Abstract
Insulators help separate active chromatin domains from silenced ones. In yeast, gene promoters act as insulators to block the spread of Sir and HP1 mediated silencing while in metazoans most insulators are multipartite autonomous entities. tDNAs are repetitive sequences dispersed throughout the human genome and we now show that some of these tDNAs can function as insulators in human cells. Using computational methods, we identified putative human tDNA insulators. Using silencer blocking, transgene protection and repressor blocking assays we show that some of these tDNA-containing fragments can function as barrier insulators in human cells. We find that these elements also have the ability to block enhancers from activating RNA pol II transcribed promoters. Characterization of a putative tDNA insulator in human cells reveals that the site possesses chromatin signatures similar to those observed at other better-characterized eukaryotic insulators. Enhanced 4C analysis demonstrates that the tDNA insulator makes long-range chromatin contacts with other tDNAs and ETC sites but not with intervening or flanking RNA pol II transcribed genes.
Keywords: chromatin, chromosome conformation capture, human, insulator, tRNA
Introduction
The differential packaging of chromatin in the eukaryotic nucleus results in differential gene regulation. Euchromatin is characterized by open, more accessible chromatin that is more likely to be transcribed while heterochromatin is more densely packed, and generally refractive to transcription (Elgin and Grewal, 2003; Huisinga et al, 2006). Regulatory elements such as enhancers positively regulate gene expression while silencers negatively affect gene expression within the context of these domains (Visel et al, 2009; Raab and Kamakaka, 2010; Sen and Grosschedl, 2010; Bulger and Groudine, 2011). Euchromatic and heterochromatic domains often reside adjacent to one another along the linear chromosome and mechanisms exist to spatially separate these domains thus aiding in proper gene expression. Regulatory elements such as enhancers and silencers are confined to the appropriate domain by several mechanisms. DNA elements called insulators are one mechanism for isolating regulatory elements (Maeda and Karch, 2003; Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006; Bushey et al, 2008).
Insulators are defined by their ability to isolate and insulate long-range regulatory elements. Enhancer-blocking insulators are DNA elements that when placed between an enhancer and a promoter block the ability of the enhancer to activate the promoter. Barrier insulators block the spread of silenced chromatin when placed between a silencing element and a gene promoter (Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006; Bushey et al, 2008). Insulators are present from yeast to man and characterization of these elements demonstrate that these elements share many common properties and mechanisms. All insulators bind specific transcription factors and localize to DNaseI hypersensitive sites. Several insulators have been shown to recruit chromatin-modifying and remodelling machines, which aid in the formation of the DNaseI hypersensitive sites. Another conserved property of insulators is that they cluster together in the nucleus forming ‘insulator bodies’ and by this process they isolate genes and their cognate regulatory elements into specific chromatin loops and sequester the genes to specific compartments in the nucleus (Maeda and Karch, 2007; Bushey et al, 2008; Raab and Kamakaka, 2010).
Silenced chromatin in Saccharomyces cerevisiae occurs at the telomeres and HM loci and involves silencers and the Sir repressors. Sir2p deacetylates histones while Sir3p and Sir4p bind the deacetylated histones in chromatin leading to gene repression (Rusche et al, 2003). In Schizosaccharomyces pombe, the centromeres, telomeres and cryptic mating type loci are packaged into silent heterochromatin. Histone deacetylases deacetylate the histones, which are then methylated by specific methylases, and the methylated histones are in turn bound by the HP1 homologue, Swi6 resulting in gene silencing (Grewal and Moazed, 2003). In vertebrates, constitutive centromeric heterochromatin is very similar to S. pombe heterochromatin with histone H3 methylated at K9 and being bound by HP1 containing protein complexes (Fodor et al, 2010). In mammals, besides constitutive heterochromatin, there is facultative intercalary heterochromatin on the chromosome arms (Trojer and Reinberg, 2007). Enzyme complexes specifically deacetylate histone H3 following which, specific enzymes methylate histone H3 at K27 and polycomb group proteins bind these modified residues resulting in the formation of silenced facultative heterochromatin. These silenced chromatin domains are separated from active chromatin domains by insulators.
Although heterochromatin is distinct in S. cerevisiae and S. pombe, specific tDNA genes function as insulators in both species suggesting conservation of function (McFarlane and Whitehall, 2009). tRNA genes are moderately repetitive, present singly or in small clusters, transcribed by RNA pol III (Dieci et al, 2007) and their expression is subject to developmental and cell-cycle regulation (Stutz et al, 1989; Fairley et al, 2003). Transcription of tRNA genes (tDNA) is mediated by the transcription factors TFIIIB and TFIIIC along with RNA pol III (Geiduschek and Kassavetis, 2001). Mammalian cells have additional factors such as OCT1, Myc, Fos/Jun and Rb that directly or indirectly regulate their transcription (Felton-Edkins et al, 2003). tDNAs have an internal promoter consisting of the A and the B box, which recruits the transcription factor TFIIIC. The B box has been shown to solely recruit TFIIIC and a single base change in the B box has been shown to completely abolish TFIIIC recruitment and tRNA transcription, replication pausing and insulation. TFIIIC mediated recruitment of TFIIIB upstream of the tDNA followed by the recruitment of RNA polymerase III culminates in transcription of the gene (Geiduschek and Kassavetis, 2001; Schramm and Hernandez, 2002).
In S. cerevisiae, a tRNA-thr gene and its flanking sequences function as an insulator and restrict the spread of silent chromatin at the cryptic mating locus HMR in either orientation (Donze et al, 1999; Donze and Kamakaka, 2001), while 30% of the tDNAs in S. pombe are found flanking centromeric heterochromatin and several of these genes have been shown to act as barrier insulators (Noma et al, 2006; Scott et al, 2006; Iwasaki et al, 2010). The insulator function of tDNAs is critically dependent upon the recruitment/binding of the transcription factors TFIIIB and TFIIIC while transcription by RNA polymerase III is not necessary for insulation (Noma et al, 2006; Simms et al, 2008; Biswas et al, 2009; Valenzuela et al, 2009). tDNA mediated insulation utilizes cohesin proteins as well as specific chromatin remodellers and histone modifiers to generate a specialized nucleosome depleted DNaseI hypersensitive site (Donze et al, 1999; Donze and Kamakaka, 2001; Damelin et al, 2002; Ng et al, 2002; Bachman et al, 2005; Gelbart et al, 2005; Jambunathan et al, 2005; Oki and Kamakaka, 2005; Dubey and Gartenberg, 2007; Parnell et al, 2008; Dhillon et al, 2009). Studies also show that TFIIIC-bound loci in S. cerevisiae and S. pombe coalesce at specific foci, which results in the formation of chromatin loops (Noma et al, 2006; D’Ambrosio et al, 2008; Haeusler et al, 2008; Duan et al, 2010; Iwasaki et al, 2010).
In Drosophila, Su(Hw) protein binds the Gypsy retrotransposon and functions as a barrier and enhancer-blocking insulator (Roseman et al, 1993). Su(Hw) mediates insulation by interacting with numerous cofactors (Gdula et al, 1996) and the protein also coalesces together forming insulator bodies that likely aid in the formation of chromatin loops (Gerasimova et al, 2000; Capelson and Corces, 2005).
The chicken HS4 site of the β-globin locus is the archetypal autonomous vertebrate insulator (Chung et al, 1993; Pikaart et al, 1998; Bell et al, 1999; Recillas-Targa et al, 2002; West et al, 2004; Huang et al, 2007; Dickson et al, 2010). Three proteins are recruited to this insulator and each is important for a specific aspect of insulator function. USF1 binds a site within cHS4 and is necessary for the recruitment of chromatin-modifying enzymes that modify histones, generate a hypersensitive site and block the spread of heterochromatin. VEZF1 binds to different sites and is important for barrier activity through its effects on DNA methylation. CTCF mediates enhancer-blocking activity and is also required to localize the insulator to specific regions of the nucleus forming chromatin loops (Yusufzai and Felsenfeld, 2004; Yusufzai et al, 2004).
The majority of functionally characterized mammalian insulators bind CTCF and CTCF is necessary for enhancer-blocking insulation in vertebrates (Bell and Felsenfeld, 1999; Magdinier et al, 2004; Filippova et al, 2005; Mishiro et al, 2009; Phillips and Corces, 2009) and data also indicate that cohesins are required for CTCF mediated insulation (Wendt et al, 2008; Mishiro et al, 2009). Besides CTCF, very few other proteins required for enhancer blocking have been identified in mammalian cells and the barrier activity of mammalian insulators has also not been well characterized either (Burgess-Beusse et al, 2002; Kim et al, 2009; Ottaviani et al, 2009a, 2009b).
One CTCF-independent mammalian enhancer-blocking insulator is an RNA polymerase III transcribed Short Interspersed Nuclear Element (SINE) (Lunyak et al, 2007; Roman et al, 2011). SINEs are transposable elements that bind RNA pol III transcription factors TFIIIC and TFIIIB and are transcribed by RNA polymerase III or RNA polymerase II (Rowold and Herrera, 2000). However, it is currently unknown if RNA pol III transcribed genes as opposed to SINE elements have the ability to function as enhancer-blocking insulators in vertebrates and it is also unclear if human pol III transcribed loci function as barrier insulators (Kim et al, 2009; Roman et al, 2011) like the recently identified mouse tDNA insulator (Ebersole et al, 2011).
Given the observation that TFIIIC-bound loci function as barrier insulators in the distantly related yeast, S. cerevisiae and S. pombe, as well as enhancer-blocking insulators in mice (Donze et al, 1999; Noma et al, 2006; Scott et al, 2006; Lunyak et al, 2007; Biswas et al, 2009), we inquired if tRNA genes could function as insulators in human cells. Here, we identify fragments in human cells that contain tRNA genes and are capable of insulator activity. We show that tRNA genes are often found in close proximity to the boundaries of repressed chromatin domains. We also find that many tRNA gene locations with respect to neighbouring RNA pol II transcribed genes are conserved through evolution, suggesting a location-specific functional role for these genes. Using a functional assay in S. pombe, we demonstrate that human tRNA genes are capable of blocking heterochromatin. We further show that the human tRNA genes also block repression mediated by the polycomb group proteins in human cells and protect transgenes from position effects. Finally, we show that, like SINE elements and other mammalian insulators, tRNA genes can efficiently block enhancer mediated transcription activation. Mapping studies of the chromatin environment at the native locus of the 3-kb putative tRNA insulator fragment has a signature found at other insulators and we also show that the tDNA-containing fragment interacts with other tDNA-containing fragments via long-range interactions thereby clustering in the nucleus and possibly forming chromatin loops and these data collectively suggest that tRNA genes likely function as insulators in human cells.
Results
In silico identification of putative tDNA insulators
In light of previous observations that DNA elements bound by the RNA pol III transcription factors TFIIIC and TFIIIB function as insulators in yeast and mice (Raab and Kamakaka, 2010), we wished to determine the locations of potential RNA pol III insulators in human cells. Given that TFIIIC recruitment is critical for these elements to function we initially mapped TFIIIC binding site containing DNA sequences (B boxes) throughout the human genome using a TFIIIC consensus sequence-based weight matrix based on a 11-bp consensus sequence (Pavesi et al, 1994). Following clustering analyses and a cutoff score of more than −9.3 and a window size of 2 kbp, we determined B-box counts and B-box probabilities and ranked >600 000 putative TFIIIC binding sites. The distribution of these putative sites is shown in Supplementary Figure S1A. In all, 608 out of the 631 human tDNA and tDNA pseudo genes (as defined by tRNAdb; http://gtRNAdb.ucsc.edu) were present in our B-box predictions with 42% of all tDNAs in the top 1% of our predictions.
Since clusters or tandem arrays of TFIIIC bound sites function as better insulators in yeast (Donze and Kamakaka, 2001; Noma et al, 2006; Valenzuela et al, 2009), we reasoned that clusters of these sites are more likely to function as insulators in humans. We therefore focused on identifying loci where multiple TFIIIC sites (two or more over a 2-kb distance) clustered together. The strongest clustered sites resided at or near tDNAs and we therefore chose to focus further analysis on clusters of tDNAs rather than B boxes alone. The top three clusters of tRNA genes were located on chromosomes 1, 6 and 17. ETC loci bind TFIIIC but not RNA pol III and interestingly these loci are not present in repetitive clusters along human chromosomes.
