Abstract
F1-ATPase is the smallest mechanical motor known. Tentoxin, a cyclic peptide produced by phytopathogenic fungi, inactivates the F1 motor in sensitive plants at nanomolar to micromolar concentrations, whereas higher concentrations surpass the natural activity of the enzyme. Single molecule studies now have clarified the molecular steps involved in both processes. Inactivation delays the dwell time of a single step in the complete 360° turn and results in an asymmetric rotation of the central rotor subunit. In contrast, rotation in the stimulated F1 particle is smooth and accompanied by strongly reduced ADP inhibition. Our study provides for the first time the direct observation of a noncompetitively inhibited state of the enzyme and directly visualizes the regulation of the molecular motor by an external natural compound. In addition, the ADP release step during catalysis was revealed by analysis of the single molecule rotation behavior. Hence, tentoxin is a sophisticated molecular tool to mark and control certain catalytic steps within the reaction pathway of the molecular F1 motor.
The catalytic moiety of the ATP synthase (F1)2 works like a molecular motor, and ATPase activity is coupled to rotation of the central γ subunit in the catalytic α3β3 core (1, 2). Rotation catalysis of F1-ATP synthase was first suggested by P. D. Boyer in his “binding-change mechanism” concept (3), and was strengthened by the x-ray crystal structure of mitochondrial F1 (4). Direct observation of the γ rotation in single F1 particles revealed the orientation of the central γ subunit in relation to the ambient three β subunits, which are taking different individual conformations at a given time during catalysis (1). The rotation was found to proceed in 120° steps, each driven by the consumption of one ATP molecule (5). The individual steps could be further divided into 80° and 40° substeps that are related to ATP binding/ADP release and phosphate release, respectively (6).
Tentoxin, a phytotoxin produced by fungi of the Alternaria species, blocks ATP hydrolysis in certain chloroplast F1 (CF1) (7–10) and causes chlorosis of the sensitive plants. The tentoxin-binding site is located at the contact surface between an α and a β subunit close to the N-terminal β-barrel structure on the α3β3 hexagon. The crystal structure of spinach chloroplast F1 complexed with tentoxin showed that the catalytic site on the αβ pair with bound toxin is in a closed conformation (11). Thus, it was deduced that tentoxin inhibits the activity of the enzyme by blocking the conformational shifts at the catalytic sites. However, which of the catalytic processes is affected could not be determined in detail so far. The stimulatory effect of tentoxin is even less understood on the molecular level, although it is accepted that binding of a second or maybe third toxin molecule induces the stimulation (9, 10). Previous single molecule experiments using genetically engineered F1 from thermophilic Bacillus PS3 (TF1) suggest that the high stimulatory concentrations of the toxin correlate with a reduced inhibition of the enzyme by ADP (12). However, the mutant TF1 showed only a partial relief from tentoxin inhibition even at high toxin concentrations (13).
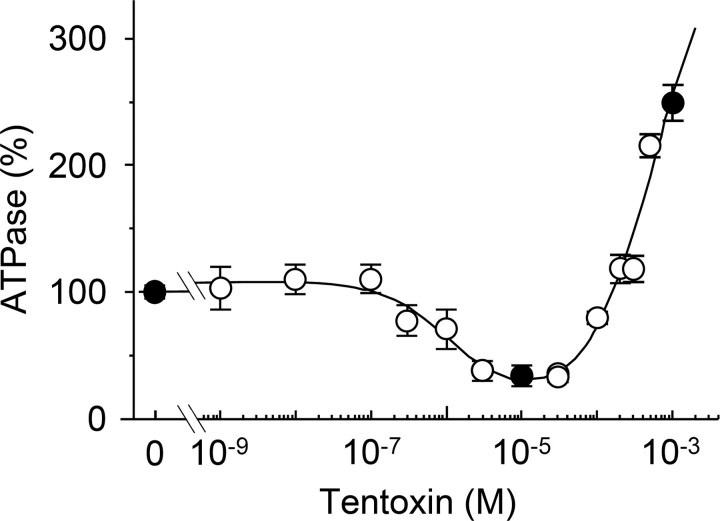
To clarify both the inhibitory and stimulatory effects of tentoxin on the single molecule level, we adopted the F1 of the cyanobacterium Thermosynechococcus elongatus BP-1 (14), which is inhibited at 5–10 μm tentoxin and substantially stimulated above 100 μm (see Fig. 1). The magnitude of inhibition and stimulation agrees well with data obtained for spinach chloroplast F1 (15) but showed a higher Ki (0.8 ± 0.5 μm) as reported for other cyanobacterial F1 complexes (16). The observed difference in the inhibition constant might be attributed to a Ser-Ala substitution in the tentoxin-binding site of the cyanobacterial enzymes, so that the hydrogen bonding with β D83 weakens the tentoxin-enzyme interaction (8, 11).
FIGURE 1.
