Abstract
While recent years have witnessed dramatic advances in our capacity to identify and quantify an ever-increasing number of plant metabolites, our understanding of how metabolism is spatially regulated is still far from complete. In an attempt to partially address this question, we studied the storage metabolome of the barley (Hordeum vulgare) vacuole. For this purpose, we used highly purified vacuoles isolated by silicon oil centrifugation and compared their metabolome with that found in the mesophyll protoplast from which they were derived. Using a combination of gas chromatography-mass spectrometry and Fourier transform-mass spectrometry, we were able to detect 59 (primary) metabolites for which we know the exact chemical structure and a further 200 (secondary) metabolites for which we have strong predicted chemical formulae. Taken together, these metabolites comprise amino acids, organic acids, sugars, sugar alcohols, shikimate pathway intermediates, vitamins, phenylpropanoids, and flavonoids. Of the 259 putative metabolites, some 12 were found exclusively in the vacuole and 34 were found exclusively in the protoplast, while 213 were common in both samples. When analyzed on a quantitative basis, however, there is even more variance, with more than 60 of these compounds being present above the detection limit of our protocols. The combined data were also analyzed with respect to the tonoplast proteome in an attempt to infer specificities of the transporter proteins embedded in this membrane. Following comparison with recent observations made using nonaqueous fractionation of Arabidopsis (Arabidopsis thaliana), we discuss these data in the context of current models of metabolic compartmentation in plants.
Technological developments in the last decade have rendered metabolomics, the comprehensive study of small molecule metabolites, an important functional genomic tool (Fiehn, 2002; Sumner et al., 2003; Weckwerth, 2003; Farré et al., 2006; Matsuda et al., 2009; Saito and Matsuda, 2010). It is currently widely applied for a variety of purposes, ranging from simple diagnostic applications (Catchpole et al., 2005; Tikunov et al., 2005; Baker et al., 2006; Scherling et al., 2009) and gene functional annotation (Tohge et al., 2005, 2007) to being an important component in systems biology-oriented research (Fernie et al., 2004; Saito and Matsuda, 2010). Metabolomics has additionally found great utility in the characterization of the metabolic response of plant cells to a range of biotic and environmental stresses (Cook et al., 2004; Hirai et al., 2004; Kaplan et al., 2004; Ishizaki et al., 2005; Urbanczyk-Wochniak and Fernie, 2005; Roessner et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Lehmann et al., 2009). However, despite the many important breakthroughs facilitated by this technology, we remain somewhat hampered in our understanding by our inability to gain high-resolution spatial data on metabolism (Stitt and Fernie, 2003; Fernie, 2007; Lunn, 2007).
Generally, metabolism is treated in a tissue-specific manner but cells within a tissue are treated as homogenous. Given that there are around 40 different cell types in plants (Goldberg, 1988; Martin et al., 2001), this is clearly an oversimplification. Recent technical advances and the widespread adoption of the GUS fusion protein expression approach (Sundaresan et al., 1995) as well as the increasing availability of resources such as Genevestigator and the BAR databases (Zimmermann et al., 2004; Brady and Provart, 2009), however, have made the identification of cell type specificity of gene expression facile. These developments have been paralleled by the increasing use of so-called single cell techniques, allowing microdissection of tissues into their constituent cell types (Schad et al., 2005; Cai and Lashbrook, 2006; Obel et al., 2009). These advances hold great promise for being able to better untangle metabolism at the cell type level, and a metabolite interface for the BAR gene expression atlas has indeed recently been published (Matsuda et al., 2009).
Going to even higher levels of resolution allows the analysis of subcellular and even suborganellar aspects of metabolism. Generally, the spatial distribution of metabolites mirrors that of the proteins involved in their metabolism. Given the large number of reports concerning plant organellar proteomes (Igamberdiev et al., 2001; Millar et al., 2001; Carter et al., 2004; Kleffmann et al., 2004; Pendle et al., 2005; Reumann et al., 2007) and the availability of the SUBA database of protein localization (Heazlewood et al., 2007), it should be relatively easy to predict the location and operation of metabolic pathways. However, the facts of organelle-specific isoforms and dual targeting of proteins often result in the duplication of metabolic reactions within different compartments of the cell (Carrari et al., 2003; Millar et al., 2006). Furthermore, several instances exist in which proteins are found in locations other than where they were thought to operate; for example, hexokinase proteins have been found in the nucleus, while the Suc synthase has been localized to the mitochondria (Subbaiah et al., 2006; Cho et al., 2007). These observations thus render such predictions of metabolite localization, on the basis of protein localization alone, liable to error. Moreover, for studies aimed at understanding the function of the vacuole, which is largely metabolically inert, such an association-based approach is likely to be highly difficult. At present, there are three different approaches for the direct determination of subcellular metabolite contents: the expression of genetically encoded marker proteins (Fehr et al., 2002), the nonaqueous fractionation technique (Gerhardt et al., 1983), and silicon oil centrifugation of protoplasts (Igamberdiev et al., 2001). The first of these requires the construction and expression of several variants of a marker protein for each individual metabolite and thus is not tractable at the metabolome level. The second approach has already been coupled to plant cells and proved to be highly informative in characterizing transgenic potato (Solanum tuberosum) plants exhibiting alterations in the Suc-to-starch pathway (Farré et al., 2001, 2006). For the third approach, several proteome studies of the vacuole have been presented for Arabidopsis (Arabidopsis thaliana; Carter et al., 2004; Shimaoka et al., 2004; Jaquinod et al., 2007). Our study, however, is focused on barley (Hordeum vulgare), since it is our experience that the purity of vacuoles isolated from this species exceeds that from any other species. To this end, we isolated pure vacuoles from protoplasts derived from barley leaves and compared the metabolome of these organelles with that of the protoplast population from which they were derived using a combination of gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-Fourier transform-mass spectrometry (LC-FT-MS). We were able to provide information concerning 61 metabolites of confirmed chemical structure and a further 135 putative metabolites on the basis of their chemical formulae and assess their partitioning between the vacuolar and extravacuolar parts of the cell. Finally, the data were compared with literature values obtained by the nonaqueous fractionation approach and in relation to tonoplast transporters described in the literature (Martinoia et al., 2000, 2007; Kaspar et al., 2010) as well as with data recently published concerning the vacuolar ionome (Smart et al., 2011).
RESULTS
Isolation of the Barley Vacuole from Protoplast
To investigate vacuole metabolomics, barley vacuoles were purified from barley protoplast. Barley mesophyll protoplasts were prepared using fresh barley leaves grown in a greenhouse for 8 d. Isolation and purification of mesophyll protoplasts and vacuoles were performed as described previously (Endler et al., 2009). In order to get rid of the isolation solution, an additional centrifugation step, as described in “Materials and Methods,” was included. Protoplast samples were prepared as three replicates (P1– P3) and vacuole samples were prepared as six replicates (V1–V6) to evaluate biological and experimental variance. Purified vacuole fractions were assessed by α-mannosidase enzymatic activity, which is known as vacuole-specific enzyme (Boller and Kende, 1979), and chlorophyll content for evaluation of the contamination of chloroplast. Four independent sets of vacuole preparations resulted in 70 ± 29 milliunits mL−1 (vacuole fractions) and 22 ± 4 milliunits mL−1 (protoplast fractions) of α-mannosidase enzymatic activity and no (vacuole fractions) and 293 ± 52 μg mL−1 (protoplast fractions) of chlorophyll content, respectively. The liquid samples were lyophilized and extracted by 80% methanol for preliminary check by GC-MS analyses. These preparations rendered high reproducibility of metabolite data between experiments. To obtain enough sample for split use in both MS-based analyses, the fractions derived from a single sample set were frozen overnight, and after defreezing, 45 μL of the aqueous solution from each tube (50–60 tubes per experiment) was recovered and pooled (vacuole fraction, 77 milliunits mL−1 of α-mannosidase enzymatic activity, no chlorophyll content; protoplast fraction, 23 milliunits mL−1 and 272 μg mL−1, respectively). The pooled fractions were lyophilized and used for the metabolomic analysis. The variance of dry weight between replicates was 8.07 ± 0.36 mg (vacuole samples; n = 6) and 8.13 ± 0.60 mg (protoplast; n = 3). For the metabolomic studies, lyophilized samples were extracted by 80% methanol for split use to both MS-based analyses.
