Abstract
The genomes of two Bacillus cereus strains (ATCC 10987 and ATCC 14579) have been sequenced. Here, we report the specificities of type II/III restriction (R) and modification (M) enzymes. Found in the ATCC 10987 strain, BceSI is a restriction endonuclease (REase) with the recognition and cut site CGAAG 24-25/27-28. BceSII is an isoschizomer of AvaII (G/GWCC). BceSIII cleaves at ACGGC 12/14. The BceSIII C terminus resembles the catalytic domains of AlwI, MlyI, and Nt.BstNBI. BceSIV is composed of two subunits and cleaves on both sides of GCWGC. BceSIV activity is strongly stimulated by the addition of cofactor ATP or GTP. The large subunit (R1) of BceSIV contains conserved motifs of NTPases and DNA helicases. The R1 subunit has no endonuclease activity by itself; it strongly stimulates REase activity when in complex with the R2 subunit. BceSIV was demonstrated to hydrolyze GTP and ATP in vitro. BceSIV is similar to CglI (GCSGC), and homologs of R1 are found in 11 sequenced bacterial genomes, where they are paired with specificity subunits. In addition, homologs of the BceSIV R1-R2 fusion are found in many sequenced microbial genomes. An orphan methylase, M.BceSV, was found to modify GCNGC, GGCC, CCGG, GGNNCC, and GCGC sites. A ParB-methylase fusion protein appears to nick DNA nonspecifically. The ATCC 14579 genome encodes an active enzyme Bce14579I (GCWGC). BceSIV and Bce14579I belong to the phospholipase D (PLD) family of endonucleases that are widely distributed among Bacteria and Archaea. A survey of type II and III restriction-modification (R-M) system genes is presented from sequenced B. cereus, Bacillus anthracis, and Bacillus thuringiensis strains.
INTRODUCTION
Bacillus cereus is a Gram-positive, soil-dwelling, aerobic bacterium. Food contaminated with some B. cereus strains can cause nausea, vomiting, and diarrhea in humans. Certain B. cereus strains produce a pore-forming toxin called CytK (CytK-1 and CytK-2), which is hemolytic and toxic toward human intestinal Caco-2 cells and Vero cells (16). The genomic DNA sequences of Bacillus cereus strains ATCC 10987 and ATCC 14579 (nonpathogenic, biosafety level 1 strains) have been published (32). Genomic analysis indicated that B. cereus ATCC 10987 is more similar to Bacillus anthracis Ames than to B. cereus ATCC 14579 (19, 29, 32). In addition, nine more Bacillus cereus genomes of pathogenic and nonpathogenic strains have been sequenced (GenBank genome database). An additional 22 B. cereus strains have been sequenced by the shotgun sequencing method (GenBank). Bioinformatic analysis of the 11 fully sequenced genomes indicated that B. cereus strain ATCC 10987 harbors the most restriction-modification (R-M) systems among the B. cereus strains, with four predicted complete R-M systems, including one partially characterized type III R-M system, one orphan 5-cytosine methyltransferase (C5 methyltransferase [MTase]), and one ParB-methylase fusion (see the REBASE database) (34). Only one type II R-M system was predicted for the ATCC 14579 strain.
The primary biological function of restriction endonucleases (REases) is to attack (restrict) foreign invading DNA during viral infections, conjugative plasmid DNA transfer, and natural DNA transformation from the environment. Phage restriction by REases can be highly efficient and is of the order of 103- to 107-fold restriction by different type of R-M systems (43). REases are classified into four major types based on their subunit architecture, NTP requirement, sequence specificity, and DNA cleavage mechanism (33). Type I R-M systems were originally discovered by studying phage-plating efficiency among different Escherichia coli hosts (3). Type I restriction enzymes are multisubunit complexes consisting of M2R2S subunits that recognize an asymmetric, bipartite sequence and require ATP hydrolysis to direct cleavage at a distant site following DNA translocations (43) (4). Type II REases with 4- to 8-bp recognition sequences and over 300 unique specificities are widely used in creating recombinant DNA molecules. Like type I R-M systems, type III R-M systems are multisubunit complexes with a Res2Mod2 configuration and require ATP hydrolysis and DNA translocation for endonuclease activity on unmodified DNA (39). Modification-dependent REases are presently grouped into type IIM if the target sequence recognition and cleavage are very specific and precise, for example, DpnI (GN6mATC) and MspJI (5mCNNR) (11, 24) (modifications are underlined), or into type IV if the cleavage is nonspecific and variable. Examples of type IV systems are EcoK McrBC (38) and SauUSI (44), which cleaves 5mC-modified DNA, and GmrSD that attacks glucosylated-hmC (glc-5hmC) T4 DNA (5).
The goal of this work was to determine the specificities of the type II and III R-M systems and the orphan MTase encoded by two B. cereus genomes (ATCC 10987 and ATCC 14579) in vitro. Understanding these R-M systems will likely facilitate the genetic study (gene transfer) of Bacillus, since similar systems (isoschizomers) also occur in other sequenced Bacillus strains. The availability of cloned MTase genes also makes it possible to modify DNA in vivo or in vitro for biological DNA transformation into other Bacillus strains with similar restriction systems (the modification of plasmid DNA increases transformation efficiency). In addition, newly discovered specificities can add a new tool to the “tool box” used by molecular biologists in creating recombinant DNA molecules.
5mC, 5hmC, and N6mA modifications of DNA bases have been shown to affect gene expression, gene silencing, and DNA repair (8, 9, 21, 23, 26, 42). Type II and III R-M systems have been implicated in epigenetic gene regulation in prokaryotes and in human disease (7, 14). The study of restriction-modification systems (iceA1 [CATG], HpyIII [GATC], M.HpyIII, and hrgA) has shed light on the pathogenesis of the disease-causing pathogen Helicobacter pylori that causes gastritis, ulcers, and gastric cancers (1, 2, 17). Phase variation in methyltransferase expression associated with type III R-M systems has been identified for a variety of pathogenic bacteria (7, 13). It was previously demonstrated that a phase-variable type III mod gene (CGAAT) in the Haemophilus influenzae Rd strain coordinates the random switching of the expression of multiple genes and constitutes a phase-variable regulon. It is not clear whether 5mC or N6mA base modification of DNA is involved in the regulation of gene expression or in the pathogenesis in Bacillus strains, although GATC methyltransferase (MTase), cytosine MTase, and amino-MTase are found among 10 sequenced B. cereus genomes (see below). It is clear, however, that pathogenic bacterial strains can take up foreign DNA by horizontal gene transfer (HGT) by a variety of mechanisms, especially in multidrug-resistant strains with a survival advantage in hospital settings. HGT is the driving force behind bacterial evolution, adaptation to niche environments, and generation of microbial diversity. Restriction enzymes (type I to IV) provide genetic barriers to such promiscuous DNA exchanges, and in certain cases, anti-restriction proteins encoded by phages and mobile genetic elements provide antagonists by inhibition of type I restriction systems (28).
Here, we report the preliminary characterization of the four restriction enzymes BceSI to -IV, one multispecificity 5-cytosine (C5) MTase (M.BceSV), one nonspecific nicking enzyme from the ATCC 10987 strain, and Bce14579I from the ATCC 14579 strain by gene expression in a heterologous host, protein purification, and enzymatic assays. A survey of putative type II and III R-M genes is also presented from sequenced B. cereus, B. anthracis, and Bacillus thuringiensis strains.
MATERIALS AND METHODS
B. cereus ATCC 10987 and ATCC 14579 genomic DNAs (gDNAs) were obtained from ATCC (Manassas, VA). Plasmid and phage DNAs (pBR322, pUC19, pTYB1, pAII17, phage λ), restriction and modification enzymes, E. coli competent cells, and chitin beads were from New England BioLabs, Inc. (NEB; Ipswich, MA). T7 expression vector pET21a was purchased from Novagen (Madison, WI). Plasmid DNA was prepared by Qiagen mini-spin columns (Qiagen, Valencia, CA). PCR primers were synthesized by IDT (Coralville, IA).
