Abstract
Background
Taxonomy and phylogeny of subclass Heterodonta including Tellinoidea are long-debated issues and a complete agreement has not been reached yet. Mitochondrial (mt) genomes have been proved to be a powerful tool in resolving phylogenetic relationship. However, to date, only ten complete mitochondrial genomes of Heterodonta, which is by far the most diverse major group of Bivalvia, have been determined. In this paper, we newly sequenced the complete mt genomes of six species belonging to Heterodonta in order to resolve some problematical relationships among this subclass.
Principal Findings
The complete mt genomes of six species vary in size from 16,352 bp to 18,182. Hairpin-like secondary structures are found in the largest non-coding regions of six freshly sequenced mt genomes, five of which contain tandem repeats. It is noteworthy that two species belonging to the same genus show different gene arrangements with three translocations. The phylogenetic analysis of Heterodonta indicates that Sinonovacula constricta, distant from the Solecurtidae belonging to Tellinoidea, is as a sister group with Solen grandis of family Solenidae. Besides, all five species of Tellinoidea cluster together, while Sanguinolaria diphos has closer relationship with Solecurtus divaricatus, Moerella iridescens and Semele scaba rather than with Sanguinolaria olivacea.
Conclusions/Significance
By comparative study of gene order rearrangements and phylogenetic relationships of the five species belonging to Tellinoidea, our results support that comparisons of mt gene order rearrangements, to some extent, are a useful tool for phylogenetic studies. Based on phylogenetic analyses of multiple protein-coding genes, we prefer classifying the genus Sinonovacula within the superfamily Solenoidea and not the superfamily Tellinoidea. Besides, both gene order and sequence data agree that Sanguinolaria (Psammobiidae) is not monophyletic. Nevertheless, more studies based on more mt genomes via combination of gene order and phylogenetic analysis are needed to further understand the phylogenetic relationships in subclass Heterodonta.
Introduction
Mitochondrial DNA (mtDNA) is the only extranuclear genome in animal cytoplasm [1]. Most metazoan mitochondrial genomes are covalently closed circular molecules which range from 14 to 42 kb in length [2], but see [3]. The typical mitochondrial genome contains the same 37 genes: 13 for protein subunits of oxidative phosphorylation enzymes (atp6, atp8, cox1–3, cob, nad1–6 and nad4l), two for mitochondrial ribosomal RNAs [small and large subunit ribosomal RNA (rrnS and rrnL)] and 22 for the transfer RNA genes (tRNAs) genes necessary for translating these 13 proteins [4]. In general, there are few intergenic nucleotides except for a single large non-coding region generally thought to contain elements that control the initiation of replication and transcription [4]. Owing to abundance of mitochondria in cells, lack of recombination, maternal inheritance (except for [5]), absence of introns, and higher evolutionary rates, mtDNA sequences are extensively used for comparative and evolutionary genomics, molecular evolution, population genetics, species identification, and phylogenetic relationships at various taxonomic levels [6]–[9].
For some phyla of animal, mitochondrial gene arrangements seem seldom to have changed. With few notable exceptions, those vertebrates studied, for instance, have identical gene arrangements [10]. However, mollusks, especially bivalves, which display an extraordinary amount of variation in gene arrangement, challenge this rule. Gene arrangement has been shown to be very powerful characters for reconstructing evolutionary relationships, and the rapidity of rearrangement within a lineage determines the level at which rearrangements are likely to be phylogenetically informative [10], [11].
In recent studies, phylogenetic analysis based on complete mt sequence data have proved to enhance resolution and statistical confidence of inferred phylogenetic trees when compared with analyses based only on small portions of the mtDNA [12]–[15]. With technological and methodological advances, and associated decreasing costs of DNA sequencing, the amplification and sequencing of whole mt genomes has become routine [16]. Consequently, there have been significant increases in the number of complete mitochondrial sequences available during the last ten years. Nevertheless, to date, only ten complete mitochondrial genomes of Heterodonta, which is by far the most diverse major group of Bivalvia [17], have been determined.
Heterodonta, encompassing richly speciose families such as the Cardiidae, Tellinidae, Veneridae and Lucinidae, and including major economic groups such as clams, cockles, geoducks and razor shells, can be hugely abundant in both marine and freshwater systems, and of considerable ecological importance in community structure as well as a trophic resource [17]–[19]. However, taxonomy and phylogeny of Heterodonta are long-debated issues, and a complete agreement has not been reached yet, even if this subclass has a rich fossil history extending from the Lower Palaezoic, with major radiations in the Late Mesozoic [20]–[22]. In particular, the morphologically-inferred phylogenies of subclass Heterodonta were challenged by recent phylogenetic studies based on molecular data. For example, the Gastrochaenidae and Hiatellidae do not form a monophyletic group with the other families of the order Myoida [23]–[24]. Monophyly of the Lucinoidea is not supported, with the families Thyasiridae and Ungulinidae not closely related to the Lucinidae [25].
The superfamily Tellinoidea of the subclass Heterodonta consists of five families (Tellinidae, Donacidae, Psammobiidae, Semelidae, Solecurtidae) [26]. Based on the information of paleontology and morphology of Tellinoidea, large numbers of research works have been performed to study the evolutionary history and taxonomy within this superfamily over a long time [27]–[30]. Howbeit, compared with molecular analyses carried out to investigate relationships within individual families of Heterodonta (e.g., Veneridae [31], [32], Sphaeriidae [33], Thyasiridae [34]), there have been few attempts to make comprehensive analysis of phylogenetic relationships of Tellinoidea on molecular level so far, not to mention analysis based on complete mitochondrial genome.
In this paper, we newly sequenced the complete mt genomes of six heterodont bivalves, including five from four families (Tellininae, Psammobiidae with two species, Semelidae, Solecurtidae) within superfamily Tellinoidea and one from superfamily Solenoidea which was ever classified in Tellinoidea, and compared their different gene arrangements. In addition, the six newly determined sequences, together with the heterodont mt genomes available in GenBank, were used to recover the phylogeny of Heterodonta in order to resolve some problematical relationships among this subclass.
Results and Discussion
Genome composition
The main structural features of the six newly sequenced mt genomes in this study are summarized in Table 1. The complete mitochondrial genomes of MOIR-0101 Moerella iridescens [JN398362], SADI-0111 Sanguinolaria diphos [JN398363], SAOL-0112 Sanguinolaria olivacea [JN398364], SESC-0121 Semele scaba [JN398365], SICO-0201 Sinonovacula constricta [JN398366] and SODI-0131 Solecurtus divaricatus [JN398367] vary in size from 16,352 bp (S. diphos) to 18,182 bp (S. olivacea). Length differences are mostly in virtue of the variation in tandem repeats within the non-coding region. The placement of all coding genes on the same strand and the lack of one protein coding gene atp8 are the most distinctive features of marine bivalve mt genomes [35], [36], without exceptions for six studied species. The overall A+T content of six newly sequenced complete mt genomes ranges from 59.19% (S. scaba) to 67.08% (S. constricta). In addition, overlapping genes, which are presumably to help prevent rearrangements of gene order and loss of genes during evolution in mammalian [37], are a common phenomenon in all newly sequenced mt genomes (Table 1).
