Abstract
Insulin-degrading enzyme (IDE) (insulysin) is a zinc metallopeptidase that metabolizes several bioactive peptides, including insulin and the amyloid β peptide. IDE is an unusual metallopeptidase in that it is allosterically activated by both small peptides and anions, such as ATP. Here, we report that the ATP-binding site is located on a portion of the substrate binding chamber wall arising largely from domain 4 of the four-domain IDE. Two variants having residues in this site mutated, IDEK898A,K899A,S901A and IDER429S, both show greatly decreased activation by the polyphosphate anions ATP and PPPi. IDEK898A,K899A,S901A is also deficient in activation by small peptides, suggesting a possible mechanistic link between the two types of allosteric activation. Sodium chloride at high concentrations can also activate IDE. There are no observable differences in average conformation between the IDE-ATP complex and unliganded IDE, but regions of the active site and C-terminal domain do show increased crystallographic thermal factors in the complex, suggesting an effect on dynamics. Activation by ATP is shown to be independent of the ATP hydrolysis activity reported for the enzyme. We also report that IDEK898A,K899A,S901A has reduced intracellular function relative to unmodified IDE, consistent with a possible role for anion activation of IDE activity in vivo. Together, the data suggest a model in which the binding of anions activates by reducing the electrostatic attraction between the two halves of the enzyme, shifting the partitioning between open and closed conformations of IDE toward the open form.
Keywords: Amyloid, Crystal Structure, Enzyme Mechanisms, Insulin, Kinetics, Peptidases, Peptide Hormones, Structural Biology, X-ray Crystallography, Yeast Metabolism
Introduction
Insulin-degrading enzyme (IDE)6 is a zinc metallopeptidase from the M16 family that has been extensively studied because of its role in regulating cellular insulin (1–3). More recently, IDE has received considerable attention for its role in degrading amyloid β (Aβ) peptides (4–7), and a genetic link between IDE and late onset Alzheimer disease has been reported (8). IDE degrades a number of other physiological peptides in vitro, including IGF-1 and IGF-2 (9), glucagon (10), atrial natriuretic peptide, TGF-α (11), and γ-endorphin (11). In addition to its metabolic role in peptide degradation, IDE has been reported to play a role in the inhibition of proteasome function by insulin (12), to degrade oxidized proteins in peroxisomes (13), to serve as a receptor accessory factor that enhances androgen and glucocorticoid receptor binding to DNA (14), and to serve as a receptor for the Varicella-Zoster virus (15, 16). Thus, IDE has a central role in mammalian physiology, and modulating its function may prove valuable for the treatment of disease states, most notably diabetes and Alzheimer disease.
Both the liganded and unliganded structures of human IDE have been recently reported (17, 18). The tertiary structure resembles a clamshell composed of four structurally similar domains arranged to enclose a large central chamber. Substrates must enter the chamber, which has a volume of ∼13,000 A3, and it is believed that a hinge-like conformational change allows substrate binding and product release. Mutations that destabilize the closed conformation increase IDE activity, suggesting that substrate binding/product release may be rate-limiting. The initial structural work (17) as well as our recent studies (19) identify two sites where substrates interact with the enzyme, one at the active site, located in domain 1, and the other at an equivalent site in the noncatalytic domain 2.
IDE exists as an equilibrium mixture of monomers, dimers, and tetramers, with dimers thought to be the predominant and most active state (11, 20). IDE is unusual among zinc metallopeptidases in that it exhibits allosteric kinetic behavior, with small peptide substrates increasing the activity of the enzyme toward the same or other small peptides (20). In addition, IDE has been shown to be activated heterotropically by polyphosphate anions (21, 22). In particular, ATP, other nucleotides, and triphosphate (PPPi) activate the enzyme toward hydrolysis of certain substrates 100–200-fold (21, 23). More recently, it has been reported that IDE has ATPase activity, which is also dependent upon a site distinct from the active site (24). In the studies presented here, we identify the site where ATP (and likely other anions) binds to IDE using x-ray crystallography and show that mutating enzyme residues in this site sharply decreases ATP/anion activation.
EXPERIMENTAL PROCEDURES
Preparation of IDE Mutants
Rat IDE (residues 41–1019), bearing the mutations K898A,K899A,S901A (IDEK898A,K899A,S901A), or R429S (IDER429S) were prepared from the pFastBac HTb-IDE plasmid (20, 25) using the QuikChange mutagenesis kit (Stratagene). Primers used for mutagenesis are listed as follows with the base changes in boldface and underlined: 1) Lys-898, Lys-899, and Ser-901 changed to alanine- IDEK898A,K899A,S901A: K898A forward, 5′-CGACTCGACAAACCAGCGAAACTCTCTGCAGAG-3′, and reverse, 5′-CTCTGCAGAGAGTTTCGCTGGTTTGTCGAGTCG-3′; K899A forward, 5′-CTCGACAAACCAGCGGCACTCTCTGCAGAGTGC-3′, and reverse, 5′-GCACTCTGCAGAGAGTGCCGCTGGTTTGTCGAG-3′; S901A forward, 5′-AAACCAGCGGCACTCGCTGCAGAGTGCGCGAAG-3′, and reverse, 5′-CTTCGCGCACTCTGCAGCGAGTGCCGCTGGTTT-3′; 2) Arg-429 converted to serine IDER429S, R429S forward, 5′-TTTAAAGATAAAGAGAGCCCACGAGGCTACACA-3′, and reverse, 5′-TGTGTAGCCTCGTGGGCTCTCTTTATCTTTAAA-3′. Bacmid and virus production were as discussed previously (25, 26).
Purification of IDE
IDE and the IDEK898A,K899A,S901A and IDER429S mutants were expressed as hexahistidine fusion proteins (hexahistidine sequence and a linker containing a tobacco etch virus protease cleavage site) in Sf9 cells and purified as described previously (25, 26). Briefly, Sf9 cells were suspended in 100 mm potassium phosphate buffer, pH 7.3, sonicated, and centrifuged at 75,000 × g for 20 min to pellet the membrane fraction. Supernatant was loaded on a His-select nickel affinity gel (Sigma) column and washed extensively with the potassium phosphate buffer containing 20 mm imidazole. Enzyme was eluting from the metal affinity resin by proteolytic removal of the hexahistidine sequence with added tobacco etch virus protease. IDE activity is known to be similar before and after cleavage of the hexahistidine fusion sequence (27). Purity was estimated by SDS-polyacrylamide gel analysis, and protein concentration was determined using the Coomassie Blue reagent with BSA as a standard.
Crystallization of IDE in Presence of ATP
Co-crystallization of wild type IDE and ATP was performed using IDE at 8 mg/ml in 50 mm Tris, pH 7.4, 1 mm DTT, 50–100 mm NaCl, 5 mm ATP, and 1 mm EDTA. The enzyme was maintained for 4 days under the above conditions before being used for crystallization trials employing the sitting drop vaporization method with Compact Clover plates (Emerald Biosystems). EDTA was used primarily to sequester any free Mg2+, which has been shown to reduce activation by ATP (21, 23). The protein/ATP solution was mixed 1:1 with well solution containing 100 mm sodium citrate, pH 6.5, 100 mm ammonium acetate, and 20% PEG 4000, and crystals were grown at 22 °C, usually accompanied by heavy precipitation. Crystals were dehydrated (28) by a brief (1–5 s) transfer to 50% PEG 4000 prior to flash cooling in liquid nitrogen (29).
