Abstract
Cysteine is one of the most versatile molecules in biology, taking over such different functions as catalysis, structure, regulation and electron transport during evolution. Research on Arabidopsis has contributed decisively to the understanding of cysteine synthesis and its role in the assimilatory pathways of S, N and C in plants. The multimeric cysteine synthase complex is present in the cytosol, plastids and mitochondria and forms the centre of a unique metabolic sensing and signaling system. Its association is reversible, rendering the first enzyme of cysteine synthesis active and the second one inactive, and vice-versa. Complex formation is triggered by the reaction intermediates of cysteine synthesis in response to supply and demand and gives rise to regulation of genes of sulfur metabolism to adjust cellular sulfur homeostasis. Combinations of biochemistry, forward and reverse genetics, structural- and cell-biology approaches using Arabidopsis have revealed new enzyme functions and the unique pattern of spatial distribution of cysteine metabolism in plant cells. These findings place the synthesis of cysteine in the centre of the network of primary metabolism.
INTRODUCTION
Cysteine has a multitude of functions in biology due to the chemical properties of reduced sulfur. Sulfide is a strong nucleophile, making it a highly reactive mediator of redox reactions. These include catalysis, activation of reactive groups (e.g. coenzyme A), disulfide bonds with their structural and regulatory functions and, in particular, electron transfer via iron-sulfur clusters. Complexes of transition metals such as iron with sulfide might have acted as catalysts of essential organosynthetic chemistry during the evolution of protolife (Martin et al., 2003; Wächtershauser, 2006). Today, they are indispensable parts of the energy-generating processes in mitochondria and chloroplasts. Cysteine is the donor of sulfide for all of these compounds in all cells. Cysteine in the proteomes of fully-sequenced genomes of members of the archea, eubacteria and eukaryotes is underrepresented compared to other amino acids (Pe'er et al., 2004), but its frequency in proteins correlates positively with increasing complexity of organisms, reflecting the risk of non-specific oxidation and an advantage of increased use of cysteine residues (Miseta and Csutora, 2000). Indeed, the evolution of life passed through geological times of largely anoxic and highly sulfidic environments until today's oxygenic atmosphere, requiring adaptation and specialization of functions of sulfide residues in proteins and in metabolism.
The biosynthesis of cysteine and its central position in primary sulfur metabolism has been extensively investigated in Arabidopsis thaliana which has delivered insight into novel and unexpected mechanisms of enzyme regulation, metabolite sensing and spatial distribution of amino acid metabolism in plant cells. The integration of reduced sulfur from assimilatory sulfate reduction in plastids into cysteine is the exclusive point of entry into downstream metabolism. The reaction is comparable in its importance to ammonia fixation by glutamine synthetase and forms an interface between sulfur, nitrogen and carbon metabolism. Accordingly, supply of sulfide for cysteine synthesis and demand for cysteine itself for protein biosynthesis or anabolic processes such as formation of methionine, glutathione, iron-sulfur clusters and many others require regulatory coordination between sulfate uptake, assimilatory reduction, cysteine synthesis and distribution of reduced sulfur into downstream metabolism. In addition, intracellular transport and whole plant allocation of primary sulfur compounds are components of this dynamic network. Analysis of Arabidopsis in recent years has indeed revealed that the synthesis of cysteine is a major regulator of the upstream reactions in primary sulfur metabolism. Cysteine synthesis occurs almost constitutively and ubiquitously in plant cells and occurs in the plastids, the cytosol and the mitochondria. For a more comprehensive overview of the general sulfur metabolism in phototrophic organisms, the reader is referred to Hell et al. (2008). In-depth reviews of plant primary sulfur metabolism are provided by Schmidt and Jäger (1992), Leustek (2000) and Takahashi et al. (2011).
Pathways leading to cysteine: sulfate uptake and assimilation
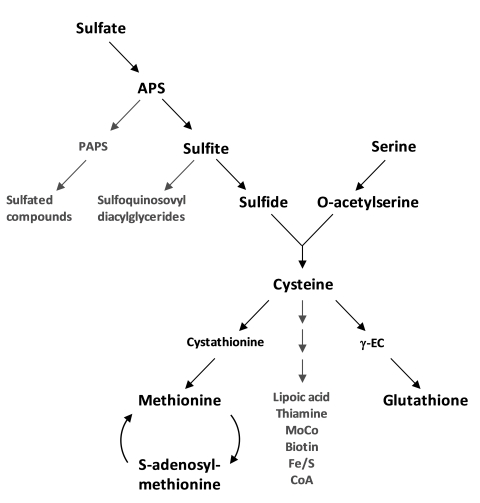
The sulfur substrate for cysteine synthesis in plants is provided by sulfate uptake and assimilatory sulfate reduction, while the amino acid backbone comes from serine metabolism (Figure 1). Sulfur in plants is generally taken up as sulfate by roots from the soil. Sulfate transporters (Sultr) form a family of 14 proteins in Arabidopsis (Hawkesford, 2003) and operate as cotransporters with 2–3 H+/SO42- transported. High affinity Sultr proteins show substrate affinities of app. Km = 10 µM and low affinity Sultr proteins of app. Km=100 µM when heterologously expressed in yeast (Takahashi et al., 2000). The cell type-specific expression of Sultr genes in the root epidermis, root cortex, root vasculature up to shoot vasculature and leaf lamina is orchestrated in complex patterns throughout the plant and driven by external supply and internal demand (see Takahashi et al., 2011, for review). Cysteine itself and metabolites upstream and downstream in the pathway are candidates for sensing or signaling of these conditions towards gene regulation (Vidmar et al., 2000; Vauclare et al., 2002; Rouached et al., 2008). In the root epidermis the response of the genes Sultr1;1 and Sultr1;2 (Shibagaki et al., 2002) to sulfate deprivation results in enhanced uptake rates within hours in Arabidopsis (Smith et al., 1997; Clarkson et al., 1999). Resupply of sulfate to roots equally rapidly represses the uptake systems. This response is archetypical in all plants and algae and is often used as a marker for conditions of sulfate deficiency and even for screens for identifying regulatory factors (Takahashi et al., 2011).
Figure 1.
Biochemical pathway of primary sulfur metabolism in Arabidopsis thaliana.
Co-substrates and products have been omitted. Endproducts are indicated in grey letters. Arrows can represent more than one reaction.
The activity of sulfate uptake and reduction can be regulated by supply and demand, i.e. genes are repressed at high sulfate availability and low demand and become de-repressed by sulfate limitation and enhanced demand for reduced sulfur. These mechanisms, however, depend on organ type, development and environmental constraints and are interwoven with the primary pathways of carbon and nitrogen (Davidian and Kopriva, 2010). De-repression of sulfate uptake in the presence of external sulfate can be mimicked by feeding of O-acetylserine (OAS), the activated form of serine and reaction intermediate of cysteine synthesis. This leads to the induction of numerous genes, in particular Sultr genes, but also genes encoding enzymes of sulfate reduction such as adenosine phosphosulfate reductase (APR) and many others (Koprivova et al., 2000; Hirai et al., 2003) including genes encoding enzymes for the breakdown of glucosinolates, i.e. secondary compounds with high sulfur content to regain internally stored sulfur. In contrast, few genes encoding enzymes of cysteine synthesis respond this way. Whether cellular contents of sulfate, sulfide OAS, cysteine and glutathione are the true primary signals for controlling sulfate uptake and reduction or rather act indirectly is still a matter of debate (Buchner et al., 2004; Hopkins et al., 2005; Davidian and Kopriva, 2010). A detailed discussion of the current status of sulfate transporter regulation in Arabidopsis and crop plants can be found in Hawkesford, (2008) and Takahashi et al., (2011).
Arabidopsis gene expression databases show, in agreement with physiological studies from a number of researchers (see Brunold, 1990; Schmidt and Jäger, 1992; Leustek et al., 2000 for review), that the genes and encoded enzymes of sulfate assimilation are expressed not only in photosynthetic but also in heterotrophic organs. Thus, root, seed and specialized non-photosynthetic cell types share a varying degree of autonomy with respect to assimilation of sulfate into cysteine. In Arabidopsis, all enzymes of this pathway including cysteine synthesis are encoded by at least three gene copies except for sulfite reductase (SiR) which is encoded by a single gene (Bork et al., 1998; Leustek, 2002). A comparative overview on the orthologous and paralogous evolution of the genes families related to primary sulfur metabolism in bacteria and eukaryotes with emphasis on Arabidopsis is available (Patron et al., 2008).
ATP sulfurylase isoforms are present in the cytosol and plastids and form activated sulfate as adenosine 5′-phosphosulfate (APS). APS is located at the first branching point of the pathway (Figure 1). A small amount of APS can be further activated to 3′-phosphoadenosine 5′-phosphosulphate (PAPS) by APS kinase in both compartments (Mugford et al., 2009). Sulfotransferases use PAPS to form sulfate esters with hydroxylgroups of various substrates (glucosinolates, flavonoids, peptides, hormons; Hernández-Sebastià et al., 2008) but reside exclusively in the cytosol. APS in plastids is reduced to sulfite by APS reductase which accepts and transfers electrons from glutathione (Gutierrez-Marcos et al., 1996; Setya et al., 1996). APS reductase gene expression responds even more strongly than that of APS kinase genes to sulfate deficiency but also to daylight and other stress conditions (reviewed in Kopriva, 2006). In addition, the APS reductase protein is regulated post-translationally in response to oxidative stress (Bick et al., 2001). Sulfite is positioned at the second branching point of the reduction sequence (Figure 1). Plastid-localized sulfolipid biosynthesis requires sulfite to form sulfoquinovosyldiacylglycerol, a lipid which is exclusively found in photoautotrophic organisms (Sanda et al., 2001; Benning et al., 2008). The UDP-SQ enzyme (SQD1) competes with SiR that reduces the bulk of sulfite to sulfide in chloroplasts using electrons from ferredoxin and a siroheme as ligand (Yonekura-Sakakibara et al., 2000). In contrast to ATP sulfurylase and APS reductase genes, the SiR gene shows little transcriptional regulation. Nevertheless, the SiR single copy gene At5g04590 is not only essential for survival, the encoded SiR activity can become limiting for the flux through the reduction pathway (Khan et al., 2010). Sulfite and sulfide are chemically very reactive and are assumed to be present in only small steady-state amounts. Among the few measurements available are concentrations from Arabidopsis leaf cytosol (55 µM sulfide) and chloroplast (125 µM sulfide) (Krüger et al., 2009) and from Arabidopsis whole leaf extracts ranging from 5 µM (Tsakraklides et al., 2002) to 25 µM (Khan et al., 2010) of sulfite.
Pathways leading to cysteine: step 1 — synthesis of OAS by serine acetyltransferase
The cysteine synthesis pathway in plants starts with serine acetyltransferase (SAT; Serat, EC 2.3.1.30) and activation of serine by acetyl transfer from acetyl coenzyme A to form O-acetylserine (OAS). OAS and sulfide are the substrates for direct sulfhydration by O-acetylserine (thiol) lyase (OAS-TL) (Figure 2). SAT and OAS-TL form the hetero-oligmeric cysteine synthase (CS) complex. In whole protein extracts the entire SAT activity is always associated with OAS-TL in the CS complex, but an excess of free and active OAS-TL is always present (see section CS complex below). This two-step pathway is conserved between archea, eubacteria and plants (Hell and Wirtz, 2008). Fungi such as Saccharomyces cerevisiae, Schizosaccharomyces pombe or Neurospora crassa are taxonomically and physiologically very heterogenous and can, depending on species and nutrient availability, use direct sulfhydration to form either cysteine or homocysteine, the latter being synthesized from O-acetylhomoserine and homoserine. Homocysteine is then either methylated to methionine or transsulfurylated via cystathionine to cysteine. While methionine and cysteine are interconvertible in fungi, the key regulator of sulfate uptake and assimilation including sulfur amino acid synthesis is methionine (Marzluf, 1997; Thomas and Surdin-Kerjan, 1997). Animals cannot assimilate sulfate but need to take up cysteine or methionine which are interconverted by transsulfurylation via cystathionine. Methionine is the essential amino acid for adult humans and excess dietary methionine can entirely replace cysteine intake (Reeds, 2000). It is important to stress that cysteine from plants is, directly or indirectly, by far the major dominating source of reduced sulfur for the nutrition of mammals and humans.