Global Difference and Fraction-Specific Peaks
Metabolite profiles were analyzed by GC-time of flight-MS for primary metabolites (Lisec et al., 2006) and LC-MS for secondary metabolites (Tohge and Fernie, 2010). The resultant data were combined into a single data matrix for multivariate analyses. First, we performed principal component analysis to visualize the extent of metabolomic changes between protoplast samples and vacuole samples (Supplemental Fig. S1A). Since a clear separation between the protoplast samples and vacuole samples was observed in PC1 (74.9%), we subsequently performed a Venn diagram analysis to illustrate the levels of commonality and difference between samples (Supplemental Fig. S1A). In total, 259 peaks were detected by GC-MS and LC-MS. Twelve peaks detected by LC-MS were exclusively detected in the vacuole samples (Supplemental Table S1). On the other hand, 34 peaks were observed as protoplast-specific peaks, meaning that 213 peaks were detected in both fractions. Given that the subcellular vacuole volume of barley leaves has been defined to be 60.5% of the total cell volume (Winter et al., 1993), we defined those compounds that were abundant at percentages above 60% in vacuole as “vacuolar accumulated.” Ninety-nine peaks were detected at high relative abundance (more than 1.5-fold change [FC]) in the vacuole samples, while 62 peaks were detected at low relative abundance (less than 0.5 FC). It is important to bear in mind here that vacuole-specific peaks could, in principle, also be detected in protoplast samples. For those that were solely detected in the vacuole, it thus seems likely that their levels were under the limit of detection in our protoplast samples. By contrast, protoplast-specific peaks suggest that these metabolites are not transported into the vacuole.
Metabolite Profiling of Primary Metabolites by GC-MS
As stated above, measurements of the relative levels of primary metabolites were performed by GC-MS (Lisec et al., 2006), and the absolute levels of 59 compounds were estimated based on concentration curves of authentic standard that were run side by side with the samples (Table I). The results show that the vacuole accumulates a large variety of different compounds: sugars, sugar alcohols, organic acids, and amino acids. A total of 22 amino acids and their related compounds were determined in vacuolar and protoplast samples. Three of them had higher vacuolar than protoplast levels (more than 90% of compound in the vacuole, more than 1.5 FC), seven were invariant (30%–90%, 0.5–1.5 FC), and seven displayed a lower vacuolar level (less than 30%, less than 0.5 FC; Table I; Fig. 1). Interestingly, His and Ala were highly more abundant in the barley vacuole than in the protoplast, followed by Trp (1.53 FC), Met (1.43 FC), and Ser (1.24 FC). The relative amounts of Glu (less than 0.02 FC), Pro (0.09 FC), Gln (0.10 FC), and Asp (0.12 FC) were very low inside the vacuole. The levels of other amino acids, such as Orn, Tyr, Ile, tyramine, Val, Phe, pyro-Glu, and Gly, were invariant. Additionally, we were also able to determine γ-aminobutyric acid amounts in both samples, but at higher levels in the protoplast samples (Table I; Fig. 1).
Table I. Metabolite contents of barley protoplasts and vacuole samples.
Values are presented as means ± sd of determinations on six replicates of vacuole fractions and three protoplast fractions. Percentage of content levels in vacuole was calculated by α-mannosidase activity. Given that the subcellular vacuole volume of barley leaves has been defined to be 60.5% of the total cell volume (Winter et al., 1993), we defined those compounds that were abundant at percentages above 60% in vacuole as vacuolar accumulated.
| Compound Name | FC | In Vacuole | Protoplast | Vacuole |
| % | nmol L−1 protoplast | |||
| Amino acids and derivatives | ||||
| His | >1.65 | >100 | 5.6 ± 1.7 | 21.9 ± 9.8 |
| Ala | >1.65 | >100 | 1,981.6 ± 127.6 | 4,789.2 ± 1,415.7 |
| Trp | 1.53 | 92.4 | 47.5 ± 3.9 | 43.9 ± 7.6 |
| Met | 1.43 | 86.7 | 22.4 ± 2.6 | 19.4 ± 2.1 |
| Ser | 1.24 | 75.2 | 4,190.2 ± 392.4 | 3,150.2 ± 1069.6 |
| Orn | 0.79 | 47.6 | 15.9 ± 0.6 | 7.6 ± 1.6 |
| Tyr | 0.78 | 47.4 | 1,612.4 ± 46.5 | 765.0 ± 92.7 |
| Ile | 0.78 | 46.9 | 1,848.5 ± 111.8 | 867.5 ± 150.4 |
| Tyramine | 0.65 | 39.4 | 12.8 ± 1.4 | 5.0 ± 0.8 |
| Val | 0.56 | 33.6 | 4,063.4 ± 442.1 | 1,364.0 ± 304.9 |
| Phe | 0.55 | 33.1 | 734.0 ± 29.0 | 242.9 ± 38.8 |
| Pyro-Glu | 0.51 | 30.6 | 1,583.2 ± 160.2 | 484.6 ± 63.5 |
| Gly | 0.43 | 26.2 | 1,949.2 ± 258.2 | 509.8 ± 131.9 |
| Asn | 0.35 | 21.4 | 980.2 ± 176.1 | 209.4 ± 103.4 |
| Thr | 0.33 | 20.2 | 4,169.7 ± 233.1 | 842.4 ± 99.1 |
| Lys | 0.31 | 18.9 | 1,028.3 ± 54.6 | 193.9 ± 42.7 |
| β-Ala | 0.31 | 18.6 | 143.6 ± 10.0 | 26.6 ± 2.4 |
| Asp | 0.12 | 7.5 | 1,358.7 ± 882.7 | 102.5 ± 17.9 |
| Gln | 0.10 | 5.8 | 990.8 ± 97.8 | 57.1 ± 9.5 |
| Pro | 0.09 | 5.2 | 62.2 ± 11.3 | 3.3 ± 0.8 |
| γ-Aminobutyric acid | <0.02 | <1 | 1,3381.1 ± 784.5 | 101.4 ± 7.9 |
| Glu | <0.02 | <1 | 98.8 ± 15.8 | 0.4 ± 0.1 |
| Organic acids | ||||
| Isocitric acid | >1.65 | >100 | 114.0 ± 1.9 | 1,080.0 ± 900.3 |
| Malic acid | >1.65 | >100 | 2,974.1 ± 78.5 | 6,352.5 ± 834.2 |
| Succinic acid | >1.65 | >100 | 3,229.1 ± 126.5 | 3,709.9 ± 1,126.6 |
| Gluconic acid | 1.52 | 92.2 | 2,911.7 ± 388.1 | 2,685.9 ± 534.6 |
| Citric acid | 0.56 | 33.9 | 126.1 ± 16.6 | 42.7 ± 7.3 |
| Threonic acid | 0.46 | 27.8 | 20.5 ± 2.4 | 5.7 ± 1.3 |
| Aconitic acid | 0.44 | 26.6 | 9.5 ± 1.6 | 2.5 ± 0.6 |
| Fumaric acid | 0.40 | 23.9 | 179.4 ± 35.4 | 42.9 ± 12.2 |
| Benzoic acid | 0.39 | 23.6 | 61.5 ± 14.2 | 14.5 ± 4.9 |
| Pyruvic acid | 0.39 | 23.3 | 23.5 ± 3.0 | 5.5 ± 0.9 |
| Glutaric acid | 0.31 | 18.9 | 0.5 ± 0.2 | 0.1 ± 0.1 |
| Maleic acid | 0.23 | 14.0 | 65.4 ± 17.3 | 9.1 ± 2.4 |
| Shikimic acid | 0.21 | 12.9 | 2.2 ± 0.3 | 0.3 ± 0.1 |
| GalUA | 0.12 | 7.3 | 351.5 ± 54.6 | 25.6 ± 7.2 |
| 2-Oxoglutarate | 0.11 | 6.9 | 3.3 ± 1.1 | 0.2 ± 0.1 |
| Glyceric acid | 0.04 | 2.7 | 544.7 ± 58.8 | 14.8 ± 1.6 |
| Sugars | ||||
| Ara | 1.44 | 87.2 | 7.9 ± 0.9 | 6.9 ± 1.2 |
| Glc | 1.01 | 61.4 | 385.1 ± 46.7 | 236.4 ± 41.2 |
| Xyl | 0.94 | 57.0 | 6.4 ± 0.8 | 3.6 ± 0.6 |
| Fru | 0.84 | 51.0 | 28.3 ± 3.4 | 14.4 ± 2.7 |
| Gal | 0.65 | 39.1 | 4.7 ± 1.7 | 1.9 ± 0.9 |
| Maltose | 0.50 | 30.5 | 13.9 ± 1.8 | 4.2 ± 0.6 |
| Gentiobiose | 0.43 | 26.2 | 11.9 ± 2.9 | 3.1 ± 0.5 |
