Abstract
Meiotic recombination initiates via programmed double-strand breaks (DSBs). We investigate whether, at a given initiation site, DSBs occur independently among the four available chromatids. For a single DSB “hot spot”, the proportions of nuclei exhibiting zero, one, or two (or more) observable events were defined by tetrad analysis and compared with those predicted by different DSB distribution scenarios. Wild-type patterns are incompatible with independent distribution of DSBs among the four chromatids. In most or all nuclei, DSBs occur one-per-pair of chromatids, presumptively sisters. In many nuclei, only one DSB occurs per four chromatids, confirming the existence of trans inhibition where a DSB on one chromosome interactively inhibits DSB formation on the partner chromosome. Several mutants exhibit only a one-per-pair constraint, a phenotype we propose to imply loss of trans inhibition. Signal transduction kinases Mec1 (ATR) and Tel1 (ATM) exhibit this phenotype and thus could be mediators of this effect. Spreading trans inhibition can explain even spacing of total recombinational interactions and implies that establishment of interhomolog interactions and DSB formation are homeostatic processes. The two types of constraints on DSB formation provide two different safeguards against recombination failure during meiosis.
Keywords: homeostasis, tetrads, spatial patterning
Meiotic recombination initiates via programmed double-strand breaks (DSBs), catalyzed by Spo11 transesterase. DSBs occur preferentially at “hot spots” (e.g., ref. 1) and are governed both locally (e.g., by absence of nucleosomes) and domainally (e.g., by global base composition) (2, 3). DSB formation is also modulated by communication along and between homologs. In cis, the presence of a strong hot spot at one position suppresses DSB formation in the vicinity over a distance of ∼25 kb, by mechanisms unknown (4–7). In trans, two effects occur: (i) Increased activity of the site on one homolog can decrease DSB formation on the partner homolog at the same and nearby positions (“trans inhibition”) [refs. 5 (case C) and 6]. This effect can occur at an artificial hot spot where DSB formation is independent of Spo11 and other factors and thus is probably a direct effect of DSB formation per se, with a DSB on one homolog disfavoring DSB formation on its partner. (ii) Alternatively, increased or decreased activity at a site on one homolog can coordinately increase or decrease DSB formation on the homolog [refs. 4, 5 (case C) 8, and 9]. This effect likely occurs before DSB formation via trans modulation of chromatin/chromosome structure.
A DSB on one chromatid identifies a homologous region, usually on a (nonsister) chromatid of the homolog, giving rise to a nascent interhomolog d-loop. These initial interhomolog interactions then undergo regulated differentiation, with a subset designated for maturation into crossover (CO) recombination products and the remainder fated for maturation into noncrossover (NCO) products (10–13). COs are evenly spaced along homolog pairs. This pattern may reflect the combined effects of a driving force for CO designation and the fact that CO designation concomitantly sets up a zone of “interference” that disfavors occurrence of further events nearby (12, 14, 15). DSB-initiated interhomolog interactions also tend to be evenly spaced along each homolog pair (e.g., ref. 16), potentially by an analogous logic.
When DSBs occur, after DNA replication, a diploid meiotic cell contains four versions of each chromosome, two sisters for each of the two (maternal and paternal) homologs. We investigate here whether, at a single given initiation site, DSBs occur independently on the four available chromatids and, if not, what constraints might apply.
Results
Theoretically Possible DSB Distributions.
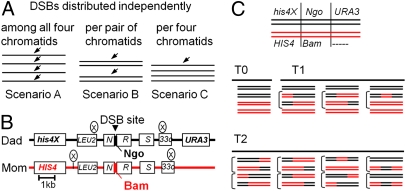
For any given DSB site, in the absence of programmed constraints, breaks would occur independently on the four chromatids. Alternatively, DSBs might be constrained to occur once per pair of chromatids, presumptively once per pair of sisters, or once per four chromatids, i.e., once per pair of homologs (Fig. 1A, scenarios A–C). These scenarios might occur singly or in combination. For all cases, the probability that a meiotic nucleus will acquire zero, one, two, or (for scenario A) more than two DSBs, at a single given site, can be calculated, as a function of the population average per-chromatid frequency of DSBs, by a suitable version of the binomial distribution (Fig. S1A).
Fig. 1.
Theory and analysis of DSB distribution among four chromatids. (A) Possible distributions of DSBs at a single site among four chromatids. (B) Map of the HIS4LEU2 locus. (C) Representative examples of nuclei exhibiting zero, one, or two (or more; text) events (T0, T1, or T2, respectively).
Per-Nucleus Analysis of in Vivo Event Distributions.
