Abstract
The Nuclear Factor kappa B (NFkB) pathway is essential for many human cancers. Therapeutics such as bortezomib (Velcade™), which interfere with nuclear factor NF-kappa-B(NFkB)signaling are of great clinical interest. NFkB signaling, however, is multifaceted and variable among tissues, developmental, and disease entities. Hence, targeted biomarkers of NFkB pathways are of prime importance for clinical research. We developed a novel real-time qPCR-based NFkB array. Only mechanistically validated NFkB targets were included. We then used random-forest classification to define individual genes and gene combinations within the NFkB pathways that define viral lymphoma subclasses as well as Kaposi sarcoma (KS). Few NFkB targets emerged that were universally present in all tumor types tested, underscoring the need for additional tumor-type specific biomarker discovery. (i) We uncovered tissue of origin-specific tumor markers, specifically CD69, CSF-1, and complement factor B (C1QBP)for PEL; IL1-beta, cyclinD3 and CD48for KS. We found that IL12, jun-B, msx-1 and thrombospondin 2 were associated with EBV co-infection in PEL. (ii) We defined the NFkB signature of Epstein-Barr virus (EBV)positive AIDS-associated Burkitt lymphoma(BL). This signature identified CCR5 as the key marker. (iii) This signature differed from EBV negative BL consistent with the idea that EBV not only activates NFkB activity but that this virus also reprograms NFkB signaling towards different targets.
Keywords: Kaposi sarcoma, EBV, KSHV, Burkitt lymphoma, random forest, tree, real-time qPCR, ATL
Introduction
Nuclear factor NF-kappa-B (NFkB) is a sequence-specific transcription factor. NFkB is crucial for the survival of B cell lymphoma. Further studies established its importance as a pro-survival factor in many cancers, not just those of lymphoid origin (reviewed in 1). NFkB is a mixed multimer of NFkB1(p50), NFKB2(p52), Rel A(p65), Rel B, and c-rel. Subunit composition, posttranscriptional modifications and cooperating transcription factors determine target gene specificity beyond recognition of the NFkB consensus motif. Hence, mRNA profiling of NFkB responsive genes can be used to stratify malignancies, and to characterize cellular responses to natural (viral infection, growth-factor signaling) or artificial (chemotherapy drugs) stimuli.
Chemotherapy induces NFkB as part of the pro-survival response to DNA damage 2. Several therapeutics directly target NFkB or NFkB regulating factors. Velcade™/bortezomib inhibit the proteasome and thereby prevents degradation of IkB. This inhibits NFkB signaling, resulting in cell death (though bortezomib’s clinical efficacy may be due to other effects as well). Bay 11-7082 has emerged as an experimental NFkB inhibitor, which causes rapid cell death in NFkB-dependent cancer models, including primary effusion lymphoma (PEL)3 and Epstein-Barr virus (EBV) infected lymphoblastoid cell lines 4.
Kaposi sarcoma (KS) is the leading cancerin people living with HIV/AIDS followed by forms of non-Hodgkin lymphoma (NHL), including Burkitt lymphoma (BL). Hodgkin lymphoma(HL) is an AIDS-associated cancer that has increased in the post-HAART era 5. Other, so-called common cancers have also become more prevalent. These AIDS-associated lymphomas and KS are associated with gamma-herpesvirus infection. Gamma-herpesvirus infection, reactivation (and subsequent replication) is regulated by NFkB 6–12. This relationship seems almost self-evident, since gamma herpesviruses with the exception of herpesvirus saimiri, establish lifelong latency in B cells, for which NFkB is the central regulator of survival and differentiation.
Results
NFkB signaling in Kaposi sarcoma
To investigate NFkB signaling we developed a targeted real-time qPCR array (Supplemental Table 1). We obtained validated NFkB target genes through literature review. We designed primer pairs such that these share a common melting temperature, similar amplicon length, and amplification efficiency.
