Abstract
The bacterial outer membrane (OM) is an exceptional biological structure with a unique composition that contributes significantly to the resiliency of Gram-negative bacteria. Since all OM components are synthesized in the cytosol, the cell must efficiently transport OM-specific lipids and proteins across the cell envelope and stably integrate them into a growing membrane. In this review, we discuss the challenges associated with these processes and detail the elegant solutions that cells have evolved to address the topological problem of OM biogenesis. Special attention will be paid to the Bam machine, a highly conserved multiprotein complex that facilitates OM β-barrel folding.
Keywords: outer membrane biogenesis, β-barrel, Bam complex, LPS
1. Introduction
Perhaps the most distinguishing feature of the typical Gram-negative bacterium is the remarkable structure referred to as the outer membrane (OM). This robust bilayer accommodates a unique complement of OM-specific proteins and lipids that perform specialized cellular functions; as such, the OM can be regarded as an extracytoplasmic organelle. This membrane functions primarily as a highly selective permeability barrier that insulates the cell against a variety of potentially cytotoxic agents in the extracellular milieu. Incorporated in the OM are membrane proteins that participate in a diverse array of processes including transport (both passive and active), proteolysis, secretion, adhesion, signaling, homeostatic control, and biogenesis and reinforcement of the OM itself [1]
The targeting and assembly of membrane proteins in general is a complex process that requires multiple folding factors. OM protein biogenesis is further complicated by the fact that these proteins must traverse one lipid bilayer and then integrate specifically into another; the cell must be capable of discriminating between inner membrane (IM) and OM proteins in addition to coordinating the assembly of each.
A comprehensive review of membrane protein folding must include a discussion of integral β-barrel protein assembly in the Gram-negative OM, particularly because this system serves as a model for understanding mitochondrial and chloroplast biogenesis (as these organelles are the products of an ancient bacterial symbiosis; [2]). Because an appreciation of the topological problem of integral OM protein biogenesis requires some familiarity with OM anatomy and physiology, we preface our discussion of the topic with a primer on the individual components of the OM.
2. Description of OM and components
2.1 OM Lipids
With respect to lipid content, the outer membrane in most Gram-negative bacteria is peculiar in that it is a composite of two major lipidic species (phospholipid and LPS) that are asymmetrically distributed in the membrane. These lipids, discussed below, have unique properties that substantially influence the fluidity and permeability of the OM.
2.1.1 Phospholipids
The majority species of phospholipid in E. coli is the zwitterionic phosphatidylethanolamine (PE), which represents roughly 75–80% of the total phospholipid content in the cell. Negatively charged phosphatidylglycerol (PG) comprises much (15–20%) of the remainder, and a PG condensate known as cardiolipin (CL) is present in small amounts. This ratio is slightly altered in the OM, where PE represents an even larger proportion of total membrane phospholipid (~90%). The relatively high concentration of phospholipids with saturated acyl chains in the OM, along with the associated increase in membrane rigidity, further distinguishes the OM from the IM [3, 4].
2.1.2 LPS
The barrier property of the OM can largely be attributed to the presence and asymmetrical distribution of the well-known bacterial glycolipid called lipopolysaccharide (LPS). This surface-molecule-turned-antigen is specifically targeted to the OM and is normally found exclusively on the outer surface of the cell [5]. LPS monomers in a bilayer exhibit strong lateral interactions with one another; this in combination with the aforementioned enrichment of fully saturated phospholipids in the inner leaflet greatly reduces the fluidity of the OM, which is rigid and gel-like in comparison to the IM [6].
LPS can be subdivided into three distinct fractions: lipid A (which secures LPS in the OM via a hexa-acylated sugar moiety), core oligosaccharide, and the distal O-antigen (which is not produced in derivatives of E. coli K-12). Synthesis of lipid A and addition of the core oligosaccharide occur at the cytoplasmic face of the IM, whereas O-antigen ligation only takes place after the nascent glycolipid is flipped to the periplasmic face of the IM [7]. LPS is shuttled to and integrated into the OM by a dedicated transport system that spans the E. coli cell envelope [7].
2.2 OM Proteins
Despite the robustness of the OM, Gram-negative bacteria are capable of selective uptake of essential nutrients, toxin secretion (pathogenicity) or efflux (multidrug resistance), and assembly of complex surface-exposed macromolecules (as well as the cell envelope itself). This is accomplished through the regulated synthesis, assembly, and activity of OM proteins [6]. Although exceptions to the following generalization have been described, the vast majority of OM proteins present in Gram-negative bacteria belong to one of two major classes:
lipoproteins, which are tethered to the periplasmic face of the OM via N-terminal lipid modifications, and
β-barrels (OMPs), which span the OM and enable the cell to interact and mediate exchange with its environment.
2.2.1 Lipoproteins
Many periplasmic proteins likely display diffuse localization patterns within the periplasm, as has been reported for periplasmic green fluorescent protein (GFP) [8]. However, a host of periplasmic proteins execute functions specifically at the inner or outer membrane. The N-terminal lipidation of membrane-associated periplasmic proteins serves to spatially restrict their activity, presumably enabling them to function more efficiently. Over 90 lipoproteins have been identified in E. coli [9], and these factors have been shown to participate in a host of processes including envelope biogenesis [10–14], cell division [15], secretion [16], and signaling [17]. Lipoproteins may interact with periplasmic proteins, integral membrane proteins, the murein layer, or other lipoproteins, and are sometimes found as vital components of transmembrane complexes.
Membrane anchoring of lipoproteins is not necessarily a prerequisite for function. Indeed, multiple essential lipoproteins in E. coli are known to function properly even when the N-terminal lipobox (required for lipidation) is deleted, so long as they are produced in excess [18, 19]. This suggests that lipidation of some periplasmic proteins is simply a matter of economy. However, mislocalization of certain envelope lipoproteins abrogates function and in some cases causes toxicity. For example, ectopic IM localization of the peptidoglycan-associated lipoprotein Lpp leads to cell lysis [20], and rerouting of the OM-associated (penicillin-binding protein) PBP cofactor LpoA to the IM renders it unable to participate in peptidoglycan biosynthesis [13]. Therefore, the proper localization of lipoproteins, generally speaking, is critical for growth and viability.
2.2.2 OMPs
In addition to the presence of LPS and the asymmetrical lipid distribution of the OM, another prominent characteristic that distinguishes it from the IM is the presence of integral membrane proteins that almost exclusively adopt a β-barrel conformation. The β-barrel structure can be imagined as an antiparallel β-sheet that wraps around a central pore to form a cylinder stabilized by an inter-strand main chain hydrogen bonding network; the barrel is closed by the noncovalent pairing of the first and last beta strands [21].
As with lipoproteins, membrane-integral β-barrel proteins carry out diverse functions in the outer membranes of Gram-negative bacteria (as well as in OMs of mitochondria and chloroplasts). The most abundant OMPs in E. coli serve as passive, relatively non-specific transporters that permit the diffusion of sugars, ions, and small hydrophilic molecules smaller than ~700 Da across the OM. Examples of these porins include OmpF, OmpC, and PhoE, all of which form highly stable trimeric complexes in the membrane. Other OMPs, such as the maltodextrin transporter LamB or the sucrose channel ScrY, facilitate diffusion of specific substrates. While the general and specific porins were among the earliest to be characterized, OMPs have since been shown to participate in an wide array of cellular processes including energy-dependent efflux (e.g. TolC), active transport (e.g. FhuA or BtuB), adhesion (e.g. Ag43), secretion (e.g. so-called autotransporters), pilus biogenesis (e.g. FimD), OM biogenesis (e.g. BamA or LptD), proteolysis (e.g. OmpT), peptidoglycan binding (e.g. OmpA), and the stress response (e.g. OMPLA) [22].
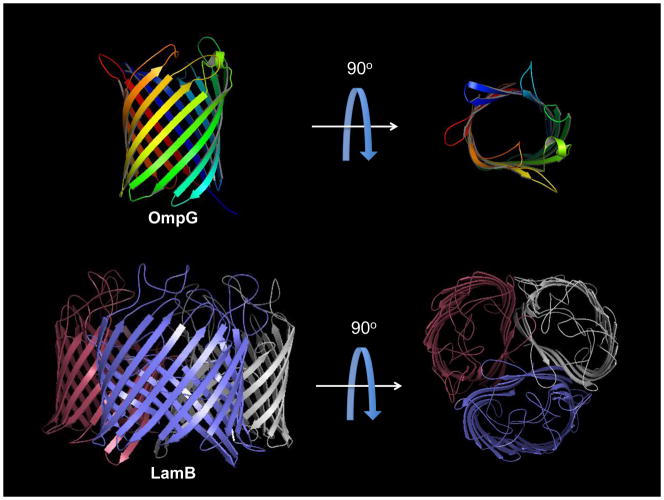
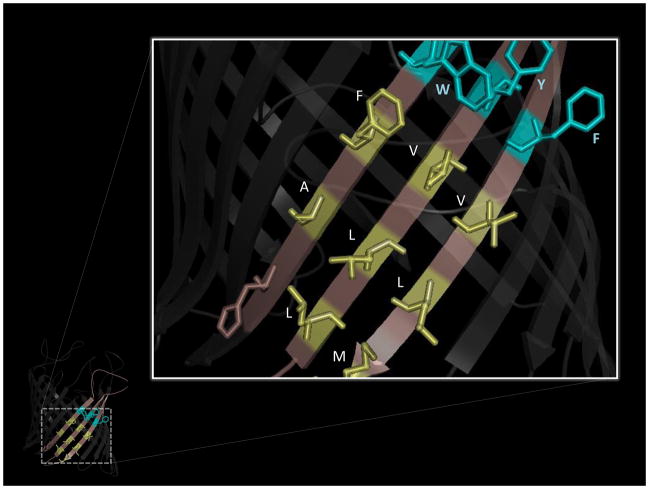
Although OM β-barrels can have widely different functions, available structural information reveals a number of physical characteristics that typify this family of proteins (Fig. 1). Every known bacterial OMP contains an even number of transmembrane (TM) β-strands such that the N- and C-termini of the barrel reside in the periplasm (it is worth noting that this is not necessarily the case for mitochondrial β-barrels). The β-strands are tilted relative to the transmembrane axis, and the length of each β-strand is correlated with the degree of the tilt. As the TM surface of an OM β-barrel is lipid-exposed, it is not surprising that hydrophobic residues are present in a dyad repeat pattern in the TM β-strands (Fig. 2A); this pattern ensures a continuous hydrophobic surface around the barrel exterior, while permitting positioning of polar residues within the interior, often resulting in formation of a water-filled pore [22]. Short turns connect the β-strands at the periplasmic face of the OM, whereas long loops bridge the strands at the extracellular face. These loops may fold into and constrict (or occlude) the barrel pore, or they may interact with LPS; in certain cases, loops are critical for protein function (e.g. [23, 24]).
Figure 1.
The general β-barrel fold. These E. coli OMPs exhibit characteristic typical of bacterial OM β-barrels, including short periplasmic turns, long extracellular loops (some of which fold back into the barrel lumen in the case of LamB, and an even number of β-strands. Note that OmpG (top, PDB ID: 2X9K) is monomeric, whereas LamB (bottom, PDB ID: 1AF6) forms stable trimers in the OM.
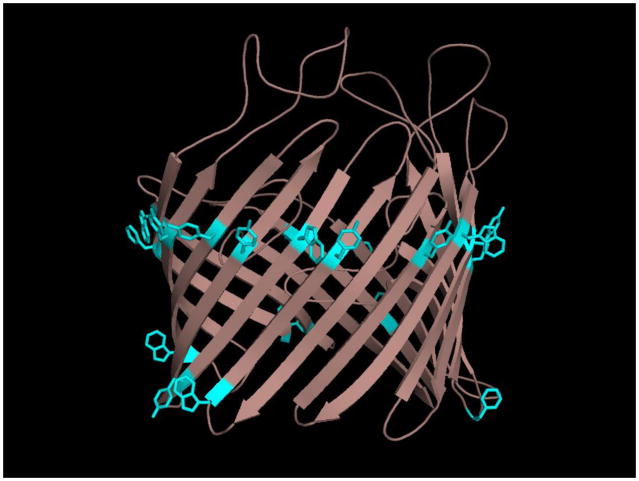
Figure 2.
Figure 2A. The alternating (dyad repeat) pattern for hydrophobic residues in TM strands 8–10 of the E. coli OM beta-barrel protein LamB (PDB ID: 1AF6). Non-polar, membrane-exposed residues are highlighted in yellow, and the aromatic side chains forming part of the aromatic girdle (see Fig. 2B) are highlighted in cyan.
Figure 2B. Residues defining the “aromatic girdle”, shown in cyan on this structure of the LamB monomer from E. coli (PDB ID: 1AF6), demarcate the membrane boundaries.
Some OMPs are hybrid proteins that contain sizeable periplasmic domains. Such domains may extend the function(s) of individual OMPs and serve as receptors, cell wall anchors, or platforms for multiprotein complex assembly [10, 12].
2.2.3 Accessory proteins & surface structures
In addition to membrane lipids and integral proteins, the OM of E. coli accommodates a variety of complex surface-exposed accessory structures. The membrane serves as a substratum for assembly of appendages such as pili, flagella, and the injectisome, and it also anchors the capsular exopolysaccharide to the cell surface. It is not surprising, therefore, that defects in OM biogenesis can have deleterious effects on processes such as virulence, motility, and biofilm formation, which can be dependent on surface appendages or capsules. Recent comprehensive reviews of the biogenesis of these structures have been presented elsewhere (cf. [25–27]). Consequently, they will not be discussed in detail here, except to say that many of the factors required to build these appendages are themselves OM proteins (e.g. FimD, the type I pilus usher, is an OM β-barrel); therefore, proper assembly of surface structures can depend indirectly on proper assembly and folding of OM proteins.
2.3 Peptidoglycan (a disclaimer)
The peptidoglycan cell wall is an elastic mesh-like network of rigid glycan strands cross-linked by flexible peptide linkers. This essential organelle is required to resist turgor pressure, promote cell growth and septation, and maintain cell shape. The fact that this critical structural element is situated between the inner and outer membranes of Gram-negative bacteria might suggest that it represents a physical barrier for nascent OM components in transit through the periplasm; however, no available evidence supports this conclusion. Additionally, attempts to determine the porosity of the murein sacculus suggest that the peptidoglycan layer is permeable to globular proteins up to 100 kDa [28]. Although the role(s) of the peptidoglycan layer in OM biogenesis (if any) has generally been difficult to characterize, the available evidence suggests that involvement of the sacculus in transport or assembly of OM components is minimal at best. Accordingly, with all due reverence for this vital cellular factor and our colleagues who study it, we will largely ignore the peptidoglycan layer in our discussion.
