Background: The cyclic nucleotide phosphodiesterases PDE10 and PDE11 contain putatively regulatory GAF domains with unknown function.
Results: Synthetic GAF domain ligands can activate both PDEs.
Conclusion: PDE10 is activated by cAMP, whereas the physiological ligand of the PDE11 GAF domains remains unknown.
Significance: This is the first demonstration of a functional role of the PDE10 and PDE11 GAF domains.
Keywords: Cyclic AMP (cAMP), Cyclic GMP (cGMP), Cyclic Nucleotide Analogues, Cyclic Nucleotides, Phosphodiesterases
Abstract
The most recently identified cyclic nucleotide phosphodiesterases, PDE10 and PDE11, contain a tandem of so-called GAF domains in their N-terminal regulatory regions. In PDE2 and PDE5, the GAF domains mediate cGMP stimulation; however, their function in PDE10 and PDE11 remains controversial. Although the GAF domains of PDE10 mediate cAMP-induced stimulation of chimeric adenylyl cyclases, cAMP binding did not stimulate the PDE10 holoenzyme. Comparable data about cGMP and the PDE11 GAF domains exist. Here, we identified synthetic ligands for the GAF domains of PDE10 and PDE11 to reduce interference of the GAF ligand with the catalytic reaction of PDE. With these ligands, GAF-mediated stimulation of the PDE10 and PDE11 holoenzymes is demonstrated for the first time. Furthermore, PDE10 is shown to be activated by cAMP, which paradoxically results in potent competitive inhibition of cGMP turnover by cAMP. PDE11, albeit susceptible to GAF-dependent stimulation, is not activated by the native cyclic nucleotides cAMP and cGMP. In summary, PDE11 can be stimulated by GAF domain ligands, but its native ligand remains to be identified, and PDE10 is the only PDE activated by cAMP.
Introduction
Phosphodiesterases (PDE)2 play important roles in the control of cyclic nucleotide signaling. To date, 11 families of PDEs differing in regulation and catalytic properties have been identified. With the exception of the photoreceptor PDE6, all PDEs are homodimers with conserved C-terminal catalytic domains and different N-terminal regulatory regions (for reviews, see Refs. 1–3).
PDE10 and PDE11 are the most recently discovered PDE families, and both hydrolyze cAMP and cGMP (4–9). Each of these enzymes is encoded by one gene giving rise to several splice variants. For PDE10, 12 splice variants characterized by unique N- and C-terminal sequences have been described (10). The predominant forms are the membrane-bound PDE10A2 and a cytosolic form, PDE10A1 or PDE10A3 in humans or rats, respectively (11). For PDE11, four splice variants differing in the length of their N termini were found (9). Only PDE11A4 contains a complete tandem of GAF domains (see below); the other splice variants are truncations thereof (8).
Kinetically, PDE10 exerts a higher affinity but a 2–5-fold lower Vmax for cAMP compared with cGMP, whereas PDE11 has little preference for either nucleotide (4–9, 12). Both PDEs display a rather restricted expression pattern. Immunofluorescence studies of brain found PDE10A predominantly in medium spiny neurons of the striatum (13). Relatively high levels of PDE10 mRNA were also found in testis (4, 6). PDE11 has been suggested to be prominent in skeletal muscle and prostate; however, because of the lack of appropriate antibodies against PDE11, the tissue distribution of this isoform is a matter of debate (7, 14–16). In situ hybridization demonstrated expression in the hippocampus, subiculum, and amygdalohippocampal area (17).
Several PDEs (PDE2, -5, -6, -10, and -11) contain a tandem of so-called GAF domains in their N-terminal regions. GAF domains are small ligand binding domains identified in many, mostly bacterial proteins (18–22). The acronym GAF is deduced from the proteins in which GAF domains were first discovered (cGMP-specific and -stimulated phosphodiesterases, Anabaena adenylyl cyclases, and Escherichia coli FhlA). In mammals, PDEs are the only proteins containing GAF domains. PDE2 and -5 are stimulated by binding of cGMP to their GAF domains (23–26).
PDE10 and PDE11 also contain tandem GAF domains. However, stimulation of PDE10 and PDE11 by cAMP or cGMP has not been demonstrated yet. Moreover, a recent publication claimed that binding of cyclic nucleotides to the PDE10 and PDE11 GAF domains does not stimulate catalytic activity (27). In another experimental approach, the PDE10 and -11 GAF domains fused to the catalytic domain of a bacterial adenylyl cyclase were studied. In these chimeric constructs, cAMP increased adenylyl cyclase activity of the PDE10 GAF fusion protein, whereas the PDE11 GAF-containing construct was responsive to cGMP (28).
Here, we used fluorophore-tagged tandem GAF domains of PDE10 and PDE11 to identify synthetic GAF ligands. Using these ligands, GAF-mediated stimulation of PDE10 and PDE11 is demonstrated. Moreover, our results show that cAMP activation of PDE10 enhances competitive inhibition of cGMP turnover by cAMP and that PDE11, despite its activation by synthetic GAF ligands, is not stimulated by the physiological cyclic nucleotides cAMP and cGMP.
EXPERIMENTAL PROCEDURES
Expression of PDE10 and PDE11 Holoenzymes and GAF Domain FRET Constructs
The open reading frames of human PDE10A1 (GenBank gi 4894715; Ref. 5) and PDE11A4 (gi 10716052; Ref. 9) were amplified by PCR from human cDNA and subcloned to pcDNA3 (Invitrogen). Using these constructs, fragments encoding the tandem GAF domains of PDE10 (Lys61–Tyr428) and PDE11 (Ala174–Val580) were amplified by PCR and subcloned into pcDNA3 between the open reading frames of CFP and YFP as described previously (29). HEK 293 cells were grown in 75-cm2 flasks (30) and transfected as described (31). Two or 3 days post-transfection, cells were harvested and lysed by sonication (Branson) in 500 μl of buffer (either 50 mm NaCl, 1 mm EDTA, 50 mm triethanolamine/HCl, pH 7.4 for expression of PDE holoenzymes or 25 mm triethanolamine/HCl, pH 7.4 for FRET constructs with both buffers containing 2 mm DTT and mammalian protease inhibitor cocktail (Sigma-Aldrich)). Lysates were cleared by centrifugation (100,000 × g for 40 min at 4 °C), and protein contents were determined by the Bradford method (Bio-Rad).
FRET Analysis of Isolated GAF Domains
FRET constructs of PDE10 tandem GAF domains obtained as described above (5 μl of cleared lysates) were analyzed in a total volume of 100 μl (25 mm triethanolamine/HCl, pH 7.4 containing 2 mm DTT and 10 mm MgCl2) in white half-area 96-well plates (Greiner) using a Cary Eclipse spectrofluorometer and a microplate accessory (Varian). For screening, nucleotides were added at a 100 μm concentration, and fluorescence was recorded for 30 min (excitation, 436 nm; emissions, 475 and 525 nm corresponding to CFP and YFP, respectively; excitation and emission slits, 5 nm). Subsequently, 0.5 μm cAMP was added to identify potential GAF domain antagonists, and fluorescence was again recorded for 30 min. Values of a water-containing well were subtracted for background correction, and ratios of emissions at 525 and 475 nm were calculated. EC50 values and 95% confidence intervals were obtained by recording nucleotide effects at concentrations between 0.1 and 100 μm.
Smaller FRET changes of the PDE11 tandem GAF domain FRET constructs necessitated analysis in more sensitive cuvettes instead of microplates; moreover, the larger volume of the cuvettes required reduction of the nucleotide concentrations because of limited availability. Cleared lysates (30 μl) containing the FRET constructs of the PDE11 GAF domains were analyzed in a Cary Eclipse spectrofluorometer (Varian) in cuvettes containing a total volume of 800 μl (25 mm triethanolamine/HCl, pH 7.4 containing 2 mm DTT and 10 mm MgCl2). Fluorescence was continuously recorded (values as above); after establishing a base line, nucleotides were added to a final concentration of 10 μm, fluorescence was recorded for at least 5 min, and subsequently 0.1 and 1 μm cGMP were added to identify potential GAF domain antagonists followed by further recording for 5 min. EC50 values and 95% confidence intervals were obtained by recording cumulative concentration response curves between 0.1 and 100 μm nucleotides.