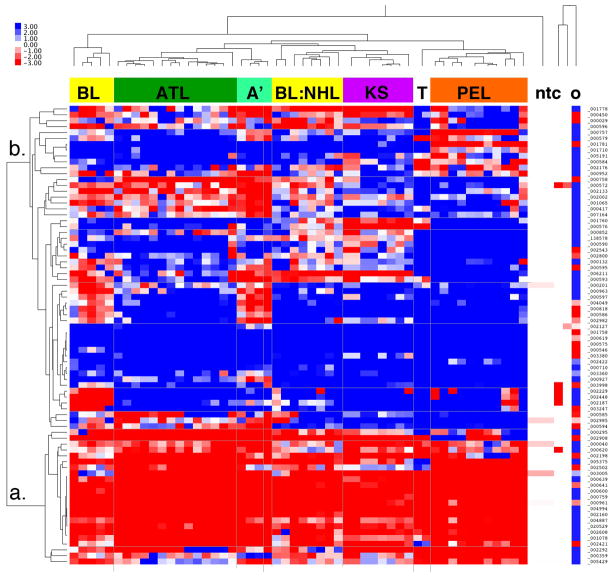
To test the hypothesis that there exist common NFkB responsive genes among AIDS defining cancers, we performed unsupervised clustering (Figure 1). We included two AIDS-associated post germinal center (GC) NHL, namely BL (n=10) and PEL (n=4 in biological replicates). We included primary KS biopsies(n=8) and KSHV-infected cell lines (n=2) as a gamma herpesvirus-dependent non-hematological cancer. We included non-HIV associated Adult T cell leukemia (ATL) (n=17) and AIDS-associated, virus-negative, NHL(n=4) as controls. In addition we included replicates of non-template control (ntc, n=5). The different AIDS-tumor types segregated according to tissue of origin.
Figure 1.
A. Heatmap representation of unsupervised clustering of all study data. Normalized (dCT) and median-centered relative mRNA levels were clustered by gene and by sample. “Blue” indicates reduction and “Red” indicates induction relative to the median for each sample. “a.” and “b.” indicate the two main clusters of mRNAs. The clusters of samples matched their presumptive origin: BL, Burkitt lymphoma; ATL, Adult T cell leukemia, A’: a novel sub-type of ATL; BL:NHL, a novel subtype of BL, which clustered with non-BL NHL samples; KS, Kaposi sarcomabiopsies, T: KS-like cell lines; PEL, primary effusion lymphoma; ntc, non-template control,; o, outlier ATL sample. Relative scale is indicated on the top left(The data points for the ntc appear white in figure 1A since the normalized data were median-centered by sample).
This demonstrates that profiling for just NFkB responsive genes sufficed to classify these tumor types. All KS biopsies, which are of endothelial lineage 13–15, clustered together. All PEL, which are of post GC B cell lineage 16, 17, clustered together. All ATL, which are of T cell lineage, clustered together; although we identified two subtypes(labeled ATL and A’ in figure 1). Two independently derived, KSHV-infected cell lines 18 clustered together (labeled T in figure 1), but separate from KS biopsies. One ATL sample (labeled “o” in Figure 1) appeared as outlier and was removed from subsequent analysis.
We identified two novel sub-types of BL: those that co-clustered with other NHL as well as a Hodgkin’s lymphoma case (HL), i.e. other virus negative B cell lymphomas(middle of Figure 1, labeled BL:NHL) and those that clustered separately(left cluster in Figure 1, labeled BL). The latter represent the EBV+ BL samples. BL group membership in these subtypes correlated perfectly with EBV status, even though EBV mRNAs levels were not included in the unsupervised clustering. This demonstrates that the NFkB pathway in BL differs based upon the presence of EBV.
