Background: Post-translational modifications generate functional heterogeneity of viral regulatory factors.
Results: Viral chromatin association by Kaposi sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) is modulated by protein arginine methyltransferase 1 (PRMT1)-directed methylation.
Conclusion: Methylation of KSHV LANA antagonizes viral reactivation.
Significance: Protein methylation contributes to the functional properties of viral regulatory proteins, including KSHV LANA.
Keywords: Chromatin Immunoprecipitation (ChIP), Herpesvirus, Histones, Protein Methylation, Virology, KSHV, Reactivation, Viral Latency
Abstract
The Kaposi sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) is a multifunctional protein with roles in gene regulation and maintenance of viral latency. Post-translational modification of LANA is important for functional diversification. Here, we report that LANA is subject to arginine methylation by protein arginine methyltransferase 1 in vitro and in vivo. The major arginine methylation site in LANA was mapped to arginine 20. This site was mutated to either phenylalanine (bulky hydrophobic, constitutive methylated mimetic) or lysine (positively charged, non-arginine methylatable) residues. The significance of the methylation in LANA function was examined in both the isolated form and in the context of the viral genome through the generation of recombinant KSHV. In addition, authentic LANA binding sites on the KSHV episome in naturally infected cells were identified using a whole genome KSHV tiling array. Although mutation of the methylation site resulted in no significant difference in KSHV LANA subcellular localization, we found that the methylation mimetic mutation resulted in augmented histone binding in vitro and increased LANA occupancy at identified LANA target promoters in vivo. Moreover, a cell line carrying the methylation mimetic mutant KSHV showed reduced viral gene expression relative to controls both in latency and in the course of reactivation. These results suggest that residue 20 is important for modulation of a subset of LANA functions and properties of this residue, including the hydrophobic character induced by arginine methylation, may contribute to the observed effects.
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV3/human herpesvirus 8) is a γ-herpesvirus linked to Kaposi sarcoma (KS) and at least two rare lymphoproliferative disorders, primary effusion lymphoma (or body cavity-based lymphoma (BCBL)) and a subset of Multicentric Castleman disease (1–4). Like all herpesviruses, KSHV has distinct latent and lytic phases whose transition is regulated primarily at the transcriptional level by transactivation, silencing, and chromatin remodeling of the viral genome (5–10). KSHV latency-associated nuclear antigen (LANA/ORF73) is a key regulatory protein that is essential for the establishment and maintenance of viral latency. LANA is a DNA-binding protein that binds to the viral latent origin of replication located at the terminal repeat sequence of the KSHV genome, is highly expressed in all KSHV-associated disorders (11–13), and is a functional homologue of Epstein-Barr virus transcription factor EBNA1 (14–17). LANA functions as both a transcriptional activator and a repressor depending on the context of promoters and cell line interrogated (18, 19). Accordingly, LANA has been shown to associate with a broad range of transcriptional regulators such as RBP-Jκ, CBP, Daxx, BRD2, RB, p53, and Sp-1 (20–26). In addition to influences via protein partner interaction, LANA function has also been reported to be regulated by post-translational modifications, including phosphorylation (27, 28), acetylation (29), poly(ADP-ribosyl)ation (30), and sumoylation.4 Among these post-translational modifications, phosphorylation, acetylation, and poly(ADP-ribosyl)ation have been reported to antagonize LANA function, measured by either genomic copy number alterations or derepression of LANA transcriptional targets such as ORF50 (28–30).
Protein arginine methylation is a post-translational modification frequently observed in nucleic acid-binding proteins. The mammalian PRMT family consists of nine highly conserved members that catalyze distinct types of methyl group addition to the guanidino group of protein arginine residues (31). Substrates include many proteins involved in RNA metabolism (32); however, the list of targets has become increasingly broad, encompassing transcriptional coactivators, corepressors, DNA repair factors, and signaling molecules (31, 33). Viral proteins are also modified by PRMTs. This class of substrates includes a variety of viral proteins including adenovirus type 5 100K (34, 35), Epstein-Barr virus EBNA1 (36), herpes simplex virus type 1 ICP27 (37, 38), human immunodeficiency virus type 1 Tat (39), hepatitis C virus NS3 (40), and hepatitis δ virus small form antigen (41). The consequences imparted by arginine methylation on viral protein function include alterations in target localization (35, 36, 38), transcriptional activation (39), viral gene expression (38), and viral replication (34, 38, 39, 42).
In this report we demonstrate that KSHV LANA is arginine-methylated in vitro and in vivo by PRMT1. Using a strategy of amino acid substitution mutants of the primary arginine methylation site in LANA in the context of the KSHV genome as well as in its isolated form, our results suggest that methylation of LANA modulates the transcriptional control of viral gene targets, mediated in part by a mechanism that involves modulation of the histone binding function of LANA.
EXPERIMENTAL PROCEDURES
Plasmids
The cloning strategy employed throughout these studies involved cloning of fragments of interest into several vectors that contain a CpoI site inserted into each polylinker. This strategy has been described previously (6, 43–46). Fragments were digested with CpoI, desalted (Qiagen) and cloned into CpoI-digested vectors for expression in mammalian cells (pCDNA3-CpoI), lentiviral transduction (pLTRExBGHpA-CpoI), baculoviral expression (pFastBac-CpoI), and for GST pulldown assays (pGEX-2T-CpoI). The resultant clones from all procedures possess an N-terminal FLAG, HA, or GST tag. A full-length wt-LANA expression vector was constructed as described above using cDNA prepared from BCBL-1 cells. This plasmid served as the template for mutagenesis and for construction of truncated LANA fragments by PCR. Oligonucleotide sequences used throughout this study are available upon request.
Protein Methylsome Library
Details of the methylase/demethylase library will be described elsewhere.5 Pertinent to this publication, PRMTs 1–8 were cloned by PCR using Pfu Turbo (Agilent Technologies), and cDNA was prepared from a pool of RNAs from several cell lines.
Mutagenesis
Mutations were introduced into wt-LANA using QuikChange II kit (Agilent Technologies). LANA amino acids 1–70 was inserted into plasmid pGex-2T-CpoI. This plasmid served as the template for mutagenesis of the individual N-terminal arginine residues of LANA.
Cell Lines
293T (SV40 large T/human embryonic kidney epithelial) and SLK cells (human endothelial cells) (47, 48) were grown in monolayer culture in DMEM, supplemented with 10% fetal bovine serum (complete DMEM) in the presence of 5% CO2. 293T BAC stable cell lines were selected and maintained in complete DMEM containing 100 μg/ml hygromycin. SLK lentiviral stable cells were selected and maintained in complete DMEM containing 1 μg/ml puromycin.
Immunoprecipitation and Immunoblotting
The procedures used have been described (43). Immunoprecipitations (IP) utilized 2 μg of preimmune IgG or 2 μg of specific IgG per immunoprecipitation. For immunoblotting, the final concentration of all primary antibodies was 1 μg/ml. Primary antibodies used were anti-FLAG-M2 (Sigma), anti-HA (Covance), anti-lamin A/C (Cell Signaling Technology), anti-β actin (Sigma), anti-PRMT1 (Sigma), or anti-H2B (Millipore). Secondary antibodies were obtained from Santa Cruz Biotechnologies.
Methylation Assays
Purified recombinant proteins were incubated for 3 h at 37 °C with 500 nm recombinant PRMT in 30 μl of methylation buffer (20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, and 1 mm PMSF) supplemented with 1 μl (0.5 μCi) of S-adenosyl-l-[methyl-3H]methionine (Amersham Biosciences). Reactions were stopped by the addition of 2× SDS-PAGE sample buffer and heating. Samples were analyzed by SDS-PAGE and autoradiography. For in vivo methylation, cells were labeled for 3 h at 37 °C with 50 μCi of l-[methyl-3H]methionine in the presence of 100 μg/ml cycloheximide. After labeling, the cells were processed for immunoprecipitation using anti-FLAG antibody. After electrophoresis of the immunoprecipitates, the gels were subjected to fluorography with Amplify (GE Healthcare) and exposed to film at −80 °C.
