Abstract
Over the last decade, the introduction of microarray technology has had a profound impact on gene expression research. The publication of studies with dissimilar or altogether contradictory results, obtained using different microarray platforms to analyze identical RNA samples, has raised concerns about the reliability of this technology. The MicroArray Quality Control (MAQC) project was initiated to address these concerns, as well as other performance and data analysis issues. Expression data on four titration pools from two distinct reference RNA samples were generated at multiple test sites using a variety of microarray-based and alternative technology platforms. Here we describe the experimental design and probe mapping efforts behind the MAQC project. We show intraplatform consistency across test sites as well as a high level of interplatform concordance in terms of genes identified as differentially expressed. This study provides a resource that represents an important first step toward establishing a framework for the use of microarrays in clinical and regulatory settings.
Recently, pharmacogenomics and toxicogenomics have been identified both by the US Food and Drug Administration (FDA) and the US Environmental Protection Agency (EPA) as key opportunities in advancing personalized medicine1,2 and environmental risk assessment3. These agencies have issued guidance documents to encourage scientific progress and to facilitate the use of these data in drug development, medical diagnostics and risk assessment (http://www.fda.gov/oc/initiatives/criticalpath/; http://www.fda.gov/cder/guidance/6400fnl.pdf; http://www.fda.gov/cdrh/oivd/guidance/1549.pdf; http://www.epa.gov/osa/genomics.htm). However, although DNA microarrays represent one of the core technologies for this purpose, concerns have been raised regarding the reliability and consistency, and hence potential application of microarray technology in the clinical and regulatory settings. For example, a widely cited study reported little overlap among lists of differentially expressed genes derived from three commercial microarray platforms when the same set of RNA samples was analyzed4. Similar low levels of overlap have been reported in other interplatform and/or cross-laboratory microarray studies5–8.
Although similar results continue to appear in peer-reviewed journals9,10, raising doubts about the repeatability, reproducibility and comparability of microarray technology11–13, several studies have also been recently published showing increased reproducibility of microarray data generated at different test sites and/or using different platforms14–18. It follows that before this technology can be applied in clinical practice and regulatory decision making, microarray standards, quality measures and consensus on data analysis methods need to be developed2,19–21.
Here we describe the MAQC project, a community-wide effort initiated and led by FDA scientists involving 137 participants from 51 organizations. In this project, gene expression levels were measured from two high-quality, distinct RNA samples in four titration pools on seven microarray platforms in addition to three alternative expression methodologies. Each microarray platform was deployed at three independent test sites and five replicates were assayed at each site. This experimental design and the resulting data set provide a unique opportunity to assess the repeatability of gene expression microarray data within a specific site, the reproducibility across multiple sites and the comparability across multiple platforms. Objective assessment of these technical metrics is an important step towards understanding the appropriate use of microarray technology in clinical and regulatory settings. This study also addresses many other needs of the scientific community pertaining to the use and analysis of microarray data (see MAQC goals in Supplementary Data online).
The MAQC project has generated a rich data set that, when appropriately analyzed, reveals promising results regarding the consistency of microarray data between laboratories and across platforms. In this article, we detail the study design, describe its implementation and summarize the key findings of the MAQC main study. The accompanying set of articles22–26 provides additional analyses and related data sets. Although the sample types used in this study are not directly representative of a relevant biological study, the study provides technical insights into the capabilities and limitations of microarray technology. Similar levels of concordance in cross-laboratory and interplatform comparisons have been independently reported using a toxicogenomics study26.
RESULTS
Experimental design
The MAQC project (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/) repeatedly assayed four pools comprised of two RNA sample types on a variety of gene expression platforms and at multiple test sites. The two RNA sample types used were a Universal Human Reference RNA (UHRR) from Stratagene and a Human Brain Reference RNA (HBRR) from Ambion. The four pools included the two reference RNA samples as well as two mixtures of the original samples: Sample A, 100% UHRR; Sample B, 100% HBRR; Sample C, 75% UHRR:25% HBRR; and Sample D, 25% UHRR75% HBRR. This combination of biologically different RNA sources and known titration differences provides a method for assessing the relative accuracy of each platform based on the differentially expressed genes detected. A unique feature of the MAQC project is that both sample type A and sample type B are commercially available to the community for a few years to come in the exact batches as those used by the MAQC project.
Six commercially available microarray platforms were tested: Applied Biosystems (ABI); Affymetrix (AFX); Agilent Technologies (AGL for two-color and AG1 for one-color); GE Healthcare (GEH); Illumina (ILM) and Eppendorf (EPP). In addition, scientists at the National Cancer Institute (NCI) generated spotted microarrays using oligonudeotides obtained from Operon. The RNA sample types were also tested on three alternative gene expression platforms: TaqMan Gene Expression Assays from Applied Biosystems (TAQ TaqMan is a registered trademark of Roche Molecular Systems, Inc.); StaRT-PCR from Gene Express (GEX) and QuantiGene assays from Panomics (QGN).
Each microarray platform provider selected three sites for testing. In most cases, five replicate assays for each of the four sample types were processed at each of the test sites. Six of the microarray providers used one-color protocols where one labeled RNA sample was hybridized to each microarray (Table 1). The Agilent two-color and NCI microarrays were tested using a two-color protocol so that two differently labeled RNA samples were simultaneously hybridized to the same microarray. The Eppendorf assay contained two identical microarrays on one glass slide, which were independently hybridized to two samples. Although only a single fluorescent dye was used, the Eppendorf data are presented in a ratio format.
Table 1.
Gene expression platforms and data analyzed in the MAQC main study
| Manufactuer | Code | Protocol | Platform | Number of probesa |
Number of test sites |
Number of samples |
Number of replicates |
Total number of microarraysb |
|---|---|---|---|---|---|---|---|---|
| Applied Biosystems | ABI | One-color microarray | Human Genome Survey Microarray v2.0 | 32,878 | 3 | 4 | 5 | 58 |
| Affymetrix | AFX | One-color microarray | HG-U133 Plus 2.0 GeneChip | 54,675 | 3 | 4 | 5 | 60 |
| Agilent | AGL | Two-color microarrayc | Whole Human Genome Oligo Microarray, G4112A | 43,931 | 3 | 2 | 10 | 56 |
| AG1 | One-color microarray | Whole Human Genome Oligo Microarray, G4112A | 43,931 | 3 | 4 | 5 | 56 | |
| Eppendorf | EPP | One-color microarray | DualChip Microarray | 294 | 3 | 4 | 5 | 60 |
| GE Healthcare | GEH | One-color microarray | CodeLink Human Whole Genome, 300026 | 54,359 | 3 | 4 | 5 | 60 |
| Illumina | ILM | One-color microarray | Human-6 BeadChip,48K v1.0 | 47,293 | 3 | 4 | 5 | 59 |
| NCI_Operon | NCI | Two-color microarray | Operon Human Oligo Set v3 | 37,632 | 2 | 4 | 5 | 33 |
| Applied Biosystems | TAQ | TaqMan assays | >200,000 assays available | 1,004 | 1 | 4 | 4 | N/A |
| Panomics | QGN | QuantiGene assays | ~ 2,600 assays available | 245 | 1 | 4 | 3 | N/A |
| Gene Express | GEX | StaRT-PCR assays | ~ 1,000 assays available | 207 | 1 | 4 | 3 | N/A |
| Total | 442 |
A global definition of probes is used to include individual probes, probe sets or primer pairs depending on the gene expression platform. The numbers listed in this table are derived from product literature and may include some platform duplication. Alternative figures for the number of probes analyzed are provided as Table S5 in Supplementary Data online.
Maximum number of microarrays per one-color protocol is 60 (3 sites × 4 sample types × 5 replicates). As described in the text, replacement hybridizations but not outlier hybridizations are included in the main study data analysis. Only data from 386 microarrays were analyzed in this article. Additional data sets are described in Table S4 in Supplementary Data online.
Although not presented in this paper, the Agilent two-color data (56 microarrays) are discussed elsewhere24. In the remaining figures, test sites and sample types are referenced using the following nomenclature: “platform code_test site_sample ID”. Sample A = 100% UHRR; Sample B = 100% HBRR; Sample C = 75% UHRR: 25% HBRR; and Sample D = 25% UHRR: 75% HBRR.
Each microarray provider used its own software to generate a quantitative signal value and a qualitative detection call for each probe on the microarray. This attention to the qualitative calls of each platform resulted in our using a potentially different number of genes in each calculation. It also had an impact on data analysis, because some, but not all, of the platforms removed suspect or low intensity data. In addition, 11 hybridizations were removed from further analysis due to quality issues. Table 1 notes the final number of hybridizations used in the final data analysis for each microarray platform. Further details are presented in Methods and Tables S1–S4 in Supplementary Data online. Pre-hybridization and post-hybridization quality information of samples is available as Supplementary Table 1 online.