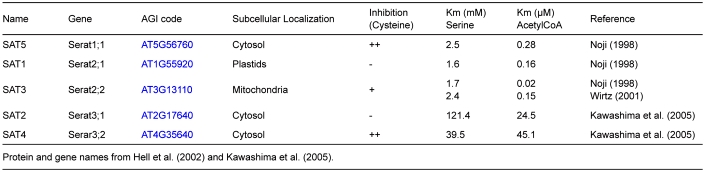
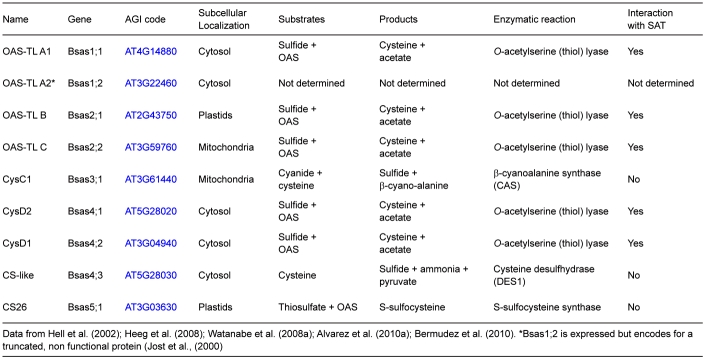
In Arabidopsis five genes (Serat) encode SAT enzymes for OAS synthesis (Table 1). Analysis of the evolution of amino acid sequences suggests that the ancestral Serat gene was of host origin and was not derived of the cyanobacterial endosymbiont (Kopriva et al., 2008). SAT1 and SAT3 are localized in plastids and mitochondria respectively, while SAT2, SAT4 and SAT5 are cytosolic (Table 1). SAT5 is most similar in sequence to SAT1 and SAT3, notwithstanding their organellar transit peptides and less related to SAT2 and SAT4 which have C-terminal extensions and low affinities to serine and acetyl coenzymeA. The genes expressing SAT2 and SAT4 are transcribed 10–100-times less than the other three. It is most likely that SAT2 and SAT4 are unable to bind OAS-TLs in the CS complex and have therefore been suspected of carrying out other or additional metabolic functions (Kawashima et al., 2005). However, a comprehensive study with SAT (serat) mutants lacking at least one isoenzyme showed that all quadruple mutant combinations expressing only one of the five SATs are viable, indicating that SAT2 and SAT4 are sufficient for survival. Quadruple mutants expressing only SAT1, SAT2 or SAT4 are strongly retarded in growth at the seedling stage while those expressing only SAT3 or SAT5 show only slight growth retardation (Watanabe et al., 2008b). These phenotypes are consistent with the insignificant reductions in total SAT activity in single mutants lacking only SAT1 or SAT2 or SAT4 and the measurable reduction of total SAT activity in the roots of the single mutant lacking only cytosolic SAT5 (Watanabe et al., 2008b; Krüger et al., 2009). Remarkably, complete loss of mitochondrial SAT3 due to insertional mutation of the corresponding gene causes the loss of 70–80% of total SAT activity but results in only slight growth retardation (Watanabe et al., 2008b). In contrast, the down-regulation of SAT3 activity by using a gene-specific artificial miRNA results in stunted growth. The severity of this phenotype correlates strictly with the degree of reduction in gene expression (Haas et al., 2008). The flux of 35S-sulfate into cysteine is strongly decreased in these plants (Haas et al., 2008) but can be completely restored by genetic complementation of the amiRNAi effect by targeting a heterologous SAT from tobacco to mitochondria (F. Haas, M. Wirtz, R. Hell, unpublished). These contradictory findings for the complete absence versus low abundance of mitochondrial SAT3 protein may be explained if it is assumed that the CS complex has a regulatory function beyond modulation of SAT enzyme activity. Genetic analysis of the contributions of the predominant SAT enzymes, SAT1 in plastids, SAT3 in mitochondria and SAT5 in the cytosol, to cysteine synthesis in leaf cells is confirmed by the only currently available biochemical measurement of compartment-specific SAT activities in leaves of pea (Pisum sativum). It is estimated that about 80–90% of total SAT activity resides in mitochondria and about 5–10% each is present in the cytosol and chloroplasts (Figure 3A) (Ruffet et al., 1995; Droux, 2003).
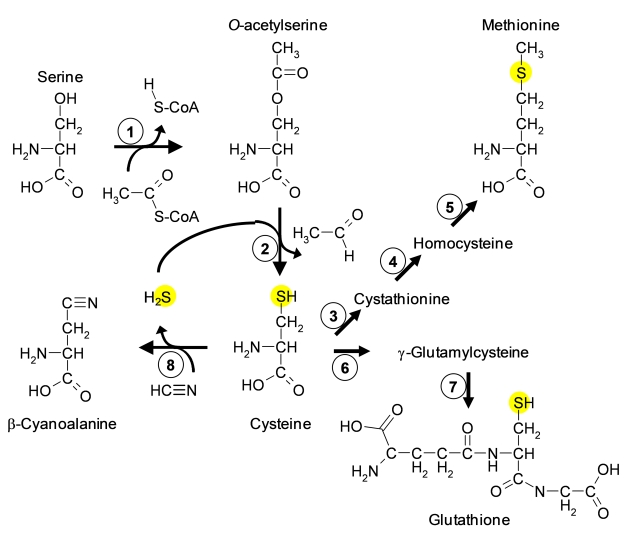
Figure 2.
Chemical structures and biosynthesis of sulfur containing metabolites.
Arrows indicate reactions. Enzymes are shown as circles with numbers. 1, serine acetyltransferase, 2, O-acetylserine(thiol)lyase, 3, cystathionine-γ-synthase, 4, cystathionine-β-lyase, 5, methionine synthase, 6, glutamate cysteine ligase, 7, glutathione synthetase, 8, β-cyanoalanine synthase. Yellow spheres highlight the reduced sulfur atom, which is fixed by cysteine synthesis.
Serat genes are expressed throughout the entire life-cycle of the plant, albeit with varying degrees of tissue-dependent and developmental patterns. Changes in transcription in response to environmental factors are generally minor. Remarkably, expression of the gene encoding SAT4 responds to sulfate deprivation and to cadmium exposure despite presumably contributing little SAT activity in vivo, pointing to a function in thiol-mediated stress response (Kawashima et al, 2005). The abundance of SAT1 and SAT3 transcript in root increases up to 10-fold after treatment with the oxidizing reagent menadione (Lehmann et al., 2009). Transcripts encoding SAT1 also increase 10-fold when a catalase2-deficient mutant of Arabidopsis is transferred from high CO2 concentrations to ambient air, presumably to provide more cysteine for glutathione synthesis to be used in the detoxification of H2O2 by the ascorbate/glutathione cycle (Queval et al., 2009). In both cases, neither SAT1 protein level nor SAT activity were determined, but in the latter case the increase in SAT1 mRNA is accompanied by an increase in total levels of cysteine and glutathione. It seems therefore that the expression of gene encoding plastidic SAT1 is triggered by oxidative stress even though plastidic SAT activity contributes only about 10% to the total SAT activity in non-stressed leaves, and that metabolic regulation of the CS complexes in the three compartments is responsible for increased production and contents of cysteine in response to environmental challenges. The concept of regulation of SAT activity in plastids is further discussed in the CS complex section below.
Table I.
The SAT gene and protein family in Arabidopsis.
The only known metabolic function of OAS in plants is in cysteine synthesis. But with respect to regulation, OAS has at least two more functions: it can induce genes of sulfur metabolism and dissociate the cysteine synthase complex. OAS is a labile compound that spontaneously converts into N-acetylserine in a pH-dependent manner. The conversion rate is 1% min-1 at pH 7.6 (Flavin and Slaughter, 1965). N-acetylserine has no known function in plant metabolism, but is the inducer of the cysteine regulon in bacteria (Kredich, 1996). OAS can be applied to Arabidopsis leaves or roots and is readily taken up. Concentrations of OAS in leaves of unstressed Arabidopsis plants show a wide range from 0.5 to 15 nmol g-1 fresh weight-1; Cysteine concentrations show less variation and range from 7 to 22 nmol g-1 fresh weight-1 (Hirai et al., 2004; Heeg et al., 2008; Watanabe et al., 2008a; Watanabe et al., 2008b; Krüger et al., 2009; Khan et al., 2010). The differences in OAS content may either reflect higher biological responsiveness to environmental status or technical deviations due to different HPLC methods used in comparison to cysteine determinations. Upon sulfate deprivation, the level of OAS increases as part of the typical sulfur deficiency response. This long term increase is mostly viewed as a passive effect due to the lack of sulfide for cysteine synthesis. Depending on the experimental conditions the OAS levels begin to increase several hours or even days after onset of external sulfate depletion, but since the mRNA contents of Sultr genes encoding high affinity transporters appear to increase earlier, the role of OAS as primary inducer of the sulfate deficiency response is questionable (Hirai et al., 2003; Buchner et al., 2004; Wirtz et al., 2004; Hopkins et al., 2005; Rouached et al., 2008). However, a high resolution study with respect to time scale, perceiving root tissue and co-determination of OAS and sulfide concentrations together with Sultr gene expression profiles upon transfer of plants from high to low sulfate containing medium has not yet been carried out. Application of OAS to Arabidopsis roots orchestrates the expression of a sulfate deficiency gene response in microarray analyses, suggesting that OAS may be either a strictly cellular or a later, secondary signal (Hirai et al., 2003; Hesse et al., 2004). Currently, the role of OAS is seen by several authors as a mediator that links the assimilation pathways of sulfate with those of nitrogen and carbon (Hawkesford and De Kok, 2006; Schachtman and Shin, 2007; Amtmann and Armengaud, 2009).
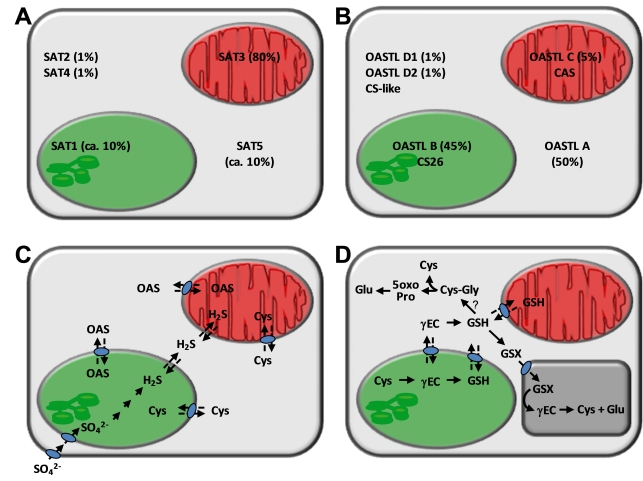
Figure 3.
Compartmentation of cysteine and glutathione synthesis in Arabidopsis thaliana.
A, B. Data in % refer to approximate relative contributions of SAT (from Pisum sativum) and OASTL (from Arabidopsis) activities, respectively, in the three compartments according to different experimental approaches (Droux, 2003; Ruffet et al., 1995; Heeg et al., 2008; Watanabe et al., 2008a). Other enzymatic functions refer to CS-like (cysteine desulfhydrase, DES1; Alvarez et al., 2010), CS26 (S-sulfocysteine synthase; Bermudez et al., 2010), CAS (β-cyanoalanine synthase, CysC1; Hatzfeld et al., 2000, Yamaguchi et al., 2000). C. Transport processes in cysteine synthesis as evidenced by knock-out mutant analyses (Heeg et al., 2008; Watanabe et al., 2008) that have not yet been identified at the molecular level. D. Synthesis and degradation of glutathione (GSH) and glutathione conjugates (GSX); γ-glutamylcysteine (γEC), 5-oxoproline (5oxoPro). γ-Glutamyltransferase activities at the plasmalemma and phytochelatin synthase activity in the cytosol are not shown. The predicted activity of γ-glutamycyclotransferase is indicated by a question mark
The enzymatic activity of SAT in Arabidopsis crude protein extracts is in the low nmol mg-1 protein min-1 range (Haas et al., 2008; Heeg et al., 2008). The in vivo activity first depends on the availability of the two substrates. Serine can be assumed to be less limiting for SAT1, SAT3 and SAT5, if at all, especially in the light and in mitochondria due to photorespiration. Reports of acetyl-coenzyme A concentrations are rare and in the order of 5 nmol g-1 fresh weight (Roughan, 1997; Tumaney et al., 2004). When compared to Km values of the major SATs 1, 3, and 5 (KmSer=1.6–2.7 mM; KmAcCoA=20–280µM; Kawashima et al., 2005), it seems more likely that acetyl coenzyme A rather than serine availability is limiting SAT activity. However, acetyl-coenzyme A feeding experiments have not been carried out to date. Feedback sensitivity of SAT to cysteine probably modulates the rate of cysteine synthesis at least in some subcellular compartments. In S. typhimurium SAT is strictly feedback inhibited by cysteine (Ki = 1 µM; Kredich, 1996) as part of the regulatory circuit of the Cys regulon. Plants have higher cellular cysteine concentrations and in Arabidopsis cytosolic SAT5 shows 50% inhibition (IC50) at 1.8 µM cysteine. Free mitochondrial SAT3 is only inhibited at higher cysteine concentrations (IC50=53 µM), while plastidic SAT1 does not show any apparent inhibition by cysteine at all (Noji et al., 1998; Kawashima et al., 2005; reviewed in Wirtz and Hell, 2006). SAT2 and SAT4 have different kinetic constants with very high Km values for serine and acetyl-coenzyme A (24 to 121 mM), complete insensitivity to cysteine in the case of SAT2 and an IC50 of 0.8 µM cysteine of SAT4 (Kawashima et al., 2005). Feedback sensitivity of cytosolic SAT5 and low or no sensitivity of the organellar SAT3 and SAT1 would point to the cytosol as the major regulatory site (Noji et al., 1998). However, this observation would not explain how mitochondrial SAT3, which constitutes the bulk of cellular SAT activity, is efficiently controlled. The assumed subcellular concentrations of cysteine in Arabidopsis leaves (Krüger et al., 2009) suggest that the feedback-sensitive cytosolic SAT5 would be completely inhibited by high cysteine concentrations, whereas the insensitive plastidic SAT1 would operate in an environment with generally low cysteine concentrations. The cysteine concentration in mitochondria carrying SAT3 with intermediate feedback sensitivity is unknown. In pea leaves, the cytosolic SAT fraction is not inhibited by cysteine, while the plastidic and mitochondrial fraction is inhibited strongly and weakly, respectively (Droux, 2003). In spinach and tobacco, organelle-localized SAT isoforms are inhibited by cysteine in a physiologically relevant range. Thus, evolution may have developed species-specific solutions for the feedback regulatory role of SATs (Droux, 2004; Wirtz and Hell, 2006). It should be cautioned, however, that these feedback analyses of the Arabidopsis SATs have been carried out with recombinant proteins in the absence of OAS-TL and thus the CS complex. The significance of feedback regulation of SAT by cysteine is further discussed in the CS complex section below.