Gene Knockdown
The shRNA directed against PRMT1 was obtained from Open Biosystems. Cells were transfected using FuGENE HD (Roche Applied Science). The shRNA expression plasmid was cotransfected with FLAG-LANA, and the knockdown was started 72 h before the methylation labeling procedure. After the labeling step, cell lysates were prepared and analyzed for LANA methylation as described above.
Cellular Fractionation
SLK, SLK-LANA, and SLK-LANA Arg-20 mutants were fractioned into nuclear and cytoplasmic pools using the NE-PER kit (Pierce). The nuclear matrix fractionation protocol has been described (49).
Recombinant Protein Production
Spodoptera frugiperda Sf9 cells were maintained in EX-CELL 420 medium (JRH Biosciences), and recombinant baculoviruses were generated as previously described (43). Recombinant baculovirus bacmid DNA was transfected into Sf9 cells by using FuGENE 6 (Roche Applied Science), and recombinant viruses were subsequently amplified twice. Expression of recombinant proteins was confirmed by immunoblotting with anti-FLAG monoclonal antibody. Large scale cultures of Sf9 cells (100 ml) were infected with recombinant baculovirus at a multiplicity of infection of 0.1–1.0, and cells were harvested 48 h after infection. Recombinant proteins were purified as described previously (6). The purity and amount of protein was measured by SDS-PAGE and Coomassie Blue staining with BSA as a standard. The preparation and purification of GST-fusion proteins has been described (6).
Recombineering Protocols
Mutagenesis of a bacterial artificial chromosome containing the entire KSHV genome (BAC-16) was performed by using a recombineering system (58). The strains and vectors used in the recombineering procedure were obtained from the National Cancer Institute, National Institutes of Health. The host strain for λ-mediated recombination was a BAC-16/SW105 transformant. A LANA targeting vector was constructed using modified pCD-LANA that contains a 5′ KSHV ∼200-bp homology arm followed by an ampicillin resistance (Bla) cassette upstream of the LANA coding region. The Bla cassette is flanked by site-specific recognition (FRT) sequences for the FLPe recombinase (50). The linearized targeting cassette contains LANA homology arms of 0.2 and 1 kb on the 5′ and 3′ side of the ampicillin resistance marker. The targeting cassette was released from the vector sequences by digestion with PstI and BamHI. The fragment was gel-purified and electroporated into induced (recombination +) SW105/BAC-16 cells. Ampicillin-resistant transformants were selected and analyzed by PCR for insertion of the targeting cassette. Two or three positive clones of each intermediate were then grown at 32 °C with removal of the targeting cassette accomplished by arabinose induction of the FLPe recombinase. Ampicillin-sensitive clones were screened by PCR for removal of the targeting cassette. Clones positive by PCR for removal of the cassette were then further verified by sequencing the targeted region of LANA. A similar approach was used for the Arg-20 mutants that were cloned into the targeting construct. The introduced Arg-20 mutations were confirmed by sequencing, and the integrity of each recombinant BAC was examined by restriction enzyme digestions and Southern blotting.
LANA Wild-type Revertant Constructs
LANA Arg-20 mutant alleles in BAC-16 were reverted to wild-type using the recombineering protocols described above. For these constructions, the mutant LANA allele in the KSHV bacmid was replaced with wild-type LANA using the wt-LANA targeting cassette. Reversion of each mutant LANA allele to wild-type was confirmed by sequencing. 293T BAC stable cell lines containing each revertant bacmid were then created as described below.
Construction of BAC Stable Cell Cultures
293T cells were transfected with 5 μg of recombinant BAC DNA using HEKfectin (Bio-Rad). After 2 days of culture, the cells were expanded to a single T175 flask, and hygromycin (100 μg/ml) selection was started. After approximately 2 weeks, the hygromycin-resistant colonies that emerged from a single flask were trypsinized and pooled to establish each culture. Early passage pools were used throughout the experiments described herein.
ChIP, KSHV Tiling Array, and ChIP-on-chip
ChIP assays were performed following the protocol described by the Farnham laboratory at the University of California-Davis. 293T BAC stable cell cultures were used in the ChIP experiments, and Dox-inducible K-Rta-BCBL-1 cells were used in ChIP-on-chip experiments (51). Antibodies used for LANA immunoprecipitation were rat anti-LANA (Advanced Biotechnologies, Inc.) and rat non-immune serum IgG (Alpha Diagnostic International). The KSHV tiling array was designed across KSHV genome sequence (NC_009333.1) and manufactured by Agilent Technologies. The probes were spotted on 8 × 15 K array format. The details of this array have been described recently (10). Labeling, hybridization, scanning, and analyzing of the arrays were done at UC Davis Cancer Center Genomic Shared Resource. The enrichment of wt-LANA binding to the KSHV genome was calculated by the intensity ratio of immunoprecipitated and input DNA for each array spot, and the probe signals were further normalized by blank subtraction and the Whitehead error model using Genomic Workbench software. Each ChIP-on-chip experiment was performed in triplicate. For real-time PCR analysis of ChIP samples, the quantity of DNA amplified corresponding to specific KSHV loci was compared with the matching input samples. Results are expressed as the ratio of the ChIP sample amplification signal normalized to the signal obtained using the sample input control.
RT-qPCR
RNA was prepared from 293T BAC stable cell lines after an induction period of 48 h. Non-reactivated 293T BAC stable cells grown in parallel served as the latent cultures. For viral reactivation, the final concentrations of the chemical-inducing agents were 3 mm sodium butyrate and 20 ng/ml phorbol myristate acetate (Sigma). RNA was prepared using RNeasy plus kits (Qiagen), and cDNA was prepared using SuperScript III first-strand synthesis reagents (Invitrogen). Gene expression was analyzed by qPCR using real-time coding region primers against the ORFs denoted by arrows in Fig. 6A. KSHV gene-specific expression was normalized to the LANA signal for each sample. LANA expression was normalized to the corresponding β-actin control for each sample.
FIGURE 6.
LANA Arg-20 mutations affects KSHV chromatin binding during latency. A, KSHV wt-LANA whole genome tiling array is shown. LANA ChIP DNAs were prepared from K-Rta-inducible BCBL-1 cells during latency. The immunoprecipitated DNA was probed against a KSHV tiling array as described under “Experimental Procedures.” The graph depicts the ratio of LANA/input signal across the KSHV genome. The entire KSHV genome is annotated at the bottom of the figure. The arrows denote ORFs evaluated by ChIP and/or RT-qPCR in the 293T BAC stable cell cultures. Several regions of the KSHV genome are indicated above the graph for reference. Ba, validation of the anti-LANA antibody in LANA ChIP assay is shown. ChIP DNA samples were prepared as described under “Experimental Procedures” and immunoprecipitated with anti-LANA antibody or normal rat IgG. DNA associated with each immunoprecipitate was analyzed by qPCR using primer pairs against the ORF73 promoter. Values are reported as the ratio of the Ct values of the amplified DNA immunoprecipitated relative to the amount of input DNA amplified. b, ChIP analysis shown is. ChIP DNA was prepared from each BAC stable cell line during latency. ChIP DNA associated with each immunoprecipitate was analyzed by qPCR using primer pairs directed against the ORFs listed. Values are reported as the ratio of the Ct values of the amplified DNA immunoprecipitated with anti-LANA antibody relative to the amount of input DNA amplified.
In Vitro Histone Binding
Binding reactions were carried as described (6, 52). Binding buffer contained 20 mm Hepes (pH 7.9), 150 mm NaCl, 1 mm EDTA, 4 mm MgCl2, 0.1% Nonidet P-40, 10% glycerol, and 1 mm PMSF. Histone octamers were formed by resuspension of lyophilized calf thymus histones (Roche Applied Science) in 2 m NaCl followed by stepwise dialysis against PBS at 4 °C overnight. Binding reactions (20 μl) contained LANA GST fusion protein and 0.5 μg/μl histone octamer. Reactions were incubated at 4 °C for 90 min. Wash buffers contained binding buffer supplemented with 125–500 mm NaCl. After extensive washing, the GST beads were resuspended in 2× sample buffer, boiled, and loaded on SDS-PAGE-15% gels. The gels were subjected to immunoblotting as described above and probed for H2B (43).