Next, we inspected individual genes. One group of genes was highly transcribed in all tumor classes, B cell lymphoma, T cell lymphoma and endothelial lineage KS alike (Figure 1, cluster a.). The least variable were SelectinP/CD62, FasL, IL11, IL6, CSF3, prostacyclin synthase, MMP9, tenascin-C, MIP-2gamma, mad-3, PDGF-beta, VCAM-1. Others in cluster a. showed variation within either the PEL (serpinA1, Rel) or within the ATL and KS (Lamb2, TGM1, VEGF-C) classes. The remainder exhibited tumor type and sample specific variation (Figure 1, cluster b). Several mRNA were highly transcribed in KS biopsies, but not exclusively so: CyclinD3, CD48, IGF-BP1, IL1-beta, proenkephalin, TAP1. Conversely, several mRNAs were highly transcribed in PEL and others, but not KS: CD69-early activation antigen, CSF-1-colony stimulating factor 1, CFB-complement factor B, which was expected as these are lymphoid-associated NFB target genes.
We performed extensive quality control assessments. Supplemental figure S1, panel A shows the distribution of CT levels, i.e. the distribution of mRNA levels on a log2 scale across all samples and all genes. Undetectable or absent mRNAs account for approximately 50% of the data(single bar at CT = 40). This is consistent with the idea that many NFkB target genes are tissue specific and tightly regulated. They are transcriptionally silent in one tumor type and induced in another. Our targeted array design enriched for this type of mRNA because many of the mRNAs in our array were initially identified by less sensitive, semi-quantitative methods, such as Northern-blot.
The remaining mRNAs varied broadly in their level of transcription. Based on the density distribution we estimate a range of ≥2(40–20) = 210 or three orders of magnitude. This large range suggests that these mRNAs are highly variable and thus may be developed in to useful diagnostic markers. Supplemental figure S1, panel B shows the distribution of CT levels for the n=470 individual non-template control reactions. With the exception of very few outliers, we did not detect any signal in the absence of template (mean = 39.81, median = 40.0, with a median absolute deviation(m.a.d.) of 0.00cycle units).
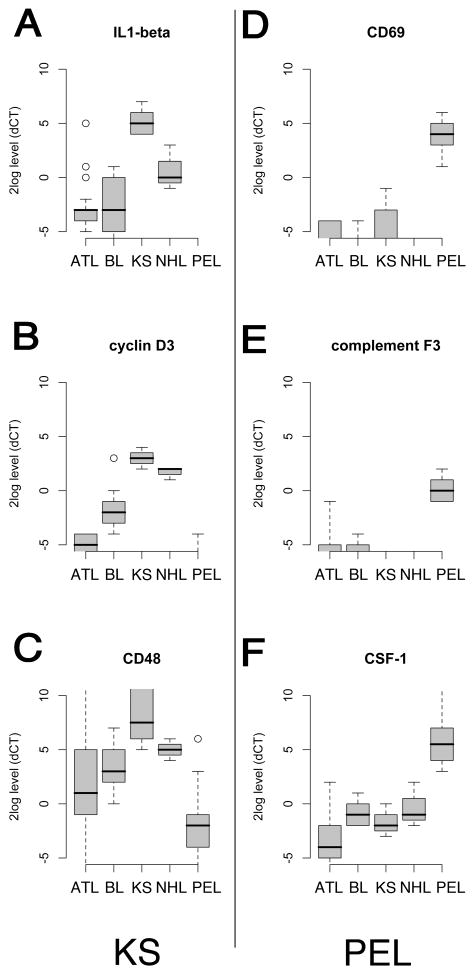
To investigate the levels of these candidate mRNAs between grossly different tumor classes we conducted pair-wise comparisons for the differentially transcribed mRNAs as identified by unsupervised clustering. We ranked the mRNAs by their differential expression (significance after adjustment for false discovery rate). For this and subsequent analyses we removed ntc, the outlier “o” and the single class samplesVG1, ATL#31, DLBCL, and BL#3. KS, an endothelial tumor, was easily distinguished from the lymphomas by any one of three single mRNAs: IL1-beta, cyclinD3 and CD48 were present at higher levels in KS than in any of the other tumors (Figure 2, A–C). These three markers were virtually absent in PEL(q< 0.001). PEL stood out on the basis of high-level expression for CD69, complement factor 3, and CSF-1(q< 0.001), which were absent from the other samples (Figure 2, D–E).