PDE Assays
Cleared lysates of HEK cells expressing either PDE10A1 or PDE11A4 (0.1–16 μg; enzyme amount adjusted to obtain maximally 20% substrate conversion) were incubated for 5 min at 37 °C with the indicated concentrations of either [32P]cGMP or [32P]cAMP (approximately 3 kBq), 3 mm MgCl2, and 1 unit of calf intestinal alkaline phosphatase (Sigma-Aldrich) in a total volume of 100 μl of 50 mm triethanolamine/HCl, pH 7.4 containing 0.5 g × liter−1 bovine serum albumin and 3 mm DTT. Incubations were stopped by addition of 900 μl of charcoal suspension (30% (v/v) in 50 mm KH2PO4, pH 2.3), and following centrifugation, formed 32P was determined as Čerenkov radiation in the supernatant.
Generation of PDE10 and PDE11 Antibodies
PCR-amplified fragments encoding the PDE10 and PDE11 tandem GAF domains (see above; without the fluorophores) were subcloned into pPR-IBA (IBA, Göttingen Germany) for bacterial expression of N-terminally Strep-tagged GAF domains. E. coli BL21(DE3)pLysS (Invitrogen) were transformed with the respective plasmid and grown to an A600 of 0.6 (12 × 250 ml). Expression was induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and carried out for 20 h at 20 °C. Bacteria were harvested by centrifugation, frozen at −70 °C or in liquid nitrogen, and resuspended after thawing in 0.25 g × liter−1 lysozyme (Boehringer) in 100 ml of 50 mm NaCl, 50 mm triethanolamine/HCl, pH 8.5 containing 1 mm EDTA and 2 mm DTT. After lysis, DNase I (Roche Applied Science) and MgCl2 (final concentrations, 0.02 g × liter−1 and 100 mm, respectively) were added, and lysates were cleared by centrifugation (100,000 × g for 40 min at 4 °C) and applied to 10 ml of Strep-Tactin-Sepharose (IBA). After elution with desthiobiotin, size exclusion chromatography was performed (Hiload 26/60 Superdex, 200 pg, Amersham Biosciences; running buffer, 25 mm triethanolamine/HCl, pH 7.4), and fractions containing the GAF domains were concentrated (Amicon centrifugal filter units, 10 kDa, Millipore) and stored at −70 °C. Polyclonal antisera were obtained by immunizing rabbits with purified GAF domains using standard protocols.
Analysis of PDE10 and PDE11 Tissue Distribution
Mouse organs and brain regions were homogenized in 4–10 volumes (w/v) of 50 mm NaCl, 2 mm DTT, 1 mm EDTA, 50 mm triethanolamine/HCl, pH 7.4 containing mammalian protease inhibitor cocktail (Sigma-Aldrich) using a glass/Teflon Potter-Elvehjem homogenizer. After centrifugation to remove nuclei (800 × g for 10 min at 4 °C), protein concentrations were determined by the Bradford method (Bio-Rad), and 20 μg of proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Protran BA-85, Schleicher & Schuell/Whatman). After blocking (Roti-Block, Carl Roth, Karlsruhe, Germany), PDE10 and PDE11 antisera (see above) were applied in a 1:10,000 dilution and detected by secondary peroxidase-coupled anti-rabbit antibodies (Pierce). Chemiluminescence detection was performed using SuperSignal West Dura chemiluminescent substrate (Pierce) and a charge-coupled device (CCD) camera-based detection system (GDS 8000, UVP). The specificity of signals was checked by addition of purified antigen (4 μg/ml) to the primary antibody solution.
Preparation of Leydig Cells and Measurement of Testosterone Release
Adult mice were sacrificed, and testes were removed and stripped from connective tissue (tunica albuginea). Tissue was suspended in Dulbecco's modified Eagle's medium and mechanically disrupted by aspirating three times into a 50-ml syringe. Seminiferous tubes and non-disrupted cells were allowed to settle for 10 min followed by centrifugation of the remaining supernatant (10 min at 250 × g at room temperature). The pellet was resuspended in 1 ml of buffer L (10 mm HEPES, 154 mm NaCl, 5.6 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 3.6 mm NaHCO3, 5.6 mm glucose, pH 7.4), layered onto 1 ml of Percoll (1.124 g × ml−1; Biochrom, Berlin, Germany), and centrifuged (15 min at 900 × g at room temperature). The Leydig cell layer remaining on top of the Percoll layer was resuspended in 1 ml of buffer L, and cell number (usually ∼4 × 107 cells/mouse) was determined using an improved Neubauer hemocytometer.
Testosterone release was measured by incubating 106 Leydig cells in 400 μl of buffer L for 3 h at 34 °C with agitation (1000 rpm; Thermomixer 5436, Eppendorf) with luteinizing hormone (National Hormone and Peptide Program) and TP-10 (Pfizer) as indicated. Subsequently, cells were removed (900 × g for 15 min), and testosterone was measured in the supernatant by radioimmunoassay (Diagnostic Systems Laboratories).
Measurement of PDE10 in Striatal Homogenates
Striata of two mice were isolated and immediately homogenized by 15 strokes with a glass/glass Potter-Elvehjem homogenizer using 10 volumes of ice cold buffer (50 mm triethanolamine/HCl, pH 7.4, 50 mm NaCl, 1 mm EDTA, 2 mm DTT, Sigma mammalian protease inhibitor cocktail). The homogenate was centrifuged at 800 × g for 10 min at 4 °C to remove cellular debris and nuclei. Of the resulting supernatant, 0.05 μl was used for PDE assays.
Fractionation of Mouse Hippocampi
Hippocampi of 10 mice were homogenized in 3 ml of buffer A (25 mm NaCl, 25 mm triethanolamine/HCl, pH 7.4 containing 1 mm EDTA, 2 mm DTT, 0.4 mm phenylmethylsulfonyl fluoride, 0.2 mm benzamidine, and 1 μm pepstatin A) using 20 strokes of a glass/Teflon Potter-Elvehjem homogenizer. After centrifugation (100,000 × g for 40 min at 4 °C), the cytosolic fraction was applied to an anion exchange column (0.5-cm inner diameter × 5 cm = 1-ml column volume; Source Q, GE Healthcare) and eluted using a 40-ml gradient to 300 mm NaCl in 2-ml fractions. 20 μl of each fraction and the cytosol were applied to Western blots; 10 μl/incubation were used for phosphodiesterase assays performed as described above.
RESULTS
Five of the 11 PDE families contain a tandem of regulatory so-called GAF domains. The GAF-containing PDE2 and PDE5 are allosterically activated by cGMP. In contrast, GAF-dependent activation of PDE10 and PDE11 has not been demonstrated; this may be partially due to the fact that the nucleotide serving as the GAF ligand is also degraded as a substrate. Therefore, we decided to screen for synthetic GAF ligands using FRET constructs in which the respective tandem of GAF domains is sandwiched between CFP and YFP.
