Abstract
Two high-resolution maps of meiotic recombination initiation sites across the genomes of budding yeast and mice illuminate broad similarities in the control of meiotic recombination in these diverse species but also highlight key differences. These studies offer new insights into the relationships between recombination, chromosome structure, and genome evolution.
Sexual reproduction in eukaryotes involves meiosis, a specialized cell division in which diploid cells differentiate into haploid gametes. The halving of the genome content is accomplished by a single round of DNA replication, followed by two rounds of chromosome segregation. This requires connecting homologous chromosomes of different parental origin called homologs before the first meiotic division. In most eukaryotes, these connections are established by the programmed formation of DNA double-strand breaks (DSBs) and their subsequent repair by interhomolog recombination, leading to reciprocal exchanges between homologs called crossovers (Hunter, 2007). Crossovers ensure proper homolog segregation and generate new allele combinations that increase genome diversity. Recombination between dispersed repeated sequences can cause genome instability, chromosome rearrangement, and, thus, genetic disorders (Sasaki et al., 2010). Therefore, meiotic DSB formation and repair must be tightly regulated to ensure precise genome division among gametes while avoiding deleterious genome modifications.
To understand this regulation, we must characterize components that control the location and frequency of meiotic DSBs. Several approaches involving either direct DSB detection or comprehensive analysis of recombination patterns have shown that recombination events cluster in discreet regions called hot spots (Pan and Keeney, 2007). Each hot spot is active in a minor fraction of cells (typically less than 10%–0.01%), whereas non-hot spot regions display even lower or undetectable recombination activity. Identification of factors that contribute to this regulation requires determining DSB locations across the genome and at high resolution, which is technically challenging. Two recent papers have made significant progress toward understanding meiotic DSB distributions. The first, in Saccharomyces cerevisiae (Pan et al., 2011), provides the first nucleotide resolution DSB map in any species. The second, in Mus musculus (Smagulova et al., 2011), presents the first genome-wide DSB map in a mammal.
Mapping DSBs in Yeast and Mice
Meiotic DSB formation occurs through a conserved mechanism involving Spo11, a homolog of the catalytic subunit of topoisomerase VI with no or little DNA sequence specificity (Keeney, 2008) (Figure 1A). After initial cleavage, Spo11 remains covalently linked to the 5′ ends of DSBs. A nick in the 5′ end releases the Spo11-oligonucleotide complex. DSB ends then undergo 5′-to-3′ resection, generating long 3′ overhangs that are bound by the Rad51 and Dmc1 recombinases (homologs of E. coli RecA) (Hunter, 2007).
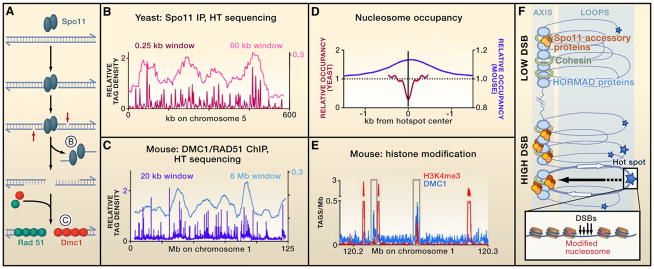
Figure 1. Genome-wide Features of Meiotic Double-Strand Breaks.
(A) Mechanisms of meiotic DNA double-strand break (DSBs) formation, showing the substrates that Pan et al. (2011) and Smagulova et al. (2011) used to map DSBs in yeast and mouse, respectively.
(B and C) DSB distributions on yeast and mouse chromosomes, with smoothing windows to show individual DSB hot spots and DSB domains.
(D) Nucleosome occupancy differs at yeast and mouse hot spots.
(E) Mouse hot spots are enriched for the trimethylation on lysine 4 of histone H3 (H3K4me3).
(B–E) These panels are plotted using data of Pan et al. (2011) and Smagulova et al. (2011). In (B) and (C), tag densities are normalized to relative genome size.
(F) Chromatin and chromosome organization influence DSB formation. DSBs occur within loops, within or outside genes (white arrows), in accessible chromatin where nucleosomes carry specific modifications and at sites where Spo11 is recruited (stars indicate different frequencies of DSBs). Some Spo11 accessory proteins (brown/orange ovals) are localized in axes, suggesting that DSB formation can involve the movement of loop sites to the axis.
Pan et al. mapped meiotic DSBs at nucleotide resolution using Spo11 immunoprecipitation, followed by amplification and then high-throughput sequencing of Spo11-linked oligonucleotides from wild-type yeast (Figure 1A). They identified 3,604 DSB hot spots, with a median hot spot width of ~190 bp and a wide range (~400-fold) in DSB activity (Figure 1B). The number of hot spots is somewhat arbitrary, as it is based on setting a threshold—here, a density of Spo11-bound oligonucleotides approximately twice that of the genome-wide average. With the criteria used, most Spo11 activity occurs within the 3,604 hot spots, and a minor but significant fraction (11%) of DSBs occurred in regions not defined as hot spots. Many features of this map agree with previous lower-resolution maps, but the higher-resolution data offer new insights, especially regarding the role of chromatin structure in DSB formation.