| Fuc | 0.42 | 25.5 | 15.9 ± 2.2 | 4.0 ± 0.8 |
| Man | 0.39 | 23.3 | 1.4 ± 0.3 | 0.3 ± 0.0 |
| Isomaltose | 0.38 | 23.1 | 3.9 ± 0.4 | 0.9 ± 0.2 |
| Rib | 0.37 | 22.3 | 6.0 ± 0.9 | 1.3 ± 0.2 |
| Suc | 0.13 | 7.8 | 54.9 ± 6.2 | 4.3 ± 0.7 |
| Fatty acids | ||||
| 18:00 | 0.57 | 34.7 | 316.9 ± 81.4 | 110.1 ± 43.0 |
| 16:00 | 0.53 | 32.0 | 305.3 ± 82.7 | 97.6 ± 40.6 |
| Others | ||||
| Mannitol | 0.48 | 29.0 | 106.8 ± 21.4 | 30.9 ± 6.5 |
| Phosphoric acid | 0.48 | 28.8 | 1,687.5 ± 283.4 | 485.8 ± 170.4 |
| Sorbitol/galactitol | 0.44 | 26.9 | 1,698.3 ± 443.6 | 456.2 ± 174.9 |
| Maltitol | 0.44 | 26.9 | 1.4 ± 0.5 | 0.4 ± 0.2 |
| Putrescine | 0.18 | 11.0 | 20.5 ± 3.1 | 2.3 ± 0.5 |
| Myoinositol | 0.08 | 4.7 | 466.7 ± 18.3 | 21.8 ± 2.8 |
| Glycerol-3-phosphate | 0.04 | 2.2 | 35.6 ± 2.7 | 0.8 ± 0.1 |
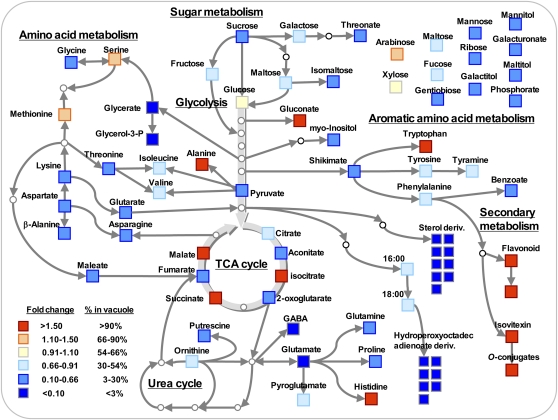
Figure 1.
Ratio of metabolite abundance in vacuole to protoplast represented on metabolic pathways. The metabolic level in the vacuole fraction was calculated against the level in the protoplast fraction. Values used for both sample types were normalized by α-mannosidase activity. The intensity of differential abundance is illustrated by a false-color scale as described in the key. GABA, γ-Aminobutyric acid.
Several of the organic acids measured had higher vacuolar than protoplast levels (Table I). Higher levels of the tricarboxylic acid (TCA) cycle intermediates isocitrate, malate, succinate, and gluconate were observed in vacuoles. The levels of aconitate, glycerate, 2-oxoglutarate, galacturonate, shikimate, maleate, glutarate, pyruvate, benzoate, fumarate, aconitate, and threonate, however, were lower in the vacuole samples (Table I; Fig. 1).
We were additionally able to detect several sugars in the vacuole samples (Table I; Fig. 1). Monosaccharides such as Ara exhibited higher levels in vacuolar than protoplast samples. Interestingly, the level of Suc was less abundant in vacuole samples. Several other sugars, namely Glc, Xyl, Fru, Gal, and maltose, were also found at abundant levels within the vacuole.
Additional nitrogen-containing metabolites were also detected in the vacuole samples, including putrescine (Table I; Fig. 1). The purine derivative adenine displayed higher levels in protoplast samples, whereas putrescine and urea levels were invariant between sample types. Among the other compounds analyzed, it is interesting that the levels of intermediates involved in membrane biogenesis, such as C16:0 and C18:0 fatty acids, were lower in vacuole samples (Table II).
Table II. Comparison of the content of secondary metabolites detected and annotated by UPLC-FT-MS between protoplasts and purified vacuole fractions.
| Retention Time | m/z | Formula | Molecular Weight | FC | In Vacuole | Peak Annotation |
| min | % | |||||
| Rt5.12 | 609.1455 | C27H30O16 | 610.1534 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt9.62 | 631.2768 | C33H38N4O6 | 586.2791 | >1.65 | High in V | Putative tetrapyrrole related |
| Rt9.77 | 631.2768 | C33H38N4O6 | 586.2791 | >1.65 | High in V | Putative tetrapyrrole related |
| Rt5.12 | 655.1513 | C28H32O18 | 656.1589 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt7.22 | 665.2819 | C32H44O12 | 620.2833 | >1.65 | High in V | Putative phenylpropanoid |
| Rt4.59 | 759.2351 | C32H42O18 | 714.2371 | >1.65 | High in V | Putative phenylpropanoid |
| Rt6.71 | 799.2083 | C38H40O19 | 800.2164 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt6.27 | 815.2038 | C38H40O20 | 816.2113 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt7.36 | 431.0979 | C21H20O10 | 432.1057 | >1.65 | High in V | Flavonoid:apigenin-6-C-glucoside |
| Rt4.07 | 693.2353 | C40H38O11 | 694.2434 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt7.52 | 475.1814 | C21H32O12 | 476.1894 | >1.65 | High in V | Putative phenyl diglycoside |
| Rt6.78 | 875.2251 | C40H44O22 | 876.2324 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt8.43 | 455.1916 | C21H30O8 | 410.1941 | >1.65 | High in V | Putative sesquiterpene glycoside |
| Rt7.97 | 475.2113 | C29H32O6 | 476.2199 | >1.65 | High in V | Putative sesquiterpene glycoside |
| Rt3.96 | 407.1719 | C21H28O8 | 408.1784 | >1.65 | High in V | Putative sesquiterpene glycoside |
| Rt6.19 | 523.1659 | C21H32O15 | 524.1741 | >1.65 | High in V | Putative iridoide |
| Rt11.14 | 637.3587 | C33H52O9 | 592.3611 | >1.65 | High in V | Putative triterpene derivatives related |
| Rt11.88 | 649.3587 | C34H52O9 | 604.3611 | >1.65 | High in V | Putative triterpene derivatives related |
| Rt6.63 | 523.1658 | C21H32O15 | 524.1741 | >1.65 | High in V | Putative iridoide |
| Rt7.48 | 631.2237 | C27H38O14 | 586.2262 | >1.65 | High in V | Putative iridoide |
| Rt5.31 | 459.1501 | C20H28O12 | 460.1581 | >1.65 | High in V | Putative phenylpropanoid |
| Rt5.78 | 507.2073 | C21H34O11 | 462.2101 | >1.65 | High in V | Putative iridoide |
| Rt7.05 | 845.2144 | C39H42O21 | 846.2219 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt6.55 | 222.0402 | C10H9NO5 | 223.0481 | >1.65 | High in V | Putative Trp derivative |
| Rt7.77 | 485.1658 | C21H28O10 | 440.1683 | >1.65 | High in V | Putative sesquiterpene glycoside |
| Rt7.10 | 609.1454 | C27H30O16 | 610.1534 | >1.65 | High in V | Putative flavonoid glycoside |
| Rt6.95 | 769.1975 | C37H38O18 | 770.2058 | 1.59 | 95.6 | Putative flavonoid glycoside |
| Rt5.88 | 669.1665 | C29H34O18 | 670.1745 | 1.54 | 92.6 | Putative flavonoid glycoside |
| Rt3.96 | 249.0875 | C11H12N2O2 | 204.0899 | 1.24 | 74.7 | Trp |
| Rt6.36 | 307.1394 | C12H22O6 | 262.1416 | 1.04 | 62.7 | Putative fatty acid glycoside |
| Rt2.97 | 210.0767 | C9H11NO2 | 165.0790 | 0.59 | 35.8 | Phe |
| Rt5.66 | 593.1504 | C27H30O15 | 594.1585 | 0.59 | 35.8 | Sapnarin, flavonoid |
| Rt3.59 | 259.1293 | C10H18N2O3 | 214.1317 | <0.02 | <1.0 | Putative dethiobiotin related |
| Rt2.69 | 283.0678 | C10H12N4O6 | 284.0757 | <0.02 | No in V | Putative xanthosine |
| Rt13.50 | 293.2116 | C18H30O3 | 294.2195 | <0.02 | No in V | Cyclopentaneoctanoic acid related |
| Rt13.74 | 293.2116 | C18H30O3 | 294.2195 | <0.02 | No in V | Cyclopentaneoctanoic acid related |
| Rt13.81 | 293.2116 | C18H30O3 | 294.2195 | <0.02 | No in V | Cyclopentaneoctanoic acid related |
| Rt12.97 | 311.2222 | C18H32O4 | 312.2301 | <0.02 | No in V | HODE related |
| Rt13.48 | 311.2222 | C18H32O4 | 312.2301 | <0.02 | No in V | HODE related |