Screening for GAF Ligands of PDE10 and PDE11 with FRET Constructs
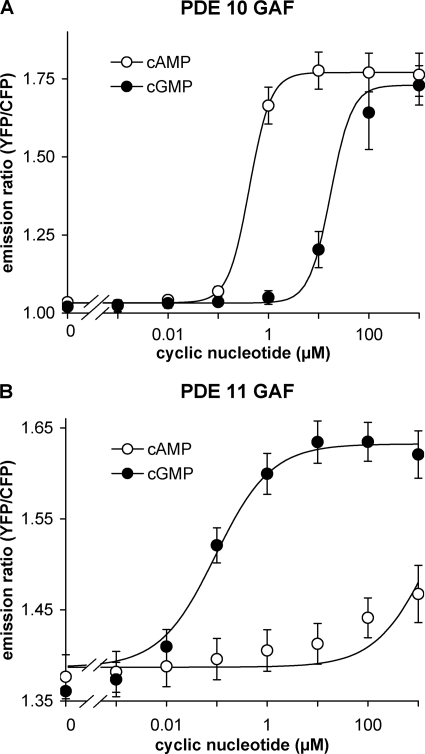
Fig. 1 depicts the cAMP and cGMP binding properties of the FRET constructs containing the tandem GAF domains of PDE10 or PDE11. The PDE10 GAF domains bound cAMP with an EC50 of ∼0.3 μm but displayed a much lower affinity for cGMP (∼30 μm). The PDE11 GAF domains almost exclusively bound cGMP (EC50 ∼ 0.1 μm); cAMP only elicited an ∼30% response even at a concentration of 1 mm.
FIGURE 1.
PDE10 GAF domains bind cAMP, and PDE11 GAF domains bind cGMP. FRET constructs containing the GAF domains of either PDE10 (A) or PDE11 (B) sandwiched between the fluorescent proteins CFP and YFP were expressed in HEK 293 cells, and changes of emission ratios between YFP and CFP elicited by the indicated concentrations of cAMP and cGMP were recorded in vitro as outlined in detail under “Experimental Procedures.” Means ± standard error of the mean of at least three experiments are depicted.
Because of the clear preference of the PDE10 FRET construct for cAMP, 33 cAMP analogues (100 μm; Table 1) were tested for their ability to elicit cAMP-like FRET changes indicating agonistic properties. Subsequently, potential antagonistic properties of the analogues were analyzed by adding a half-maximally effective cAMP concentration (0.5 μm). However, none of the tested substances blocked or reduced the cAMP-induced changes, and therefore, no antagonists were identified. Of the identified agonists, two displayed an EC50 of less than 1 μm; for three further analogues, an EC50 of less than 10 μm was determined.
TABLE 1.
Characteristics of cAMP analogues as tested on isolated PDE10 GAF domains as FRET construct
Half-maximally effective concentrations (EC50) of different cAMP analogues to elicit conformational changes of the PDE10 GAF domains are given. Conformational changes of the GAF domains were determined by FRET measurements in vitro as outlined under “Experimental Procedures.” EC50 values are from at least three independent determinations performed in duplicate; 95% confidence intervals are given in parentheses. —, no FRET change detectable at 100 μm. Full chemical names and structures of the analogues are listed in supplemental Table 1 and supplemental Fig. 1.
| Analogue | EC50 |
|---|---|
| μm | |
| cAMP | 0.28 (0.14–0.58) |
| 1-NO-cAMP | 17 (11–28) |
| 2-DMA-cAMP | >100 |
| Rp-cAMPS | 40 (25–64) |
| Sp-cAMPS | 40 (23–71) |
| 8-N3-cAMP | — |
| 2-Aza-ϵ-cAMP | — |
| 2-AHA-cAMP | >100 |
| Ara-cAMP | 4.0 (2.0–7.7) |
| 8-N3-ϵ-cAMP | — |
| 2′-NH2-cAMP | 37 (27–50) |
| 8-Br-cAMP | 54 (32–93) |
| 6-Bn-cAMP | — |
| 6-Bnz-cAMP | >100 |
| 8-BT-cAMP | — |
| cCMP | — |
| 6-Cl-cPuMP | 10 (6–18) |
| 8-Cl-cAMP | 41 (30–57) |
| 8-CPT-cAMP | — |
| 2-Cl-cAMP | 1.1 (0.6–2.0) |
| 2-Cl-MA-cAMP | — |
| 2′-dcAMP | 0.95 (0.51–1.77) |
| 7-CH-cAMP | 0.59 (0.22–1.5) |
| 2′-F-cAMP | — |
| ϵ-cAMP | 5.5 (3.4–9.0) |
| 8-OH-cAMP | — |
| 8-MA-cAMP | — |
| 6-MB-cAMP | 10 (5–20) |
| 2′-O-Me-cAMP | — |
| 6-DMA-cPuMP | — |
| 6-MA-cPuMP | — |
| 6-Phe-cAMP | — |
| cTMP | — |
| cUMP | >100 |
The PDE11 tandem GAF construct displayed smaller FRET changes than the PDE10 construct. Therefore, the screen had to be performed in cuvettes instead of microplates, which required 10-fold higher assay volumes. Because of limited availability, the analogue concentration was therefore reduced to 10 μm. Of the 31 cGMP analogues tested for agonistic properties, eight elicited cGMP-like FRET changes with EC50 values between 1 and 10 μm. None of the analogues displayed antagonistic features (tested in the presence of 0.1 and 1 μm cGMP).
PDE10 Stimulation by GAF Domain Ligands
Next, we used the identified GAF ligands to study activation of PDE10 recombinantly expressed in HEK cells. Measurements were performed with freshly prepared cytosolic fractions because cGMP stimulation of PDE5 has been reported to be impaired after storage or purification (24).
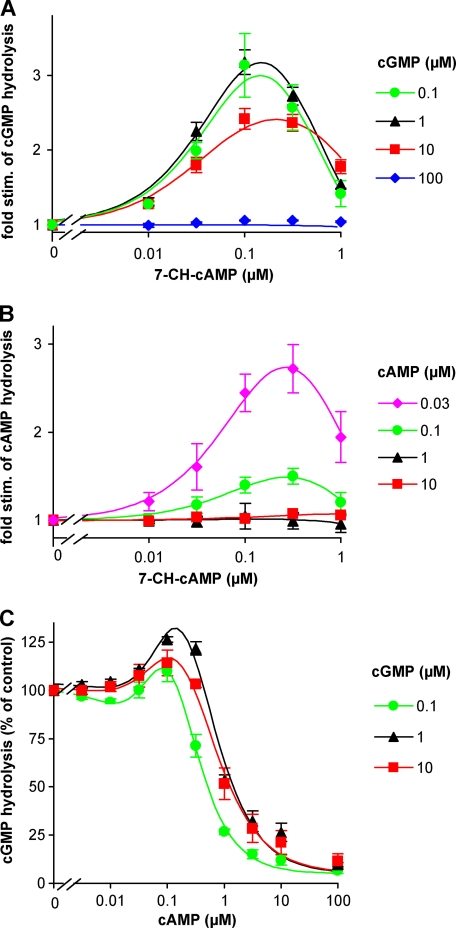
First, 7-CH-cAMP was chosen as the most promising agonist for PDE10 as it displayed the lowest EC50 value in the FRET analyses. The PDE10 GAF domains bind cAMP with high affinity (see above). To avoid an effect of the substrate on the GAF domains, we first analyzed cGMP-hydrolyzing PDE activity. At 0.1 and 1 μm cGMP, 7-CH-cAMP activated PDE10 3-fold (Fig. 2A). At higher cGMP concentrations of 10 and 100 μm, stimulation by 7-CH-cAMP was reduced or abolished, respectively, indicating that cGMP in high concentrations also binds to the GAF domains, thereby prestimulating the enzyme and reducing the maximally achievable stimulation factor. Next, the stimulatory effect of 7-CH-cAMP on cAMP-hydrolyzing activity was analyzed. At a substrate concentration of 0.03 μm cAMP, the analogue stimulated PDE10 ∼3-fold (Fig. 2B), which is comparable with the maximal stimulation observed with cGMP as substrate and demonstrates that cAMP binding to the GAF domains was negligible at the lowest cAMP concentration. However, already at 0.1 μm cAMP, the 7-CH-cAMP-induced activation was reduced (to 1.5-fold), indicating that the GAF domains were half-saturated by cAMP at this concentration. Activation by 7-CH-cAMP was abolished at 1 and 10 μm cAMP. The EC50 value for 7-CH-cAMP stimulation of the holoenzyme was about 0.05 μm, which is 10-fold lower than the value observed in the FRET constructs. The concentration-response curves were bell-shaped, indicating that higher concentrations of 7-CH-cAMP also bound to the catalytic domains and thereby inhibited substrate turnover. Other GAF domain ligands with a reasonable affinity in the FRET assays, such as 2′-dcAMP, were tested with regard to their stimulatory properties on the holoenzyme; however, none of them displayed higher stimulation factors.