A direct comparison of results across platforms was challenging because of inherent differences in protocols, number of data points per platform and data preprocessing methods. Whenever possible, all platforms were included in any comparisons, but occasionally results from one or two platforms were excluded from an analysis because the data comparison was untenable and forced contrivance that was ultimately uninformative. Although some data from the alternative platforms are presented in this article, a more thorough discussion is included elsewhere22.
Probe mapping
Microarray experiments generally rely on a hybridization intensity measurement for an individual probe to infer a transcript abundance level for a specific gene. This relationship raises several difficult issues, including: which gene corresponds to which probe, and how sensitive and specific is the probe. Previous publications have suggested that some of the variability in cross-platform studies was due to annotation problems that made it difficult to reconcile which genes were measured by specific probes27–30. Despite the fact that the human genome sequence is complete, the final list of actual genes has yet to be determined. All identifiers are moving targets, and even the NCBI hand-curated reference sequences are often modified. Another issue is that a gene expression assay designed to measure a given RNA target may unknowingly detect multiple alter-natively spliced transcripts, which may have different functions and expression patterns. Thus, the number of genes or transcripts detected with a gene expression platform is inherently difficult to define and quantify.
A unique advantage of the MAQC project is that most of the sequence information for the probes used in each gene expression technology was provided by the manufacturers. We mapped the probes (see Supplementary Methods online and Supplementary Notes online) to the RefSeq human mRNA database31 (http://www.ncbi.nlm.nih.gov/RefSeq) and to the AceView database32 (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly), a less curated but more comprehensive database, which includes all the RefSeq, GenBank and dbEST human cDNA sequences. Although the total number of probes varied across platforms, the six high-density microarray platforms assayed similar numbers of Entrez genes (15,429–16,990) and had similar percentages of probes (68–84%) that aligned to AceView transcripts (see Table S5 in Supplementary Data online). We found that 23,971 of the 24,157 RefSeq NM Accessions from the March 8, 2006 release were assayed by at least one platform (Supplementary Table 2 online) and that 15,615 Accessions were assayed by all high-density microarray platforms used in the MAQC study. Because of alternative splicing, each platform mapped to roughly four RefSeq transcripts per three Entrez genes.
To simplify the interplatform comparison, we condensed the complex probe-target relationships to a ‘one-probe-to-one-gene’ list. The 15,615 RefSeq entries on all of the high-density microarray platforms represented 12,091 Entrez genes. For each gene, we selected a single RefSeq entry (Supplementary Table 4 online), primarily the one annotated by TaqMan assays, or secondarily the one targeted by the majority of platforms. When a platform contained multiple probes matching the same RefSeq entry, only the probe closest to the 3′ end was included in the common set (Supplementary Table 3 online). In this way, we selected for each high-density platform 12,091 probes matching a common set of 12,091 reference sequences from 12,091 different genes (Supplementary Table 5 online).
Intraplatform data repeatability and reproducibility
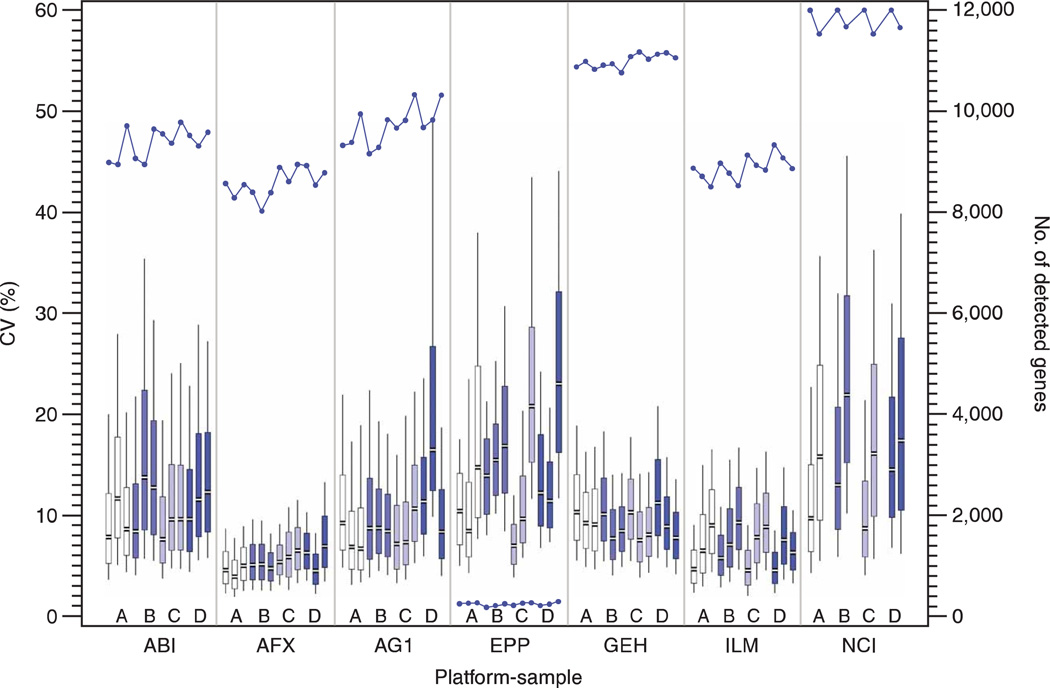
We examined microarray data for consistency within each platform by reviewing both the intrasite repeatability and the intersite reproducibility at two levels: the quantitative signal values and the qualitative detection calls. Only genes that were detected in at least three of the five sample replicates (or generally detected genes) were included in most of these calculations. This filter accounts for the different manner in which the microarray platforms identified genes below their quality thresholds, and directs our research away from the less confident, noisy results. The number of generally detected genes for each sample type at each site varied from 8,000 to 12,000 for the high-density microarray platforms, but was relatively consistent between test sites using the same platform (Fig. 1).
Figure 1.
Repeatability of expression signal within test sites. For the one-color platforms, the CV of the expression signal values between site replicates of the same sample type was calculated for all generally detected genes. The distributions of these replicate CVs are presented in a series of twelve box and whiskers plots for each microarray platform: one for each of the four sample types at the three test sites. The plots are highlighted to distinguish the sample replicates: sample A (white), sample B (light blue), sample C (light purple) and sample D (dark blue). The twelve plots showing results from the platforms with three test sites are presented in the following order from left to right: A1, A2, A3, B1, B2, B3, C1, C2, C3, D1, D2 and D3. For the two-color NCI platform, the CV of the expression Cy3/Cy5 ratios between site replicates of the same sample type was similarly calculated. The distributions of these replicate CVs are presented in a series of eight box and whiskers plots from the two NCI test sites in the following order from left to right: A1, A2, B1, B2, C1, C2, D1, and D2. The median (gap), interquartile range as well as the 10th and 90th percentile values are indicated in each plot. Only genes from the 12,091 common set that were detected in at least three of the replicates were included in the box plots and CV calculations. This number varies by platform/sample/test site and is noted as the line plot with the secondary axis and as Table S6 in Supplementary Data online. The platforms and sample types are labeled according to the nomenclature presented in Table 1.
The coefficient of variation (CV) of the quantitative signal values between the intrasite replicates was calculated using the generally detected subset from the 12,091 common genes for each sample type at every test site. The distribution of the replicate CV measures across the set of detected genes is displayed in a series of box and whiskers plots in Figure 1. Most of the one-color microarray platforms and test sites demonstrated similar replicate CV median values of 5–15%, although the distributions of replicate CV results differed between platforms. For the two-color NCI microarrays, the replicate CVs were calculated using the Cy3/Cy5 ratios. (Sample type A was used as the Cy5 reference in all NCI hybridizations.) These values were only slightly larger than the one-color signals for the same sample type.