No available method can evaluate the number of DSBs occurring at a particular site on a per-nucleus basis. However, at a strong DSB hot spot, nearly all recombination events that occur in the immediate vicinity of that position have been initiated by DSBs at that site. At such a locus, the number of recombinational interactions in a single nucleus, detected after meiosis, can serve as a proxy for the number of DSBs. In budding yeast, such analysis is possible by classical tetrad analysis. Given suitable marker differences between maternal and paternal homologs at the locus of interest, the number and nature of recombination events occurring in each individual meiotic cell are manifested in the genotypes of its four resulting haploid spores. The four haploid spores of each tetrad are separated, placed on growth medium, and germinated under fully permissive conditions, and genotypes of the cells in each spore clone determined by physical and genetic methods. Frequencies of nuclei exhibiting different numbers of recombinational interactions, and thus different numbers of in vivo DSBs, can, after accommodating certain complexities (below), be compared with the frequencies predicted by theoretical scenarios for DSB distributions among the four chromatids.
This approach is illustrated for the HIS4LEU2 hot spot, where DSBs occur within a single ∼100-bp region (5, 17). Recombination events initiated by these DSBs were detected using strains in which the two parents differ by three markers: a pair of heterozygosities at nearby flanking positions and a single base pair heterozygosity within the ∼100-bp DSB site itself (Fig. 1B). Individual chromatid genotypes were determined (SI Materials and Methods) and tetrads sorted into categories according to the number of recombination events detected (full tetrad descriptions in Fig. S2). Tetrads with zero events exhibit two chromatids of each parental genotype (type 0; Fig. 1C, Middle Left). Tetrads with one event exhibit either two nonparental chromatids whose marker arrangements reflect a single CO or one nonparental chromatid reflecting non-Mendelian segregation at the central marker without any evidence of a CO, i.e., a single “noncrossover” event (non-Mendelian segregation or “NCO+”) (type 1; Fig. 1C, Middle Right). Tetrads exhibiting other marker combinations must have undergone two or more events, with various combinations of COs and/or NCO+ events possible (type 2; Fig. 1C, Bottom). Analysis of a sufficiently large number of tetrads yields experimentally derived frequencies of type 0, type 1, and type 2 tetrads that describe the probabilities that a nucleus will contain zero, one, or two (or more) experimentally detectable recombinational interactions (T0, T1, and T2; Table 1).
Table 1.
Per-nucleus recombination patterns
| Frequencies of different tetrad types* |
Frequency of events per chromatid |
|||||
| Strain | No. tetrads analyzed | T0 | T1 | T2 | Observed | Total† |
| HIS4LEU2 | ||||||
| Wild type | 343 | 0.20 | 0.61 | 0.19 | 0.25 | 0.29 |
| sml1Δ | 288 | 0.23 | 0.60 | 0.17 | 0.23 | 0.28 |
| tel1Δ | 326 | 0.18 | 0.46 | 0.36 | 0.30 | 0.35 |
| sgs1-ΔC795 | 239 | 0.27 | 0.52 | 0.21 | 0.23 | 0.33 |
| sgs1-ΔC795‡ | 239 | 0.27 | 0.57 | 0.16 | 0.22 | 0.31 |
| ndj1Δ | 312 | 0.34 | 0.50 | 0.16 | 0.21 | 0.26 |
| mec1Δ sml1Δ§ | 240 | 0.37 | 0.47 | 0.17 | 0.20 | 0.29 |
| dmc1ΔRAD54OP | 284 | 0.46 | 0.33 | 0.21 | 0.19 | 0.22 |
| spo11HA | 412 | 0.38 | 0.51 | 0.11 | 0.18 | 0.22 |
| HIS4LEU2old | 347 | 0.31 | 0.60 | 0.09 | 0.20 | 0.29 |
| his4::URA3-arg4 | 388 | 0.40 | 0.48 | 0.11 | 0.18 | 0.21 |
*As in Fig. 1 except data for his4::URA3-arg4 are from ref. 18. Margins of error at 95% confidence levels are ≤0.06 in all cases (Table S1). Spore viability frequencies are in Table S1.
†“Total” represents the sum of the frequencies of observed events plus invisible events and corresponds to the total population average level of DSBs per chromatid, defined as parameter “X” in binomial distribution equations (text and Fig. S1). Total = X = [Obs/(1 − inv)], where inv is the fraction of DSBs that give an invisible event (text). Values of inv are specified experimentally for all strains: inv = 0.15 except for sgs1-ΔC795 and mec1Δ sml1Δ, inv = 0.3; ndj1Δ, inv = 0.2; and HIS4LEU2old, inv = 0.3 (SI Materials and Methods).
‡Adjusted for increase in two-event DSBs (ref. 21 and SI Materials and Methods).
§sml1Δ suppresses the inviability of mec1Δ. sml1Δ alone has no detectable effect on recombination (Table S1 and text).
T0, T1, and T2 were defined at three hot spots in wild-type (WT) meiosis: HIS4LEU2 (above); a second allele of this same locus, “HIS4LEU2old”, where DSBs occur at two different sites separated by ∼2 kb (site I and site II) (5); and an unrelated hot spot, his4::URA3-arg4, created by molecular insertion of ARG4 sequences at HIS4 (18, 19). T0, T1, and T2 were also defined for HIS4LEU2 in seven mutant strains known to affect recombination without dramatically affecting spore viability (required for this analysis). Table 1 shows T0, T1, and T2 and the population-average frequency of experimentally observed events (“Obs”), for all 10 strains.