RESULTS
LANA Is Arginine-methylated in Vitro
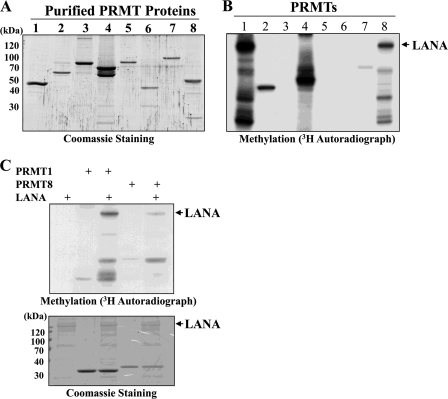
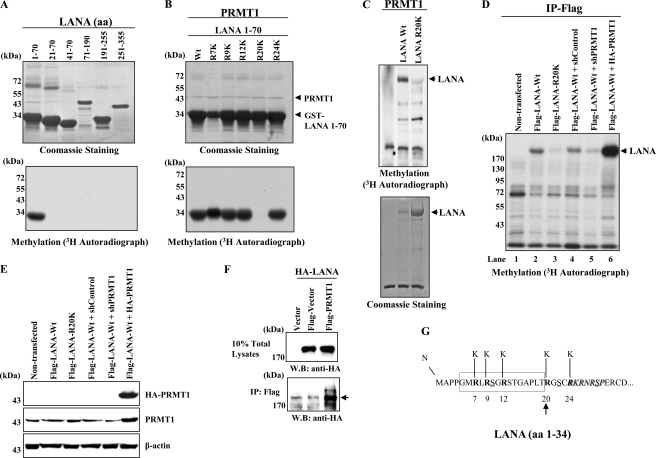
Our laboratory has developed a methylase/demethylase library that contains nearly the entire repertoire of human PRMTs, lysine methyltransferases, and lysine demethylases.5 Included in this library are eight of the nine mammalian PRMTs described to date. As part of a search for novel protein modifications directed by PRMTs, we screened a panel of recombinant viral proteins for their ability to serve as substrates for methylation in vitro. This panel included several KSHV proteins including LANA, ORF K8 (K-bZIP), ORF50 (K-Rta), and viral DNA replication enzymes. Although several important KSHV regulatory factors such as K-bZIP and K-Rta did not score in this assay, one strong positive hit that emerged from this screen was LANA. An example of the methylation reactions utilized in this screen is shown in Fig. 1, B and C. Each reaction contained purified, recombinant substrates (i.e. LANA), individual PRMTs (Fig. 1A), and S-adenosyl-l-[methyl-3H]methionine. A methylated protein with a molecular weight consistent with the migration of LANA was only observed in reactions containing PRMT1 and PRMT8 as the enzymatic activity (Fig. 1B, lanes 1 and 8). As PRMT8 expression is largely restricted to the brain and is localized to the cell membrane (53), we have focused our studies on the relationship between PRMT1 and LANA. Control reactions using histone substrates demonstrated that all of the PRMTs were enzymatically active (data not shown), although there were differences in specific activity. Using a series of LANA deletion proteins as substrates, we narrowed the methylation site(s) to the N-terminal ∼350 amino acids of LANA (data not shown). Further deletion analysis of this region of LANA mapped the methylation site(s) to the N-terminal 20 amino acid residues of LANA (Fig. 2A). This region of LANA contains a single RG motif, characteristic of the arginine methylation site glycine and arginine-rich patches (54). The first 5 N-terminal arginine residues of LANA (amino acids 1–70) were individually mutated to lysine, and the mutant proteins were purified as GST fusions (Fig. 2B). Using these substrates, a single PRMT1 methylation site was mapped in vitro to LANA residue Arg-20 (Fig. 2B). To examine the effect of the Arg-20 mutation on PRMT1-directed arginine methylation in the context of full-length LANA, the R20K mutation was inserted into a baculovirus vector, which expressed full-length LANA. When compared against wt-LANA protein, the methylation signal obtained using LANA R20K was greatly diminished (Fig. 2C). Taken together, these results map the major in vitro PRMT1 methylation site in LANA to residue Arg-20 (Fig. 2, B and C).
FIGURE 1.
LANA is arginine-methylated in vitro. A, purified PRMTs produced in baculovirus-infected cells were electrophoresed on 8% SDS-PAGE and stained with Coomassie Brilliant Blue. Numbers at the top of each lane of the gel signify PRMT designation. B, PRMT screening against LANA is shown. An analysis of methylation reactions using purified recombinant LANA and individual PRMTs is shown. Numbers at the top of each lane of the gel signify PRMT designation. Reaction products were electrophoresed on 8% SDS-PAGE and processed as described under “Experimental Procedures.” The arrow denotes arginine-methylated LANA. C, confirmatory methylation reactions demonstrate methyltransferase activity of PRMT1 and PRMT8 on LANA substrate.
FIGURE 2.
Mapping of the LANA PRMT1 arginine methylation site. A, LANA deletion fragments (amino acids 1–355) in Coomassie Brilliant Blue stain (top panel) and methylation reaction products (lower panel) of GST-LANA deletions electrophoresed on 8% SDS-PAGE are shown. The numbers above the Coomassie-stained gel indicate LANA amino acids. B, fine mapping of the LANA PRMT1 arginine methylation site is shown. Coomassie Brilliant Blue stain (top panel) and methylation reaction products (lower panel) of N-terminal Arg to Lys substitution mutants of GST-LANA amino acids 1–70 electrophoresed on 8% SDS-PAGE are shown. C, shown is the methylation of full-length LANA. In vitro methylation reactions using the indicated proteins, Coomassie Brilliant Blue stain (lower panel), and methylation reaction products (top panel) electrophoresed on 8% SDS-PAGE is shown. The arrow in the autoradiograph indicates arginine-methylated LANA. D, shown is in vivo methylation analysis of lysates prepared from 293T cells transfected with the indicated constructs. Cells were labeled, harvested, and analyzed as described under “Experimental Procedures.” Reaction products were electrophoresed on 8% SDS-PAGE. The arrow indicates arginine-methylated LANA. IP, immunoprecipitate. E, shown is confirmation of partial cellular PRMT1 knockdown and overexpression of HA-PRMT1. F, LANA-PRMT1 interaction is shown. 293T cells were transfected with the indicated plasmids and probed with the indicated antibodies. The arrow indicates the HA-LANA present in anti-FLAG-PRMT1 immunoprecipitates. WB, Western blot. G, the N-terminal 34 amino acids of LANA are shown. The LANA N-terminal histone binding domain (52) is boxed, the N-terminal amino acid cluster of the LANA bipartite NLS is italicized, and putative phosphorylation sites (55) are underlined (Ser-10, Ser-22 protein kinase C sites; Ser-29, protein kinase A/cdc-2-type kinase sites). The individual R/K mutants utilized in the mapping experiments are in bold. The arrow denotes the LANA arginine methylation site.
LANA Is Arginine-methylated in Vivo
As a next step, arginine methylation of LANA was evaluated in vivo. 293T cells were transfected with expression vectors expressing full-length FLAG-tagged wt-LANA or LANA R20K. Two days post-transfection, the cells were labeled for 3 h with [3H]methionine in the presence of a high concentration (100 μg/ml) of cycloheximide. Lysates were prepared, and LANA was immunoprecipitated using anti-FLAG antibody. Methylation of wt-LANA was readily detectable (Fig. 2D, lanes 2 and 4). In contrast, the signal was markedly reduced in immunoprecipitates prepared from LANA R20K lysates (Fig. 2D, lane 3). These results suggest that Arg-20 is the major methylation site in LANA but that there may be additional, minor methylation sites yet to be defined. Additional experiments demonstrated that partial depletion of cellular PRMT1 expression by shPRMT1, but not shControl, greatly reduced the LANA methylation signal (Fig. 2D, lanes 4 and 5). Conversely, transient overexpression of PRMT1 significantly increased LANA methylation (Fig. 2D, lane 6). Control blots confirm the partial knockdown and overexpression of PRMT1 in these experiments (Fig. 2E). Importantly, LANA physically associated with PRMT1 in transfected 293T cells, suggesting the specificity of the methylation (Fig. 2F). These results established that PRMT1 interacts with and methylates LANA in vivo. A summary of the methylation site mapping experiments in relationship to the N-terminal LANA histone binding domain, nuclear localization signal (NLS), and potential phosphorylation sites is shown in Fig. 2G (52, 55).
