Abstract
Protein recognition and binding, which result in either transient or long-lived complexes, play a fundamental role in many biological functions, but sometimes also result in pathologic aggregates. We use a simplified simulation model to survey a range of systems where two highly flexible protein chains form a homodimer. In all cases, this model, which corresponds to a perfectly funneled energy landscape for folding and binding, reproduces the macroscopic experimental observations on whether folding and binding are coupled in one step or whether intermediates occur. Owing to the minimal frustration principle, we find that, as in the case of protein folding, the native topology is the major factor that governs the choice of binding mechanism. Even when the monomer is stable on its own, binding sometimes occurs fastest through unfolded intermediates, thus showing the speedup envisioned in the fly-casting scenario for molecular recognition.
Analyzing the orchestration of the cell's activity requires understanding how the proteins and nucleic acids in the cell recognize and bind to each other. The homo- or hetero-oligomers (1–3), which are formed when proteins bind, play an essential role in many biological processes, but sometimes also result in disease (4). Deciphering the dynamic principles of protein association is crucial for the understanding of protein networking, protein function, and malfunction and to design more stable complexes as pharmacological inhibitors. Here we show that the ability of globular proteins to assemble themselves into well defined structures is well understood through energy landscape theory.
Efficient and robust folding has been achieved by the evolution of protein sequences that satisfy the principle of minimal frustration and that therefore have a landscape that can be described as a partially rugged funnel (5–9). The optimization of the energetic interactions for these small and intermediate size proteins is apparently so good that the effects of energetic frustration are hard to discern quantitatively in wild-type proteins but are easily seen in some designed molecules. Once energetic frustration is sufficiently small, topology becomes the key determinant of folding mechanism. In a perfectly funneled landscape, the structural heterogeneity observed in folding transition states and the partially folded ensemble are determined by geometrical constraints reflecting the tradeoff between chain entropy and folding stabilization energy, and can be inferred reasonably when the protein native structure alone is known (8). Comparisons between theoretical predictions and experiments have confirmed this hypothesis. The structures of transition states (6, 7), as measured by Φ value analysis (10), and the existence of folding intermediates (6, 11) are well predicted in models where energetic frustration is completely absent and that contain topological information alone (Gō models; ref. 12). This strongly supports minimal frustration as the mechanism used by evolution for protein design.
The configurational search space in folding is large, thus forcing proteins to be selected to obey the minimal frustration principle to simplify the search. The search space in molecular recognition is smaller than for folding, but it is still vast when we consider the large number of possible dimers that could form in a cell. We must then ask whether binding processes also have funneled landscapes. Several papers have recently addressed this qualitatively for real proteins (13–15). Quantitative simulation studies of lattice model proteins have also given insights about the polymer physics aspects (16). Here, we ask about the kinetic consequences of such a funneled binding landscape for the detailed mechanism of recognition. Several different mechanisms for protein recognition and association have been proposed, varying in the degree to which subunits rearrange during recognition. The simplest proposed mechanism is rigid-body docking guided by chemical and structural complementarities (the venerable “lock-and-key” mechanism). Other scenarios emphasize that plasticity may be fundamental for recognition. Koshland's proposal of “induced fit” (17) took the first step by suggesting that flexible recognition after binding can optimize binding interactions formed initially in an encounter complex. Another view, the recently proposed “conformational selection” mechanism, hypothesizes that recognition takes place between preselected conformers that are optimized for binding (18, 19). These mechanisms assume the existence of nearly completely folded subunits before association, but some proteins fold only upon association (15, 20, 21). These proteins often participate in regulatory activities and are believed to be intrinsically unstructured (22). One selective advantage of folding only at the time of binding is the possibility to achieve high specificity with low affinity (23). Coupled folding and binding also allows a single molecule to have the capability to bind to several different targets, and thereby function as a nonlinear element in control pathways (20, 24). A kinetic advantage for being initially unfolded before binding has been postulated through the “fly-casting mechanism” (25), which has been quantified in one case by using free energy functional methods. An unstructured protein can have a greater capture radius for a specific binding site than a folded state with its restricted conformational freedom, and thereby results in an enhanced speed of recognition. This advantage may be even greater when one accounts for the kinetic difficulty of desolvating a large rigid protein–protein interface (26).