Experimental vs. Predicted Event Distributions.
T0, T1, and T2 (above) cannot be compared directly with the predicted probabilities of nuclei exhibiting zero, one, two (or more) DSBs given by binomial distribution analysis, for two reasons:
i) Some DSB-provoked recombinational interactions are “invisible” to experimental detection because they do not alter the array of assayed markers along the chromatids. Such interactions include (i) recombinations between a DSB and its sister chromatid and (ii) recombinations between a DSB and a homolog chromatid that are resolved without yielding either a CO or non-Mendelian segregation at the central marker (“NCO−” events). Theoretical equations for scenarios A–C were thus modified to include a parameter, “inv”, which is the fraction of DSBs that give invisible events (Fig. S1D). The resulting equations yield the theoretically predicted probabilities that a nucleus will exhibit zero, one, or two experimentally observable events (P0, P1, P2), as a function of both the population average DSB frequency per chromatid (X) and inv.
ii) In scenario A (but not scenarios B and C), a nucleus might acquire three or four DSBs (Fig. 1). Over the range of DSB levels relevant in vivo (below) the predicted possible frequencies of such nuclei are sufficiently small that they can be safely ignored. Nonetheless, we assume that such nuclei survive but exhibit multiple events, thus appearing as type 2 tetrads (Fig. S1B). Indistinguishable results are obtained by assuming that such nuclei do not yield four viable spores and are thus not represented in the experimental dataset (Fig. S1B legend).
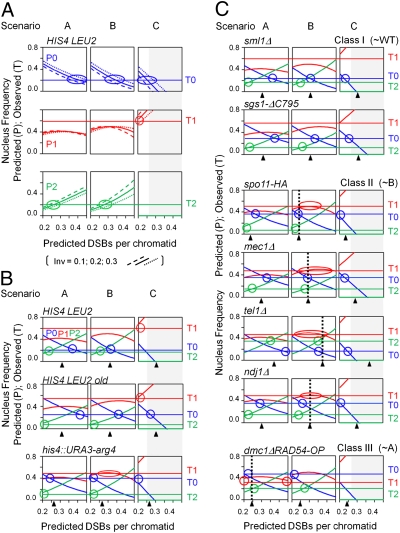
The possibility of a match between the experimental data for any given strain and any particular DSB distribution scenario can be evaluated by comparing T0, T1, and T2 with P0, P1, and P2 for that scenario over appropriate ranges of total DSB levels and frequencies of invisible events (X and inv), as illustrated for HIS4LEU2 in WT meiosis (Fig. 2A). For each scenario (A, B, or C), predicted probability curves for each nucleus type (P0, P1, and P2 in blue, red, and green, respectively) are shown for values of X and inv that span experimentally observed values (Table 1). The three different scenarios give qualitatively different predicted patterns, all of which are relatively insensitive to variations in inv. Values of T0, T1, and T2 are shown, relative to predicted patterns, as horizontal lines in Fig. 2A. An individual match (T0/P0, T1/P1, or T2/P2) occurs when the horizontal line intersects a predicted curve (Fig. 2A, circles). When a given dataset matches a given theoretical scenario, (i) matches occur for all three nucleus types, (ii) the three matches all occur at the same single value of X, and (iii) this common value of X corresponds to the value of X observed experimentally (Table 1 and Fig. 2, black triangles). By these criteria, the distributions of nucleus types for HIS4LEU2 in WT meiosis are not compatible with any of the three basic scenarios (A–C). The same results are seen more compactly in Fig. 2 B and C, with matches evaluated at the experimentally defined value of inv for each strain (Table 1).
Fig. 2.
Probabilities of tetrads with zero, one, or two events as a function of total DSB levels as predicted by scenarios A–C (P0, P1, and P2; curves) (Fig. 1) are compared with observed frequencies of such tetrads (T0, T1, and T2; horizontal lines) (Table 1). Black triangles indicate experimentally derived frequencies of total DSBs (Table 1, “Total”). Possible matches are indicated by vertical dashed lines. (A) HIS4LEU2 in WT meiosis. Theoretical distributions are shown for three values of inv. (B and C) Comparisons of indicated strains at appropriate corresponding values of inv (Table 1).
Event Distributions in WT Meiosis.
If DSBs were distributed independently among the four chromatids (scenario A; Fig. 2 A and B, Left), increasing DSB level would be accompanied by a decreased probability of nuclei with zero observable events (P0) and an increased probability of nuclei exhibiting two observable events (P2). These two effects roughly balance such that the predicted probability of nuclei exhibiting one observable event (P1) remains relatively constant.
In WT meiosis, for all three hot spots examined, the observed probabilities of nucleus types (T0, T1, and T2) do not match those predicted by scenario A (P0, P1, and P2) (Fig. 2 A and B). In all three cases, (i) there is no match between observed and predicted probabilities of one-event nuclei at any DSB level because the observed frequency is always significantly higher than the predicted frequency, and (ii) matches are possible for zero- and two-event nuclei, but occur at nonoverlapping ranges of DSB levels that, also, are higher and lower than the experimental value, respectively. Thus, meiotic DSBs at a single hot spot are not distributed independently among the four chromatids.