| Rt11.30 | 327.2171 | C18H32O5 | 328.2250 | <0.02 | No in V | HODE related |
| Rt11.64 | 327.2171 | C18H32O5 | 328.2250 | <0.02 | No in V | HODE related |
| Rt11.94 | 327.2171 | C18H32O5 | 328.2250 | <0.02 | No in V | HODE related |
| Rt10.86 | 327.2172 | C18H32O5 | 328.2250 | <0.02 | No in V | HODE related |
| Rt12.18 | 329.2328 | C18H34O5 | 330.2406 | <0.02 | No in V | HODE related |
| Rt15.37 | 477.3215 | C28H46O6 | 478.3294 | <0.02 | No in V | Sterol glycoside |
| Rt15.53 | 491.3372 | C29H48O6 | 492.3451 | <0.02 | No in V | Sterol glycoside |
| Rt12.81 | 619.4210 | C35H58O6 | 574.4233 | <0.02 | High in V | Putative flavonoid glycoside |
| Rt13.09 | 619.4211 | C35H58O6 | 574.4233 | <0.02 | High in V | Putative tetrapyrrole related |
| Rt12.81 | 621.4371 | C36H62O8 | 622.4445 | <0.02 | High in V | Putative tetrapyrrole related |
Metabolite Profiling of Secondary Metabolites by LC-MS
Next, ultra-performance liquid chromatography (UPLC)-FT-MS analysis was performed for the determination of secondary metabolite contents in the different samples. Detected peaks were identified and annotated by a combined usage of standard compounds, database surveys on the basis of accurate mass-to-charge ratio (m/z) values as analyzed by FT-MS (Giavalisco et al., 2009; Horai et al., 2010), and comparison between coelution analysis with well-analyzed plant extracts such as Arabidopsis (Tohge et al., 2005; Nakabayashi et al., 2010) and tomato (Solanum lycopersicum) fruits (Moco et al., 2006; Iijima et al., 2008). By UPLC-FT-MS analysis, 200 peaks were detected in total. Twelve of these peaks were undetected in the protoplast samples, whereas 34 peaks were undetected in the vacuolar samples (Supplemental Table S1). Both Phe and Trp displayed similar relative levels as analyzed by GC-MS. Of 12 vacuole-specific peaks, four peaks were annotated as glycoflavone derivatives (Table II). The major flavonoid isovitexin (Kaspar et al., 2010), which was identified by standard compound confirmation, was also highly detected in vacuolar samples. In addition, analysis of the other six glycoflavone derivatives annotated by UPLC-FT-MS revealed that all putative glycoflavone derivatives were more highly abundant in the vacuolar than the protoplast samples. Sterol derivatives represent one of the most important (and abundant) classes of secondary metabolite in barley (Bush et al., 1971; Hübke et al., 2005). In total, nine peaks were annotated as sterol derivatives, including both sitosterol and cholesterol derivatives (Table II). These putative sterol derivatives (Bush et al., 1971; Konig and Seifert, 1998), however, were not detected in vacuolar samples. In addition, seven putative hydroperoxyoctadecadienoic acid (HODE)-related peaks (Hübke et al., 2005) were also only detected in protoplast samples. Indeed, sterol derivatives and HODE were not even detected in the vacuolar samples in their glycosidic forms.
Integration of Metabolomic Data Sets with a Subcellarly Targeted Coexpression Analysis
In an attempt to predict the underlying cytosolic-vacuolar transport system, we next evaluated tonoplast proteins predicted to have a transport function within the context of the data collected here. This integration analysis was performed by comparing the data obtained here with published data. For this purpose, we evaluated 88 proteins reported to be tonoplast proteins of barley (Endler et al., 2006). As an initial step, we bioinformatically evaluated the possibility that individuals within either set of proteins represented transporter proteins. In order to interpret these combined data sets, an integrative analysis was performed utilizing coexpression analysis of publicly available microarray data. In order to convert GenBank identifiers to those of the Affymetrix probe, an amino acid BLAST search was performed using PLEXdb BLAST (http://www.plexdb.org/modules/tools/plexdb_blast.php). In this process, the 88 proteins were converted to 128 probe sets (since the numbers are not identical, comments on cross-hybridization are given in the list in Supplemental Table S1).
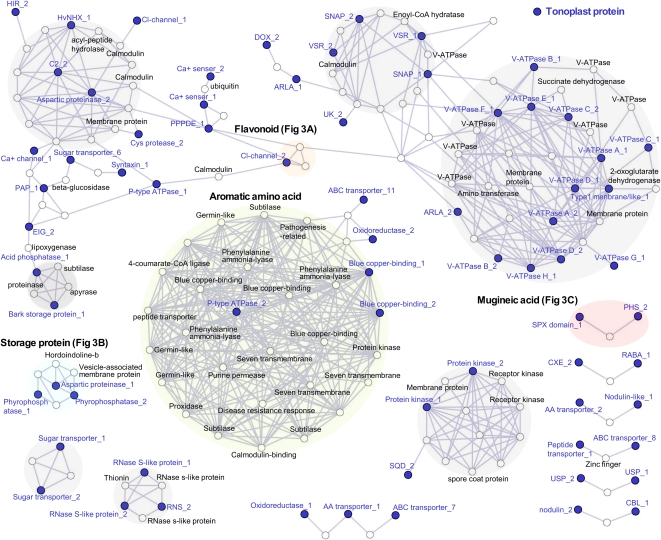
To classify gene expression clusters, we performed coexpression analyses with the 128 probe sets using PlaNet for Barley (Mutwil et al., 2011; http://aranet.mpimp-golm.mpg.de/barnet) to facilitate this process (Fig. 2). The framework of the coexpression network was constructed by the connections between vacuolar probe sets using the PlaNet database. Coexpressed network genes encoding tonoplast proteins were separated into 13 subgroups (Fig. 2; Supplemental Fig. S2), which were constructed by 70 vacuolar probe sets and 112 genes (Supplemental Table S2). The dense cluster group (middle group in Fig. 2) is well correlated with aromatic amino acid-related genes such as Phe ammonia lyase and generally expressed in photosynthetic tissues (PlaNet barley expression profiling; http://aranet.mpimp-golm.mpg.de/barnet). By contrast, a second dense cluster (top right group in Fig. 2) showed a cluster of vacuolar ATP synthase proteins and TCA cycle-related genes, such as 2-oxoglutarate dehydrogenase and succinate dehydrogenase.
Figure 2.
Frameworks of the coexpression network of barley vacuolar probe sets. The quality-checked coexpression network for barley was obtained from the PlaNet server (http://aranet.mpimp-golm.mpg.de/download/). Genes (probe sets) are depicted as nodes, while significant coexpression similarity between any two probe sets is represented by an edge. Probe sets corresponding to tonoplast-located proteins (depicted by blue nodes) were used as queries for searching the coexpression networks (Supplemental Table S2). The analysis extracted all probe sets connected to at least two vacuolar probe sets (i.e. strongly associated with vacuolar processes). Annotation of the probe sets and PageMan analysis of ontology terms can be found in Supplemental Table S2. ARLA, ADP-ribosylation factor; CBL, calcineurin B; CXE, carboxyesterase; DOX, lipoxygenase; EIG, elicitor-inducible protein; GST, glutathione S-transferase; PAP, purple acid phosphatase; PHS, propyzamide-hypersensitive protein; RNS, RNase; SNAP, soluble N-ethylmaleimide-sensitive factor attachment protein; SQD, sulfoquinovosyldiacylglycerol glycosyltransferase; UK, unknown protein; USP, universal stress protein; V-ATPase, vacuolar ATP synthase; VSR, vacuolar targeting receptor.