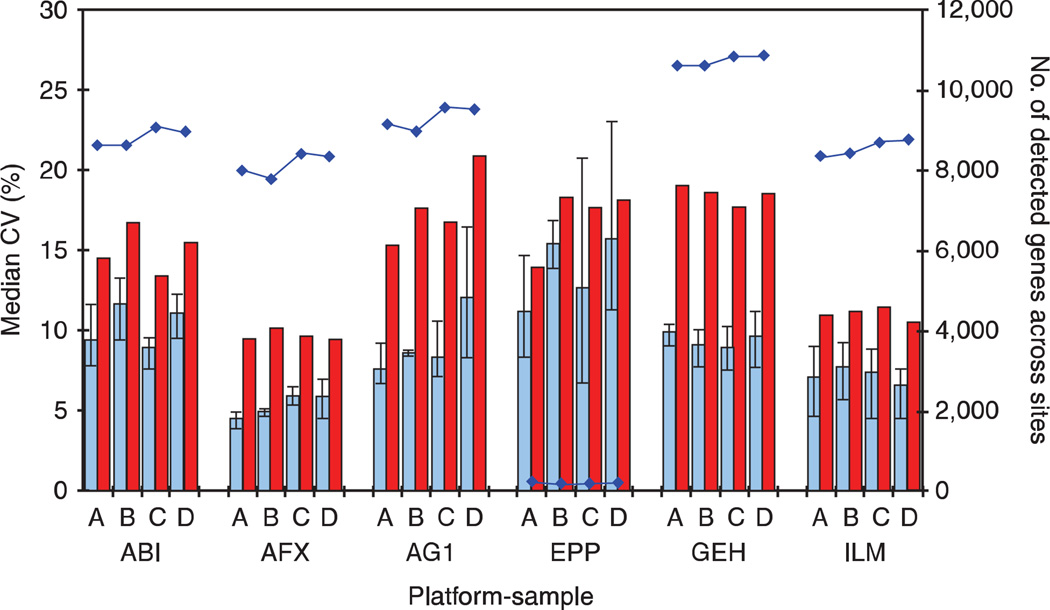
We next examined the total CV of the quantitative signal, which included both the intrasite repeatability as well as variation due to intersite differences. By definition, the total CV measure (n ≤ 15) will be larger than the replicate CV measures (n ≤ 5). Median values for the total CV distribution and the average of three replicate CV medians for each platform are presented in Figure 2. Overall, the total CV median was very consistent across all platforms, ranging from 10% to just over 20% and not dramatically higher than the replicate CV median values. In general, the total CV median was up to twice as large as the replicate CV median, but this result is not unexpected and simply implies that site-related effects should be taken into account when combining data from multiple sites using the same platform.
Figure 2.
Signal variation within and between test sites. For each of the four sample types, the replicate CV of signal within a test site (blue bar) and the total CV of signal across and within sites (red bar) are presented. As in Figure 1, genes detected in at least three of the replicates of a sample type at a single test site are included in the replicate CV calculation. Genes present in the intersection of these gene lists are included in the total CV calculation. (These gene lists are therefore slightly different than those in Figure 1.) The number of such genes within each platform and sample type is noted by blue dots connected by lines and is read on the secondary axis. It is also reported as Table S6 in Supplementary Data online. Intrasite normalization was performed according to default settings for each manufacturer, and intersite normalization was performed by scaling between sites (see main text). The NCI platform is omitted because data from only two test sites was available in the main study so intersite reproducibility measures may not be representative. The platforms and sample types are labeled according to the nomenclature presented in Table 1.
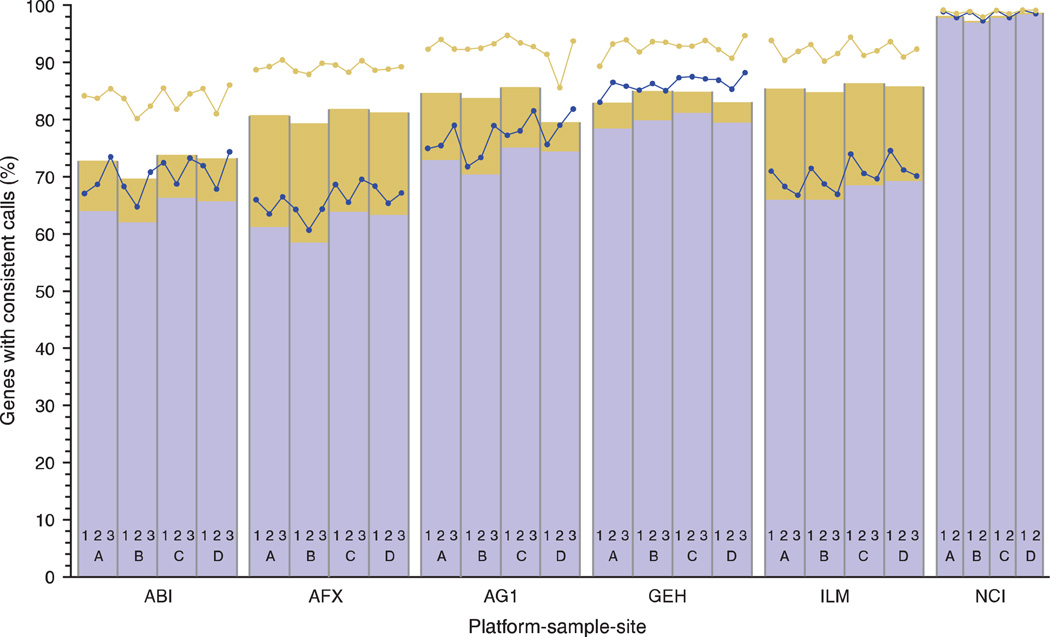
To assess variation in the qualitative measures, the percentage of the 12,091 common genes with concordant detection calls between replicates of the same sample type was calculated for each of the four sample types on each platform (Fig. 3). These figures include either all sample replicates at a single site (n ≤ 5) or all sample replicates across the test sites (n ≤ 15). Most one-color test sites demonstrated 80–95% concordance in the qualitative calls for the sample replicates within their facility. The value dropped to 70–85% concordance for the reproducibility of the qualitative calls across all three test sites. It is not surprising that platforms with more detected calls (Fig. 1) generally had higher concordance percentages. For example, the NCI microarrays detected almost all of the 12,091 common genes and had concordance percentages near 100% between test sites. Microarray platforms that had lower numbers of detected genes generally had reduced concordance percentages. Interestingly, the GE Healthcare platform had both a large number of genes detected (~ 11,000 per hybridization) and approximately 85% concordance between test sites.
Figure 3.
Concordance of detection calls within and between test sites. For the 12,091 common genes, detection calls within each platform were categorized as either ‘detected’ or ‘not detected.’ For each sample type within each platform, the percentage of genes with calls that were perfectly concordant as ‘detected’ within the replicates for a given site is plotted as blue dots, and the corresponding percentage of genes with calls perfectly concordant as ‘detected’ across all sites are plotted as the blue bars. The total percentage of genes with perfectly concordant calls (detected and not detected) within a site is plotted as the yellow dots, and the corresponding percentage of genes with calls perfectly concordant across all sites is plotted as the top of the yellow bars. The bars are split between perfectly detected genes (blue portion) and perfectly not detected genes (yellow portion) across all test sites. It is not expected that detected genes are concordant across sample types. The number of perfectly detected genes for each test site is provided as Table S6 in Supplementary Data online. As described in the main text, the stringency with which individual platforms determine that the data for a gene is sufficiently reliable to be called detected has different manufacturer defaults, leading to altered concordance percentages. Changes in the settings for sensitivity/specificity may shift the proportion of the bar assigned to each detection category. Because reliability depends on platform-specific details, detected calls do not correspond directly to relative abundance and may vary between platforms. Note: as some platforms have removed outlier hybridizations, the number of replicates within (n ≤ 5) and between sites (n ≤ 15) varies for determining concordance.
Interplatform data comparability
Expression values generated on different platforms cannot be directly compared because unique labeling methods and probe sequences will result in variable signals for probes that hybridize to the same target. Alternatively, the relative expression between a pair of sample types should be maintained across platforms. For this reason, we examined the microarray data for comparability between platforms by reviewing sample type B relative to sample type A expression values with three different metrics: differential gene list overlap, log ratio compression and log ratio rank correlation. For log ratio compression and rank correlation, only generally detected genes from the common 12,091 gene list were included in the analysis. For the gene list overlap, all 12,091 common genes were considered.