Pathways leading to cysteine: step 2 — integration of sulfide by O-acetylserine (thiol) lyase
The second step of cysteine synthesis is catalyzed by OAS-TL (EC 2.5.1.47). It belongs to the superfamily of pyridoxal phosphate-dependent enzymes and carries out a β-substitution of the acetyl group of OAS by sulfide, hence the alternative name Bsas for β-substituted alanine synthase and the older name cysteine synthase (Figure 2; Hatzfeld et al., 2000; Watanabe et al., 2008a). In bacteria and plants, the maximal specific activity of OAS-TL in whole protein extracts is one to two orders of magnitude higher than that of SAT. In Arabidopsis leaves, the ratio is approximately 60:1 on a per protein basis (ca. 600 nmol mg-1 min-1 OAS-TL activity compared to ca. 10 nmol mg-1 min-1 SAT activity; Heeg et al., 2008). SAT activity is therefore generally considered as rate limiting for cysteine synthesis. The ancestral Oastl gene in chlorophytes and plants was derived from cyanobacteria (Kopriva et al., 2008). In Arabidopsis, there are nine Oastl-like genes (Table 2) including OastlA2 (Bsas1;2; AT3G22460), a pseudogene which is transcribed but encodes a truncated, non-functional protein (Jost et al., 2000). Thus, the term OAS-TL A always refers to only OAS-TL A1 in the subsequent text. The three most highly expressed genes encode the major isoenzymes in the cytosol, plastids and mitochondria: OAS-TL A (AT4G14880, Bsas1;1), B (AT3G22460, Bsas2;1), C (AT2G43750, Bsas2;2), respectively. These three proteins and the low abundant D1 (AT3G04940, Bsas4;2) and D2 (AT5G28020, Bsas4;1) proteins are enzymatically true OAS-TLs (Yamaguchi et al., 2000; Wirtz et al., 2004; Heeg et al., 2008). According to analyses of several T-DNA knock-out lines, the cytosolic OAS-TL A contributes about 55% of enzyme activity and most of the flux of labelled sulfur in non-stressed leaves. Chloroplast OAS-TL B and mitochondrial OAS-TL C contributes approximately 45% and 5% of the total activity in leaves respectively (Figure 3B; Heeg et al., 2008; Watanabe et al., 2008a). This distribution matches findings from cellular fractionation experiments with other plant species (Lunn et al., 1990; Kuske et al., 1996). The Km-values for OAS of OAS-TL A, B and C are in the range of 310–690 µM and thus 10–60-times higher than OAS concentrations in the cytosol and plastids (Wirtz et al., 2004; Krüger et al., 2009). In contrast, Km-values for sulfide range from 3 to 6 µM (Wirtz et al., 2004) and are thus 2–4-times lower compared to total tissue sulfide concentrations under sufficient and limiting sulfur supply (Krueger et al., 2010). Subcellular sulfide concentrations have been reported to be even higher (Krüger et al., 2009). These compartments differ in their average pH values. The preferred substrate form of OAS-TL is HS- that shows the least changes in abundance of the H2S <-> HS- <-> S2- equilibrium between pH 7 and 8 in a dissociation graph (Wirtz et al., 2004). These kinetic data indicate that OAS but not sulfide is rate-limiting for cysteine synthesis, at least under sufficient sulfur supply.
Table II.
The OAS-TL gene and protein family in Arabidopsis.
In contrast to these OAS-TL isoforms, AtCysC1 (AT3G61440, Bsas3;1) is a β-cyanoalanine synthase. The gene is strongly expressed and the encoded protein preferentially catalyzes the formation of β-cyanoalanine and sulfide from the substrates cyanide and cysteine in mitochondria (Hatzfeld et al., 2000; Jost et al., 2000). Cyanide may be stored as γ-glutamyl-β-cyanoalanine or degraded by nitrilase activity to asparagines (Watanabe et al., 2008a). This detoxification process is assumed to play an important role in protecting complex IV of the respiratory chain from cyanide inhibition. OAS-TL and CAS can only be distinguished by their reaction kinetics and substrate preferences, since they are able to catalyse both reactions in vitro (Warrilow and Hawkesford, 2000). Infact, an in-gel staining method for OAS-TLs is based on the CAS reaction of cysteine and KCN-Pb reagent to β-cyanoalanine and insoluble PbS (Warrilow and Hawkesford, 1998). Considering the biochemical characteristics of the Arabidopsis proteins it appears unlikely that OAS-TLs and CAS can operate in the respective reverse directions in vivo (Jost et al., 2000). Kinetic analyses based on these biochemical observations suggest that spinach leaves possess only a mitochondrial CAS but no OAS-TL protein (Warrilow and Hawkesford, 2000). Whether cytosolic OAS-TLs D1 and D2 contribute to cysteine synthesis in Arabidopsis at all is disputable, but a detailed analysis is missing to date. Both are only weakly expressed and OAS-TL D1 shows 10-times higher Km values for sulfide and OAS at lower vmax rates when compared to OAS-TLs A, B and C, at least when expressed as fusion proteins (Yamaguchi et al., 2000). Although low OAS-TL activity was reported for OAS-TL D2 at saturating substrate conditions, it is unlikely to function as an efficient OAS-TL in vivo. OAS-TL D2 has a substitution of Ser at position 88 in the Asn loop in the OAS-TL catalytic centre (Rabeh and Cook, 2004). Mutation of this residue in OAS-TL A eliminated catalytic activity (Bonner et al., 2005), suggesting that the observed OAS-TL activity of OAS-TL D2 may be residual or a side activity of another enzymatic function. On the other hand, OAS-TL D1 and D2 were purified by SAT affinity chromatography, indicating their ability to form a CS complex, a feature that is regarded as hallmark of OAS-TLs (Heeg et al., 2008). Thus, OAS-TL D1 and D2 must be considered as true OAS-TL enzymes in a biochemical sense but have no currently known functions in vivo. The OAS-TL-like protein family further comprises cytosolic CS-like (Bsas4;3, AT5G28030) and plastidic CS26 (Bsas5;1, AT3G03630; Table 2). In both enzymes the Ser at position 88 is also substituted. As expected from the serine exchange in the catalytic center, CS-like protein has no OAS-TL activity but is able to cleave cysteine to form sulfide, ammonia and pyruvate (Hatzfeld et al., 2000; Ãlvarez et al., 2010a). It represents a cysteine desulfhydrase (termed DES1) and therefore a reaction that leads away from cysteine. The CS26 isoform can act as a S-sulfocysteine synthase accepting thiosulfate instead of sulphide as a donor of reduced sulfur for incorporation into OAS (Bermudez et al., 2010).
All true OAS-TL proteins are assumed to associate with SAT in the CS complex, but not AtCysC1, CS26 and CS-like. This is shown by affinity purification of the OAS-TL proteins from Arabidopsis leaves using cytosolic SAT as an immobilized anchor (Heeg et al., 2008). Only OAS-TL A, B, C, D1 and D2 have been isolated. Their relative abundance corresponded to the loss of OAS-TL activity in single T-DNA insertion mutants lacking either oastlA, oastlB or oastlC. This indicates no compensatory changes in the abundance of isoforms and a remarkable similarity of the OAS-TL patterns between the loss-of-functions mutants, in other plant tissues and under stress and is in line with nearly unchanged gene expression profiles in publicly available microarray databases (Heeg et al., 2008; Wirtz et al., 2010a). The intensities of OAS-TL proteins in 2-dimensional gels also correlate with transcription patterns reported in Genevestigator (Zimmermann et al., 2004). Expression and translation of the major OAS-TLs in different tissues appear to be in a linear relationship. According to databases and numerous reports, OAS-TL genes are semi-constitutively expressed during development from seedling to silique (Wirtz et al., 2010a), i.e. they are ubiquitously expressed with varying abundance in different cells types. The only reported significant change of expression is the induction of OastlA gene on exposure to cadmium which is consistent with the cadmium sensitivity of oastlA mutants (Dominguez-Solis et al., 2001). Overexpression of OastlA is reported to enhance resistance to cadmium (Dominguez-Solis et al., 2004). The microarray-based gene expression databases (AtGenExpress, Genevestigator) report no increase of OastlA mRNA during cadmium stress but those experiments may have been carried out using different experimental conditions with respect to metal chelators present in the growth media.
The abundance of the major OAS-TL isoforms in leaves is further confirmed by determination of absolute protein levels. Using calibration by recombinant antigens and polyclonal antisera the following concentrations were determined: OAS-TL A at 3.2 ng µg-1 soluble protein, OAS-TL B at 1.0 ng µg-1 soluble protein and OAS-TL C at 0.38 ng µg-1 soluble protein (Wirtz et al., 2010a). Interestingly, these isoforms are represented by up to seven different protein species in two-dimensional gels. Mass spectrometry indicates that OAS-TL A, B and probably also C are amino-terminally acetylated and have other unresolved modifications (Wirtz et al., 2010a). N-terminal acetylation is a frequent modification in eukaryotic cells and has been connected to protein stability in yeast (Hwang et al., 2010). When the kinetic properties of acetylated native and non-acetylated recombinant OAS-TL A are compared, this modification appears to have no detectable influence, leaving its function in cysteine synthesis open (Wirtz et al., 2010a). Post-translational modifications of OAS-TL A may have regulatory functions, especially with respect to interaction with SAT. However, there is so far no direct evidence suggesting that a moderate increase or decrease of the already high cytosolic activity can have an impact on cysteine synthesis. OAS-TL A was shown to be S-nitrosylated via transnitrosylation from naturally-occurring substrates such as S-nitrosoglutathione (Lindermayr et al., 2005). However, the activity of recombinant OAS-TL was not affected by S-nitrosylation but was highly sensitive to heavy metals (Wirtz et al., 2005). OAS-TL A was reported to be particularly sensitive to treatment with peroxynitrite, resulting in 90% inhibition of the activity of recombinant enzyme. The modified enzyme carries a 3-nitroTyr302 residue near the OAS binding site (Ãlvarez et al., 2010b). This mechanism may represent a way of coupling OAS-TL activity to oxidative stress by nitric oxide, although this physiological function still needs to be verified in vivo.
The cysteine synthase complex
The CS complex was first discovered in S. typhimurium (Kredich et al., 1969; Kredich, 1996). It shares a number of features with the plant CS complexes: upon extraction of protein from cells, all SAT activity is associated with OAS-TL activity; the complex spontaneously forms in vitro, and is stabilized by sulfide and dissociated by OAS; OAS-TL is almost inactive in the complex but becomes re-activated when released; the intermediate, OAS, is not channeled but freely leaves the CS complex; a molar excess of OAS-TL to SAT protein is required to achieve optimal cysteine production (reviewed in Kredich, 1996; Droux, 2003; Wirtz and Hell, 2006; Hell and Wirtz, 2008). The molecular properties of bacterial and plant CS complexes are presented in the section on structure-function relations below. The bacterial CS complex has not been connected to regulation of cysteine synthesis, whereas the CS complex is in the center of regulating cellular sulfur homoeostasis in plants. Enterobacteria rely on a classical nutrient-controlled regulon with a DNA binding protein (CysB) as activator. When cysteine is absent in the surrounding environment, the feedback-inhibition of SAT encoded by the CysE gene is relieved and the bacterial cell begins to produce OAS. OAS converts to NAS which binds to CysB and activates transcription of the Cys regulon. All genes of primary sulfur metabolism are regulated by this activator except CysE. This guarantees sufficient OAS is always produced, providing a mechanism for sensing the availability of cysteine (Kredich, 1996).
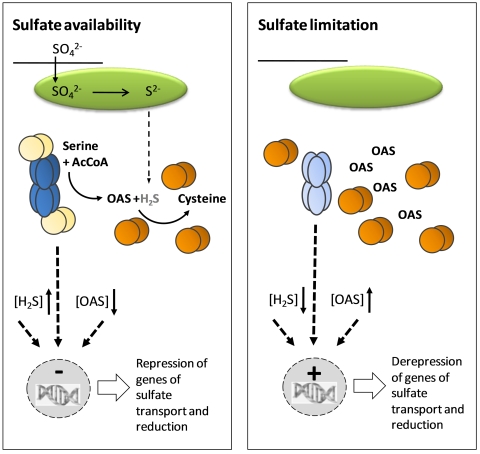
Work with Arabidopsis CS complexes in recent years has revealed a metabolic sensing system for cellular sulfur homeostasis (Figure 4). Since SAT is more active when associated with OAS-TL, the amount of SAT inside the complex determines the rate of OAS production and cysteine synthesis (Droux et al., 1998). SATs in the cytosol, plastid and mitochondrion interact with OAS-TL via its C-terminus (Bogdanova and Hell, 1997; Jost et al., 2000). Modulation of SAT activity inside and outside the CS complex may be achieved in different ways: (1) excess OAS-TL is required to stabilize SAT similar to a chaperon function (Droux et al., 1998); (2) modification of substrate affinities or vmax of SAT upon complex formation (no significant changes observed yet); (3) protection of SAT by OAS-TL from targeted degradation (not analyzed yet); (4) decreased feedback sensitivity of SAT to cysteine inside the complex (Kumaran et al., 2009; Wirtz et al., 2010b). Indeed, cytosolic SAT5 and mitochondrial SAT3 are less sensitive to feedback inhibition when associated with their respective OAS-TL partners compared to their free form (Wirtz et al., 2010b). A cytosolic SAT from soybean (AF452452.1) shows similar properties in this respect (Kumaran et al., 2009). Another soybean SAT with dual cytosolic/plastidic localization becomes less feedback sensitive to cysteine after phosphorylation by a calcium-dependent protein kinase (Li et al., 2006). Modification of feedback-sensitivity, albeit by different mechanisms, therefore seems to be a common feature to control SAT activity and the flux of reduced sulfur into cysteine.
