Abstract
The level and pattern of nucleotide variation in duplicate gene provide important information on the evolutionary history of polyploids and divergent process between homoeologous loci within lineages. Kengyilia is a group of allohexaploid species with the StYP genomic constitutions in the wheat tribe. To investigate the evolutionary dynamics of the Pgk1 gene in Kengyilia and its diploid relatives, three copies of Pgk1 homoeologues were isolated from all sampled hexaploid Kengyilia species and analyzed with the Pgk1 sequences from 47 diploid taxa representing 18 basic genomes in Triticeae. Sequence diversity patterns and genealogical analysis suggested that (1) Kengyilia species from the Central Asia and the Qinghai-Tibetan plateau have independent origins with geographically differentiated P genome donors and diverged levels of nucleotide diversity at Pgk1 locus; (2) a relatively long-time sweep event has allowed the Pgk1 gene within Agropyron to adapt to cold climate triggered by the recent uplifts of the Qinghai-Tibetan Plateau; (3) sweep event and population expansion might result in the difference in the dN/dS value of the Pgk1 gene in allopatric Agropyron populations, and this difference may be genetically transmitted to Kengyilia lineages via independent polyploidization events; (4) an 83 bp MITE element insertion has shaped the Pgk1 loci in the P genome lineage with different geographical regions; (5) the St and P genomes in Kengyilia were donated by Pseudoroegneria and Agropyron, respectively, and the Y genome is closely related to the Xp genome of Peridictyon sanctum. The interplay of evolutionary forces involving diverged natural selection, population expansion, and transposable events in geographically differentiated P genome donors could attribute to geographical differentiation of Kengyilia species via independent origins.
Introduction
Duplication is a prominent feature of plant genomic architecture. Genome duplication or polyploidy provides a reservoir of duplicate genes as substrates for potential evolutionary innovation [1]. Analysis of the levels of diversity and the patterns of substitution in duplicate gene not only traces evolutionary history of polyploids [2], but also provides insight into how the evolutionary process differs between lineages and between homoeologous loci within lineages [3], [4]. Theoretical and empirical investigation suggested that the diversity of duplicate gene is unlikely equivalent, and may arise from various forms of natural selection [3], [5]–[7], population size and history [8], introgression [9], mating system [10], recombination [11], mutation rate [6], and gene conversion [12]. It has been reported that transposable element indels shaped the homoeologous loci, which was responsible for the patterns of diversity of duplicate gene [13]. In addition, forces acting on the levels and patterns of diversity also arise from the domestication bottlenecks [14]. Therefore, differences in the levels and patterns of nucleotide diversity of duplicate gene may reflect numerous forcing factors. To segregate the effects of various forcing factors, it is necessary to obtain evolutionary dynamic data from additional homoeologous loci within a given phylogenetic framework [3].
Kengyilia Yen et J. L. Yang, a polyploid perennial genus in the wheat tribe (Poaceae: Triticeae), includes about 22 perennial species distributed in a different range of natural habitats over the upper and middle mountain ranges of Central Asia and the Qinghai-Tibetan Plateau [15]. Cytogenetic evidence suggested that Kengyilia species arose from two hybridization events followed by genome doubling of three ancestral diploid species with different genomes St, Y and P [15]–[19]. The St and P genomes are derived from Pseudoroegneria (Nevski) Á Löve and Agropyron Gaertn., respectively [20]. It is unknown where the Y genome originates, although it is a fundamental Kengyilia genome [19]. Dewey [21] considered that the Y genome has its origin in Central Asia or the Himalaya region, and may be extinct. Analysis of some StY genome species using β-amylase gene sequences yielded distinct presumed Y-genome starch synthase sequences [22]. Based on ITS sequence analysis, Liu et al. [23] suggested that the Y genome might originate from the St genome. However, data presented by Sun et al. [24] suggested that the Y genome is sister to the W and P genomes. Therefore, the origin of Y genome is open for further study.
Previous studies based on RAPD (Random amplified polymorphic DNA polymorphism) [25], RAMP (Random Amplified Microsatellite Polymorphism) [26], C-banded karyotypes [27], and ITS sequence [28] suggested that the pattern of evolutionary differentiation of Kengyilia species associated with geographical origin from Central Asia and the Qinghai-Tibetan plateau. Zhou et al. [25] speculated that the pattern of evolutionary differentiation of Kengyilia species might genetically arise from its parental lineages with two different geographical origins (Central Asia and The Qinghai-Tibetan plateau). Based on the cytogenetic and geographic data, Yen et al. [19] hypothesized that the biological factors from diploid Agropyron (P genome) species might play an important role in influencing the genetic differentiation of Kengyilia species. While these studies add to our understanding of phylogeny and genetic differentiation of Kengyilia, little is known about the evolutionary forces acting on the geographical differentiation of Kengyilia, and further information on whether the biological factors from the P genome influences the patterns of genetic diversity of Kengyilia species is still outstanding.
Phosphoglycerate kinase (Pgk1), a key ATP-generating enzyme in the glycolytic pathway, catalyzes the conversion of 1, 3-diphosphoglycerate to 3-phosphoglycerate. Analysis of the Pgk1 gene showed that it is present as a single copy per diploid chromosome in grass [29]. The Pgk1 gene has been successfully used to study the phylogeny and evolutionary history of Triticum/Aegilops complex [30], [31]. In this study, three homoeologous copies the Pgk1 gene were isolated from each the fifteen sampled Kengyilia species and analyzed with those from 47 diploid taxa representing 18 basic genomes in Triticeae. The objectives were to: (1) document the patterns of molecular evolutionary divergence among homoeologues of the Pgk1 gene in hexaploid StYP Kengyilia and between polyploidy and its diploid genome donor; (2) determine whether the patterns of Pgk1 sequence variation within the P genome lineages reflects the geographical differentiation of Kengyilia species; (3) explore evolutionary forces acting on the Kengyilia species with different geographical region; (4) identify the possible origin of the Y genome.
Materials and Methods
Taxon sampling
Fifteen Kengyilia species were included in this study and were analyzed together with 47 diploid taxa representing 18 basic genomes in the tribe Triticeae (Table S1). Pgk1 sequences for 9 accessions representing the S, D, I, R and A genomes were obtained from published data [30]. The remaining Pgk1 sequences are new data and have been deposited in GenBank. Bromus inermis L. was used as outgroup. The seed materials with PI and W6 numbers were kindly provided by American National Plant Germplasm System (Pullman, Washington, USA), while the seed materials with ZY and Y numbers were collected by ourselves, which no specific permit is required. The plants and voucher specimens are deposited at Herbarium of Triticeae Research Institute, Sichuan Agricultural University, China (SAUTI).
