S514N, G607D, G629H, and T693M); homomeric interactions as well as interactions with FASN, ACLY, and FATP4 were determined. BRET ratios are given as percent WT of n = 4 independent experiments. No BRET ratio is given in cases of luciferase signals below a method-specific cutoff of 13,000. B, schematic illustration of the consequences of disease-causing mutations in ABCD1 on the PPI network established by wild-type ALDP (yellow lines). Missense mutations can induce distinct molecular phenotypes affecting individual PPI (edgetic perturbations, gray dashed line) or the availability of the ALDP protein (node removal, gray node).
Taken together, missense mutations in the ABCD1 gene can induce edgetic perturbations of the PPI relevant to the fatty acid synthesis-transport machinery at the peroxisomal membrane described herein (Fig. 5B).
DISCUSSION
Because loss of the biological function of ALDP alone is not sufficient to explain the onset of demyelination and the broad spectrum of disease severity in X-ALD, it is highly likely that a set of other proteins modulates the disease expression. To further clarify the subject, we searched for interaction partners of this ABC transporter. Using an approach combining GST pulldown, co-immunoprecipitation, and BRET, we identified a protein complex consisting of two peroxisomal ABC half-transporters (ALDP and PMP70) interacting with the fatty acid metabolism enzymes ACC1, ACLY, FASN, and FATP4.
Applying GST pulldown and co-immunoprecipitation experiments, we demonstrated the interactions of ALDP with ACLY and FATP4 as well as the interactions of PMP70 with FASN and FATP4. To circumvent potential interference with solubilization procedures that might disrupt PPI or promote interactions not present in the physiological cell context, we performed BRET measurements in intact living cells. This led to the confirmation of the already well established homo- and heterodimerization of ALDP and PMP70. Furthermore, we substantiated PPI of ALDP and PMP70 with ACLY and FASN identified by pulldown and co-immunoprecipitation experiments in this study, respectively, as well as the known hetero-oligomerization of ACLY and FASN. The identification of a binary PPI network of the homomeric ABC full-transporters ALDP and PMP70, or a heteromeric combination thereof, with ACLY and FASN indicated the formation of a protein complex.
ACLY catalyzes the formation of acetyl-CoA from citrate and CoA and thus provides the first building block for de novo lipid synthesis (28, 29). FASN is a multifunctional enzyme that performs seven sequential reactions to convert acetyl-CoA and malonyl-CoA to palmitate (30–32). The fact that ACLY and FASN are both known to act in the cytoplasm points to the assembly of this complex at the cytoplasmic side of the peroxisomal membrane. This was substantiated by the observation that the cytoplasmic parts of both PMP70 and ALDP interact with ACLY and FASN. In addition, ACC1 was shown to interact with ACLY but not with PMP70, ALDP, or FASN.
From these results we deduced the existence of a fatty acid synthesis-transport machinery composed of ACLY and FASN and either an ALDP homo-oligomer, a PMP70 homo-oligomer, or a PMP70-ALDP hetero-oligomer. As it was proposed that ALDP and PMP70 transport fatty acids of different chain lengths into the peroxisome (17–19), varying compositions of the machinery may be responsible for different synthesis and transport pathways, e.g. for fatty acids of different chain lengths or classes.
Moreover, the observation of different binding affinities of the proteins to each other revealed that the likelihood for the occurrence of the single complex compositions differs. The ABC transporter within the complex is composed mainly of ALDP or PMP70 homo-oligomers, which is consistent with reports from earlier studies (16). However, we again confirmed that ALDP also has the capacity to bind PMP70 in vivo. So far, mutations in the PMP70 gene have not been associated with X-ALD (33). However, it has been proposed that PMP70 influences the inflammatory phenotype in X-ALD (34). Furthermore, overexpression of PMP70 in X-ALD fibroblasts corrects VLCFA β-oxidation (35). This raises the hypothesis that PMP70 transporters partially take over the functions of ALDP transporters in X-ALD and that ACLY therefore may have an impact on the course of the disease. As heterodimerization of ALDP with PMP70 might allow for the transport of fatty acids of different chain lengths, the formation of the ALDP-PMP70 heterotransporter may attenuate the clinical phenotype in patients with ALDP deficiency, where an ALDP-PMP70 transporter has a better function than the mutated ALDP-ALDP transporter. It is conceivable that a varying proportion of homo- and heterotransporters in individual patients contributes to the high phenotypic variability observed in X-ALD patients.