LANA Arginine Methylation and Subcellular Localization
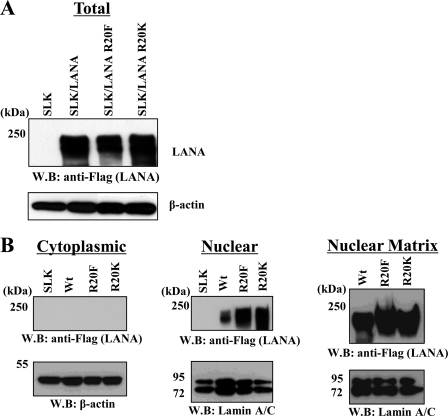
Our mapping studies positioned the arginine methylation site adjacent to (i) the LANA major histone binding domain and (ii) the LANA NLS (52, 55). Arginine methylation has been shown to regulate subcellular localization of proteins (36). Therefore, the effect of methylation on LANA localization was evaluated using cellular fractionation procedures on lysates prepared from SLK cells engineered to overexpress wt-LANA or LANA Arg-20 mutants. We substituted the positively charged arginine 20 methylation site residue of LANA with either of two amino acids in an attempt to infer the effect of methylation on LANA function. In one case, a bulky hydrophobic residue (Phe) was introduced; this residue has been proposed to mimic the constitutively arginine-methylated state (56). SLK cells were transduced with lentiviral vectors encoding FLAG-tagged versions of LANA or mutant LANA and selected with puromycin. To avoid clonal variation, mass cultures of puromycin-resistant pools of cells were screened for LANA wild-type or Arg-20 mutant expression by Western blotting. The immunoblots shown in Fig. 3A demonstrate that the steady-state level of LANA protein was similar among the cell lines, suggesting similar protein stability among the mutants. Lysates from these cell lines were then fractionated into cytoplasmic and nuclear pools, and each fraction was probed for LANA by Western blotting. The results showed that LANA protein is entirely nuclear regardless of the status of Arg-20 (Fig. 3B). Moreover, probing of nuclear matrix fractions from this same series of SLK cells demonstrated that LANA or mutant LANA was preferentially associated with the nuclear matrix fraction (Fig. 3B). Taken together, these results suggest that methylation is not involved in either LANA protein stability or nuclear localization, effects attributed to arginine methylation that have been observed with other viral proteins (34, 40).
FIGURE 3.
Arginine methylation does not influence LANA cellular localization. A, Western analysis (WB) of total wt-LANA/mutant LANA expression in SLK lentiviral stable cell lines is shown. B, Western analysis of cytoplasmic/nuclear and nuclear matrix-associated LANA fractionation from extracts prepared from SLK/lentiviral LANA/mutant LANA stable cell lines is shown. Blots were probed with the indicated antibodies.
LANA Arg-20 Mutation Shows Increased LANA-Histone Octamer Interaction
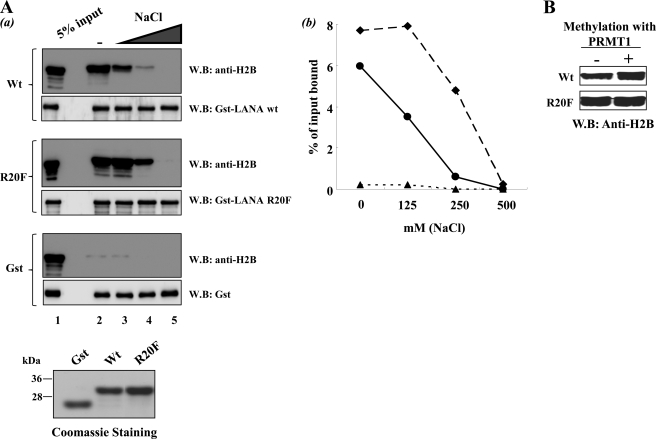
As a next step we examined effects on histone binding. In contrast to the lack of effect of the Arg-20 methylation site on LANA subcellular localization (Fig. 3), this site did influence LANA-histone interaction. An elevated degree of in vitro histone octamer binding by the R20F methylation mimic LANA protein was observed under our standard binding conditions (Fig. 4Aa, lane 2). Moreover, when preformed LANA-histone octamer complexes were washed with binding buffers containing increasing amounts of sodium chloride, the LANA R20F mutant protein showed increased binding stability at higher salt concentrations relative to wild-type LANA (Fig. 4A, a, lanes 3–5, and b). In addition, prior methylation of wild type, but not R20F mutant LANA, by PRMT1 also resulted in increased histone binding in vitro (Fig. 4B). Note that in this latter series of experiments, PRMT1 was removed from the reactions subsequent to methylation by washes with high salt buffer (500 mm NaCl) with 5% glycerol before histone binding. As the protein substrates used in these experiments were produced in Escherichia coli, which do not encode arginine methylases, GST-wt LANA represents a purely non-methylated protein. These results suggest that methylation of LANA increased its affinity to histone octamers in vitro.
FIGURE 4.
Enhanced histone octamer binding by methylation mimic LANA R20F and by arginine methylation of wt-LANA. Aa, in vitro histone binding by LANA and LANA R20F mutant is shown. H2B immunoblot analysis (WB) of in vitro histone binding reactions is shown. GST-LANA 1–70, GST-LANA 1–70 R20F mutant, or GST control beads were incubated with histone octamer as described under “Experimental Procedures.” Binding reactions were then washed with binding buffer (lane 2) or binding buffer supplemented with 125 mm (lane 3), 250 mm (lane 4), or 500 mm (lane 5) NaCl. 5% of input histone octamer is shown for each. Loading controls for each reaction are shown by GST immunoblot. Coomassie Brilliant Blue staining of each GST-LANA species used in the reactions is also shown. b, shown is quantitation of the H2B binding immunoblots in a (circles, wt-LANA; diamonds, LANA R20F; triangles, GST). B, coupled in vitro methylation/histone binding reactions are shown. GST-LANA 1–70 and GST-LANA 1–70 R20F mutant were incubated with recombinant PRMT1 (2 μm) and S-adenosyl-l-methylmethionine (4 mm) for 3 h at 37 °C. Control reactions were similarly set up without PRMT1. After incubation, the beads were washed several times with PBS containing 500 mm NaCl plus 5% glycerol. The beads were then equilibrated with histone binding buffer and assayed for histone octamer binding under standard conditions as described under “Experimental Procedures”.