DNA amplification, homoeologous sequence isolate, and sequencing
DNA extraction followed a standard CTAB protocol [32]. The Pgk1 gene was amplified with the Pgk1-specific primers PgkF1 (5′-TCGTCCTAAGGGTG TTACTCCTAA-3′) and PgkF2 (5′-AAGCTCGCGCCACCACCAGTTGAG-3′). PCR was conducted under cycling conditions reported previously [29]. PCR products were cloned into the pMD18-T vector (TaKaRa) following the manufacture's instruction.
PCR amplicons of single-copy nuclear genes from allopolyploid species will produce a heterogeneous mix of homoeologues. To separate the homoeologues of the Pgk1 gene from each Kengyilia accession, we performed the following process. Firstly, approximately 30 positive clones from each accession were screened by direct PCR using primer PgkF1 and M13R (on the side of the cloning site in the plasmid). Secondly, St-type (5′-GGTATTCTTGTGTTCCACACCA-3′) and P-type (5′-ATCZAGACYTCTAATCAAGCA-3′) Pgk1-specific primers were designed and used each together with the reverse primer PgkF2 to screen the St- and P-type Pgk1 sequences from above 30 positive clones with Pgk1 inserts, respectively. The positive clones containing the Y-type Pgk1 sequences were also obtained. The cloned PCR products were commercially sequenced in both directions by TaKaRa Biotechnology Co. Ltd. (Dalian, China), and an additional internal primer (5′-GATGGAGCTGTTTCAAACC-3′) was used to sequence the internal portion of the cloned PCR products. All the sequences from Kengyilia species were determined based on at least five independent St-, Y- and P-type clones, respectively.
Data analysis
Multiple sequences were aligned using ClustalX [33] followed by manual adjustment. To reduce the size of the matrixes and the possible impact of PCR artifacts, unique substitutions in single clones were ignored and several identical sequences were represented by a single sequence in alignments. Following an initial phylogenetic analysis, the number of sequences used for alignment was reduced by keeping only one sequence if more sequences of the same accession formed a monophyletic group.
To assess the divergence and genetic relationships between allopolyploids and its diploid progenitors, nucleotide diversity was estimated by Tajima's π [34], Watterson's θ [35], the number of fixed differences (SF) and the numbers of shared polymorphisms (SS) [36]. Tests of neutrality including Tajima's, and Fu and Li's D statistic were performed as described by Tajima [34], and Fu and Li [37]. Significance of D-values was estimated with the simulated distribution of random samples (1000 steps) using a coalescence algorithm assuming neutrality and population equilibrium [38].
To detect selective constraints on the coding portions (the introns were excluded) of the homoeologous Pgk1 gene, the ratio of nonsynonymous to synonymous substitution (dN/dS) were computed using the modified Nei-Gojobori method in MEGA 4.0 [39] and the single likelihood ancestor counting (SLAC) approach implemented by the Datamonkey analysis [40]. In the modified Nei-Gojobori analysis, the significance of difference between d N and d S was estimated using the Z statistics, with standard errors based on 1000 bootstrap replicates using MEGA 4.0 [39]. In SLAC analysis, a 95% confidence interval (95% C.I.) for the ratio of d N to d S was estimated using profile likelihood [40]. We also performed the McDonald–Kreitman (1991) test on the coding portions of the homoeologous Pgk1 gene using DnaSP 4.10.9 [41]. Significance of the test was determined by a Fisher exact test [42].
Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI). The evolutionary model used for the phylogenetic analysis was determined using ModelTest v3.0 with Akaike information criterion (AIC) [43]. The optimal model identified was GTR+G+I. ML analysis was performed using PAUP*4.0b10 (Swofford D L, Sinauer Associates, http://www.sinauer.com). ML heuristic searches were performed with 100 random addition sequence replications and TBR branch swapping algorithm. The robustness of the trees was estimated by bootstrap support (BS) [44]. BI analysis was performed using MrBayes v3.0 [45]. Four MCMC (Markov Chain Monte Carlo) chains (one cold and three heated) were run for 1,000,000 generations. The first 2500 trees were stationary discarded as “burn-in”. The remaining trees were used to construct the 50%-majority rule consensus trees. The statistical confidence in nodes was evaluated by posterior probabilities (PP).
Clock-like evolution of Pgk1 sequences within Kengyilia and its putative diploid species was evaluated with a likelihood ratio test comparing the likelihood scores from the unconstrained and clock-constrained analyses, implemented in PAUP*4.0b10. Substitution rates were significantly heterogeneous (χ2 = 173.44, df = 67, P<0.0001), implying a very poor fit to the molecular clock. Therefore, divergence times with 95% confidence intervals were estimated using Bayesian relaxed molecular clock method, implemented in BEAST v1.4.6 [46]. The lack of fossils for Triticeae precluded a direct calibration of tree topologies. Instead, molecular dating was based on the intron region of the Pgk1 gene clock of 0.0051 substitutions per site per MY (million year) [29]. Calibration points were performed using a relaxed uncorrelated lognormal molecular clock. A Yule speciation tree prior was furthermore specified, which assumes a constant speciation rate among lineages, with a log-normal prior for birth rate. MCMC searches were run for 10,000,000 generations under GTR+I model (with the associated parameters specified by ModelTest as the priors). Tracer 1.4 [47] was used to ensure the convergence of the mixing in terms of the effective sample size (ESS) values and the coefficient rate. Resulting trees were analyzed using TreeAnnotator available in BEAST where the burn-in (2000 trees) was removed and a maximum credibility tree was constructed. Trees were then viewed in FigTree v. 1.3.1 (http://tree.bio.ed.ac.uk/).
Results
Sequence analysis
Following the screen of Pgk1 homoeologues, three distinct types of Pgk1 sequences (St-, P- and Y-type) were obtained from all 15 Kengyilia species. At least 15 positive clones (including 5 St-type, 5 P-type and 5 Y-type clones) were sequenced from each accession. In cases when multiple identical sequences resulted from cloned PCR products of one accession, only one sequence was included in the data set. Consequently, 45 unique sequences were obtained and analyzed together with those from 47 diploid taxa representing 18 basic genomes in Triticeae.
