Abstract
The accumulation of protein adducts caused by carbonyl stress (CS) is a hallmark of cellular aging and other diseases, yet the detailed cellular effects of this universal phenomena are poorly understood. An understanding of the global effects of CS will provide insight into disease mechanisms and can guide the development of therapeutics and lifestyle changes to ameliorate their effects. To identify cellular functions important for the response to carbonyl stress, multiple genome-wide genetic screens were performed using two known inducers of CS. We found that different cellular functions were required for resistance to stress induced by methylglyoxal (MG) and glyoxal (GLY). Specifically, we demonstrate the importance of macromolecule catabolism processes for resistance to MG, confirming and extending known mechanisms of MG toxicity, including modification of DNA, RNA, and proteins. Combining our results with related studies that examined the effects of ROS allowed a comprehensive view of the diverse range of cellular functions affected by both oxidative and carbonyl stress. To understand how these diverse cellular functions interact, we performed a quantitative epistasis analysis by creating multimutant strains from those individual genes required for glyoxal resistance. This analysis allowed us to define novel glyoxal-dependent genetic interactions. In summary, using multiple genome-wide approaches provides an effective approach to dissect the poorly understood effects of glyoxal in vivo. These data, observations, and comprehensive dataset provide 1) a comprehensive view of carbonyl stress, 2) a resource for future studies in other cell types, and 3) a demonstration of how inexpensive cell-based assays can identify complex gene-environment toxicities.
Keywords: glyoxal, carbonyl stress, chemogenomics, yeast deletion collection
A hallmark of aging and its attendant ailments is the accumulation of DNA lesions, oxidized proteins, and carbonylated proteins and lipids (Beckman and Ames 1998; Harman 1981; Kujoth et al. 2005; Oliver et al. 1987; Stadtman 1992). It is well-documented that these defects are caused by reactive molecules such as superoxide anions (O2−), hydrogen peroxide (H2O2), and hydroxy radicals (OH) formed as by-products of cellular metabolism (Barrera et al. 2008; Bertram and Hass 2008; Shringarpure and Davies 2002; Stadtman 2006).
Less well-studied but no less consequential are the effects of carbonyl stress created by reactive carbonyl compounds (RCC), such as glyoxal, 3DG (3-deoxyglucosone) and methylglyoxal (MG). Glyoxal is formed by lipid and DNA oxidative degradation as well as via autoxidation of glycolaldehyde (Benov and Fridovich 1998). MG is a by-product of metabolic processes, including threonine catabolism (Murata et al. 1986) and lipid peroxidation (Poli et al. 1985). MG can also arise enzymatically during glycolysis (O’Brien et al. 2005; Thornalley 1996). In addition, diverse environmental sources, such as cigarette smoke and automobile exhaust, are abundant sources of carbonyls (O’Brien et al. 2005; Saint-Jalm and Moree-Testa 1980; Zervas et al. 2002). The widespread thermal processing of food can result in MG and other aldehydes (Nemet et al. 2006; Wells-Knecht et al. 1995).
Advanced glycation end products (AGE) arising from carbonyl stress are thought to contribute to chronic diseases, such as diabetes, chronic obstructive pulmonary disease, ischemia/reperfusion, and Alzheimer’s disease (Ellis 2007). Understanding the result of carbonyl stress is an essential first step to characterize the impacts of CS stress on cell physiology.
The well-conserved glyoxalase system is the cell’s principal defense against AGEs and aldehydes, detoxifying MG and glyoxal in the presence of glutathione (GSH) to glycolate and D-lactate, respectively (Thornalley 1998). Both can also be detoxified by NADPH-dependent aldose reductases (Vander Jagt et al. 1992). Previous work using yeast has focused primarily on MG (Aguilera et al. 2005; Maeta et al. 2005) and showed that the conserved HOG MAP kinase pathway is important for the induction of MG-responsive genes. However, no comprehensive, genome-wide analysis of the biological consequences of these modifications exists. Identifying the cellular functions necessary for providing resistance to these toxic molecules will provide insight into the molecular mechanisms that underlie diseases associated with CS and could suggest therapeutic interventions.
Materials and Methods
Reagents
Methylglyoxal, glyoxal, nicotinamide, isonicotinamide, and aminoguanidine were purchased from Sigma-Aldrich (St. Louis, MO). Nicotinamide and isonicotinamide were dissolved in sterile H2O and filtered sterilized.
Yeast strains, plasmids, and growth conditions
Yeast deletion strains were obtained from the yeast deletion collection. ORF-containing plasmids, listed in Table S5, were obtained from Charlie Boone or constructed by gap-repair (Oldenburg et al. 1997). Yeast transformations were performed using the standard lithium acetate method (Gietz et al. 1995) and selected synthetic complete medium lacking uracil (SCM URA−). For growth curve analysis, individual strains were inoculated into 100 μl of YPD or SCM URA− and grown to saturation for ∼20 h at 30°C. Overnight cultures were resuspended by shaking for 15 min, diluted into 100 μl of media in 96-well plates, and grown in Tecan GENios microplate readers for 24 h. The growth rate of each culture was monitored by measuring the OD600 every 15 min as previously described (Giaever et al. 2004). Doubling-time calculations and area under growth curve (AUGC) analysis were performed as previously described (Hoon et al. 2008; Lee et al. 2005b). We used AUGC analysis for Figure 1B, as we found that this method more accurately captured the fitness of strains under severe growth inhibition (>50% inhibition). AUGC was calculated uniformly over a 24 h window from the start of the experiment. For lower levels of growth inhibition, doubling time during the exponential growth phase was used for fitness comparisons.
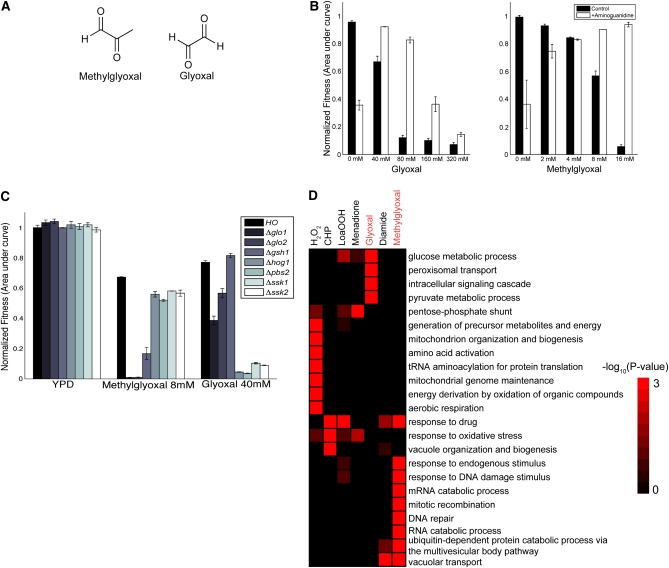
Figure 1 .
