Abstract
The temperature-sensitive swoH1 mutant of Aspergillus nidulans was previously identified in a screen for mutants with defects in polar growth. In the present work, we found that the swoH1 mutant swelled, lysed, and did not produce conidia during extended incubation at the restrictive temperature. When shifted from the permissive to the restrictive temperature, swoH1 showed the temperature-sensitive swelling phenotype only after 8 h at the higher temperature. The swoH gene was mapped to chromosome II and cloned by complementation of the temperature-sensitive phenotype. The sequence showed that swoH encodes a homologue of nucleoside diphosphate kinases (NDKs) from other organisms. Deletion experiments showed that the swoH gene is essential. A hemagglutinin-SwoHp fusion complemented the mutant phenotype, and the purified fusion protein possessed phosphate transferase activity in thin-layer chromatography assays. Sequencing of the mutant allele showed a predicted V83F change. Structural modeling suggested that the swoH1 mutation would lead to perturbation of the NDK active site. Crude cell extracts from the swoH1 mutant grown at the permissive temperature had ∼20% of the NDK activity seen in the wild type and did not show any decrease in activity when assayed at higher temperatures. Though the data are not conclusive, the lack of temperature-sensitive NDK activity in the swoH1 mutant raises the intriguing possibility that the SwoH NDK is required for growth at elevated temperatures rather than for polarity maintenance.
Spores of the filamentous fungus Aspergillus nidulans break dormancy and expand isotropically before sending out a germ tube and switching to polar-tip growth. Further growth occurs exclusively at the hyphal tip (45). The temperature-sensitive swoH1 mutant of A. nidulans was originally isolated in a screen for polarity maintenance defects (46) based on the fact that the mutant hyphae swell shortly after germ tube emergence at the restrictive temperature. The sequence of the swoH gene reveals that it encodes a homologue of yeast YNK1, a nucleoside diphosphate kinase (NDK). NDKs catalyze the transfer of the γ-phosphate from a nucleoside triphosphate to a nucleoside diphosphate and are important in nucleotide metabolism (2, 28).
NDK null mutants of Escherichia coli and Pseudomonas aeruginosa are viable (27, 87). NDK null mutants of Saccharomyces cerevisiae and Schizosaccharomyces pombe are normal in vegetative growth, sporulation, mating, and morphology, though they have much lower NDK activity than the wild types (10 and 20%, respectively) (19, 27). Enzymes other than NDK are assumed to furnish these low levels of NDK activity (19, 32, 38, 55, 77, 84). The only reported NDK mutation in a filamentous fungus, a P72H change in Neurospora crassa, causes reduced NDK activity and deficient light response for perithecial polarity (57, 58). In plants, NDK interacts with phytochromes, photoreceptors that relay environmental light signals to nuclear genes (13, 26, 73). Higher organisms contain multiple isoforms of NDK, some of which have been shown to be expressed in a tissue-specific manner and to have different subcellular localizations (3, 7, 18, 21, 35, 43, 52, 63, 82). In Drosophila melanogaster, Caenorhabditis elegans, and Xenopus laevis, NDKs are essential for development (12, 40, 62, 63). In mammals, NDKs are involved in differentiation, cell survival, tumor metastasis, and proliferation (5, 35, 36, 56, 61, 70, 71, 74, 75). Both prokaryotic and eukaryotic NDKs form oligomers. NDK oligomerization has been correlated with its ability to interact with other molecules and carry out its function (4, 15, 20, 41, 42, 67, 81).
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. The identification of the temperature-sensitive swoH1 mutant has been previously described (46). The media used were as previously reported (46). Strain construction and genetic analysis used standard A. nidulans techniques (24, 30, 39).
TABLE 1.
Strains used in this paper
| Strain | Genotype |
|---|---|
| A104a | yA2 adE20 acrA1 phenA2 pyroA4 lysB5 sB3 nicB8 coA1 |
| A254a | biA1 AcrA1 wA3 ileA3 cnxE16 adD3 |
| A773a | pyrG89 wA3 pyroA4 |
| A850a | biA1 ΔargB::trpCΔB methG1 veA1 trpC801 |
| A852a | biA1 ΔargB::trpCΔB methG1 veA1 trpC801 pabaA1 yA2 ΔargB::trpCΔB veA1 trpC801 |
| APW13b | swoH1 (ts−) pabaA char |
| AXL20c | swoH1 (ts−) pyroG89 pyroA4 |
| AXL30d | swoH::argB biA1 ΔargB::trpCΔB methG1 veA1 trpC801 pabaA1 yA2 ΔargB::trpCΔB veA1 trpC801 |
Available from Fungal Genetics Stock Center, Department of Microbiology, University of Kansas Medical Center, Kansas City.
Isolated by Patrick Westfall (Momany laboratory).
Isolated by crossing APW13 with A773.
Diploid A852 with one copy of swoH gene replaced by argB marker.
Growth conditions and microscopic observation.
The conditions for growth and preparation of cells were as previously reported (24). Briefly, spores were inoculated on coverslips in liquid medium in a petri dish. After incubation, the cells were fixed and the nuclei were stained with Hoechst 33258 (Sigma, St. Louis, Mo.). Microscopic observations were made using a Zeiss (Thornwood, N.Y.) Axioplan microscope, and digital images were acquired using an Optronics (Goleta, Calif.) digital imaging system. Images were prepared using Photoshop version 5.5 (Adobe, Mountain View, Calif.).
DNA isolation.
DNA was isolated from A. nidulans using previously described methods (24).
Cloning by complementation and plasmid rescue.
A random genomic plasmid library carrying the pyr4 marker and sequences for autonomous replication provided by Greg May (M. D. Anderson Cancer Center, University of Texas, Houston) (60) was transformed into protoplasts of the swoH1 mutant AXL20 using standard protocols (86). DNA was purified from non-temperature-sensitive (ts+) pyrG prototrophs and used to transform E. coli XL1-blue. Three plasmids were recovered, and restriction mapping showed that they contained the same genomic DNA inserts.
Identification and sequencing of the complementing gene by transposon tagging.
Transposons were randomly inserted into the complementing plasmid using the GPS-1 system (New England Biolabs, Beverly, Mass.). The resulting plasmids, each containing one copy of the transposon at random sites, were sequenced using primers unique for the transposon ends on an ABI 3700 DNA Analyzer (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. The sequences were assembled and analyzed using the Phred (version 0.000925c), Phrap (version 0.990319), and Consed (version 11.0) computer programs (http://depts.washington.edu/ventures/uwtech/license/express/ppcombo.htm#consed) as previously described (76). The assembled contig was used to search the National Center for Biotechnology Information databases (http://www.ncbi.nlm.nih.gov) using the BLAST program to identify open reading frames (ORFs). Plasmids with transposons inserted within ORFs were transformed into the swoH1 mutant. Plasmids that failed to rescue the swoH1 mutant at the restrictive temperature had transposon insertions within a single ORF.
Mitotic mapping.
The swoH1 mutant strain APW13 was fused with the mitotic-mapping strain A104 by standard methods (39). Conidia of the heterozygous diploid were incubated on complete medium with proper supplements containing 60 μg of benomyl/ml for 2 days and transferred to complete medium with supplements for 2 weeks. The genotypes of the resulting haploid sectors were scored.
Meiotic mapping.
The swoH1 mutant strain APW13 (swoH1 pabaA char) was crossed with the chromosome II meiotic-mapping strain A254 (biA1 AcrA1 wA3 ileA3 cnxE16 adD3). Individual cleistothecia were isolated under a stereomicroscope, and the genotypes of ascospores released from cleaned cleistothecia were scored. There were 23, 28, 43, 45, and 50 map units between swoH1 and markers AcrA1, wA3, ileA3, cnxE16, and adD3, respectively.
Southern blotting.