FIGURE 2.
PDE10 stimulation by cAMP analogue 7-CH-cAMP. cGMP- (A) and cAMP (B)-hydrolyzing activities of recombinantly expressed PDE10 holoenzyme were determined in the presence of increasing concentrations of the cAMP analogue 7-CH-cAMP at the indicated substrate concentrations. cAMP concentrations greater than 0.03 μm stimulate the enzyme, thereby hiding 7-CH-cAMP-mediated stimulation. C, cAMP effects on cGMP-hydrolyzing activity of PDE10 in the presence of different substrate concentrations were measured at the indicated cAMP concentrations. cAMP at 0.1 μm stimulates the enzyme by up to 25%. Values are means ± standard error of the mean of three determinations performed in duplicate, normalized to the activity determined in the absence of 7-CH-cAMP (A and B) or cAMP (C) at the indicated substrate concentration. stim., stimulation.
cAMP Stimulates and Inhibits PDE10-catalyzed cGMP Hydrolysis
The 50% reduction of 7-CH-cAMP-mediated stimulation at a substrate concentration of 0.1 μm cAMP demonstrated that cAMP activated PDE10 with an EC50 of ∼0.1 μm (see Fig. 2B). To directly assess cAMP stimulation of the PDE10 holoenzyme, the effect of cAMP on cGMP turnover was measured (Fig. 2C). cAMP elicited a biphasic effect on cGMP turnover: a low cAMP concentration (0.1 μm) caused a very slight (up to 25%) albeit significant stimulation of cGMP turnover. However, 1 μm cAMP already led to a pronounced inhibition of cGMP turnover. Interestingly, the cAMP-induced inhibition of cGMP turnover was nearly independent of the cGMP concentration as it was almost indistinguishable at the different cGMP substrate concentrations (0.1–10 μm) used. The results thus demonstrate that PDE10 nearly exclusively hydrolyzes cAMP independently of the cGMP concentration present in case cAMP reaches low micromolar concentrations.
PDE11 Stimulation by GAF Domain Ligands
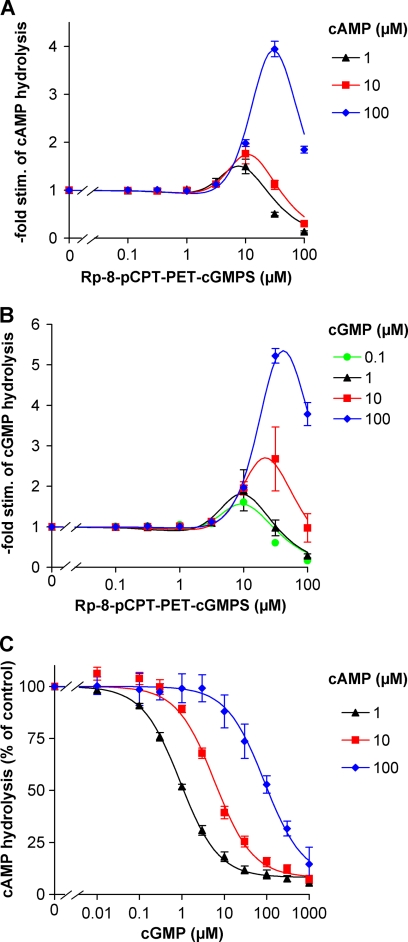
The bulky cGMP analogue Rp-8-pCPT-PET-cGMPS that displayed a low micromolar EC50 in the FRET screens (see Table 2) was analyzed with regard to its stimulatory properties on the PDE11 holoenzyme. The analogue at a concentration of 30 μm stimulated PDE11-catalyzed cAMP or cGMP turnover up to 5-fold (Fig. 3, A and B). Unexpectedly, maximal stimulation by the analogue was only observed at the highest cAMP or cGMP substrate concentration of 100 μm. At lower cAMP or cGMP concentrations (0.1–10 μm), the stimulation factor concentration-dependently declined to between 2.5- and 1.5-fold stimulation. The reduced stimulation factors can be explained by competition of the analogue with the substrate at the catalytic domain that is overcome at high substrate concentrations. Accordingly, 100 μm analogue caused robust inhibition of substrate turnover. Other analogues with similar, low micromolar EC50 values in the FRET screen (see Table 2) were tested regarding their stimulatory properties but did not cause higher stimulation factors (data not shown).
TABLE 2.
Characteristics of cGMP analogues as tested on isolated PDE11 GAF domains as FRET construct
Half-maximally effective concentrations (EC50) of different cGMP analogues to elicit conformational changes of the PDE11 GAF domains are given. Conformational changes of the GAF domains were determined by FRET measurements in vitro. EC50 values are from at least three independent determinations; 95% confidence intervals are given in parentheses. —, no FRET change detectable at 10 μm. Full chemical names and structures of the analogues are listed in supplemental Table 2 and supplemental Fig. 2.
| Analogue | EC50 |
|---|---|
| μm | |
| cGMP | 0.32 (0.15–0.65) |
| 2-NH2-cPuMP | >10 |
| 8-AET-cGMP | — |
| 8-APT-cGMP | — |
| 1-NH2-cGMP | — |
| 2′-AHC-cGMP | — |
| Sp-2′AHC-cGMPS | — |
| 8-Br-cGMP | 4.4 (2.5–7.9) |
| Rp-8-Br-cGMPS | — |
| Sp-8-Br-cGMPS | — |
| 8-pCPT-cGMP | — |
| Rp-8-pCPT-cGMPS | — |
| Sp-8-pCPT-cGMPS | — |
| Rp-8-pCPT-PET-cGMPS | 5.4 (2.8–11) |
| Sp-8-pCPT-PET-cGMPS | 2.2 (0.9–5.1) |
| 5,6-DM-cBIMP | — |
| DB-cGMP | — |
| 5,6-DCl-cBIMP | — |
| Sp-5,6-DCl-cBIMPS | — |
| 2′-dcGMP | 6 (1.6–11) |
| Rp-cGMPS | 4.9 (1.6–15) |
| Sp-cGMPS | — |
| MANT-cGMP | — |
| 2′-O-MS-cGMP | 1.1 (0.4–3.2) |
| 2′-O-MS-TME-cGMP | 1.2 (0.4–2.9) |
| 2′-O-ME-cGMP | >10 |
| PET-cGMP | 3.6 (1.2–11) |
| 8-Br-PET-cGMP | — |
| Rp-8-Br-PET-cGMPS | — |
| Sp-8-Br-PET-cGMPS | — |
| cPuMP | — |
| cXMP | — |
FIGURE 3.
PDE11 is stimulated by cGMP analogue but not by cGMP. Stimulation (stim.) of recombinant PDE11 holoenzyme by the cGMP analogue Rp-8-pCPT-PET-cGMPS was assessed at the indicated concentrations of the substrates cAMP (A) and cGMP (B). High cAMP or cGMP concentrations (100 μm) did not impair the 4–5-fold stimulation by the GAF domain ligand. C, measurement of cAMP hydrolysis at the indicated substrate concentrations in the presence of increasing concentrations of cGMP reveals no stimulation but only competitive inhibition. Means ± standard error of the mean of three independent experiments performed in duplicates are shown after normalization to the activity observed in the absence of Rp-8-pCPT-PET-cGMPS (A and B) or cGMP (C) at the substrate concentrations indicated.
