Abstract
Genetic nonself recognition systems such as vegetative incompatibility operate in many filamentous fungi to regulate hyphal fusion between genetically dissimilar individuals and to restrict the spread of virulence-attenuating mycoviruses that have potential for biological control of pathogenic fungi. We report here the use of a comparative genomics approach to identify seven candidate polymorphic genes associated with four vegetative incompatibility (vic) loci of the chestnut blight fungus Cryphonectria parasitica. Disruption of candidate alleles in one of two strains that were heteroallelic at vic2, vic6, or vic7 resulted in enhanced virus transmission, but did not prevent barrage formation associated with mycelial incompatibility. Detailed characterization of the vic6 locus revealed the involvement of nonallelic interactions between two tightly linked genes in barrage formation, heterokaryon formation, and asymmetric, gene-specific influences on virus transmission. The combined results establish molecular identities of genes associated with four C. parasitica vic loci and provide insights into how these recognition factors interact to trigger incompatibility and restrict virus transmission.
ENDOGENOUS mycoviruses have been reported to alter the ability of plant pathogenic fungi to cause disease (reviews by Nuss 2005, 2010; Ghabrial and Suzuki 2009; Pearson et al. 2009). These mycovirus infections generally result in reduced virulence, termed hypovirulence, and offer the potential for development of biological control strategies for a range of pathogenic fungi. Unlike viruses of plants and animals, viruses of fungi uniformly lack an extracellular phase in their replication cycle (Wickner 2001). Transmission is limited to intracellular mechanisms such as cytoplasmic exchange during hyphal anastomosis (fusion of hyphae) or transmission through asexual spores. While mycoviruses are able to move freely through the hyphal network that comprises a fungal colony, transmission between different strains of the same fungal species is often regulated by a genetic nonself recognition system governed by vegetative incompatibility (Glass and Kuldau 1992; Leslie 1993; Glass et al. 2000; Saupe 2000). Vegetative incompatibility may be evident when mycelia of genetically distinct conspecific strains grow together and form a barrage, or demarcation line, along the zone of contact. It can also be assayed by allowing conspecific strains to fuse to form a heterokaryon (cells having more than one nuclear type); incompatible heterokaryons grow slowly and with an abnormal morphology or exhibit no growth. While often used interchangeably, barrage formation and heterokaryon incompatibility are not necessarily concordant, but both represent manifestations of vegetative incompatibility (review in Smith et al. 2006) wherein incompatible interactions result in localized programmed cell death (PCD) (Jacobson et al. 1998; Biella et al. 2002) that restricts exchange of cellular contents (Leslie and Zeller 1996) and transmission of viruses, transposable elements, and senescence plasmids (Caten 1972; Hartl et al. 1975).
The influence of fungal vegetative incompatibility on the transmission and efficacy of virulence-attenuating mycoviruses for biological control has been studied most extensively for hypoviruses responsible for hypovirulence of the chestnut blight fungus Cryphonectria parasitica. Results of field studies conducted in both Europe and North America generally indicate that hypovirus transmission and biological control are more effective in C. parasitica populations that exhibit lower vegetative incompatibility diversity (Anagnostakis et al. 1986; Heiniger and Rigling 1994; Robin et al. 2000, 2009; Milgroom and Cortesi 2004). The vegetative incompatibility system in C. parasitica is controlled by at least six genetic vegetative incompatibility (vic) loci with only two alleles known at each locus (Anagnostakis 1982b; Huber 1996; Cortesi and Milgroom 1998). Hyphae of C. parasitica strains that contain the same alleles at all vic loci freely fuse and support virus transmission. Hyphae of strains that contain different alleles at one or more vic loci undergo incompatible reactions that result in cell death and restriction of virus transmission. Genotypes corresponding to all combinations of alleles at six vic loci have been identified and are represented by a collection of corresponding tester stains (Cortesi and Milgroom 1998). This has allowed analysis of the influence of differences at specific vic loci on virus transmission, leading to the confirmation of allele-specific influences on the frequency and symmetry of virus transmission (Huber and Fulbright 1994; Cortesi et al. 2001). These differences are correlated with allele-specific differences in the rate of PCD associated with incompatible interactions, in which delayed PCD allows greater virus transmission (Biella et al. 2002).
Although vegetative incompatibility is a common phenomenon in filamentous ascomycete fungi, only a limited number of genes controlling this process have been characterized at the molecular level in two species: Neurospora crassa and Podospora anserina (reviewed in Saupe et al. 2000; Glass and Dementhon 2006; Pinan-Lucarre et al. 2007; Paoletti and Saupe 2009). Heterokaryon incompatibility (het) genes in these two species are characterized by significant allelic polymorphism and generally encode proteins that contain a conserved HET domain (Pfam:PF06985). The variable protein domains determine specificity while the HET domain may transduce the recognition signal to activate PCD (Paoletti and Clavé 2007). The variable and HET domains can be incorporated into the same protein or reside on tightly linked gene products (Glass and Dementhon 2006; Kaneko et al. 2006; Micali and Smith 2006; Paoletti and Saupe 2009).
Five of the C. parasitica vic loci have been linked to molecular markers on a genetic linkage map (Kubisiak and Milgroom 2006). We hypothesized that, if the C. parasitica vic system resembles the nonself recognition systems that operate in N. crassa and P. anserina, then the C. parasitica vic loci should be identifiable as regions of hypervariability located near the linked markers. We confirmed this prediction by identifying seven candidate polymorphic genes associated with four vic loci through comparative analysis of the genome sequences of two C. parasitica strains, EP155 (reference genome sequence) and EP146, that were genetically determined to have allelic differences at vic2, vic4, vic6, and vic7. A role in restriction of virus transmission was demonstrated by disruption of the polymorphic candidate genes associated with the vic loci previously implicated by genetic analysis as restricting virus transmission, vic2, vic6, and vic7 (Cortesi et al. 2001). Nonallelic interactions between two tightly linked genes at the vic6 locus were shown to trigger incompatibility and influence the frequency and symmetry of virus transmission. RNA silencing was recently shown to serve as an effective antiviral defense mechanism in C. parasitica (Segers et al. 2007; Zhang et al. 2008; Sun et al. 2009b). The results of this study also strengthen an emerging view of the complementary nature of RNA silencing and the vic system in fungal antiviral defense at the cellular and population levels, respectively.
Materials and Methods
Fungal strains and growth conditions
The C. parasitica strains used in this study (Table 1) were maintained on potato dextrose agar (PDA; Difco, Detroit) at 22°–24° with a 12-h/12-h light/dark cycle and a light intensity of 1300–1600 lx. EP155 (ATCC 38755), used for generation of the reference C. parasitica genome sequence (http://genome.jgi-psf.org/Crypa2/Crypa2.home.html), is a virulent, hypovirus-free orange pigment-producing strain isolated by Sandra Anagnostakis (Connecticut Agricultural Experiment Station, New Haven, CT) in 1977 from a canker on Castanea dentata (Marshall) Borkh. in a field plot in Bethany, Connecticut. Strain EP146 (ATCC 64671) is a virulent, hypovirus-free, brown pigment-producing strain isolated in 1976 from the George Washington National Forest near Franklin, West Virginia by William MacDonald (West Virginia University, Morgantown, WV). This strain was chosen for resequencing because of mating type and pigmentation differences with the reference genome strain EP155 and previous use in laboratory and field studies. (Chen et al. 1993; Root et al. 2005).
Table 1 . Cryphonectria parasitica strains used in this study.
| Strain | Characteristic(s) | Source/reference |
|---|---|---|
| EP155 | Mating type MAT-2; vic genotype 2211-22 | ATCC 38755 |
| EP146 | Mating type MAT-1; vic genotype 2112-11 | ATCC 64671 |
| DK80 | EP155 genetic background; Δcpku80 | Lan et al. (2008) |
| DK80hygR | DK80 with hygromycin resistance | This study |
| DK80neoR | DK80 with G418 resistance | This study |
| DK80 Δvic2-2 | DK80 background; vic2-2::hygR | This study |
| DK80 Δvic6-2 | DK80 background; vic6-2::hygR | This study |
| DK80 Δvic7-2 | DK80 background; vic7-2::hygR | This study |
| DK80 Δpix6-2 | DK80 background; pix6-2::hygR | This study |
| DK80 Δpix6-2 Δvic6-2 | DK80 background; pix6-2::neoR, vic6-2::hygR | This study |
| EU-5 | Tester strain for vc type EU-5 (isolate P1-11), vic genotype 2211-22 | Cortesi and Milgroom (1998) |
| EU-6 | Tester strain for vc type EU-6 (isolate P1-6), vic genotype 2111-22 | Cortesi and Milgroom (1998) |
| EU-18 | Tester strain for vc type EU-18 (isolate P24-33), vic genotype 2211-21 | Cortesi and Milgroom (1998) |
| EU-21 | Tester strain for vc type EU-21 (isolate P10-18), vic genotype 2211-12 | Cortesi and Milgroom (1998) |
| EU-21hygR | EU-21 with hygromycin resistance | This study |
| EU-21neoR | EU-21 with G418 resistance | This study |
| EU-21 Δvic6-1 | EU-21 background; vic6-1::neoR | This study |
| EU-21 Δpix6-1 | EU-21 background; pix6-1::neoR | This study |
vic genotypes are abbreviated as the alleles at each of the six known vic loci, e.g., 2211-22 is the abbreviation for genotype vic1-2, vic2-2, vic3-1, vic4-1, vic6-2, vic7-2 (Cortesi and Milgroom 1998).