The NDK gene was radiolabeled using the Redi-prime II DNA-labeling system (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's instructions and was used to probe a chromosome-specific genomic library (9) (available from the Fungal Genetics Stock Center [http://www.fgsc.net/]). Four cosmids from chromosome II hybridized to the NDK gene: W3A06, W12E04, W14F07, and W3C12.
Sequencing of the swoH1 mutant allele.
The swoH1 mutant allele was amplified from AXL20 genomic DNA by three independent PCRs using the Expand High Fidelity PCR system (Roche Diagnostics). The primers used for PCR amplifications were 5′-CGTACTAGATTGACTTCCCTGTC and 5′-GTGACGCAGTTTCCTAGAGATG. After purification with the Qiagen (Valencia, Calif.) PCR cleanup kit, the PCR products were sequenced on both strands by primer walking using an ABI310 sequencer (Applied Biosystems) according to the manufacturer's instructions. The sequences obtained were compared with the wild-type allele using GeneDoc (version 2.6.001) (http://www.psc.edu/biomed/genedoc) with default parameters. All three reactions gave the same G561T mutation.
Construction of the swoH null allele.
Flanking sequences from upstream and downstream of the swoH gene (1.3 kb each) were amplified by high-fidelity PCR. The primers used to amplify the 5′flanking sequence of swoH with the addition of PstI restriction sites were 5′-GAATTCCGAAATAGAAGCCGAGCAG and 5′-AACTGCAGAACCAATCCATTGGGTACGTTTGAGAAGAGGG. The primers used to amplify the 3′ flanking sequence of swoH with the addition of SacII restriction sites were 5′-ACCCCGCGGGGACCCTCAGAGTTCATCTCTAG and 5′-ACCCCGCGGGGAGCTGGTGGGTTTTTGTTCGG. After double digestion with EcoRI and PstI, the 5′ flanking sequence was inserted into pargBC-1 (44) bearing the argB gene as the selectable marker. The resulting plasmid, pargBC-5F, was then ligated with 3′ flanking sequence after digestion with SacII. The correct insert direction of the 3′ flanking sequence was confirmed by restriction mapping and PCR using the primers 5′-CGCCAGCTCAACATCAGC and 5′-CTCTGCATCTGTGCGGTC. The plasmid pargBC-5F-3F was then digested with KpnI and separated on a 1% agarose gel. The 4.4-kb fragment containing argB with swoH gene flanking sequences on both sides (5F-argB-3F) was purified using the Qiagen gel purification kit.
A linear fragment containing 5F-argB-3F was transformed into the A. nidulans haploid strain A850 (ΔargB), and transformants were selected for growth on minimal medium lacking arginine. All transformants were screened by PCR and verified by Southern blotting to distinguish ectopic from homologous integration. Of 72 haploid transformants screened, none had homologous integration (data not shown). Therefore the fragment 5F-argB-3F was transformed into the A. nidulans diploid strain A852 (ΔargB), and transformants were selected for arginine prototrophy. Genomic DNAs from 72 diploid transformants were isolated and digested with BamHI. A Southern blot was performed with 5′ flanking sequence of swoH as the probe. Only one transformant was confirmed to have a homologous integration on one chromosome and a wild-type copy on the other. The transformant was then treated with benomyl to induce haploidization as described for mitotic mapping. In total, 315 haploid sectors were scored for genotype. None of the haploid sectors grew on minimal medium without arginine supplement.
Protein sequence alignment.
Sequences of NDKs from different organisms were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) and were aligned using GeneDoc (version 2.6.001) with default parameters.
Homology modeling.
The SwoHp model was prepared by homology modeling using the SWISS-MODEL automated homology-modeling program (http://www.expasy.org/swissmod/) (23, 65, 66). Five template models from the protein data bank, 1BE4 (1), 1NUE (49), 1NSQ, 1B99 (22), and 1NHK (85) (with 70.5, 69.6, 66.9, 61.6, and 49.3% sequence identity with SwoHp, respectively), were submitted in “First Approach” mode after reduction of the coordinate files to monomeric subunits lacking ligands. SWISS-MODEL incorporated all protein sequences in the model, so no additional loop building or energy minimization was performed. All residues in the protein except residue Glu4 were reasonably modeled based on geometry and energetics. In both the wild-type and mutant structures, the side chain carboxyl group of Glu4 was oriented so that it would bridge the main chain nitrogen of Val84 (a favorable contact) and the main chain carbonyl of Arg6 (an unfavorable contact). Since this configuration is unfavorable unless the glutamic acid is uncharged, and since Glu6 did not have this orientation in any of the crystal structures, the side chain was manually repositioned to direct the carboxyl group toward the surface of the protein, consistent with other NDK structures. To evaluate whether the V83F swoH1 mutation could affect NDK oligomerization, SwoHp oligomers were generated by fitting multiple SwoHp monomers to the bacterial NDK homotetrameric and eukaryotic NDK homohexameric structures using Swiss-PDB Viewer (http://www.expasy.org/spdbv/) (23). The NDK oligomers were generated from the crystallographic and noncrystallographic operators listed in the respective protein data bank files.
Generation of three-HA-tagged SwoHp.
To construct the three-hemagglutinin (HA)-tagged SwoHp, the swoH gene was modified to contain a NotI restriction site at the N terminus by high-fidelity PCR amplification. The primers used were 5′-CGGGATCCCGATGACTAAGGAAAAAAGCGGCCGCAAAACTAATTCTGAGCAGACGTAAGTGCAT and 5′-ACATGCATGCATGTTTATTCCTTCTCATAGATCCAGCCG. The product (swoH-NotI) was inserted into the pGR3-AMA1-Not1 vector behind the bidirectional niiA-niaD promoter (provided by Greg May) (59), creating the pGR3-AMA1-swoH-NotI plasmid. The three-HA tag was obtained by digesting plasmid pBSE66 (provided by Scott Erdman, Department of Biology, Syracuse University) with NotI, and the 100-bp fragment was gel purified with the Qiagen gel purification kit. The three-HA fragment was ligated into pGR3-AMA1-swoH-NotI. The resulting plasmid, pXL1, which has a correct three-HA insertion site and direction, was confirmed by sequencing using the primers 5′-CGGGATCCCGATGACT and 5′-GTTGGGAAACTGTGCTGC.
Purification of three-HA-tagged SwoHp.
Approximately 107 conidia were inoculated into 100 ml of complete medium with proper supplements and incubated at 30°C overnight with shaking. Hyphae were harvested by filtering the conidia through two layers of cheesecloth and were washed with cold H2O. The cells were ground with mortar and pestle in liquid nitrogen. The powder was suspended in 2 volumes of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40) containing a Complete Protease Inhibitor Cocktail tablet (Roche). The suspension was centrifuged at 14,000 × g for 30 min. This crude cell extract of the supernatant was used for affinity purification on an anti-HA affinity matrix (Roche) according to the manufacturer's instructions. Purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The final purified HA-SwoHp was confirmed by silver staining and Western blotting with anti-HA monoclonal antibody (Abcam Ltd., Cambridge, United Kingdom).
Phosphate transferase activity by TLC assay.
Reactions were carried out essentially as previously described (37), except buffer D (25 mM HEPES, pH 7.0, 50 mM NaCl, 10 mM MgCl2, 10 μM dithiothreitol [DTT]) was used. ATP (1.8 mM) and GDP (1.8 mM) were added as substrates. The purified fusion protein was added to start the reaction, and the mixture was incubated at 37°C for 30 min. A control reaction with the addition of EDTA was always performed. An aliquot from each reaction mixture was spotted onto a 20- by 20-cm polyethyleneimine- cellulose-F thin-layer chromatography (TLC) plate and was developed in a saturated tank containing 0.75 M KH2PO4 (pH 3.6). The TLC plate was air dried, and the nucleotides were visualized under UV light at a wavelength of 254 nm and photographed.