Steady state ATP hydrolysis activity of the biotinylated α3β3γCys125/222 complex in the presence of tentoxin. ATP hydrolysis activity was measured with an ATP-regenerating system coupled to the oxidation of NADH. The samples were preincubated with the indicated toxin concentrations at 22 °C for at least 30 min, and tentoxin was also included in the reaction buffer. The reaction was measured at 25 °C for 5 min in the presence of 2 mm ATP and calculated relatively to the conditions without tentoxin (2.6 ± 0.2 μmol of Pi released per mg of protein/min), which was set as 100%. Black circles indicate the toxin concentrations used in our single molecule experiments. The curve shown in the figure was obtained by the least square analysis of the data using an equation proposed by Santolini et al. for the analysis of tentoxin binding (Equations 6–8 in Ref. 10). From this analysis, an apparent dissociation constant of 1.08 μm was obtained for tentoxin inhibition and of 565 μm for tentoxin stimulation.
By using the cyanobacterial F1 subcomplex, we studied the inhibition and stimulation modes of F1 by tentoxin at the single molecule level and successfully clarified the molecular steps involved in both processes. Inhibiting concentrations of tentoxin affect the ADP release step at the catalytic sites, whereas rotation of the γ subunit was smooth in stimulated F1 particles because of a reduced ADP inhibition state, but nevertheless showed a slightly decreased rotating speed compared with the noninhibited enzyme. Thus the phytotoxin tentoxin is certainly a valuable tool to study catalytic steps within the reaction pathway of the molecular F1 motor.
EXPERIMENTAL PROCEDURES
Materials—Biotin-PEAC5-maleimide was purchased from Dojindo (Kumamoto, Japan). Tentoxin, ATP, phosphoenolpyruvate, and bovine serum albumin were obtained from Sigma. Pyruvate kinase, lactate dehydrogenase, and NADH were purchased from Roche Diagnostics. Other chemicals were of the highest grade commercially available.
Expression Plasmid—Expression plasmid for the α3β3γ complex of T. elongates BP-1 was constructed as described (14). For selective labeling of the γ subunit, suitable for single molecule experiments, all natural cysteine residues were substituted by serine using the Mega-primer method (17). Two additional cysteines were introduced in position 125 and 222. Substitution of A125C was done using the following mutation primer, 5′-GGGATAGTCACGGCGCTGGAAATATTGGCATGCCTTGCGACCC-3′, and Val-222 was changed to Cys by applying mutation primer 5′-CATTTGGAAGTCAACCGCGAGAAAACCTCGACGCTGCCCGCTCTGC-3′. The plasmid was named pTR19FWR and was used for the expression of α3β3γCys125/222.
Protein Preparation—Expression, purification, and labeling of α3β3γCys125/222 with biotin-PEAC5-maleimide were done as described before (14) and confirmed by PAGE in the presence of 0.1% (w/v) SDS (SDS-PAGE) and following immunoblot analysis (supplemental Fig. S5).
ATPase Activity Measurement—ATPase activity was monitored in the presence of an ATP-regenerating system (18) in 50 mm HEPES-KOH, pH 8.0, 100 mm KCl, 2 mm MgCl2, 2 mm ATP, 50 μg/ml pyruvate kinase, 50 μg/ml lactate dehydrogenase, 2 mm phosphoenolpyruvate, and 0.2 mm NADH. The assay was carried out at 25 °C. The rate of ATP hydrolysis was determined by monitoring the decrease in NADH absorption at 340 nm after addition of purified α3β3γCys125/222 using a spectrophotometer V-550 (Jasco, Tokyo, Japan). When the substrate ATPγS (5 mm) was used or in measurements in the presence of ADP (5 mm), the activity was monitored as the liberation of phosphate determined by a colorimetric method (19). Preincubation of the enzyme with tentoxin was performed for 30 min in 50 mm HEPES-KOH, pH 8.0, 100 mm KCl, 2 mm MgCl2 with the indicated toxin concentrations.
Rotation Assay—Rotation assays were done as described (14) with minor adjustments. Biotinylated complexes (10 μl) in 50 mm HEPES-KOH, pH 8.0, 100 mm KCl, and 1% (w/v) bovine serum albumin were infused into a flow chamber and were incubated for 2 min at room temperature. The flow chamber was then washed with 50 μl of 50 mm HEPES-KOH, pH 8.0, 100 mm KCl to remove unattached complexes. Streptavidin-coated beads (100, 210, or 340 nm diameter) in 50 mm HEPES-KOH, pH 8.0, 100 mm KCl, and 1% (w/v) bovine serum albumin were infused into the flow chamber and were incubated for 15 min. Rotation was initiated by addition of 80 μl of assay buffer (50 mm HEPES-KOH, pH 8.0, 100 mm KCl, 0.8 mm MgCl2, 0.25 or 20 μm ATP, 100 μg/ml pyruvate kinase, and 2 mm phosphoenolpyruvate) after washing the flow chamber with 50 μl of 50 mm HEPES-KOH, pH 8.0, and 100 mm KCl. In experiments with tentoxin, the indicated concentration of the toxin was included in the assay buffer. Rotation of the duplex beads attached on the γ subunit was monitored with a conventional optical microscope type IX 71 (Olympus, Tokyo, Japan) with a 100× objective lens. Images were recorded with a digital video recorder. For fast recording we used a dark-field microscope (IX 71, Olympus) equipped with a mercury lamp and a high speed camera (Hi-DcamII, NAC Image Technology, Tokyo) at 2,000 frames/s. Recorded images were analyzed by custom software as described (20).