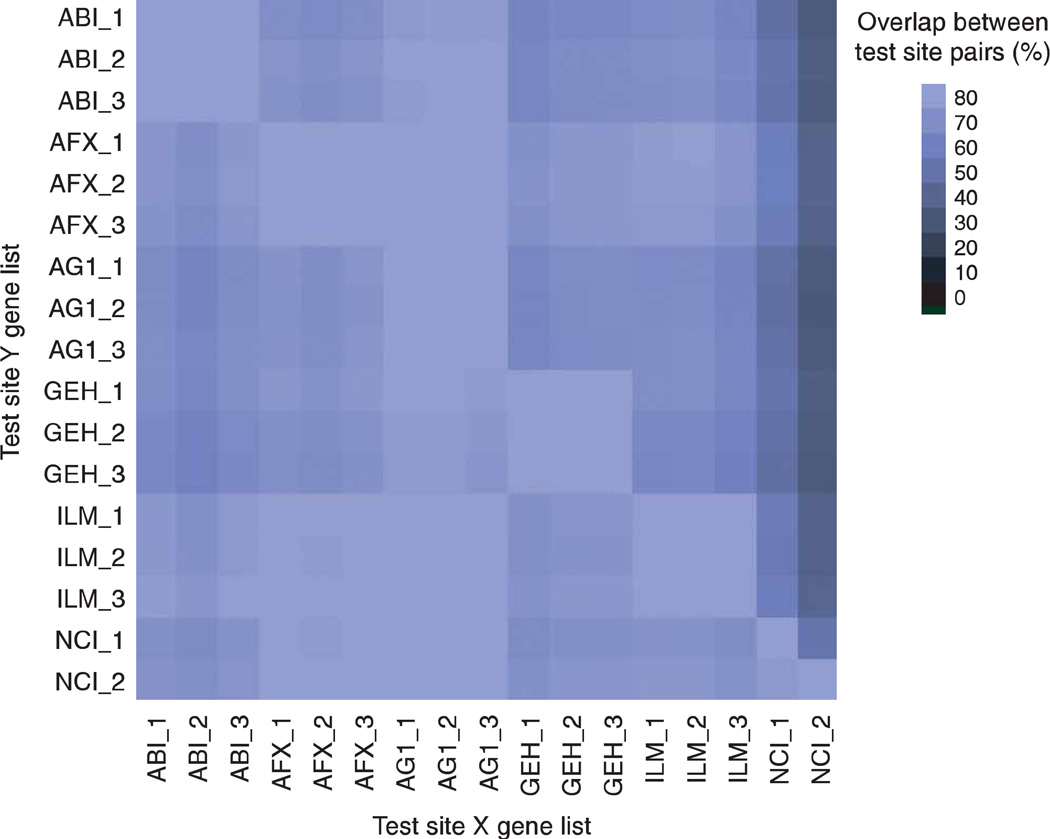
A list of differentially expressed genes was generated for each test site and compared to lists from other test sites using the same platform and those using a different platform. A percent score was calculated to indicate the number of genes in common between each pair of test site lists. The percentage of overlap for each comparison is displayed in Figure 4. Note the graphic comparisons are asymmetrical indicating the analysis is performed in two directions. That is, the percentage of test site Y genes on the list from test site X can be different from the percentage of test site X genes on the test site Y list. For all but the NCI test sites, the gene list overlap is at least 60% for each test site comparison (both directions) with many site pairings achieving 80% or more between platforms and 90% within platforms. Typically, the genes that the NCI microarray platform identified as differentially expressed were also identified on the other platforms, suggesting a low false positive rate for this platform. However, the converse was not necessarily true, most likely due to more log ratio compression observed in the NCI platform and the use of a stringent P-value threshold.
Figure 4.
Agreement of gene lists. This graph indicates the concordance of genes identified as differentially expressed for pairs of test sites, labeled as X and Y. A list of differentially expressed genes between sample type A replicates versus sample type B replicates was generated for each test site (using the 12,091 common genes with ≥ twofold change and P < 0.001 thresholds) and compared for commonality to other test sites. The size of these gene lists is reported as Table S7 in Supplementary Data online. No filtering related to the qualitative detection call was performed. The color of the square in the matrix reflects the percent overlap of genes on the list for the test site Y (listed in row) that are also present on the list for the test site X (listed in column). A light-colored square indicates a high percent overlap between the gene lists at both test sites. A dark-colored square indicates a low percent overlap, suggesting that most genes identified in site Y were not identified in site X. Numerical values for the percent overlap are presented as Table S9 in Supplementary Data online. Note: the graph is asymmetric and not complementary. Only the six high-density microarray platforms are presented. As described in the text, data from some platforms were omitted from these calculations because of quality issues. The platforms and sample types are labeled according to the nomenclature presented in Table 1.
The _1, _2 and _3 suffixes refer to test site location.
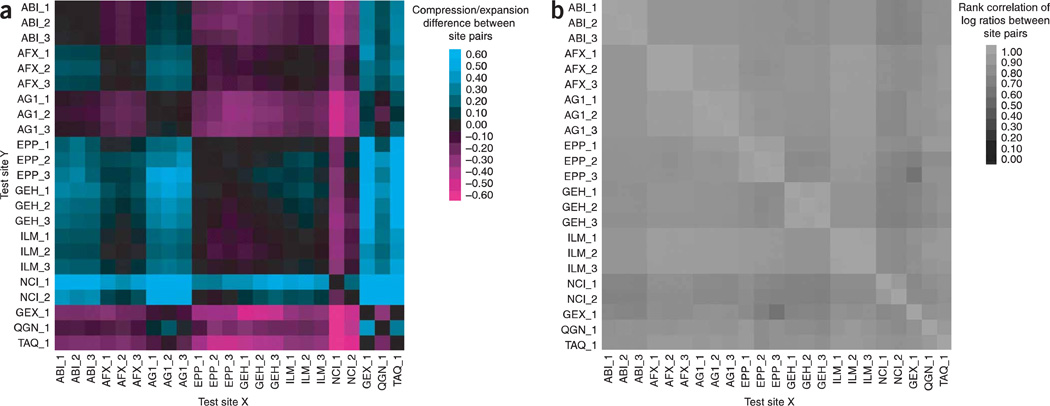
Each microarray platform has a defined background correction method and dynamic range of signal detection, which can lead to over- or underestimates of log ratios and fold changes in expression between sample types. To examine the level of compression or expansion in log ratios, we determined the best fitted line for the log ratio estimates between pairs of test sites. The percent difference of the slope for each comparison is displayed in Figure 5a. An ideal slope of 1 would result in a percent difference of 0; negative or positive percent differences in the slope of the ideal line indicate compression or expansion of the log ratios in one test site relative to the other. For each commercial one-color platform, good agreement was observed between its three test sites. Most of the interplatform test site comparisons also showed little compression or expansion. Test site 1 for the NCI microarrays produced consistently different results from the other test sites, both within and between platforms.
Figure 5.
Agreement of log ratios across platforms and test sites. (a) Log ratio compression/expansion. This graph indicates the percent difference from equivalency between platform/sites (corresponding to a slope value 1 for the best fitted line using orthogonal regression) of the log ratio differential expression using A and B replicates. A dark spot implies equivalency (slope = 1 → percent difference = 0). A positive percent difference in slope from the ideal line (aqua) indicates compression of log signal for test site Y relative to test site X. A negative percent difference in the ideal line (magenta) indicates expansion. Read as “What is the difference from equivalence in slope (m = 1) for the test site Y versus test site X ?” Only genes detected by both test sites in at least three replicates of sample type A and three replicates of sample type B are included in the calculation, and the number for each pair is reported as Table S8 in Supplementary Data online. Numerical values for the percent difference are presented as Table S10 in Supplementary Data online. Note: the graph is asymmetric, but approximately complementary. As described in the text, data from some platforms were omitted from these calculations due to quality issues. The platforms and sample types are labeled according to the nomenclature presented in Table 1. The _1, _2 and _3 suffixes refer to test site location, (b) Rank correlation of log ratios. This graph indicates the correlation of the log ratio differential expression values (using A versus B replicates) when we examine their rank. Large positive log ratio values would be ranked high and large negative log ratio values would be ranked low. Read as “What is the correlation of the rank log ratio values between the test site Y and the test site X?” Only genes generally detected in both sample types A and B and by both test sites are included in the calculation, and the number for each pair is reported as Table S8 in Supplementary Data online. Numerical values for the rank correlation are presented as Table S11 in Supplementary Data online. Note: the graph is symmetric. As described in the text, data from some platforms were omitted from these calculations due to quality issues. The platforms and sample types are labeled according to the nomenclature presented in Table 1.
The _1, _2 and _3 suffixes refer to test site location.
The comparability of results across platforms was also examined using a rank correlation metric. Log ratios for the differential expression observed between sample B replicates and sample A replicates were calculated for the generally detected common genes and then compared between test sites and across platforms. The rank correlations of the log ratios are displayed visually in Figure 5b. Good agreement was observed between all sites, even those using different microarray platforms. In fact, the median rank correlation was 0.87 and the smallest rank correlation value was 0.69 between the microarray platforms.
Assessing relative accuracy
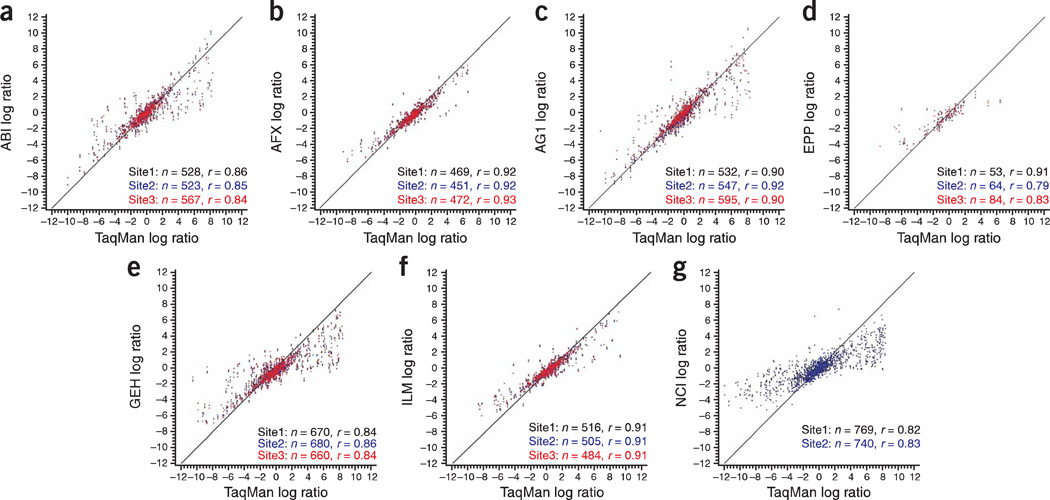
The relative accuracy of the microarray platforms can be assessed using either the titrated mixtures of the RNA samples23 or gene abundance measurements collected with alternative technologies22. Figure 5, as well as Tables S12 and S13 in Supplementary Data online, illustrate the relative rank correlation and compression/expansion values for log (B/A) between microarray-based and alternative gene expression technologies. Further comparisons between each microarray platform relative to the TaqMan assays are presented as scatter plots in Figure 6.