Interestingly, ACLY is more likely to associate with the PMP70 than with the ALDP transporter in the complex, even though ALDP homodimers have been described as predominating (16). The complex occurring with the lowest probability would be ALDP-PMP70-FASN-ACLY, indicating that its formation is rare but possible. We showed that ALDP and PMP70 bind FASN with comparable affinity. Thus, the synthesis of fatty acids by FASN might be closely linked to the transport of the respective VLCFA and LCFA into the peroxisome by both the homo- and heteromeric ALDP/PMP70 transporters. It is of note that BRET experiments for FASN were performed exclusively using the C-terminal thioesterase domain (BC007909). This demonstrates that beyond the interdomain region (36), the thioesterase domain on the one hand has the capacity to self-associate and on the other hand may be involved in establishing interactions inside the PPI network. Furthermore, the interaction of ALDP with ACLY and FASN is of special interest in view of the involvement of the latter proteins in the lipid metabolism of myelin formation and integrity and thus brain functions, which are impaired in patients with X-ALD. Studies involving heterozygous ACLY knock-out mice show high ACLY expression in the neural tube and brain during development and in glial cells and cholinergic neurons in adult mice (37). Palmitate synthesized by FASN can be elongated to VLCFAs that are components of the ceramides, sphingolipids, and glycerolipids needed for brain development including myelination, neurological functions, and cell signaling (38).
Moreover, we have shown that ACC1 and the acyl-CoA synthetase FATP4 are associated with the protein complex identified here. ACC1 carboxylates acetyl-CoA to malonyl-CoA, the second building block for lipid synthesis by FASN. FATP4, a member of the family of the fatty acid transporters (39), transports LCFAs and VLCFAs. In addition, FATP4 is capable of activating both LCFAs and VLCFAs to their CoA derivatives (26, 40, 41). However, elongation of LCFAs is catalyzed by the fatty-acid elongase machinery, an endoplasmic reticulum-bound enzyme complex (42). Thus, ACC1 and FATP4 constitute the missing links in this multimodal unit of fatty acid synthesis and transport, providing metabolites for FASN enzymatic action as well as transesterification of its product to acyl-CoA for subsequent transport across the peroxisomal membrane. FATP4 interacts with three members of the complex, namely ALDP, PMP70, and FASN, but not with ACLY. These PPI imply that FATP4 is needed to activate fatty acids synthesized by FASN, which then are transported by ALDP and/or PMP70 across the peroxisomal membrane into the peroxisomal matrix for subsequent β-oxidation. Interesting findings with respect to FATP4 dysfunction inside the complex arise from experiments carried out in skin cells lacking FATP4 (43). Here, the activation of signaling pathways, where most probably the accumulated free unesterified fatty acids served as ligands, triggered STAT3 and cytokines, leading to up-regulation of STAT3 targets by EGFR signaling. The connection of ACC1 to the complex is restricted to a single PPI with ACLY. Both, ACC1 and ACLY provide the building blocks for fatty acid synthesis by FASN and, with respect to a directed functional sequence of the machinery described, could be classified as the raw material supply unit. This unit is linked to the central fatty acid synthesis enzyme, FASN, but is only loosely connected to the transport unit at the end of the functional sequence (FATP4 and ABC transporters). These findings together with the notion of similar structural conformations of the ternary complexes, as indicated by comparable BRET50 values of individual PPI, may indicate an intended and not an accidental occurrence of the complex in evolution.