Data Collection and Structure Determination
Final data were collected at Beamline-X4C of the National Synchrotron Light Source at Brookhaven National Laboratory, and the data were processed using HKL2000 (30). The structure of IDE crystallized with ATP was determined by molecular replacement with PHASER (31) using the native rat IDE coordinates (19) and refined using REFMAC5 within CCP4 (Collaborative Computational Project, No. 4) with subsequent model building performed using COOT (32). Water molecules were assigned using PHENIX (33), CCP4, and COOT. The quality of the final structure was analyzed using the Molprobity server (34), and figures were made using PyMOL (PyMOL Molecular Graphics System) and Chimera (35). Crystallographic data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Crystallographic summary of data collection and structure determination for the IDE-ATP complex crystal structure
| Data collection and structure refinement | IDE-ATP |
|---|---|
| Resolution | 50.0–2.27 Å |
| Space group | C2 |
| mol/asymmetric unit | 1 |
| a | 115.0 Å |
| b | 70.7 Å |
| c | 114.1 Å |
| α | 90.00° |
| β | 92.63° |
| γ | 90.00° |
| λ | 0.9794 Å |
| Completenessa | 92.0% (88.7%) |
| Redundancya | 3.3 (2.7) |
| Rsyma | 0.09 (0.48) |
| I/σa | 11.0 (1.9) |
| R/Rfree | 0.22/0.27 |
| Bond lengths | 0.007 Å |
| Bond angles | 1.04° |
| No. of water molecules | 226 |
| Ramachandran favored | 96.8% |
| Ramachandran outliers | 0.0% |
a Data indicate statistics for the last resolution shell (2.33–2.27 Å) as shown in parentheses.
Enzyme Activity Assay
IDE activity was determined by measuring the increase in fluorescence that occurs when the enzyme cleaves the internally quenched fluorogenic substrate Abz-GGFLRKHGQ-EDDnp at the Arg–Lys bond (26). The reaction was followed on a SpectraMax Gemini XS fluorescence plate reader using excitation and emission wavelengths of 318 and 419 nm, respectively. Initial velocities were determined using SoftMax Pro 4.0 and kinetic constants calculated using GraphPad Prism 4.0. ATPase activity was measured by following Pi release with acid molybdate as described by Harder et al. (36).
TNP-ATP Fluorescence Measurements
TNP-ATP fluorescence was monitored at excitation and emission wavelengths of 403 and 547 nm, respectively, using an LS55 luminescence spectrometer (PerkinElmer Life Sciences) at 20 ± 0.1 °C. TNP-ATP (Molecular Probes, Inc.) as supplied was diluted to 4 mm with 50 mm Tris-HCl, pH 7.4, and added to achieve the indicated concentrations. Competition experiments used to assess triphosphate affinity for wild type and mutant IDE were carried out with 5 μm enzyme in 50 mm Tris-HCl, pH 7.4, with constant 10 μm TNP-ATP and PPPi varying from 0 to 60 mm. Decrease in fluorescent enhancement at 547 nm was followed.
IDE Activity Assay in Yeast
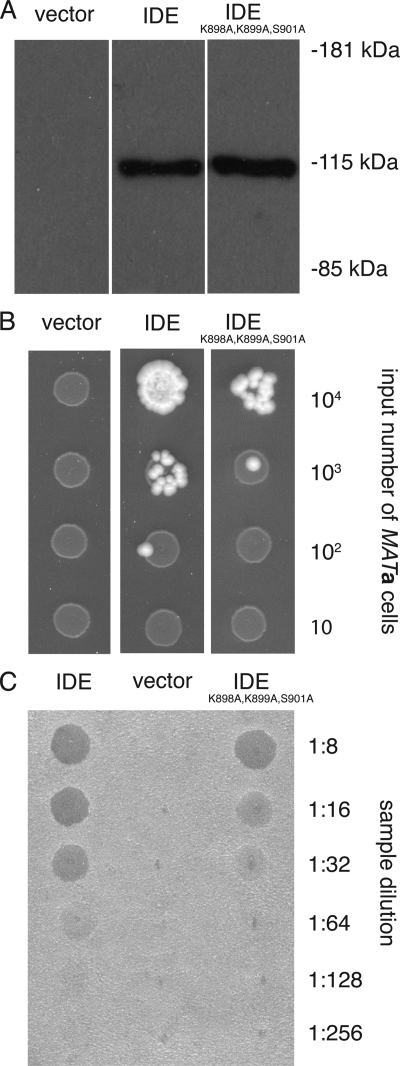
Wild type IDE and IDEK898A,K899A,S901A activities were compared in Saccharomyces cerevisiae using both a serial dilution mating test and a spot halo assay as described previously (37, 38). The assays take advantage of the ability of IDE, in lieu of the yeast M16 metallopeptidases Axl1p and Ste23p, to participate in the proteolytic maturation of yeast a-factor mating pheromone. In the mating test, IH1793 cells (MATα lys1 (39)) were mixed with decreasing numbers of strain Y272 cells (MATa trp1 leu2 ura3 his4 can1 axl1::LEU2 ste23::LEU2) bearing expression plasmids encoding IDE (pWS839; 2μ URA3 PPGK-HIS6n::TEV::IDE(41–1019)), IDEK898A,K899A,S901A (pWS841; 2μ URA3 PPGK-HIS6n::TEV::IDE(41–1019)K898A,K899A,S901A), or an empty expression vector (pRS316 (40)). The initial ratio of MATα to MATa cells was 9:1 in a 100-μl volume, with subsequent mixtures having sequentially 10-fold less MATa cells. Portions of mating mixtures (10 μl) were spotted on minimal media. Growth of diploid colonies, indicative of mating, was scored 3 days after incubation of plates at 30 °C. For the halo assay, yeast bearing the expression plasmids listed above were cultured in 5 ml of selective media for 3 days at 30 °C in 15-ml conical polypropylene tubes, which adsorb a-factor on their walls. The adsorbed a-factor was extracted by voiding the contents of the culture, washing the tubes several times with deionized water, and desorbing a-factor with methanol. Recovered a-factor samples were dried by speed-vac, and samples were reconstituted in methanol (50 μl). 2-Fold serial dilutions of the samples were prepared in YEPD liquid media, and a portion (2 μl) of each dilution was spotted onto a lawn of RC757 cells (MATα lys1 sst2-1 (41)) that had been spread on YEPD solid media; RC757 yeast undergo a strong growth arrest phenotype in the presence of a-factor (42). Halo formation was allowed to proceed for 18 h at 30 °C. Results of mating tests and halo assays were recorded by scanning plates with a Microtek flatbed scanner and importing digitized images into Photoshop and PowerPoint.