Figure 4.
Model for regulation of cysteine synthesis by the CS complex
The simplified regulatory model illustrates the processes in the cytosol. Sulfate is transported from the extracellular space via the cytosol into the plastids (green oval), where it is reduced to sulfide. Sulfide can leave the plastids to serve in the cytosol as substrate for cysteine synthesis by free OAS-TL dimers (orange spheres). The carbon- and nitrogen-containing backbone of cysteine, OAS, is synthesized by SAT within the CS complex (dark blue ellipses). OAS leaves the complex because OAS-TL dimers within the CS complex (yellow spheres) are catalytically inactive. Limitation of sulfate results in the decrease of sulfide and increase of OAS concentrations, resulting in dissociation of the CS complex. SAT activity is down-regulated upon dissociation of the CS complex, because free SAT hexamers (light blue ellipses) are sensitive to inhibition by cysteine. The dissociation of the CS complex and the effectors, OAS and sulfide, may act as signals (dotted arrows), which trigger transcription of genes involved in sulfate transport and reduction, allowing the resupply of sulfide.
Further evidence for a regulatory role of the CS complex is derived from the effectors sulfide and OAS. Sulfide stabilizes the CS complex (Wirtz and Hell, 2006). Dissociation of SAT and OAS-TL in vitro in the presence of sulfide requires OAS concentrations in the millimolar range which can probably never be reached in vivo (Wirtz et al., 2004). In contrast, in the absence of sulfide, i.e. during sulfate deprivation, OAS effectively dissociates the CS complex. The dissociation of 50% of the complex is achieved in the presence of 50 to 70 µM OAS and exhibits positive cooperativity: below the 50 to 70 µM OAS threshold, the CS complex is fully associated; above the OAS threshold, it is fully dissociated (Berkowitz et al., 2002). Since OAS concentrations in the cytosol in non-starved plants are in the range of the threshold (Krüger et al., 2009) and are known to increase while sulfide levels decrease during sulfate deprivation, the system provides a perfect molecular switch to modify SAT activity and adjust it to the metabolic situation of the cell via CS complex association and dissociation (Hell and Hillebrand, 2001). The inactivation of OAS-TL in the CS complex (Droux et al., 1998; Berkowitz et al., 2002) is the reason why OAS is not channeled but is released into the surrounding solution. The role of OAS as a potential signal for induction of genes of sulfate uptake and reduction has been discussed in the sections above. OAS would therefore not only serve as a substrate for cysteine synthesis but act as an effector for CS complex dissociation and a signal for gene expression, at least after prolonged sulfate deprivation, to recover cellular sulfur homeostasis.
Evidence for the metabolic regulation by the CS complex is indirectly provided by the analysis of T-DNA mutants (Heeg et al., 2008). An oastlAB double mutant that has only about 5% of wildtype OAS-TL activity (provided by mitochondrial OAS-TL C) shows only slight growth reduction and has even higher cysteine and glutathione levels at elevated OAS contents. These observations suggest that more sulfide from chloroplasts reaches the mitochondria compared to wildtype plants, where it stabilizes the mitochondrial CS complex to provide cysteine for the requirements of the rest of the cell. Increased sulfide concentrations in mitochondria of oastlAB plants would promote enhanced SAT activity and more OAS is produced, resulting in a flux of 35S-sulfate into cysteine that is comparable to wildtype (Heeg et al., 2008). Further proof of the CS complex function is derived from de-regulation of cysteine synthesis by overexpression of a dominant negative mutant of SAT in the cytosol of tobacco (Wirtz and Hell, 2007). Mitochondrial SAT3 can be completely inactivated by mutation of a histidine residue in the catalytic center but still retains its ability to interact with OAS-TL proteins from Arabidopsis (Wirtz et al., 2001). When expressed in the cytosol of transgenic tobacco, the inactive SAT interacts with endogenous OAS-TL and outcompetes tobacco SAT. The dramatic increase in total cysteine and glutathione in these tobacco plants is interpreted as over-compensation of mitochondrial and/or plastidic CS complexes for the down-regulated cysteine formation in the cytosol (Wirtz and Hell, 2007).
In summary, the CS complex acts as a sensor for the availability of sulfide which reflects the sulfate supply of the cell (Figure 4). Sufficient level of sulfide stabilizes the CS complex in which SAT is active and produces OAS. Since OAS-TL in the CS complex is inactive, OAS is released from the complex and is used by free OAS-TL for cysteine synthesis. Decreased sulfide concentration due to sulfur deprivation allows OAS to accumulate because limited sulfide is available for cysteine formation. The accumulation of OAS above its cellular threshold level causes the CS complex to dissociate. SAT activity stalls and produces OAS at a lower rate to sustain acetyl coenzyme A until OAS triggers the de-repression of genes controlling sulfate transport and reduction. When the level of sulfide is restored, the CS complex can re-associate and SAT activity resumes. Since this regulatory model was first fully outlined (Hell and Hillebrand, 2001; Wirtz and Hell, 2006), it has not been contradicted by any published experiments investigating the regulation of cysteine synthesis. However, it should be cautioned that several aspects need further investigation. The mechanisms of SAT inactivation are not fully understood, the experimentally determined structure of the CS complex with its effector binding sites is unknown and the link between the CS complex as a fast sulfide sensor for sulfur homoeostasis and the comparatively slow trigger function of OAS on gene expression needs to be further explored.
The properties of the CS complex may be further extended by interaction with additional partners. A yeast-two-hybrid screen described the interaction of plastid SAT1 with the cyclophilin, CYP20-3 (Dominguez-Solis et al., 2008). This protein family has peptidyl-prolyl-cis-trans-isomerase activity and may assist the association of SAT in the CS complex. Lack of CYP20-3 apparently results in decreased synthesis of cysteine and glutathione in chloroplasts under light and oxidative stress and consequently in a severe stress phenotype in Arabidopsis thaliana (Dominguez-Solis et al., 2008). Since SAT and OAS-TL associate very efficiently in vitro (Droux et al., 1998; Berkowitz et al., 2002) and plastidic SAT contributes to cellular OAS formation only to a minor extent (Figure 3), the role of CYP20-3 needs further investigation. If SAT1 activation is required for enhanced thiol production to achieve resistance to oxidative stress, the serat2;1 mutant lacking SAT1 (Watanabe et al., 2008b) should be more sensitive under such stress conditions. This, however, seems not to be the case (A. Speiser, M. Wirtz, R. Hell, unpublished data). Another observation describes the interaction of OAS-TL A with the C-terminal STAS domain of sulfate transporter, Sultr1;2, in the yeast-two-hybrid system (Shibagaki and Grossman, 2010). The analysis of sulfate transport in yeast cells expressing Sultr1;2 and OAS-TL A together suggests that the interaction has a negative impact on transporter activity, whereas the activity of OAS-TL measured in vitro was enhanced by co-incubation with the STAS domain. This finding would provide an elegant link between cysteine synthesis in the cytosol and regulation of sulfate uptake at the plasmalemma (Shibagaki and Grossman, 2010). It is currently unclear how the abundant, highly active and almost constitutively expressed OAS-TL A can mediate such changes in the physiological context. However, these initial reports point to possible additional regulatory functions that depend on the presence of SAT and OAS-TL proteins or the CS complex.
Compartmentation of cysteine metabolism
An important consequence of the extensive analysis of knock-out mutants of the Sat (Serat) and Oastl genes and some of their combinations is the intracellular exchange of metabolites in the cysteine metabolic pathway. Comparison of the serat and oastl mutants has led to the hypothesis that serine, sulfide, OAS and cysteine may also cross organellar membranes (Figure 3C). For instance, mutants lacking cytosolic OAS-TL A are fully viable, indicating that plastids and possibly mitochondria are able to provide cysteine for the requirements of the cytosol (Heeg et al., 2008; Lopez-Martin et al., 2008; Watanabe et al., 2008a). Furthermore, the viability of a mutant lacking plastidic OAS-TL B suggests sulfide is able to exit the plastid to the cytosol and, as evidenced by an oastlAB double mutant, to enter mitochondria (Heeg et al., 2008). Since mitochondria are the only site of cysteine production in this double mutant, cysteine must be able to reach the cytosol and plastids. Likewise, different mutant combinations lacking SATs provide evidence for the transport of OAS into and out of the organelles (Watanabe et al., 2008b; Krüger et al., 2009). To date, mechanisms for transporting other intermediates necessary for cysteine metabolism are yet to be identified. With sulfide in equilibrium with H2S, it is generally assumed that this uncharged form is able to penetrate lipid membranes in a way similar to water (Jacques, 1936). Indeed, in the context of analysis of aquaporin substrate specificities, it becomes evident that H2S can pass biological membranes more readily than water without the requirement of any facilitator (Mathai et al., 2009).
It should be noted that possibly not all plant species are able to freely exchange OAS and cysteine between sub-cellular compartments. The strong accumulation of cysteine and glutathione in tobacco with dominant-negative overexpression of an enzymatically inactive SAT in the cytosol has been interpreted as inactivation of the cytosolic CS complex by competition between endogenous and transgenic SAT. The strong accumulation of thiols in the transgenic tobacco lines has been suggested to result from insufficient exchange of OAS or cysteine between the cytosol and organelles where the respective CS complexes react by over-compensatory cysteine production (Wirtz and Hell, 2007).
The consideration of glutathione biosynthesis and degradation in Arabidopsis adds complexity to this subcellular exchange of pathway intermediates but is particularly important with respect to cysteine demand and recycling (Figure 3D). The first enzyme of glutathione biosynthesis, glutamate cysteine ligase (GCL; GSH1, AT4G23100), forms γ-glutamylcysteine (γ-EC) and is exclusively localized in plastids. The second enzyme, glutathione synthetase (GS; GSH2, AT5G27380), is concluded to be present in plastids but preferentially in the cytosol from the abundance of mRNA splice variants (Wachter et al., 2005). GS null mutants are seedling lethal but can be complemented by expression of GS only in the cytosol (Pasternak et al., 2008). The identification of a gene family encoding CLT transporters in the plastid inner membrane that are related to chloroquine resistance proteins from the malaria parasite, Plasmodium falciparum, revealed transport of γ-glutamylcysteine and glutathione between the plastid and the cytosol (Maughan et al., 2010). In the oastlAB double mutant (Heeg et al., 2008) that lacks cysteine synthesis in the cytosol and the plastids, the following scenario may be envisaged: Sulfide is generated from sulfate in the plastid, diffuses through the cytosol to the mitochondrion where it is integrated into cysteine. Transport of cysteine from the mitochondrion to the plastid allows its integration into γ-glutamylcysteine which may then be transported to the cytosol as the substrate for glutathione biosynthesis. This example of metabolite shuffling illustrates how complex the seemingly simple cysteine to glutathione pathway can be and presumably how important intracellular communication is.
Structure-function relations of the cysteine synthase complex
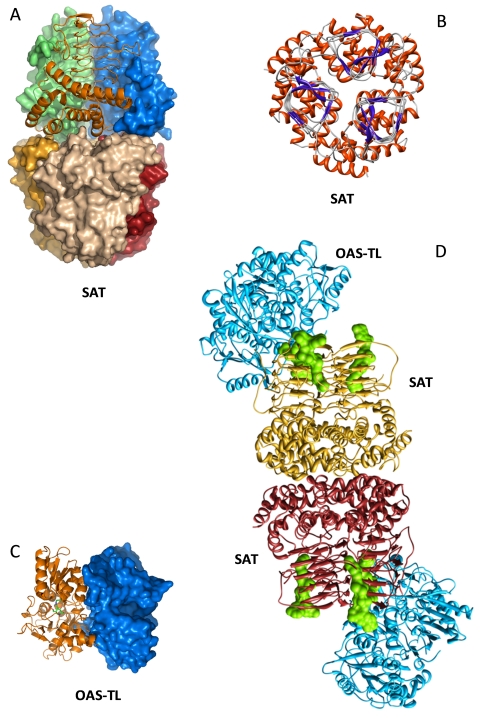
The reversible association and dissociation with concomitant activation and deactivation of the subunits of the cysteine synthase complex must be based on its structural properties. At present crystal structures of SAT (Gorman and Shapiro, 2004; Olsen et al., 2004; Pye et al., 2004) and OAS-TL (Burkhard et al., 1998; Burkhard et al., 1999) from bacteria and of cytosolic OAS-TL A from Arabidopsis (Bonner et al., 2005) are known. Amino acid sequence similarities between enterobacterial and plant SAT are in the order of 34% to 44 %. They are sufficient for reliable protein modeling of the plant orthologues that predicts a pronounced domain structure of plant SATs: (1) transit peptide in case of organellar isoforms; (2) N-terminal α-helix cluster of mature protein; (3) central β-sheet cluster; (4) C-terminus (Bogdanova and Hell, 1997; Noji et al., 1998). The β-sheet cluster shows the highest homology between plants and bacteria and places SATs in a family of acyltransferases with an unusual left-handed parallel β-helix which carries the catalytically active domain (Vuorio et al., 1991; Vaara, 1992). Computational modeling of this domain shows a prism-like structure that is composed of β-sheets of a hexapeptide described as [LIV]-[GAED]-X2-[STAV]-X (Vaara, 1992; Raetz and Roderick, 1995) and arranged in a left-handed helix of four complete rounds. Site-directed mutagenesis of Arabidopsis mitochondrial SAT3 showed a histidine on a loop that protrudes from the prism-like structure is involved in catalysis, presumably by binding acetyl-coenzyme A (Wirtz et al., 2001). By analogy to E. coli SAT (Olsen et al., 2004), the modeled SAT3 β-helix domain is arranged as a trimer with three catalytic sites located in the clefts between each subunit (Feldman-Salit et al., 2009). In bacteria, two SAT trimers form a hexamer (Hindson et al., 2000; Gorman and Shapiro, 2004; Pye et al., 2004) and, indeed, the α-helical cluster in SAT3 is responsible for SAT-SAT interaction in yeast two-hybrid assays (Bogdanova and Hell, 1997). The C-terminus consists of a region close to the β-sheet domain that is involved in feedback-inhibition by cysteine in both E. coli and plant SATs (Noji et al., 1998; Inoue et al., 1999) and a flexible end of about 10 amino acids with a terminal isoleucine. The structure of E. coli SAT shows that cysteine binds in the same pocket as serine, explaining the competitive inhibition mechanism of SAT (Olsen et al., 2004). Deletion analysis shows that the C-terminus is responsible for interaction with OAS-TL (Bogdanova and Hell, 1997; Mino et al., 1999). Although the final 10 amino acids are highly divergent between bacterial and plant SAT, the interaction is largely dependent on the C-terminal isoleucine in both cases (Campanini et al., 2005; Francois et al., 2006). Detailed comparisons of structure and reaction mechanism of Arabidopsis and bacterial SAT and OAS-TLs are available (Wirtz and Droux, 2005; Hell and Wirtz, 2008; Yi et al., 2010).