Mutation-induced alterations in protein conformation and/or abundance of ALDP at the peroxisomal membrane are expected to impair the formation and function of the newly identified fatty acid synthesis-transport machinery, which may have an influence on the clinical course of X-ALD patients. To verify this hypothesis, we analyzed the effect of five disease-causing missense mutations in the ABCD1 gene on the PPI, supposed to be crucial for the formation and function of the protein complex. The impact of mutations on protein complexes or higher order interaction networks can be determined with respect to the abundance of the affected proteins (nodes) and to the ability of the nodes to establish interactions (edges). We showed that three of these mutations, G116R, S514N, and G607D, induced edgetic perturbations of PPI by exerting partly countervailing effects on ALDP homo-oligomerization and the interaction between ALDP and FASN and ALDP and ACLY, respectively. This confirmed that mutations can alter the ALDP interaction network, a mechanism that is expected to contribute to the phenotypic variability in X-ALD patients. However, it has to be taken into account that the majority of ABCD1 gene mutations are associated with a total lack of ALDP. In these cases, the mutation induces a loss of all interactions with this protein, a process called node removal, which would lead to even more deleterious consequences for the formation and function of the fatty acid synthesis-transport machinery and in turn for the metabolic function in X-ALD patients (Fig. 5B).
As cellular functions are mediated through complex systems of macromolecules and metabolites linked through biochemical and physical interactions, disease-causing mutations may also affect entire systems or interactome properties (44). In this context, altered interactions with further proteins associated with this network may have an impact on other metabolic or signaling pathways. To address this issue, we linked the fatty acid synthesis-transport machinery to the cellular interactome and identified 20 PPI with proteins involved in four different functional entities (peroxisomal, other organelles, fatty acid metabolism, or cell cycle and signaling pathways).
Besides the interaction of ALDP and PMP70 with PEX19, which is essential for the cellular housekeeping of the peroxisomal function, hetero-oligomerization of these ABC half-transporters with ALDR points to the potential existence of further ABC full-transporter subspecies. In view of the hypothesized variation in the composition of complexes described here, this may expand the cellular set of different fatty acid synthesis-transport machineries available for specific metabolic conditions. The PPI of PMP70 with MEST (membrane of the endoplasmic reticulum) and CLN3 (lysosomal membrane) represents an interesting link of the fatty acid synthesis-transport machinery to disease-relevant lipid metabolism pathways occurring in other cellular organelles. A possible extension of the ALDP-PMP70-ACC1-ACLY-FASN-FATP4 network arises from the interaction of ACLY with AMPKb, a protein kinase that phosphorylates acetyl-CoA carboxylase 1. The biosynthesis of malonyl-CoA by ACC1 linked to the protein complex through a possible PPI would provide the second building block for de novo lipid synthesis. All other interactions take place with proteins that are involved in the processes of cell cycle and cell signaling, e.g. PELO (chromosome segregation), GSK-3b (various signaling pathways), and IKBKE (host defense signaling pathway). The functional impact arising from some of these PPI is in line with and may further extend the relationship of the fatty acid synthesis-transport machinery to cell growth and inflammatory processes. Because of the broad spectrum of cellular functions of this interactome, it is likely that perturbations of one component, e.g. of ALDP in X-ALD, result in remote cellular dysfunctions. Detailed analyses of the pathways involved may in future reveal epigenetic mechanisms linked to the pathogenesis of X-ALD.
In conclusion, we propose a model of ABC full-transporters, ACC1, ACLY, FASN, and FATP4, as forming a thus far unknown peroxisomal fatty acid synthesis-transport machinery. The varying binding affinities of the proteins involved and the hypothesized different substrate specificities of ABC full-transporter subspecies may indicate the existence of alternative complexes for specific demands at fluctuating metabolic needs.
Our findings provide a new insight into lipid metabolism at a molecular level and contribute novel aspects to the understanding of the pathogenesis in X-ALD. Future work will extend the knowledge on additional proteins involved in this process and investigate the remote functions of the machinery and its single components.
Supplementary Material
Acknowledgments
We thank Jürgen G. Haas for providing the Gateway entry clones of FASN and PMP70 from his copy of the Mammalian Gene Collection; Anja Schultze, Anna Heckel- Pompey, and Ilona Dahmen for excellent technical assistance; and Mathias Woidy for technical assistance and critical input on data evaluation.
This work was supported by Deutsche Forschungsgemeinschaft Grant GA354/7-1 (to J. G.) and by the Bavarian Genome Research Network (BayGene) and LMUexcellent Grant 42595-6 (to A. C. M.).

This article contains supplemental Tables 1–5.