Methylgloxal and glyoxal inhibit yeast growth. A) Chemical structure of methylglyoxal and glyoxal. B) Growth curve analysis for measuring the fitness of wild-type (HO) strain in glyoxal (left) and methylglyoxal (right) in the presence (black bars) and absence of 20 mM aminoguanidine (white bars). Fitness was quantified using area under growth curve and normalized to growth in YPD ± SD (n = 3). Growth inhibition caused by aminoguanidine in the absence of glyoxal and methylglyoxal is discussed in the text. C) Fitness of deletion strains in methylglyoxal and glyoxal ± SD (n = 3) analyzed by growth curve analysis. Strains selected were deficient in genes previously shown to be important for resistance to methylglyoxal stress in yeast. D) Heatmap representing significant GO terms of strains identified to be significantly sensitive (false discovery rate < 0.05) to methylglyoxal and glyoxal by pooled fitness screens. For comparison, we included strains identified to be sensitive to oxidants hydrogen peroxide (H2O2), cumene hydroperoxide (CHP), linoleic acid 13-hydroperoxide (LoaOOH), menadione, and diamide by a previous study (Thorpe et al. 2004). The heatmap is colored according to the P value of each GO term.
Genome-wide screening and analysis
MSP and DSP screens were performed as previously described (Hoon et al. 2008) with the modification that only the homozygous deletion strains were screened against both methylglyoxal and glyoxal. For DSP, glyoxal was screened at 20 mM and methylglyoxal was screened at 8 mM for 5 and 20 generations. For MSP, yeast genomic library transformants were grown for 20 generations in glyoxal (40 mM) and supplemented with isonicotinamide (25 mM) where indicated. Methods for pooled growth, OD monitoring, automatic cell dilution, and cell harvesting were performed as previously described (Pierce et al. 2006). Competitive screens were designed such that strains that are not resistant should get diluted out over the course of four dilutions every 5 generations. There was variation in terms of the number of starting cells for each strain in the pool, but we normalized this by taking the ratio of the microarray intensities between the control and treated pools. Both DSP and MSP were analyzed using a high-density oligonucleotide tag array manufactured by Affymetrix (Pierce et al. 2006). For DSP, barcode probe intensities were extracted and processed as previously described (Pierce et al. 2006). Each array was mean-normalized and the fold change (log2 control/treatment) was calculated by comparison with a set of control arrays. At least two biological replicates were carried out for each treatment condition. The log2 ratios of both tags were averaged to generate a single score for each gene for use in the DSP-MSP plots. We used rank product analysis (Breitling et al. 2004) to identify strains that were significantly sensitive to either methylglyoxal or glyoxal. A false discovery rate cutoff of 0.05 was used. The list of sensitive strains can be found in Table S1 and Table S2. For the glyoxal deletion resistance screen, the homozygous deletion pools were screened and processed as described above. The results are presented in Table S3. For MSP, ORF probe intensities were extracted and processed in the same way as the barcode probes. Each ORF is represented by at least two probes, and the log2 ratios of each probe were averaged to generate a single score for each gene. To identify each suppressor locus, the log2 ratio of intensities were ordered by genomic location of each ORF and analyzed using a sliding window to identify loci that have at least two adjacent ORFs with log2 ratios ≥ 1.6. GO analysis was performed using GOstats (Falcon and Gentleman 2007), a bioconductor package written in R.
Double-mutant strain construction
We assessed genetic interactions among a subset of genes that confer resistance to glyoxal by generating multiple combinations of double-deletion strains for quantitative fitness analysis (St. Onge et al. 2007). MATa haploids deletion strains were obtained from the yeast deletion collection, and MATα haploids deletion strains were obtained from Charlie Boone. Double-deletion strains were constructed by the synthetic genetic array (SGA) protocol with minor modifications using a Singer RoToR HDA (Singer Instruments, Somerset, UK) (Stansfield and Stark 2007; Tong and Boone 2006). Approximately 800 double-deletion mutants were constructed among 15 deletion strains that were sensitive to glyoxal and 24 deletion strains that were resistant to glyoxal. Single-deletion strains with the same drug resistance cassette (Kanr-Natr and Natr-Kanr) as the double-mutant strains were constructed using the HO-deletion strain as a query strain. In MATa haploids, genes were replaced with a kanamycin resistance marker gene (Kanr), and in MATα haploids, genes were replaced with a nourseothricin resistance marker gene (Natr). This method allowed each double-deletion gene pair to be constructed twice (Kanr-Natr and Natr-Kanr) independently. Fitness values between each gene pair were highly correlated (R2 = 0.92). The fitness of strains was determined in the presence and absence of glyoxal. One double mutant (Δhog1Δccw12) could not be constructed because the genes in the pair were genetically linked. Fitness values between each gene pair were highly correlated (Figure 4B, R2 = 0.92), demonstrating the reproducibility of the assay. Strains for which fitness values of Kanr-Natr double-deletion strains differed from that of the Natr- Kanr deletion strains (|Wxy-Wyx|/ > 0.2) were filtered and not used in the analysis.
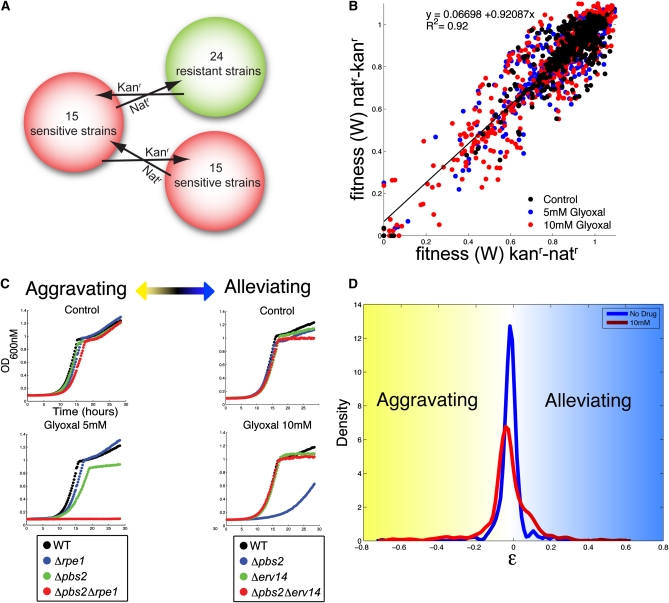
Figure 4 .
Quantitative epistasis analysis. A) Double deletion strains were constructed between 15 glyoxal sensitive strains and 24 glyoxal resistant strains. Each double mutant was constructed independently twice using two different markers (Kanr and Natr). B) Fitness correlation between reciprocal double-deletion mutants for each gene pair in the presence and absence of glyoxal. The correlation coefficient (R) and the best fitting line are shown. C) Examples of double-deletion mutants displaying aggravating and alleviating genetic interaction in the presence of glyoxal. Δpbs2Δrpe1 mutants were strongly aggravating in 5 mM glyoxal with ε = −0.66, and Δpbs2Δerv14 were strongly alleviating (suppression) in 10 mM glyoxal with ε = 0.44. D) Distribution of ε values for all double mutant pairs grown in YPD (blue) or 10 mM glyoxal (red).
Growth assay for epistasis analysis
Deletion strains arrayed on YPD/agar were inoculated into 96-well plates containing 100 μl of YPD using a Singer RoToR HDA (Singer Instruments). Cultures were grown to saturation for 20 h at 30°C and stored at 4°C for 24–48 h. The cells were then resuspended by shaking for 15 min. Cultures were diluted into 100 μl volumes in 96-well plates using the Singer RoToR HDA and grown in Tecan GENios microplate readers for 30 h at 30°C. The growth rate of each culture was monitored by measuring the OD600 every 15 min. The doubling time (D) was calculated exactly as previously described (St. Onge et al. 2007). The fitness of each deletion strain was calculated as the ratio of the doubling time of the parental wild-type strain divided by that of the mutant. We quantified the genetic interaction between each gene pair using a multiplicative model. If a strain deleted for gene x has a fitness of Wx and a strain deleted for gene y has a fitness of Wy, then the double mutant strain is expected to have a fitness (Wxy) of Wx × Wy. We measured the deviation εxy from this expectation, where εxy = Wxy – Wx × Wy. For each gene pair, the εxy values for each Kanr-Natr and Natr-Kanr were averaged and used to generate the heatmap.