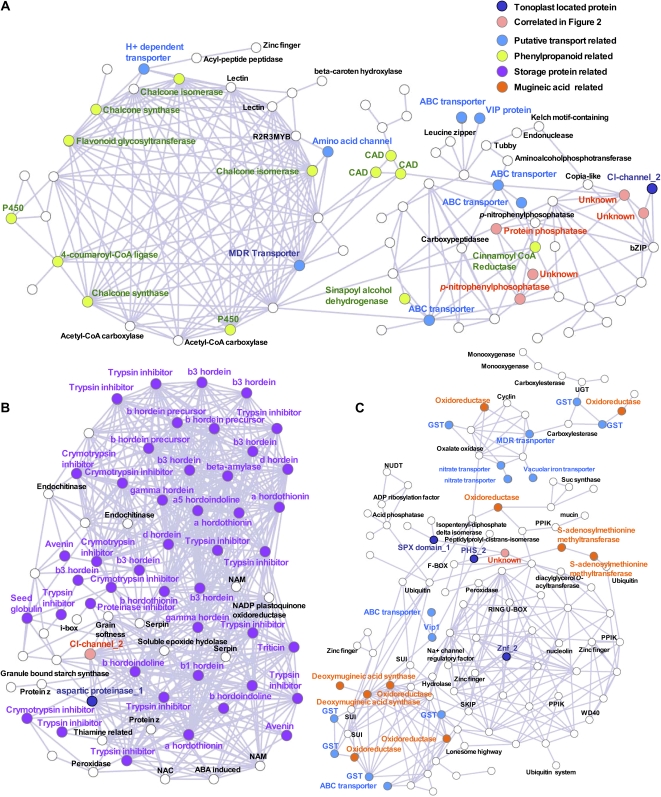
As a next step, we evaluated the barley coexpressed genes in each cluster group independently to facilitate the prediction of related metabolic pathways. In order to analyze these networks in more detail, some subclusters were focused using the PlaNet database. A gene of interest was queried to search a specific subcluster classified by PlaNet based on the framework described in Figure 2. We were able to identify three examples (Fig. 3). First, we focused on a subcluster (orange in Fig. 2) between groups, because this is a hub cluster to connect to the vacuolar ATP synthase cluster to calmodulin clusters (Fig. 2). Figure 3A shows a correlation network of a gene encoding a chloride channel protein (Contig6783_at) and unknown proteins (Contig3508_at and Contig3508_s_at). This network revealed a network connection to stress-inducible secondary metabolites such as flavonoid, because chalcone synthase (Contig7356_at and Contig7358_at), chalcone isomerase (Contig9047_at and Contig9048_at), 4-coumaroyl-CoA ligase (Contig4677_at), P450 (HVSCEb0006P14r2_at and HVSCEb0006O20r2_at), and flavonoid glycosyltransferase (Contig11602_at) were observed in this cluster. In this network, analysis showed the candidates for transporter genes that are putatively related to secondary metabolism, such as multidrug-resistant-type transporter (HVSMEn0015M0015Mo1r2_s_at) and H+-dependent transporter (Contig26311_at). Other examples indicate a subcluster that showed a small dense network with many genes encoding storage proteins (Fig. 3B). The network in Figure 3B was correlated with many storage protein genes. In this network, many genes encoding storage proteins and related proteins, such as α-β-γ-hordein, hordoindoline, prolamine, avenin, and a trypsin inhibitor, were observed. These proteins were predicted to be transporters for storage protein, as mentioned above, and were highly expressed during seed development and correlated with the genes hordein and avenin. The tonoplast-located proteins aspartic protease (Contig3144_at) and Cl− channel protein (Contig7078_at) were strongly connected to the storage protein cluster. Again, we next performed gene expression tree analysis using only the small cluster that was characterized by “contains similarity to a tetracycline resistance efflux protein” (Contig13913_at) and putative protein phosphatase (Contig9964_at). The network showed a correlation network with mugineic acid biosynthesis, such as deoxymugineic acid synthase (Contig3519_at and HVSMEa0014J15r2_x_at) and oxidoreductase (Contig6722_at, Contig9968_at, Contig9968_at, Contig10877_at, Contig11890_at, and HV_CEa0006E02r2_s_at; Fig. 3C). In addition, this network also showed strong connection with many genes encoding transport-related protein, such as ATP-binding cassette (ABC) transporter (Contig2625_at and Contig7226_at) and glutathione S-transferase (Contig4137_at, Contig9632_at, HA03B19u_s_at, and HVSMEa0014H14r2_s_at), because mugineic acid is a phytosiderophore and a ligand compound that is a metal chelator in graminaceous plants.
Figure 3.
Subclusters of coexpression network analysis to predict the functions of tonoplast-located proteins. Subclusters of the frameworks of the coexpression network in Figure 2 indicated flavonoid- and phenylpropanoid-related biosynthesis (A), storage protein (B), and mugineic acid biosynthesis (C). Genes (probe sets) are depicted as nodes, while significant coexpression similarity between any two probe sets is represented by an edge. Probe sets corresponding to tonoplast-located proteins (depicted by blue nodes; Supplemental Figure S3) were used as queries for searching the coexpression networks (Supplemental Table S2). Annotation of the probe sets and PageMan analysis of ontology terms can be found in Supplemental Table S2. CAD, Cinnamoyl-alcohol dehydrogenase; GST, glutathione S-transferase; MDR, multidrug-resistant-type transporter; NAM, no apical meristem; NUDT, nudix hydrolase homolog; PHS, propyzamide-hypersensitive protein; PPIK, phosphatidylinositol 3- and 4-kinase; SKIP, SKP-interacting partner; UGT, UDP-glycosyltransferase; VIP, vanabin-interacting protein.
DISCUSSION
The process of homeostasis requires that the levels of certain metabolite pools be maintained at near-constant levels. One mechanism to achieve this is the sequestration of metabolites into a relatively metabolically inert environment such as the vacuole. Our current knowledge of the vacuole comes largely from studies of isolated vacuoles as well as cell biological studies of exocytosis, endocytosis, and autophagy (Phan et al., 2008; Liu et al., 2009). These approaches have revealed a wide range of metabolites including sugars, organic acids, amino acids as well as chlorophyll catabolites, glutathione conjugates, bile acids, and sulfate conjugates (Martinoia et al., 2000; Farré et al., 2001); however, until recently, this was rather fragmentary. The application of the nonaqueous fractionation method in potato tubers when coupled to GC-MS analysis represented a first attempt to address this more systematically (Farré et al., 2001). This has been further refined by improvements in the resolution of the nonaqueous technique due to the ability to assess ever-smaller sample volumes in addition to the additional use of FT-ion cyclotron resonance (ICR)-MS, which has a far greater mass accuracy and thus potential for compound prediction. This combined approach was recently applied to Arabidopsis (Krueger et al., 2011) and produced an unprecedented amount of information concerning spatial resolution. Here, we utilized both GC-MS and FT-ICR-MS but instead applied them to silicon oil-centrifuged vacuoles from protoplast preparations of barley. Following this approach, we were able to detect some 259 metabolites and to define that some 12 of them were preferentially localized to the vacuole, 34 were preferentially localized outside of the vacuole, and 52 were distributed equally in the cell.
Primary metabolites generally displayed the partition that would be anticipated with organic acids and sugars as well as a few amino acids clearly being stored here. Of particular note are the high accumulations of isocitrate, citrate, and aconitate as well as Glc and Fru. That said, there were some clear exceptions to that statement, with the organic acids fumarate and 2-oxoglutarate being essentially absent in the vacuole, as were Suc and shikimate. Barley was additionally characterized by the accumulation of His, Ala, Trp, Met, and Ser in the vacuole; however, many of the other amino acids preferentially accumulate outside the vacuole. The pattern of organic acid partitioning suggests that 2-oxoglutarate and fumarate do not represent storage compounds in barley. In the case of 2-oxoglutarate, this is perhaps unsurprising given the crucial role of this metabolite in nitrate assimilation and the TCA cycle (Araújo et al., 2008, 2011). The fact that fumarate appears not to be a storage metabolite clearly contrasts with the situation in Arabidopsis, where, like malate, it appears to play a crucial role as a carbon store (Fahnenstich et al., 2007; Zell et al., 2010). The difference between these species may well be explained by the presence of a cytosolic fumarase (Pracharoenwattana et al., 2010) and the fact that fumarate is an important transport form of carbon in Arabidopsis (Chia et al., 2000). The relative levels of sugars is consistent with a very high vacuolar invertase activity in barley, as has been described for other species including potato, maize (Zea mays), carrot (Daucus carota), and tobacco (Nicotiana tabacum; Büssis et al., 1997; Tang et al., 1999; Trouverie et al., 2004; Junker et al., 2006). Furthermore, it is consistent with the presence of a large number of sugar-conjugated secondary metabolites in this compartment.