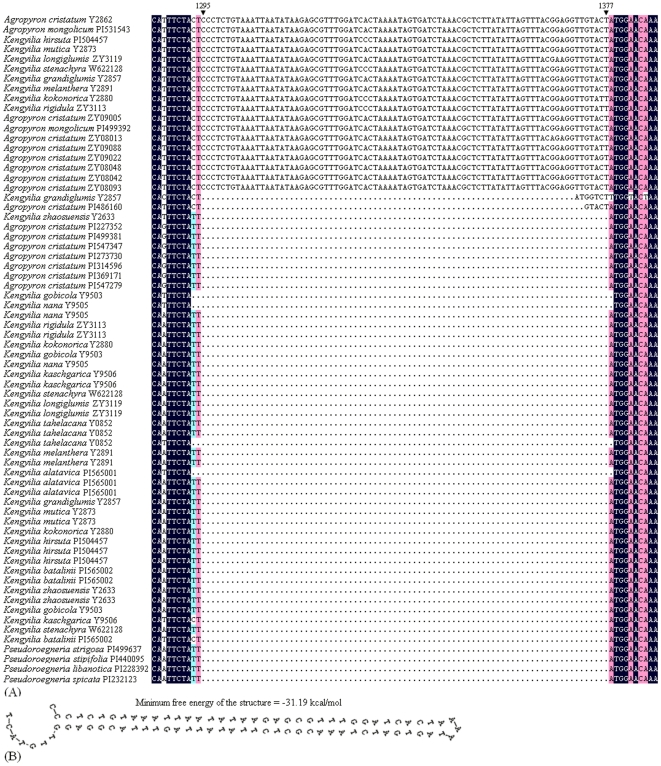
The DNA sequence of the Pgk1 gene includes 5 exons and 4 introns, which was in agreement with previous studies [29], [30]. The sequence comparison from all the species studied here showed that the length of DNA sequences ranged from 1341 bp to 1484 bp, and the DNA sequences in most accessions were ∼1390 bp. Pgk1 sequence matrix including both exons and introns contains 1522 characters, of which 20.04% (305/1522) were variable, and 8.74% (133/1522) were parsimony informative. Sequence alignment showed that an 83-bp insertion was detected for the P-type sequences at position 1295–1377 in the intron region from eight Kengyilia species (K. longiglumis, K. mutica, K. melanthera, K. hirsuta, K. stenachyra, K. rigidula, K. kokonorica and K. grandiglumis), two Agropyron mongolicum accessions (PI 531543 and PI 499392) and eight Agropyron cristatum accessions (Y2862, ZY08013, ZY08042, ZY09022, ZY08048, ZY09088, ZY08093 and ZY09005) (Figure 1, A). Secondary structure analysis indicated an inverted-repeat region in the 83-bp insertion (Figure 1, B). BLAST search against the transposable elements (TEs) stored in the TREP (Triticeae Repeat) showed that the 83-bp insertion belongs to MITE stowaway element.
Figure 1. Pgk1 gene sequence analysis.
(A) Partial alignment of the amplified sequences of the Pgk1 gene from Kengylia and its putative diploid species. (B) Secondary structure of MITE elements.
Phylogenetic analyses
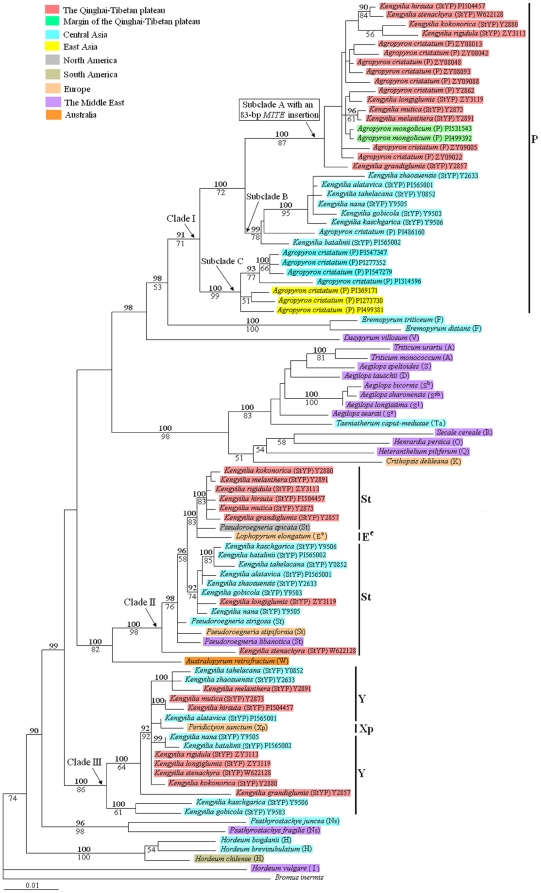
To reveal the putative genome donors of Kengyilia, the Pgk1 sequences of all the polyploid species were included in the phylogenetic analyses (ML and BI), together with 47 diploid taxa representing 18 genomes in Triticeae. ML analysis yielded a single phylogenetic tree (−Lnlikelihood = 9139.1240), with the following estimated ML parameters: the assumed nucleotide frequencies A: 0.2682, C: 0.1918, G: 0.2332, T: 0.3068, the proportion of invariable sites = 0.2519, gamma shape parameter = 0.6693. ML and Bayesian analyses recovered the same topology. The tree illustrated in Figure 2 was the ML tree with posterior probabilities (PP) above and bootstrap support (BS) below branches.
Figure 2. Maximum-likelihood tree inferred form the Pgk1 sequence of Kengyila and its affinitive species within Triticeae.
Numbers with bold above nodes are Bayesian posterior probability values ≥90% numbers below nodes are bootstrap values ≥50%. The numbers after species names refer to the distinct homoeologous copy of the Pgk1 gene. The capital letters in bracket indicate the genome type of the species. Different color labeled the geographic information of Kengyilia species and its affinitive donors.
The phylogenetic tree showed that the St-, P- and Y-type sequences from Kengyilia species were split into three well supported clades (Figure 2). The Clade I included the P-type sequences of Kengyilia and the sequences of Agropyron (91% PP and 71% BS). Three subclades (A, B and C subclade) with high statistical support were recognized in this clade. Subclade A included all the Kengyilia species and Agropyron cristatum accessions from the Qinghai-Tibetan Plateau and Agropyron mongolicum from the Alashan (margin of the Qinghai-Tibetan Plateau)(100% PP and 87% BS). It was worth mentioning that the sequence in subclade A had an 83-bp MITE stowaway insertion at position 1295–1377. Subclade B contained all the Kengyilia species from Central Asia and one A. cristatum accession from Central Asia (PI 486160) (99% PP and 78% BS). Subclade C consisted of four A. cristatum accessions from Central Asia and three A. cristatum accessions from East Asia (100% PP and 99% BS). The Clade II comprised all the St-type sequences of Kengyilia and the sequences from Pseudoroegneria species and Lophopyrum elongatum (100% PP and 98% BS). In this Clade, Pseudoroegneria spicata and Lo. elongatum were grouped with six Kengyilia species from the Qinghai-Tibetan Plateau (100% PP and 83% BS). Kengyilia longiglumis from the Qinghai-Tibetan Plateau was clustered with all the sampled Kengyilia species from Central Asia (92% PP and 74% BS). Three Pseudoroegneria species formed paraphyletic, and Kengyilia stenachyra was placed at the base of the Clade II. The Clade III included all the Y-type sequences of Kengyilia and the sequence from Peridictyon sanctum (100% PP and 86% BS). In Clade III, Kengyilia kaschgarica was grouped with Kengyilia gobicola with 100% PP and 61% BS, while the remaining Kengyilia species formed one subclade (100% PP and 64% BS).