3. The topological problem of OM biogenesis
None of the constituent parts of the OM are synthesized in situ. As a consequence, OM lipids and proteins must overcome a series of obstacles in order to traverse the cell envelope from their site of synthesis in the cytoplasm or the inner leaflet of the IM to their final location in the membrane. Following synthesis, OM-directed components must first be translocated across the IM. This is an energetically unfavorable process for both lipids and proteins, so this step must be coupled to an exergonic reaction [29]. As membrane proteins and lipids necessarily contain highly hydrophobic domains, they must then be at least partially shielded from the aqueous periplasm in order to prevent aggregation and/or irreversible misfolding due to hydrophobic collapse. Additionally, these components must be specifically targeted to the OM, as the presence of OM components in the IM can cause disastrous effects [30, 31]. As such, it is imperative that targeting of OM factors be specific and unidirectional. Lastly, OM lipids and proteins must be properly assembled into the OM, and in the case of OMPs, locally folded. To complicate matters further, OM integration must occur during growth in the absence of any obvious energy source (the periplasm is devoid of ATP and the OM is not energized by an ion gradient), all while maintaining a robust permeability barrier [32].
The Gram-negative cell has evolved elegant solutions to the topological problem of OM biogenesis. Although each OM constituent (with the possible exception of phospholipid) has a dedicated periplasmic transport and OM assembly system, the targeting process for all components can be generally described as follows:
OM components are synthesized in the cytosol (or at the cytosolic face of the IM),
energy derived from ATP hydrolysis promotes translocation across (and, in some cases, release from) the IM,
periplasmic chaperones shield hydrophobic patches and direct the components to the OM for assembly,
a dedicated OM protein (or protein complex) coordinates energy-independent substrate transfer and OM integration.
Each transport system accomplishes the steps above through the concerted efforts of envelope factors present in each cellular compartment. In this review, we will describe these systems in detail, with special attention paid to the transport and OM assembly of β-barrel proteins.
4. Solutions to the topological problem of OM biogenesis
4.1 Lipids
Every OM lipid is an IM emigrant. LPS and OM-destined phospholipids are synthesized at the cytoplasmic face of the IM, flipped to the periplasmic face by IM proteins, and directed to the OM [33]. The lipopolysaccharide transport (Lpt) has been extensively characterized, and recent studies (discussed below) have contributed significantly to our understanding of LPS biogenesis. In stark contrast, the mechanism of transmembrane phospholipid transport remains mysterious.
4.1.1 LPS Transport
The journey to the OM begins for LPS following synthesis at the cytoplasmic face of the IM (for review see [34]). “Rough” LPS (lipid A + core) is first flipped to the periplasmic face of the IM by the ATP-dependent translocase MsbA, which can also promote the translocation of phospholipids. Upon entering the periplasm, O-antigen is distally ligated onto rough LPS to form mature (“smooth”) LPS. The IM ABC transporter LptBFG likely energizes the release of LPS from the IM [35, 36], and the bitopic membrane protein LptC binds LptA at the IM [37] and promotes transfer of the lipid A moiety of LPS to the soluble periplasmic factor LptA [38].
Until recently, it was not known whether LptA forms a soluble periplasmic intermediate with LPS or whether it serves as the periplasmic component of a transenvelope protein bridge, although results of previous studies were more consistent with the latter model [39]. Recent reports provide genetic and biochemical evidence that strongly supports the IM-OM bridge model for LPS transport [37, 40].
Following IM extraction, the remaining steps in LPS transport and assembly proceed in an energy-independent fashion (Fig. 3). This likely involves the affinity-driven directional transfer of LPS monomers from the LPS binding factor at the IM (LptC), across the periplasmic transmembrane bridge (formed by LptA), to a two-protein OM complex composed of the β-barrel LptD and the OM lipoprotein LptE. The manner in which LptDE promotes the cell surface assembly of LPS remains to be determined, though an attractive model proposed by the Kahne group [18, 41] posits that LPS is directly inserted into the OM outer leaflet though a lateral opening in the LptD barrel (perhaps gated by LptE). Such a model is especially appealing in light of the fact that OMPs permitting lateral diffusion of lipids into the OM have already been described [42].
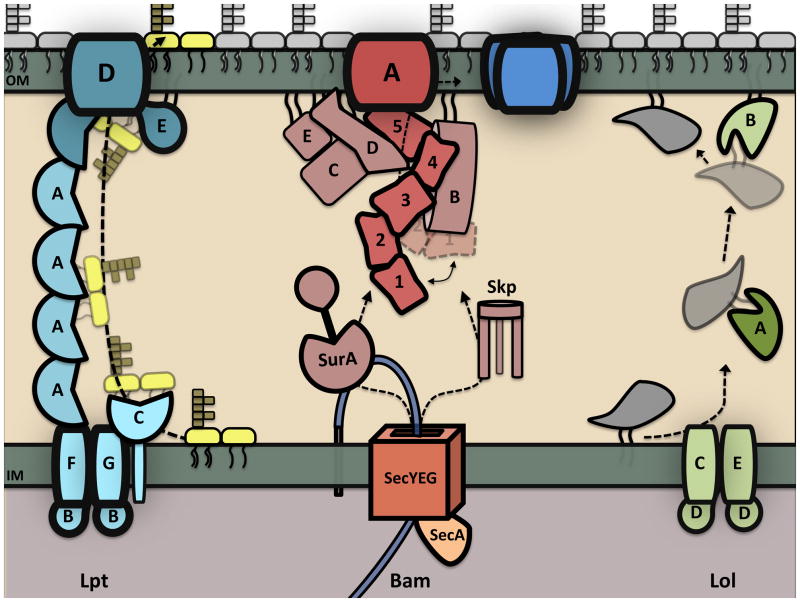
Figure 3.
OM biogenesis in E. coli. All components of the bacterial OM are synthesized in the cytoplasm and translocated across the IM into the periplasmic space. Once in the periplasm, each OM constituent traverses the envelope and integrates into the OM via a dedicated transport system (shown above). The Lpt components form a transenvelope protein bridge that shields the lipid A moiety from the aqueous environment and funnels LPS through LptDE to the cell surface. OM β-barrels (OMPs) cross the IM through the Sec translocon and associate with periplasmic chaperones that target OMPs to the Bam machine for assembly. OM lipoproteins are extracted from the IM in an ATP-dependent fashion and transferred to an OM receptor (LolB) via a periplasmic carrier protein (LolA). Phospholipid transport is not shown, as the mechanism by which they are trafficked to the OM is not known.
4.1.2 Phospholipid Transport
Nothing is known about how phospholipids are shuttled from the IM to the OM. The translocation (‘flip-flop’) of phospholipids to the periplasmic face of the IM can be mediated actively by MsbA or passively by α-helical TM peptides [43–45], although the details of translocation by either route are not understood. Beyond this point, while it is clear that OM phospholipids originate in the IM [46], the mechanism of intermembrane transport remains elusive. Periplasmic lipid vesicles have not been observed visually, and it is unclear whether the peptidoglycan layer would permit the passage of vesicles across the periplasm [4]. Zones of adhesion between the IM and OM have been postulated [47, 48], though their existence remains controversial [49]. It is known that phospholipid shuttling does not require ATP or protein synthesis, but is heavily dependent on the proton motive force [50]. If the bulk of cellular phospholipid transport occurred through zones of membrane fusion or sites of membrane collision [51], it is not obvious why the proton motive force (PMF) would be required (unless IM-OM fusion itself requires the PMF).
Unlike protein and LPS transport, phospholipid transport is bidirectional [52]. A conserved system mediating retrograde phospholipid transport (OM to IM) has been identified in E. coli and was shown to contribute to the maintenance of lipid asymmetry (Mla) at the OM [53]. While the Mla system participates in phospholipid trafficking, it must not be the only means for phospholipid transport, as mla mutant phenotypes are subtle and the system is dispensable for viability [53].
Numerous mechanisms of nonvesicular lipid transport have been observed or proposed in eukaryotes [51], and lipid trafficking in diderm prokaryotes could occur by one or more of these mechanisms in an analogous fashion. Elucidation of the lipid trafficking process(es) employed by bacteria will clearly require further inquiry.
4.2 Proteins
All cellular proteins are synthesized in the cytoplasm, and proteins destined for the OM are no exception. Because export of lipoproteins and OMPs requires the Sec translocase, which only accommodates unfolded substrates [29], the earliest challenges facing nascent OM proteins involve stabilization of unfolded, aggregation-prone polypeptides in the cytoplasm and targeting of these preproteins to Sec for translocation.
Proteins bound for the cell envelope of E. coli can either be translocated during (SRP-dependent) or following (SecB-dependent) translation. In general, integral IM proteins are co-translationally targeted to Sec via the signal recognition particle (SRP)-dependent pathway, whereas exported proteins are maintained in an unfolded, translocation-competent state after translation by the cytoplasmic chaperone SecB, which binds regions of the mature, unfolded protein [54]. These secreted proteins are synthesized with an N-terminal signal sequence that serves as a recognition domain for SecA, the catalytic component of the translocon, which harnesses energy derived from ATP hydrolysis to drive polypeptide translocation through the channel formed by SecYEG [29].
Interestingly, OMPs are not strictly dependent on the post-translational, SecB-dependent targeting pathway for export. An increase in the hydrophobic character of OmpA or LamB signal sequences results in the co-translational, SecB-independent export of these OMPs, which are still correctly targeted to and assembled into the OM following translocation [55, 56]. This suggests that post-translational secretion, while characteristic of all OM proteins, is not important for successive maturation steps following translocation.
Upon entry into the periplasmic compartment, the paths of lipoproteins and OMPs diverge. In this section, we discuss the OM targeting and assembly pathways for the two major types of OM protein after IM translocation.
4.2.1 Lipoproteins
A lipoprotein is not born that way. The maturation process involves a series of post-translational modifications made to nascent lipoproteins following IM translocation. Specifically, prolipoproteins bearing a conserved lipobox sequence (present at the boundary between the signal sequence and cleavage product) are acylated at a conserved N-terminal cysteine residue both prior to and immediately following removal of the signal sequence. The resulting covalently affixed lipid moiety securely anchors the mature lipoprotein to the membrane [57]. Some lipoproteins are retained in the IM and function there, but the majority (90%) are targeted to the OM by a dedicated periplasmic transport system called Lol [58].
Before a mature lipoprotein can be targeted to the OM it must first be liberated from the IM. Since lipoproteins are securely attached to membranes, it is not surprising that extraction of the lipid moiety from the IM is an energy-dependent process. This extraction is catalyzed by the concerted efforts of LolCDE (the constituent members of an IM-associated ATPase) and LolA (the periplasmic lipoprotein release factor). LolCDE energizes the transfer of the lipoprotein acyl chains from the IM to LolA, which forms a soluble periplasmic complex with the free lipoprotein. LolA then transfers the lipid moiety in a “mouth-to-mouth” fashion [59] to the structurally similar lipoprotein acceptor LolB, itself an OM lipoprotein, at which time the LolA-lipoprotein complex is dissociated and the lipid transferred to the inner leaflet of the OM via LolB. Thus, the localization of LolB at the OM and LolCDE at the IM ensures that lipoprotein transport is unidirectional and irreversible.
How is the Lol transport system able to distinguish OM lipoproteins from those that function at the IM? In E. coli, the identity of the residue(s) immediately following the acylated N-terminal cysteine specifies the fate of the lipoprotein. The presence of an aspartate at this +2 position causes IM retention of lipoproteins, and the presence of most other amino acids at +2 generally leads to OM localization. Asp(+2) serves as a “Lol avoidance signal” that prevents the release of lipoproteins from the IM, perhaps by promoting the formation of a tight lipoprotein-phospholipid complex that does not interact with LolCDE [60]. An elegant genetic selection conducted by the Pugsley group [61] led to the identification of alternative Lol avoidance +2 residues (e.g. Pro and Trp), however, these residues are not found in endogenous lipoboxes in E. coli. Additional studies have demonstrated a role for the residue at the +3 position in Lol avoidance [62], suggesting that the +2 residue is necessary but not sufficient for IM retention.
It is interesting to note that the Lol system has recently been implicated in the biogenesis of a non-lipoprotein [16]. The secretion of pullulanase is facilitated by the Pul secretin, a homododecamer of the OM protein PulD [63], and OM localization of PulD requires the OM lipoprotein PulS [64]. Indirect evidence for a LolA-PulS-PulD periplasmic complex suggests that PulD might ‘piggy-back’ on PulS to co-opt Lol for OM localization [16]. Although the Lol-Pul heterotrimer could not be observed directly, its putative existence represents a novel targeting mechanism for membrane-spanning OM proteins.
4.2.2 OMPs
4.2.2.1 Late translocation steps
Nascent β-barrel proteins enter the periplasm in an N- to C-terminal fashion during transit through the Sec translocon [29]. Since unfolded OMPs have a tendency to aggregate in solution, it is conceivable that interactions with periplasmic chaperones occur during or immediately following translocation. Indeed, a number of periplasmic factors have been implicated in facilitating late steps in translocation and/or release from the translocon. A recent report of the crystal structures of SecDF, two accessory components of the Sec machine, implies a role for these factors in mediating late steps in translocation in a manner dependent on the PMF [65]. Beyond the secretion machinery, there is evidence suggesting that the IM-associated periplasmic chaperone PpiD positively influences translocation of OMPs across the IM via early interactions with unfolded polypeptides exiting the Sec translocon [66]. This finding is intriguing insofar as it offers an explanation for the phenotype(s) associated with deletion of ppiD in vivo; however, all of the evidence supporting a role for PpiD in late steps of translocation was obtained in an in vitro system, potentially complicating interpretation of the results.