Structural Classification of Homodimers
A first step in experimentally characterizing the mechanism of protein assembly is to find out whether association starts from unfolded subunits (two-state dimers) or folded subunits (three-state dimers) (27–29). For a so-called two-state dimer [also described as a permanent (or obligatory) dimer (1)], the monomer is intrinsically unstructured and folds only upon binding. Binding with a three-state mechanism usually has as an intermediate the individual folded monomers but some homodimers formed via a three-state mechanism in which the intermediate actually corresponds to a dimeric structure rather than a folded monomer.
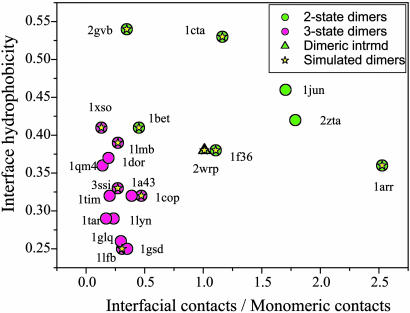
Eleven homodimers, which have been experimentally studied, were selected for theoretical study. Of these, five have been experimentally classified as two-state dimers, five are classified as three-state dimers with a folded monomeric intermediate, and one is a three-state dimer but with a dimeric intermediate. The dimers selected for the survey span a range of topology, secondary structure content, and interface geometry. Fig. 1 shows a “phase diagram” correlating the association mechanism found experimentally with a structural classification of two- and three-state homodimers based on the number of intramonomeric and interfacial native contacts as well as the hydrophobicity of the interface. Two-state dimers are characterized by a higher ratio of interfacial contacts to monomeric contacts. The interface is extensive and crucial for stabilizing such two-state dimers. There is also a tendency for two state dimers to have a more hydrophobic interface. Previous analyses (1, 30), suggest that, when folding and association obligatorily occur together, the resulting interface is hydrophobic, similar to the core of a single domain protein, but that for hydrophilic interfaces, recognition occurs commonly when association follows folding. Recognition in the latter case is perhaps guided by long-range electrostatic interactions (31, 32). Although, this “phase diagram” differentiates between the two types of homodimers, three of the large two-state dimers (1f36, 2gvb, and 1bet) are characterized by low/intermediate values of the ratio between interfacial to monomeric native contacts, suggesting the possibility of a more complicated association than that for smaller two-state dimers.
Fig. 1.
A phase diagram that correlates the association mechanism of the homodimers with their structural properties. The two- and three-state homodimers are structurally classified based on the number of interfacial and intramonomeric native contacts as well as the hydrophobicity of the interface (the 11 selected homodimers for simulation are designated by a star). The interface hydrophobicity was calculated based on the normalized occurrence of each amino acid in interfacial contacts multiplied by its hydrophobicity factor (50). The classification as two-state, three-state, and existence of dimeric intermediate is based on experimental data. In general, a two-state dimer is characterized by higher ratio of interfacial native contacts to monomeric native contacts, and a more hydrophobic interface, in comparison to a three-state dimer. The two-state homodimers include 1cta (Troponin C site III) (40), 1arr (Arc repressor) (33, 41), 2gvb (gene V protein) (34, 42), 1f36 (factor for inversion stimulation) (36), 1bet (β nerve growth factor) (43). The three-state homodimers include: 1lmb (λ repressor) (44), 1cop (λ Cro repressor) (45), 1lfb (LFB1 transcription factor) (46), 3ssi (S. subtilisn inhibitor) (47), 1xso (superoxide dismutase) (48). The class of three-state homodimers with dimeric intermediate is represented by 2wrp (Trp repressor) (49) and denoted by the same color as the two-state dimers because its dimerization does not involve a preexisting folded monomer. For references on other dimers in the phase diagram, see supporting information.