Table 1. Main structural features of the six newly sequenced mt genomes in this study.
| Moerella iridescens | Sanguinolaria diphos | Sanguinolaria olivacea | Semele scaba | Sinonovacula constricta | Solecurtus divaricatus | |
| Total size | 16799 | 16352 | 18182 | 17117 | 17224 | 16749 |
| A+T % | 65.72 | 63.36 | 65.27 | 59.19 | 67.08 | 60.15 |
| rrnS | 863 | 876 | 949 | 861 | 909 | 887 |
| rrnL | 1268 | 1343 | 1346 | 1330 | 1228 | 1380 |
| No. of tRNA | 22 | 21 | 21 | 20 | 22 | 22 |
| No. of gene overlapping | 3 | 6 | 9 | 2 | 2 | 4 |
| Size range of gene overlapping | 1 | 1 to 44 | 1 to 44 | 1 to 20 | 1 to 3 | 1 |
| cox1 | 1671 | 1755 | 1758 | 1692 | 1692 | 1725 |
| (ATA/TAA) | (ATA/TAG) | (ATA/TAA) | (ATG/TAA) | (ATT/TAA) | (ATA/TAG) | |
| cox2 | 861 | 873 | 858 | 1206 | 843 | 867 |
| (ATG/TAG) | (ATG/TAG) | (ATG/TAA) | (ATG/TAG) | (ATG/TAG) | (ATG/TAA) | |
| cox3 | 867 | 936 | 936 | 894 | 804 | 894 |
| (ATA/TAA) | (ATG/TAG) | (ATA/TAG) | (ATG/TAG) | (ATG/TAG) | (ATG/TAG) | |
| nad1 | 924 | 924 | 939 | 921 | 930 | 927 |
| (GTG/TAG) | (ATT/TAG) | (ATA/TAA) | (ATG/TAG) | (ATA/TAG) | (ATA/TAA) | |
| nad2 | 1014 | 1071 | 1077 | 1056 | 1056 | 1062 |
| (ATA/TAA) | (ATG/TAG) | (ATG/TAA) | (TTG/TAA) | (ATG/TAG) | (ATG/TAG) | |
| nad3 | 360 | 363 | 378 | 360 | 366 | 360 |
| (ATT/TAA) | (ATT/TAA) | (ATG/TAG) | (ATA/TAG) | (ATG/TAG) | (ATG/TAA) | |
| nad4 | 1335 | 1303 | 1362 | 1365 | 1392 | 1347 |
| (ATA/TAG) | (ATA/T) | (ATA/TAA) | (GTG/TAG) | (ATG/TAA) | (TTG/TAG) | |
| nad4L | 291 | 315 | 312 | 291 | 288 | 291 |
| (ATG/TAG) | (ATG/TAA) | (ATG/TAA) | (GTG/TAA) | (ATG/TAA) | (GTG/TAG) | |
| nad5 | 1746 | 1788 | 1800 | 1737 | 1761 | 1734 |
| (TTG/TAA) | (ATA/TAA) | (ATT/TAG) | (ATG/TAA) | (ATT/TAA) | (ATG/TAG) | |
| nad6 | 537 | 567 | 555 | 633 | 531 | 567 |
| (ATT/TAA) | (ATA/TAG) | (ATA/TAG) | (ATA/TAA) | (ATG/TAG) | (ATA/TAG) | |
| cob | 1233 | 1233 | 1242 | 1176 | 1146 | 1248 |
| (ATA/TAA) | (ATG/TAG) | (ATG/TAG) | (ATT/TAG) | (ATG/TAA) | (GTG/TAA) | |
| atp6 | 777 | 720 | 687 | 711 | 702 | 846 |
| (ATG/TAA) | (ATA/TAG) | (ATG/TAA) | (ATG/TAA) | (ATA/TAG) | (ATG/TAA) |
For each, total size of the mt genome, the percent of overall A+T content, size of rrnS and rrnL, number of tRNA, number of gene overlapping and its size range, and size of the protein coding genes (start and stop codons in parentheses) are presented. Gene lengths are in bp.
Protein coding genes
Table 1 shows the initiation and termination codons for the 12 protein-coding genes (PCGs) encoded by the six mt genomes. Most of PCGs (64/72) appear to start with the conventional codon ATN (ATG, N = 34; ATA, N = 22; ATT, N = 8), which is typical for metazoan mt genomes [2]. There are also TTG (N = 3) and GTG (N = 5) acted as start codons, which are not unusual start codons in molluscan mt genomes but in several gastropod mt genomes [38]. All 12 PCGs of six mt genomes end in full termination codon (TAG, N = 37; TAA, N = 34), except for nad4 gene of S. diphos ending with the incomplete stop codon T which may be modified to a complete TAA stop codon via posttranscriptional polyadenylation [39]. In contrast to the available heterodont bivalves mt genomes from GenBank, the mt genome of S. olivacea has the longest cox1 (1758 bp) and nad2 (1077 bp) genes, S. scaba has the longest cox2 (1206 bp) and nad6 (633 bp) genes, and S. diphos has the shortest nad4 (1303 bp) gene.
Transfer and ribosomal RNA genes
In the mt genomes of metazoan, almost all amino acids but leucine and serine are decoded by only one tRNA each [40]. Without exception, there are 22 typical tRNAs interspersed throughout the mt genome of M. iridescens, S. divaricatus, and S. constricta. The trnF is missing in both S. diphos and S. olivacea, and S. scaba lacks trnY and trnS1. Deficiencies of tRNA genes are often observed in protozoans, fungi, algae, plants and low metazoans [41], [42]. In this study, most of tRNAs can be folded into the typical secondary structures (not shown).
BLAST searches indicated approximate locations of the two rRNA genes, but their exact boundaries can not been determined [43]. The size of rrnL flanked by nad6 and atp6 in all six mt genomes ranges from 1228 bp (S. constricta) to 1380 bp (S. divaricatus). The length of rrnS varies from 861 bp (S. scaba) to 949 bp (S. olivacea). The rrnS genes of M. iridescens, S. diphos and S. divaricatus position in between trnG and major non-coding region. However, rrnS genes of S. olivacea, S. scaba, and S. constricta lie in between cox2 and trnS1, between trnW and cox2, and between trnM and cox3, respectively. Both lengths of rrnL and rrnS are within the range of genome sizes of already sequenced molluscan mtDNAs.
Non-coding regions
There are a large number of non-coding regions (NCR) including in the six mt genomes each. The number of NCR varies from 16 (S. diphos and S. olivacea) to 25 (S. constricta). The total length of unassignable nucleotides ranges from 1022 bp (6.25% of the genome) in S. diphos to 2730 bp (15.01% of the genome) in S. olivacea (Table 2).
