Abstract
Though highly efficient at fermenting hexose sugars, Saccharomyces cerevisiae has limited ability to ferment five-carbon sugars. As a significant portion of sugars found in cellulosic biomass is the five-carbon sugar xylose, S. cerevisiae must be engineered to metabolize pentose sugars, commonly by the addition of exogenous genes from xylose fermenting fungi. However, these recombinant strains grow poorly on xylose and require further improvement through rational engineering or evolutionary adaptation. To identify unknown genes that contribute to improved xylose fermentation in these recombinant S. cerevisiae, we performed genome-wide synthetic interaction screens to identify deletion mutants that impact xylose utilization of strains expressing the xylose isomerase gene XYLA from Piromyces sp. E2 alone or with an additional copy of the endogenous xylulokinase gene XKS1. We also screened the deletion mutant array to identify mutants whose growth is affected by xylose. Our genetic network reveals that more than 80 nonessential genes from a diverse range of cellular processes impact xylose utilization. Surprisingly, we identified four genes, ALP1, ISC1, RPL20B, and BUD21, that when individually deleted improved xylose utilization of both S. cerevisiae S288C and CEN.PK strains. We further characterized BUD21 deletion mutant cells in batch fermentations and found that they produce ethanol even the absence of exogenous XYLA. We have demonstrated that the ability of laboratory strains of S. cerevisiae to utilize xylose as a sole carbon source is suppressed, which implies that S. cerevisiae may not require the addition of exogenous genes for efficient xylose fermentation.
Keywords: recombinant yeast, ethanol, xylose, functional genomics, chemical genomics
Cellulosic fermentation for the production of fuels and chemicals has many advantages as cellulose, the main component of plant cell walls, is very abundant in agricultural and forestry waste and this feedstock does not compete with valuable food source (reviewed in Lynd et al. 2008; Rubin 2008). Though cellulose is abundant and renewable, numerous hurdles remain in making cellulosic ethanol production an economically viable industry. In contrast to cane sugar or starch fermentation, due to the complex nature of the carbohydrate present in cellulosic biomass, a significant amount of xylose and arabinose (5-carbon sugars derived from the hemicellulose portion of the lignocellulose) is present in the biomass hydrolysates (Saha 2003). Indeed, after glucose, D-xylose is the second most abundant sugar in hemicelluloses. Therefore, in order to maximize the potential for ethanol production, ethanologenic fermentation strains must be capable of utilizing both pentose and hexose sugars present in the lignocellulose.
Though Saccharomyces cerevisiae has an exceptional ability for rapid anaerobic growth and fermentation of hexose sugars, it has been generally reported that laboratory strains exhibit only a negligible metabolism of xylose (Chiang et al. 1981; Gong et al. 1983; Wang et al. 1980). This phenotype is in spite of the fact that S288C laboratory yeast has endogenous genes that appear to encode a putative xylose utilization pathway (Figure 1). The present strategy to improve the ability of S. cerevisiae to ferment xylose has been the introduction of exogenous genes from xylose-fermenting fungi (reviewed in Hahn-Hagerdal et al. 2007; Matsushika et al. 2009; Van Vleet and Jeffries 2009). The most common strategy involves the introduction of xylose reductase (XR) and xylitol dehydrogenase (XDH) genes and the overexpression of the endogenous xylulokinase gene (XKS1) (Hallborn et al. 1991; Ho et al. 1998; Jin et al. 2002; Kotter et al. 1990). As the XR/XDH pathway can result in a cofactor imbalance that has been shown to negatively impact metabolic flux (reviewed in Hahn-Hagerdal et al. 2007; Matsushika et al. 2009; Van Vleet and Jeffries 2009), a second strategy has emerged that introduces a bacterial xylose isomerase (XI) gene into yeast, which allows for the slow metabolism of xylose via the endogenousXks1 (Karhumaa et al. 2005; Kuyper et al. 2003; Madhavan et al. 2009; Walfridsson et al. 1996). Considerable effort has been made to improve pentose fermentation, including engineering or optimizing xylose enzyme activity (XR, XK, XDH, and XI), xylose transport, and the pentose phosphate pathway; reducing redox imbalances; and other strategies (reviewed in Hahn-Hagerdal et al. 2007; Matsushika et al. 2009). Despite significant directed efforts focusing on known proteins or pathways impacting xylose utilization and fermentation, ethanol production from xylose remains inefficient in recombinant S. cerevisiae strains and suggests that novel strategies should be considered.
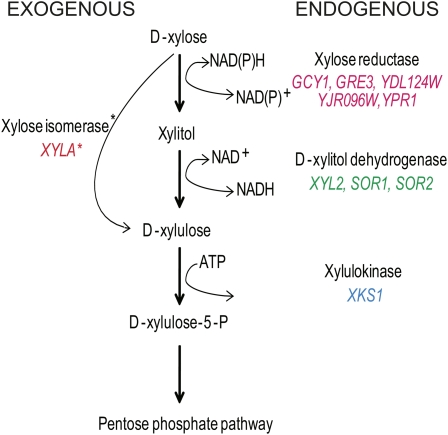
Figure 1 .
The xylose utilization pathway in S. cerevisiae, including xylose isomerase strategy to improve xylose utilization. Putative endogenous S. cerevisiae genes of the xylose utilization pathway are listed on the right and marked in different colors: pink for genes involved in xylose reducatase pathways; green for xylitol dehydrogenase; and blue for xylulokinase. The exogenous xylose isomearse gene XYLA is listed on the left, highlighted in red, and marked by *. For this study, XYLA was isolated from Piromyces Sp E2. Adapted from Wenger et al. (2010).
It is clear that despite the common belief that S. cerevisiae cannot use xylose as a carbon source, many wild and industrial wine yeast strains are in fact capable of xylose utilization (Attfield and Bell 2006; Wenger et al. 2010). The ability of some industrial yeast to utilize xylose was recently mapped to a putative xylitol dehydrogenase gene, XDH1 that is not present in the laboratory S288C strain (Wenger et al. 2010). Importantly, Wenger et al. (2010) determined that XDH1 requires the endogenous XR genes GRE3 and YPR1 and the endogenous XK gene XKS1 to allow for xylose utilization. In contrast, they also found that three putative XDH genes, SOR1, SOR2, and XYL2, suppressed the ability of XDH1 expressing strains to utilize xylose (Wenger et al. 2010). This work not only illustrated that S. cerevisiae is genetically “primed” to ferment xylose but also suggested that additional endogenous proteins may either positively or negatively impact the ability of S. cerevisiae to utilize xylose. In an attempt to isolate such proteins, we performed genome-wide synthetic genetic array (SGA) screens (Tong et al. 2001) to identify deletion mutants that impact the xylose utilization of strains expressing the XI gene XYLA from Piromyces sp. E2 (herein referred to as pXYLA) and strains expressing both XYLA and an additional copy of the endogenous XK gene XKS1 (herein referred to as pXYLA,XKS1). To compliment this effort, we screened the deletion mutant array to identify mutants whose growth is either negatively or positively impacted by xylose. Our genetic network reveals that more than 80 nonessential genes from a diverse range of cellular processes impact xylose utilization. Surprisingly, we identified four deletion mutants that improved xylose consumption of laboratory S288C, even in the absence of exogenous XYLA. Next, we deleted these genes in an ethanol-tolerant CEN.PK strain and confirmed that all four mutants improved xylose utilization and that the deletion of BUD21 improves xylose fermentation. These data suggest that the natural ability of S. cerevisiae to recognize and utilize xylose was suppressed, and they validate the use of systems biology approaches using S288C as a means to identify novel pathways contributing to xylose fermentation.