Bimolecular Folding: Association Simulations with an Energetically Minimally Frustrated Model
The effects of the monomer and interface geometry on the association mechanism of the 11 selected homodimers were studied by simulating two identical monomeric chains interacting via a Gō model which takes into account only contact interactions found in the folded dimer. This model lacks energetic frustration and, accordingly, corresponds to a perfectly funneled energy landscape. Each residue is represented by a single bead centered at the Cα position by using a force field similar to that used by Clementi et al. (6). In the framework of the model, all native contacts are represented by the 10-12 Lennard Jones form without any discrimination between the various chemical types of interaction. Moreover, both the intra- and intermonomeric contacts (interfacial contacts) are treated in the same way without any bias toward separate folding or toward binding (a detailed description of this energy function can be found in supporting information, which is published on the PNAS web site). Nonspecific binding is not contained in this model because only native contacts are included. A survey of two-state folders has been made by Koga and Takada using Gō model (7). In most cases, the experimental Φ values were reproduced by this model that contains topological information alone. In some cases where the protein's symmetric topology allows two degenerate patterns of residues to function as critical folding nuclei, the degeneracy can be broken by the residual energetic heterogeneity. The predictions of perfectly funneled landscapes for folding with intermediates have been highlighted in several other studies (6, 11). Although the structure and presence of partially folded intermediate ensembles is well predicted by these simple models, the absolute values of barriers and stabilities are a fine balance that is sensitive to details of the potential.
To enhance the sampling of association events, the two identical subunits of each homodimer are linked by a glycine chain, which acts mainly to hold the two unbound subunits (folded or unfolded) in close proximity during their dynamics. The linker's length was determined by the distance between the C terminus of subunit A and the N terminus of subunit B, and it was designed not to interfere with any intra- or intersubunit contacts that stabilize the folded dimer. Covalently linked Arc repressor (33) and gene V protein (34) were experimentally found to be fully functional and with an enhanced folding rate and stability. Accordingly, these experiments suggest that the linker plays a passive, largely entropic role by keeping the unbound monomers at high local concentrations during folding.
For each fused homodimer, few tens constant temperature simulations were performed (each includes >4 × 107 integration time steps) starting from a dimeric conformation or unfolded monomers. The set of trajectories was combined by using the weighted histogram analysis method (35) to provide the folding temperature, Tf, (defined as the temperature where the free energy of the unfolded and folded states are identical) from the peak of the specific heat versus temperature and to calculate the thermodynamic properties of the systems.
Association of Obligatory Dimers: Folding by Binding
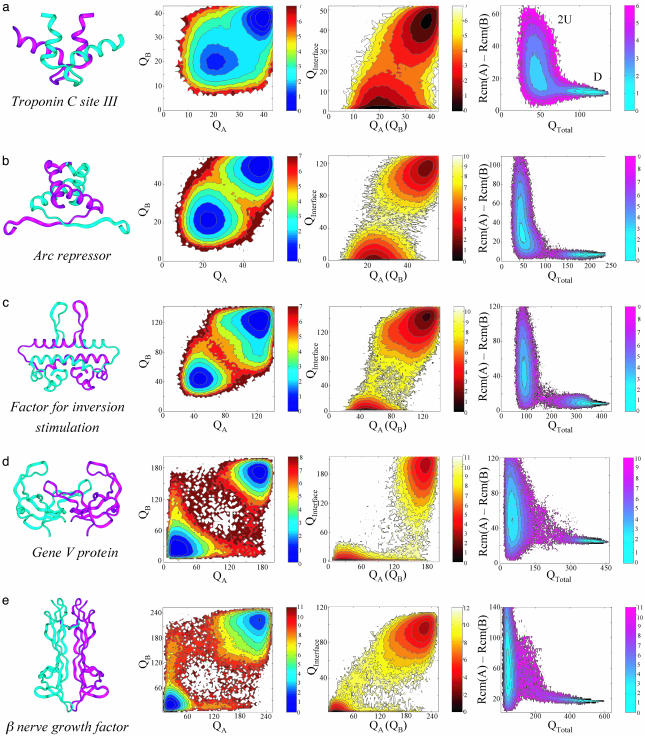
