Abstract
How does matrix metalloproteinase-12 (MMP-12 or metalloelastase) degrade elastin with high specific activity? NMR suggested soluble elastin to cover surfaces of MMP-12 far from its active site. Two of these surfaces have been found, by mutagenesis guided by the BINDSIght approach, to affect degradation and affinity for elastin substrates but not a small peptide substrate. Main exosite 1 has been extended out to Asp124 that binds calcium. Novel exosite 2 comprises residues from the II–III loop and β-strand I near the back of the catalytic domain. The high exposure of these distal exosites may make them accessible to elastin made more flexible by partial hydrolysis. Importantly, combination of a lesion at each of exosites 1 and 2 and active site decreased catalytic competence towards soluble elastin by 13- to 18-fold to the level of MMP-3, homologue and poor elastase. Double mutant cycle analysis of conservative mutations of Met156 (exosite 2) and either Asp124 (exosite 1) or Ile180 (active site) had additive effects. Compared to polar substitutions observed in other MMPs, Met156 enhanced affinity and Ile180 kcat for soluble elastin. Both residues detracted from the higher folding stability with polar mutations. This resembles the trend in enzymes of an inverse relationship between folding stability and activity. Restoring Asp124 from combination mutants enhanced kcat for soluble elastin. In elastin degradation, exosites 1 and 2 contributed independently of each other and Ile180 at the active site, but with partial coupling to Ala182 near the active site. The concept of weak, separated interactions coalescing somewhat independently can be extended to this proteolytic digestion of a protein from fibrils.
Keywords: elastin degradation, protein-protein interactions, steady-state enzyme kinetics, double mutant cycle analysis
Exosites are pivotal in thrombin recognition of therapeutic inhibitors (1) and peptide substrates from receptors by its catalytic domain (2). Exosites of MMPs‡ have normally been regarded as residing instead in the non-catalytic C-terminal hemopexin-like domain and fibronectin II-like modules inserted in MMP-2 and 9 (3, 4). The additional domains support binding and specificity for substrates from protein fibrils. The hemopexin domain enhances binding and proteolysis of collagen triple helices by several MMPs (5–11). The fibronectin II-like modules are required for MMP-2 and 9 to bind and digest elastin (12), gelatin (8, 13), and collagens (10, 14). In contrast, MMP-12 requires only its catalytic domain to degrade elastin. (Mature MMP-12 in vivo retains only its catalytic domain due to autolytic removal of its hemopexin domain (15).) What is it about the catalytic domain of MMP-12 making it so active upon elastin? NMR suggested surfaces of MMP-12 remote from its active site can be covered by soluble α-elastin, such as a strip across the β-sheet and a bowl from helix A to helix C (16). We investigated whether any of these may function as exosites to support elastin degradation.
Elastin constitutes half the protein mass of the aorta (17) and provides the recoil that dampens the high pressure waves from heart contraction (18). Heavy desmosine cross-linking (19) could account for why elastin can last a lifetime. Elastin degradation is characteristic of atherosclerosis (20), aortic aneurysms, and emphysema (COPD). MMP-12, also known as the metalloelastase secreted by macrophages, has been implicated in each disease (21–24). Proteolytic release of elastin fragments strongly signals for repair and chronic inflammation by potently attracting leukocytes as in emphysema (25), stimulating proliferation of lymphocytes, fibroblasts and smooth muscle cells, and promoting angiogenesis (24). These are characteristic of age-related arterial aneurysm formation (24), plus proteolytic clearance of elastin from the aneurysms (22).
Compared to its closest homolog of MMP-3 catalytic domain (MMP-3cd), MMP-12 has a 12- to 13-fold advantage in elastin degradation in terms of catalytic efficiency, turnover number (kcat), and extent of peptide release (16, 26). Subtleties accounting for this difference have been a topic of investigation. MMP-3cd has nearly 3 kcal/mol greater folding stability (27), in accord with the hypothesis that enzyme variants typically trade stability for activity (28–30). Millisec timescale backbone dynamics enveloped the active site of MMP-12 and much more so in MMP-3cd (27).
Among several elastases compared, MMP-12 exhibited the highest affinity for insoluble elastin fibrils (22). Moreover, most of MMP-12 remained bound, even with competition by the TIMP-1 inhibitor of MMPs. MMP-12 was the main MMP observed bound to elastin undergoing digestion in human aortic aneurysms (22). The notably high affinity for elastin was hypothesized to result from multiple interfaces between MMP-12 and α-elastin that were suggested by NMR (16).
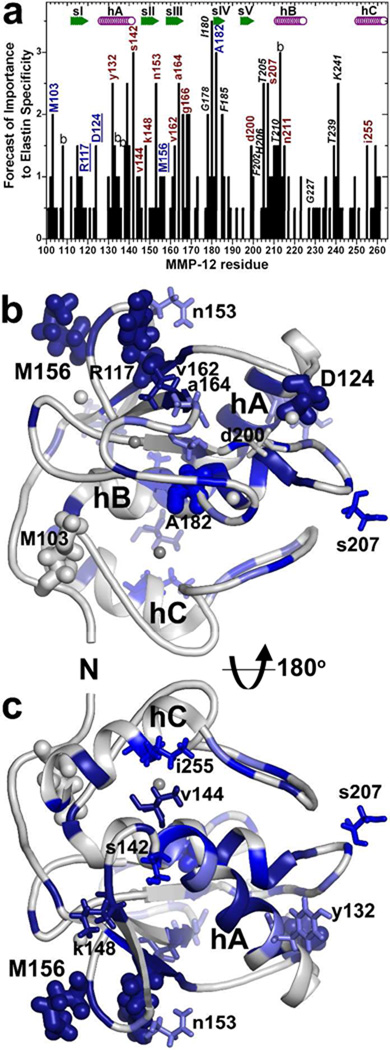
Within these putative interfaces with α -elastin, the BINDSIght strategy (referring to bioinformatics and NMR discovery of specificity of interactions) predicted at least 28 sequence positions to be potentially distinctive (16) (Fig. 1). Ten of these residues from five loops on the perimeter of the active site cleft were investigated and implicated in enhanced specificity for elastin substrates (16). The remaining sequence positions distant from the active site are considered herein. These candidates are located at N-terminus through β-strand III, the beginning of the S-shaped III–IV loop, and helix C (Fig. 1). Elastin is too amorphous and heterogeneous to crystallize and too repetitious in sequence and too prone to self-association for NMR in solution (31). Given the barriers to structure determination of complexes with elastin, mutagenesis and catalytic characterization have been attractive for evaluating positions in MMP-12 affecting elastin degradation.
Fig. 1.
MMP-12 residues distal from the active site chosen for mutagenesis based on putative contacts with 20 kD α-elastin species (Eln-20) and BINDSIght forecast of importance (16): (a) Residues were selected for mutagenesis based on this BINDSIght plot suggesting interfaces that could be distinctive. Those with blue and red-brown labels were characterized in this study of remote surfaces and those in black italics in the prior study of the active site periphery (16). The score in the plot accumulated with NMR evidences of surfaces that are covered or perturbed in chemical shift by the Eln-20 binding partner and distinctions in sequence (16). Uppercase labels mark residues found to support higher catalytic efficiency toward fEln-100 than a conservative mutation, and lowercase for those that do not. Those underlined are consequential positions distant from the active site. “b” denotes a buried residue. In the front view (b) and back view (c) of the catalytic domain, dark blue, medium blue, and light blue represent, respectively, strong, medium, and mild protection of the surface by Eln-20 from an exogenous Gd·EDTA probe. Side chains are plotted for residues mutated and characterized in this study. Side chains that affected digestion of 100 kD α-elastin (fEln-100) are drawn with thick sticks, and those that did not with thin sticks. Zinc ions are darker gray and calcium lighter gray.
Two tools were used in testing if remote residues participate in elastolysis. One tool is the fluor-labeled 100 kDa fraction of α-elastin designated fEln-100. This soluble substrate introduced the ability to compare quantitatively the steady-state kinetics of proteolysis of an elastin substrate (16, 32). fEln-100 retains the heavy desmosine cross-linking of intact elastin fibrils (32–34) and the capacity to self-assemble at physiological temperature (35). Considering the progression of elastin degradation to clearance (22) via three dozen or more sites of hydrolysis by MMP-12 (36), proteolysis of fEln-100 was suggested to represent later stages of elastolysis when elastin has already been nicked and the fibrils partly disrupted (16).
The second tool is double mutant cycle analysis of mutational effects on catalysis. It clarifies how additive the effects of mutations may be. Double mutant cycle analysis applied to enzyme transformation of small molecules showed that well-separated lesions independently impaired the transition-state stabilization, i.e. ΔΔGT‡ is additive (37). Closely neighboring lesions, on the other hand, have lacked independence and were not energetically additive (37). When applied to protein-protein interfaces, the additive character of impairments to affinity increased with distance of separation of the lesions (38). Fully additive behavior was observed for lesions separated far enough apart to reside within independent interfacial clusters (39) or to be free of electrostatic coupling (38). Mildvan and coworkers described different scenarios involving double mutations and insights regarding enzyme mechanism. The energetics of the mutational interactions, defined relative to the wild type as reference, can be additive, partially additive, synergistic, absent, or antagonistic (40). Switching to the double mutant as the reference point for comparing mutational effects, called “inverse thinking”, simplifies description of non-additive effects (41).