Construction of Yeast Expression Vectors Encoding IDE
pWS839 and pWS841 were created by plasmid-based PCR-directed recombination (43). In brief, the IDE encoding sequences were amplified by PCR from the pFastBac HTb vectors described above (IDE and IDEK898A,K899A,S901A) such that the ends of the PCR products contained the desired coding region and 39-bp extensions on either end that were homologous to the expression vector (pSM703 (44), 2μ URA3 PPGK) at sites on opposite sides of the polylinker. The PCR product was co-transformed into Y272 yeast along with pSM703 that had been digested with XmaI and NotI. Plasmids were isolated from yeast colonies surviving selection and screened for the presence of an appropriate insert by restriction digest analysis.
Preparation of Yeast Cell Extracts and Immunoblotting
Yeast extracts for immunoblot analysis were generated by alkaline lysis and TCA precipitation as described previously (45). The recovered protein precipitates were resuspended in hot (100 °C) urea sample buffer (250 mm Tris, pH 8.0, 6 m urea, 4% SDS, and 0.01% bromphenol blue) and clarified by centrifugation before use (16,000 × g, 1 min). An equivalent percentage amount of each sample was subject to SDS-PAGE followed by immunoblot analysis using an IDE-specific antibody, an appropriate secondary antibody, and chemiluminescence detection methods (BM Chemiluminescence Blotting Substrate, Roche Applied Science).
Docking of Triphosphate
Docking of triphosphate to the inner surface of domains 3 and 4 was done with AutoDock Vina (46) and AutoDockTools (47). The search region was defined interactively, and rotational flexibility was allowed for all ligand backbone bonds. Surface views were prepared with the program Chimera (35) using APBS and PDB 2PQR for electrostatic calculations (48).
RESULTS
IDE-ATP Complex Crystal Structure
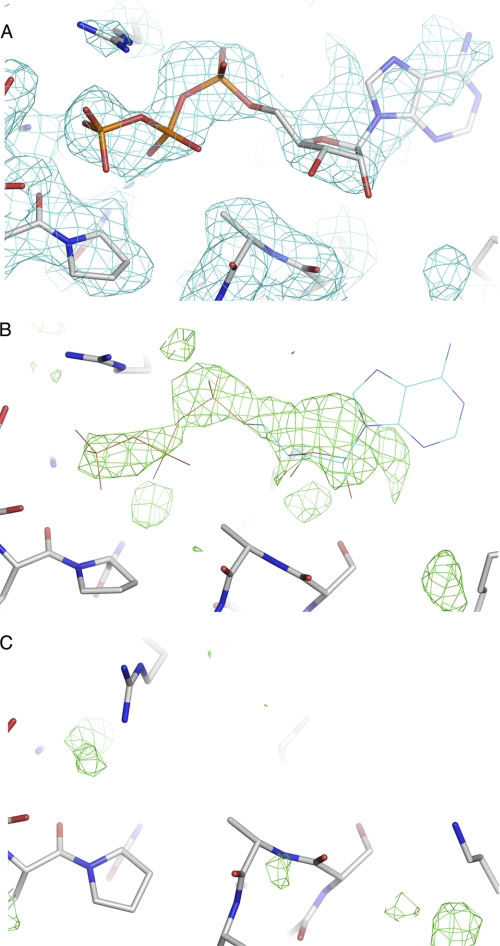
Previous studies have shown that the polyphosphate anions ATP and PPPi act in vitro as heterotropic activators of IDE toward the hydrolysis of small peptides, including the fluorogenic substrate Abz-GGFLRKHGQ-EDDnp (21, 26). To determine the interactions important for ATP binding in IDE, we crystallized rat IDE in the presence of ATP and determined the structure of the enzyme at 2.27 Å by molecular replacement using our previously determined native rat IDE coordinates (19). A difference electron density map revealed the presence of two bound ligands. The first region of strong positive difference density was at the previously described distal binding site (17). Attempts to model in the triphosphate moiety of ATP into the positive electron density at this site resulted in several unfavorable contacts with the enzyme. We have found that cleavage of the N-terminal hexahistidine tag from inactive IDE leads to binding of cleaved peptide derived from the fusion sequence at the distal site (19), and a three-residue polyalanine peptide modeled into this site accounted for most of the observed electron density.
The second ligand site is located on the inner surface of domain 4 (the loop between helices α31 and α32) near the interfaces with domains 2 and 3. The observed difference electron density was consistent with bound ATP, and a model for the ligand was placed at this site (Fig. 1, A and B). Electron density for the adenine ring of the ATP molecule was absent, indicating that this portion of the molecule is disordered. This observation agrees with previous studies suggesting that ATP binding occurs exclusively through the triphosphate moiety (21, 23). In prior work, crystal structures of rat IDE were determined under the same crystallization and data collection conditions except that ATP (and EDTA) were not present (19). Importantly, these structures do not show strong connected difference density in the proposed ATP-binding site (Fig. 1C), indicating that the density seen in this region is due to ATP and not another component of the buffer.
FIGURE 1.
ATP density. A, SIGMAA weighted 2Fo − Fc electron density map (blue) in the region of the ATP-binding site is displayed at 0.8 of the root mean square deviation of the map. The final model is shown in a stick representation. B, difference electron density (Fo − Fc) used for initial placement of ATP. The density (displayed with a cutoff of 2.3 times the root mean square deviation of the map) was phased from the IDE model after molecular replacement and initial refinement. The final ATP model is shown in a line representation. C, simulated annealing omit difference electron density (Fo − Fc, 2.3 times the root mean square deviation of the map) in the region of the ATP-binding site from an IDE crystal structure determined in the absence of ATP (19). Crystallization and data collection conditions were the same as used for the ATP-bound structure except ATP and EDTA were not included in the final soaking solution.
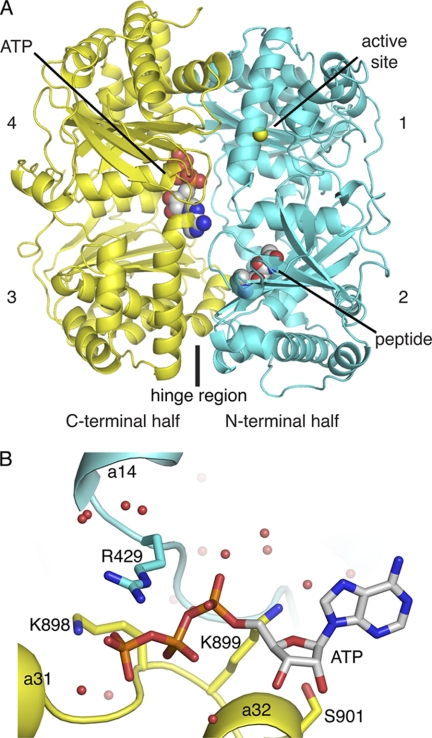
The model of ATP binding suggests several potential interactions with the enzyme (Fig. 2). The side chains of residues Arg-429, Lys-898, Lys-899, and Ser-901 are in position to potentially make hydrogen bond interactions with the modeled ATP ligand. Also the amide nitrogen of Lys-898 is well positioned to hydrogen bond with the phosphate groups in the model. Arg-429 is the only domain 2 residue that might contribute to ATP binding. It may be somewhat analogous to an arginine finger found in some other GTP- and ATP-hydrolyzing proteins (49, 50), where the charge on the guanidinium group stabilizes additional negative charge development during the transition state of ATP hydrolysis. It should be noted that the high micromolar affinity of IDE for ATP/polyphosphates suggests that these residues do not make strong contacts with the bound ligand.