Perhaps surprisingly, a large number of the secondary metabolites we detected were not found to be preferentially accumulated in the vacuole; however, there are some clear exceptions to this. Among the 49 peaks that were annotated, a chemical formula by accurate m/z value, 26 of the secondary metabolites were detected to be at higher abundance in the vacuole. Characterization of the chemical composition of secondary metabolites in barley has not been extensively reported, although there are some publications concerning flavonoids (Norbaek et al., 2000; Kaspar et al., 2010), sterol derivatives (Bush et al., 1971; Konig and Seifert, 1998), and HODE (Hamberg and Hamberg, 1996; Hübke et al., 2005). In our metabolite profiling by FT-MS with accurate m/z value for chemical formula prediction, we confirmed a formula for these compounds. All peaks we listed in Table II except Phe and Trp were not observed in Arabidopsis or tomato tissues, but similar peaks such as glycoflavone glycosides were detected in monocot plants such as rice (Oryza sativa) and maize (Besson et al., 1985; Snook et al., 1995). In our measurement of barley secondary metabolites, 12 putative flavonoids, seven putative HODE-related compounds, three cyclopentaneoctanoic acid-related compounds, and nine putative sterol derivatives were annotated. The compounds in each class displayed similar behavior; for example, all flavonoids were higher in vacuole, whereas HODE-related compounds and sterol-related compounds were not detected in vacuole samples. The sterol-related compounds are likely located at the tonoplast membrane (Yoshida and Uemura, 1986). In addition, the cycloartenol synthase protein (AAT38887 homolog, Contig11705_at; Supplemental Table S2), which is involved in sterol biosynthesis, was previously identified as a “membrane fusion and remodeling protein” (Endler et al., 2006).
In the case of dicot plants such as Arabidopsis leaves, flavonol-O-glycoside, anthocyanin, and glucosinolates have been identified as major secondary metabolites (Tohge et al., 2005; Maruyama-Nakashita et al., 2006; Hirai et al., 2007; Matsuda et al., 2009), whereas other dicots such as Solanaceae species, including tomato and tobacco, accumulate glycoalkaloids, quinate derivatives, and acylated sugars (Moco et al., 2006; Butelli et al., 2008; Iijima et al., 2008; Adato et al., 2009). On the other hand, in leaves of monocot plant species such as rice, maize, and barley, flavonol-O-glycoside and flavonol-C-glycoside (glycoflavone) are reported (Norbaek et al., 2000; Kaspar et al., 2010). Typical flavonoids such as flavonol and anthocyanin are transported to the vacuole after their conversion to glycosides and in some cases acylates, either in exchange for protons or by direct energization by ABC transporters. In Arabidopsis research, multidrug and toxic compound extrusion-type transporter (Debeaujon et al., 2001; Marinova et al., 2007) and the soluble glutathione S-transferase proteins (Kitamura et al., 2004, 2010) are characterized as key players of flavonoid transport into the vacuole. In maize, genetic evidence was provided that ABC transporters are implicated in anthocyanin transport (Goodman et al., 2004). Based on this assumption, an integration analysis was performed utilizing coexpression analysis of publicly available microarray data. Generally, coexpression analysis does not work well for transport-related proteins, since the expression of genes encoding transport-related proteins such as ABC transporters is generally low, and they are not usually stress responsive (Klein et al., 2006). Despite this, gene coexpression analysis using data obtained for different developmental stages has previously revealed that TT12 (a multidrug and toxic compound extrusion transporter for seed pigment proanthocyanidin in Arabidopsis) was well correlated with enzymatic genes of proanthocyanidin metabolism, such as BAN leucocyanidin leucopelargonidin (At1g61720) and laccase-like polyphenol oxidase (AtLAC15; At5g48100; Tohge and Fernie, 2010). In the other example of transporters for glucosinolate that accumulated in the vacuole of Brassica species, BAT5 (At4g12030), which is required for the biosynthesis of Met-derived glucosinolates, was well correlated in the coexpression network analysis (Hirai et al., 2007; Gigolashvili et al., 2009). Our trial of coexpression network analysis gave us many hints that may aid in understanding of the vacuolar transport system for flavonoid, storage proteins, and mugineic acid. The case of flavonoid acts as a positive control, given that this accumulation in the vacuole was experimentally annotated in several plant species (Song et al., 2010). Intriguingly, it also suggested a transport system for phytosiderophores, and it would seem likely that such a system plays a role in metal chelators under metal stress and nutrient-deficient conditions. As we showed in Figures 2 and 3, some metabolites that need to be transported into the vacuole revealed correlation in gene expression between those genes involved in their biosynthesis and those involved in their transport system. In this study, we used barley as an example of the power of combining data from metabolite profiling, subcellular proteomics, and in silico coexpression analysis in order to better understand the storage metabolome. Using this approach, we were able to characterize the storage metabolome of this species in spite of the fact that the genome sequence of barley is not yet available.
In summary, the results presented here provide the basis for a more comprehensive understanding of the vacuolar systems of transport and metabolism. While the partitioning of a number of these metabolites is in keeping with what would be anticipated, we additionally provide information on a number of other metabolites. That said, comparison between the barley vacuole data presented here and that found in Arabidopsis suggests that while many factors are conserved between species, there are subtle differences also. While it is possible that some of these differences are due to the different approaches used between the two studies, it would appear likely that the vacuolar metabolome, like the cellular metabolome (Fiehn, 2007; Krueger et al., 2011), is context dependent. With this in mind, it will be highly interesting in future studies to compare the storage metabolomes of a broad range of tissues types and its dynamic response to cellular circumstances, including exposure to salt stress, drought, and conditions that induce senescence.
MATERIALS AND METHODS
Plant Materials and Harvest
Barley (Hordeum vulgare var Baraka) was grown in vermiculite (Vermica) in a controlled-environment chamber (16 h of light/8 h of dark, 300 mE m–2 s–1, 227°C, 60% relative humidity) and watered with Luwasa hydroculture nutrient solution (Interhydro). The primary leaves were harvested between 8 and 9 am as described by Rentsch and Martinoia (1991) and Endler et al. (2006).
Protoplast and Vacuole Isolation for Metabolomics
Barley protoplasts and vacuoles for metabolomics were isolated by a slight modification of the procedure described by Rentsch and Martinoia (1991). Briefly, for protoplast isolation, after digestion of the leaves, protoplasts were collected by centrifugation at 1,200g for 10 min on a cushion constituted of digestion medium (500 mm sorbitol, 1 mm CaCl2, and 10 mm MES-KOH, pH 6) containing 30% Percoll (v/v). The supernatant was removed, the protoplasts were mixed with the Percoll cushion, and additional osmotically dissolved Percoll (digestion medium solved in Percoll) was added to give a final Percoll concentration of about 40%. For the metabolomics of the protoplasts, the protoplast solution was overlaid with medium C (500 mm sorbitol, 20% Percoll, pH 6, 1 mm CaCl2, and 10 mm MES-KOH, pH 5.6) and medium D (500 mm betaine, 1 mm CaCl2, and 10 mm MES-KOH, pH 5.6). The volume used depended on the quantity of protoplasts isolated. After centrifugation for 10 min at 1,200g, the protoplasts were recovered from the upper interphase. For the metabolomic studies of protoplasts, 200 μL of a silicon oil mixture (AR200:AR20, 8:2) was placed at the bottom of polyethylene microcentrifugation tubes (400-μL capacity). Two milliliters of protoplasts was mixed with 2 mL of medium D, and 200 μL of this mix was placed on the top of the silicon oil mixture. Protoplasts were sedimented by a 2 × 20-s centrifugation at 10,000g. The polyethylene tubes (50 per experiment) were frozen overnight. The bottoms of the frozen tubes were cut with a razor blade and collected. One milliliter of water was added and vortexed, and the aqueous fraction containing a tiny amount of silicon oil but no tube tips was transferred in a fresh tube. The mixture was centrifuged at 4,000g for 10 min. The water phase was removed, lyophilized, and used for the metabolomic analysis.