Figure 2.
Supervised class comparison of study data represented as Box plots. The box covers the 25 to 75 percentile of the data, i.e. the inter quartile range (IQR). The horizontal line indicates the median. Whiskers demarcate the minimum and maximum of the data. Open circles denote outliers (1.5 * IQR from the 1st and 3rd quartiles). A, B, C; shown are the top most discriminating mRNAs that distinguish KSHV-associated KS from AIDS-associated lymphoma. D, E, F; shown are the top most discriminating mRNAs that distinguish PEL from AIDS-associated lymphoma and ATL. For these markers p < 0.001 after adjustment for false discovery rate. Relative levels on a log10 scale for the indicated mRNAs are shown on the vertical and classes on the horizontal axis.
NFkB pathway profiling distinguishes AIDS-associated lymphoma
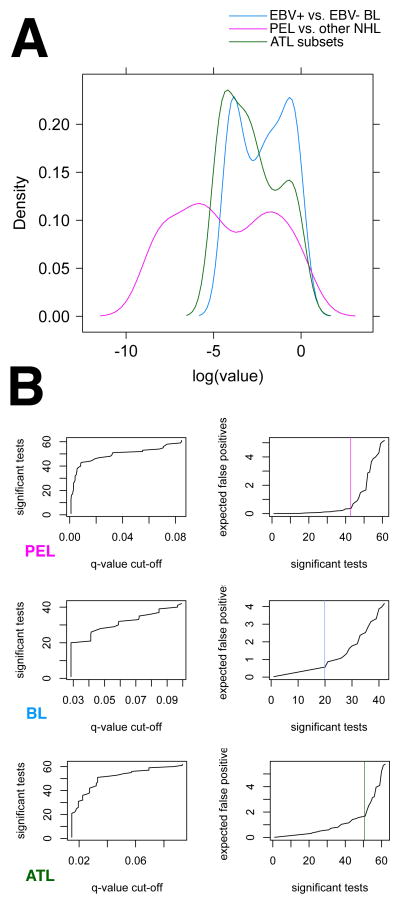
The result of our unsupervised analyses was that we could distinguish PEL for BL, and that we could establish subsets of BL and subsets of ATL on the basis of target NFkB responsive gene profiling. We used supervised, pair-wise comparison to establish significance levels for each gene in the array. Figure 3 A plots the p value distribution for each of the three comparisons: PEL to BL(purple line), the two novel BL sub sets to each other(blue line) and the two novel ATL subsets to each other (green line). The distribution for each comparison set was bi-modal, i.e. some of the genes showed a significant difference in gene expression(lower log(p-value), or left peak), others did not(log(pvalue) centered around 0, or right peak). As expected the differences between the two histological variant B cell lymphomas PEL and BL were much greater (log(p value) for the left peak at (−5...−10) compared to differences with in the BL or the ATL subsets. It is important to remember that each tumor within the BL and ATL subsets carried the same histological diagnosis and these sub-sets could only be identified after profiling. The exception is BL. BL can also be subdivided based on EBV status. This analysis thus identified a set of novel biomarkers, which differed between subsets of BL and a set of biomarkers, which differed between two novel subsets of ATL. Conversely, this supervised analysis demonstrates that there exist two different subtypes of BL and two different subtypes of ATL.
Figure 3.
Tree-based classification of PEL, BL subsets and ATL subsets. (A) Density plot of p values for each individual mRNA in the array that result from two-sample non-parametric comparison of PEL to all other lymphomas (PEL, purple), the two subsets of BL (BL, blue) and the two subsets of ATL (ATL, green). (B) Corresponding all other NHL diagnostic plots for q-value calculation. For each comparison the left panel plots the number of significant tests, i.e. genes at a given q-value cut-off level. Note that for PEL ~ 40 genes were significant to q≤ 0.02, for BL and ATL subsets only ~ 20. For each comparison the right panel plots the expected number of false positive markers among the top most significant tests on the vertical and the number of significant tests on the horizontal axis. Note that for PEL < 1 false positive is expected within the top 40 most differentially regulated genes, for BL subsets 3–4 false positives are expected and for ATL subsets also < 1.
