Abstract
Cytochrome P450 3A4 (CYP3A4) metabolizes more than 50% of prescribed drugs. The expression of CYP3A4 changes during liver development and may be affected by the administration of some drugs. Alternative mRNA transcripts occur in more than 90% of human genes and are frequently observed in cells responding to developmental and environmental signals. Different mRNA transcripts may encode functionally distinct proteins or contribute to variability of mRNA stability or protein translation efficiency. The purpose of this study was to examine expression of alternative CYP3A4 mRNA transcripts in hepatocytes in response to developmental signals and drugs. cDNA cloning and RNA sequencing (RNA-Seq) were used to identify CYP3A4 mRNA transcripts. Three transcripts were found in HepaRG cells and liver tissues: one represented a canonical mRNA with full-length 3′-untranslated region (UTR), one had a shorter 3′-UTR, and one contained partial intron-6 retention. The alternative mRNA transcripts were validated by either rapid amplification of cDNA 3′-end or endpoint polymerase chain reaction (PCR). Quantification of the transcripts by RNA-Seq and real time quantitative PCR revealed that the CYP3A4 transcript with shorter 3′-UTR was preferentially expressed in developed livers, differentiated hepatocytes, and in rifampicin- and phenobarbital-induced hepatocytes. The CYP3A4 transcript with shorter 3′-UTR was more stable and produced more protein compared with the CYP3A4 transcript with canonical 3′-UTR. We conclude that the 3′-end processing of CYP3A4 contributes to the quantitative regulation of CYP3A4 gene expression through alternative polyadenylation, which may serve as a regulatory mechanism explaining changes of CYP3A4 expression and activity during hepatocyte differentiation and liver development and in response to drug induction.
Introduction
Significant interindividual variation in response to drugs, metabolized by the cytochrome P450 3A4 (CYP3A4), exists among both adult and pediatric patients. However, the causative factors are still not fully identified. CYP3A4 is highly expressed in adult liver and small intestine and catalyzes the metabolism of a variety of xenobiotics and endobiotics, including drugs, environmental chemicals, carcinogens, and steroid hormones (Lamba et al., 2002). Expression of CYP3A4 increases during liver development from very low levels in the prenatal and neonatal stages, with a gradual increase in childhood, to become the major P450 enzyme in adult liver (Stevens et al., 2003). This ontogenic expression pattern may influence the CYP3A4-dependent drug metabolism among neonates, infants, children, and adults (Zhou, 2008). In addition, many drugs can up- or down-regulate CYP3A4 expression (Luo et al., 2004). The induction or inhibition of CYP3A4 expression by some drugs is a major clinical concern for drug-drug interactions in patients receiving multiple CYP3A4-metabolizing drugs. Therefore, understanding the regulatory mechanisms of CYP3A4 expression in response to developmental and environmental signals is critically important for drug therapy. Many studies have focused on the regulatory mechanisms of nuclear receptors, including pregnane X receptor (PXR) (Kliewer et al., 1998), constitutive androstane receptor (Martínez-Jiménez et al., 2007), and glucocorticoid receptor (Khan et al., 2009) on CYP3A4 expression. However, little is known about alternative transcription and RNA processing of CYP3A4 and its implication on regulation of CYP3A4 expression during liver development and drug administration.
A single gene can produce multiple mRNA transcripts by alternative transcription and RNA processing by using alternative transcription start sites, splicing, or polyadenylation. A genome-wide study has indicated that more than 90% of human genes produce more than one mRNA transcripts and approximately 86% have a frequency of 15% or more for the minor transcripts (Wang et al., 2008). Various mRNA transcripts may translate into proteins with different efficiency or produce proteins with different structures or functions (Pan et al., 2008). Utilization of alternative transcription start sites, splicing, or polyadenylation is often regulated by developmental stages, differentiation signals (Castle et al., 2008), environmental changes, or disease status (Cáceres and Kornblihtt, 2002). Hence, alternative transcription and RNA processing as versatile processes can be integrated with other regulatory mechanisms to modulate cellular responses to developmental and environmental signals and to fine tune the functions of gene products (Licatalosi and Darnell, 2010).
Up to now, alternative splicing events have not been reported for the CYP3A4 gene. Multiple CYP3A4 mRNA transcripts with alternative polyadenylation were found when cDNA sequences related to cytochrome P450 nifedipine oxidase in human liver were identified (Beaune et al., 1986; Molowa et al., 1986; Bork et al., 1989). Northern blot analysis with the cDNA probes revealed two hybridization bands in some human liver samples, particularly in the samples from patients who received dexamethasone treatment (Molowa et al., 1986). The mRNA transcript with a shorter 3′-UTR had much stronger signal intensity than the mRNA transcript with a longer 3′-UTR (Molowa et al., 1986; Bork et al., 1989). However, in NCBI GenBank database, the mRNA transcript with the longer 3′-UTR is considered the canonical transcript for CYP3A4. The expression of alternative CYP3A4 mRNA transcripts in response to developmental signals or drug administration is unknown.
Various technologies have been developed to identify mRNA transcripts, including cDNA cloning and RNA sequencing-based approaches. cDNA cloning combined with conventional sequencing is a traditional method to identify novel mRNA transcripts. However, this method is generally not quantitative and is labor extensive. RNA sequencing (RNA-Seq) uses next generation sequencing technology (Mortazavi et al., 2008; Nagalakshmi et al., 2008; Pan et al., 2008; Wang et al., 2008), which has low levels of background noise, no upper limit for quantification, and a high degree of reproducibility for both technical and biological replicates. More importantly, RNA-Seq has the power to identify novel mRNA transcripts very efficiently (Wang et al., 2008, 2009; Trapnell et al., 2010).
The HepaRG cell line we used in our investigation is a newly derived human liver cell line (Gripon et al., 2002). With a change in culture conditions, undifferentiated cells are capable of differentiating into hepatocyte-like and biliary epithelial-like cells (Gripon et al., 2002). Compared with other human liver cell lines, such as HepG2 cells, HepaRG cells express various drug-metabolizing enzymes and nuclear receptors, comparable with those found in cultured primary human hepatocytes (Hart et al., 2010) and hence are more suitable for studies of drug-metabolizing enzymes, such as CYP3A4 (Guillouzo et al., 2007). A previous study showed that CYP3A4 mRNA expression and enzyme activity in HepaRG cells increased in response to prototypical CYP3A4 inducers, such as rifampicin and phenobarbital (Anthérieu et al., 2010).