Screening of BINDSIght-proposed sequence positions by mutagenesis has enlarged the remote frontier of the main exosite (now exosite 1) and revealed distant exosite 2 that support elastolysis by MMP-12. Strategic combination of a mutation in the active site with remote and novel positions in exosites 1 and 2 is enough to diminish selectively the elastolytic activity of MMP-12 by at least its 12-fold margin of superiority to the homologous MMP-3cd negative control. This reached a milestone in engineering elastin specificity out of metalloelastase and strongly suggests specific combinations of well-separated sites of weak interaction with α-elastin that boost the catalytic efficiency of this proteolysis.
EXPERIMENTAL PROCEDURES
Preparation of MMP-12 variants
The mutations in human MMP-12 catalytic domain (aa100–263) were generated by QuikChange mutagenesis using PCR master mix (Stratagene). Mutated enzymes were purified as described (42) but with an improved folding procedure (43). Mutant enzymes were stored in small aliquots at −80 ºC in 20 mM Tris-HCl (pH 7.5), 10 mM CaCl2, 0.1 mM ZnCl2, 100 mM NaCl, 50% w/v glycerol.
Enzymatic assays
The concentration of intact enzyme active sites was determined by inhibitor titration of initial velocities (44) with galardin (GM6001, EMD Biosciences) and soluble Knight’s peptide substrate FS-6 (EMD) (45). Kinetics of proteolytic digestion of FS-6 and fEln-100 (32) were monitored as progress curves at 25 ºC with a BioTek Synergy MX plate reader. Fluorescence excitation and emission of FS-6 were at 328 and 393 nm, respectively. Excitation and emission of fEln-100 were at 490 and 512 nm, respectively. All kinetic experiments were performed in TNC buffer of 20 mM Tris-HCl (pH 7.5), 10 mM CaCl2, 0.1 mM ZnCl2, 100 mM NaCl, 0.035% (w/v) Brij-35.
Fitting of steady-state enzyme kinetics
Individual kcat/Km, Km, and kcat were obtained by nonlinear regression and global fitting of two progress curves collected at appropriate concentrations as described in detail previously (46). The modification of the procedure for fEln-100 of fitting the first kinetic phase has been illustrated (32).
Peptide release from insoluble elastin fibrils
Mutational variants of human MMP-12 were compared by measuring the release of fluorescent peptides from insoluble elastin-fluorescein particles (75-37 µm, Elastin Products Co.). 80 nM enzyme variants were incubated with 2 mg/ml elastin-fluorescein in TNC buffer with gentle shaking at 37 ºC for 15 hrs. The supernatants containing the fluorescent peptides released were diluted 10-fold for fluorescence measurements using the BioTek Synergy MX plate reader with excitation at 490 nm and emission at 520 nm. Measurements of fluorescence from proteolytic release of peptides were first normalized by the fluorescence of substrate-only samples and then expressed as ratios against wild type MMP-12 controls. Values are given as mean ± S.E.M.
Double Mutant Cycle Analysis
This was performed in the inverse mode in order to make non-additive behavior simpler and more intuitive. Inverse thinking uses the double (or triple) mutation as the energetic reference point in order to highlight the gains from restoring each mutated position to the wild type sequence (41). Consequently, changes in transition state stabilization (47) were calculated from the catalytic efficiencies expressed as:
| (eq. 1) |
Likewise, changes in free energy of substrate binding (40) were approximated as:
| (eq. 2) |
Changes in activation free energy(48) were calculated as:
| (eq. 3) |
In inverse thinking, partially additive mutational damage is seen as cooperative since the gain from restoring both mutated residues to wild type exceeds the energetic sum of restoring both residues individually. Synergistic mutational damage is regarded in inverse thinking as anti-cooperative because the gain in restoring both residues to the native sequence is less than the energetic sum of restoring them one-at-a-time. Antagonism is non-additive behavior that is more obvious in the inverse mode because restoring one residue has the opposite effect of restoring the other residue (41).
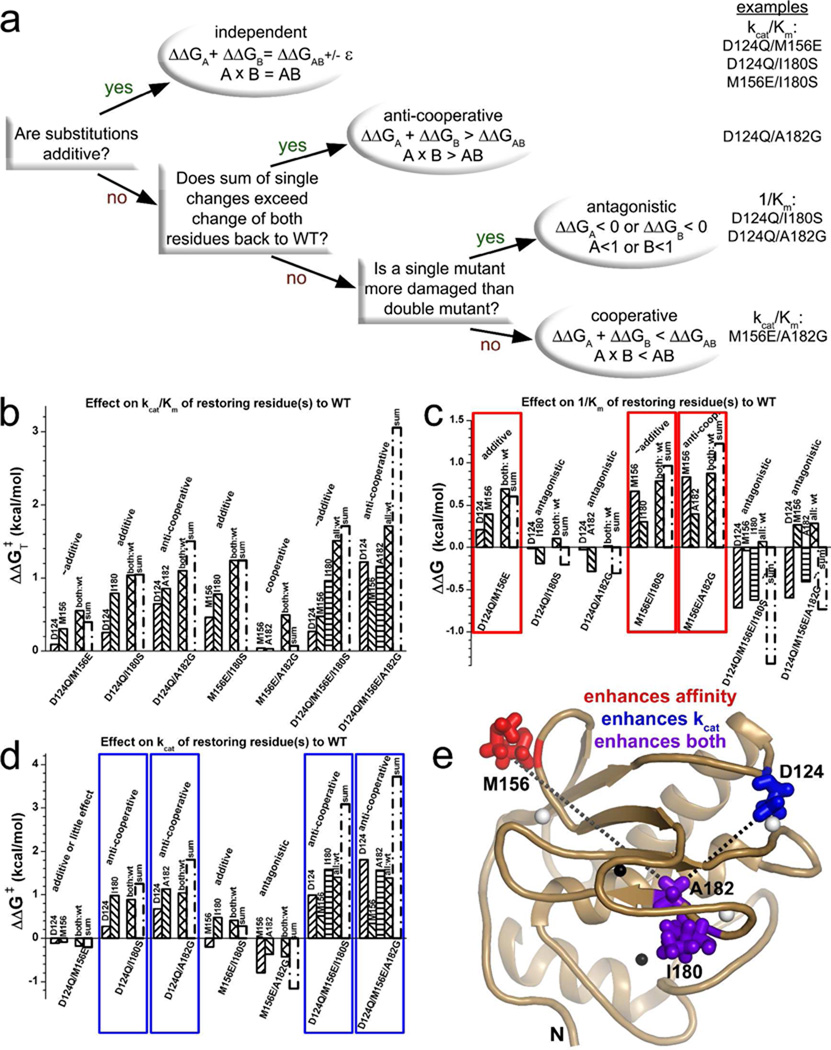
The categories of behavior of combining mutations were identified by the decision tree shown in Fig. 4a. For effects to be considered additive (independent), the sum of free energies of restoring each residue individually to wild type needed to agree with the total difference between combined mutation and wild type to within the tolerance ε of 0.2 kcal/mol, as well as within 30%. A sum of single residue changes back to wild type exceeding restoration of both or all positions back to wild type by > ε is considered anti-cooperative (41) (Fig. 4a). The sum of restoring single positions to wild type being less than the difference between combined mutation and wild type (by ε at least) is either cooperative of antagonistic. In antagonism, the variant with a combination of mutations outperforms a simpler mutant having a position restored to wild type (Fig. 4a).
Fig. 4.
Double mutant cycle analysis, in the inverse mode (41), of effects on catalytic turnover of fEln-100 of restoring mutated positions to the wild type human MMP-12 sequence. Panel (a) shows the decision tree for categorizing the behaviors. Examples of each behavior are listed at right. “A” or “B” refers to the ratio given as the argument of the ln function of eq. 1, 2, or 3 for a single mutant relative to the double mutant reference. “AB” refers to the ratio of the wild type relative to the double mutant. Free energy comparisons are shown for kcat/Km (b), 1/Km (c), and kcat (d) for fEln-100 and using eq. 1, 2, or 3, respectively. The wild type residue restored is labeled above each column. The double or triple mutant reference point is listed below the columns. For example, the first column in (b) reports the difference between M156E and D124Q/M156E variants, which restored D124 to the single mutant. The sum of each free energy from each single residue restored to wild type is marked. The measurement of fully wild type is marked in the comparisons with cross-hatched columns labeled either as “both:wt” (relative to double mutants) or “all:wt” (relative to triple mutants). Panel (e) maps major trends onto the NMR structure (50). Red and blue highlight combinations whose contributions to catalytic efficiency are mainly through affinity (1/Km) or catalytic turnover (kcat), respectively. Purple in (e) marks side chains that support kcat (d) and also enhance affinity for fEln-100 relative to double mutants with M156E (c). Dotted lines in (e) denote non-additive coupling with the Ala182 position that is anti-cooperative with Asp124 (black) or cooperative with Met156 (gray) with respect to catalytic efficiency. Omission of dotted lines between Asp124, Met156, and Ile180 indicates their independent (additive) effects on kcat/Km.
Affinity for soluble elastin
Titrations with α-elastin (Elastin Products Co.) were monitored with intrinsic tryptophan fluorescence emission spectra of MMP-12 variants excited at 280 nm. Elastin is devoid of tryptophan. Each point in titrations was measured with triplicate samples at 25 ºC using a BioTek Synergy MX plate reader within 30 min of mixing prior to substantial proteolysis. Fluorescence quenching at 323 nm was normalized by the maximum quenching at the plateau at 2 to 3 mg/ml α-elastin.