Vegetative incompatibility genotyping
Vegetative incompatibility genotyping was performed by pairing the C. parasitica strain of interest with each of the 64 European (EU) tester strains described by Cortesi and Milgroom (1998) on agar medium amended with bromocresol green pH indicator dye as described (Cortesi and Milgroom 1998).
Virus transmission assay
Virus transmission was assayed by placing a donor strain infected with hypovirus CHV-1/EP713 and a virus-free recipient strain 1 cm apart on PDA and incubating them at 22°–24° for 7 days. Virus transmission to the recipient strain can be detected by phenotypic conversion of the expanding edge of the recipient strain colony following contact of the two strains (see Figure 1 of Cortesi et al. 2001). Hypovirus CHV-1/EP713 infection was established in the different donor strains tested in this study by transfection of fungal spheroplasts with hypovirus CHV-1/EP713 transcripts generated in vitro from a full-length viral cDNA clone using the protocol developed by Chen et al. (1994).
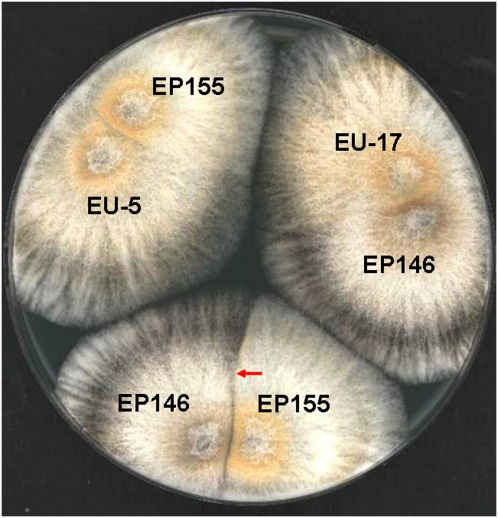
Figure 1 .
Vegetative incompatibility (vic) genotyping of C. parasitica strain EP146. Strains of C. parasitica that differ at one or more vic loci form barrage lines (a line of dead cells) when colonies of the two strains merge as observed for the pairing of strains EP155 and EP146 (barrage line indicated by red arrow at bottom of the culture plate). Cortesi and Milgroom (1998) established a collection of 64 C. parasitica tester strains that represent all possible genotypes arising from the six genetically determined vic loci. Strain EP155, the strain used for generation of the reference genome sequence (http://genome.jgi-psf.org/Crypa2/Crypa2.home.html), was previously reported (Cortesi and Milgroom 1998) to give a compatible reaction (fusion of hyphae with no barrage formation) with tester strain EU-5 (vic genotype 2211-22) as shown at the left of the culture plate. Resequenced strain EP146 was found to be compatible with tester strain EU-17, as shown at the right side of the culture plate, and thus to have the vic genotype 2112-11.
Heterokaryon incompatibility assay
Heterokaryon incompatibility assays were done as described by Smith et al. (2006) with slight modifications. Briefly, strains were paired by placing 1- to 2-mm3 blocks of agar containing actively growing mycelium ∼2 mm apart on a cellophane membrane overlaid on top of PDA. The plates were incubated at 30° for 24 hr or until the paired colony margins overlapped by ∼1 mm at which time the membrane with the paired colonies was transferred to PDA+hyg+G418 [PDA, 30 μg/ml hygromycin B (Roche, Laval, QC, Canada), and 20 μg/ml G418 (an analog of neomycin: BioShop Canada, Burlington, ON, Canada)]. The double-selection plates were incubated an additional 3–4 days at 30° before observation for heterokaryotic outgrowths. Mycelium from the edge of outgrowths or from the zone of confluence of paired colonies was subsequently transferred to PDA+hyg+G418 for growth rate measurements.
Genome sequencing and analysis
Genome sequence data for strain EP155, including gene models and predicted protein functional information, were obtained from the Joint Genome Institute of the U.S. Department of Energy (http://genome.jgi.psf.org/Crypa2/Crypa2.download.ftp.html). Data for strain EP146 were generated in the New Mexico State University (NMSU) Genome Sequencing Laboratory according to the Roche 454 GS FLX Titanium protocols for library preparation, emulsion PCR reaction, and sequencing (Margulies et al. 2005). Subsequently, quality-screened reads were mapped against the EP155 genome with Roche gsMapper v.2.5p1 software. Combined gsMapper alignments and EP155 genome data were then used to create a web-based interface using the freely available Generic Genome Browser (GBrowse) from the Generic Model Organism Database (http://gmod.org). Homology searches using specific gene sequences were performed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). GenBank accession numbers for the candidate vic genes are as follows: vic6-1, JN367268; vic6-2, JN256666; pix6-1, JN367270; pix6-2, JN367269; vic2-1, JN367272; vic2-2, JN367271; vic2a-1, JN367274; vic2a-2, JN367273; vic4-1, JN367275; and vic4-2, JN367276.
Small-scale genomic DNA isolation and disruption of candidate vic genes
C. parasitica strains were cultured on cellophane-overlaid PDA plates until the colony size became ∼1.5–2 cm in diameter. Mycelia were gently removed and transferred to 2-ml screw-capped Sarstedt tubes containing 0.5 g of 0.5-mm zirconium beads in 400 μl extraction buffer (40 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 0.5% SDS). The tubes were shaken for 30 sec at 5000 rpm in a Mini BeadBeater (Biospec Products, Bartlesville, OK), cooled in ice, and subjected to two additional rounds of shaking and cooling. The samples were extracted with phenol/chloroform/iso-amyl alcohol (25/24/1) and the aqueous phase was collected for ethanol precipitation. After centrifugation, pellets were resuspended in 10–40 μl of water depending on the pellet size.
Disruption of the candidate vic genes was performed according to the PCR-based strategy (Kuwayama et al. 2002; Sun et al. 2009a) with modifications. Three PCR products, which included a selectable marker and two flanking target gene-specific sequences, were generated in the first round of PCR. A second round of PCR was performed (30 cycles consisting of denaturation at 98° for 5 sec, annealing at 64° for 20 sec, and extension at 72° for 1.5 min) to assemble the three fragments using Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA) and two end oligonucleotide primers corresponding to the flanking gene-specific sequences. Transformation was performed according to the method of Churchill et al. (1990), followed by selection of putative transformants in the presence of 40 μg/ml of hygromycin or 20 μg/ml of G418. Putative disruptants were placed under intense constant light condition (∼4000 lx) to promote asexual sporulation (Hillman et al. 1990) followed by the selection of uninucleate single conidial isolates on antibiotic-containing PDA to eliminate heterokaryons. Disruption of the candidate vic genes was confirmed in single-spore transformants by PCR analyses.
Results
Genotyping EP146 vic genes
Cortesi and Milgroom (1998) established a collection of 64 European C. parasitica isolates or “tester strains” that represent all possible genotypes arising from the six genetically identified vic loci, each with two alleles. C. parasitica strain EP155 (see Table 1), the strain used to generate the reference genome sequence (http://genome.jgi-psf.org/Crypa2/Crypa2.home.html), was previously shown to be compatible with tester strain EU-5 and thus have the genotype 2211-22, where each number refers to the allele present at the respective vic loci, i.e., vic1-2, vic2-2, vic3-1, vic4-1, vic6-2, and vic7-2 (Cortesi and Milgroom 1998). The vic genotype for resequenced C. parasitica strain EP146 was determined by pairing with each of the 64 tester strains. Compatibility was observed with tester strain EU-17 (Figure 1) that has the vic genotype 2112-11. Thus, strain EP146 differs from the reference strain EP155 at four vic loci: vic2, vic4, vic6, and vic7.
Polymorphism-based identification of C. parasitica vic gene alleles
The EP146 genome was sequenced to ∼11× coverage (486.6 Mbp) using Roche 454 high-throughput sequencing technology. Raw reads (average length of 384 bp) were individually mapped onto the scaffolds composing the C. parasitica strain EP155 reference genome sequence within a searchable genome browser.