Next we identified individual biomarkers that can differentiate between BL and ATL subtypes. Again, we started with the easy comparison, that between the his to logically different PEL and BL. After adjustment for multiple comparisons19 approximately 40 genes were differentially transcribed (Figure 3, panel PEL). Their unadjusted p-values were < 0.00001. We calculated a false positive rate of <1. This suggests that any one of these mRNAs can be used to molecularly differentiate PEL from BL. As an alternative method we used random forest classification(reviewed in 20). This identified BcL-xl, Tap1, CSF-1, CSF-2, FasL, and complement factor B as the most robust classifiers to distinguish BL from PEL
Approximately 20 genes in our array were differentially transcribed between the two subsets of BL as identified by unsupervised clustering (Figure 3, panel BL). Their unadjusted p-values were < 0.001. As expected, subsets of BL were more closely related to each other than BL to PEL. Cox-2, TNFalpha, and CCR5 were the most robustly differentially transcribed mRNAs between the two BL subsets. High level of these three mRNAs correlated with the presence of EBV. This suggests on the on hand that these three mRNAs can be used to molecularly differentiate EBV+ BL from EBV− BL. On the other hand, if one wanted to test the efficacy of an NFkB inhibitor for BL, these would be good biomarkers only for EBV+ BL, not for EBV− BL.
Approximately 20 genes in our array were differentially transcribed between the two subsets of ATL as identified by unsupervised clustering (Figure 3, panel ATL). Their unadjusted p-values were not significant, suggesting that the heterogeneity among ATL cases was not very well captured in a targeted array based on NFkB signaling.
Since PEL can either be singly infected with KSHV alone, or dually infected with KSHV and EBV, we were interested whether any NFkB dependent genes were associated with EBV infection.
Discussion
AIDS-associated malignancies are in dire need of new therapies. Since their frequency in the general population is comparably low and since patients who develop AIDS-associated malignancies are often too sick to participate in experimental treatments, clinical trials in AIDS-associated malignancies are faced with low enrollment. How then, can we nevertheless obtain clinical data that support the efficacy of novel therapeutics? This is a general obstacle faced by all who study disease that affect marginal or especially vulnerable populations. One solution is the use of biomarkers to support studies of drug action. Here we used a targeted approach to identify biomarkers that are relevant for AIDS associated malignancies and that can be developed into biomarkers for the evaluation of drugs that affect the NFkB pathway. We focused on the NFkB pathway, because it is central to gamma-herpesvirus-associated cancers, which in turn are overrepresented among AIDS patients 21.
We identified CD69, CSF-1, and complement factor B (C1QBP) as PEL specific biomarkers that are known downstream targets of NFkB. We identified IL1-beta, cyclin D3 and CD48 as NFkB-regulated KS specific biomarkers. These NFkB targets were not expressed in BL or ATL.
We obtained evidence in support of two molecular subtypes of BL, which can be differentiated on the basis of their EBV status and we determined thatCox-2, TNFalpha, and CCR5 were robustly expressed only in EBV positive BL. This corroborates a recent study by Piccaluga et al 22, who were able to distinguish BL from other NHL, but more importantly who also distinguished three types of BL--endemic, spontaneous, and HIV-associated on the basis of mRNA profiling. Further this study found that genes in the TNF/NFkB pathway were overrepresented among the mRNAs that differentiated among BL subsets. This underscores the utility of pathway-targeted profiling if biological insights or larger scale data suggest particular biological pathways.