Stability measurements
The folding stability of selected MMP-12 variants was measured by chemical denaturation using urea at 37 ºC and detection of intrinsic tryptophan fluorescence emission. 1.3 µM working stock solutions of each variant of MMP-12 in TNC buffer and TNC buffer with 10 M urea were mixed and incubated for 10 min. to obtain equilibrated solutions with [urea] between these extremes. Each mixture was repeated in triplicate. Excitation was at 290 nm and emission at 316 nm in the BioTek Synergy MX plate reader. The data were interpreted and the linear transition region extrapolated to zero denaturant by the method of Pace (49). The folding stability at zero denaturant ΔG(H2O) is:
| (eq. 4) |
where the slope m is the dependence on denaturant concentration (49).
Normal modes analysis
Point mutations were introduced to the coordinates of the lowest energy NMR structure of human MMP-12(E219A) (50) using PyMol (51). An anisotropic network model was used to simulate modes of motion using a server online (52). The modes were compared between control and mutants by squared fluctuations, deformation energy, and trajectories of the motion.
RESULTS
To investigate further why MMP-12 is highly active in digesting proteolysis-resistant elastin substrates, the remote regions of its catalytic domain that were covered by soluble α-elastin in an NMR assay (16) are evaluated herein.
Selection of mutations
Recent BINDSIght analysis integrated NMR evidences of MMP-12 surfaces that bind the 20 kDa fraction of α-elastin (Eln-20) with sequence positions distinguishing sub-families of MMPs (16). The scores generated from those data suggest possible importance of sequence positions to elastin interactions (Fig. 1a). The study herein evaluates influences on elastolysis outside the periphery of the active site by surveying broadly the remote sites that have significant BINDSIght scores for putative interactions with Eln-20. The 18 positions not labeled with italics in Fig. 1a were mutagenized and their kinetics of digestion of fEln-100 characterized. These include a swath or residues extending from Asn153 and Met156 of the remote II–III loop and Arg117 (top left of Fig. 1b) through Val162, Ala164 and Gly166 at β-strand III over to calcium-binding Asp124, as well as to Asp-200 and Ser-207 of the V-B loop (right). Also pictured in Fig. 1b are Met103 near the N-terminus and Ala182 at the active site in the center. Flipping the nearly standard view of the enzyme to a back side view (Fig. 1c) brings to the foreground residues mutated such as Ile-255 of helix C (top), Val144 and Ser142 at the C-terminal end of helix A, Tyr132 at the opposite end of helix A, and Lys148 of β-strand II. These residues were each replaced by the corresponding side chain of a homologous MMP not known to be an elastase. Each of the single point mutants retained from 90 to 119% of wild type MMP-12 activity towards soluble Knight’s peptide substrate FS-6 (Fig. 2a, Tables 1, S1) indicating that each remained intact as an MMP.
Fig. 2.
Comparison of activities of single, double, and triple mutants. a) Catalytic efficiencies are plotted for degradation of the soluble α-elastin substrate fEln-100 and the control peptide substrate FS-6. The kcat/Km values are normalized by kcat/Km of wild type MMP-12 (Table 1). b) Release of peptides from overnight digestion of fluorescein-tagged, insoluble elastin fibrils is normalized by that of wild type MMP-12.
Table 1.
Catalytic properties (at 25 ºC, pH 7.5) of MMP-12 variants found to have consequential mutations.
| MMP-12 varianta | Location of mutation |
General Substrate FS-6 |
fEln-100 |
Relative activity of fEln-100 FS-6 normalized to |
||||
|---|---|---|---|---|---|---|---|---|
| kcat/Km,/M/s | kcat, s−1 |
Km, µM |
kcat/Km /M/s |
kcat,10 2s−1 |
Km µM |
wtMMP12c: selectivity for fEln- 100 |
||
| wt MMP-12 b | 133,800 ± 6000 | 17.3 ± 0.8 | 129 ± 4 | 10690 ± 430 | 1.06 ± 0.04 | 1.06 ± 0.04 | 100.0% | |
| wt MMP-3 b | 20,660 ± 620 | 3.2 ± 0.1 | 154 ± 5 | 830 ± 40 | 0.08 ± 0.01 | 0.94 ± 0.04 | 50.3% | |
| M103F | N-terminus | 146,300 ± 4400 | 18.7 ± 0.6 | 128 ± 4 | 8040 ± 320 | 0.92 ± 0.04 | 1.15 ± 0.04 | 68.8% |
| R117S | Stand I, next to II–III loop | 120,800 ± 3600 | 13.7 ± 0.4 | 113 ± 3 | 7130 ± 290 | 0.72 ± 0.03 | 1.15 ± 0.04 | 73.8% |
| D124Q | I-A loop, Ca2+ ligand | 136,700 ± 4100 | 17.1 ± 0.5 | 125 ± 4 | 7060 ± 280 | 1.24 ± 0.05 | 1.75 ± 0.07 | 64.6% |
| M156E | II–III loop | 135,200 ± 4100 | 16.9 ± 0.5 | 125 ± 4 | 4890 ± 200 | 1.17 ± 0.05 | 2.39 ± 0.10 | 45.3% |
| I180Sb | Bulge-edge in III–IV loop | 141,800 ± 4300 | 24.1 ± 2.2 | 170 ± 15 | 2860 ± 140 | 0.37 ± 0.06 | 1.3 ± 0.1 | 25.2% |
| A182G | Strand IV | 120,100 ± 3600 | 15.6 ± 0.5 | 130 ± 4 | 4970 ± 200 | 0.57 ± 0.02 | 1.14 ± 0.05 | 51.8% |
| D124Q/M156E | 134,700 ± 4000 | 16.7 ± 0.5 | 124 ± 4 | 4210 ± 200 | 1.43 ± 0.06 | 3.40 ± 0.14 | 39.1% | |
| D124Q/I180S | 67,230 ± 2020 | 16.2 ± 0.5 | 241 ± 7 | 1855 ± 80 | 0.24 ± 0.01 | 1.27 ± 0.05 | 34.5% | |
| D124Q/A182G | 54,610 ± 1640 | 10.6 ± 0.3 | 194 ± 6 | 1660 ± 70 | 0.18 ± 0.01 | 1.09 ± 0.04 | 38.1% | |
| M156E/I180S | 57,420 ± 1720 | 6.8 ± 0.2 | 119 ± 4 | 1310 ± 50 | 0.52 ± 0.02 | 3.99 ± 0.16 | 28.6% | |
| 145,400 ± 4400 | 17.2 ± 0.5 | 118 ± 4 | 4650 ± 190 | 2.16 ± 0.09 | 4.66 ± 0.19 | 40.0% | ||
| D124Q/M156E/I180S | 53,660 ± 1600 | 9.7 ± 0.3 | 180 ± 5 | 829 ± 33 | 0.10 ± 0.01 | 1.19 ± 0.05 | 19.3% | |
| D124Q/M156E/A182G | 41,570 ± 1250 | 8.4 ± 0.3 | 202 ± 6 | 593 ± 24 | 0.10 ± 0.01 | 1.71 ± 0.07 | 17.9% | |
| D124Q/M156E/F185Y | 133,400 ± 4000 | 18 ± 0.5 | 135 ± 4 | 3570 ± 140 | 0.48 ± 0.02 | 1.35 ± 0.05 | 33.5% | |
| D124Q/M156E/T205K | 159,700 ± 4800 | 18 ± 0.5 | 113 ± 4 | 4500 ± 180 | 0.65 ± 0.03 | 1.45 ± 0.06 | 35.3% | |
Each mutation is found at the equivalent position in a less elastolytic homologue such as MMP-3, −8, −10, −11, −27, or MT-MMP.
previously reported in ref (16).
heights from Fig. 2a of black columns divided by hatched columns, i.e. kcat/Km,fEln100/ kcat/Kmfs6 after normalizing each kcat/Km to the wild type
Mutations inconsequential to elastolysis
Point mutants of remote surfaces of MMP-12 protected by Eln-20 (16) that retained 84 to 101% of wild type activity towards fEln-100 were Y132A, S142E and V144A at either end of helix A, K148T of β-strand II, N153Y of the II–III loop, and I255V/R of helix C (Fig. 1c; supplemental Table S1). Consequently, putative contacts of elastin with this region appear unlikely to enhance elastin degradation directly, though they might support the anchoring of MMP-12 to elastin fibrils. Also inconsequential to elastolysis were V162S, A164V and G166R at and near β-strand III and D200E and S207V of the V-B loop on one flank of the active site (Fig. 1b; supplemental Table S1).