The aims of this study were to identify alternative transcripts of CYP3A4 mRNA in liver by cDNA cloning and RNA-Seq and to examine quantitative changes of different CYP3A4 mRNA transcripts during liver development, hepatocyte differentiation, and in response to drugs. To that end, expression levels of the identified CYP3A4 mRNA transcripts were quantified in human liver tissues from subjects of various ages, including fetal, neonatal/pediatric, and adult stages. Expression levels of the transcripts were also compared between undifferentiated and differentiated HepaRG cells. Finally, expression levels of the transcripts were examined in differentiated HepaRG cells and primary human hepatocytes after incubation with PXR (rifampicin) or constitutive androstane receptor (phenobarbital) activator.
Materials and Methods
HepaRG Cell Culture.
HepaRG cells and culture medium were provided by Biopredic International (Rennes, France). The undifferentiated HepaRG cells were seeded at 2 × 105 cells/well in six-well plates, and maintained in the growth medium for 2 weeks. HepaRG cells were differentiated by culturing the cells in the differentiation medium containing 2% dimethyl sulfoxide (DMSO) for 2 more weeks. For the drug treatment experiments, differentiated HepaRG cells were incubated with serum-free growth medium for 24 h, then incubated for another 24 h with solvent control (0.1% DMSO), rifampicin (10 μM), or phenobarbital (750 μM).
Primary Human Hepatocytes.
Primary human hepatocyte cultures were obtained through the Liver Tissue Cell Distribution System (Kostrubsky et al., 1999) from the University of Pittsburgh. At arrival, the cells were incubated at 37°C for 3 h, and the medium was renewed into Hepatocyte Maintenance Medium (Lonza, Basel, Switzerland) for 24 h before experiments. For the drug treatment experiments, primary human hepatocytes were incubated with solvent control (0.1% DMSO), rifampicin (10 μM), or phenobarbital (750 μM) for 24 h. DMSO, rifampicin, and phenobarbital were purchased from Sigma-Aldrich (St. Louis, MO).
Human Liver Tissues.
Anonymized fetal, neonatal/pediatric, and adult human liver tissues were obtained from tissue retrieval programs supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the University of Maryland Brain and Tissue Bank for Developmental Disorders (Baltimore, MD), and the Laboratory of Developmental Biology at the University of Washington (Seattle, WA). The collection and use of these tissues have been declared as nonhuman subject research by the University of Missouri-Kansas City Pediatric Health Sciences Review Board. The post mortem interval was <6 h, and all tissues were maintained at −80°C before use.
RNA Isolation and Reverse Transcription.
Total RNA was extracted from primary human hepatocytes or HepaRG cells using TRIzol reagent (Invitrogen, Carlsbad, CA) or from human liver tissues using an RNeasy kit (QIAGEN, Valencia, CA). The optical density at 260/280 nm values of all RNA samples were above 1.8. The 95% confidence interval of RNA quality indicator values were between 7.7 and 9.3 when analyzed on an Experion Bioanalyzer (Bio-Rad Laboratories, Hercules, CA). Total RNA (1 μg) was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and random hexamers in a final volume of 50 μl.
cDNA Cloning and Sequencing.
Thirty nanograms of cDNA from a fetal liver (gestational week 26, white donor; Clontech Laboratories, Mountain View, CA) was used for PCR amplification with a first and a nested primer sets in the 5′- and 3′-UTRs of the CYP3A4 gene (Table 1). Subsequently, the PCR products were digested with KpnI and XbaI and cloned into the pCR4-TOPO vector (Invitrogen) using a rapid DNA ligation kit (Roche Diagnostics, Indianapolis, IN). After transformation into DH5α one-shot Escherichia coli cells (Invitrogen), colonies were picked and plasmid DNA was extracted. The insert fragments were released by restriction enzyme digestion and examined on a 0.7% agarose gel. A CYP3A4 mRNA transcript was identified by DNA sequencing with a DYEnamic ET dye terminator cycle sequencing kit (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). Unincorporated fluorescent dye-terminators were removed from the sequencing reactions by solid-phase magnetic CleanSEQ beads (Agencourt, Beverly, MA) before running the reactions on a MegaBACE 500 Sequencing System (GE Healthcare). Initial sequencing was performed with the insert flanking primers specific for the pCR4-TOPO vector. CYP3A4 exon-specific primers were designed to close the gap when no sequence overlap was achieved.
TABLE 1.
The primer sequences
Accession numbers: CYP3A4, NM_017460.3; GAPDH, NM_002046.3; ACTB, NM_001101.3.
| Gene and Method | Amplification | Direction | Sequence (5′ to 3′) | PCR Product |
|---|---|---|---|---|
| bp | ||||
| CYP3A4 | ||||
| cDNA cloning | 5′-UTR | CACATAGCCCAGCAAAGAGCAACAC | ||
| 3′-UTR | ATTTATGCAGTCCATTGGATGAAGCC | |||
| Nested PCR | CGGggtaccTGAAAGGAAGACTCAGAGGAGAGAG | |||
| GCtctagaGGTCTCTGGTGTTCTCAGGCACAG | ||||
| End-point PCR | From exon 5 to intron 6 | F | TTGCTGTCTCCAACCTTCACCAGT | 255 |
| R | GTTGCATTACCACAGCCCTCCTTT | |||
| 3′RACE-PCR | cDNA synthesis | GCGAGCACAGAATTAATACGACTCACTATAGGT 12VN | ||
| Outer PCR | F | CTGCATTGGCATGAGGTTTGCTCT | ||
| R | GCGAGCACAGAATTAATACGACT | |||
| Inner PCR | F | TTGAGTCAAGGGATGGCACCGTAA | ||
| R | CGCGGATCCGAATTAATACGACTCACTATAGG | |||
| Sequencing | GTGACCAAATCAGTGTGAGGAGGT | |||
| RT-PCR | Canonical 3′-UTR | F | AATCCTAGCAGTTTGGGAGGCTGA | 142 |
| R | TGAGATTACAGGCGAGTCCACCAT | |||
| Canonical and Short 3′-UTR | F | TTGAGTCAAGGGATGGCACCGTAA | 156 | |
| R | ATGCAGTCCATTGGATGAAGCCCA | |||
| Intron-6 retention | F | GAGCTGATATTCCTGCTGTTGGGT | 85 | |
| R | TCTTGGGAGACCCATTGAAGTTGC | |||
| GAPDH | ||||
| RT-PCR | From exon 4 to exon 6 | F | AATCCCATCACCATCTTCCAGGAG | 199 |
| R | CATGGTTCACACCCATGACGAACA | |||
| ACTB | ||||
| RT-PCR | From exon 3 to exon 4 | F | ACCAACTGGGACGACATGGAGAAA | 192 |
| R | TAGCACAGCCTGGATAGCAACGTA |
F, forward; R, reverse; 3′RACE-PCR, rapid amplification of cDNA 3′ ends-PCR
Validation of CYP3A4 mRNA Transcript by Endpoint PCR.