Materials and Methods
Yeast strains
The yeast strains used in this study are listed in Table 1. The MATa deletion mutant array (DMA) was purchased from OpenBiosystems (Catalog no. YSC1053). The deletion strains generated for this study were designed using a standard PCR-mediated gene insertion technique (Longtine et al. 1998) and confirmed by PCR analysis using two sets of primer pairs (sequences available upon request). Plasmids were transformed into wild-type cells using a standard LiAc protocol (Gietz and Schiestl 2007).
Table 1 . S. cerevisiae strains used in this study.
| Strain Name | Genetic Background | Origin |
|---|---|---|
| Y7092 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ | Tong and Boone 2006 |
| YKB2179 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ pXYLA | This study |
| YKB2178 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ pXYLA,XKS1 | This study |
| YKB2069 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+alp1Δ::kanMX6 | This study |
| YKB2530 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ isc1Δ::kanMX6 | This study |
| YKB2531 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ rpl20bΔ::kanMX6 | This study |
| YKB2532 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ bud21Δ::kanMX6 | This study |
| YKB2534 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ alp1Δ::kanMX6 isc1Δ::NAT | This study |
| YKB2535 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ alp1Δ::kanMX6 rpl20bΔ::NAT | This study |
| YKB2536 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ isc1Δ::kanMX6 rpl20bΔ::NAT | This study |
| YKB2537 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ bud21Δ::kanMX6 alp1Δ::NAT | This study |
| YKB2538 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ bud21Δ::kanMX6, isc1Δ::NAT | This study |
| YKB2539 | MATα can1Δ::STE2pr-SP-his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 LYS2+ bud21Δ::kanMX6 rpl20bΔ::NAT | This study |
| YKB2684 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 (CEN.PK 113-13D) | Entian and Koetter 1998 |
| YKB2680 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 + pXYLA,XKS1 | This study |
| YKB2666 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 bud21Δ::kanMX6 | This study |
| YKB2667 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 bud21Δ::kanMX6 + pXYLA,XKS1 | This study |
| YKB2668 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 alp1Δ::kanMX6 | This study |
| YKB2670 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 isc1Δ::kanMX6 | This study |
| YKB2665 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 rpl20bΔ::kanMX6 | This study |
Plasmid construction for xylose isomerase (XYLA) and xylulokinase (XKS1)
The XYLA and XKS1 genes were cloned into the pYES2 shuttle vector (Invitrogen), each preceded by the Kozak sequence (GCCACC) and flanked by the triosephosphate isomerase constitutive promoter (TPIp) and thecytochrome C1 transcription terminator (CYC1tt). First, the GAL1 promoter sequence was removed from pYES2, and then TPIp and XYLA were amplified by PCR with genomic DNA from S. cerevisiae and Piromyces sp. E2 genomic DNA, respectively, and inserted upstream of CYC1tt, as described previously (Kuyper et al. 2003), resulting in pYES2-XYLA-CYC1tt [herein called pXYLA (pKB60)]. TPIp was obtained using the primer combination of TPI-NheI-Fwd 5′-GATCGCTAGCTGTTsTAAAGATTACGGATAT-3′ and TPI-EcoRI-Rev 5′-GATCGAATTCTTTTAGTTTATGTATGTGTTTTTTGTAG-3′. Primers for PCR amplification from S. cerevisiae were based on genomic sequence data for strain S288C, although the strain used as DNA template was CEN.PK113-13D. Phusion HF DNA polymerase (Finnzymes) was used for all PCR reactions. Next, a XKS1-CYC1tt-TPIp cassette was assembled. A second TPIp was amplified by PCR using primer pair TPI-HindIII-Fwd 5′-GATCAAGCTTTGTTTAAAGATTACGGATAT-3′ and TPI-EcoRI-Rev. CYC1tt could not be successfully PCR-amplified from the pYES2 vector, so the following strategy was taken to excise it for cloning: pYES2 was digested with NsiI and religated upon itself, reducing it to a 2.2-kbp mini-pYES2 version of itself, still containing the bla resistance gene CYC1tt with a single NsiI site just upstream of it and the pUC origin. A HindIII site was introduced 100 bp downstream of CYC1tt by PCR-amplifying mini-pYES2 with the primer combination of pYES-CYC-AvrII-Fwd 5′-GATCCCTAGGTTCGGCTGCGGCGAGCGGTA-3′ and pYES-CYC-HindIII-AvrII-Rev 5′-GATCCCTAGGAAGCTTCGACCGAGCGCAGCGAGTCA-3′. The resulting PCR product was digested with AvrII, ligated upon itself, and then transformed into Escherichia coli DH5α. The vector was reisolated and digested with NsiI and HindIII to release the CYC1tt fragment. XKS1 was amplified by PCR from S. cerevisiae genomic DNA with primer pairXKS1-EcoRI-Fwd 5′-GATCGAATTCGCCACCATGAGAGTCTTTTCCAGTTCGCTTAA-3′ andXKS1-NsiI-Rev 5′-GATCATGCATATGTTGTGTTCAGTAATTCAGAGACAG-3′. The XKS1 and new TPIp PCR products were digested with NsiI and HindIII, respectively, and in a single reaction ligated together with the CYC1tt fragment. The resulting cassette was reamplified by PCR using primersXKS1-EcoRI-Fwd and TPI-EcoRI-Rev, digested with EcoRI, and inserted at the EcoRI site between the first TPIp and XYLA in pYES2-TPIp-xylA-CYC1tt, resulting in the final pYES2-TPIp-XKS1-CYCtt-TPIp-XYLA-CYC1tt vector [herein called pXYLA,XKS1 (pKB61)].
Growth conditions and dot assay experiments
Cells were grown in standard YEP or synthetic complete (SC) media supplemented with glucose or xylose to a final concentration of 2%, unless otherwise described. To assess growth under xylose and glucose conditions, wild-type cells were grown in YEP media supplemented with either 2% glucose or xylose, and transformed cells were grown in SC-uracil (to maintain the plasmid) supplemented with either 2% xylose or glucose. Cells were grown to midlog phase in the carbon source of interest, and then either growth curve or dot assays were performed. Semi-aerobic growth curve analysis was performed in triplicate on Multiskan Ascent plate reader (Thermo Electron Corporation) in sealed plates, at 30° with shaking at 480 rpm prior to the OD600 measurements that were taken every 30 min over a 20-hr period. Dot assays were performed by spotting 5 µl of 10-fold serial dilutions (OD600 = 0.1, 0.01, 0.001, 0.0001) onto specified media, and sealed plates were incubated at 30°. All growth curve and dot assay experiments were repeated using three different isolates of each strain.
Glucose and xylose consumption assays
Independent yeast colonies were cultured in 2 ml of selective media supplemented with either 2% glucose or xylose and incubated at 30° overnight with shaking. Following overnight culturing, 500 µl of the culture was transferred into 10 ml of fresh media supplemented with 2% xylose or glucose and incubated for 24 hr at 30°. Cells were harvested by centrifugation, washed once with sterile YEP media, and pitched at 1.5 × 107 cells/ml into shaker flasks containing either 100 ml of glucose- or xylose-containing media at 13 g/l. Flasks were shaken at 250 rpm at 30°. Consumption assays were performed in triplicate, and samples were taken at 24-hr intervals to check for sugar utilization. The glucose concentration was determined using the Glucose (GO) Assay Kit (Sigma GAGO-20) per the manufacturer’s guidelines. Xylose concentration was determined using the phloroglucinol assay (Eberts et al. 1979). Briefly, the color reagent of 0.5 g of phloroglucinol (Sigma), 100 ml of glacial acetic acid, and 10 ml of concentrated HCl was freshly prepared and kept in the dark. Stock standard xylose (10 g/l) was prepared by dissolving D-xylose powder in saturated benzoic acid (Sigma) and used for preparation of the calibration curve. Samples (200 µl) were mixed with 5 ml color reagent and subsequently heated at 100° for 4 min. The reaction was rapidly cooled to room temperature in iced water, and the absorbance at 540 nm was recorded.