Distal point mutations that selectively diminish elastolysis
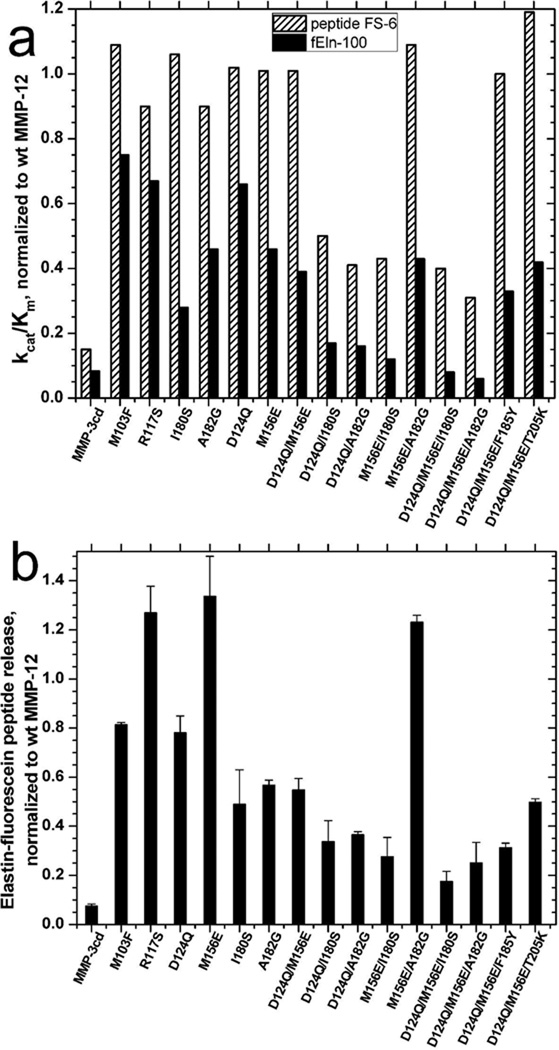
MMP-12 with lesions of M103F at the N-terminus, R117S at β-strand I, or D124Q at the C3 calcium binding site (53) in the I-A loop detracted 25%, 35%, or 35% from catalytic efficiency (kcat/Km) of digestion of fEln-100 (Fig. 2a; Table 1). Peptide release from insoluble elastin fibrils was also modestly diminished by MMP-12(M103F) and MMP-12(D124Q), but not by MMP(R117S). Adjacent to Arg117 is the M156E lesion of the II–III loop that decreased by 53% the catalytic efficiency towards fEln-100 (Fig. 2a; Table 1). It modestly enhanced peptide release from insoluble elastin fibrils (Fig. 2b). The new A182G lesion in β-strand IV at the active site served as a positive control that also diminished 53% of catalytic efficiency towards fEln-100 (Fig. 2a, Table 1), a magnitude similar to that of most lesions more peripheral to the active site (16). The D124Q or M156E lesions eroded activity upon fEln-100 by impairing Km and affinity for the substrate (Table 1; supplemental Fig. S2). The R117S and A182G lesions instead detracted from catalytic turnover kcat (Table 1).
Combinations of remote and active site mutations
Remote point mutations that selectively decreased activity for the fEln-100 protein substrate were combined with mutations closer to the active site in order to try to accumulate losses of activity towards fEln-100 without affecting activity for a control peptide substrate. Well-separated mutations that each selectively impair digestion of fEln-100 were chosen for combinations since studies of enzyme catalysis on small molecules (37) and protein-protein affinity (38, 39) found mutations to be additive when distant. Two of the three most remote mutations, D124Q (21.6 Å) and M156E (23.6 Å), were combined with mutations at the active site such as I180S (10.0 Å) and A182G (8.4 Å). (In parentheses are listed their Cα distances to the active site zinc in the free state in solution (50).) The M156E substitution is characteristic of MMP-3 (stromelysin 1). D124Q is native to MMP-8, 10, and 11. I180S is found in MMP-10 (stromelysin 2). A182G is found in MMP-26 and 27. Asp124 is 15 Å from Ile180, and 13 Å from Ala182. Met156 is 27 Å from Ile180 and 21 Å from Ala182.
All but one combination of two of these well-separated mutations further reduced activity towards both fEln-100 (Fig. 2a) and elastin-fluorescein (Fig. 2b) beyond the single mutations, i.e. D124Q/M156E, D124Q/I180S, D124Q/ A182G, M156E/I180S, and M156E/A182G. However, MMP-12(M156E/A182G) was like MMP-12(M156E) in retaining wild type or more activity towards insoluble elastin-fluorescein fibrils than did MMP-12(A182G) (Fig. 2b). MMP-12(M156E/A182G) is also unique in that its kcat for fEln-100 was twice that of wild type (Table 1). Non-additive behaviors of combinations with A182G are described further below. Combination of the M156E lesion with either the remote D124Q or active site A182G lesion retained wild type activity toward the peptide substrate FS-6 while the three other double mutations did not (Fig. 2a). These latter three (D124Q/I180S, D124Q/A182G, and M156E/I180S) still selectively diminished activity towards the fEln-100 substrate more than towards the FS-6 peptide substrate. This selectivity was beyond that of single D124Q, M156E, or A182G mutations, as evident from the decreased ratios of heights of black bars to hatched bars in Fig. 2a, expressed as percentages in the last column of Table 1.
MMP-12 variants with apparent Km for fEln-100 increased by more than two-fold (Table 1) were checked for accompanying decreases in affinity for α-elastin. (α-elastin has been recommended for elastase assays (54)). Titrations monitoring intrinsic tryptophan fluorescence emission of MMP-12 show saturable binding of α-elastin (supplementary Fig. S2a). Each of these MMP-12 mutants (M156E, D124Q/M156E, M156E/I180S, and M156E/A182G) displayed decreased affinity relative to wild type that qualitatively agrees with the fitted 2.3- to 4.4-fold increases in apparent Km for fEln-100 (supplementary Fig. S2b,c). The shapes of the association plots do not conform to the shapes of simple 1:1 binding isotherms (supplementary Fig. S2d). This is consistent with the complexities of a collection of binding events at the three dozen sites of hydrolysis in elastin (36), variability in affinity for these sites suggested by fast and slow phases of hydrolysis (32), and the widely varied masses within α-elastin. The M156E lesion appears sufficient to account for the decreased affinity. Addition of D124Q appeared to further decrease the affinity for α-elastin slightly (supplementary Fig. S2b,c).
Exosites 1 and 2
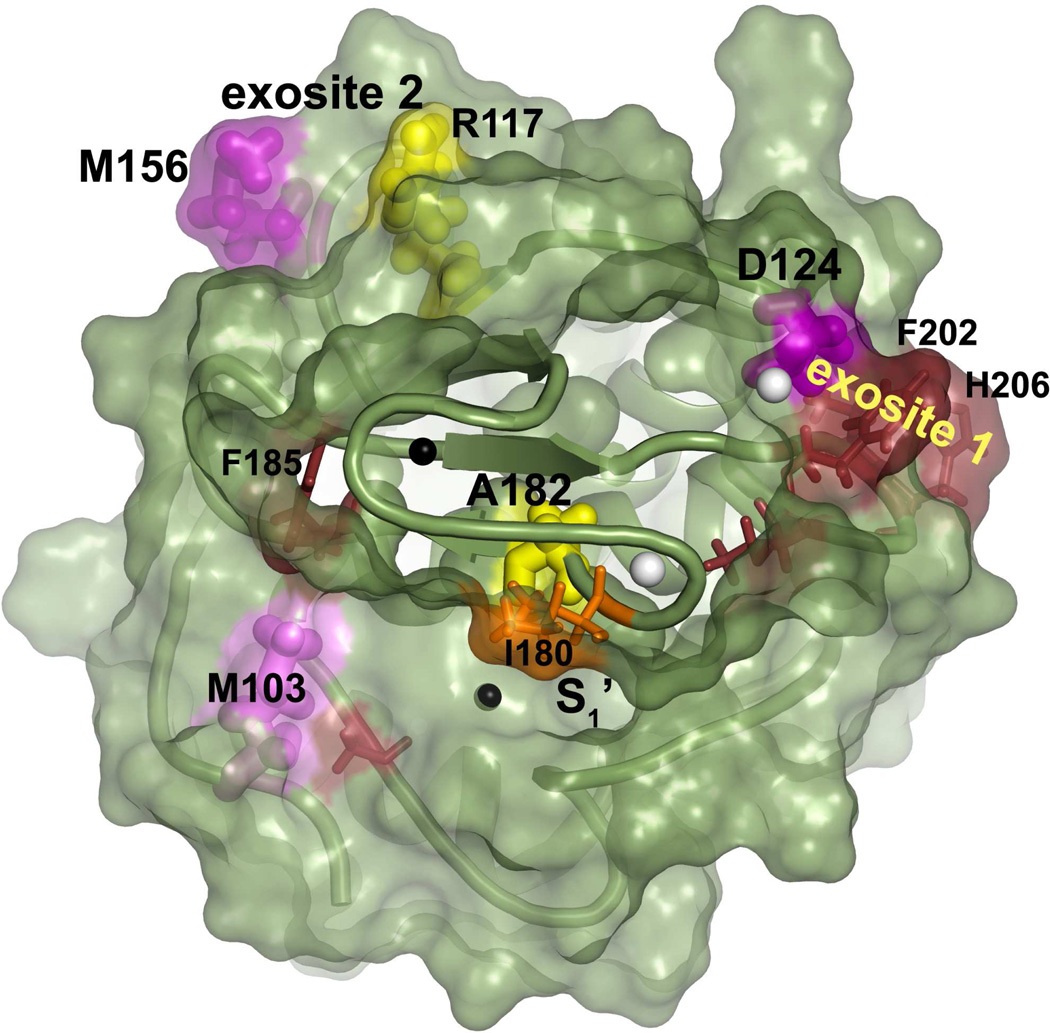
MMP-12(D124Q) and each of the combination mutants that include this mutation had their activities toward fEln-100 and elastin-fluorescein selectively diminished (Fig. 2 and Table 1). Asp124 is assigned to principal exosite 1 in elastin degradation by the rationale given in Discussion. Reductions in elastolysis by MMP-12(M156E/I180S) and MMP-12 (D124Q/M156E) strengthened the case for contributions arising from Met156 and neighboring Arg117 (Fig. 2, Table 1), which form a distinct patch designated exosite 2 (Fig. 3).