Coupled enzyme assay of NDK activity.
Approximately 107 conidia were inoculated into 100 ml of complete medium with proper supplements at 30°C and incubated for 24 h with shaking. The hyphae were harvested by filtering the conidia through two layers of cheesecloth and were washed with cold H2O. The cells were ground with mortar and pestle in liquid nitrogen. The powder was suspended in 2 volumes of buffer A (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5% glycerol, 0.02% NaZ3, 0.1 M (NH4)2SO4, 1 mM DTT, 3 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride). The suspension was centrifuged at 10,000 × g for 10 min and again at 18,000 × g for 30 min. The ammonium sulfate-precipitated fraction between 45 and 70% was isolated by centrifugation at 40,000 × g for 40 min. The pellet was dissolved in buffer B (50 mM Tris-HCl, pH 7.5, 0.02% NaZ3, 5 mM MgC12, 1 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride, 5 mM 2-mercaptoethanol) and dialyzed against the same buffer overnight, aliquoted into small tubes, quickly frozen in liquid nitrogen, and stored at −80°C. The protein concentration was quantified using the Bio-Rad protein calibration kit with bovine serum albumin as a standard.
NDK activity was determined using a modification of the continuous spectrophotometric assay of Mourad and Parks (50, 51). ATP (2 mM) and TDP (0.6 mM) were added to 0.75 ml of buffer C and preincubated at assay temperature. The reaction was initiated by the addition of cell extract containing 15 μg of total protein. The reaction was terminated after 10 min by rapidly heating the mixture to 100°C for 5 min. The tubes were cooled by plunging them into ice. The precipitated protein was removed by centrifugation at 14,000 × g for 4 min. The supernatant (0.6 ml) was taken into tubes containing phosphoenolpyruvate (2.5 mM), NADH (1 mM), and pyruvate kinase (10 U/ml)- lactate dehydrogenase (20.5 U/ml) in 0.4 ml of buffer C (100 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2). The absorbance was measured at 340 nm at room temperature in a spectrophotometer (DU640B; Beckman). TDP was converted to TTP at the expense of ATP, and the amount of ADP formed was estimated by measuring NADH disappearance. Negative control assays without TDP and without enzyme NDK were always performed. The assay was performed three times, and the average was considered the NDK activity.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited with the GenBank Data Libraries under accession no. AAL23684. The genomic sequence of the complementing gene, its intron locations based on protein alignment and consensus splice sequences (6), and the predicted protein sequence have been deposited in GenBank (accession no. AY057453).
RESULTS AND DISCUSSION
The swoH1 mutant swells and lyses at restrictive temperature.
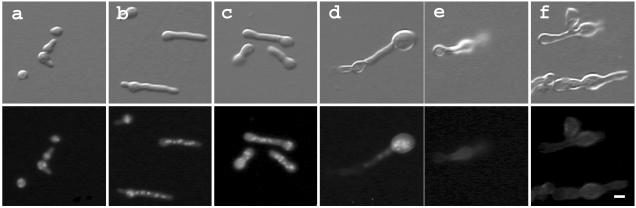
In wild-type A. nidulans, spores break dormancy and grow isotropically before sending out a germ tube and growing by polar-tip extension (45). Polar growth requires two genetically separable steps: polarity establishment (marking the spot from which the germ tube will emerge) and polarity maintenance (directing the cellular machinery to deposit new material so that the germ tube emerges and continues to extend). Previous work showed that with overnight incubation at the restrictive temperature, swoH1 mutant cells stop tip elongation and swell shortly after germ tube emergence (46). To determine the phenotype with longer incubation at the restrictive temperature, we cultured swoH1 and wild-type cells for up to 3 days. At the restrictive temperature, swoH1 cells began to swell at their tips at 10 h (Fig. 1c) and to lyse at 12 h (Fig. 1d). By 38 h, >90% of the cells had lysed, leaving ghost cells without nuclei (Fig. 1f). Wild-type cells showed normal growth under the same conditions.
FIG. 1.
swoH1 mutant cells swell and lyse at restrictive temperature. Conidia of the swoH1 mutant AXL20 were inoculated onto coverslips in complete liquid medium and incubated at the restrictive temperature (42°C). The cells were fixed and stained with Hoechst 33258 to visualize the nuclei. The upper and lower rows show differential interference contrast (DIC) and fluorescent images of the same fields. (a) 6 h, (b) 8 h, (c) 10 h, (d) 12 h, (e) 24 h, (f) 38 h. Bar, 5 μm.
SwoHp is necessary for hyphal growth.
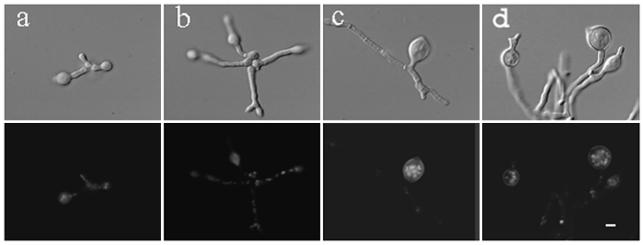
In previous studies, swoH1 cells incubated at the permissive temperature for 10 h and shifted to the restrictive temperature for 2 h showed normal hyphal extension (46). This was interpreted to mean that SwoHp is not needed for polarity maintenance. However, if SwoHp were required for polarity maintenance but was very stable, germ tubes would be expected to remain normal for some time after the shift from permissive to restrictive temperature. To better determine the timing of SwoHp function, conidia of the wild type and the swoH1 mutant were incubated at the permissive temperature for 2, 4, 6, and 8 h and then shifted to the restrictive temperature for 12 h. Wild-type cells showed normal hyphal extension. The swoH1 mutant cells swelled in all cases (Fig. 2). Growth at the permissive temperature for up to 17 h before being shifted to the restrictive temperature gave similar results (data not shown). However, swollen hyphae were not obvious when the swoH1 mutant was shifted to the restrictive temperature for <8 h (data not shown). The observation that the swollen phenotype was observed only after >8 h of incubation at the restrictive temperature regardless of whether the cell had yet sent out a germ tube suggested two possible explanations. The first is that SwoHp made at the permissive temperature is functional while SwoHp made at the restrictive temperature is not. In this scenario, a reservoir of functional protein supports normal growth for several hours after the shift to the restrictive temperature. The second possible explanation is that the mutant SwoHp is functional, but not fully so, and that more SwoHp activity is required for growth at the restrictive temperature. In this scenario, the mutant protein can meet the needs of the cell growing at the permissive temperature but not at the restrictive temperature. If the first explanation were correct, one would expect SwoHp to be very active at 30°C and inactive at 42°C. If the second explanation were correct, one would expect that SwoHp activity levels would be nearly equivalent at the two temperatures.
FIG. 2.
The swoH1 mutant cannot maintain polar growth upon shift from permissive to restrictive temperature. Conidia of the swoH1 mutant AXL20 were grown in liquid complete medium at the permissive temperature (30°C) for 2, 4, 6, and 8 h and then shifted to restrictive temperature (42°C) for 12 h (a, b, c, and d, respectively). The cells were fixed and stained with Hoechst 33258 to visualize the nuclei. The upper and lower rows show DIC and fluorescent images of the same fields. Bar, 5 μm.
SwoHp is necessary for conidiation.
A. nidulans produces asexual spores (conidia) on aerial hyphae (conidiophores). To determine if SwoHp is needed for asexual reproduction, we tested the ability of the swoH1 mutant to produce conidia at the restrictive temperature. Since the swoH1 mutant stopped growing during early vegetative growth at the restrictive temperature, we cultured the swoH1 mutant at the permissive temperature in liquid medium with shaking for 8, 12, and 20 h and then plated hyphae on solid medium to induce conidiation at the restrictive temperature. The plates were examined by stereoscope after incubation for 1, 2, 3, and 4 days. While the swoH1 mutant incubated at the permissive temperature conidiated normally, no conidia were formed in cultures shifted to the restrictive temperature, indicating that SwoHp is necessary for conidiation at the restrictive temperature (data not shown).