RESULTS AND DISCUSSION
Low Concentrations of Tentoxin Inhibit the ADP Release Step—For our single molecule experiments, we attached streptavidin-coated polystyrene beads (diameter ∼210 nm) via the biotinylated γ subunit to the α3β3γ complex and observed the ATP-driven rotation of the γ subunit at 250 nm ATP using a phase contrast microscope. Rotation was stepwise, and three stop positions could be clearly distinguished (Fig. 2A and supplemental movies 1 and 3). These positions were separated by 120° and correspond to the positions where the enzyme waits for the substrate ATP (20–22). The influence of tentoxin on the rotation pattern was studied in buffer exchange experiments. A rotating particle was then observed twice for an interval of 5 min before and after tentoxin addition to examine the influence of the toxin. Exchange of the buffer solution on its own did not alter the rotation pattern (supplemental Fig. S1). In contrast, rotation of γ became asymmetric, and one dwell time in a complete 360° turn became substantially delayed (Fig. 2A and supplemental movies 2 and 4) when inhibitory concentrations of tentoxin (10 μm) were supplied. This is in contrast to rotation experiments performed with the engineered TF1, which related tentoxin inhibition to a complete stop of the γ rotation (12). Rotation of the cyanobacterial enzyme continued after infusion of inhibitory concentrations of tentoxin, but the rotation speed significantly decreased (31.8 ± 7.9% compared with the enzyme without tentoxin). Bulk ATPase activity measurements showed almost the same decrease in activity (32.6 ± 2.6%) at this tentoxin concentration. To answer the question whether a fast dissociation and rebinding of the inhibitor could be in charge of the partial inactivation, we determined the kinetic properties for the first tentoxin-binding site in bulk ATPase activity measurements (supplemental Fig. S2). At 10 μm tentoxin, the time constant for binding was 52 s, whereas the time constant for dissociation was 714 s. The resulting kinetic constants were kon = 1.9 × 103 m-1 s-1 and koff = 1.4 × 10-3 s-1. The kon was 1 order of magnitude slower compared with those of the spinach CF1, whereas koff was faster than that for CF1 (10). Nevertheless, binding was fast enough to proceed during rotation experiments, and dissociation was too slow to explain the suppressed rotation. Moreover, 9 of 14 particles stopped at the same mostly inhibited site, and four changed their position once during the observation interval (300 s). One particle showed two equal inhibited positions and was not included for further analysis. The three resting positions observed during tentoxin inhibition matched well with the ATP waiting positions assigned before buffer exchange (Fig. 2, A, insets, and B). For calculation of angle differences, data of 13 independent particle observations were merged and aligned at the most inhibited position. No significant change or shift was detected for all three positions as follows: -1.6 ± 10.7° at 0° (set arbitrarily for the most inhibited position), -0.6 ± 12.8° at 120°, and 0.5 ± 11.3° at 240° (Fig. 2B). In the most recent model of F1 rotation (6), ATP binding and ADP release are the events that take place at these positions. Our precise determination of the resting positions eliminated the possibility that other catalytic processes such as ATP hydrolysis or phosphate release are affected by tentoxin. Bulk phase activity measurements of F1 using the slowly hydrolyzable ATP analogue, ATPγS, were performed to confirm this conclusion. Under these conditions ATPγS hydrolysis was the rate-limiting step, and no inhibitory (108.3 ± 23.3% activity) or stimulatory (111.1 ± 15.0%) effect of tentoxin could be detected when 10 μm tentoxin for inhibition or 1 mm for stimulation were supplied. Furthermore, detailed analysis of the rotation speed of γ between dwell periods at 20 μm ATP showed that torque generation is not affected by tentoxin (supplemental Fig. S3). To further specify which catalytic process is affected by the toxin, we determined the Km value for ATP in the presence of different toxin concentrations. In accordance with reports on spinach F1 (23), the inhibitor acts noncompetitively with respect to ATP, and the Km value for ATP did not change in response to the tentoxin concentrations (Km values for our enzyme were 115.2 ± 35.1 μm ATP without tentoxin; 97.3 ± 14.6 μm ATP at 2 μm and 92.3 ± 4.3 μm ATP at 10 μm toxin; see supplemental Fig. S4). This suggests that not ATP binding but another event such as ADP release is affected by the toxin. This interpretation is supported by the results of Hu et al. (24), who found that tentoxin inhibits the release of ADP after binding of a nonhydrolyzable ATP analogue.
FIGURE 2.