Fig. 3.
Residues from exosites that enhance elastin degradation. Newly identified residues are pink or yellow and labeled with larger type. Adjoining residues identified previously are dark red or orange and labeled with smaller type. Residues found to enhance Km are purple or kcat are yellow, both with side chains plotted with large bonds. Residues previously found to enhance Km are dark red or kcat are orange (16), both with small bonds. Zinc ions are black and calcium white. The S-shaped III–IV loop is in the foreground.
Triple mutations
In an effort to reduce the elastase activity of MMP-12 to the level of its closest homologue of MMP-3cd, third mutations were added to the lesions in exosites 1 and 2 of MMP-12(D124Q/M156E). Importantly, the triple mutations combining a lesion at each of exosites 1 and 2 with one bordering the central region of the active site, namely D124Q/M156E/I180S and D124Q/M156E/A182G, each conferred a significantly greater loss of catalytic efficiency in digesting fEln-100 than the parental double mutant of D124Q/M156E (Fig. 2). Elastin digestion by these triple mutants was impaired to greater extent than towards the FS6 peptide substrate, as evident by at least two-fold drops in selectivity (Table 1). MMP-12(D124Q/M156E/I180S) and MMP-12(D124Q/M156E/A182S) acting on fEln-100 each had kcat decreased by almost 11-fold and kcat/Km by 13-fold and 18-fold, respectively (Table 1). While these two triple mutants displayed less activity towards fEln-100 than the negative control of MMP-3cd, they retained 2- to 2.6-fold more activity towards the FS-6 peptide (Table 1), i.e. competence as an MMP. These results suggested the combined importance of exosites 1 and 2 and active site in the specific activity of MMP-12 for elastin-derived substrates.
In contrast, adding to MMP-12(D124Q/M156E) a third mutation on the periphery of the active site, whether F185Y near unprimed subsites or T205K beyond primed subsites at the opposite end of the cleft, only slightly further decreased hydrolysis of either fEln-100 or elastin-fluorescein (Fig. 2, Table 1). MMP-12(D124Q/M156E/F185Y) and MMP-12 (D124Q/M156E/T205K) were much less compromised (kcat/Km down by 3-fold and 2.4-fold) than those with the third lesion (I180S and A182Q) closer to the catalytic center. The triple mutations having F185Y or T205K substitutions experienced only modest decreases in selectivity for the fEln-100 (Table 1) and elastin-fluorescein (Fig. 2b).
Double mutant cycle analysis
This tested the hypothesis that elastin interaction and degradation results from a sum of weak interactions. Potentially modular, weak interactions can be tested by simple combinations of two mutations. The inverse mode of double mutant cycle analysis was employed. It reckons the free energies of single mutations and wild type relative to the reference point of the double mutant. Single mutations are regarded as restoring the wild type residue (41) (listed above the columns in Fig. 4b–d). Combinations of Asp124, Met156, and Ile180 had additive effects on transition-state stabilization (ΔΔGT‡), i.e. they independently promoted catalytic efficiency (kcat/Km) of hydrolysis of fEln-100 (Fig. 4a,b). Met156 did so by enhancing affinity (1/Km) for fEln-100 compared with double mutants (Fig. 4c, red boxes). Met156 combined with either Asp124 or Ile180 was essentially independent and additive with respect to 1/Km and kcat (Fig. 4c,d). The enhancements of catalytic efficiency by restoring Asp124 relative to double or triple mutants mainly increased catalytic turnover (blue boxes in Fig. 4d and Table 1). Increases of kcat/Km by Ile180 resulted from enhancements of both affinity and kcat (Fig. 4c–e).
In contrast, combinations of mutations involving Ala182 show non-additive effects on catalytic efficiency (ΔΔGT‡) (Fig. 4b). This implies functional coupling of Ala182 with the Asp124 and Met156 positions (signified by dashed lines in Fig. 4e). Like Ile180, Ala182 improved kcat and ΔΔG‡ (blue boxes in Fig. 4d and Table 1) relative to two of three multiple mutations. Two double mutants and two triple mutants each containing the D124Q lesion and either I180S or A182G shared the pattern of antagonism of affinity (1/Km, Fig. 4c) and anti-cooperative influences on catalytic turnover (Fig. 4d). Among the I180S-containing combination mutants, the effects on affinity and kcat were compensatory, achieving additive effects on transition-state stabilization, i.e. independent effects on catalytic efficiency (Fig. 4b). The A182-containing combination mutants fell short of this independence.
F185Y and T205K are effectual point mutations on the perimeter of the active site (16). Among the triple mutants, D124Q/M156E/F185Y and D124Q/M156E/T205K exhibited comparatively weaker and nearly additive effects upon catalytic efficiency and turnover number with fEln-100. Their activation free energies ΔΔG‡ were additive and transition-state stabilization ΔΔG‡T nearly additive (supplementary Fig. S1). MMP-12(D124Q/M156E/T205K) was, however, no more impaired in catalytic efficiency upon fEln-100 than MMP-12(D124Q/M156E) (Table 1, Fig. 2a, supplementary Fig. S1). The mild antagonism implies coupling between T205K and another lesion, probably D124Q nearby.
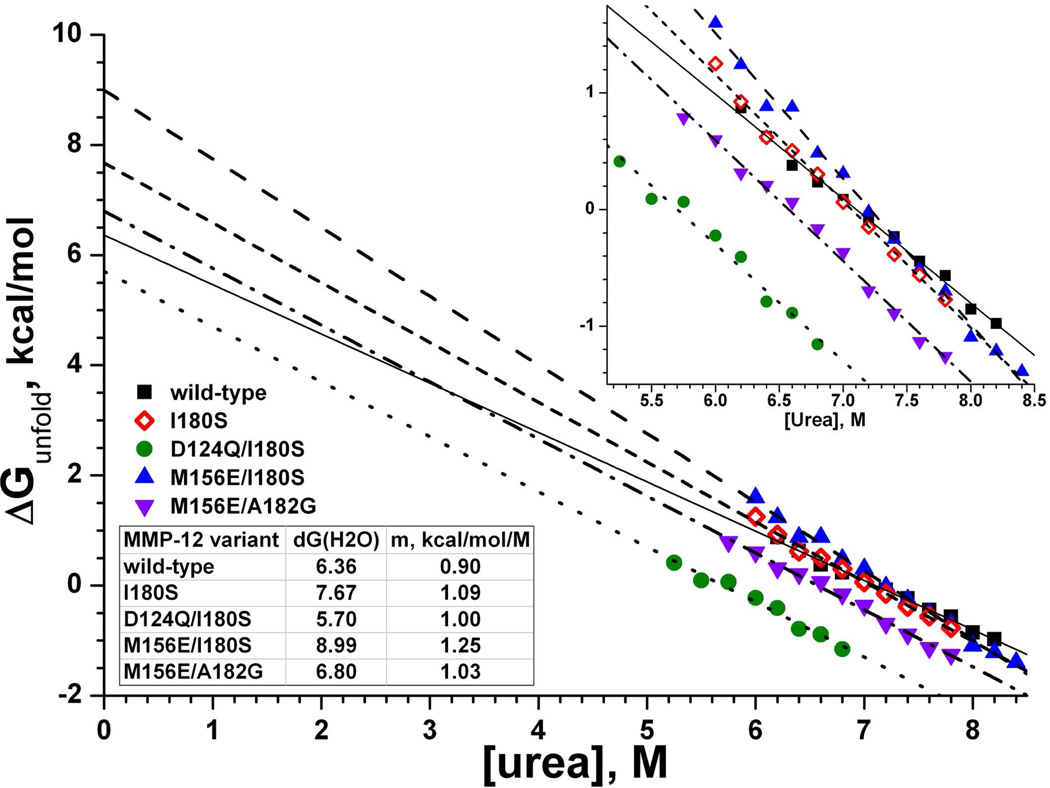
Folding stabilities
Whether effects on protein folding could indirectly affect catalysis was probed in selected variants of MMP-12 by chemical denaturation using urea. The folding stabilities measured at 37 ºC by the linear extrapolation method (49) were 6.4 kcal/mol for wild type, 7.7 kcal/mol for MMP-12(I180S), 5.7 kcal/mol for MMP-12(D124Q/I180S), 9.0 kcal/mol for MMP-12(M156E/I180S), and 6.8 kcal/mol for MMP-12(M156E/A182G) (Fig. 5). These stabilities strongly suggest that MMP-12 remained folded in the presence of D124Q, M156E, I180S, and A182G substitutions. Introduction of the D124Q lesion to MMP-12(I180S) destabilized it by 2 kcal/mol (Fig. 5), consistent with loss of a carboxylate side chain from binding a calcium ion at the C3 site. MMP-12(I180S) was 1.3 kcal/mol more stable than wild type. MMP-12(M156E/I180S) was 2.6 kcal/mol more stable than wild type, while loosing 8-fold of its activity towards fEln-100, 3.6-fold towards elastin-fluorescein fibrils, and 2.3-fold toward the general peptide substrate FS-6. This behavior follows a trend among variants of diverse enzymes where trades between activity and stability were observed (28–30).