An NDK gene complements the swoH1 mutant.
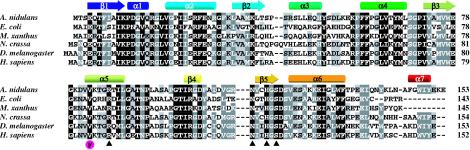
A genomic plasmid library carrying a pyr4 marker was transformed into swoH1 pyrG mutant protoplasts. Transformants were selected on minimal medium at the restrictive temperature. Total DNA from transformants was introduced into E. coli, and complementing plasmids were isolated. Restriction mapping showed that all of the complementing plasmids contained the same genomic DNA insert. One complementing plasmid, pH42, was sequenced using a transposon tag strategy. Five ORFs were found in pH42. The complementing gene was identified based on the fact that two transposon insertions within this single ORF disrupted the plasmid's ability to complement the swoH1 mutant, while transposons inserted into other ORFs had no effect (data not shown). A search of the National Center for Biotechnology Information database showed that the ORF that complemented swoH1 encodes a homologue of an NDK with 67% amino acid identity with the S. cerevisiae YNK1 and 69% identity with the Homo sapiens NDK NM23-H2. NDKs convert nucleoside diphosphates to nucleoside triphosphates and are a highly conserved family across prokaryotes and eukaryotes (Fig. 3). Higher organisms contain several isoforms of NDK, while yeasts and bacteria contain only one. A BLAST search of the Whitehead Institute Center for Genome Research A. nidulans genome database (http://www-genome.wi.mit.edu) with the swoH gene sequence yielded no additional hits, suggesting that there is only one NDK gene in A. nidulans. In addition, Southern blots of the A. nidulans genomic DNA with the swoH gene showed only one band (data not shown).
FIG. 3.
Multiple alignment of NDK sequences. Protein sequences of NDKs from different organisms were obtained from GenBank (accession numbers AAL23684 [NDK from A. nidulans], NP_417013 [NDK from E. coli], A35539 [NDK from M. xanthus), CAD37041 [NDK from N. crassa], P08879 [NDK from D. melanogaster], P22392 [NDK B from H. sapiens]). The solid boxes represent identical or highly similar residues; dark and light shading indicate 75 and 50% similar residues, respectively. Secondary structures are shown above the corresponding amino acid sequences as arrows (β-strands) or cylinders (α-helices) coded blue (N terminus) to red (C terminus). The A. nidulans swoH1 V83F mutation is indicated by the purple circled F below the sequence. Amino acids important for catalytic phosphate transferase activity are indicated by arrowheads below the sequence, based on Morera et al. and Postel et al. (48, 69).
Mapping of swoH1.
Mitotic mapping was used to identify the chromosome on which the swoH gene lies. A heterozygous diploid was made by fusing the swoH1 mutant strain APW13 with the mitotic-mapping strain A104, which has a marker on each chromosome. Chromosome loss was induced by benomyl treatment of the heterozygous diploid. The resulting haploid sectors have a set of chromosomes that are a random mixture from either parent. The genotypes of these haploid sectors were scored. The acrAr marker segregated in repulsion to the swoH1 ts− phenotype, indicating that swoH is on chromosome II (Table 2).
TABLE 2.
Mitotic mapping of the swoH genea
| CHRb | Marker | Segregation in repulsion (%)c |
|---|---|---|
| I | ts− | 100 |
| adE20− | 35 | |
| II | acrAr | 96 |
| III | phenA2 | 71 |
| IV | pyroA4− | 63 |
| V | lysB5− | 60 |
| VI | sB3− | 57 |
| VII | nicB8− | 59 |
Diploid was generated by fusing swoH1 mutant strain APW13 with mitotic-mapping strain A104. The diploid was treated with benomyl to induce haploidization. Genotypes of haploid sectors were scored.
CHR, chromosome on which marker is located.
Percentage of conidia that have markers segregated in repulsion to swoH1 ts− phenotype. The marker of chromosome VIII in the mitotic-mapping strain A104 is also ts− (compact morphology), which makes it impossible to score at the same time as swoH1 (ts−). Out of 350 haploid sectors, 99 were ts−, which may be caused by reduced viability of the ts− strain.
The location of the swoH1 locus on chromosome II was determined by meiotic mapping. The swoH1 mutant strain APW13 was crossed with the meiotic-mapping strain A254. Genotypes of the progeny ascospores were scored (n = 315), and the distances between the swoH1 locus and other markers on chromosome II were determined by recombination frequencies. Based on the genetic map of chromosome II (available from the Fungal Genetics Stock Center [http://www.fgsc.net/]), the swoH gene is located on the left arm of chromosome II and is 23 map units away from AcrA1.
Southern blotting experiments showed that the NDK plasmid which complemented the swoH1 mutant phenotype hybridized to four cosmids from a chromosome II-specific library (9). The positions of these cosmids on the A. nidulans physical map are consistent with our genetic mapping of the swoH gene (14) (http://www.gla.ac.uk/Acad/IBLS/molgen/aspergillus/maps.html). The fact that the physical position of the complementing plasmid is consistent with the genetic position of the swoH1 locus indicated that NDK represents the authentic swoH gene and not a high-copy-number suppressor.
Deletion of the swoH gene is lethal.
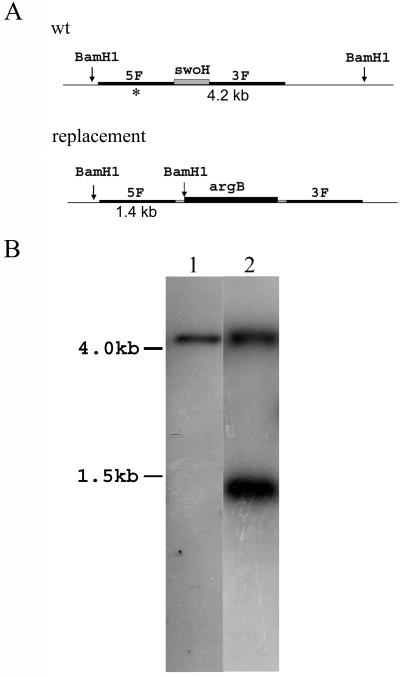
To determine the effect of its loss, we attempted to replace swoH with the argB marker by homologous recombination in a haploid. Only ectopic integrations were found in 72 haploid transformants screened, suggesting that the swoH gene may be essential. To determine if this explanation is correct, the disruption construct was transformed into the A. nidulans diploid stain A852 (ΔargB). Transformants that grew on medium without arginine were tested by Southern blotting. Only one diploid transformant of 72 screened was heterozygous, with one chromosome retaining the wild-type copy of swoH and one with the homologously integrated argB (Fig. 4). This diploid transformant (AXL30) was induced to haploidize, and the genotypes of the resulting haploid sectors were scored. None of the 315 haploid sectors scored were argB+, indicating that only those haploid sectors with the intact swoH gene survived. This result indicated that the swoH gene is essential in A. nidulans.
FIG. 4.
Homologous replacement of swoH with argB to create a heterozygous diploid. (A) BamHI sites near swoH loci for wild-type (wt) and replacement alleles. The asterisk indicates 5′ flanking sequence used to probe the Southern blot. The distances between BamHI sites are indicated below. (B) DNA of the heterozygous diploid transformant was digested with BamHI and probed with 5′ flanking sequence of the swoH gene. Lane 1, wild type; lane 2, heterozygous diploid AXL30. The sizes are indicated on the left.
