Abstract
Expression of c-Myb is required for normal hematopoiesis and for proliferation of myeloid leukemia blasts and a subset of T cell leukemia but its role in B-cell leukemogenesis is unknown.
We tested the role of c-Myb in p190BCR/ABL-dependent B-cell leukemia in mice transplanted with p190BCR/ABL-transduced marrow cells with a c-Myb allele (Mybf/d) and in double transgenic p190BCR/ABL/Mybw/d mice. In both models, loss of a c-Myb allele caused a less aggressive B-cell leukemia.
In p190BCR/ABL expressing human B-cell leukemia lines, knockdown of c-Myb expression suppressed proliferation and colony formation.
Compared to c-Mybw/f cells, expression of Bmi1, a regulator of stem cell proliferation and maintenance, was decreased in pre-B cells from Mybw/d p190BCR/ABL transgenic mice. Ectopic expression of a mutant c-Myb or Bmi1 enhanced the proliferation and colony formation of Mybw/d p190BCR/ABL B-cells; by contrast, Bmi1 downregulation inhibited colony formation of p190BCR/ABL-expressing murine B cells and human B-cell leukemia lines. Moreover, c-Myb interacted with a segment of the human Bmi1 promoter and enhanced its activity.
In blasts from nineteen Ph1 adult ALL patients, levels of c-Myb and Bmi1 showed a positive correlation. Together, these findings support the existence of a c-Myb-Bmi1 transcription regulatory pathway required for p190BCR/ABL leukemogenesis.
Keywords: Oncogene, Transcription Factor, Stem Cells, Leukemia
Introduction
In addition to chronic myeloid leukemia (CML), the Philadelphia (Ph1) chromosome is also detected in 20–40% of acute lymphoblastic leukemia (ALL), a poor- prognosis disease subtype (1). The Ph1 chromosome is the result of the t(9;22) reciprocal translocation which generates three BCR/ABL fusion genes each containing different 5′ portions of BCR fused to exons 2 – 11 of ABL with the breakpoint in BCR determining size, activity of the gene product and disease phenotype (2). p190BCR/ABL is the result of head-tail fusion of BCR exon 1 with ABL exon 2 (e1-a2 fusion) and encodes a 7.0 kb mRNA which is translated into a 190 kDa protein with constitutive tyrosine kinase activity, the highest among the three forms of BCR/ABL (3,4).
Mechanisms implicated in BCR/ABL-dependent transformation of hematopoietic cells include activation of the RAS, STAT5 and PI-3 kinase pathways which provide proliferative and anti-apoptotic signals thought to be essential for the process of leukemogenesis (5–7). These pathways are activated by all BCR/ABL variants (8); however, certain pathways are activated by p190BCR/ABL in a B-cell specific manner (9–11), possibly accounting for its distinct role in B-cell transformation.
Expression of p210BCR/ABL induces also changes in the levels and activity of several transcription factors; besides members of the STAT family, p210BCR/ABL enhances the expression of c-Myc (12) and the activity of the B-catenin/LEF-1 and the Hedgehog pathways, both of which may regulate the self-renewal of normal and BCR/ABL-transformed mouse and human progenitor subsets (13–15). BCR/ABL can also repress the expression of C/EBPα and JunB, two transcription factors causally linked to development of leukemia and myeloproliferative disorders in mice and humans (16–18). Expression of c-Myb is also regulated by p210BCR/ABL (19), and AML and CML blasts rely on its expression for proliferation and survival more than the normal counterpart (20, 21). Using a genetic approach, we recently demonstrated that optimal levels of c-Myb are required for p210BCR/ABL-dependent transformation of hematopoietic progenitors and leukemogenesis (22).
Duplication or translocation of the c-Myb gene has recently been identified in a cohort of pediatric T-ALL patients (23,24) and knockdown of c-Myb expression in T-ALL cell lines promoted differentiation (23), suggesting that it may have a pathogenic role at least in a subset of T-ALL patients. Less is known about the role of c-Myb in B-ALL and in mouse models of B-cell leukemia. Using in vivo and in vitro assays of p190BCR/ABL-dependent leukemogenesis and transformation of B-cell progenitors, we show here that c-Myb hemizygous B-cells are less leukemogenic and clonogenic than the normal counterpart. Downregulation of c-Myb expression also reduced proliferation and cluster formation of human p190BCR/ABL Z-181 and SUP-B15 B-ALL cell lines. By hybridization of oligonucleotide arrays with RNA from p190BCR/ABL/Myb+/− pre-B cells, we identified differentially expressed genes with potential roles in B-cell leukemogenesis. One such gene, the hematopoietic stem cell regulator Bmi1 (25), is a direct transcriptional target of c-Myb. Ectopic expression of Bmi1 rescued the defective colony-forming potential of p190BCR/ABL/Myb+/− B-cells; by contrast, knockdown of Bmi1 expression suppressed colony formation of p190BCR/ABL-expressing Myb+/+ murine B cells and human ALL cells. Expression of c-Myb and Bmi1 was correlated in blast cells from nineteen BCR/ABL-positive adult ALL patients.
Together, these studies suggest that c-Myb regulates the process of p190BCR/ABL-dependent B-cell leukemogenesis, at least in part, through its effect on the levels of Bmi1; since Bmi1 is involved in stem cell self renewal, c-Myb-regulated abnormal expression of Bmi1 might represent an important mechanism for transformation and maintenance of p190BCR/ABL-expressing pre-B cells.
Materials and methods
Mice
Mybf/f and Mybf/d mice (26) were maintained by brother-sister mating. C57BL/6J mice were obtained from the Jackson Laboratory. ICR-SCID mice were obtained from Taconic (Hudson, NY, USA). B6;CBA-Tg(BCR-ABL) 623Hkp mice were obtained from the NCI Mouse Repository (27). BCR-ABL+Mybw/f and BCR-ABL+Mybw/d mice were generated through crosses of B6;CBA-Tg(BCR-ABL) 623Hkp and Mybf/d mice. Strains were genotyped as described (26,27). Experiments were performed according to the Animal Care and Use Committee’s guidelines.
Plasmids
MSCVp190wt-IRES/eGFP (MigRI-p190) was provided by Dr Richard Van Etten (Tufts University, Boston, MA). MigRI-Δ(358–452)Myb-GFP was previously described (19). MSCV Bmi1-IRES-eYFP was obtained by removing full length Bmi1 from the MSCV-Bmi1-pgk-EGFP plasmid provided by Dr. Guy Sauvageau (Clinical Research Institute of Montreal, Montreal, Quebec, Canada) and cloning it at the EcoRI-XhoI sites of the MSCV vector. Two pLKO.1-based lentiviral shRNA vectors, TRCN0000040058 and TCRN0000012565 (Open Biosystems, Huntsville, AL, USA), were used for Myb and Bmi1 knockdown, respectively. A scrambled lentivirus shRNA (#1864, Addgene, Cambridge, MA) was used as control. pLVTSH-Myb which encodes a doxycycline (DOX)-inducible c-Myb shRNA targeting nucleotides 1,161–1,187 (from the AUG start site) of the human c-Myb mRNA was kindly provided by Dr. Thomas Gonda (University of Queensland, Queensland, Australia).