PCR cloning of restriction-modification genes.
Phusion DNA polymerase was used to amplify restriction (R or res) and modification (M or mod) genes in PCR.
(i) Cloning of BceSI R and M genes.
The BceSI mod and res genes (Bce_1018 and Bce_1019) were amplified together by PCR, and both genes were inserted into a pUC19-based vector pRRS (SalI/CIP [calf intestinal alkaline phosphatase]). A 6×His tag was engineered at the N terminus of the Mod subunit. After nickel column purification of the Res/Mod subunits from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced cell extracts, restriction activity was analyzed using phage λ and pBR322 DNAs.
(ii) Cloning of the BceSII restriction gene.
The bceSIIR gene (flanked by HindIII and SphI sites) was amplified by PCR and inserted into pUC19 with compatible ends and transferred into E. coli strain ER2566 carrying the AvaII MTase (pLG-avaIIM) for expression.
(iii) Cloning of the BceSIII restriction gene.
The bceSIIIR gene (flanked by NdeI and BamHI sites) was amplified by PCR and inserted into pET21a with compatible ends and transferred into E. coli strain ER2566 carrying the two BceAI MTases (pACYC-bceAIM1M2).
(iv) Cloning of the BceSIV MTase (bceSIVM) gene.
The bceSIVM gene (flanked by BamHI sites) was amplified by PCR and inserted into pUC19 with the same cohesive ends. Plasmid DNA with the correct insert orientation was prepared and challenged with BbvI (GCAGC) or Fnu4HI (GCNGC).
(v) Cloning of the BceSIV R1 (bceSIVR1) and R2 (bceSIVR2) genes.
Open reading frame (ORF) Bce_368 (predicted DNA helicase, ATPase/GTPase, R1 subunit, large subunit) (flanked by NdeI and BamHI sites) was amplified by PCR and inserted into a T7 expression vector pAII17 and transformed into an E. coli expression host T7 Express lacIq (NEB). Similarly, a PCR fragment carrying ORF Bce_369 (R2 subunit with a phospholipase D [PLD] endonuclease catalytic domain and specificity function, small subunit) (flanked by NdeI and BamHI sites) was inserted into a T7 expression vector pET21a and transformed into E. coli expression host T7 Express lacIq. E. coli T7 Express lacIq (pET21a-bceSIVR2) formed very small colonies, indicating toxicity of the inserted gene; a plasmid carrying a protective M.BbvI, pACYC-bbvIM (GCAGC), was cotransformed into the expression strain to form E. coli T7 Express lacIq (pET21a-bceSIVR2, pACYC-bbvIM), which formed colonies of normal size.
(vi) Cloning of the parB-M gene and the BceSV MTase (bceSVM) genes.
A PCR fragment carrying the parB-M fusion gene with a 6×His tag (∼1,260 bp, flanked by NdeI and BamHI sites) was inserted into pAII17 and transformed into a T7 expression strain ER2566 (also called T7 Express). Another PCR fragment (3.4 kb flanked by BamHI sites), carrying both parB-M and bceSVM genes, was inserted into pAII17 (BamHI/CIP treated) in the correct orientation or opposite orientation relative to the T7 promoter. The overexpressed ParB-M protein was found in the insoluble fraction when IPTG induction was carried out at 37°C. Lower temperature induction, at 25°C to 30°C, increased the protein solubility.
(vii) Cloning of the Bce14579I R1 and R2 genes.
ORF BC_0939 (the bce14579IR1 gene) (flanked by NdeI and XhoI sites) was amplified by PCR from gDNA and inserted into a pTYB1 expression vector in fusion with an intein and chitin-binding domain (CBD). ORF BC_0940 (the bce14579IR2 gene with a TAA stop codon) (flanked by restriction sites NdeI and EcoRI) was amplified by PCR and inserted into pTYB1 as a nonfusion construct. To protect the E. coli DNA, M.Fnu4HI (GCNGC) was coexpressed in the expression host ER2566 (pACYC-fnu4HIM, pTYB1-bce14579IR2).
Preparation of cell extracts and protein purification.
IPTG induction of protein production was carried out for 3 to 5 h by the addition of a final concentration of 0.5 mM IPTG at the late log phase of cultures. For small-scale enzyme production at the initial screening stage, 10 ml of cell culture was sufficient. For medium-scale enzyme production, 1 to 2 liters of IPTG-induced cell culture was used. Cell lysis was carried out by the freeze-thaw method followed by sonication (microtip for a 0.5- to 1-ml volume, medium tip for a 30- to 60-ml volume). A Ni-nitrilotriacetic acid (Ni-NTA) spin kit and fast flow Ni-NTA resin from Qiagen were used for protein purification. The Impact protein expression and purification system (NEB) using CBD and chitin beads was employed to purify BceSIV R1, the R1/R2 complex, Bce14579I R1, or Bce14579I R1/R2 subunits by following the manufacturer's protocol. In vitro synthesis of the ParB-M fusion protein was carried out using a PurExpress protein synthesis kit from NEB.
DNA cleavage assays.
DNA cleavage assays by BceSIV or Bce14579I were carried out using 1× NEB buffer 4 supplemented with ATP or GTP at 37°C for 1 h. One μg of DNA and various amounts of the indicated crude lysate or purified protein was used. Reactions were stopped by the addition of 10× stop buffer. Cleavage products were resolved on 0.8% to 1% agarose gels. To determine cleavage sites, the cleaved DNA was purified by using a spin column and sequenced with appropriate primers. Runoff sequencing was carried out using a BigDye Terminator v3.1 cycle sequencing kit from Applied Biosystems (Life Technologies-ABI, Carlsbad, CA).
ATPase and GTPase assays.
Reactions were carried out as previously described (25) using the indicated amounts (micrograms) of BceSIV with either 1 mM ATP (NEB) or 1 mM GTP (Sigma G8877). Reactions were carried out in a UV-transparent clear-bottom 96-well plate at 37°C for 12 min, with data collected every 10 s. The optical density at 340 nm (OD340) was read by using a SpectraMax Plus 384 (Molecular Devices, Silicon Valley, CA) spectrophotometer. The maximum apparent velocity (Vmax) was calculated using SoftMax Pro data acquisition and analysis software within the linear range of 20 to 90 s. Control assays were performed in the absence of BceSIV.
Bioinformatic analysis of protein homologs.
The “bestfit” program of GCG software was used to compare amino acid similarity between two proteins (41). The “Gene cluster” function of the KEGG database (http://www.genome.jp/kegg/kegg2.html) was used to find homologs of R-M genes or functionally associated genes. The online server BlastP and REBASE were used to search protein homologs and isoschizomers. For phylogeny analysis, the protein sequences were aligned by ClustalW2. The evolutionary history was inferred using the neighbor-joining method (35). The evolutionary distances computed using the p-distance method were defined as the numbers of amino acid differences per site (30). The analysis included 35 amino acid sequences from four groups of known PLD family endonucleases and some putative endonucleases (hypothetical proteins with the signature HxKxD catalytic residues of PLD family endonucleases). All positions containing gaps and missing data were eliminated. There were a total of 159 positions in the final data set. Evolutionary analyses were conducted using MEGA5 (40).
RESULTS AND DISCUSSION
Cloning and expression of BceSI.