The lethality of the swoH null mutant is consistent with the lysis phenotype of the swoH1 mutant and important functions of NDKs in development and differentiation in multicellular eukaryotes but contrasts with findings in the unicellular organisms E. coli, P. aeruginosa, S. cerevisiae, and S. pombe, in which NDK is dispensable. This intriguing correlation between the complexity of the life style and the essentiality of NDKs suggests that NDK might play more important roles in multicellular than in unicellular organisms. Consistent with this idea, in Myxococcus xanthus, a bacterium with a complex multicellular life style (31, 34, 78, 80), NDK is essential (25, 53, 54).
SwoHp has phosphate transferase activity.
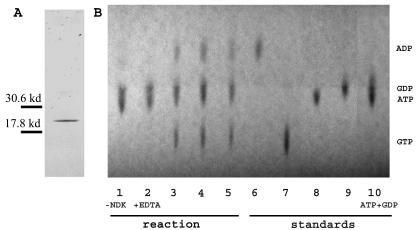
A three-HA- SwoH fusion was inserted behind the bidirectional niiA-niaD promoter in the pGR3-AMA1-Not1 vector (59). When the resulting construct was transformed into the swoH1 mutant, it complemented the mutant phenotype at the restrictive temperature in inductive medium (Fig. 5f) but not in repressive medium (Fig. 5e) (72), indicating that the three-HA N-terminally tagged SwoHp is functional. Empty vector had no effect on growth (data not shown). The HA-SwoHp fusion protein was purified on an anti-HA affinity matrix. Only one band of the appropriate size was detected on SDS-PAGE by silver staining after affinity column purification (Fig. 6A). The purified fusion protein was used to assay phosphate transferase activity with Mg2+ as a cofactor, ATP as a phosphate donor, and GDP as a phosphate acceptor. The reaction was visualized by TLC. When HA-SwoHp was not added to the reaction mixture, only the substrates ATP and GDP were seen (Fig. 6B). When HA-SwoHp was added to the reaction mixture, both the substrates ATP and GDP and the products ADP and GTP were seen (Fig. 6B). When the chelator EDTA was added along with HA-SwoHp, only the substrates ATP and GDP were seen (Fig. 6B). These results proved that SwoHp has phosphate transferase activity, consistent with its identification as an NDK.
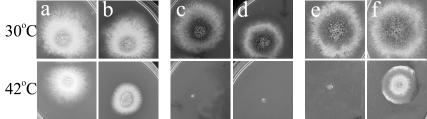
FIG. 5.
HA-SwoHp fusion complements swoH1 mutant. Shown are the wild type (a and b), a swoH1 mutant (c and d), and a swoH1 mutant transformed with the HA-SwoHp fusion under the inducible promoter niiA-niaD (e and f). Spores were inoculated on repressive medium (a, c, and e) and inductive medium (b, d, and f) and incubated at 30 or 42°C for 2 days.
FIG. 6.
HA-SwoHp shows phosphate transferase activity. (A) Affinity-purified HA-SwoHp was electrophoresed on an SDS-PAGE denaturing gel and silver stained. (B) A phosphate transferase assay was performed with ATP as the γ-phosphate donor and GDP as the acceptor. Lanes: 1, without HA-SwoHp; 2, with 100 ng of HA-SwoHp plus 100 mM EDTA; 3, with 40 ng of HA-SwoHp; 4, with 100 ng of HA-SwoHp; 5, with 140 ng of HA-SwoHp; 6, ADP; 7, GTP; 8, GDP; 9, ATP; 10, mixture of ATP and GDP. The positions of the standards are labeled on the right. −, without; +, with.
The swoH1 mutation likely distorts the NDK active site.
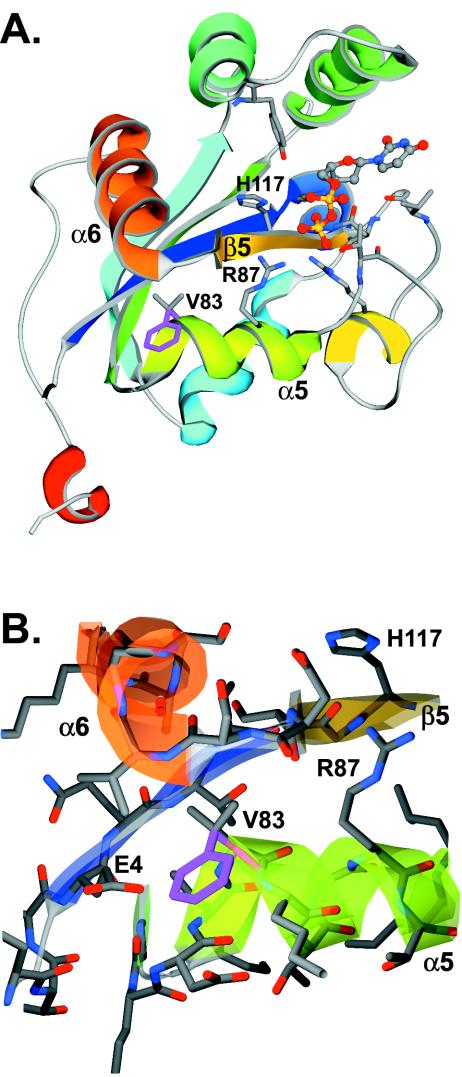
Sequencing of the swoH1 mutant allele revealed one mutation, G561T, which is predicted to change an absolutely conserved valine to phenylalanine at position 83 of the protein (Fig. 3). Since the NDK family is highly conserved both at the primary protein sequence level (Fig. 3) and at the protein structure level (4, 12, 15, 17, 21, 33, 43, 48, 68, 69), we were able to predict the protein structure of SwoHp using homology modeling. Like other NDKs, SwoHp is predicted to have a core of four internal β-strands surrounded by 6 α-helices (Fig. 7A). The predicted substrate-binding pocket is bound by α-helix 1, the N terminus of β-strand 5, and the C terminus of α-helix 5. The active-site residues R87 and H117 protrude into the pocket from α5 and β5, respectively. In other eukaryotes, NDKs form homohexamers with residues on the outer face of α-helix 5, opposite the substrate-binding pocket, forming the subunit interface. Our homology model is consistent with oligomerization of SwoHp through residues on α-helix 5.
FIG. 7.
Ribbon representation of the A. nidulans SwoHp homology model. (A) The SwoHp homology model of one subunit is portrayed in a ribbon representation, with secondary-structure features colored blue (N terminus) through red (C terminus). The substrate molecules are shown in ball-and-stick representations. The swoH1 mutation V83F occurs on α-helix 5. The side chain of wild-type valine shown in ball and stick representation is colored purple, and the swoH1 mutant phenylalanine is colored gray. The side chains of the active residues R87 on α-helix 5 and H117 on β-strand 3 are also shown in ball-and-stick representations and colored gray. The color scheme is the same as that in Fig. 3. (B) Enlargement of the region near V83F, with residues shown in ball-and-stick representations.
The swoH1 mutation (V83F) is not predicted to cause any dramatic overall structural change but likely alters the relative positions of α5 and β5, where the conserved active-site residues R87 and H117 are located (Fig. 7B). In the wild type, V83 occupies the space between the start of α5 and the start of α6, where it likely helps keep active-site residues properly positioned. The more bulky phenylalanine residue in the swoH1 mutant cannot be accommodated in the same space and must project outward, resulting in a small void in the protein. To compensate for this void, α5 and β5 would likely move closer to each other, creating suboptimal interactions between the substrate and the enzyme active-site residues and thus reduced NDK activity in the mutant. Oligomerization is likely not disturbed in the swoH1 mutant. The V83F change is predicted to lie on the face of α-helix 5 opposite the subunit interface.
swoH1 mutant cell extract has reduced NDK activity.