The simulations show that, for five of the systems, unfolded chains directly bind to form a folded dimer. This finding agrees with the experimental classification of these systems as two-state dimers. The free energy surface of the binding process of each homodimer is projected onto several candidate reaction coordinates for folding and binding: the monomeric native contacts (QA and QB), interfacial native contacts (QInterface), the total number of native contacts (QTotal), and the distance between the center of mass of the two subunits [Rcm(A)–Rcm(B)] (Fig. 2). These provide a detailed investigation of the binding mechanism and, particularly, the existence of coupling between the monomer folding as well as between folding and binding. For Troponin C site III (Arc repressor), these figures show coupling between the monomer folding as the only populated states are those where the two chains are either unfolded (low QA and QB) or folded (high QA and QB). Also, the monomer (either A or B) folding is coupled to the interface formation (i.e., binding) as indicated by the observation that a folded monomer (high QA or QB) exists only when the interface is formed (i.e., high QInterface). In the simulations, the two chains fold concurrently upon their association (time evolution of monomeric and interfacial native contacts are shown in Fig. 3a). The free energy surface projected onto the total number of dimeric native contacts (QTotal) and the separation distance (the distance between the center of mass of the two subunits) shows only two-free energy minima: one corresponding to two unfolded chains that can be very apart from each other (U), and the other to a compact folded dimer (D). For larger two-state homodimers (Fig. 2 c–e) partially folded monomers, which are marginally stable, are also detected. Although their associations are thermodynamically two-state, kinetic intermediates are involved. Consistent with equilibrium denaturation studies of FIS protein, the simulations indicate the existence of an unstable monomeric intermediate in the absence of urea (36). Consistent with the simulations, an obligatory thermodynamic dimeric intermediate has also been experimentally detected during the association of the unstable monomers of the domain of Escherichia coli Trp repressor (residues 2–66) (Figs. 3b and 4f). We find that the intermediate is rather asymmetric and is probably consists of a dimer with a single fully folded subunit, another 60% folded subunit, and ≈80% of the interfacial contacts formed.
Fig. 2.
Free energy surfaces of folding and binding of obligatory (two-state) dimers. Free energy surfaces of the simulated homodimers are plotted as a function of the intrasubunit native contacts (QA and QB), intersubunit native contacts, (Qinterface), the total number of native contacts (QTotal), and the separation distance between the two chains [Rcm(A)–Rcm(B)]. The simulations reproduce the experimentally inferred mechanisms regarding the coupling between folding and binding: the monomers constitute the dimers fold concurrently with their binding. The free energy surfaces are calculated at their transition temperatures (the folded and unfolded states have identical free energy values) defined by the peak of the specific heat profile as a function of temperature: 0.94 ε, 1.08 ε, 1.06 ε, 1.13 ε, and 1.21 ε for Troponin C site III, Arc repressor, Factor for inversion stimulation, gene V protein, and β nerve growth factor, respectively. The free energy is in units of ε. We note that an unfolded monomer is not entirely unfolded but is partially structured, containing ≈20–40% of the native contacts.
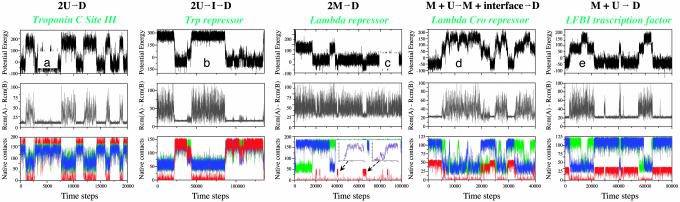
Fig. 3.
Typical trajectories of folding and association of representative homodimers presented in Figs. 2 and 4. The time evolution of the potential energy, the separation distance, as well as QA (green), QB (blue), and QInterface (red), illustrate the coupling between folding and binding [Troponin C site II (a), Arc repressor (b), and Trp repressor (c)], binding of two already folded monomers [λ repressor (d)], and the cases where recognition occurs by an unfolded subunit [λ Cro repressor (e) and LFB1 transcription factor (f)]. All of the trajectories are at the same temperatures as the corresponding free energy surfaces (Figs. 2 and 4).