Nucleotide diversity and strength of selection
Two overall measures of nucleotide diversity, π and θw, were separately calculated for the St, Y and P genomes of Kengyilia, and for Agropyron (Table S2). The estimates of nucleotide diversity in the P genome of Kengyilia from the Qinghai-Tibetan Plateau were π = 0.0105, θw = 0.0128, while in the P genome of Agropyron from the Qinghai-Tibetan Plateau and its margin region (Alashan), the estimates of nucleotide diversity were π = 0.0064, θw = 0.0093. The estimates of nucleotide diversity in the P genome of Kengyilia from Central Asia were π = 0.0088, θw = 0.0120, while in the P genome of Agropyron from Central Asia, the estimates of nucleotide diversity were π = 0.0142, θw = 0.0170. π was also separately calculated for synonymous and nonsynonymous sites. The overall number of polymorphic sites at homoeologous loci of Pgk1 sequence from Kengyilia St, Y and P genomes was 51, 85, and 82, respectively. The overall number of polymorphic sites in the P genome of Kengyilia was lower than that in the P genome of diploid Agropyron. The Tajima [34] and Fu and Li's [37] tests were conducted on each of eight data sets (Table S2). Tajima's and Fu and Li's D values of the P genome lineage from the Qinghai-Tibetan Plateau were −0.9473 (P>0.05) and −0.8423 (P>0.05) for Kengyilia, and −1.5089 (P<0. 05) and −1.9190 (P<0. 05) for Agropyron, respectively. The same parameters in the P genome lineage from Central Asia were −1.5244 (P<0. 05) and −1.5709 (P<0. 05) for Kengyilia, and −1.2570 (P<0. 05) and −1.2570 (P<0. 05) for Agropyron, respectively.
Speciation genetics suggested that hybridization or differentiation between two species can be inferred through comparisons of shared nucleotide polymorphisms with fixed differences [48]. Closely related taxa are expected to harbor a relative higher level of shared polymorphisms because the divergence event has not lasted long enough to erase all ancestral polymorphisms [36]. The number of shared and fixed differences at Pgk1 locus between Kengyilia and its putative diploid donor were shown in Table 1. Three shared polymorphisms and no fixed difference were observed between the St-type sequence of Kengyilia and that of Pseudoroegneria. Thirty-four shared polymorphisms and no fixed difference were found between the all the sampled P-type sequence of Kengyilia and that of Agropyron. For the P genome lineage of sympatric origin, the number of shared polymorphisms was higher than the number of fixed difference, while for the P genome lineage of allopatric origin, the number of shared polymorphisms was lower than the number of fixed difference.
Table 1. Estimation of shared polymorphisms and fixed differences between Kengyilia and its putative diploid genome donor based on the Pgk1 sequences.
| SS | SF | |
| St genome lineage | ||
| Kengyilia – Pseudoroegneria | 3 | 0 |
| P genome lineage | ||
| Kengyilia (QTPa) – Agropyron (QTP) | 6 | 0 |
| Kengyilia (QTP) – Agropyron (CAb) | 5 | 7 |
| Kengyilia (CA) – Agropyron (CA) | 9 | 2 |
| Kengyilia (CA) – Agropyron (QTP) | 3 | 14 |
| Kengyilia (Overall) – Agropyron (Overall) | 34 | 0 |
QTP is the abbreviation of the Qinghai-Tibetan Plateau.
CA is the abbreviation of Central Asia.
The non-synonymous to synonymous rate ratio dN/dS is indicative of the change of selective pressures. The dN/dS ratios of >1, = 1 and <1 indicate positive selection, neutral evolution and purifying selection on the coding portions, respectively. Prior to the estimation of selective constraints on the coding portions of the Pgk1 gene in Kengyilia St, Y and P genome and its putative diploid genome donor, the average non-synonymous (dN) and synonymous (dS) distances with standard errors were calculated using the modified Nei-Gojobori method (Table S3). Both Z-Test and SLAC statistics showed that almost all the dN/dS values were significantly <1, strongly indicating that the Pgk1 gene in the St, Y and P genomes of Kengyilia and its putative diploid genome donor have subjected to purifying selection. Comparison among the Pgk1 coding portions of Agropyron lineages with different geographical region revealed that the dN/dS value of the Qinghai-Tibetan Plateau Agropyron was not significantly (Z-test with P = 0.1455) below 1 and nearly 3-fold higher than that in the Central Asia Agropyron. The McDonald–Kreitman (MK) test of selective pressures was performed to compare the Qinghai-Tibetan Plateau P genome lineage with Central Asia P genome lineage. Total 19 mutations were found in the Qinghai-Tibetan Plateau Agropyron, of which 11 were nonsynonymous and eight were synonymous. Among the 19 mutations found in Central Asia Agropyron, six were nonsynonymous and 13 were synonymous. Significant departure from neutrality was detected for Agropyron (P = 0.037) from the Qinghai-Tibetan Plateau.