Fig. 5.
Folding stabilities of selected enzyme variants under chemical denaturation at 37 ºC. ΔG(H2O) is the free energy to unfold the protein in absence of denaturant accessible by linear extrapolation from the linear unfolding transition region to the Y-intercept. The fitted slope of [urea] dependence is m in eq. 4 (49). The reproducibility of extrapolated ΔG(H2O) observed was 1σ ≈ 3%. The inset shows an expanded view of the transition region.
Robustness of dynamics
Whether mutations might influence indirectly by perturbing dynamics was considered. Normal modes analysis found no differences in simulated modes of motion to result from either the D124Q or I180S substitutions. In the fourth lowest mode, a 2% increase in squared fluctuations of distant Thr154 seemed to result from the A182G mutation. Differences in squared fluctuations of Pro104 and Thr154 accompanied the M156E substitution in the sixth, eighth, and tenth lowest modes. Differences from M156E at Phe171 and Asp175 of the S-shaped loop and Ser189 of the IV–V loop were also present in mode 8 (supporting Fig. S3). Apparent changes from this mutation were two- to three-fold larger in mode 8 than in mode 6. The fluctuations of mode 8 with the M156E mutation are portrayed in supporting Movie S1. M156E could have small effects on the dynamics of neighboring loops, but these are not at the active site.
DISCUSSION
Weak, additive interactions outside active site in enzyme transformation of a protein
Protein-protein interfaces have been parsed into multiple modules that are each comparatively weak in affinity (39). Enzymes interacting specifically with small molecule substrates are thought to employ several weak interactions within their active sites. The mutagenesis herein and NMR results (16) strongly suggest that macromolecular substrates derived from elastin extend far beyond the active site in their contacts with MMP-12, thereby influencing both catalytic efficiency and Michaelis constant Km. The demonstration herein of distant sites (Fig. 3) independently supporting an enzyme action on a protein substrate comes by way of co-adding contributions of exosites 1, 2, and active site periphery in catalytic efficiency upon fEln-100 (Fig. 4). This fits conceptually alongside past demonstrations of additive behaviors in enzyme transformation of small molecules and in protein-protein interactions. Common to these diverse scenarios is evidence for separated, weak contacts conferring affinity and adjustable specificity. The modest sizes of effects on catalytic efficiency of distal exosite 1 or 2 lesions D124Q or M156E are evident as transition-state stabilizations ΔΔG‡T averaging 0.21 ± 0.08 or 0.43 ± 0.08 kcal/mol, when reversing these respective mutations relative to the multiple mutations (apart from A182G-containing mutants; see below). These impairments of elastin degradation are similar in size to single point mutations described on the periphery of the active site (16). Ile180 and Ala182 closer to the active site contribute more on average to ΔΔG‡T at 0.82 ± 0.09 and 0.63 ± 0.49 kcal/mol, respectively, relative to multiple mutants. Combination of either or both of D124 and M156 with either I180 or A182 results in more significant transition-state stabilizations of 1.04 to 1.71 kcal/mol (Fig. 4a), highlighting the phenomenon of ΔΔG‡T adding up from separate interfaces (Fig. 3).
MMP exosites
This work’s identification of functional exosites within the catalytic domain of an MMP complements past findings of exosites in other domains of MMPs for recognition of fibrillar substrates. The fibronectin II-like modules inserted within the V-B loop of MMP-2 and 9 could be viewed as corresponding to exosite 1 as an extreme elaboration of it (55). Effects of altered V-B loop sequences on collagen triple helical peptidase activity (7, 56, 57) can in retrospect be regarded as evidences for the equivalent of exosite 1 being important in collagenolytic MMPs. Exosite 1 and other locations peripheral to the active site cleft are also clearly important to MMP-12 recognition of collagen IV and V triple helices (11, 16). Mutations or removal of exosites from fibronectin II-like modules of MMP-2 or 9 (12–14) or exosites of the catalytic domain of MMP-12 (Fig. 3) (16) have analogous effects of damaging interactions with fibrillar protein substrates while retaining the catalytic turnover of small peptide substrates that do not reach the remote exosites. Consequently, selective interference in interactions of one MMP with elastin or triple helices may be achievable by targeting antibodies around the equivalent of exosite 1.
Rationale for Asp124 belonging to exosite 1 in elastolysis
Asp124 lies in close van der Waals contact with Phe202 which was previously identified as part of the main exosite (16). Both Asp124 and Phe202 are about 9 Å distant from the side chains of His206 and Thr-205 that cluster in the same exosite patch (Fig. 3). Joint binding of the C3 calcium ion by the Asp124 and Glu199 carboxyl groups and the Glu199 and Glu201 carbonyl oxygens (58) bridges Asp124 to the V-B loop containing most of the residues of exosite 1. Consequently, structural proximity and connectivity is the main argument for including Asp124 in principal exosite 1. Corroborating evidence comes from non-additive effects on fEln-100 hydrolysis of adding the T205K mutation to MMP-12(D124Q/M156E) (supplementary Fig. S1, Table 1, Fig. 2) that suggest coupling to T205K. This probably resulted from the proximity across exosite 1 of the D124Q lesion, since nearby mutations in interfaces have usually been non-additive (37–39). The presumed functional, energetic coupling of D124Q and T205K would support their connectivity within the same structural cluster within the protein-protein interface, the concept developed by ref (39). Thus, assignment of Asp124 to exosite 1 in the context of elastin degradation is based on its structural proximity to exosite 1, calcium bridging to the V-B loop, and apparent functional coupling with Thr205.
Role of Asp124
This aspartate that binds the C3 calcium ion with its carboxyl group is conserved in 18 of 23 MMPs and replaced by glutamine or lysine in five not needing it for activity (53). Alanine substitution of this aspartate in MMP-26 decreased its affinity for this calcium ion and proteolytic activity (59). The decreased activity of D124Q-containing MMP-12 mutants for large substrates from elastin (Fig. 2, 3, 4 and Table 1) implies a functional contribution of this remote aspartate-containing loop that joins β-strand I with helix A in the β-α-β motif. The functional role for this loop now appears analogous to corresponding loops of β-α-β motifs that support the active site of triose phosphate isomerase or the nucleotide binding site of the Rossman fold (60). Despite the small defect of MMP-12(D124Q) in affinity for the soluble elastin substrate (Table 1), restoration of Asp124 instead benefitted kcat for fEln-100 compared with D124Q-containing combination mutants (blue boxes in Fig. 4d). Anti-cooperative kcat behavior of Asp124 combined with either Ile180 or Ala182 in double and triple mutants (Fig. 4d) implies that these residues facilitate the same non-rate-limiting step, according to ref (40). This step probably affects catalytic turnover due to the similarity between ΔΔG‡ (for kcat) and ΔΔG‡T (for kcat/Km) in double mutant cycle analysis of most D124Q-containing multiple mutants (Fig. 4b,d). Consistent with this, changes in Km for fEln-100 are minimal for these four multiple mutants (Table 1). Since a wild type level of catalytic efficiency for the peptide substrate is maintained by four of the eight D124Q mutants (Fig. 2a, Table 1), the catalytic consequences from D124Q can be subtle enough to affect only turnover of the cross-linked and proteolysis-resistant elastin substrates. Modest destabilization of the protein fold and / or perturbation of active site structure by the D124Q mutation might have indirectly compromised enzymatic interaction, perhaps by affecting hydrogen bonding of the Ile180 to Ala182 region with the backbone of elastin substrates especially.
Independence of exosite 2
The long distances of Met156 from active site (24 Å to zinc) and exosite 1 (26 Å to Asp124) indicate that it belongs to neither. The additivity of Met156 with either Asp124 or Ile180 in digestion of fEln-100 (Fig. 4) implies the independence of their effects on the same step; see ref (40). Considering the size of the Km effects (Table 1, Fig. 4), this potentially rate-limiting step might be binding and positioning of fEln-100 to support turnover.
Functional coupling of Ala182 to Asp124 and Met156
Ala182 exhibited functional coupling to Asp124 and Met156 despite separations of 13 and 21 Å, respectively (Fig. 4e). The thermodynamic coupling of Ala182 and Asp124 could facilitate the same non-rate-limiting catalytic step in elastolysis by the argument given above. Ala182 and Met156 were cooperative in transition state stabilization (Fig. 4b), anti-cooperative in affinity for the soluble elastin (Fig. 4c), and antagonistic toward kcat (Fig. 4d). The latter two observations suggest that Ala182 and Met156 may facilitate the same step (40). Ala182 hydrogen bonding of the NH group of the scissile bond of a peptide substrate (61) and the enhancement of α-elastin affinity by Met156 further raises the question of whether these two residues might jointly position segments of α-elastin for cleavage.
There is considerable evidence for allostery acting on the pro-domain in the activation of a few MMPs (4). The functional and energetic coupling of Ala182 with the Asp124 and Met156 positions, symbolized by dashed lines in Fig. 4e, suggests the possibility of allosteric transmission across the catalytic domain of an MMP as well. A182G conferring a slight increase of simulated motion to the Thr154 neighbor of Met156 is consistent with the possibility of some transmission between active site and exosite 2. Belonging to the same large rigid structural cluster has been proposed as a source of the non-additivity of residues separated by long distances (62). This may be plausible in that Ala182 could be rigidly connected by the β-sheet to the locales of Asp124 and Met156.