Culture conditions, isolation and retroviral/lentiviral transduction of bone marrow cells
Bone marrow cells were isolated from femurs of Mybf/d, Mybf/f, BCR-ABL+Mybw/f and BCR-ABL+Mybw/d, and non-nucleated cells were removed by hypotonic lysis. Prior to transduction, Mybf/f and Mybf/d marrow cells were cultured in pre-stimulation media with murine IL-7 (10). Retroviral transductions were performed as described (22). Lentiviral transduction was performed using concentrated supernatant (http://ubik.microbiol.washington.edu/protocols/bl3/virusconcentration.htm) from 293T cells co-transfected with pLKO.1shRNA vector, pMDG and pCMVΔR874 plasmids. Lentiviral stocks were titrated on K562 cells using a 10−2–10−7dilution. Viral transduction was performed at a cell density of 2×105 cells/ml. Transduced cells were plated at 1×105 cells/ml in the presence of puromycin (2μg/ml). Percentage of cells surviving after 2 days of selection was used to calculate viral titer. Approximately 1.1×109 infectious units/mL were used in lentiviral transduction experiments. One round of co-sedimentation and incubation were carried out as described above.
Bone marrow transplantation and leukemogenesis assays
2×106BCR-ABL transduced Mybf/f and Mybf/d bone marrow cells were transplanted into lethally-irradiated (11 Gy in two split doses of 5.5 Gy each) C57BL/6J mice by tail vein injection. Presence of GFP-positive BCR/ABL-expressing cells in bone marrow and spleen was evaluated by flow cytometry. For determination of survival, mice appearing moribund were sacrificed and bone marrow and spleen were harvested for immunophenotyping of p190BCR/ABL-transduced GFP+ cells.
Flow Cytometry and Immunophenotyping
Bone marrow and spleen single cell suspensions from transplanted mice were evaluated for GFP-positivity using normal bone marrow and splenocytes as a control. Lineage of GFP-positive cells was determined using PE-Cy5-anti-mouse CD19 (Biolegend) and PE-anti-mouse Gr-1 (BD Pharmingen). Immunophenotyping of B cells was performed using anti-mouse antibodies: APC-B220 (Biolegend), PE-CD43 (BD Pharmingen), and FITC-IgM (BD Pharmingen).
Colony Formation Assay
Bone marrow cells from BCR-ABL+Mybw/f and BCR-ABL+Mybw/d mice were plated in “Complete” Methylcellulose Medium for Pre-B Cell Colony Assays (Stem Cell Technology) at 50,000 cells per plate. Cells transduced for knockdown or overexpression experiments were plated at100,000 cells/plate. Colonies were scored after 7–11 days.
RNA interference
For siRNA transfection, 1 × 106 Z-181 or SUP-B15 cells were resuspended in 100 μl of nucleofector V solution (Amaxa, Cologne, Germany) and mixed with 5 μg of ON-TARGET plus SMART pool siRNA (ThermoFisherScientific, Dharmacon, Waltham, MA). The solution was added to the Amaxa electrode cuvettes and electroporated in Amaxa Electroporator II, using program S-18 (Z-181) or X-01 (SUP-B15). Afterward, cells were diluted in 2 ml IMDM (supplemented with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM L-glutamine) at 37°C (5 × 105 cells/ml).
Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP Assay Kit (USB Corporation, Cleveland, Ohio). Briefly, 2.5 × 107 exponentially growing Z-181 cells were cross-linked with 1% formaldehyde, incubated for 10 min at 37°C and treated with glycine (final concentration125 mM; 5 minutes at room temperature). Cells were then washed with ice-cold PBS and resuspended in 1mL of lysis buffer with a protease inhibitor cocktail and sonicated at 30% power for 20 pulses of 15seconds each in a Branson Sonifier 450 (Branson Ultrasonics, Danbury, CT). Chromatin was pre-cleared with 50 μL proteinA-agarose beads for 60 min at 4°C with rotation and pre-cleared lysates were immunoprecipitated with the anti c-Myb antibody (8 μg; clone 1-1, Upstate Biotechnology, Lake Placid, NY) at 4°C overnight with rotation.
Immunoprecipitations without antibody (no antibody control) or an anti-mouse IgG (8 μg; Thermo Scientific Pierce, Rockford, IL) were included with each experiment. Immune complexes were collected with 50 μL protein A-agarose beads for 60 minutes at 4°C with rotation (except for 10μL of supernatant of the no antibody control saved as input) and washed once with 1 mL each of: low salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 150 mM NaCl), high salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 1.5 M NaCl), LiCl wash buffer (250 mM LiCl; 1% NonidetP-40 [NP-40]; 1% sodium deoxycholate; 1 mM EDTA; and 10 mM Tris-HCl, pH 8.0); and twice with 10 mM Tris-HCl, pH 8.0; and 1 mM EDTA. Immune complexes were next eluted using freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). Cross-links were reversed by heating at 65°C overnight in the presence of 0.2 M NaCl. DNA was recovered using PrepEase DNA Clean-Up Kit (USB Corporation). ChIP DNA (2 μL) was used as a template for semi-quantitative PCR (32 cycles) using the following Bmi-1 primers: (sense 5′ACGGGCCTGACTACACCGACACT 3′, antisense 5′ CCCGATCTCTGCCTCTCATA 3′ from −238 to +23), (sense 5′ GTTTCCACTCTGCCTTCAGC 3′, antisense 5′ CCCGATCTCTGCCTCTCATA 3′, from −111 to + 23),( sense 5′AGA GAG ATG GAC TGA CAA ATG C 3′, antisense 5′ GTG AGG AAA CTG TGG ATG AGG 3′ from + 5954 to + 6103).
Patients
Levels of c-Myb and Bmi1 mRNA were analyzed in peripheral blood or bone marrow blasts of nineteen newly diagnosed BCR-ABL1-positive adult acute lymphoblastic leukemia (ALL) patients. Diagnosis of ALL was made on the basis of morphologic, biochemical and immunologic characteristics of leukemic cells. Patients’ median age was 53 years (range: 18–76); median blast percentage at diagnosis was 90% (range: 49–99). Main characteristics of the patients are shown in Supplementary Table 2.