At HIS4LEU2, scenario B is excluded by absence of any possible match for one-event nuclei and scenario C is excluded because it implies that every nucleus will get either zero or one DSB, whereas two-event nuclei occur with significant probability (Fig. 2 A and B). However, scenario B gives a significantly better match than scenario A, with a lesser discrepancy for one-event nuclei (P1 vs. T1) and matches for both zero- and two-event nuclei (P0 vs. T0 and P2 vs. T2) at intermediate DSB frequencies that are approximately within the range of experimentally defined values. Thus, imposition of the one-per-pair constraint significantly improves the match between observed and predicted distributions of nucleus types relative to that obtained with scenario A. Moreover, scenarios B and C are inadequate to explain observed event distributions for complementary reasons: The observed level of one-event nuclei is too high for scenario B, but not for scenario C, whereas the observed level of two-event nuclei is too high for scenario C, but not for scenario B. Thus, the in vivo situation might be explained satisfactorily by a combination of one-per-two and one-per-four constraints. Any one-per-four constraint would imply trans inhibition.
Event distributions at the two other loci in WT meiosis support the possibility of a mixed B/C scenario. At HIS4LEU2old, predicted relationships to scenarios B and C are very closely similar to those for HIS4LEU2 (Fig. 2B and further discussion in Fig. S3) and a one-per-four constraint was identified previously at this locus by physical analysis (5). his4::URA3-arg4 exhibits the same trends, with a closer match with scenario B (Fig. 2B) specifically supporting existence of a one-per-two constraint.
Event Distributions in Mutants.
For the seven analyzed mutants, event distributions at HIS4LEU2 are also not explained by independent distribution of DSBs among four chromatids, for reasons analogous to the case for WT (Fig. 2C, scenario A). sml1Δ and sgs1-ΔC795 exhibit event distributions that are not significantly different from that of WT (Fig. 2C, class I). sml1Δ lacks a negative regulator of ribonucleotide reductase and is implicated in Mec1/ATR-mediated regulation of DNA replication (20). sgs1-ΔC795 lacks a helicase that eliminates multichromatid recombination intermediates (21). Correspondingly, Sml1 does not affect recombination and Sgs1 affects recombination at steps well after DSB formation and establishment of DSB/partner interactions (21). The other five mutants exhibit a significantly different event distribution from WT (P < 0.001 by G-test). Among these, four mutants exhibit possible matches with scenario B (Fig. 2C, class II), with scenario C excluded by the presence of significant levels of two-event nuclei. Identification of this pattern in several mutants strongly supports the existence of a one-per-pair constraint on DSB distribution. By implication, WT would exhibit a mixture of scenarios B and C and the mutants would be specifically defective in the one-per-four constraint. These four mutants are tel1Δ and mec1Δ sml1Δ (hereafter mec1Δ; Table 1, footnote §), which, respectively, lack the related chromosome-based signal transduction molecules corresponding to mammalian ATM and ATR; ndj1Δ, which lacks a telomere-specific binding protein and exhibits multiple defects, some of which are indirect consequences of regulatory surveillance controls (22–25); and spo11-HA3His6 (hereafter spo11HA), which is defective in the efficiency of Spo11-mediated DSB catalysis (15). The final mutant, dmc1Δ RAD54-OP, exhibits a possible match to scenario A (independent distribution among four chromatids) (Fig. 2C, class III). Scenario C and scenario B are both excluded by the too-low frequency of one-event nuclei and also, for scenario C, by the presence of two-event nuclei (Fig. 2C). This mutant lacks meiotic RecA homolog Dmc1 and overexpresses Rad54 helicase, which substantially suppresses dmc1Δ defects (26).
Scenario D Combines One-per-Two and One-per-Four Constraints.
The above results point strongly to a situation where a one-per-pair constraint is present in all nuclei of all analyzed strains and a one-per-quartet constraint (trans inhibition) is also present in WT and operates with reduced efficiency in certain mutants compared with WT HIS4LEU2 and in his4::URA3-arg4 compared with the two HIS4LEU2 alleles (Fig. 3A and below).
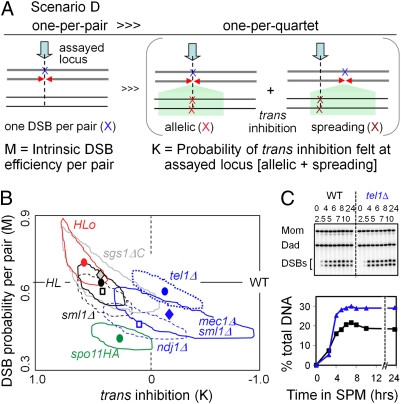
Fig. 3.