An endpoint PCR was used to validate the CYP3A4 mRNA transcript with partial intron-6 retention. A specific primer set (Table 1) was designed from exon 5 to intron 6 to amplify a product from cDNA with an expected size of 255 bp.
RNA Sequencing.
Total RNA from HepaRG cells or primary human hepatocytes was used for RNA-Seq. The RNA-Seq experiments were performed according to Illumina RNA-Seq protocol (San Diego, CA). In brief, a population of poly(A)+ RNA was selected and converted to a library of cDNA fragments (220–450 bp) with adaptors attached to both ends using an Illumina mRNA-Seq sample preparation kit. The quality of the library preparation was confirmed by analysis on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cDNA fragments were then sequenced on an Illumina HiSeq 2000 to obtain 100-bp sequences from both ends (paired end). The resulting reads were aligned to the reference human genome GRCh37.61/hg19 by TopHat (http://tophat.cbcb.umd.edu/) at settings of maximum 10 multiple hits per read, maximum three mismatches allowed, and fragment length ranged between 100 and 350. The reads were assembled into genes and transcripts by Cufflinks (http://cufflinks.cbcb.umd.edu/) (Trapnell et al., 2010). The counts of RNA-Seq fragments were used to indicate the abundance of the identified mRNA transcripts, which was presented in fragments per kilobase of exon per 106 fragments mapped (FPKM) (Trapnell et al., 2010).
Rapid Amplification of cDNA 3′-End.
For the validation of the alternative polyadenylation of CYP3A4 mRNA, a rapid amplification of cDNA 3′-end (3′RACE) was performed by using the FirstChoice RNA ligase-mediated rapid amplification of cDNA ends (RIM-RACE; Ambion Inc., Austin, TX). In brief, first-strand cDNA was synthesized from 1 μg of total RNA, using an oligo(dT) containing adapter primer in a 20-μl reaction. One microliter of the cDNA was then amplified as follows: the outer 3′RACE adaptor primer (which was complimentary to the anchored adapter) and the customized outer CYP3A4 primer were used for a first amplification, followed by a nested PCR with the inner 3′RACE adaptor primer and the inner CYP3A4-specific primer. The PCR products were separated on a 2% agarose gel. Bands of the expected sizes were excised and purified with a QIAquick gel extraction kit (QIAGEN). Subsequently, DNA was sequenced by quick single-pass DNA sequencing provided by ACGT Inc. (Wheeling, IL). Primer sequences are shown in Table 1.
Quantification of the CYP3A4 mRNA Transcripts by Real-Time PCR.
Specific primers were designed according to the CYP3A4 gene sequence (GenBank accession number NM_017460.3) using PrimerQuest and synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences, PCR product length, and other details are provided in Table 1 and Supplemental Fig. 1. The transcripts with canonical 3′-UTR and partial intron-6 retention have unique sequences that can be amplified by specific primer sets for the transcripts, but the transcript with short 3′-UTR has sequence identical to the transcript with canonical 3′-UTR; therefore, the designed primer set can only be used to detect transcripts with canonical and shorter 3′-UTR. The expression levels of the identified CYP3A4 mRNA transcripts were determined by SYBR Green (Invitrogen) real-time PCR in triplicate reactions on cDNA from human liver tissues of different ages/developmental stages, HepaRG cells at undifferentiated and differentiated stages, and HepaRG cells or primary human hepatocytes treated with rifampicin or phenobarbital. One microgram of total RNA was reverse-transcribed in a 50-μl reaction, the cDNAs were then diluted 5-fold, and 1 μl was used for a SYBR Green real-time PCR. The real-time PCRs were conducted in a 7900 HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA) starting with conditions consisting of 95°C for10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The mean Cq of glyceraldehyde-3-phosphate dehydrogenase (GenBank accession number NM_002046.3) and actin β (GenBank accession number NM_001101.3) were used as internal controls for normalization. The comparative Cq (ΔΔCq) method was employed for the relative quantification in the real time PCRs. PCR efficiencies of the primer sets and the raw data of the RT-PCRs are provided in Supplemental Tables 1 to 5. The minimum information for publication of quantitative real-time PCR experiments can be found in Supplemental Table 6. Control incubations in which the reverse transcription step was excluded resulted in Cq values greater than 35, indicating the absence of contamination from genomic DNA.
mRNA Stability Assay.
Differentiated HepaRG cells were treated with actinomycin D (2 μM) for 0, 2, 4, and 6 h. Total RNA was isolated by using TRIzol reagent and reverse-transcribed with Moloney murine leukemia virus reverse transcriptase and random primers (Invitrogen). Real-time qPCR was used to detect the expression of canonical 3′-UTR and a composite value representing the canonical + shorter 3′-UTR mRNA transcripts of CYP3A4. Fold changes of mRNA levels at 2, 4, and 6 h were compared with the mRNA level at 0 h (assigned a value of 1 for reference). The natural logarithm (ln) of fold changes were plotted with different time points (0, 2, 4, and 6 h). Half-life of mRNA was determined from logarithmic transformation (natural logarithm) of the mRNA levels at each time point (0, 2, 4, and 6 h) and calculated as t1/2 = 0.693/(lnC1-lnC2)/t, t representing the time interval between successive mRNA determination and C1 and C2 corresponding to the amount of mRNA at the initial and successive time points, respectively.
Construction of Plasmids Containing CYP3A4 3′-UTR.
Plasmids were constructed by introducing sequences of the CYP3A4 3′-UTRs into the downstream region of a firefly luciferase gene in a pMirTarget vector (OriGene, Rockville, MD). The pMirTarget vector contains a red fluorescent protein (RFP) sequence serving as an internal control to monitor and normalize the transfection efficiency. The canonical and the shorter 3′-UTRs of the CYP3A4 gene (GenBank accession number NM_017460.3, Homo sapiens) were PCR-amplified from human genomic DNA with a high-fidelity PCR enzyme and subcloned into the pMirTarget vector after the stop codon of the luciferase gene. To generate the plasmid of luciferase expression with only the canonical 3′-UTR of CYP3A4, the proximal poly (A) signal for the shorter 3′-UTR of CYP3A4 was mutated from the sequence AATAAA to ACGCAA. All clones had the expected insert sizes (Supplemental Fig. 3). The inserts were sequenced and confirmed to perfectly match to the CYP3A4 reference sequences (data not shown).