Xylose chemical genomic and XYLA and XYLA,XKS1 synthetic genetic interaction screens
Robotic manipulation of the deletion mutant array was conducted using a Singer RoToR HDA (Singer Instruments). For the genome-wide XYLA and XYLA,XKS1 SGA screens, the MATa query strain Y7092 (Tong and Boone 2006) was transformed with either pXYLA (YKB2179) or pXYLA,XKS1 (YKB2178). The resulting query strains were mated to the MATa deletion mutant array, and SGA methodology (Tong et al. 2001; Tong et al. 2004) was used with the modifications previously described to maintain selection of the plasmid (Measday et al. 2005). After the final round of pinning on SD-uracil (2% glucose), the DMA containing the plasmids were subsequently pinned onto plates containing 2% xylose (SX-uracil). To identify deletion mutants that displayed growth defects or advantages on xylose, the deletion mutant array was also screened directly on 2% xylose plates. All three genome-wide screens were performed in triplicate at 30°, and growth was visually scored for slow growth, lethality, or suppression after one day on glucose and two days on xylose. Putative genetic interactions identified in a minimum of two out of three replicates in any of the five screens (DMA on xylose, pXYLA on glucose, pXYLA,XKS1 on glucose, pXYLA on xylose, and pXYLA,XKS1 on xylose) were confirmed in all five conditions. In brief, the deletion mutant was transformed using traditional methods (Gietz and Schiestl 2007) with the vector control pRS415 (Sikorski and Hieter 1989), pXYLA, or pXYLA,XKS1, and a series of dot assays were performed on SC-uracil media with either 2% glucose or 2% xylose as the sole carbon source at 30°. Confirmed genetic interactions and xylose sensitivity are listed insupporting information,Table S1.
Aerobic xylose fermentation
Independent yeast colonies, isolated from YPD-agar plates (1% yeast extract, 2% peptone, 2% glucose, 2% agar), were first cultivated in 50 ml conical tubes containing 15 ml YEP (1% yeast extract, 2% peptone) medium supplemented with 2% (20 g/l) glucose at 30°, 200 rpm. Inoculum cultures were started by transferring 500 µl of the tube-grown cultures into 250 ml flasks containing 25 ml YEP medium supplemented with 2% (20 g/l) xylose or 2% (20 g/l) glucose (depending on the sugar to be used in the batch fermentations) and incubated for 24 hr at 30° and 200 rpm. The cells from these precultures were harvested by centrifugation at 18,000 g for 5 min at 4°, washed twice with sterile YEP media, and then used to inoculate final batch fermentations at an initial OD620nm = 0.1. Batch fermentations were performed using 250 ml flasks containing 50 ml of YEP medium supplemented with 2% (20 g/l) of the desired carbon source (xylose or glucose) as indicated and incubated in a rotary shaker at 30° and 200 rpm. Samples for determination of biomass and metabolite concentration were periodically withdrawn under sterile conditions. The batches were terminated when no further changes in OD were observed, as after the sugar has been depleted the cells can begin to consume the ethanol. The data plotted were observed by reading until the maximum concentration of ethanol was observed.
Kinetic parameters calculation
For the characterization of the strains used in this work, specific rates of growth (µ), glucose consumption (qGlc), xylose consumption (qXyl), ethanol production (qEtOH), yield of ethanol on glucose (YEtOH/Glc), and yield of ethanol on xylose (YEtOH/Xyl) were determined. The µ, qGlc, and qXyl values were calculated during exponential growth phase. Because growth rates and ethanol production kinetics differed among studied strains, qEtOH, YEtOH/Glc, and YEtOH/Xyl were calculated considering only the ethanol production phase, defined as the period from starting one sample before ethanol was detected up to the point when a sharp decrease in ethanol accumulation was observed. Following the same criteria, plots were constructed using only the data corresponding to the ethanol production phase. Flask cultures were performed at least in duplicate. The values reported represent the means of the experiments performed.
Biomass and metabolite analyses
Cell growth was followed as optical density at 620 nm (spectrophotometer GENESYS20, ThermoFisher Scientific). Biomass was determined as dry-cell weight as described previously (Parachin et al. 2010). Glucose, xylose, ethanol, xylitol, acetate, and glycerol were analyzed by high-performance liquid chromatography (HPLC) (UltiMate 3000, Dionex) with refractive index detector (Shodex). Samples were loaded onto an Aminex HPX-87H ion exchange column (BioRad) operated at 42° and eluted with 5 mm H2SO4 at a flow rate of 0.4 ml/min.
Results
Genome-wide chemical and synthetic interaction analysis identifies novel regulators of xylose utilization
Our goal was to use SGA methodology to systematically screen the laboratory S288C yeast deletion mutant array to identify genetic determinants of xylose fermentation. As S288C has limited ability to utilize xylose (Chiang et al. 1981; Gong et al. 1983; Wang et al. 1980), our first goal was to construct a SGA query strain (Tong et al. 2001) with improved growth on xylose. Two plasmids, one expressing the Piromyces sp. E2 XI gene XYLA and the other expressing the combination of XYLA and the endogenous XK gene XKS1, were introduced into the wild-type laboratory strain (A, B inFigure S1). A series of growth curve analyses determined that, although the addition of pXYLA endowed growth on xylose, those cells expressing both XYLA and additional XKS1 (pXYLA,XKS1) displayed a further improvement in growth on xylose (Figure S2). Similarly, in aerobic batch fermentations, strains containing pXYLA or pXYLA,XKS1 displayed 14-fold and 15-fold improvement in xylose consumption, respectively, compared to the wild-type strains (C inFigure S2). These results demonstrate that the introduction of XYLA and additional XKS1 improves the ability of the SGA query strain to utilize xylose as a carbon source, and they replicate previous studies using exogenous XI genes (reviewed in Van Maris et al. 2007).
We performed genome-wide SL-SGA screens with both pXYLA- and pXYLA,XKS1-containing query strains. In brief, query strains containing pXYLA or pXYLA,XKS1 were mated to the yeast deletion mutant array, and the SGA methodology was used to incorporate the plasmids into the deletion mutants (Tong et al. 2001). The growth of the deletion mutants containing the plasmids was tested on both glucose (Figure 2) and xylose (Figure 3) media. As a control, the deletion mutant array (without pXYLA or pXYLA,XKS1) was also screened for growth on xylose. Deletion mutants or deletion mutant–plasmid combinations that resulted in synthetic lethality (SL), slow growth (SG), or improved growth (suppressor) were identified (see Materials and Methods). To confirm the chemical and genetic interactions, regardless of which screen the mutant was identified in, each mutant was independently transformed with three different plasmids (vector control, pXYLA, and pXYLA,XKS1), dot assays were performed on plates containing either 2% glucose or 2% xylose as the carbon source, and growth was scored (seeTable S1 for a full list of interactions).
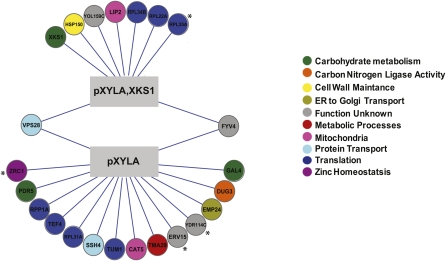
Figure 2 .
Genetic interaction network of pXYLA and pXYLA,XKS1 on glucose. Genome-wide synthetic interaction SGA screens were performed using query strains that were transformed with pXYLA (YKB2179) and pXYLA,XKS1 (YKB2178). Genes are represented by nodes that are color-coded according to their SGD cellular roles (www.yeastgenome.org) and/or assigned through review of the literature. Interactions are represented by edges. Deletion mutants that display synthetic lethal interactions are indicated by *.