We next measured the distance between each tDNA and its neighbour (Supplementary Figure S1B), plotting the cumulative frequency. Approximately 50% of human tDNAs are located within 5 kb of another tDNA and many (23%) of the tDNAs are found <1 kb from a second tDNA (Oler et al, 2010). This clustering is similar to what is observed for tDNAs in Drosophila melanogaster, Caenorhabditis elegans and S. pombe but distinct from what is observed for S. cerevisiae (Kuhn et al, 1991), suggesting that tDNA clusters may have functional significance.
Syntenic conservation of tDNAs
tDNAs are moderate copy repetitive elements, present in multiple copies within the genome (Guthrie and Abelson, 1982; Frenkel et al, 2004; Goodenbour and Pan, 2006), suggesting that the selection pressure on any individual tDNA gene should not be high and therefore its position within the genome need not be strongly conserved. Consistent with this hypothesis, in bacterial cells the position of tDNAs is not well conserved among closely related species (Withers et al, 2006; Copeland et al, 2009). Syntenic alignments highlight blocks of the human genome that are conserved in another organism's genome. We asked whether or not human tDNA locations were syntenically conserved with respect to their neighbouring pol II transcribed genes. We used syntenic alignments of the human, chimp, mouse and opossum genomes (Waterston et al, 2002; Karolchik et al, 2003; Schwartz et al, 2003) and identified the locations of tDNAs in each of these organisms (Lowe and Eddy, 1997; http://gtRNAdb.ucsc.edu). We considered a tDNA to be the same if tDNAs from both organisms had the same anticodon (this constraint was more stringent than using the same isoacceptor). Genome-wide analyses of all the human tDNAs showed that the majority of tDNAs were syntenic in chimps and nearly 50% of the human tDNAs were in syntenic positions in mouse (274/622) and ∼25% of tDNAs were syntenic in opossum (159/622) (see Table I and Figure 1A). These results are similar to recent results on the distribution of tDNAs in other eukaryotes (Bermudez-Santana et al, 2010).
Table 1. Percent of repetitive RNA in conserved positions in genome.
| tDNA | snRNA | 7SL RNA | |
|---|---|---|---|
| Chimp | 85 | 92 | 89 |
| Mouse | 44 | 3 | 1 |
| Opossum | 26 | 3 | NAa |
| a7SL RNA data not available in RepeatMasker data set. | |||
Figure 1.
Location of tDNAs suggests functional roles. (A) A schematic representation of a highly conserved genomic locus spanning ∼140 kb on chromosome 17. Gene diagrams illustrate the positions of the nine RNA pol II transcribed genes in this region. The position of tDNAs is depicted as tick marks. Synteny of the tDNAs within this locus is shown as a further tick mark in the row of the species in which it is syntenic. (B) Box and whisker plots depicting the distance between binding sites and the end points of histone H3 lysine 27 trimethylated domains in K562 cells.
As a control, we used the RepeatMasker data for other small repetitive RNA families to compare their locations within the genome (Smit, Hubley Green: RepeatMasker Open-3.0 http://www.repeatmasker.org; 1996–2007). snRNA and 7SL RNA are transcribed by RNA pol II and pol III. We determined the locations of the genes for these RNAs across different mammalian species as well and found that these genes were not conserved relative to their neighbouring pol II transcribed protein-coding genes (human–mouse snRNA that was syntenically conserved was 140/4287 and human–opossum was 115/4287. Similarly, human-mouse 7SL RNAs that were syntenic were 6/943, but human-opossum data were not available in repeat masker). The relatively high degree of location conservation of tDNAs suggests that the tDNA position within the genome may have a functional significance (though there are other interpretations such as slower turnover as well).
tDNA clusters are located at transitions of chromatin domains
Gene regulation is mediated in part by modifications of the histones in chromatin. Numerous studies have demonstrated that particular modifications correlate with specific gene activity states (Barski et al, 2007; Mikkelsen et al, 2007). Histone H3 K27 trimethylation (H3K27me3) is most often associated with silenced genes while histone H3 K36 trimethylation is most often associated with transcriptionally active genes (Schones and Zhao, 2008). Initially, we identified putative insulator tDNAs by examining their position relative to H3K27me3 domains (Barski et al, 2007). We reasoned that if some tDNAs functioned as insulators then these should be present at these transition zones.
To determine in general if tDNAs were located at transition points between active and repressed chromatin domains, we measured the distance between each tDNA and the nearest end point of an H3K27me3 repressed domain (Figure 1B). Although in yeast nearly all the tDNAs are occupied by TFIIIC, this is not the case for human cells (Harismendy et al, 2003; Roberts et al, 2003; Moqtaderi and Struhl, 2004; Barski et al, 2010; Moqtaderi et al, 2010; Oler et al, 2010; Raha et al, 2010). We reasoned that occupied and unoccupied tDNAs might vary in their position relative to H3K27me3 domains. To assess this, we used the occupancy levels for each of the tDNAs from the ENCODE project. We subdivided the list into quartiles and took the tDNAs in the first (highly occupied n=158) and last (unoccupied n=169) quartile. We determined the distance between these tDNAs and the edges of the H3K27me3 domains as defined by the ENCODE project. Active tDNAs were closer to the boundary of a repressed domain (located on average 41 kbp (median of 28 kb) from the boundary of the repressed domain) compared with inactive tDNAs (average of 127 kbp from a boundary (median of 33 kb) and located within the repressed domain), demonstrating that active tDNAs reside adjacent to an H3K27me3 domain boundary compared with the unoccupied tDNAs (P=0.002 Wilcoxon test). As a control, we also performed this analysis on putative CTCF insulators (n=18 441). CTCF, Rad21 bound DNaseI hypersensitive sites were first identified as putative CTCF insulators sites. These sites were then analysed with respect to distance from H3K27me3 domains and were found to be on average 71 kbp (median of 31 kb) from an H3K27me3 boundary. These data are consistent with mapping data showing that only a subfraction of CTCF sites flank H3K27me3 domains (Barski et al, 2007; Cuddapah et al, 2009). The random sites, as expected, were not present near the H3K27me3 domain boundaries.
Another function of chromatin insulators is to function as enhancer blockers. The expression patterns of a pair of genes separated by an insulator are less likely to be similar since the insulator will prevent co-regulation of these genes. On the other hand, pairs of genes lacking an insulator might have similar expression patterns since there is a greater likelihood of these genes sharing regulatory modules and therefore being co-regulated. If tDNAs act as enhancer blockers, then pairs of genes separated by tDNAs might have expression patterns that are less correlated than pairs of genes lacking tDNAs. We investigated 750 randomly chosen gene pairs and analysed the ENCODE expression data from K562 and HeLa cells for these genes. There was some correlation in the expression patterns for these pairs of genes (r=0.30 and 0.34 for K562 and HeLa cells, respectively), which is consistent with data showing that there is some clustering of co-regulated genes in eukaryotes (Singer et al, 2005). We next investigated all gene pairs separated by bound CTCF, a known enhancer-blocking insulator. When two genes were separated by CTCF, this correlation was lost (r=0.03 and 0.04 for K562 and HeLa cells, respectively). When gene pairs were separated by TFIIIC bound tDNAs, similarly to the CTCF data, the correlation in expression was also lost (r=0.04 and 0.09 for K562 and HeLa cells, respectively). These data are consistent with the model that tDNAs might be functioning as chromatin insulators.
We then identified specific loci where clusters of tDNA resided at the boundaries of H3K27me3 domain. Our analyses identified numerous loci where tDNAs could be functioning as barrier insulators with tDNA clusters on chromosomes 6 and 17 being the most striking. We focused our attention on the region of chromosome 17 that contains five clusters of tDNAs (Figure 1A). This is a particularly gene-rich region comprising 8 RNA pol II transcribed protein-coding genes, one dubious unannotated open reading frame and 18 tDNAs within 150 kb. This locus is highly conserved throughout mammalian evolution (Figure 1A) and the RNA pol II transcribed genes within this locus remained syntenic throughout mammalian evolution, during the 100 million years of evolutionary time between opossum and humans (Karolchik et al, 2003). Twelve of the eighteen tDNAs found in this region in humans were also found in the same position in opossum. Most importantly, the position of the tDNAs relative to the pol II transcribed protein-coding genes is conserved.
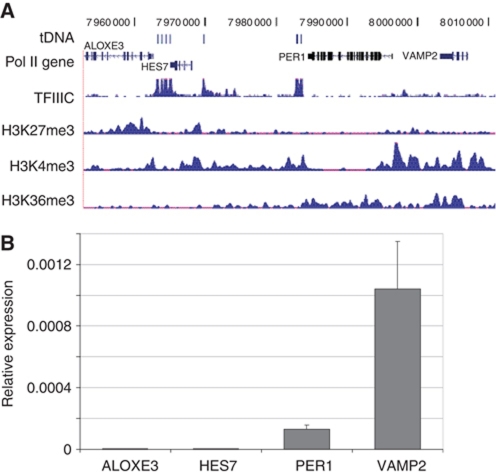
In CD4+ T cells at this locus on chromosome 17, high levels of H3K27me3 encompass the arachidonate lipoxygenase gene (ALOXE3) but the levels of H3K27me3 reduce near the cluster of four tDNAs, and drops further at the second cluster of tDNAs adjacent to the hairy and enhancer of split 7 (HES7) gene. Adjacent to this domain is a large domain enriched in H3K36me3, a mark of active chromatin (Barski et al, 2007).
Recent chromatin profiling in nine different human cell lines echoes this distribution pattern and adds further details (Ernst et al, 2011; see Figure 2A). In K562 cells, H3K27me3 is high over the ALOXE3 gene, decreases over the HES7 gene and finally disappeared over the Period homologue 1 (PER1) gene. In contrast, H3K4 trimethylation is very low over the ALOXE3 gene, increases at the HES7 gene and is highly enriched at genes beyond this region. Finally, H3K36me3 is not observed on the ALOXE3 or HES7 genes but is present over the coding regions PER1 and the other genes in this cluster. Based on this distribution of histone marks, it was proposed that ALOXE3 is transcriptionally silent, HES7 was transcriptionally poised but inactive while PER1 was transcriptionally active. We have also mapped the distribution of H3K27me3 and H3K36me3 across this region using qChIP and the distribution pattern of these histone marks across most of our probes show similar distribution patterns as the ENCODE genome-wide patterns (see Supplementary Figure S2).
Figure 2.
Identification of chromatin domains in K562 cells. (A) ENCODE ChIP-seq data for the distribution of TFIIIC, histone H3K27me3, H3K4me3 and H3K36me3 are plotted across the chromosome 17 domain. The position of tDNAs and RNA pol II transcribed genes is shown. (B) Transcription status of genes at the chromosome 17 locus. Quantitative RT–PCR analyses of RNA isolated from K562 cells. RNA was measured from three independent biological replicates and error bars represent standard deviation. Schematic representation showing the locations of genes is present above the graph.
tDNAs demarcate differentially transcribed functional domains on chromosome 17
The ChIP data show that tDNAs are present at the transition between the K27me3 bound (potentially silenced) ALOXE3 gene and the H3K27me3 and H3K4me bound (potentially inactive but poised) HES7 gene. The data further show that tDNAs are present at the second transition between the potentially inactive HES7 and the H3K36me3 and H3K4me3 bound (potentially active) PER1 genes. If indeed there are distinct chromatin domains juxtaposed with each other, then a prediction is that genes within the H3K27me3 domain should be transcriptionally silent while the genes enriched for H3K36me3 should be active. We measured the transcriptional states of the genes within these two domains on chromosome 17 by RT–PCR performed on total RNA purified from actively dividing K562 cells (Figure 2B). ALOXE3, the gene with the highest levels of H3K27me3 was highly repressed in our cell line, as was HES7, the gene flanked by tDNAs and which contained both H3K27me3 and H3K4me3 confirming the prediction that HES7 is an inactive gene. In contrast, both PER1 and the vesicle-associated membrane protein 2 (VAMP2) genes, which were enriched for H3K4me3 and H3K36me3, were correspondingly highly expressed in this cell line demonstrating that the tDNAs reside in a transition zone separating both structural and functional domains. The AurKB gene, which is located further downstream and is enriched for H3K4me3 and H3K36me3, is also active in K562 cells (data not shown).