Arabidopsis OAS-TL A and S. typhimurium and H. influenza OAS-TLs also share 30 to 40 % amino acid sequence identity and are structurally very similar (Burkhard et al., 1998; Burkhard et al., 1999; Bonner et al., 2005). OAS-TLs form stable homodimers of predicted molecular masses of 68 to 75 kD. In crystal structures, the two active sites of the dimer face each other, each carrying a tightly bound pyridoxal phosphate as cofactor. The active site is placed in a cleft deep in the center of each monomer (Rabeh and Cook, 2004), a feature that is important for understanding the inhibition of OAS-TL by SAT (see below). The β-replacement reaction of the acetyl group of OAS by elimination and substitution by sulfide is one of the best described enzyme reactions in general. OAS binds first with a very low dissociation constant (<1µM OAS), but the low affinity (>300µM OAS) limits the overall reaction but not the irreversible substitution by sulfide. These kinetic properties form the basis for the regulatory function of the CS complex (Wirtz et al., 2004). The mechanism of enzymatic inhibition of OAS-TL upon association of the CS complex is based on the binding of the flexible C-terminal end of SAT to the catalytic center of OAS-TL. Evidence for this elegant explanation comes from co-crystallization of OAS-TLs from H. influenza and Arabidopsis OAS-TL A with decapeptides of the corresponding SATs (Huang et al., 2005; Francois et al., 2006). Association and dissociation of the CS complex is based on competition of OAS with the C-terminal end of SAT for the substrate binding site of OAS-TL.
No crystal structure of the CS complex from any organism is currently available which is mainly due to the lability of SAT proteins. However, biochemical analyses reveal the mechanisms of SAT and OAS-TL interaction. Binding constants of C-terminal SAT decapeptides from H. influenza and S. typhimurium to OAS-TL from H. influenza are in the same range, suggesting a general binding mechanism in bacteria. However, with KD = 0.6 to 1 µM, they are 250-times lower compared to the affinity of full-length bacterial SAT (Mino et al., 2000; Campanini et al., 2005). The decapeptide of Arabidopsis cytosolic SAT5 shows much higher affinity to OAS-TL A (KD=5–100 nM; Kumaran and Jez, 2007) and is in the same range of affinities of the full-length SATs from the cytosol and mitochondria of Arabidopsis to OAS-TLs (KD=25–40nM; Droux et al., 1998; Berkowitz et al., 2002; Wirtz et al., 2010b). The different binding constants of γ-proteobacterial and Arabidopsis CS complexes may support the regulatory function of the plant complex which requires dissociation by OAS. It should be cautioned that these data rely on the existence of only one binding site between SAT and OAS-TL that is identical with the binding site for OAS. At present, the binding site(s) of sulfide leading to complex stabilization is unknown.
The quaternary composition of the CS complex is therefore important for understanding its function. The pioneering work of Nicholas Kredich with S. typhimurium describes a molecular mass of 309 kD for the hetero-oligomeric CS complex: 160 kD for the SAT hexamer and 68 kD for the OAS-TL dimer. A 6:4 ratio of SAT to OAS-TL subunits in the assembled complex is deduced from these data (Kredich et al., 1969; Kredich, 1996). The core of this complex is formed by the dimer of trimers (hexamer; Hindson et al., 2000) of SAT which would recruit two OAS-TL dimers. This stoichiometry is supported by data from fluorescence titration (Campanini et al., 2005). The molecular masses of CS complexes from spinach, tobacco, Arabidopsis (Droux et al., 1992; Wirtz and Hell, 2007; Wirtz et al., 2010b) and also soybean (Gycine max; Wirtz et al., 2010b) are in the same range which initially suggests a similar composition as in bacteria. Since the C-termini of SAT subunits in one trimer point to the same direction, they all potentially can bind an OAS-TL dimer at one catalytic site next to each other and possibly in a sequential manner. The same arrangement would be possible on the second trimer of the SAT hexamer. However, the presence of only two OAS-TL dimers in the CS complex points to binding of only one dimer at each of the two distal ends of the SAT hexamer. This organization would fulfill a sense for symmetry but still leaves open the question of binding by one or two SAT C-termini (Campanini et al., 2005; Wirtz and Hell, 2006) (Figure 5). Indeed, plant and bacterial SAT decapeptides bind to OAS-TL dimers in a 2:1 ratio, but full SAT trimers rather than decapeptides could exert steric hindrance and prevent more than one OAS-TL dimer from binding. Addition of excess decapeptides to the assembled bacterial CS complex results in no detectable binding of the decapeptide (Campanini et al., 2005). This suggests that complex-bound OAS-TL has no accessible catalytic center and would be consistent with the almost complete inactivation of the enzyme in plant and bacterial CS complexes (Kredich et al., 1969; Droux et al., 1998; Wirtz et al., 2001). The resulting absence of substrate channeling of OAS is another cornerstone of the metabolic regulatory model of the plant CS complex (Hell and Hillebrand, 2001). The experimental data are corroborated by modeling the mitochondrial SAT3/ OAS-TL C complex of Arabidopsis (Feldman-Salit et al., 2009). When the so far elusive and not crystallized C-terminal tail of SAT is modeled and included in docking studies based on diffusional encounter complexes of SAT and OAS-TL that are generated by Brownian dynamics simulation, a stoichiometric ratio of six OAS-TL dimers to one SAT hexamer in the CS complex is geometrically possible. However, binding energy calculations suggest that a ratio of only two OAS-TL dimers to one SAT hexamer is more likely (Feldman-Salit et al., 2009).
The comparable quaternary composition of bacterial and both cytosolic and mitochondrial Arabidopsis CS complexes is confirmed by several biochemical methods that all indicate a 6:4 ratio of SAT:OAS-TL monomer composition. These studies include the OAS-triggered dissociation of the Arabidopsis cytosolic CS complex in native leaf extracts (Wirtz et al., 2010b). Moreover, compared to bacterial CS complexes, the Arabidopsis SAT-OASTL binding constant of KD=30 µM is ten times higher and the calculated binding energies are generally lower, pointing to an energetically easier dissociation than association process, as would be expected for a regulatory function (Wirtz et al., 2010b). However, the spontaneous formation of the Arabidopsis CS complexes is well supported by a negative Gibbs free energy (ΔG = -33 kcal mol-1) with negative values of ΔS and ΔH. The organization of a CS complex from the cytosol of soybean has been reported to be an exception consisting of a single SAT homotrimer with each subunit binding one OAS-TL dimer in sequentially decreasing affinity (Kumaran et al., 2009). The 3:6 ratio of monomeric subunits would result in a total molecular mass in the same range as the Arabidopsis CS complexes with a 6:4 ratio (Kumaran et al., 2009). However, this study used a cDNA sequence (AF452452.1) that does not correspond to the available soybean genomic DNA database. A subsequent investigation using a soybean SAT cDNA that matches the genomic database showed a hexameric SAT and likely 6:4 ratio of subunits (Wirtz et al., 2010b).
The most important feature for the regulatory function of the plant CS complex known at present is the modification or activation of SAT activity in the complex compared to free SAT. The cytosolic soybean CS complex reported by Kumaran et al., (2009) exhibits a two-fold increase in SAT activity and a decreased feedback sensitivity from 2 to 70 µM cysteine compared to free SAT. SAT activity remains unchanged in the Arabidopsis cytosolic and mitochondrial CS complexes but feedback sensitivity to cysteine is significantly reduced in both complexes, although to a much lower extent compared to soybean (Wirtz et al., 2010b). The release of feedback inhibition of another soybean SAT isoform with cytosolic and plastid localization is achieved by a calcium-dependent protein kinase (Liu et al., 2006). The corresponding phosphorylation site is not conserved in Arabidopsis SATs. Taken together, these results point to an important role of the modification of SAT feedback sensitivity to cysteine, either by CS complex formation or phosphorylation, to control the rate of cysteine synthesis.
Pathways downstream of cysteine: methionine synthesis, glutathione relations, and cysteine degradation
Cysteine is the direct or indirect donor for all reduced sulfur in a plant cell. The most important ones in Arabidopsis include methionine, glutathione, iron-sulfur cluster, molybdenum cofactor, vitamins (coenzyme A, lipoic acid, thiamine, biotin) and secondary compounds such as camalexin and glucosinolates. The biosynthesis and metabolism of these compounds have been discussed in other chapters of The Arabidopsis Book or special review articles (Balk and Lobreaux, 2005; Schwarz and Mendel, 2006; Hell et al., 2008; Rouhier et al., 2008; Noctor et al., 2011; Takahashi et al., 2011). Only the most nearby reactions of cysteine in this network will be presented here. A substantial fraction of cellular cysteine delivers reduced sulfur for methionine synthesis. To improve the content of the essential amino acid methionine in the diets of humans and non-ruminant animals is one important reason why plant biotechnology attempts to enhance cysteine synthesis in Arabidopsis and subsequently in crop plants (Galili et al., 2005; Khan and Hell, 2008). Cystathionine-γ-synthase in the plastids catalyzes the first committed step of cystathionine synthesis (Figure 2). Its activity is rate-limiting and feedback-regulated by S-adenosylmethionine (SAM) at the level of mRNA stability in Arabidopsis (Ravanel et al., 1998; Chiba et al., 2003). Cystathionine-β-lyase then catalyzes the release of homocysteine which is subsequently methylated to methionine by methionine synthase. Methionine belongs to the aspartate family of amino acids and is co-regulated with the biosynthesis of the other essential amino acids threonine, branched chain amino acids and lysine but not with sulfate assimilation or cysteine synthesis (Wirtz and Droux, 2005; Jander and Joshi, 2009). That the regulation of methionine biosynthesis is separated from cysteine synthesis causes considerable problems for the biotechnological improvement of crops (Galili et al., 2005). As for cysteine the steady-state content of methionine is rather low (about 1 nmol g-1 fresh weight; Ranocha et al., 2001), and the turnover must be assumed to be high to meet the demands for protein synthesis. Most of the free methionine is actually bound in SAM and the methionine transport form S-methylmethionine (Ranocha et al., 2001). The Yang cycle regenerates methionine and SAM for ethylene synthesis. In fact it is mostly the sulfur group (as thioketobutyric acid) that is recycled, stressing the requirement for reduced sulfur in this process. As a consequence severe sulfate starvation can negatively affect the efficiency of the Yang cycle in Arabidopsis (Bürstenbinder et al., 2007).
Figure 5.
Three-dimensional model structures of components of the mitochondrial CS complex from A. thaliana.
A. Surface presentation of SAT consisting of a dimer of two trimers. Every subunit has a different color, one SAT subunit (orange) is shown as ribbon model. B. SAT trimer viewed from top. Red, α -helices clustered at the bottom of each SAT subunit form the interaction sites with the α—helices of the second SAT trimer in the CS complex. Blue and grey, domain of β-sheets and turns forming a prism-like shape in each SAT subunit. A loop extends from the prism of each subunit into the space towards the clockwise following subunit as part of the catalytic centre formed by both neighboring subunits. C, OAS-TL dimer is shown as surface (blue) and ribbon (gold) model with the pyridoxal phosphate cofactor (green) inside. The cleft in the centre of the surface model of one subunit shows the catalytic centre and binding site of the SAT carboxy terminus. D, CS complex model of a SAT (golden) hexamer with two OAS-TL dimers (blue, purple) at distal ends binding to one of the carboxy termini of a SAT trimer. Acetyl Coenzyme A (cyan) is positioned at each of the six possible catalytic sites. Figures were generated using Chimera by courtesy of A. Feldman-Salit and R. Wade as described in Feldman-Salit et al. (2009).