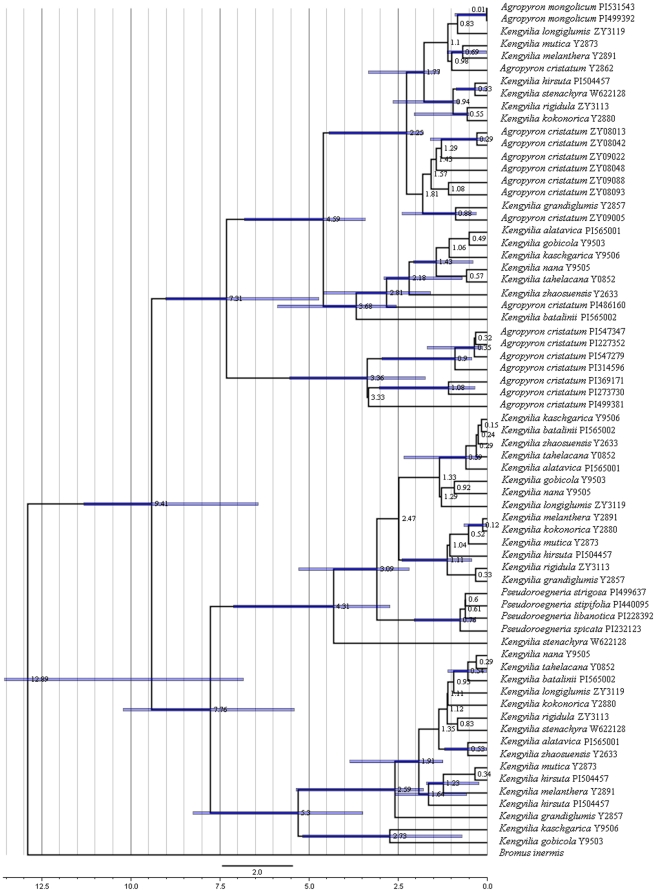
The BEAST analyses of the intron region of the Pgk1 sequences from Kengyilia and its putative diploid species generated a time-calibrated tree (Figure 3). Under a lognormal relaxed clock, the coefficient of rate variation was estimated to be 0.985 (95% C.I., 0.653–1.352), indicating that relaxed clock was appropriate. The birth rate indicated by the Yule prior is 0.474 (95% C.I., 0.323–0.622). The mean ages with 95% confidence intervals were indicated in the chronogram (Figure 3). Time calibration analysis demonstrated that the divergence time of the St, Y, and P genome lineages was 4.31 MYA (95% C.I., 2.72–7.12), 5.30 MYA (95% C.I., 3.48–8.25), and 7.31 MYA (95% C.I., 4.72–9.01), respectively. The split between the P genome lineages from the Qinghai-Tibetan Plateau and from Central Asia took place about 4.59 MYA (95% C.I., 3.42–6.81).
Figure 3. A time-calibrated tree inferred form the intron region of the Pgk1 sequence of Kengyila and its putative donors using a Bayesian relaxed clock method in BEAST.
Discussion
Pgk1 homoeologues and evolutionary history of Kengyilia
Cytogenetic analyses have concluded that all the Kengyilia species contain the StYP genomic constitutions [15]–[19]. Three homoeologous types of the Pgk1 gene, St-, Y- and P-type, were obtained from all the polyploid Kengyilia species in the present study. Phylogenetic analysis showed that the St-type homoeologous sequences were grouped with the sequences of Pseudoroegneria with 100% PP and 98% BS, and the P-type homoeologous sequences were clustered with the sequences of Agropyron with 91% PP and 71% BS. Wakeley and Hey [36] pointed out that closely related species are expected to harbor a relative higher level of shared polymorphisms than fixed differences. Our analysis of shared/fixed polymorphisms showed that more shared polymorphisms than fixed differences were observed between the St-type sequences and the sequences of Pseudoroegneria and between the P-type sequences and the sequences of Agropyron. Phylogenetic and diversity analysis thus indicate that Kengyilia is closely related to Pseudoroegneria and Agropyron. Combined with previous cytogenetic studies [16], [20], it can be concluded that the Pseudoroegneria and Agropyron species served as the St and P genome donors during the polyploid speciation of the Kengyilia species.
The Y genome is represented in all the Kengyilia species and many Asiatic tetraploid and some hexaploids in Triticeae [21]. No diploid species containing Y have been identified [49]. Chromosome pairing analysis indicated low affinities between the St and Y genomes [50]. ITS sequence data of Liu et al. [23] showed that the Y genome may be originated from the St genome. RPB2 [24] and EF-G [51] data suggested that the Y genome was sister to the W genome and has a different origin from the St genome. Chromosome pairing suggested that the W genome has very low homoeology with the St and Y genome [52]. Considering the suggestion of Torabinejad and Mueller [52], Sun et al. [51] pointed out that more sequence data are needed to reveal the relationship of the Y genome with other genomes in Triticeae. In this study, phylogenetic analysis indicated that the Y-type Pgk1 sequences were distinct from the St- and P-type sequences, which provides additional support for the independent origin of the Y genome. In phylogenetic tree (Figure 2), the Y-type sequences were grouped with the sequences from Peridictyon sanctum (Xp genome donor). This is in agreement with recent genealogical analysis of single-copy nuclear gene Acc1 sequence in the species with Y genome in Triticeae (Sha et al., unpublished data), where the Y-type Acc1 homoeologues were clustered with the sequences from Dasypyrum (V genome) species, Heteranthelium piliferum (Q genome) and Peridictyon sanctum (Xp genome). Therefore, it suggested that the Y genome may be closely related to the Xp genome in Peridictyon sanctum.
Geographical differentiation of P genome
Genus Agropyron is the P haplome donor to Kengyilia and contains approximately eight diploid (PP) or tetraploid (PPPP) or hexaploid (PPPPPP) species. Agropyron cristatum and A. mongolicum are the only two diploid species within Agropyron [21]. Phytogeographically, A. cristatum is widely distributed in Eurasian temperate region, while A. mongolicum is restricted in some regions of northern China. The present Pgk1 gene data showed that the sampled A. cristatum from East Asia, the Qinghai-Tibetan plateau, and Central Asia did not form monophyletic group but were scattered into three distinct subclades (subclade A, B, and C). Genetic differentiation among A. cristatum population based on pairwise FST estimates was relatively high, ranging from 14.93% to 65.74% (East Asia – Central Asia: 14.93%; the Qinghai-Tibetan plateau – Central Asia: 51.26%; East Asia – the Qinghai-Tibetan plateau: 65.74%). These results indicated that A. cristatum populations from different geographical origins were genetically heterogeneity. High level of genetic differentiation and divergent population structure of A. cristatum could be attributed to restricted gene flow caused by geographical isolation.
Data from RAPD [25], RAMP [26], C-banded karyotypes [27], and ITS sequence [28] suggested that Kengyilia species were geographical differentiated. Our phylogenetic analysis of Pgk1 sequences demonstrated that the separation of the P genomes in Kengyilia species is in good agreement with their geographical origins – Central Asia and the Qinghai-Tibetan plateau distinction. The accessions of Kengyilia species and A. cristatum from the Qinghai-Tibetan Plateau, and A. mongolicum from Alashan is differentiated from the accessions of Kengyilia species and one A. cristatum accession from Central Asia. Second, more shared polymorphisms than fixed differences (Kengyilia (CA) – Agropyron (CA): 9 vs. 2; Kengyilia (QTP) – Agropyron (QTP): 6 vs. 0) were observed between the sympatric P genome lineage, while less shared polymorphisms than fixed differences (Kengyilia (CA) – Agropyron (QTP): 3 vs. 14; Kengyilia (QTP) – Agropyron (CA): 5 vs. 7) were found between the allopatric P genome lineage. Third, an 83 bp MITE element insertion in the Pgk1 gene were found in the P genome of the Qinghai-Tibetan Plateau Kengyilia species and their sympatric diploid donors, while this element was absent in the same position of the sequence alignment in the P genome lineages from Central Asia. Finally, time-calibrated phylogeny suggested that speciation event of Central Asia Kengyilia species (about 3.68 MYA) may be prior to that of the Qinghai-Tibetan Plateau Kengyilia species (about 2.25 MYA). These different lines indicated that the P genome in Kengyilia species is highly differentiated according to their geographical origin. The Central Asia and the Qinghai-Tibetan Plateau Kengyilia species thus have independent origins.