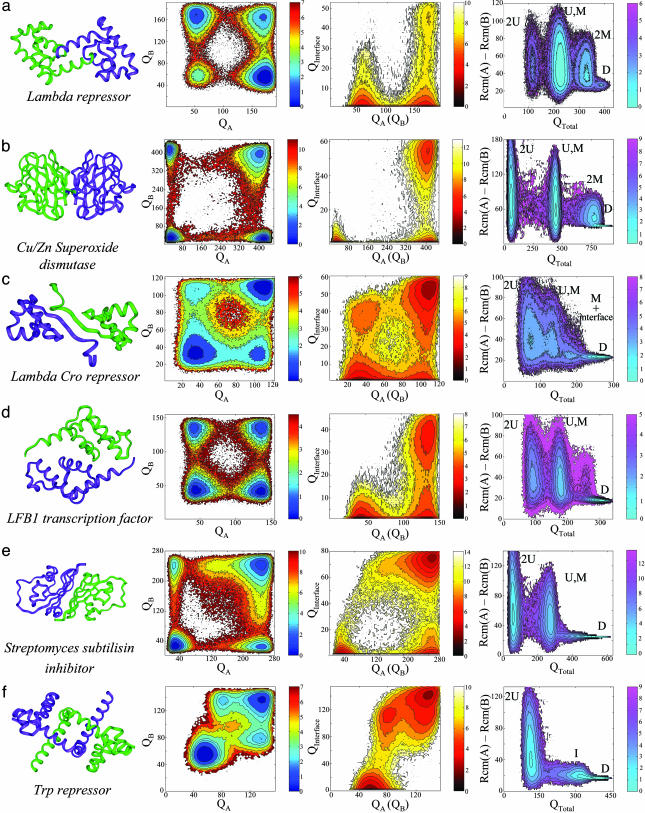
Fig. 4.
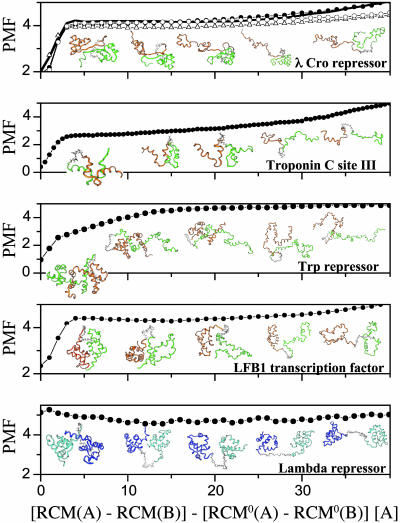
Free energy surfaces of folding and binding of nonobligatory (three-state) dimers. An unbound folded monomer exist for the three-state dimers (a–e) [except for Trp repressor (f) with a dimeric intermediate]. The formation of some three-state dimers [λ Cro repressor (c), LFB1 transcription factor (d), and S. subtilisin inhibitor (e)] preferentially occurs by binding between folded and unfolded monomers and not by binding two already folded chains as found for λ repressor (a) and Cu/Zn superoxide dismutase (b). For Trp repressor, a coupling between folding and binding with a dimeric intermediate is observed. For three-state homodimers (may have more than a single peak in the specific heat curve), the free energy surfaces were plotted at temperatures in which the unfolded and folded states have the same free energy: 0.99 ε, 1.27 ε, 0.99 ε, 1.20 ε, 0.96 ε, and 1.04 ε for λ repressor, Cu/Zn superoxide dismutase, λ Cro repressor, LFB1 transcription factor, S. subtilisin inhibitor, and Trp repressor, respectively. The free energy is in units of ε.
Association of Nonobligatory Dimers: Induced-Fit and Beyond
The dimers (three-state) that, in the laboratory, bind from folded monomers are also found to follow a three-state mechanism in the simulation. However, on closer inspection, the formation of some of these homodimers cannot be described completely by the traditional mechanisms. Instead, association occurs often between folded and unfolded monomers. In these simulations, the formation of dimeric λ repressor from two unfolded chains is decoupled from the monomer folding, and several different configurations are observed along a folding trajectory. Among these, conformations with a single folded monomer (high QA and low QB and vice versa) or with two folded monomers (high QA and QB) were detected (Figs. 3c and 4a). Moreover, conformations with two folded monomers exist with incompletely formed interfaces (high QA and QB together with low QInterface), reflecting the fact that the monomers are autonomous entities. The free energy surface for λ repressor projected onto QTotal and the separation distance (Fig. 4a) indicates the existence of four states: two unfolded subunits (U), a single folded monomer (M), two folded monomers (2M), and a folded dimer (D). A similar association mechanism was also found for Cu/Zn superoxide dismutase (Fig. 4b). The gradual increase of the number of interfacial native contacts during a binding event (Fig. 3c) indicates that dimer formation follows binding when two already folded monomers successfully collide and later adjust their relative orientation to optimize the interface.