The finding of highest analogue-mediated stimulation in the presence of high substrate concentrations demonstrates that neither cGMP nor cAMP acted as ligands at the analogue binding site. This is in stark contrast to the observations made for PDE10 (see above) and for PDE2 and PDE5 (31). Here, analogue-mediated stimulation is observed only at low concentrations of the physiological nucleotides; higher concentrations of the physiological nucleotides led to stimulation of the enzymes by themselves, thereby masking stimulation by the respective analogue. Because it is reasonable that the analogue Rp-8-pCPT-PET-cGMPS acted via the GAF domain, binding of cGMP to the GAF domain appears unlikely. On the other hand, the analogue might theoretically stimulate PDE11 by a site different from a putative cGMP stimulatory site. However, no evidence for cGMP stimulation of PDE11 has been obtained.
cGMP Inhibits PDE11-catalyzed cAMP Turnover
Because cGMP bound to the PDE11 FRET construct and has been shown to activate chimeric adenylyl cyclases containing the PDE11 GAF domains (28), the effect of cGMP on PDE11 cAMP turnover was analyzed. The experiments displayed a classical competitive inhibition of cAMP hydrolysis by cGMP with a rightward shift of the cGMP concentration-response curves at increasing cAMP concentrations. These results are compatible with the assumption that the cyclic nucleotides compete at the catalytic domain. Both nucleotides displayed a similar affinity to the catalytic domains as equimolar cGMP concentrations (1, 10, and 100 μm) led to an ∼50% inhibition of cAMP turnover at the different cAMP substrate concentrations (1, 10, and 100 μm), respectively.
Distribution and Properties of Native PDE10
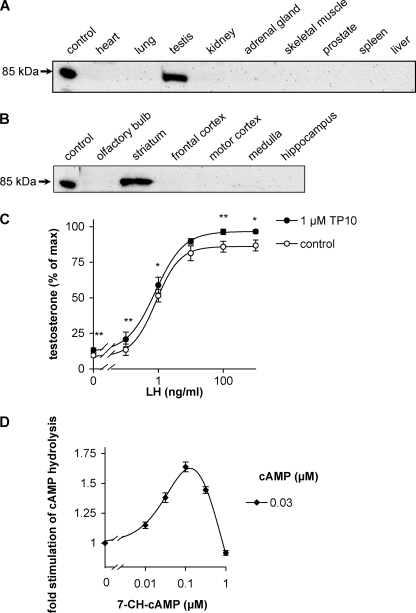
As the current knowledge on the tissue distribution of PDE10 is mostly based on mRNA distribution, we analyzed PDE10 protein expression in Western blots. For generation of antibodies, the tandem GAF domains of the FRET constructs but without the fluorophores were expressed in E. coli, purified, and used for immunization of rabbits. Subsequently, distribution of PDE10 in mice was analyzed in Western blots. The antibody recognized an 85-kDa protein compatible with the size of PDE10 in testis (Fig. 4A) and brain (Fig. 4B). Analysis of the distribution in brain revealed a prominent expression in striatum. The results confirm earlier results obtained using Northern blots, in situ hybridization, and immunohistochemistry (4–6, 13).
FIGURE 4.
PDE10 expression is confined to testis and striatum. Mouse organs (A) and brain regions (B) were homogenized and analyzed in Western blots using a newly generated antiserum raised against the GAF domains of PDE10. Recombinant human PDE10A1 was applied as a positive control. Shown is one representative blot of three. Larger sections of Western blots are shown in supplemental Fig. 3. C, primary murine Leydig cells (106) were stimulated by the indicated concentrations of luteinizing hormone in the absence or presence of the PDE10 inhibitor TP-10, and testosterone release (3 h at 34 °C) was measured by radioimmunoassay. (*, p < 0.05; **, p < 0.01.) D, stimulation of cAMP-degrading activity in homogenates of murine striatum by increasing concentrations of the GAF domain ligand 7-CH-cAMP was assessed at a substrate concentration of 0.03 μm cAMP. Values are means ± standard error of the mean.
Because of the prominent expression of PDE10 in testis, we asked whether the enzyme is involved in regulation of testosterone production. Testosterone production by Leydig cells is stimulated by luteinizing hormone (LH), and this stimulatory effect is mediated by cAMP (32). First, the PDE10 inhibitor TP-10 (33) was used to analyze the relative contribution of PDE10 to cAMP hydrolysis. In homogenates of murine Leydig cell preparations, cAMP-hydrolyzing activity was inhibited by approximately 30% by 1 μm TP-10 when measured at substrate concentrations between 10 nm and 1 μm cAMP (data not shown). Next, the effect of PDE10 inhibition on LH stimulation of testosterone production was assessed. TP-10 increased testosterone production, although the effect was not very pronounced (Fig. 4C). Especially in the absence of LH or in the presence of a low LH concentration (0.1 ng/ml), PDE10 inhibition increased testosterone production by approximately 50%.
To exclude the possibility that stimulation by the GAF agonist 7-CH-cAMP is solely a property of the recombinantly expressed enzyme, homogenates of mouse brain striatum that displayed a relatively high PDE10 content in Western blots were prepared. Therein, 7-CH-cAMP caused a 1.6-fold stimulation of cAMP-degrading activity as compared with the 3-fold stimulation observed with the recombinant enzyme under the same conditions (Fig. 4D). The EC50 value in the range of 30 nm 7-CH-cAMP was comparable with that of the recombinant enzyme. These findings demonstrate that the native enzyme from mouse brain striatum is subject to GAF domain-triggered stimulation and show that PDE10 represents a substantial portion of cAMP-degrading activity in this brain region at the tested nanomolar cAMP concentrations.
PDE11 Displays Predominant Expression in Hippocampus
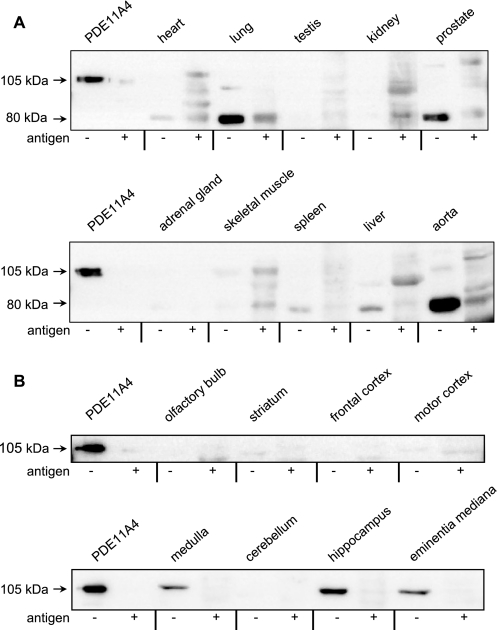
To analyze tissue distribution of PDE11, antibodies were generated by using purified PDE11 GAF domains. The obtained antibody recognized two major proteins (Fig. 5), one at the expected molecular mass of PDE11A4 at 105 kDa and a smaller protein at 80 kDa (see below). PDE11A4 (105 kDa) was found almost exclusively in brain (Fig. 5B). Detailed analysis of brain regions demonstrated highest expression levels in hippocampus and slightly lower amounts in medulla and median eminence. The smaller protein at 80 kDa produced much stronger signals and was found in lung and aorta. The size of this protein is compatible with the predicted molecular mass of PDE11A3 (8). However, on the mRNA level, PDE11A3 has been exclusively found in testis and has never been described to occur in lung or aorta (9). On the other hand, the signals obtained in lung and aorta seemed to be specific as tested by competition with the purified GAF domains used for immunization. Hence, the respective protein was purified from mouse lung by immunoaffinity chromatography using the antiserum, but the purified protein neither showed cyclic nucleotide-degrading activity nor yielded any peptide related to phosphodiesterases in mass spectrometry analyses. The 80-kDa signal thus has to be considered to be nonspecific. Taken together, substantial levels of PDE11 were only found in brain especially in hippocampus.
FIGURE 5.