TFIIIC binds to tDNAs on chr17
In yeast, tDNA mediated insulator activity is dependent in part upon the binding of the transcription factor TFIIIC to the tDNA promoter and in part on flanking sequences. Unlike yeast, where all of the tDNAs are occupied by TFIIIC, recent mapping data for the RNA pol III transcriptome have clearly shown that not all tDNAs in the human genome are occupied by TFIIIC (Barski et al, 2010; Moqtaderi et al, 2010; Oler et al, 2010; Raha et al, 2010). If our model that tDNAs bound by TFIIIC act as insulators is correct, then we would expect at a minimum that the tDNAs on chromosome 17 to be bound by TFIIIC. Due to the repetitive nature of tDNA and despite our best efforts, we were unable to design specific primers for all of the tDNAs across this region but were able to study TFIIIC binding across some of the tDNAs. Using these primers, we mapped the distribution of TFIIIC by ChIP coupled with qPCR along chromosome 17 (Figure 3A–B; Supplementary Figure S3). TFIIIC was only bound to tDNA-containing loci (probes 4, 7 and 14) but was absent from regions of chromosome17 that lacked a tDNA (remaining probes). The recent genome-wide mapping data for the RNA pol III transcriptome in numerous human cells (Barski et al, 2010; Moqtaderi et al, 2010; Oler et al, 2010; Raha et al, 2010) also show that these tRNA genes on chromosome 17 are bound by TFIIIC (Figure 2A). Similarly to the results obtained in yeast (Valenzuela et al, 2009), the levels of TFIIIC occupancy at the human tDNAs varied though the underlying reason for the variation are not known.
Figure 3.
Distribution of proteins on chromosome 17. (A) Diagram of the tDNA clusters and surrounding RNA pol II transcribed genes on chromosome 17. Numbered black bars (1–14) depict the location of PCR probes (sequences present in supplementary tables). All chromatin IP experiments were performed in K562 cells. (B) Quantitative chromatin immunoprecipitations using antibodies against a TFIIIC subunit (GTF3C5) were performed to determine the distribution of these proteins across a region of chromosome 17. (C) Acetylated histone H3 distribution across a region of chromosome 17. (D) CTCF was mapped across this region by quantitative chromatin immunoprecipitation. (E) The protein Rad21 distribution was mapped across the tDNA cluster on chromosome 17 using quantitative ChIP and antibodies against Rad21. All qChIP experiments are the average of at least two independent crosslinkings, with two immunoprecipitations from each crosslinking. Error bars represent standard error.
Chromatin structure at the tDNA is ‘insulator-like’
Insulator activity in yeast, Drosophila and chicken cells utilizes additional factors such as cohesins, histone acetylases and chromatin remodellers (Donze et al, 1999; West et al, 2004; Oki and Kamakaka, 2005; Gaszner and Felsenfeld, 2006; Huang et al, 2007; McNairn and Gerton, 2008; Dhillon et al, 2009; Dorsett, 2009; Wendt and Peters, 2009; Bose and Gerton, 2010; Wood et al, 2010). We wished to know if the DNA fragments containing human tDNA clusters had a chromatin signature similar to that observed at insulators in yeast and chicken cells. We mapped the distribution of acetylated histones by qChIP using PCR amplicons across the tDNAs on chromosome 17. Peaks of histone acetylation were observed at specific sites and these peaks were in the vicinity of the TFIIIC peaks (Figure 3C) though these peaks were shifted relative to the TFIIIC peaks possibly because tDNAs are nucleosome free and position nucleosomes adjacent to the gene (Morse et al, 1992).
Most mammalian insulators utilize CTCF for insulation, which recruits cohesin proteins. Cohesin proteins have been shown to function in both barrier and enhancer-blocking insulator activity in yeast, Drosophila and mammalian cells (Donze et al, 1999; Laloraya et al, 2000; Glynn et al, 2004; Dubey and Gartenberg, 2007; Gause et al, 2008; Dorsett, 2009; Wendt and Peters, 2009; Zlatanova and Caiafa, 2009; Bose and Gerton, 2010; Wood et al, 2010). We therefore mapped the distribution of CTCF and the Rad21 subunit of the cohesin protein complex across the region of interest on chromosome 17 (Figure 3D and E). We identified a peak of CTCF binding adjacent to the cluster of four tDNA genes upstream of ALOXE3. The binding site identified maps to the site previously shown to bind CTCF by the ENCODE consortium. We also observed peaks of cohesin binding and some of these peaks are coincide with CTCF peaks consistent with data that CTCF recruits cohesins to specific sites in the genome (Gause et al, 2008; Parelho et al, 2008; Rubio et al, 2008; Stedman et al, 2008; Mishiro et al, 2009; Wendt and Peters, 2009). Taken together, our mapping studies show the chromatin environment near the tDNA fragment and the trans-factors recruited to the fragment that contains the tDNAs are characteristic of other known insulators. At this point, the dependency relationships between TFIIIC, CTCF and cohesins and their individual role in insulation are not known.
tDNAs function as enhancer-blocking insulators
The presence of tDNAs at a chromatin state transition zone could simply be a chance occurrence. Alternatively, the tDNA genes could be functioning as insulators. Long-range gene regulation involves enhancers and silencers that positively or negatively regulate a promoter of a gene. Insulators restrict the action of these elements in a position-dependent manner (Raab and Kamakaka, 2010). We first inquired if a human tDNA-containing fragment had an ability to block communication between an enhancer and a promoter when inserted between these two elements. While tDNAs function as barrier insulators in yeast, they have never been tested for enhancer-blocking activity since classic enhancers do not exist in these organisms. We used the mammalian enhancer-blocking assay developed by the Felsenfeld laboratory (Chung et al, 1993). The human tDNA-containing fragments were cloned between the murine HS2 enhancer of the globin locus and a neomycin reporter driven by the human γ-globin promoter. This enhancer–promoter pair is active in K562 cells. Chicken HS4, as well as many other insulators, have been shown to robustly block the activation of the neomycin gene by HS2. The constructs generated were linearized and equal amounts of these constructs were used to transfect K562 cells. Neomycin-resistant colonies were allowed to grow in soft agar on plates and the number of neomycin-resistant colonies was counted after staining.
A construct containing the enhancer–promoter cassette was used as a control and all data were normalized to this cassette (Figure 4B, lane 1). A construct containing just the promoter was used as the negative control (Figure 4B, lane 2). We used a construct with a fragment containing two copies of the 1.2-kb long full chicken HS4 insulator inserted between the enhancer and promoter and used this construct as a control for insulation (Figure 4B, lane 3). The duplicated cHS4 insulator reduced the number of neomycin-resistant colonies to ∼40% of the uninsulated construct. We next tested six human DNA containing fragments from chromosome 17 (see Figure 4A for relative locations of these genes). One fragment contained a single tDNA that is present immediately upstream of HES7 (Figure 4B, lane 5). One fragment contained two tDNAs that are present immediately downstream of PER1 (Figure 4B, lane 6). One fragment contained three tDNAs located upstream of AURKB (Figure 4B, lane 9) and three DNA fragments contained four tDNAs (Figure 4B, lanes 4, 7 and 8). In addition, we tested three DNA fragments from chromosomes 6 and 19, each of which contained between one and two tDNA genes (Figure 4B, lanes 10–12).
Figure 4.
Human tDNAs possess enhancer-blocking activity. (A) Schematic representation of the chromosome 17 locus analysed in the enhancer-blocking assay with the tDNA clusters shown as bars. (B) Enhancer-blocking constructs containing murine HS2 enhancer of the β-globin gene and the human γ-globin promoter drive expression of a neomycin-resistant gene. A positive control for insulator activity contains two copies of the cHS4 insulator. Constructs lacking the enhancer or lacking an insulator fragment were used as controls for expression of the neomycin resistance gene. tDNA-containing fragments from chromosomes 6, 17 and 19 were cloned between the enhancer and the promoter. The arrowheads indicate the number of tDNAs present in each test fragment. K562 cells were transfected and the number of neomycin-resistant colonies growing in soft agar were quantified. The data are the average of three transfection experiments and error bars are standard error. (C) Analysis of enhancer-blocking ability of the first two clustered tDNAs from upstream of the ALOXE3 gene. Arrowheads indicate the number and orientation of the DNA fragments (asterisks denote P<0.02).
With regards to the tDNAs on chromosome 17, the fragment (labelled HES7) containing the single tDNA was unable to block the enhancer. The fragment containing two tDNAs (labelled PER1) has moderate enhancer-blocking activity (50% of the uninsulated control) while the fragment with four tDNAs (labelled ALOXE3) has robust enhancer-blocking activity (40% of the uninsulated control), and was equivalent to that observed for the duplicated chicken HS4 insulator. Similarly, the TMEM and AURK1 fragments have strong insulator activity while the AURK2 is a weak insulator. We see similar variability in insulation for tDNA-containing fragments from chromosomes 6 and 19 with the fragment on chromosome 19 being very robust in enhancer blocking.
We next focused our attention on the cluster of four tDNAs upstream of ALOXE3. Besides binding TFIIIC, this cluster has binding sites for CTCF. We focused on the two telomere-proximal tDNAs from this cluster and asked if just these two tDNAs (lacking CTCF) could function in enhancer blocking (Figure 4C). Inserting this 1.2 kb fragment between the enhancer and the promoter demonstrated that these two tDNAs could block the enhancer but the activity was not as robust as that observed for the entire cluster of four tDNAs with the CTCF site (∼60% of the control) (Figure 4C, lane 13).
The yeast tDNA fragment can function as a barrier in either orientation (Donze and Kamakaka, 2001). Similarly, the chicken HS4 fragment can function as an enhancer blocker in either orientation (Pikaart et al, 1998; Bell et al, 1999; Recillas-Targa et al, 1999). We therefore tested the tDNA-containing fragment in both orientations. Like chicken HS4, the tDNA fragment functions in an orientation-independent manner to block the enhancer from communicating with the promoter (compare lanes 13 and 14 in Figure 4).
It was possible that proteins binding flanking sequences mediated the insulation observed. tDNAs have an internal promoter composed of two binding sites called the A and B boxes. The tDNA B box is only known to bind the transcription factor TFIIIC is critical for TFIIIC binding to the tDNA promoter and for tDNA transcription (Geiduschek and Tocchini-Valentini, 1988). In addition, insulator activity of tDNAs in yeast requires the binding of TFIIIC to the tDNA B-box promoter (Donze and Kamakaka, 2001) and mutations in the B box abolish binding of TFIIIC, which results in loss of gene transcription and insulation. To determine if TFIIIC was necessary for enhancer-blocking activity in human K562 cells, we made three base pair mutations in the highly conserved tDNA B box. We assayed the ability of this mutated tDNA to block communication between the enhancer and the promoter. The results from this experiment show that the mutated tDNA-containing fragment had significantly reduced ability to block the enhancer (compare lanes 1, 14 and 15 in Figure 4). This result also indicates that the reduction in number of colonies in constructs containing an insulator was not due to an increase in distance between the enhancer and the promoter. Furthermore, since this fragment does not contain any CTCF binding sites it is likely that enhancer blocking is mediated in a CTCF-independent manner.
In yeast, a single tDNA that is a weak insulator can be converted to a strong insulator by multimerization (Donze and Kamakaka, 2001; Noma et al, 2006; Valenzuela et al, 2009). The chicken HS4 insulator also has the same property (Pikaart et al, 1998). Since the two tDNAs were not as robust in insulation as the entire native cluster of four tDNAs at ALOXE3, we asked if duplicating these two tDNAs would improve insulation. The duplicated fragment led to a further decrease in the number of neomycin-resistant colonies to ∼35% of constructs containing no insulator (compare lanes 14 and 16 in Figure 4C). These data show clearly that duplication increased activity to that observed for the native fragment containing four tDNAs (Figure 4B, lane 4). Mutagenizing the four B boxes in the duplicated fragment significantly eliminated its ability to insulate (compare lanes 16 and 17 in Figure 4C).
One possibility was that the tDNAs were functioning not only by blocking the enhancer from communicating with the promoter but also by simply repressing the enhancer. To address this issue, we cloned the tDNA-containing fragment immediately upstream of the enhancer. Our data show that in this configuration (lane 18 in Figure 4C) the enhancer was still able to activate the promoter, demonstrating that the tDNA was functioning as a bona fide enhancer blocker and not simply as a silencer of the enhancer, though these may be related phenomenon (Petrykowska et al, 2008).