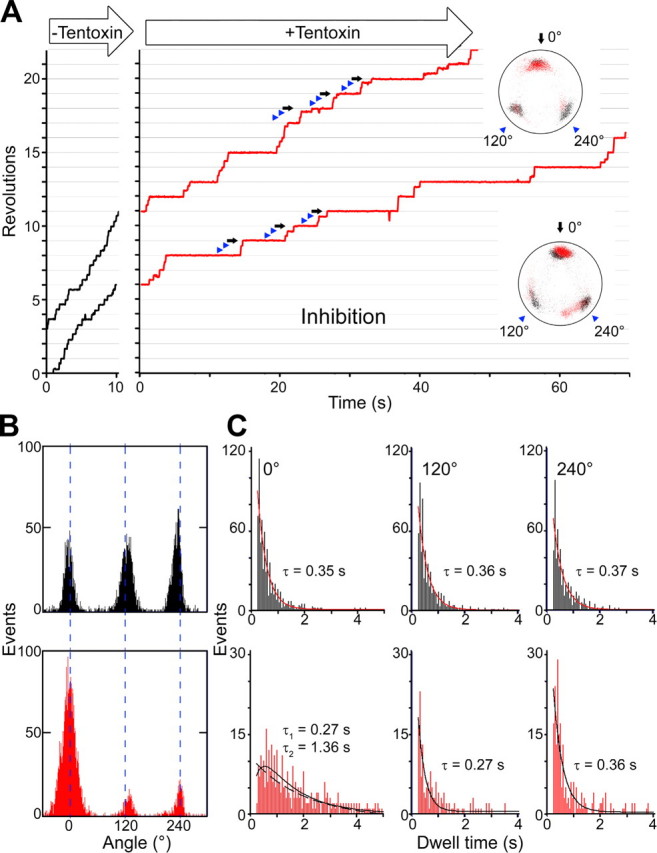
Rotation under inhibitory concentrations of tentoxin. A, particles (diameter 210 nm) were observed at 250 nm ATP for 5 min. After solution exchange with buffer containing 10 μm tentoxin, the same particles were observed for a further 5 min. Insets show the trace of the bead centroid before (black dots) and after tentoxin infusion (red dots). Under inhibitory conditions, revolution was asymmetric with one long pause (black arrows) and two short pauses (blue arrowheads) per 360° turn. B, angle distribution of one rotating particle at 250 nm ATP before (black bars) and after (red bars) tentoxin infusion was analyzed. Change or shift of positions was as follows: -1.6 ± 10.7° at 0° (set arbitrarily for the most inhibited position), -0.6 ± 12.8° at 120°, and 0.5 ± 11.3° at 240°. C, dwell times at 0, 120, and 240° before (black bars) and after (red bars) tentoxin infusion are plotted and analyzed. Total number of pauses were 736 for 0°, 629 for 120°, and 603 for 240° before solution exchange and 342 for 0°, 219 for 120°, and 215 for 240° after solution exchange (data from nine particles were aligned at the most inhibited position, which was set as 0°). All dwell time distributions could be fitted with a single exponential decay, besides the 0° position after tentoxin infusion, which shows a double exponential decay with R = 0.81 (dashed line represent a single exponential decay with R = 0.78).
Thereafter, the dwell times at all three positions obtained with and without tentoxin were determined (Fig. 2C). The distribution of pause length always followed a single exponential decay with time constants similar to the ATP binding rate of TF1 (25). The dwell time distribution at the mostly inhibited position (0°), however, gave a more accurate fit with a double exponential equation. The first time constant (0.27 s) was similar to that of the other resting positions or those of the noninhibited enzyme, whereas the second constant (1.36 s) was significantly slower and clearly attributed to tentoxin. This strengthened our idea that tentoxin affects an independent event like ADP release but not the ATP-binding event. This newly identified event was not reported by dwell time analysis of single molecule experiments so far. The two independent events located at 0° that were observed in our single molecule experiments do not fit the recent mechanistic model proposed by Mao and Weber (26), which attributes the ADP release step to the 40° substep. However, our data comply with a recent rotation model of Adachi et al. (6). Irrespective of their fit to a mechanistic model, the data demonstrate that tentoxin can be a useful tool to analyze specific steps during the rotation and to gain further insight into the basic mechanistic operation mode of F1-ATPases.