Generation of LANA Arg-20 Mutants Using Recombineering
The influence of the LANA arginine methylation site was examined in the context of the KSHV genome. This newly developed bacmid is called KSHV BAC-16. BAC-16 is derived from rKSHV.219; this is a recombinant KSHV in which GFP is constitutively expressed (e.g. in latency) (57). BAC-16 also contains the selectable resistance marker hygromycin. Details of the construction of the parental KSHV BAC-16 will be published elsewhere.6 Using this new BAC system in conjunction with recombineering protocols (58), we incorporated the LANA Arg-20 mutants into the KSHV genome, individually replacing the endogenous wt-LANA allele with the Arg-20 substitutions. A LANA wild-type control was also constructed in which the wt-LANA allele on the targeting vector was used to replace the corresponding LANA wild-type allele on BAC-16. In addition, revertant LANA alleles in each Arg-20 substitution BAC were constructed in which the wt-LANA allele on the targeting vector was used to replace the mutant LANA Arg-20 allele that had been previously introduced into BAC-16. These constructs served as additional controls for potential influences of unintended mutations introduced into the bacmids during the course of recombineering. Construction of each recombinant BAC utilized a two-step procedure as described under “Experimental Procedures” and is shown schematically in Fig. 5A. Recombineering at the LANA locus was carried out using a SW105 transformant carrying BAC-16. This strain carries the BAC of interest as well as inducible systems for λ-mediated homologous recombination (step 1) and FLPe-mediated excision (step 2) of the targeting cassette. In step one, a linearized targeting cassette containing the LANA substitution mutation of interest and a selectable marker (ampicillin) was introduced into the recipient BAC cells by electroporation. Recombination between the linear targeting cassette and the LANA locus on the BAC proceeded using λ-mediated homologous recombination machinery that is present in SW105 (Fig. 5A). Drug-resistant intermediates were then screened by PCR for the presence of the targeting cassette. In step 2, the targeting cassette was excised from 1–3 individual positive intermediates using induced FLPe recombinase (Fig. 5A). Colonies (now ampicillin-sensitive) were screened by PCR for FRT-dependent excision of the selectable marker. The introduced mutation in the BAC was then sequence-verified. Final validation of the recombineered BAC was confirmed by restriction enzyme digestions and Southern blotting. Revertant LANA alleles were constructed in a similar fashion, except that recombination utilized individual SW105 strains carrying LANA Arg-20 mutant BACs and a wt-LANA targeting cassette. A Southern blot analysis of the LANA R20K mutant construction is shown in Fig. 5C. Probing recombinant BAC DNA clones with a LANA probe generated a band pattern consistent with an insertion of the targeting cassette at the LANA locus. BamHI-digested DNA purified from BAC intermediates was predicted to show a ∼1.1-kb band shift upon successful recombination of the targeting cassette (Fig. 5C, right panel, lanes B′ and C′) and, similarly, return to the position of the BAC-16 parental band (Fig. 5C, right panel, lane A′) upon FLPe-mediated removal of the targeting cassette (Fig. 5C, right panel, lanes D′, E′, and F′). When probed with an FRT-Bla fragment, only the BAC intermediates (Fig. 5C, left panel, lanes B and C) and not the final recombinants (Fig. 5C, left panel, lanes D, E, and F) or parental BAC (Fig. 5C, left panel, lane A) contain the corresponding fragment, as predicted. The band patterns obtained by Southern blot analysis of R20F and wt-LANA control bacmid constructs were identical to those listed in Fig. 5C. Final confirmation of each allele replacement strategy was conducted by sequence analysis of PCR products generated using primer pairs that bracket the mutation site in the recombinant BAC DNA. Fig. 5D depicts a small portion of the nucleotide sequence of the 5′ end of the LANA locus near the LANA ATG and compares this sequence with the residual nucleotides that remain in the final BAC recombinants. The recombinant BAC DNAs were then transfected into 293T cells, and mass cultures of hygromycin-resistant BAC stable cells (Fig. 5B) were created containing each of the Arg-20 mutants, the wt-LANA control, or the LANA revertants. After Hirt DNA extraction (59) and qPCR using LANA-specific primers, each culture was found to contain 1–5 genome copies per cell using a BAC-16 bacmid standard curve (data not shown). The BAC cultures were then used to study the effect of LANA methylation at position Arg-20 on KSHV viral gene expression.
FIGURE 5.
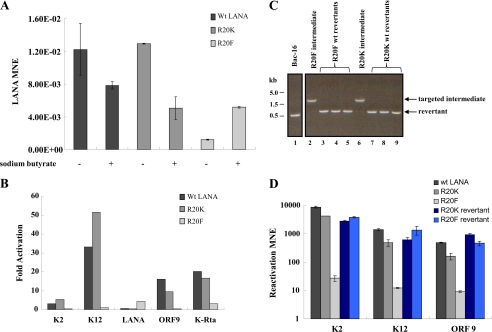
KSHV BAC-16 recombineering. A, shown is a schematic depiction of the recombineering procedures used for construction of BAC-16 LANA mutants. B, establishment of 293T BAC stable cell line shows >90% GFP-positive cells after hygromycin selection. FITC channel (GFP, left) and phase (right) images are shown. C, Southern analysis used recombinant KSHV bacmid DNA that was digested with BamHI and probed as indicated. Analysis of BAC-16 LANA R20K allele is shown. D, shown is the nucleotide sequence near the LANA initiator methionine of the parental BAC-16 (upper sequence) compared with a bacmid that had undergone successful recombineering (lower sequence). The LANA ATG is underlined. The single residual 34-bp FRT site plus 12 bp derived from the targeting vector are shown at the insertion site between nucleotides −1 and +1 relative to the LANA ATG.
LANA Occupancy of KSHV Genome during Latency
We wished to examine the effects of Arg-20 methylation on KSHV gene expression profiles of several select viral genes during latency and viral reactivation using the BAC stable cell lines. Before undertaking these studies, we needed to determine the LANA target viral genes throughout the KSHV genome, as we envisioned using the stable cell lines to measure KSHV gene expression of viral ORFs, whose gene expression was directly regulated by LANA. To achieve this, we constructed a KSHV tiling array and conducted a LANA ChIP on KSHV chip analyses with K-Rta-inducible BCBL-1 cells (51). Surprisingly, although there are peaks and troughs in the LANA binding pattern, the array depicts LANA occupancy throughout the entire latent KSHV genome when we considered 2-fold enrichment as significant (Fig. 6A). ChIP-KSHV-chip analyses were verified by qPCR with primers specific for the LANA promoter region (Fig. 6Ba). Interestingly, during viral reactivation there is an immediate (i.e. within 4 h) disassociation of LANA from the KSHV genome after K-Rta induction (supplemental Fig. 1). These results propose a model in which LANA may play a key role in maintaining the entire latent KSHV epigenome during latency and rapidly disassociates from the genome during the early stages of reactivation.
Increased Chromatin Interaction in Vivo by LANA R20F Mutation during Latency
After major LANA binding sites were in hand, we examined the effects of arginine methylation on recruitment of LANA to select target regions. LANA ChIP analysis was used to examine the degree of LANA wt or Arg-20 mutant association with the KSHV genome. We examined several regions where LANA was significantly enriched in naturally infected cells. Two regions containing lower levels of LANA enrichment (vIRF3 and ORF64) were also included in the analysis. As shown in Fig. 6Bb, the ChIP analysis demonstrated an increase in LANA R20F occupancy within 7/7 LANA recruitment sites in viral genes analyzed during latency, which is consistent with the observed increase in affinity toward histone octamers in vitro. In contrast, the degree of LANA R20K occupancy at this same panel of viral genes was reduced (5/7 genes) or equivalent (2/7 genes) to wt-LANA binding (Fig. 6Bb). Consistent with the LANA ChIP-KSHV-chip results, using Bac stable LANA ChIP DNA, non-enriched genes (vIRF3 and ORF64) also showed low levels of LANA recruitment during latency when analyzed by qPCR (Fig. 6Bb).
LANA Arginine Methylation and KSHV Gene Expression
If LANA plays a role in gene repression during reactivation (60) and LANA R20F differs in recruitment, one may expect to see the effects on viral gene expression at LANA target genes. RNA was prepared from 293T BAC stable cell cultures during latency and at 48 h post-reactivation. Reactivation was accomplished by sodium butyrate and phorbol myristate acetate treatment, and the gene expression profiles of KSHV genes were determined by quantitative RT-PCR. Genes examined included immediate early (ORF50), early (K2, ORF9), and latent (K12 and ORF73) (Fig. 7, A and B). LANA expression in each cell culture was normalized to β-actin (Fig. 7A). The expression of each additional KSHV gene under evaluation was normalized relative to the corresponding LANA value to minimize the effects of episomal copy number for each cell line. When 293T cells harboring BAC-16 episomes with the LANA R20F allele were examined, KSHV gene expression was reduced under both latent and reactivated conditions relative to 293T wt-LANA cells (Fig. 7A and data not shown). When viewed as -fold changes in response to reactivation, the gene expression results in the R20F culture were markedly reduced relative to the response of the wt control or R20K cell lines (Fig. 7B). Importantly, this profile of gene expression correlated with recruitment of LANA, where higher enrichment of LANA showed reduced target gene expression. ORF73 expression was modestly stimulated in LANA R20F Bac stable cells, whereas wt- and R20K LANA expression declined slightly during reactivation (Fig. 7A). Similar results to those described above were obtained when we conducted this experiment using (i) K-Rta overexpression as a means of reactivation and (ii) chemical reactivation using an independently derived series of 293T BAC stable cells (data not shown).