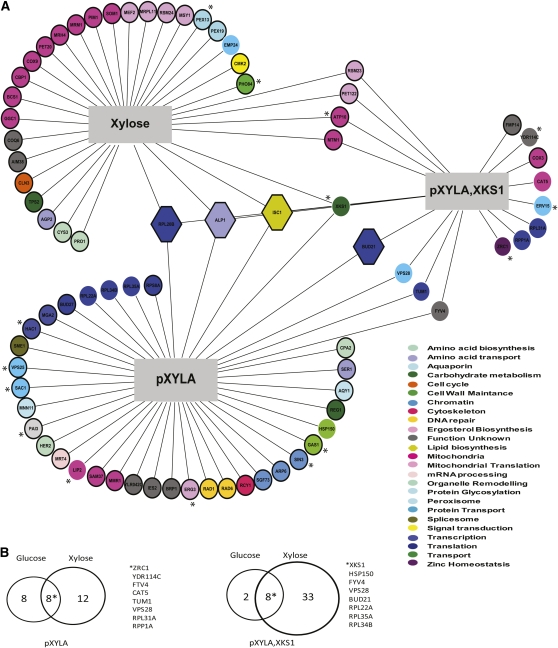
Figure 3 .
Xylose sensitivity and genetic interaction network of pXYLA and pXYLA,XKS1 on xylose. (A) Genome-wide synthetic interaction SGA screens were performed using query strains that were transformed with pXYLA (YKB2179) and pXYLA,XKS1 (YKB2178) and screened xylose plates. In addition, a chemical genomic screen was performed screening the deletion mutant array directly on xylose plates. Genes are represented by nodes that are color-coded according to their SGD cellular roles (www.yeastgenome.org) and/or assigned through review of the literature. Interactions are represented by edges. For deletion mutants displaying SG or SL interactions, the nodes are circles. For the deletion mutants that display improved growth phenotype (suppressors), the nodes are hexagons. Genes that interact with pXYLA or pXYLA,XKS1 only on xylose have their nodes outlined in black. Deletion mutants that display synthetic lethal interactions are indicated by *. (B) Venn diagrams showing the number of overlapping genes in each of the screens performed: pXYLA on xylose and glucose and pXYLA,XKS1 on xylose and glucose.
The addition of pXYLA or pXYLA,XKS1 negatively affected the growth of 23 deletion mutants on glucose, and each plasmid had largely distinct genetic interactions (Figure 2). The addition of pXYLA resulted in slow growth in 13 deletion mutants and was lethal to deletion mutants of the vacuole zinc transporter ZRC1 (MacDiarmid et al. 2000) and the uncharacterized genes ERV15 and YDR114c. While additional XKS1 relieved the lethality in these three strains, it caused lethality in deletion mutants of the large 60S ribosomal subunit RPL35A (Venema and Tollervey 1999) and slow growth in 8 additional deletion mutants. Only deletion mutants of the ESCRT-I component VPS28 and the uncharacterized gene FYV4 were sensitive to both plasmids. This result suggests that, although the addition of the exogenous gene XYLA and increased XKS1 expression did not detectably impact the growth rate or consumption of glucose in a S288C wild-type background (Figure S2), their addition is not neutral and has cellular consequences in certain deletion mutant backgrounds. Although there have been contradictory reports regarding the impact of XKS1 overexpression on growth rate and xylose consumption (Ho et al. 1998; Johansson et al. 2001; Matsushika and Sawayama 2008; Richard et al. 2000; Rodriguez-Pena et al. 1998; Toivari et al. 2001), it is clear that XKS1 driven by the TPI1 promoter not only increases xylose consumption (C inFigure S2) but also results in a synthetic dosage effect in many mutant backgrounds.
The majority of strains in the deletion mutant array could grow, albeit slowly, on media where the sole carbon was 2% xylose. We found that 4 deletion mutants were not viable (ATP10, PEX13, PHO84, and XKS1) on xylose and 26 deletion mutants displayed slow growth on xylose (Figure 3,Table S1). Remarkably, the vast majority of xylose-sensitive mutants could be rescued by the addition of pXYLA or pXYLA,XKS1 plasmid. The exception to this was XKS1. As expected, xks1Δ cells could not grow on 2% xylose, but they could be partially rescued by pXYLA or pXYLA,XKS1. As pXYLA,XKS1 could only partially rescue xks1Δ on xylose, it suggests that both copies of XKS1, the endogenous genomic copy and the plasmid-borne copy, are necessary to promote maximum xylose utilization S288C.
Similar to the results on glucose, pXYLA and pXYLA,XKS1 displayed largely distinct genetic interactions on xylose (Figure 3A). While pXYLA negatively affected the growth of 16 deletion mutants on xylose (3 SL and 13 SG), pXYLA,XKS1 negatively affected the growth of 37 deletion mutants on xylose (8 SL and 29 SG). For both plasmids, the genetic interactions they displayed on glucose were not all replicated on xylose media (Figure 3B). This result indicates that the detrimental effects of XYLA or XKS1 expression on glucose in certain deletion mutant backgrounds can be alleviated by growth on xylose. The pXYLA and pXYLA,XKS1 xylose interaction network is not limited to known players in carbohydrate metabolism; rather, the network implies that genes associated with diverse cellular pathways are required for XI-mediated xylose metabolism, including translation, DNA repair, and protein transport. For example, of the 53 genes identified between the two plasmids, FUNSPEC analysis (Robinson et al. 2002) indicates that a significant number encode components of the cytoplasmic ribosomal large subunit (P = 4.1e−5) or are implicated in the GO biological process of translation elongation (P = 0.003805). This chemical and functional genomics study dramatically expands our knowledge of proteins and pathways that are required for xylose utilization and optimal function of the XI/XK pathway.
Xylose utilization is inhibited by suppressors in S288C
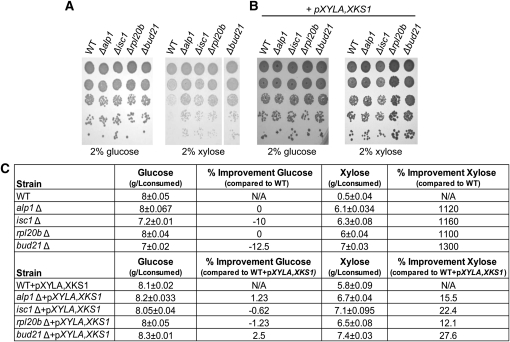
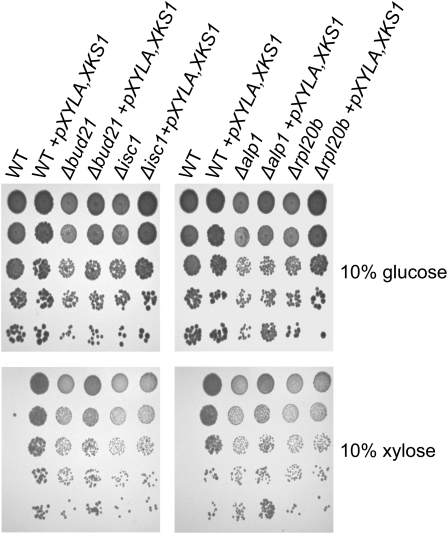
The genome-wide chemical and genetic screens identified four deletion mutants that improved growth on xylose: RPL20B, ALP1, ISC1, and BUD21. All four putative suppressors, which to date have not been linked to pentose metabolism, are implicated in three diverse biological processes.Alp1 is an arginine transporter (Regenberg et al. 1999),Isc1 is a mitochondrial membrane–localized inositol phosphosphingolipid phospholipase C (Betz et al. 2002),Rpl20B is a component of the large (60S) ribosomal subunit (Lecompte et al. 2002; Planta and Mager 1998; Venema and Tollervey 1999), andBud21 is a component of the small ribosomal subunit (SSU) processosome (Dragon et al. 2002). To confirm that the null mutants of these genes confer advantageous growth on xylose, the genes were directly knocked out of a S288C laboratory strain, and a series of growth analyses and sugar consumption assays were performed (Figure 4). Deletion of these genes did not detectably impact growth on glucose (Figure 4A), and only deletion of ISC1 and BUD21 resulted in minor decreases in glucose consumption (Figure 4C). However, when the suppressor strains were grown on xylose media, they grew better than the wild-type strain (Figure 4A) and had dramatic improvements in xylose consumption (Figure 4C). We next asked if these suppressors could improve the consumption of xylose over and above that conferred by pXYLA,XKS1. Though the suppressors in combination with pXYLA,XKS1 did not dramatically improve growth on xylose, the deletion of BUD21 and RPL20B displayed reproducibly minor improvements of growth on xylose (Figure 4B). The suppressors of all these genes improved the consumption of xylose of strains transformed with pXYLA,XKS1 from a modest 12% for RPL20B to 27.5% for BUD21. We next asked if a combination of the suppressors might result in a further improvement of xylose utilization. Though no double mutant was deleterious to growth on glucose or xylose media (data not shown), improvements in xylose consumption were either negligible or modest at best (Table S2). This finding suggests that the proteins encoded by these genes, although not presently linked, may be working through the same pathway or mechanisms to inhibit xylose consumption in S288C S. cerevisiae.