The 1.2-kb tDNA-containing fragment functions as an enhancer blocker only in the presence of a functional B-box promoter and insulation is lost upon mutating the B box. To confirm that the effect is due to loss of TFIIIC binding, we decided to map the distribution of TFIIIC in the wild-type and mutant fragment. We subcloned the wild-type or mutant 1.2 kb fragments into an episome (pCEP9) and stably transfected K562 cells with the construct (which was autonomously maintained by the EBNA1 protein and the OriP origin present on the episome) (Supplementary Figure S4A). In order to distinguish the episomal tDNAs from the chromosomal tDNAs, we incorporated 29 bp unique tags flanking the two tDNAs. ChIP with antibodies against TFIIIC showed binding to the wild-type tDNA Gln (though not the tDNA Lys) and TFIIIC binding to the Gln tDNA was lost in the 3-bp B-box mutant (Supplementary Figure S4B).
Human tDNAs function as barrier insulators in S. pombe
Some insulators function solely as enhancer blockers or as barrier insulators while other insulators are composite insulators that possess both activities (Raab and Kamakaka, 2010). Having shown for the first time that tDNAs can function as enhancer-blocking insulators, we next turned our attention to ask whether human tDNA-containing DNA fragments could also function as barrier insulators and had the ability to block silenced heterochromatin. In S. pombe, the RNAi machinery works in conjunction with transcription factors to recruit histone deacetylases and methylases to generate chromatin depleted of acetylated histones and enriched for histone H3K9me3. The HP1 sequence homologue Swi6p binds the methylated chromatin to mediate silencing of genes (Grewal and Moazed, 2003; Grewal and Elgin, 2007). This heterochromatin is very similar to constitutive heterochromatin in mammals (Trojer and Reinberg, 2007). Furthermore, tDNA transcription factors are conserved between yeast and mammals such that yeast factors can transcribe mammalian tDNAs (Huang and Maraia, 2001). In addition, the binding sites for S. pombe TFIIIC is very similar to the binding site for human TFIIIC (Hamada et al, 2001; Moqtaderi et al, 2010). We therefore chose to begin our barrier analyses of human tDNAs by using an assay for barrier activity in S. pombe.
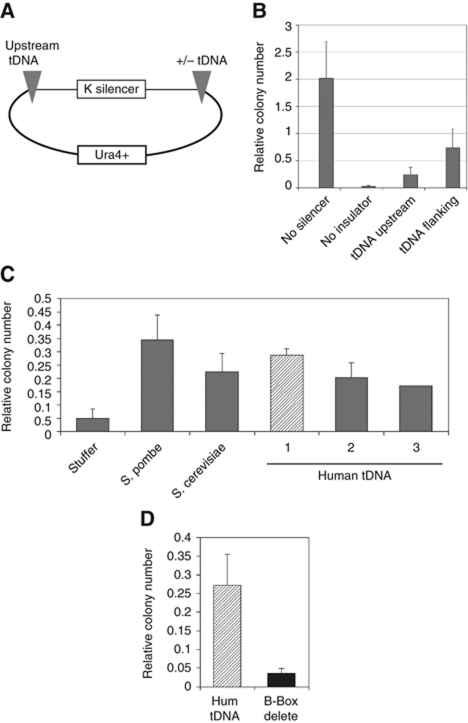
We developed a barrier assay in S. pombe that is similar to the barrier assay in S. cerevisiae. When an S. pombe replicating plasmid with an Ura4+ reporter gene is transformed into an S. pombe uracil auxotroph, it enables the strain to grow on medium lacking uracil. However, incorporation of an S. pombe silencer such as the 6.4-kb centromeric K repeat from chromosome 3 into this replicating plasmid allows the silencer to recruit various histone deacetylases, methylases and repressor proteins to generate heterochromatin which spreads to silence the Ura4+ reporter. Thus, strains transformed with this plasmid are unable to grow on media lacking uracil and appear untransformed (Partridge et al, 2002). As a test of the assay, we first determined if flanking the silencer with an S. pombe tDNA insulator would block the spread of heterochromatin and allow growth of the transformants on medium lacking uracil. We generated plasmids where we inserted an S. pombe tDNA insulator upstream of, or flanking the K repeat silencer (Figure 5). As controls we had plasmids either lacking the silencer or plasmids containing the silencer but lacking a putative tDNA insulator. Equal amounts of these plasmids were used to independently transform an ura4− leu1− S. pombe auxotroph. In order to control for transformation efficiency, we co-transformed the S. pombe strain with a second S. pombe replicating plasmid that contained the Leu1+ gene but no silencer sequences. Half of the transformation was plated on S. pombe minimal media plates lacking uracil, while the remaining transformants were plated on minimal media plates lacking leucine. Colonies were counted and the number of Ura+ colonies was normalized to the Leu+ colonies for each transformation. A plot of the number of transformants using this assay is shown in Figure 5B. Ura4+ plasmids containing no silencer generated a large number of colonies, while plasmids with the silencer (no insulator) generated very few colonies. Interestingly, insertion of an S. pombe tDNA upstream of the silencer (tDNA upstream) resulted in little insulation of the reporter whereas flanking the silencer with the tDNA (tDNA flanking) led to higher levels of insulation.
Figure 5.
Human tDNAs function as barrier insulators in S. pombe. (A) Schematic depiction of the plasmid construct used to test for barrier insulation in S. pombe. (B) Equal amounts of various plasmids containing S. pombe tDNAs were used along with a Leu1+ plasmid to transform an S. pombe strain. Transformants were plated on minimal media plates lacking either uracil or leucine and the number of colonies able to grow on media lacking uracil or leucine were counted. Data shown is the average with standard error of three independent transformation experiments. (C) Human tDNA pairs from the ALOXE3 and PER1 cluster (horizontal bars) on chromosome 17 were cloned into the plasmid shown in (A). Insulator activity of recombinant plasmids containing human tDNAs was determined as described for (B). Data shown is the average with standard error of three independent transformation experiments. Colony number was normalized to the number of colonies in a construct lacking a silencer. (D) A 3 bp mutation in the human tDNAs in the strongest fragment (hatched column in C) was mutated and the insulator assay was repeated as described.
Having developed this simple assay we next asked if human tDNAs can function as insulators in S. pombe to block heterochromatin. We also tested the highly characterized S. cerevisiae tDNA insulator for its ability to block silencing in S. pombe. PCR fragments containing tDNAs were cloned between the S. pombe silencer and the Ura4+ gene (Figure 5A). The S. cerevisiae HMR tDNA insulator was able to function as an insulator in S. pombe, albeit less strongly than the S. pombe tDNA (Figure 5C). Analyses of the human tDNAs from chromosomes 6 and 17 indicated that several human tDNA fragments were able to protect the Ura4+ gene from silencing (Figure 5C; Supplementary Figure S4). Interestingly, not all human tDNA fragments had robust insulation abilities. This is consistent with the enhancer-blocking data and data in S. cerevisiae where some tDNAs are able to function as insulators while other tDNAs are not (Donze and Kamakaka, 2001). Among the human tDNAs, there appears to be a range of abilities to protect the Ura4+ gene from silencing, which do not strictly fall into yes or no categories, but instead appear to show a continuum of ability. Similar results were seen using tDNA genes from other loci on chromosome 6 (see Supplementary Figure S4). Some of the human tDNA fragments with robust insulation abilities were similar in strength to the known S. pombe tDNA barrier insulator. The most effective human tDNA fragment in this assay contained two out of the four tDNAs that are the most telomere-proximal tDNAs in the chromosome 17 cluster and separate the ALOXE3 gene from the HES7 gene.
To determine if the B-box sequence is required for human tDNAs to act as insulators in S. pombe, we made 3 bp mutations in the highly conserved residues in the B box in the tDNAs contained within the strongest insulator in Figure 5C (dark bar). These mutations significantly reduced the ability of this fragment to block silencing (Figure 5D). These data suggest that insulation by the human tDNAs in S. pombe was most likely mediated by the binding of S. pombe TFIIIC to the promoter of these genes (Huang and Maraia, 2001).
tDNAs can block repression in human cells
We chose to further characterize the DNA fragment containing these two tDNAs (ALOXE3 tDNA1). These two genes are in the same relative position in the opossum genome as they are in human genome (Figure 1) and are located near the transition between the active and repressed chromatin domains (Figure 2) and displayed robust insulation activity in the human enhancer blocking as well as the S. pombe barrier assay.
Tethering a repressor protein (polycomb protein CBX4) upstream of a reporter gene has been shown to induce a repressed state at the reporter gene (Smallwood et al, 2007; Vincenz and Kerppola, 2008). We decided to adapt this assay to further test the ability of the tDNA to function as an insulator. We developed a system to determine if tDNAs could block repression originating from a specific source. Nine Gal4 binding sites were cloned upstream of a luciferase reporter gene that was driven by the CMV promoter and we inserted the 1.2-kb DNA fragment containing the ALOXE3 tDNAs (but lacking CTCF) or one copy of the 1.2-kb chicken HS4 insulator between the Gal4 binding sites and the luciferase gene (Figure 6). These reporters were integrated into the genome of HEK-293 cells at a specific site using recombination mediated cassette exchange (Bode et al, 2000). The cell lines were analysed for proper integration by PCR. We then transiently transfected these cell lines with expression plasmids expressing Gal4–CBX4 or the Gal4 DNA binding domain (DBD) along with a plasmid expressing eGFP to mark transfected cells.
Figure 6.
Human tDNAs can block repression. Tethering of a repressor protein can silence genes. ALOXE3 tDNA pair1 or a cHS4 insulator-containing fragment were cloned between 9 × Gal4 binding sites and the luciferase gene and integrated into an FRT site in HEK-293 cells. Gal4–CBX4 or Gal4 DBD alone was transfected along with a GFP expressing plasmid and GFP+ cells were first sorted and then replated. Expression of the luciferase gene was measured 24 h later. The graph depicts percent expression by comparing Gal4–DBD transfected lines with GBD-CBX4 lines from three experiments. Asterisk denotes P<0.05.
We sorted eGFP-positive cells by fluorescence activated cell sorting (FACS), equal numbers of these transfected cells were replated, and after 24 h luciferase activity was quantified. In three independent experiments, we found that when cell lines containing no insulator were transfected with Gal4–CBX4, expression of luciferase was reduced 50% relative to the same cell line transfected with Gal4–DBD alone. In cell lines containing a tDNA insulator, expression of the luciferase reporter was two-fold higher than in the no insulator line (Figure 6). We also tested a single copy of the chicken HS4 insulator, and in this experiment, this well-established insulator functioned intermediately such that the expression of luciferase was 1.5-fold that of the no insulator cell line (Figure 6).
We also generated a cell line where the B box in the tDNA insulator had a 3-bbp mutation. When these lines were tested for their ability to block Gal4–CBX4 mediated repression of the luciferase gene, we found that the mutations in the tDNA insulator's B box disrupted the ability of the tDNA to act as an insulator such that the expression of luciferase was nearly identical to cell lines with no insulator (Figure 6).
tDNAs can protect a randomly integrated transgene from silencing in human cells
While the previous experiment demonstrated that tDNAs could block the spread of polycomb-mediated repression from a synthetic silencer, the effect was not very robust due to the narrow dynamic range of this assay. To directly assess the ability of tDNA-containing fragments to act as a barrier insulator in human cells, we performed a classical transgene protection assay (Pikaart et al, 1998; Emery et al, 2000). In mammalian cells, transgenes randomly integrate into the genome either as single or as multiple tandem copies. The expression of the transgene is dependent upon the number of copies integrated as well as the site of integration. Most often, single copy transgenes become silenced after a few days in culture due to the spread of heterochromatin from neighbouring sequences. Flanking the transgenes with insulators has previously been shown to slow the rate of silencing of transgenes, protecting them from the neighbouring heterochromatic silencing (Pikaart et al, 1998; Emery et al, 2000). We built a cassette where an eGFP reporter gene was placed under the control of the human γ-globin promoter (Figure 7A). Upon random integration into the human genome, the eGFP is usually silenced. We flanked the eGFP gene with either a single copy of the 1.2-kb fragment (containing two tDNAs) from chromosome 17 or with one copy of the well-characterized 1.2 kb chicken insulator from the β-globin locus, cHS4. We also tested the tDNA-containing DNA fragment where the B boxes were mutated. Human K562 cells were co-transfected with the linear plasmid containing the transgene as well as a plasmid expressing the puromycin resistance gene. We initially selected for puromycin-resistant transfectant colonies and then analysed the expression of the eGFP reporter after the drug was withdrawn. This allowed us to select for stable transfectants without any bias towards sites that are permissive to transcription. Once we had obtained stable transfectants, we determined the copy number of the eGFP reporter cassette that had integrated by qPCR of genomic DNA isolated from various stable cell lines. We investigated cell lines where the eGFP was in 1–3 copies in the genome (since K562 cells are effectively triploid cells; Naumann et al, 2001). We monitored expression of eGFP in many independent cell lines for each reporter construct by FACS over a period of 11 weeks.