Figure 6.
Correlation between microarray and TaqMan data. The scatter plots compare the log ratio differential expression values (using A versus B replicates) from each microarray platform relative to values obtained by TaqMan assays. Each point represents a gene that was measured on both the microarray and TaqMan assays. The spot coloring indicates whether the data were generated in test site 1 (black), test site 2 (blue) or test site 3 (red) for the microarray platform. Only genes that were generally detected in sample type A replicates and sample type B replicates were used in the comparisons. The exact number of probes analyzed for each test site and its correlation to TaqMan assays are listed in the bottom right corner of each plot. As described in the text, data from some platforms were omitted from these calculations because of quality issues. The platforms and sample types are labeled according to the nomenclature presented in Table 1. The line shown is the ideal 45° line.
The log ratios of sample type B to sample type A expression detected on the TaqMan assays were compared to the log ratios obtained for the same genes on the microarray assays. Only genes that were generally detected in both sample A and B replicates on the TaqMan assays and on the microarray were included in this analysis. The relative accuracy of each high-density platform to the TaqMan assay data was generally higher for those microarray platforms with fewer genes detected as indicated by number and magnitude of deviations from the ideal 45° line indicated in Figure 5a and Figure 6.
Correlation with alternative platforms
Similarly, the Affymetrix, Agilent, and Illumina platforms displayed high correlation values of 0.90 or higher with TaqMan assays based on comparisons of ~ 450–550 genes, whereas the GE Healthcare and NCI platforms had a reduced average correlation of 0.84, but included almost 30% more genes in the data comparisons. These additional genes were not identified as ‘not detected’ during the data review process, but may represent less confident results due to lower signals exhibiting greater variance. Thus, much of the difference in comparability metrics may be a reflection of the algorithm used to assign detection calls. Similar correlation values for the microarray platforms were observed relative to each of the other alternative platforms, StaRT-PCR, and QuantiGene22.
DISCUSSION
The results of the MAQC project provide a framework for assessing the potential of microarray technologies as a tool to provide reliable gene expression data for clinical and regulatory purposes. All one-color microarray platforms had a median CV of 5–15% for the quantitative signal (Fig. 1) and a concordance rate of 80–95% for the qualitative detection call (Fig. 3) between sample replicates. This variation increased when data from different test sites using the same platform were included (Figs. 2 and 3). However, lists of differentially expressed genes averaged ~89% overlap between test sites using the same platform and ~74% overlap across one-color microarray platforms (Fig. 4). Importantly, the ranks of log ratios were highly correlated among the microarrays (minimum R = 0.69; Fig. 5b), indicating that all platforms were detecting similar changes in gene abundance. These results indicate that, for these sample types and these laboratories, microarray results were generally repeatable within a test site, reproducible between test sites and comparable across platforms, even when the platforms used probes with sequence differences as well as unique protocols for labeling and expression detection.
Within the MAQC study, there were notable differences in various dimensions of performance between microarray platforms. Some platforms had better intrasite repeatability overall (e.g., Illumina), better intersite reproducibility (e.g., Affymetrix), or more consistency in the detection calls (e.g., GE Healthcare). Likewise, some platforms were more comparable to TaqMan assays (e.g., Applied Biosystems and Agilent one-color), whereas others demonstrated signal compression (e.g., NCI_Operon). Some of these differences were manifest in the apparent power analyses (see Figure SI in Supplementary Data online) as test sites with smaller CV values (Fig. 1) typically had more power to discriminate differences between groups, as would be expected. Other differences might have been related to the platform’s signal-to-analyte response characteristics22. It is important to note that 11 (2.4%) of the 453 microarray hybridizations were removed from the analysis due to quality issues (listed as Table SI in Supplementary Data online). The relative performance of some platforms might have been altered if this data filter had not been applied.
Each microarray platform has made different trade-offs with respect to repeatability, sensitivity, specificity and ratio compression. One interesting result was that platforms with divergent approaches to measuring expression often generated comparable results. For example, data from Affymetrix test sites, which use multiple short oligonudeotide probes per target with perfect match and mismatch sequences, and Illumina test sites, which use plasma-etched silicon wafers containing beads with long oligonudeotide probes, were remarkably similar in the numbers of genes detected and the detection consistency, gene list overlap and ratio compression analyses. In other words, the expression patterns generated were reflective of biology regardless of the differences in technology.
Some of the results were affected by differences in data analysis and detection call algorithms. This effect is most noticeable in the fold-change compression observed in the two-color results from the NCI microarrays, which generally included low intensity probes resulting in over 95% detection call rate. The comparability of the NCI microarrays relative to the other platforms improves when background is based on ‘alien’ or negative control sequences. This alternative method reduces the detection call rate to 60–70%, while generally increasing the absolute fold changes in up- and down-regulated genes (E.S.K., unpublished data). Interestingly, the NCI platform had lower intrasite repeatability (Fig. 1), but demonstrated comparable rankings in log ratios when compared to the other platforms (Fig. 5b).
Additional analyses of the MAQC data are provided in the accompanying articles. For example, the microarray platforms detected known differences in gene abundance between defined RNA mixtures23 and generated differential expression results that were comparable with other gene expression platforms22–24. The comparability of the gene expression results increased when the microarrays and other methodologies analyzed overlapping sequences from the same gene22. Furthermore, external RNA controls included in some microarray platforms were useful predictors of technical performance25.
Direct comparison of different microarray platforms is neither a new nor an original idea in the realm of high-throughput biology. However, the data set generated by the MAQC project is unique in both its size and content. The main study compares seven different microarray platforms and includes ~ 60 hybridizations per platform using well-characterized, commercially available RNA sample types. Including the reagents used in the two pilot studies and the toxicogenomics validation study26, 1,327 microarrays have been used for this project (see Table S4 in Supplementary Data online). Moreover, the availability of the probe sequences in the MAQC project enabled us to approach the interplatform comparisons with greater scientific rigor. We performed detailed probe mapping to confirm identity and reveal potential sequence- or target-based differences between the gene expression platforms. This analysis confirmed that the great majority of probes were very carefully chosen and of high quality.
Most of the results in this report are based on a set of 12,091 common genes that are represented on six high-density microarray platforms, but which generally use different probe sequences for detection. Our probe selection procedure may have introduced a bias in the study because the imposed criteria neither reflect the platform design philosophies nor does it account for the very rich underlying biology. More than one probe per target can be a highly desirable feature on microarray platforms because a single probe may not capture all tissue-specific effects. We also found a number of probes that were not gene specific, suggesting a strategy of targeting multigene families.
The MAQC data set captures intrasite, intersite and interplatform differences. However, it does not address protocol, time or other technical variables within a test site because all test sites used the same protocol and generated replicate data at approximately the same time (except as noted under data filtering). The effect and levels of these sources of variation have been described in other studies15'33. Furthermore, our analysis does not include performance metrics based on ‘biology’ (e.g., Gene Ontology terms or pathways)26. Though a relatively high level of concordance of differentially expressed gene lists were observed in this study, it is possible that a higher level of agreement would be detected using these other methods of gene list concordance34, or that a lower level would be observed with sample types that were more realistically similar.
It should be noted that the results presented in this paper in terms of log ratios and overlap of lists of differentially expressed genes were derived from comparing sample types A and B, which exhibited the greatest differences among the four sample types used in the MAQC project. In practical applications, the expected differences between sample types (e.g., treated versus control animals) are usually much smaller compared to those seen between sample types A and B. Therefore, the comparability of microarray data reported in this paper does not necessarily mean that the same level of consistency would be achieved in toxicogenomic or pharmacogenomic applications. This difference can be seen from the relatively lower power and smaller overlap of gene lists (see Figures S1–S2 in Supplementary Data online) when comparing sample types C and D, where the maximum fold change is three.
The MAQC data set can be used to compare normalization methods23 and data analysis algorithms26 (see Figure S2 in Supplementary Data online), similar to a currently available website (http://affycomp.biostat.jhsph.edu) which illustrates the impact of the different data analysis methods on expression results30–34. It is our hope that future studies will add to the MAQC data set. For example, microarray providers could submit gene expression results from new microarrays with updated probe content and then use the MAQC data set to confirm consistency with older versions of the microarray. In an effort to equally represent all platforms and to present results in a timely manner, this publication analyzed only 386 microarray hybridizations from 20 test sites. However, additional data sets from the MAQC main study are available (listed as Tables S1–S4 in Supplementary Data online). Although most sites generated quality results, some differences were detected between test sites using the same platform. Thus, microarray studies need unified metrics and standards, which can be used to identify suboptimal results and monitor performance in microarray facilities.