The periplasmic chaperone Skp (seventeen kilodalton protein) was also shown to interact with translocation intermediates of model OMPs at the periplasmic face of the IM [67]. This early interaction seems to require release of the nascent polypeptide from ribosomes, which may suggest that some degree of secondary structure formation is required in order for Skp to bind an emerging OMP. It has been proposed that Skp promotes the release of newly translocated OmpA from the IM [67], but the fact that such evidence is derived from spheroplast experiments, combined with the demonstrated inability of Skp to enhance release of PhoE from Sec in a similar system [68] makes further inquiry necessary before firm conclusions can be drawn. Still, the processing delay observed for OMPs in the absence of certain periplasmic or OM-associated OMP biogenesis factors [69] may signify a role for extracytoplasmic proteins in accepting substrates before translocation is complete.
Following IM translocation, the mature OMP is liberated from its IM-associated signal sequence by periplasmic peptidases. Retarding or altogether preventing signal sequence cleavage by mutation of the processing site interferes with the targeting, assembly, and oligomerization of the trimeric OMP LamB, demonstrating the importance of signal sequence processing for OMP biogenesis. Additionally, the release of this processing-defective LamB variant from the IM is delayed, which may explain the observed activation of the phage shock (Psp) response [30], which is known to be sensitive to IM stress in general and disruption of the PMF in particular [70]. Assuming that Psp activation is an indicator of IM perturbation, it is possible that IM-tethered LamB may assemble aberrantly into the cytoplasmic membrane; such an event is likely to be toxic to the cell, and the reported toxicity associated with high-level production of processing-defective LamB further supports this notion.
4.2.2.2 Interactions with periplasmic chaperones
The importance of OMP-chaperone interactions in the periplasm is well established. Left to its own devices, a wayward beta barrel in the aqueous periplasmic space can wreak havoc in several respects: a) the surface hydrophobicity and insolubility of OMPs quickly leads to the toxic accumulation of misfolded aggregates, b) aberrant association with and assembly into the IM perturbs the membrane and causes stress response activation, and c) failure to correctly target chaperone-dependent OMPs (such as the essential LPS assembly factor LptD) compromises the OM permeability barrier and negatively affects cell viability. Periplasmic chaperones serve to maintain nascent OMPs in a folding-competent state while preventing off-pathway misfolding and aggregation, and they are critically important for specific targeting of OMPs to the OM [1].
In E. coli, the periplasmic protein SurA (so called because it was initially identified in a screen for genes required for stationary phase survival [71]) is regarded as the primary periplasmic OMP chaperone [72]. A role for SurA in extracytoplasmic folding (and OMP biogenesis in particular) was proposed simultaneously by several groups in the mid 1990’s [73–75], and subsequent genetic, biochemical, and structural inquiry has led to a refined understanding of the function of this chaperone.
SurA was initially supposed to function in envelope protein folding in light of several in vivo observations:
Overexpression of surA suppresses OM permeability defects and stress response induction associated with impaired OM biogenesis [75],
Deletion of surA results in activation of the σE extracytoplasmic stress response [74].
These findings implicated SurA in the targeting and/or folding of β-barrel proteins, raising the possibility that SurA is an OMP chaperone. Consistent with a role in OMP biogenesis, SurA was shown to interact specifically with unfolded (but not folded) OMPs [76, 77], as well as with model peptides containing motifs that are enriched in OM β-barrels [77–82].
An unexpected structural motif found in all SurA homologs is the peptidyl-prolyl cis-trans isomerase (PPIase) domain of the parvulin type. The surA ORF encodes two PPIase domains referred to as P1 and P2 [73, 75]. Both domains were shown to be dispensable for the chaperone activity of SurA, which instead resides in a molecular cradle formed by the association of the N- and C-terminal domains [76, 83, 84]. It remains unclear at this time what the importance of the PPIase domains might be, although the fact that deleting either parvulin domain from SurA causes novobiocin sensitivity in UPEC strains of E. coli [84] may imply a specific, parvulin-dependent function of SurA. It is conceivable that these domains are required to catalyze proline isomerization in some specific substrate (or substrates), thereby accelerating folding; indeed, P1 and P2 do endow SurA with the ability to isomerize peptidyl-prolyl bond [76, 83]. However, mutation of catalytic site residues in P2 effectively abolishes the PPIase activity of SurA without affecting chaperone activity [76]. In addition, the P1 domain of SurA is conserved in the leptospiral homolog of SurA, but this domain is devoid of PPIase activity [85]. Taken together, these findings show that the contributions of the PPIase domains to the chaperone function of SurA are probably minimal. Still, the extraordinary conservation of at least one parvulin domain across SurA homologs implies a role for PPIase domains in somehow increasing the efficiency of OMP targeting/assembly, or in catalyzing an as-yet-unidentified secondary function of SurA. It is worth noting that the ability of SurA to interact with model peptides rich in aromatic residues is dependent on the P1 domain, which binds these peptides well even in isolation [82]. As such, the parvulin domains may influence substrate binding by promoting specific interactions.
SurA is known to enhance the rate of OMP monomer folding [69, 73, 74]. Additionally, the deletion or depletion of surA causes strong induction of the σE stress response (which is activated by unfolded OMPs), implying that β-barrel folding is compromised in the absence of SurA. This could reflect a general role for this chaperone in OMP assembly. To determine the substrate range of SurA, Collet and colleagues compared the OM proteome of a wild-type strain of E. coli to that of a ΔsurA mutant [86]. Surprisingly, they found that many OMPs (15 of the 23 observed) are not affected by the absence of SurA. Furthermore, many OMPs of reduced abundance in a ΔsurA strain are controlled at the level of synthesis by σE; consequently, the relative contribution of SurA to folding of those OMPs could not be unambiguously determined, as depletion of surA effectively results in the downregulation of major OMP genes.
Despite this, proteomic analysis did lead to the identification of two OMPs whose decreased abundance in a ΔsurA mutant could not be explained by decreased synthesis: FhuA, a ferrichrome transporter, and LptD/Imp, the integral OM component of the Lpt transport system required for assembly of LPS into the OM. Levels of LptD in the membrane are reduced by an order of magnitude in the absence of SurA, despite the fact that lptD transcription is increased upon σE activation [86]. This suggests that LptD is a (perhaps the) primary substrate of SurA; it is certainly the most important substrate, given the significant OM permeability and growth defects associated with impaired LptD biogenesis [87]. In light of this, it is interesting to note that lptD and surA are organized into an operon [86, 88].
It has been known for some time that defects in LPS biogenesis can have downstream effects on OMP assembly. For example, reduced OMP levels [89, 90] and slowed kinetics of porin folding and trimerization [91] are observed in LPS truncation mutants, and the arrest of lipid synthesis by cerulenin treatment causes a block in OmpF assembly [92]. In addition, the increase in OM permeability associated with reduced expression [93] or mutation [94] of LptD has been described in detail (indeed, the original lptD gene name is imp, reflecting the increased membrane permeability of lptD mutants). Some have gone so far as to blame most of the pleiotropic effects associated with loss of SurA on reduced levels of LptD in a ΔsurA background [86]. This might suggest that the function of SurA in OMP biogenesis is secondary to its role as a bona fide LptD chaperone, at least in E. coli. However, the ability of SurA to greatly increase the efficiency of OmpT folding in an in vitro β-barrel assembly system [95] and the accumulation of unfolded porin observed upon SurA depletion and change in OM density [72] clearly indicate a role for SurA as a general OMP chaperone. Furthermore, in addition to LptD, a requirement for SurA has been described for a number of specific OMPs, including the OM components of the chaperone/usher pilus assembly systems for both P (PapC) and type I (FimD) pili [84, 96], the autotransporters EspP [97, 98], Hbp [99], and IcsA [100], and the adhesin intimin [101].
Crystallographic studies on the tertiary structure of SurA across various species have highlighted the modular nature of SurA and homologous chaperones (including the cytoplasmic ribosome-associated chaperone Trigger Factor) [83, 85, 102]. The first solution structure of E. coli SurA revealed a core chaperone domain tightly associated with the P1 domain, whereas the ‘satellite’ P2 domain is loosely tethered to the core module by a flexible linker [83][93]. However, a subsequent structure of SurA lacking the P2 domain (SurAΔP2) and bound to a model peptide demonstrated a striking conformational rearrangement: in the presence of the aromatic-rich, α-helical C-peptide, the P1 domain completely dissociates from the chaperone core to bind the peptide, and the chaperone domains of each SurAΔP2 monomer directly interact such that an extensive dimer interface is formed [82]. It should be noted that a structural homolog of SurA present in Bordetella (Par27) also crystallizes as a dimer, although the dimer interface differs from that observed for SurAΔP2 [102]. It is tempting to speculate that SurA can drastically alter its conformation or oligomeric state in order to interact with specific substrates, however, there is as yet no evidence that oligomers of SurA are physiologically relevant.
In addition to SurA, the other chaperone believed to play an important role in OMP biogenesis is Skp. Skp has a long and complicated history, having been originally identified as a DNA-binding factor [103], a dubious role for an extracytoplasmic protein. Skp was later implicated in late steps of IM translocation [104, 105]; although Skp is not involved in translocation, this was not the last time that such a role for this chaperone was proposed (see section 4.2.2.1).
It was not until eight years after the discovery of Skp that its primary function was realized. Skp was isolated in an elegant biochemical screen for periplasmic proteins that selectively bind unfolded OMPs [106], and was independently identified in a genetic screen for transposition events that cause activation of the σE stress response [75]. Skp was later shown to promote formation of soluble periplasmic intermediates of OmpA [67]. Together, these findings strongly supported a role for Skp in OMP biogenesis.
The importance of Skp was fully appreciated when it was shown that it is essential for viability in a ΔsurA mutant background; that is, surA and skp constitute a synthetically lethal pair [72, 107]. Deletion of skp causes slight stress response induction, but no significant effect on OMP assembly was observed [72]. Additionally, proteomic analysis suggests that no OMP in E. coli depends on Skp for assembly (J. Collet, personal communication). However, depletion of skp in a ΔsurA mutant leads to accumulation of unfolded OMPs, strong induction of the σE stress response, and cell death. Whereas the abundance of individual OMPs is not significantly decreased in a Δskp mutant, levels of all OMPs are reduced upon simultaneous disruption of surA and skp (J. Collet, personal communication). The synthetic lethality of the skp surA pair strongly suggests that these chaperones have OMP substrates in common (i.e. Skp and SurA are partially redundant), and that together they are responsible for transporting the bulk of E. coli OMPs to the OM. It should be noted that chaperone requirements in different organisms are quite diverse. For example, no synthetic phenotypes are observed when a ΔsurA Δskp double mutant is constructed in N. meningitidis; this may reflect the reduced importance of SurA in this organism relative to E. coli [108].
The structure of Skp has been aptly compared to the body plan of a jellyfish [109]. Skp is a homotrimer comprising a core “body” domain and three α-helical “tentacles” that extend from it [110], bearing some resemblance to the structures of the archeal chaperone prefoldin [109] and the mitochondrial intermembrane space chaperone Tim9.10 [111][93]. The tentacle helices form the walls of a deep hydrophobic pit that is thought to bind unfolded substrate proteins via a combination of hydrophobic and electrostatic interactions [112]. Indeed, NMR and site-directed fluorescence spectroscopy indicate that the OmpA β-barrel domain is buried deep within the cavity formed by the tentacles [113], whereas the hydrophilic periplasmic domain remains outside of the Skp cavity and assumes its native conformation [114]. The flexibility of the tentacles [109] theoretically enables Skp to accommodate OMPs of diverse sizes by tolerating large fluctuations in cavity volume. This would explain the ability of Skp to form 1:1 complexes with OMPs ranging in size from 19–89 kDa and containing 8–16 β-strands [112]. In vivo, Skp exhibits a broad substrate spectrum that includes porins [68, 106, 112, 115], OmpA [116], autotransporters [97, 98, 117], and intimin [101]. Skp has also been implicated in virulence in Salmonella, and indirect evidence suggests that it is important for cell-cell spread in Shigella [100, 118].
Some evidence to support a role for minor periplasmic chaperones in OM biogenesis is available in the literature. For example, the IM-tethered, parvulin-like periplasmic protein PpiD was initially identified in a screen for genes that, when overexpressed, suppressed the OM permeability defect of a surA mutant. The results of ensuing experiments showed that ppiD could not be deleted in a strain lacking surA, leading the authors to conclude that these factors are at least partially redundant [119]. While these findings have been called into question [96, 120], evidence is mounting that PpiD does indeed serve as a periplasmic chaperone, although its substrates may not necessarily be OMPs [120, 121]. It has been proposed that PpiD functions as a “gatekeeper”, interacting with nascent envelope proteins as they are translocated into the periplasm [66] (see 4.2.2.1). The observed IM localization and in vitro chaperone activity of PpiD [120] are consistent with this hypothesis; however, because all evidence for involvement of PpiD in translocation (or translocon release) was obtained using spheroplasted cells [66][93], it remains to be seen whether or not PpiD facilitates late steps of translocation in vivo.
The chaperone function of FkpA, an FKBP-like [122], dimeric [123] periplasmic PPIase, has also been interrogated genetically and biochemically. Like SurA, FkpA exhibits chaperone activity in vitro [124, 125] and in vivo [126, 127] that is independent of its PPIase activity [125]. It is reportedly involved in cell invasion and virulence in various Gram-negatives [128–130], although the relative importance of FkpA in these processes has been disputed [131]. FkpA is also known to be essential for the toxicity of colicin M in a manner that does require PPIase activity [132], and while it has been implicated in maturation of the autotransporter EspP [133], colicin M remains the sole confirmed in vivo substrate of FkpA. Although periplasmic chaperone activity for this PPIase has been demonstrated, there is currently no evidence that FkpA is involved in OMP maturation in any direct way [134, 135].
Considerably less is known about PpiA, the fourth and final PPIase present in the E. coli periplasm. Although the regulation of this factor by the CpxAR [136], which suggests a role for periplasmic PPIases in the envelope stress response, no in vivo substrates for PpiA have been identified, and no OMP biogenesis defects are observed in its absence [96]. Accordingly, PpiA is not thought to participate in OMP folding in any direct way [135].
The periplasmic ATP- independent serine protease DegP is a key housekeeping factor that can promote both refolding and proteolysis of unfolded, misfolded, or aggregated proteins in the envelope [137]. Particularly important during times of stress, the synthesis of DegP is activated in response to a wide variety of extracytoplasmic stresses [138]. This protease is also specifically induced by the presence of periplasmic OMPs and responds by degrading dead-end, off-pathway folding intermediates and/or forming massive molecular cages around folding-competent off-pathway substrates to promote proper assembly [139].