Results
Methyglyoxal and glyoxal inhibit yeast growth
MG is a potent inhibitor of yeast growth (Aguilera and Prieto 2001; Aguilera et al. 2005; Bito et al. 1997; Inoue and Kimura 1996; Inoue et al. 1998; Maeta et al. 2005). Using a quantitative fitness assay, we quantified the growth inhibition for MG and glyoxal (Figure 1A). The IC50 for MG is 8.6 mM (for S. cerevisiae strain BY4743), approximately 5-fold less than that for glyoxal (IC50 of 45.5 mM) (Figure 1B). Aminoguanidine (AG), a α,β-dicarbonyl scavenging agent (Cameron et al. 2005) and known suppressor of carbonyl stress, rescues the growth inhibition caused by both compounds (Figure 1B), suggesting that the cellular effects are specific. In the absence of methylglyoxal or glyoxal treatment with AG (20 mM) inhibits yeast growth, likely due to AG reacting with pyruvate to form a hydrazone adduct at high concentrations (Thornalley 2003); in contrast; the carbonyl compounds modulate this effect.
HOG pathway components are both required for resistance to MG and glyoxal
In yeast, the high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway regulates osmotic homeostasis (O’Rourke et al. 2002). Previous work has determined that the HOG pathway mediates MG resistance, presumably via promoting the expression of genes involved in MG metabolism (Aguilera et al. 2005; Inoue et al. 1998). One such gene is GLO1, encoding glyoxalase I, an enzyme that converts MG into S-D-lactoylgluthathione in the presence of glutathione (Inoue and Kimura 1996). In contrast, we found glyoxal and MG exert different effects on mutants involved in HOG signaling and glyoxalase activity, suggesting distinct mechanisms of action. We confirmed this with individual growth tests of selected strains in glyoxal and methylglyoxal. For example, strains missing GLO1, GLO2, or GSH1 were more sensitive to MG stress than strains missing HOG1, PBS2, SSK1, or SSK2, whereas the opposite pattern of sensitivities was observed for glyoxal stress (Figure 1C).
Genome-wide fitness profiling demonstrates physiologically distinct responses to MG and glyoxal
We used two genome-wide assays: 1) yeast deletion collection to identify genes that confer sensitivity, and 2) an overexpression assay to define genes that confer resistance to CS. Genome-wide fitness profiling showed that glyoxal and MG affect cells very differently. We screened both compounds against a collection of 4700 homozygous deletion strains (Giaever et al. 2002; Giaever et al. 2004; Hillenmeyer et al. 2008; Hoon et al. 2008; Lee et al. 2005b) and identified 458 deletion strains that were significantly sensitive (false discovery rate < 0.05) to either MG or GLY (supporting information, Table S1 and Table S2). Functional enrichment analysis using Gene Ontology (GO) annotations of sensitive strains showed distinct requirements for resistance to each compound (Figure 1D). MG-sensitive genes were enriched for protein, mRNA, and DNA metabolic processes, in agreement with the suggested mechanism of action of MG in forming DNA, RNA, and protein adducts (Kalapos 1999). In contrast, glyoxal resistance required genes involved in glucose metabolism and peroxisomal and signal transduction processes. Our study corroborates, on a genome-wide level, previous studies in which MG and glyoxal were shown to exert different effects in vivo [e.g., MG and glyoxal induced distinct signals for MAP family kinases in human endothelial cells (Akhand et al. 2001)].
The effects of MG and glyoxal were also distinct from that observed for other oxidants. We compared GO annotations for genes identified following treatment with other oxidants in a previous study (Thorpe et al. 2004) and found that the effects of oxidative stress are typically broad but specific to each oxidant (Figure 1D). Here, we found that additional functions, beyond those associated with ROS, were important for CS resistance.
Carbonyl stress leads to DNA, RNA, and protein dysfunction
We found that multiple processes involved in the repair and degradation of damaged cellular macromolecules factor in the CS response. Glyoxal and MG are potent arginine-directed glycating agents and covalently cross-link proteins via modification of arginine residues, leading to protein dysfunction (Rabbani and Thornalley 2008). Protein modification by MG activates ubiquitin/proteasome-dependent proteolysis (Du et al. 2006). Consistent with this observation was our finding that strains deleted for genes involved in ubiquitin-dependent protein degradation (∆shp1, ∆bsd2, ∆stp22, ∆doa4, ∆swm1, ∆bst1, ∆cdc26, ∆rad6, ∆eps1, ∆ rtt101, ∆grr1, ∆vps25, ∆def1, ∆doa1, ∆ snf7, ∆ srn2, ∆vps36, ∆ubx2, ∆ubx4, ∆vps20, ∆ydj1, ∆snf8, ∆vps28, and ∆bro1) were sensitive to MG. For glyoxal, the requirement for ubiquitin-related pathways was less pronounced, with fewer identified strains (∆reg1, ∆doa4, ∆ubc8, ∆ jem1, and ∆ydj1). Together, the MG and GLY results demonstrate that protein catabolism plays a major role in detoxifying glycated proteins.
Besides producing protein adducts, methylglyoxal also reacts with guanine bases in RNA and DNA (Kang et al. 1996; Murata-Kamiya and Kamiya 2001). Because messenger RNA (mRNA) quality control is essential, we expected to uncover gene deletion strains involved in mRNA metabolism. The first step in mRNA decay is deadenylation, followed by mRNA degradation either by decapping followed by 5′→3′ decay or by 3′→5′ decay (Garneau et al. 2007). As expected, several strains deficient in deadenylation (∆ccr4, ∆pop2, and ∆not5), decapping and 5′→3′ decay (∆dhh1,∆pat1,∆lsm6, ∆lsm1, and ∆lsm7) and 3′→5′ decay (∆ski8, ∆ski7, ∆ski3, and ∆ski2) were sensitive to methylglyoxal. Furthermore, multiple strains involved in DNA repair (∆mms4, ∆rad18, ∆rpn4, ∆rad59, ∆rad57, ∆rad55, ∆hpr1, ∆mus81, ∆rad51, ∆rad4, ∆rad6, ∆rpb9, ∆rad54, ∆srs2, ∆def1, ∆doa1, ∆rad5, ∆mms22, ∆rad52, ∆rad14, ∆mre11, ∆tho2, ∆rad50, ∆snf2, ∆rad1, and ∆mms1) were also sensitive to MG. In contrast, DNA repair and mRNA decay pathways were not required for glyoxal resistance, even at higher doses (data not shown). Together, these results demonstrate that MG damages cellular macromolecules more potently than glyoxal.
Glyoxal-resistant deletion strains
Having identified numerous CS-sensitive deletion strains, we asked which deletion strains manifest CS resistance. Such genes can be interpreted as negative regulators of stress resistance. Pooled fitness profiling assays have focused on identifying deletion strains with reduced fitness in experimental conditions, in part because the assay’s dynamic range for assessing resistance is poor [i.e., modest perturbations are used (∼IC10)] and in part because a large amount of the sample is hybridized. To identify resistant strains, we developed an optimized “resistance screen” in which the entire pool of deletion strains is treated with a high dose of compound (e.g., IC50) over 20 generations of growth (Figure 2A and Materials and Methods), and a smaller amount of material is hybridized (Figure 2A). This new assay identified bona fide resistant deletion strains. Following treatment with glyoxal at an IC50 concentration, most of the strain-specific TAG intensities were at background levels compared to controls, while those strains that remained constant or increased during the course of the experiment represented glyoxal-resistant strains.