For the vacuole isolation, protoplasts were purified using a similar gradient but replacing medium C by C′ (400 mm sorbitol, 20% Percoll, pH 7.2, 30 mm KCl, 20 mm HEPES-KOH, pH 7.2, 0.1% bovine serum albumin, and 0.2 mm dithiothreitol) and medium D by D′ (400 mm betaine, 30 mm KCl, 20 mm HEPES-KOH, pH 7.2, and 0.2 mm dithiothreitol). Vacuoles were released from protoplasts by forcing the protoplasts through a syringe and purified by flotation as described by Rentsch and Martinoia (1991). This procedure occurred at 4°C. In order to get rid of the isolation medium, 30 μL of vacuoles was added to 70 μL of a medium containing 22% Percoll, pH 7.2, 400 mm sorbitol, 30 mm KCl, 20 mm HEPES-KOH, pH 7.2, 0.1% bovine serum albumin, and 0.2 mm dithiothreitol. The samples were overlaid with 200 μL of silicone oil AR200 and 60 μL of water. After mixing for 6 min, the vacuoles were floated by centrifugation at 10,000g for 20 s. Tubes were frozen overnight, and after defreezing, 45 μL of the aqueous solution from each tube (50–60 per experiment) was recovered and pooled. The pooled fraction was lyophilized and used for the metabolomic analysis.
Extraction for Metabolite Analysis
Lyophilized sample was homogenized using a ball mill precooled with liquid nitrogen and extracted in 1,400 μL of methanol, and 60 μL of internal standard (0.2 mg ribitol mL–1 water) was subsequently added as a quantification standard. Samples were homogenized by the Retsch Muhle mixer mill for 2 min at 25 L s–1. One thousand two hundred microliters of supernatant after centrifugation at 14,000 rpm for 10 min was transferred to a glass vial. After mixing with 750 μL of CHCl3, 1,500 μL of water was added and vortexed for 15 s. After centrifugation for 15 min at 4,000 rpm, supernatant was taken (50 and 100 μL for GC-MS analysis and 1,000 μL for LC-MS analysis) from the upper polar phase into fresh Eppendorf tubes (also 2 μL). Sample dried by the SpeedVac for at least 2 h without heating was used for metabolite analysis.
Derivatization and Analysis of Primary Metabolites Using GC-MS
Metabolite extraction for GC-MS was performed by a method modified from that described by Roessner-Tunali et al. (2003). The extraction, derivatization, standard addition, and sample injection were exactly as described previously (Lisec et al., 2006). Both chromatograms and mass spectra were evaluated using either TAGFINDER (Luedemann et al., 2008) or the MASSLAB program (ThermoQuest), and the resulting data were prepared and presented as described (Roessner et al., 2001).
UPLC-FT-MS of Secondary Metabolites
UPLC separation of secondary metabolites was performed according to a previously published protocol (Giavalisco et al., 2009) using a Waters Acquity UPLC system. The UPLC apparatus was equipped with an HSS T3 C18 reverse-phase column (100 × 2.1 mm i.d., 1.8-μm particle size; Waters), which was operated at a temperature of 40°C. The mobile phases consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The flow rate of the mobile phase was 400 μL min–1, and 2 μL of sample was loaded per injection. The following gradient profile was applied: after a 1-min isocratic run at 99% A, a linear 12-min gradient was applied to 65% A; this was immediately followed by a 1.5-min gradient to 30% A before a 1-min gradient to 1% A; then, a 1.5-min isocratic period at 1% A followed, before switching back to 99% A to reequilibrate the column for 2.5 min, before the next sample could be injected. The UPLC device was connected to the FT-ICR apparatus via a TriVersa NanoMate (Advion). The UPLC flow rate at 400 μL min–1 was split 1:1,000 with a T-valve (Advion). One-tenth of 1% (400 nL min–1) was directly loaded to the FT-ICR-MS device, while 99.9% was discarded. The sample was infused into the mass spectrometer via a nanospray chip (type A; Advion) by applying a voltage of 1.8 kV in the positive and 1.9 kV in the negative ionization mode. Spray sensing was used between 1 and 17 min of the UPLC gradient. The mass spectra were acquired using the LTQ FT-ICR-Ultra mass spectrometer (Thermo-Fisher). The spectra were recorded using full-scan mode, covering a mass range from m/z 100 to 1,500. Resolution was set to 50,000, and maximum loading time for the ICR cell was set to 500 ms. The transfer capillary temperature was set to 200°C, and the MS spectra were recorded from 0 to 19 min of the UPLC gradient. Molecular masses, retention times, and associated peak intensities were extracted from the raw files using the RefinerMS software (version 5.3; GeneData). Peak prediction and annotation in the data matrix by LC-MS was conducted by databases (for review, see Tohge and Fernie, 2009) such as MASSBANK (Horai et al., 2010) and KNApSAcK (Shinbo et al., 2006), based on accurate m/z analyzed by FT-MS.
Coexpression Analysis via Proteome Data
The coexpression network analysis of wheat (Triticum aestivum) was performed by PlaNet (Mutwil et al., 2011) using Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Global comparison of all detected compounds by GC-MS and LC-MS in protoplast and vacuole.
Supplemental Figure S2. Probe identifiers of genes shown in the framework of the coexpression network in Figure 2.
Supplemental Figure S3. Probe identifiers of genes shown in the framework of the coexpression network in Figure 3.
Supplemental Table S1. Secondary metabolite-related peaks detected by UPLC-FT-MS
Supplemental Table S2. The barley vacuole proteins used for coexpression analysis.
References
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Dominguez E, Wang ZH, De Vos RCH, Jetter R, et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SIMYB12-regulated transcriptional network. PLoS Genet 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulfur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Trenkamp S, Bunik VI, Fernie AR. (2008) Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, Beale MH. (2006) A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol J 4: 381–392 [DOI] [PubMed] [Google Scholar]
- Besson E, Dellamonica G, Chopin J, Markham KR, Kim M, Koh HS, Fukami H. (1985) C-Glycosylflavones from Oryza sativa. Phytochemistry 24: 1061–1064 [Google Scholar]
- Boller T, Kende H. (1979) Hydrolytic enzymes in the central vacuole of plant cells. Plant Physiol 63: 1123–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Provart NJ. (2009) Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush PB, Grunwald C, Davis DL. (1971) Changes in sterol composition during greening of etiolated barley shoots. Plant Physiol 47: 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssis D, Heineke D, Sonnewald U, Willmitzer L, Raschke K, Heldt HW. (1997) Solute accumulation and decreased photosynthesis in leaves of potato plants expressing yeast-derived invertase either in the apoplast, vacuole or cytosol. Planta 202: 126–136 [DOI] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Cai SQ, Lashbrook CC. (2006) Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J 48: 628–637 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR. (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan SQ, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole GS, Beckmann M, Enot DP, Mondhe M, Zywicki B, Taylor J, Hardy N, Smith A, King RD, Kell DB, et al. (2005) Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc Natl Acad Sci USA 102: 14458–14462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DW, Yoder TJ, Reiter WD, Gibson SI. (2000) Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 211: 743–751 [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. (2007) Glucose signaling through nuclear hexokinase1 complex in Arabidopsis. Plant Signal Behav 2: 123–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Reiland S, Gerrits B, Schmidt UG, Baginsky S, Martinoia E. (2009) In vivo phosphorylation sites of barley tonoplast proteins identified by a phosphoproteomic approach. Proteomics 9: 310–321 [DOI] [PubMed] [Google Scholar]
- Fahnenstich H, Saigo M, Niessen M, Zanor MI, Andreo CS, Fernie AR, Drincovich MF, Flügge UI, Maurino VG. (2007) Alteration of organic acid metabolism in Arabidopsis overexpressing the maize C4 NADP-malic enzyme causes accelerated senescence during extended darkness. Plant Physiol 145: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Tech S, Trethewey RN, Fernie AR, Willmitzer L. (2006) Subcellular pyrophosphate metabolism in developing tubers of potato (Solanum tuberosum). Plant Mol Biol 62: 165–179 [DOI] [PubMed] [Google Scholar]
- Farré EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L. (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr M, Frommer WB, Lalonde S. (2002) Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc Natl Acad Sci USA 99: 9846–9851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR. (2007) The future of metabolic phytochemistry: larger numbers of metabolites, higher resolution, greater understanding. Phytochemistry 68: 2861–2880 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Fiehn O. (2002) Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol 48: 155–171 [PubMed] [Google Scholar]
- Fiehn O. (2007) Cellular metabolomics: the quest for pathway structure. In Lindon JC, Nicholson JK, Holmes E, eds, The Handbook of Metabonomics and Metabolomics, Chapter 2. Elsevier BV, Amsterdam, pp 35–54 [Google Scholar]
- Gerhardt R, Stitt M, Heldt HW. (1983) Subcellular metabolite determination in spinach leaves through non-aqueous fractionation. Physiol Chem 364: 1130–1131 [Google Scholar]