Although DegP is a potent protease, it also exhibits considerable chaperone activity even when proteolytic activity is abrogated by mutation of the active site serine [138]. The chaperone activity of DegP is thought to predominate at lower temperatures, whereas a temperature-dependent conformational switch favors protease activity at higher temperatures [140]. DegP is often regarded as a periplasmic chaperone primarily because the simultaneous deletion of surA and degP is not tolerated [107]; this suggests that DegP is an essential component of a secondary OMP assembly pathway [72]. While the importance of DegP in maintaining homeostasis and responding to stress is clear, less is understood about how it might contribute to OM assembly of β-barrels. Overproduction of a protease-deficient variant of DegP (DegPS210A) has been shown to suppress the lethal effects associated with the expression of folding-defective OMPs [141, 142], but the DegPS210A chaperone can only bind and sequester these OMPs in the periplasm; it cannot return them to the folding pathway. While DegP may be important for preventing toxic aggregation of misfolded or mistargeted OMPs through a combination of proteolysis and sequestration, it is not clear at this time whether it represents a dead-end for OMPs or whether they can re-enter the folding pathway after a productive association with DegP.
4.2.2.3 OM targeting/Substrate recognition
The targeting of secretory proteins to the Sec translocon is known to depend on the presence of an N-terminal signal peptide that is recognized by components of the Sec apparatus. An important question in the field of OMP biogenesis involves the signal(s) and recognition factor(s) required for β-barrel assembly in vivo. The existence of a sorting signal for OMPs was initially proposed based on the observation that the C-terminal amino acid of β-barrels is absolutely required for OM assembly [143]. Phylogenetic analysis revealed a high degree of sequence conservation at this position; the C-terminal residue is almost always aromatic, and a phenylalanine side chain is found most often at this position [144]. While mutation or deletion of this C-terminal phenylalanine essentially blocks assembly and causes periplasmic accumulation in vivo [142], such OMP variants can still fold properly into bilayers in vitro [145], implying that these C-terminal mutants are folding-competent but are not recognized by an assembly factor.
Although the putative OMP recognition factor has yet to be identified in bacteria, a recent investigation into the targeting of mitochondrial β-barrels convincingly demonstrated the involvement of an OM peripheral membrane protein in selective recognition of β-barrels via the C-terminal β-strand, which contains a conserved sequence motif termed the “β-signal” [146]. This mitochondrial OMP recognition factor, Sam 35, is an essential component of a highly conserved multiprotein complex that is required for β-barrel assembly across phylogeny. These results strongly suggest that the cellular machinery that facilitates OMP assembly is able to identify and bind substrates by recognizing a conserved motif in the final β-strand [146]. The manner in which OMPs are assembled into a biological membrane by the bacterial homolog of this machine will be discussed in the following section.
4.2.2.4 OM assembly
Incorporation of peptides into the low-dielectric-constant lipid phase of a bilayer requires complete saturation of main-chain hydrogen bonding potential within membrane-spanning regions. In α-helical TM proteins, the hydrogen bonding potential of the backbone is satisfied by the sequential formation of internal hydrogen bonds that stabilize the helix and permit partitioning into the bilayer (assuming the side chains to be exposed to the lipid phase are nonpolar). In β-barrel membrane proteins, however, the backbone polar groups are neutralized by hydrogen bonding between neighboring β-strands. As a consequence, whereas the TM α-helices of IM proteins can be individually released into the membrane, the folding and membrane integration of OMPs are predicted to occur simultaneously, such that OMP assembly can be described as an “all-or-none” folding event [21]. This fundamental difference between IM and OM protein assembly suggests that the folding pathways of these membrane protein families are distinct.
In contrast to IM protein assembly, the mechanisms of OMP folding and membrane integration in vivo are poorly understood, although in vitro analysis of β-barrel folding into membranes affords some insight into the general characteristics of β-barrels that promote membrane folding. Since the exterior surface of the barrel is buried within a lipid bilayer, it is not surprising that side chains exposed to the hydrophobic core of the membrane are primarily short-chain aliphatic residues with high Kyle-Doolittle values [147]. Indeed, in vitro folding experiments with a model OMP showed that the majority of lipid-exposed β-barrel residues must be hydrophobic, and that prolines and charged residues are generally not tolerated among membrane-facing residues [148]. Aromatic side chains are overrepresented in OM β-barrels, particularly at the membrane-solvent interface [21], where they occur in rings around the barrel rims to form so-called “aromatic girdles” [6], presumably influencing the stability of the folded OMP within the membrane (Fig. 2B). It has also been shown that β-hairpins comprising a β-sheet need not necessarily be covalently linked, as fragments of a single β-barrel structure are still assembled properly into the membrane as a β-barrel monomer [149]. This implies that the minimal structural unit required for β-barrel assembly is shorter than the full-length OMP.
In silico modeling of OMP unfolding highlights the contribution of various structural features of β-barrel that can impact stability [150, 151]. In addition to a barrel domain and extracellular loops, some OMPs exhibit unique features such as barrel plugs (which exhibit significant secondary structure with the barrel lumen), or helical “out-clamps” (which may reinforce weakly stable transmembrane β-strands at the membrane-solvent interface; [151]). Additionally, a number of OMPs oligomerize in the membrane, shielding high-energy interaction surfaces and stabilizing membrane strands [151]. The stability of some membrane-integral OMP trimers is striking; dissociation of porin trimers, for example, only occurs after heating at temperatures about 70°C in the presence of detergent (ex. [69]).
While experimental systems devised to study assembly of β-barrels into biological membranes have shown the process to occur spontaneously (see [152]), the rate of OMP folding in vitro occurs far too slowly to be physiologically relevant. This implies the existence of folding factors at the OM that act to increase the rate of OMP assembly so as to support rapid growth. These folding factors have been recently identified [10, 153, 154], and a great deal of information about the so-called β-barrel assembly machine (Bam) has been generated since its discovery.
It is now clear that OMP assembly is dependent on Bam in vivo in every organism in which it has been tested [10, 153–158]. Furthermore, Bam is extremely well conserved; functionally and structurally homologous cousins of BamA/Omp85, the most evolutionarily ancient Bam component, have been identified in mitochondria (Sam50/Tob55), chloroplasts (Toc75), chromalveolates [156] and all diderm bacteria. This suggests that Bam catalyzes a fundamental biological process, and that membrane β-barrels across phylogeny assemble by a nearly universal mechanism.
Bam is an oligomeric membrane-associated protein complex composed of the OM β-barrel BamA and a variable number of accessory OM lipoproteins that physically interact with it [10, 159–163]. The complex was initially discovered in E. coli [10, 87] and was found to comprise four lipoproteins (BamBCDE) that stably bind to the sizeable N-terminal periplasmic domain of BamA [10, 161, 162]. Attempts to determine the stoichiometry of Bam suggest that each complex contains one of each Bam component, although the low molecular weight of BamE makes it difficult to unambiguously determine whether one or two BamE molecules are present per complex [95].
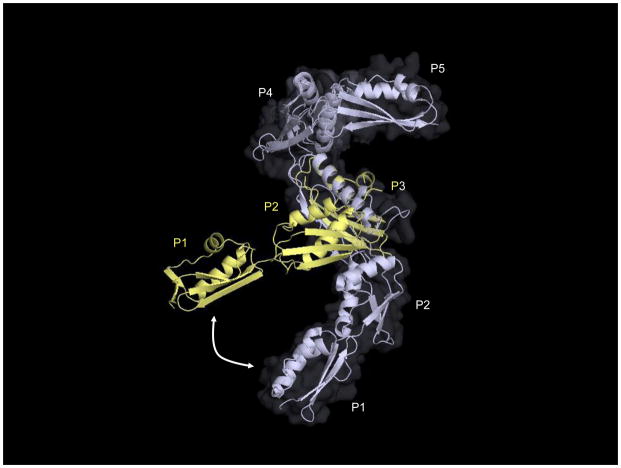
The hub of the physical complex is the periplasmic domain of BamA, which is a helical beaded chain comprising five structurally homologous POTRA (polypeptide translocation associated) domains [161, 164, 165]. These domains, numbered P1-P5 from the N-terminus, each display a β-α-α-β-β architecture and assume a characteristic fold. Although the BamA POTRA domains are strikingly similar with respect to structure (they can essentially be superimposed), they have surprisingly little primary sequence identity and domain deletion experiments imply distinct functional roles for individual POTRAs [161]. The BamA barrel and at least a subset of its POTRA domains are essential for viability [161, 166]. The POTRAs, then, must participate in some critical step(s) in the OMP assembly process. Interestingly, diverse bacteria have different requirements for the periplasmic domain of BamA. In E. coli, for example, deletion of even a single POTRA domain leads to impaired β-barrel assembly and reduced viability [161]. The cell is able to tolerate deletion of P1 and/or P2; but removal of P3, P4, or P5 is lethal even in the presence of P1 and P2; the minimal BamA is P3-5 plus the C-terminal membrane domain [161]. However, Neisseria meningitidis is viable after deletion of P1-4 with only marginal effects on viability, suggesting that the majority of the periplasmic domain of BamA is dispensable in this species [166]. It is conceivable that this reflects a difference in the relative importance of particular Bam substrates in E. coli compared to Neisseria. It has not escaped our notice that the LPS assembly factor LptD, a Bam substrate and essential protein in E. coli [88], is dispensable for viability in Neisseria [167]. Although there are many possible explanations for the species-specific difference in the relative importance of the POTRAs, it is tempting to suggest that P1-P4 are, to different degrees, required for the assembly of LptD; this hypothesis correlates the dispensability of P1-P4 in Neisseria with the dispensability of LptD in that organism. In any case, the precise contribution of each individual POTRA domain to the assembly process remains to be determined.
Although it is unclear how the periplasmic domain of BamA interacts directly with substrates, quite a bit is known about how the POTRAs mediate Bam complex formation. The periplasmic domain of BamA is required for the physical associations between BamA and the remaining complex members. The stable association of BamA with BamB requires most of the BamA periplasmic domain; Removal of any POTRA except P1 disrupts the BamA-B connection [161]. In addition, point mutations or small insertions in P3 weaken or altogether disrupt the BamA-B interaction, perhaps indicating a particular requirement for P3 in nucleating or maintaining this association [161].
In contrast, only P5 is necessary for the physical association of BamA with BamC, D, and E, which form a lipoprotein subcomplex that interacts with BamA following BamCDE heterotrimerization [95, 161]. From biochemical data presently available it is difficult to resolve the nature of the physical interactions between BamA and the individual members of the BamCDE subcomplex. It is tempting to conclude that the interactions between BamC/E and BamA occur indirectly through BamD, as C-terminal truncations of BamD prevent the association of BamA with both BamC and BamE [11, 162]. This model is probably overly simplistic, however, as C-terminal BamD truncations (as well as bamE null mutations) also compromise the BamA-D interaction [162]. It is possible that BamC and/or BamE do associate directly with BamD, and in so doing stabilize a direct BamA-D interaction. While it is clear that our understanding of the Bam physical interaction network would benefit greatly from additional biochemical inquiry, it can at least be said (with some certainty) that the primary physical determinants for the BamA-CDE interaction(s) are contained within P5, the barrel-proximal POTRA domain. bamD, like bamA, encodes an essential member of the Bam complex. Because BamA and BamD, as the only essential members of an essential machine, likely interact in order to facilitate OMP assembly [11], it is not surprising that P5 is specifically required for viability in distantly related Gram-negatives. The exception that proves the rule is the mitochondrial BamA homolog Sam50, which contains just one POTRA domain in addition to its C-terminal β-barrel domain [165]. Unexpectedly, the lone POTRA domain of Sam50 is dispensable for viability, and β-barrel assembly appears to proceed unperturbed in its absence [146]. This may be a consequence of the fact that the essential accessory protein in this system (the peripheral membrane protein Sam35) associates with Sam50 not via the POTRA domain but rather through the membrane-embedded β-barrel [146, 168]. It will be interesting to determine whether the minimal requirement for P5 in bacteria reflects its importance as the complex assembly platform.
In addition to their discrete POTRA requirements, it is important to note that BamB and the BamCDE subcomplex interact with BamA independently; that is, mutations in or deletion of BamB does not affect the physical association between BamA and BamCDE, and vice versa [11, 95, 161, 162]. This may suggest that BamB and BamCDE fulfill discrete functions and/or participate in separate steps of OMP assembly, although what these functions might be remains unclear.
The precise roles of the accessory lipoproteins have not been conclusively determined, but genetic analysis of the complex has yielded some insight into the relative importance of these factors nonetheless. As mentioned above, BamD is an essential complex member (and the only essential lipoprotein); its depletion, much like the depletion of BamA [10], causes OMP assembly to stall altogether [11]. The nearly identical effects of depleting bamA and bamD implies that they either work together to catalyze an essential step in β-barrel assembly, or that they are each required to effect distinct but equally important sequential steps.
The remaining three lipoproteins are individually dispensable [10, 11, 162]. Deletion of either bamC or bamE has minimal effects on β-barrel assembly at best; these single mutants grow normally and exhibit subtle reductions in steady state OMP levels (if any). Null mutations in bamB cause more appreciable OM biogenesis defects, including decreased OMP levels, increased OM permeability, and activation of extracytoplasmic stress responses [10, 87, 162, 169, 170]. However, it is unlikely that BamB is important for the assembly of all Bam substrates, as deletion of this lipoprotein enhances the assembly of several OMPs, including the efflux pump TolC [169] and LptD4213, a folding-defective variant of the OMP that mediates surface assembly of LPS [87]. This may signify a more specialized role for BamB in assembly of some as-yet-undetermined subset of OM β-barrels.
Although single deletions of the non-essential Bam lipoproteins are tolerated, deleterious synthetic effects are observed when these deletions are combined with mutations in various OMP assembly factors. For example, null mutations in bamC or bamE (discussed above) have only mild effects on OM biogenesis, but simultaneous deletion of both factors causes significant induction of the σE stress response and a sharp reduction in OMP levels [162]. Even more striking is the effect of combining a bamE deletion with a null mutation in bamB - this double mutant is not viable [162]. These genetic interactions may imply partial functional redundancy between various Bam components, or they may reflect the inability of Bam to support growth when the efficiency of OMP assembly is reduced below some critical threshold. In any case, these synthetic effects demonstrate critically important roles for the non-essential lipoproteins, despite their individual dispensability.