RT-PCR analysis of c-Myb and Bmi1 expression in patient samples
Total RNA was extracted from mononuclear cells separated by Ficoll-Hypaque density gradient centrifugation using the RNeasy total RNA isolation kit (Qiagen, Valencia, CA) according to manufacturer’s instructions; 1μg of RNA was used for cDNA synthesis with Moloney murine leukemia virus reverse transcriptase (Invitrogen) in the presence of dNTPs and Random Hexamer primers (Perkin-Elmer, Vaterstetten, Germany).
MYB and BMI1 expression was assessed using Syber Green PCR master Mix (Applied Biosystems, Foster City, CA) and primers described in Supplementary Table 3. Taqman Syber Green assays were performed in duplicate using a 7900 Real-Time PCR system and 7900 System Software (Applied Biosystems, Foster City, CA) and a thermal protocol consisting of: 50°C for 2 min, followed by 95°C for 10 min, then 40 cycles of 95°C for 1 min and 60°C for 1 min. Results were normalized to the level of ubiquitously expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Standard curves were generated for experiments measuring absolute (copy number) expression values and the results were expressed as ratio MYB/GAPDH and BMI1/GAPDH.
Statistical Analyses
Due to small sample size, exact methods were used for survival’s analyses using StatXact version 8 by Cytel. The Wilcoxon-Gehan test for censored data was computed for each study. The Wilcoxon-Gehan test is similar to the log rank test for survival data, but it assigns a higher weight to early observations and gradually reduces the weight over time. Kaplan-Meier plots were generated using R software. All p-values reported are based on exact 2-sided tests. All other statistical analyses were conducted using unpaired Student’s t-test.
Results
Frequency of B cell subsets in heterozygous c-Myb knockout mice
To test the requirement of c-Myb in normal B-cells and in p190BCR/ABL-dependent leukemogenesis, we used heterozygous c-Myb knockout mice (referred here as Mybf/d) because deletion of both Myb alleles induces a marked decrease of mature B cells by a block at the pro-B to pre-B transition (28).
We first assessed whether loss of a c-Myb allele correlated with reduced c-Myb expression in marrow mononuclear and CD19+ B cells. Compared to cells with two functional Myb alleles (Mybf/f), levels of c-Myb were reduced approximately 50% in both cell fractions from mice with only a functional Myb allele (Mybf/d) (Supplementary Figure 1A). We then assessed the frequency of B cell subsets; consistent with previous studies using adult mice on a mixed genetic background (26,28), the proportion of pro-B, immature and mature B cells was similar in bone marrow and spleen of Mybf/f and Mybf/d mice while there was a slight but not significant (p= 0.307) decrease of pre-B cells in the Mybf/d mice (Supplementary Figure 1B).
Mybf/f and Mybf/d bone marrow cells have a similar capacity to reconstitute B cell subsets in recipient mice
We further investigated the requirement of c-Myb expression in B cell lymphopoiesis by assessing B cell reconstitution in lethally-irradiated B6. SJL-Ptprca Pepcb/BoyJ congenic mice (CD45.1+) used as recipients of Mybf/f or Mybf/d bone marrow cells (CD45.2+). Mybf/f and Mybf/d donor marrow cells, identified by expression of the CD45.2 antigen, generated similar proportions of total and B cell subsets in bone marrow and spleen of recipient mice (Supplementary Figure 1C–E and Supplementary Figure 2). The pre-B-cell subset in bone marrow (1.15% greater in Mybf/f than in Mybf/d recipients) (Supplementary Figure 1D) and the immature B cell subset in the spleen (3.74% greater in Mybf/f than in Mybf/d recipients) (Supplementary Figure 1E) showed minor, not significant differences (p=0.280 and 0.310, respectively).
Requirement of c-Myb in two models of p190BCR/ABL- dependent leukemogenesis
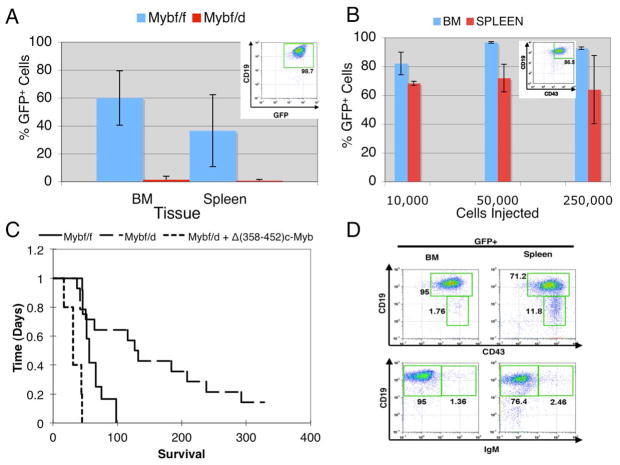
We first assessed whether c-Myb expression is necessary for p190BCR/ABL -dependent leukemogenesis using bone marrow cells from Mybf/f and Mybf/d mice transduced with the MigRI/GFP-p190BCR/ABL retrovirus and transplanted into lethally-irradiated recipient mice. Transplanted mice (six/group) were sacrificed 8 weeks post-injection; mice injected with Mybf/f cells showed extensive infiltration of GFP-positive cells into bone marrow and spleen; by contrast, mice transplanted with Mybf/d bone marrow cells had very few GFP-positive cells in these tissues (Figure 1A). Nearly all GFP-positive cells in the bone marrow and spleen of Mybf/f recipient mice were CD19+ (Figure 1A, inset), indicative of B-cell leukemia.
Figure 1.
c-Myb hemizygous bone marrow cells are less efficiently transformed by the p190BCR/ABL oncogene. (A) Infiltration of p190BCR/ABL-transduced Mybf/f and Mybf/d bone marrow cells into bone marrow and spleen of lethally-irradiated recipient mice. GFP expression from the MigRI/GFP bicistronic retrovirus was used to detect BCR/ABL-expressing donor cells. Recipient mice were sacrificed 8 weeks after transplantation with 2×106 MigRI p190BCR/ABL-transduced bone marrow cells. GFP+ cells were detected by flow cytometry; inset shows that nearly all GFP+ cells were CD19+; (B) Different numbers of CD19+CD43+ GFP+ cells (inset) from mice transplanted with p190BCR/ABL-transduced Mybf/f marrow cells were injected into sub-lethally-irradiated secondary recipient mice and GFP-positive cells analyzed by flow cytometry in bone marrow and spleen once moribund mice were sacrificed (2–3 weeks post-injection). (C) Kaplan-Meier plot shows survival of mice injected with 2 × 106 p190BCR/ABL-transduced Mybf/f or Mybf/d bone marrow cells transduced with p190BCR/ABL alone with p190BCR/ABL and Δ(358–452) c-Myb. (D) Representative immunophenotype of GFP+ cells from bone marrow and spleen of mice injected with p190BCR/ABL-transduced Mybf/f cells [CD19+CD43+ (top panel); CD19+IgM+ (bottom panel)].