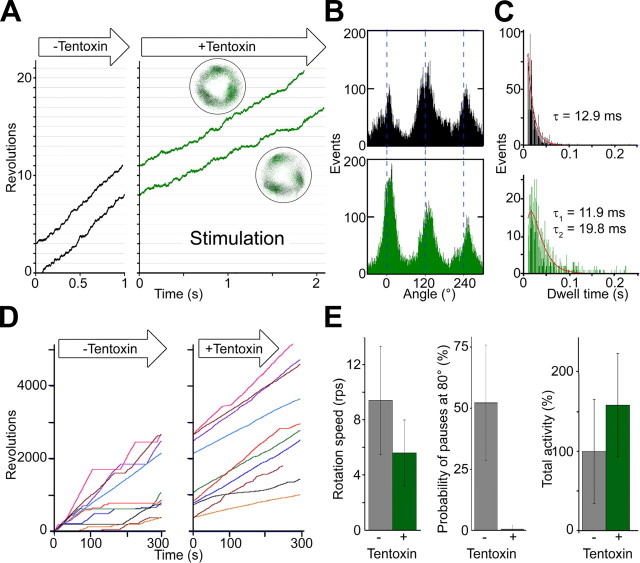
High Concentrations of Tentoxin Relieve the ADP Inhibition—Next, we studied the stimulation of the F1 complex by tentoxin. To this end, rotation experiments were carried out at 20 μm ATP because the time required for the binding of the second or maybe the third toxin molecule depends on the catalytic state of the enzyme (10), and stimulation was not observed at concentrations below 5 μm ATP (not shown). At 20 μm ATPase activity was stimulated to 140–190%. According to the kon ∼1 × 107 m-1 s-1 determined by the dwell time analysis in Fig. 2C before tentoxin infusion, the substrate binding at this ATP concentration takes about 5 ms. To visualize the stepwise ATP-driven rotation, we used a combination of smaller beads (100 nm diameter) and a high speed camera (time resolution 2,000 frames/s). The rotation speed at 1 mm tentoxin increased compared with those of the inhibited enzyme, but achieved only 60–70% of the speed obtained with the noninhibited enzyme (without toxin) (Fig. 3A). Most remarkably the rotation of stimulated F1 was smooth, and no clear long pauses could be detected. The three resting positions observed with the high speed camera did not shift (3.2 ± 12.7°) after tentoxin infusion (Fig. 3B). During long observation periods, we sometimes found a transition to a slower speed with a single distinct resting position (not shown). For the spinach enzyme, binding and dissociation of stimulatory tentoxin molecule(s) to the binding site(s) take only a few seconds in the presence of ATP (10). We therefore assume that the observed transition was because of release and rebinding of stimulatory tentoxin molecules.
FIGURE 3.
Rotation under stimulatory concentrations of tentoxin. A, close-up views of rotation observed at 20 μm ATP before (left part) and after buffer exchange with 1 mm tentoxin (right part) are indicated. Insets show the trace of the beads centroid before (black dots) and after tentoxin infusion (green dots). B, angle distribution of one rotating particle at 20 μm ATP before (black bars) and after (green bars) tentoxin infusion are indicated. C, dwell times before (black bars) and after (green bars) tentoxin infusion are plotted and analyzed. Total number of pauses was 1,070 before and 667 after solution exchange (data from three merged particles). Dwell time distribution before tentoxin infusion could be fitted with a single exponential decay. Stimulated particles show a double exponential decay. D, long term observation of F1 rotation with and without stimulatory tentoxin concentrations (1 mm) are indicated. Rotating particles were observed twice at 20 μm ATP for 5 min. Before tentoxin infusion, rotation of the beads often showed long pauses above 5 s at 80°. Stimulated particles show slower rotation speed but virtually no long pauses (>5 s), which lead to an increased net activity calculated as total revolutions per total observation time (summarized in E).
Because the stimulated F1 showed smooth rotation, we could not distinguish differences among the three resting positions (Fig. 3B). Hence, all observed dwell times were merged and fitted with an exponential equation (Fig. 3C). The dwell time distribution without tentoxin followed a single exponential decay with a time constant of 12.9 ms. Because we could detect three distinct resting positions separated by 120° and catalytic dwell periods (the process of ATP hydrolysis at the catalytic site) are still faster than the ATP-binding event, we assign these pauses as ATP waiting dwell periods. As in tentoxin inhibition, the dwell time distribution changed after toxin infusion and followed a double exponential decay. The first time constant (11.9 ms) was similar to the constant before buffer exchange (12.9 ms), whereas the second (19.8 ms) was slightly slower (Fig. 3C). Taken together, high concentrations of tentoxin seem to partially relieve the inhibition event after substrate binding. However, it is still unclear how the binding of a second or third tentoxin molecule may alter the conformation or relief of the inhibition as a high resolution structure of a stimulatory F1-tentoxin complex was not resolved so far.