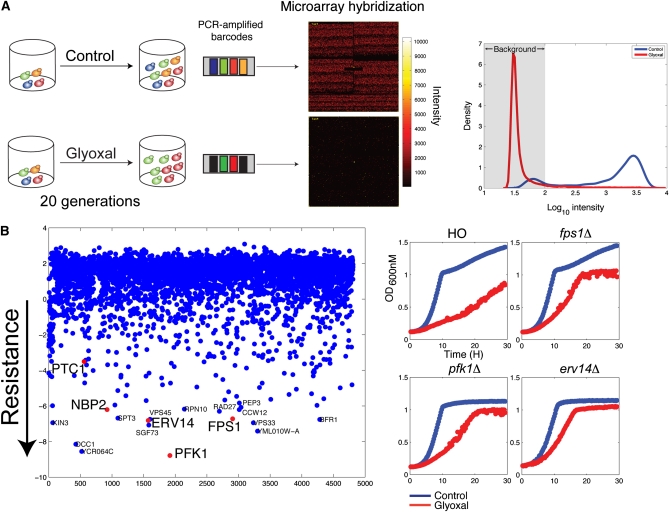
Figure 2 .
Identifying deletion strains resistant to glyoxal stress. A) (left) Schematic of screen used for identifying deletion strains resistant to glyoxal stress. A homozygous deletion pool was grown for 20 generations in 80 mM glyoxal. Barcodes were amplified and hybridized to TAG4 arrays. (right) Strong selection in the presence of glyoxal resulted in selection for strains highly resistant to glyoxal. TAG intensities are mostly at background (gray region) in cells treated with glyoxal compared to control. B) (left) Log2 fold ratio of control/treatment of TAG array results. Negative fold ratios indicate tags that are over-represented in glyoxal selection. Genes mentioned in the text are highlighted in red. (right) Confirmation growth curves of deletion strains resistant to glyoxal.
Multiple strains were significantly resistant to glyoxal, and the majority of these strains were confirmed individually (Figure 2B, Table S3). Among these genes, PTC1, a type 2C protein phosphatase that negatively regulates the HOG pathway by dephosphorylating Hog1 (Warmka et al. 2001), and Nbp2, a protein that recruits Ptc1 to the Pbs2-Hog1 complex, provide additional evidence that upregulation of the HOG pathway is required for glyoxal resistance (Mapes and Ota 2004). GO analysis of these resistant deletion strains showed enrichment for intracellular/endosome transport protein metabolic processes and organelle organization (Table S4).
A sensitized suppressor screen to understand glyoxal stress
To further characterize the effects of glyoxal, we employed a third genome-wide assay. Specifically, we identified multicopy suppressors of glyoxal sensitivity using multicopy suppressor profiling (MSP) to identify genes that confer resistance to compounds when over-represented (Hoon et al. 2008; Rine et al. 1983).
During our experiments with deletion resistant screens, we discovered that sir2 deletion mutants displayed resistance to glyoxal. This observation, combined with the well-characterized relationship between Sir2 and NAD metabolism (Blander and Guarente 2004), inspired us to test if nicotinamide, an endogenous inhibitor of Sir2 (Bitterman et al. 2002), would act as a chemical modifier in the glyoxal MSP assay. We found that nicotinamide conferred resistance to glyoxal (Figure 3A) whereas isonicotinamide, an antagonist of nicotinamide inhibition and an activator of Sir2 deacetylase activity (Sauve and Schramm 2004), conferred sensitivity to glyoxal (Figure 3A). Therefore, we mimicked activation of Sir2 using isonicotinamide to amplify the sensitivity of the MSP screen (Figure 3B). We identified multiple suppressors containing genomic DNA that were over-represented in pools grown in glyoxal compared with control (Table 1), and isonicotinamide administration identified additional suppressors. One of the most resistant loci identified was GLO1, the enzyme that metabolizes glyoxal. This observation confirms that this experimental approach can identify relevant genes involved in glyoxal resistance. Several suppressors were confirmed by isogenic tests with wild-type yeast harboring plasmids containing individually cloned ORFs (Figure S1).
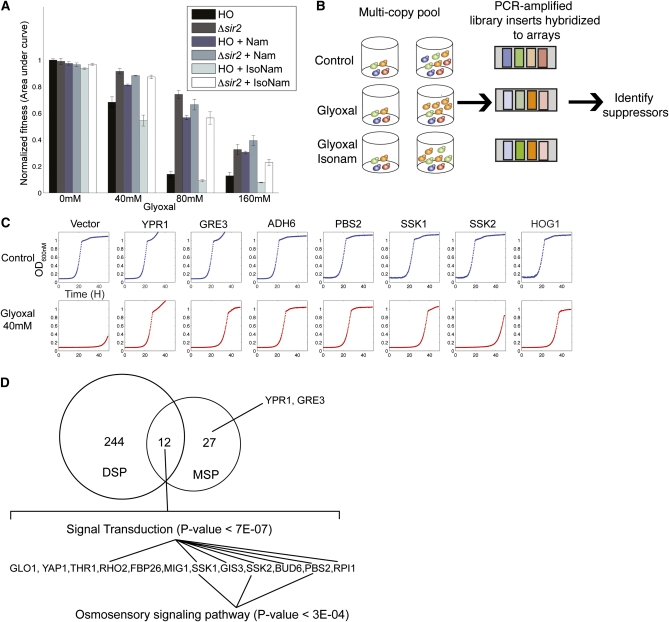
Figure 3 .
Sensitized glyoxal suppressor screen. A) Fitness ± SD (n = 3) of wild-type and SIR2 mutants grown in different glyoxal concentrations with and without supplementing nicotinamide (NAM) and isonicotinamide (IsoNAM) at 5 mM and 25 mM, respectively. B) Schematic of multicopy suppressor screen with glyoxal and isonicotinamide. A pool of yeast strains harboring a genomic library was grown competitively in the presence of control, glyoxal, and glyoxal supplemented with isonicotinamide. Plasmids were isolated and inserts were amplified by PCR and hybridized to ORF probes present on the TAG4 array. Over-represented ORFs in treatment vs. control are identified as candidate suppressors. C) Confirmatory growth curves of individually cloned suppressors identified by MSP. D) Overlap of genes identified by MSP and DSP. MSP genes include genes found in suppressor loci in Table 1. In cases where individual genes were not identified by subcloning, all genes in the suppressor loci were included. Genes that were found in both assays were enriched for components in the osmosensory signaling pathway by Gene Ontology (GO) analysis.