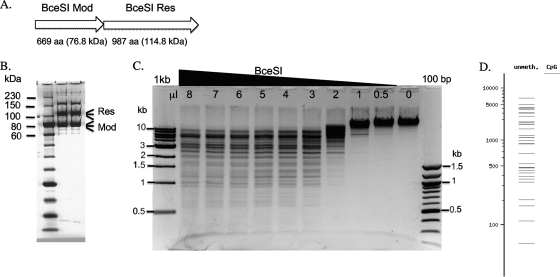
The BceSI R-M genes have been cloned previously in an 11-kb DNA fragment and expressed in E. coli (20). BceSI was shown to be a type III REase requiring ATP as a cofactor. But the BceSI recognition sequence and cleavage sites were not determined previously. The gene organization of the BceSI R-M system is shown in Fig. 1A. We amplified the mod and res genes (Bce_1018 and Bce_1019) together using PCR and cloned both genes into a pUC19-based vector pRRS. A 6×His tag was engineered at the N terminus of the Mod subunit. After nickel column purification of the Res/Mod subunits from IPTG-induced cell extracts, partial restriction activity was detected (data not shown). Analysis of the partially purified BceSI enzyme by SDS-PAGE indicated that the Mod expression was higher than the Res expression. Close inspection of the ribosome binding site and the spacer before the res gene start codon revealed that the spacer is 12 nucleotides (nt) long (GGAGGT [S/D]-agtgaatataaa [spacer]-ATG [start codon]), and the long spacer (shown in lowercase letters) was probably the cause of poor Res expression. Therefore, PCR mutagenesis was carried out to delete 6 nt out of 12 nt in the spacer region before the ATG start codon. The final translation signal contains the GGAGGT-tataaa-ATG sequence of the res gene, which resulted in elevated res gene expression. BceSI was partially purified by a nickel-agarose column with equally expressed Res and Mod subunits. It can be further purified from a Hi-Trap heparin column (Fig. 1B), although a few contaminating bands are still visible in the partially purified BceSI protein preparation. BceSI is active in all four NEB restriction buffers (data not shown). The addition of 1 mM sinefungin (an S-adenosylmethionine [SAM] analog) slightly stimulated BceSI activity (data not shown). A titration of the BceSI enzyme in digestion of λ DNA is shown in Fig. 1C. The real digestion pattern did not perfectly match the digestion pattern predicted in silico (NEB cutter), presumably reflecting the cleavage preference of two sites arranged in a head-to-head or tail-to-tail orientation (the 3′ end of 5′-CGAAG-3′ is defined as the “head” [see below]) as observed with other type III REases.
Fig 1.
BceSI gene organization and characterization of BceSI restriction enzyme. (A) Gene organization of the BceSI R-M system. Mod, modification subunit; Res, restriction subunit. (B) SDS-PAGE analysis of BceSI after two-column purification (nickel agarose and Hi-Trap heparin Sepharose). (C) BceSI digestion of phage λ DNA. One μg of λ DNA was digested by various amounts of BceSI endonuclease (0.5 μg/μl) in NEB buffer 4 at 37°C for 1 h. (D) BceSI digestion pattern generated by NEB cutter. The real digestion and hypothetical digestion patterns do not completely match, presumably due to BceSI requiring two sites in a head-to-head or tail-to-tail orientation for efficient cleavage. unmeth., unmethylated; CpG, CpG dinucleotide.
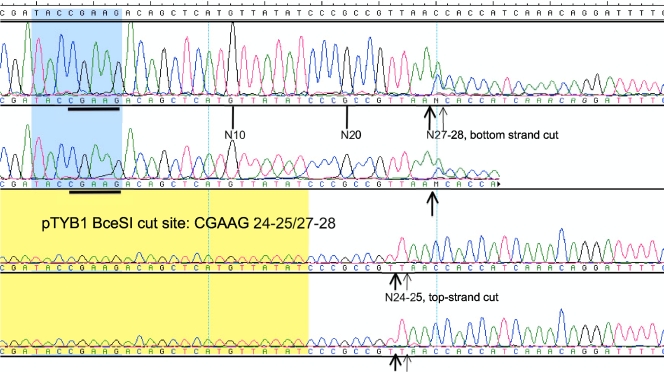
Restriction mapping of the BceSI recognition sequence on pTYB1 and runoff sequencing of the cleavage products indicated that BceSI cleaves the sites CGAAG 24-25/27-28 (top-strand major cut at N24 and minor cut at N25; bottom-strand major cut at N27 and minor cut at N28 as shown in Fig. 2). Additional mapping of the cleavage site (CGAAG 24/27-28) in pBR322 indicated that the top-strand cleavage is rather fixed at N24 (see Fig. S1 in the supplemental material) and the bottom-strand cut is somewhat variable at N27 and N28 (data not shown), probably reflecting the flanking sequence effects. To further confirm the cleavage distance of the bottom strand, BceSI-digested DNA fragments were blunted by T4 DNA polymerase and cloned into pUC19, and the cloning junctions were sequenced. CGAAG BceSI recognition site, and the bottom strand cuts at N27 or N28 were confirmed from five independent isolates. One star site with one base off, TGAAG, was also detected in a cloned fragment, indicating that the CGAAG specificity can be relaxed into YGAAG (Y, C or T). It is likely that the BceSI Mod subunit modifies the 4th nt in the CGAAG sequence to yield CGA[N6mA]G. M.BceSI and M.StyLTI (CAGAG) share 55% and 62% amino acid sequence identity and similarity, respectively, and both recognition sites can be defined as CRRAG (R, A or G) with a common adenine at the 4th base (12). However, it is still unknown which adenine is modified by the BceSI Mod subunit. BlastP analysis shows that over 20 close homologs of M.BceSI and M.StyLTI with more than 45% amino acid sequence identity are present in sequenced microbial genomes (more than 100 homologs with >30% amino acid [aa] sequence identity), reflecting the widespread distribution of BceSI close relatives (12). A crystallography study of BceSI in complex with DNA is in progress (Gupta and Aggarwal, unpublished).
Fig 2.
Runoff sequencing to determine the cut site on cleaved pTYB1. Sequencing reactions were carried out using two identical templates with different concentrations. The top two panels show sequencing results from the cleaved bottom strand; the bottom two panels show sequencing results from the cleaved top strand. The determined cut site is CGAAG 24-25/27-28 (top-strand cut at N24-25 and bottom-strand cut at N27-28).
Cloning and expression of BceSII in E. coli.
The predicted amino acid sequence of BceSII shows 29% to 30% amino acid sequence identity and 42% to 43% sequence similarity to HgiBI, HgiEI, and HgiCII REases with the recognition sequence G/GWCC (REBASE) (15). In addition, the bceSIIM gene sequence is very similar to the avaIIM gene sequence (27), with the known target site GGWCC. The bceSIIR gene-induced cell extracts produced a partial digestion pattern similar to that seen with AvaII digestion (see Fig. S2B in the supplemental material) or with mixed digestion by AvaII/BceSII (see Fig. S2C). The BceSII enzyme produced by in vitro transcription and translation (PurExpress) also generated the AvaII cleavage pattern (data not shown). By bioinformatic analysis and enzymatic assays, we demonstrated that BceSII is an isoschizomer of HgiBI, HgiEI, HgiCII, and AvaII. Although M.AvaII and M.BceSII show strong sequence similarity (49% amino acid sequence identity/59% sequence similarity by the “bestfit” program of GCG software), their companion REase AvaII and BceSII do not, suggesting that either the BceSII R-M genes evolved at a different mutational rate due to different genetic selection pressure or the two genes were acquired as separate events by HGT.
Cloning and expression of BceSIII in E. coli.