Figure 7.
tDNAs can protect a randomly integrated transgene from silencing. (A) Schematic representation of the constructs used in the transgene protection assay. (B) A box plot of the percent of GFP expressing cells is plotted as a function of number of days. A single copy of chicken HS4 is compared with a single copy of the ALOXE3 tDNA pair1 insulator and an uninsulated transgene. The box includes the 25th–75th percent quartiles, the line is the median and the whiskers are 1.5 × the interquartile range. Open circles denote data points that are outside the whiskers (n=13 cell lines for no insulator; 15 cell lines for chicken HS4, 13 cell lines for tDNA and 15 cell lines for B-box delete).
The difference between the insulated and uninsulated lines is most apparent by looking at a box plot of cells expressing eGFP at a given day among all cell lines combined (Figure 7B). Our results showed that in most cell lines, cassettes with no flanking insulators were rapidly silenced (yellow box plot), while cassettes which were flanked with either the 1.2-kb tDNA-containing fragment (green box plot) or a single copy of the 1.2-kb chicken HS4 element (blue box plot) were partially protected from silencing in many cell lines over an 80-day period. Seven of the ten uninsulated lines (single copy integrants) were silenced by day 40. However, for both cHS4 and the tDNA-flanked lines, the majority of lines were still expressing at moderate-to-high levels at day 40 and even at day 80 (see Supplementary Figure S6A–D, for data on individual cell lines). These data show that both cHS4 and the human tDNA fragment were similarly effective in insulation. Interestingly, silencing did not appear to be a gradual decrease in expression of the reporter but an all or none phenomenon generating two populations of cells—either expressing eGFP or silenced and not expressing eGFP at all. To determine if the B-box sequence is required for the human tDNA-containing fragment to act as an insulator, we made 3 bp mutations in the highly conserved residues in the B box in the tDNAs. Mutations in the B box severely reduced the ability of this fragment to protect the transgenes. Mutations in the B box do not abolish insulation, suggesting that either this mutation does not completely eliminate TFIIIC binding or other factors that bind this DNA fragment play a role in insulation. These results do suggest that a single copy of the 1.2-kb tDNA-containing fragment was equivalent to a single copy of the well-characterized chicken insulator HS4 in the transgene protection assay and show that this effect is partially dependent upon TFIIIC binding to the B-box element present within the tDNA.
tDNAs coalesce together in human cells
Studies in yeast (Noma et al, 2006; Ruben et al, 2011), Drosophila (Gerasimova et al, 2000; Capelson and Corces, 2005), chicken (Yusufzai and Felsenfeld, 2004; Yusufzai et al, 2004) and human cells (Bartolomei, 2009; Mishiro et al, 2009) have shown that insulators cluster together and/or localize to special compartments in the nucleus separating distinct functional domains into separate chromatin domains. Since we show that tDNA-containing fragments have the ability to insulate genes, one prediction would be that these putative insulators would coalesce together in the nucleus forming distinct chromatin domains thereby sequestering different genes and their regulatory elements in different nuclear subcompartments. We decided to focus our attention on the DNA fragment containing four tDNAs immediately upstream of ALOXE3 on chromosome 17. We were curious about the three-dimensional organization of this region and decided to perform a 4C experiment (Zhao et al, 2006). We were interested in identifying interactions between the tDNA cluster and other sites in the genome in K562 cells in an unbiased manner. We performed a modified e4C-Seq (see Materials and methods; Schoenfelder et al, 2010b) using a Csp6I restriction fragment present in the ALOXE3 tDNA cluster as bait. Following deep sequencing, the prey sequences were mapped across the reference genome. This analysis was repeated using independently crosslinked samples and the hits from the two biological replicates were compared (Figure 8, sample 1 and sample 2). Very similar long-range interaction patterns were observed in both experiments (correlation coefficient for all sequenced prey fragments was 0.85), suggesting that the interactions are robust and reproducible. We focused our attention on interactions between the bait fragment and prey fragments on chromosome 17 and have not tried to comprehensively identify interactions between this tDNA cluster and sites on other chromosomes. Peaks were identified and analysed (see Supplementary Figure S7). The tRNA genes on chromosome 17 are evolutionarily conserved with respect to gene organization and contain many different RNA pol II transcribed genes that play different roles in the cell. Our analysis showed that the tDNA cluster physically contacted the neighbouring tDNA clusters on chromosome 17 but not the promoters of the RNA pol II transcribed genes even though these genes were closer to the bait. Interestingly, the ALOXE3 cluster makes physical contact with the PER1 tDNA cluster, the TMEM tDNA cluster and the AURK1 tDNA cluster. At this point, it is not clear if these contacts are simultaneous and long lasting or occur transiently in pair-wise fashion. Surprisingly, even though the second AURK2 tDNA cluster is only 3 kbp from the first AURK1 tDNA cluster, the ALOXE3 tDNA cluster did not make significant contact with this second cluster but the reason for this is unclear and will require additional experiments. We also detect small but reproducible interactions with ETC loci (that bind TFIIIC but not RNA pol III) present between PER1 and VAMP2 as well as between PFAS and C17orf68 (see asterisks in Figure 8A). This property of TFIIIC bound sites interacting with other TFIIIC bound sites is not solely a property of tDNAs on chromosome 17. An independent 4C experiment using a different bait sequence containing tDNAs on chromosome 6 also showed specific contacts between the bait tDNA and other tDNAs on that chromosome (data not shown).
Figure 8.
tDNAs coalesce together in human cells. (A) 4C-Seq analysis of the ALOXE3 tDNA cluster with associated loci on the human chromosome 17. A schematic representation of the RNA pol II and pol III transcribed genes around the bait fragment is shown. 4C reads are shown for the two biological replicates (sample 1 and sample 2). The bait fragment is marked by a red line. Asterisks mark ETC sites. (B) Quantitative 3C analysis of the ALOXE3 DNA fragment and its interactions with other Pst1 fragments across a 150-kb region in K562 cells. Relative interactions are plotted in arbitrary units. tDNA-containing fragments are marked by red bars at the top of the figure. Error bars are standard error of three reactions.
Having identified numerous interactions between the tDNA-containing fragments on chromosome 17 we confirmed these interactions using 3C (Dekker et al, 2002). To perform this analysis, we designed primers across the region of interest on chromosome 17 with all primers oriented in a single direction (Supplementary Table 4). The primers were first tested for amplification efficiencies using a BAC clone encompassing the 150-kb domain from chromosome 17 (data not shown) and all primers had similar binding/amplification efficiencies. K562 cells were then crosslinked, digested with PstI, ligated and the ligation products were analysed by PCR using these primers followed by quantitation as described (Miele et al, 2006). Major interactions were observed for primers present in restriction fragments containing tDNAs (Figure 8B) while intervening fragments showed low to no interactions. These data confirm the 4C data demonstrating that restriction fragments containing tDNAs interact with one another in the nucleus.
Discussion
Insulators are able to perform at least one of two functions: they can block the spread of silenced chromatin emanating from silencers and/or they can disrupt enhancer mediated gene activation. In small eukaryotes like S. cerevisiae and S. pombe, insulators are primarily promoters of highly active RNA pol II and RNA pol III transcribed genes that block the spread of silencing. In metazoans, insulators are primarily autonomous elements though there are some examples of gene promoters functioning as insulators (Raab and Kamakaka, 2010). In vertebrates, almost all insulators identified to date have been autonomous elements that bind the transcription factor CTCF (Phillips and Corces, 2009; Zlatanova and Caiafa, 2009). In this report, we have used a bioinformatic approach to initially identify putative tDNA insulators in human cells. We analysed the chromatin at these putative sites using available genome-wide data and extended the analysis at one of the potential tDNA insulator sites by quantitative ChIP. Our qChIP analyses are consistent with the prediction that DNA sequences containing tDNAs most likely aid in the partitioning of chromosomes. Using several different functional assays, we show that these tDNAs could function as insulators—both as barriers that restrict the spread of repressed chromatin and as enhancer blockers in human cells. Insulation mediated by the tDNA-containing fragment was dependent in part on the internal tDNA promoter, suggesting that it may be dependent upon the transcription factor TFIIIC. Our data show that promoters of genes can function as insulators in mammalian cells just as they do in yeast and the data suggest that the ability of tDNAs to insulate genes may be an inherent property of tDNAs. While tDNA bound TFIIIC likely plays a role in insulation in human cells, additional experiments will be necessary to determine which other factors that bind in the vicinity of these tDNAs also play a role in insulation.
tDNA clustering and insulation
Another key property of insulators is their tendency to cluster at specific sites in the genome. The scs and scs’ insulators interact directly with each other while the Gypsy insulator which binds Su(Hw) and other proteins forms insulator bodies (Gerasimova et al, 2000; Capelson and Corces, 2005; Dorman et al, 2007; Wallace and Felsenfeld, 2007; Bushey et al, 2008; Dorsett, 2009; Wendt and Peters, 2009; Zlatanova and Caiafa, 2009; Wood et al, 2010). tDNAs in yeast also tend to localize and cluster at specific sites in the genome via the recruitment of cohesins and condensins (Haeusler and Engelke, 2006; Dubey and Gartenberg, 2007; D’Ambrosio et al, 2008; Haeusler et al, 2008; Duan et al, 2010; Ruben et al, 2011). Our 4C data show that tDNA genes cluster in the human nucleus with other tDNA genes. We have analysed two different loci, one on chromosome 6 (data not shown) and the second on chromosome 17 using restriction fragments that contain tDNA genes as bait. Analysis of the interacting partners primarily identified other tDNAs and ETC sites. While the tDNA clusters used as bait had CTCF binding sites in its vicinity, the bait fragment preferentially bound to prey fragments containing TFIIIC binding sites and not CTCF binding sites. This is most obvious when one analyses DNA sequences on either side of the bait fragment on chromosome 17. The bait fragment adjacent to the ALOXE3 gene interacted with the tRNA genes to its right (centromere proximal) over a 150-kb region rather than the numerous CTCF sites to its left (telomere proximal). While the tDNA fragments that interact with the bait fragment have CTCF binding sites in their vicinity, the ETC loci that interact with the bait fragment lack CTCF sites. This is especially evident at the ETC locus between C17orf68 and the PFAS gene, suggesting that CTCF may not be the sole determinant for long-range interactions in human cells and suggests that other factors or TFIIIC alone may be playing a role in these interactions. These data collectively suggest that numerous proteins possibly including CTCF, TFIIIC as well as other unidentified proteins may regulate long-range interactions.
Many transcriptionally active RNA pol II promoters coalesce together at transcription factories (West and Fraser, 2005). Our 4C data show that the tDNAs on chromosome 17 interact with other tDNAs in three-dimensional space in the nucleus but do not interact with neighbouring RNA pol II enhancers and promoters, suggesting the presence of specific RNA pol III transcription factories. Consistent with this observation is immunofluorescence data demonstrating RNA pol III nuclear foci (Pombo et al, 1999).
It is believed that the mechanism by which enhancer-blockers function is by modulating long-range interactions in the nucleus. Two main models to explain enhancer-blocking insulators have been proposed. One model, based on the observation that enhancers interact with promoters, suggests that the enhancer blocker acts as a decoy by directly interacting with the enhancer, thereby precluding functional interactions between the enhancer and the promoter. A second model, based on the observation that insulators interact with one another to form insulator bodies, suggests that insulators interact with each other to partition the chromatin fiber into loops, such that enhancers in one loop do not or cannot activate promoters in a different loop (Geyer, 1997; Gerasimova and Corces, 2001). While our data do not formally preclude either insulator model, it is possible that clustering of tDNAs at RNA pol III transcription factories (West and Fraser, 2005; Bartlett et al, 2006; Xu and Cook, 2008; Schoenfelder et al, 2010a) could lead to enhancer blocking by sequestering RNA pol II promoters in a nuclear compartment deficient in pol II and enriched for pol III. This raises the possibility that insulator bodies may simply be specific transcription factories. Sequestering a specific gene promoter to one specific factory over another may result in either gene activation or repression/inactivation depending upon the transcription factors present at that factory. Enhancers and insulators could be visualized as DNA elements in a competition for recruitment of a promoter to a specific factory in the nucleus.