Table 1 . Multicopy suppressors identified by MSP.
| Locus | Log2 Ratio | Average Ratio | Confirmed Suppressor |
|---|---|---|---|
| Glyoxal | |||
| GLO1;YML003W | 2.52,3.25 | 2.88 | GLO1 |
| YDR366C;YDR367W;YPR1;XRS2 | 3.29,2.07,2.20,2.37 | 2.48 | YPR1 |
| YMR317W;ADH6 | 2.86,2.04 | 2.45 | ADH6 |
| PBN1;LRE1 | 2.13,2.35 | 2.24 | ND |
| Glyoxal + isonicotinamide | |||
| PBN1;LRE1 | 4.81,4.74 | 4.78 | ND |
| YDR366C;YDR367W;YPR1;XRS2 | 5.05,4.14,4.53,4.73 | 4.61 | YPR1 |
| YMR317W;ADH6 | 4.73,4.49 | 4.61 | ADH6 |
| MRPL28;STP1 | 3.07,4.24 | 3.66 | STP1 |
| MED8;SOY1;MSI1;PGI1 | 4.25,4.09,3.68,2.60 | 3.65 | MED8 |
| YML007C-A;YAP1;GIS4;[tS(AGA)M];TRM12;GLO1;YML003W | 2.40,2.46,2.33,0.00,4.90,5.80,6.43 | 3.47 | GLO1 |
| THR1;PPA1;RPN1 | 3.16,3.83,3.11 | 3.37 | THR1 |
| ZDS2;YML108W;PML39 | 3.60,3.48,2.86 | 3.31 | ZDS2 |
| NST1;RHO2 | 2.88,3.58 | 3.23 | RHO2 |
| YGR125W;YGR126W | 2.42,3.66 | 3.04 | YGR126W |
| FBP26;VPS35 | 2.95,2.96 | 2.96 | ND |
| CCC2;[YDR271C];GLO2;DON1 | 2.94,0.00,4.38,4.04 | 2.84 | GLO2 |
| YGL036W;MIG1 | 3.38,2.26 | 2.82 | ND |
| LCP5;YER128W | 2.94,2.65 | 2.79 | ND |
| SSL1;SSK1 | 2.50,3.09 | 2.79 | SSK1 |
| MIG2;SIP2 | 2.56,3.00 | 2.78 | MIG2 |
| GIS3;IOC2 | 2.14,3.39 | 2.77 | GIS3 |
| BUD22;ERG5;SOK2 | 2.40,2.76,2.91 | 2.69 | ND |
| STB1;KRI1 | 2.91,2.40 | 2.66 | ND |
| YKR023W;DBP7 | 2.73,2.45 | 2.59 | ND |
| TRR2;CDC12 | 2.55,2.52 | 2.54 | ND |
| NIP7;SRP72 | 2.57,2.44 | 2.50 | ND |
| ALG12;SSK2 | 2.00,2.79 | 2.40 | ND |
| RBA50;[snR84];HLR1;QCR7;APA2 | 3.36,0.00,3.30,3.04,2.19 | 2.38 | HLR1 |
| YGR016W;YGR017W | 2.42,2.18 | 2.30 | ND |
| BSP1;YPR172W;VPS4 | 2.14,2.38,2.39 | 2.30 | ND |
| DDP1;YOR164C | 2.27,2.29 | 2.28 | ND |
| CBS1;[YDL068W];COX9 | 2.00,0.00,2.48 | 1.49 | ND |
| Singletons | |||
| DAM1 | 3.55 | ND | |
| MTH1 | 3.10 | ND | |
| CDC34 | 3.00 | ND | |
| BUD6 | 2.90 | ND | |
| PBS2 | 2.90 | PBS2 | |
| RPI1 | 2.87 | ND | |
| SEC6 | 2.85 | ND | |
| YPT7 | 2.83 | ND | |
| DNA2 | 2.82 | ND | |
| GRE3 | 2.70 | GRE3 | |
Syntenic genes identified by MSP screen. Singletons are genes that do not have neighboring genes with high log2 ratios. ORFs in brackets [] are not present on the microarray and thus not assessed by MSP. ND, not tested by single ORF analysis.
Deletion- and overexpression-based assays are complementary
Surprisingly, only 4% (12/287) of genes were identified in both the overexpression-resistance and deletion-sensitivity assays (Figure 3D). There are multiple reasons for this lack of overlap, both biological and technical. These assays were designed to yield orthologous data; e.g., deletion screens are unlikely to identify genes with overlapping or redundant function (Tong et al. 2001). Overexpression screens can identify suppressor genes that are functionally redundant because expression of a single gene can confer resistance. Indeed, we identified several aldehyde reductases that, when overexpressed, behaved this way. GRE3 is an aldehyde reductase that is capable of metabolizing MG and is regulated by the HOG pathway (Aguilera and Prieto 2001; Aguilera et al. 2005). YPR1 is another aldehyde reductase that has high specificity for 2-methylbutyraldehyde (Ford and Ellis 2002). ADH6 is an alcohol dehydrogenase able to detoxify aldehydes in vivo (Larroy et al. 2002). All three genes strongly suppressed glyoxal toxicity in single-strain confirmations (Figure 3C). Although each of these genes was identified by overexpression profiling, none was sensitive to glyoxal as a single-gene deletion (Table S2), reflecting potential functional redundancy and underscoring the benefit of combining the results of multiple assays.
Despite the small overlap between loss-of-function and gain-of-function assays, 12 genes where identified in both assays (Figure 3D). These genes were enriched for signal transduction; e.g., three components of the HOG pathway (PBS2, SSK1, and SSK2) were identified. Overexpression of HOG1 conferred resistance to glyoxal (Figure 3C), even though it was not identified by MSP. On the basis of our earlier work, genes that ranked high in both assays were enriched for functions directly related to drug mechanism of action (Hoon et al. 2008); accordingly, we concluded that the HOG pathway is critical for glyoxal resistance, and because mutants in the glyoxalase pathway were much less sensitive (Figure 1C), we suggest that additional effectors of glyoxal resistance are regulated by the HOG pathway.
Integrating genome-wide glyoxal sensitivity and resistance data
Using the data from the deletion- and overexpression-based screens, we generated a ranked list of sensitive and resistant strains and performed epistasis analysis by systematically creating double mutants between genes involved in CS resistance and measuring their fitness in the presence of glyoxal using high resolution growth curve analysis on the approximately 800 multimutants. Epistasis, defined as the influence of a mutation in one gene on the phenotype of another, can be formalized so that any deviation from the predicted effects of combined mutations can be quantified to derive genetic interaction networks and dissect cellular pathways (Boone et al. 2007; Mani et al. 2008) (St. Onge et al. 2007). All possible combinations of double-deletion strains were constructed for 15 deletion strains sensitive to glyoxal, and all possible combinations between the same 15 sensitive deletion strains and 24 resistant deletion strains were constructed for a total of ∼800 double-deletions strains that were screened for fitness in the presence and absence of glyoxal (Figure 4A).We performed the screens at two glyoxal concentrations (5 mM and 10 mM) that were lower than the pooled screen concentration to increase the likelihood of identifying both alleviating and aggravating interactions. Each double mutant was constructed twice, and we found that fitness values between reciprocal gene pairs were highly correlated (Figure 4B). Using fitness values for double- and single-deletion strains, we quantified the genetic interaction (expressed as ε) between gene pairs, where εxy = Wxy – Wx × Wy. By performing our screens at multiple drug doses with both resistant and sensitive mutant strains (using high-resolution growth curves), we could identify subtle genetic interactions ranging from aggravation (ε < 0) to suppression (ε > 0). For example, in 5 mM glyoxal, loss of RPE1 severely aggravated glyoxal sensitivity of a Δpbs2 strain, whereas in 10 mM glyoxal, loss of ERV14 completely suppressed glyoxal sensitivity of a Δpbs2 strain (Figure 4C). The distribution of ε values showed that a greater number of genetic interactions were uncovered when the screen was performed in the presence of stress (St. Onge et al. 2007) (Figure 4D).