The predicted amino acid sequence of BceSIII (Bce_5605) shows 97% amino acid sequence identity to the type IIS REase BceAI (ACGGC 12/14) (31) (C. Nkenfou and R. Morgan, unpublished data) (see REBASE) (34). BceSIII is also identical to an ORF found in the genome of B. cereus AH1271 (genome coordinate 5031127.0.5032902; ORF, bcere0028_51390) (GenBank accession number CM000739; Protein Data Bank identification code EEL79209). Therefore, it is predicted that BceAI, BceSIII, and BceAH1271-ORF bcere0028_51390 (EEL79209) are isoschizomers. Purified BceSIII shows a cleavage pattern and cut sites identical to those of BceAI by gel electrophoresis (see Fig. S3 in the supplemental material) and by DNA runoff sequencing of the cleavage products (data not shown). In the BlastP analysis, the C terminus of BceSIII is moderately similar to the C terminus of AlwI (GGATC 4/5), with ∼30% amino acid sequence similarity. Although BceSIII and AlwI do not recognize similar DNA sites (ACGGC versus GGATC), the similarity at the C terminus of the two proteins suggests that they may share similar catalytic domains. The predicted catalytic domains of four REases/NEases, AlwI, BceSIII, MlyI, and Nt.BstNBI, are shown in Fig. 3. This catalytic domain differs in amino acid sequence from the typical type IIS catalytic domain found in FokI, which contains PD-X-D/EXK catalytic residues. The AlwI/BceSIII catalytic residues (E-X-PD-X12-E-X8-9-Q-X3-E-X6-H) are further variations of the catalytic site PD-X6-E-X14-QR that is found in the bottom-strand catalytic site of BtsCI, BsmI, Mva1269I, Nb.BsrDI (B subunit), and Nb.BtsI (B subunit) (45). The predicted catalytic residues of BceSIII remain to be confirmed experimentally.
Fig 3.
Multiple amino acid sequence alignment (ClustalW) in the catalytic domains of Nt.BstNBI, MlyI, AlwI, and BceSIII. Nt.BstNBI, the large subunit of BstNBI, a top-strand nicking enzyme (GAGTCN4↓). The recognition sequences of MlyI, AlwI, and BceSIII are GAGTC 5/5, GGATC 4/5, and ACGGC 12/14, respectively. The putative catalytic residues of BceSIII (E—PD-X12-E—Q-X8-9-E-X6-H) are shown in bold and underlined. Asterisk, identical amino acid residues; colon or period, amino acid residues with similar properties.
Cloning and expression of BceSIV in E. coli.
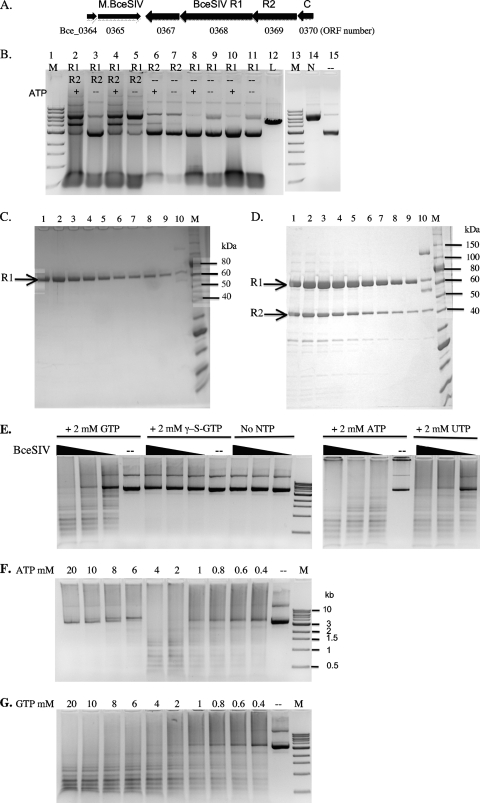
The BceSIV R-M system is carried on an operon with perhaps six genes (Fig. 4A). M.BceSIV (Bce_0365) is predicted to be a C5 MTase displaying significant sequence similarity to C5 MTases that modify GCWGC, such as M.Lsp1109I (66% and 74% aa sequence identity and similarity, respectively) (34). The bceSIVM gene (flanked by BamHI sites) was amplified by PCR and inserted into pUC19. Plasmid DNA with the correct insert orientation was prepared and digested by BbvI (GCAGC) or Fnu4HI (GCNGC). The plasmid DNA was mostly resistant to BbvI digestion (about 10% of plasmid DNA was linearized) and sensitive to Fnu4HI digestion (data not shown). It was concluded that M.BceSIV modifies GCWGC sites and rendered the sites resistant to BbvI digestion. One putative transcription repressor (Bce_364) is located upstream of M.BceSIV. There is one ORF (Bce_367) immediately downstream of M.BceSIV which had no detectable endonuclease activity when it was expressed in E. coli using IPTG-induced cell extracts or as expressed protein from an in vitro transcription and translation system (data not shown). The function of ORF Bce_367 is still unknown. Another small ORF (Bce_370) shows amino acid sequence similarity to the C (controller) protein of R-M systems, which might control the transcription of Bce_367, 368 (R1), and 369 (R2) in the BceSIV operon.
Fig 4.
Characterization of BceSIV endonuclease. (A) Schematic diagram of six ORFs of the BceSIV operon. The gene identifier (Bce_0364 to Bce_0370) is indicated below the ORFs. Bce_0364, a possible transcription regulator, similar to phage repressors; Bce_0365, M.BceSIV (GCWGC); Bce_0367, unknown function, no apparent endonuclease activity; Bce_0368, R1 subunit of BceSIV, NTPase/helicase; Bce_0369, BceSIV endonuclease catalytic subunit, R2 subunit; Bce_0370, putative transcription regular, similar to the controller (C) protein of other R-M systems. (B) Digestion of pBR322 DNA by cell extracts containing BceSIV R1, R2, and R1+R2 in the presence or absence of ATP (2 mM). Lanes 1 and 13, 1-kb DNA ladder; lanes 2 to 11 contain either R1, R2, or R1+R2 as indicated; lane 12, linear DNA; lane 14, nicked circular DNA; lane 15, uncut pBR322 DNA. (C) Purified BceSIV R1 subunit from a chitin column. The predicted molecular mass of R1 is 69.6 kDa. Lanes 1 to 9, eluted fractions containing R1; lane 10, total protein from the chitin column (R1-intein-CBD fusion protein, R1, and intein-CBD). (D) Purified BceSIV R1 and R2 subunits from a chitin column (lanes 1 to 9). The predicted molecular mass of R2 is 42.7 kDa. Lane 10, total protein from the chitin column. (E) BceSIV digestion of pBR322 in the presence of GTP, γ-S-GTP (nonhydrolyzable GTP analog), ATP, and UTP. (F) BceSIV digestion of pBR322 in the presence of ATP in the range of 0.4 mM to 20 mM. M, 1 kb DNA ladder. (G) BceSIV digestion of pBR322 in the presence of GTP in the range of 0.4 mM to 20 mM. Lanes M, 1-kb DNA ladder.
Cell extracts containing ORF Bce_368 (predicted DNA helicase, ATPase/GTPase, BceSIV R1 subunit, large subunit) displayed no apparent REase activity, although a low level of nicking activity was detected but may have been due to nonspecific nuclease activity from the crude extracts (Fig. 4B, lanes 8 to 11). However, we found that cells expressing ORF Bce_369 (BceSIV R2 subunit with catalytic and specificity function, small subunit) formed very small colonies, indicating toxicity of the inserted gene. To alleviate toxicity, we coexpressed M.BbvI (GCAGC), and these cells grew normally. Not surprisingly, we found that R2 alone has a low level of endonuclease activity (Fig. 4B, lane 6), and its activity was stimulated by the addition of R1 cell extract (Fig. 4B, lanes 2 and 4). The activity stimulation is dependent on the presence of ATP (see below for the NTP requirement).