Luciferase Activity Assay.
For functional analyses of translation efficiency of alternative polyadenylation mRNA transcripts of CYP3A4, luciferase assays were performed using differentiated HepaRG cells. The cells were seeded at 2 × 104 cells/well in 96-well plates and cultured for 24 h. Plasmid DNAs (0.2 μg) were transfected into cells by Lipofectamine 2000 (Invitrogen) for 48 h (DNA/Lipofectamine 2000 ratio = 1:2.5). The expression of RFP was analyzed in situ on an adherent cell cytometer (Celigo cytometer; Cyntellect Inc., San Diego, CA) to obtain cell images in both bright field and red fluorescence (Supplemental Fig. 4). The ratios of cell densities from bright field to red fluorescence in each entire well were analyzed by Celigo Software and used for normalization of transfection efficiency. The firefly luciferase activities were measured with a luciferase assay system (Promega, Madison, WI) and normalized by transfection ratios measured by RFP expression.
Results
Identification of Alternative CYP3A4 mRNA Transcripts.
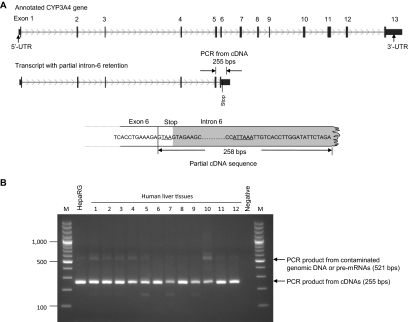
A novel CYP3A4 mRNA transcript with partial intron-6 retention was identified in a fetal liver (gestational week 26, white donor) cDNA clone (Fig. 1A). A translational stop codon (TAA) was found at the beginning of intron 6. A potential polyadenylation signal sequence (ATTAAA) is located downstream of the stop codon. The transcript with partial intron-6 retention was validated by endpoint PCR from cDNAs of HepaRG cells and 12 human liver samples with a primer set binding to exon 5 and intron 6, which produced an amplicon with the expected size of 255 bp in all tested samples (Fig. 1B).
Fig. 1.
Identification of an alternative CYP3A4 mRNA transcript with partial intron-6 retention. A, detection of the CYP3A4 mRNA transcript by cDNA cloning and sequencing. TAA, translational stop codon; ATTAAA, polyadenylation signal. B, validation of the transcript by endpoint PCR using cDNAs of HepaRG cells and human liver tissues with primers binding to exon 5 and intron 6. Human liver tissues: 1 to 4, fetal; 5 to 8, neonatal/pediatric; and 9 to 12, adult. M, DNA marker (base pairs).
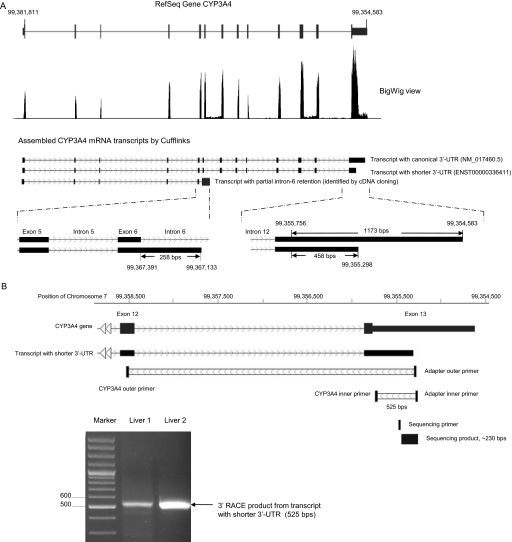
RNA-Seq confirmed the transcript with partial intron-6 retention and identified an alternative polyadenylated mRNA transcript in the 3′-UTR region of CYP3A4 (Fig. 2A). The RefSeq CYP3A4 gene in the human assembly of GRCh37/hg19 is located on the minus strand of chromosome 7 between 99,354,583 and 99,381,811 with 13 exons. Sequencing reads from an mRNA library constructed from total RNA of HepaRG cells were mapped to the reference genome by TopHat. Distribution of the sequencing reads within the CYP3A4 gene region is displayed in Fig. 2A by upload a BigWig file of the sample to the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). Thirteen peaks of the sequencing reads were observed along the RefSeq CYP3A4 gene. The first twelve exons were covered by a single peak only. However, in exon 13, a region with high-level reads followed by a region with low-level reads was found. A sharp decrease of the sequencing reads occurred at chromosome position 99,355,298 in the 3′-UTR of CYP3A4. The RNA-Seq reads ended at coordinate 99,354,583 of chromosome 7, which matched with the termination of the RefSeq full-length (canonical) 3′-UTR of CYP3A4 in the reference sequence. Faint peaks were also observed in introns 6 and 11. After assembly by Cufflinks, three CYP3A4 mRNA transcripts with alternative polyadenylation and partial intron-6 retention were identified (Fig. 2A). The 3′-UTR of the transcript with canonical 3′-UTR had a length of 1173 bp, whereas the 3′-UTR of the shorter 3′-UTR transcript had a length of only 458 bp. The transcript with partial intron-6 retention contained 258 bp of intron-6 sequence. However, Cufflinks did not assemble a transcript with intron-11 retention. The canonical transcript corresponds to the GenBank accession number NM_017460.5 in the NCBI GenBank and RefSeq CYP3A4 gene in the UCSC Genome Browser. The CYP3A4 transcript with shortened 3′-UTR is same as CYP3A4-001 (ENST00000336411) in Ensembl Gene Predictions (Ensembl 62). The transcript with partial intron-6 retention is annotated in neither the NCBI nor the Ensembl database.
Fig. 2.
Identification of an alternative CYP3A4 mRNA transcript with a shorter 3′-UTR by RNA-Seq and validation by 3′RACE-PCR. A, expression of the CYP3A4 gene in HepaRG cells analyzed by RNA-Seq. Short-paired 100-bp RNA sequencing reads were aligned to the reference human genome hg19 by TopHat and viewed as distribution peaks by the UCSC genome browser. Cufflinks assembled the reads to the canonical and the shorter 3′-UTR transcripts and calculated their expression levels presented as FPKM. B, validation of the shorter 3′-UTR transcript by 3′RACE-PCR. First, total RNA of two liver tissues were reverse-transcribed with the poly(T)-containing adapter primer, subsequently followed by 3′RACE-PCR amplification with the adapter outer primer and the CYP3A4 outer primer and a nested PCR amplification with the adapter inner primer and the CYP3A4-specific inner primer. The PCR products were separated on a 2% agarose gel.