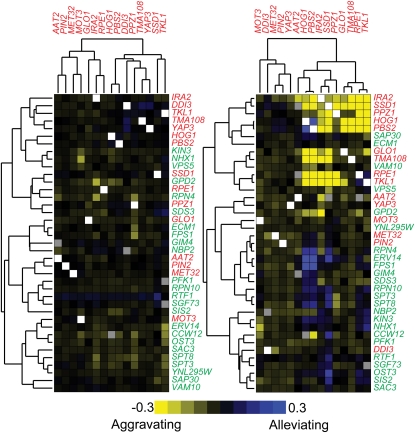
We summarized all the epistatic tests as a heatmap, with strains ordered by hierarchical clustering of their ε values (Figure 5). Genes of related function clustered together; e.g., HOG1 and PBS2 were highly correlated (r = 0.98) in both YPD and YPD + glyoxal. In contrast, other pairs of genes shared genetic interactions only in the presence of glyoxal. RPN10, which encodes the 19S regulatory particle (RP) of the 26S proteasome, clusters with SPT3 and SPT8, members of the SAGA transcriptional regulatory complex. It was recently shown that the 19S RP alters SAGA and stimulates its interaction with transcription activators (Lee et al. 2005a). We also note that a strain deleted for GCN5, the catalytic subunit of SAGA, was resistant to glyoxal (Table S3) and RPE1, an epimerase with roles in the non-oxidative part of the pentose phosphate pathway that has been shown to display increased sensitivity to hydrogen peroxide (Juhnke et al. 1996). In the presence of glyoxal, RPE1 clusters with TKL1, a transketolase that functions in the same pathway (Walfridsson et al. 1995), and both are involved in NADPH production, important for protection against reactive oxidative stress. Because alleviating interactions arise when a mutation in one gene impairs the function of a pathway, thereby masking the effects of mutations in other members of the same pathway (Mani et al. 2008), the alleviation observed between RPE1 and TKL1 (ε = 0.15) suggests that the two genes operate in the same pathway. In contrast, mutants of both RPE1 and TKL1 shared aggravating interactions with mutants of IRA2, SSD1, PPZ1, HOG1, and PBS2, indicating that they buffer glyoxal resistance via distinct pathways. Thus, our epistasis analysis of over 800 multimutants detected known relationships between genes in the same pathway and uncovered novel relationships between pathways that were detected only under glyoxal-induced stress.
Figure 5 .
Genetic interaction profiles predict pathways. Genes hierarchically clustered (Pearson correlation) according to similar patterns of genetic interaction (ε) for growth in YPD (left) and in 10 mM glyoxal (right). The heatmap is colored accordingly to the ε value of each double mutant: yellow for aggravating interactions (ε < 0) and blue for alleviating interactions (ε > 0). Genes identified as sensitive to glyoxal when deleted singly are highlighted in red, and genes identified as resistant to glyoxal when deleted singly are highlighted in green.
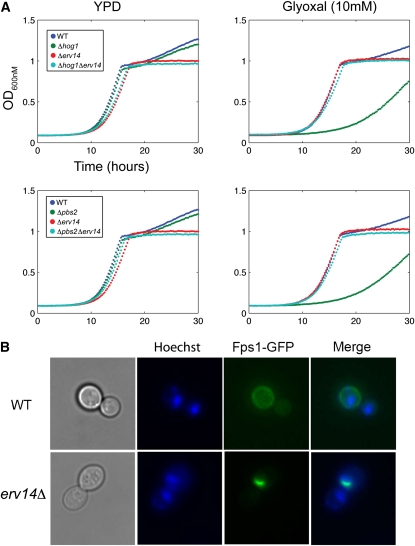
Loss of Fps1 and Erv14 abolishes the requirement for the HOG pathway
Erv14 is a transmembrane protein found in the ER and the early Golgi compartment involved in COPII cargo selection (Nakanishi et al. 2007; Powers and Barlowe 1998; Powers and Barlowe 2002). Our genetic interaction analysis showed that Erv14 clusters with Fps1, a membrane channel involved in glycerol export. Deletion of both genes resulted in complete suppression of glyoxal sensitivity of Δhog1 and Δpbs2 mutants, demonstrating that both FPS1 and ERV14 function downstream of the HOG pathway (Figure 6A). Fps1 is known to mediate the uptake of acetic acid, arsenite, and antimonite, and downregulation of Fps1 activity via Hog1 confers resistance to the same toxins (Mollapour and Piper 2007; Thorsen et al. 2006; Wysocki et al. 2001). On the basis of the degree of resistance conferred by loss of FPS1 to both Δhog1 and Δpbs2 mutants, we speculate that glyoxal may enter the cell via the Fps1 plasma membrane channel. Because loss of Fps1 also suppressed glyoxal sensitivity in other mutants (e.g., Δtma108, Δtkl1, and Δppz1) at much higher glyoxal concentrations (Figure S2), it is also possible that Erv14 regulates transport of Fps1 to the plasma membrane. To test the hypothesis that loss of Erv14 renders cells resistant to glyoxal by disrupting plasma membrane localization of Fps1, we examined a GFP-tagged allele of Fps1 and found that, in fact, localization of Fps1 to the plasma membrane was disrupted in Δerv14 mutants (Figure 6B). This observation confirmed that epistatic studies derived from the genome-wide screens can form the basis of specific, testable hypotheses.
Figure 6 .
Erv14 deletion abolishes requirement for HOG pathway. A) Growth curve of double mutants Δhog1Δerv14 (top row) and Δpbs2Δerv14 (bottom row) in YPD and glyoxal (10 mM). B) Live cell imaging of Fps1 distribution. Wild-type (BY4741) and Δerv14 cells expressing C-terminally GFP-tagged Fps1 from the pUG23-FPS1GFP plasmid were grown in synthetic media lacking methionine. DNA staining was performed with Hoescht stain 33258.
Discussion
Characterization of chemical stresses, such as MG and glyoxal, is challenging because numerous pathways (e.g., stress response and chemical detoxification) are involved. To address these challenges, we employed several unbiased chemical genomic assays. First, using competitive fitness measurements, we defined the genetic determinants of CS resistance and then determined the relationships between these genes in the presence of CS using multimutant analysis (St. Onge et al. 2007). The result is a comprehensive view of the physiology that underlies the cellular response to CS. We show that the genetic requirements for MG and glyoxal resistance are distinct and that different carbonyl stresses require distinct genetic cohorts for cell survival. We demonstrate the importance of macromolecule catabolism for resistance to MG, confirming and extending known mechanisms of MG toxicity. Downregulation of the glucose repression pathway was also implicated in the control of glyoxal resistance, specifically MIG1, SNF1, and MIG2 (Table 1).