For dimeric λ Cro repressor, LFB1 transcription factor, and Streptomyces subtilisin inhibitor, binding dominantly takes place between unfolded and folded chains (Figs. 4 c–e). For λ Cro repressor, the four states that constitute its free energy surface correspond to unfolded and folded dimer, a single folded monomer, and a folded monomer with a formed interface with an unfolded monomer. The transient complex between folded and unfolded chains can either result in a folded dimer by a folding of the bound unfolded subunit or be lead to separation of folded and unfolded chains (Fig. 3d). The free energy surfaces for λ Cro repressor were studied also by using longer linkers of 12, 20, and 30 glycine residues and without a linker, where the distance between the monomer center of mass is constrained, giving the same results (see supporting information). Just as for λ Cro repressor, symmetric LFB1 transcription factor (Fig. 4d) and S. subtilisin inhibitor (Fig. 4e) dimerize by a partially coupled binding and folding with an asymmetric association pathway. In contrast to λ Cro repressor, the transient complex between folded and unfolded chains is hardly populated because its formation is accompanied by a fast folding of the unfolded subunit (accordingly, only three states with low free energy constitute their free energy surfaces). The dimerization of these homodimers illustrates that, although a single monomer may fold irrespective of each other, folding is faster after its binding because the already-folded subunit acts as a template for folding of the unfolded chain. The folding of an isolated monomer has a larger free energy barrier than folding as a fused homodimer (supporting information). The high local concentration manifested here by the linker allows folding to be assisted by other subunits. The enhanced folding is reminiscent of catalytic folding via prosequences (37).
The Fly-Casting Mechanism
The applicability of the fly-casting speedup (25) for association scenarios was examined by plotting the free energy as a function of separation distance between the two subunits. A gradual decrease of the free energy indicates a long-range attraction that involves partially unfolded monomer. Fig. 5 demonstrates the fly-casting speedup for four homodimers that are experimentally classified as either two- or three-state dimers, but in our study, all are found to involve speedier recognition by unfolded chain. An attraction is observed when the separation distance between the two subunits is ≈30 Å larger than the native separation distance. The strongest fly-casting effect is detected for Trp repressor where an unfolded chain folds after swapping a helix and forming a stable interface. We predict that a similar recognition mechanism is common to other domain-swapped oligomers where unfolding of the folded monomers is a prerequisite for the oligomer formation (38), presumably via a fly-casting mechanism. For λ repressor where association occurs between already folded subunits that recognize each other upon collision, the fly-casting mechanism was not observed and no significant decrease in the free energy was found for any separation distance. Presumably, the difference from the free energy functional study (25) arises because the DNA is left out of the present calculation.
Fig. 5.
Free energy profiles (units of ε) as a function of the separation distance between the two chains. Note that for each dimer the separation distance was shifted by subtracting the separation distance in the native dimer. A gradual decrease of the free energy indicates a weak interaction between at least a single unfolded chain and its target, showing that binding occurs by the fly-casting mechanism [for λ Cro repressor, the profiles are shown for linkers of 12 (filled circles), 20 (triangle), and 30 (open circles) glycine residues and without a linker (solid line), indicating similar effect starting at separation distance of 20 Å]. On the other hand, a flat free energy profile indicates a collision between two folded chains. For each dimer, the snapshots illustrate conformations with a shifted separation distance of 30, 20, 10, 5, and 0 Å (for clearer representation, the backbone was added to each Cα conformation).
Conclusions
Our simulation survey of the formation of 11 homodimers in all cases reproduces their experimentally binding mechanisms by using a simple model. In particular, the agreement between the binding mechanisms found in experiment and from simulations with energetically minimally frustrated models (corresponding to a perfectly funneled landscape) is a strong indication that binding processes have funneled landscapes. Our study shows that the binding mechanism is robust and is governed by protein topology. As for protein folding, topology is an important factor for the protein binding mechanism, and the degree of topological frustration of a monomer determines whether the binding will occur between two unfolded or folded chains. Although, as the phase diagram (Fig. 1) indicates, the two-state or three-state mechanism of the dimer binding/folding is determined by a simple count of contacts, along with their fractional hydrophobic character, the detailed mechanisms are more subtle but apparently follow from topology alone. Several proteins with few interfacial contacts exhibit fly-casting (e.g., λ Cro repressor), whereas others bind via induced fit (e.g., λ repressor), and a higher-order characterization of the topology of the network of interactions at the surface would be needed to quantify this distinction. Moreover, our survey complements the experiments because it predicts that, for some dimers, the unfolded monomer may play an important role in binding the other subunit even when the monomer is stable on its own. This observation supports the accumulating evidence for the critically important role of unfolded proteins in cell biology (20, 39). The asymmetric pathway of association of two identical chains through an unfolded intermediate shows the speedup postulated in the fly-casting mechanism, which provides a leading explanation for the biological advantage, and therefore prevalence, of unfolded proteins. This energy landscape framework may be applicable to a wide range of cellular binding processes. The distinct binding mechanisms observed here for homodimers depends not only on the protein topology, but also on the concentrations, which are relatively high in the present dimer simulations because of the constraint that is applied on the two monomers. Similar behavior has been recently reported for protein aggregation by using lattice simulations (16). Dynamics and plasticity are indispensable for bimolecular recognition, and there is a much broader spectrum of binding scenarios than previously imagined.