Table 2. A comparison of non-coding regions (NCR) within the six mt genomes.
| Largest NCR | ||||||
| Species | No. of NCR | Total lenth (bp) | Proportion of themt genome (%) | Lenth (bp) | A+T % | Location |
| Moerella iridescens | 23 | 1604 | 9.55 | 1200 | 70.00 | rrnS - trnM |
| Sanguinolaria diphos | 16 | 1022 | 6.25 | 674 | 70.77 | rrnS - trnM |
| Sanguinolaria olivacea | 16 | 2730 | 15.01 | 2272 | 69.67 | trnG - trnI |
| Semele scaba | 23 | 1567 | 9.15 | 1166 | 58.92 | cob - trnG |
| Sinonovacula constricta | 25 | 2134 | 12.39 | 1492 | 66.89 | nad2 - trnK |
| Solecurtus divaricatus | 22 | 1160 | 6.93 | 775 | 65.81 | rrnS - trnM |
Due to lacking discrete conserved sequence blocks, the control regions of invertebrates' mt genomes, unlike those of vertebrates, are not well characterized [44]. In general, the mt genome contains one major non-coding region with some peculiar patterns (e.g. AT-rich, hairpin structures, T-stretch, C-rich,tandem repeats), believed to play a role in initiating and/or regulating mitochondrial transcription and replication [2], [45]–[48]. The largest non-coding region (MNR) of the six mt genomes with all the patterns mentioned above is identified as a putative control region (CR). As highly rearranged gene order in bivalves, the MNR is not conserved at the same location among bivalves [42]. In this study, four different locations (between rrnS and trnM, trnG and trnI, cob and trnG, nad2 and trnK) of MNR occur.
Among the six mt genomes, the MNRs vary in size from 674 bp (S. diphos) to 2272 bp (S. olivacea) (Table 2). Moreover, the A+T content of the putative CR in each mt genome is higher or slightly lower than that of the whole mt genome. There are some sections of nucleotide sequence existed in MNR, all of which can form a typical hairpin-like secondary structures (Figure S1). The conserved flanking sequences around the hairpin structures exhibit conserved motifs: 5′-flanking sequences show a TATA element (except for S. divaricatus) which has also been reported in Crustacea [49], while 3′-flanking sequences possess a TTTAT element in M. iridescens, S. scaba, S. olivacea and S. divaricatus. It is assumed that these structures are of functional importance involving in the origin of the replication of mtDNA [48]. Long T-stretches of 18 bp, 15 bp, 18 bp and 13bp are observed in the MNR of S. diphos, S. olivacea, S. divaricatus and S. constricta, respectively, which may provide essential signals for the replication initiation of mtDNA [46]. In addition, the C-rich sequences, predicted to facilitate formation of the D-loop by decelerating the extension of heavy-strand synthesis at this location in some vertebrates [45], exist in the MNR of M. iridescens, S. scaba and S. olivacea.
Tandem repeats are also detected in MNR of five mt genomes (Figure S2; Figure S3; Figure S4; Figure S5; Figure S6; Figure S7), but that of S. diphos. Especially in MNR of M. iridescens mt genome, three distinct tandem repeat units are found, one of which comprises 14.4 nearly identical copies of a 54 bp unit. Besides, S. olivacea has 2 copies of 98 bp and S. scaba has 2.8 copies of 109 bp. Such large tandem repeat units are also reported in the bivalves Acanthocardia tuberculata [50] and Placopecten magellanicus [51]. Further study on tandem repeats in the control region is needed, as it is important to illuminate the functional implications of the repeats and the molecular mechanisms that generate the repeats [52].
Gene order rearrangements
In contrast with other metazoans, the phylum Mollusca has long been known to exhibit an exceptionally variable arrangement of genes within mitochondrial DNA [53]–[55]. So far, due to coding on one strand probably, all bivalves whose mt genomes have been presented display enormous gene rearrangements (but see oysters [56]). It is suspected that coding on both strands may be inhibitory to mt genome rearrangement, because rearranging a genome with dual-strand coding may be more complicated and cause more harm than that codes on one strand [57].
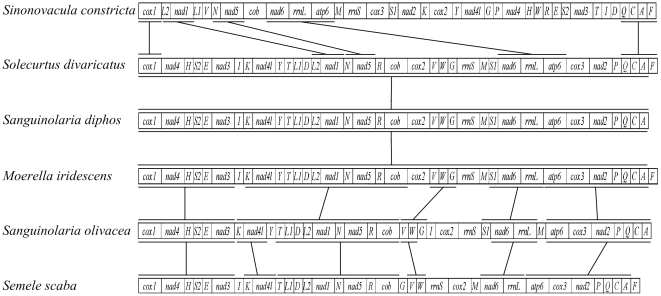
In this study, we compare the gene order rearrangements of six newly sequenced mt genomes (Figure 1). The six heterodont bivalves exhibit five different gene orders, among which M. iridescens and S. divaricatus have the identical gene order. Furthermore, the five patterns of gene arrangement differ from any gene order ever reported in molluscs. The gene order of S. constricta is remarkably unlike that of five other species, even excluding the tRNA, which might indicate the relatively distant relationship as also revealed in the phylogenetic analysis in this study (see below). And five other complete mt genomes differ primarily in the position of tRNA genes, whose secondary structures allow them to translocate more frequently than either rRNAs or protein coding genes [58], [59]. When disregarding tRNA genes, all five species belonging to superfamily Tellinoidea show the same gene arrangements except for translocations of genes rrnS and cox2 in S. scaba. The relatively high level of conservation in the gene order may verify the close lineage relationship. In addition, there are three small blocks, trnK-nad4l, trnV-trnW and nad6-rrnL, and three large blocks, cox1-nad4-trnH-trnS2-trnE-nad3, trnT-trnL1-trnD-trnL2-nad1-trnN-nad5-trnR-cob and atp6-cox3-nad2-trnP-trnQ-trnC-trnA, shared by S. olivacea and S. scaba.
Figure 1. Linear representation of the mitochondrial gene arrangement in six newly sequenced bivalves.
As is the standard convention for metazoan mt genomes, cox1 has been designated the start point for all genomes. All genes are transcribed from left-to-right. The bars indicate identical gene blocks. The non-coding regions are not presented and gene segments are not drawn to scale.