To test for a physical interaction of BceSIV R1 and R2 subunits, the bceSIVR1 gene was inserted into a pTYB1 vector (NdeI/XhoI sites) as a fusion with an intein and a chitin-binding domain (CBD). The BceSIV R1 subunit was purified from a chitin column (Fig. 4C). IPTG-induced cell extracts containing BceSIVR1-intein-CBD were also mixed with BceSIV R2 crude extracts and loaded onto a chitin column. After extensive washing, the R1/R2 complex was eluted from the column by the addition of dithiothreitol (DTT) in an overnight intein cleavage reaction. Figure 4D shows the copurified R1/R2 subunits from the eluted fractions. The partially purified BceSIV complex shows REase activity in the presence of 2 mM ATP, GTP, and UTP (Fig. 4E). The nonhydrolyzable GTP analog, γ-S-GTP, does not stimulate BceSIV activity (Fig. 4E), suggesting that GTP hydrolysis is required for BceSIV activity (see further experimental evidence below for GTP hydrolysis to GDP). The stimulative effects by ATP and GTP appear to be variable at different nucleotide concentrations. ATP concentrations between 0.4 mM and 4 mM stimulated BceSIV activity with optimal stimulation at 2 to 4 mM (ATP). High concentrations of ATP (6 to 20 mM), however, inhibit BceSIV cleavage activity (Fig. 4F). The range of GTP concentrations in stimulating BceSIV activity is wider, as BceSIV is active at 0.4 to 20 mM (GTP). dATP and dGTP also have a stimulative effect on BceSIV activity (data not shown). Protein crystals have been grown from purified BceSIV (Y. Gupta and A. Aggarwal, unpublished results). We concluded that BceSIV is composed of R1 and R2 subunits. The R1 subunit alone does not show detectable endonuclease activity. BceSIV activity is stimulated by the addition of GTP, ATP, UTP, or dNTP, with a preference for a wide range of GTP concentrations in stimulating endonuclease activity.
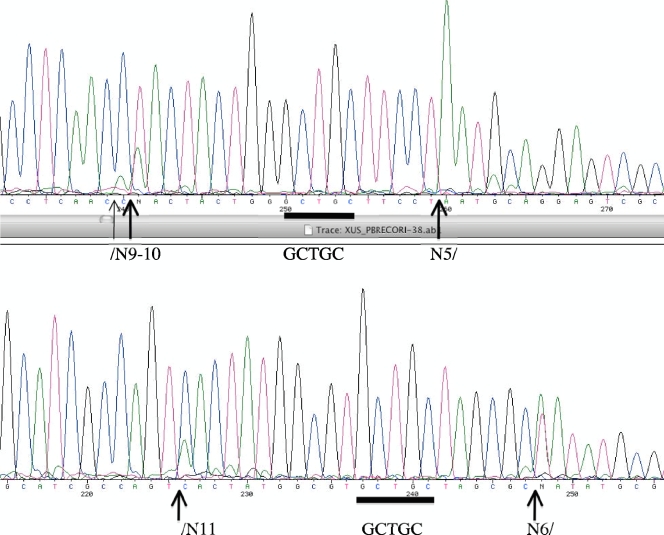
To determine the cut sites of BceSIV, pBR322 digested by BceSIV was sequenced. Figure 5 shows two such runoff sequencing results. The extra A peaks (double peaks or an extremely high A peak) in the sequence read indicate DNA breaks on the template strand. The cleavage sites are located on both sides of GCAGC (GCTGC) recognition sequences but are somewhat asymmetrical (↓N9-11 GCWGC N5-7↓). The digested DNA banding pattern by BceSIV is nearly identical to that of BbvI, but the cleavage sites of BceSIV are different from that of BbvI (BbvI cleaves at a fixed distance, GCAGC 8/12).
Fig 5.
DNA runoff sequencing of BceSIV-cleaved pBR322. Arrows indicate a mixed peak or high adenine (A) peak where the templates have been cleaved by BceSIV. The recognition sequence GCWGC is shown by a blue bar. The variable cleavage sites by BceSIV (↓N9-11 GCWGC N5-7↓) are indicated.
Type I and III REases requires ATP hydrolysis, and specifically, the type IV restriction enzyme EcoK McrBC requires GTP hydrolysis for restriction activity. The cofactor requirement by BceSIV is reminiscent of type I, III, and IV REases. Only CglI restriction activity has been shown to have its activity stimulated by ATP. In the previously characterized CglI R-M system from Corynebacterium glutamicum ATCC 13032, gpORF1 (homolog of BceSIV R2 subunit) of CglI recognizes the sequence GCSGC (cut sites unknown) (37). By measuring the restriction efficiency of a conjugative plasmid transfer, it was determined that CglI ORF1 encodes the major restriction activity, and CglI ORF2 contributes to a small but significant level of restriction by complementation analysis. CglI gpORF2 contains conserved motifs A and B of NTPases. Interestingly, CglI restriction activity can be alleviated by heat treatment, since heating CglIR-positive cells prior to mating can dramatically increase the conjugation efficiency, suggesting that the CglI R-M system is probably encoded by a mobile genetic element such as a prophage. Thus, CglI is considered to encode a “stress-sensitive” restriction system that can be “cured” by heat shock (36, 37).
Table 1 lists the seven homologs of BceSIV R1/R2 restriction systems in sequenced microbial genomes. In some cases, a very short patch repair gene (VSR) is associated with the R-M systems. In addition to the homologs encoded by two adjacent genes, there are over 95 homologs encoded by fused R1-R2 genes in the range of ∼700 to ∼1,049 aa long. In GenBank and protein databases, these proteins are annotated as the Z1 domain-containing endonucleases or superfamily II DNA helicases. The NTPase domains in the R1-R2 fusion proteins share sequence homology with eukaryotic MORC ATPases in DNA repair enzymes and enzymes involved in chromatin structure (22). Presumably, the fused endonucleases from microbes recognize the DNA sequence GCWGC, GCSGC, or GCNGC (or possibly evolve into a methylation-dependent REase with the recognition site G5mCNGC if the cognate MTase is missing or inactive). In summary, the BceSIV-like enzymes could consist of two subunits or monomeric peptides with a NTPase/helicase fusion with a PLD endonuclease catalytic domain and a target recognition domain (TRD); they may cleave outside their recognition sequences and in some cases require NTP hydrolysis for DNA translocation and enhanced restriction activity; and cut sites may be variable.
Table 1.
BceSIV R1 and R2 homologs or R1-R2 fusion homologsa
| Bacterial strain | Homolog |
VSR | |
|---|---|---|---|
| R1 (NTPase) | R2 (PLD-TRD) | ||
| Bacillus cereus ATCC 10987 | BCE_0368 | BCE_0369 | |
| Lactobacillus delbrueckii ATCC BAA-365 | LBUL_1144 | LBUL_1143 | LBUL_1145 |
| Neisseria gonorrhoeae FA 1090 | NGO0363 | NGO0364 | NGO0300 |
| Neisseria gonorrhoeae NCCP11945 | NGK_0520 | NGK_0521 | NGK_0449 |
| Haemophilus somnus 2336 | HSM_0598 | HSM_0597 | |
| Corynebacterium glutamicum ATCC 13032 | NCgl1705 | NCgl1704 | |
| Corynebacterium glutamicum (Bielefeld) | Cg1998 | Cg1997 | |
| Neisseria meningitidis 053442 | NMCC_0745 | NMCC_0746 | |
| Corynebacterium resistens | CRES_0473 | CRES_0474 | |
| R1-R2 fusion (NTPase-R) |
|||
| Sulfurovum sp. NBC37-1 | SUN_2431 (923 aa) | SUN_2429 | |
| Aromatoleum aromaticum EbN1 | EbA6352 (860 aa) | ||
| Rhodopseudomonas palustris BisA53 | RPE_3251 (891 aa) | RPE_3253 | |
| Ruminococcus albus | Rumal_0705 (927 aa) | Rumal_0709 | |
| “Candidatus Koribacter versatilis” | Acid345_4267 (899 aa) | ||
More than 95 R1-R2 fusion homologs (∼700 to 1,000 aa long) are found in sequenced microbial genomes (BlastP total score, 1,923 to 232). VSR, very short patch repair endonuclease for T/G mismatch.