Figure 4 .
Deletion of ALP1, BUD21, ISC1, or RPL20B improve the xylose utilization of S288C yeast even in the absence of exogenous XI. Wild-type (WT, Y7092), alp1Δ (YKB2069), isc1Δ (YKB2530), rpl20bΔ (YKB 2531), and bud21Δ (YKB2532) cells without (A) or with pXYLA,XKS1 (B) were plated in 10-fold serial dilutions onto YEP plates containing glucose or xylose. Plates were incubated at 30° for 1 day on glucose and 2 days on xylose. (C) Analysis of the consumption of glucose and xylose of the four suppressor mutants, alp1Δ (YKB2069), isc1Δ (YKB2530), rpl20bΔ (YKB 2531), and bud21Δ (YKB2532), compared to wild-type (WT, Y7092), either alone or transformed with pXYLA,XKS1. Growth cultures were started in either 13% glucose or xylose media, pitched with 1.5 × 107 cells/ml and carried out at 30° with for 168 hr. The calculation for g/l of sugars consumed corresponds to the concentrations of sugars observed at the end of the batches compared to the starting amount.
Bud21 suppresses xylose utilization and fermentation in CEN.PK 113-3D
We next wanted to determine whether the suppressors identified in S288C also functioned as suppressors of xylose utilization in other yeast strains. For this purpose, we utilized the ethanol-tolerant MATa CEN.PK 113-3D strain (Van Dijken et al. 2000) that has previously been used in numerous xylose fermentation studies (Eliasson et al. 2000; Johansson et al. 2001; Van Vleet et al. 2008). Deletion mutants in CEN.PK 133-3D (hereafter called CEN.PK) were generated for BUD21, ISC1, ALP1, and RPL20b and subsequently transformed with pXYLA,XKS1. As seen for S288C, deletion of the suppressors resulted in the dramatic improvement of the ability of CEN.PK to grow on xylose plates, even in the absence of pXYLA,XKS1 (Figure 5).
Figure 5 .
Deletion of ALP1, BUD21, ISC1, or RPL20B improve the xylose utilization of CEN.PK yeast. CEN.PK 133-13D wild-type (WT, YKB2684), alp1Δ (YKB2668), isc1Δ (YKB2670), rpl20bΔ (YKB2665), and bud21Δ (YKB2666) cells, alone or transformed with pXYLA,XKS1, were plated in 10-fold serial dilutions onto YEP plates containing glucose or xylose. Plates were incubated at 30° for 1 day on glucose and 2 days on xylose.
We next performed a series of controlled aerobic batch fermentation studies to assess whether one of the suppressors could improve xylose fermentation. BUD21 was selected as its deletion resulted in the greatest improvement in xylose consumption in the S288C background with and without the addition pXYLA,XKS1 (Figure 4). The fermentations were performed using CEN.PK and CEN.PK bud21Δ with and without pXYLA,XKS1 in both glucose (Table 2,Figure S3) and xylose (Table 3, Figure 6). For xylose and glucose fermentation, the concentrations of xylose or glucose, ethanol, acetate, and glycerol were measured, along with the cell biomass. In addition, xylitol levels were measured in the xylose fermentation. During glucose fermentation, the addition of pXYLA,XKS1 or deletion of BUD21 resulted in an increment in the specific growth rate and final biomass levels compared with the control strain; however, this rise in biomass production resulted in nearly identical decreases in both ethanol and glycerol production (Table 2,Figure S3). Surprisingly, the CEN.PK strain containing both the bud21Δ and pXYLA,XKS1 not only caused a further reduction in ethanol and glycerol production but also a dramatic increase in acetate production. This increase may reflect the increase in growth rate of this strain and the associated energetic demand.
Table 2 . Kinetic parameters from Saccharomyces cerevisiae CEN.PK 113-13D derivative strains grown aerobically in YEP medium supplemented with glucose 20 g/l.
| Strain | Biomassa (gDCW/l) | µ (h−1) | qGlc (gGlc/gDCW⋅h) | qEtOH (gEtOH/gDCW−h) | Acetatea (g/l) | Glycerola (g/l) | Ethanola (g/l) | YEtOH/Glc (gEtOH/gGlc) |
|---|---|---|---|---|---|---|---|---|
| WT | 5.018 ± 0.184 | 0.354 ± 0.041 | 0.236 ± 0.019 | 0.144 ± 0.018 | 0.495 ± 0.022 | 1.429 ± 0.313 | 10.586 ± 1.683 | 0.535 ± 0.071 |
| WT pXYLA,XKS1 | 6.791 ± 0.345 | 0.420 ± 0.004 | 0.238 ± 0.020 | 0.128 ± 0.002 | 0.732 ± 0.032 | 0.637 ± 0.016 | 7.609 ± 0.121 | 0.443 ± 0.011 |
| bud21Δ | 6.915 ± 0.170 | 0.416 ± 0.002 | 0.329 ± 0.005 | 0.134 ± 0.006 | 0.670 ± 0.055 | 0.705 ± 0.080 | 7.647 ± 0.121 | 0.473 ± 0.031 |
| bud21Δ pXYLA,XKS1 | 7.978 ± 0.046 | 0.549 ± 0.009 | 0.131 ± 0.003 | 0.052 ± 0.003 | 2.810 ± 0.120 | 0.201 ± 0.055 | 5.809 ± 0.308 | 0.326 ± 0.001 |
Aerobic batch fermentations were performed in YEP supplemented with 2% glucose as described in the Materials and Methods using CEN.PK 133-13D wild-type (WT, YKB2684), WT transformed with pXYLA,XKS1 (YKB2680), bud21Δ (YKB2666), and bud21Δ transformed with pXYLA,XKS1 (YKB2667). Values listed are the average ± SE of duplicate experiments. Specific rates of growth (µ), glucose consumption (qGlc), ethanol production (qEtOH), and yield of ethanol on glucose (YEtOH/Glc) were determined. Values of µ and qGlc were calculated during exponential growth phase.
Values obtained at the end of each fermentation.