The phenotype of transformed (GFP+) cells in recipient mice was analyzed by flow cytometry based on CD19 and CD43 expression, and double-positive CD19/CD43 cells (Figure 1B, inset) from mice transplanted with p190BCR/ABL-transduced marrow cells from Mybf/f mice were sorted by GFP expression and injected into sublethally-irradiated recipient mice. Two mice each were transplanted with 2.5×105, 5×104, or 1×104 pro-B cells. Within 18 days, all secondary recipient mice became moribund and were sacrificed. Each showed massive infiltration of bone marrow and spleen with p190BCR/ABL-transduced GFP-positive cells (Figure 1B). To further assess whether the pro-B cell leukemia was transplantable, tertiary recipient mice were injected with GFP+CD19+CD43+ cells sorted from secondary recipients. As few as 100 cells were sufficient to cause leukemia within 24 days of injection (data not shown).
To determine whether the reduced infiltration of GFP-positive leukemic cells in mice transplanted with p190BCR/ABL-transduced Mybf/d bone marrow cells correlated with longer survival, mice injected with p190BCR/ABL -transduced bone marrow cells from Mybf/f and Mybf/d mice were sacrificed when terminally ill and bone marrow and spleen evaluated by flow cytometry for the presence of GFP-positive leukemic cells. Median survival of mice (n=12) transplanted with p190BCR/ABL-transduced marrow cells from Mybf/f mice was 56 days, whereas that of mice (n=14) inoculated with p190BCR/ABL-transduced marrow cells from Mybf/d mice was 130 days (Figure 1C). The difference in survival of the two groups was statistically significant (p=0.007; Log-rank test).
The reduced leukemogenic potential of p190BCR/ABL-transduced Mybf/d marrow cells was rescued by co-infection with a retrovirus encoding a mutant form of c-Myb: mice (n=5) injected with doubly-transduced Mybf/d marrow cells survived even less of those injected with p190BCR/ABL-transduced Mybf/f marrow cells (Figure 1C). GFP+ cells taken from the bone marrow and spleen of leukemic mice exhibited a pro-B cell phenotype (Figure 1D).
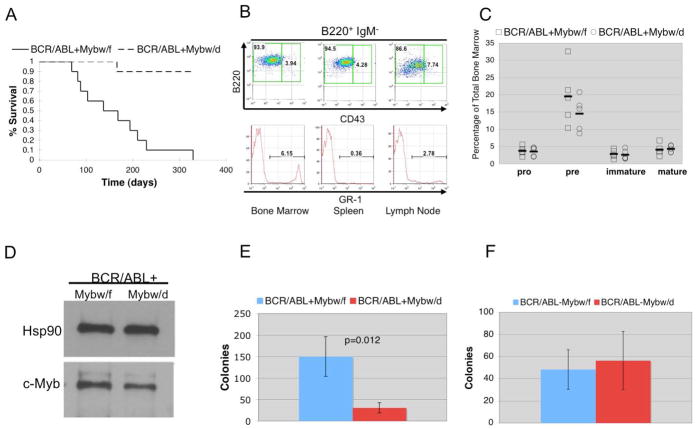
The role of c-Myb expression in p190BCR/ABL-dependent leukemogenesis was also assessed using intercrosses of p190BCR/ABL transgenic mice which typically develop a B cell leukemia (28) and c-Myb heterozygous knockout mice (Mybf/d) to obtain mice with one (BCR/ABL+Mybw/d) or two (BCR/ABL+Mybw/f) functional c-Myb alleles. These F1 mice exhibited significantly different survivals (p=0.0006): all ten BCR/ABL+Mybw/f mice were dead 11 months after birth (median survival, 153 days) while only one of ten mice died in the BCR/ABL+Mybw/d group (median survival, > 300 days) (Figure 2A). Hematopoietic tissues of moribund mice were infiltrated with B220+CD43low/−IgM− cells whereas only few GR-1-positive myeloid cells were detected (Figure 2B), consistent with the development of pre-B cell leukemia.
Figure 2.
Loss of one copy of c-Myb suppresses the leukemogenic potential of B cells from p190BCR/ABL-transgenic mice.(A) Survival of BCR/ABL+Mybw/d and BCR/ABL+Mybw/f mice; (B) Frequency of B lymphocyte and myeloid cells in hematopoietic tissues of terminally ill mice evaluated using B cell (B220, CD43, IgM) and myeloid (GR-1) markers; (C) Frequency of B cell subsets in the bone marrow of 8 week old BCR/ABL+ Mybw/f and BCR/ABL+Mybw/d mice; (D) Expression of c-Myb (anti-Myb, clone 1-1, Upstate Biotechnology) in pre-B cells from BCR-ABL+Mybw/f and BCR-ABL+Mybw/d mice. Levels of Hsp90 were measured as control of equal loading; Colony formation of bone marrow cells from BCR-ABL+Mybw/f and BCR-ABL+Mybw/d mice (E) and BCR-ABL−Mybw/f and BCR-ABL−Mybw/d mice (F). Cells were plated at 5×104 cells/plate in methylcellulose supplemented with IL-7 and colonies were scored seven days after plating. Panels represent combined results from 3 experiments, each performed in triplicate.
Low levels of c-Myb reduce colony formation of p190BCR/ABL-transformed B cell progenitors
To further test the effects of reduced c-Myb levels in p190BCR/ABL-expressing B cells, we performed additional in vitro assays using cells from 8-week old BCR/ABL+Mybw/f and BCR/ABL+Mybw/d mice. The frequency of pro-B, immature B, and mature B cells was essentially identical in the bone marrow of BCR/ABL+Mybw/f and BCR/ABL+Mybw/d mice, while the difference in pre-B cell frequencies was not significant (Figure 2C); pre-B cells from BCR/ABL+Mybw/d mice showed decreased levels of c-Myb (approximately 50% by densitometry) compared to the pre-B cell counterpart from the bone marrow of BCR/ABL+Mybw/f mice (Figure 2D). Compared to BCR/ABL+Mybw/f bone marrow, colony formation of BCR/ABL+Mybw/d in “complete” methylcellulose medium for pre-B cell colony assays was significantly reduced (Figure 2E; p=0.012). Such difference appears to be due to increased colony formation of cells expressing p190BCR/ABL and carrying two functional copies of the c-Myb gene, as bone marrow cells from BCR/ABL−Mybw/d littermates formed the same number of colonies of BCR/ABL−Mybw/f cells (p=0.551) (Figure 2F) and BCR/ABL+Mybw/d bone marrow cells (Supplementary Table 1). The reduced colony formation of BCR/ABL+Mybw/d pre-B cells was rescued by expression of a mutant form of c-Myb with longer half-life (19) (Supplementary Figure 3A and B).
c-Myb-regulated Bmi1 expression is required for p190BCR/ABL-dependent leukemogenesis
Since c-Myb functions as a transcription factor, the reduced leukemogenic potential of p190BCR/ABL-expressing Myb+/− B cells may be caused by changes in the expression of transcriptionally-regulated downstream targets of c-Myb.