Scenario D. (A) DSBs occur with an intrinsic probability of M per pair of chromatids (Left), irrespective of any trans effect. In addition, some fraction of the nuclei, K, that would otherwise have given rise to DSBs on both homologs give rise, instead, to a DSB on only one of the two homologs (Right). K reflects trans inhibition in which, at the assayed site, a DSB on one chromosome is inhibited by a DSB either at the exact allelic site on the homolog (allelic inhibition) or in the vicinity of that allelic site (spreading inhibition). M and K are thus “intrinsic DSB efficiency per pair of chromatids” and “strength of trans inhibition,” respectively. Scenario D is describable by the binomial distribution F(0) = F(0)B = (1 − M)2; F(1) = F(1)B + M2K = 2M(1 − M) + M2K; F(2) = M2(1 − K). And predicted probabilities of nuclei exhibiting zero, one, or two recombination events (F0, F1, and F2) can be converted to frequencies of observable events (P0, P1, and P2) as a function of inv (defined as in scenarios A–C) as for other scenarios (text and Fig. S1 C and D). (B) By scenario D, each experimental dataset (T0, T1, or T2) is described by unique values of K and M (symbols; surrounding areas = 95% confidence levels from Table 1 and Table S1) (text). HIS4LEU2, M = 0.68, K = 0.44; HIS4LEU2 old, M = 0.73, K = 0.65; ndj1Δ, M = 0.53, K = 0.12; mec1Δ, M = 0.56, K = −0.15; tel1Δ, M = 0.67, K = −0.1; sgs1-ΔC795, M = 0.72, K = 0.48; and spo11HA, M = 0.46, K= 0.27. (C) DSB levels in WT and tel1Δ at HIS4LEU2 in a rad50S background over time in meiosis. (Upper) One-dimensional gels; (Lower) quantification.
This combination of constraints, “scenario D”, is again describable by the binomial distribution (Fig. 3 A and B). Here, DSBs occur with an intrinsic probability of “M” per pair of chromatids, irrespective of any trans effect. In addition, some fraction of the nuclei, “K”, that would otherwise have given rise to DSBs on both homologs give, instead, a DSB on only one of the two homologs. M and K thus correspond to “intrinsic DSB efficiency per pair of chromatids” and “strength of trans inhibition,” respectively. Predicted probabilities of nuclei exhibiting zero, one, or two recombination events (P0, P1, and P2) are a function of these two variables plus inv (defined above).
For any experimental array of nucleus types, at a specified inv, scenario D defines specific unique values of both M and K at which T0, T1, and T2 match P0, P1, and P2 (Fig. 3B).
The predicted level of trans inhibition for HIS4LEU2 and HIS4LEU2old in WT meiosis, and for the two mutants with WT-like event patterns (class I above; sml1Δ and sgs1-ΔC795), is very high, K = 0.45–0.65 (Fig. 3B and Fig. S4). That is, in any nucleus where a first DSB occurs on one homolog, the probability that a DSB will also occur on the partner chromosome is reduced by ∼50% of what it would have been in the absence of trans inhibition. All four of these strains also exhibit very high predicted intrinsic DSB probabilities, M = 0.6–0.7. That is, a given homolog (pair of chromatids) has an intrinsic probability of DSB formation of 60–70% in the absence of trans inhibition. Both parameter values match other indications that HIS4LEU2 is an unusually robust and tightly controlled DSB hot spot (e.g., refs. 15 and 27).
A previous study showed that trans inhibition can spread for some distance along the target chromosome in the vicinity of the position exactly allelic to the site that is the source of the effect (5) (Fig. 3A, Right). Thus, at any particular assayed position, the amount of trans inhibition as described by the parameter K of scenario D actually reflects the sum of two components (Fig. 3A): (i) “allelic inhibition”, i.e., inhibition of DSB formation at the assayed site by a DSB at the exact allelic site on the homolog “donor” chromosome, and (ii) “spreading inhibition”, reflecting the fact that DSB formation at the assayed position is also inhibited by DSBs that occur in the vicinity of the allelic site on the homolog (DSB donor).
The relative contributions of “allelic” and “spreading” effects (e.g., at HIS4LEU2) are not known. However, the two effects are differentiable. A mutation that globally reduces the efficiency of DSB formation per se, M, will have no effect on allelic trans inhibition but will reduce the strength of spreading trans inhibition because it will reduce the number of (nearby) DSBs that are contributing to this effect at the assayed locus. Thus, by scenario D, such a mutation will reduce K as well as M. Correspondingly, the spo11HA mutation, which is thought to specifically reduce DSB formation (15), exhibits not only a predicted reduction of ∼35% in the intrinsic probability of DSB formation (M), but also a reduction of ∼39% in trans inhibition (K) (Fig. 3B).