FIGURE 2.
Overview of IDE structure and details of proposed ATP-binding site. A, backbone trace of IDE crystallized in the presence of ATP is shown in a ribbon representation with the four structurally related domains numbered, and the N- and C-terminal halves of the molecule colored in cyan and yellow, respectively. The expected position of the active site zinc ion is shown as a yellow sphere, but the metal is absent in the crystal structure. The region believed to act as a hinge for a proposed conformational change is also indicated. Modeled ATP and a 3-mer polyalanine peptide are shown in a space-filling representation and indicated by labels. B, proposed ATP-binding site is shown with selected side chains in a stick representation and ordered water as red spheres. ATP modeled into difference density present in the site is also show in stick representation.
A structural alignment of the IDE-ATP complex with the native IDE crystal structure (19) did not reveal any significant changes in average conformation at the ATP-binding site. However, a comparison of the thermal factors for native IDE and the IDE-ATP crystal structures (which are essentially isomorphous and were determined at nearly the same resolution) suggest a global change in dynamics of IDE upon binding ATP (supplemental Fig. S1). In particular, higher thermal factors are observed for many of the residues within the catalytic domain of IDE. The average thermal factor on all atoms of the catalytic domain (residues 42–285) of IDE is 29.5 Å2 as compared with a value of 49.9 Å2 for the same residues of IDE-ATP. Overall, the two models have similar thermal factors, with an average value of 39.4 for all unliganded IDE protein atoms and 42.4 for all IDE-ATP protein atoms, indicating that the differences in the catalytic domain are not the result of an overall difference in the two crystal structures.
Mutagenesis and Biochemical Analysis of ATP-binding Site of IDE
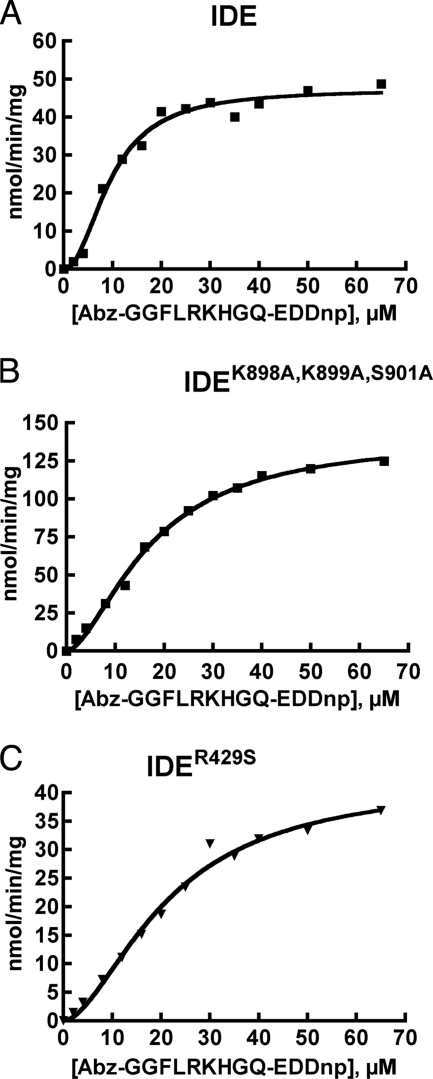
Site-directed mutagenesis was used to eliminate potential binding interactions identified in the crystal structure. Two mutant forms of IDE were generated, the first with mutations K898A,K899A,S901A (IDEK898A,K899A,S901A), and the second with the single mutation R429S (IDER429S). With the synthetic substrate Abz-GGFLRKHGQ-EDDnp, both mutants retained the sigmoidal substrate versus velocity response observed with IDE (Fig. 3), which is characteristic of allosteric homotropic activation (20). Hill coefficients of 1.7 were calculated for both mutants, which is slightly less than the value of 2.1 obtained for IDE. The IDEK898A,K899A,S901A mutant showed a 3-fold increase in kcat relative to IDE, whereas IDER429S had essentially the same kcat as IDE (Table 2). Both mutants had a roughly 2-fold higher Km values for the fluorogenic substrate compared with IDE.
FIGURE 3.
Comparison of kinetics of IDE and IDE containing ATP-binding site mutations. Reactions were conducted in 50 mm Tris-HCl, pH 7.4, with IDE (0.5 μg) (A), IDEK898A,K899A,S901A (0.5 μg) (B), or IDER429S (1.0 μg) (C) using Abz-GGFLRKHGQ-EDDnp as the substrate.
TABLE 2.
Summary of kinetic properties of IDE, IDER429S, and IDEK898A,K899A,S901A with Abz-GGFLRKHGQ-EDDnp as a substrate
| IDE | IDER429S | IDEK898A,K899A,S901A | |
|---|---|---|---|
| Vmax (nmol/min/mg) | 47.2 ± 1.7 | 42.3 ± 3.2 | 140.8 ± 5.5 |
| kcat (min−1) | 5.2 ± 0.2 | 4.7 ± 0.3 | 15.5 ± 0.6 |
| Km (mm) | 9.6 ± 0.7 | 21.3 ± 2.4 | 17.3 ± 1.0 |
| Hill coefficient | 2.1 ± 0.3 | 1.7 ± 0.2 | 1. 7 ± 0.1 |
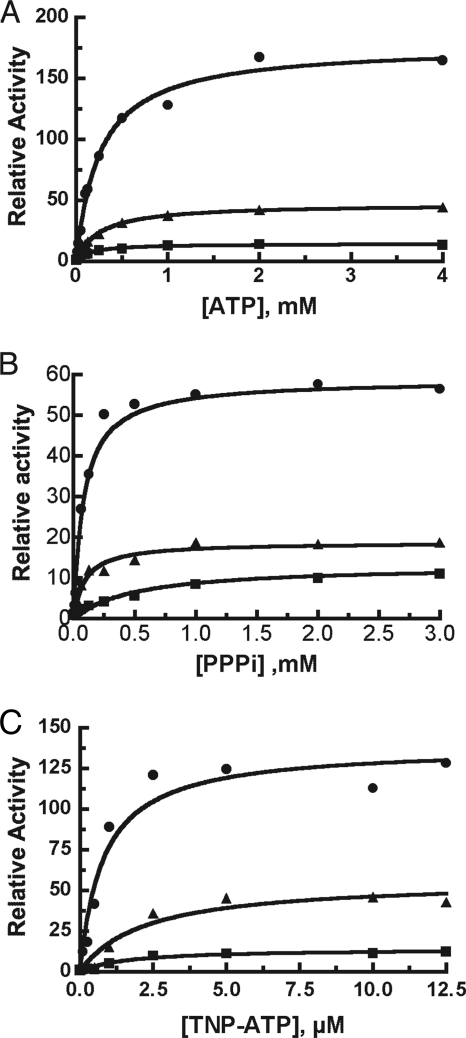
The ability of ATP, triphosphate, and TNP-ATP to activate both IDE mutants is dramatically reduced relative to the wild type enzyme, however (Fig. 4). ATP activates IDE more than 100-fold with Abz-GGFLRKHGQ-EDDnp as the substrate. The level of IDER429S activation is about 35% of the wild type enzyme value, and there is even less activation of IDEK898A,K899A,S901A by ATP, ∼15%. The concentration of ATP needed to achieve 50% activation (Ka) appears to be ∼2-fold lower for the mutants than the wild type enzyme (Ka = 1.6 mm for IDE and 0.6 mm for both IDEK898A,K899A,S901A and IDER429S). However as noted by Dixon and Webb (51) these Ka values do not directly reflect the affinity of the enzyme for ATP as they are dependent of the Km for the substrate, which is ∼2-fold higher for each of the mutants. In addition, the steady-state equation contains terms in [A]2 and [S]2 making the apparent Ka a complex constant. Activation by triphosphate showed essentially the same apparent Ka or wild type IDE and the IDER429S mutant (0.08 and 0.09 mm, respectively) but a higher apparent Ka for IDEK898A,K899A,S901A (0.5 mm). Activation by TNP-ATP showed an increased apparent Ka for IDEK898A,K899A,S901A and IDER429S mutant (2.2 μm) compared with a Ka of 0.9 μm for the wild type enzyme.