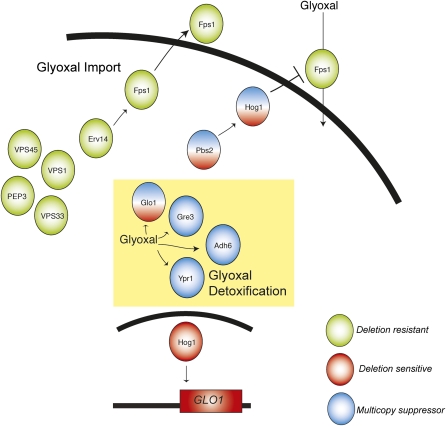
The effects of glyoxal on the cell are pervasive and deleterious, yet poorly understood. Integrating our observations, we suggest a model of glyoxal resistance in yeast (Figure 7) with the intention of generating testable hypotheses for follow-up studies. In this model, glyoxal detoxification is mediated via multiple reductases, GRE3, ADH6, YPR1 and GLO1, which were identified via multicopy suppression to confer glyoxal resistance. A functioning HOG pathway is critical for glyoxal resistance by regulating the expression of GLO1, which encodes a glyoxal detoxifying enzyme. In addition to GLO1, our data suggest that other downstream effectors participate in mediating glyoxal resistance. One candidate effector is FPS1, a plasma membrane channel that is targeted for endocytic degradation by Hog1 phosphorylation (Mollapour and Piper 2007). Deletion of FPS1 abolishes the sensitivity of HOG pathway mutants to glyoxal, similar to that observed for acetic acid, arsenite, and antimonite (Mollapour and Piper 2007; Thorsen et al. 2006; Wysocki et al. 2001). Several mutants responsible for ER and early Golgi transport were observed to be resistant to glyoxal. In particular, loss of ERV14 also abolishes sensitivity of HOG pathway mutants. We speculate that this is due to defective localization of Fps1 to the cell surface with a concomitant reduction in glyoxal levels. One way to test this model would be to measure the intracellular glyoxal concentrations during treatment of wild-type and mutant strains.
Figure 7 .
Model for glyoxal resistance. We highlight four main pathways necessary for glyoxal resistance. The HOG pathway positively regulates the expression of GLO1 and negatively regulates FPS1 in conferring glyoxal resistance. We hypothesize that the HOG pathway targets Fps1 for degradation, thereby reducing glyoxal import. Moreover, we hypothesize that transport of Fps1 to the cell surface is mediated by Erv14 and associated proteins. We also identified two other mechanisms by which glyoxal resistance is managed: 1) downregulation of the Ras/cAMP/PKA pathway and 2) glyoxal metabolism.
In summary, we show the broad range of specific cellular functions that are important for carbonyl stress resistance. Using multiple genome-wide approaches, we were able to comprehensively identify multiple genetic requirements for glyoxal stress resistance. Furthermore, a quantitative epistasis analysis comprising ∼800 strains allowed us to uncover glyoxal-dependent genetic interactions. These data comprise the first genome-wide assessment of the role of gene dose and of epistatic relationships with respect to their role in the response to carbonyl stress. Because many of the pathways affected by carbonyl stress are well conserved, our observations and their underlying datasets provide a resource for future studies in yeast and other cell types. For example, aging, while not typically thought of as a stress-related disorder, manifests many of the features of carbonyl accumulation–related toxicity. Accordingly, comprehensive gene dose studies may serve as a cost-effective and comprehensive approach for toxicity assessment toward achieving the goal of transforming both general and environmental health (Collins et al. 2008).
Supplementary Material
Acknowledgments
We thank members of the chemogenomics lab at the Stanford Genome Technology Center for discussions. Charlie Boone kindly supplied yeast strains and plasmids. S. Hoon is supported by a graduate fellowship from the Agency for Science Technology and Research (Singapore). R.W. Davis is supported by grants from the National Institutes of Health (NIH). G. Giaever and C. Nislow are supported by grants from the NIH and Canadian Institutes of Health Research (CIHR) (MOP-81340 to G.G.; MOP-84305 to C.N.).
Literature Cited
- Aguilera J., Prieto J. A., 2001. The saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 39: 273–283 [DOI] [PubMed] [Google Scholar]
- Aguilera J., Rodriguez-Vargas S., Prieto J. A., 2005. The HOG MAP kinase pathway is required for the induction of methylglyoxal-responsive genes and determines methylglyoxal resistance in saccharomyces cerevisiae. Mol. Microbiol. 56: 228–239 [DOI] [PubMed] [Google Scholar]
- Akhand A. A., Hossain K., Mitsui H., Kato M., Miyata T., et al. , 2001. Glyoxal and methylglyoxal trigger distinct signals for map family kinases and caspase activation in human endothelial cells. Free Radic. Biol. Med. 31: 20–30 [DOI] [PubMed] [Google Scholar]
- Barrera G., Pizzimenti S., Dianzani M. U., 2008. Lipid peroxidation: control of cell proliferation, cell differentiation and cell death. Mol. Aspects Med. 29: 1–8 [DOI] [PubMed] [Google Scholar]
- Beckman K. B., Ames B. N., 1998. The free radical theory of aging matures. Physiol. Rev. 78: 547–581 [DOI] [PubMed] [Google Scholar]
- Benov L., Fridovich I., 1998. Superoxide dependence of the toxicity of short chain sugars. J. Biol. Chem. 273: 25741–25744 [DOI] [PubMed] [Google Scholar]
- Bertram C., Hass R., 2008. Cellular responses to reactive oxygen species-induced DNA damage and aging. Biol. Chem. 389: 211–220 [DOI] [PubMed] [Google Scholar]
- Bito A., Haider M., Hadler I., Breitenbach M., 1997. Identification and phenotypic analysis of two glyoxalase II encoding genes from saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 272: 21509–21519 [DOI] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A., 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277: 45099–45107 [DOI] [PubMed] [Google Scholar]
- Blander G., Guarente L., 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73: 417–435 [DOI] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B. J., 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P., 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Cameron N. E., Gibson T. M., Nangle M. R., Cotter M. A., 2005. Inhibitors of advanced glycation end product formation and neurovascular dysfunction in experimental diabetes. Ann. N. Y. Acad. Sci. 1043: 784–792 [DOI] [PubMed] [Google Scholar]
- Collins F. S., Gray G. M., Bucher J. R., 2008. Toxicology. transforming environmental health protection. Science 319: 906–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Zeng J., Ou X., Ren X., Cai S., 2006. Methylglyoxal downregulates raf-1 protein through a ubiquitination-mediated mechanism. Int. J. Biochem. Cell Biol. 38: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Ellis E. M., 2007. Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol. Ther. 115: 13–24 [DOI] [PubMed] [Google Scholar]
- Falcon S., Gentleman R., 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Ford G., Ellis E. M., 2002. Characterization of Ypr1p from saccharomyces cerevisiae as a 2-methylbutyraldehyde reductase. Yeast 19: 1087–1096 [DOI] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J., 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Giaever G., Flaherty P., Kumm J., Proctor M., Nislow C., et al. , 2004. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA 101: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Harman D., 1981. The aging process. Proc. Natl. Acad. Sci. USA 78: 7124–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., et al. , 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., et al. , 2008. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4: 498–506 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Kimura A., 1996. Identification of the structural gene for glyoxalase I from saccharomyces cerevisiae. J. Biol. Chem. 271: 25958–25965 [PubMed] [Google Scholar]
- Inoue Y., Tsujimoto Y., Kimura A., 1998. Expression of the glyoxalase I gene of saccharomyces cerevisiae is regulated by high osmolarity glycerol mitogen-activated protein kinase pathway in osmotic stress response. J. Biol. Chem. 273: 2977–2983 [DOI] [PubMed] [Google Scholar]