To determine NDK activity levels, crude cell extracts from the wild type and the swoH1 mutant incubated overnight at the permissive temperature (30°C) were analyzed at 25, 42, and 54°C by a coupled enzyme assay (Table 3). Because the swoH1 mutant is temperature sensitive (the phenotype is observed at 42°C but not at 30°C), we expected that the NDK levels from swoH1 would be roughly equivalent to those from the wild type when assayed at lower temperatures and would fall relative to the wild type at higher temperatures. However, at 25°C, the NDK activity of the swoH1 mutant was only 20% of that of the wild type. At 42°C, the NDK activity of the swoH1 mutant increased to 24% of that of the wild type, and at 54°C, it increased to 30% of that of the wild type.
TABLE 3.
swoH1 mutant crude cell extract has reduced NDK activitya
| Assay temp (°C) | Activity (μmol ADP/ng of protein) |
Ratio (%)b | |
|---|---|---|---|
| swoH1 mutant | Wild type | ||
| 25 | 0.18 ± 0.03 | 0.93 ± 0.04 | 19.3 |
| 42 | 0.24 ± 0.02 | 0.98 ± 0.08 | 24.2 |
| 54 | 0.31 ± 0.03 | 1.05 ± 0.09 | 29.5 |
The same amount of protein was added to each reaction mixture. The results from three independent experiments were averaged.
swoH1 mutant/wild type.
Because our NDK activity assays were performed on crude cell extracts, caution must be used in interpreting the results. Other proteins have been shown to possess phosphate transferase activity like NDK and to synthesize nucleoside triphosphates (19, 32, 38, 55, 77, 84). Indeed, NDK null mutants of S. cerevisiae and S. pombe have 10 and 20% of wild-type NDK activity, respectively (19, 27). Enzymes other than NDK are assumed to furnish these low levels of NDK activity (19, 32, 38, 55, 77, 84). It is unclear whether the low levels of NDK activity assayed in crude cell extracts of the swoH1 mutant are from the mutated SwoHp or other phosphate transferase enzymes. Regardless, the NDK activity levels in mutant cell extracts compared to those in wild-type cell extracts suggest that the V83F mutation in the swoH1 mutant does not confer temperature sensitivity on the SwoH NDK protein. Though failure to detect thermosensitive NDK activity in our swoH ts mutant was surprising, it is consistent with the perturbation of the active-site pocket predicted by structural modeling. The suboptimal interactions predicted between the substrate and the enzyme active-site residues would be expected to reduce NDK activity regardless of the temperature.
Our NDK activity assays raise the intriguing possibility that reduced phosphate transferase activity in swoH1 is sufficient to support normal growth at the permissive temperature but not at the higher restrictive temperature, i.e., that there is a requirement for wild-type levels of NDK at elevated temperatures. The idea that more NDK activity might be required for sustained growth at elevated temperatures is consistent with our temperature shift experiments, in which the swoH1 swollen phenotype was never seen with <8 h of incubation at the restrictive temperature. If there is an increased need for NDK with other kinds of stress, it might explain our recovery of only a single heterozygous swoH+/ΔswoH::argB diploid among 72 diploids screened. Our transformation procedure uses protoplasting—certainly a stress for the cell.
There is evidence that NDKs might be involved in response to stress in other systems (16, 25, 47, 79, 83). Human and fish NDKs coimmunoprecipitate with the heat shock protein HSP70 (37). The S. cerevisiae NDK is transcriptionally upregulated under cell-damaging conditions (29). E. coli NDK copurifies with HSP70 proteins, including DnaK, an important modulator of heat stress response that is required for growth at high temperatures (8, 10, 11, 64). The precise role of NDKs in the stress response, and whether this role involves any change in NTP levels, is unclear.
Even though swoH1 was isolated in a screen for temperature-sensitive polar-growth mutants, our data raise the possibility that the SwoH NDK might actually be required for sustained growth at higher temperatures rather than polarity maintenance. All fungal growth after the initial isotropic-to-polar switch is by tip extension. Thus, the cessation of polar growth and tip swelling in the swoH1 mutant at the restrictive temperature may actually be the death throes of a mutant unable to cope with elevated temperature.
Acknowledgments
This work was sponsored by Department of Energy Biosciences grant DE-FG02-97ER20275 to M.M.
We thank Brian Shaw for assistance in sequencing and identification of the complementing plasmid, Patrick Westfall for APW13 strain construction, and Greg May for providing the plasmid library and niiA-niaD promoter used in this work.
REFERENCES
- 1.Abdulaev, N. G., G. N. Karaschuk, J. E. Ladner, D. L. Kakuev, A. V. Yakhyaev, M. Tordova, I. O. Gaidarov, V. I. Popov, J. H. Fujiwara, D. Chinchilla, E. Eisenstein, G. L. Gilliland, and K. D. Ridge. 1998. Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure. Biochemistry 37:13958-13967. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, R. P., B. Robison, and R. E. Parks, Jr. 1978. Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol. 51:376-386. [DOI] [PubMed] [Google Scholar]
- 3.Agou, F., S. Raveh, S. Mesnildrey, and M. Veron. 1999. Single strand DNA specificity analysis of human nucleoside diphosphate kinase B. J. Biol. Chem. 274:19630-19638. [DOI] [PubMed] [Google Scholar]
- 4.Almaula, N., Q. Lu, J. Delgado, S. Belkin, and M. Inouye. 1995. Nucleoside diphosphate kinase from Escherichia coli. J. Bacteriol. 177:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amendola, R., R. Martinez, A. Negroni, D. Venturelli, B. Tanno, B. Calabretta, and G. Raschella. 2001. DR-nm23 expression affects neuroblastoma cell differentiation, integrin expression, and adhesion characteristics. Med. Pediatr. Oncol. 36:93-96. [DOI] [PubMed] [Google Scholar]
- 6.Ballance, D. J. 1986. Sequences important for gene expression in filamentous fungi. Yeast 2:229-236. [DOI] [PubMed] [Google Scholar]
- 7.Barraud, P., L. Amrein, E. Dobremez, S. Dabernat, K. Masse, M. Larou, J. Y. Daniel, and M. Landry. 2002. Differential expression of nm23 genes in adult mouse dorsal root ganglia. J. Comp. Neurol. 444:306-323. [DOI] [PubMed] [Google Scholar]
- 8.Barthel, T. K., and G. C. Walker. 1999. Inferences concerning the ATPase properties of DnaK and other HSP70s are affected by the ADP kinase activity of copurifying nucleoside-diphosphate kinase. J. Biol. Chem. 274:36670-36678. [DOI] [PubMed] [Google Scholar]
- 9.Brody, H., J. Griffith, A. J. Cuticchia, J. Arnold, and W. E. Timberlake. 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19:3105-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukau, B. 1993. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 9:671-680. [DOI] [PubMed] [Google Scholar]
- 11.Bukau, B., and G. C. Walker. 1990. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiadmi, M., S. Morera, I. Lascu, C. Dumas, G. Le Bras, M. Veron, and J. Janin. 1993. Crystal structure of the Awd nucleotide diphosphate kinase from Drosophila. Structure 1:283-293. [DOI] [PubMed] [Google Scholar]
- 13.Choi, G., H. Yi, J. Lee, Y. K. Kwon, M. S. Soh, B. Shin, Z. Luka, T. R. Hahn, and P. S. Song. 1999. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401:610-613. [DOI] [PubMed] [Google Scholar]