Table 3 . Kinetic parameters from Saccharomyces cerevisiae CEN.PK 113-13D derivative strains grown aerobically in YEP medium supplemented with xylose 20 g/l.
| Strain | Biomassa (gDCW/l) | µ (h−1) | qXyl (gXyl/gDCW⋅h) | qEtOH (gEtOH/gDCW−h) | Xylitola (g/l) | Acetatea (g/l) | Glycerola (g/l) | Ethanola (g/l) | YEtOH/Xyl (gEtOH/gXyl) |
|---|---|---|---|---|---|---|---|---|---|
| WT | 0.298 ± 0.014 | 0.018 ± 0.02 | ND | ND | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | ND |
| WT pXYLA,XKS1 | 11.567 ± 0.199 | 0.132 ± 0.006 | 0.031 ± 0.001 | 0.010 ± 0.000 | 0.580 ± 0.164 | 0.525 ± 0.038 | 0.264 ± 0.048 | 5.295 ± 0.425 | 0.352 ± 0.003 |
| bud2Δ1 | 12.922 ± 0.117 | 0.143 ± 0.006 | 0.031 ± 0.001 | 0.006 ± 0.001 | 1.350 ± 0.083 | 0.301 ± 0.042 | 0.399 ± 0.070 | 3.189 ± 0.170 | 0.201 ± 0.017 |
| bud21Δ pXYLA,XKS1 | 2.841 ± 0.04 | 0.265 ± 0.004 | 0.061 ± 0.005 | 0.017 ± 0.004 | 0.294 ± 0.048 | 0.297 ± 0.019 | 0.000 ± 0.000 | 1.033 ± 0.041 | 0.315 ± 0.018 |
Aerobic batch fermentations were performed in YEP supplemented with 2% xylose as described in Materials and Methods using CEN.PK 113-13D wild-type (WT, YKB2684), WT transformed with pXYLA,XKS1 (YKB2680), bud21Δ (YKB2666), and bud21Δ transformed with pXYLA,XKS1 (YKB2667). Values are the average ± SE of triplicate experiments. Specific rates of growth (µ), xylose consumption (qXyl), ethanol production (qEtOH), and yield of ethanol on xylose (YEtOH/Xyl) were determined. Values of µ and qXyl were calculated during exponential growth phase.
Values obtained at the end of each fermentation.
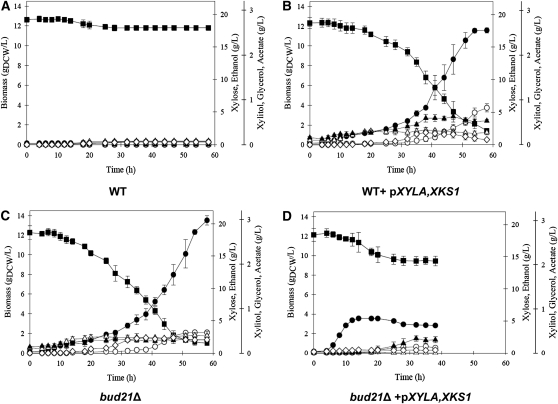
Figure 6 .
Deletion of BUD21 improves the xylose fermentation of CEN.PK yeast. Fermentation profile of the S. cerevisiae CEN.PK 113-13D derivative strains during aerobic batch cultivation in xylose (20 g/l). (A) CEN.PK 113-13D wild-type (WT, YKB2684), (B) WT transformed with pXYLA,XKS1 (YKB2680), (C) bud21Δ (YKB2666), and (D) bud21Δ transformed with pXYLA,XKS1 (YKB2667). Biomass (●), xylose (▪), ethanol (○), glycerol (△), acetate (▴), and xylitol (⋄). The batches were terminated when no further changes in OD were observed. Each data point represents the mean ± SD from triplicate experiments.
As expected, the control CEN.PK strain showed no increase in biomass or ethanol production in xylose (Figure 6, Table 3). In case of the CEN.PK strain containing pXYLA,XKS1, there was a significant increase in biomass and ethanol production (5.295 g/l), and a low level of xylitol was detected in the cells at the end of the fermentation (Figure 6, Table 3). The greatest biomass increase was observed for the suppressor-deficient strain bud21Δ, once again confirming that the strain is capable of sustained growth in xylose media. Although the bud21Δ strain was also able to produce ethanol (3 g/l), its ethanol production may be hindered due to a buildup of xylitol in the cells (Table 3), which may in turn be the result of a redox imbalance in the cells. Unfortunately, the combination of bud21Δ and pXYLA,XKS1 did not result in an improvement in fermentative capacity in xylose (Figure 6D). Despite the fast initial burst in growth (increased μ), biomass did not increase over the course of the experiment and ethanol production was low (1 g/l), although its overall ethanol yield (YEtOH/Xyl) was nearly equivalent to the CEN.PK pXYLA,XKS1 strain (Table 3). Although combining the XI with the deletion of the suppressor BUD21 did not lead to additive ethanol production, the SGA screen identified a deletion mutant that can consume xylose and produce ethanol at production levels and kinetics similar to strains containing exogenous xylose pathway genes (Matsushika et al. 2009). Not only does this result justify the continued use of functional genomics as a tool for identifying novel genetic strategies to improve industrial yeast strains but it also implies that S. cerevisiae may not require the addition of exogenous genes for efficient xylose fermentation.
Discussion
The majority of studies to date on improving xylose utilization in S. cerevisiae have taken a candidate gene or directed approach (reviewed in Van Vleet and Jeffries 2009). Unfortunately, these approaches are limited by our knowledge of the pathways/proteins regulating xylose utilization. To our knowledge, this is the first study to apply genome-wide chemical and synthetic genetic interaction screens to systematically assess the contribution of nonessential genes to xylose utilization via both endogenous and exogenous XI pathways. Indeed, despite the wealth of genetic interactions generated by large-scale SGA screening (Costanzo et al. 2010; Tong et al. 2001; Tong et al. 2004), only four genetic interactions have been identified for XKS1 (Costanzo et al. 2010; Fiedler et al. 2009), which may reflect the fact that most SGA screens have been conducted on standard glucose media.
We found that, in spite of the numerous reports indicating that laboratory S. cerevisiae strains cannot utilize xylose (Batt et al. 1986; Chiang et al. 1981; Gong et al. 1983; Wang et al. 1980), the vast majority of deletion mutant strains could survive when xylose was the sole carbon source (Figure 3A). This is not unexpected as the S. cerevisiae genome encodes a putative xylose utilization pathway (Figure 1) and many members of the Saccharomyces sensu stricto group have the ability to utilize xylose as a carbon source (Attfield and Bell 2006; Wenger et al. 2010). While XKS1 was identified as being essential for growth on xylose, no other genes encoding components of endogenous xylose pathway (Figure 1) were identified in any of the screens. This is likely because there are multiple putative redundant genes that encode for xylose reductase and D-xylitol dehydrogenase, whereasXks1 may be the sole xylulokinase. That said, pXYLA alone could rescue xks1Δ growth on xylose, which suggests that there may be alternative proteins/pathways in yeast that can convert D-xylulose and that there may be alternative noncanonical XR and XDH genes, too. Many of the xylose-sensitive deletion mutants are, as expected, implicated in mitochondrial functions because, during growth on xylose, the redox cofactors NADH and NADPH are involved in catabolism and biosynthesis (Mo et al. 2009; Salusjarvi et al. 2003; Shi et al. 2002). Remarkably, the vast majority of xylose-sensitive mutants could be rescued by the addition of pXYLA or pXYLA,XKS1 plasmid. One interpretation of this result is that the proteins encoded by the xylose-sensitive genes are essential for or contribute to S288C’s endogenous xylose utilization pathway but that they do not detectably impact xylose utilization mediated when XI is present. Alternatively, as mitochondrial mutants display fitness defects on nonfermentable carbons sources (Steinmetz et al. 2002), the detected growth defect may not be specific to xylose but apply to any nonpreferred carbon source.
Using the SGA methodology, we systematically screened the contribution of nonessential yeast genes to XI/XK-mediated xylose metabolism (Figure 3A,Table S1). One striking feature of the network was the abundance of deletion mutants identified that encode proteins implicated in various aspects of translation. Similarly, genes implicated in translation were upregulated in XDH1 xylose-positive strains (Wenger et al. 2010). This result suggests that translation plays a key role in remodeling the cell for xylose utilization, which is understandable in light of the dramatic changes in the transcriptome (Bengtsson et al. 2008; Jin et al. 2002; Jin et al. 2004; Kuyper et al. 2005; Runquist et al. 2009; Salusjarvi et al. 2006; Wenger et al. 2010) and proteome (Karhumaa et al. 2009; Salusjarvi et al. 2008; Salusjarvi et al. 2003) that occurred upon xylose exposure. This result may also explain the identification of genes implicated in chromatin remodeling or transcription and protein transport. Understanding why these proteins are required for XYLA,XKS1-dependent utilization on xylose may reveal novel genetic mechanisms to improve xylose utilization of industrial strains.