Interestingly, one assayed mutant, tel1Δ, exhibits no alteration in the intrinsic efficiency of DSB formation with an essentially complete absence of trans inhibition (Fig. 3B), implying that these two features of DSB formation are functionally distinct and implicatingTel1 as required for trans inhibition per se. Two other assayed mutants, mec1Δ sml1Δ and ndj1Δ, are similar to spo11HA in exhibiting reductions in both M and K; however, compared with spo11HA, both mutants exhibit lesser reductions in DSB efficiency but greater defects in trans inhibition (Fig. 3B). Thus, in both mutants, the reduction in DSB efficiency cannot (fully) account for the defect in trans inhibition. Because sml1Δ exhibits no difference from WT, the mec1Δ sml1Δ defect is attributable to the mec1Δ mutation. Correspondingly, Mec1 and Ndj1 are implicated as being required, separately, for both efficient DSB formation and efficient trans inhibition. We note that the loss of a one-per-four constraint in these mutants could alternatively be interpreted as reflecting a loss of the one-per-pair-of-sisters constraint but retention of trans inhibition. We cannot exclude this possibility but consider it to be less obvious.
For the final assayed mutant, dmc1Δ RAD54-OP, scenario D predicts a value of K much less than zero (K = −1.2). This prediction might mean that occurrence of a DSB on one chromosome increases (rather than decreases) the probability of a DSB on the partner chromosome, e.g., by abrogating the one-per-pair constraint. Alternatively, promiscuous action of Rad51 in this mutant might yield discoordination of the two DSB ends such that one DSB produces interactions with more than one partner (21).
We also note that because the effect of trans inhibition is to preclude formation of a subset of DSBs, any mutation that reduces trans inhibition will tend to increase the population-average levels of DSBs (and thus DSB-initiated recombinational interactions). Correspondingly, tel1Δ, which decreases trans inhibition without affecting DSB efficiency (Fig. 3B), exhibits a higher frequency of recombination events than WT (Table 1). This increase, not previously known, is also seen by physical analysis of total levels of DSBs (Fig. 3C) at HIS4LEU2. Similarly, spo11HA, mec1Δ sml1Δ, and ndj1Δ mutants, although reduced for the efficiency of DSB formation, nonetheless exhibit a higher frequency of recombination events than would be predicted from their DSB efficiencies alone (Fig. S5). Thus, functions involved in trans inhibition will appear as negative regulators of DSB formation (Discussion).
Trans Inhibition at Weaker DSB Sites.
By scenario D, the intrinisic activity of a DSB site and its efficiency in effecting trans inhibition are unrelated. Thus, DSBs occurring at a weak DSB site might still be perfectly efficient at inhibiting DSB formation at the allelic position on the homolog. Distribution analysis would detect such a feature if it were possible to specifically identify the events arising only from one particular weak site. In contrast, physical analysis of population average DSB levels will detect trans inhibition only at very hot sites. Trans inhibition reduces DSB formation only the minority subset of nuclei that would otherwise have undergone two DSBs (one on each homolog), which is a significant fraction of the total only for the very hottest DSB sites. This feature explains why, in previous physical analyses of DSB levels, trans inhibition was detected only for the two hottest analyzed sites [refs. 5 (case D) and 6]. Even at his4::URA3-arg4, a still quite active DSB hot spot, trans inhibition is readily detected by scenario D analysis (M = 0.43 and K = 0.18) but would never be detected by physical analyses, where presence/absence of inhibition is predicted to alter DSB/event levels by only ∼1% (Fig. S6). This consideration has a further implication: The fact that another type of trans effect could be detected by physical analysis even at weaker sites (introductory section at the beginning of this paper) implies that this second process is affecting many or all nuclei, not a minority subset, in accord with its occurrence before DSB formation (introductory section and refs. 5, 8, and 27).
Discussion
The presented results demonstrate that meiotic DSBs at a single initiation site are not independently distributed among the four available chromatids. Logically, DSBs could be constrained to occur one-per-pair of chromatids and/or one-per-quartet of chromatids. Experimentally observed patterns of nucleus types in WT meiosis are not compatible with either of these constraints alone but are well explained by a combination of the two. It appears that DSB formation (i) is always constrained to occur one-per-pair of chromatids, presumptively one-per-pair of sisters, and (ii) is constrained in some nuclei to occur one-per-four chromatids. The latter constraint implies communication between homologs in trans. This trans inhibition spreads for some distance along the target chromosome. Mutant phenotypes raise the possibility that signal transduction kinases Mec1 (ATR) and Tel1 (ATM) are potential direct effectors of trans inhibition.
Molecular Basis for Constraints on DSB Distribution Among Chromatids.
A one-per-pair constraint on DSB formation presumptively reflects a situation in which a DSB can occur on only one of the two sister chromatids. This constraint, not previously appreciated, could reflect features built into pre-DSB recombination complexes, which contain both sisters (11, 17, 28), via core recombinosome features and/or surrounding chromosome structural determinants (13, 17, 28). A one-per-four constraint was previously implied by physical analysis of DSBs. That analysis also suggested that this trans inhibition effect is directly mediated by DSBs, with occurrence of a DSB on one chromosome actively disfavoring formation of a DSB on the homolog partner, and showed that this effect spreads some distance away (5, 6). The present study confirms and extends these conclusions.