The AspCNI endonuclease also carries a PLD family endonuclease domain and a TRD with the recognition sequence GCCGC and variable cut sites (GenBank accession number HQ446229; R. Morgan, unpublished data). The AspCNI endonuclease and BceSIV R2 share 24% and 42% amino acid sequence identity and similarity, respectively. The PLD family endonuclease catalytic domain is located at the N terminus of AspCNI (1 to ∼160 aa), and the TRD is predicted to be located at the C terminus. No further sequence information is available for downstream of the AspCNI restriction gene, so the possible existence of an R1 gene encoding NTPase is not yet confirmed. So far, this group of enzymes (group 1) includes BceSIV, Bce14579I, CglI, NgoFVII, and some other two-subunit REases. Group 2 enzymes include many uncharacterized REases that are R1-R2 fusions. Enzymes in groups 1 and 2 are closely related and differ in gene organization. The PLD family catalytic domain has also been found in the type IIS enzymes BfiI and BmrI (ACTGGG 5/4) (6, 10, 18), and a few homologs of BfiI/BmrI are found in sequenced bacterial genomes. Here, we tentatively assigned BfiI/BmrI and close homologs to enzyme group 3. Interestingly, the PLD endonuclease catalytic domain and ATPase/DNA helicase domain have also been found in another group of methylation-dependent type IV REases represented by SauUSI (S5mCNGS; S, C or G) (44). Thus, during the evolution of R-M systems in prokaryotes, the PLD catalytic domain and the NTPase/helicase domain have been adopted by and evolved in different types of REases. We tentatively assigned SauUSI-like enzymes to group 4 of PLD-family endonucleases. In addition, we also included four putative PLD-family proteins with unknown functions (hypothetical proteins with PLD family signature catalytic sites HxKxD) in the phylogeny analysis. Fig. S4 in the supplemental material shows the phylogenetic analysis based on the amino acid sequences of the four groups of PLD family endonucleases mentioned above. As expected, group 1 and group 2 enzymes (R1-R2 fusion) form a closely related clade. Group 3 enzymes BfiI, BmrI, and a close relative form another clade. SauUSI-like group 4 enzymes from Gram-positive bacteria with PLD-catalytic/DNA helicase/5mC TRD domains also form a clade, but they are more divergent, possibly reflecting diverse specificities. Two putative PLD family endonucleases from a Thioalkalivibrio sp. and Planctomyces maris form a small outlier that is distantly related to SauUSI. The four hypothetical proteins with PLD family signature HxKxD catalytic sites are more related to group 1 enzymes. Overall, the PLD family REases may be the second-largest groups of restriction enzymes (>600) next to the PDxD/ExK-containing REases.
GTPase and ATPase assays for BceSIV (R1+R2).
We next determined if BceSIV had ATPase or GTPase activity using NADH as a substrate. In this reaction, BceSIV first hydrolyzes ATP/GTP to ADP/GDP. The second reaction step is catalyzed by pyruvate kinase (PK), which transfers a phosphate group from phosphoenolpyruvate (PEP) to ADP/GDP, yielding one molecule of pyruvate and one molecule of ATP/GTP. The third reaction step is controlled by lactate dehydrogenase (LDH), which catalyzes the interconversion of pyruvate and lactate with a concomitant interchange of NADH and NAD+ (25). The difference between the reduced form (339 nm) of NADH and the oxidized form (259 nm) is read using UV absorption spectra at OD340 (see Fig. S5 in the supplemental material). Fig. S5B in the supplemental material shows representative raw data of UV absorption at 340 nm with various amounts of BceSIV with either 1 mM GTP or 1 mM ATP added in the presence of 0.5 μg DNA. We found that with increasing amounts of BceSIV, NADH was converted to NAD+ at increasing rates. The Vmax (apparent velocity) values from multiple experiments were calculated and averaged from the NADH conversion curve and plotted against the amount of BceSIV to yield the relative ATPase and GTPase activity in term of milliunits (mUnits)/min as shown in Fig. S5C and S5D in the supplemental material. We found that BceSIV has higher relative ATPase activity than GTPase activity. The R1 subunit alone has no GTPase activity in the same enzymatic assay (data not shown).
Cloning and expression of M.BceSV, a multispecificity C5 MTase.
M.BceSV encoded by ORF Bce_0393 was predicted to be a C5 MTase with GGCC specificity (HaeIII isoschizomer) in REBASE. An upstream gene, ORF Bce_0392, was predicted to be a ParB-like nuclease fused to an amino-methyltransferase. The ParB-methylase (ParB-M) fusion is similar in amino acid sequence to a hypothetical protein from Streptococcus phage phi-m46.1, whose homologs can be found in over 100 sequenced bacterial and phage/prophage genomes. We created three plasmids using pAII17 that expressed the parB-M fusion (6×His tagged) either alone or with the bceSVM genes. The plasmids containing both the parB-M and bceSVM genes (pAII17-parB-M-bceSVM) inserted the genes into the vector in the correct orientation or opposite orientation relative to the T7 promoter. By SDS-PAGE analysis of the IPTG-induced total cell extracts, coexpression of ParB-M (∼47 kDa) and M.BceSIV (∼78 kDa) was detected; however, the majority of the expressed ParB-M protein was found in the insoluble fraction (data not shown). IPTG induction at 30°C increased the protein solubility to a certain extent, leading to approximately 1/3 of the ParB-M protein being soluble in the supernatant (data not shown). Partially purified ParB-M appears to have nonspecific nuclease (nicking) activity on pBR322 (see Fig. S6A, lane 6) and pUC19 (see Fig. S6B, lane 6). In addition, ParB-M appears to have a DNA nicking-associated concatenation activity on pBR322 and pUC19. A high-molecular-weight DNA (a possible dimer) accumulated after ParB-M treatment (see Fig. S6A to C in the supplemental material). The activity was not caused by other E. coli proteins, since control extracts in the absence of ParB-M do not show such a strong activity (see Fig. S6C in the supplemental material). The dimer DNA can be resolved into a linear form after restriction digestion with HindIII or PstI, indicating no major DNA deformation by the ParB-M protein (see Fig. S6A and B, lanes 2 and 3, in the supplemental material). Further E. coli exonuclease III treatment removed all linear and nicked circular plasmid DNA (see Fig. S6A, lane 5, and Fig. S6B, lane 4). The smearing of pBR322 and pUC19 was most likely caused by the nonspecific nicking endonuclease activity of ParB-M.
The biological function of ParB-M fusion is unknown. We produced a small amount of the ParB-M fusion protein using an in vitro transcription and translation system (PurExpress) and examined the methyltransferase activity on phage λ DNA and pXbaI DNA (pUC19 with a large XbaI fragment from adenovirus DNA). However, we were unable to detect the transfer of [H3]SAM to DNA (data not shown). Either the DNA substrates used here do not carry the true target site for ParB-M or the protein has no MTase activity.