The tripeptide γ-glutamylcysteinylglycine (glutathione) can be regarded as a chemically modified cysteine molecule. The flanking amino acids (glutamate and glycine) result in a shift in redox potential for the cysteine residue and protect the sulfhydryl group against oxidation (Meyer and Hell, 2005; Meyer, 2008), while the γ-peptide bond prevents proteolysis (Figure 2). It is tempting to speculate that during evolution glutathione became necessary to stabilize when the oxygen concentration on the earth increased due to photosynthesis. Alternatively, the stabilization of cysteine in glutathione may have been necessary in early life, since all organisms synthesize glutathione in the same way or carry glutathione-related reductants (Meyer and Hell, 2005; Meyer, 2008). It is therefore not surprising that the entry reaction catalyzed by GCL is highly regulated by transcriptional and post-translational mechanisms and thus rate-liming for glutathione synthesis (Yi et al., 2010). Throughout the life cycle of modern land plants, glutathione functions in scavenging and detoxifying reactive oxygen species (ROS), heavy metals and xenobiotics, in redox signaling, glutathionylation, electron transfer in catalytic processes (e.g. APS reduction by APR) (Foyer and Noctor, 2009; Noctor et al., 2011), and in other more specialized tasks such as cofactor in glucosinolate synthesis (Geu-Flores et al., 2009). Most of these tasks are mediated by glutathione reductase, γ-glutamyltransferase, glutathione-S-transferase and glutaredoxin. The glutathione redox functions in protection and ROS signaling seem to be so vital that cytosol and mitochondria of Arabidopsis have a glutaredoxin-based complementary system in case of insufficient glutathione reduction (Marty et al., 2009). Another example for the role of glutathione-mediated redox homeostasis is the prevention of cotranslational proteolytic processing defects (Frottin et al., 2009). In Arabidopsis, glutathione operates as reservoir for cysteine. When cells are supplied with OAS or cysteine, the intracellular excess of cysteine is transiently saved as glutathione. If plants are sulfate-starved, levels of glutathione decrease before those of cysteine (Koprivova et al., 2000; Tsakraklides et al., 2002; Hirai et al., 2003). Breakdown of glutathione is initiated in four possible ways: First, γ-glutamlytransferases (GGT1, AT1G23310 and GGT2, AT1G70580) associated with the outer side of the plasmalemma catalyse the degradation of oxidized glutathione to free cysteine and glutamate in the apoplast, possibly as part of a extracellular glutathione retrieval system e.g. during long distance transport (Martin et al., 2007; Ohkama-Ohtsu et al., 2007b; Destro et al., 2010). Second, an unidentified γ-glutamylcyclotransferase activity releases cysteine and 5-oxo-proline which is converted to glutamate by 5-oxoprolinase. According to this hypothesis γ-glutamlytransferases are hardly involved in degradation of free glutathione in the cell (Ohkama-Ohtsu et al., 2008). Third, glutathione-S-transferase activities conjugate the cysteine moiety to endogenous or xenobiotic substrates (Dixon and Edwards, 2011). The conjugates are rapidly transported by multidrug resistance protein transporters of the ABC-type family (Sanchez-Fernandez et al., 2001) to the vacuole where GGT4 (AT4G39650) initiates the degradation via γ-EC to cysteine (Grzam et al., 2007; Martin et al., 2007; Ohkama-Ohtsu et al., 2007a). Whether cysteine can be recovered from the vacuole is currently unknown. Finally, breakdown of glutathione conjugates may be initiated through the removal of the carboxyterminal glycine residue by a side activity of phytochelatin synthase to form a γ-EC conjugate (Grzam et al., 2006; Blum et al., 2007).
The direct pathway leading to the degradation of cysteine has been well investigated in animals, where cysteine is degraded and the sulfhydryl group oxidised to sulfite and sulfate in mitochondria (Kisker et al., 1997). Although sulfite oxidase is present in Arabidopsis, it is exclusively localized in peroxisomes and appears not to function in cysteine degradation but acts as a salvage enzyme (Hänsch et al., 2007). So far no clear degradation pathway of cysteine has been identified in plants. At least three groups of cysteine desulfurylases can cleave the sulfhydryl group and thus potentially contribute to cysteine homeostasis (Papenbrock et al., 2007; Hell and Wirtz, 2008). Cytosolic OAS-TL isoform CS-like catalyzes the desulfhydration of L-cysteine to sulfide, ammonia and pyruvate and is therefore a L-cysteine desulfhydrase. CS-like null mutants exhibit decreased total cysteine desulfhydrase activity, indicating the presence of additional enzymes with similar activities (Ãlvarez et al., 2010a). Next, D-Cys desulfhydrases (EC 4.4.1.15) with high specificity for this enantionmer have been identified (Riemenschneider et al., 2005; Papenbrock et al., 2007). This reaction would be important in combination with a cysteine amino-acid racemase which still needs to be demonstrated (Gördes et al., 2010), leaving the role of D-cysteine unclear (Papenbrock et al., 2010). However, both desulfhydrase reactions result in the release of sulfide. Free sulfide could potentially be recycled by an OAS-TL or released to the atmosphere as H2S. The third option is the desulfuration reaction of cysteine to alanine and elemental sulfur or sulfide. The catalyzing enzymes belong to the NifS-like proteins and provide sulfur for the biosynthesis of iron-sulfur clusters (Balk and Lobreaux, 2005), molybdenum cofactor (Bittner and Mendel, 2010) and may also be involved in biosynthesis of other sulfur compounds in plastids such as thiamine (Van Hoewyk et al., 2007). Another degradation pathway could begin with the deamination of cysteine by an aminotransferase to yield mercaptopyruvate. The reduced sulfur moiety could then be transferred to nucleophilic acceptors by members of the sulfurtransferase/rhodanese family which has yet to be characterized extensively in plants (Papenbrock et al., 2010). Finally, reduced sulfur could be generated from the degradation of cysteine. This is possibly carried out by a cystine lyase which catalyzes the cleavage of the β-carbon-sulfide link of L-cystine and releases thiocysteine, pyruvate and ammonia. Thiocysteine might be further metabolized to thiocyanate or hydrogen sulfide or channeled into other pathways. In contrast to animals (Cooper, 1983), such degradation pathway still needs to be identified in plants. Cystine lyase has been identified from Brassica oleracea and Arabidopsis (Jones et al., 2003) where it may contribute to cysteine degradation, secondary metabolism or pathogen defense.
Concluding Remarks
The synthesis of cysteine connects the assimilatory pathways of carbon, nitrogen and sulfur. The cysteine synthase complex of SAT and OAS-TL resides in the cytosol, plastids and mitochondria where independent protein biosynthesis takes place. The pathway intermediates are produced in a compartment-specific manner in leaves, producing sulfide in the plastid, the bulk of OAS in the mitochondrion and the majority of cysteine in the cytosol, at least under non-stressed conditions. These intermediates appear to be freely exchangeable between compartments as evidenced by the analysis of insertion mutants. Cysteine synthesis in the three compartments may have additional functions in response to stress and developmental cues. The cysteine synthase complex is a sensor for OAS and sulfide and regulates the activity of SAT in response to the actual cysteine supply. Its dynamic dissociation contributes to the regulation of cysteine homeostasis in the different compartments with respect to synthesis of proteins, methionine, glutathione, iron-sulfur clusters and many other compounds. If the cysteine synthase complex has additional functions, besides regulation of SAT activity, e.g. in regulation of OAS inducible genes needs further exploration. The role of cysteine degradation in higher plants is currently unclear. Research in Arabidopsis is expected to contribute most to answer these questions.
Footnotes
Citation: Rüdiger Hell and Markus Wirtz (2011) Molecular Biology, Biochemistry and Cellular Physiology of Cysteine Metabolism in Arabidopsis thaliana The Arabidopsis Book 9:e0154. doi:10.1199/tab.0154
elocation-id: e0154
First published on December 16, 2011: e0154. doi: 10.1199/tab.0154
REFERENCES
- Ãlvarez C., Calo L., Romero L.C., Garcia I., Gotor C. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010a;152:656–669. doi: 10.1104/pp.109.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ãlvarez C., Lozano-Juste J., Romero L.C., Garcia I., Gotor C., Leon J. Inhibition of Arabidopsis O-Acetylserine(thiol)lyase A1 by Tyrosine Nitration. J. Biol. Chem. 2010b;286:578–586. doi: 10.1074/jbc.M110.147678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A., Armengaud P. Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr. Opin. Plant Biol. 2009;12:275–283. doi: 10.1016/j.pbi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Balk J., Lobreaux S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Benning C., Garavito R.M., Shimojima M. Sulfolipid biosynthesis and function in plants. In: Hell R., Dahl C., Leustek T., editors. Sulfur metabolism in phototrophic organisms. Dordrecht, The Netherlands: Springer; 2008. pp. 189–204. [Google Scholar]
- Berkowitz O., Wirtz M., Wolf A., Kuhlmann J., Hell R. Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J. Biol. Chem. 2002;277:30629–30634. doi: 10.1074/jbc.M111632200. [DOI] [PubMed] [Google Scholar]
- Bermudez M.A., Paez-Ochoa M.A., Gotor C., Romero L.C. Arabidopsis S-Sulfocysteine Synthase Activity Is Essential for Chloroplast Function and Long-Day Light-Dependent Redox Control. Plant Cell. 2010;22:403–416. doi: 10.1105/tpc.109.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J.A., Setterdahl A.T., Knaff D.B., Chen Y., Pitcher L.H., Zilinskas B.A., Leustek T. Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry. 2001;40:9040–9048. doi: 10.1021/bi010518v. [DOI] [PubMed] [Google Scholar]
- Bittner F., Mendel R.R. Cell Biology of Molybdenum. In: Hell R., Mendel R.R., editors. Cell Biology of Metals and Nutrients. Heidelberg: Springer; 2010. pp. 119–144. [Google Scholar]
- Blum R., Beck A., Korte A., Stengel A., Letzel T., Lendzian K., Grill E. Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J. 2007;49:740–749. doi: 10.1111/j.1365-313X.2006.02993.x. [DOI] [PubMed] [Google Scholar]
- Bogdanova N., Hell R. Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 1997;11:251–262. doi: 10.1046/j.1365-313x.1997.11020251.x. [DOI] [PubMed] [Google Scholar]
- Bonner E.R., Cahoon R.E., Knapke S.M., Jez J.M. Molecular basis of cysteine biosynthesis in plants: structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. J. Biol. Chem. 2005;280:38803–38813. doi: 10.1074/jbc.M505313200. [DOI] [PubMed] [Google Scholar]
- Bork C., Schwenn J.D., Hell R. Isolation and characterization of a gene for assimilatory sulfite reductase from Arabidopsis thaliana. Gene. 1998;212:147–153. doi: 10.1016/s0378-1119(98)00155-3. [DOI] [PubMed] [Google Scholar]
- Brunold C. Reduction of sulfate to sulfide. In: Rennenberg H., Brunold C., De Kok LJ., Stulen E., editors. Sulfur nutrition and sulfur assimilation in higher plants. SPB Academic Publisher; 1990. pp. 13–32. [Google Scholar]
- Buchner P., Stuiver C.E.E., Westerman S., Wirtz M., Hell R., Hawkesford M.J., De Kok L.J. Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol. 2004;136:3396–3408. doi: 10.1104/pp.104.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Tai C.H., Ristroph C.M., Cook P.F., Jansonius J.N. Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol. 1999;291:941–953. doi: 10.1006/jmbi.1999.3002. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Rao G.S., Hohenester E., Schnackerz K.D., Cook P.F., Jansonius J.N. Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol. Biol. 1998;283:121–133. doi: 10.1006/jmbi.1998.2037. [DOI] [PubMed] [Google Scholar]
- Bürstenbinder K., Rzewuski G., Wirtz M., Hell R., Sauter M. The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 2007;49:238–249. doi: 10.1111/j.1365-313X.2006.02942.x. [DOI] [PubMed] [Google Scholar]
- Campanini B., Speroni F., Salsi E., Cook P.F., Roderick S.L., Huang B., Bettati S., Mozzarelli A. Interaction of serine acetyltransferase with O-acetylserine sulfhydrylase active site: evidence from fluorescence spectroscopy. Protein Sci. 2005;14:2115–2124. doi: 10.1110/ps.051492805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003;100:10225–10230. doi: 10.1073/pnas.1831512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson D.T., Eugénio D., Sara A. Uptake and assimilation of sulphate by sulphur deficient Zea mays cells: the role of O-acetyl-L-serine in the interaction between nitrogen and sulphur assimilatory pathways. Plant Physiology and Biochemistry. 1999;37:283–290. [Google Scholar]
- Cooper A.J.L. Biochemistry of Sulfur-Containing Amino Acids. Annu. Rev. Biochem. 1983;52:187–222. doi: 10.1146/annurev.bi.52.070183.001155. [DOI] [PubMed] [Google Scholar]
- Davidian J.C., Kopriva S. Regulation of Sulfate Uptake and Assimilation--the Same or Not the Same? Mol. Plant. 2010;3:314–325. doi: 10.1093/mp/ssq001. [DOI] [PubMed] [Google Scholar]