Smagulova et al. mapped DSBs formed during mouse spermatogenesis by high-throughput sequencing of single-strand DNA (ssDNA) bound to DMC1 or RAD51. To overcome the low abundance of recombination intermediates in wild-type cells, they used a mutant (Hop2−/−) that lacks a strand invasion accessory protein and that accumulates intermediates bound to DMC1 and RAD51. Approximately 10,000 hot spots were identified (p < 10−4 and a false-discovery rate of 6.7%). Most hot spots were separated by 60–330 kb, with a wide range in hot spot activity (Figure 1C). The DSB map from Hop2−/− mutants was confirmed by a similar study with wild-type mice and by direct molecular assays at a few hot spots. The DSB map presented by Smagulova et al. displays reasonable agreement with crossover maps of chromosome I obtained in a different genetic background (Paigen et al., 2008).
Do Nucleosomes Protect or Provide Accessibility for Recombination?
Past studies in S. cerevisiae and S. pombe highlighted the role of chromatin in the localization of DSBs and uncovered two specific findings: (1) DSBs occur in regions of accessible chromatin that are present in mitotic as well as in meiotic cells and (2) the binding of transcription factors and chromatin modifiers influences DSB levels (Lichten, 2008). The high-resolution data of Pan et al. provide the clearest picture to date of DSB localization with respect to nucleosomes. Most DSBs (88.2%) map within the nucleosome-depleted regions (NDR) of promoters, upstream of transcription start sites and flanked by positioned nucleosomes (Figure 1D). Comparing DSB patterns with genome-wide patterns of transcription factor binding in mitotic cells suggests that some transcription factors prevent Spo11 access to their binding site, whereas other transcription factors do not block DSB formation. DSBs do occur within transcribed regions of some genes, but these show low-nucleosome occupancy or poorly positioned nucleosomes. Overall, it appears that DSB formation in yeast is opportunistic, occurring where DNA is sufficiently exposed to allow access by Spo11 (Lichten, 2008).
In contrast, when Smagulova et al. analyzed global patterns of nucleosome positioning in mouse testis chromatin, they found a modest enrichment in nucleosome occupancy at hot spot centers (Figure 1D). This apparent difference from yeast may indicate that, in mice, DSB formation involves interactions between DSB-forming proteins and nucleosomes. It is also possible that the chromatin organization at a given hot spot in the fraction of the cells where it is active differs from the overall population, with a nucleosome being displaced only in the active fraction, for example.
H3K4me3: Different Strategies for a Common Task?
Histone H3 lysine 4 trimethylation (H3K4me3) is associated with sites of active transcription, and recent studies have shown that H3K4me3 is enriched at hot spots in both yeast (Borde et al., 2009) and mouse (Buard et al., 2009). In yeast, a single methyl-transferase, Set1, catalyzes the H3K4me3 modification. Set1 is part of the COMPASS complex, which is recruited to the 5′ ends of genes through its interaction with RNA polymerase II. Cells mutant for Set1 (set1D) show reduced DSBs and major changes in DSB localization, suggesting that H3K4me3 is important for normal DSB formation, but not absolutely required.
Smagulova et al. showed that 94% of mouse DSB hot spots overlap with peaks of H3K4me3 enrichment, but most of these enriched regions are not transcription promoters, which show greater H3K4me3 levels (Figure 1E). Recent studies have identified the histone methyl transferase PRDM9 as an important determinant of hot spot localization in mammals (Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010). PRDM9 has DNA binding specificity, which is determined by a DNA-binding domain with multiple zinc fingers. This domain is highly variable both within and among species. Interestingly, Smagulova et al. found a motif enriched at hot spots that matches the predicted PRDM9 binding sequence of the mouse strain analyzed. The DSB machinery may thus be recruited by PRDM9, proteins interacting with PRDM9, and features of the chromatin established by PRDM9.
Because many transcription promoters active in spermatocytes are marked by H3K4me3 but are not DSB hot spots, H3K4me3 is clearly insufficient to promote DSB formation in mice, and other chromatin modifications may be involved in providing specificity. Mice lacking Prdm9 (Prdm9−/−) do form meiotic DSBs, but their levels and location have not been determined. Paradoxically, these mutant mice appear to have DSB repair defects (Hayashi et al., 2005), suggesting that PRDM9 may be important for later steps in recombination.