Kubisiak and Milgroom (2006) reported the construction of a C. parasitica linkage map based on a cross between Japanese isolate JA17 and Italian isolate P17-8 and identified a number of markers linked to five of the known vic loci: vic1, vic2, vic4, vic6, and vic7. In this study, we updated the published linkage map by adding simple sequence repeat (SSR) markers mined from the EP155 reference genome sequence (Supporting Information, Table S1), using methods described in Kubisiak et al. (2007). We used the vic-linked markers generated in the JA17 × P17-8 mapping population as starting reference positions on the EP155 reference genome sequence assembly to screen for sequence polymorphisms with the 454-generated EP146 genome sequence that could correspond to candidate vic alleles.
vic2:
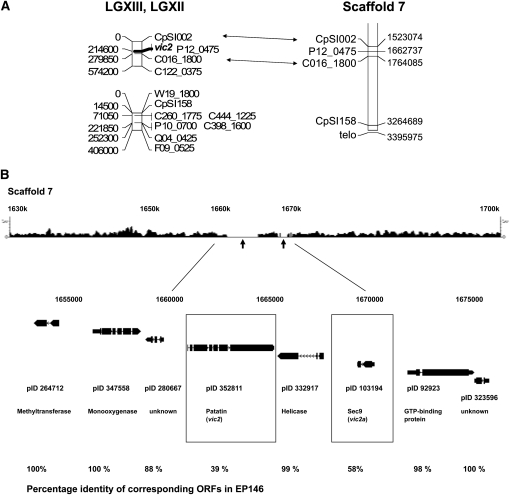
Sequences corresponding to two markers in linkage group (LG)XIII shown previously to be closely linked to the vic2 locus were identified on Scaffold 7 of the EP155 genome sequence assembly, version 2. The vic2 locus was mapped 4.5 cM from marker C016_1800 on one side and 14.8 cM from SSR marker CpSI002 on the other side and found to cosegregate with marker P12_0475 (Figure 2A). Only one region of significant sequence polymorphism was identified between the CpSI002 and C016_1800 linkage markers on Scaffold 7. This consisted of a 10-kbp region that extended from position 1,661,000 to 1,671,000 and contained the vic2 cosegregating marker P12_0475 at position 1,662,737. This 10-kbp polymorphic region contained two open reading frames (ORFs), one encoding a member of the patatin-like phospholipase family [Pfam 01734 (Andrews et al. 1988)] and the other a protein related to a fungal plasma membrane SNARE Sec9 protein (reviewed in Hong 2005; Jahn and Scheller 2006) (Figure 2B). Remarkably, a comparison of the predicted amino acid sequences for these ORFs from strains EP155 and EP146 revealed that both were highly polymorphic [39% and 58% identity, respectively (Figure S1 and Figure S2)]. In contrast, the 2.7-kbp sequence separating the two ORFs showed only a moderate level of single-nucleotide polymorphisms and contained a 444-codon ORF encoding a helicase-like protein that was 99% identical in EP155 and EP146; high levels of sequence similarity were also found for proteins flanking this region. The patatin-like and sec9-like genes in strain EP155 were designated vic2-2 and vic2a-2, respectively, while these genes in strain EP146 were designated candidate vic alleles vic2-1 and vic2a-1, respectively.
Figure 2 .
The candidate vic2 locus. (A) Alignment of LGXII and LGXIII of the C. parasitica genetic linkage map generated by Kubisiak and Milgroom (2006), the latter of which contains the position of the vic2 locus, with Scaffold 7 of version 2 of the C. parasitica genome sequence. The estimated physical distance (base pairs) of each marker from the end of the linkage group (shown on the left side of the linkage map) was calculated using the estimate of 1 cM = 14.5 kbp (Kubisiak and Milgroom 2006). Markers linking the physical and genetic maps are connected by double arrows. (B) The region of sequence polymorphism for C. parasitica strains EP155 and EP146 and machine-annotated ORFs located near the predicted vic2 locus. The top track shows the density plot of the 454 sequence reads (scale indicated at both ends) that match a corresponding portion of Scaffold 7 of the EP155 reference genome sequence assembly. The 10-kbp region of polymorphism between the EP155 and EP146 genome sequences in the region mapped for the vic2 locus is indicated by the absence of matching 454 sequence reads (arrows). The ORFs located within a 23-kbp region containing the polymorphic locus, with accompanying protein ID numbers, are shown below the 454 sequence read track. The candidate vic2 genes vic2-2 (patatin-like protein, pID 352811) and vic2a-2 (sec9-like protein, pID103194) are indicated by boxes. The percentages of amino acid identity for the corresponding EP155 and EP146 ORFs are indicated at the bottom.
vic4:
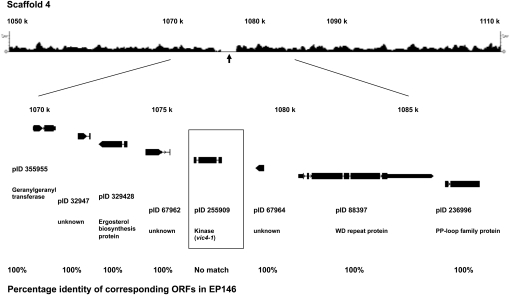
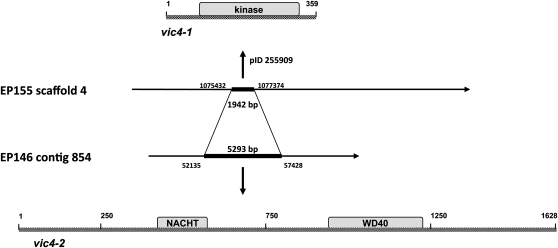
The vic4 locus was mapped between EP155 genome-specific SSR linkage markers CPG3 and CpSI116 on LGI that corresponded to positions 1,897,839 and 345,343, respectively, on Scaffold 4 (Figure S3), ∼42.4 cM from the latter. Using the same strategy as described for the vic2 locus, we identified a polymorphic region extending from position 1,075,432 to 1,077,374 that narrowly defined a single annotated ORF of 359 aa that contained a protein kinase c-like domain in the EP155 sequence (Figure 3). Interestingly, this sequence was replaced in the corresponding EP146 sequence contig by a larger ORF of 1628 aa that contained the classic NACHT-NTPase and WD repeat domains characteristic of a number of fungal heterokaryon incompatibility genes (Figure 4) (reviewed in Chevanne et al. 2010). The idiomorphic genes encoding the ORF with the protein kinase c-like domain in strain EP155 and the corresponding NACHT-NTPase/WD repeat-encoding gene in strain EP146 were designated candidate vic genes vic4-1 and vic4-2, respectively.
Figure 3 .
The candidate vic4 locus: the region of sequence polymorphism for C. parasitica strains EP155 and EP146 and machine-annotated ORFs located near the predicted vic4 locus. The ∼2-kbp region of polymorphism between the EP155 and EP146 genome sequences in the region in Scaffold 4 mapped for the vic4 locus (Figure S3) is indicated by the absence of 454 sequence reads generated from EP146 DNA that match the corresponding EP155 reference genome sequence (arrow). The open reading frames (ORFs) located within the 19-kbp region containing the polymorphic locus, with accompanying protein ID numbers and amino acid residue lengths, are shown below the 454 sequence read track. The boxed candidate vic4 allele, vic4-1 (pID 255909), in strain EP155 is absent in strain EP146, while other surrounding predicted proteins show perfect matches, as indicated at the bottom.
Figure 4 .
The candidate vic4-1 allele in EP155, which contains a fructosamine kinase/phosphotransferase enzyme family domain, is replaced by an unrelated ORF of 1628 amino acids in length in the corresponding EP146 sequence. This candidate vic4-2 allele contains NACHT and WD40 motifs reported for het genes from P. anserina (reviewed in Paoletti and Saupe 2009).
vic6:
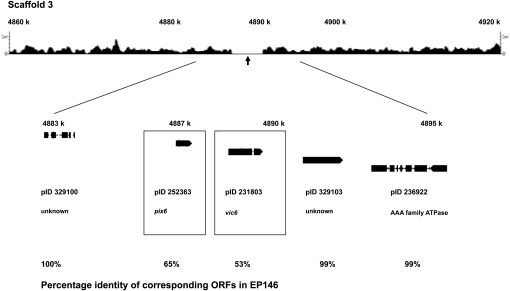
The vic6 locus was mapped by Kubisiak and Milgroom (2006) to reside 13.8 cM and 15.2 cM from linkage group XIV SSR linkage markers CpSI135 and CpSI136, respectively, which were located at positions 5,015,225 and 5,033,338, respectively, on Scaffold 3 (Figure S4). A 3710-bp region of sequence polymorphism was identified (Figure 5), extending from position 4,887,200 to 4,890,200 that coincided primarily with a single coding region of 705 amino acids with sequence similarity to N. crassa heterokaryon incompatibility gene pin-c2 on the basis of BLAST analysis (5e-43) and containing a clearly defined HET domain (PF06985) (Kaneko et al. 2006). Analysis of the EP146 sequence contig corresponding to this region of Scaffold 3 revealed an ORF at the position of nucleotide polymorphism that encoded a 729-amino-acid ortholog of the EP155 protein with a conserved HET domain but high levels of amino acid polymorphism in the N-terminal and C-terminal flanking regions (53% overall identity) (Figure S5). These ORFs in strains EP155 and EP146 were designated candidate alleles vic6-2 and vic6-1, respectively. The region of polymorphism extended upstream from vic6 into the C-terminal portion (65% identity) of a small gene of 177 codons (Figure S6). The allelic forms of this small ORF were designated pix6-2 and pix6-1 (Figure 5, partner of vicsix, based on functional analyses described below) in strains EP155 and EP146, respectively. Note that a high level of nucleotide identity is restored in the regions flanking the stretch of intense polymorphism associated with the candidate vic6 locus.
Figure 5 .