One of our noteworthy finding during this study is that S. diphos and S. olivacea, which belong to the same genus Sanguinolaria, have different gene arrangements with three translocations of trnI, trnV-trnW-trnG and trnM. The case that differences in the gene arrangement occur in the same genus is seldom reported, yet in genus Dendropoma [16] and genus Crassostrea [57]. Besides, M. iridescens, S. divaricatus and S. diphos have a completely identical gene order, if lacking a trnF in S. diphos is ignored. As described above, it is surprising that the gene arrangement of S. diphos is more similar with that of M. iridescens and S. divaricatus than that of S. olivacea, while S. diphos and S. olivacea should have close relatives according to traditional taxonomy. Meanwhile, this result is consistent with the conclusion from the phylogenetic analysis (see below). Early analyses of mtDNA had led to the proposition that a comparative analysis of mt gene order could proved to be a useful phylogenetic tool [11], [60], [61]. By mt gene order comparisons, Smith et al. provided evidence that two echinoderm classes, sea stars and brittle stars, form a monophyletic group to the exclusion of two others, sea cucumber and sea urchins [60]. Akasaki et al. examined the relationships of subclass Coleoidea via comparing extensive mt gene arrangements, and concluded that order Octopoda might be the most ancestral among this subclass Coleoidea in accordance with the phylogenetic tree [61]. In this study, the results obtained here support that comparisons of mt gene order rearrangments, to some extent, are a useful tool for phylogenetic studies.
Phylogenetic analyses of Heterodonta
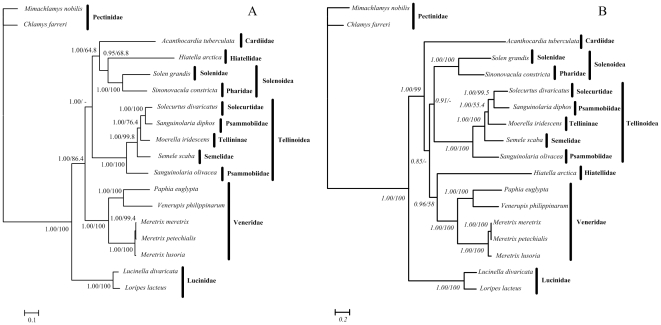
ML and Bayesian trees based on amino acid and nucleotide sequences of 12 concatenated protein-coding genes (except atp8 gene) were performed to reconstruct phylogenetic relationships within heterodont bivalves (Figure 2). The tree topologies based on amino acid and nucleotide sequences were largely congruent and received high supports in most nodes.
Figure 2. Phylogenetic trees of heterodont bivalves based on the concatenated amino acid (A) and nucleotide sequences (B) of 12 protein-coding genes (except atp8 gene).
Numbers in the nodes correspond to Bayesian posterior probabilities (left) and ML bootstrap proportions (right). Dashes indicate support values below 50%.
Using mt genome data, the KH and SH tests were performed to evaluate the alternative morphology-based hypotheses and previous molecular studies. All 7 alternative topologies including 2 topologies presented in our study were summarized in Table S1. The best topology was phylogenetic analysis based on amino acid data shown in Figure 2A (KH and SH tests p = 1.000), while the second best one was phylogenetic analysis based on nucleotide sequences in Figure 2B (KH test p = 0.005 and SH test p = 0.210). All other topologies were significantly rejected (KH and SH P<0.001) which confirmed the phylogenetic analyses based on our data.
Our analysis shows the two species of family Lucinidae form a single clade as a sister group to other heterodont bivalves, indicating that Lucinidae is monophyletic, in accordance with previous viewpoint of Taylor [17]. Five species belonging to family Veneridae, including Paphia euglypta, Venerupis philippinarum, Meritrix meretrix, Meritrix petechialis and Meretrix lusoria cluster together, supporting the monophyly of family Veneridae [62], [63]. In the phylogenetic trees based on the amino acid data, the rock-boring and nestling Hiatella arctica belonging to the order Myoida is as a sister group with Solenidae+Pharidae (superfamily Solenoidea) questioning the subdivision of the subclass Heterodonta into two orders Veneroidea and Myoida, which corroborates the finding of previous molecular analysis [17], [64]. It should be noticed that A. tuberculata evidenced as a member of superfamily Cardioidea based on molecular analysis [17], [50], is placed in a long branch as a sister group to Hiatelloidea+Solenoidea, which is robustly supported by BI but ML based on 12-protein amino acid data (Figure 2A). While in the phylogentic analyses based on nucleotide data, the position of this species is as sister group with the other heterodont bivalves except for Lucinidae (Figure 2B). Both of results are incompatible with a sister relationship of Cardioidea to Tellinoidea, reported by Campbell [23], Steiner and Hammer [65], Dreyer et al. [66], and Taylor et al. [34] based on short fragments of nuclear gene or mt DNA. Therefore, more phylogenetic analyses based on more molecular data, in addition to morphological characters, are required in order to resolve the relationship among the subclass Heterodonta in the future.
The position of genus Sinonovacula has been debated over a long time. Using morphological evidences such as the formation of the siphons and so on, Sinonovacula had previously been suggested by Yonge [67], [68] that it should be removed from the family Solenidae where it was placed by Ghosh [69] and transferred to the Tellinoidea. This decision was accepted and followed by Keen [70], Habe [71] and Vokes [72] who transferred Sinonovacula to Solecurtidae within the Tellinoidea. Subsequently, using shell and anatomical characters, von Cosel [73] retransferred Sinonovacula to Solenoidea. And then, the result of phylogenetic analyses of heterodont bivalves based on rRNA genes by Taylor [17] was in agreement with von Cosel's decision. Our phylogenetic analyses of multiple protein-coding genes not only show that S. constricta is distant from the Solecurtidae belonging to Tellinoidea, but also indicate that S. constricta is as a sister group with Solen grandis of family Solenidae. In other words, we prefer classifying the genus Sinonovacula within the superfamily Solenoidea and not the superfamily Tellinoidea.
All five species of superfamily Tellinoidea from four different families form a clade strongly supported by three trees (except for ML tree based on amino acid sequence), which corroborates the monophyly of Tellinoidea. This result is also reported by Taylor et al. [17]. However, according to the phylogenetic trees in this study, S. diphos has a closer relationship with S. divaricatus, M. iridescens and S. scaba rather than with S. olivacea, highly supported by BI and ML, implying that two species presently classified into Sanguinolaria (Psammobiidae) do not form monophyletic groups. These unexpected findings suggesting that the current taxonomy should be brought into question and a careful review of this genus is required. Similar conclusions that Semelidae, Donacidae and Tellinidae are not monophyletic were ever made when Taylor et al. analyzed familial relationships within Tellinoidea [17]. In fact, to date, there has no special research for the taxonomy of this superfamily based on molecular data. Thereby, in the future study, more detailed analyses with a larger taxon sampling and more rapidly evolving molecular markers including mt genome are still necessary in order to test the taxonomy of superfamily Tellinoidea based on morphology and clarify familial relationships within this superfamily.
Conclusions
In this study, we newly determined the complete mt genomes of six bivalves, increasing the number of complete mt genomes sequenced within subclass Heterodonta from 10 to 16. By comparative study of the gene order rearrangements and phylogenetic relationships of the species belonging to Tellinoidea, our results support that comparisons of mt gene order rearrangements, to some extent, are a useful tool for phylogenetic studies. Based on phylogenetic analyses of multiple protein-coding genes, we prefer classifying the genus Sinonovacula within the superfamily Solenoidea and not the superfamily Tellinoidea. Besides, both gene order and sequence data agree that Sanguinolaria (Psammobiidae) is not monophyletic. Nevertheless, more studies based on more mt genomes via combination of gene order and phylogenetic analysis are needed to further understand the phylogenetic relationships in subclass Heterodonta including superfamily Tellinoidea and Solenoidea.