FIGURE 4.
Effect of ATP-binding site mutations on the ability of ATP, PPPi, and TNP-ATP to increase the rate of Abz-GGFLRKHGQ-EDDnp hydrolysis. Activity was determined in 50 mm Tris-HCl, pH 7.4, with 10 μm Abz-GGFLRKHGQ-EDDnp as substrate at the indicated concentrations of ATP (A), PPPi (B), or TNP-ATP (C). Each reaction contained 0.5 μg of enzyme. Relative activity is the ratio of substrate cleaved at a given concentration of polyphosphate species to that cleaved in the absence of the polyphosphate species. IDE, closed circles; IDER429S, closed triangles; IDEK898A,K899A,S901A, closed squares.
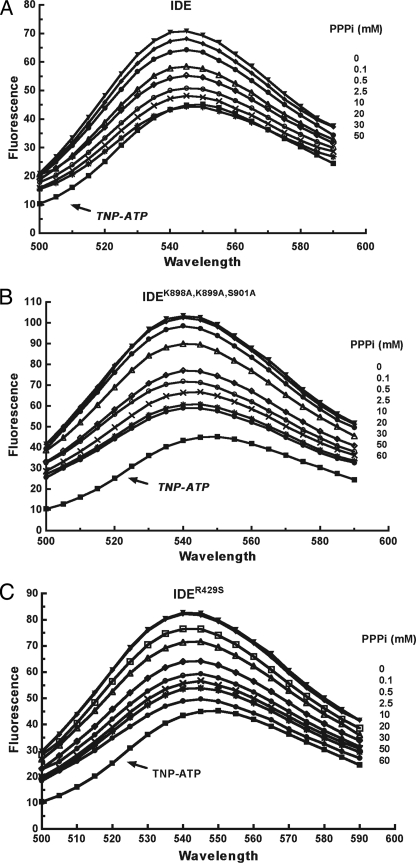
Both mutants bound TNP-ATP with a greater fluorescent enhancement than IDE (Fig. 5, top spectrum in each panel). We measured the ability of PPPi to displace bound TNP-ATP because high concentrations of ATP interfered with the TNP-ATP fluorescence spectra. As seen in Fig. 5, PPPi can completely displace bound TNP-ATP from IDE with half-maximal displacement at 0.6 mm PPPi. For the two NEP mutants, we found that TNP-ATP displacement was incomplete with half-maximal displacement at ∼1.3 mm PPPi for IDEK898A,K899A,S901A and ∼1.0 mm for IDER429S. These higher half-maximal displacement concentrations suggest a weaker binding to the mutant IDE forms. The inability of PPPi to completely displace TNP-ATP from the mutants further suggests an alteration in the local environment of the binding site. In particular, it suggests a less electrostatic/polar character to the interaction. Taken together, these kinetic and analog binding results are consistent with the IDEK898A,K899A,S901A and IDER429S mutations affecting the binding of ATP, likely through a decrease in affinity as well as a change in the orientation or location of the bound molecule.
FIGURE 5.
Displacement of bound TNP-ATP by triphosphate. Fluorescence emission spectra were recorded in 50 mm Tris-HCl, pH 7.4, with 10 μm TNP-ATP, 5 μm IDE, or IDE mutant and the indicated concentration of PPPi.
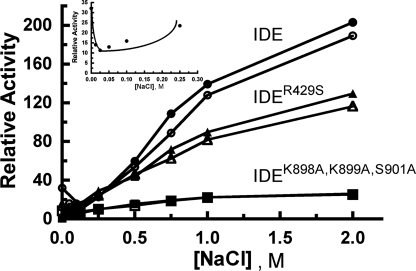
The activity of wild type IDE with Abz-GGFLRKHGQ-EDDnp as substrate is increased by high concentrations of NaCl to an even greater extent than by ATP (Fig. 6). As observed with ATP activation, the two mutants are also activated but to a lesser extent (Fig. 6). In the presence of 0.2 mm ATP (producing an ∼30-fold activation of IDE), ATP-dependent heterotropic activation is first reduced by NaCl, but then activation is observed (Fig. 6, inset). As shown in Table 2, activation by NaCl is about 2-fold greater than activation by ATP for IDE and IDEK898A,K899A,S901A but close to 4-fold greater with IDER429S.
FIGURE 6.
Effect of NaCl on IDE-dependent hydrolysis of Abz-GGFLRKHGQ-EDDnp hydrolysis and on activation by ATP. Activity was determined in 50 mm Tris-HCl, pH 7.4, with 10 μm Abz-GGFLRKHGQ-EDDnp as substrate, IDE, IDEK898A,K899A,S901A, or IDER429S, and the indicated concentration of NaCl. The effect of NaCl was determined in the absence (closed symbols) or presence (open symbols) of 0.2 mm ATP. The inset shows that NaCl initially inhibited but then activated the reaction in the presence of ATP.
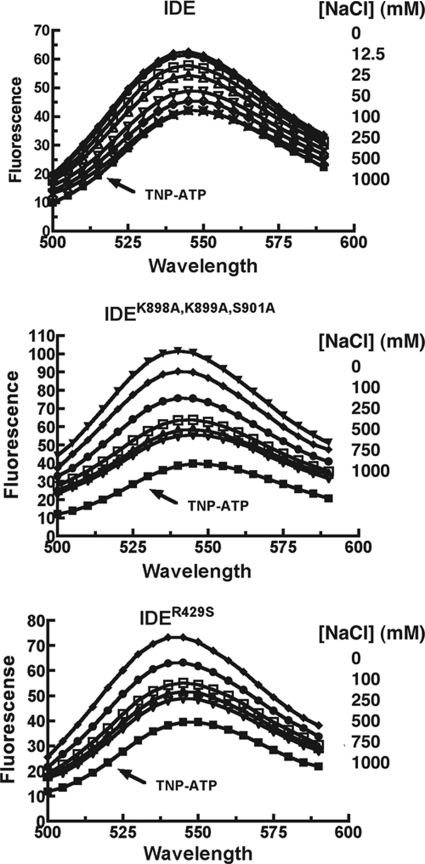
That NaCl can completely displace ATP (TNP-ATP) from IDE is shown in Fig. 7. As observed with PPPi, TNP-ATP displacement from IDEK898A,K899A,S901A and IDER429S is incomplete. As also seen with PPPi (Fig. 5), displacement of TNP-ATP from IDEK898A,K899A,S901A was the least complete. These effects of salt concentration on IDE activity are supportive of an electrostatic mechanism for anion activation.