PDE11 is restricted to brain and predominantly expressed in hippocampus. A newly generated antiserum against the PDE11 GAF domains was used to identify PDE11 expression in organs (A) and brain regions (B) of mice in Western blots. Human PDE11A4 (105 kDa) served as a positive control. Preabsorption of the antiserum with purified GAF domains (+ antigen; 4 μg × ml−1) was performed to check signal specificity and abolished PDE11A4 signals in hippocampus, medulla, and eminentia mediana. No indication was found that the 80-kDa signal in aorta and lung is related to PDE11 (see “Results”). Shown is one representative blot of three. Larger sections of Western blots are shown in supplemental Figs. 4 and 5.
Activation of Native PDE11 from Hippocampus
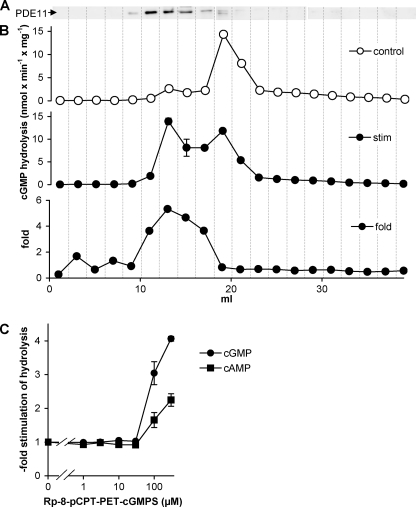
The PDE11 enzyme expressed in HEK cells was not stimulated by either cAMP or cGMP but only by a heavily modified cyclic nucleotide analogue. We wondered whether the native PDE11 in hippocampus might contain a post-translational modification or an interaction/dimerization partner that enables cGMP stimulation. Therefore, we tested whether the analogue stimulates cyclic nucleotide-degrading activity in hippocampal homogenates. However, the analogue did not have any effect in homogenates probably due to the minor contribution of PDE11 to total PDE activity in hippocampus. Hence, mouse hippocampi were fractionated by anion exchange chromatography to separate the PDEs. Fractions were analyzed in Western blots, and cGMP PDE activity was measured in the absence and presence of the GAF ligand Rp-8-pCPT-PET-cGMPS (Fig. 6A). As judged by Western blot, PDE11 eluted between 10 and 16 ml. These fractions displayed a rather low cGMP-hydrolyzing activity in the absence of the GAF ligand but the highest PDE activity in the presence of the GAF ligand with a maximal stimulation factor of 5. In concentration-response curves for the GAF ligand (Fig. 6C), cAMP- and cGMP-degrading activities were stimulated slightly less than observed with the recombinant enzyme (compare with Fig. 3, A and B), and ∼10-fold higher concentrations of the analogue were required. Nevertheless, also the native PDE11 enzyme was stimulated by the GAF domain ligand in the presence of 100 μm cAMP or cGMP, reinforcing the conclusion that the native cyclic nucleotides did not activate the enzyme.
FIGURE 6.
GAF-mediated stimulation of PDE11 enriched from hippocampus. A, hippocampi from mice were fractionated by anion exchange chromatography, and fractions were analyzed in Western blots. B, cGMP-hydrolyzing activity in the fractions was measured at a substrate concentration of 100 μm cGMP in the absence (control) and the presence (stim) of the GAF domain agonist Rp-8-pCPT-PET-cGMPS (100 μm). Highest stimulation factors (5-fold) were observed in the fractions containing PDE11 in Western blots. C, in the fraction with the highest PDE11 content, concentration-response curves for the GAF domain ligand were obtained by measuring cAMP- and cGMP-degrading activities at substrate concentrations of 100 μm. Values are means ± standard error of the mean.
DISCUSSION
The GAF domain-containing PDE10 and PDE11 were identified nearly 10 years ago, but the only experimental data in favor of cyclic nucleotides acting as GAF domain ligands of these PDEs were derived from chimeric proteins composed of PDE tandem GAF domains and the catalytic domains of Anabaena adenylyl cyclases. In these chimeric proteins, the PDE10 GAF domains conferred cAMP stimulation, whereas the PDE11 GAF domains mediated cGMP stimulation (28). In contrast, a later study of the PDE10 and PDE11 holoenzymes claimed that binding of cAMP and cGMP does not stimulate PDE10 and PDE11, respectively (27). In our study, synthetic GAF domain ligands were identified using FRET constructs of the tandem GAF domains of PDE10 or PDE11. With the identified agonists, GAF-mediated stimulation of these enzymes is demonstrated for the first time.
Stimulation of PDE10
The most potent PDE10 GAF agonist (7-CH-cAMP) stimulated the holoenzyme 3-fold. Full stimulation of the enzyme was only observed at the lowest substrate concentration (30 nm) at which allosteric binding of the substrate to the GAF domains can be considered to be negligible. More importantly, the finding that stimulation by the analogue was abolished by higher cAMP substrate concentrations demonstrates stimulation of PDE10 by cAMP itself (EC50 ∼ 0.1 μm).
A stimulation factor of 3 does not appear very impressive at first glance. On the other hand, the GAF agonist inhibited catalytic turnover at slightly higher concentrations than those required for stimulation. Thus, the observed 3-fold stimulation by the analogue probably does not represent the stimulation factor of PDE10 but is limited by competition at the catalytic domain.
In an earlier study, 7-CH-cAMP was reported to bind to PDE10 without activating the enzyme (27). However, even the positive control displayed an only 4-fold activation (PDE2 activation by 5,6-DM-cBIMP), which is much lower than the 40-fold activation observed in our laboratory (31). This indicates severely impaired enzymes and/or suboptimal assay conditions; possibly the addition of N-terminal His tags or storage at −20 °C impaired stimulation of the holoenzymes while preserving their ability to bind cyclic nucleotides. In analogy, PDE5 was termed “cGMP-binding, cGMP-specific PDE” for decades because cGMP stimulation of the enzyme was obviously lost during storage or purification (24).
Because cAMP activated PDE10, it was interesting to analyze how cAMP affects cGMP breakdown by PDE10. cAMP slightly stimulated cGMP hydrolysis (by 25% at 0.1 μm cAMP); however, slightly higher cAMP concentrations completely inhibited cGMP turnover (IC50 ∼ 0.7 μm). Interestingly, the inhibitory potency of cAMP was almost independent of the cGMP substrate concentration; this is apparently in contrast to plain competition at the catalytic domain and suggests that cAMP binding to the GAF domains increased the preference of the catalytic domain for cAMP. However, the measured cAMP inhibition curves can roughly be obtained when cGMP catalytic rates are calculated from the published km and Vmax values of PDE10.3 On the other hand, the affinity of cAMP to the catalytic domains cannot be determined without stimulating the enzyme via its GAF domains, and the published enzymatic constants already account for the GAF-mediated cAMP effects. The similarity of the EC50 for GAF-mediated cAMP stimulation (0.1 μm) and the published km values suggests that the published km values are substantially determined by the underlying GAF-mediated cAMP stimulation.
PDE10 binds cAMP at its second GAF domain, termed GAF-B (27, 28), which is reminiscent of PDE2 in which cGMP is bound to GAF-B as well (19). Based on the crystal structure of PDE2, a general mechanism for GAF-dependent PDE activation has been proposed (34). In the basal, non-activated state, the catalytic pockets of the dimerized catalytic domains are packed against each other. Binding of cGMP to GAF-B rotates the catalytic domains with further subtle conformational changes (“swing-out of the H loop”), thereby enabling substrate access. It is tempting to speculate that such a conformational change in PDE10 may also increase the preference of the catalytic domain for cAMP. However, as structural information about PDE10 is limited to isolated catalytic or GAF domains (35, 36), definitive conclusions await resolution of the stimulated and unstimulated holoenzyme structures, which may be facilitated by the GAF agonist identified in the present study.
PDE10 is predominantly expressed in testis and the striatum. In testis, the isobutylmethylxanthine-insensitive PDE8 has been shown to be a major regulator of LH-induced testosterone production (37), and the small effect elicited by the PDE10 inhibitor in our study suggests that PDE10 accounts for a fraction of the remaining isobutylmethylxanthine-sensitive PDEs in this pathway. In striatum, the stimulation of cAMP-degrading activity by the cAMP analogue 7-CH-cAMP demonstrates that GAF-mediated stimulation is a feature of the native PDE10 too and that PDE10 is responsible for a substantial amount of cAMP-degrading activity in striatum.