While periplasmic chaperones should not be regarded as bona fide Bam complex members, the physical interaction between periplasmic factors and the Bam complex has been demonstrated. Specifically, binding of SurA to BamA can be detected in the presence of a chemical crosslinker, which traps this presumably transient association [171]. The SurA-BamA interaction is thought to facilitate the transfer of substrate from the chaperone to the assembly machine. The physical determinants of this association are not entirely clear, but there is biochemical evidence that support a role for P1 of BamA in SurA docking: point mutations within this POTRA domain diminish binding to the chaperone [171].
Interestingly, the SurA-BamA interaction does not require BamB [170]. This is somewhat unexpected given the apparent functional relationship between SurA and BamB: these assembly factors identically affect the kinetics of LamB assembly when they are deleted [69], and affect the folding of an overlapping subset of OMPs [86]. Furthermore, BamB was shown to be important for the assembly of Bam substrates delivered by SurA [95]. Since loss of BamB does not affect the ability of SurA to interact with Bam, it is unlikely that SurA binds BamB directly (or cooperatively to BamA in the presence of BamB). BamB may instead exert some undetermined effect on BamA to promote assembly of SurA substrates [172].
Although attempts have been made to crosslink Skp and DegP to Bam, no such interactions have been observed [72]. It remains to be seen whether this reflects the transient nature of this interaction; it is formally possible that, in contrast with SurA, these chaperones do not directly interact with the Bam machine, but instead deliver substrate by some transfer mechanism that does not require docking to Bam.
5. A first glimpse at the physical Bam complex: Structural insights
5.1 BamA
As an essential and nearly ubiquitous protein, BamA (along with its homologs) has received a tremendous amount of attention since its relatively recent discovery as a membrane biogenesis factor. While extensive genetic, biochemical, electrophysiological, structural, and phylogenetic analyses have been conducted on this primeval protein, a detailed understanding of its function in β-barrel folding remains elusive. Still, recent investigations into the structure and function of BamA have yielded some insight into its activity.
The first glimpse of BamA came in the form of two atomic resolution structures of most of the periplasmic domain [161, 173]. Bioinformatic analysis conducted prior to the solutions of the structure strongly suggested that the periplasmic domain of BamA is subdivided into five POTRA domains [164, 165], but the low sequence homology between the POTRAs left open the possibility that these domains would be structurally distinct. In light of this, the first structures revealed something quite unexpected: the POTRAs present in the structures (P1-P4), despite lack of sequence homology, are so structurally homologous that the individual POTRA domains can be neatly superimposed with very little deviation (<1.80 Å r.m.s.d.) from one POTRA to the next [173].
Despite the striking similarity between POTRA domains, there is one domain that displays some unique features. P3 differs from the others in two important respects: 1) the loop between the two α-helices is significantly longer (10 additional residues) in P3 than in the remaining POTRAs, and 2) the second β-strand contains a “β-bulge” which exposes a surface for binding of additional β-strands by β-augmentation [161, 173]. The P1-P4 fragment crystallized as a dimer, and the dimer interface is created by the P3 β-bulge binding to a short segment of P5 present in the construct which templates β-strand formation and extends the P3 β-sheet. Interestingly, the observed β-augmentation occurred in a parallel fashion in one structure [161] and an antiparallel fashion in the other [173], suggesting that POTRAs may tolerate opposite binding orientations so as to accommodate a wide variety of OMP substrates. Whether or not this reflects the ability of BamA to associate with substrate in this way in vivo is far from certain, but these interactions provide an attractive mechanism for the binding of OMP β-strands (or β-hairpins) to POTRAs.
Comparing the two P1-P4 crystal structures revealed one of the most intriguing features of the periplasmic domain of BamA. While the relative orientations of the P1-P2 and P3-P4 domains are virtually identical in both structures, suggesting rigidity at these interfaces, a significant difference at the interdomain angle between P2 and P3 was readily apparent. Whereas the Sousa group observed a 130° angle between these POTRAs, placing the periplasmic domain in an extended conformation, the structure solved by Kahne and colleagues showed this angle to be smaller by 30°, resulting in a “bent” conformation relative to the Sousa structure (Fig. 4). There is extensive hydrogen bonding between P1-P2 and P3-P4, yet there are exceedingly few observed polar contacts between P2 and P3 [173]. This may imply a significant degree of conformational flexibility at the “hinge” between these two POTRA domains; indeed, this notion is supported by NMR, PELDOR, and SAXS data that confirm inflexibility at the P1-P2 joint [174, 175] as well as the relative conformational freedom observed between P2 and P3 [176]. EOM analysis of the SAXS data further suggests that the two observed conformational states are not simply randomly sampled conformations that were trapped in the crystal lattice, but rather two preferred conformations assumed by BamA in solution [176]. The additional solution structures of the P4-P5 tandem pair, together with analysis of the behavior of that pair in solution, suggests that the P4-P5 joint is also rigid [176, 177]. Taken together, the periplasmic domain of BamA can potentially be thought of as two rigid “arms” (comprising P1-P2 and P3-P5) that are connected by a flexible linker.
Figure 4.
The periplasmic domain of BamA exhibits conformation flexibility about the hinge between POTRA domains 2 and 3. Two independently structures are superimposed at POTRA 3. The yellow structure (PDB ID: 2QDF) represents the “bent” conformation, and the purple conformation (PDB ID: 3EFC, 3OG5) represents the “extended” conformation.
The functional relevance of this conformational flexibility, if any, is unclear. However, several related pieces of evidence may provide a clue. Aside from the difference in the P2-P3 angle observed between the Sousa and Kahne structures, there is an additional divergence that is apparent: the L2 loop between the two α-helices in P3 is disordered in the Kahne structure (in which BamA assumes the “bent” conformation), but well-ordered and resolved in the Sousa structure. It is appealing to suggest that this may indicate a conformational switch between the bent and extended states that is regulated by a disorder-to-order transition at L2 of P3. While there is no available data to support such a notion, we note with interest the isolation of a partial loss-of-function mutation in BamA that maps precisely to the L2 loop mentioned above. This mutant, bamA6, exhibits a mild decrease in the steady state levels of OMPs, but enhances the folding of an OMP with a complex biogenesis pathway [178]. Whether or not the bamA6 mutation biases the conformation of BamA towards one of the two observed states remains to be seen.
Although the structure of the complete periplasmic domain of BamA has been solved, there is no structural data available for the C-terminal β-barrel domain. However, the structure of the two-partner secretion transporter FhaC, a distantly related Omp85 homolog, was solved concurrently with BamA [179]. This structure reveals a 16-stranded β-barrel that contains an unusually long extracellular loop (L6). This loop, a common motif among Omp85 family members, is conserved in BamA, and is predicted to be even longer than in FhaC; assuming the loop extends into the lumen of the barrel, as the FhaC structure predicts, it is more than long enough to reach the periplasmic space [180]. This loop contains a highly conserved tetrad motif found in all Omp85 homologs, including BamA. Mutation or deletion of this motif in FhaC renders the transporter unable to secrete its passenger, the FHA adhesin [179, 181]. No reports regarding the functional importance of the tetrad motif (or L6 in general) for BamA are available. It will indeed be interesting to see whether or not this conserved extracellular loop plays a role in OM biogenesis.
5.2 BamB
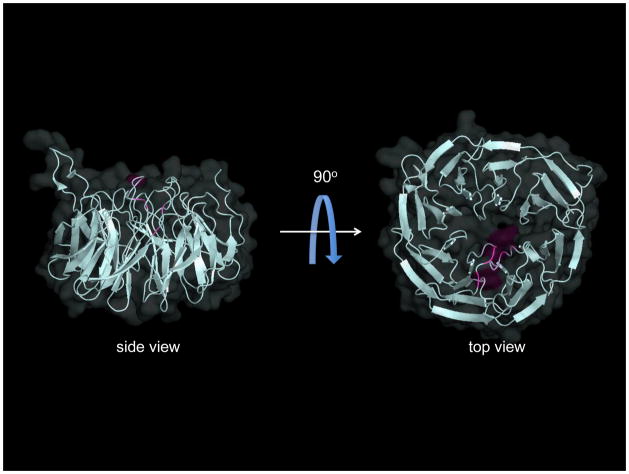
2011 proved to be a red-letter year for structural biology of the Bam complex. Four independent crystal structures of BamB were reported [182, 183] in as many months! Unlike the previous structural characterization of BamA, the independently-solved structures of BamB did not reveal significant conformational or architectural differences between the structures - structural comparisons between them show that they deviate very little from one another (r.m.s.d. = 0.5–0.7 Å; [182]). Even so, the individual groups contributing uniquely to our understanding of BamB and its contribution(s) to Bam-mediated OMP assembly.
In silico modeling of the BamB structure predicted that this lipoprotein assumes a β-propeller fold in the periplasm [160, 170]; this was confirmed in each of the recently solved crystal structures. The BamB propeller has eight blades, each comprising four antiparallel β-strands. The blades are joined by long interconnecting loops (ILs) that form a surface at the narrower rim (“top”) of the propeller. A highly electronegative deep groove at the “bottom” of the propeller has been proposed to participate in protein-protein interactions [184].
In light of the BamB structure, previous identification of BamB residues critical for BamA binding (and vice versa) suggests a putative BamA-B interaction surface. Mutational analysis of BamB revealed five residues required for the physical association with BamA [170] that cluster to two adjacent β-blade interconnecting loops (IL4 and IL5) on the top of propeller (see Fig. 5). Mutation of these residues results in a bamB null phenotype and prevents the stable association of BamB and BamA (without compromising the stability of BamB itself; [170]). The BamB crystal shows that these residues are closely apposed and implies that they specify a BamA binding site [182, 183]. Consistent with this, a deletion in a propeller blade connected to IL5 abrogates BamB function [87]. Truncation of this blade may cause IL5 to be buried within the central funnel, thus preventing this loop from accessing the solvent-exposed surface and interacting with BamA.
Figure 5.
The solution structure of BamB (PDB ID: 3P1L). Residues shown previously to be involved in the BamA-BamB interaction are highlighted in magenta (Vuong et al 2008).
The reordering of residues in the unique β-bulge present in BamA-P3 was previously shown to disrupt the interaction between BamA and BamB, implying the presence of a binding site for BamB at the edge of the P3 β-sheet [161]. Since an interaction between the β-bulge and a short fragment of P5 (comprising a single β-strand) was observed in both P1-P4 crystal structures, it was proposed that this mode of binding (β-augmentation) may be relevant for interactions between BamA and its substrates and/or interaction partners [161, 173]. Intriguingly, a simulation of the BamA-B interaction using a protein docking algorithm suggests an attractive model for this association in which the β-bulge in P3 binds to IL4 of BamB by β-augmentation [172]. This model is satisfying because both BamB IL4 and the BamA β-bulge are known to be important for the BamA-B interaction, and because the β-bulge is known to bind β-strands by β-augmentation [161, 170, 173]. Additional evidence supporting this model includes the observed surface electronegativity of BamB in the region surrounding IL4-5, which is relevant in light of the high positive nature of P3 specifically. Electrostatic interactions at these domains may further stabilize the interaction between BamB and BamA P3 [172].
The hydrophobic pockets formed between neighboring BamB propeller blades have also been suggested to serve as OMP binding sites [183]. However, this proposal is based on intermolecular contacts observed between BamB monomers in the crystal lattice, and while BamB does increase the efficiency of OMP assembly [95], there is no evidence that it directly interacts with OMPs. The involvement of the P3 β-bulge and IL4/5 in the BamA-B interaction was confirmed genetically and biochemically [161, 170]; thorough dissection of the proposed role of BamB in OMP binding must also be performed before a direct OMP-BamB interaction can be confirmed.
5.3 BamC
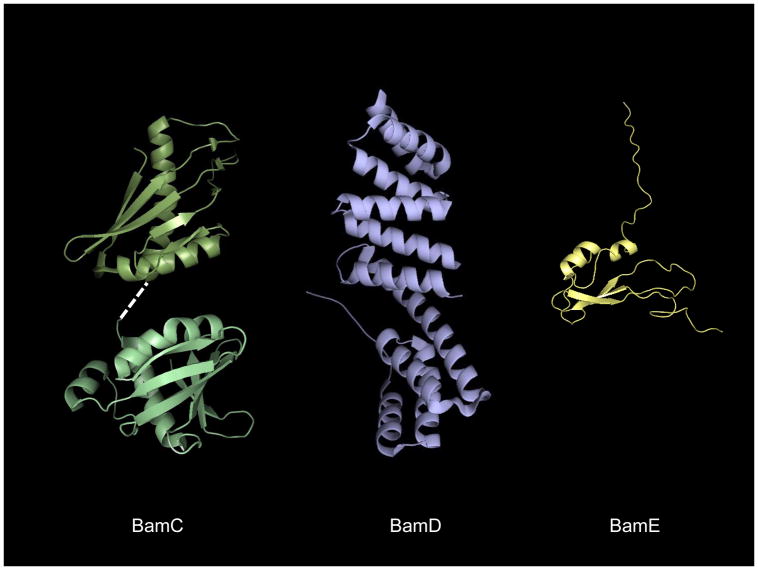
Two separate groups solved the structure of BamC independently [182, 185]. Both report a disordered N-terminus (comprising the first ~70–100 residues) that could not be resolved in either case. What remains are two structurally homologous helix grip domains (Fig. 6) connected by a predicted α-helical linker that was also not resolved in the crystal structure.
Figure 6.
The solution structures of E. coli BamCDE. The BamD (PDB ID: 2YHC) and BamE (PDB ID: 2KM7) structures are oriented with the N-termini pointing toward the top of the page. The structurally homologous helix grip domains of BamC (PDB ID: 2LAE, 2LAF) are shown on the left, with the extreme C-terminal domain at bottom. A dashed white line indicates the unresolved helix linking the two domains.
Unfortunately, the structures provide little insight into the function of BamC or the physical interaction(s) between BamC and other members of the complex. Firstly, unlike BamA and BamB, no BamC mutants have ever been reported (with the exception of a complete bamC deletion), making it difficult to identify residues or domains involved in function and/or protein-protein interactions. Furthermore, the intrinsic disorder of the extreme N-terminus leaves a significant gap in our structural understanding of this minor OMP assembly factor. Genetic and biochemical determination of functionally important BamC domains must be obtained before the relevance of its 3D structure can be appreciated.