Thus, in both yeast and mice, several important questions remain. Does H3K4me3 have a direct role in DSB formation, or is it a collateral result of the histone-methyl transferase activity of PRDM9 and Set1? If the DSB machinery is recruited by a direct or indirect interaction with PRDM9 or Set1, how are DSBs formed in their absence?
The Big Picture: Features of Chromosome Organization that Shape Recombination
Although the factors that determine individual yeast and mouse DSB hot spots may differ, the overall patterns and intensities of DSB are remarkably similar in the two organisms, in particular when regions corresponding to similar fractions of the genome are compared (see window sizes and tag densities in Figures 1B and 1C). Thus, despite an ~200-fold difference in genome size, yeast and mouse form meiotic DSBs at similar levels (~160/cell in yeast, 200–300/cell in mice) (Pan et al., 2011; Kauppi et al., 2011). In both organisms, irregularly alternating domains of greater and lower DSB activity are evident at multiple levels of resolution, with varying hot spot density and individual DSB activity both contributing to domain patterns. Interestingly, these higher-order patterns appear to be conserved among related but diverged strains and species in both yeast and mouse (Mancera et al., 2008; Paigen et al., 2008), as if DSB domains reflect higher-order chromosome features that diverge less rapidly than underlying sequences.
Factors responsible for DSB domains remain unknown, but studies point toward a role for chromosome structure and organization. At the time of DSB formation, chromosomes are organized into chromatin loops anchored to an axis, a linear protein structure enriched for cohesins and for meiosis-specific proteins with HORMA domains (Hunter, 2007). A remarkable study by Panizza et al. (2011) recently showed that a subset of the Spo11 accessory proteins required for DSB formation are also axis associated. However, most DSBs form in loop sequences (Blat et al., 2002), raising the possibility that DSB formation involves recruitment of Spo11-bound sequences to chromosome axes through interactions with Spo11 accessory proteins (Figure 1F).
DSB levels could be potentially affected by regional differences in loop size, in the ratio of loop-associated to axis-associated segments, or in axis protein composition. Consistent with this, mutating condensins in Caenorhabditis elegans increases axis length and also leads to increased DSB formation (Mets and Meyer, 2009). Moreover, in many organisms, including mice and humans, meiotic chromosome axes are longer and recombination frequencies greater in females than in males. Most strikingly, the short region of homology between the X and Y sex chromosomes, also called the pseudoautosomal region, has the greatest DSB activity density in the mouse genome (Smagulova et al., 2011) and also has a longer than normal axis and shorter chromatin loops (Kauppi et al., 2011).
In addition to the alternating domains seen in most of the genome, specific chromosome elements show reduced levels of recombination and DSBs. Centromeres and pericentric regions display reduced meiotic recombination in many organisms (Lichten, 2008), and Pan et al. show reduced DSB formation in an ~5 kb region around yeast centromeres and in an ~20 kb region adjacent to telomeres. Mouse centric and telomeric regions contain highly repetitive sequences, and Smagulova et al. did not analyze these regions. It will be of interest to establish whether DSBs still form at reduced levels in these regions in mammals, as they do in yeast, or whether DSB formation is truly silenced in these regions.
Both yeast and mouse genomes contain dispersed repeated elements that are at risk for deleterious genome rearrangement through nonallelic recombination (Sasaki et al., 2010). Pan et al. found DSBs to be substantially reduced (~5- to 10-fold) in Ty elements, the major repeat family in yeast, as would be expected from their closed chromatin conformation (Lichten, 2008). By contrast, some human repeats, such as retrotransposon THE-1A LTRs, are highly recombinogenic in correlation with the presence of predicted PRDM9 binding sequences, and Smagulova et al. report that the related MTC and MTD LTRs of mouse are also enriched for PRDM9-binding sites and DSB hot spots. Given the dynamic nature of mammalian hot spots and the diversity of PRDM9, it will be interesting to evaluate the generality of these findings. In any case, selective pressure for sequence-directed DSB formation must be strong enough in mammals to overcome disadvantages of DSB formation in sequences at risk for nonallelic recombination.
Implications for the Role of Meiotic Recombination in Homolog Pairing
Although the mechanism of meiotic homolog pairing is not completely understood, it is known to involve multiple interactions along chromosomes. DSB ends are thought to mediate this process in yeast and mice, with recombinase-mediated strand exchange stabilizing early interhomolog interactions (Storlazzi et al., 2010). However, the need to avoid nonallelic interactions may constrain DSB distributions along chromosomes, and the potential flexibility of Set1 and PRDM9, in terms of target site specificity, may be favorable in the face of such challenges. Another specific challenge involves the need for homologous interaction between the X and Y sex chromosomes in the relatively small pseudoautosomal region, and this challenge may be met by the specific organization of this region and targeting of Spo11 activity (Kauppi et al., 2011). It is interesting to note that some organisms, such as Drosophila melanogaster and C. elegans, use mechanisms independent of DBSs to pair homologs. The number of DSBs in these organisms is strikingly lower than that in yeast or mouse (Gerton and Hawley, 2005), and meiotic recombination may function simply to create crossover connection between homologs. Thus, it will be interesting to know how the levels and distributions of DSBs are controlled in these species.