The candidate vic6 locus: the region of sequence polymorphism for C. parasitica strains EP155 and EP146 and machine-annotated ORFs located near the predicted vic6 locus (Figure S4). The ∼3.7-kbp region of polymorphism between the EP155 and EP146 genome sequences on Scaffold 3 in the region mapped for the vic6 locus is indicated by the absence of 454 sequence reads generated from EP146 DNA that match the corresponding EP155 reference genome sequence (arrow). The ORFs located within a 13.4-kbp region containing the polymorphic locus, with accompanying protein ID numbers, are shown below the 454 sequence read track. The boxed candidate vic6 gene (pID 231803), which contains a HET domain, and adjacent ORF pix6 (pID 252363) showed a greater degree of sequence polymorphism than surrounding ORFs, which showed near perfect matches.
vic7:
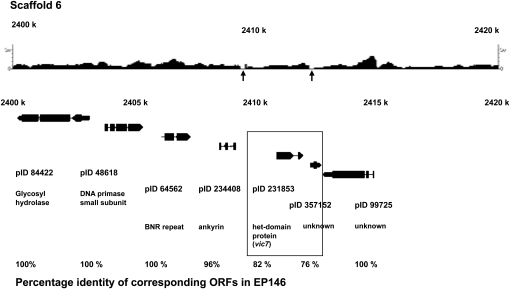
The vic7 locus was mapped 13.6 cM (∼197,200 bp) from SSR marker CpSI006, which is located on Scaffold 6 at position 2,553,152 (Figure S7). Inspection of the EP155/EP146 sequence difference browser track revealed only two small (∼200–300 bp) closely spaced regions of polymorphism located near positions 2,410,778 and 2,412,159, respectively (Figure 6). Neither region of polymorphism resided within an annotated ORF, but the first was adjacent to an ORF encoding a protein containing an ankyrin motif and the second was adjacent to a protein containing a HET motif. Further inspection determined that the ORF containing the HET domain extended in the C-terminal direction to give a predicted protein consisting of >600 amino acid residues. The corresponding alleles in strains EP155 and EP146 were 87% identical with a highly conserved HET domain and significant polymorphism limited primarily to the C-terminal domain (Figure S8). The different alleles of this candidate vic gene in strains EP155 and EP146 were designated vic7-2 and vic7-1, respectively.
Figure 6 .
The candidate vic7 locus: the region of sequence polymorphism for C. parasitica strains EP155 and EP146 and machine-annotated ORFs located near the predicted vic7 locus. Polymorphisms between the EP155 and EP146 genome sequences on Scaffold 6 in the region mapped for the vic7 locus (Figure S7) are indicated by the absence of 454 sequence reads generated from EP146 DNA that match the corresponding EP155 reference genome sequence (arrows). The ORFs located within a 20-kbp region containing the polymorphic locus, with accompanying protein ID numbers, are shown below the 454 sequence read track. The two boxed EP155 ORFs, pID 231853 and pID 357152, showed a greater degree of sequence divergence than the surrounding ORFs, which showed near perfect or perfect matches. When manually annotated, these two ORFs became one HET domain-containing ORF corresponding to candidate vic7 allele vic7-2. The percentages of amino acid identity for the corresponding EP155 and EP146 ORFs surrounding the putative vic7 locus are indicated at the bottom.
Association between polymorphism of candidate vic alleles and vic genotypes of C. parasitica field isolates
The polymorphisms associated with the putative vic loci provided the basis for PCR-based differentiation of the individual alleles for all candidate vic genes (primer information in Table S2). Allele specificity for the vic4, vic6 (both vic6 and pix6), and vic7 loci was easily determined by the relative size of the allele-specific PCR products visualized in agarose gels (Figure S9) and confirmed by sequence analysis. Nucleotide sequence differences were used to distinguish the candidate vic2-1 and vic2-2 alleles and the adjacent sec9-like gene alleles vic2a-1 and vic2a-2 associated with the vic2 locus. These PCR-based diagnostic tools were then used to compare the specificity of candidate vic allele with vic genotyping results for 26 field strains isolated from Finzel, Maryland (MD), Crevoladossala, Italy (VO), Bartow, West Virginia (BRU), and Depot Hill, New York (DU) (Milgroom and Cortesi 1999). As indicated in Table 2, complete concordance was observed between PCR-based vic allele specificity and vic tester strain-based genotyping for isolates from these four independent populations.
Table 2 . Correspondence between barrage-based vic genotype and PCR-based vic genotype in C. parasitica field isolates.
| vic2 locus | vic6 locus | ||||||
|---|---|---|---|---|---|---|---|
| Isolatea | Genotype | vic2(ptn)b | vic2a(sec9)b | vic4c | vic6c | pix6c | vic7c |
| EP155 | 2211-22 | 2 | 2 | 1 | 2 | 2 | 2 |
| EP146 | 2112-11 | 1 | 1 | 2 | 1 | 1 | 1 |
| MD3 | 2112-11 | 1 | 1 | 2 | 1 | 1 | 1 |
| MD8 | 2212-12 | 2 | 2 | 2 | 1 | 1 | 2 |
| MD11 | 2121-11 | 1 | 1 | 1 | 1 | 1 | 1 |
| MD16 | 1212-22 | 2 | 2 | 2 | 2 | 2 | 2 |
| MD18 | 2211-11 | 2 | 2 | 1 | 1 | 1 | 1 |
| MD19 | 1122-11 | 1 | 1 | 2 | 1 | 1 | 1 |
| MD22 | 2112-12 | 1 | 1 | 2 | 1 | 1 | 2 |
| MD28 | 1212-11 | 2 | 2 | 2 | 1 | 1 | 1 |
| MD33 | 2211-22 | 2 | 2 | 1 | 2 | 2 | 2 |
| MD46 | 2122-11 | 1 | 1 | 2 | 1 | 1 | 1 |
| MD52 | 1211-12 | 2 | 2 | 1 | 1 | 1 | 2 |
| MD55 | 1112-21 | 1 | 1 | 2 | 2 | 2 | 1 |
| VO1 | 2112-22 | 1 | 1 | 2 | 2 | 2 | 2 |
| VO21 | 2111-22 | 1 | 1 | 1 | 2 | 2 | 2 |
| VO59 | 2111-11 | 1 | 1 | 1 | 1 | 1 | 1 |
| BRU2 | 2212-21 | 2 | 2 | 2 | 2 | 2 | 1 |
| BRU10 | 2222-12 | 2 | 2 | 2 | 1 | 1 | 2 |
| BRU16 | 2212-22 | 2 | 2 | 2 | 2 | 2 | 2 |
| BRU17 | 2112-21 | 1 | 1 | 2 | 2 | 2 | 1 |
| BRU19 | 2211-11 | 2 | 2 | 1 | 1 | 1 | 1 |
| BRU69 | 1211-11 | 2 | 2 | 1 | 1 | 1 | 1 |
| DU14 | 1212-11 | 2 | 2 | 2 | 1 | 1 | 1 |
| DU29 | 2212-22 | 2 | 2 | 2 | 2 | 2 | 2 |
| DU63 | 2111-11 | 1 | 1 | 1 | 1 | 1 | 1 |
| DU72 | 2121-11 | 1 | 1 | 1 | 1 | 1 | 1 |
| DU74 | 2211-12 | 2 | 2 | 1 | 1 | 1 | 2 |
vic genotypes are abbreviated as the alleles at each of six known vic loci, as defined in Table 1. The PCR products for vic2(ptn) and vic2a(sec9) are both associated with the vic2 locus while the PCR products for vic6 and pix6 are both associated with the vic6 locus. PCR-based markers are not yet available for vic1 or vic3.
MD isolates are from Finzel, Maryland; VO isolates are from Crevoladossola, Italy; BRU isolates are from Bartow, West Virginia; DU isolates are from Depot Hill, New York. These isolates were used in a previous population study (Milgroom and Cortesi 1999).
vic2 candidates vic2 patatin-like protein (ptn) and vic2a Sec9-like protein (sec9): PCR products were directly sequenced to assign allele 1 or 2.
vic4, vic6, pix6, and vic7: alleles assigned on the basis of the PCR product size. In addition, for all MD isolates, vic6 and vic7 PCR products were sequenced along with several isolates from the rest of the list.
Disruption and functional analysis of candidate vic gene alleles
To define the functional role of the polymorphic C. parasitica candidate vic gene alleles in vegetative incompatibility, candidate vic alleles vic2-2, vic6-2, and vic7-2 were disrupted by transforming strain DK80, a mutant of the standard C. parasitica laboratory strain EP155 containing a disruption of the nonhomologous end-joining DNA repair pathway ku80 gene homolog, to promote homologous recombination (Lan et al. 2008). Analysis of multiple independent disruption mutant strains for each candidate vic allele failed to find any obvious changes in growth, colony morphology, sporulation, or virulence (data not shown). Multiple selected disruption mutant strains for vic2-2 (DK80 Δvic2-2), vic6-2 (DK80 Δvic6-2), and vic7-2 (DK80 Δvic7-2) were subsequently examined for the effect of allele disruption on mycelial incompatibility, characterized by the formation of barrages or lines of demarcation (Powell 1995) and mycovirus transmission (Huber and Fulbright 1994; Cortesi et al. 2001). The candidate vic4 allele was not disrupted because heteroallelism at the vic4 locus, while associated with barrage formation, does not negatively affect virus transmission (Cortesi et al. 2001).
Mycelial incompatibility was not altered by disruption of candidate vic2-2, vic6-2, or vic7-2 alleles. That is, barrage formation still occurred when these mutants were paired with the corresponding heteroallelic tester strains EU-6 (2111-22), EU-21 (2211-12), and EU-18 (2211-21), respectively (not shown).