5.4 BamD
The crystal structure of BamD was also reported independently by two groups. One group reported the structure of E. coli BamD [182], whereas the other recently solved structure is that of BamD from the distantly related Rhodothermus marinus [186]. Both show that BamD is composed of multiple tetratricopeptide (TPR) repeats packed together into a long superhelical structure (Fig. 6). Many TPR-repeat-containing proteins are known to scaffold multiprotein complexes [186], and structural homologs of BamD that serve as chaperone receptors have been shown to bind substrate via recognition of C-terminal targeting signals in a conserved binding pocket [187]. Since residues at the C-terminus of OMPs are known to be critically important for assembly [143], it has been suggested that one function of BamD may be to selectively bind OM β-barrels in the periplasm via recognition of their C-termini [182, 186]. This is consistent with the finding that Sam35, an essential component of the OM complex that assembles β-barrels in mitochondria, directly binds nascent β-barrels in a manner that is entirely dependent on the presence of a C-terminal recognition signal [146]. It is tempting to speculate that BamD serves a similar function in the bacterial OM, selectively recognizing and binding OMPs prior to membrane integration.
A clear difference between the two available BamD structures is the shortened C-terminus of E. coli BamD relative to that of Rhodothermus. Generally speaking, successive TPR motifs pack against one another to stabilize each repeat, and the interaction surface of the terminal TPR is capped off by an α-helix [186]. Whereas this capping helix is present in Rhodothermus BamD (where it terminates the TPR tandem array after the fifth TPR repeat), this helix is missing altogether in E. coli BamD [186]. Since C-terminal truncations of BamD in E. coli destabilize the interaction(s) of BamD with all other Bam complex members, it has been suggested that this domain is required for the association of BamD with the complex [11]. The variable conservation of the C-terminal capping helix may reflect variations in Bam complex constitution or binding interactions between complex members.
5.5 BamE
As with BamC, no partial loss-of-function mutations in BamE have been reported. Still, the recently solved structures of BamE [182, 188] provide insight into function by highlighting potential protein-protein and protein-membrane interaction surfaces (Fig. 6). A thorough analysis of functionally important BamE residues was conducted by glycine, cysteine, and alanine-scanning mutagenesis [189], which revealed a cluster of surface-exposed residues that, when mutated, compromise the OM permeability barrier in a manner resembling a ΔbamE mutant. A subset of these residues was further shown to influence binding to BamD (supposedly the only Bam component that interacts with BamE in vitro) and/or PG, an anionic minor OM phospholipid. The demonstrated interaction between BamE and PG in vitro may imply a fascinating and unexpected role for this lipoprotein: association of the Bam complex with PG-rich regions of the OM [189]. In light of the fact that PG self-associates in the membrane [190] and that anionic phospholipids can promote protein translocation and drive interactions between membrane proteins and the bilayer [191], it is tempting to suggest that localizing OMP assembly to regions of the OM replete with negatively-charged PG may enhance the rate of β-barrel folding or membrane integration in vivo. It will be interesting to see whether the binding preference for PG exhibited by BamE is relevant in the periplasm.
Several groups reported the in vitro dimerization [188] or hexamerization [182] of BamE, and expression of BamE in the cytoplasm of E. coli also resulted in dimer formation [189]. While stoichiometric analysis of the purified Bam complex may be consistent with dimerization of BamE [95], analysis of the oligomeric state of BamE in the periplasm suggested that BamE is exclusively present as a monomer in its native environment [189], calling into question the physiological relevance of the BamE dimer.
6. Probing the function of the Bam complex
6.1 In vivo assembly kinetics
Prior to the discovery of the Bam complex, analysis of the β-barrel assembly mechanism was more or less limited to in vitro model OMP refolding studies (see [152], identification of intramolecular determinants of efficient β-barrel folding (ex.[134, 148]), and elucidation of the folding pathway for individual OMPs by detection of folding intermediates (ex. [145, 192–195]), although visualization of such intermediates was sometimes complicated by the fact that OMP assembly happens too quickly in vivo to reliably trap intermediates [69]. Identification and characterization of the Bam machine has contributed tremendously to our general understanding of β-barrel targeting and folding, and the results of several seminal studies have shed light on the manner in which OMP folding factors operate coordinately to facilitate OMP assembly.
The maturation of the abundant E. coli OMP LamB from cytoplasmic synthesis to OM assembly can be tracked in vivo by pulse-chase analysis [195]. Ureta et al discovered that slowing cell growth by reducing the incubation temperature increased the half-life of LamB assembly intermediates, thereby permitting more accurate detection and quantification of the conformations LamB assumes during the normal assembly pathway. To probe the function of β-barrel folding factors and assess their contributions to OMP assembly, the kinetics of LamB folding were determined in strains deleted for various envelope proteins that had been previously implicated in OMP biogenesis [69].
In a wild-type cell grown at low temperature, LamB (along with the abundant trimeric porins) proceeds through a folding pathway in which the precursor is processed to a mature form in the periplasm, folded into a monomeric species, and finally converted to a stable LPS-associated trimer. In the absence of the periplasmic chaperone SurA or the Bam complex accessory lipoprotein BamB, the conversion of unfolded mature LamB to folded LamB monomer is dramatically delayed, whereas the trimerization of folded LamB occurs at a normal rate [69].
The importance of this kinetic analysis is underscored by the fact that assessing steady-state OMP levels in surA or bamB mutants only shows that OMP assembly is compromised without revealing why. Ureta et al not only identified the LamB folding step that is accelerated by SurA and BamB (unfolded → folded monomer), but also showed that surA and bamB null mutations have identical effects on LamB maturation. This finding suggests that these two factors promote the same step in OMP folding [69]; said differently, both SurA and BamB are needed for efficient folding assembly of a highly abundant OM protein.
6.2 Accommodating diverse substrates
OMPs come in a wide variety of shapes and sizes, and Bam must be sufficiently robust to accommodate the structural variation that functional differentiation may require. OmpA, for example, is among the ‘lightweight’ OMPs, its membrane domain comprising just eight β-strands [152]. This OMP functions primarily not as a diffusion channel but as a reinforcement factor, binding the murein sacculus and anchoring the cell wall to the OM [22]. In stark contrast, the wide-mouthed iron transporter FecA is required for energy-dependent import of an iron chelate through the gated pore formed by its large 22-strand β-barrel domain [22]. Clearly, the Bam complex must tolerate diversity among its substrates and adapt to facilitate the folding of unique and, in some cases, challenging substrates.
The OM LPS transporter LptD is one such substrate. Assembly of this essential OMP is complicated by several factors: 1) LptD contains a periplasmic N-terminal domain, which is involved in formation of an essential transenvelope Lpt protein bridge [37], 2) the OM lipoprotein LptE forms a plug in the LptD barrel [18, 41] and is absolutely required for the biogenesis of LptD [196, 197], 3) disulfide bonds linking the N-terminal periplasmic domain of LptD to the β-barrel must form properly in order for the protein to function [196], and 4) LptD assembly is exquisitely dependent on the chaperone SurA [86]. Clearly, folding and function of LptD depend heavily on multiple interactions with various envelope factors, including BamA.
The tight transmembrane association of LptDE and the requirement for LptE in LptD folding raise the question of whether BamA coordinates the assembly of the LptDE heterodimer. A recent report describes the isolation of bamA mutations that suppress the LptDE biogenesis defects associated with a partial loss-of-function mutation in lptE (lptE6, [197]). The LptE6 variant interferes with LptDE complex assembly; however, once formed, the LptDE6 complex is stable. Suppressor mutations that alter a specific proline residue in BamA P2 restore efficient LptDE6 complex assembly [197]. Although it is not yet clear exactly how BamA influences LptDE complex formation, the identification of lptE suppressors in bamA strongly implies a fascinating new role for Bam: coordinating the temporally and spatially regulated folding and heterooligomerization of an OMP together with its structural lipoprotein partner.
Autotransporters (ATs) provide another example of complex Bam substrates. ATs are OMPs that are thought to co-opt Bam for OM translocation of covalently-linked N-terminal passenger domains [97, 198, 199]. Compelling biochemical evidence from the recent elegant analysis of Bam-mediated AT assembly suggests that both the passenger and barrel domains of an AT interact with periplasmic chaperones and BamA, whereas the barrel domain additionally interacts with BamB and BamD [97, 99, 198]. Characterization of the temporal association between the E. coli AT EspP and Bam complex members shows that the association between Bam lipoproteins and EspP outlast the association between EspP and BamA [198]. It is tempting to speculate that the prolonged interactions between BamB/BamD and EspP reflect the late/terminal function(s) of these lipoproteins; since they are the last folding factors to contact EspP, they may well catalyze a final assembly step [198]. It will be very interesting to see if the Bam lipoproteins function in this fashion during the assembly of OM β-barrels in general.
The Bernstein group also proposed the existence of a checkpoint mechanism that prevents late steps in assembly of the AT barrel domain from occurring until the passenger domain has been completely secreted; this is supported by the finding that stalling of passenger domain translocation causes folding of the partially assembled β-barrel domain to stall as well [198]. This is an intriguing result in and of itself, as it suggests that the rate of Bam-mediated β-barrel assembly can be determined by a coupled event (in this case, OM translocation).
6.3 In vitro characterization of Bam function
For nearly twenty years [200], in vitro analysis of OMP folding into a lipid bilayer has been conducted in order to study the folding kinetics and physical determinants of β-barrel assembly, to determine the effects of variations in environmental conditions or phospholipid content, or to dissect the interaction(s) between an OMP and a chaperone. Analysis of this kind yielded significant insight into the molecular events that occur during β-barrel folding and the conditions that favor it [152].
In vivo, β-barrel assembly requires the Bam complex. Does the in vitro folding pathway of a particular OMP resemble the folding pathway of the same OMP in vivo? This is a critically important question to consider if in vitro studies are to aid in our understanding of OMP folding in living cells. Ideally, to permit a mechanistic understanding of β-barrel folding in living cells, in vitro analysis of OMP assembly would include the purified Bam complex; such reconstitution systems have contributed greatly to the present understanding of other transport processes, including Sec-dependent IM translocation [201], maltose transport [202], and lipoprotein sorting [203]. To that end, the Kahne group at Harvard University recently reported a significant achievement: the in vitro reconstitution of the E. coli Bam machine [95].
The Kahne group found that the Bam holocomplex could be efficiently reconstituted by first purifying native BamAB and BamCDE and then combining these subcomplexes to form the heteropentamer. Proteoliposomes containing the five-member Bam complex were able to increase the rate of in vitro OMP folding by several orders of magnitude [95]. Efficient OMP folding in this system requires the complete Bam complex and the SurA chaperone, but does not require LPS.
The beauty of the Bam reconstitution lies in the ingenious choice of substrate. Accurate kinetic analysis of OMP assembly in vitro requires a sensitive assay that permits detection of newly-folded substrate over time. To achieve this, the Kahne group took advantage of the protease activity of the OM β-barrel OmpT. A fluorogenic peptide added to proteoliposomes along with the unfolded OMP essentially serves as a reporter of OmpT folding: only folded OmpT cleaves the peptide, therefore the rate of fluorescence production is a function of the rate of OmpT folding [95]. This highly sensitive system affords real-time, quantitative analysis of Bam-assisted OMP folding kinetics.
The reconstitution of Bam in vitro is pivotal in part because it clearly demonstrates that the complete Bam complex is necessary for efficient β-barrel assembly. Moreover, it shows that the process occurs spontaneously and does not require an energy source. The rate of the folding reaction in this reconstituted system (on the order of seconds to minutes) is comparable to assembly rates observed for model OMPs in vivo, which suggests that Bam is sufficient for assembly of β-barrels on a physiologically relevant timescale [95].
Now that facilitated OMP assembly has been successfully reconstituted in vitro, it will be interesting to employ this system to ask additional mechanistic questions about the Bam apparatus. What experimental conditions favor increased folding efficiency? Does Bam turn over in vitro (i.e. can a single Bam complex process cycle through multiple rounds of substrate assembly)? Is the reconstituted machine competent to assemble a variety of OMPs? Can the folding of complex substrates (e.g. LptD or ATs) also be reconstituted in vitro, and are additional factors are required? Omission of any one Bam component in vitro greatly reduces folding efficiency of OmpT [95]; is this generally true for OMPs in this system, or are individual Bam components dispensable for certain substrates? Deletion of the C-terminal phenylalanine of PhoE abrogates assembly in vivo without affecting in vitro refolding whatsoever [145]; does a mutation of this kind prevent OMP recognition by Bam in vitro? The reconstitution system makes the problem of β-barrel assembly far more tractable. The combined use of this system together with mutant OMP biogenesis factors that exhibit altered activity in vivo will likely provide mechanistic insight into this fascinating biological problem.
HIGHLIGHTS.
OM biogenesis requires exquisitely regulated and dedicated transport systems.
OMP folding is catalyzed by a conserved multiprotein complex.
Structural characterization of Bam has afforded significant insight into function.