We obtained evidence in support of molecular subtypes of ATL that were not linked to histological stage. However, we did not have the statistical power to identify any one specific biomarker in our targeted arrays, which could differentiate between these two subtypes.
Lymphomas have been the subject of innumerous profiling studies. Studies on AIDS-associated lymphomas or KS are rarer 13, 14, 16, 17, 23. Some of the new biomarkers that we discovered here, were previously investigated:C1QBP had been detected in PEL, but the other two markers (CD69, CSF-1) were not previously associated with PEL. CD69 was originally identified as a T cell activation marker, but is also highly transcribed in follicular lymphoma 24. This also holds true for CSF-1 and C1QBP. In regard to KS profiling data are even more limited. CyclinD3 is a bona fide endothelial cell marker, expression of which correlates with CD31levels. CD48 is expressed in endothelial cells as well as leukocytes and can be up regulated by LPS (via NFkB) and VEGF-1.
Cox-2, TNFalpha, and CCR5 were the most robustly differentially transcribed NFkB targets in our two BL subsets. Higher mRNAlevels correlated with the presence of EBV. In PEL, high levels of IL12, jun-B, msx-1 and thrombospondin 2 were best correlated with the presence of EBV. This suggests an important generalization: whereas EBV modulates NFkB signaling, it is the tumor tissue of origin that restricts which NFkB target gene repertoire that is open to regulation by the virus.
Some NFkB target genes emerged that were universally present and that could be used to follow drug effects in a large variety of different cancers. This is not entirely unexpected and underscores the importance of multi-analyte assays in cancer research. Unfortunately this would also suggest that single biomarker responses can not be compared between tumor types or even between lymphoma subtypes.
These biomarkers are clinically relevant because many agents that target NFkB in cancer are in advanced clinical testing. These biomarkers are robust, because they can be measured individually by real-time qPCR. We hope that they facilitate the testing of novel NFkB inhibitors for AIDS-associated malignancies.
Methods
Sample processing
Frozen biopsy samples were obtained under informed consent at the participating institutions. Diagnosis was confirmed by pathology review and de-identified samples were used for analysis. RNA was isolated following our published methods 25 and quality established using an Agilent Bioanalyzer (Supplemental Figure S2).
Analysis
Multiple housekeeping genes were included in the qPCRarray. If > 50% of housekeeping genes failed to yield a signal, the entire sample was excluded from further analysis. If<50% of housekeeping genes failed, the individual data points were omitted from further analysis. Next the median CT of all housekeeping genes was calculated for each sample and subtracted from the CT for all other genes (dCT = CTgene − medianCThouse). The dCT data were standardized by sample and subjected to unsupervised clustering. We divided the ATL and BL each into two subsets according to Figure 1 and rounded our real-time qPCRdata (dCT) to the nearest integer. We next calculated individual Wilcox-Rank tests with continuity correction as implemented in R version 2.8.0 for each gene in the array between KS and all non-KS samples. We adjusted for multiple comparison using q-value 19. This yielded 10 genes with q ≤0.001. We used random forest classifiers to identify the most robust markers for each sub-set 20.
Supplementary Material
Acknowledgments
This work was supported by the AIDS malignancy clinical trials consortium (AMC), the University cancer research grant (UCRF), the center for AIDS research (CfAR), NIH (CA112217, CA121935, DE018304, CA019014)and the Leukemia and lymphoma Society.