Previous reports have relied heavily on the statistical significance (P value) rather than on the actual measured quantity of differential expression (fold change or ratio) in identifying differentially expressed genes. This strict reliance on P values alone has resulted in the apparent lack of agreement between sites and microarray platforms20,26. Our results from analyzing the MAQC human data sets (see Figure S2 in Supplementary Data online) and the rat toxicogenomics data set26 indicate that a straightforward approach of fold-change ranking plus a nonstringent P cutoff can be successful in identifying reproducible gene lists, whereas ranking and selecting differentially expressed genes solely by the t-test statistic predestine a poor concordance in results, in particular for shorter gene lists, due to the relatively unstable nature of the variance (noise) estimate in the t-statistic measure. More robust methods such as ranking using the test statistic from the Significance Analysis of Microarrays (SAM)35 did not generate more reproducible results compared to fold-change ranking in our cross-laboratory and interplatform comparisons. Our results are consistent with previously published data20. Furthermore, the impact of normalization methods on the reproducibility of gene lists becomes minimal when the fold change, instead of the P value, is used as the ranking criterion for gene selection24,26.
Two initiatives for microarray reference materials are currently in progress. A group led by FDA’s Center for Drug Evaluation and Research (CDER) developed two mixed-tissue RNA pools with known differences in tissue-selective genes that can be used as rat reference materials36, whereas the External RNA Controls Consortium (ERCC) is testing polyadenylated transcripts that can be added to each RNA sample before processing to monitor the technical performance of the assay37. The MAQC project complements these efforts by establishing several commercially available human reference RNA samples, and an accompanying large data set, which can be used by the scientific community to compare results generated in their own laboratories for quality control and performance validation efforts. In fact, the commercial availability of the MAQC reference sample types allowed several laboratories to generate and submit additional gene expression data to the MAQC project after the official deadline (listed as Table S4 in Supplementary Data online).
Repeated intersite comparisons, such as a proficiency testing, are required three times a year for many Clinical Laboratory Improvement Amendments (CLIA) assays and may also be useful in microarray facilities to monitor the comparability and consistency of data sets generated over time38. For example, a proficiency testing program evaluated the performance over a 9-month period of 18 different laboratories by repeatedly hybridizing three replicates of the same two RNA sample types to Affymetrix microarrays (L.H.R. and W.D.J., unpublished results). This study revealed the range of quality metrics and the impact of protocol differences on the microarray results. The MAQC human reference RNA sample types could be used in this kind of intersite proficiency testing program.
In summary, the technical performance of microarrays as assessed in the MAQC project supports their continued use for gene expression profiling in basic and applied research and may lead to their use as a clinical diagnostic tool as well. International organizations such as ERCC37, the Microarray Gene Expression Data Society39 and this MAQC project are providing the microarray community with standardization of data reporting, common analysis tools and useful controls that can help provide confidence in the consistency and reliability of these gene expression platforms.
METHODS
Probe mapping
Affymetrix, Agilent, GE Healthcare, Illumina and Operon oligonucleotides used by the NCI provide publicly available probe sequences for their microarray platforms in a spreadsheet format (websites listed in Supplementary Data online). The probe sequences for Applied Biosystems microarrays can be individually obtained through the Panther database (http://www.pantherdb.org) and the sequences of the intended regions for QuantiGene (Panomics) assays are available upon request. Probe sequences for Eppendorf microarrays are not yet publicly available, but were provided to the MAQC project for confidential analysis. Gene Express provided annotation and approximate forward and reverse primer locations for the StaRT-PCR assays, which were sufficient to localize the intended target. For TaqMan assays, Applied Biosystems provided Assay ID, amplicon size, assay location on the RefSeq and a context sequence (exact 25-nt sequence that includes the TaqMan assay detection probe). The MAQC probe mapping (Supplementary Methods online and Supplementary Notes online) used the March 8, 2006 RefSeq release containing 24,000 curated accessions to which we subjectively added 157 entries that were recently either withdrawn or retired from the NCBI curation. AceView comparisons were based on the August 2005 database32.
An exact match of the sequence of the probe to the database entry was required. Probes matching only the reverse strand of a transcript were excluded as well as probes matching more than one gene. An exact match of 80% of the probes within a probe set (usually 9 probes out of 11) was required for Affymetrix. The results based on these stringent criteria are provided as Supplementary Tables 2–5 online and summarized as Table S5 in Supplementary Data online. The counts for the StaRT-PCR and TaqMan assays were based on the annotation provided by Gene Express and Applied Biosystems. In the AceView analysis, the mapping was tolerant to low levels of noncentral mismatches, but applied a stringent gene-specific filter so that probes which potentially cross-hybridize were removed even if they had a single exact match.
RNA preparation
The total RNA sources were tested and selected based on the results of 160 microarrays from Pilot Project I (data not shown). The Universal Human Reference RNA (catalog no. 740000) and Human Brain Reference RNA (catalog no. 6050) were generously donated by Stratagene and Ambion, respectively. The four titration mixtures of the samples were selected based on the results of 254 microarrays from Pilot Project II (data not shown) and prepared as described elsewhere23. The titration pools were mixed at the same time at one site using a documented protocol (MAQC_RNA_Preparation_SOP.doc) available at the MAQC website (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/). Each test site received 50-µg aliquots of the four sample types and confirmed the RNA quality using a Bioanalyzer (Agilent) before initiating target preparation.
Target preparation and quality assessments
Every test site was provided with instructions (MAQC_Sample_Processing_Overview_SOP.doc) on the processing of RNA samples, conducting quality assessment of RNA reference samples, target preparations and replication guidelines, standardized nomenclature for referencing samples and a template for reporting quality assessment data (MAQC_RNA_Quality_Report_Template.xls). The gene expression vendors generously provided all reagents to the test sites. Each microarray test site assessed cRNA yields using a spectrophotometer and determined the median transcript sizes using a Bioanalyzer (Agilent). Pre-hybridization and post-hybridization quality metrics are presented as Supplementary Table 1 online. Some statistically significant differences were observed in these quality metrics between sites (data not shown).
Affymetrix, Agilent, Applied Biosystems and Eppendorf test sites added platform-specific external RNA controls to the samples before processing25. Data were submitted to the FDA’s National Center for Toxicological Research (FDA/NCTR) directly from each test site and distributed to the eleven official analysis sites for review. Lists of the gene expression test sites and data analysis centers are available as Tables S1 and S2 in Supplementary Data online. All test sites for one vendor used the same target preparation protocols and processed all replicates at approximately the same time, with two exceptions: (i) Microarray slides at the NCI test sites were scanned at 100% laser power, but the photomultiplier setting varied from slide to slide and (ii) some outlier hybridizations were repeated at a later date as described below. Exact protocols for sample processing are available at the MAQC website (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/) and are briefly described in Supplementary Data online.
Data filters
Outlier hybridizations were repeated or removed from the analysis after the original data submission deadline in October 2005. One site each for the NCI and GE Healthcare platforms repeated all sample types in the MAQC study (NCI_2 and GEH_2) due to protocol issues. One Illumina site (ILM_2) repeated two samples in the MAQC study due to low cRNA yield, and another Illumina site (ILM_1) did not hybridize one sample replicate due to the same reason. Data quality from 11 hybridizations at seven test sites (ABI_2, ABI_3, AG1_1, AG1_2, AG1_3, AGL_1 and AGL_2) was not satisfactory. More details are provided as Table S3 in Supplementary Data online.
Data processing
The platform-specific methods used for background subtraction, data normalization and the optional incorporation of offset values are described in Supplementary Data online. Each test site submitted its data (including image files) to the FDA/NCTR. All data were imported into the ArrayTrack database system40,41 and preprocessed and normalized according to the manufacturer’s suggested procedures. Each gene was reviewed for quality and marked with a detection call, using the manufacturer’s protocol. Data in a uniform format were distributed to all test sites and official data analysis sites for independent study.
Data analysis
Data analyses were performed on either all of the 12,091 common genes or a subset of this group based on the qualitative detection call reported for each hybridization. The size of these subsets in each of the test sites for each sample type is reported as Table S6 in Supplementary Data online.
Signal repeatability and reproducibility
The coefficient of variation (CV) of the signal or Cy3/Cy5 values (not log transformed) between the intrasite replicates (n ≤ 5) was calculated for genes that were detected in at least three replicates of the same sample type within a test site. The distributions of these replicate CV values are displayed in Figure 1. The replicate CV medians from three test sites are included in Figure 2. A total CV (Fig. 2) of the signal values was calculated for all replicates across three test sites (n ≤ 15) using the intersection of the generally detected gene lists (that is, genes detected in at least three replicates at all three sites). A global scaling normalization is inherently applied to data from the GE Healthcare and Agilent platforms, but is not part of data extraction and normalization on the Applied Biosystems, Affymetrix (using PLIER+16) and Illumina platforms. To account for these differences, Applied Biosystems, Affymetrix and Illumina provided scaling factors for each test site that were included when measuring the total CV.