FIGURE 7.
Displacement of bound TNP-ATP by NaCl. Fluorescence emission spectra were recorded as in Fig. 6 in 50 mm Tris-HCl, pH 7.4, with 10 μm TNP-ATP, 5 μm IDE or IDE mutant, and the indicated concentration of NaCl.
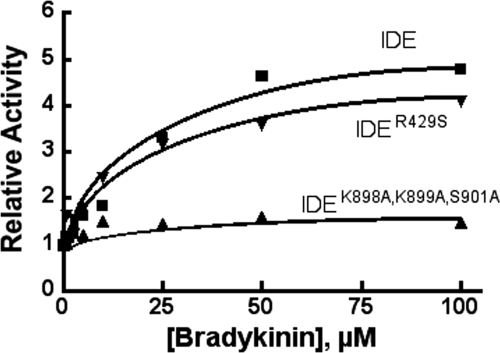
We have previously shown that small peptides like bradykinin can act as activators of Abz-GGFLRKHGQ-EDDnp hydrolysis, and interactions between peptide and anion activation have been suggested (20). We thus determined the effect ATP-binding site mutations would have on the ability of bradykinin to increase the rate of Abz-GGFLRKHGQ-EDDnp hydrolysis. Similar rates were observed for IDER429S and wild type IDE, although the observed rate for IDEK898A,K899A,S901A was significantly reduced, suggesting that ATP- and peptide-dependent activation may be linked (Fig. 8).
FIGURE 8.
Effect of bradykinin on Abz-GGFLRKHGQ-EDDnp hydrolysis by IDE, IDEK898A,K899A,S901A, or IDER429S. Activity was determined in 50 mm Tris-HCl, pH 7.4, with 10 μm Abz-GGFLRKHGQ-EDDnp as substrate and indicated concentrations of bradykinin. Reactions contained 0.5 μg of protein.
IDE has been reported to exhibit ATPase activity (24). Thus the question arises as to whether activation of IDE by ATP is coupled to its hydrolysis. We found that the nonhydrolyzable ATP analog γ-methylene ATP is able to activate Abz-GGFLRKHGQ-EDDnp hydrolysis nearly as well as ATP (supplemental Fig. S2), demonstrating that ATP hydrolysis is not necessary for IDE activation. IDE-dependent ATPase activity was estimated to be less than 1 nmol/min/mg IDE using 1 mm ATP as substrate, and this rate did not change in the presence of Abz-GGFLRKHGQ-EDDnp substrate. The much higher rate of peptide cleavage (see Table 3) over ATP hydrolysis also supports the conclusion that ATPase activity is not coupled to activation.
TABLE 3.
Comparison of activation by ATP to activation by NaCl
| Enzyme form | Rate increase at 5 mm ATP | Rate increase at 2 m NaCl |
|---|---|---|
| IDE | 102 | 228 |
| IDE-AAA | 16 | 27 |
| IDER429S | 37 | 130 |
Effect of ATP-binding Site Mutation on Intracellular Activity of IDE
An established yeast system for functional studies of IDE (37, 38) was used to assess the significance of anion activation on IDE activity in a cellular environment. The assay is based on the ability of IDE to support production of mature a-factor mating pheromone in the absence of yeast metallopeptidases (i.e. Axl1p and Ste23p) that normally promote its maturation. When expressed in this yeast system, IDE and the ATP-binding site mutant IDEK898A,K899A,S901A were produced at approximately the same protein levels (Fig. 9A). Wild type IDE, however, promoted mating to a greater extent than IDEK898A,K899A,S901A (Fig. 9B). Similarly, a spot halo assay revealed greater production of a-factor by yeast expressing wild type IDE relative to yeast expressing the ATP-binding site mutant (Fig. 9C). The results indicate that an intact polyphosphate/anion-binding site is required for maximal IDE activity in yeast, suggesting that heterotropic activation by anions likely influences IDE activity in vivo.
FIGURE 9.
Activity of wild type IDE and ATP-binding site mutant IDEK898A,K899A,S901A in the yeast S. cerevisiae. A, immunoblot of yeast-expressed IDE and IDEK898A,K899A,S901A. Equivalent percentage amounts of total cellular extracts from yeast containing an empty expression vector or plasmids encoding IDE or the IDEK898A,K899A,S901A mutant were analyzed by SDS-PAGE and immunoblotting using an IDE-specific antibody. B, mating test for IDE activity. An excess of MATα yeasts were mixed with decreasing numbers of MATa cells deficient for Axl1p and Ste23p that were transformed with either an empty expression vector or a vector encoding IDE or IDEK898A,K899A,S901A as indicated. Mating, which leads to colony growth, is dependent on production of mature a-factor pheromone by plasmid-encoded IDE. C, spot halo assay for a-factor production. Concentrated samples of a-factor recovered from yeast bearing the indicated expression vector were spotted at the indicated dilutions on a lawn of RC757 MATα cells, which undergo growth arrest upon exposure to a-factor pheromone. The zones of growth arrest are the dark circular regions within the yeast lawn.
DISCUSSION
ATP-binding Site of IDE
A number of observations suggest that the site identified by crystallography is indeed the site where ATP and presumably other anions bind. Mutating nearby basic residues, as well as Ser-901, in the IDER429S and IDEK898A,K899A,S901A variants profoundly affects the level of heterotropic activation by ATP. In addition, these mutations affect the interaction of TNP-ATP with the enzyme. The fluorescent yield from the TNP group is altered in the mutant IDE constructs, suggesting a change in the environment local to the binding site. Also, we find that PPPi and NaCl can completely compete off the bound TNP-ATP from the wild type enzyme but not with the two IDE mutants, particularly IDEK898A,K899A,S901A. This result again suggests a change in the local environment, in this case one that increases the nonpolar nature of the TNP interaction, which would then not be disrupted by an increase in ionic strength. The conversion of nearby charged and polar residues to nonpolar alanine in the IDEK898A,K899A,S901A mutant could arguably result in just that effect. Finally, a structural similarity search (DaliLite version 3) using domains 3 and 4 of IDE identifies a number of related enzymes that have nucleotide substrates (supplemental Fig. S3A). In particular, IDE has structural similarity with polymerases and other nucleotidyltransferases, adenylyl cyclase, and GTP cyclohydrolase III, all palm domain-containing enzymes (52), and the nucleotide-binding positions in these enzymes at least partially overlap the identified ATP-binding site in IDE.