- ALDP
- adrenoleukodystrophy protein
- aa
- amino acid(s)
- ABC
- ATP-binding cassette
- ACC
- acetyl-CoA carboxylase
- ACLY
- ATP-citrate lyase
- BRET
- bioluminescence resonance energy transfer
- FASN
- fatty-acid synthase
- LCFA
- long chain fatty acid
- PMP70
- peroxisomal membrane protein 70 kDa
- PPI
- protein-protein interaction(s)
- Rluc
- Renilla reniformis luciferase
- VLCFA
- very long chain fatty acid
- X-ALD
- X-linked adrenoleukodystrophy.
REFERENCES
- 1. Mosser J., Douar A. M., Sarde C. O., Kioschis P., Feil R., Moser H., Poustka A. M., Mandel J. L., Aubourg P. (1993) Nature 361, 726–730 [DOI] [PubMed] [Google Scholar]
- 2. Kamijo K., Taketani S., Yokota S., Osumi T., Hashimoto T. (1990) J. Biol. Chem. 265, 4534–4540 [PubMed] [Google Scholar]
- 3. Gärtner J., Moser H., Valle D. (1992) Nat. Genet. 1, 16–23 [DOI] [PubMed] [Google Scholar]
- 4. Lombard-Platet G., Savary S., Sarde C. O., Mandel J. L., Chimini G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1265–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Geel B. M., Assies J., Weverling G. J., Barth P. G. (1994) Neurology 44, 2343–2346 [DOI] [PubMed] [Google Scholar]
- 6. Moser H. W. (1997) Brain 120, 1485–1508 [DOI] [PubMed] [Google Scholar]
- 7. Gärtner J., Braun A., Holzinger A., Roerig P., Lenard H. G., Roscher A. A. (1998) Neuropediatrics 29, 3–13 [DOI] [PubMed] [Google Scholar]
- 8. Higgins C. F. (1992) Br. Med. Bull. 48, 754–765 [DOI] [PubMed] [Google Scholar]
- 9. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gottesman M. M., Ambudkar S. V. (2001) J. Bioenerg. Biomembr. 33, 453–458 [DOI] [PubMed] [Google Scholar]
- 11. Germann U. A., Pastan I., Gottesman M. M. (1993) Semin Cell Biol. 4, 63–76 [DOI] [PubMed] [Google Scholar]
- 12. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., et al. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 13. Liu L. X., Janvier K., Berteaux-Lecellier V., Cartier N., Benarous R., Aubourg P. (1999) J. Biol. Chem. 274, 32738–32743 [DOI] [PubMed] [Google Scholar]
- 14. Smith K. D., Kemp S., Braiterman L. T., Lu J. F., Wei H. M., Geraghty M., Stetten G., Bergin J. S., Pevsner J., Watkins P. A. (1999) Neurochem. Res. 24, 521–535 [DOI] [PubMed] [Google Scholar]
- 15. Tanaka A. R., Tanabe K., Morita M., Kurisu M., Kasiwayama Y., Matsuo M., Kioka N., Amachi T., Imanaka T., Ueda K. (2002) J. Biol. Chem. 277, 40142–40147 [DOI] [PubMed] [Google Scholar]
- 16. Hillebrand M., Verrier S. E., Ohlenbusch A., Schäfer A., Söling H. D., Wouters F. S., Gärtner J. (2007) J. Biol. Chem. 282, 26997–27005 [DOI] [PubMed] [Google Scholar]
- 17. van Roermund C. W., Visser W. F., Ijlst L., van Cruchten A., Boek M., Kulik W., Waterham H. R., Wanders R. J. (2008) FASEB J. 22, 4201–4208 [DOI] [PubMed] [Google Scholar]
- 18. Imanaka T., Aihara K., Takano T., Yamashita A., Sato R., Suzuki Y., Yokota S., Osumi T. (1999) J. Biol. Chem. 274, 11968–11976 [DOI] [PubMed] [Google Scholar]
- 19. Kemp S., Theodoulou F. L., Wanders R. J. (2011) Br. J. Pharmacol. 164, 1753–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ewart G. D., Cannell D., Cox G. B., Howells A. J. (1994) J. Biol. Chem. 269, 10370–10377 [PubMed] [Google Scholar]