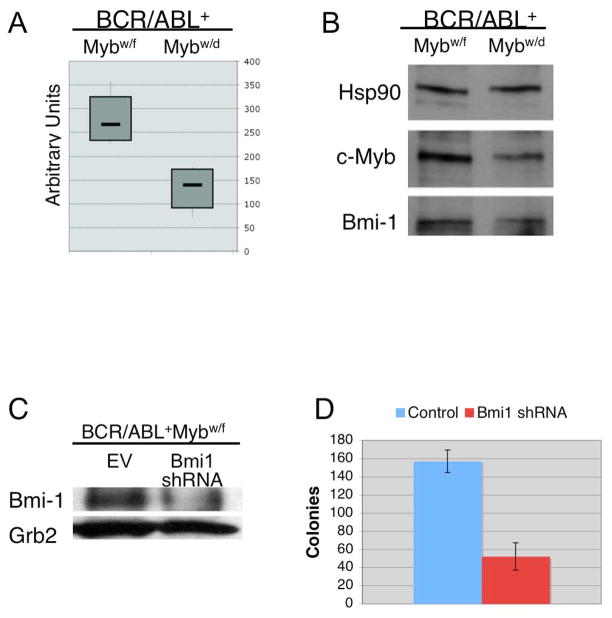
Thus, we probed oligonucleotide microarrays with pre-B cell RNA from pre-leukemic BCR/ABL+Mybw/f and BCR/ABL+Mybw/d mice (5–7 weeks of age; n=4 each). This cell subset was chosen because of the propensity of BCR/ABL+Mybw/f mice to develop BALL with a pre-B cell phenotype (Figure 2B). Global gene expression profiles show distinct patterns between BCR/ABL+Mybw/f and BCR/ABL+Mybw/d pre-B cells (Supplementary Figure 4A and B). Known c-Myb regulated genes such as Bcl-2, c-Kit, Rag2, adenosine receptor 2B (activated) and GATA-2 (repressed) were identified in our array data; we also identified other candidate c-Myb targets with a potential involvement in transformation such as members of the WNT/β-catenin family and the polycomb group gene Bmi1. We decided to investigate Bmi1 further because of its role in hematopoietic stem cell self-renewal and leukemogenesis (25). Bmi1 mRNA expression is lower (p=0.0396) in BCR/ABL+Mybw/d than in BCR/ABL+Mybw/f pre-B cells (Figure 3A; Supplementary Figure 4A and B) and correlates with reduced protein levels (approximately 40% by densitometry) (Figure 3B).
Figure 3.
Requirement of Bmi1 expression in p190BCR/ABL-transformed pre-B cells. (A) Box plot shows levels of Bmi1 mRNA in BCR/ABL+Mybw/f and BCR/ABL+Mybw/d pre-B cells (n=4); (B) Western blot shows c-Myb and Bmi1 expression (anti- Bmi1 antibody, Millipore, clone F6) in pre-B cells (representative of 4 samples) from 5–7 week old BCR/ABL+Mybw/f and BCR/ABL+Mybw/d mice. Levels of Hsp90 were measured as control of equal loading; (C) Western blot shows Bmi1 expression in BCR/ABL+Mybw/f bone marrow cells lentivirally transduced with the empty vector (EV) or a Bmi1-specific shRNA. Levels of Grb2 (anti Grb2, BD Transduction Laboratories) were measured as loading control; (D) Pre-B colony formation of Bmi1 shRNA-transduced BCR/ABL+Mybw/f bone marrow cells from 8-week old mice. Bone marrow cells were transduced with the EV or Bmi1 shRNA lentivirus for 24h and plated (100,000 cells/plate) in methylcellulose supplemented with IL-7. Plates (n=6/each experiment) were scored after 10 days. 98% cells from plucked colonies were CD19+.
To evaluate whether Bmi1 is required for p190BCR/ABL-dependent transformation, its expression was knocked-down in BCR/ABL+Mybw/f bone marrow cells lentivirally transduced with a pre-tested Bmi1 shRNA (Figure 3C). Bmi1 shRNA-transduced BCR/ABL+Mybw/f bone marrow cells (from an 8-week old mouse) showed a significant reduction in pre-B colony formation compared to empty vector-transduced cells (p=0.001) (Figure 3D).
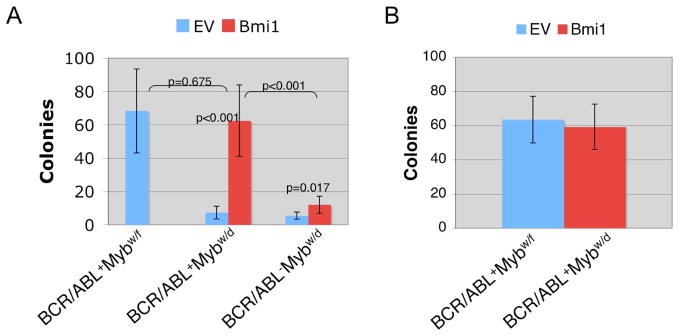
We also overexpressed Bmi1 in BCR/ABL+Mybw/d bone marrow via retroviral transduction, and transduced cells were utilized for colony formation assays. Upon plating on IL-7-supplemented methylcellulose plates, Bmi1-transduced bone marrow cells were markedly more clonogenic than empty vector (EV)-transduced cells (p<0.001) (Figure 4A). Because Bmi1 can promote B cell transformation (29), we transduced BCR/ABL−Mybw/d bone marrow cells with the Bmi1 or the control vector to assess whether expression of Bmi1 alone was sufficient to increase colony numbers. As expected, expression of Bmi1 alone did increase the number of IL-7-dependent colonies (p=0.017) (Figure 4A); however, colonies were significantly fewer than those derived from cells co-expressing BCR/ABL and Bmi1 (p< 0.001) (Figure 4A), suggesting that Bmi1 cooperates with p190BCR/ABL in B cell transformation. Since Bmi1 promotes the self renewal of normal and leukemic hematopoietic stem cells (28), we assessed whether Bmi1-transduced BCR/ABLMybw/d cells isolated from colonies formed in IL-7-supplemented primary plates would form secondary colonies. EV-transduced BCR/ABL+Mybw/d cells formed no colonies (data not shown), while Bmi1-transduced BCR/ABL+Mybw/d cells formed secondary colonies as effectively as BCR/ABL+Mybw/f cells (Figure 4B), indicating that Bmi1 expression is important for self-renewal of p190BCR/ABL-transformed pre-B cells.
Figure 4.