To further validate the shorter 3′-UTR transcript, we performed 3′RACE-PCR with RNA samples from two liver tissues. Two primer sets were designed as shown in Fig. 2B and Table 1. After the 3′RACE-PCR, a single band was observed on an agarose gel with a size in agreement with the shorter 3′-UTR transcript length (525 bp) as predicted by 3′ RACE. Sequencing of this product revealed a poly(A)-containing sequence (Supplemental Fig. 2A) that was uniquely mapped to the 3′-UTR of CYP3A4 (Supplemental Fig. 2B). Transcription termination of the short 3′-UTR occurred at chromosomal position 99,355,302, which differed by only four bases from the termination position (99,355,298) detected by RNA-Seq. In addition, an AATAAA sequence is located 15 bases (99,355,317) upstream of the termination site (99,355,302), which may serve as a recognition signal for the poly(A) polymerase complex.
Differential Expression of the CYP3A4 mRNA Transcripts during HepaRG Cell Differentiation.
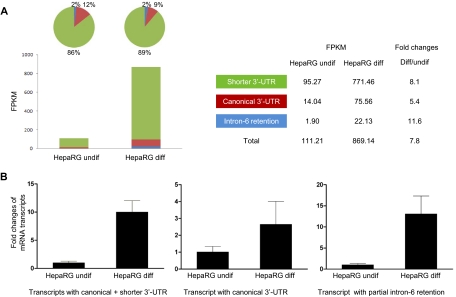
Differential expression of the alternative CYP3A4 mRNA transcripts was observed during hepatocyte differentiation in cultured HepaRG cells (Fig. 3). RNA-Seq quantified expression levels of these three transcripts in both undifferentiated and differentiated HepaRG cells (Fig. 3A). FPKM was used to present abundance of the sequenced reads assembled to each transcript by Cufflinks. The transcript with the shorter 3′-UTR was the dominant transcript and represented 86% of total CYP3A4 transcripts in undifferentiated HepaRG cells, whereas the transcripts with canonical 3′-UTR and intron-6 retention comprised 12 and 2%, respectively. A 7.8-fold increase in FPKM value was found for three transcripts together in differentiated HepaRG cells compared with undifferentiated HepaRG cells; however, the increase was higher for the shorter 3′-UTR (8.1-fold) or intron-6 retention transcript (11.6-fold) than for the canonical 3′-UTR transcript (5.4-fold). A preference for the shorter 3′-UTR transcript relative to the canonical transcript existed in the differentiated HepaRG cells.
Fig. 3.
Expression of the CYP3A4 mRNA transcripts determined by RNA-Seq (A) (n = 1) and RT-qPCR (B) (n = 3) in HepaRG cells before and after differentiation. FPKM is used to present expression levels of each CYP3A4 mRNA transcript in RNA-Seq experiments. In RT-qPCR experiments, the average expression level in undifferentiated HepaRG cells is set as 1. Fold changes of expression levels of each transcript in differentiated HepaRG cells are compared with the values in undifferentiated HepaRG cells. Black column, mean; bar, S.D. of biological triplicate samples.
A similar result was found when the result from RNA-Seq was verified by RT-qPCR (Fig. 3B). Compared with undifferentiated HepaRG cells, expression of the canonical transcript in differentiated HepaRG cells increased by an average of only 2.6-fold in three triplicate samples, but expression of the canonical transcript and the shorter 3′-UTR transcripts together increased by 10.0-fold, implying relatively greater up-regulation of the shorter transcript during differentiation. Expression of the intron-6 retention transcript was increased by 13.1-fold after HepaRG cells were differentiated.
Differential Expression of the CYP3A4 mRNA Transcripts in HepaRG Cells or Primary Human Hepatocytes Treated with Different Drugs.
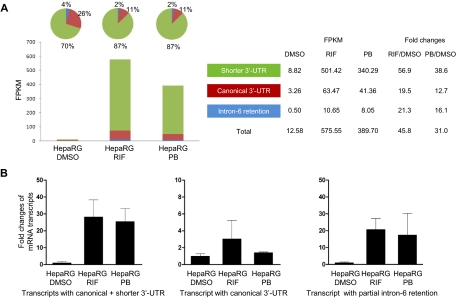
Expression levels of the CYP3A4 mRNA transcripts increased in differentiated HepaRG cells treated with rifampicin (RIF) or phenobarbital (PB) compared with HepaRG cells treated with 0.1% DMSO (control) (Fig. 4). In a RNA-Seq measurement (Fig. 4A), the transcript with shorter 3′-UTR was the dominant transcript consisting of 70%, whereas the transcripts with canonical 3′-UTR and intron-6 retention comprised 26 and 4%, respectively, in HepaRG cells treated with 0.1% DMSO control. The FPKM values were increased by 45.8- and 31.0-fold for three transcripts together in HepaRG cells treated with RIF and PB, respectively, compared with that treated with DMSO control. However, the fold increase was higher for the shorter 3′-UTR (56.9 and 38.6 for RIF and PB, respectively) than canonical 3′-UTR transcript (19.5 and 12.7) or intron-6 retention transcript (21.3 and 16.1). A preference for the shorter 3′-UTR transcript relative to the canonical or intron-6 retention transcripts existed in differentiated HepaRG cells after RIF and PB treatment.
Fig. 4.
Expression of the CYP3A4 mRNA transcripts determined by RNA sequencing (A) (n = 1) and RT-qPCR (B) (n = 3) in differentiated HepaRG cells after drug treatments. The differentiated HepaRG cells were treated with DMSO solvent control (0.1%), RIF (10 μM), or PB (750 μM) for 24 h. In the RT-qPCR experiment, the average expression level treated with DMSO is set as 1. Fold changes of expression levels of the transcripts after drug treatment are compared with the values in DMSO. Black column, mean; bar, S.D. of biological triplicate samples.
RT-qPCR confirmed the RNA-Seq result in HepaRG cells after drug treatment (Fig. 4B). The fold induction for the canonical transcript was 3.0 and 1.4 for RIF- and PB-treated groups, respectively. When RT-PCR was performed with primers detecting both the canonical and the shorter 3′-UTR transcripts, the induction increased to 28.2- and 25.5-fold in RIF- and PB-treated groups, respectively.