- Destro T., Prasad D., Martignago D., Bernet I.L., Trentin A.R., Renu I.K., Ferretti M., Masi A. Compensatory expression and substrate inducibility of γ-glutamyl transferase GGT2 isoform in Arabidopsis thaliana. J. Exp. Bot. 2010;62:805–814. doi: 10.1093/jxb/erq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D.P., Edwards R. Glutathione Transferases. The Arabidopsis Book. 2010;8:e0131. doi: 10.1199/tab.0131. doi: 10.1199/tab.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis J.R., Gutierrez-Alcala G., Vega J.M., Romero L.C., Gotor C. The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J. Biol. Chem. 2001;276:9297–9302. doi: 10.1074/jbc.M009574200. [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis J.R., Lopez-Martin M.C., Ager F.J., Ynsa M.D., Romero L.C., Gotor C. Increased cysteine availability is essential for cadmium tolerance and accumulation in Arabidopsis thaliana. Plant Biotech. J. 2004;2:469–476. doi: 10.1111/j.1467-7652.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis J.R., He Z., Lima A., Ting J., Buchanan B.B., Luan S. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. USA. 2008;105:16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M. Plant serine acetyltransferase: new insights for regulation of sulphur metabolism in plant cells. Plant Physiol. Biochem. 2003;41:619–627. [Google Scholar]
- Droux M. Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynth. Res. 2004;79:331–348. doi: 10.1023/B:PRES.0000017196.95499.11. [DOI] [PubMed] [Google Scholar]
- Droux M., Martin J., Sajus P., Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch. Biochem. Biophys. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- Droux M., Ruffet M.L., Douce R., Job D. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants-structural and kinetic properties of the free and bound enzymes. Eur. J. Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Feldman-Salit A., Wirtz M., Hell R., Wade R.C. A Mechanistic Model of the Cysteine Synthase Complex. J. Mol. Biol. 2009;386:37–59. doi: 10.1016/j.jmb.2008.08.075. [DOI] [PubMed] [Google Scholar]
- Flavin M., Slaughter C. Synthesis of the succinic ester of homoserine, a new intermediate in the bacterial biosynthesis of methionine. Biochemistry. 1965;4:1370–1375. doi: 10.1021/bi00883a022. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. Redox Regulation in Photosynthetic Organisms: Signaling, Acclimation, and Practical Implications. Antioxid. Redox. Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Francois J.A., Kumaran S., Jez J.M. Structural Basis for Interaction of O-Acetylserine Sulfhydrylase and Serine Acetyltransferase in the Arabidopsis Cysteine Synthase Complex. Plant Cell. 2006;18:3647–3655. doi: 10.1105/tpc.106.047316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Espagne C., Traverso J.A., Mauve C., Valot B., Lelarge-Trouverie C., Zivy M., Noctor G., Meinnel T., Giglione C. Cotranslational proteolysis dominates glutathione homeostasis to support proper growth and development. Plant Cell. 2009;21:3296–3314. doi: 10.1105/tpc.109.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Amir R., Hoefgen R., Hesse H. Improving the levels of essential amino acids and sulfur metabolites in plants. Biol. Chem. 2005;386:817–831. doi: 10.1515/BC.2005.097. [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Nielsen M.T., Nafisi M., Moldrup M.E., Olsen C.E., Motawia M.S., Halkier B.A. Glucosinolate engineering identifies a [γ]-glutamyl peptidase. Nat. Chem. Biol. 2009;5:575–577. doi: 10.1038/nchembio.185. [DOI] [PubMed] [Google Scholar]
- Gördes D., Kolukisaoglu U., Thurow K. Uptake and conversion of D-amino acids in Arabidopsis thaliana. Amino Acids. 2010;40:553–563. doi: 10.1007/s00726-010-0674-4. [DOI] [PubMed] [Google Scholar]
- Gorman J., Shapiro L. Structure of serine acetyltransferase from Haemophilus influenzae Rd. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1600–1605. doi: 10.1107/S0907444904015240. [DOI] [PubMed] [Google Scholar]
- Grzam A., Martin M., Hell R., Meyer A. γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett. 2007;581:3131–3138. doi: 10.1016/j.febslet.2007.05.071. [DOI] [PubMed] [Google Scholar]
- Grzam A., Tennstedt P., Clemens S., Hell R., Meyer A.J. Vacuolar sequestration of glutathione S-conjugates outcompetes a possible degradation of the glutathione moiety by phytochelatin synthase. FEBS Lett. 2006;580:6384–6390. doi: 10.1016/j.febslet.2006.10.050. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J.F., Roberts M.A., Campbell E.I., Wray J.L. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc. Natl. Acad. Sci. USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas F.H., Heeg C., Queiroz R., Bauer A., Wirtz M., Hell R. Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol. 2008;148:1055–1067. doi: 10.1104/pp.108.125237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch R., Lang C., Rennenberg H., Mendel R.R. Significance of plant sulfite oxidase. Plant Biol. 2007;9:589–595. doi: 10.1055/s-2007-965433. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Maruyama A., Schmidt A., Noji M., Ishizawa K., Saito K. β-Cyanoalanine synthase is a mitochondrial cysteine cynthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1172. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford M.J. Transporter gene families in plants: the sulphate transporter gene family redundancy or specialization? Physiol. Plant. 2003;117:155–163. [Google Scholar]
- Hawkesford M.J. Uptake, distribution and subcellular transport of sulfate. In: Hell R., Dahl C., Leustek T., editors. Sulfur metabolism in phototrophic organisms. Dordrecht, The Netherlands: Springer; 2008. pp. 17–32. [Google Scholar]
- Hawkesford M.J., De Kok L.J. Managing sulphur metabolism in plants. Plant, Cell Environ. 2006;29:382–395. doi: 10.1111/j.1365-3040.2005.01470.x. [DOI] [PubMed] [Google Scholar]
- Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell. 2008;20:168–185. doi: 10.1105/tpc.107.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R., Hillebrand H. Plant concepts for mineral acquisition and allocation. Curr. Opin. Biotechnol. 2001;12:161–168. doi: 10.1016/s0958-1669(00)00193-2. [DOI] [PubMed] [Google Scholar]
- Hell R., Wirtz M. Metabolism of cysteine in plants and phototrophic bacteria. In: Hell R., Dahl C., Leustek T., editors. Sulfur metabolism in phototrophic organisms. Dordrecht, The Netherlands: Springer; 2008. pp. 61–94. [Google Scholar]
- Hell R., Jost R., Berkowitz O., Wirtz M. Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids. 2002;22:245–257. doi: 10.1007/s007260200012. [DOI] [PubMed] [Google Scholar]
- Hell R., Dahl C., Knaff D.B., Leustek T. Sulfur Metabolism in Phototrophic Organisms. Doodrecht: Springer; 2008. [Google Scholar]
- Hernández-Sebastià C., Varin L., Marsolais F. Sulfotransferases from plants, algae and phototrophic bacteria. In: Hell R., Dahl C., Leustek T., editors. Sulfur metabolism in phototrophic organisms. Dordrecht, The Netherlands: Springer; 2008. pp. 113–132. [Google Scholar]
- Hindson V.J., Moody P.C., Rowe A.J., Shaw W.V. Serine acetyltransferase from Escherichia coli is a dimer of trimers. J. Biol. Chem. 2000;275:461–466. doi: 10.1074/jbc.275.1.461. [DOI] [PubMed] [Google Scholar]
- Hirai M., Fujiwara T., Awazuhara M., Kimura T., Noji M., Saito K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003;33:651–663. doi: 10.1046/j.1365-313x.2003.01658.x. [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., Yano M., Goodenowe D.B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins L., Parmar S., Blaszczyk A., Hesse H., Hoefgen R., Hawkesford M.J. O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol. 2005;138:433–440. doi: 10.1104/pp.104.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Vetting M.W., Roderick S.L. The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase. J Bacteriol. 2005;187:3201–3205. doi: 10.1128/JB.187.9.3201-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C.S., Shemorry A., Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Noji M., Saito K. Determination of the sites required for the allosteric inhibition of serine acetyltransferase by L-cysteine in plants. Eur. J. Biochem. 1999;266:220–227. doi: 10.1046/j.1432-1327.1999.00847.x. [DOI] [PubMed] [Google Scholar]
- Jacques A.G. The kinetics of penetration: XII. Hydrogen sulfide. J. Gen. Physiol. 1936;19:397–418. doi: 10.1085/jgp.19.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Joshi V. Aspartate-Derived Amino Acid Biosynthesis in Arabidopsis thaliana. The Arabidopsis Book. 2009;7:e0121. doi: 10.1199/tab.0121. doi: 10.1199/tab.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.R., Manabe T., Awazuhara M., Saito K. A new member of plant CS-lyases. A cystine lyase from Arabidopsis thaliana. J. Biol. Chem. 2003;278:10291–10296. doi: 10.1074/jbc.M212207200. [DOI] [PubMed] [Google Scholar]
- Jost R., Berkowitz O., Wirtz M., Hopkins L., Hawkesford M.J., Hell R. Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene. 2000;253:237–247. doi: 10.1016/s0378-1119(00)00261-4. [DOI] [PubMed] [Google Scholar]
- Kawashima C.G., Berkowitz O., Hell R., Noji M., Saito K. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol. 2005;137:220–230. doi: 10.1104/pp.104.045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.S., Hell R. A Future Crop Biotechnology View of Sulfur and Selenium. In: Jez J.M., editor. Sulfur: A Missing Link between Soils, Crops, and Nutrition. Madison: 2008. pp. 293–312. [Google Scholar]
- Khan M.S., Haas F.H., Allboje Samami A., Moghaddas Gholami A., Bauer A., Fellenberg K., Reichelt M., Hansch R., Mendel R.R., Meyer A.J., Wirtz M., Hell R. Sulfite Reductase Defines a Newly Discovered Bottleneck for Assimilatory Sulfate Reduction and Is Essential for Growth and Development in Arabidopsis thaliana. Plant Cell. 2010;22:1216–1231. doi: 10.1105/tpc.110.074088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C., Schindelin H., Rees D.C. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu. Rev. Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- Kopriva S. Regulation of Sulfate Assimilation in Arabidopsis and Beyond. Ann. Bot. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S., Patron N., Keeling P., Leustek T. Phylogenetic analysis of sulfate assimilation and cysteine biosynthesis in phototrophic organisms. In: Hell R., Dahl C., Leustek T., editors. Sulfur metabolism in phototrophic organisms. Dordrecht, The Netherlands: Springer; 2008. pp. 33–60. [Google Scholar]
- Koprivova A., Suter M., den Camp R.O., Brunold C., Kopriva S. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 2000;122:737–746. doi: 10.1104/pp.122.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich N.M. Biosynthesis of cysteine. In: Neidhardt F.C., Curtiss R., Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umberger E., editors. Escherichia coli and Salmonella typhimurium. Cellular and molecular biology. Washington D.C: ASM Press; 1996. pp. 514–527. [Google Scholar]
- Kredich N.M., Becker M.A., Tomkins G.M. Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J. Biol. Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- Krueger S., Donath A., Lopez-Martin M.C., Hoefgen R., Gotor C., Hesse H. Impact of sulfur starvation on cysteine biosynthesis in T-DNA mutants deficient for compartment-specific serine-acetyltransferase. Amino Acids. 2010;39:1029–1042. doi: 10.1007/s00726-010-0580-9. [DOI] [PubMed] [Google Scholar]
- Krüger S., Niehl A., Martin M.C.L., Steinhauser D., Donath A., Hildebrandt T., Romero L.C., Hoefgen R., Gotor C., Hesse H. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopis. Plant, Cell Environ. 2009;32:349–367. doi: 10.1111/j.1365-3040.2008.01928.x. [DOI] [PubMed] [Google Scholar]
- Kumaran S., Jez J.M. Thermodynamics of the interaction between O-acetylserine sulfhydrylase and the C-terminus of serine acetyltransferase. Biochemistry. 2007;46:5586–5594. doi: 10.1021/bi7001168. [DOI] [PubMed] [Google Scholar]
- Kumaran S., Yi H., Krishnan H.B., Jez J.M. Assembly of the cysteine synthase complex and the regulatory role of protein-protein interactions. J. Biol. Chem. 2009;284:10268–10275. doi: 10.1074/jbc.M900154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuske C.R., Hill K.K., Guzman E., Jackson P.J. Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia Mill. Plant Physiol. 1996;112:659–667. doi: 10.1104/pp.112.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Schwarzlander M., Obata T., Sirikantaramas S., Burow M., Olsen C.E., Tohge T., Fricker M.D., Moller B.L., Fernie A.R., Sweetlove L.J., Laxa M. The Metabolic Response of Arabidopsis Roots to Oxidative Stress is Distinct from that of Heterotrophic Cells in Culture and Highlights a Complex Relationship between the Levels of Transcripts, Metabolites, and Flux. Mol. Plant. 2009;2:390–406. doi: 10.1093/mp/ssn080. [DOI] [PubMed] [Google Scholar]
- Leustek T. Sulfate metabolism. The Arabidopsis Book. 2002;1:e0017. doi: 10.1199/tab.0017. doi: 10.1199/tab.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M.N., Bick J.-A., Davies J.P. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- Li L., Kim B.G., Cheong Y.H., Pandey G.K., Luan S. A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Yoo B.-C., Lee J.-Y., Pan W., Harmon A.C. Calcium-regulated phosphorylation of soybean serine acetyltransferase in response to oxidative stress. J. Biol. Chem. 2006;281:27405–27415. doi: 10.1074/jbc.M604548200. [DOI] [PubMed] [Google Scholar]
- Lopez-Martin M.C., Becana M., Romero L.C., Gotor C. Knocking Out Cytosolic Cysteine Synthesis Compromises the Antioxidant Capacity of the Cytosol to Maintain Discrete Concentrations of Hydrogen Peroxide in Arabidopsis. Plant Physiol. 2008;147:562–572. doi: 10.1104/pp.108.117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E., Droux M., Martin J., Douce R. Localization of ATP-sulfurylase and O-acetylserine(thiol)lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.N., Saladores P.H., Lambert E., Hudson A.O., Leustek T. Localization of Members of the {γ}-Glutamyl Transpeptidase Family Identifies Sites of Glutathione and Glutathione S-Conjugate Hydrolysis. Plant Physiol. 2007;144:1715–1732. doi: 10.1104/pp.106.094409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Rotte C., Hoffmeister M., Theissen U., Gelius-Dietrich G., Ahr S., Henze K. Early cell evolution, eukaryotes, anoxia, sulfide, oxygen, fungi first (?), and a tree of genomes revisited. IUBMB Life. 2003;55:193–204. doi: 10.1080/1521654031000141231. [DOI] [PubMed] [Google Scholar]