- Juhnke H., Krems B., Kotter P., Entian K. D., 1996. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. 252: 456–464 [DOI] [PubMed] [Google Scholar]
- Kalapos M. P., 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 110: 145–175 [DOI] [PubMed] [Google Scholar]
- Kang Y., Edwards L. G., Thornalley P. J., 1996. Effect of methylglyoxal on human leukaemia 60 cell growth: modification of DNA G1 growth arrest and induction of apoptosis. Leuk. Res. 20: 397–405 [DOI] [PubMed] [Google Scholar]
- Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., et al. , 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484 [DOI] [PubMed] [Google Scholar]
- Larroy C., Fernandez M. R., Gonzalez E., Pares X., Biosca J. A., 2002. Characterization of the saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem. J. 361: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., et al. , 2005a. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436 [DOI] [PubMed] [Google Scholar]
- Lee W., St. Onge R. P., Proctor M., Flaherty P., Jordan M. I., et al. , 2005b. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeta K., Izawa S., Inoue Y., 2005. Methylglyoxal, a metabolite derived from glycolysis, functions as a signal initiator of the high osmolarity glycerol-mitogen-activated protein kinase cascade and calcineurin/Crz1-mediated pathway in saccharomyces cerevisiae. J. Biol. Chem. 280: 253–260 [DOI] [PubMed] [Google Scholar]
- Mani R., St. Onge R. P., Hartman J. L., 4th, Giaever G., Roth F. P., 2008. Defining genetic interaction. Proc. Natl. Acad. Sci. USA 105: 3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapes J., Ota I. M., 2004. Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J. 23: 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M., Piper P. W., 2007. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell. Biol. 27: 6446–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Saikusa T., Fukuda Y., Watanabe K., Inoue Y., et al. , 1986. Metabolism of 2-oxoaldehydes in yeasts. Possible role of glycolytic bypath as a detoxification system in L-threonine catabolism by saccharomyces cerevisiae. Eur. J. Biochem. 157: 297–301 [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N., Kamiya H., 2001. Methylglyoxal, an endogenous aldehyde, crosslinks DNA polymerase and the substrate DNA. Nucleic Acids Res. 29: 3433–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Suda Y., Neiman A. M., 2007. Erv14 family cargo receptors are necessary for ER exit during sporulation in saccharomyces cerevisiae. J. Cell Sci. 120: 908–916 [DOI] [PubMed] [Google Scholar]
- Nemet I., Varga-Defterdarovic L., Turk Z., 2006. Methylglyoxal in food and living organisms. Mol. Nutr. Food Res. 50: 1105–1117 [DOI] [PubMed] [Google Scholar]
- O’Brien P. J., Siraki A. G., Shangari N., 2005. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit. Rev. Toxicol. 35: 609–662 [DOI] [PubMed] [Google Scholar]
- Oldenburg K. R., Vo K. T., Michaelis S., Paddon C., 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25: 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C. N., Ahn B. W., Moerman E. J., Goldstein S., Stadtman E. R., 1987. Age-related changes in oxidized proteins. J. Biol. Chem. 262: 5488–5491 [PubMed] [Google Scholar]
- O’Rourke S. M., Herskowitz I., O’Shea E. K., 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18: 405–412 [DOI] [PubMed] [Google Scholar]
- Pierce S. E., Fung E. L., Jaramillo D. F., Chu A. M., Davis R. W., et al. , 2006. A unique and universal molecular barcode array. Nat. Methods 3: 601–603 [DOI] [PubMed] [Google Scholar]
- Poli G., Dianzani M. U., Cheeseman K. H., Slater T. F., Lang J., et al. , 1985. Separation and characterization of the aldehydic products of lipid peroxidation stimulated by carbon tetrachloride or ADP-iron in isolated rat hepatocytes and rat liver microsomal suspensions. Biochem. J. 227: 629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., Barlowe C., 1998. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the drosophila cornichon gene product. J. Cell Biol. 142: 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., Barlowe C., 2002. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol. Biol. Cell 13: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani N., Thornalley P. J., 2008. The dicarbonyl proteome: proteins susceptible to dicarbonyl glycation at functional sites in health, aging, and disease. Ann. N. Y. Acad. Sci. 1126: 124–127 [DOI] [PubMed] [Google Scholar]
- Rine J., Hansen W., Hardeman E., Davis R. W., 1983. Targeted selection of recombinant clones through gene dosage effects. Proc. Natl. Acad. Sci. USA 80: 6750–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jalm Y., Moree-Testa P., 1980. Study of nitrogen-containing compounds in cigarette smoke by gas chromatography-mass spectrometry. J. Chromatogr. A 198: 188–192 [DOI] [PubMed] [Google Scholar]
- Sauve A. A., Schramm V. L., 2004. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr. Med. Chem. 11: 807–826 [DOI] [PubMed] [Google Scholar]
- Shringarpure R., Davies K. J., 2002. Protein turnover by the proteasome in aging and disease. Free Radic. Biol. Med. 32: 1084–1089 [DOI] [PubMed] [Google Scholar]
- St. Onge R. P., Mani R., Oh J., Proctor M., Fung E., et al. , 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E. R., 1992. Protein oxidation and aging. Science 257: 1220–1224 [DOI] [PubMed] [Google Scholar]
- Stadtman E. R., 2006. Protein oxidation and aging. Free Radic. Res. 40: 1250–1258 [DOI] [PubMed] [Google Scholar]
- Stansfield I., Stark M. J. R., 2007. Yeast Gene Analysis. Elsevier/Academic Press, Amsterdam andBoston [Google Scholar]
- Thornalley P. J., 2003. Use of aminoguanidine (pimagedine) to prevent the formation of advanced glycation endproducts. Arch. Biochem. Biophys. 419: 31–40 [DOI] [PubMed] [Google Scholar]
- Thornalley P. J., 1998. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact. 111–112: 137–151 [DOI] [PubMed] [Google Scholar]
- Thornalley P. J., 1996. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification–a role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 27: 565–573 [DOI] [PubMed] [Google Scholar]
- Thorpe G. W., Fong C. S., Alic N., Higgins V. J., Dawes I. W., 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 101: 6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M., Di Y., Tangemo C., Morillas M., Ahmadpour D., et al. , 2006. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol. Biol. Cell 17: 4400–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array analysis in saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Vander Jagt D. L., Robinson B., Taylor K. K., Hunsaker L. A., 1992. Reduction of trioses by NADPH-dependent aldo-keto reductases. aldose reductase, methylglyoxal, and diabetic complications. J. Biol. Chem. 267: 4364–4369 [PubMed] [Google Scholar]
- Walfridsson M., Hallborn J., Penttila M., Keranen S., Hahn-Hagerdal B., 1995. Xylose-metabolizing saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61: 4184–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmka J., Hanneman J., Lee J., Amin D., Ota I., 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells-Knecht K. J., Zyzak D. V., Litchfield J. E., Thorpe S. R., Baynes J. W., 1995. Mechanism of autoxidative glycosylation: Identification of glyoxal and arabinose as intermediates in the autoxidative modification of proteins by glucose. Biochemistry 34: 3702–3709 [DOI] [PubMed] [Google Scholar]
- Wysocki R., Chery C. C., Wawrzycka D., Van Hulle M., Cornelis R., et al. , 2001. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in saccharomyces cerevisiae. Mol. Microbiol. 40: 1391–1401 [DOI] [PubMed] [Google Scholar]
- Zervas E., Montagne X., Lahaye J., 2002. Emission of alcohols and carbonyl compounds from a spark ignition engine. influence of fuel and air/fuel equivalence ratio. Environ. Sci. Technol. 36: 2414–2421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.