Methods
Taxon sampling and DNA extraction
In this study, each of the six bivalve complete mt genomes sequenced was obtained from a single specimen. M. iridescens was collected from Leqing (Zhejiang province of China), S. diphos was from Beihai (Guangxi province of China), S. scaba was from Lingao (Hainan province of China), and meanwhile, all three were preserved in EtOH 95% in 2008. S. olivacea was sampled in Rizhao (Shandong province of China) and preserved frozen at −80°C in 2009. S. constricta and S. divaricatus were collected in Qingdao (Shandong province of China) and Rizhao in 2011, respectively.
The total genomic DNA was extracted from adductor muscle by a modification of standard phenol–chloroform procedure that has been described by Li et al. [74] and visualized on 1.0% agarose gel.
PCR amplification and sequencing
In order to design long-PCR primers, we first obtained partial cox1 and rrnL gene sequences, with the universal primers of LCO1490/HCO2198 [75], and 16SF/16SR [76], respectively. Polymerase chain reaction (PCR) was performed in a total volume of 50 µl including 2 U Taq DNA polymerase (Takara), about 100 ng template DNA, 1 µM forward and reverse primers, 200 µM of each dNTP, 1×PCR buffer and 2 mM MgCl2. The PCR reaction was carried out in TaKaRa PCR Thermal Cycler Dice Model TP600 (Takara Bio Inc.) under the following conditions: an initial denaturation for 3 min at 94°C, then 35 cycles of denaturation for 45 s at 94°C, annealing for 45 s at 52°C, extension for 1 min at 72°C, and final extension for 5 min at 72°C.
Subsequently, each mitochondrial genome was amplified by long-PCR technique [77] based on the two specific primer pairs, which were designed from the obtained partial sequences using Primer Premier 5.0 (http://www.premierbiosoft.com/). PCR reactions were carried out in 50 µl reaction mixtures containing 33.5 µl of sterile distilled H2O, 5 µl of 10×LA PCR buffer II (Mg2+ plus, Takara), 8 µl of dNTP (10 mM each), 1 µl of each primer (10 µM), 0.5 µl of LA Taq polymerase (5 U/µl, Takara), and 1 µl of DNA template (50 ng). The long PCR cycling was set up with an initial denaturation step at 94°C for 2 min, followed by 35 cycles comprising denaturatin at 94°C for 20 s, annealing at 60°C for 35 s and extension at 68°C for from 7 to 15 min depending on the expected length of the PCR products. The process was completed with a final extension at 72°C for 10 min. PCR products were purified using EZ-10 spin column DNA gel extraction kit (Sangon Biotech), and then directly sequenced with the primer walking method. The sequencing was conducted on an ABI PRISM 3730 (Applied Biosystems) automatic sequencer.
Sequence analysis and gene annotation
All sequence data were analysed and arranged to create the full genome using the Seqman program from DNASTAR (http://www.DNASTAR.com). Protein coding genes were analysed by ORF Finder [78] using the invertebrate mitochondrial code. The rRNA genes were identified with DOGMA [79] and BLAST searches [80]. The boundaries of each gene were determined with multiple alignments of other published bivalve mitochondrial sequences. The tRNA genes were identified by DOGMA and tRNAscan-SE Search Server [81] with a COVE score cutoff of 1.0 and the invertebrate mitochondrial genetic code for secondary structure prediction. The whole mt genome sequence was tested for potentially tandem repeats by TANDEM REPEAT FINDER, Version 4.0 [82].
Phylogenetic analyses
Eighteen molluscan mt genomes including six newly determined mt genomes, as well as those of all other heterodont bivalves, were used to illustrate the phylogenetic relationship of Heterodonta (Table 3). Chlamys farreri and Mimachlamys nobilis from the subclass Pteriomorphia served as outgroups. Owing to the fact that most bivalve species lack the atp8 gene, amino acid sequences of 12 concatenated protein-coding genes were used in phylogenetic analysis. The alignment of the amino acid sequences of each 12 mitochondrial PCGs was aligned with Clustal X [83] using default settings, followed by manual correction. After areas of dubious alignment were isolated using Gblocks [84] (default settings) and excluded from the analysis, the 12 separate amino acid sequence alignments were concatenated to a single multiple sequence alignment, which consisted of 2026 sites. The nucleotide sequence was substituted from the concatenated amino acid alignment and the final sequence consisted of 7076 sites.
Table 3. List of the species whose mt genome sequences were used in phylogenetic analysis in present paper.
| Species | Classification | Accession Number | Reference |
| Paphia euglypta | Bivalvia; Heteroconchia; Veneroida; Veneroidea; Veneridae | GU269271 | [93] |
| Venerupis philippinarum | Bivalvia; Heteroconchia; Veneroida; Veneroidea; Veneridae | AB065375 | Okazaki et al., unpublished |
| Meretrix meretrix | Bivalvia; Heteroconchia; Veneroida; Veneroidea; Veneridae | GQ463598 | [94] |
| Meretrix lusoria | Bivalvia; Heteroconchia; Veneroida; Veneroidea; Veneridae | GQ903339 | [95] |
| Meretrix petechialis | Bivalvia; Heteroconchia; Veneroida; Veneroidea; Veneridae | EU145977 | [96] |
| Lucinella divaricata | Bivalvia; Heteroconchia; Veneroida; Lucinoidea; Lucinidae | EF043342 | Dreyer et al., unpublished |
| Loripes lacteus | Bivalvia; Heteroconchia; Veneroida; Lucinoidea; Lucinidae | EF043341 | Dreyer et al., unpublished |
| Acanthocardia tuberculata | Bivalvia; Heteroconchia; Veneroida; Cardioidea; Cardiidae | DQ632743 | [50] |
| Hiatella arctica | Bivalvia; Heteroconchia; Myoida; Hiatelloidea; Hiatellidae | DQ632742 | [50] |
| Solen grandis | Bivalvia; Heteroconchia; Veneroida; Solenoidea; Solenidae | HQ703012 | [97] |
| Sinonovacula constricta | Bivalvia; Heteroconchia; Veneroida; Solenoidea; Pharidae | JN398366 | This study |
| Moerella iridescens | Bivalvia; Heteroconchia; Veneroida; Tellinoidea; Tellinidae | JN398362 | This study |
| Sanguinolaria diphos | Bivalvia; Heteroconchia; Veneroida; Tellinoidea; Psammobiidae | JN398363 | This study |
| Sanguinolaria olivacea | Bivalvia; Heteroconchia; Veneroida; Tellinoidea; Psammobiidae | JN398364 | This study |
| Semele scaba | Bivalvia; Heteroconchia; Veneroida; Tellinoidea; Semelidae | JN398365 | This study |
| Solecurtus divaricatus | Bivalvia; Heteroconchia; Veneroida; Tellinoidea; Solecurtidae | JN398367 | This study |
| Chlamys farreri | Bivalvia; Pteriomorphia; Pectinoida; Pectinoidea; Pectinidae | EU715252 | [36] |
| Mimachlamys nobilis | Bivalvia; Pteriomorphia; Pectinoida; Pectinoidea; Pectinidae | FJ415225 | Xu et al., unpublished |
Two methods: Maximum likelihood (ML) and Bayesian inference (BI) were used to reconstruct phylogenetic relationships of heterodont bivalves. For phylogenetic analyses based on amino acid data, MtArt+I+G evolutionary model was chosen as the best-fit model of amino acid evolution by ProtTest version 2.4 [85] at the default setting based on Akaike Information Criterion (AIC). As the MtArt evolutionary model is not available in MrBayes, the WAG model (the second best-fit model according to ProtTest) was used in Bayesian analysis. For phylogenetic analyses based on nucleotide data, the most appropriate model GTR+I+G was selected by MODELTEST [86] using the Akaike information criterion. The ML analysis was conducted with PHYML 3.0 program [87] and 1000 bootstraps were used to estimate the node reliability. BI was performed on combined database using MrBayes 3.1 [88] In the case of the Bayesian analysis, the Markov chain Monte Carlo (MCMC) were run for 5,000,000 generations (sampling every 100 generations) to allow adequate time for convergence. After omitting the first 25,000 “burnin” tree, the remaining 25,000 sampled trees were used to estimate the 50% of majority rule consensus tree and the Bayesian posterior probabilities. All phylogenetic parameters were checked with Tracer v 1.5 [89]. Alternative phylogenetic hypotheses from previous morphological and molecular studies were tested using the Kishino-Hasegawa (KH) test [90] and Shimodaira-Hasegawa (SH) test [91] implemented in CONSEL [92].