The effect of disruption of vic allele candidates vic2-2, vic6-2, and vic7-2 on virus transmission is presented in Table 3. Hypovirus transmission occurred successfully when EP155 and the derived DK80 strains were paired as either virus donor or virus recipient, showing that the deletion of the ku80 gene homolog does not affect hypovirus transmission. Similar to previous reports (Cortesi et al. 2001), allelic differences at the vic2 locus [DK80 (2211-22) vs. EU-6 (2111-22)] significantly reduced hypovirus transmission frequency to levels of 0/20–1/20 depending on whether the vic2-2 allele is in the donor or the recipient strain, respectively. Disruption of the candidate vic2-2 allele (patatin-like gene) in strain DK80 resulted in no significant increase in transmission when the mutant strain [DK80 Δvic2-2 (2211-22), where disruption is signified by a strike, e.g., 2] was the virus donor and EU-6 (2111-22) was the recipient, but resulted in 100% transmission when the mutant strain was the recipient.
Table 3 . Hypovirus transmission frequency for disruption mutants of candidate vic alleles at vic2, vic6, and vic7.
| Donor | Recipient | Transmission | P-valuea |
|---|---|---|---|
| DK80 (2211-22) | EP155 (2211-22) | 20/20 | |
| EP155 (2211-22) | DK80 (2211-22) | 20/20 | |
| DK80 (2211-22) | EU-6 (2111-22) | 0/20 | 0.245 |
| DK80 Δvic2-2 (2211-22) | EU-6 (2111-22) | 2/20 | |
| EU-6 (2111-22) | DK80 (2211-22) | 1/20 | <0.001 |
| EU-6 (2111-22) | DK80 Δvic2-2 (2211-22) | 20/20 | |
| DK80 (2211-22) | EU-21 (2211-12) | 1/20 | 0.171 |
| DK80 Δvic6-2 (2211-22) | EU-21 (2211-12) | 4/20 | |
| EU-21 (2211-12) | DK80 (2211-22) | 3/20 | <0.001 |
| EU-21 (2211-12) | DK80 Δvic6-2 (2211-22) | 20/20 | |
| DK80 (2211-22) | EU-18 (2211-21) | 2/20 | 0.015 |
| DK80 Δvic7-2 (2211-22) | EU-18 (2211-21) | 9/20 | |
| EU-18 (2211-21) | DK80 (2211-22) | 20/20 | ND |
| EU-18 (2211-21) | DK80 Δvic7-2 (2211-22) | 20/20 | |
| DK80 Δvic6-2 (2211-22)b | EU-21 (2211-12) | 4/20 | <0.001 |
| DK80 Δvic6-2 (2211-22) | EU-21 Δvic6-1 (2211-12) | 20/20 | |
| EU-21 (2211-12)b | DK80 (2211-22) | 3/20 | <0.001 |
| EU-21 Δvic6-1 (2211-12) | DK80 Δvic6-2 (2211-22) | 20/20 |
P-values for Fisher’s exact tests comparing the frequency of virus transmission when a vic allele in one isolate is disrupted. ND, not determined.
Repeated from above for comparison with double-allele disruption mutant parings.
Similar results for virus transmission were observed for disruption of candidate vic allele vic6-2. In this case, allelic differences at the vic6 locus [DK80 (2211-22) vs. EU-21 (2211-12)] resulted in a reduction in hypovirus transmission frequency to levels of 1/20 if the recipient strain contains the vic6-1 allele and to 3/20 if the recipient strain contains the vic2-2 allele. As observed for disruption of vic2-2, disruption of the candidate vic6-2 allele resulted in no significant increase in virus transmission when the mutant strain was the donor, but resulted in 100% transmission when the mutant strain was the virus recipient.
As reported by Cortesi et al. (2001) and observed in Table 3, strains heteroallelic at the vic7 locus [DK80 (2211-22) vs. EU-18 (2211-21)] exhibit an asymmetric virus transmission pattern with 100% transmission occurring when the recipient contained the vic7-2 allele and much lower transmission occurring when the recipient contained vic7-1. Since virus transmission to a vic7-2–containing strain of a vic7 heteroallelic pair is already 100%, disruption of the candidate vic7-2 allele cannot increase acceptance of virus. However, there was a significant increase in virus transmission frequency from 2/20 to 9/20 (Table 3) when the vic7-2 mutant strain served as the virus donor.
The increased frequency of virus transmission observed for vic2-2 and vic6-2 mutant strains when serving as a recipient and the vic7-2 mutant when serving as the virus donor strain is consistent with a role for these candidate vic alleles in vegetative incompatibility, even though barrage formation is still observed. This raised the question of whether disruption of both candidate vic alleles of a heteroallelic pair would abolish barrage formation and eliminate the barrier to virus transmission.
Disruption of vic6-2 and vic6-1
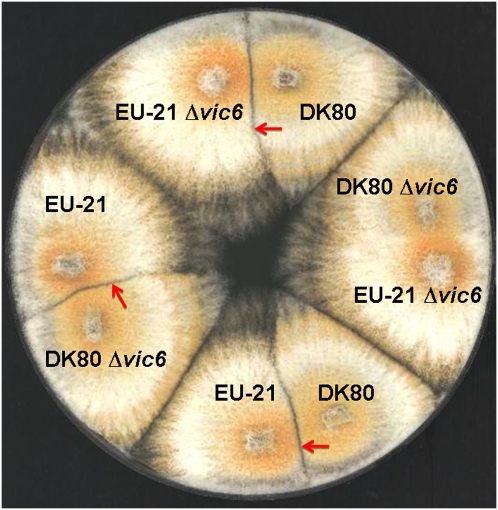
The candidate vic6-1 allele was disrupted in tester strain EU-21(2211-12) that differs in vic alleles from the DK80 strain only at the vic6 locus (Cortesi and Milgroom 1998). Disruption of either the candidate vic6-1 or the vic6-2 alleles independently resulted in no change in mycelial incompatibility (Figure 7). The DK80 Δvic6-2 mutants formed barrages when paired with the EU-21 tester strain and retained compatibility with the DK80 parent strain and EP155 (see above), while the EU-21 Δvic6-1 mutants formed barrages when paired with EP155 or DK80 and were compatible with the EU-21 parent strain (results not shown). However, barrage formation was abolished when the two mutant strains were paired (Figure 7). Additionally, the frequency of virus transmission increases to 100% when the vic6 alleles are disrupted in both DK80 and EU-21 irrespective of which mutant strain is donor or recipient (Table 3). Thus, both barrage formation and restrictions to virus transmission were eliminated when both alleles were disrupted.
Figure 7 .
Mycelial incompatibility assay for strains disrupted in candidate vic alleles vic6-1 and vic6-2 in incompatible isolates DK80 (2211-22) and tester strain EU-21 (2211-12) (barrage lines indicated by red arrows). Barrage formation resulting from an incompatible interaction occurred when vic6-1 disruption mutants [EU-21 Δvic6-1 (2211-12)] were paired with strain DK80 as shown at the top or when vic6-2 disruption mutants [DK80 Δvic6-2 (2211-22)] were paired with tester strain EU-21 as shown at the left. In contrast, barrage formation was abolished when both mutants were paired as shown at the right [DK80 Δvic6-2 (2211-22) vs. EU-21 Δvic6-1 (2211-12)].
We next tested whether vic6 disruption mutant strains could form stable heterokaryons using methods described previously (Smith et al. 2006). Heterokaryon formation results in rapidly growing sectors on double-antibiotic medium when each individual strain carries resistance to only one antibiotic. In this study, we used PDA containing hygromycin and G418 (PDA+hyg+G418) as the double-selection medium. As indicated in Table 4, heterokaryotic outgrowths were observed with strain pairs that had identical alleles at all six vic loci, e.g., DK80hygR and DK80neoR. Upon transfer of hyphae from these outgrowths to fresh (PDA+hyg+G418) medium, a sustained radial growth rate of between 6.5 and 8.0 mm per day was observed. Outgrowths were not observed for the vic6 heteroallelic pairs [DK80neoR (2211-22) + EU-21hygR (2211-12)] or [DK80hygR (2211-22) + EU-21neoR (2211-12)] or for heteroallelic pairs deleted in either vic6-1 or vic6-2 alone. In contrast, heterokaryotic sectors did develop for the pairing of EU-21 Δvic6-1 (2211-12) and DK80 Δvic6-2 (2211-22) that have their respective allelic forms (vic6-1 and vic6-2) disrupted. Hyphal transfers from the [DK80 Δvic6-2 + EU-21 Δvic6-1] outgrowths to PDA+hyg+G418 medium grew equivalently to compatible heterokaryons (7.4 ± 0.4 mm per day, SE, n = 5) and significantly faster than transfers from vic6-incompatible pairings (<0.3 mm per day). Thus, stable heterokaryon formation is possible only when both vic6 gene allelic forms are deleted in the paired strains. This confirms a role for the candidate polymorphic vic6 alleles as determinants of vegetative incompatibility.