Concordance of detection call
Analyses were performed on all 12,091 common genes using the feature quality metrics provided by the manufacturers. All calls were resolved to a Detected or Not Detected status. Details on each platform’s method of determining qualitative calls are provided in Supplementary Data online. In general, the results are provided regarding the consistency of the resolved detection calls. If the call was missing because the microarray was absent, then the detection value was not considered. Otherwise, the qualitative call was considered, including those cases where the signal value was missing.
Gene list agreement
A list of differentially expressed genes was identified for each test site using the usual two group t-test that assumes equal variances between groups resulting in a pooled estimate of variance. This calculation is based on log signal. The criteria were P value < 0.001 and a mean difference greater than or equal to twofold. No filtering related to gene detection was performed. For each pair of test sites, the number of genes in both lists was identified. Percent overlap (Fig. 4) was calculated as the number of genes in common divided by number of genes on the list from one test site. For example, the agreement score for test site Y relative to test site X equals the number of genes on both lists divided by the number of genes on the test site Y list.
Log ratio comparability
The log ratio of each gene is defined as the average of log signal for all sample B replicates minus the average of log signal of all sample A replicates. (This value is the equivalent of the log of the ratio of the geometric average of signal for all sample A replicates to the geometric average of signal for all sample B replicates.) Only genes that were detected in at least three sample A replicates and detected in at least three sample B replicates for both test sites were included. To detect compression or expansion (Fig. 5a), the slope (m) was calculated for each pair of test sites using orthogonal regression due to the potential measurement error in both sites. This analysis is based on the formula y = mx + b, where y is the log ratio from test site Y and×is the log ratio from test site X. As the ideal slope is 1, the percent difference from ideal is simply m − 1. Comparability between a pair of test sites was also examined using Spearman rank correlations of the log ratios (Fig. 5b). This value compares the relative position of a gene in the test site X rank order of the log ratio (fold change) values against its position in test site Y rank order. Scatter plots of the log ratios from all sites against the log ratios generated with the TaqMan assays are presented in Figure 6.
Accession numbers
All data are available through GEO (series accession number: GSE5350), ArrayExpress (accession number: E-TABM-132), ArrayTrack (http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/), and the MAQC web site (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/).
Supplementary Material
ACKNOWLEDGMENTS
All MAQC participants freely donated their time and reagents for the completion and analysis of the MAQC project. Participants from the National Institutes of Health (NIH) were supported by the Intramural Research Program of NIH, Bethesda, Maryland. D.H. thanks Ian Korf for BLAST discussions. This study utilized a number of computing resources, including the high-performance computational capabilities of the Biowulf PC/Linux cluster at the NIH (http://biowulf.nih.gov/) as well as resources at the analysis sites.
AUTHORS
The following authors contributed to project leadership:
Leming Shi1, Laura H Reid2, Wendell D Jones2, Richard Shippy3, Janet A Warrington4, Shawn C Baker5, Patrick J Collins6, Francoise de Longueville7, Ernest S Kawasaki8, Kathleen Y Lee9, Yuling Luo10, Yongming Andrew Sun9, James C Willey11, Robert A Setterquist12, Gavin M Fischer13, Weida Tong1, Yvonne P Dragan1, David J Dix14, Felix W Frueh15, Federico M Goodsaid15, Damir Herman16, Roderick V Jensen17, Charles D Johnson18, Edward K Lobenhofer19, Raj K Puri20, Uwe Scherf21, Jean Thierry-Mieg16, Charles Wang22, Mike Wilson12,18, Paul K Wolber6, Lu Zhang9,23, William Slikker, Jr1, Leming Shi1, Laura H Reid2
Project leader: Leming Shi1
Manuscript preparation team leader: Laura H Reid2
MAQC Consortium:
Leming Shi1, Laura H Reid2, Wendell D Jones2, Richard Shippy3, Janet A Warrington4, Shawn C Baker5, Patrick J Collins6, Francoise de Longueville7, Ernest S Kawasaki8, Kathleen Y Lee9, Yuling Luo10, Yongming Andrew Sun9, James C Willey11, Robert A Setterquist12, Gavin M Fischer13, Weida Tong1, Yvonne P Dragan1, David J Dix14, Felix W Frueh15, Federico M Goodsaid15, Damir Herman16, Roderick V Jensen17, Charles D Johnson18, Edward K Lobenhofer19, Raj K Puri20, Uwe Scherf21, Jean Thierry-Mieg16, Charles Wang22, Mike Wilson12,18, Paul K Wolber6, Lu Zhang9,23, Shashi Amur15, Wenjun Bao24, Catalin C Barbacioru9, Anne Bergstrom Lucas6, Vincent Bertholet7, Cecilie Boysen25, Bud Bromley25, Donna Brown26, Alan Brunner3, Roger Canales9, Xiaoxi Megan Cao27, Thomas A Cebula28, James J Chen1, Jing Cheng29, Tzu-Ming Chu24, Eugene Chudin5, John Corson6, J Christopher Corton14, Lisa J Croner30, Christopher Davies4, Timothy S Davison18, Glenda Delenstarr6, Xutao Deng22, David Dorris12, Aron C Eklund17, Xiao-hui Fan1, Hong Fang27, Stephanie Fulmer-Smentek6, James C Fuscoe1, Kathryn Gallagher31, Weigong Ge1, Lei Guo1, Xu Guo4, Janet Hager32, Paul K Haje33, Jing Han20, Tao Han1, Heather C Harbottle34, Stephen C Harris1, Eli Hatchwell35, Craig A Hauser36, Susan Hester14, Huixiao Hong27, Patrick Hurban19, Scott A Jackson28, Hanlee Ji37, Charles R Knight38, Winston P Kuo39, J Eugene LeClerc28, Shawn Levy40, Quan-Zhen Li41, Chunmei Liu4, Ying Liu42, Michael J Lombardi17, Yunqing Ma10, Scott R Magnuson43, Botoul Maqsodi10, Tim McDaniel4, Nan Mei1, Ola Myklebost44, Baitang Ning1, Natalia Novoradovskaya13, Michael S Orr15, Terry W Osborn38, Adam Papallo17, Tucker A Patterson1, Roger G Perkins27, Elizabeth H Peters38, Ron Peterson45, Kenneth L Philips19, P Scott Pine15, Lajos Pusztai46, Feng Qian27, Hongzu Ren14, Mitch Rosen14, Barry A Rosenzweig15, Raymond R Samaha9, Mark Schena33, Gary P Schroth23, Svetlana Shchegrova6, Dave D Smith47, Frank Staedtler45, Zhenqiang Su1, Hongmei Sun27, Zoltan Szallasi48, Zivana Tezak21, Danielle Thierry-Mieg16, Karol L Thompson15, Irina Tikhonova32, Yaron Turpaz4, Beena Vallanat14, Christophe Van7, Stephen J Walker49, Sue Jane Wang15, Yonghong Wang8, Russ Wolfinger24, Alex Wong6, Jie Wu27, Chunlin Xiao9, Qian Xie27, Jun Xu22, Wen Yang10, Liang Zhang29, Sheng Zhong50, Yaping Zong51, William Slikker, Jr1
Scientific management (National Center for Toxicological Research, US Food and Drug Administration): Leming Shi, Weida Tong, Yvonne P. Dragan, William Slikker, Jr.
