Abstract
A DNA fragment encoding Ribonuclease H (EC 3. 1.26.4) was isolated from an extreme thermophilic bacterium, Thermus thermophilus HB8, by its ability to complement the temperature-sensitive growth of an Escherichia coli rnhA deficient mutant. The primary amino acid sequence showed 56% similarity to that of E. coli RNase HI but little or no homology to E. coli RNase HII. Enzymes derived from thermophilic organisms tend to have fewer cysteines than their bacterial counterparts. However, T. thermophilus RNase H has one more cysteine than its E. coli homologue. Stability of the RNase H in extracts of T. thermophilus to elevated temperatures was the same for the protein expressed in E. coli. T. thermophilus RNase H should, therefore, be a useful tool for editing RNA-DNA hybrid molecules at higher temperatures and may also be stable enough to be used in a cyclical process. It was suggested that regulation of expression of the RNase H may be different from that of E. coli. RNase HI.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bockrath R., Wolff L., Farr A., Crouch R. J. Amplified RNase H activity in Escherichia coli B/r increases sensitivity to ultraviolet radiation. Genetics. 1987 Jan;115(1):33–40. doi: 10.1093/genetics/115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. DNA sequence and coding properties of mutD(dnaQ) a dominant Escherichia coli mutator gene. J Mol Biol. 1986 Jul 5;190(1):113–117. doi: 10.1016/0022-2836(86)90080-x. [DOI] [PubMed] [Google Scholar]
- Dasgupta S., Masukata H., Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987 Dec 24;51(6):1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Foster P. L., Sullivan A. D., Franklin S. B. Presence of the dnaQ-rnh divergent transcriptional unit on a multicopy plasmid inhibits induced mutagenesis in Escherichia coli. J Bacteriol. 1989 Jun;171(6):3144–3151. doi: 10.1128/jb.171.6.3144-3151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C. O., Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990 Jun 26;29(25):5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Mellert W., Moelling K. Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J. 1988 Jan;7(1):239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase Y., Inoue H., Ohtsuka E. Secondary structure in formylmethionine tRNA influences the site-directed cleavage of ribonuclease H using chimeric 2'-O-methyl oligodeoxyribonucleotides. Biochemistry. 1990 Sep 18;29(37):8793–8797. doi: 10.1021/bi00489a041. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hollingsworth H. C., Nossal N. G. Bacteriophage T4 encodes an RNase H which removes RNA primers made by the T4 DNA replication system in vitro. J Biol Chem. 1991 Jan 25;266(3):1888–1897. [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. RNase H-defective mutants of Escherichia coli: a possible discriminatory role of RNase H in initiation of DNA replication. Mol Gen Genet. 1984;195(1-2):17–22. doi: 10.1007/BF00332717. [DOI] [PubMed] [Google Scholar]
- Itaya M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Crouch R. J. DNA sequence of the gene coding for Escherichia coli ribonuclease H. J Biol Chem. 1983 Jan 25;258(2):1276–1281. [PubMed] [Google Scholar]
- Kanaya S., Kimura S., Katsuda C., Ikehara M. Role of cysteine residues in ribonuclease H from Escherichia coli. Site-directed mutagenesis and chemical modification. Biochem J. 1990 Oct 1;271(1):59–66. doi: 10.1042/bj2710059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miura Y., Sekiguchi A., Iwai S., Inoue H., Ohtsuka E., Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990 Mar 15;265(8):4615–4621. [PubMed] [Google Scholar]
- Kane C. M. Renaturase and ribonuclease H: a novel mechanism that influences transcript displacement by RNA polymerase II in vitro. Biochemistry. 1988 May 3;27(9):3187–3196. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
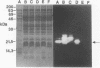
- Keller W., Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama Y., Furukawa K. Cloning and sequence analysis of tryptophan synthetase genes of an extreme thermophile, Thermus thermophilus HB27: plasmid transfer from replica-plated Escherichia coli recombinant colonies to competent T. thermophilus cells. J Bacteriol. 1990 Jun;172(6):3490–3495. doi: 10.1128/jb.172.6.3490-3495.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M. S., Berger S. L. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc Natl Acad Sci U S A. 1989 May;86(10):3539–3543. doi: 10.1073/pnas.86.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G., Crouch R. J., Post K., Hu S. C., McKelvin D., Zweig M., Court D. L., Gerwin B. I. Functional organization of the murine leukemia virus reverse transcriptase: characterization of a bacterially expressed AKR DNA polymerase deficient in RNase H activity. J Virol. 1988 Nov;62(11):4376–4380. doi: 10.1128/jvi.62.11.4376-4380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Horiuchi T., Sekiguchi M. Structure and expression of the dnaQ mutator and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7137–7141. doi: 10.1073/pnas.80.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193(2):231–237. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pickett G. G., Kogoma T., Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T., Imahori K. Physiochemical properties of deoxyribonucleic acid from an extreme thermophile. J Biochem. 1974 Jan;75(1):179–183. doi: 10.1093/oxfordjournals.jbchem.a130372. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Quiñones A., Kücherer C., Piechocki R., Messer W. Reduced transcription of the rnh gene in Escherichia coli mutants expressing the SOS regulon constitutively. Mol Gen Genet. 1987 Jan;206(1):95–100. doi: 10.1007/BF00326542. [DOI] [PubMed] [Google Scholar]
- Rong Y. W., Carl P. L. On the molecular weight and subunit composition of calf thymus ribonuclease H1. Biochemistry. 1990 Jan 16;29(2):383–389. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawano N., Oshima T. Cloning of 3-isopropylmalate dehydrogenase gene of an extreme thermophile and partial purification of the gene product. J Biochem. 1981 Feb;89(2):677–682. doi: 10.1093/oxfordjournals.jbchem.a133245. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonwirth H., Frank P., Kedinger C., Büsen W. Class II ribonuclease H comigrates with, but is distinct from, the third largest subunit of calf thymus RNA polymerase I. Biochim Biophys Acta. 1990 Sep 10;1087(1):31–38. doi: 10.1016/0167-4781(90)90117-k. [DOI] [PubMed] [Google Scholar]
- Wintersberger U., Kühne C., Karwan R. Three ribonucleases H and a reverse transcriptase from the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 1988 Dec 20;951(2-3):322–329. doi: 10.1016/0167-4781(88)90102-9. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]