Multimerization and insulation
In S. cerevisiae, the ability to function as an insulator resides in the ability of the tDNA-containing fragment to stably bind its transcription factors TFIIIC and TFIIIB. Weak tDNA insulators become robust insulators when they are multimerized and data show that binding of the tDNA by the transcription factors is in competition with spreading repressor proteins (Donze and Kamakaka, 2001; Valenzuela et al, 2009). Similarly in S. pombe, tDNA insulators are present in arrays thus providing robust insulation (Noma et al, 2006; Iwasaki et al, 2010). In Drosophila and vertebrates, many insulators are compound DNA elements that bind multiple factors to mediate robust insulation (Gaszner and Felsenfeld, 2006; Bushey et al, 2008). We have identified tDNA insulators at specific loci in human cells. The locus we have studied has multiple tDNA genes clustered together. We have not deleted individual genes within the cluster to determine if they all function together in insulation. However, we have found that multimerizing the tDNAs results in an increase in enhancer blocking in human cells. It is, therefore, reasonable to suggest that the cluster of tDNAs together have a greater ability to insulate compared with a single tDNA gene. The clustering would increase the probability of stable binding of transcription factors and lead to successful insulation and could be a property that was selected for at specific sites in the genome.
We have characterized one locus in human cells where tDNAs cluster and we have found that the tDNAs from this locus can insulate genes. There are numerous others sites in the human genome where tDNAs cluster. tDNA also seem to cluster in other species such as C. elegans and Drosophila (Supplementary Figure S1B). It is possible that these sites are also functioning as insulators and future experiments should help answer this question. At this stage, we are unable to definitively say that all clustered tDNAs are functional insulators in all human cell types. It is entirely possible that some tDNA clusters function as insulators in one cell type but not another in a regulated manner regulated by other transcription factors.
tDNA conservation and insulation
tRNA genes are some of the most ancient genes in the genome and are thought to be present prior to the separation of the three domains of life (Eigen et al, 1989). Unlike paralogous copies of protein-coding genes, paralogous copies of tDNAs do not diverge in their sequences. Eukaryotic tDNAs have RNA polymerase III type 2 promoters (Geiduschek and Kassavetis, 2001; Schramm and Hernandez, 2002; Dieci et al, 2007). This promoter is present within the body of the gene and consists of two conserved regions named the A and B boxes. The promoter element recruits TFIIIC, TFIIIB and RNA polymerase III, which are conserved among eukaryotes (Huang and Maraia, 2001) such that many of the RNA polymerase III transcription factors are interchangeable within yeast and between humans and yeast (Teichmann et al, 1997; Proshkina et al, 2006).
This raises the question of why tDNAs were selected as insulators in diverse species. One interesting property of tDNAs is the observation that the sequences of tDNAs are highly conserved and the substitution rate of sequences within the gene is very low (compared with protein-coding genes; Dujon et al, 2004; Dujon, 2006; Withers et al, 2006). This property could have played a pivotal role in the selection of tDNAs as insulators. The requirement to maintain a precise tRNA structure (for protein translation) likely places limits to the changes possible within tRNA genes. Since the promoters of these genes lie within the body of the gene, alterations to these elements would consequently also be under severe selection pressure and these sequences would also not alter rapidly. This could help explain the conservation of tDNA promoters and their consequent selection as insulators in varied species.
Most RNA pol II transcribed genes rarely multimerize into arrays while tDNAs are often present as arrays. Multimerization also would aid in insulation simply as a function of probability of occupancy of the tDNAs by their cognate factors. This would result in a greater probability of chromatin disruption and thus greater probability of insulation. The selective advantage of insulation provided to RNA pol II transcribed genes by neighbouring tDNAs would aid in maintaining the tDNAs at that particular site thus the tDNA genes would remain syntenic with the surrounding genome. In agreement with this hypothesis, we found many tDNAs remained in syntenic positions throughout mammalian evolution. Analyses also show that many tDNAs remained syntenic across species and often locate at the edges of syntenic blocks in other organisms besides humans including Drosophila and yeast (Bermudez-Santana et al, 2010)
tDNAs and pol II genes
In human cells, many tDNAs reside adjacent to RNA pol II promoters and there is a clear positive correlation between the activity of the RNA pol II protein-coding gene and the activity of tDNA genes that reside immediately adjacent to the promoters of pol II transcribed genes (Moqtaderi et al, 2010; Oler et al, 2010; Raha et al, 2010). It is likely that RNA pol II genes are activated by promoter-specific transcription factors and the opening of chromatin by these factors could aid TFIIIC/TFIIIB to bind and transcribe adjacent tDNAs at some sites in the genome. It should be pointed out that while the tDNAs adjacent to the ALOXE3 promoter are most likely active since they bind RNA pol III, ALOXE3 is not active (see Figure 2).
In the context of tDNA-mediated enhancer blocking, we can speculate as to how pol II transcribed genes might affect and be affected by pol III transcribed genes. In one scenario, the effect could be unidirectional such that pol II-specific transcription factors regulate tDNA transcription but not vice versa. Alternatively, pol II-specific transcription factors could stimulate binding of TFIIIC/TFIIIB to tDNAs which in turn could lead to enhancer blocking, the consequences of which would be to generate a more stable transcription state where the RNA pol II transcribed gene activity state would not switch very often. The stabilization of gene activity states is critical for normal development and differentiation (Weintraub, 1985; Bernstein et al, 2006). Stabilization could be a key role for tDNAs and other insulators and may explain the observation that many tDNAs remain syntenic with their pol II transcribed genes. Future experiments dissecting multiple tDNA clusters in human cells should clarify these questions.
How do human tDNAs function as barriers?
Most insulators are DNaseI hypersensitive sites that are bound by sequence-specific transcription factors (Oki and Kamakaka, 2005; Xi et al, 2007; Boyle et al, 2008). Barrier insulators including tDNA barriers bind specific transcription factors that in turn recruit chromatin remodelling and modifying machines to generate and maintain a hypersensitive site. This disruption in the chromatin fiber is thought to play an important role in insulation (Gaszner and Felsenfeld, 2006; Raab and Kamakaka, 2010). The data with human tDNAs suggest a similar mechanism of action. Besides the observation that human tDNAs are nucleosome free and histones flanking tDNAs have active chromatin marks (Barski et al, 2010; Moqtaderi et al, 2010; Oler et al, 2010) preliminary data from our laboratory also show that p300 is present in the immediate vicinity of the tDNAs. This acetylase has been shown to interact with TFIIIC (Mertens and Roeder, 2008) and acetylates histones in chromatin and it is therefore likely that this acetylase recruited in the vicinity of the tDNA insulator (either directly or indirectly) sets up a chromatin state amenable to pol III factor binding and insulation.
While heterochromatin in S. cerevisiae, S. pombe and human cells is quite distinct, tDNAs function as insulators in all three systems, suggesting that the mechanism of insulation is conserved. What is likely to be the core mechanism that allows tDNAs to insulate different forms of heterochromatin? Based on the studies in yeast, we have suggested that stable binding of tDNA transcription factors coupled with histone acetylation are drivers for insulation (Oki and Kamakaka, 2005). If the histones are acetylated then they are not going to be deacetylated and by definition they will also not be methylated. The presence of active acetylation marks would preclude binding of repressor proteins such as the polycomb group proteins and the HP1 family of proteins thus terminating the spread of repressed chromatin. Furthermore, tDNA transcription is directly regulated by various transcription factors such as OCT, Fos/Jun and Myc and possibly even CTCF that bind in the vicinity of tDNAs (Hoeffler and Roeder, 1985; Felton-Edkins et al, 2003; Woiwode et al, 2008; Oler et al, 2010) and play a role in generating a chromatin environment amenable for tDNA transcription, the consequences of which could be insulation of RNA pol II transcribed genes. The fact that tDNAs are transcriptionally active likely allows these sites to remain in an open nucleosomal-free configuration aiding in their function as insulators. Thus, while there are additional complexities in silenced chromatin in different organisms, the basic mechanism by which tDNAs are transcribed allows them to simultaneously act as insulators to all different forms of heterochromatin and tDNAs may have inherent molecular properties that allow them to be suborned as insulators.
Materials and methods
Cell culture
K562 cells and HEK-293 cells were obtained from ATCC and HEK-293-FRT cells were obtained from Graeme Cottrell (Cottrell et al, 2005). HEK-293 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM)+10% fetal bovine serum (FBS) with penicillin/streptomycin. K562 cells were cultured in Iscove's modification of DMEM containing 10% FBS and penicillin/streptomycin. All cells were maintained in a 37°C incubator in 5% CO2.
Synteny analysis
Synteny chains of human/mouse and human/possum alignments were downloaded from the UCSC genome browser hg18 build (Karolchik et al, 2003). A tDNA was considered to be syntenic with its surroundings if it satisfied at least one of the following conditions. If the tDNA was located in a top-level chain it was considered to be syntenic, since this showed that the tDNA was located in a fully conserved region of the genome. If the tDNA in humans was located in a gap in the top-level chain, the gap in the next species (which corresponded to the human gap) was searched for a tDNA that contained the identical anticodon using http://gtRNAdb.ucsc.edu or tRNAscan-SE (Lowe and Eddy, 1997). If such a second-species tDNA was found, then the human tDNA was considered to be syntenic, else it was considered not to be syntenic. Similar analysis was performed for other repetitive RNA families using location data obtained from RepeatMasker tracks for the snRNA family of repeats and the srpRNA (7SL RNA) family also downloaded from hg18 annotations on http://genome.ucsc.edu.
RT–PCR
RNA was collected from K562 cells grown as described above using Trizol (Invitrogen, USA) per manufacturer's recommendations. In all, 5 μg RNA was converted to cDNA using Superscript III reverse transcriptase per manufacturer's instructions (Invitrogen). Equal amounts of cDNA were used in qPCRs to determine if gene-specific products were present. Primers used for qPCR are available in supplementary table.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were done as previously described (Fujita and Wade, 2004). Briefly, K562 cells were grown in IMDM supplemented with 10% FBS and penicillin and streptomycin. In all, 1 × 108 cells were pelleted and fixed in 1% formaldehyde in PBS for 10 min at room temperature. Fixation was stopped by adding glycine to 0.125 M and incubating for 5 min at room temperature. Cells were washed twice in PBS and lysed in a solution of 1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.0 and 1 mM PMSF for 10 min. Chromatin was sheared using a Diagenode BioRuptor for eight cycles of 30 s at high power with 2 min rest after each cycle. Chromatin was spun at 4°C for 10 min. Chromatin was diluted in 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.0, 167 mM NaCl, 1 mM PMSF to a concentration of 1 × 107 cells/ml. Chromatin was then precleared using protein A beads (Millipore, Billerica, MA) by adding 25 μl beads/ml and rotating for 30 min at 4°C. Chromatin was briefly spun to pellet the beads and cleared chromatin was aliquoted (2 ml per tube). One to four micrograms of antibodies (H3K36me3 ab9050, Abcam, Cambridge, MA; H3K27me3-17-622, acetylated H3-06-599, CTCF-07-729, Millipore; GTF3C5-BP301-242A, Bethyl Laboratories, Montgomery, TX) were added to tubes and incubated overnight at 4°C. In all, 50 μl protein A beads were added to each sample and rotated for 1 h at 4°C. Beads were washed once each with low salt (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA and 0.1% SDS), high salt (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA and 0.1% SDS), LiCl (10 mM Tris–HCl pH 8.0 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate and 1 mM EDTA), and twice with TE (10 mM Tris–HCl pH 7.5, 1 mM EDTA). DNA/protein complexes were eluted from the beads using a solution of 1% SDS and 100 mM sodium bicarbonate by rotating at room temperature for 15 min two times. Eluates were pooled, 25 μl of 5 M NaCl was added and the eluate was incubated at 65°C overnight to reverse the crosslinks. In all, 10 μl of 0.5 M EDTA, 10 μl Tris–HCl pH 6.5 and 2 μl proteinase K (10 mg/ml) was added to each sample and incubated 1 h at 42°C. After phenol–chloroform extraction, DNA was ethanol precipitated and quantified using the Picogreen dsDNA quantitation kit (Invitrogen) using a fluorescent spectrophotometer. Equal amounts of DNA were used in qPCRs with SybrGreen. Fold enrichment was calculated as follows: 2ΔCt (between input and immunoprecipitated material). This was further normalized to a negative control locus within the same genomic region as indicated in the figure legends. All primers used in qPCRs are available in Supplementary Table 2.