When the bceSVM gene was inserted and expressed from plasmid pAII17-parB-M-bceSVM, the resulting M.BceSV modifies target sites on the plasmid and renders the plasmid resistant to restriction digestion with the same or overlapping specificities. Plasmid DNA prepared from IPTG-induced overnight cultures with the correct insert orientation (T7 promoter_parB-M_bceSVM) was resistant to digestion by EaeI (YGGCCR), HaeIII (GGCC), Fnu4HI (GCNGC), HpaII (CCGG), NlaIV (GGNNCC), and BssHII (GCGCGC) (see Fig. S6D in the supplemental material). The plasmid was also partially resistant to digestion by HhaI (GCGC), MspI (CCGG), and PspGI (CCWGG). If the insert was in the opposite orientation of the T7 promoter, the plasmid DNA was only partially resistant to digestion by EaeI, HaeIII, Fnu4HI, HpaII, NlaIV, and BssHII (data not shown), presumably due to the low level of expression of the multispecificity MTase. The vector pAII17 prepared from an IPTG-induced overnight culture was sensitive to digestion by these REases (ER2566 is a Dcm-deficient E. coli B strain) (see Fig. S6E in the supplemental material). Plasmid pAII17-parB-M was also cleaved by these REases, indicating that the expressed M.BceSV was solely responsible for the plasmid modification and resistance (data not shown). It was concluded that BceSV is a multispecificity MTase that carries at least five specificities: GGCC, GCNGC, CCGG, GGNNCC, and GCGCGC/GCGC (predicted modified cysteine is underlined). To test the genomic DNA modification by the multispecificity MTase M.BceSV, B. cereus genomic DNA (gDNA) was digested by AvaII, BbvI, BceAI, EaeI, HaeIII, Fnu4HI, HpaII, NlaIV, and BssHII. The gDNA was resistant to AvaII, BbvI, and BceAI digestion (data not shown), most likely due to the modifications conferred by the expressed MTases M.BceSII (GGWCC), M.BceSIV (GCWGC), and M.BceSIII (ACGGC) in the native strain. The gDNA was partially digested by EaeI, HaeIII, Fnu4HI, HpaII, and NlaIV (data not shown), indicating that M.BceSIV expression was incomplete in the native cells.
Cloning, expression, and purification of Bce14579I R1 and R2 subunits.
The Bce14579I R1 gene was cloned in fusion with an intein and chitin-binding domain, and the R2 gene was cloned as nonfusion. The R1 subunit was purified from a chitin column following intein cleavage (see Fig. S7A, lane 6). The R1 and R2 complex was also partially purified by mixing the R1 and R2 cell extracts and loading onto a chitin column. SDS-PAGE analysis indicated that only a small amount of R2 subunit protein was copurified with R1 (molecular mass, 66.1 kDa). An induced band of R2 protein approximately 50 kDa (predicted mass, 52.8 kDa) was detected in the total proteins (see Fig. S7A, lanes 2 and 3), but the R2 protein was not abundant in the soluble fraction (supernatant cell extract following centrifugation) (lanes 4 and 5). Lowering the temperature of IPTG induction to 25°C overnight increased the soluble R2 subunit protein (data not shown). Nevertheless, the partially purified R1/R2 enzyme was active in digestion of DNA. Fig. S7B in the supplemental material shows that R1 alone is inactive in DNA cleavage (lanes 1 to 3). The Bce14579I R1/R2 complex is active in digesting DNA in the presence of ATP or GTP (lanes 4 to 11). However, a high concentration of ATP (10 mM) inhibits restriction activity. The addition of dATP, dCTP, dGTP, or dTTP (5 mM) also has some stimulatory effects on Bce14579I activity. The R1/R2 enzyme has minimal activity in DNA cleavage in the absence of NTP or in the presence of nonhydrolyzable γ–S-ATP (5 mM). TseI (GCWGC) digestion or TseI/Bce14579I double digestions gave rise to a similar partial digestion patterns (lanes 18 and 19), suggesting that TseI and Bce14579I share similar sequence specificities. To determine the cleavage sites of Bce14579I, partially digested pBR322 DNA was used in runoff sequencing. The DNA incision takes place near the GCTGC site with variable cleavage distance (GCTGC N3-6↓); long-distance cuts at ↓N14 GCAGC, ↓N31 GCAGC, and ↓N44 GCAGC were also detected from cloned DNA fragments following Bce14579I digestion of λ DNA (data not shown), suggesting that Bce14579I is capable of translocation on DNA.
The properties of the four active BceSI-IV R-M systems, the multispecificity M.BceSV orphan MTase, and Bce14579I are summarized in Table 2. The putative type I and type IV restriction systems have not been studied yet.
Table 2.
Restriction and modification enzymes from Bacillus cereus strains ATCC 10987 and ATCC 14579
| Bacillus cereus strain | Restriction-modification enzyme (type) | Sequence specificity (description) |
|---|---|---|
| ATCC 10987 | BceSI (type III REase) | CGAAG 24-25/27-28 (unique specificity) |
| BceSII (type II) | G/GWCC (AvaII isoschizomer) | |
| BceSIII (type IIS) | ACGGC 12/14 (BceAI isoschizomer) | |
| BceSIV (type IIT) | GCWGC (TseI neoschizomer) | |
| BceSV, multispecificity C5 MTase, orphan MTase | GGCC, GCNGC, CCGG, GGNNCC, and GCGCGC (GCGC) | |
| ParB-methylase, DNA-nicking enzyme fused with an amino-methyltransferase | M specificity unknown, nonspecific DNA nicking activity | |
| ATCC 14579 | Bce14579I (type IIT) | GCWGC (TseI neoschizomer) |
Some putative type II and III R-M systems from the other nine B. cereus genomes are summarized in Table S1 in the supplemental material. The B. cereus 03BB102 strain carries a putative heterodimeric REase (predicted sequence specificity, GCNGC or GCSGC) encoded by Bce03_0978 and 0979. But the cognate MTase is missing. Instead, a predicted M specificity (CCCGC) is located nearby. It would be interesting to see whether this MTase can be rapidly evolved into GCCGC in vitro or in vivo to match the heterodimeric REase GCSGC. The B. cereus 03BB102 strain encodes a putative multispecificity C5M with predicted GGCC, GCGC, CCGG, and other specificities. B. cereus AH820 carries a BceSIV-like R-M system encoded by BCAH820_1008 (M), BCAH820_1009 (R1) and BCAH820_1011 (R2), with a predicted GCNGC specificity. B. cereus G9842 carries at least five putative C5 MTases: M1 (GCNGC or GCSGC), M2 (GCNNGC), and three MTases encoded by a plasmid, M3 (amino-MTase), M4 (GGNCC, C5M), and M5 (TCGA, C5M). A BceSIV-like REase is encoded by BCG9842_B4335 (R1) and BCG9842_B4334 (R2). B. cereus Q1 carries an orphan adenine MTase (GATC). The cytotoxic NVH 391-98 strain carries a BbvI-like type II R-M system (GCAGC) and a R-M system with GATC specificity. In summary, 10 out of 11 sequenced B. cereus strains carry type II or III R-M systems with predicted recognition of 4 to 6 bp. A BceSIV-like restriction system can be found in five B. cereus genomes. Multispecificity C5 MTases can be found in two B. cereus genomes. A number of orphan C5 MTases are also found in the B. cereus genomes. These M genes may have been acquired recently or be the remains of inactive genes after DNA deletion and rearrangement. In addition, horizontal gene transfer may also occur between Bacillus species. A close homolog of BceSIII and BceAI is found in Bacillus thuringiensis (BthB ORF05523). An orphan C5 MTase with predicted sequence specificity of YGGCCR (M.EaeI isoschizomer) was found in the whole-genome shotgun sequence of B. cereus AH603 (GenBank accession number CM000737, gene identifier Bcere0026_58650). A BceSIV-like heterodimeric REase can also be found in the whole-genome shotgun sequence of B. cereus AH603 with the predicted GCWGC specificity. However, the protein size of R1/R2 changes considerably; the R1 (ATPase, bcere0026_8030) and R2 (endonuclease, bcere0026_8040) subunits are 494 aa and 869 aa, respectively. There are an additional 22 WGS sequences of B. cereus genomes in GenBank, and the putative type II R-M systems remain to be analyzed in detail.