One of the most interesting findings of this study was the identification of four genes, BUD21, ALP1, ISC1, and RPL20B, which suppress the natural ability of yeast to utilize xylose. Deletion of these genes resulted in increased xylose consumption and growth on xylose media to levels similar to yeast containing the XI/XK system (Figures 4 and 5). Hence, there may be fitness disadvantages in retaining the ability to consume xylose that laboratory yeast has evolved to inhibit. How are the suppressors inhibiting xylose utilization? Of the four suppressors identified, bud21Δ had the greatest impact on xylose consumption (Figure 4).Bud21 is a component of the small ribosomal subunit (SSU) processosome that contains U3 snoRNA (Dragon et al. 2002), and expression of BUD21 mRNA is upregulated in high-sucrose media, possibly due a strong osmotic response (Ando et al. 2006; Perez-Torrado et al. 2010).Bud21 is also involved in ribosomal protein biogenesis and processing, functions transiently repressed by stress conditions, such as those experienced during a fermentation environment (Hirasawa et al. 2010). Therefore, deletion of certain aspects of the stress response may be advantageous, allowing for bypassing some of the initial stress conditions that occur during xylose fermentation. Alternatively, both the XR/XDH and XI/XK xylose utilization pathways cause dramatic transcriptional remodeling of the cell (Bengtsson et al. 2008; Fendt and Sauer 2010; Hector et al. 2009; Jin et al. 2004; Pitkanen et al. 2003; Runquist et al. 2009; Salusjarvi et al. 2008; Salusjarvi et al. 2003; Wenger et al. 2010). Therefore, the suppressors may all be regulating some aspect of transcription or translation, which is likely the case ofRpl20b andBud21. Of course, these two possibilities are not mutually exclusive as the stress response is largely mediated by remodeling of the cells’ transcriptome and proteome. As robust stress-tolerance is essential for yeast to efficiently ferment cellulosic hydrolysates at an industrial scale, if the suppressors are functioning by modulating aspects of stress response, they may not be an appropriate genetic strategy for the improvement of industrial yeast strains. The mechanism by which these proteins inhibit xylose utilization in yeast will require further study to assess their value for the cellulosic biofuel industry.
We found that deletion of the suppressor BUD21 improved growth on xylose and that CEN.PK bud21Δ cells were able to ferment xylose and produce ethanol (Figure 6, Table 3). Unfortunately, the combination of bud21Δ and pXYLA,XKS1 did not result in further increases in ethanol production; rather, it was detrimental in the batch fermentations (Figure 6, Table 3). Though there was an initial fast burst in biomass accumulation, CEN.PK bud21Δ pXYLA,XKS1 cells plateaued in growth much sooner than CEN.PK wild-type cells. This is surprisingly as neither S288C nor CEN.PK bud21Δ transformed with pXYLA,XKS1 displayed growth defects on xylose plates (Figures 4 and 5). One possibility is that, although the cultures were aerobic, oxygen may be limiting at the high cellular concentrations of batch fermentations, effects that do not occur on plates. The exact reason for the differences in growth on plates and culture require further investigation. However, it is clear that, at least in the case of deletion of BUD21, activation of S. cerevisiae’s natural ability to utilize and ferment xylose is not synergistic with exogenous pXYLA,XKS1 in aerobic xylose fermentations. It may be beneficial to exploit the molecular “barcodes” of yeast mutant collections and microarray-based methods (reviewed in Smith et al. 2010) to identify mutants that improve either recombinant XI/XK or XR/XDH/XK xylose fermentation in anaerobic batch cultures that better replicate industrial-scale biofuel production. Remarkably, the performance of the CEN.PK bud21Δ strain in batch fermentation in media in which xylose is the sole carbon source was similar or better than many recombinant yeast strains (Matsushika et al. 2009). This finding suggests the addition of exogenous genes may not be the sole method of improving the xylose fermentation efficiency of Saccharomyces; rather, efforts should focus on identifying and optimizing the endogenous xylose utilization pathway in yeast.
Supplementary Material
Acknowledgments
The authors thank Dr. P. Kötter, Institute of Microbiology, J.W. Goethe Universität, Frankfurt, Germany, for the kind gift of the CEN.PK 113-13D yeast strain. This work was supported by a grant to K.B., R.M., and V.J.J.M. by the Agricultural Bioproducts Innovation Program of Agriculture and Agri-Food Canada and by grants to K.B. from the National Sciences and Engineering Research Council of Canada (NSERC) and an Early Researcher Award from the Ontario Government. K.B. is a Canada Research Chair (CRC) in Chemical and Functional Genomics, and V.M is a CRC in Microbial Genomics and Engineering. P.Q. was supported by a NSERC Undergraduate Student Research Award.
Footnotes
Supporting information is available online athttp://www.g3journal.org/lookup/suppl/doi:10.1534/g3.111.000695/-/DC1.
Literature Cited
- Ando A., Tanaka F., Murata Y., Takagi H., Shima J., 2006. Identification and classification of genes required for tolerance to high-sucrose stress revealed by genome-wide screening of Saccharomyces cerevisiae. FEM. Yeast Res. 6: 249–267 [DOI] [PubMed] [Google Scholar]
- Attfield P. V., Bell P. J., 2006. Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEM. Yeast Res. 6: 862–868 [DOI] [PubMed] [Google Scholar]
- Batt C. A., Caryallo S., Easson D. D., Jr, Akedo M., Sinskey A. J., 1986. Direct evidence for a xylose metabolic pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 28: 549–553 [DOI] [PubMed] [Google Scholar]
- Bengtsson O., Jeppsson M., Sonderegger M., Parachin N. S., Sauer U., et al. , 2008. Identification of common traits in improved xylose-growing Saccharomyces cerevisiae for inverse metabolic engineering. Yeast 25: 835–847 [DOI] [PubMed] [Google Scholar]
- Betz C., Zajonc D., Moll M., Schweizer E., 2002. ISC1-encoded inositol phosphosphingolipid phospholipase C is involved in Na+/Li+ halotolerance of Saccharomyces cerevisiae. Eur. J. Biochem. 269: 4033–4039 [DOI] [PubMed] [Google Scholar]
- Chiang L. C., Gong C. S., Chen L. F., Tsao G. T., 1981. d-Xylulose fermentation to ethanol by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 42: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., et al. , 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberts T. J., Sample R. H., Glick M. R., Ellis G. H., 1979. A simplified, colorimetric micromethod for xylose in serum or urine, with phloroglucinol. Clin. Chem. 25: 1440–1443 [PubMed] [Google Scholar]
- Eliasson A., Christensson C., Wahlbom C. F., Hahn-Hagerdal B., 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66: 3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Koetter P., 1998. Yeast mutant and plasmid collections, pp. 431–449 Yeast Gene Analysis, edited by Brown A., Tuite M. Academic Press, San Diego [Google Scholar]
- Fendt S. M., Sauer U., 2010. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biol. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D., Braberg H., Mehta M., Chechik G., Cagney G., et al. , 2009. Functional organization of the S. cerevisiae phosphorylation network. Cell 136: 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 35–37 [DOI] [PubMed] [Google Scholar]
- Gong C. S., Claypool T. A., McCracken L. D., Maun C. M., Ueng P. P., et al. , 1983. Conversion of pentoses by yeasts. Biotechnol. Bioeng. 25: 85–102 [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B., Karhumaa K., Jeppsson M., Gorwa-Grauslund M. F., 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108: 147–177 [DOI] [PubMed] [Google Scholar]
- Hallborn J., Walfridsson M., Airaksinen U., Ojamo H., Hahn-Hagerdal B., et al. , 1991. Xylitol production by recombinant Saccharomyces cerevisiae. Biotechnology (N. Y.) 9: 1090–1095 [DOI] [PubMed] [Google Scholar]