In contrast to the reduced rotation speed of stimulated F1 complexes, the bulk ATPase activity is significantly higher than that without tentoxin (Fig. 1). Rotation experiments with a tentoxin-sensitive TF1 mutant (12) demonstrated that high toxin concentrations reduce the probability of long rotation pauses (>20 s), which were attributed to ADP inhibition. ADP inhibition is a common feature of F1-ATPases (27–29) and occurs when the hydrolysis product ADP remains in one catalytic nucleotide-binding site and arrests the γ subunit 80° distinct from the ATP waiting position (30). During the long observation experiments in the absence of tentoxin, we could confirm this inhibition for the cyanobacterial enzyme (Fig. 3D, left panel, and see Fig. 7B of Ref. 14). On average, the complex stayed in such pauses for 52% of the observation time. The situation dramatically changed upon infusion of high tentoxin concentrations. After buffer exchange rotation speed was reduced (Fig. 3A), but the enzyme rotated continuously and stayed in the ADP inhibition pauses less than 1% (Fig. 3D, right panel, and E). This effect counteracted the reduced rotation speed of tentoxin-stimulated complexes and resulted in a higher net activity calculated as total revolutions per observation time (Fig. 3E). Also the magnitude of the stimulation on the single molecule level was very similar (∼160%) compared with bulk ATPase activity measurements. We verified the correlation of tentoxin stimulation and reduced ADP inhibition in ATPase activity measurements. It is known that the magnitude of ADP inhibition is enhanced by external ADP (30) and azide (31, 32), whereas phosphate (33) and the nonionic detergent lauryldimethylamine oxide (27) diminish the inhibition. Therefore, the stimulatory effect of high tentoxin concentrations should be pronounced at high ADP inhibition, whereas the stimulation should be suppressed when ADP inhibition is diminished. Measurements of the ATPase activity with or without 1 mm tentoxin confirmed this assumption. Under these conditions, ATPase activity was stimulated to 174 ± 7%, whereas addition of ADP (5 mm) and azide (95 μm) increased the stimulatory effect of tentoxin to 289 ± 16 and 249 ± 7%, respectively. On the other hand, no stimulation was observed when 50 mm phosphate was added (97 ± 7%).
Molecular Processes of Inhibition and Stimulation Caused by Tentoxin—Although it is well accepted that a single tentoxin molecule can completely inhibit the CF1-ATPase activity, there is still a debate on the number of the tentoxin molecules involved in the reactivation process.
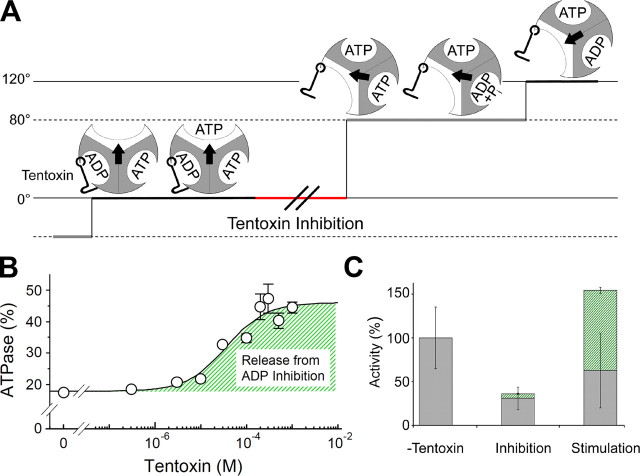
In this study, we visualized the effects of different tentoxin concentrations on the activity of the F1-ATPase and confirmed that binding of a single tentoxin molecule is sufficient for complete inhibition of the enzyme (Fig. 2). Based on our single molecule studies, we propose a model of inhibition and stimulation of F1 complexes by tentoxin (Fig. 4). We showed that tentoxin inhibition induced an asymmetric rotation by extending only one dwell time in the complete 360° turn (Fig. 2A). The corresponding event takes place directly after ATP binding and stops the reaction pathway before the 80° substep proceeds, which was observed as long pauses in Fig. 2A (τ = 1.36 s). Thus we could clarify the ADP release step at 0° during rotation of the γ subunit. This means that a single tentoxin molecule bound to one binding site acts like a clamp as shown in Fig. 4A and inhibits the transition of the related β subunit from a closed to an open conformation as suggested by the crystal structure of the spinach chloroplast F1 complexed with tentoxin (11). This particular feature could be useful to mark a native enzyme at one position within a complete 360° turn. A similar feature was recently reported using a TF1 complex carrying a mutation at the catalytic sites (34). However, tentoxin would enable us to gain more easily further insights into the catalytic properties of each catalytic site separately using a native enzyme. In addition, the inhibitory effect is partially relieved by binding of stimulatory tentoxin molecules breaking the asymmetry of rotation and restoring the rotation speed to some extent.
FIGURE 4.
Molecular processes of inhibition and stimulation caused by tentoxin. A, model of tentoxin inhibition is indicated. The 120° step with the prolonged pause at 0° is shown. The three catalytic sites are colored in gray for closed and white for open conformations. The orientation of the central γ subunit is symbolized by an arrow, and the tentoxin molecule by a hook. Tentoxin inhibits the reaction sequence after substrate binding to an open catalytic site by keeping the catalytic site with the bound inhibitor in a closed conformation. B, bulk ATPase activities of the F1 complex at different tentoxin concentrations were measured at 2 mm ATP. The ratio of activity in the presence of 95 μm azide divided by that in the absence of azide was calculated. The resulted ratio followed a sigmoid binding curve with a dissociation constant of 36.3 μm, which was calculated using the equation published previously (Equation 8 in Ref. 10). C, inhibitory and stimulatory tentoxin effects on ATP hydrolysis are combined. Inhibition by tentoxin is because of a strongly reduced rotation speed (gray bars), which is partially relieved in case of the stimulated enzyme. The activity at high tentoxin concentration is moreover boosted by a strongly reduced ADP inhibition (green hatched bar). Both processes generate an enzymatic complex with a highly enhanced activity, explaining the curious regulation pattern shown in Fig. 1.