Supporting Information
Hairpin-like secondary structures in the mitochondrial putative control regions of M. iridescens , S. scaba , S. divaricatus , S. diphos , S. olivacea and S. constricta . Conserved motifs in 5′- and 3′-flanking sequences are underlined.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Moerella iridescens A. Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Moerella iridescens B. Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Sanguinolaria olivacea . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Semele scaba . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Sinonovacula constricta . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Solecurtus divaricatus . Symbol “-” indicates an insertion or deletion.
(DOC)
Tests of alternative topologies.
(DOC)
Acknowledgments
We are grateful to Jun Chen for providing the species of Moerella iridescens, Sanguinolaria diphos, Semele scaba. We also thank Xiaodong Zheng and Yaosen Qian for collecting Sanguinolaria olivacea, Solecurtus divaricatus.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the grants from the 973 Program (2010CB126406) and National Natural Science Foundation of China (31072207). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 3.Bridge D, Cunningham CW, Schierwater B, Desalle R, Buss LW. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci. 1992;89:8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 5.Doucet-Beaupre H, Breton S, Chapman EG, Blier PU, Bogan AE, et al. Mitochondrial phylogenomics of the bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10:50. doi: 10.1186/1471-2148-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert P, Cywinska A, Ball S, Waard J. Biological identification through DNA-barcodes. Proc R Soc Lond Ser B. 2002;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moritz C, Brown W. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci. 1987;84:7183–7187. doi: 10.1073/pnas.84.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curole JP, Kocher TD. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. 1999;14:394–398. doi: 10.1016/s0169-5347(99)01660-2. [DOI] [PubMed] [Google Scholar]
- 9.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 10.Boore JL, Medina M, Rosenberg LA. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol Biol Evol. 2004;21:1492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- 11.Boore JL, Brown WM. Big tree from little genomes: mitochondrial gene order as a phylogenetic tool. Cuur Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- 12.Zardoya R, Meyer A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol. 1996;13:933–942. doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
- 13.Russo CA, Takezaki N, Nei M. Efficiencies of different tree-building methods in recovering a known vertebrate phylogeny. Mol Biol Evol. 1996;13:525–536. doi: 10.1093/oxfordjournals.molbev.a025613. [DOI] [PubMed] [Google Scholar]
- 14.Mueller RL. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Sys Biol. 2006;55:289–300. doi: 10.1080/10635150500541672. [DOI] [PubMed] [Google Scholar]
- 15.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2001;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 16.Rawlings TA, Maclnnis MJ, Bieler R, Boore JL, Collins TM. Sessile snails, dynamic genomes: gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BMC Genomics. 2010;11:440. doi: 10.1186/1471-2164-11-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor JD, Williams ST, Glover EA, Dyal P. Molecular phylogeny of heterodont bivalves (Mollusca: Bivalvia: Heterodonta): new analyses of 18 S and 28 S rRNA genes. Zool Scr. 2007;36:587–606. [Google Scholar]
- 18.Prezant RS. Beesley PL, Ross GJB, Wells A, editors. Heterodonta: Introduction. Mollusca: The southern synthesis. Fauna of Australia. 1998. pp. 289–294. Melbourne, CSIRO Publishing.
- 19.Lewis TL, Esler D, Boyd WS. Effects of predation by sea ducks on clam abundance in soft-bottom intertidal habitats. Mar Ecol Prog Ser. 2007;329:131–144. [Google Scholar]
- 20.Cope JCW. The early phylogeny of the Class Bivalvia. Palaeontology. 1997;40:713–746. [Google Scholar]
- 21.Crame JA. Evolution of taxonomic diversity gradients in the marine realm: evidence from the composition of recent bivalve faunas. Paleobiology. 2000;26:188–214. [Google Scholar]
- 22.Crame JA. Evolution of taxonomic diversity gradients in the marine realm: a comparison of Late Jurassic and recent bivalve faunas. Paleobiology. 2002;28:184–207. [Google Scholar]
- 23.Campbell DC. Harper EM, Taylor JD, Crame JA, editors. Molecular evidence on the evolution of the Bivalvia. The evolutionary biology of the Bivalvia. 2000. pp. 31–46. London, Geological Society.
- 24.Giribet G, Distel DL. Lydeard C, Lindberg D, editors. Bivalve phylogeny and molecular data. Molecular systematics and phylogeny of Mollusks. 2003. pp. 45–90. Washigton DC, Smithsonian Institution Press.