- Hector R. E., Bowman M. J., Skory C. D., Cotta M. A., 2009. The Saccharomyces cerevisiae YMR315W gene encodes an NADP(H)-specific oxidoreductase regulated by the transcription factor Stb5p in response to NADPH limitation. New Biotechnol. 26: 171–180 [DOI] [PubMed] [Google Scholar]
- Hirasawa T., Furusawa C., Shimizu H., 2010. Saccharomyces cerevisiae and DNA microarray analyses: what did we learn from it for a better understanding and exploitation of yeast biotechnology? Appl. Microbiol. Biotechnol. 87: 391–400 [DOI] [PubMed] [Google Scholar]
- Ho N. W., Chen Z., Brainard A. P., 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. S., Jones S., Shi N. Q., Jeffries T. W., 2002. Molecular cloning of XYL3 (D-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl. Environ. Microbiol. 68: 1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. S., Laplaza J. M., Jeffries T. W., 2004. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 70: 6816–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B., Christensson C., Hobley T., Hahn-Hagerdal B., 2001. Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl. Environ. Microbiol. 67: 4249–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa K., Hahn-Hagerdal B., Gorwa-Grauslund M. F., 2005. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22: 359–368 [DOI] [PubMed] [Google Scholar]
- Karhumaa K., Pahlman A. K., Hahn-Hagerdal B., Levander F., Gorwa-Grauslund M. F., 2009. Proteome analysis of the xylose-fermenting mutant yeast strain TMB 3400. Yeast 26: 371–382 [DOI] [PubMed] [Google Scholar]
- Kotter P., Amore R., Hollenberg C. P., Ciriacy M., 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18: 493–500 [DOI] [PubMed] [Google Scholar]
- Kuyper M., Harhangi H. R., Stave A. K., Winkler A. A., Jetten M. S., et al. , 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEM. Yeast Res. 4: 69–78 [DOI] [PubMed] [Google Scholar]
- Kuyper M., Hartog M. M., Toirkens M. J., Almering M. J., Winkler A. A., et al. , 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEM. Yeast Res. 5: 399–409 [DOI] [PubMed] [Google Scholar]
- Lecompte O., Ripp R., Thierry J. C., Moras D., Poch O., 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30: 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lynd L. R., Laser M. S., Bransby D., Dale B. E., Davison B., et al. , 2008. How biotech can transform biofuels. Nat. Biotechnol. 26: 169–172 [DOI] [PubMed] [Google Scholar]
- MacDiarmid C. W., Gaither L. A., Eide D., 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19: 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan A., Tamalampudi S., Ushida K., Kanai D., Katahira S., et al. , 2009. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl. Microbiol. Biotechnol. 82: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Matsushika A., Inoue H., Kodaki T., Sawayama S., 2009. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 84: 37–53 [DOI] [PubMed] [Google Scholar]
- Matsushika A., Sawayama S., 2008. Efficient bioethanol production from xylose by recombinant saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J. Biosci. Bioeng. 106: 306–309 [DOI] [PubMed] [Google Scholar]
- Measday V., Baetz K., Guzzo J., Yuen K., Kwok T., et al. , 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102: 13956–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M. L., Palsson B. O., Herrgard M. J., 2009. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachin N. S., Bengtsson O., Hahn-Hagerdal B., Gorwa-Grauslund M. F., 2010. The deletion of YLR042c improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Yeast 27: 741–751 [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R., Panadero J., Hernandez-Lopez M. J., Prieto J. A., Randez-Gil F., 2010. Global expression studies in baker's yeast reveal target genes for the improvement of industrially-relevant traits: the cases of CAF16 and ORC2. Microb. Cell Fact. 9: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen J. P., Aristidou A., Salusjarvi L., Ruohonen L., Penttila M., 2003. Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab. Eng. 5: 16–31 [DOI] [PubMed] [Google Scholar]
- Planta R. J., Mager W. H., 1998. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14: 471–477 [DOI] [PubMed] [Google Scholar]
- Regenberg B., During-Olsen L., Kielland-Brandt M. C., Holmberg S., 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36: 317–328 [DOI] [PubMed] [Google Scholar]
- Richard P., Toivari M. H., Penttila M., 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190: 39–43 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Grigull J., Mohammad N., Hughes T. R., 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pena J. M., Cid V. J., Arroyo J., Nombela C., 1998. The YGR194c (XKS1) gene encodes the xylulokinase from the budding yeast Saccharomyces cerevisiae. FEMS Microbiol. Lett. 162: 155–160 [DOI] [PubMed] [Google Scholar]
- Rubin E. M., 2008. Genomics of cellulosic biofuels. Nature 454: 841–845 [DOI] [PubMed] [Google Scholar]
- Runquist D., Hahn-Hagerdal B., Bettiga M., 2009. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell Fact. 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccharomyces Genome Database, 2011 Saccharomyces Genome Database.http://www.yeastgenome.org
- Saha B. C., 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30: 279–291 [DOI] [PubMed] [Google Scholar]
- Salusjarvi L., Kankainen M., Soliymani R., Pitkanen J. P., Penttila M., et al. , 2008. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salusjarvi L., Pitkanen J. P., Aristidou A., Ruohonen L., Penttila M., 2006. Transcription analysis of recombinant saccharomyces cerevisiae reveals novel responses to xylose. Appl. Biochem. Biotechnol. 128: 237–261 [DOI] [PubMed] [Google Scholar]
- Salusjarvi L., Poutanen M., Pitkanen J. P., Koivistoinen H., Aristidou A., et al. , 2003. Proteome analysis of recombinant xylose-fermenting Saccharomyces cerevisiae. Yeast 20: 295–314 [DOI] [PubMed] [Google Scholar]
- Shi N. Q., Cruz J., Sherman F., Jeffries T. W., 2002. SHAM-sensitive alternative respiration in the xylose-metabolizing yeast Pichia stipitis. Yeast 19: 1203–1220 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., Ammar R., Nislow C., Giaever G., 2010. A survey of yeast genomic assays for drug and target discovery. Pharmacol. Ther. 127: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Scharfe C., Deutschbauer A. M., Mokranjac D., Herman Z. S., et al. , 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31: 400–404 [DOI] [PubMed] [Google Scholar]
- Toivari M. H., Aristidou A., Ruohonen L., Penttila M., 2001. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab. Eng. 3: 236–249 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- van Dijken J. P., Bauer J., Brambilla L., Duboc P., Francois J. M., et al. , 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26: 706–714 [DOI] [PubMed] [Google Scholar]
- van Maris A. J., Winkler A. A., Kuyper M., de Laat W. T., van Dijken J. P., et al. , 2007. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv. Biochem. Eng. Biotechnol. 108: 179–204 [DOI] [PubMed] [Google Scholar]
- Van Vleet J. H., Jeffries T. W., 2009. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol. 20: 300–306 [DOI] [PubMed] [Google Scholar]
- Van Vleet J. H., Jeffries T. W., Olsson L., 2008. Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on D-xylose. Metab. Eng. 10: 360–369 [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D., 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Walfridsson M., Bao X., Anderlund M., Lilius G., Bulow L., et al. , 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62: 4648–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Y., Shopsis C., Schneider H., 1980. Fermentation of a pentose by yeasts. Biochem. Biophys. Res. Commun. 94: 248–254 [DOI] [PubMed] [Google Scholar]
- Wenger J. W., Schwartz K., Sherlock G., 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.