In summary, PDE10 is potently stimulated by cAMP, and the GAF-mediated stimulation alters the enzymatic parameters such that the enzyme acts as a cAMP-specific phosphodiesterase. Only in cells with extremely low cAMP concentrations, PDE10 can contribute to cGMP degradation. In those cells, a small increase in cAMP to 1 μm could inhibit cGMP breakdown by PDE10.
Stimulation of PDE11
The GAF domains of PDE11 mediate stimulation of the enzyme. With the cGMP analogue Rp-8-pCPT-PET-cGMPS, an up to 5-fold stimulation of cGMP or cAMP hydrolysis is observed. Interestingly, the stimulatory effect of Rp-8-pCPT-PET-cGMPS occurs at high substrate concentrations (100 μm cAMP or cGMP). The data therefore imply that the enzyme is not activated by the physiological nucleotides cAMP and cGMP. If cAMP or cGMP had activated PDE11 via the GAF domains, stimulation by the GAF agonist should not be detectable at such high substrate concentrations as was shown for PDE2, PDE5, and PDE10 (Ref. 31 and above). In accordance with the conclusion that cGMP does not activate PDE11, cAMP hydrolysis was not activated by cGMP. Rather, a plain competitive inhibition of cAMP breakdown by equimolar cGMP concentrations was observed. Why did the GAF ligand of PDE11 yield lower stimulation factors at lower substrate concentrations? Obviously, the GAF ligand also has some affinity to the catalytic domain as can be seen from the bell-shaped concentration-response curves and inhibits substrate turnover more efficiently at lower substrate concentrations, thereby diminishing the stimulation factor (see Fig. 3, A and B).
The identification of an analogue stimulating PDE11 only at high substrate concentrations is reminiscent of the stimulation of the photoreceptor PDE6 by the PDE inhibitors dipyridamole and M&B 22,948 at high substrate concentrations (38). However, the paradoxical stimulatory effect of PDE inhibitors under millimolar substrate conditions observed for PDE6 required preincubation of PDE6 with the inhibitors in the absence of cGMP, whereas the presence of cGMP during the preincubation abolished the stimulatory effect. This is in contrast to the effect of the cGMP analogue on PDE11, which did not require access to the enzyme in the absence of cGMP but exerted its action in the presence of cGMP or cAMP during the PDE assays. Hence, the paradoxical stimulation of PDE6 by competitive inhibitors is a distinct phenomenon that is unrelated to the Rp-8-pCPT-PET-cGMPS-mediated stimulation of PDE11.
Why did cGMP bind to the FRET constructs with a reasonably high affinity (EC50 ∼ 0.3 μm) but fail to stimulate PDE11 holoenzyme? PDE11A4 contains a relatively long N terminus preceding the GAF domains (197 amino acids). In chimeric proteins composed of the N-terminal part of PDE11A4 including the GAF domains and the catalytic domain of Anabaena adenylyl cyclases, the N-terminal part preceding the GAF domains had a tremendous effect on the ability of cGMP to stimulate the chimera. Truncation of 196 N-terminal amino acids increased the cGMP affinity 20-fold (39). However, deletion of these 196 amino acids in the PDE11A4 holoenzyme abolished stimulation by the cGMP analogue but did not enable cGMP stimulation (data not shown). Irrespective of the uncertainty resulting from conclusions based on mutant proteins, it stands to reason that the different protein environments of the GAF domains in Anabaena adenylyl cyclases, FRET constructs, or the PDE11 holoenzyme strongly determine the GAF domain function.
The observation that the GAF domains are capable of stimulating PDE11 but do not mediate stimulation by cGMP raises the question whether the enzyme is regulated by the GAF domains in vivo. On the one hand, GAF domains of other proteins have been reported to mediate regulation by other small molecules (21); therefore, PDE11 may well be activated by another so far unidentified small molecule. On the other hand, a post-translational modification or an additional interaction partner as a prerequisite for PDE11 activation by cGMP cannot be ruled out, although even the native PDE11A4 enriched from mouse hippocampi was not stimulated by cGMP but only by the cGMP analogue identified in this study. In summary, GAF domains, despite their homology, confer a variety of regulatory properties to PDEs: fast and sustained cGMP-induced activation to PDE2 and PDE5, respectively; cAMP-mediated inhibition of cGMP turnover to PDE10; and hypothetically activation by non-cyclic nucleotide ligands to PDE11.
Supplementary Material
Acknowledgment
We gratefully acknowledge the technical assistance of Ulla Krabbe, Arkadius Pacha, Erika Mannheim, and Caroline Vollmers.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ko1157 and the Kommission für Finanzautonomie und Ergänzungsmittel of the Medical Faculty (KOFFER). H. G. G. is the chief executive officer and F. S. is the head of research and development of BIOLOG Life Science Institute, which sells cyclic nucleotide analogues for research purposes.

This article contains supplemental Tables 1 and 2 and Figs. 1–5.
km (cGMP) ∼ 10 μm; km (cAMP) ∼ 0.1 μm; Vmax (cGMP) ∼ 5 × Vmax (cAMP).
- PDE
- phosphodiesterase
- CFP
- cyan fluorescent protein
- LH
- luteinizing hormone
- YFP
- yellow fluorescent protein
- DTT
- dl-dithiothreitol.
REFERENCES
- 1. Bender A. T., Beavo J. A. (2006) Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol. Rev. 58, 488–520 [DOI] [PubMed] [Google Scholar]
- 2. Conti M., Beavo J. (2007) Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 3. Francis S. H., Corbin J. D., Bischoff E. (2009) Cyclic GMP-hydrolyzing phosphodiesterases. Handb. Exp. Pharmacol. 191, 367–408 [DOI] [PubMed] [Google Scholar]
- 4. Fujishige K., Kotera J., Michibata H., Yuasa K., Takebayashi S., Okumura K., Omori K. (1999) Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A). J. Biol. Chem. 274, 18438–18445 [DOI] [PubMed] [Google Scholar]
- 5. Loughney K., Snyder P. B., Uher L., Rosman G. J., Ferguson K., Florio V. A. (1999) Isolation and characterization of PDE10A, a novel human 3′,5′-cyclic nucleotide phosphodiesterase. Gene 234, 109–117 [DOI] [PubMed] [Google Scholar]
- 6. Soderling S. H., Bayuga S. J., Beavo J. A. (1999) Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc. Natl. Acad. Sci. U.S.A. 96, 7071–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fawcett L., Baxendale R., Stacey P., McGrouther C., Harrow I., Soderling S., Hetman J., Beavo J. A., Phillips S. C. (2000) Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc. Natl. Acad. Sci. U.S.A. 97, 3702–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hetman J. M., Robas N., Baxendale R., Fidock M., Phillips S. C., Soderling S. H., Beavo J. A. (2000) Cloning and characterization of two splice variants of human phosphodiesterase 11A. Proc. Natl. Acad. Sci. U.S.A. 97, 12891–12895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuasa K., Kotera J., Fujishige K., Michibata H., Sasaki T., Omori K. (2000) Isolation and characterization of two novel phosphodiesterase PDE11A variants showing unique structure and tissue-specific expression. J. Biol. Chem. 275, 31469–31479 [DOI] [PubMed] [Google Scholar]