- 25.Williams ST, Taylor JD, Glover EA. Molecular phylogeny of the Lucinoidea (Bivalvia): non-monophyly and separate acquisition of bacterial chemosymbiosis. J Mollus Stud. 2004;70:187–202. [Google Scholar]
- 26.Bieler R, Mikkelsen PM. Bivalvia-a look at the branches. Zool J Linn Soc. 2006;148:223–235. [Google Scholar]
- 27.Trueman ER. The cardinal ligament of the Tellinacea. J Mollusc Stud. 1966;37:111–117. [Google Scholar]
- 28.Taylor JD. The structure evolution of the bivalve Mollusca. Paleontology. 1973;16:519–534. [Google Scholar]
- 29.Pohlo R. Evolution of the Tellinacea. J Mollusc Stud. 1982;48:245–256. [Google Scholar]
- 30.Simone LRL, Wilkinson S. Comparative morphological study of some Tellinidae from Thailand (Bivalvia: Tellinoidea). Raffles B Zool. 2008;18:151–190. [Google Scholar]
- 31.Kappner I, Bieler R. Phylogeny of venus clams (Bivalvia: Venerinae) as inferred from nuclear and mitochondrial gene sequences. Mol Phylogenet Evol. 2006;40:317–331. doi: 10.1016/j.ympev.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Mikkelsen PM, Bieler R, Kappner I, Rawlings TA. Phylogeny of Veneroidea (Mollusca: Bivalvia: Heterodonta) based on morphology and molecules. Zool J Linn Soc. 2006;2006, 148:439–521. [Google Scholar]
- 33.Lee T, Ó Foighil D. Phylogenetic structure of the Sphaeriinae, a global clade of freshwater bivalve molluscs, inferred from nuclear (ITS-1) and mitochondrial (16 S) ribosomal gene sequences. Zool J Linn Soc. 2003;137:245–260. [Google Scholar]
- 34.Taylor JD, Williams ST, Glover EA. Evolutionary relationships of the bivalve family Thyasiridae (Mollusca; Bivalvia), monophyly and superfamily status. J Mar Biol Assoc UK. 2007;87:565–574. [Google Scholar]
- 35.Yu ZN, Wei ZP, Kong XY, Shi W. Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis - a case of “Tandem duplication-random loss” for genome rearrangement in Crassostrea? BMC Genomics. 2008;9:477. doi: 10.1186/1471-2164-9-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu KF, Kanno M, Yu H, Li Q, Kijima A. Complete mitochondrial DNA sequence and phylogenetic analysis of Zhikong scallop Chlamys farreri (Bivalvia: Pectinidae). Mol Biol Rep. 2010;38:3067–3074. doi: 10.1007/s11033-010-9974-8. [DOI] [PubMed] [Google Scholar]
- 37.Uda K, Komeda Y, Koyama H, Koga K, Fujita T, et al. Complete mitochondrial genomes of two Japanese precious corals, Paracorallium japonicum and Corallium konojoi (Cnidaria, Octocorallia, Coralliidae): Notable differences in gene arrangement. Gene. 2011;476:27–37. doi: 10.1016/j.gene.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Grande C, Templado J, Zardoya R. Evolution of gastropod mitochondrial genome arrangements. BMC Evol Biol. 2008;8:61. doi: 10.1186/1471-2148-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ojala D, Montoya J, Attardi G. TRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 40.Podsiadlowski L, Braband A, Mayer G. The complete mitochondrial genome of the onychophoran Epiperipatus biolleyi reveals a unique transfer RNA set and provides further support for the ecdysozoa hypothesis. Mol Biol Evol. 2008;25:42–51. doi: 10.1093/molbev/msm223. [DOI] [PubMed] [Google Scholar]
- 41.Schneider A, Marechal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 42.Ren JF, Shen X, Jiang F, Liu B. The mitochondrial genomes of two scallops, Argopecten irradians and Chlamys farreri (Mollusca: Bivalvia): the most highly rearranged gene order in the family Pectinidae. J Mol Evol. 2010;70:57–68. doi: 10.1007/s00239-009-9308-4. [DOI] [PubMed] [Google Scholar]
- 43.Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ. Mitochondrial DNA sequence and gene organization in the Australian Blacklip Abalone Haliotis rubra (Leach). Mar Biotechnol. 2005;7:645–658. doi: 10.1007/s10126-005-0013-z. [DOI] [PubMed] [Google Scholar]
- 44.Serb JM, Lydeard C. Complete mtDNA Sequence of the north American freshwater mussel, Lampsilis ornata (Unionidae): An Examination of the Evolution and Phylogenetic Utility of Mitochondrial Genome Organization in Bivalvia (Mollusca). Mol Biol Evol. 2003;20:1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- 45.Kumazawa Y, Ota H, Nishida M, Ozawa T. The complete nucleotide sequence of a snake (Dinodon semicarinatus) mitochondrial genome with two identical control regions. Genetics. 1998;150:313–329. doi: 10.1093/genetics/150.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito S, Tamura K, Aotsuka T. Replication origin of mitochondrial DNA in insects. Genetics. 2005;171:1695–1705. doi: 10.1534/genetics.105.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faber EJ, Stepien AC. Tandemly repeated sequences in the mitochondrial DNA control region and phylogeography of the pike-perches Stizostedion. Mol Phylogenet Evol. 1998;10:310–322. doi: 10.1006/mpev.1998.0530. [DOI] [PubMed] [Google Scholar]
- 48.Zhang DX, Szymura JM, Hewitt GM. Evolution and structural conservation of the control region of insect mitochondrial DNA. J Mol Evol. 1995;40:382–391. doi: 10.1007/BF00164024. [DOI] [PubMed] [Google Scholar]
- 49.Kilpert F, Podsiadlowski L. The complete mitochondrial genome of the common sea slater, Lagia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics. 2006;7:241. doi: 10.1186/1471-2164-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreyer H, Steiner G. The complete sequences and gene organization of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica – and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 2006;3:13. doi: 10.1186/1742-9994-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.La Roche J, Snyder M, Cook DI, Fuller K, Zouros E. Molecular characterization of a repeat element causing large-scale size variation in the mitochondrial DNA of the sea scallop Placopecten magellanicus. Mol Biol Evol. 1990;7:45–64. doi: 10.1093/oxfordjournals.molbev.a040586. [DOI] [PubMed] [Google Scholar]
- 52.Mundy NI, Winchell CS, Woodruff DS. Tandem Repeats and Heteroplasmy in the Mitochondrial DNA Control Region of the Loggerhead Shrike (Lanius ludovicianus). J Hered. 1996;87:21–26. doi: 10.1093/oxfordjournals.jhered.a022948. [DOI] [PubMed] [Google Scholar]
- 53.Boore JL, Brown WM. Mitochondrial genomes and the phylogeny of mollusks. Nautilus. 1994;108:61–78. [Google Scholar]
- 54.Boore JL, Daehler LL, Brown WM. Complete sequence, gene arrangement, and genetic code of mitochondrial DNA of the cephalochordate Branchiostoma floridae (Amphioxus). Mol Biol Evol. 1999;16:410–418. doi: 10.1093/oxfordjournals.molbev.a026122. [DOI] [PubMed] [Google Scholar]
- 55.Kurabayashi A, Ueshima R. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: systematic implication the genome organization. Mol Biol Evol. 2000;17:266–277. doi: 10.1093/oxfordjournals.molbev.a026306. [DOI] [PubMed] [Google Scholar]