Table S2 in the supplemental material lists the putative type II R-M genes and their predicted specificities among sequenced B. anthracis and B. thuringiensis strains. A C5M with the predicted target site YGGCCR was present in five out of six sequenced B. anthracis genomes. The YGGCCR C5 MTase can also been found in sequenced B. cereus and B. thuringiensis strains, suggesting HGT among the closely related Bacillus species. There are possible type IV methylation-dependent restriction systems (McrBC-like or Mrr-like) among the B. anthracis strains (not listed in the table). Among the sequenced B. thuringiensis genomes, five strains carry five possible cytosine MTase specificities. Two plasmid-encoded MTases carry frameshifts within the M gene (TRD region), and C5M motifs IX and X may be produced as separate peptides. The B. thuringiensis A1 Hakam strain may encode a type IIG restriction enzyme with R-M-S fusion. In the B. thuringiensis BMB171 strain, there are two putative Z1-domain containing endonucleases with similar predicted specificities of GCWGC or GCSGC: BMB171_C0823 is next to a cognate MTase (GCWGC) and Bth171ORF4698 is a stand-alone endonuclease. The two Z1 domain-containing putative endonucleases share amino acid sequence similarity to the BceSIV R1 protein (NTPase/helicase). Additional bioinformatic analysis and experimentation are needed to confirm the specificities of these putative R-M genes in the Bacillus genomes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Don Comb and Jim Ellard for support and encouragement. We are grateful to Rich Roberts, Bill Jack, Lise Raleigh, Geoff Wilson, and Siu-Hong Chan for discussions and critical comments. We appreciate NEB's DNA sequencing lab for the runoff sequence.
The publication cost was paid by New England BioLabs, Inc.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print 28 October 2011
REFERENCES
- 1. Ando T, Aras RA, Kusugami K, Blaser MJ, Wassenaar TM. 2003. Evolutionary history of hrgA, which replaces the restriction gene hpyIIIR in the hpyIII locus of Helicobacter pylori. J. Bacteriol. 185:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ando T, et al. 2002. A Helicobacter pylori restriction endonuclease-replacing gene, hrgA, is associated with gastric cancer in Asian strains. Cancer Res. 62:2385–2389 [PubMed] [Google Scholar]
- 3. Arber W. 1974. DNA modification and restriction. Prog. Nucleic Acid Res. Mol. Biol. 14:1–37 [DOI] [PubMed] [Google Scholar]
- 4. Arber W, Linn S. 1969. DNA modification and restriction. Annu. Rev. Biochem. 38:467–500 [DOI] [PubMed] [Google Scholar]
- 5. Bair CL, Black LW. 2007. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 366:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bao Y, et al. 2008. Expression and purification of BmrI restriction endonuclease and its N-terminal cleavage domain variants. Protein Expr. Purif. 58:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belland RJ, Morrison SG, Hogan D. 1996. A phase-variable type III restriction-modification system in Neisseria gonorrhoeae, poster 117, p. 360–361. In Zollinger WD, Frasch CE, Deal CD. (ed), Abstr. 10th Int. Pathogenic Neisseria Conf, Baltimore, MD http://neisseria.org/ipnc/1996/Neis1996-chap6.pdf [Google Scholar]
- 8. Berger J, Bird A. 2005. Role of MBD2 in gene regulation and tumorigenesis. Biochem. Soc. Trans. 33:1537–1540 [DOI] [PubMed] [Google Scholar]
- 9. Bucci C, et al. 1999. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell 3:435–445 [DOI] [PubMed] [Google Scholar]
- 10. Chan SH, Bao Y, Ciszak E, Laget S, Xu SY. 2007. Catalytic domain of restriction endonuclease BmrI as a cleavage module for engineering endonucleases with novel substrate specificities. Nucleic Acids Res. 35:6238–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen-Karni D, et al. 2011. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U. S. A. 108:11040–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dartois V, De Backer O, Colson C. 1993. Sequence of the Salmonella typhimurium StyLT1 restriction-modification genes: homologies with EcoP1 and EcoP15 type-III R-M systems and presence of helicase domains. Gene 127:105–110 [DOI] [PubMed] [Google Scholar]
- 13. De Bolle X, et al. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35:211–222 [DOI] [PubMed] [Google Scholar]
- 14. de Vries N, et al. 2000. Phase variation in a type III restriction-modification system of Helicobacter pylori. Gastroenterology 118:A736 [Google Scholar]
- 15. Dusterhoft A, Erdmann D, Kroger M. 1991. Isolation and genetic structure of the AvaII isoschizomeric restriction-modification system HgiBI from Herpetosiphon giganteus Hpg5: M. HgiBI reveals high homology to M.BanI. Nucleic Acids Res. 19:3207–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fagerlund A, Ween O, Lund T, Hardy SP, Granum PE. 2004. Genetic and functional analysis of the cytK family of genes in Bacillus cereus. Microbiology 150:2689–2697 [DOI] [PubMed] [Google Scholar]
- 17. Figueiredo C, et al. 2000. Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori. Gene 246:59–68 [DOI] [PubMed] [Google Scholar]
- 18. Grazulis S, et al. 2005. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl. Acad. Sci. U. S. A. 102:15797–15802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han CS, et al. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188:3382–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hegna IK, Bratland H, Kolsto AB. 2001. BceS1, a new addition to the type III restriction and modification family. FEMS Microbiol. Lett. 202:189–193 [DOI] [PubMed] [Google Scholar]
- 21. Iqbal K, Jin SG, Pfeifer GP, Szabo PE. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 108:3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iyer LM, Abhiman S, Aravind L. 2008. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jelinic P, Shaw P. 2007. Loss of imprinting and cancer. J. Pathol. 211:261–268 [DOI] [PubMed] [Google Scholar]
- 24. Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B. 1986. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell 46:993–1000 [DOI] [PubMed] [Google Scholar]
- 25. Li Z, Galvin BD, Raverdy S, Carlow CK. 2011. Identification and characterization of the cofactor-independent phosphoglycerate mutases of Dirofilaria immitis and its Wolbachia endosymbiont. Vet. Parasitol. 176:350–356 [DOI] [PubMed] [Google Scholar]
- 26. Marinus MG. 2010. DNA methylation and mutator genes in Escherichia coli K-12. Mutat. Res. 705:71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matveyev AV, Young KT, Meng A, Elhai J. 2001. DNA methyltransferases of the cyanobacterium Anabaena PCC 7120. Nucleic Acids Res. 29:1491–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McMahon SA, et al. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 37:4887–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mols M, de Been M, Zwietering MH, Moezelaar R, Abee T. 2007. Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ. Microbiol. 9:2933–2944 [DOI] [PubMed] [Google Scholar]
- 30. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 31. Nkenfou C. 2002. Cloning and studying of the cleavage flexibility of some type IIs restriction endonucleases. Ph.D. thesis University of Yaounde I, Yaounde, Cameroon [Google Scholar]
- 32. Rasko DA, et al. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts RJ, et al. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 36. Schäfer A, Tauch A, Droste N, Puhler A, Kalinowski J. 1997. The Corynebacterium glutamicum cglIM gene encoding a 5-cytosine methyltransferase enzyme confers a specific DNA methylation pattern in an McrBC-deficient Escherichia coli strain. Gene 203:95–101 [DOI] [PubMed] [Google Scholar]
- 37. Schäfer A, Schwarzer A, Kalinowski J, Pühler A. 1994. Cloning and characterization of a DNA region encoding a stress-sensitive restriction system from Corynebacterium glutamicum ATCC 13032 and analysis of its role in intergeneric conjugation with Escherichia coli. J. Bacteriol. 176:7309–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sutherland E, Coe L, Raleigh EA. 1992. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225:327–358 [DOI] [PubMed] [Google Scholar]
- 39. Szczelkun MD. 2011. Translocation, switching and gating: potential roles for ATP in long-range communication on DNA by type III restriction endonucleases. Biochem. Soc. Trans. 39:589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thompson SM. 2003. Constructing and refining multiple sequence alignments with PileUp, SeqLab, and the GCG suite. Curr. Protoc. Bioinformatics Chapter 3: Unit 3.6 [DOI] [PubMed] [Google Scholar]
- 42. Walsh CP, Xu GL. 2006. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 301:283–315 [DOI] [PubMed] [Google Scholar]
- 43. Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585–627 [DOI] [PubMed] [Google Scholar]
- 44. Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. 2011. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39:5597–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu SY, et al. 2007. Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res. 35:4608–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.