- Giavalisco P, Köhl K, Hummel J, Seiwert B, Willmitzer L. (2009) 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal Chem 81: 6546–6551 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Rollwitz I, Humphry M, Gershenzon J, Flügge UI. (2009) The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21: 1813–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB. (1988) Plants: novel developmental processes. Science 240: 1460–1467 [DOI] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. (2004) A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16: 1812–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Hamberg G. (1996) Peroxygenase-catalyzed fatty acid epoxidation in cereal seeds: sequential oxidation of linoleic acid into 9(S),12(S),13(S)-trihydroxy-10(E)-octadecenoic acid. Plant Physiol 110: 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45: 703–714 [DOI] [PubMed] [Google Scholar]
- Hübke H, Garbe LA, Tressl R. (2005) Characterization and quantification of free and esterified 9- and 13-hydroxyoctadecadienoic acids (HODE) in barley, germinating barley, and finished malt. J Agric Food Chem 53: 1556–1562 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Romanowska E, Gardestrom P. (2001) Photorespiratory flux and mitochondrial contribution to energy and redox balance of barley leaf protoplasts in the light and during light-dark transitions. J Plant Physiol 158: 1325–1332 [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, Suzuki T, Suzuki H, Okazaki K, Kitayama M, et al. (2008) Metabolite annotations based on the integration of mass spectral information. Plant J 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ. (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Wuttke R, Nunes-Nesi A, Steinhauser D, Schauer N, Büssis D, Willmitzer L, Fernie AR. (2006) Enhancing vacuolar sucrose cleavage within the developing potato tuber has only minor effects on metabolism. Plant Cell Physiol 47: 277–289 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar S, Matros A, Mock HP. (2010) Proteome and flavonoid analysis reveals distinct responses of epidermal tissue and whole leaves upon UV-B radiation of barley (Hordeum vulgare L.) seedlings. J Proteome Res 9: 2402–2411 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Yamazaki M, Saito K, Narumi I. (2010) Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J 62: 549–559 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Klein M, Burla B, Martinoia E. (2006) The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett 580: 1112–1122 [DOI] [PubMed] [Google Scholar]
- Konig M, Seifert K. (1998) Uptake and translocation of exogenously applied 7-oxysterols in barley. Phytochemistry 49: 1257–1263 [Google Scholar]
- Krueger S, Giavalisco P, Krall L, Steinhauser M-C, Büssis D, Usadel B, Flügge UI, Fernie AR, Willmitzer L, Steinhauser D. (2011) A topological map of the compartmentalized Arabidopsis thaliana leaf metabolome. PLoS ONE 6: e17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, et al. (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu YM, Xiong Y, Bassham DC. (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Lunn JE. (2007) Compartmentation in plant metabolism. J Exp Bot 58: 35–47 [DOI] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Duplessis S, Ditengou F, Lagrange H, Voiblet C, Lapeyrie F. (2001) Developmental cross talking in the ectomycorrhizal symbiosis: signals and communication genes. New Phytol 151: 145–154 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Massonneau A, Frangne N. (2000) Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol 41: 1175–1186 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. (2009) MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57: 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. (2006) Recent surprises in protein targeting to mitochondria and plastids. Curr Opin Plant Biol 9: 610–615 [DOI] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CHR. (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Yamazaki M, Saito K. (2010) A polyhedral approach for understanding flavonoid biosynthesis in Arabidopsis. N Biotechnol 27: 829–836 [DOI] [PubMed] [Google Scholar]
- Norbaek R, Brandt K, Kondo T. (2000) Identification of flavone C-glycosides including a new flavonoid chromophore from barley leaves (Hordeum vulgare L.) by improved NMR techniques. J Agric Food Chem 48: 1703–1707 [DOI] [PubMed] [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M. (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ. (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan NQ, Kim SJ, Bassham DC. (2008) Overexpression of Arabidopsis sorting nexin AtSNX2b inhibits endocytic trafficking to the vacuole. Mol Plant 1: 961–976 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Zhou WX, Keech O, Francisco PB, Udomchalothorn T, Tschoep H, Stitt M, Gibon Y, Smith SM. (2010) Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J 62: 785–795 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Martinoia E. (1991) Citrate transport into barley mesophyll vacuoles: comparison with malate-uptake activity. Planta 184: 532–537 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma CL, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O. (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Patterson JH, Forbes MG, Fincher GB, Langridge P, Bacic A. (2006) An investigation of boron toxicity in barley using metabolomics. Plant Physiol 142: 1087–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR. (2001) High-resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems: identification of phenocopies. Plant Physiol 127: 749–764 [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F. (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Schad M, Mungur R, Fiehn O, Kehr J. (2005) Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods 1: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherling C, Ulrich K, Ewald D, Weckwerth W. (2009) A metabolic signature of the beneficial interaction of the endophyte Paenibacillus sp. isolate and in vitro-grown poplar plants revealed by metabolomics. Mol Plant Microbe Interact 22: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T. (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45: 672–683 [DOI] [PubMed] [Google Scholar]
- Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. (2006) KNApSAcK: a comprehensive species-metabolite relationship database. Plant Metabolomics 57: 165–181 [Google Scholar]
- Smart KE, Smith JA, Kilburn MR, Martin BG, Hawes C, Grovenor CR. (2011) High-resolution elemental localization in vacuolate plant cells by nano-scale secondary ion mass spectrometry. Plant J 63: 870–879 [DOI] [PubMed] [Google Scholar]
- Snook ME, Widstrom NW, Wiseman BR, Byrne PF, Harwood JS, Costello CE. (1995) New C-4″-hydroxy derivatives of maysin and 3′-methoxymaysin isolated from corn silks (Zea mays). J Agric Food Chem 43: 2740–2745 [Google Scholar]
- Song WY, Park J, Mendoza-Cózatl DG, Suter-Grotemeyer M, Shim D, Hörtensteiner S, Geisler M, Weder B, Rea PA, Rentsch D, et al. (2010) Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci USA 107: 21187–21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Fernie AR. (2003) From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol 14: 136–144 [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Palaniappan A, Duncan K, Rhoads DM, Huber SC, Sachs MM. (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem 281: 15625–15635 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA. (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tang GQ, Lüscher M, Sturm A. (1999) Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. (2005) A novel approach for nontargeted data analysis for metabolomics: large-scale profiling of tomato fruit volatiles. Plant Physiol 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. (2009) Web-based resources for mass-spectrometry-based metabolomics: a user’s guide. Phytochemistry 70: 450–456 [DOI] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. (2010) Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tohge T, Yonekura-Sakakibara K, Niida R, Watanabe-Takahashi A, Saito K. (2007) Phytochemical genomics in Arabidopsis thaliana: a case study for functional identification of flavonoid biosynthesis genes. Pure Appl Chem 79: 811–823 [Google Scholar]
- Trouverie J, Chateau-Joubert S, Thévenot C, Jacquemot MP, Prioul JL. (2004) Regulation of vacuolar invertase by abscisic acid or glucose in leaves and roots from maize plantlets. Planta 219: 894–905 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Fernie AR. (2005) Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J Exp Bot 56: 309–321 [DOI] [PubMed] [Google Scholar]
- Weckwerth W. (2003) Metabolomics in systems biology. Annu Rev Plant Biol 54: 669–689 [DOI] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW. (1993) Subcellular volumes and metabolite concentrations in barley leaves. Planta 191: 180–190 [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Uemura M. (1986) Lipid composition of plasma membranes and tonoplasts isolated from etiolated seedlings of mung bean (Vigna radiata L.). Plant Physiol 82: 807–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell MB, Fahnenstich H, Maier A, Saigo M, Voznesenskaya EV, Edwards GE, Andreo C, Schleifenbaum F, Zell C, Drincovich MF, et al. (2010) Analysis of Arabidopsis with highly reduced levels of malate and fumarate sheds light on the role of these organic acids as storage carbon molecules. Plant Physiol 152: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]