DSB Hot Spot Evolution
DSB repair by recombination leads to the replacement of sequences on the broken chromatid by gene conversion from the repair donor. If hot spot activity is controlled in cis by sequences near DSB sites, then gene conversion should replace “hot” alleles with “cold” alleles over evolutionary time. Extending this logic leads to the paradoxical conclusion that DSB hot spots and, thus, meiotic recombination should no longer exist.
Several arguments have the potential to invalidate this paradox. DSB activity at a site may be determined in cis by elements located outside of normal gene conversion tracts. In addition, cis-acting elements that make a site “hot” might have other functions that select for their retention. Alternatively, the loss of hot spots through gene conversion may be countered either by a restoring activity at the same sites or by forming hot spots at new locations. It appears that yeast and mammals have found distinct solutions to this apparent problem.
In yeast, diverged S. cerevisiae strains and Saccharomyces species show a high degree of conservation of recombination patterns (Mancera et al., 2008; Tsai et al., 2010). Given that yeast DSB hot spots do not require specific sequence motifs and are mostly located in transcription promoter regions, maintenance of hot spot activity in yeast may be linked to the maintenance of chromatin structure through the selection for functional promoters.
Hot spot dynamics are strikingly different in mammals, in which hot spot locations are highly polymorphic between individuals and between closely related species (Baudat et al., 2010; Myers et al., 2010). This remarkable plasticity in hot spot distribution has important consequences for the use of genetic linkage as a tool, such as in genome-wide association studies, because linkage between a trait and a scored sequence marker can potentially vary widely in different populations. Diversity in hot spot distribution is correlated with the high diversity and rapid evolution of minisatellite repeats that encode the zinc finger array in PRDM9 (Ponting, 2011), leading to changes in DNA binding specificity and, thus, changes in hot spot localization. A major unsolved issue is how selection acts on Prdm9 and whether the selection is through PRDM9’s role in promoting recombination or other roles, such as a hypothetical control of gene expression, especially given that PRDM9 belongs to the PRDM family of transcription factors.
The Landscape in the Future
These studies highlight the impressionistic nature of DSB landscapes in yeast and mice: they have marked differences when brushstrokes are examined in close detail but have striking similarities when one steps back and takes a broader view (Figures 1B and 1C). Several key questions remain to be answered about the different layers controlling DSB activity (Figure 1F). For instance, features that influence DSB activity at the scale of chromosomal domains, such as chromatin loop organization, are still unknown. At higher resolution, further analysis of the relationship between DSB locations and genes in the mouse and other genomes will be important to glean mechanistic insights and to understand the evolutionary impact of meiotic DSB repair on gene linkage patterns.
One major implication of these recent studies is that, given the conservation of the molecular mechanism of meiotic DSB formation among diverse organisms, similar approaches can potentially be developed to study DSB landscapes in other species. Mechanistic understanding will also be more powerful when DSB patterns are coupled with landscapes of DSB repair outcomes (e.g., patterns of gene conversion and crossover), as well as with other features of chromosome organization, including the three-dimensional organization in the nucleus. Given the various strategies that are used in different organisms for meiotic recombination and the impact of this process on genome evolution, it will be exciting to compare the molecular machines that perform this remarkable and key step of sexual reproduction across diverse species.
Acknowledgments
We thank Dan Camerini-Otero, Galina Pethukova, Scott Keeney, and members of their groups for data used in Figure 1. We apologize to many colleagues whose work was not cited here due to space limits. M.L. is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH; B.d.M. is supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (09-BLAN-0269-01), the Association pour la Recherche contre le Cancer, and the Fondation pour la Recherche Médicale.
References
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthès P, Grey C, de Massy B. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, Hawley RS. Nat Rev Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Hunter N. Curr Topics Genet. 2007;17:381–442. [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. Science. 2011;331:916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M. Genome Dyn Stab. 2008;3:165–193. [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Keeney S. PLoS Biol. 2007;5:e333. doi: 10.1371/journal.pbio.0050333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Trends Genet. 2011;27:165–171. doi: 10.1016/j.tig.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Nat Rev Mol Cell Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. Cell. 2010;141:94–106. doi: 10.1016/j.cell.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Burt A, Koufopanou V. Proc Natl Acad Sci USA. 2010;107:7847–7852. doi: 10.1073/pnas.0908774107. [DOI] [PMC free article] [PubMed] [Google Scholar]