FIGURE 7.
LANA Arg-20 mutations affects KSHV gene expression. A, shown is an analysis of LANA expression by RT-qPCR among 293T BAC-16 stable cell cultures during latency and at 48 h reactivation. LANA expression was normalized to β-actin. Values represent the mean ±S.D. of three determinations (MNE, mean normalized expression). B, -fold expression is shown. RNA was harvested from each culture during latency and at 48 h reactivation. cDNA was prepared, and gene expression was analyzed by qPCR using primer pairs directed against the ORFs listed. The -fold change in expression (reactivated/latent) for each ORF is shown among each culture of BAC stable cells. Latent and reactivated expression was normalized to LANA. C, LANA Arg-20 revertant alleles are shown. Ethidium bromide staining of 1.2% agarose gel analysis of PCR products amplified from BAC DNA using primers that bracket the Arg-20 site of LANA. Indicated PCR products are the targeted intermediate (1600 bp) and the recombined allele (500 bp). BAC-16 parental (lane 1), targeted LANA R20F (lane 2), and targeted R20K (lane 6) are shown. Cultures containing the targeted intermediates were grown and treated with arabinose for 1 h to induce FLPe-mediated recombination. BAC DNA was prepared from three individual revertants derived from targeted intermediates of R20F (lanes 3–5) and R20K (lanes 7–9) and screened by PCR as described above. Each of the final recombinants positive by PCR were verified as wild-type revertants by sequence analysis. D, gene expression analysis of Arg-20 wild-type revertant alleles is shown. BAC stable cell lines were created for each revertant allele, and KSHV gene expression was analyzed during latency or at 48 h post-chemical reactivation. Gene expression was analyzed by RT-qPCR using primers specific for the KSHV ORFs listed (MNE, mean normalized expression × 1000, reference LANA).
Finally, to show that the phenotype we observed is not due to nonspecific mutations, we constructed wild-type revertant alleles of each LANA Arg-20 mutant (Fig. 7C). 293T BAC stable cell lines of each revertant were established, and the gene expression phenotype of each culture was examined as described in the previous section. The revertant cultures contained amounts of KSHV genomic DNA copy numbers similar to the Arg-20 mutant cultures (data not shown). When compared with the gene expression results for the Arg-20 mutant cultures, all revertant BAC-containing cultures exhibited changes in KSHV gene expression (Fig. 7D) that approached the values obtained with the wt-LANA control BAC culture.
DISCUSSION
In this report we demonstrate that the KSHV LANA protein is subject to arginine methylation. The major arginine methylation site in LANA was mapped to residue Arg-20. This modification further expands the LANA repertoire of post-translational modifications including phosphorylation (28), acetylation (29), poly(ADP-ribosyl)ation (30), and sumoylation.4 Importantly, we could detect arginine methylated LANA in vivo, and we have preliminary estimates of this value as ≤2% LANA methylation relative to the total amount of immunoprecipitated LANA present in latent BCBL-1 cell extracts (supplemental Fig. 2A). A higher value of ∼5% methylation was obtained analyzing lysates from LANA-transfected 293T cells (supplemental Fig. 2B). One issue that remains is when and where LANA arginine methylation occurs and the relative levels of arginine-methylated versus unmethylated LANA under different conditions. Indeed, the question of the relative levels of modified versus unmodified species applies to all post-translational protein modifications, including LANA modifications. It is tempting to speculate that methylation of LANA may be a signal for recruiting protein complexes, which recognizes protein methylation as seen in cellular histone binding proteins (for a recent review, see Ref. 61). Specific tools such as modified LANA-specific antibodies may be needed to answer those questions. The exact nature and location of the methyl mark(s) on LANA molecules present in BCBL-1 cells is still not known. However, because PRMT1 is classified as a type I PRMT (31) and our knockdown data indicate that PRMT1 is the predominant enzymatic activity responsible for methylation of transfected LANA in 293T cells, the arginine methyl mark on LANA at position Arg-20 is most likely to be asymmetric dimethylation. Our detailed mutagenesis studies, which focused on the N terminus of LANA, suggest that Arg-20 is the major arginine methylation site. However, our results using the full-length LANA R20K mutant indicate that there may be other minor methylation sites in LANA. The role and nature of these additional methylation sites remain to be determined. We have conducted large-scale LANA immunoprecipitations using nuclear extracts prepared from HeLa-LANA-overexpressing cells and have also identified PRMT1 as a LANA interacting partner.5 This observation taken together with the ability of LANA to be methylated in vitro and in vivo by PRMT1 suggests that LANA is a genuine PRMT1 substrate and not a physically interacting but non-methyl accepting partner, as has been observed for other PRMT-interacting partners (56, 62, 63).
Our mapping of the LANA arginine methylation site to residue Arg-20 is very intriguing given the fact that this site is interposed between the dominant LANA histone binding domain (52) and the LANA NLS (55). Moreover, this region of LANA contains several predicted protein phosphorylation sites, suggesting the possibility for modulation of LANA function via phospho-methyl modification cassettes or switches as has been seen in other proteins (64, 65). Mutation of LANA residue Arg-20 did not influence LANA protein stability, nuclear localization, or nuclear matrix association; however, this residue did affect LANA histone binding activity in vitro and LANA-KSHV chromatin association in vivo. Importantly, cell lines harboring LANA R20F mutant BACs responded to reactivation signals very poorly. Inspection of the ChIP and gene expression data for a given KSHV locus suggests an inverse correlation between LANA association during latency and the -fold response to chemical reactivation. A high relative level of LANA binding within a specific KSHV gene resulted in a lower -fold response to chemical reactivation, whereas a low or intermediate level of binding had the opposite effect. ORF73 was an exception to this trend, where expression was modestly stimulated in LANA R20F BAC stable cells, possibly the result of hyperstimulation of the LANA promoter (66) by increased R20F binding. Taken together, we interpret these results to suggest that arginine methylation of LANA results in a LANA molecule that is more effective at KSHV chromatin binding and subsequent silencing of certain regions of the KSHV genome. Importantly, the arginine methylation site is conserved in Rhesus monkey rhadinovirus ORF73. In fact, Rhesus monkey rhadinovirus ORF73 contains five RG motifs in the N-terminal region of ORF73, indicating a possible conserved function of arginine methylation. Consistent with our data suggesting that Arg-20 methylation strengthens the LANA-chromatin interaction, LANA R20K (a non-arginine-methylatable residue) exhibited the lowest relative LANA occupancy at seven of nine KSHV loci examined and a higher-fold response at two loci where this parameter was assayed (K2 and K12). These results suggest that arginine methylation, in addition to other factors such as target DNA sequence and other DNA binding proteins, may influence LANA chromatin binding. Interestingly, a recent report showed that methylation of EBNA2 is required for interaction with DNA-bound RBP-Jκ to modulate gene expression (67). Because LANA has been shown to associate with RBP-Jκ, this function may be conserved between Epstein-Barr virus and KSHV (20).
The replacement of arginine with a lysine residue does preserve the positive charge of the position and renders this site refractory to arginine methylation; however, this introduction might be potentially complicated due to the ability of lysine to be subjected to other post-translational modifications such as lysine methylation, acetylation, and ubiquitylation. Importantly, the analysis of LANA Arg-20 revertant alleles suggests that the observed gene expression phenotype among the LANA Arg-20 mutants is LANA-specific and not due to the introduction of nonspecific mutations outside of the targeted region. Nonetheless, we acknowledge that the notion of whether or not the R20F mutation accurately mimics methylation is subject to debate, and therefore, it can be argued that if Arg-20 plays an important role in LANA function in the unmethylated state, then any mutation of this residue would impact LANA function.