Human enhancer-blocking assay
Putative insulators were cloned into the AscI site of pNI (Gift of Gary Felsenfeld). Enhancer-blocking assays were performed as previously described with modifications (Chung et al, 1993). Briefly, 800 ng of linearized plasmid was transfected into 4 × 105 K562 cells using Lipofectamine 2000 as per manufacturer's instructions (Invitrogen). Two days after transfection, cells were split into triplicate and plated in IMDM+10% FBS+800 μg/ml G418 with 0.3% Noble agar. After 2 weeks, plates were stained for 1 h with 0.005% crystal violet and washed to remove excess stain. Pictures of plates were taken using a gel documentation system (Fluorchem, Alpha Innotech). Numbers of colonies were counted using ImageJ software by converting to a binary image and using the Analyze particles tool. Colony numbers for each experiment were normalized to number of colonies from pNI (equal to 1).
S. pombe barrier assay
A fragment containing two tDNAs from the S. pombe centromere was cloned into the SacI site of pUR19K (gift from K Scott and R Allshire; Partridge et al, 2002). Putative insulator fragments containing tDNAs were PCR amplified (see Supplementary Table 3 for cloned sequences) and cloned into the PstI site of pUR19K between the K repeat silencer (ClaI fragment) and the Ura4+ gene. Location and information about each tDNA fragment are in supplementary table. Equal amounts of pUR19K and a plasmid containing the Leu1+ gene (Adams et al, 2005) were transformed into an S. pombe strain (ROP53-ura−, leu−) and plated on yeast minimal media plates lacking uracil or leucine. After 5 days growth at 30°C, the number of colonies was counted on each plate and the number of uracil-positive colonies was reported relative to the number of leucine-positive colonies.
Human repressor-blocking assay
Stable HEK-293 cell lines were made using site-specific recombination. An HEK-293 cell line containing an FRT site (gift of Graeme Cottrel) was transfected with constructs containing nine Gal4 binding sites upstream of a luciferase reporter driven by the CMV promoter and the putative insulators shown in the figure. Insulators were cloned as an AscI–FseI fragment between the binding sites and the luciferase gene driven by the CMV promoter. Expression of flp recombinase by co-transfection of pOG44 allowed single integrations of these constructs into a specific locus. These stable cell lines were subsequently co-transfected with Gal4–CBX4 or Gal4–DBD alone (Vincenz and Kerppola, 2008) and a GFP plasmid. GFP-positive cells were sorted on a BD FacsAria cell sorter (BD Biosciences, San Jose, CA) and 1 × 105 cells were plated. Supernatant was harvested 24 h later and assayed for luciferase expression using a Ready-to-Glow Secreted Luciferase Kit (Clontech, Mountain View, CA). Percent expression was determined as the amount of luciferase expression of the cell line transfected with the repressor normalized to the cell line transfected with the Gal4 DNA binding domain alone.
Human transgene protection assay
Constructs were linearized with ScaI and co-transfected with pBABE-puro into 4 × 105 K562 cells using Lipofectamine 2000 per manufacturer's conditions at a 3:1 ratio. Forty-eight hours post transfection, cells were plated in 2% Methocel 4000 (Sigma-Aldrich, St. Louis, MO) containing 2 μg/ml puromycin. After 7 days, single colonies were isolated and re-plated individually in 96-well plates containing K562 conditioned media with 2 μg/ml puromycin. After a further 5 days, colonies were expanded in 24-well plates, grown 3 more days and then transferred to 6-well plates. At this point, selection was removed (day 0 is removal of selection), and GFP expression was measured by FACS at the time points indicated. Total selection time was 15 days. DNA was collected from these cell lines and qPCR was performed to identify copy number. FACS was performed by washing cells two times in PBS and was carried out on a BD LSRII (BD Biosciences). For some early time points, we were unable to collect enough cells and these plots may appear blank. Data were analysed using FlowJo software (Treestar, Inc, Ashland, OR).
4C analysis
K562 cells were cultured in Iscove's DMEM, supplemented with 10% FBS, and the cells harvested at 400 g for 8 min at 4°C. Approximately 1 × 108 cells were fixed for 5 min in 2% formaldehyde in DMEM/10% FBS at room temperature, and then quenched by the addition of ice-cold glycine to 125 mM. The cells were harvested at 400 g for 8 min at 4°C, washed in cold PBS, and then nuclei were extracted by a 10–15-min incubation on ice in lysis buffer (10 mM Tris–HCl, pH 8, 10 mM NaCl, 0.2% NP-40, 1 × complete EDTA-free protease inhibitor cocktail; Roche). The nuclei were harvested at 800 g for 5 min at 4°C and separated into 1 × 107 aliquots in 500 μl Buffer B (Fermentas). SDS was added to 0.3%, and the nuclei were incubated for 1 h at 37°C, 950 r.p.m. Triton X-100 was added to 1.8% and the nuclei were incubated for a further hour at 37°C, 950 r.p.m. Csp6I (600 U; Fermentas) was added and the nuclei were digested overnight at 37°C, 950 r.p.m. The restriction enzyme was inactivated and the chromatin solubilized by the addition of SDS to 1.6% and incubation for 30 min at 65°C, 950 r.p.m. The chromatin was diluted into 7 ml of 1.1 × T4 DNA ligase buffer (New England Biolabs), Triton X-100 was added to 1%, and the chromatin was incubated at 37°C for 1 h. T4 DNA ligase (800 U; New England Biolabs) was added and ligation was carried out at 16°C for 4 h, followed by 30 min at room temperature. Proteinase K was added to 100 μg/ml and crosslinks were reversed overnight at 65°C. 3C DNA was subsequently purified by RNase A digestion (50 μg/ml; 37°C for 30 min), phenol extraction, chloroform extraction and ethanol precipitation. DNA was quantified by a PicoGreen assay (Invitrogen).
Generation of 3C material for confirmation of e4C peaks
3C was done as previously described (Dekker, Current Protocols). DNA was digested with PstI (New England Biolabs). PCR was carried out using MangoMix (Bioline) using 250–300 ng DNA and the following conditions: 95°C for 5 min; 38 cycles of 95°C for 30 s, 68°C for 45 s, 70°C for 1 min; 70°C for 5 min and products were separated on a 1.5% agarose gel stained with 0.5 μg/μl ethidium bromide. Images of gels were captured using a gel doc and band intensities quantified using ImageJ. Interaction frequencies were calculated relative to a control template made from BAC RP11-1D5, which contained all expected ligation products in equimolar amounts (Childrens Hospital Oakland Research Institute).
High-resolution e4C-Seq
In all, 20 μg aliquots of 3C DNA were diluted in 10 mM Tris–HCl pH 8.5 and fragmented using a probe sonicator (MISONIX Sonicator 3000) to generate 3C DNA fragments in the size range of 100–500 bp. In all, 20 μg of sonicated 3C DNA from two independent batches of K562 cells was used as template for 50 μl primer extension reactions with 5 pmol each of R1, R3 and R4 biotinylated primers separately, 200 μM dNTPs and 2 U Vent (exo-) DNA polymerase (New England Biolabs) in manufacturer-supplied buffer, under the following conditions: 95°C, 4 min; 60°C, 2 min; 72°C, 10 min. The primer extension reactions were terminated by rapid chilling on ice. Unincorporated biotinylated primers were removed using QiaQuick PCR purification columns (Qiagen). In all, 50 μl of the primer-extended material was bound to 200 μg magnetic streptavidin-coated beads (Dynabeads M-280 Streptavidin; Invitrogen) with 50 μl binding buffer (provided with Dynabeads kilobase BINDER kit; Invitrogen) for 4 h at 25°C, 1200 r.p.m. The beads were washed twice with 500 μl wash buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 2 M NaCl) and once with 500 μl 10 mM Tris–HCl, pH 8, before repairing DNA ends using 0.6 U T4 DNA Polymerase (New England Biolabs) in 110 μl NEB buffer 2 with 100 μM dNTPs, 100 μg/ml BSA at 16°C for 30 min. The beads were washed as previously and the bound DNA ligated to 200 pmol of custom-made blunt ended versions of the Illumina P1 adapter with 2000 U high-concentration T4 DNA ligase (New England Biolabs) in 40 μl 1 × T4 ligase buffer (New England Biolabs) overnight at 37°C. PE P1 adapter is the Illumina P1 adapter without the 5′-T to allow blunt-ended ligation in the generation libraries. GCG was introduced at the 3′-end of the forward strand of the adapters to allow for multiplexing with other samples.
The beads were washed as previously and used as template in a 50-μl PCR with 400 nM of Illumina PE P2-linked nested primer, 400 nM Illumina P1 sequencing primer (which is identical to the PE P1 adapter forward strand), 200 μM dNTPs, and 5 U HotStar Taq DNA polymerase (Qiagen) in 1 × manufacturer-supplied buffer, and the following cycle conditions: 95°C, 15 min; 23 cycles of 94°C, 30 s, 55°C, 30 s, 72°C, 1 min; 55°C, 2 min; 72°C, 10 min. e4C products (R1, R3 and R4) were taken as the supernatant of the PCR, and purified with a QiaQuick PCR purification column (Qiagen).
Primers used in generation of this library are listed below.
Sequencing e4C-Seq libraries
Products were size fractionated by electrophoresis on a 2% agarose gel and the appropriate-sized DNA was purified using a Qiaquick Gel Extraction Kit (Qiagen). The size-selected material was amplified with Illumina PCR amplification bridging primers (PE PP1 primer and PE PP2 primer for paired-end libraries) for 10 cycles as per the manufacturer's instructions. Libraries were sequenced as paired-end reads using a Genome Analyzer IIa (Illumina). The first three sequenced nucleotides correspond to the indexed ‘barcode’ that was incorporated. The average DNA fragment sizes for the paired-end sequenced libraries were 200–300 bp including all adapter sequences.
Sequence alignment and mapping
Reads were aligned to the human reference genome (Hg18) using bowtie. Start positions of reads were extracted and were assigned to the corresponding restriction fragment for visualization on the UCSC genome browser. Replicate 1 generated 0.33 million uniquely mapped reads and Replicate 2 generated 4.83 million uniquely mapped reads. The replicates were highly correlated within the 1-MB region surrounding the bait region, but not elsewhere in the genome, leading us to conclude we cannot reliably analyse the region distant from the bait or on other chromosomes using this data set. To calculate peaks within this region, the number of reads for each restriction fragment was plotted with respect to number of restriction fragments from the bait point. We plotted a loess smoothing line (span=0.4) and an approximation of the 95% confidence interval. Peaks above this confidence interval suggest a real long-range interaction. Sequences and bed and bam files generated from mapping can be found under GEO accession GSE31105
Primers and adapters
The sequences of all primers and oligonucleotides used in this study are given below. All Illumina adapter sequences were procured from the Illumina technical service (Oligonucleotide sequences© 2006 and 2008 Illumina Inc. All rights reserved) and were commercially synthesized with HPLC purification. Adapters were made by mixing equimolar amounts of forward and reverse strand oligonucleotides, heating to 90°C for 2 min, and then cooling slowly using a thermal cycler.
Biotinylated Bait: 5′-/5BioTEG/GTCCCGCCCAAGTCCCTTA-3′;
PE GCG-P1 adapter forward: ACACTCTTTCCCTACACGACGCTCTTCCGATCTGCG;
PE GCG-P1 adapter reverse: P-CGCAGATCGGAAGAGCGTCGTGTAGGGAAAGA-Am;
PE P2-linked R1 nested primer: TCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTCCTGCCCCATTCCCACTCTA;
PE PP1 primer: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGAT CT;
PE PP2 primer: CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT.
Supplementary Material
Acknowledgments
We would like to thank Adam West, Mike Bulger and Frederique Magdinier for protocols and advice during the course of this research. We would like to thank Gary Felsenfeld, Nigel Bunnett, Robin Allshire, Kristin Scott, Michael Carey, Claudius Vincenz and Naoko Tanese for plasmids and reagents. We would also like to acknowledge the technical help of Serdar Kasakyan. This work was supported by a grant from the NIH (GM078068) and the University of California Cancer Research coordinating committee to RTK and (T32-GM008646, and the GREAT Training Program of the UC System wide Biotechnology Research and Education Program, grant # 2008–16) to JR. Integrated DNA technologies synthesized the wild-type and mutant 1.2 kb fragment analysed in Supplementary Figure S4.