- 56.Wu XY, Xu XD, Yu ZN, Wei ZP, Xia JJ. Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol Phylogenet Evol. 2010;57:448–454. doi: 10.1016/j.ympev.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 57.Ren JF, Liu X, Jiang F, Guo XM, Liu B. Unusual conservation of mitochondrial gene order in Crassostrea oyster: evidence for recent speciation in Asia. BMC Evol Biol. 2010;10:394. doi: 10.1186/1471-2148-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boore JL, Brown WM. The complete DNA sequence of the mitochondrial genome of the black chiton Katharina tunicate. Genetics. 1994;138:423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cantatore P, Gadaleta MN, Roberti M, Saccone C, Wilson AC. Duplication and remolding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature. 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- 60.Smith MJ, Arndt A, Gorski S, Fajber E. The phylogeny of echinoderm classes based on mitochondrial gene arrangements. J Mol Evol. 1993;36:545–554. doi: 10.1007/BF00556359. [DOI] [PubMed] [Google Scholar]
- 61.Akasaki T, Nikaido M, Tsuchiya K, Segawa S, Hasegawa M, et al. Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Mol Phylogenet Evol. 2006;38:648–658. doi: 10.1016/j.ympev.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 62.Mikkelsen PM, Bieler R, Kappner I, Rawlings TA. Phylogeny of Veneroidea (Mollusca: Bivalvia: Heterodonta) based on morphology and molecules. Zool J Linn Soc. 2006;148:439–521. [Google Scholar]
- 63.Giribet G, Wheeler W. On bivalve phylogeny: a high-level analysis of the Bivalvia (Mollusca) based on combined morphology and DNA sequence data. Invertebr Biol. 2002;121:271–324. [Google Scholar]
- 64.Canapa A, Barucca M, Marinelli A, Olmo E. A molecular phylogeny of Heterodonta (Bivalvia) based on small ribosomal subunit RNA sequences. Mol Phylogenet Evol. 2001;21:156–161. doi: 10.1006/mpev.2001.0992. [DOI] [PubMed] [Google Scholar]
- 65.Steiner G, Hammer S. Harper EM, Taylor JD, Crame JA, editors. Molecular phylogeny of the Bivalvia inferred from 18 S rDNA sequences with particular reference to the Pteriomorphia. The evolutionary biology of the Bivalvia. 2000. pp. 11–29. London, Geological Society.
- 66.Dreyer H, Steiner G, Harper EM. Molecular phylogeny of Anomalodesmata (Mollusca: Bivalvia) inferred from 18 S rRNA sequences. Zool J Linn Soc. 2003;139:229–246. [Google Scholar]
- 67.Yonge CM. On the structure and adaptations of the Tellinacea, deposit-feeding Eulamellibranchia. Phil Trans B. 1949;234:29–76. [Google Scholar]
- 68.Yonge CM. On the structure, biology and systematic position of Pharus legumen (L.). J Mar Biol Assoc UK. 1959;38:277–290. [Google Scholar]
- 69.Ghosh E. Taxonomic studies on the soft parts of the Solenidae. Rec Indian Mus. 1920;19:47–78. [Google Scholar]
- 70.Keen AM. Moore RC, editor. Superfamily Tellinacea. Treatise on invertebrate paleontology. 1969. Kansas, The Geological Society of America and the University of Kansas Press.
- 71.Habe T. Systematics of Mollusca in Japan: Bivalvia and Scaphopoda. 1977. Tokyo, Zukan-no-Hokuryukan Press.
- 72.Vokes HE. Genera of the Bivalvia: a systematic and bibliographic catalogue. 1980. New York, Paleontological Research Institution Press.
- 73.von Cosel R. Morton B, editor. An introduction to the razor shells (Bivalvia: Solenacea). The Bivalvia - Proceedings of a Memorial Symposium in Honour of Sir Charles Maurice Yonge, Edinburgh, 1986. 1990. Hong Kong, Hong Kong University Press.
- 74.Li Q, Park C, Kijima A. Isolation and characterization of microsatellite loci in the Pacific abalone, Haliotis discus hannai. J Shellfish Res. 2002;21:811–815. [Google Scholar]
- 75.Folmer O, Black M, Hoeh R, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 76.Simon C, Frati F, Beckenback A, Crespi B, Liu H, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primers. Annals Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 77.Cheng S, Chang SY, Gravitt P, Respress R. Long PCR. Nature. 1994;369:684–685. doi: 10.1038/369684a0. [DOI] [PubMed] [Google Scholar]
- 78. NCBI Open Reading Frame Finder (NCBI website. Available: http://www.ncbi.nlm.nih.gov/gorf/gorf.html. Accessed 2011 Oct 1.)
- 79.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 80. The Basic Local Alignment Search Tool (BLAST) (NCBI website. Available: http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 2011 Oct 1.)
- 81.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 85.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 86.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 87.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 88.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 89.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Honinoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 91.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 92.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 93.Xu XD, Wu XY, Yu ZN. The mitogenome of Paphia euglypta (Bivalvia: Veneridae) and comparative mitogenomic analyses of three venerids. Genome. 2010;53:1041–1052. doi: 10.1139/G10-096. [DOI] [PubMed] [Google Scholar]
- 94.He CB, Wang J, Gao XG, Song WT, Li HJ, et al. The complete mitochondrial genome of the hard clam Meretrix meretrix. Mol Biol Rep. 2011;38:3401–3409. doi: 10.1007/s11033-010-0449-8. [DOI] [PubMed] [Google Scholar]
- 95.Wang HX, Zhang SP, Li Y, Liu BZ. Complete mtDNA of Meretrix lusoria (Bivalvia: Veneridae) reveals the presence of an atp8 gene, length variation and heteroplasmy in the control region. Comp Biochem Physiol D. 2010;5:256–264. doi: 10.1016/j.cbd.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 96.Ren JF, Shen X, Sun M, Jiang F, Yu Y, et al. The complete mitochondrial genome of the clam Meretrix petechialis (Mollusca: Bivalvia: Veneridae). Mitochondr DNA. 2009;20:78–87. doi: 10.1080/19401730902964425. [DOI] [PubMed] [Google Scholar]
- 97.Yuan Y, Li Q, Kong LF, Yu H. The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): a novel gene order and unusual non-coding region. Mol Biol Rep. 2012;39:1287–1292. doi: 10.1007/s11033-011-0861-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hairpin-like secondary structures in the mitochondrial putative control regions of M. iridescens , S. scaba , S. divaricatus , S. diphos , S. olivacea and S. constricta . Conserved motifs in 5′- and 3′-flanking sequences are underlined.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Moerella iridescens A. Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Moerella iridescens B. Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Sanguinolaria olivacea . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Semele scaba . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Sinonovacula constricta . Symbol “-” indicates an insertion or deletion.
(DOC)
Alignment of the tandem repeats in the largest non-coding region (MNR) of mitochondrial genome of Solecurtus divaricatus . Symbol “-” indicates an insertion or deletion.
(DOC)
Tests of alternative topologies.
(DOC)