Exosite 2 in advanced elastin digestion
Hydrolysis of insoluble fluorescein-labeled elastin may represent early stages of digestion of rigid fibrils while digestion of fEln-100 could represent later stages of digestion that make cross-linked chains more flexible (16). Thus, retention of high peptide release from elastin fibrils by MMP-12(R117S), MMP-12(M156E), and MMP-12(M156E/A182G) (Fig. 2b) may suggest contacts with these residues to be unimportant initially in elastin degradation when the fibrils remain too rigid to bend around to reach exosite 2. Since Met156, Arg117, and Asp124 are most exposed, they may be the distal contacts most easily reached by elastin made flexible enough by partial hydrolysis.
Trades of stability for activity
The gains in folding stability of 1.3 kcal/mol by MMP-12(I180S) and 2.6 kcal/mol by MMP-12(M156E/I180S) (Fig. 5) vis-à-vis their losses of activity (Fig. 2) are consistent with the trend of enzyme variants trading away thermal stability for more activity (28–30). Such a tradeoff was also observed when comparing MMP-3 and 12 (26). Both Met156 and Ile180 place a bulky hydrophobic side chain on the surface of MMP-12, something that is typically destabilizing to a protein structure. Substitutions by glutamate and serine at these two positions (native to MMP-3 and 10, respectively) presumably alleviate the energetically unfavorable ordering of waters around the exposed hydrophobic side chains. Natural selection of Met156 and Ile180 appears to have favored high activity of MMP-12 over its stability. Remote Met156 enhances affinity for soluble elastin while central Ile180 enhances catalytic turnover (Table 1).
Similarity of MMP-12 triple mutants to MMP-3cd
The 2.6 kcal/mol of stabilization of MMP-12(M156E/I180S) is reminiscent of the 2.8 kcal/mol stabilization of MMP-3cd relative to MMP-12 (26). More importantly, both MMP-12 (D124Q/M156E/I180S) and MMP-12 (D124Q/ M156E/A182G) resemble MMP-3cd in terms of catalytic efficiency, turnover number, and Km for fEln-100 (Table 1, Fig. 2a). Furthermore, these two triple mutants also resemble MMP-3cd activity towards the peptide substrate, particularly in terms of Km but also within a factor of two to three for turnover number and catalytic efficiency (Table 1). These combinations of three mutations (among 65 differences in sequence) were enough to devolve MMP-12 into MMP-3-like behavior.
Analogy with exosites of thrombin
Thrombin’s Exosite I is accessed by peptide substrates from protease-activated receptors that bend from the active site around to reach it. Affinity for the peptide was disrupted by mutations at either Exosite I or the active site (2). The polypeptide inhibitors hirudin, ornithodorin, and rhodnin each extend from the active site and place their basic and hydrophobic groups across Exosite I (2). The concept of various polypeptide partners bending from active site around to exosite I seems to generalize to MMP-12. Its catalytic turnover of and affinity for substrates from collagen triple helices and α-elastin is clearly enhanced by exosite 1, which is suggestive of bending of otherwise rigid triple helices in order for them to reach exosite 1 in the Michaelis complex (16). This could be analogous to peptide substrate positions P4’ and P5’ modulating turnover by a serine protease, a cysteine protease, and MMP-2 (63).
Summary
MMP-12 digestion of substrates derived from elastin now appears to be enhanced by a second and more remote exosite centered around Met156 of the II–III loop and Arg117 of β-strand I. Main exosite I at the V-B loop should be expanded to include calcium-binding Asp124 of the I-A loop in the context of elastolysis. The importance of exosite 1 to elastolysis by MMP-12 may have analogies with digestion of proteins from fibrils by collagenolytic MMPs and with peptidase activities of some serine and cysteine proteases. Increased catalytic efficiency for soluble elastin results from affinity enhanced by Met156 and catalytic turnover enhanced by Asp124. Met156 sacrifices folding stability, whereas Asp124 enhances it. Ile180 trades away some stability for higher kcat for substrates from protein fibrils. Combining one lesion at each of Asp124 (exosite I), Met156 (exosite II), and either Ile180 or Ala182 at the β-strand IV edge of the active site significantly and selectively decreased catalytic efficiency of turnover of soluble α-elastin down to less than or equal to that of negative control MMP-3cd, while preserving more activity on a peptide substrate. The net deterioration of elastolysis by these lesions, except for A182G, is additive and gives specific examples of weak interactions widely distributed from active site to its periphery and exosites 1 and 2 supporting elastolysis. Thus, the concept of additive combinations of weak, separated interactions has now been demonstrated in a proteolytic enzyme transformation of a protein substrate, broadening the range of examples of additive behavior. The importance of the two novel exosites in elastin degradation is underscored by the achievement of MMP-3-like catalytic behavior via only three distantly separated point mutations of MMP-12.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Mark Palmier for expert advice on fitting of steady-state kinetics to enzyme progress curves and insightful comments on the manuscript.
Footnotes
This work was supported by NIH grants R01GM57289, GM57289-08S1, and American Heart Association grant 0855714G.
The abbreviations used are: BINDSIght, bioinformatics; NMR discovery of specificity of interactions; cd, catalytic domain as in MMP-3cd; Eln-20, α-elastin species of 20 kDa; fEln-100, fluorescently labeled α-elastin species of 100 kDa; MMP, matrix metalloproteinase; S.E.M., standard error of the mean; TIMP-1, tissue inhibitor of matrix metalloproteinases-1; TNC, Tris, NaCl, and CaCl2; wt, wild type
SUPPORTING INFORMATION AVAILABLE
Table S1 describes mutants with activities nearly intact. Fig. S1 depicts “double” mutant cycle analysis for triple mutations with a point mutation on either extreme of the active site cleft. Fig. S2 compares affinity for α-elastin of mutants with apparent Km increased by at least two-fold. Fig. S3 compares amplitude of motions in normal mode 8 before and after the M156E mutation. Movie S1 illustrates normal mode 8 with M156E present. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bode W. Structure and interaction modes of thrombin. Blood Cells Mol Dis. 2006;36:122–130. doi: 10.1016/j.bcmd.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, Di Cera E. Molecular mapping of thrombin-receptor interactions. Proteins. 2001;45:107–116. doi: 10.1002/prot.1130. [DOI] [PubMed] [Google Scholar]
- 3.Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 4.Sela-Passwell N, Rosenblum G, Shoham T, Sagi I. Structural and functional bases for allosteric control of MMP activities: Can it pave the path for selective inhibition? Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2010;1803:29–38. doi: 10.1016/j.bbamcr.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Clark IM, Cawston TE. Fragments of human fibroblast collagenase. Purification and characterization. Biochem J. 1989;263:201–206. doi: 10.1042/bj2630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Connell JP, Docherty AJ. The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- 7.Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H. Identification of the (183)RWTNNFREY(191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 8.Patterson ML, Atkinson SJ, Knauper V, Murphy G. Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS Lett. 2001;503:158–162. doi: 10.1016/s0014-5793(01)02723-5. [DOI] [PubMed] [Google Scholar]
- 9.Knauper V, Cowell S, Smith B, Lopez-Otin C, O’Shea M, Morris H, Zardi L, Murphy G. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J. Biol. Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- 10.Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 11.Bhaskaran R, Palmier MO, Lauer-Fields JL, Fields GB, Van Doren SR. MMP-12 Catalytic Domain Recognizes Triple Helical Peptide Models of Collagen V with Exosites and High Activity. J. Biol. Chem. 2008;283:21779–21788. doi: 10.1074/jbc.M709966200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shipley JM, Doyle GA, Fliszar CJ, Ye QZ, Johnson LL, Shapiro SD, Welgus HG, Senior RM. The structural basis for the elastolytic activity of the 92-kDa and 72- kDa gelatinases. Role of the fibronectin type II-like repeats. J. Biol. Chem. 1996;271:4335–4341. doi: 10.1074/jbc.271.8.4335. [DOI] [PubMed] [Google Scholar]
- 13.Collier IE, Krasnov PA, Strongin AY, Birkedal-Hansen H, Goldberg GI. Alanine scanning mutagenesis and functional analysis of the fibronectin-like collagen-binding domain from human 92-kDa type IV collagenase. J. Biol. Chem. 1992;267:6776–6781. [PubMed] [Google Scholar]
- 14.O’Farrell TJ, Pourmotabbed T. The fibronectin-like domain is required for the type V and XI collagenolytic activity of gelatinase B. Arch Biochem Biophys. 1998;354:24–30. doi: 10.1006/abbi.1998.0662. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro SD, Kobayashi DK, Ley TJ. Cloning and characterization of a unique elastolytic metalloproteinase produced by human alveolar macrophages. J. Biol. Chem. 1993;268:23824–23829. [PubMed] [Google Scholar]