- Marty L., Siala W., Schwarzlander M., Fricker M.D., Wirtz M., Sweetlove L.J., Meyer Y., Meyer A.J., Reichheld J.-P., Hell R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:9109–9114. doi: 10.1073/pnas.0900206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G.A. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- Mathai J.C., Missner A., Kugler P., Saparov S.M., Zeidel M.L., Lee J.K., Pohl P. No facilitator required for membrane transport of hydrogen sulfide. Proc. Natl. Acad. Sci. USA. 2009;106:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan S.C., Pasternak M., Cairns N., Kiddle G., Brach T., Jarvis R., Haas F., Nieuwland J., Lim B., Muller C., Salcedo-Sora E., Kruse C., Orsel M., Hell R., Miller A.J., Bray P., Foyer C.H., Murray J.A., Meyer A.J., Cobbett C.S. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. USA. 2010;107:2331–2336. doi: 10.1073/pnas.0913689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.J. The integration of glutathione homeostasis and redox signaling. J. Plant Physiol. 2008;165:1390–1403. doi: 10.1016/j.jplph.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth. Res. 2005;86:435–457. doi: 10.1007/s11120-005-8425-1. [DOI] [PubMed] [Google Scholar]
- Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. Purification and characterization of serine acetyltransferase from Escherichia coli partially truncated at the C-terminal region. Biosci. Biotechnol. Biochem. 1999;63:168–179. doi: 10.1271/bbb.63.168. [DOI] [PubMed] [Google Scholar]
- Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci. Biotechnol. Biochem. 2000;64:1628–1640. doi: 10.1271/bbb.64.1628. [DOI] [PubMed] [Google Scholar]
- Miseta A., Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000;17:1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- Mugford S.G., Yoshimoto N., Reichelt M., Wirtz M., Hill L., Mugford S.T., Nakazato Y., Noji M., Takahashi H., Kramell R., Gigolashvili T., Flugge U.-I., Wasternack C., Gershenzon J., Hell R., Saito K., Kopriva S. Disruption of Adenosine-5′-Phosphosulfate Kinase in Arabidopsis Reduces Levels of Sulfated Secondary Metabolites. Plant Cell. 2009;21:910–927. doi: 10.1105/tpc.109.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C.H. Glutathione. The Arabidopsis Book. 2011;9:e0142. doi: 10.1199/tab.0142. doi: 10.1043/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji M., Inoue K., Kimura N., Gouda A., Saito K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Zhao P., Xiang C., Oliver D. Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 2007a;49:878–888. doi: 10.1111/j.1365-313X.2006.03005.x. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Oikawa A., Zhao P., Xiang C., Saito K., Oliver D.J. A γ-Glutamyl Transpeptidase-Independent Pathway of Glutathione Catabolism to Glutamate via 5-Oxoproline in Arabidopsis. Plant Physiol. 2008;148:1603–1613. doi: 10.1104/pp.108.125716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Radwan S., Peterson A., Zhao P., Badr A., Xiang C., Oliver D. Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 2007b;49:865–877. doi: 10.1111/j.1365-313X.2006.03004.x. [DOI] [PubMed] [Google Scholar]
- Olsen L., Huang B., Vetting M., Roderick S. Structure of serine acetyltransferase in complexes with CoA and its cysteine feedback inhibitor. Biochemistry. 2004;43:6013–6019. doi: 10.1021/bi0358521. [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Guretzki S., Henne M. Latest news about the sulfurtransferase protein family of higher plants. Amino Acids. 2010;41:43–57. doi: 10.1007/s00726-010-0478-6. [DOI] [PubMed] [Google Scholar]
- Papenbrock J., Riemenschneider A., Kamp A., Schulz-Vogt H.N., Schmidt A. Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants - from the field to the test tube and back. Plant Biol. 2007;9:582–588. doi: 10.1055/s-2007-965424. [DOI] [PubMed] [Google Scholar]
- Pasternak M., Lim B., Wirtz M., Hell R., Cobbett C.S., Meyer A.J. Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 2008;53:999–1012. doi: 10.1111/j.1365-313X.2007.03389.x. [DOI] [PubMed] [Google Scholar]
- Patron N.J., Durnford D.G., Kopriva S. Sulfate assimilation in eukaryotes: fusions, relocations and lateral transfers. BMC Evol. Biol. 2008;8:39. doi: 10.1186/1471-2148-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I., Felder C.E., Man O., Silman I., Sussman J.L., Beckmann J.S. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- Pye V.E., Tingey A.P., Robson R.L., Moody P.C. The structure and mechanism of serine acetyltransferase from Escherichia coli. J. Biol. Chem. 2004;279:40729–40736. doi: 10.1074/jbc.M403751200. [DOI] [PubMed] [Google Scholar]
- Queval G., Thominet D., Vanacker H., Miginiac-Maslow M., Gakiere B., Noctor G. H2O2-Activated Up-Regulation of Glutathione in Arabidopsis Involves Induction of Genes Encoding Enzymes Involved in Cysteine Synthesis in the Chloroplast. Mol. Plant. 2009;2:344–356. doi: 10.1093/mp/ssp002. [DOI] [PubMed] [Google Scholar]
- Rabeh W.M., Cook P.F. Structure and mechanism of O-acetylserine sulfhydrylase. J. Biol. Chem. 2004;279:26803–26806. doi: 10.1074/jbc.R400001200. [DOI] [PubMed] [Google Scholar]
- Raetz C.R., Roderick S.L. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- Ranocha P., McNeil S.D., Ziemak M.J., Li C., Tarczynski M.C., Hanson A.D. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575–584. doi: 10.1046/j.1365-313x.2001.00988.x. [DOI] [PubMed] [Google Scholar]
- Ravanel S., Gakiere B., Job D., Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeds P.J. Dispensable and Indispensable Amino Acids for Humans. J. Nutr. 2000;130:1835S–1840S. doi: 10.1093/jn/130.7.1835S. [DOI] [PubMed] [Google Scholar]
- Riemenschneider A., Wegele R., Schmidt A., Papenbrock J. Isolation and characterization of a D-cysteine desulfhydrase protein from Arabidopsis thaliana. Febs. J. 2005;272:1291–1304. doi: 10.1111/j.1742-4658.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- Rouached H., Wirtz M., Alary R., Hell R., Arpat A.B., Davidian J.-C., Fourcroy P., Berthomieu P. Differential Regulation of the Expression of Two High-Affinity Sulfate Transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 2008;147:897–911. doi: 10.1104/pp.108.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan P.G. Stromal concentrations of coenzyme A and its esters are insufficient to account for rates of chloroplast fatty acid synthesis: evidence for substrate channelling within the chloroplast fatty acid synthase. Biochem. J. 1997;327:267–273. doi: 10.1042/bj3270267. (Pt 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N., Lemaire S.D., Jacquot J.-P. The Role of Glutathione in Photosynthetic Organisms: Emerging Functions for Glutaredoxins and Glutathionylation. Annu. Rev. Plant Biol. 2008;59:143. doi: 10.1146/annurev.arplant.59.032607.092811. [DOI] [PubMed] [Google Scholar]
- Ruffet M.L., Lebrun M., Droux M., Douce R. Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur. J. Biochem. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez R., Davies T.G., Coleman J.O., Rea P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Sanda S., Leustek T., Theisen M.J., Garavito R.M., Benning C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 2001;276:3941–3946. doi: 10.1074/jbc.M008200200. [DOI] [PubMed] [Google Scholar]
- Schachtman D.P., Shin R. Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 2007;58:47–69. doi: 10.1146/annurev.arplant.58.032806.103750. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Jäger K. Open questions about sulfur metabolism in plants. Annu. Rev. Plant Physiol Plant. Mol. Biol. 1992;43:325–349. [Google Scholar]
- Schwarz G., Mendel R.R. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- Setya A., Murillo M., Leustek T. Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylsulfate reductase. Proc. Natl. Acad. Sci. USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Grossman A.R. Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J. Biol. Chem. 2010;285:25094–25102. doi: 10.1074/jbc.M110.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Rose A., McDermott J.P., Fujiwara T., Hayashi H., Yoneyama T., Davies J.P. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002;29:475–486. doi: 10.1046/j.0960-7412.2001.01232.x. [DOI] [PubMed] [Google Scholar]
- Smith F.W., Hawkesford M.J., Ealing P.M., Clarkson D.T., Vanden Berg P.J., Belcher A.R., Warrilow A.G. Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J. 1997;12:875–884. doi: 10.1046/j.1365-313x.1997.12040875.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000;23:171–182. doi: 10.1046/j.1365-313x.2000.00768.x. [DOI] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G., Martin M., Chalam R., Tarczynski M.C., Schmidt A., Leustek T. Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J. 2002;32:879–889. doi: 10.1046/j.1365-313x.2002.01477.x. [DOI] [PubMed] [Google Scholar]
- Tumaney A.W., Ohlrogge J.B., Pollard M. Acetyl coenzyme A concentrations in plant tissues. J. Plant Physiol. 2004;161:485–488. doi: 10.1078/0176-1617-01258. [DOI] [PubMed] [Google Scholar]
- Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol. Lett. 1992;76:249–254. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D., Pilon M., Pilon-Smits E.A. The functions of NifS-like proteins in plant sulfur and selenium metabolism. Plant Science. 2007;174:117–123. [Google Scholar]
- Vauclare P., Kopriva S., Fell D., Suter M., Sticher L., von Ballmoos P., Krahenbuhl U., den Camp R.O., Brunold C. Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 2002;31:729–740. doi: 10.1046/j.1365-313x.2002.01391.x. [DOI] [PubMed] [Google Scholar]
- Vidmar J.J., Tagmount A., Cathala N., Touraine B., Davidian J.E. Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett. 2000;475:65–69. doi: 10.1016/s0014-5793(00)01615-x. [DOI] [PubMed] [Google Scholar]
- Vuorio R., Hirvas L., Vaara M. The Ssc protein of enteric bacteria has significant homology to the acyltransferase Lpxa of lipid A biosynthesis, and to three acetyltransferases. FEBS Lett. 1991;292:90–94. doi: 10.1016/0014-5793(91)80841-p. [DOI] [PubMed] [Google Scholar]
- Wachter A., Wolf S., Steininger H., Bogs J., Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 2005;41:15–30. doi: 10.1111/j.1365-313X.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Wachtershauser G. From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya. Philos Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1787–1806. doi: 10.1098/rstb.2006.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow A., Hawkesford M. Separation, subcellular location and influence of sulphur nutrition on isoforms of cysteine synthase in spinach. J. Exp. Bot. 1998;49:1625–1636. [Google Scholar]
- Warrilow A.G., Hawkesford M.J. Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J. Exp. Bot. 2000;51:985–993. doi: 10.1093/jexbot/51.347.985. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kusano M., Oikawa A., Fukushima A., Noji M., Saito K. Physiological Roles of the β-Substituted Alanine Synthase Gene Family in Arabidopsis. Plant Physiol. 2008a;146:310–320. doi: 10.1104/pp.107.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Mochida K., Kato T., Tabata S., Yoshimoto N., Noji M., Saito K. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell. 2008b;20:2484–2496. doi: 10.1105/tpc.108.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M., Droux M. Synthesis of the sulfur amino acids: cysteine and methionine. Photosynth. Res. 2005;86:345–362. doi: 10.1007/s11120-005-8810-9. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J. Plant Physiol. 2006;163:273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Hell R. Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell. 2007;19:625–639. doi: 10.1105/tpc.106.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M., Droux M., Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J. Exp. Bot. 2004;55:1785–1798. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Berkowitz O., Droux M., Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur. J. Biochem. 2001;268:686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Lindermayr C., Durner J., Hell R. Post-translational modification of cytosolic OAS-TL from Arabidopsis thaliana. In: Saito K., Kok L.J. De, Stulen I., Hawkesford M.J., Schnug E., Sirko A., Rennenberg H., editors. Sulfur Transport and Assimilation in Plants in the Post Genomic Era. Leiden: Backhuys Publishers; 2005. pp. 91–94. [Google Scholar]
- Wirtz M., Heeg C., Samami A.A., Ruppert T., Hell R. Enzymes of cysteine synthesis show extensive and conserved modifications patterns that include Nα-terminal acetylation. Amino Acids. 2010a;39:1077–1086. doi: 10.1007/s00726-010-0694-0. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Birke H., Heeg C., Mueller C., Hosp F., Throm C., Koenig S., Feldman-Salit A., Rippe K., Petersen G., Wade R.C., Rybin V., Scheffzek K., Hell R. Structure and function of the hetero-oligomeric cysteine synthase complex in plants. J. Biol. Chem. 2010b;285:32810–32817. doi: 10.1074/jbc.M110.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Nakamura T., Kusano T., Sano H. Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and β-cyanoalanine synthase. Plant Cell Physiol. 2000;41:465–476. doi: 10.1093/pcp/41.4.465. [DOI] [PubMed] [Google Scholar]
- Yi H., Galant A., Ravilious G.E., Preuss M.L., Jez J.M. Sensing Sulfur Conditions: Simple to Complex Protein Regulatory Mechanisms in Plant Thiol Metabolism. Mol. Plant. 2010;3:269–279. doi: 10.1093/mp/ssp112. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Onda Y., Ashikari T., Tanaka Y., Kusumi T., Hase T. Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 2000;122:887–894. doi: 10.1104/pp.122.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]