By the analysis presented, several mutants exhibit only a one-per-pair constraint. Scenario D analysis envisions that this phenotype implies loss of trans inhibition. Mutants lacking Mec1 or Tel1, related chromosome-based signal transduction kinases homologous to ATR and ATM, respectively, are two such mutants. Direct involvement of these particular molecules in this process would be attractive, given their diverse central roles in complex global and local regulation of chromosomal processes in normal cellular programs, including CO interference during meiosis in Drosophila, which is also a “spreading trans inhibition effect” (29–31). Also, a recent mouse study identified ATM as a negative regulator of DSB formation, with an increase in DSB levels relative to WT, analogous to the findings reported here (32). Thus, abrogation of trans inhibition could explain all or part of ATM's role in that organism. This analysis further suggests that Mec1/ATR (but not Tel1/ATM) is also required for efficient DSB formation per se, irrespective of its role in trans inhibition, implying two mechanistically distinct functions for this molecule during early meiotic prophase. Absence of Ndj1 confers defects in both DSB formation and trans inhibition similar to those conferred by absence of Mec1/ATR. Because Ndj1 appears to localize specifically at telomeres (24, 25), its roles may be indirect, e.g., via triggering of regulatory effects that alter Mec1 activity.
DSB studies have also revealed that an increase in DSB activity at one position tends to inhibit DSBs in the vicinity in cis (Introduction). This effect, not visible in the present analysis, could potentially operate by a common mechanism to trans inhibition. Alternatively, cis and trans inhibition might be distinct, occurring by differentiable mechanisms, e.g., on the donor and “recipient” homologs and/or before and after DSB formation, respectively.
Implications for Patterning of DSBs/Interhomolog Interactions.
In any given nucleus, DSBs occur at a subset of all available possible positions, differently in different nuclei. When a DSB contacts its homolog partner and sets up an interhomolog interaction, it concomitantly sets up trans inhibition of DSB formation on the homolog. This effect necessarily also sets up trans inhibition of additional interhomolog interactions. Such inhibition occurs not only at the allelic position on the target chromosome but also over some distance in the vicinity of that position along the target chromosome, thus disfavoring formation of interhomolog interactions nearby. The existence of spreading trans inhibition will confer two important effects on spatial patterning of DSBs and, most critically, interhomolog interactions (Fig. 4 A and B).
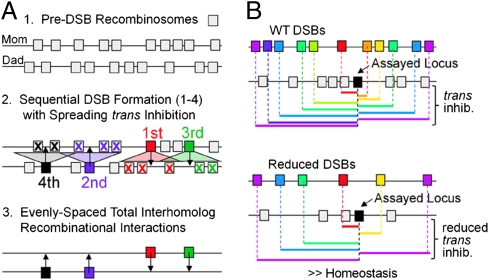
Fig. 4.
Spreading trans inhibition confers even spacing (A) and homeostasis (B) of DSBs and resultant interhomolog interactions. (A) Starting with a random array of pre-DSB recombination complexes (1, gray boxes), sequential DSBs and interhomolog interactions will tend to fill in the holes between prior events (2), conferring a tendency for even spacing (3). In 2, pre-DSB recombinosomes unaffected by trans inhibition could be inactivated by cis inhibition (text). (B) (Upper) Given an array of pre-DSB recombination complexes (colored boxes), formation of DSBs and interhomolog interactions at any particular assayed locus (black box) will be inhibited by spreading trans inhibition emanating from DSBs at nearby loci on the homolog (colored lines). (Lower) Given a global decrease in the intrinsic probability of DSB formation throughout the genome, e.g., by a defect in Spo11-mediated catalysis (and thus a reduced probability of DSBs along any given region of a particular nucleus, indicated by X's), all regions will concomitantly exhibit a decrease in the probability that those processes will be inhibited by spreading trans inhibition (seen for a given assayed locus by fewer colored lines). Thus, at all loci throughout the genome, the effect of reduced catalysis efficiency will be partially compensated for by the reduction in spreading trans inhibition, resulting in “homeostasis” for both DSBs and, more importantly, interhomolog interactions.
Even spacing.
Occurrence of a DSB and ensuing interhomolog interaction at one position disfavors subsequent occurrence of other such events in the immediate vicinity of that first interaction. Then, as the DSB/interhomolog interaction process continues, subsequent events will tend to fill in the holes between previous events. As a result DSB/interhomolog interactions will tend to be evenly spaced along the chromosome (Fig. 4A) (ref. 12 and discussion in ref. 16), thus explaining the experimentally observed pattern (Introduction).
Homeostasis.
When the intrinsic probability of DSB formation at a particular locus decreases, the level of spreading trans inhibition emanation from that position will also decrease, thus resulting in an increase in the overall probability of DSBs (on the homolog) and interhomolog interactions in the vicinity (Fig. 4B). By extension, if there is a global decrease in the probability of DSB formation throughout the genome, every locus will be similarly affected. As a result, throughout the genome, the levels of both DSBs and interhomolog interactions will be decreased less than they would otherwise have been. Put another way, because of spreading trans inhibition, both DSB formation and establishment of interhomolog interactions are homeostatic processes: A local or global reduction is partially compensated by a concomitant reduction in resulting spreading inhibition.