- 10. O'Connor V., Genin A., Davis S., Karishma K. K., Doyère V., De Zeeuw C. I., Sanger G., Hunt S. P., Richter-Levin G., Mallet J., Laroche S., Bliss T. V., French P. J. (2004) Differential amplification of intron-containing transcripts reveals long term potentiation-associated up-regulation of specific Pde10A phosphodiesterase splice variants. J. Biol. Chem. 279, 15841–15849 [DOI] [PubMed] [Google Scholar]
- 11. Kotera J., Sasaki T., Kobayashi T., Fujishige K., Yamashita Y., Omori K. (2004) Subcellular localization of cyclic nucleotide phosphodiesterase type 10A variants, and alteration of the localization by cAMP-dependent protein kinase-dependent phosphorylation. J. Biol. Chem. 279, 4366–4375 [DOI] [PubMed] [Google Scholar]
- 12. Yuasa K., Ohgaru T., Asahina M., Omori K. (2001) Identification of rat cyclic nucleotide phosphodiesterase 11A (PDE11A): comparison of rat and human PDE11A splicing variants. Eur. J. Biochem. 268, 4440–4448 [DOI] [PubMed] [Google Scholar]
- 13. Seeger T. F., Bartlett B., Coskran T. M., Culp J. S., James L. C., Krull D. L., Lanfear J., Ryan A. M., Schmidt C. J., Strick C. A., Varghese A. H., Williams R. D., Wylie P. G., Menniti F. S. (2003) Immunohistochemical localization of PDE10A in the rat brain. Brain Res. 985, 113–126 [DOI] [PubMed] [Google Scholar]
- 14. Yuasa K., Kanoh Y., Okumura K., Omori K. (2001) Genomic organization of the human phosphodiesterase PDE11A gene. Evolutionary relatedness with other PDEs containing GAF domains. Eur. J. Biochem. 268, 168–178 [DOI] [PubMed] [Google Scholar]
- 15. Loughney K., Taylor J., Florio V. A. (2005) 3′,5′-Cyclic nucleotide phosphodiesterase 11A: localization in human tissues. Int. J. Impot. Res. 17, 320–325 [DOI] [PubMed] [Google Scholar]
- 16. D'Andrea M. R., Qiu Y., Haynes-Johnson D., Bhattacharjee S., Kraft P., Lundeen S. (2005) Expression of PDE11A in normal and malignant human tissues. J. Histochem. Cytochem. 53, 895–903 [DOI] [PubMed] [Google Scholar]
- 17. Kelly M. P., Logue S. F., Brennan J., Day J. P., Lakkaraju S., Jiang L., Zhong X., Tam M., Sukoff Rizzo S. J., Platt B. J., Dwyer J. M., Neal S., Pulito V. L., Agostino M. J., Grauer S. M., Navarra R. L., Kelley C., Comery T. A., Murrills R. J., Houslay M. D., Brandon N. J. (2010) Phosphodiesterase 11A in brain is enriched in ventral hippocampus and deletion causes psychiatric disease-related phenotypes. Proc. Natl. Acad. Sci. U.S.A. 107, 8457–8462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aravind L., Ponting C. P. (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22, 458–459 [DOI] [PubMed] [Google Scholar]
- 19. Martinez S. E., Beavo J. A., Hol W. G. (2002) GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol. Interv. 2, 317–323 [DOI] [PubMed] [Google Scholar]
- 20. Hurley J. H. (2003) GAF domains: cyclic nucleotides come full circle. Sci. STKE 2003, PE1. [DOI] [PubMed] [Google Scholar]
- 21. Zoraghi R., Corbin J. D., Francis S. H. (2004) Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol. Pharmacol. 65, 267–278 [DOI] [PubMed] [Google Scholar]
- 22. Schultz J. E. (2009) Structural and biochemical aspects of tandem GAF domains. Handb. Exp. Pharmacol. 191, 93–109 [DOI] [PubMed] [Google Scholar]
- 23. Beavo J. A., Hardman J. G., Sutherland E. W. (1971) Stimulation of adenosine 3′,5′-monophosphate hydrolysis by guanosine 3′,5′-monophosphate. J. Biol. Chem. 246, 3841–3846 [PubMed] [Google Scholar]
- 24. Rybalkin S. D., Rybalkina I. G., Shimizu-Albergine M., Tang X. B., Beavo J. A. (2003) PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J. 22, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mullershausen F., Friebe A., Feil R., Thompson W. J., Hofmann F., Koesling D. (2003) Direct activation of PDE5 by cGMP: long-term effects within NO/cGMP signaling. J. Cell Biol. 160, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corbin J. D., Blount M. A., Weeks J. L., 2nd, Beasley A., Kuhn K. P., Ho Y. S., Saidi L. F., Hurley J. H., Kotera J., Francis S. H. (2003) [3H]Sildenafil binding to phosphodiesterase-5 is specific, kinetically heterogeneous, and stimulated by cGMP. Mol. Pharmacol. 63, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 27. Matthiesen K., Nielsen J. (2009) Binding of cyclic nucleotides to phosphodiesterase 10A and 11A GAF domains does not stimulate catalytic activity. Biochem. J. 423, 401–409 [DOI] [PubMed] [Google Scholar]
- 28. Gross-Langenhoff M., Hofbauer K., Weber J., Schultz A., Schultz J. E. (2006) cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J. Biol. Chem. 281, 2841–2846 [DOI] [PubMed] [Google Scholar]
- 29. Russwurm M., Mullershausen F., Friebe A., Jäger R., Russwurm C., Koesling D. (2007) Design of fluorescence resonance energy transfer (FRET)-based cGMP indicators: a systematic approach. Biochem. J. 407, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russwurm C., Zoidl G., Koesling D., Russwurm M. (2009) Dual acylation of PDE2A splice variant 3: targeting to synaptic membranes. J. Biol. Chem. 284, 25782–25790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jäger R., Schwede F., Genieser H. G., Koesling D., Russwurm M. (2010) Activation of PDE2 and PDE5 by specific GAF ligands: delayed activation of PDE5. Br. J. Pharmacol. 161, 1645–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Habert R., Lejeune H., Saez J. M. (2001) Origin, differentiation and regulation of fetal and adult Leydig cells. Mol. Cell. Endocrinol. 179, 47–74 [DOI] [PubMed] [Google Scholar]
- 33. Schmidt C. J., Chapin D. S., Cianfrogna J., Corman M. L., Hajos M., Harms J. F., Hoffman W. E., Lebel L. A., McCarthy S. A., Nelson F. R., Proulx-LaFrance C., Majchrzak M. J., Ramirez A. D., Schmidt K., Seymour P. A., Siuciak J. A., Tingley F. D., 3rd, Williams R. D., Verhoest P. R., Menniti F. S. (2008) Preclinical characterization of selective phosphodiesterase 10A inhibitors: a new therapeutic approach to the treatment of schizophrenia. J. Pharmacol. Exp. Ther. 325, 681–690 [DOI] [PubMed] [Google Scholar]
- 34. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., Menniti F. S. (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the x-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H., Liu Y., Hou J., Zheng M., Robinson H., Ke H. (2007) Structural insight into substrate specificity of phosphodiesterase 10. Proc. Natl. Acad. Sci. U.S.A. 104, 5782–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Handa N., Mizohata E., Kishishita S., Toyama M., Morita S., Uchikubo-Kamo T., Akasaka R., Omori K., Kotera J., Terada T., Shirouzu M., Yokoyama S. (2008) Crystal structure of the GAF-B domain from human phosphodiesterase 10A complexed with its ligand, cAMP. J. Biol. Chem. 283, 19657–19664 [DOI] [PubMed] [Google Scholar]
- 37. Vasta V., Shimizu-Albergine M., Beavo J. A. (2006) Modulation of Leydig cell function by cyclic nucleotide phosphodiesterase 8A. Proc. Natl. Acad. Sci. U.S.A. 103, 19925–19930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gillespie P. G., Beavo J. A. (1989) Inhibition and stimulation of photoreceptor phosphodiesterases by dipyridamole and M&B 22,948. Mol. Pharmacol. 36, 773–781 [PubMed] [Google Scholar]
- 39. Gross-Langenhoff M., Stenzl A., Altenberend F., Schultz A., Schultz J. E. (2008) The properties of phosphodiesterase 11A4 GAF domains are regulated by modifications in its N-terminal domain. FEBS J. 275, 1643–1650 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.