Another key function of LANA is episomal maintenance during latency. Because protein methylation has been reported to affect both protein-protein and protein-nucleic acid interactions (31, 54), it is possible that arginine methylation of LANA may influence KSHV episomal maintenance. However, our experiments suggest that this may not be the case. After transient transfections of BAC DNA into 293T cells without incorporation of a selection procedure, GFP fluorescence declined precipitously for all BAC transfectants regardless of the LANA Arg-20 status (supplemental Fig. 3).
When combined with recombineering protocols, the KSHV bacmid used in these experiments allows one to readily construct and evaluate mutations and the corresponding revertant alleles within the context of the KSHV genome. The procedure adapted herein is very efficient and yields a very low background of non-recombinants. We have utilized our current mutagenesis protocol to introduce mutations into multiple KSHV loci including K-Rta and K-bZIP. The BAC-16 system could also be amenable to recombineering procedures that utilize single-stranded oligonucleotides (68) or short denatured PCR fragments as targeting vectors (69) to further facilitate BAC-16 manipulation.
In the course of conducting these experiments, we carried out a LANA ChIP-chip experiment using a customized, whole genome KSHV tiling array that we designed. Our results demonstrate for the first time that LANA is associated with essentially the entire latent KSHV genome in BCBL-1 cells with several highly enriched regions. This may suggest that LANA has additional roles as a structural element coordinating the binding, arrangement, and modulation of factors that interact with KSHV chromatin. This is consistent with the finding that LANA function can be partially replaced by histone H1 (70). Interestingly, the LANA-episome interaction rapidly disassociates upon K-Rta-mediated reactivation such that at 4 h post-reactivation, LANA association with the KSHV episome is markedly reduced (supplemental Fig. 1). Because this study utilized samples derived from early time points, this phenotype is not likely due to encapsidation of the KSHV genome. A previous report had observed the rapid loss of LANA from the ORF50 region during reactivation (29); our results now expand this loss to the entire episome. We are currently investigating mechanisms of LANA dissociation from the KSHV genome. Importantly, our LANA ChIP-chip result complements recent reports that have provided comprehensive maps of active and repressive histone marks along the entire KSHV chromosome (8, 9), as well as a map of the binding sites across the KSHV genome of an associated cellular histone demethylase (10). These findings together with our LANA tiling array data may provide new insights into epigenetic regulation via viral proteins. Moreover, the results reported herein underscore the potential of relatively understudied methylation/demethylation events to influence the function of viral proteins.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA147791 (to Y. I.) and P01-DE19085 (to H.-J. K., J. U. J., and Y. I.). Additional funding was provided by University of California Davis Cancer Center Support Grant P30 CA93373.

This article contains supplemental Figs. 1–3.
Y. Izumiya, unpublished results.
K. Kim and Y. Izumiya, unpublished information.
J. U. Jung, personal communication.
- KSHV
- Kaposi sarcoma (KS)-associated herpesvirus
- LANA
- latency-associated nuclear antigen
- BCBL-1
- body cavity-based lymphoma
- PRMT
- protein arginine methyltransferase
- BAC
- bacterial artificial chromosome
- FRT
- Flp recombinase target
- FLPe
- enhanced Flp recombinase
- qPCR
- quantitative PCR.
REFERENCES
- 1. Cesarman E., Knowles D. M. (1999) The role of Kaposi sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin Cancer Biol. 9, 165–174 [DOI] [PubMed] [Google Scholar]
- 2. Wong E. L., Damania B. (2005) Linking KSHV to human cancer. Curr. Oncol. Rep. 7, 349–356 [DOI] [PubMed] [Google Scholar]
- 3. Schulz T. F. (2006) The pleiotropic effects of Kaposi sarcoma herpesvirus. J. Pathol. 208, 187–198 [DOI] [PubMed] [Google Scholar]
- 4. Ganem D. (2010) KSHV and the pathogenesis of Kaposi sarcoma. Listening to human biology and medicine. J. Clin. Invest. 120, 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun R., Lin S. F., Gradoville L., Yuan Y., Zhu F., Miller G. (1998) A viral gene that activates lytic cycle expression of Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U.S.A. 95, 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Izumiya Y., Ellison T. J., Yeh E. T., Jung J. U., Luciw P. A., Kung H. J. (2005) Kaposi sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79, 9912–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang P. C., Fitzgerald L. D., Van Geelen A., Izumiya Y., Ellison T. J., Wang D. H., Ann D. K., Luciw P. A., Kung H. J. (2009) Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 69, 5681–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toth Z., Maglinte D. T., Lee S. H., Lee H. R., Wong L. Y., Brulois K. F., Lee S., Buckley J. D., Laird P. W., Marquez V. E., Jung J. U. (2010) Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6, e1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Günther T., Grundhoff A. (2010) The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6, e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang P. C., Fitzgerald L. D., Hsia D. A., Izumiya Y., Wu C. Y., Hsieh W. P., Lin S. F., Campbell M., Lam K. S., Luciw P. A., Tepper C. G., Kung H. J. (2011) Histone demethylase JMJD2A regulates Kaposi sarcoma-associated herpesvirus replication and is targeted by a viral transcriptional factor. J. Virol. 85, 3283–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kedes D. H., Lagunoff M., Renne R., Ganem D. (1997) Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi sarcoma-associated herpesvirus. J. Clin. Invest. 100, 2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mesri E. A., Cesarman E., Boshoff C. (2010) Kaposi sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verma S. C., Lan K., Robertson E. (2007) Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312, 101–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim C., Sohn H., Lee D., Gwack Y., Choe J. (2002) Functional dissection of latency-associated nuclear antigen 1 of Kaposi sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76, 10320–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grundhoff A., Ganem D. (2003) The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77, 2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J., Garber A. C., Renne R. (2002) The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76, 11677–11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han S. J., Hu J., Pierce B., Weng Z., Renne R. (2010) Mutational analysis of the latency-associated nuclear antigen DNA binding domain of Kaposi sarcoma-associated herpesvirus reveals structural conservation among γ-herpesvirus origin-binding proteins. J. Gen. Virol. 91, 2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelley-Clarke B., De Leon-Vazquez E., Slain K., Barbera A. J., Kaye K. M. (2009) Role of Kaposi sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J. Virol. 83, 4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An F. Q., Compitello N., Horwitz E., Sramkoski M., Knudsen E. S., Renne R. (2005) The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280, 3862–3874 [DOI] [PubMed] [Google Scholar]
- 20. Lan K., Kuppers D. A., Robertson E. S. (2005) Kaposi sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 79, 3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Platt G.M., Simpson G. R., Mittnacht S., Schulz T. F. (1999) Latent nuclear antigen of Kaposi sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73, 9789–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Radkov S. A., Kellam P., Boshoff C. (2000) The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 23. Verma S. C., Borah S., Robertson E. S. (2004) Latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78, 10348–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim C., Gwack Y., Hwang S., Kim S., Choe J. (2001) The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi sarcoma-associated herpesvirus. J. Biol. Chem. 276, 31016–31022 [DOI] [PubMed] [Google Scholar]
- 25. Murakami Y., Yamagoe S., Noguchi K., Takebe Y., Takahashi N., Uehara Y., Fukazawa H. (2006) Ets-1-dependent expression of vascular endothelial growth factor receptors is activated by latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus through interaction with Daxx. J. Biol. Chem. 281, 28113–28121 [DOI] [PubMed] [Google Scholar]
- 26. Friborg J., Jr., Kong W., Hottiger M. O., Nabel G. J. (1999) p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402, 889–894 [DOI] [PubMed] [Google Scholar]