Supplementary Material
Acknowledgments
This work has been funded by the National Science Foundation-sponsored Center for Theoretical Biological Physics (Grants PHY-0216576 and 0225630) with additional support from MCB-0084797. Y.L also acknowledges support from the Rothschild and Fulbright foundations.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jones, S. & Thornton, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wodak, S. & Janin, J. (2002) Adv. Protein Chem. 61, 9–72. [DOI] [PubMed] [Google Scholar]
- 3.Ofran, Y. & Rost, B. (2003) J. Mol. Biol. 325, 377–387. [DOI] [PubMed] [Google Scholar]
- 4.Solomon, B., Taraboulos, A. & Katchalski-Katzir, E. (2001) Conformational Diseases—A Compendium (Bialik, Jerusalem).
- 5.Onuchic, J. N., Socci, N. D., Luthey-Schulten, Z. & Wolynes, P. G. (1996) Fold. Des. 1, 441–450. [DOI] [PubMed] [Google Scholar]
- 6.Clementi, C., Nymeyer, H. & Onuchic, J. N. (2000) J. Mol. Biol. 298, 937–953. [DOI] [PubMed] [Google Scholar]
- 7.Koga, N. & Takada, S. (2001) J. Mol. Biol. 313, 171–180. [DOI] [PubMed] [Google Scholar]
- 8.Baker, D. (2000) Nature 405, 39–42. [DOI] [PubMed] [Google Scholar]
- 9.Lee, J. C., Chang, I. J., Gray, H. B. & Winkler, J. R. (2002) J. Mol. Biol. 320, 159–164. [DOI] [PubMed] [Google Scholar]
- 10.Fersht, A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman, New York).
- 11.Clementi, C., Jennings, P. A. & Onuchic, J. N. (2000) Proc. Natl. Acad. Sci. USA 97, 5871–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda, Y., Taketomi, H. & Go, N. (1978) Biopolymers 17, 1531–1548. [Google Scholar]
- 13.Tsai, C.-J., Kumar, S., Ma, B. & Nussinov, R. (1999) Protein Sci. 8, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkhivker, G. M., Bouzida, D., Gehlhaar, D. K., Rejto, P. A., Freer, S. T. & Rose, P. W. (2002) Curr. Opin. Struct. Biol. 12, 197–203. [DOI] [PubMed] [Google Scholar]
- 15.Papoian, G. A. & Wolynes, P. G. (2003) Biopolymers 68, 333–349. [DOI] [PubMed] [Google Scholar]
- 16.Dima, R. I. & Thirumalai, D. (2002) Protein Sci. 11, 1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshland, D. E. J. (1958) Proc. Natl. Acad. Sci. USA 44, 98–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma, B., Kumar, S., Tsai, C.-J. & Nussinov, R. (1999) Protein Eng. 12, 713–720. [DOI] [PubMed] [Google Scholar]
- 19.Bosshard, H. R. (2001) News Physiol. Sci. 16, 171–173. [DOI] [PubMed] [Google Scholar]
- 20.Dyson, H. J. & Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60. [DOI] [PubMed] [Google Scholar]
- 21.Dunker, A. K., Lawson, J. D., Brown, C. J., Williams, R. M., Romero, P., Oh, J. S., Oldfield, C. J., Campen, A. M., Ratliff, C. M., Hipps, K. W., et al. (2001) J. Mol. Graphics Model. 19, 26–59. [DOI] [PubMed] [Google Scholar]
- 22.Uversky, V. N. (2002) Protein Sci. 11, 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spolar, R. & Record, M. (1994) Science 263, 777–784. [DOI] [PubMed] [Google Scholar]