Affiliations:
1National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Road, Jefferson, Arkansas 72079, USA; 2Expression Analysis, Inc., 2605 Meridian Parkway, Durham, North Carolina 27713, USA; 3GE Healthcare, 7700 S. River Parkway, Suite 2603, Tempe, AZ 85284, USA; 4Affymetrix, Inc., 3420 Central Expressway, Santa Clara, California 95051, USA; 5Illumina, Inc. 9885 Towne Centre Drive, San Diego, California 92121, USA; 6Agilent Technologies, Inc., 5301 Stevens Creek Blvd., Santa Clara, California 95051, USA; 7Eppendorf Array Technologies, rue du Séminaire 20a, 5000 Namur, Belgium; 8NCI Advanced Technology Center, 8717 Grovemont Circle, Bethesda, Maryland 20892, USA; 9Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA; 10Panomics, Inc., 6519 Dumbarton Circle, Fremont, California 94555, USA; 11Medical University of Ohio, 3000 Arlington Avenue, Toledo, Ohio 43614, USA; 12Ambion, An Applied Biosystems Business, 2130 Woodward Street, Austin, Texas 78744, USA; 13Stratagene Corp., 11011 North Torrey Pines Road, La Jolla, California 92130, USA; 140ffice of Research and Development, US Environmental Protection Agency, 109 TW Alexander Drive, Research Triangle Park, North Carolina 27711, USA; 15Center for Drug Evaluation and Research, US Food and Drug Administration, 10903 New Hampshire Avenue, Silver Spring, Maryland 20993, USA; 16National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, Maryland 20894, USA; 17University of Massachusetts-Boston, 100 Morrissey Boulevard, Boston, Massachusetts 02125, USA; 18Asuragen, Inc., 2150 Woodward, Austin, Texas 78744, USA; 19Cogenics™, A Division of Clinical Data, Inc., 100 Perimeter Park Drive, Suite C, Morrisville, North Carolina 27560, USA; 20Center for Biologies Evaluation and Research, US Food and Drug Administration, 29 Lincoln Drive, Bethesda, Maryland 20892, USA; 21Center for Devices and Radiological Health, US Food and Drug Administration, 2098 Gaither Road, Rockville, Maryland 20850, USA; 22UCLA David Geffen School of Medicine, Transcriptional Genomics Core, Cedars-Sinai Medical Center, 8700 Beverly Boulevard, Los Angeles, California 90048, USA; 23Solexa, Inc., 25861 Industrial Boulevard, Hayward, California 94545, USA; 24SAS Institute, Inc., 100 SAS Campus Drive, Cary, North Carolina 27513, USA; 25Vialogy Corp., 2400 Lincoln Avenue, Altadena, California 91001, USA; 26Operon Biotechnologies, 2211 Seminole Drive, Huntsville, Alabama 35805, USA; 27Z-Tech Corp., 3900 NCTR Road, Jefferson, Arkansas 72079, USA; 28Center for Food Safety and Applied Nutrition, US Food and Drug Administration, 8401 Muirkirk Road, Laurel, Maryland 20708, USA; 29CapitalBio Corp., 18 Life Science Parkway, Changping District, Beijing 102206, China; 30Biogen Idec, 5200 Research Place, San Diego, California 92122, USA; 31US Environmental Protection Agency, Office of the Science Advisor, 1200 Pennsylvania Avenue, NW, Washington, DC 20460, USA; 32Yale University, W.M. Keck Biotechnology Resource Laboratory, Microarray Resource, 300 George Street, New Haven, Connecticut 06511, USA; 33TeleChem Arraylt, 524 E. Weddell Drive, Sunnyvale, California 94089, USA; 34Center for Veterinary Medicine, US Food and Drug Administration, 8401 Muirkirk Road, Laurel, Maryland 20708, USA; 35Cold Spring Harbor Laboratory, 500 Sunnyside Boulevard, Woodbury, New York 11797, USA; 36Burnham Institute, 10901 North Torrey Pines Road, La Jolla, California 92037, USA; 37Stanford University School of Medicine, 318 Campus Drive, Stanford, California 94305, USA; 38Gene Express, Inc., 975 Research Drive, Toledo, Ohio 43614, USA; 39Harvard School of Dental Medicine, Department of Developmental Biology, 188 Longwood Avenue, Boston, Massachusetts 02115, USA; 40Vanderbilt University, 465 21st Avenue South, Nashville, Tennessee 37232, USA; 41University Texas Southwestern Medical Center, 6000 Harry Hines Boulevard/ND6.504, Dallas, Texas 75390, USA; 42University of Texas at Dallas, Department of Computer Science, MS EC31 Richardson, Texas 75083, USA; 43GenUs BioSystems, Inc., 1808 Janke Drive Unit M, Northbrook, Illinois 60062, USA; 44Norwegian Microarray Consortium, Rikshospitalet - Radiumhospitalet Health Centre, Montebello, N0310 Oslo, Norway; 45Novartis, 250 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA; 46MD Anderson Cancer Center, Breast Medical Oncology Department-Unit 1354, 1155 Pressler Street, Houston, Texas 77230, USA; 47Luminex Corp., 12212 Technology Boulevard, Austin, Texas 78727, USA; 48Harvard Medical School, Children's Hospital Informatics Program at the Harvard-MIT Division of Health Sciences and Technology (CHIP@HST), Boston, Massachusetts 02115, USA; 49Wake Forest University School of Medicine, Department of Physiology and Pharmacology, Medical Center Boulevard, Winston-Salem, North Carolina 27157, USA; 50University of Illinois at Urbana-Champaign, Department of Bioengineering, 1304 W. Springfield Avenue, Urbana, Illinois 61801, USA; 51Full Moon Biosystems, Inc., 754 N. Pastoria Avenue, Sunnyvale, California 94085, USA.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
DISCLAIMER
This work includes contributions from, and was reviewed by, the FDA, the EPA and the NIH. This work has been approved for publication by these agencies, but it does not necessarily reflect official agency policy. Certain commercial materials and equipment are identified in order to adequately specify experimental procedures. In no case does such identification imply recommendation or endorsement by the FDA, the EPA or the NIH, nor does it imply that the items identified are necessarily the best available for the purpose.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests (see the Nature Biotechnology website for details).
References
- 1.Lesko LJ, Woodcock J. Translation of pharmacogenomics and pharmacogenetics: a regulatory perspective. Nat. Rev. Drug Discov. 2004;3:763–769. doi: 10.1038/nrd1499. [DOI] [PubMed] [Google Scholar]
- 2.Frueh FW. Impact of microarray data quality on genomic data submissions to the FDA. Nat. Biotechnol. 2006;24:1105–1107. doi: 10.1038/nbt0906-1105. [DOI] [PubMed] [Google Scholar]
- 3.Dix DJ, et al. A framework for the use of genomics data at the EPA. Nat. Biotechnol. 2006;24:1108–1111. doi: 10.1038/nbt0906-1108. [DOI] [PubMed] [Google Scholar]
- 4.Tan PK, et al. Evaluation of gene expression measurements from commercial micro-array platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 6.Ivanova NB, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 7.Miller RM, et al. Dysregulation of gene expression in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-lesioned mouse substantia nigra. J. Neurosci. 2004;24:7445–7454. doi: 10.1523/JNEUROSCI.4204-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortunel NO, et al. Comment on ‘“Sternness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. author reply 393. [DOI] [PubMed] [Google Scholar]
- 9.Miklos GL, Maleszka R. Microarray reality checks in the context of a complex disease. Nat. Biotechnol. 2004;22:615–621. doi: 10.1038/nbt965. [DOI] [PubMed] [Google Scholar]
- 10.Frantz S. An array of problems. Nat. Rev. Drug Discov. 2005;4:362–363. doi: 10.1038/nrd1746. [DOI] [PubMed] [Google Scholar]
- 11.Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 12.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 13.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl. Acad. Sci. USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen D, et al. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6:63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobbin KK, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin. Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- 16.Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nat. Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 17.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat. Methods. 2005;2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 18.Kuo WP, et al. A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat. Biotechnol. 2006;24:832–840. doi: 10.1038/nbt1217. [DOI] [PubMed] [Google Scholar]
- 19.Shi L, et al. QA/QC: challenges and pitfalls facing the microarray community and regulatory agencies. Expert Rev. Mol. Diagn. 2004;4:761–777. doi: 10.1586/14737159.4.6.761. [DOI] [PubMed] [Google Scholar]
- 20.Shi L, et al. Cross-platform comparability of microarray technology: intra-platform consistency and appropriate data analysis procedures are essential. BMC Bioinformatics. 2005;6 Suppl. 2:S12. doi: 10.1186/1471-2105-6-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji H, Davis RW. Data quality in genomics and microarrays. Nat. Biotechnol. 2006;24:1112–1113. doi: 10.1038/nbt0906-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canales RD, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 23.Shippy R, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat. Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 25.Tong W, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat. Biotechnol. 2006;24:1132–1139. doi: 10.1038/nbt1237. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat. Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 27.Mecham BH, et al. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004;32:e74. doi: 10.1093/nar/gnh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6:107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22:789–794. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- 31.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thierry-Mieg D, J TM. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biology. 2006;7 Suppl. 1:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bammler T, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat. Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 34.Harr B, Schlotterer C. Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res. 2006;34:e8. doi: 10.1093/nar/gnj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson KL, et al. Use of a mixed tissue RNA design for performance assessments on multiple microarray formats. Nucleic Acids Res. 2005;33:el87. doi: 10.1093/nar/gni186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker SC, et al. The External RNA Controls Consortium: a progress report. Nat. Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 38.Reid LH. The value of a proficiency testing program to monitor performance in microarray laboratories. Pharm. Discov. 2005;5:20–25. [Google Scholar]
- 39.Ball CA, et al. Standards for microarray data. Science. 2002;298:539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 40.Tong W, et al. ArrayTrack–supporting toxicogenomic research at the U.S. Food and Drug Administration National Center for Toxicological Research. Environ. Health Perspect. 2003;111:1819–1826. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong W, et al. Development of public toxicogenomics software for microarray data management and analysis. Mutat. Res. 2004;549:241–253. doi: 10.1016/j.mrfmmm.2003.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.