We anticipated that mutating IDE residues in the polyphosphate-binding site would greatly decrease affinity and thus alter the concentration dependence of activation by ATP. The mutations do decrease triphosphate binding, another observation that supports correct identification of the site, but the affinity loss is less than anticipated. One explanation for this unexpected observation is that there may be alternative binding interactions that compensate for these mutations. As noted, the proposed ATP/anion-binding site is located in a larger positively charged inner surface formed by domains 3 and 4 (supplemental Fig. S3B). Given the size of the electropositive patch, there are a number of potential sites for ATP/polyphosphate binding in addition to the one identified crystallographically, and this likely holds true for other anions as well. The presence of alternative sites is supported by computational docking of triphosphate to the surface, which finds a number of binding modes clustered not only at the identified site but also at two nearby locations (supplemental Fig. S3C). The estimated interaction strengths differ by less than 0.7 kcal/mol for all 18 docked triphosphate groups shown, indicating that other sites might have affinities close to that of the primary site.
We propose then that we have identified a preferred binding site that under crystallization solution conditions has higher occupancy than other possible sites. Binding to this site would reduce affinity for the nearby sites by making the surrounding surface less electropositive, resulting in the observed one-to-one binding stoichiometry (21). Mutating this site, however, would likely allow alternative binding to nearby sites at a not greatly diminished affinity, giving activation over a similar concentration range. In this model, binding at the specific site would activate IDE to a greater extent than binding at nonspecific sites. This would account for the sharp decrease in the extent of activation when the identified site is ablated. Activation would still occur but only to the lower extent supported by binding to an alternative site or sites.
The greater activity of wild type IDE over IDEK898A,K899A,S901A in the yeast assays suggests that anions are important in determining the level of IDE activity in vivo. Typical intracellular nucleoside triphosphate concentrations (53) match the observed activation range for IDE (21, 22), but most nucleotides in the cell are complexed with divalent cations, likely reducing their effectiveness in activating the enzyme (21). Also, the low millimolar chloride concentrations in the cytosol are insufficient to play a significant role in IDE activation based on our results. Other small anionic species present in cells (54), including inositol and other sugar phosphates, small glycosaminoglycans, metabolic intermediates (pyruvate, lactate, malate, oxaloacetate, etc.), and peptides, may contribute, however. Glutathione, which carries a net negative charge, may be a contributor to activation depending on binding affinity, because it is present in the cytosol at millimolar concentrations. Activation of the insulin receptor transiently increases the intracellular concentration of glucose 6-phosphate in animal cells, and a role for this anion in IDE activation would be attractive from a regulatory perspective. Preliminary assays indicate that 1 mm glucose 6-phosphate produces a 2-fold activation of IDE on a small substrate, suggesting it may indeed contribute to intracellular IDE activity. It should be noted, however, that anionic activation of IDE may be constitutive and not regulatory. Further work will be required to determine the anion or anions primarily responsible for IDE activation in cells and whether their levels vary sufficiently to modulate IDE activity.
Determining the relevance of enhanced IDE activity will also require additional studies, particularly because IDE is known to be involved in a number of physiological functions. In vitro, ATP/polyphosphate does not greatly enhance IDE activity toward two important large substrates, insulin and Aβ (21, 22, 55). However, identified activators that appear to bind at the ATP/anion site have been shown to stimulate Aβ hydrolysis by IDE when another competing peptide is present (55). It may be that in the complex multisubstrate environment of the cell, anions increase hydrolysis of insulin and Aβ in the same manner.
Potential Activation Mechanism
The location of the ATP/anion-binding site determined in this work suggests a possible mechanism for activation by polyphosphate/polyanion binding. Previous studies by Tang and co-workers (17, 18) have indicated that the proposed hinge-like conformational change necessary for substrate entry/product release is likely rate-limiting. Heterotropic activation may occur through a shift in the conformational equilibrium toward the open state of the enzyme, because ATP binding is accompanied by an increase in the hydrodynamic radius of IDE (18). The inner surface of the IDE substrate-binding chamber contributed by domains 3 and 4 carries a strong positive electrostatic potential, whereas the inner surface contributed by domains 1 and 2 is strongly negative (see supplemental Fig. S3B). Thus binding of anions to the positively charged half of the chamber likely reduces the electrostatic attraction between the two chamber walls, increasing the open state population. This electrostatic mechanism is consistent with the activation of IDE by salt, because higher ionic strength would also reduce the electrostatic attraction between the two walls of the binding chamber. In fact, high concentrations of the small chloride ion may shield the attractive interactions of the two surfaces more effectively than larger polyanions, resulting in the observed greater activation. This mechanism also may explain why IDEK898A,K899A,S901A, which reduces the overall charge on the positively charged half of the chamber, shows an ∼3-fold higher kcat than the wild type enzyme. In addition, the increased thermal motion seen for the catalytic domain in the crystal structure may result from a loosening of the interface between the two halves of IDE. The lack of polyphosphate/polyanion activation for large substrates may reflect their ability to shield electrostatic attraction between the chamber walls. Larger substrates may also block polyphosphate/anion binding to the specific site.
The conformation of the structurally related enzyme pitrilysin is of interest regarding the proposed activation mechanism. The two inner chamber walls of this enzyme (Protein Data Bank accession code 1Q2L) both carry an overall negative surface charge, and it has been crystallized only in the open conformation. Thus, in pitrilysin repulsion between two negatively charged surfaces likely plays a role in stabilizing the open form, just as attraction between two oppositely charged surfaces may stabilize the closed form of IDE.
Im et al. (18) reported that ATP induced a conformational change in human IDE, specifically an increase in the β-sheet character as determined by CD spectroscopy. Although we noted increased thermal factors largely in the catalytic domain, we did not detect any significant change in the average IDE structure when ATP was bound. Interestingly, triphosphate, which is as effective as ATP at increasing IDE activity (Fig. 4) (21), did not produce the same conformational changes as ATP (18). This suggests that the conformational effects produced by ATP as observed by Im et al. (18) are likely not related to its ability to increase substrate hydrolysis.
Supplementary Material
Acknowledgments
We acknowledge RapiData 2007 and the staff at Beamline X4 at NSLS at Brookhaven National Laboratory for their assistance with the x-ray data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants DA02243, NS38041, DA016176, T32 DA016176, GM067092, and P20 RR20171.

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (code 3TUV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IDE
- insulin-degrading enzyme (or insulysin)
- Aβ
- amyloid β peptide
- TNP-ATP
- 2′,3′-O-(2,4,6-trinitrophenyl)adenosine-5′-triphosphate
- Abz
- ortho-aminobenzoic acid
- EDDnp
- N-(ethylenediamine)-2,4-dinitrophenyl.