Recent studies using genetic markers in many genera suggested that multiple origins (including independent origin) of polyploid species are the rule rather than the exception [2], [53]. A better understanding of the potential evolutionary outcomes for polyploid populations of independent origin is of particular evolutionary interest. Symonds et al. [53] emphasized that the fates of polyploid populations of independent origins varied depending on the amount of genetic variation initially contributed by the diploid progenitors. The present Pgk1 gene genealogical structure and patterns of shared/fixed polymorphisms indicated the occurrence of independent origins of Kengyilia species. This offers an opportunity to address the potential evolutionary outcomes of independent origins within Kengyilia. On the basis of Pgk1 sequences of the P genome lineage from the Qinghai-Tibetan Plateau, the level of nucleotide diversity in Kengyilia (π = 0.0105; θ w = 0.0128) was higher than that in diploid Agropyron (π = 0.0064; θ w = 0.0093), and Tajima's and Fu and Li's D statistic suggests a departure from the equilibrium neutral model at this locus, with an excess of rare sequence variants in Kengyilia species. Greater diversity could reflect gene flow from diploid Agropyron population. For the P genome lineage from Central Asia, the level of nucleotide diversity in Kengyilia (π = 0.0088; θ w = 0.0120) was lower than that in diploid Agropyron (π = 0.0142; θ w = 0.0170), and the values of Tajima's and Fu and Li's D statistic in Kengyilia were significantly negative and lower than the values of same parameter calculated from Agropyron. This is compatible with a genetic bottleneck created by recent polyploidization. Diverged levels of nucleotide suggested that the P genome lineages of Kengyilia with independent origin have distinct evolutionary potentials.
Evolution of Pgk1 sequences in Kengyilia
Since gene duplication results in functional redundancy, divergent selective pressure may act on the duplicated copies that are critical for the subsequent variation, retention or loss of the duplicated genes [54]. Isolation and characterization of three divergent Pgk1 homoeologues from all the hexaploid Kengyilia species studied here suggested the retention of triplicated Pgk1 homoeologues in Kengyilia species. The dN/dS ratio of three divergent Pgk1 homoeologues was significantly below 1 (Z-test with P<0.05; SLAC with 95% C.I.: 0–1), suggesting that the Pgk1 sequences are selectively constrained as most mutations in functional genes are expected to be disadvantageous.
The dN/dS value of the Qinghai-Tibetan Plateau Agropyron was nearly 3-fold higher than that in Central Asia Agropyron. Significant MK test (P = 0.037) between them suggested an excess of nonsynonymous substitutions, which is traditional viewed as an outcome of positive selection. This appears paradoxical in light of the strong signature of purifying selection in Pgk1 sequences. Relaxed purifying selection, selective sweep, and population expansion may explain this paradox. The relaxed purifying selection hypothesis is very unlikely because it did not explain our result of less significant Tajima's D values in the high dN/dS group (P = 0.044) than in the low dN/dS group (P = 0.017). The deficiency of synonymous polymorphisms in the high dN/dS group in comparison with the low dN/dS group was also not the result of relaxed purifying selection. The following evidences support the hypothesis of selective sweep. First, the dN/dS value of the Qinghai-Tibetan Plateau Agropyron was significantly higher than that in Central Asia Agropyron. Significant difference in dN/dS ratio was considered to be a result of selective sweep [55]. Second, our result showed less levels of nucleotide diversity in the high dN/dS group than in the low dN/dS group, which is consistent with the expectation that sweep results in reduced polymorphism in the high dN/dS group [55]. Third, Palmé et al. [56] emphasized that selective sweep would cause more negative Tajima's D and lower silent diversity in the high dN/dS group than in the low dN/dS group. Corresponding to this suggestion, the present result showed that the Tajima's D value in the high dN/dS group (−1.5089) is more negative than that in the low dN/dS group (−1.2570), and the levels of silent diversity in the high dN/dS group (π = 0.0089) is less than that in the low dN/dS group (π = 0.0236). Finally, given that the Pgk1 gene in the Qinghai-Tibetan Plateau Agropyron population has undergone selective sweep, it is suggested that the sweep event for the Pgk1 gene in Plateau may be associated with an evolutionary adaptation to local cold climate conditions. Recent study focusing on the responses of plant to cold stress has revealed that Phosphoglycerate kinase (Pgk1) is an up-regulated response protein to cold stress, a feature of adaptive evolution [57]. Palaeoclimatic evidence indicated that the cold climate effects of the Qinghai-Tibetan Plateau resulted from its large-scale uplifting and consequent glacial cycles during the Quaternary (2.4 MYA to the present) [58], [59]. The present molecular dating suggested that the divergence time of the P genome lineage including the high dN/dS Agropyron population from the Qinghai-Tibetan Plateau was dated to 2.25 MYA, and the divergence time of the P genome lineage including the low dN/dS Agropyron population from Central Asia was dated to 3.38 MYA. Considering the conversion of low to high dN/dS value resulting from selective sweep, the age of selective sweep might have happened ≈2.3–3.4 MYA. Therefore, it is possible that a relatively long-time sweep event allow the Pgk1 gene within Agropyron to adapt to cold climate triggered by the recent uplifts of the Qinghai-Tibetan Plateau. Because demographic processes such as range expansion can have a similar impact on DNA variation to that caused by a selective sweep in a population [60]. It can also not rule out the possibility that population expansion might have contributed to the present significant MK tests and difference in the dN/dS ratio. Analysis of diversity showed that the levels of diversity in the Qinghai-Tibetan Plateau Agropyron population (π = 0.0089; θw = 0.0093) is significant lower than that in Central Asia population (π = 0.0236; θw = 0.0170), indicating a recent expansion in the Qinghai-Tibetan Plateau Agropyron population. This suggestion is further determinative of the more significant negative Tajima's and Fu and Li's D values. Population expansion in associated with Pleistocene glacial cycles might have accelerated the fixation of mildly deleterious replacement mutations that became effectively neutral in the Qinghai-Tibetan Plateau Agropyron populations.