Table 4 . Heterokaryon compatible (+) and incompatible (−) pairings with mean growth rates in parentheses (millimeters per day).
| hygR strains | ||||
|---|---|---|---|---|
| Dk80hygR (2211-22) | EU-21hygR (2211-12) | DK80 Δvic6-2 (2211-22) | DK80 Δvic7-2 (2211-22) | |
| neoR strains | ||||
| DK80neoR | + | − | + | + |
| (2211-22) | (8.0) | (<0.1) | (6.7) | (6.6) |
| EU-21neoR | − | + | − | − |
| (2211-12) | (<0.3) | (7.6) | (<0.1) | (<0.1) |
| EU-21 Δvic6-1 | − | + | + | − |
| (2211-12) | (<0.3) | (6.7) | (7.0) | (<0.2) |
Evidence for nonallelic interactions at the vic6 locus
It was of interest to test whether the vic6 alleles could functionally replace each other. To this end, the disrupted vic6 allele in mutant strain DK80 Δvic6-2 was replaced with an intact vic6-1 allele from strain EP146 linked to a neomycin resistance gene. Examination of DK80 Δvic6-1 replacement strains revealed abnormal growth characteristics and colony morphology (Figure S10) that included reduced aerial hyphae, reduced biomass production, irregular margins, and reduced conidiation. The association of abnormal phenotypic changes with the replacement of the disrupted vic6-2 allele with an intact vic6-1 allele suggested the possibility of functional interactions between the vic6-1 product and a gene product other than that encoded by vic6-2.
As indicated in Figure 5, the candidate vic6 locus also contains a small polymorphic ORF, pix6, adjacent to the candidate vic6 gene. The two pix6 alleles were found to be in linkage disequilibrium with the corresponding vic6 alleles in the natural C. parasitica populations examined (Table 2), consistent with a functional association, because repeated recombination in nature is likely to have eroded this complete association even though the two loci are tightly linked. Independent disruption of pix6-2 in strain DK80 or pix6-1 in strain EU-21 did not eliminate barrage formation (Table 5, pairings E and F). However, the pix6-2 and pix6-1 disruption mutant strains were found to very efficiently promote hypovirus transmission (100%) when serving as the donor, but exhibited no increase when serving as a recipient (Table 5). Importantly, this is completely opposite to the properties of the vic6 disruption mutant strains that promoted virus transmission only when serving as the recipient strain (summarized in Table 5, pairings B and C).
Table 5 . Summary of effects of gene disruption in the vic6 locus on barrage formation and hypovirus transmission.
| Virus movementa | |||||
|---|---|---|---|---|---|
| Pairing | Strains | Genotype | Barrage | % (2 to 1) | % (1 to 2) |
| A | DK80 | pix6-2, vic6-2 | Yes | 5 (1/20) | 15 (3/20) |
| EU-21 | pix6-1, vic6-1 | ||||
| B | DK80 Δvic6b | pix6-2, Δvic6-2 | Yes | 15 (6/40) | 100 (30/30) |
| EU-21 | pix6-1, vic6-1 | ||||
| C | DK80 | pix6-2, vic6-2 | Yes | 100 (20/20) | 17 (5/30) |
| EU-21 Δvic6 | pix6-1, Δvic6-1 | ||||
| D | DK80 Δvic6 | pix6-2, Δvic6-2 | No | 100 (20/20) | 100 (20/20) |
| EU-21 Δvic6 | pix6-1, Δvic6-1 | ||||
| E | DK80 Δpix6 | Δpix6-2, vic6-2 | Yes | 100 (32/32) | 0 (0/30) |
| EU-21 | pix6-1, vic6-1 | ||||
| F | DK80 | pix6-2, vic6-2 | Yes | 7 (2/30) | 100 (20/20) |
| EU-21 Δpix6 | Δpix6-1, vic6-1 | ||||
| G | DK80 Δpix6 | Δpix6-2, vic6-2 | No | 100 (20/20) | 100 (20/20) |
| EU-21 Δpix6 | Δpix6-1, vic6-1 | ||||
| H | DK80 Δpix6 | Δpix6-2, vic6-2 | Yes | 100 (32/32) | 10 (3/30) |
| EU-21 Δvic6 | pix6-1, Δvic6-1 | ||||
| I | DK80 Δvic6 | pix6-2, Δvic6-2 | Yes | 13 (4/30) | 100 (20/20) |
| EU-21 Δpix6 | Δpix6-1, vic6-1 | ||||
| J | DK80 Δpix6Δvic6 | Δpix6-2, Δvic6-2 | No | 100 (20/20) | 100 (20/20) |
| EU-21 | pix6-1, vic6-1 | ||||
“2 to 1” means virus transmission from a strain with a vic6-2 genotype, e.g., DK80, to a strain with a vic6-1 genotype, e.g., EU-21. The inverse holds for “1 to 2”. Transmission data are reported as percentage of independent trials, with numbers in parentheses showing the number of successes over the number of trials.
All pix6-1 and vic6-1 gene disruption mutants were made in the strain EU-21 background. All pix-2 and vic6-2 disruption mutants were made in the strain DK80 background. Disrupted alleles are indicated in bold.
As observed when pairing disruption mutant Δvic6-1 and Δvic6-2 strains (Tables 3 and 5, pairing D), the paired Δpix6-1 and Δpix6-2 mutant strains failed to form barrages and exhibited no resistance to virus transmission (Table 5, pairing G). Thus, potential allelic interactions either between vic6-1 and vic6-2 (pairing G) or between pix6-1 and pix6-2 (pairing D) alone are insufficient to cause barrage formation or restrict virus transmission (100% transmission in both directions). In contrast, barrage formation was observed for all paired mutant strains in which one of the two possible nonallelic interactions between pix6 and vic6 alleles remained intact (Table 5, pairings B, C, E, and F). Moreover, elimination of both potential allelic interactions and one, but not both, possible pix6-vix6 nonallelic interaction by disruption of pix6-2 and vic6-1 or pix6-1 and vic6-2 alleles still allowed barrage formation but resulted in an asymmetric, allele-specific, loss of resistance to virus transmission (Table 5, pairings H and I). Virus transmission was unrestricted from the DK80 Δpix6-2 vic6-2 strain into the EU-21 pix6-1 Δvic6-1 strain (pairing H), but was restricted in this pairing when the EU-21–derived strain was the virus donor. The reciprocal situation was observed in pairing I: virus transmission was restricted from DK80 pix6-2 Δvix6-2 into EU-21 Δpix6-1 vic6-1 but was unimpeded in the opposite direction. Importantly, disruption of pix6-2 and vic6-2 in the same strain (Table 5, paring J) eliminated barrage formation (Figure S11) as well as resistance to virus transmission in both directions (Table 5).
The combined results provide strong evidence that vegetative incompatibility for strains heteroallelic at the vic6 locus requires nonallelic rather than allelic interactions between two tightly linked genes, pix6 and vic6. Nonallelic interactions involving both sets of pix6 and vic6 alleles are required for a robust incompatibility reaction that severely restricts two-way virus transmission. Disruption of one of the two nonallelic interactions weakens the incompatibility reaction as indicated by enhanced virus transmission. Moreover, the asymmetric, gene-specific nature of the resulting enhancement in virus transmission suggests an element of mechanistic directionality underlying the incompatible reaction.
Discussion
The C. parasitica vegetative incompatibility system has been the subject of considerable interest for >3 decades due to its reported role in limiting virus transmission and consequently, the effectiveness of virus-mediated control of chestnut blight (Van Alfen et al. 1975; Anagnostakis 1982a; Nuss 1992; MacDonald and Fulbright 1991; Heiniger and Rigling 1994; Milgroom and Cortesi 2004). Using a comparative genomics approach, we report here the identification of a total of seven candidate incompatibility genes associated with four C. parasitica vic loci. Subsequent functional analysis confirmed that candidate genes at the vic2, vic6, and vic7 loci do contribute to restriction of virus transmission. These studies provide molecular and functional confirmation for a role of the vic loci in regulating virus transmission, based previously on genetic evidence only. Detailed analysis of the candidate vic6 locus revealed a gene complex consisting of two tightly linked but distinct genes and provided evidence for the contribution of nonallelic interactions to the restrictions placed by the C. parasitica vegetative incompatibility system on mycovirus transmission.
The approach used to identify the candidate vic genes took advantage of the general polymorphic nature of nonself recognition genes (reviewed in Richman 2000). It also relied on the collection of 64 vic tester strains (Cortesi and Milgroom 1998) to determine the differences in vic genotypes for the reference and resequenced strains. Additionally, it was guided by the predicted positions of the vic loci on a genetic linkage map (Kubisiak and Milgroom 2006; Kubisiak et al. 2007) and corresponding genome sequence assembly (Department of Energy/Joint Genome Institute and Cryphonectria genome consortium, unpublished results). Reliance on the presence of HET domains (PF06985) associated with the majority of het gene complexes in N. crassa and P. anserina alone for identification of C. parasitica vic genes was not an option. Blast analysis with the HET domain gave 96 hits in the C. parasitica genome assembly (our unpublished results). This compares with 120 HET domains for P. anserina (Paoletti et al. 2007), 55 for N. crassa, and 38 for Aspergillus oryzae (Fedorova et al. 2005). Moreover, neither the vic2 nor the vic4 gene candidates were found to contain HET domains.