General Implications for Spatial Patterning.
The logic described above for DSB formation and establishment of interhomolog interactions is the same as that previously proposed for CO patterning, with a driving force for CO designation, and each CO designation event nucleates a spreading zone of inhibition that disfavors further CO designation events nearby (CO interference). As more and more events occur, they will fill in the holes between prior events, yielding a tendency for even spacing of COs along a chromosome (12, 14). Additionally, CO designation has been shown to exhibit homeostasis (15, 33). We therefore propose that meiotic recombination-mediated interhomolog interactions are spatially patterned by (at least) two successive rounds of patterning that involve precisely the same logic, first for DSBs and then for COs. The basic features of even spacing and homeostasis will apply to any spatial patterning process that involves the basic logic of a “driving force” for occurrence of an event plus coupling of event occurrence to spreading inhibition and to any specific mechanism that operates by imposition and relief of mechanical stress (12).
Biological Significance(s) of One-per-Two and One-per-Four Constraints on DSB Distribution.
Constraining DSB formation to one-per-pair of sisters ensures that, if ensuing interhomolog recombination goes awry, the sister chromatid will always be present as an intact template for repair of the DSB. Constraining DSB formation to one-per-pair of homologs ensures that the complex events involved in establishment of an interhomolog interaction will tend not to occur twice at the same (allelic) chromosomal position or, given spreading of trans inhibition, at two nearby chromosomal positions. Given that interhomolog recombinational interactions ultimately develop into discrete physical linkages between the structural axes of homologs (e.g., ref. 16), it may be particularly important not to try to have two such linkages in close proximity because attempted formation of two interaxis “bridges” at the same or nearby positions is likely to be deleterious. Because DSB-mediated interhomolog interactions mediate whole-chromosome juxtaposition (“pairing”) of homologs, even spacing will be advantageous in ensuring that this process occurs regularly along the entire length of a chromosome. Homeostasis should also be advantageous because, as pointed out previously for CO homeostasis (15), it provides a buffering system that protects meiotic chromosome dynamics from local, regional, and/or global defects in DSB formation and/or establishment of total interhomolog interactions.
Materials and Methods
Strains are derived from SK1 (Table S2). Tetrad asci were dissected onto YPD plates. Spore phenotypes were determined by replica plating and colony PCR or Southern blot (SI Materials and Methods). The theoretical DSBs distributions at a single DSB site were described by a set of binomial equations (Fig. S1) and the level of invisible events (inv) was specified for each strain as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Keeney, N. Hunter, and D. Bishop for strains/plasmids. Support (for L.Z.) and partial support (for A.S. and K.P.K.) for this research were provided by National Institutes of Health Grant GM044794 (to N.E.K.). A.S. was also supported by Consiglio Nazionale delle Ricerche and by Italian Ministry of Foreign Affairs (2011 Grant “Con il contributo del Ministero degli Affari Esteri, Direzione Generale per la Promozione del Sistema Paese”). K.P.K. was also supported by a National Research Foundation of Korea grant funded by the government of the Republic of Korea (Ministry of Education, Science and Technology, no. 20110029504).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117937108/-/DCSupplemental.
References
- 1.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau DH, editors. Recombination and Meiosis: Models, Means and Evolution (Genome Dynamics and Stability 3) New York: Springer; 2008. pp. 165–193. [Google Scholar]
- 3.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 4.Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res. 2008;36:984–997. doi: 10.1093/nar/gkm1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan QQ, Xu F, White MA, Petes TD. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocco V, Nicolas A. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 9.Bullard SA, Kim S, Galbraith AM, Malone RE. Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 11.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Heidelberg: Springer; 2006. pp. 381–442. [Google Scholar]
- 12.Kleckner N, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleckner N, Zhang L, Weiner B, Zickler D. Meiotic chromosome dynamics. In: Karsten R, editor. Genome Organization and Function in the Cell Nucleus. Weinheim, Germany: Wiley-VCH; 2011. pp. 487–533. [Google Scholar]
- 14.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 15.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storlazzi A, et al. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010;141:94–106. doi: 10.1016/j.cell.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KP, et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosaka H, Shinohara M, Shinohara A. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 2008;4:e1000196. doi: 10.1371/journal.pgen.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu HY, Burgess SM. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol. 2006;16:2473–2479. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 25.Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 26.Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney S, Kleckner N. Communication between homologous chromosomes: Genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996;1:475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 28.Panizza S, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Barchi M, et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carballo JA, Cha RS. Meiotic roles of Mec1, a budding yeast homolog of mammalian ATR/ATM. Chromosome Res. 2007;15:539–550. doi: 10.1007/s10577-007-1145-y. [DOI] [PubMed] [Google Scholar]
- 31.Carpenter AT. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma. 1979;75:259–292. doi: 10.1007/BF00293472. [DOI] [PubMed] [Google Scholar]
- 32.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci USA. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.