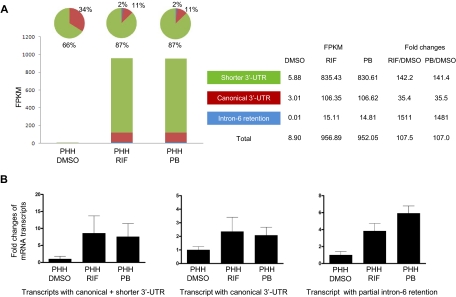
Furthermore, a preference for the shorter 3′-UTR transcript relative to the canonical transcript was also found in primary human hepatocytes (PHH) after drug treatments (Fig. 5). By RNA-Seq (Fig. 5A), the transcript with shorter 3′-UTR represented 66% of total CYP3A4 transcripts in PHH treated with 0.1% DMSO control, with the increased percentage to 87% in PHH treated with RIF or PB for 24 h. FPKM values were increased by 107.5- and 107.0-fold for the three transcripts together in PHH treated with RIF and PB, respectively, compared with that with treated DMSO control. Furthermore, the increase was higher for the shorter 3′-UTR (142.1- and 141.4-fold for RIF and PB, respectively) than canonical 3′-UTR transcript (35.4- and 35.5-fold). The intron-6 retention transcript is increased most in PHH after drug treatment (1511- and 1481-fold for RIF and PB, respectively), but the FPKM is 0.01 (<1%) in the DMSO control group.
Fig. 5.
Expression of the CYP3A4 mRNA transcripts determined by RNA sequencing (A) (n = 1) and RT-qPCR (B) (n = 3) in primary human hepatocytes after drug treatments. The primary human hepatocytes were treated with DMSO solvent control (0.1%), RIF (10 μM), or PB (750 μM) for 24 h. Three primary human hepatocytes were isolated from male donors at age of 55, 56, and 58. In the RT-qPCR experiment, the average expression level treated with DMSO is set as 1. Fold changes of expression levels of the transcripts after drug treatment are compared with the values in DMSO. Black column, mean; bar, S.D. of biological triplicate samples.
A similar result was obtained measured by RT-qPCR when primary human hepatocytes were treated with RIF or PB (Fig. 5B). Expression of the canonical transcript increased by 2.4- and 2.1-fold after RIF and PB treatment, respectively, but expression of the canonical and shorter 3′-UTR transcripts together increased by 8.6- and 7.6-fold in RIF- and PB-treated groups, respectively. The intron-6 retention transcript increased by 3.8- and 5.9-fold, respectively.
Differential Expression of the CYP3A4 mRNA Transcripts in Human Livers from Different Aged Donors.
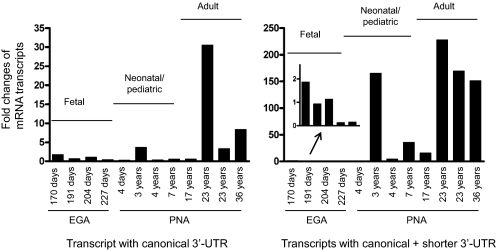
Expression of the alternative CYP3A4 mRNA transcripts was quantified in liver tissues of humans of various ages, including four fetal samples at estimated gestational ages of 170, 191, 204, and 227 days; four neonatal/pediatric at postnatal ages of 4 days and 3, 4, and 7 years; and four adults aged 17, 23, 23, and 36 years (Fig. 6). The expression of the canonical CYP3A4 mRNA transcript increased through age up to 31-fold. In contrast, both canonical and shorter 3′-UTR transcripts together increased with increasing age up to approximately 228-fold. A preference for the shorter 3′-UTR transcript compared with the canonical transcript was observed during human liver development. Significant interindividual variation existed within each age group.
Fig. 6.
Relative fold changes of the CYP3A4 mRNA transcripts determined by RT-qPCR in liver tissues from humans of various ages. Average expression level in fetal liver is set as 1. Fold changes of expression levels of mRNA transcripts in each sample are compared with the fetal average value. Fetal livers include four samples with estimated gestational ages (EGA) from 170 to 227 days. Neonatal/pediatric livers contain four samples from postnatal ages (PNA) 4 days to 7 years; the age of four adult livers ranges from 17 to 36 years.
mRNA Stability of the CYP3A4 mRNA Transcripts.
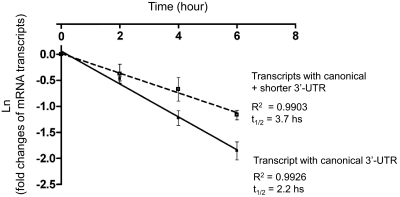
HepaRG cells were treated with actinomycin D to inhibit transcription at different time points, and the decay of the mRNA transcripts was investigated by real-time PCR (Fig. 7). A composite half-life of 3.7 h was observed for the combined canonical and shorter 3′-UTR mRNA transcripts, whereas the half-life of the mRNA transcript with the canonical 3′-UTR alone was 2.2 h. These data indicate that mRNA transcript with the shorter 3′-UTR was more stable than the mRNA transcript with the canonical 3′-UTR.
Fig. 7.
mRNA stability of the CYP3A4 transcripts. Levels of the CYP3A4 mRNA transcripts were determined by RT-qPCR in differentiated HepaRG cells at 0, 2, 4, and 6 h after the treatment of a transcription inhibitor, actinomycin D (2 μM) (n = 3). Fold changes of the mRNA transcript levels at 2, 4, and 6 h were compared with the levels at 0 h (considered as 1). Fold changes (ln) were plotted with the different time points (0, 2, 4, and 6 h). Half-life of mRNA was calculated at t1/2 = 0.693/(lnC1-lnC2)/t, with t = time interval between C1 and C2; C1 = amount of mRNA at t = 0, and C2 = amount of mRNA at t = 2, 4, or 6 h, respectively.
Influence of Alternative CYP3A4 3′-UTRs on Translation Revealed by Luciferase Assay.
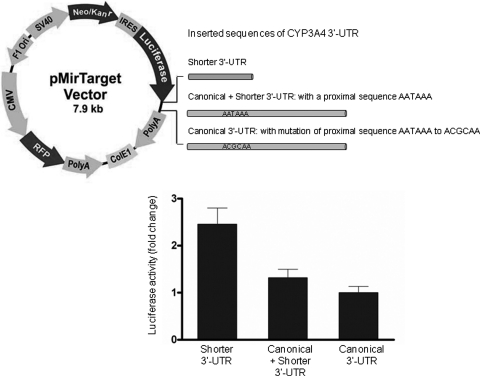
Luciferase reporter system was used to measure protein synthesis for three plasmid constructs containing different CYP3A4 3′-UTR in differentiated HepaRG cells. The shorter 3′-UTR of CYP3A4 produced more luciferase than the canonical 3′-UTR (Fig. 8). The protein expression derived from the shorter 3′-UTR of CYP3A4 was 2.4-fold higher than that from the canonical 3′-UTR.