- 14.Clutterbuck, A. J. 1997. The validity of the Aspergillus nidulans linkage map. Fungal Genet. Biol. 21:267-277. [DOI] [PubMed] [Google Scholar]
- 15.Dumas, C., I. Lascu, S. Morera, P. Glaser, R. Fourme, V. Wallet, M. L. Lacombe, M. Veron, and J. Janin. 1992. X-ray structure of nucleoside diphosphate kinase. EMBO J. 11:3203-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaton, P., H. L. Byers, N. Leeds, M. A. Ward, and M. J. Shattock. 2002. Detection, quantitation, purification, and identification of cardiac proteins S-thiolated during ischemia and reperfusion. J. Biol. Chem. 277:9806-9811. [DOI] [PubMed] [Google Scholar]
- 17.Erent, M., P. Gonin, J. Cherfils, P. Tissier, G. Raschella, A. Giartosio, F. Agou, C. Sarger, M. L. Lacombe, M. Konrad, and I. Lascu. 2001. Structural and catalytic properties and homology modelling of the human nucleoside diphosphate kinase C, product of the DRnm23 gene. Eur. J. Biochem. 268:1972-1981. [DOI] [PubMed] [Google Scholar]
- 18.Fournier, H. N., S. Dupe-Manet, D. Bouvard, M. L. Lacombe, C. Marie, M. R. Block, and C. Albiges-Rizo. 2002. Integrin cytoplasmic domain-associated protein 1α (ICAP-1α) interacts directly with the metastasis suppressor nm23-H2, and both proteins are targeted to newly formed cell adhesion sites upon integrin engagement. J. Biol. Chem. 277:20895-20902. [DOI] [PubMed] [Google Scholar]
- 19.Fukuchi, T., J. Nikawa, N. Kimura, and K. Watanabe. 1993. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene 129:141-146. [DOI] [PubMed] [Google Scholar]
- 20.Galvis, M. L., S. Marttila, G. Hakansson, J. Forsberg, and C. Knorpp. 2001. Heat stress response in pea involves interaction of mitochondrial nucleoside diphosphate kinase with a novel 86-kilodalton protein. Plant Physiol. 126:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilles, A. M., E. Presecan, A. Vonica, and I. Lascu. 1991. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J. Biol. Chem. 266:8784-8789. [PubMed] [Google Scholar]
- 22.Gonin, P., Y. Xu, L. Milon, S. Dabernat, M. Morr, R. Kumar, M. L. Lacombe, J. Janin, and I. Lascu. 1999. Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography. Biochemistry 38:7265-7272. [DOI] [PubMed] [Google Scholar]
- 23.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 24.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey, C., and P. W. French. 2000. Effects on protein kinase C and gene expression in a human mast cell line, HMC-1, following microwave exposure. Cell Biol. Int. 23:739-748. [DOI] [PubMed] [Google Scholar]
- 26.Hasunuma, K., and N. Yabe. 1998. Early events occurring during light signal transduction in plants. Tanpakushitsu Kakusan Koso 43:1443-1452. (In Japanese.) [PubMed]
- 27.Izumiya, H., and M. Yamamoto. 1995. Cloning and functional analysis of the ndk1 gene encoding nucleoside-diphosphate kinase in Schizosaccharomyces pombe. J. Biol. Chem. 270:27859-27864. [DOI] [PubMed] [Google Scholar]
- 28.Janin, J., and D. Deville-Bonne. 2002. Nucleoside-diphosphate kinase: structural and kinetic analysis of reaction pathway and phosphohistidine intermediate. Methods Enzymol. 354:118-134. [DOI] [PubMed] [Google Scholar]
- 29.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20:8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 31.Kuner, J. M., and D. Kaiser. 1982. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J. Bacteriol. 151:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroda, A., and A. Kornberg. 1997. Polyphosphate kinase as a nucleoside diphosphate kinase in Escherichia coli and Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 94:439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladner, J. E., N. G. Abdulaev, D. L. Kakuev, M. Tordova, K. D. Ridge, and G. L. Gilliland. 1999. The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina. Acta Crystallogr. D 55:1127-1135. [DOI] [PubMed] [Google Scholar]
- 34.LaRossa, R., J. Kuner, D. Hagen, C. Manoil, and D. Kaiser. 1983. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J. Bacteriol. 153:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H. Y., and H. Lee. 1999. Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett. 145:93-99. [DOI] [PubMed] [Google Scholar]
- 36.Lee, I. H., S. I. Chang, K. Okada, H. Baba, and H. Shiku. 1997. Transcription effect of nm23-M2/NDP kinase on c-myc oncogene. Mol. Cells 7:589-593. [PubMed] [Google Scholar]
- 37.Leung, S. M., and L. E. Hightower. 1997. A 16-kDa protein functions as a new regulatory protein for Hsc70 molecular chaperone and is identified as a member of the Nm23/nucleoside diphosphate kinase family. J. Biol. Chem. 272:2607-2614. [DOI] [PubMed] [Google Scholar]
- 38.Lu, Q., and M. Inouye. 1996. Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism. Proc. Natl. Acad. Sci. USA 93:5720-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, G. C., and E. Kafer. 1974. Genetic analysis of the reciprocal translocation T2(I;8) of Asperigillus using the technique of mitotic mapping in homozygous translocation diploids. Genetics 77:11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda, I., Y. Kohara, M. Yamamoto, and A. Sugimoto. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11:171-176. [DOI] [PubMed] [Google Scholar]
- 41.Mesnildrey, S., F. Agou, A. Karlsson, D. D. Bonne, and M. Veron. 1998. Coupling between catalysis and oligomeric structure in nucleoside diphosphate kinase. J. Biol. Chem. 273:4436-4442. [DOI] [PubMed] [Google Scholar]
- 42.Mesnildrey, S., F. Agou, and M. Veron. 1997. The in vitro DNA binding properties of NDP kinase are related to its oligomeric state. FEBS Lett. 418:53-57. [DOI] [PubMed] [Google Scholar]
- 43.Milon, L., P. Meyer, M. Chiadmi, A. Munier, M. Johansson, A. Karlsson, I. Lascu, J. Capeau, J. Janin, and M. L. Lacombe. 2000. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 275:14264-14272. [DOI] [PubMed] [Google Scholar]
- 44.Momany, M., and J. E. Hamer. 1997. The Aspergillus nidulans septin encoding gene, aspB, is essential for growth. Fungal Genet. Biol. 21:92-100. [DOI] [PubMed] [Google Scholar]
- 45.Momany, M., and I. Taylor. 2000. Landmarks in the early duplication cycles of Aspergillus fumigatus and Aspergillus nidulans: polarity, germ tube emergence and septation. Microbiology 146:3279-3284. [DOI] [PubMed] [Google Scholar]
- 46.Momany, M., P. J. Westfall, and G. Abramowsky. 1999. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon, H., B. Lee, G. Choi, D. Shin, D. T. Prasad, O. Lee, S. S. Kwak, D. H. Kim, J. Nam, J. Bahk, J. C. Hong, S. Y. Lee, M. J. Cho, C. O. Lim, and D. J. Yun. 2003. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA 100:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morera, S., M. Chiadmi, G. LeBras, I. Lascu, and J. Janin. 1995. Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium. Biochemistry 34:11062-11070. [PubMed] [Google Scholar]
- 49.Morera, S., M. L. Lacombe, Y. Xu, G. LeBras, and J. Janin. 1995. X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 Å resolution. Structure 3:1307-1314. [DOI] [PubMed] [Google Scholar]
- 50.Mourad, N., and R. E. Parks, Jr. 1966. Erythrocytic nucleoside diphosphokinase. III. Studies with free and phosphorylated enzyme and evidence for an essential thiol group. J. Biol. Chem. 241:3838-3844. [PubMed] [Google Scholar]