Duplicate genes in polyploid lineages are often preserved in function by more strongly purifying selection [61]. Comparative analyses showed that the dN/dS value in the P genome of the Qinghai-Tibetan Plateau Kengyilia species was lower than that in their sympatric diploid relatives, indicating that more strongly purifying selection acts to conserve the function of the Pgk1 gene in Kengyilia. However, a slightly elevated dN/dS value in the P genome of the Central Asia Kengyilia species compared to their sympatric diploid relatives might suffer from polyploidization bottleneck as suggested by the present estimate of diversity, because population bottleneck may result in reduced selection [62].
It is worth mentioning that the dN/dS value of the P genome of Kengyilia lineage from the Qinghai-Tibetan Plateau (dN/dS = 0.2645) was higher than that from Central Asia (dN/dS = 0.2018). Non-significantly negative Tajima's D value coupled with greater diversity in the high dN/dS group excluded the possibility that selective sweep could resulted in the difference in the dN/dS value of the P genome of Kengyilia. A highly non-significant difference between this two dN/dS ratios (P = 0.499 by Fisher's exact test) based on the McDonald-Kreitman test was found, rejecting the hypothesis of relaxed purifying selection. Considering the difference in the dN/dS value of two allopatric Agropyron lineages, it is possible that the difference in the dN/dS value of the P genome of two allopatric Kengyilia lineages may be genetically from geographically differentiated P genome donors via independent origins. This difference is not completely erased, although polyploidization bottleneck might occur in Central Asia Kengyilia lineages and strong purifying selection might act on the Pgk1 gene in the Qinghai-Tibetan Plateau Kengyilia lineages.
Supporting Information
Kengyilia species and other related genera in Triticeae used in this study.
(DOC)
Estimates of nucleotide diversity and test statistics at Pgk1 locus in Kengyilia St, Y and P genome and its putative diploid genome donor.
(DOC)
Detection of selection pressure on the coding portions of the Pgk1 gene in Kengyilia St, Y and P genome and its putative diploid genome donor.
(DOC)
Acknowledgments
We are very grateful to American National Plant Germplasm System (Pullman, Washington, USA) providing the part seed materials. We also thank two anonymous reviewers for their very useful comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Trangenic Major Program (Nos. 20112X08009-001), the National Natural Science Foundation of China (Nos. 30900087, 30870154), Special Fund for Agro-Scientific Research in the Public Interest of China (Nos. 201003021), and the Science and Technology Bureau (Nos. 2060503) and Education Bureau of Sichuan Province. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hovav R, Udall JA, Chaudhary B, Rapp R, Flagel L, et al. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci U S A. 2008;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since Plant Speciation. New Phytol. 2003;161:173–191. [Google Scholar]
- 3.Small RL, Wendel JF. Differential evolutionary dynamics of duplicated paralogous Adh loci in allotetraploid cotton (Gossypium). Mol Biol Evol. 2002;19:597–607. doi: 10.1093/oxfordjournals.molbev.a004119. [DOI] [PubMed] [Google Scholar]
- 4.Aagaard JE, Willis JH, Phillips PC. Relaxed selection among duplicate floral regulatory genes in Lamiales. J Mol Evol. 2006;63:493–503. doi: 10.1007/s00239-005-0306-x. [DOI] [PubMed] [Google Scholar]
- 5.Moore RC, Purugganan MD. The early stages of duplicate gene evolution. Proc Natl Acad Sci U S A. 2003;100:15682–15687. doi: 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo WZ, Cai CP, Wang CB, Zhao L, Wang L, et al. A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics. 2008;9:314. doi: 10.1186/1471-2164-9-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, et al. Evolutionary history of GS3, a gene conferring grain size in rice. Genetics. 2009;182:1323–34. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan CZ, Xiang QYJ, Remington DL, Purugganan MD, Wiegmann BM. Evolutionary patterns in the antR-Cor gene in the dwarf dogwood complex (Cornus, Cornaceae). Genetica. 2007;130:19–34. doi: 10.1007/s10709-006-0016-3. [DOI] [PubMed] [Google Scholar]
- 9.Slotte T, Huang HR, Lascoux M, Ceplitis A. Polyploid speciation did not confer instant reproductive isolation in Capsella. Mol Biol Evol. 2008;25:1472–1481. doi: 10.1093/molbev/msn092. [DOI] [PubMed] [Google Scholar]
- 10.Gong PC, Bao Y. Nucleotide diversity of the homoeologous adh 1 loci in the American allotetraploid Oryza species. Plant Syst Evol. 2008;276:243–253. [Google Scholar]
- 11.Salmon A, Flagel L, Bao Y, Udal JA, Wendel JF. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 2010;186:123–134. doi: 10.1111/j.1469-8137.2009.03093.x. [DOI] [PubMed] [Google Scholar]
- 12.Takuno S, Nishio T, Satta Y, Innan H. Preservation of a pseudogene by gene conversion and diversifying selection. Genetics. 2008;180:517–531. doi: 10.1534/genetics.108.091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun GL, Daley T, Ni Y. Molecular evolution and genome divergence at RPB2 gene of the St and H genome in Elymus species. Plant Mol Biol. 2007;64:645–655. doi: 10.1007/s11103-007-9183-6. [DOI] [PubMed] [Google Scholar]
- 14.Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol. 2007;24:1506–1517. doi: 10.1093/molbev/msm077. [DOI] [PubMed] [Google Scholar]
- 15.Yang JL, Yen C, Baum BR. Kengyilia: synopsis and key to species. Hereditas. 1992;116:25–28. [Google Scholar]
- 16.Jensen KB. Genome analysis of Eurasisan Elymus thoroldianus, E. melantherus, and E. kokonoricus (Poaceae: Triticeae). Int J Plant Sci. 1996;157:136–141. [Google Scholar]
- 17.Zhou YH. Study on karyotypes of 5 species of Kengyilia. Guihaia. 1994;14:163–169. [Google Scholar]
- 18.Zhang XQ, Yang JL, Yen C, Zheng YL, Zhou YH. Cytogenetic and systematic analysis of Kengyilia gobicola, K. zhaosuensis and K. batalinii var. nana (Poaceae). Genet Resour Crop Evol. 2000;47:451–454. [Google Scholar]
- 19.Yen C, Yang JL, Baum BR. Biosystematics of Triticeae. 2006. Volume 3. Chinese Agriculture Press, Beijing.
- 20.Löve A. Conspectuse of Triticeae. Feddes Rep. 1984;95:425–524. [Google Scholar]
- 21.Dewey DR. Gustafson JP, editor. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. Gene Manipulation in Plant Improvement. 1984. pp. 209–279. Plenem Press, New York.