Fig. 8.
Luciferase activity from plasmid constructs containing different CYP3A4 3′-UTR sequences. The plasmids were constructed by inserting different CYP3A4 3′-UTR sequences into the downstream region of a firefly luciferase gene in a pMir-Target Vector. Luciferase activity was used for detecting protein synthesis with the shorter CYP3A4 3′-UTR, canonical + shorter 3′-UTR, and canonical 3′-UTR alone (n = 5). Red fluorescence produced by an RFP was used for normalizing the transfection efficiency. In the pMir-Target vector: CMV, cytomegalovirus promoter; PolyA (downstream of Luciferase), human growth hormone poly(A) sequence (586 bp); SV40, simian vacuolating virus 40 promoter; Neo/Kanr, neomycin/kanamycin resistant; IRES, internal ribosome entering sequence; Luciferase, firefly luciferase; PolyA (downstream of RFP), HSV thymidine kinase poly(A) sequence (419 bp); F1 Ori, F1 phage origin of replication.
Discussion
In the current study, we applied cDNA cloning and RNA-Seq methods to identify two alternative CYP3A4 mRNA transcripts, one of which contained partial intron-6 retention and the other with a shorter 3′-UTR.
The CYP3A4 mRNA transcript with partial intron-6 retention can potentially be translated into a novel protein with a shortened amino acid sequence as a result of a translational stop codon in intron 6. However, the absence of a heme-binding signature in the encoded polypeptide precludes a catalytically active protein. Proportion of this transcript in all examined samples is less than 2% of total CYP3A4 mRNA. Therefore, its influence on CYP3A4 function is considered to be limited; as a result, we did not investigate function of this transcript in the current study. Although RNA-Seq also showed faint peaks in intron 11, Cufflinks did not assemble a transcript with intron-11 retention. Thus, the possible existence of intron-11 retention cannot be excluded based on the current evidence, and this remains an issue for further investigation.
We have verified an additional polyadenylation site in the CYP3A4 gene, which produced an transcript with a shorter 3′-UTR than the canonical transcript. The canonical full-length 3′-UTR is 1173 bp long and contains two AAUAAA sequences at positions 440 and 1156 bp of the 3′-UTR. The AAUAAA sequence at 1156 serves as the polyadenylation signal for the canonical transcript. In addition, 3′RACE sequencing data demonstrated that CYP3A4 pre-mRNA could also be cleaved 15 nucleotides downstream of the AAUAAA sequence at the 440-bp position to generate a transcript with a shorter 3′-UTR. Furthermore, a GU/U-rich region, including a UUUUUGGU sequence, is located downstream of this AAUAAA sequence. Both the AAUAAA sequence and the GU/U-rich accessory sequence could conceivably provide binding sites for the polyadenylation machinery to produce the alternative transcript with the shorter 3′-UTR.
Although both the canonical and shorter 3′-UTR transcripts are expected to produce an identical protein, the latter has less regulatory sequence elements in the 3′-UTR for post-transcriptional and translational regulation. RNA polyadenylation, including cleavage of nascent pre-mRNA and the addition of a 3′ poly(A) tail, is required for maturation of most mRNA transcripts (Millevoi and Vagner, 2010). Cleavage of nascent pre-mRNA is usually guided by a polyadenylation signal sequence in the 3′-UTR of a pre-mRNA, typically containing the nucleotides AAUAAA, AUUAAA, or other AU rich hexamers. A pre-mRNA is often cleaved 10 to 30 nucleotides downstream of the polyadenylation signal sequence, and a poly(A) tail of 200 to 250 nucleotides is added. Transcripts without 3′-end processing will be degraded or not transported efficiently to the cytoplasm. In addition, 3′-UTR regions can harbor many cis-acting elements involved in regulating the polyadenylation process (Hu et al., 2005), being recognized by RNA binding proteins, or interacting with microRNAs (miRNAs) (Legendre and Gautheret, 2003).
Studies have shown that alternative polyadenylation is a common biological phenomenon and occurs in more than 50% of human genes (Tian et al., 2005). The complexity of RNA 3′-end processing provides the potential for extensive regulation of gene expression in addition to transcription initiation (Zhao et al., 1999; Beaudoing et al., 2000; Gilmartin, 2005). In fact, different stimuli, including physiological conditions (cell proliferation, differentiation, and development) (Sandberg et al., 2008; Ji et al., 2009), and pathological states (cancer, inflammation, and infection) (Flavell et al., 2008; Mayr and Bartel, 2009), can influence the relative usage of an alternative poly(A) site and determine which transcripts need to be processed. Differential lengths of 3′-UTRs produced by alternative polyadenylation are a coordinated mechanism for altered expression of many genes during T-cell activation (Sandberg et al., 2008), embryonic development (Ji et al., 2009), neuronal activation (Flavell et al., 2008), and somatic cell reprogramming (Ji and Tian, 2009).
Our findings also demonstrated that physiological conditions (hepatocyte differentiation and liver development) as well as external stimuli (drugs) may influence alternative polyadenylation in the CYP3A4 gene. The expression of the shorter 3′-UTR transcript is preferentially increased over its canonical counterpart during hepatocyte differentiation, liver development, and in response to drug induction. A previous study also showed that the transcript with the shorter 3′-UTR was the major transcript in human liver in response to CYP3A4 activity of midazolam hydroxylation (He et al., 2006).
The 3′-UTR lengthening and shortening can affect mRNA stability, transport, and protein production. It has been reported that mRNAs with shorter 3′-UTR mediated by alternative polyadenylation allowed oncogenes to produce 10-fold more proteins in cancer cell lines (Mayr and Bartel, 2009). Cell proliferation across diverse cell types and tissues is associated with widespread reductions in the 3′-UTR based regulatory capacity of mRNAs (Sandberg et al., 2008). In this study, we provide evidence in an mRNA decay assay that the CYP3A4 mRNA transcript with the shorter 3′-UTR is more stable than the mRNA transcript with the canonical 3′UTR. When the shorter CYP3A4 3′-UTR is inserted into a plasmid containing a luciferase reporter system, this construct can produce more luciferase protein than a construct containing the canonical 3′-UTR. These results suggest that, in addition to the regulation of CYP3A4 transcription by nuclear receptors, differential usage of alternative polyadenylation sites may also contribute to the complexity of CYP3A4 mRNA expression and ultimate CYP3A4 activity in human liver. It is tempting to speculate that if the shorter 3′-UTR is subject to less regulatory influence than the canonical transcript, it may provide an adaptive mechanism to respond to perturbations of the system and facilitate just-in-time regulation to meet the changing demands of a developing organism or changes in its environment. For a better understanding of CYP3A4 gene regulation, it is important to investigate the physiological conditions as well as the inductive states that are related to the increased use of the alternative polyadenylation producing the shorter 3′-UTR.