Author contributions: JR performed the experiments, analysed the experimental and bioinformatic data and wrote the manuscript. John Chiu helped generate constructs described in Figures 4 and 6. JZ performed the bioinformatic analyses described in Supplementary Figure S1A. SK analysed the 4C genomic data described in Figure 8. SK performed the experiments described in Figure 8A. Early chromatin immunoprecipitation experiments were performed under the supervision of Dr Wade in his laboratory. DH supervised the bioinformatic analyses described. Almost all of the work was performed in the laboratory of RK who also initiated and supervised the project and co-wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams C, Haldar D, Kamakaka RT (2005) Construction and characterization of a series of vectors for Schizosaccharomyces pombe. Yeast 22: 1307–1314 [DOI] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD (2005) TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev 19: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K (2010) Pol II and its associated epigenetic marks co-localize throughout the genome with pol III-transcribed non-coding RNA genes. Nat Struct Mol Biol 17: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartlett J, Blagojevic J, Carter D, Eskiw C, Fromaget M, Job C, Shamsher M, Trindade IF, Xu M, Cook PR (2006) Specialized transcription factories. Biochem Soc Symp 73: 67–75 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS (2009) Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 23: 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (1999) Stopped at the border: boundaries and insulators. Curr Opin Genet Dev 9: 191–198 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bermudez-Santana C, Attolini CS, Kirsten T, Engelhardt J, Prohaska SJ, Steigele S, Stadler PF (2010) Genomic organization of eukaryotic tRNAs. BMC Genomics 11: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Biswas M, Maqani N, Rai R, Kumaran SP, Iyer KR, Sendinc E, Smith JS, Laloraya S (2009) Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae. Mol Cell Biol 29: 2889–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J, Schlake T, Iber M, Schubeler D, Seibler J, Snezhkov E, Nikolaev L (2000) The transgeneticist’s toolbox: novel methods for the targeted modification of eukaryotic genomes. Biol Chem 381: 801–813 [DOI] [PubMed] [Google Scholar]
- Bose T, Gerton JL (2010) Cohesinopathies, gene expression, and chromatin organization. J Cell Biol 189: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G (2002) The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA 99 (Suppl 4): 16433–16437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG (2008) Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20: 105–116 [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Copeland CS, Marz M, Rose D, Hertel J, Brindley PJ, Santana CB, Kehr S, Attolini CS, Stadler PF (2009) Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genomics 10: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell GS, Roosterman D, Marvizon JC, Song B, Wick E, Pikios S, Wong H, Berthelier C, Tang Y, Sternini C, Bunnett NW, Grady EF (2005) Localization of calcitonin receptor-like receptor and receptor activity modifying protein 1 in enteric neurons, dorsal root ganglia, and the spinal cord of the rat. J Comp Neurol 490: 239–255 [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA (2002) The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell 9: 563–573 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dhillon N, Raab J, Guzzo J, Szyjka SJ, Gangadharan S, Aparicio OM, Andrews B, Kamakaka RT (2009) DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J 28: 2583–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson J, Gowher H, Strogantsev R, Gaszner M, Hair A, Felsenfeld G, West AG (2010) VEZF1 elements mediate protection from DNA methylation. PLoS Genet 6: e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23: 614–622 [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT (1999) The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT (2001) RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J 20: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman ER, Bushey AM, Corces VG (2007) The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol 18: 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2009) Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res 17: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS (2010) A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey RN, Gartenberg MR (2007) A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev 21: 2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B (2006) Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet 22: 375–387 [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E et al. (2004) Genome evolution in yeasts. Nature 430: 35–44 [DOI] [PubMed] [Google Scholar]
- Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, Larionov V (2011) tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle 10: 2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M, Lindemann BF, Tietze M, Winkler-Oswatitsch R, Dress A, von Haeseler A (1989) How old is the genetic code? Statistical geometry of tRNA provides an answer. Science 244: 673–679 [DOI] [PubMed] [Google Scholar]
- Elgin SC, Grewal SI (2003) Heterochromatin: silence is golden. Curr Biol 13: R895–R898 [DOI] [PubMed] [Google Scholar]
- Emery DW, Yannaki E, Tubb J, Stamatoyannopoulos G (2000) A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc Natl Acad Sci USA 97: 9150–9155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairley JA, Scott PH, White RJ (2003) TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J 22: 5841–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ (2003) Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2: 181–184 [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM (2005) Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8: 31–42 [DOI] [PubMed] [Google Scholar]
- Fodor BD, Shukeir N, Reuter G, Jenuwein T (2010) Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol [DOI] [PubMed] [Google Scholar]
- Frenkel FE, Chaley MB, Korotkov EV, Skryabin KG (2004) Evolution of tRNA-like sequences and genome variability. Gene 335: 57–71 [DOI] [PubMed] [Google Scholar]
- Fujita N, Wade PA (2004) Use of bifunctional cross-linking reagents in mapping genomic distribution of chromatin remodeling complexes. Methods 33: 81–85 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gause M, Schaaf CA, Dorsett D (2008) Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays 30: 715–718 [DOI] [PubMed] [Google Scholar]
- Gdula DA, Gerasimova TI, Corces VG (1996) Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA 93: 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA (2001) The RNA polymerase III transcription apparatus. J Mol Biol 310: 1–26 [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Tocchini-Valentini GP (1988) Transcription by RNA polymerase III. Annu Rev Biochem 57: 873–914 [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T (2005) Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev 19: 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet 35: 193–208 [DOI] [PubMed] [Google Scholar]
- Geyer PK (1997) The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev 7: 242–248 [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour JM, Pan T (2006) Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34: 6137–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Abelson J (1982) Organization and expression of tRNA genes in Saccharomyces cerevisiae. In The Molecular Biology of the Yeast Saccharomyces, Vol. Metabolism and Gene Expression, Strathern JN, Jones EW, Broach JR (eds), pp 487–528. Cold Spring Harbor, NY: Cold Spring Harbor Labs [Google Scholar]
- Haeusler RA, Engelke DR (2006) Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res 34: 4826–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR (2008) Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar D, Kamakaka RT (2006) tRNA genes as chromatin barriers. Nat Struct Mol Biol 13: 192–193 [DOI] [PubMed] [Google Scholar]
- Hamada M, Huang Y, Lowe TM, Maraia RJ (2001) Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol Cell Biol 21: 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O (2003) Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J 22: 4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler WK, Roeder RG (1985) Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell 41: 955–963 [DOI] [PubMed] [Google Scholar]
- Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G (2007) USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol 27: 7991–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Maraia RJ (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res 29: 2675–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Brower-Toland B, Elgin SC (2006) The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma 115: 110–122 [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K (2010) Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D (2005) Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171: 913–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ (2003) The UCSC genome browser database. Nucleic Acids Res 31: 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Ebersole T, Kouprina N, Noskov VN, Ohzeki J, Masumoto H, Mravinac B, Sullivan BA, Pavlicek A, Dovat S, Pack SD, Kwon YW, Flanagan PT, Loukinov D, Lobanenkov V, Larionov V (2009) Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Res 19: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Clarke L, Carbon J (1991) Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc Natl Acad Sci USA 88: 1306–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, Yung Y, Montoliu L, Glass CK, Rosenfeld MG (2007) Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251 [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F (2003) Ensuring enhancer fidelity. Nat Genet 34: 360–361 [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F (2007) Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev 17: 394–399 [DOI] [PubMed] [Google Scholar]
- Magdinier F, Yusufzai TM, Felsenfeld G (2004) Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J Biol Chem 279: 25381–25389 [DOI] [PubMed] [Google Scholar]
- McFarlane RJ, Whitehall SK (2009) tRNA genes in eukaryotic genome organization and reorganization. Cell Cycle 8: 3102–3106 [DOI] [PubMed] [Google Scholar]
- McNairn AJ, Gerton JL (2008) The chromosome glue gets a little stickier. Trends Genet 24: 382–389 [DOI] [PubMed] [Google Scholar]
- Mertens C, Roeder RG (2008) Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol 28: 5764–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J (2006) Mapping chromatin interactions by chromosome conformation capture. Curr Protoc Mol Biol, Chapter 21: Unit 21.11 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M (2009) Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K (2004) Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol 24: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K (2010) Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH, Roth SY, Simpson RT (1992) A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol 12: 4015–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann S, Reutzel D, Speicher M, Decker HJ (2001) Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk Res 25: 313–322 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16: 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI (2006) A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Oki M, Kamakaka RT (2005) Barrier function at HMR. Mol Cell 19: 707–716 [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hegedorn CH, Graves BJ, Cairns BR (2010) Human RNA polymerase III transcriptomes and relationships to Pol II promoters, enhancer-binding factors and chromatin domains. Nat Struct Mol Biol 17: 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Rival-Gervier S, Boussouar A, Foerster AM, Rondier D, Sacconi S, Desnuelle C, Gilson E, Magdinier F (2009a) The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet 5: e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Schluth-Bolard C, Rival-Gervier S, Boussouar A, Rondier D, Foerster AM, Morere J, Bauwens S, Gazzo S, Callet-Bauchu E, Gilson E, Magdinier F (2009b) Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J 28: 2428–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR (2008) RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S (1994) Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res 22: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrykowska HM, Vockley CM, Elnitski L (2008) Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res 18: 1238–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev 12: 2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR (1999) Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J 18: 2241–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkina GM, Shematorova EK, Proshkin SA, Zaros C, Thuriaux P, Shpakovski GV (2006) Ancient origin, functional conservation and fast evolution of DNA-dependent RNA polymerase III. Nucleic Acids Res 34: 3615–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Kamakaka RT (2010) Insulators and promoters: closer than we think. Nat Rev Genet 11: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M (2010) Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci USA 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Bell AC, Felsenfeld G (1999) Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc Natl Acad Sci USA 96: 14354–14359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G (2002) Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA 99: 6883–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR (2003) The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA 100: 14695–14700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, Ballestar E, Gomez-Skarmeta JL, Vavrova-Anderson J, White RJ, Montoliu L, Fernandez-Salguero PM (2011) Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res 21: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Pirrotta V, Geyer PK (1993) The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J 12: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowold DJ, Herrera RJ (2000) Alu elements and the human genome. Genetica 108: 57–72 [DOI] [PubMed] [Google Scholar]
- Ruben GJ, Kirkland JG, Macdonough T, Chen M, Dubey RN, Gartenberg MR, Kamakaka RT (2011) Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS One 6: e21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P (2010a) The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P (2010b) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Zhao K (2008) Genome-wide approaches to studying chromatin modifications. Nat Rev Genet 9: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W (2003) Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16: 119–129 [DOI] [PubMed] [Google Scholar]
- Sen R, Grosschedl R (2010) Memories of lost enhancers. Genes Dev 24: 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, Edwards DJ, Donze D (2008) TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell 7: 2078–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH (2005) Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol Biol Evol 22: 767–775 [DOI] [PubMed] [Google Scholar]
- Smallwood A, Esteve PO, Pradhan S, Carey M (2007) Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 21: 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Gouilloud E, Clarkson SG (1989) Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes Dev 3: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Teichmann M, Dieci G, Huet J, Ruth J, Sentenac A, Seifart KH (1997) Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J 16: 4708–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Reinberg D (2007) Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28: 1–13 [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Kamakaka RT (2009) Transcription independent insulation at TFIIIC-dependent insulators. Genetics 183: 131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT (2006) Chromatin insulators. Annu Rev Genet 40: 107–138 [DOI] [PubMed] [Google Scholar]
- Vincenz C, Kerppola TK (2008) Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci USA 105: 16572–16577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA (2009) Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G (2007) We gather together: insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Weintraub H (1985) Assembly and propagation of repressed and depressed chromosomal states. Cell 42: 705–711 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Peters JM (2009) How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res 17: 201–214 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- West AG, Fraser P (2005) Remote control of gene transcription. Hum Mol Genet 14 Spec No 1: R101–R111 [DOI] [PubMed] [Google Scholar]
- West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G (2004) Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell 16: 453–463 [DOI] [PubMed] [Google Scholar]
- Withers M, Wernisch L, dos Reis M (2006) Archaeology and evolution of transfer RNA genes in the Escherichia coli genome. RNA 12: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL (2008) PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol 28: 4204–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ (2010) Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet 11: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Cook PR (2008) The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta 1783: 2155–2160 [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Felsenfeld G (2004) The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc Natl Acad Sci USA 101: 8620–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P (2009) CTCF and its protein partners: divide and rule? J Cell Sci 122(Part 9): 1275–1284 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.