Overexpression of Bmi1 rescues pre-B colony formation of BCR/ABL+Mybw/d bone marrow cells. (A) Pre-B cell colony formation of BCR/ABL+Mybw/d and BCR/ABL−Mybw/d total bone marrow retrovirally-transduced with Bmi1 or the empty vector (EV) (n=6 each). Cells were plated (100,000/plate) in IL-7 supplemented methylcellulose and scored 7 days later. Colony formation of EV-transduced BCR/ABL+Mybw/f colony formation was included for comparison; (B) Secondary colony formation of Bmi1-transduced BCR/ABL+Mybw/d and EV-transduced BCR/ABL+Mybw/f cells. Primary plates were solubilized and single cells replated (20,000 cells/plate) in IL-7-supplemented methylcellulose. Secondary colonies were counted after 10 days.
Bmi1 is a direct c-Myb target
Because reduced c-Myb expression correlated with lower Bmi1 levels (Figure 3A and B), we investigated whether expression of Bmi1 was directly regulated by c-Myb. Search of the murine and human Bmi1 promoter using transcription factor binding site databases (i.e. www.gene-regulation.com) indicated the presence of putative Myb binding sites, suggesting that Bmi1 might be a direct Myb target.
Using a human Bmi1 5′ flanking sequence-LUC plasmid (30), we performed luciferase assays in 293T cells co-transfected with the MigRI empty vector or the MigRI-Myb full length cDNA. In three independent experiments, Bmi1-driven LUC activity was transactivated approximately two-fold (Supplementary Figure 5A, left); a similar effect was observed using the Bmi1-LUC plasmid with a mutated c-Myc binding site (30) (Supplementary Figure 5A, right); since c-Myc has been reported to be a c-Myb target (31), the effect of Myb on the Bmi1 promoter might be direct. Chromatin immunoprecipitation (ChIP) assays carried out in the Ph1-positive Z-181 B-cell leukemia line (32) demonstrated that the −238 to +43 or the −111 to +43 segment of the Bmi1 5′ flanking region (but not nucleotides +5954 to + 6103 in the coding sequence) was amplified from anti-c-Myb ChIPs (Supplementary Figure 6C), suggesting that the interaction occurs in correspondence of the underlined putative c-Myb binding site at nucleotides +3 to +8 (Supplementary Figure 6B). To assess the role of this putative binding site, a Myb-binding mutant 5′ Bmi1-Luc plasmid was generated and used in promoter transactivation assays. The promoter with a mutated Myb-binding site was no longer transactivated by c-Myb (Supplementary Figure 5B, right), while the corresponding wild-type promoter showed a two-fold transactivation (Supplementary Figure 5B, left).
c-Myb and Bmi1 expression is required for proliferation and survival of human p190BCR/ABL-positive leukemic cells
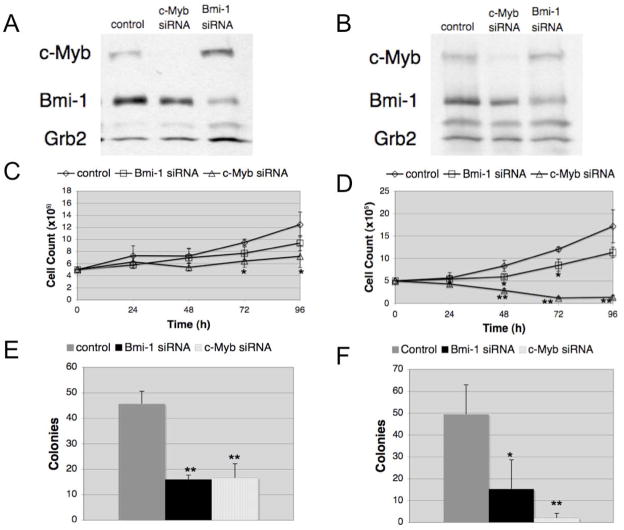
To assess whether levels of c-Myb also have an important role in Ph1ALL, we knocked-down c-Myb expression in Z-181 and SUP-B15 B-ALL cell lines and examined the effects on proliferation and colony formation. c-Myb siRNA-transfected cells showed markedly decreased c-Myb expression and proliferated less (p<0.05, p≤0.002; Z-181 and SUP-B15 cells, respectively) than scrambled siRNA-transfected cells (Figure 5A–D). The effect of reduced c-Myb expression was also tested upon plating Z-181 and SUP-B15 cells in IL-7-supplemented methylcellulose to allow growth of pre-B cells. c-Myb siRNA-transfected Z-181 and SUP-B15 cells exhibited significantly reduced (p≤0.002) colony formation compared to control cells (Figure 5E and F). Colony formation of Z-181 and SUP-B15 cells transduced with a lentivirus expressing a doxycycline-inducible Myb shRNA was also markedly suppressed (Supplementary Figure 6). Downregulation of Myb expression was associated with decreased levels of Bmi1 (approximately 50 and 25% in siRNA-transfected Z-181 and SUP-B15 cells and 50 and 75% in shRNA-transduced Z-181 and SUP-B15 cells, respectively) (Figure 5A and B, and Supplementary Figure 7).
Figure 5.
Expression of c-Myb and Bmi1 is required for proliferation and colony formation of Ph1 Z-181 B-ALL and SUP-B15 cells. Western blots show c-Myb and Bmi1 expression in scrambled, c-Myb or Bmi1 siRNA-transfected Z-181 (A) or SUP-B15 (B) cells; levels of Grb2 were measured for normalization. Proliferation of Z-181 (C) or SUP-B15 (D) cells after transfection with scrambled, c-Myb or Bmi1 siRNA. Transfected cells were plated at 100,000 cells/ml (in triplicate) and counted every 24 hours. Colony formation of Z-181 cells (E) or SUP-B15 (F) cells transfected with scrambled, c-Myb, or Bmi1 siRNA. Cells were plated at 100,000 (Z-181) or 75,000 cells/plate (SUP-B15 cells) in rhIL-7-containing methylcellulose. Colonies were scored after 10 days. (*p-value< 0.05, ** p-value p≤0.002).