- 24.Kirwacki, R., Hengst, L., Tennant, L., Reed, S. & Wright, P. (1996) Proc. Natl. Acad. Sci. USA 93, 11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoemaker, B. A., Portman, J. J. & Wolynes, P. G. (2000) Proc. Natl. Acad. Sci. USA 97, 8868–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung, M. S., Garcia, A. E. & Onuchic, J. N. (2002) Proc. Natl. Acad. Sci. USA 99, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neet, K. E. & Timm, D. E. (1994) Protein Sci. 3, 2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, D., Tsai, C.-J. & Nussinov, R. (1998) Protein Sci. 7, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakhnovich, E. I. (1999) Nat. Struct. Biol. 6, 99–102. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, C.-J., Xu, D. & Nussinov, R. (1997) Protein Sci. 6, 1793–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber, G. & Fersht, A. (1996) Nat. Struct. Biol. 3, 427–431. [DOI] [PubMed] [Google Scholar]
- 32.Sheinerman, F. B., Norel, R. & Honig, B. (2000) Curr. Opin. Struct. Biol. 10, 153–159. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, C. R. & Sauer, R. T. (1996) Biochemistry 35, 13878–13884. [DOI] [PubMed] [Google Scholar]
- 34.Liang, H., Sandberg, W. S. & Terwilliger, T. C. (1993) Proc. Natl. Acad. Sci. USA 90, 7010–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrenberg, A. M. & Swendsen, R. H. (1989) Phys. Rev. Lett. 63, 1195–1198. [DOI] [PubMed] [Google Scholar]
- 36.Hobart, S. A., Ilin, S., Moriarty, D. F., Osuna, R. & Colon, W. (2002) Protein Sci. 11, 1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryan, P. N. (2002) Chem. Rev. 102, 4805–4816. [DOI] [PubMed] [Google Scholar]
- 38.Rousseau, F., Schymkowitz, J. W. H. & Itzhaki, L. S. (2003) Structure (London) 11, 243–251. [DOI] [PubMed] [Google Scholar]
- 39.Matouschek, A. (2003) Curr. Opin. Struct. Biol. 13, 98–109. [DOI] [PubMed] [Google Scholar]
- 40.Monera, O. D., Shaw, G. S., Zho, B.-Y., Sykes, B. D., Kay, C. M. & Hodges, R. S. (1992) Protein Sci. 1, 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowie, J. U. & Sauer, R. T. (1989) Biochemistry 28, 7139–7143. [DOI] [PubMed] [Google Scholar]
- 42.Liang, H. & Terwilliger, T. C. (1991) Biochemistry 30, 2772–2782. [DOI] [PubMed] [Google Scholar]
- 43.Timm, D. E., Haseth, P. L. D. & Neet, K. E. (1994) Biochemistry 33, 4667–4676. [DOI] [PubMed] [Google Scholar]
- 44.Huang, G. & Oas, T. (1995) Proc. Natl. Acad. Sci. USA 92, 6878–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jana, R., Hazbun, T. R., Mollah, A. K. M. M. & Mossing, M. C. (1997) J. Mol. Biol. 273, 402–416. [DOI] [PubMed] [Google Scholar]
- 46.Leiting, B., Francesco, R. D., Tomei, L., Cortese, R., Otting, G. & Wuthrich, K. (1993) EMBO J. 12, 1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akasaka, K., Fujii, S., Hayashi, F., Rokushika, S. & Hatano, H. (1982) Biochem. Int. 5, 637–642. [Google Scholar]
- 48.Mei, G., Rosato, N., Silva, N., Rusch, R., Gratton, E., Savini, I. & Finazzi-Agro, A. (1992) Biochemistry 31, 7224–7230. [DOI] [PubMed] [Google Scholar]
- 49.Gloss, L. M. & Matthews, C. R. (1998) Biochemistry 37, 16000–16010. [DOI] [PubMed] [Google Scholar]
- 50.Pacios, L. (2001) J. Chem. Inf. Comput. Sci. 41, 1427–1435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.