- 51.Mourad, N., and R. E. Parks, Jr. 1966. Erythrocytic nucleoside diphosphokinase. II. Isolation and kinetics. J. Biol. Chem. 241:271-278. [PubMed] [Google Scholar]
- 52.Munier, A., C. Feral, L. Milon, V. P. Pinon, G. Gyapay, J. Capeau, G. Guellaen, and M. L. Lacombe. 1998. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 434:289-294. [DOI] [PubMed] [Google Scholar]
- 53.Munoz-Dorado, J., M. Inouye, and S. Inouye. 1990. Nucleoside diphosphate kinase from Myxococcus xanthus. I. Cloning and sequencing of the gene. J. Biol. Chem. 265:2702-2706. [PubMed] [Google Scholar]
- 54.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1990. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J. Biol. Chem. 265:2707-2712. [PubMed] [Google Scholar]
- 55.Noguchi, T., and T. Shiba. 1998. Use of Escherichia coli polyphosphate kinase for oligosaccharide synthesis. Biosci. Biotechnol. Biochem. 62:1594-1596. [DOI] [PubMed] [Google Scholar]
- 56.Nosaka, K., M. Kawahara, M. Masuda, Y. Satomi, and H. Nishino. 1998. Association of nucleoside diphosphate kinase nm23-H2 with human telomeres. Biochem. Biophys. Res. Commun. 243:342-348. [DOI] [PubMed] [Google Scholar]
- 57.Ogura, Y., Y. Yoshida, K. Ichimura, C. Aoyagi, N. Yabe, and K. Hasunuma. 1999. Isolation and characterization of Neurospora crassa nucleoside diphosphate kinase NDK-1. Eur. J. Biochem. 266:709-714. [DOI] [PubMed] [Google Scholar]
- 58.Ogura, Y., Y. Yoshida, N. Yabe, and K. Hasunuma. 2001. A point mutation in nucleoside diphosphate kinase results in a deficient light response for perithecial polarity in Neurospora crassa. J. Biol. Chem. 276:21228-21234. [DOI] [PubMed] [Google Scholar]
- 59.Osherov, N., J. Mathew, and G. S. May. 2000. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 31:181-188. [DOI] [PubMed] [Google Scholar]
- 60.Osherov, N., and G. May. 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otsuki, Y., M. Tanaka, S. Yoshii, N. Kawazoe, K. Nakaya, and H. Sugimura. 2001. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc. Natl. Acad. Sci. USA 98:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ouatas, T., B. Abdallah, L. Gasmi, J. Bourdais, E. Postel, and A. Mazabraud. 1997. Three different genes encode NM23/nucleoside diphosphate kinases in Xenopus laevis. Gene 194:215-225. [DOI] [PubMed] [Google Scholar]
- 63.Ouatas, T., M. Selo, Z. Sadji, J. Hourdry, H. Denis, and A. Mazabraud. 1998. Differential expression of nucleoside diphosphate kinases (NDPK/NM23) during Xenopus early development. Int. J. Dev. Biol. 42:43-52. [PubMed] [Google Scholar]
- 64.Paek, K. H., and G. C. Walker. 1987. Escherichia coli dnaK null mutants are inviable at high temperature. J. Bacteriol. 169:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 66.Peitsch, M. C. 1995. Protein modeling by E-mail. Bio/Technology 13:658-660. [Google Scholar]
- 67.Polosina, Y., K. F. Jarrell, O. V. Fedorov, and A. S. Kostyukova. 1998. Nucleoside diphosphate kinase from haloalkaliphilic archaeon Natronobacterium magadii: purification and characterization. Extremophiles 2:333-338. [DOI] [PubMed] [Google Scholar]
- 68.Postel, E. H. 1998. NM23-NDP kinase. Int. J. Biochem. Cell Biol. 30:1291-1295. [DOI] [PubMed] [Google Scholar]
- 69.Postel, E. H., B. A. Abramczyk, S. K. Gursky, and Y. Xu. 2002. Structure-based mutational and functional analysis identify human NM23-H2 as a multifunctional enzyme. Biochemistry 41:6330-6337. [DOI] [PubMed] [Google Scholar]
- 70.Postel, E. H., B. M. Abramczyk, M. N. Levit, and S. Kyin. 2000. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc. Natl. Acad. Sci. USA 97:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Postel, E. H., S. J. Berberich, J. W. Rooney, and D. M. Kaetzel. 2000. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 32:277-284. [DOI] [PubMed] [Google Scholar]
- 72.Punt, P. J., J. Strauss, R. Smit, J. R. Kinghorn, C. A. van den Hondel, and C. Scazzocchio. 1995. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 15:5688-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quail, P. H. 2000. Phytochrome-interacting factors. Semin. Cell Dev. Biol. 11:457-466. [DOI] [PubMed] [Google Scholar]
- 74.Roymans, D., K. Vissenberg, C. De Jonghe, R. Willems, G. Engler, N. Kimura, B. Grobben, P. Claes, J. P. Verbelen, C. Van Broeckhoven, and H. Slegers. 2001. Identification of the tumor metastasis suppressor Nm23-H1/Nm23-R1 as a constituent of the centrosome. Exp. Cell Res. 262:145-153. [DOI] [PubMed] [Google Scholar]
- 75.Roymans, D., R. Willems, K. Vissenberg, C. De Jonghe, B. Grobben, P. Claes, I. Lascu, D. Van Bockstaele, J. P. Verbelen, C. Van Broeckhoven, and H. Slegers. 2000. Nucleoside diphosphate kinase beta (Nm23-R1/NDPKβ) is associated with intermediate filaments and becomes upregulated upon cAMP-induced differentiation of rat C6 glioma. Exp. Cell Res. 261:127-138. [DOI] [PubMed] [Google Scholar]
- 76.Shaw, B. D., C. Momany, and M. Momany. 2002. Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot. Cell 1:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiba, T., K. Tsutsumi, K. Ishige, and T. Noguchi. 2000. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry 65:315-323. [PubMed] [Google Scholar]
- 78.Shimkets, L. J., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song, E. J., Y. S. Kim, J. Y. Chung, E. Kim, S. K. Chae, and K. J. Lee. 2000. Oxidative modification of nucleoside diphosphate kinase and its identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biochemistry 39:10090-10097. [DOI] [PubMed] [Google Scholar]
- 80.Spormann, A. M., and D. Kaiser. 1999. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 181:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uno, T., M. Ueno, M. Kikuchi, and Y. Aizono. 2002. Purification and characterization of nucleoside diphosphate kinase from the brain of Bombyx mori. Arch. Insect Biochem. Physiol. 50:147-155. [DOI] [PubMed] [Google Scholar]
- 82.Venturelli, D., R. Martinez, P. Melotti, I. Casella, C. Peschle, C. Cucco, G. Spampinato, Z. Darzynkiewicz, and B. Calabretta. 1995. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proc. Natl. Acad. Sci. USA 92:7435-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wada, Y., T. Kayo, and A. Koizumi. 2002. Characterization of gene expression profile associated with energy restriction-induced cold tolerance of heart. Microsc. Res. Tech. 59:313-316. [DOI] [PubMed] [Google Scholar]
- 84.Wheeler, L. J., N. B. Ray, C. Ungermann, S. P. Hendricks, M. A. Bernard, E. S. Hanson, and C. K. Mathews. 1996. T4 phage gene 32 protein as a candidate organizing factor for the deoxyribonucleoside triphosphate synthetase complex. J. Biol. Chem. 271:11156-11162. [DOI] [PubMed] [Google Scholar]
- 85.Williams, R. L., D. A. Oren, J. Munoz-Dorado, S. Inouye, M. Inouye, and E. Arnold. 1993. Crystal structure of Myxococcus xanthus nucleoside diphosphate kinase and its interaction with a nucleotide substrate at 2.0 Å resolution. J. Mol. Biol. 234:1230-1247. [DOI] [PubMed] [Google Scholar]
- 86.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaborina, O., N. Misra, J. Kostal, S. Kamath, V. Kapatral, M. E. El-Idrissi, B. S. Prabhakar, and A. M. Chakrabarty. 1999. P2Z-independent and P2Z receptor-mediated macrophage killing by Pseudomonas aeruginosa isolated from cystic fibrosis patients. Infect. Immun. 67:5231-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]