We also assessed proliferation and colony formation of Bmi1 siRNA-transfected Z-181 and SUP-B15 cells; both cell lines proliferated less and formed fewer colonies (p<0.05 and p<0.002, respectively) than the scramble siRNA-transfected counterpart (Figures 5C–F); of interest, inhibition of colony formation was more pronounced than inhibition of proliferation.
c-Myb and Bmi1 expression shows a positive correlation in BCR-ABL1+ ALL patients
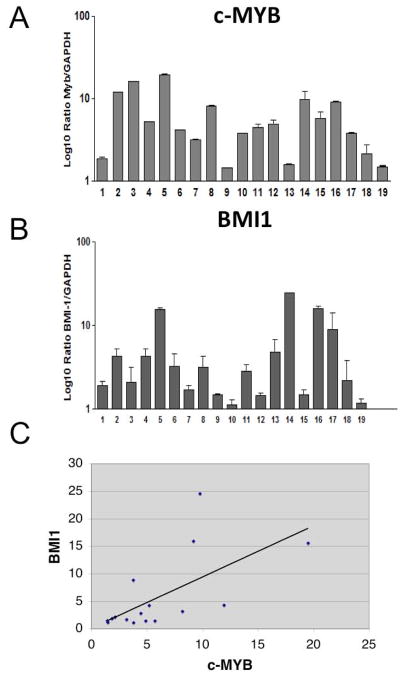
Expression of c-Myb and Bmi1 mRNA was assessed in blast cells of nineteen newly diagnosed BCR-ABL1-positive adult ALL patients (Figure 6A and B). The median value of the Myb/GAPDH ratio in ALL blasts was 4.46 (range, 1.43–19.9) whereas the median value of the Bmi1/GAPDH ratio was 2.835 (range, 1.12–24.57). Overall, expression of c-Myb and Bmi1 showed a trend toward positive correlation when considering the totality of the ALL samples (p=0.0478; Spearman correlation analysis). The correlation in the ALL group was even stronger when limiting the analysis to the p190BCR/ABL-positive samples (p= 0.0132) (Figure 6C)
Figure 6.
c-Myb (A) and BMI1 (B) mRNA levels in BCR-ABL1-positive ALL patients. Absolute levels of c-Myb and Bmi1 mRNA were detected in blasts of BCR-ABL1-positive ALL patients by reverse transcription and SYBR green-based real-time PCR. Relative c-Myb and Bmi1 transcript levels were calculated using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression to normalize for equivalent cDNA levels and results were expressed as ratio between c-Myb and GAPDH and Bmi1 and GAPDH. The bar graph shows the average numbers of c-Myb mRNA copies from duplicates. Error bars represent standard deviations. (*) denotes p210BCR/ABL-positive ALL samples. (C) Scatter plot represents the correlation of Myb and Bmi1 mRNA levels in p190BCR/ABL-positive ALL samples.
Discussion
In this study, we investigated the role of c-Myb in B-cell leukemogenesis induced by p190BCR/ABL because, in spite of its essential role in B cell development and its abundant expression in B-ALL (including few cases of Ph1 ALL) (33, 34), no data exist in support of an important role of c-Myb in models of oncogene-induced B cell malignancies. We utilized: i) lethally-irradiated mice transplanted with wild-type (f/f) or Myb+/− (f/d) marrow cells transduced with the p190BCR/ABL retrovirus; and ii) 190BCR/ABL transgenic mice intercrossed with knockout mice carrying one functional c-Myb allele (w/d). In both models, reduced c-Myb expression caused longer disease latency and survival of leukemic mice. In the bone marrow transplantation/leukemogenesis model, p190BCR/ABL induced a pro-B cell leukemia (CD19+ CD43+) while p190BCR/ABL transgenic mice developed a leukemia with a predominant pre-B cell (B220+CD43low/−IgM−) phenotype; pre-B cells from p190BCR/ABL/Mybw/d transgenic mice were less clonogenic than the Mybw/f counterpart, suggesting that optimal levels of c-Myb are required for proliferation and survival of p190BCR/ABL-transformed pre-B cells. Direct proof that this effect is specific for p190BCR/ABL-transformed cells stems from the observation that no differences in colony formation were noted using Mybw/f and Mybw/d pre-B cells from p190BCR/ABL-negative littermates. To address potential mechanisms that may explain the selective effect of c-Myb dosage in p190BCR/ABL-transformed cells, we performed microarray analyses using RNA from p190BCR/ABL-expressing and normal Mybw/f and Mybw/d pre-B cells; levels of Myb most affected the gene expression profile of p190BCR/ABL-transformed cells, but a subset of genes was also modulated in normal B cells; we reasoned that among the genes differentially expressed in Mybw/f and Mybw/d p190BCR/ABL-transformed pre-B cells there may be one or more whose change in expression is mechanistically linked to the impaired leukemogenic potential of Mybw/d p190BCR/ABL-expressing pre-B cells.
One such gene is Bmi1 which is an attractive candidate as a relevant c-Myb target because of its role in self-renewal and maintenance of normal and leukemic stem cells (25, 35) and its cooperation with classical oncogenes in neoplastic transformation (36–39). Through perturbation of gene expression, luciferase, and ChIP assays we demonstrated that Bmi1 is a direct target of Myb; moreover, its ability to restore the defective colony formation of c-Mybw/d p190BCR/ABL pre-B cells to levels comparable to those of the p190BCR/ABL-transformed counterpart with basal c-Myb levels supports the notion that Bmi1 is a functionally relevant c-Myb target for p190BCR/ABL-dependent transformation of pre-B cells.
Of equal importance, secondary colony formation of Bmi1-transduced c-Mybw/d p190BCR/ABL pre-B cells was also undistinguishable from that of Mybw/f p190BCR/ABL-transformed pre-B cells, suggesting that high expression of Bmi1 is necessary for their self-renewal.
Since normal pre-B cells possess limited self-renewal potential, p190BCR/ABL would not be able to maintain their transformed state had these cells not acquired self-renewal properties through a regulatory pathway dependent, in part, on increased Myb-Bmi1 expression.
Indeed, expression of c-Myb and Bmi1 was higher in p190BCR/ABL-expressing pro-B and pre-B cells than in their normal counterpart (Supplementary Figure 8).
While Bmi1 expression appears to be important for p190BCR/ABL-dependent leukemogenesis, it is unlikely that it is the only relevant c-Myb target for maintenance of the transformed phenotype of p190BCR/ABL-expressing B cells.
Bmi1-silenced p190BCR/ABL marrow cells from transgenic mice formed fewer B cell colonies than the control counterpart; although not directly comparable, c-Mybw/d p190BCR/ABL B-cells were even less clonogenic than Bmi1-silenced p190BCR/ABL-expressing cells which would indicate that the consequences of reduced c-Myb expression for p190BCR/ABL transformation of pre-B cells do not depend entirely on decreased Bmi1 levels.
The role of c-Myb and Bmi1 was also tested in p190BCR/ABL-expressing Ph1 ALL lines; proliferation and colony formation of Z-181 and SUP-B15 cells was suppressed by c-Myb or Bmi1 RNA interference and Bmi1 levels were decreased in c-Myb-silenced cells whereas c-Myb levels did not change in Bmi1-silenced cells.
Together, these findings suggest that expression of c-Myb and Bmi1 is functionally related and that the c-Myb-Bmi1 pathway plays an important role in regulating proliferation and survival of Ph1 ALL cells, although the effects of c-Myb may be, in part, Bmi1-independent and expression of Bmi1 may be regulated by other oncogenic proteins including c-Myc (30).