The complete correspondence between the results of PCR-based vic allele specificity and vic-tester strain-aided genotyping for 26 C. parasitica field isolates (Table 3) provides strong correlative evidence linking the polymorphic candidate vic alleles with the vic system. The strains tested to date represent the two sequenced laboratory strains and field isolates from three independent North American populations and one European population. The field isolates are valuable for establishing an association between the vegetative incompatibility phenotype and specific alleles because they are derived from natural populations in which repeated recombination would have broken down linkage disequilibrium that might have resulted from spurious correlations in laboratory strains. Analysis of the amplified and sequenced PCR fragments identified only two alleles per locus and no evidence for additional alleles. A much wider survey of allelic diversity in field isolates is warranted, including in isolates from Japan and China, where C. parasitica is native and from where it was introduced into North America and Europe, causing devastating chestnut blight epidemics (reviewed in Milgroom and Cortesi 2004). Analysis of vic allele sequences would allow a determination of whether they are under positive selection and polymorphism is maintained by balancing selection, as found for N. crassa and P. anserina (Saupe et al. 1995; Wu et al. 1998; Micali and Smith 2006; Chevanne et al. 2010; Hall et al. 2010). The PCR-based test for vic allele specificity also provides a valuable tool for determining the vic diversity profiles of C. parasitica forest and orchard populations. The molecular identification of vic alleles opens new approaches for constructing a more complete picture of the influence of vic diversity on hypovirus transmission at the population level and a better understanding of how mycoviruses influence the evolution of the vic system.
A functional role in the restriction of virus transmission was demonstrated for three of the candidate vic genes through gene disruption analysis. Disruption of the candidate vic2 allele encoding the patatin-like protein resulted in a dramatic increase in virus transmission from 5 to 100% when the mutant strain served as the recipient, but resulted in little change if the mutant served as the donor (Table 3). A similar dramatic asymmetric effect on virus transmission was observed when the vic6 disruption mutant served as a recipient in virus transmission assays (Table 3). Analysis of the HET domain-containing candidate vic7 gene was complicated by the fact that virus transmission is not inhibited when the recipient strain has the vic7-2 allele. Consequently, disruption of this allele present in the DK80 strain would not result in any increase. However, the trend toward higher transmission rates (from 2/20 to 9/20) observed when the mutant strain served as the donor is consistent with a contribution of the candidate vic7 to restriction of virus transmission.
The fact that the increase in virus transmission observed for the vic mutant strains was not accompanied by a loss of barrage formation was initially surprising. However, our results reinforce previous observations that heteroallelism at the vic4 locus results in barrage formation but does not prevent heterokaryon formation (Smith et al. 2006) or, more importantly for this study, does not restrict hypovirus transmission (Cortesi et al. 2001); i.e., barrage formation and restriction of virus transmission are not necessarily tightly coupled. One interpretation is that disruption of one of the alleles for a heteroallelic pair retards PCD to an extent that virus transmission is increased, especially when the mutant serves as the recipient strain, but not to an extent that barrage formation is prevented. A slight delay in the initiation of PCD following fusion of the donor and recipient hyphae would increase the frequency with which virus can escape into adjacent recipient cells and then move unrestricted through the recipient mycelium, infecting the mycelium in the growing margin of the colony (Biella et al. 2002).
Genetic mechanisms of vegetative incompatibility can be placed into two categories. First, allelic interactions are triggered by interplay between different alleles of a single incompatibility gene. Second, nonallelic interactions involve different linked or unlinked incompatibility genes. Examples of nonallelic incompatibility gene pairs include un-24/het-6 and het-c/pin-c in N. crassa (review in Dementhon et al. 2006) and het-c/het-e and het-c/het-d in P. anserina (review in Saupe 2000).
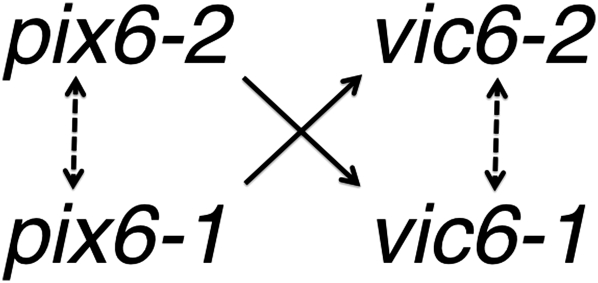
Strong evidence for nonallelic interactions was obtained for the two polymorphic genes, pix6 and vic6, contained within the candidate vic6 locus. As indicated in Figure 8, pairing of strains heteroallelic at the vic6 locus can result in a limited set of possible interactions between these tightly linked genes that include two allelic interactions (pix6-1 with pix6-2 and vic6-1 with vic6-2) and two nonallelic interactions (pix6-1 with vic6-2 and pix6-2 with vic6-1). Since mutant strain DK80 Δpix6-2 vic6-2 was compatible with strain EU-21 Δpix6-1 vic6-1 and strain DK80 pix6-2 Δvic6-2 was compatible with strain EU-21 pix6-1 Δvic6-1, it is clear that the potential pix6 or vic6 allelic interactions alone are insufficient to cause barrage formation (Figure 8 and summarized in Table 5).
Figure 8 .
Model for interactions between the linked genes pix6 and vic6 at the vic6 locus. Nonallelic interactions supported by data summarized in Table 5 are indicated by solid arrows. Arrow direction reflects how the pix6 gene product potentially acts in trans with the cis-acting vic6 gene product to effect asymmetric virus transmission. No evidence was obtained in this study for potential allelic interactions (dashed double arrows).
Elimination of the potential vic6 allelic interaction and either of the two possible pix6–vic6 nonallelic interactions by disruption of a single vic6 allele allowed barrage formation but resulted in asymmetric loss of resistance to virus transmission by the Δvic6 strain (Table 5, pairings B and C). In contrast, elimination of the potential pix6 allelic interaction and either of the two pix6–vic6 nonallelic interactions by disruption of a single pix6 allele allowed barrage formation but resulted in asymmetric decreased resistance to virus transmission by the strain that has vic6 intact (Table 5, parings E and F). This pattern of barrage formation but asymmetric enhancement of virus transmission holds when Δpix6 and Δvic6 mutant strains were paired (Table 5, pairings H and I), in which case both potential allelic interactions and one potential nonallelic interaction are eliminated. Thus, barrage formation was observed for every mutant pairing in which one of the possible nonallelic interactions remained intact. However, the strength of the incompatible reaction was decreased when one nonallelic interaction was disrupted, as witnessed by enhanced virus transmission.
The observation that a strain with pix6 disrupted has enhanced virus donor capability while a strain disrupted at vic6 is more susceptible to virus infection is intriguing. It will be interesting to determine whether this gene-dependent asymmetry in enhanced virus transmission correlates with asymmetric delay in PCD for the two interacting strains and whether the asymmetry is dependent on the presence of virus. It is conceivable that the pix6 gene product is mobile and acts in trans to trigger PCD initially or preferentially by interacting with the gene product of an anchored, cis-acting incompatible gene product of vic6. For example, disruption of the pix6-1 allele, the corresponding vic6-2 allele, or both would then result in an asymmetric delay in PCD in the vic6-2–containing strain, leading to the opportunity for increased virus transmission early on in the interaction, without eliminating eventual PCD, heterokaryon incompatibility, and barrage formation. Under this scenario, a similar situation would hold for disruption of the pix6-2 and/or vic6-1 alleles.
Multiple polymorphic genes were also identified at the candidate vic2 and vic7 loci, raising the possibility for nonallelic interactions at these loci as well. For example, nonallelic interactions parallel to those observed for the vic6 and pix6 alleles could be envisioned for the closely linked vic2 and vic2a alleles. Future studies will focus on the physical interactions, cellular distribution, and movement of candidate vic gene products to provide a mechanistic understanding of how these interactions drive PCD and resistance to virus transmission. Strategies that disrupt all or most allelic and nonallelic interactions would be expected to enhance hypovirus spread and biological control potential.
An inducible RNA silencing surveillance system was recently shown to serve as a cellular antiviral defense response in C. parasitica to target hypovirus RNA for degradation (Segers et al. 2007; Zhang et al. 2008; Sun et al. 2009b). Hypoviruses, in turn, encode a suppressor of RNA silencing, p29. Hypovirus mutants that lack p29 accumulate to a much lower level in infected mycelia, resulting in a reduced level of virus transmission through asexual spores (Suzuki et al. 2003). That is, p29 is able to promote virus transmission by suppressing the cellular RNA silencing antiviral defense response. Thus, the RNA silencing surveillance system and the vic genetic system could be viewed as having evolved as complementary cellular and population-level antiviral defense mechanisms. Interestingly, Biella et al. (2002) reported that hypovirus infection influences the frequency of vic-associated programmed cell death, with an additional potential mechanism for promoting its own transmission. This raises the possibility of a mechanistic link between the interactions of hypoviruses with the two principal antiviral defense strategies of vegetative incompatibility and RNA silencing.
Supplementary Material
Acknowledgments
The authors thank Haofeng Chen and Peter Houde of the New Mexico State University Genome Sequencing Laboratory for technical assistance. Thomas Kubisiak, U.S. Department of Agriculture Forest Service, Southern Research Station, Southern Institute of Forest Genetics, Saucier, Mississippi, made major contributions to assembly of C. parasitica genome sequence scaffolds and the alignment of linkage group markers with the scaffolds. This work was supported by National Science Foundation awards DBA-0821806 (to P. Houde), MCB-1051453 (to A.L.D.), and MCB-1051331 (to D.L.N.). The authors declare that they have no conflict of interest.