The molecular structure of the reactivated enzyme and the conformational shifts related to the stimulation process need to be clarified by further structural studies on a tentoxin-stimulated chloroplast F1. Yet our single molecular analysis has resolved that the activity of the stimulated enzyme is boosted because of the strong reduction of ADP inhibition. To quantify the liberation of ADP inhibition, we monitored the bulk ATPase activity with and without azide at different tentoxin concentrations (Fig. 4B). The difference in the apparent Kd value for tentoxin stimulation obtained from Fig. 1 (565 μm) and that from Fig. 4B (36.3 μm) is because of the difference in the experimental conditions; the assay reported in Fig. 1 is the observation of the direct stimulatory effect of tentoxin, whereas the plots in Fig. 4B are calculated indirectly from the ratio of the activity in the presence and in the absence of azide to evaluate the contribution of ADP inhibition for the stimulation process. Without tentoxin, the enzyme activity was severely reduced to less than 20% in the presence of azide. With increasing concentrations of tentoxin, the activity was less affected and dropped only to 45% at 1 mm. Noteworthy, the azide effect at 10 μm tentoxin (inhibition condition) was similar to the enzyme without toxin. This implies that the reduced ADP inhibition is an exquisite feature of the stimulated enzyme caused by the binding of a second or third toxin molecule (Fig. 4C). In addition, this tentoxin effect, which is directed against the ADP inhibition, can easily explain the observed differences in the stimulation level of the tentoxin-sensitive ATPases (i.e. CF1, the mutant TF1, and the F1 complex from T. elongatus) at high concentrations of the toxin. The purified CF1 from spinach chloroplasts contains one to two tightly bound ADP molecules (35–37), and its activity is normally very low because of the ADP inhibition at steady state conditions. The activity of the enzyme can be stimulated more than 10-fold by the addition of high concentrations of the toxin (7–10). In contrast, purified TF1 does not contain any bound nucleotides (38, 39), and the incubation with Mg-ADP easily induces the strong ADP inhibition state (40). This specific property of the TF1-ATPase explains why the tentoxin-sensitive mutant prepared from TF1 did not show the overactivation even at high tentoxin concentrations (13).
Supplementary Material
Acknowledgments
We thank M. Tsumuraya and T. Fuse-Murakami for technical assistance and F. Boris, M. Yoshida, H. Strotmann, and P. Gräber for reading the manuscript and for fruitful discussions.
This work was supported by Grant-in-aid for Scientific Research on Priority Areas 18074002 (to T. H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the German Academic Exchange Service (DAAD) and the German Research Foundation Grant GR1616/8 (to G. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Movies S1–S4.
Footnotes
The abbreviations used are: F1 catalytic moiety of the ATP synthase; CF1, chloroplast F1; TF1, F1 from thermophilic Bacillus PS3; ATPγS, adenosine 5′-O-(thiotriphosphate).
References
- 1.Noji, H., Yasuda, R., Yoshida, M., and Kinosita, K., Jr. (1997) Nature 386 299-302 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, M., Muneyuki, E., and Hisabori, T. (2001) Nat. Rev. Mol. Cell Biol. 2 669-677 [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. D. (1993) Biochim. Biophys. Acta 1140 215-250 [DOI] [PubMed] [Google Scholar]
- 4.Abrahams, J. P., Leslie, A. G., Lutter, R., and Walker, J. E. (1994) Nature 370 621-628 [DOI] [PubMed] [Google Scholar]
- 5.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., Jr., and Itoh, H. (2001) Nature 410 898-904 [DOI] [PubMed] [Google Scholar]
- 6.Adachi, K., Oiwa, K., Nishizaka, T., Furuike, S., Noji, H., Itoh, H., Yoshida, M., and Kinosita, K., Jr. (2007) Cell 130 309-321 [DOI] [PubMed] [Google Scholar]
- 7.Steele, J. A., Uchytil, T. F., Durbin, R. D., Bhatnagar, P., and Rich, D. H. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 2245-2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avni, A., Anderson, J. D., Holland, N., Rochaix, J. D., Gromet-Elhanan, Z., and Edelman, M. (1992) Science 257 1245-1247 [DOI] [PubMed] [Google Scholar]
- 9.Mochimaru, M., and Sakurai, H. (1997) FEBS Lett. 419 23-26 [DOI] [PubMed] [Google Scholar]