A significant correlation between c-Myb and Bmi1 expression was noted in comparing c-Myb and Bmi1 mRNA levels in BCR/ABL-expressing ALL blasts; this finding is interesting because the expression of both genes is likely to be subjected to multiple regulatory mechanisms and their levels might be more closely related in clonogenic cells.
In summary, optimal levels of c-Myb expression are required for p190BCR/ABL-dependent transformation and leukemogenesis and the effects of c-Myb depend, in part, on its ability to enhance the expression of Bmi1, a gene that regulates the self-renewal of hematopoietic stem cells (25, 35), endowing p190BCR/ABL-transformed pro- and pre-B cells with “stem cell properties” essential for leukemia maintenance.
Supplementary Material
Acknowledgments
We thank Dr Dimri (Department of Cancer Biology, Evanston Northwestern Healthcare Research Institute, Evanston, IL) for the Bmi1-Luc plasmid. We thank Dr. Sankar Addya (Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA) for analysis of microarray data.
This work was supported by National Cancer Institute grants CA95111 and P0178890 (B.C) and European LeukemiaNet, AIRC, AIL and Fondazione Del Monte di Bologna e Ravenna (G.M. and I.I.) and NIAID grant AI059294 (T.P.B). A.R.S. was supported by a fellowship of the American-Italian Cancer Foundation (AICF) and is currently supported by a fellowship from AIRC.TW was supported by a National Institutes of Health predoctoral fellowship (T32-CA09683-14).
Footnotes
This work is dedicated to the memory of Dr Alan M. Gewirtz
Conflict of interest
The authors declare no conflict of interest
Authorship contributions
T. Waldron performed most experiments and wrote first draft of the manuscript. M. De Dominici performed the experiments with human ALL lines. A.R. Soliera performed Luciferase and ChIP assays on the Bmi-1 promoter. I. Iacobucci, A. Leonetti and G. Martinelli performed real time PCR studies on ALL samples. Y Zhang and R. Martinez performed microarray hybridization analyses. T. Hyslop performed statistical analyses. T.P. Bender provided Mybf/d mice and help for the immunophenotype analyses. B. Calabretta designed experiments and wrote manuscript.
References
- 1.Gleissner B, Gokbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW, et al. Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002;99:1536–1543. doi: 10.1182/blood.v99.5.1536. [DOI] [PubMed] [Google Scholar]
- 2.Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- 3.Clark SS, McLaughlin J, Timmons M, Pendergast AM, Ben-Neriah Y, Dow LW, et al. Expression of a distinctive BCR-ABL oncogene in Ph1-positive acute lymphocytic leukemia (ALL) Science. 1988;239:775–777. doi: 10.1126/science.3422516. [DOI] [PubMed] [Google Scholar]
- 4.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 5.Goga A, McLaughlin J, Afar DE, Saffran DC, Witte ON. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 6.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 7.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okuda K, Golub TR, Gilliland DG, Griffin JD. p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene. 1996;13:1147–1152. [PubMed] [Google Scholar]
- 9.Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Liu Y, Pelletier S, Buchdunger E, Warmuth M, Fabbro D, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet. 2004;36:453–461. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- 12.Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene. 2002;21:7137–7146. doi: 10.1038/sj.onc.1205942. [DOI] [PubMed] [Google Scholar]
- 13.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 17.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell. 2001;104:21–32. doi: 10.1016/s0092-8674(01)00188-x. [DOI] [PubMed] [Google Scholar]
- 18.Yang MY, Liu TC, Chang JG, Lin PM, Lin SF. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood. 2003;101:3205–3211. doi: 10.1182/blood-2002-05-1598. [DOI] [PubMed] [Google Scholar]
- 19.Corradini F, Cesi V, Bartella V, Pani E, Bussolari R, Candini O, Calabretta B. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant Myb mutants. J Biol Chem. 2005;280:30254–30262. doi: 10.1074/jbc.M504703200. [DOI] [PubMed] [Google Scholar]
- 20.Calabretta B, Sims RB, Valtieri M, Caracciolo D, Szczylik C, Venturelli D, et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of Myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci U S A. 1991;88:2351–2355. doi: 10.1073/pnas.88.6.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratajczak MZ, Hijiya N, Catani L, DeRiel K, Luger SM, McGlave P, et al. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of Myb antisense oligodeoxynucleotides. Blood. 1992;79:1956–1961. [PubMed] [Google Scholar]
- 22.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 2008;111:4771–4779. doi: 10.1182/blood-2007-08-105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 24.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayeuela JM, Dik WA, et al. The MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 25.Lessard J, Sauvageau G. Bmi1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 26.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, Groffen J. Acute leukaemia in bcr/abl transgenic mice. Nature. 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- 28.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 29.Alkema MJ, Jacobs H, van Lohuizen M, Berns A. Pertubation of B and T cell development and predisposition to lymphomagenesis in Emu Bmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 1997;15:899–910. doi: 10.1038/sj.onc.1201262. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns, van Lohuizen M. Bmi1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536–546. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans JL, Moore TL, Kuehl WM, Bender T, Ting JP. Functional analysis of Myb protein in T-lymphocytic cell lines shows that it trans-activates the c-myc promoter. Mol Cell Biol. 1990;10:5747–5752. doi: 10.1128/mcb.10.11.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Estrov Z, Talpaz M, Zipf TF, Kantarjian HM, Ku S, Ouspenskaia MV, et al. Role of granulocyte-macrophage colony-stimulating factor in Philadelphia (Ph1)-positive acute lymphoblastic leukemia: studies on two newly established Ph1-positive acute lymphoblastic leukemia cell lines (Z-119 and Z-181) J Cell Physiol. 1996;166:618–630. doi: 10.1002/(SICI)1097-4652(199603)166:3<618::AID-JCP17>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Mavilio F, Sposi NM, Petrini M, Bottero L, Matinucci M, De Rossi G, et al. Expression of cellular oncogenes in primary cells from human acute leukemias. Proc Natl Acad Sci USA. 1986;83:4394–4398. doi: 10.1073/pnas.83.12.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 35.Rizo A, Olthof S, Han L, Vellenga E, de Haan G, Schuringa JJ. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood. 2009;114:1498–1505. doi: 10.1182/blood-2009-03-209734. [DOI] [PubMed] [Google Scholar]
- 36.Haupt Y, Bath ML, Harris AW, Adams JM. Bmi1 transgene induces lymphomas and collaborates with Myc in tumorigenesis. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- 37.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns, van Lohuizen M. Bmi1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warner JK, Wang JC, Takenaka K, Doulatov S, McKenzie JL, Harrington L, Dick JE. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia. 2005;19:1794–1805. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- 39.Datta S, Hoenerhoff MJ, Bommi P, Sainger R, Guo WJ, Dimri M, et al. Bmi1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer Res. 2007;67:10286–10295. doi: 10.1158/0008-5472.CAN-07-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.