REFERENCES
- 1. Goldstein B. J., Livingston J. N. (1981) Metabolism 30, 825–835 [DOI] [PubMed] [Google Scholar]
- 2. Yonezawa K., Yokono K., Yaso S., Hari J., Amano K., Kawase Y., Sakamoto T., Shii K., Imamura Y., Baba S. (1986) Endocrinology 118, 1989–1996 [DOI] [PubMed] [Google Scholar]
- 3. Hari J., Shii K., Roth R. A. (1987) Endocrinology 120, 829–831 [DOI] [PubMed] [Google Scholar]
- 4. McDermott J. R., Gibson A. M. (1997) Neurochem. Res. 22, 49–56 [DOI] [PubMed] [Google Scholar]
- 5. Kurochkin I. V., Goto S. (1994) FEBS Lett. 345, 33–37 [DOI] [PubMed] [Google Scholar]
- 6. Miller B. C., Eckman E. A., Sambamurti K., Dobbs N., Chow K. M., Eckman C. B., Hersh L. B., Thiele D. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6221–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E. A., Frosch M. P., Eckman C. B., Tanzi R. E., Selkoe D. J., Guenette S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vepsäläinen S., Parkinson M., Helisalmi S., Mannermaa A., Soininen H., Tanzi R. E., Bertram L., Hiltunen M. (2007) J. Med. Genet. 44, 606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Misbin R. I., Almira E. C., Duckworth W. C., Mehl T. D. (1983) Endocrinology 113, 1525–1527 [DOI] [PubMed] [Google Scholar]
- 10. Reiser M., Bernstein H. G., Ansorge S., Dorn A. (1987) Acta Histochem. 82, 35–39 [DOI] [PubMed] [Google Scholar]
- 11. Safavi A., Miller B. C., Cottam L., Hersh L. B. (1996) Biochemistry 35, 14318–14325 [DOI] [PubMed] [Google Scholar]
- 12. Bennett R. G., Fawcett J., Kruer M. C., Duckworth W. C., Hamel F. G. (2003) J. Endocrinol. 177, 399–405 [DOI] [PubMed] [Google Scholar]
- 13. Morita M., Kurochkin I. V., Motojima K., Goto S., Takano T., Okamura S., Sato R., Yokota S., Imanaka T. (2000) Cell Struct. Funct. 25, 309–315 [DOI] [PubMed] [Google Scholar]
- 14. Kupfer S. R., Wilson E. M., French F. S. (1994) J. Biol. Chem. 269, 20622–20628 [PubMed] [Google Scholar]
- 15. Li Q., Ali M. A., Cohen J. I. (2006) Cell 127, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q., Krogmann T., Ali M. A., Tang W. J., Cohen J. I. (2007) J. Virol. 81, 8525–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen Y., Joachimiak A., Rosner M. R., Tang W. J. (2006) Nature 443, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Im H., Manolopoulou M., Malito E., Shen Y., Zhao J., Neant-Fery M., Sun C. Y., Meredith S. C., Sisodia S. S., Leissring M. A., Tang W. J. (2007) J. Biol. Chem. 282, 25453–25463 [DOI] [PubMed] [Google Scholar]
- 19. Noinaj N., Bhasin S. K., Song E. S., Scoggin K. E., Juliano M. A., Juliano L., Hersh L. B., Rodgers D. W. (2011) PLoS One 6, e20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song E. S., Juliano M. A., Juliano L., Hersh L. B. (2003) J. Biol. Chem. 278, 49789–49794 [DOI] [PubMed] [Google Scholar]
- 21. Song E. S., Juliano M. A., Juliano L., Fried M. G., Wagner S. L., Hersh L. B. (2004) J. Biol. Chem. 279, 54216–54220 [DOI] [PubMed] [Google Scholar]
- 22. Camberos M. C., Pérez A. A., Udrisar D. P., Wanderley M. I., Cresto J. C. (2001) Exp. Biol. Med. 226, 334–341 [DOI] [PubMed] [Google Scholar]
- 23. Yao H., Hersh L. B. (2006) Arch. Biochem. Biophys. 451, 175–181 [DOI] [PubMed] [Google Scholar]
- 24. Del Carmen Camberos M., Cresto J. C. (2007) Exp. Biol. Med. 232, 281–292 [PubMed] [Google Scholar]
- 25. Song E. S., Daily A., Fried M. G., Juliano M. A., Juliano L., Hersh L. B. (2005) J. Biol. Chem. 280, 17701–17706 [DOI] [PubMed] [Google Scholar]
- 26. Song E. S., Mukherjee A., Juliano M. A., Pyrek J. S., Goodman J. P., Jr., Juliano L., Hersh L. B. (2001) J. Biol. Chem. 276, 1152–1155 [DOI] [PubMed] [Google Scholar]
- 27. Song E. S., Rodgers D. W., Hersh L. B. (2011) J. Biol. Chem. 286, 13852–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heras B., Martin J. L. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 29. Rodgers D. W. (1997) Methods Enzymol. 276, 183–203 [DOI] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 34. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins Struct. Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 35. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 36. Harder K. W., Owen P., Wong L. K., Aebersold R., Clark-Lewis I., Jirik F. R. (1994) Biochem. J. 298, 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim S., Lapham A. N., Freedman C. G., Reed T. L., Schmidt W. K. (2005) J. Biol. Chem. 280, 27481–27490 [DOI] [PubMed] [Google Scholar]
- 38. Alper B. J., Nienow T. E., Schmidt W. K. (2006) Biochem. J. 398, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michaelis S., Herskowitz I. (1988) Mol. Cell. Biol. 8, 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chan R. K., Otte C. A. (1982) Mol. Cell. Biol. 2, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marcus S., Xue C. B., Naider F., Becker J. M. (1991) Mol. Cell. Biol. 11, 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. (1997) Nucleic Acids Res. 25, 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., Brodsky J. L. (2001) Mol. Biol. Cell 12, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujimura-Kamada K., Nouvet F. J., Michaelis S. (1997) J. Cell Biol. 136, 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trott O., Olson A. J. (2010) J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Unni S., Huang Y., Hanson R. M., Tobias M., Krishnan S., Li W. W., Nielsen J. E., Baker N. A. (2011) J. Comput. Chem. 32, 1488–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahmadian M. R., Stege P., Scheffzek K., Wittinghofer A. (1997) Nat. Struct. Biol. 4, 686–689 [DOI] [PubMed] [Google Scholar]
- 50. Nadanaciva S., Weber J., Wilke-Mounts S., Senior A. E. (1999) Biochemistry 38, 15493–15499 [DOI] [PubMed] [Google Scholar]
- 51. Dixon M., Webb E. C. (1979) The Enzymes, 3rd Ed., pp. 381–399, Academic Press, New York [Google Scholar]
- 52. Beese L. S., Derbyshire V., Steitz T. A. (1993) Science 260, 352–355 [DOI] [PubMed] [Google Scholar]
- 53. Beis I., Newsholme E. A. (1975) Biochem. J. 152, 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones L. S., Yazzie B., Middaugh C. R. (2004) Mol. Cell. Proteomics 3, 746–769 [DOI] [PubMed] [Google Scholar]
- 55. Cabrol C., Huzarska M. A., Dinolfo C., Rodriguez M. C., Reinstatler L., Ni J., Yeh L. A., Cuny G. D., Stein R. L., Selkoe D. J., Leissring M. A. (2009) PLoS One 4, e5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.