- 16.Palmier MO, Fulcher YG, Bhaskaran R, Duong VQ, Fields GB, Van Doren SR. NMR and bioinformatics discovery of exosites that tune metalloelastase specificity for solubilized elastin and collagen triple helices. J. Biol. Chem. 2010;285:30918–30930. doi: 10.1074/jbc.M110.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starcher BC, Galione MJ. Purification and comparison of elastins from different animal species. Anal Biochem. 1976;74:441–447. doi: 10.1016/0003-2697(76)90224-4. [DOI] [PubMed] [Google Scholar]
- 18.Wagenseil JE, Mecham RP. Vascular extracellular matrix and arterial mechanics. Physiol Rev. 2009;89:957–989. doi: 10.1152/physrev.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81:229–240. doi: 10.1002/bdrc.20111. [DOI] [PubMed] [Google Scholar]
- 20.Robert L, Robert AM, Jacotot B. Elastin-elastase-atherosclerosis revisited. Atherosclerosis. 1998;140:281–295. doi: 10.1016/s0021-9150(98)00171-3. [DOI] [PubMed] [Google Scholar]
- 21.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 22.Curci JA, Liao S, Huffman MD, Shapiro SD, Thompson RW. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J Clin Invest. 1998;102:1900–1910. doi: 10.1172/JCI2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JL, George SJ, Newby AC, Jackson CL. Divergent effects of matrix metalloproteinases 3, 7, 9, and 12 on atherosclerotic plaque stability in mouse brachiocephalic arteries. Proc Natl Acad Sci U S A. 2005;102:15575–15580. doi: 10.1073/pnas.0506201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonicelli F, Bellon G, Debelle L, Hornebeck W. Elastin-elastases and inflamm-aging. Curr Top Dev Biol. 2007;79:99–155. doi: 10.1016/S0070-2153(06)79005-6. [DOI] [PubMed] [Google Scholar]
- 25.Hunninghake GW, Davidson JM, Rennard S, Szapiel S, Gadek JE, Crystal RG. Elastin fragments attract macrophage precursors to diseased sites in pulmonary emphysema. Science. 1981;212:925–927. doi: 10.1126/science.7233186. [DOI] [PubMed] [Google Scholar]
- 26.Liang X, Arunima A, Zhao Y, Bhaskaran R, Shende A, Byrne TS, Fleeks J, Palmier MO, Van Doren SR. Apparent tradeoff of higher activity in MMP-12 for enhanced stability and flexibility in MMP-3. Biophys J. 2010;99:273–283. doi: 10.1016/j.bpj.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang J, Liu E, Yu Y, Kitajima S, Koike T, Jin Y, Morimoto M, Hatakeyama K, Asada Y, Watanabe T, Sasaguri Y, Watanabe S, Fan J. Macrophage metalloelastase accelerates the progression of atherosclerosis in transgenic rabbits. Circulation. 2006;113:1993–2001. doi: 10.1161/CIRCULATIONAHA.105.596031. [DOI] [PubMed] [Google Scholar]
- 28.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold FH, Wintrode PL, Miyazaki K, Gershenson A. How enzymes adapt: lessons from directed evolution. Trends in Biochemical Sciences. 2001;26:100–106. doi: 10.1016/s0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- 30.Beadle BM, Shoichet BK. Structural bases of stability-function tradeoffs in enzymes. J Mol Biol. 2002;321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 31.Tamburro AM, Bochicchio B, Pepe A. The dissection of human tropoelastin: from the molecular structure to the self-assembly to the elasticity mechanism. Pathol Biol (Paris) 2005;53:383–389. doi: 10.1016/j.patbio.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Palmier MO, Fulcher YG, Van Doren SR. Solubilized elastin substrate for continuous fluorimetric assay of kinetics of elastases. Anal Biochem. 2011;408:172–174. doi: 10.1016/j.ab.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partridge SM, Davis HF. The chemistry of connective tissues. 3. Composition of the soluble proteins derived from elastin. Biochem J. 1955;61:21–30. doi: 10.1042/bj0610021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mecham RP, Lange G. Measurement by radioimmunoassay of soluble elastins from different animal species. Connect Tissue Res. 1980;7:247–252. doi: 10.3109/03008208009152360. [DOI] [PubMed] [Google Scholar]
- 35.Cox BA, Starcher BC, Urry DW. Coacervation of alpha-elastin results in fiber formation. Biochim Biophys Acta. 1973;317:209–213. doi: 10.1016/0005-2795(73)90215-8. [DOI] [PubMed] [Google Scholar]
- 36.Taddese S, Weiss AS, Neubert RH, Schmelzer CE. Mapping of macrophage elastase cleavage sites in insoluble human skin elastin. Matrix Biol. 2008;27:420–428. doi: 10.1016/j.matbio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber G, Fersht AR. Energetics of protein-protein interactions: Analysis ofthe Barnase-Barstar interface by single mutations and double mutant cycles. Journal of Molecular Biology. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 39.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mildvan AS, Weber DJ, Kuliopulos A. Quantitative interpretations of double mutations of enzymes. Arch Biochem Biophys. 1992;294:327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- 41.Mildvan AS. Inverse thinking about double mutants of enzymes. Biochemistry. 2004;43:14517–14520. doi: 10.1021/bi048052e. [DOI] [PubMed] [Google Scholar]
- 42.Parkar AA, Stow MD, Smith K, Panicker AK, Guilloteau JP, Jupp R, Crowe SJ. Large-scale expression, refolding, and purification of the catalytic domain of human macrophage metalloelastase (MMP-12) in Escherichia coli. Protein Expr Purif. 2000;20:152–161. doi: 10.1006/prep.2000.1280. [DOI] [PubMed] [Google Scholar]
- 43.Zheng X, Ou L, Tong X, Zhu J, Wu H. Over-expression and refolding of isotopically labeled recombinant catalytic domain of human macrophage elastase (MMP-12) for NMR studies. Protein Expr Purif. 2007;56:160–166. doi: 10.1016/j.pep.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Knight CG. Active-site titration of peptidases. Methods Enzymol. 1995;248:85–101. doi: 10.1016/0076-6879(95)48008-0. [DOI] [PubMed] [Google Scholar]
- 45.Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem. 2004;328:166–173. doi: 10.1016/j.ab.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 46.Palmier MO, Van Doren SR. Rapid determination of enzyme kinetics from fluorescence: overcoming the inner filter effect. Anal Biochem. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson AJ, Fersht AR, Blow DM, Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983;22:3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- 48.Klimacek M, Nidetzky B. The oxyanion hole of Pseudomonas fluorescens mannitol 2-dehydrogenase: a novel structural motif for electrostatic stabilization in alcohol dehydrogenase active sites. Biochemical Journal. 2009;425:455–463. doi: 10.1042/BJ20091441. [DOI] [PubMed] [Google Scholar]
- 49.Pace CN, Shaw KL. Linear Extrapolation Method of Analyzing Solvent Denaturation Curves. Proteins: Structure, Functionand Genetics Suppl. 2000;4:1–7. doi: 10.1002/1097-0134(2000)41:4+<1::aid-prot10>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 50.Bhaskaran R, Palmier MO, Bagegni NA, Liang X, Van Doren SR. Solution structure of inhibitor-free human metalloelastase (MMP-12) indicates an internal conformational adjustment. J Mol Biol. 2007;374:1333–1344. doi: 10.1016/j.jmb.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLano WL. The PyMOL Molecular Graphics System DeLano Scientific. CA: Palo Alto; 2002. [Google Scholar]
- 52.Eyal E, Yang L-W, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22:2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 53.Maskos K. Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie. 2005;87:249–263. doi: 10.1016/j.biochi.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 54.Keller S, Mandl I. Solubilized Elastin as a Substrate for Elastase and Elastase Inhibitor Determinations. Biochemical Medicine. 1971;5:342–347. [Google Scholar]
- 55.Van Doren SR. Structural Basis of Extracellular Matrix Interactions with Matrix Metalloproteinases. In: Parks WC, Mecham RP, editors. Extracellular Matrix Degradation. Berlin: Springer-Verlag; 2011. pp. 123–144. [Google Scholar]
- 56.Pelman GR, Morrison CJ, Overall CM. Pivotal molecular determinants of peptidic and collagen triple helicase activities reside in the S3’ subsite of matrix metalloproteinase 8 (MMP-8): the role of hydrogen bonding potential of ASN188 and TYR189 and the connecting cis bond. J. Biol. Chem. 2005;280:2370–2377. doi: 10.1074/jbc.M409603200. [DOI] [PubMed] [Google Scholar]
- 57.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. The roles of substrate thermal stability and P2 and P1’ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 2006;281:38302–38313. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
- 58.Lang R, Kocourek A, Braun M, Tschesche H, Huber R, Bode W, Maskos K. Substrate specificity determinants of human macrophage elastase (MMP- 12) based on the 1.1 A crystal structure. J Mol Biol. 2001;312:731–742. doi: 10.1006/jmbi.2001.4954. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Park HI, Sang QX. Calcium regulates tertiary structure and enzymatic activity of human endometase/matrilysin-2 and its role in promoting human breast cancer cell invasion. Biochem J. 2007;403:31–42. doi: 10.1042/BJ20061390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Branden C, Tooze J. Introduction to Protein Structure. 2nd ed. New York: Garland Publishing; 1999. [Google Scholar]
- 61.Bertini I, Calderone V, Fragai M, Luchinat C, Maletta M, Yeo KJ. Snapshots of the reaction mechanism of matrix metalloproteinases. Angew Chem Int Ed Engl. 2006;45:7952–7955. doi: 10.1002/anie.200603100. [DOI] [PubMed] [Google Scholar]
- 62.Istomin AY, Gromiha MM, Vorov OK, Jacobs DJ, Livesay DR. New insight into long-range nonadditivity within protein double-mutant cycles. Proteins Structure, Functionand Bioinformatics. 2008;70:915–924. doi: 10.1002/prot.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.