Acknowledgments
We thank Nate Rigel for helpful discussion. This work was supported by National Institute of General Medical Sciences Grant GM34821
Abbreviations
- LPS
lipopolysaccharide
- IM
inner membrane
- OM
outer membrane
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- OMP
OM β-barrel protein
- GFP
green fluorescent protein
- PMF
proton motive force
- ATP
adenosine triphosphate
- Psp
phage shock protein
- NMR
nuclear magnetic resonance
- PELDOR
pulsed-electro double resonance
- SAXS
small-angle X-ray scattering
- rmsd
root-mean-square deviation
- AT
autotransporter
- Bam
β-barrel assembly machine
- POTRA
polypeptide translocation-associated
- TPR
tetratricopeptide repeat
- IL
interconnecting loop
- EOM
ensemble optimization method
- FHA
filamentous hemagglutinin
- Mla
maintenance of lipid assymetry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Lugtenberg EJ, Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim Biophys Acta. 1976;441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 4.Huijbregts RP, de Kroon A, de Kruijff B. Topology and transport of membrane lipids in bacteria. Biochim Biophys Acta. 2000;1469:43–61. doi: 10.1016/s0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 5.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annual review of biochemistry. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullineaux CW, Nenninger A, Ray N, Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol. 2006;188:3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyadai H, Tanaka-Masuda K, Matsuyama S-i, Tokuda H. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem. 2004;279:39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- 10.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Typas A, Banzhaf M, Saparoea BvdBv, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, Rechenberg Mv, Breukink E, Blaauwen Td, Gross CA, Vollmer W. Regulation of Peptidoglycan Synthesis by Outer-Membrane Proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collin S, Guilvout I, Nickerson NN, Pugsley AP. Sorting of an integral outer membrane protein via the lipoprotein-specific Lol pathway and a dedicated lipoprotein pilotin. Mol Microbiol. 2011;80:655–665. doi: 10.1111/j.1365-2958.2011.07596.x. [DOI] [PubMed] [Google Scholar]
- 17.Laubacher ME, Ades SE. The Rcs Phosphorelay Is a Cell Envelope Stress Response Activated by Peptidoglycan Stress and Contributes to Intrinsic Antibiotic Resistance. Journal of Bacteriology. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chng S-S, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukahara J, Mukaiyama K, Okuda S, Narita S, Tokuda H. Dissection of LolB function--lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 2009;276:4496–4504. doi: 10.1111/j.1742-4658.2009.07156.x. [DOI] [PubMed] [Google Scholar]
- 20.Yakushi T, Tajima T, Matsuyama S, Tokuda H. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. Journal of Bacteriology. 1997;179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 22.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Delattre AS, Clantin B, Saint N, Locht C, Villeret V, Jacob-Dubuisson F. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 277:4755–4765. doi: 10.1111/j.1742-4658.2010.07881.x. [DOI] [PubMed] [Google Scholar]
- 24.Kramer RA, Zandwijken D, Egmond MR, Dekker N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease ompT. Eur J Biochem. 2000;267:885–893. doi: 10.1046/j.1432-1327.2000.01073.x. [DOI] [PubMed] [Google Scholar]
- 25.Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annual review of biochemistry. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 28.Vázquez-Laslop N, Lee H, Hu R, Neyfakh AA. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. Journal of Bacteriology. 2001;183:2399–2404. doi: 10.1128/JB.183.8.2399-2404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.du Plessis DJF, Nouwen N, Driessen AJM. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Carlson JH, Silhavy TJ. Signal sequence processing is required for the assembly of LamB trimers in the outer membrane of Escherichia coli. Journal of Bacteriology. 1993;175:3327–3334. doi: 10.1128/jb.175.11.3327-3334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–5249. doi: 10.1038/sj.emboj.7601402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagan CL, Silhavy TJ, Kahne DE. ?-Barrel Membrane Protein Assembly by the Bam Complex. Annual review of biochemistry. 2010 doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 33.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 34.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Narita S-i, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS LETTERS. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandeo P, Villa R, Martorana AM, Samalikova M, Grandori R, Dehò G, Polissi A. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. Journal of Bacteriology. 2011;193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem. 2010;285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suits MDL, Sperandeo P, Dehò G, Polissi A, Jia Z. Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. Journal of molecular biology. 2008;380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 40.Chng S-S, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freinkman E, Chng S-S, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci USA. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Berg B. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem. 2010;11:1339–1343. doi: 10.1002/cbic.201000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doerrler WT. An Escherichia coli Mutant Defective in Lipid Export. Journal of Biological Chemistry. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 44.Kol MA, de Kroon AIPM, Killian JA, de Kruijff B. Transbilayer movement of phospholipids in biogenic membranes. Biochemistry. 2004;43:2673–2681. doi: 10.1021/bi036200f. [DOI] [PubMed] [Google Scholar]
- 45.Kol MA, van Dalen A, de Kroon AIPM, de Kruijff B. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J Biol Chem. 2003;278:24586–24593. doi: 10.1074/jbc.M301875200. [DOI] [PubMed] [Google Scholar]
- 46.Osborn MJ, Rick PD, Lehmann V, Rupprecht E, Singh M. Structure and biogenesis of the cell envelope of gram-negative bacteria. Ann N Y Acad Sci. 1974;235:52–65. doi: 10.1111/j.1749-6632.1974.tb43256.x. [DOI] [PubMed] [Google Scholar]
- 47.Bayer ME. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- 48.Bayer ME. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J Struct Biol. 1991;107:268–280. doi: 10.1016/1047-8477(91)90052-x. [DOI] [PubMed] [Google Scholar]
- 49.Kellenberger E. The ‘Bayer bridges’ confronted with results from improved electron microscopy methods. Mol Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 50.Donohue-Rolfe AM, Schaechter M. Translocation of phospholipids from the inner to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:1867–1871. doi: 10.1073/pnas.77.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prinz WA. Lipid trafficking sans vesicles: where, why, how? Cell. 2010;143:870–874. doi: 10.1016/j.cell.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem. 1977;252:7405–7412. [PubMed] [Google Scholar]
- 53.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proceedings of the National Academy of Sciences. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilly AA, Crane JM, Randall LL. Export chaperone SecB uses one surface of interaction for diverse unfolded polypeptide ligands. Protein Sci. 2009;18:1860–1868. doi: 10.1002/pro.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowers CW, Lau F, Silhavy TJ. Secretion of LamB-LacZ by the signal recognition particle pathway of Escherichia coli. Journal of Bacteriology. 2003;185:5697–5705. doi: 10.1128/JB.185.19.5697-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee HC, Bernstein HD. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci USA. 2001;98:3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 58.Okuda S, Tokuda H. Lipoprotein Sorting in Bacteria. Annu Rev Microbiol. 2011 doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 59.Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hara T. Mechanism Underlying the Inner Membrane Retention of Escherichia coli Lipoproteins Caused by Lol Avoidance Signals. Journal of Biological Chemistry. 2003;278:40408–40414. doi: 10.1074/jbc.M307836200. [DOI] [PubMed] [Google Scholar]
- 61.Seydel A, Gounon P, Pugsley AP. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol. 1999;34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 62.Gennity JM, Kim H, Inouye M. Structural determinants in addition to the amino-terminal sorting sequence influence membrane localization of Escherichia coli lipoproteins. Journal of Bacteriology. 1992;174:2095–2101. doi: 10.1128/jb.174.7.2095-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley AP. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardie KR, Lory S, Pugsley AP. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011 doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antonoaea R, Fürst M, Nishiyama K-I, Müller M. The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry. 2008;47:5649–5656. doi: 10.1021/bi800233w. [DOI] [PubMed] [Google Scholar]
- 67.Schäfer U, Beck K, Müller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 68.Harms N, Koningstein G, Dontje W, Muller M, Oudega B, Luirink J, de Cock H. The early interaction of the outer membrane protein PhoE with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem. 2001;276:18804–18811. doi: 10.1074/jbc.M011194200. [DOI] [PubMed] [Google Scholar]
- 69.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MPH, Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiology Reviews. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 71.Tormo A, Almiron M, Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. Journal of Bacteriology. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rouviere PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes & Development. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 75.Missiakas D, Betton JM, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 76.Behrens S, Maier R, de Cock H, Schmid FX, Gross CA. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 2001;20:285–294. doi: 10.1093/emboj/20.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bitto E, McKay DB. Binding of phage-display-selected peptides to the periplasmic chaperone protein SurA mimics binding of unfolded outer membrane proteins. FEBS LETTERS. 2004;568:94–98. doi: 10.1016/j.febslet.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 78.Bitto E, McKay DB. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- 79.Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem. 2005;280:23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- 80.Stymest KH, Klappa P. The periplasmic peptidyl prolyl cis-trans isomerases PpiD and SurA have partially overlapping substrate specificities. FEBS J. 2008;275:3470–3479. doi: 10.1111/j.1742-4658.2008.06493.x. [DOI] [PubMed] [Google Scholar]
- 81.Webb HM, Ruddock LW, Marchant RJ, Jonas K, Klappa P. Interaction of the periplasmic peptidylprolyl cis-trans isomerase SurA with model peptides. The N-terminal region of SurA id essential and sufficient for peptide binding. J Biol Chem. 2001;276:45622–45627. doi: 10.1074/jbc.M107508200. [DOI] [PubMed] [Google Scholar]
- 82.Xu X, Wang S, Hu Y-X, McKay DB. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. Journal of molecular biology. 2007;373:367–381. doi: 10.1016/j.jmb.2007.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bitto E, McKay DB. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure. 2002;10:1489–1498. doi: 10.1016/s0969-2126(02)00877-8. [DOI] [PubMed] [Google Scholar]
- 84.Watts KM, Hunstad DA. Components of SurA required for outer membrane biogenesis in uropathogenic Escherichia coli. PLoS ONE. 2008;3:e3359. doi: 10.1371/journal.pone.0003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giuseppe PO, Atzingen MV, Nascimento ALTO, Zanchin NIT, Guimarães BG. The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA. Journal of Structural Biology. 2011;173:312–322. doi: 10.1016/j.jsb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet J-F. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–2443. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 88.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 89.Ames GF, Spudich EN, Nikaido H. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J Bacteriol. 1974;117:406–416. doi: 10.1128/jb.117.2.406-416.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koplow J, Goldfine H. Alterations in the outer membrane of the cell envelope of heptose-deficient mutants of Escherichia coli. J Bacteriol. 1974;117:527–543. doi: 10.1128/jb.117.2.527-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laird MW, Kloser AW, Misra R. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. Journal of Bacteriology. 1994;176:2259–2264. doi: 10.1128/jb.176.8.2259-2264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bolla JM, Lazdunski C, Pagès JM. The assembly of the major outer membrane protein OmpF of Escherichia coli depends on lipid synthesis. EMBO J. 1988;7:3595–3599. doi: 10.1002/j.1460-2075.1988.tb03237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abe S, Okutsu T, Nakajima H, Kakuda N, Ohtsu I, Aono R. n-Hexane sensitivity of Escherichia coli due to low expression of imp/ostA encoding an 87 kDa minor protein associated with the outer membrane, Microbiology (Reading. Engl) 2003;149:1265–1273. doi: 10.1099/mic.0.25927-0. [DOI] [PubMed] [Google Scholar]
- 94.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hagan CL, Kim S, Kahne D. Reconstitution of Outer Membrane Protein Assembly from Purified Components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Justice SS, Hunstad DA, Harper JR, Duguay AR, Pinkner JS, Bann J, Frieden C, Silhavy TJ, Hultgren SJ. Periplasmic peptidyl prolyl cis-trans isomerases are not essential for viability, but SurA is required for pilus biogenesis in Escherichia coli. Journal of Bacteriology. 2005;187:7680–7686. doi: 10.1128/JB.187.22.7680-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. Journal of Bacteriology. 2009;191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sauri A, Soprova Z, Wickström D, de Gier J-W, Van der Schors RC, Smit AB, Jong WSP, Luirink J. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease, Microbiology (Reading. Engl) 2009;155:3982–3991. doi: 10.1099/mic.0.034991-0. [DOI] [PubMed] [Google Scholar]
- 100.Purdy GE, Fisher CR, Payne SM. IcsA Surface Presentation in Shigella flexneri Requires the Periplasmic Chaperones DegP, Skp, and SurA. Journal of Bacteriology. 2007;189:5566–5573. doi: 10.1128/JB.00483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bodelon G, Marin E, Fernandez LA. Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of intimin from enteropathogenic Escherichia coli strains. J Bacteriol. 2009;191:5169–5179. doi: 10.1128/JB.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clantin B, Leyrat C, Wohlkönig A, Hodak H, Ribeiro EdA, Martinez N, Baud C, Smet-Nocca C, Villeret V, Jacob-Dubuisson F, Jamin M. Structure and plasticity of the peptidyl-prolyl isomerase Par27 of Bordetella pertussis revealed by X-ray diffraction and small-angle X-ray scattering. Journal of Structural Biology. 2010;169:253–265. doi: 10.1016/j.jsb.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 103.Aasland R, Coleman J, Holck AL, Smith CL, Raetz CR, Kleppe K. Identity of the 17-kilodalton protein, a DNA-binding protein from Escherichia coli, and the firA gene product. Journal of Bacteriology. 1988;170:5916–5918. doi: 10.1128/jb.170.12.5916-5918.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thome BM, Hoffschulte HK, Schiltz E, Muller M. A protein with sequence identity to Skp (FirA) supports protein translocation into plasma membrane vesicles of Escherichia coli. FEBS Lett. 1990;269:113–116. doi: 10.1016/0014-5793(90)81132-8. [DOI] [PubMed] [Google Scholar]
- 105.Thome BM, Muller M. Skp is a periplasmic Escherichia coli protein requiring SecA and SecY for export. Mol Microbiol. 1991;5:2815–2821. doi: 10.1111/j.1365-2958.1991.tb01990.x. [DOI] [PubMed] [Google Scholar]
- 106.Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 107.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. Journal of Bacteriology. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Volokhina EB, Grijpstra J, Stork M, Schilders I, Tommassen J, Bos MP. Role of the periplasmic chaperones Skp, SurA, and DegQ in outer membrane protein biogenesis in Neisseria meningitidis. Journal of Bacteriology. 2011;193:1612–1621. doi: 10.1128/JB.00532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 110.Korndörfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11:1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- 111.Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 112.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J Mol Biol. 2007;374:91–105. doi: 10.1016/j.jmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 113.Qu J, Behrens-Kneip S, Holst O, Kleinschmidt JH. Binding regions of outer membrane protein A in complexes with the periplasmic chaperone Skp. A site-directed fluorescence study. Biochemistry. 2009;48:4926–4936. doi: 10.1021/bi9004039. [DOI] [PubMed] [Google Scholar]