- 27. Varjosalo M., Björklund M., Cheng F., Syvänen H., Kivioja T., Kilpinen S., Sun Z., Kallioniemi O., Stunnenberg H. G., He W. W., Ojala P., Taipale J. (2008) Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 133, 537–548 [DOI] [PubMed] [Google Scholar]
- 28. Cheng F., Weidner-Glunde M., Varjosalo M., Rainio E. M., Lehtonen A., Schulz T. F., Koskinen P. J., Taipale J., Ojala P. M. (2009) KSHV reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathog. 5, e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu F., Day L., Gao S. J., Lieberman P. M. (2006) Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi sarcoma-associated herpesvirus lytic transcription. J. Virol. 80, 5273–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohsaki E., Ueda K., Sakakibara S., Do E., Yada K., Yamanishi K. (2004) Poly(ADP-ribose) polymerase 1 binds to Kaposi sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 78, 9936–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bedford M. T., Clarke S. G. (2009) Protein arginine methylation in mammals. Who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gary J. D., Clarke S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 33. Boisvert F. M., Côté J., Boulanger M. C., Richard S. (2003) A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 34. Koyuncu O. O., Dobner T. (2009) Arginine methylation of human adenovirus type 5 L4 100-kilodalton protein is required for efficient virus production. J. Virol. 83, 4778–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iacovides D. C., O'Shea C. C., Oses-Prieto J., Burlingame A., McCormick F. (2007) Critical role for arginine methylation in adenovirus-infected cells. J. Virol. 81, 13209–13217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shire K., Kapoor P., Jiang K., Hing M. N., Sivachandran N., Nguyen T., Frappier L. (2006) Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 80, 5261–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mears W. E., Rice S. A. (1996) The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA binding activity and determines in vivo methylation. J. Virol. 70, 7445–7453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hibbard M. K., Sandri-Goldin R. M. (1995) Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol. 69, 4656–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie B., Invernizzi C. F., Richard S., Wainberg M. A. (2007) Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J. Virol. 81, 4226–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rho J., Choi S., Seong Y. R., Choi J., Im D. S. (2001) The arginine 1493 residue in QRRGRTGR1493G motif IV of the hepatitis C virus NS3 helicase domain is essential for NS3 protein methylation by the protein arginine methyltransferase 1. J. Virol. 75, 8031–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y. J., Stallcup M. R., Lai M. M. (2004) Hepatitis δ virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78, 13325–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu J., Shin B., Park E. S., Yang S., Choi S., Kang M., Rho J. (2010) Protein arginine methyltransferase 1 regulates herpes simplex virus replication through ICP27 RGG-box methylation. Biochem. Biophys. Res. Commun. 391, 322–328 [DOI] [PubMed] [Google Scholar]
- 43. Izumiya Y., Izumiya C., Hsia D., Ellison T. J., Luciw P. A., Kung H. J. (2009) NF-κB serves as a cellular sensor of Kaposi sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jκ coactivator. J. Virol. 83, 4435–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hamza M. S., Reyes R. A., Izumiya Y., Wisdom R., Kung H. J., Luciw P. A. (2004) ORF36 protein kinase of Kaposi sarcoma herpesvirus activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 279, 38325–38330 [DOI] [PubMed] [Google Scholar]
- 45. Izumiya Y., Lin S. F., Ellison T. J., Levy A. M., Mayeur G. L., Izumiya C., Kung H. J. (2003) Cell cycle regulation by Kaposi sarcoma-associated herpesvirus K-bZIP. Direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 77, 9652–9661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izumiya Y., Lin S. F., Ellison T., Chen L. Y., Izumiya C., Luciw P., Kung H. J. (2003) Kaposi sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta. Physical association and promoter-dependent transcriptional repression. J. Virol. 77, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herndier B. G., Werner A., Arnstein P., Abbey N. W., Demartis F., Cohen R. L., Shuman M. A., Levy J. A. (1994) Characterization of a human Kaposi sarcoma cell line that induces angiogenic tumors in animals. AIDS 8, 575–581 [DOI] [PubMed] [Google Scholar]
- 48. Siegal B., Levinton-Kriss S., Schiffer A., Sayar J., Engelberg I., Vonsover A., Ramon Y., Rubinstein E. (1990) Kaposi sarcoma in immunosuppression. Possibly the result of a dual viral infection. Cancer 65, 492–498 [DOI] [PubMed] [Google Scholar]
- 49. Ohsaki E., Suzuki T., Karayama M., Ueda K. (2009) Accumulation of LANA at nuclear matrix fraction is important for Kaposi sarcoma-associated herpesvirus replication in latency. Virus Res 139, 74–84 [DOI] [PubMed] [Google Scholar]
- 50. Buchholz F., Angrand P. O., Stewart A. F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16, 657–662 [DOI] [PubMed] [Google Scholar]
- 51. Nakamura H., Lu M., Gwack Y., Souvlis J., Zeichner S. L., Jung J. U. (2003) Global changes in Kaposi sarcoma-associated virus gene expression patterns after expression of a tetracycline-inducible Rta transactivator. J. Virol. 77, 4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barbera A. J., Chodaparambil J. V., Kelley-Clarke B., Joukov V., Walter J. C., Luger K., Kaye K. M. (2006) The nucleosomal surface as a docking station for Kaposi sarcoma herpesvirus LANA. Science 311, 856–861 [DOI] [PubMed] [Google Scholar]
- 53. Lee J., Sayegh J., Daniel J., Clarke S., Bedford M. T. (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896 [DOI] [PubMed] [Google Scholar]
- 54. Bedford M. T., Richard S. (2005) Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 55. Piolot T., Tramier M., Coppey M., Nicolas J. C., Marechal V. (2001) Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75, 3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mostaqul Huq M. D., Gupta P., Tsai N. P., White R., Parker M. G., Wei L. N. (2006) Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 25, 5094–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vieira J., O'Hearn P. M. (2004) Use of the red fluorescent protein as a marker of Kaposi sarcoma-associated herpesvirus lytic gene expression. Virology 325, 225–240 [DOI] [PubMed] [Google Scholar]
- 58. Copeland N. G., Jenkins N. A., Court D. L. (2001) Recombineering. A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 [DOI] [PubMed] [Google Scholar]
- 59. Arad U. (1998) Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques 24, 760–762 [DOI] [PubMed] [Google Scholar]
- 60. Li Q., Zhou F., Ye F., Gao S. J. (2008) Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yun M., Wu J., Workman J. L., Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robin-Lespinasse Y., Sentis S., Kolytcheff C., Rostan M. C., Corbo L., Le Romancer M. (2007) hCAF1, a new regulator of PRMT1-dependent arginine methylation. J. Cell Sci. 120, 638–647 [DOI] [PubMed] [Google Scholar]
- 63. Lei N. Z., Zhang X. Y., Chen H. Z., Wang Y., Zhan Y. Y., Zheng Z. H., Shen Y. M., Wu Q. (2009) A feedback regulatory loop between methyltransferase PRMT1 and orphan receptor TR3. Nucleic Acids Res. 37, 832–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rust H. L., Thompson P. R. (2011) Kinase consensus sequences. A breeding ground for cross-talk. ACS Chem. Biol. 6, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fischle W., Wang Y., Allis C. D. (2003) Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 [DOI] [PubMed] [Google Scholar]
- 66. Renne R., Barry C., Dittmer D., Compitello N., Brown P. O., Ganem D. (2001) Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus. J. Virol. 75, 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gross H., Barth S., Palermo R. D., Mamiani A., Hennard C., Zimber-Strobl U., West M. J., Kremmer E., Grässer F. A. (2010) Asymmetric arginine dimethylation of Epstein-Barr virus nuclear antigen 2 promotes DNA targeting. Virology 397, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Swaminathan S., Ellis H. M., Waters L. S., Yu D., Lee E. C., Court D. L., Sharan S. K. (2001) Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis 29, 14–21 [DOI] [PubMed] [Google Scholar]
- 69. Yang Y., Sharan S. K. (2003) A simple two-step, “hit and fix” method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 31, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shinohara H., Fukushi M., Higuchi M., Oie M., Hoshi O., Ushiki T., Hayashi J., Fujii M. (2002) Chromosome binding site of latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76, 12917–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.