- 21. Gloeckner C. J., Mayerhofer P. U., Landgraf P., Muntau A. C., Holzinger A., Gerber J. K., Kammerer S., Adamski J., Roscher A. A. (2000) Biochem. Biophys. Res. Commun. 271, 144–150 [DOI] [PubMed] [Google Scholar]
- 22. Halbach A., Lorenzen S., Landgraf C., Volkmer-Engert R., Erdmann R., Rottensteiner H. (2005) J. Biol. Chem. 280, 21176–21182 [DOI] [PubMed] [Google Scholar]
- 23. Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfleger K. D., Eidne K. A. (2006) Nat. Methods 3, 165–174 [DOI] [PubMed] [Google Scholar]
- 25. Kashiwayama Y., Asahina K., Morita M., Imanaka T. (2007) J. Biol. Chem. 282, 33831–33844 [DOI] [PubMed] [Google Scholar]
- 26. Vizoso Pinto M. G., Villegas J. M., Peter J., Haase R., Haas J., Lotz A. S., Muntau A. C., Baiker A. (2009) Proteomics 9, 5303–5308 [DOI] [PubMed] [Google Scholar]
- 27. Jia Z., Moulson C. L., Pei Z., Miner J. H., Watkins P. A. (2007) J. Biol. Chem. 282, 20573–20583 [DOI] [PubMed] [Google Scholar]
- 28. Kornacker M. S., Ball E. G. (1965) Proc. Natl. Acad. Sci. U.S.A. 54, 899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Linn T. C., Srere P. A. (1979) J. Biol. Chem. 254, 1691–1698 [PubMed] [Google Scholar]
- 30. Smith S. (1994) FASEB J. 8, 1248–1259 [PubMed] [Google Scholar]
- 31. Wakil S. J. (1989) Biochemistry 28, 4523–4530 [DOI] [PubMed] [Google Scholar]
- 32. Wakil S. J., Stoops J. K., Joshi V. C. (1983) Annu. Rev. Biochem. 52, 537–579 [DOI] [PubMed] [Google Scholar]
- 33. Kemp S., Wanders R. J. (2007) Mol. Genet. Metab. 90, 268–276 [DOI] [PubMed] [Google Scholar]
- 34. Di Benedetto R., Denti M. A., Salvati S., Attorri L., Di Biase A. (2009) Neurochem. Int. 54, 37–42 [DOI] [PubMed] [Google Scholar]
- 35. Braiterman L. T., Zheng S., Watkins P. A., Geraghty M. T., Johnson G., McGuinness M. C., Moser A. B., Smith K. D. (1998) Hum. Mol. Genet. 7, 239–247 [DOI] [PubMed] [Google Scholar]
- 36. Chirala S. S., Jayakumar A., Gu Z. W., Wakil S. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3104–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beigneux A. P., Kosinski C., Gavino B., Horton J. D., Skarnes W. C., Young S. G. (2004) J. Biol. Chem. 279, 9557–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hannun Y. A., Obeid L. M. (2002) J. Biol. Chem. 277, 25847–25850 [DOI] [PubMed] [Google Scholar]
- 39. Hirsch D., Stahl A., Lodish H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8625–8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herrmann T., Buchkremer F., Gosch I., Hall A. M., Bernlohr D. A., Stremmel W. (2001) Gene 270, 31–40 [DOI] [PubMed] [Google Scholar]
- 41. Hall A. M., Wiczer B. M., Herrmann T., Stremmel W., Bernlohr D. A. (2005) J. Biol. Chem. 280, 11948–11954 [DOI] [PubMed] [Google Scholar]
- 42. Ohno Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y., Sassa T., Kihara A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin M. H., Chang K. W., Lin S. C., Miner J. H. (2010) Dev. Biol. 344, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong Q., Simonis N., Li Q. R., Charloteaux B., Heuze F., Klitgord N., Tam S., Yu H., Venkatesan K., Mou D., Swearingen V., Yildirim M. A., Yan H., Dricot A., Szeto D., Lin C., Hao T., Fan C., Milstein S., Dupuy D., Brasseur R., Hill D. E., Cusick M. E., Vidal M. (2009) Mol. Syst. Biol. 5, 321 (abstr.) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.