- 114.Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci USA. 2009;106:1772–1777. doi: 10.1073/pnas.0809275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jarchow S, Lück C, Görg A, Skerra A. Identification of potential substrate proteins for the periplasmic Escherichia coli chaperone Skp. Proteomics. 2008;8:4987–4994. doi: 10.1002/pmic.200800288. [DOI] [PubMed] [Google Scholar]
- 116.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 117.Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J Bacteriol. 2009;191:815–821. doi: 10.1128/JB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rowley G, Skovierova H, Stevenson A, Rezuchova B, Homerova D, Lewis C, Sherry A, Kormanec J, Roberts M. The periplasmic chaperone Skp is required for successful Salmonella Typhimurium infection in a murine typhoid model, Microbiology (Reading. Engl) 2011;157:848–858. doi: 10.1099/mic.0.046011-0. [DOI] [PubMed] [Google Scholar]
- 119.Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matern Y, Barion B, Behrens-Kneip S. PpiD is a player in the network of periplasmic chaperones in Escherichia coli. BMC Microbiol. 2010;10:251. doi: 10.1186/1471-2180-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weininger U, Jakob RP, Kovermann M, Balbach J, Schmid FX. The prolyl isomerase domain of PpiD from Escherichia coli shows a parvulin fold but is devoid of catalytic activity. Protein Sci. 2010;19:6–18. doi: 10.1002/pro.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Horne SM, Kottom TJ, Nolan LK, Young KD. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect Immun. 1997;65:806–810. doi: 10.1128/iai.65.2.806-810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saul FA, Arié J-P, Vulliez-le Normand B, Kahn R, Betton J-M, Bentley GA. Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity. Journal of molecular biology. 2004;335:595–608. doi: 10.1016/j.jmb.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 124.Bothmann H, Pluckthun A. The periplasmic Escherichia coli peptidylprolyl cis, trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J Biol Chem. 2000;275:17100–17105. doi: 10.1074/jbc.M910233199. [DOI] [PubMed] [Google Scholar]
- 125.Ramm K, Plückthun A. The periplasmic Escherichia coli peptidylprolyl cis, trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J Biol Chem. 2000;275:17106–17113. doi: 10.1074/jbc.M910234199. [DOI] [PubMed] [Google Scholar]
- 126.Arie JP, Sassoon N, Betton JM. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol. 2001;39:199–210. doi: 10.1046/j.1365-2958.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- 127.Hullmann J, Patzer SI, Römer C, Hantke K, Braun V. Periplasmic chaperone FkpA is essential for imported colicin M toxicity. Mol Microbiol. 2008;69:926–937. doi: 10.1111/j.1365-2958.2008.06327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Helbig JH, Konig B, Knospe H, Bubert B, Yu C, Luck CP, Riboldi-Tunnicliffe A, Hilgenfeld R, Jacobs E, Hacker J, Fischer G. The PPIase active site of Legionella pneumophila Mip protein is involved in the infection of eukaryotic host cells. Biol Chem. 2003;384:125–137. doi: 10.1515/BC.2003.013. [DOI] [PubMed] [Google Scholar]
- 129.Horne SM, Kottom TJ, Nolan LK, Young KD. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infection and Immunity. 1997;65:806–810. doi: 10.1128/iai.65.2.806-810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moro A, Ruiz-Cabello F, Fernández-Cano A, Stock RP, González A. Secretion by Trypanosoma cruzi of a peptidyl-prolyl cis-trans isomerase involved in cell infection. EMBO J. 1995;14:2483–2490. doi: 10.1002/j.1460-2075.1995.tb07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Humphreys S, Rowley G, Stevenson A, Kenyon WJ, Spector MP, Roberts M. Role of periplasmic peptidylprolyl isomerases in Salmonella enterica serovar Typhimurium virulence. Infect Immun. 2003;71:5386–5388. doi: 10.1128/IAI.71.9.5386-5388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Helbig S, Patzer SI, Schiene-Fischer C, Zeth K, Braun V. Activation of colicin M by the FkpA prolyl cis-trans isomerase/chaperone. J Biol Chem. 2011;286:6280–6290. doi: 10.1074/jbc.M110.165274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ruiz-Perez F, Henderson IR, Nataro JP. Interaction of FkpA, a peptidyl-prolyl cis/trans isomerase with EspP autotransporter protein. Gut microbes. 2010;1:339–344. doi: 10.4161/gmic.1.5.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 135.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 136.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 137.Meltzer M, Hasenbein S, Mamant N, Merdanovic M, Poepsel S, Hauske P, Kaiser M, Huber R, Krojer T, Clausen T, Ehrmann M. Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res Microbiol. 2009;160:660–666. doi: 10.1016/j.resmic.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 138.Subrini O, Betton J-M. Assemblies of DegP underlie its dual chaperone and protease function. FEMS Microbiol Lett. 2009;296:143–148. doi: 10.1111/j.1574-6968.2009.01658.x. [DOI] [PubMed] [Google Scholar]
- 139.Krojer T, Sawa J, Huber R, Clausen T. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat Struct Mol Biol. 2010;17:844–852. doi: 10.1038/nsmb.1840. [DOI] [PubMed] [Google Scholar]
- 140.Krojer T, Sawa J, Schäfer E, Saibil HR, Ehrmann M, Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 141.CastilloKeller M, Misra R. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. Journal of Bacteriology. 2003;185:148–154. doi: 10.1128/JB.185.1.148-154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Misra R, CastilloKeller M, Deng M. Overexpression of protease-deficient DegP(S210A) rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. Journal of Bacteriology. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.de Cock H, Struyvé M, Kleerebezem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. Journal of molecular biology. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 144.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. Journal of molecular biology. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 145.Jansen C, Heutink M, Tommassen J, de Cock H. The assembly pathway of outer membrane protein PhoE of Escherichia coli. Eur J Biochem. 2000;267:3792–3800. doi: 10.1046/j.1432-1327.2000.01417.x. [DOI] [PubMed] [Google Scholar]
- 146.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 147.Zeth K, Thein M. Porins in prokaryotes and eukaryotes: common themes and variations. Biochem J. 2010;431:13–22. doi: 10.1042/BJ20100371. [DOI] [PubMed] [Google Scholar]
- 148.Koebnik R. Membrane assembly of the Escherichia coli outer membrane protein OmpA: exploring sequence constraints on transmembrane beta-strands. Journal of molecular biology. 1999;285:1801–1810. doi: 10.1006/jmbi.1998.2405. [DOI] [PubMed] [Google Scholar]
- 149.Koebnik R. In vivo membrane assembly of split variants of the E.coli outer membrane protein OmpA. EMBO J. 1996;15:3529–3537. [PMC free article] [PubMed] [Google Scholar]
- 150.Adamian L, Naveed H, Liang J. Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim Biophys Acta. 2011;1808:1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Naveed H, Jackups R, Liang J. Predicting weakly stable regions, oligomerization state, and protein-protein interfaces in transmembrane domains of outer membrane proteins. Proc Natl Acad Sci USA. 2009;106:12735–12740. doi: 10.1073/pnas.0902169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kleinschmidt JH. Membrane protein folding on the example of outer membrane protein A of Escherichia coli. Cellular and Molecular Life Sciences (CMLS) 2003;60:1547–1558. doi: 10.1007/s00018-003-3170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. The Journal of Cell Biology. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 155.Arnold T, Zeth K, Linke D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. J Biol Chem. 2010;285:18003–18015. doi: 10.1074/jbc.M110.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bullmann L, Haarmann R, Mirus O, Bredemeier R, Hempel F, Maier UG, Schleiff E. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Haarmann R, Ibrahim M, Stevanovic M, Bredemeier R, Schleiff E. The properties of the outer membrane localized Lipid A transporter LptD. J Phys Condens Matter. 2010;22:454124. doi: 10.1088/0953-8984/22/45/454124. [DOI] [PubMed] [Google Scholar]
- 158.Schleiff E, Maier UG, Becker T. Omp85 in eukaryotic systems: one protein family with distinct functions. Biological Chemistry. 2011;392:21–27. doi: 10.1515/BC.2011.005. [DOI] [PubMed] [Google Scholar]
- 159.Anwari K, Poggio S, Perry A, Gatsos X, Ramarathinam SH, Williamson NA, Noinaj N, Buchanan S, Gabriel K, Purcell AW, Jacobs-Wagner C, Lithgow T. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus. PLoS ONE. 2010;5:e8619. doi: 10.1371/journal.pone.0008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gatsos X, Perry AJ, Anwari K, Dolezal P, Wolynec PP, Likić VA, Purcell AW, Buchanan SK, Lithgow T. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiology Reviews. 2008;32:995–1009. doi: 10.1111/j.1574-6976.2008.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 162.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Volokhina EB, Beckers F, Tommassen J, Bos MP. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. Journal of Bacteriology. 2009;191:7074–7085. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 165.Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 166.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stroud DA, Becker T, Qiu J, Stojanovski D, Pfannschmidt S, Wirth C, Hunte C, Guiard B, Meisinger C, Pfanner N, Wiedemann N. Biogenesis of mitochondrial {beta}-barrel proteins: the POTRA domain is involved in precursor release from the SAM complex. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Noinaj N, Fairman JW, Buchanan SK. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. Journal of molecular biology. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 175.Ward R, Zoltner M, Beer L, El Mkami H, Henderson IR, Palmer T, Norman DG. The orientation of a tandem POTRA domain pair, of the beta-barrel assembly protein BamA, determined by PELDOR spectroscopy. Structure. 2009;17:1187–1194. doi: 10.1016/j.str.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 176.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang H, Gao ZQ, Hou HF, Xu JH, Li LF, Su XD, Dong YH. High-resolution structure of a new crystal form of BamA POTRA4–5 from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 67:734–738. doi: 10.1107/S1744309111014254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol. 2006;1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 179.Clantin B, Delattre A-S, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the Membrane Protein FhaC: A Member of the Omp85-TpsB Transporter Superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 180.Jacob-Dubuisson F, Villeret V, Clantin B, Delattre A-S, Saint N. First structural insights into the TpsB/Omp85 superfamily. Biological Chemistry. 2009;390:675–684. doi: 10.1515/BC.2009.099. [DOI] [PubMed] [Google Scholar]
- 181.Delattre A-S, Clantin B, Saint N, Locht C, Villeret V, Jacob-Dubuisson F. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 2010;277:4755–4765. doi: 10.1111/j.1742-4658.2010.07881.x. [DOI] [PubMed] [Google Scholar]
- 182.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011 doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Heuck A, Schleiffer A, Clausen T. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. Journal of molecular biology. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 184.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. Journal of molecular biology. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 185.Warner LR, Varga K, Lange OF, Baker SL, Baker D, Sousa MC, Pardi A. Structure of the BamC Two-Domain Protein Obtained by Rosetta with a Limited NMR Data Set. Journal of molecular biology. 2011 doi: 10.1016/j.jmb.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal Structure of BamD. An Essential Component of the β-Barrel Assembly Machinery of Gram Negative Bacteria. Journal of molecular biology. 2011 doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kriechbaumer V, von Löffelholz O, Abell BM. Chaperone receptors: guiding proteins to intracellular compartments. Protoplasma. 2011 doi: 10.1007/s00709-011-0270-9. [DOI] [PubMed] [Google Scholar]
- 188.Kim KH, Kang H-S, Okon M, Escobar-Cabrera E, McIntosh LP, Paetzel M. Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry. 2011;50:1081–1090. doi: 10.1021/bi101659u. [DOI] [PubMed] [Google Scholar]
- 189.Knowles TJ, Browning DF, Jeeves M, Maderbocus R, Rajesh S, Sridhar P, Manoli E, Emery D, Sommer U, Spencer A, Leyton DL, Squire D, Chaudhuri RR, Viant MR, Cunningham AF, Henderson IR, Overduin M. Structure and function of BamE within the outer membrane and the β-barrel assembly machine. EMBO reports. 2011;12:123–128. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vanounou S, Parola AH, Fishov I. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. A study with pyrene-labelled phospholipids. Mol Microbiol. 2003;49:1067–1079. doi: 10.1046/j.1365-2958.2003.03614.x. [DOI] [PubMed] [Google Scholar]
- 191.Patel GJ, Behrens-Kneip S, Holst O, Kleinschmidt JH. The periplasmic chaperone Skp facilitates targeting, insertion, and folding of OmpA into lipid membranes with a negative membrane surface potential. Biochemistry. 2009;48:10235–10245. doi: 10.1021/bi901403c. [DOI] [PubMed] [Google Scholar]
- 192.Eppens EF, Nouwen N, Tommassen J. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 1997;16:4295–4301. doi: 10.1093/emboj/16.14.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fourel D, Mizushima S, Pagès JM. Dynamics of the exposure of epitopes on OmpF, an outer membrane protein of Escherichia coli. Eur J Biochem. 1992;206:109–114. doi: 10.1111/j.1432-1033.1992.tb16907.x. [DOI] [PubMed] [Google Scholar]
- 194.Reid J, Fung H, Gehring K, Klebba PE, Nikaido H. Targeting of porin to the outer membrane of Escherichia coli. Rate of trimer assembly and identification of a dimer intermediate. J Biol Chem. 1988;263:7753–7759. [PubMed] [Google Scholar]
- 195.Vos-Scheperkeuter GH, Witholt B. Assembly pathway of newly synthesized LamB protein an outer membrane protein of Escherichia coli K-12. J Mol Biol. 1984;175:511–528. doi: 10.1016/0022-2836(84)90182-7. [DOI] [PubMed] [Google Scholar]
- 196.Ruiz N, Chng S-S, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci USA. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chimalakonda G, Ruiz N, Chng S-S, Garner RA, Kahne D, Silhavy TJ. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2011;108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ieva R, Tian P, Peterson JH, Bernstein HD. PNAS Plus: Sequential and spatially restricted interactions of assembly factors with an autotransporter {beta} domain. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rossiter AE, Leyton DL, Tveen-Jensen K, Browning DF, Sevastsyanovich Y, Knowles TJ, Nichols KB, Cunningham AF, Overduin M, Schembri MA, Henderson IR. The essential BAM complex components BamD and BamA are required for autotransporter biogenesis. Journal of Bacteriology. 2011 doi: 10.1128/JB.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Surrey T, Jähnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc Natl Acad Sci USA. 1992;89:7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Watanabe M, Nicchitta CV, Blobel G. Reconstitution of protein translocation from detergent-solubilized Escherichia coli inverted vesicles: PrlA protein-deficient vesicles efficiently translocate precursor proteins. Proc Natl Acad Sci U S A. 1990;87:1960–1964. doi: 10.1073/pnas.87.5.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Davidson AL, Nikaido H. Overproduction, solubilization, and reconstitution of the maltose transport system from Escherichia coli. J Biol Chem. 1990;265:4254–4260. [PubMed] [Google Scholar]
- 203.Kanamaru K, Taniguchi N, Miyamoto S, Narita S-i, Tokuda H. Complete reconstitution of an ATP-binding cassette transporter LolCDE complex from separately isolated subunits. FEBS J. 2007;274:3034–3043. doi: 10.1111/j.1742-4658.2007.05832.x. [DOI] [PubMed] [Google Scholar]