References
- 1.Ghosh S. Handbook of Transcription Factor NF-kappaB. Boca Raton: CRC Press; 2007. [Google Scholar]
- 2.Elkon R, Rashi-Elkeles S, Lerenthal Y, Linhart C, Tenne T, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol. 2005;6:R43. doi: 10.1186/gb-2005-6-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller SA, Schattner EJ, Cesarman E. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood. 2000;96:2537–42. [PubMed] [Google Scholar]
- 4.Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A. 2000;97:6055–60. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engels EA, Biggar RJ, Hall HI, Cross H, Crutchfield A, Finch JL, Grigg R, Hylton T, Pawlish KS, McNeel TS, Goedert JJ. Cancer risk in people infected with human immunodeficiency virus in the United States. Int J Cancer. 2008;123:187–94. doi: 10.1002/ijc.23487. [DOI] [PubMed] [Google Scholar]
- 6.Krug LT, Moser JM, Dickerson SM, Speck SH. Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency. PLoS Pathog. 2007;3:e11. doi: 10.1371/journal.ppat.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossmann C, Podgrabinska S, Skobe M, Ganem D. Activation of NF-kappaB by the latent vFLIP gene of Kaposi’s sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J Virol. 2006;80:7179–85. doi: 10.1128/JVI.01603-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chugh P, Matta H, Schamus S, Zachariah S, Kumar A, Richardson JA, Smith AL, Chaudhary PM. Constitutive NF-{kappa}B activation, normalFas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc Natl Acad Sci U S A. 2005;102:12885–90. doi: 10.1073/pnas.0408577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guasparri I, Keller SA, Cesarman E. KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med. 2004;199:993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Brown HJ, Song MJ, Deng H, Wu TT, Cheng G, Sun R. NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77:8532–40. doi: 10.1128/JVI.77.15.8532-8540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol. 2004;78:4108–19. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science. 1999;286:300–3. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 13.Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, Libermann T, Dezube BJ, Fingeroth JD, Detmar M. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet. 2004;36:683–5. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 14.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–93. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 15.Sturzl M, Brandstetter H, Roth WK. Kaposi’s Sarcoma: A review of gene expression and ultrasructure of KS spindle cells in vivo. AIDS research and human retroviruses. 1992;8:1753–62. doi: 10.1089/aid.1992.8.1753. [DOI] [PubMed] [Google Scholar]
- 16.Klein U, Gloghini A, Gaidano G, Chadburn A, Cesarman E, Dalla-Favera R, Carbone A. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood. 2003;101:4115–21. doi: 10.1182/blood-2002-10-3090. [DOI] [PubMed] [Google Scholar]
- 17.Jenner RG, Maillard K, Cattini N, Weiss RA, Boshoff C, Wooster R, Kellam P. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci U S A. 2003;100:10399–404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An FQ, Folarin HM, Compitello N, Roth J, Gerson SL, McCrae KR, Fakhari FD, Dittmer DP, Renne R. Long-Term-Infected Telomerase-Immortalized Endothelial Cells: a Model for Kaposi’s Sarcoma-Associated Herpesvirus Latency In Vitro and In Vivo. J Virol. 2006;80:4833–46. doi: 10.1128/JVI.80.10.4833-4846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fielding A. Cluster and Classification Techniques for the biosciencesed. Cambridge: Cambridge University Press; 2007. [Google Scholar]
- 21.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 22.Piccaluga PP, De Falco G, Kustagi M, Gazzola A, Agostinelli C, Tripodo C, Leucci E, Onnis A, Astolfi A, Sapienza MR, Bellan C, Lazzi S, et al. Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood. 2011;117:3596–608. doi: 10.1182/blood-2010-08-301556. [DOI] [PubMed] [Google Scholar]
- 23.Fan W, Bubman D, Chadburn A, Harrington WJ, Jr, Cesarman E, Knowles DM. Distinct subsets of primary effusion lymphoma can be identified based on their cellular gene expression profile and viral association. J Virol. 2005;79:1244–51. doi: 10.1128/JVI.79.2.1244-1251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohen SP, Troyanskaya OG, Alter O, Warnke R, Botstein D, Brown PO, Levy R. Variation in gene expression patterns in follicular lymphoma and the response to rituximab. Proc Natl Acad Sci U S A. 2003;100:1926–30. doi: 10.1073/pnas.0437875100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papin J, Vahrson W, Hines-Boykin R, Dittmer DP. Real-time quantitative PCR analysis of viral transcription. Methods Mol Biol. 2004;292:449–80. doi: 10.1385/1-59259-848-x:449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.