In conclusion, hepatocytes preferentially express an alternative CYP3A4 mRNA transcript with the shorter 3′-UTR during hepatocyte differentiation, liver development, and in response to drug induction. The 3′-end processing of CYP3A4 mRNAs may also participate in the quantitative regulation of CYP3A4 gene expression through alternative polyadenylation, which may serve as a regulatory mechanism explaining changes of CYP3A4 expression and activity during liver development and xenobiotic exposure, including drug administration. Considerable interindividual variation in drug biotransformation has been observed for clinically used drugs that are metabolized by CYP3A4. However, the underlying environmental and genetic determinants are still not fully understood. Alternative polyadenylation may serve as an additional regulatory mechanism contributing to individual variation of CYP3A4-mediated drug metabolism.
Supplementary Material
Acknowledgments
We thank Biopredic International (Rennes, France) for providing HepaRG cells and culture medium for this study. We thank Drs. Curtis Klaassen and Andrea Gaedigk for critical reading of the manuscript. Human tissue was obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD).
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This study was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant 1R01-GM087376–01A1] (to X.Z.); the National Institutes of Health National Center for Research Resources [Grant 5P20-RR021940] (to X.Z.); and the National Institutes of Health National Institute for Environmental Health Sciences [Grant 1R01-ES019487–01A1] (to X.Z.) and [Grant 1R01-ES10855] (to J.S.L.). The Liver Tissue Cell Distribution System (Kostrubsky et al., 1999) from the University of Pittsburgh was funded by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (Contract N01-DK7-0004/HHSN26700700004C).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.074393.
- 3′RACE
- rapid amplification of cDNA 3′ end
- 3′-UTR
- 3′-untranslated region
- bp
- base pair(s)
- DMSO
- dimethyl sulfoxide
- FPKM
- fragments per kilobase of exon per 106 fragments mapped
- miRNA
- microRNA
- NCBI
- National Center for Biotechnology Information
- PB
- phenobarbital
- PCR
- polymerase chain reaction
- PHH
- primary human hepatocyte
- PXR
- pregnane X receptor
- qPCR
- quantitative polymerase chain reaction
- RFP
- red fluorescent protein
- RIF
- rifampicin
- RNA-Seq
- RNA sequencing
- RQI
- RNA quality indicator values
- RT-qPCR
- real time quantitative polymerase chain reaction
- svRNA
- small vault RNA
- UCSC
- University of California Santa Cruz
- UTR
- untranslated region.
Authorship Contributions
Participated in research design: Li, Gaedigk, Hart, Leeder, and Zhong.
Conducted experiments: Li and Gaedigk.
Performed data analysis: Li, Gaedigk, and Hart.
Wrote or contributed to the writing of the manuscript: Li, Gaedigk, Hart, Leeder, and Zhong.
References
- Anthérieu S, Chesné C, Li R, Camus S, Lahoz A, Picazo L, Turpeinen M, Tolonen A, Uusitalo J, Guguen-Guillouzo C, et al. (2010) Stable expression, activity, and inducibility of cytochromes P450 in differentiated HepaRG cells. Drug Metab Dispos 38:516–525 [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. (2000) Patterns of variant polyadenylation signal usage in human genes. Genome Res 10:1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaune PH, Umbenhauer DR, Bork RW, Lloyd RS, Guengerich FP. (1986) Isolation and sequence determination of a cDNA clone related to human cytochrome P-450 nifedipine oxidase. Proc Natl Acad Sci USA 83:8064–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork RW, Muto T, Beaune PH, Srivastava PK, Lloyd RS, Guengerich FP. (1989) Characterization of mRNA species related to human liver cytochrome P-450 nifedipine oxidase and the regulation of catalytic activity. J Biol Chem 264:910–919 [PubMed] [Google Scholar]
- Cáceres JF, Kornblihtt AR. (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18:186–193 [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. (2008) Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40:1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60:1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM. (2005) Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev 19:2517–2521 [DOI] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. (2002) Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. (2007) The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168:66–73 [DOI] [PubMed] [Google Scholar]
- Hart SN, Li Y, Nakamoto K, Subileau EA, Steen D, Zhong XB. (2010) A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos 38:988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Court MH, Greenblatt DJ, von Moltke LL. (2006) Factors influencing midazolam hydroxylation activity in human liver microsomes. Drug Metab Dispos 34:1198–1207 [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. (2005) Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. (2009) Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA 106:7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. (2009) Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4:e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Chow EC, van Loenen-Weemaes AM, Porte RJ, Pang KS, Groothuis GM. (2009) Comparison of effects of VDR versus PXR, FXR and GR ligands on the regulation of CYP3A isozymes in rat and human intestine and liver. Eur J Pharm Sci 37:115–125 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- Kostrubsky VE, Ramachandran V, Venkataramanan R, Dorko K, Esplen JE, Zhang S, Sinclair JF, Wrighton SA, Strom SC. (1999) The use of human hepatocyte cultures to study the induction of cytochrome P-450. Drug Metab Dispos 27:887–894 [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294 [DOI] [PubMed] [Google Scholar]
- Legendre M, Gautheret D. (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11:75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Guenthner T, Gan LS, Humphreys WG. (2004) CYP3A4 induction by xenobiotics: biochemistry, experimental methods and impact on drug discovery and development. Curr Drug Metab 5:483–505 [DOI] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, Castell JV, Gómez-Lechón MJ. (2007) Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8:185–194 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. (2010) Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38:2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molowa DT, Schuetz EG, Wrighton SA, Watkins PB, Kremers P, Mendez-Picon G, Parker GA, Guzelian PS. (1986) Complete cDNA sequence of a cytochrome P-450 inducible by glucocorticoids in human liver. Proc Natl Acad Sci USA 83:5311–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ. (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307:573–582 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63:405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF. (2008) Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 9:310–322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.