- 10.Santolini, J., Haraux, F., Sigalat, C., Moal, G., and Andre, F. (1999) J. Biol. Chem. 274 849-858 [DOI] [PubMed] [Google Scholar]
- 11.Groth, G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3464-3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlova, P., Shimabukuro, K., Hisabori, T., Groth, G., Lill, H., and Bald, D. (2004) J. Biol. Chem. 279 9685-9688 [DOI] [PubMed] [Google Scholar]
- 13.Groth, G., Hisabori, T., Lill, H., and Bald, D. (2002) J. Biol. Chem. 277 20117-20119 [DOI] [PubMed] [Google Scholar]
- 14.Konno, H., Murakami-Fuse, T., Fujii, F., Koyama, F., Ueoka-Nakanishi, H., Pack, C. G., Kinjo, M., and Hisabori, T. (2006) EMBO J. 25 4596-4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker, W. C., Du, Z., Hein, R., Gromet-Elhanan, Z., and Richter, M. L. (2001) Biochemistry 40 7542-7548 [DOI] [PubMed] [Google Scholar]
- 16.Ohta, Y., Yoshioka, T., Mochimaru, M., Hisabori, T., and Sakurai, H. (1993) Plant Cell Physiol. 34 523-529 [Google Scholar]
- 17.Landt, O., Grunert, H. P., and Hahn, U. (1990) Gene (Amst.) 96 125-128 [DOI] [PubMed] [Google Scholar]
- 18.Stiggall, D. L., Galante, Y. M., and Hatefi, Y. (1979) Methods Enzymol. 55 308-315, 319-321 [DOI] [PubMed] [Google Scholar]
- 19.Taussky, H. H., and Shorr, E. (1953) J. Biol. Chem. 202 675-685 [PubMed] [Google Scholar]
- 20.Yasuda, R., Noji, H., Kinosita, K., Jr., and Yoshida, M. (1998) Cell 93 1117-1124 [DOI] [PubMed] [Google Scholar]
- 21.Shimabukuro, K., Yasuda, R., Muneyuki, E., Hara, K. Y., Kinosita, K., Jr., and Yoshida, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14731-14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishizaka, T., Oiwa, K., Noji, H., Kimura, S., Muneyuki, E., Yoshida, M., and Kinosita, K., Jr. (2004) Nat. Struct. Mol. Biol. 11 142-148 [DOI] [PubMed] [Google Scholar]
- 23.Dahse, I., Pezennec, S., Girault, G., Berger, G., Andre, F., and Liebermann, B. (1994) J. Plant Physiol. 143 615-620 [Google Scholar]
- 24.Hu, N., Mills, D. A., Huchzermeyer, B., and Richter, M. L. (1993) J. Biol. Chem. 268 8536-8540 [PubMed] [Google Scholar]
- 25.Sakaki, N., Shimo-Kon, R., Adachi, K., Itoh, H., Furuike, S., Muneyuki, E., Yoshida, M., and Kinosita, K., Jr. (2005) Biophys. J. 88 2047-2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao, H. Z., and Weber, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18478-18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jault, J. M., Matsui, T., Jault, F. M., Kaibara, C., Muneyuki, E., Yoshida, M., Kagawa, Y., and Allison, W. S. (1995) Biochemistry 34 16412-16418 [DOI] [PubMed] [Google Scholar]
- 28.Milgrom, Y. M., and Boyer, P. D. (1990) Biochim. Biophys. Acta 1020 43-48 [DOI] [PubMed] [Google Scholar]
- 29.Guerrero, K. J., Xue, Z. X., and Boyer, P. D. (1990) J. Biol. Chem. 265 16280-16287 [PubMed] [Google Scholar]
- 30.Hirono-Hara, Y., Noji, H., Nishiura, M., Muneyuki, E., Hara, K. Y., Yasuda, R., Kinosita, K., Jr., and Yoshida, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13649-13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowler, M. W., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8646-8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasilyeva, E. A., Minkov, I. B., Fitin, A. F., and Vinogradov, A. D. (1982) Biochem. J. 202 15-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitome, N., Ono, S., Suzuki, T., Shimabukuro, K., Muneyuki, E., and Yoshida, M. (2002) Eur. J. Biochem. 269 53-60 [DOI] [PubMed] [Google Scholar]
- 34.Ariga, T., Muneyuki, E., and Yoshida, M. (2007) Nat. Struct. Mol. Biol. 14 841-846 [DOI] [PubMed] [Google Scholar]
- 35.Shoshan, V., Shavit, N., and Chipman, D. M. (1978) Biochim. Biophys. Acta 504 108-122 [DOI] [PubMed] [Google Scholar]
- 36.Carlier, M. F., and Hammes, G. G. (1979) Biochemistry 18 3446-3451 [DOI] [PubMed] [Google Scholar]
- 37.Hisabori, T., and Mochizuki, K. (1993) J. Biochem. (Tokyo) 114 808-812 [DOI] [PubMed] [Google Scholar]
- 38.Ohta, S., Tsubo, M., Oshima, T., Yoshida, M., and Kagawa, Y. (1980) J. Biochem. (Tokyo) 87 1609-1617 [DOI] [PubMed] [Google Scholar]
- 39.Hisabori, T., Muneyuki, E., Odaka, M., Yokoyama, K., Mochizuki, K., and Yoshida, M. (1992) J. Biol. Chem. 267 4551-4556 [PubMed] [Google Scholar]
- 40.Yoshida, M., and Allison, W. S. (1983) J. Biol. Chem. 258 14407-14412 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.