- 22.Mason-Gamer RJ, Burns MM, Naum M. Polyploidy, introgression, and complex phylogenetic patterns within Elymus. Czech J Genet Plant Breed. 2005;41:21–26. [Google Scholar]
- 23.Liu Q, Ge S, Tang H, Zhang X, Zhu G, et al. Phylogenetic relationships in Elymus (Poaceae, Triticeae) based on the nuclear ribosomal transcribed spacer and chloroplast trnL-F sequences. New Phytol. 2006;170:411–420. doi: 10.1111/j.1469-8137.2006.01665.x. [DOI] [PubMed] [Google Scholar]
- 24.Sun GL, Ni Y, Daley T. Molecular phylogeny of RPB2 gene reveals multiple origin,geographic differentiation of H genome, and the relationship of the Y genome to other genomes in Elymus species. Mol Phylogenet Evol. 2008;46:897–907. doi: 10.1016/j.ympev.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 25.Zhou YH, Zheng YL, Yang JL, Yen C, Jia JZ. Relationships among Kengyilia species assessed by RAPD markers. Acta Phytotaxon Sin. 2000;38:515–521. [Google Scholar]
- 26.Zhang L, Zheng YL, Wei YM, Liu SG, Zhou YH. The genetic diversity and similarities among Kengyilia species based on random amplified microsatellite polymorphism. Genet Resour Crop Evol. 2005;52:1011–1017. [Google Scholar]
- 27.Zeng J, Cao G, Liu J, Zhang HQ, Zhou YH. C-banding analysis of eight species of Kengyilia (Poaceae: Triticeae). J Appl Genet. 2008;49:11–21. doi: 10.1007/BF03195244. [DOI] [PubMed] [Google Scholar]
- 28.Zeng J, Zhang L, Fan X, Zhang HQ, Yang RW, et al. Phylogenetic analysis of Kengyilia species based on nuclear ribosomal DNA internal transcribed spacer sequences. Biol Plantarum. 2008;52:231–236. [Google Scholar]
- 29.Huang SX, Sirikhachornkit A, Faris JD, Su XJ, Gill BS, et al. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol Biol. 2002;48:805–820. doi: 10.1023/a:1014868320552. [DOI] [PubMed] [Google Scholar]
- 30.Huang SX, Sirikhachornkit A, Su XJ, Faris J, Gill B, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci U S A. 2002;12:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilian B, Özkan H, Deusch O, Effgen S, Brandolini A, et al. Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol. 2007;24:217–227. doi: 10.1093/molbev/msl151. [DOI] [PubMed] [Google Scholar]
- 32.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 33.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tajima F. Statistical method for testing the neutral mutation of hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watterson GA. On the number of segregation sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 36.Wakeley J, Hey J. Estimating ancestral population parameters. Genetics. 1997;145:847–855. doi: 10.1093/genetics/145.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson RR. Futuyma D, Antonovics J, editors. Gene genealogies and the coalescent process. Oxford Surveys in Evolutionary Biology. 1990. pp. 1–44. Oxford University Press, New York.
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 41.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DNA sequence polymorphism, version 4.10.4. 2005. DNAsp4 computer software. Barcelona Univ. Barcelona, Spain.
- 42.Sokal RR, Rohlf FJ. Biometry, 3rd edn. 1995. W.H. Freeman, New York.
- 43.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Huelsenbeck JP, Ronquist FR. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 46.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambaut A, Drummond AJ. Tracer v1.4: MCMC trace analyses tool. 2007 Available: http://beast.bio.ed.ac.uk/Tracer. Accessed 2012 Jan 4. [Google Scholar]
- 48.Noor MAF, Feder JL. Speciation genetics: evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- 49.Wang RR-C, Bothmer RV, Dvorak J, Fedak G, Linde-Laursen I, et al. Wang RR-C, Jensen KB, Jaussi C, editors. Genome symbols in the Triticeae (Poaceae). Proc 2nd Intern. 1994. pp. 29–34. Triticeae Symp, Logan, Utah, U S A.
- 50.Lu BR, Bothmer von R. Genomic constitution of Elymus parviglumis and E. pseudonutans, Triticeae (Poaceae). Hereditas. 1990;113:109–119. [Google Scholar]
- 51.Sun GL, Komatsuda T. Origin of the Y genome in Elymus and its relationship to other genomes in Triticeae based on evidence from elongation factor G (EF-G) gene sequences. Mol Phylogenet Evol. 2010;56:727–733. doi: 10.1016/j.ympev.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 52.Torabinejad J, Mueller RJ. Genome Constitution of the Australian hexaploid grass Elymus scabrus (Poaceae, Triticeae). Genome. 1993;36:147–151. doi: 10.1139/g93-018. [DOI] [PubMed] [Google Scholar]
- 53.Symonds VV, Soltis PS, Soltis DE. Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution. 2010;64:1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- 54.Chapman BA, Bowers JE, Feltus FA, Paterson AH. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiospermgenome duplication. Proc Natl Acad Sci U S A. 2006;103:2730–2735. doi: 10.1073/pnas.0507782103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser HB, Moses AM, Schadt EE. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A. 2010;107:2977–82. doi: 10.1073/pnas.0912245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmé AE, Wright M, Savolainen O. Patterns of divergence among conifer ESTs and polymorphism in Pinus sylvestris identify putative selective sweeps. Mol Biol Evol. 2008;25:2567–2577. doi: 10.1093/molbev/msn194. [DOI] [PubMed] [Google Scholar]
- 57.Hashimoto M, Komatsu S. Proteomics analysis of rice seedlings during cold stress. Proteomics. 2007;7:1293–1302. doi: 10.1002/pmic.200600921. [DOI] [PubMed] [Google Scholar]
- 58.Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 59.Zheng BX, Xu QQ, Shen YP. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quatern Int. 2002;97–98:93–101. [Google Scholar]
- 60.Rokas A, Atkinson RJ, Brown GS, West SA, Stone GN. Understanding patterns of genetic diversity in the oak gallwasp Biorhiza pallida: demographic history or a Wolbachia selective sweep? Heredity. 2001;87:294–304. doi: 10.1046/j.1365-2540.2001.00872.x. [DOI] [PubMed] [Google Scholar]
- 61.Otto SP. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–37. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 62.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–99. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kengyilia species and other related genera in Triticeae used in this study.
(DOC)
Estimates of nucleotide diversity and test statistics at Pgk1 locus in Kengyilia St, Y and P genome and its putative diploid genome donor.
(DOC)
Detection of selection pressure on the coding portions of the Pgk1 gene in Kengyilia St, Y and P genome and its putative diploid genome donor.
(DOC)