Literature Cited
- Anagnostakis S. L., 1982a Biological control of chestnut blight. Science 215: 466–471 [DOI] [PubMed] [Google Scholar]
- Anagnotakis S. L., 1982b Genetic analysis of Endothia parasitica: linkage map of four single genes and three vegetative compatibility types. Genetics 102: 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostakis S. L., Hau B., Kranz J., 1986. Diversity of vegetative compatibility groups of Cryphonectria parasitica in Connecticut and Europe. Plant Dis. 70: 536–538 [Google Scholar]
- Andrews D., Beames B., Summers M. D., Park W. D., 1988. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J. 252: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biella S., Smith M. L., Aist J. R., Cortesi P., Milgroom M. G., 2002. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. Biol. Sci. 269: 2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caten C. E., 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72: 221–229 [DOI] [PubMed] [Google Scholar]
- Chevanne D., Saupe S. J., Clave C., Paoletti M., 2010. WD-repeat instability and diversification of the Podospora anserina hnwd non-self recognition gene family. BMC Evol. Biol. 10: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Choi G. H., Nuss D. L., 1993. Mitotic stability and nuclear inheritance of integrated viral cDNA in engineered hypovirulent strains of the chestnut blight fungus. EMBO J. 12: 2991–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Choi G. H., Nuss D. L., 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264: 1762–1764 [DOI] [PubMed] [Google Scholar]
- Churchill A. C. L., Ciufetti L. M., Hansen D. R., Van Etten H. D., Van Alfen N. K., 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17: 25–31 [Google Scholar]
- Cortesi P., Milgroom M. G., 1998. Genetics of vegetative incompatibility in Cryphonectria parasitica. Appl. Environ. Microbiol. 64: 2988–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesi P., McCulloch C. E., Song H., Lin H., Milgroom M. G., 2001. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 159: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementhon K., Iyer G., Glass N. L., 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot. Cell 5: 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N. D., Badger J. H., Robson G. D., Wortman J. R., Nierman W. C., 2005. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial S. A., Suzuki N., 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47: 353–384 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Dementhon K., 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9: 553–558 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Kuldau G. A., 1992. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu. Rev. Phytopathol. 30: 201–224 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Jacobson D. J., Shiu P. K. T., 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34: 165–186 [DOI] [PubMed] [Google Scholar]
- Hall C., Welch J., Kowbel D. J., Glass N. L., 2010. Evolution and diversity of a fungal self/nonself recognition locus. PLoS ONE 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Dempster E. R., Brown S. W., 1975. Adaptive significance of vegetative incompatibility in Neuorspora crassa. Genetics 81: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiniger U., Rigling D., 1994. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 32: 581–599 [Google Scholar]
- Hillman B. I., Shapira R., Nuss D. L., 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80: 950–956 [Google Scholar]
- Hong W., 2005. SNAREs and traffic. Biochim. Biophys. Acta 1744: 493–517 [PubMed] [Google Scholar]
- Huber D. H., 1996. Genetic analysis of vegetative incompatibility polymorphisms and horizontal transmission in the chestnut blight fungus Cryphonectria parasitica. Ph.D. Thesis, Michigan State University, East Lansing, MI [Google Scholar]
- Huber D. H., Fulbright D. W., 1994. Preliminary investigations on the effect of individual genes upon the transmission of dsRNA in Cryphonectria parasitica, pp. 15–19 in Proceedings of the International Chestnut Conference, edited by Double M. L., MacDonald W. L. West Virginia University Press, Morgantown, WV [Google Scholar]
- Jacobson D. J., Beurkens K., Klomparens K. L., 1998. Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet. Biol. 23: 45–56 [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H., 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Biol. 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Kaneko I., Dementhon K., Xiang Q., Glass N. L., 2006. Nonalleic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172: 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisiak T. L., Milgroom M. G., 2006. Markers linked to vegetative incompatibility (vic) genes and a region of high heterogeneity and reduced recombination near the mating type locus (MAT) in Cryphonectria parasitica. Fungal Genet. Biol. 43: 453–463 [DOI] [PubMed] [Google Scholar]
- Kubisiak T. L., Dutech C., Milgroom M. G., 2007. Fifty-three polymorphic microsatellite loci in the chestnut blight fungus, Cryphonectria parasitica. Mol. Ecol. Notes 7: 428–432 [Google Scholar]
- Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., et al. , 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 15: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X., Yao Z., Zhou Y., Shang J., Lin H., et al. , 2008. Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 53: 59–66 [DOI] [PubMed] [Google Scholar]
- Leslie J. F., 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31: 127–150 [DOI] [PubMed] [Google Scholar]
- Leslie J. F., Zeller K. A., 1996. Heterokaryon incompatibility in fungi: more than just another way to die. J. Genet. 75: 415–424 [Google Scholar]
- MacDonald W. L., Fulbright D. W., 1991. Biological control of chestnut blight: use and limitations of transmissible hypovirulence. Plant Dis. 75: 656–661 [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., et al. , 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali C. O., Smith M. L., 2006. A nonself recognition gene complex in Neurospora crassa. Genetics 173: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgroom M. G., Cortesi P., 1999. Analysis of population structure of the chestnut blight fungus based on vegetative incompatibility genotypes. Proc. Natl. Acad. Sci. USA 96: 10518–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgroom M. G., Cortesi P., 2004. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42: 311–338 [DOI] [PubMed] [Google Scholar]
- Nuss D. L., 1992. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 56: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss D. L., 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3: 632–642 [DOI] [PubMed] [Google Scholar]
- Nuss D. L., 2010. Mycoviruses, pp. 145–152 Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC [Google Scholar]
- Paoletti M., Clave C., 2007. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot. Cell 6: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., Saupe S. J., 2009. Fungal incompatibility: evolutionary origin and pathogen defense? BioEssays 31: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Saupe S. J., Clave C., 2007. Genesis of a fungal non-self recognition repertoire. PLoS ONE 2: e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M. N., Beever R. E., Boine B., Arthur K., 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinan-Lucarre B., Paoletti M., Clave C., 2007. Cell death by incompatibility in the fungus Podospora. Semin. Cancer Biol. 17: 101–111 [DOI] [PubMed] [Google Scholar]
- Powell W. A., 1995. Vegetative incompatibility and mycelial death of Cryphonectria parasitica detected with a pH indicator. Mycologia 87: 738–741 [Google Scholar]
- Richman A., 2000. Evolution of balanced genetic polymorphism. Mol. Ecol. 9: 1953–1963 [DOI] [PubMed] [Google Scholar]
- Robin C., Anziani C., Cortesi P., 2000. Relationship between biological control, incidence of hypovirulence, and diversity of vegetative incompatibility types of Cryphonectria parasitica in France. Phytopathology 90: 730–737 [DOI] [PubMed] [Google Scholar]
- Robin C., Capdeville X., Martin M., Traver C., Colinas C., 2009. Cryphonectria parasitica vegetative compatibility type analysis of populations in south-western France and northern Spain. Plant Pathol. 58: 527–535 [Google Scholar]
- Root C., Balbalian C., Bierman R., Geletka L. M., Anagnostakis S., et al. , 2005. Multi-seasonal field release and spermatization trials of transgenic hypovirulent strains of Cryphonectria parasitica containing cDNA copies of hypovirulent CHV1–EP713. For. Pathol. 35: 277–297 [Google Scholar]
- Saupe S. J., 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64: 489–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S. J., Turcq B., Bégueret J., 1995. Sequence diversity and unusual variability at the het-c locus involved in vegetative incompatibility in the fungus Podospora anserina. Curr. Genet. 27: 466–471 [DOI] [PubMed] [Google Scholar]
- Saupe S. J., Clavé C., Bégueret J., 2000. Vegetative incompatibility in filamentous fungi: Podospora and Neurospora provide some clues. Curr. Opin. Microbiol. 3: 608–612 [DOI] [PubMed] [Google Scholar]
- Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L., 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 104: 12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. L., Gibbs C. C., Milgroom M. G., 2006. Heterokaryon incompatibility function of barrage-associated vegetative incompatibility genes (vic) in Cryphonectria parasitica. Mycologia 98: 43–50 [DOI] [PubMed] [Google Scholar]
- Sun Q., Choi G. H., Nuss D. L., 2009a Hypovirus-responsive transcription factor gene pro1 of the chestnut blight fungus Cryphonectria parasitica is required for female fertility, asexual spore development, and stable maintenance of hypovirus infection. Eukaryot. Cell 8: 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Choi G. H., Nuss D. L., 2009b A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 106: 17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Maruyama K., Moriyama M., Nuss D. L., 2003. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and verticle transmission. J. Virol. 77: 11697–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alfen N. K., Jaynes R. A., Anagnostakis S. L., Day P. R., 1975. Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science 189: 890–891 [DOI] [PubMed] [Google Scholar]
- Wickner R. B., 2001. Viruses of yeasts, fungi and parasitic microorganisms, pp. 629–658 Fields Virology, 4th Ed., edited by Knipe D. M., Howley P. M. Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Wu J., Saupe S. J., Glass N. L., 1998. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl. Acad. Sci. USA 95: 12398–12403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Segers G. C., Sun Q., Deng F., Nuss D. L., 2008. Characterization of hypovirus derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2 dependent pathway. J. Virol. 82: 2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.