Abstract
Vibrio vulnificus, the cause of septicemia and serious wound infection in humans and fishes, require iron for its pathogenesis. Hemin uptake through the outer membrane receptor, HupA, is one of its many mechanisms by which it acquires iron. We report here the identification of an additional TonB-dependent hemin receptor HvtA, that is needed in conjunction with the HupA protein for optimal hemin utilization. The HvtA protein is significantly homologous to other outer membrane hemin receptors and its expression in trans restored the uptake of hemin and hemoglobin, the latter to a weaker extent, in a mutant strain that was defective in both receptors. Quantitative RT-PCR suggested that transcription of the hvtA gene was iron regulated. The operon containing the hvtA gene is homologous to the operon in V. cholerae containing the hemin receptor gene hutR suggesting a vertical transmission of the hvtA cluster from V. cholerae to V. vulnificus.
Keywords: Iron, Hemin, Hemoglobin, Outer membrane protein
Introduction
Vibrio vulnificus is a Gram negative halophilic pathogen that is a causative agent of diseases like gastroenteritis, life-threatening septicemia in humans and eels resulting in rapid mortality rate (Amaro et al. 1995; Gulig et al. 2005). Infection can be acquired either through consumption of raw seafood or with the exposure of open wounds to aquatic environments that contain the pathogen. It is also known to be opportunistic on certain immunocompromised individuals and those with health conditions such as liver cirrhosis, hemochromatosis, and beta thalassaemia (Hlady and Klontz 1996; Morris 1988; Strom and Paranjpye 2000).
Iron, which is not readily available in the environment or in the normal vertebrate host, has been shown to be an absolute requirement for this bacterium for its pathogenesis (Wright et al. 1981). Thus, V. vulnificus has developed several mechanisms of iron sequestration either (i) through the biosynthesis and secretion of small molecular weight compounds like the catechol vulnibactin and an as yet to be characterized hydroxamate-type siderophore, that are known to have high affinity for iron and are used to scavenge and transport iron from the environment inside the cell cytosol (Litwin et al. 1996; Okujo et al. 1994; Simpson and Oliver 1983), or (ii) by making use of hemin or hemin-containing proteins from the host. The latter mechanism includes the transport of hemin via specific outer membrane receptors in a TonB-dependent manner either by acquiring hemin from hemoproteins or from hemoglobin with the aid of proteases (Litwin and Byrne 1998; Nishina et al. 1992; Wandersman and Delepelaire 2004). Many other virulence factors like the RTX (repeats in toxin) toxin (Lee et al. 2007; Liu et al. 2007; Liu et al. 2009), capsular polysaccharides (Smith and Siebeling 2003; Wright et al. 1999), flagella (Lee et al. 2004; Kim and Rhee 2003), protease (Kothary and Kreger 1987), and iron-regulated genes (Alice et al. 2008) have also been reported to play important roles in its pathogenesis.
Several aspects of iron utilization in V. vulnificus have been studied. Litwin and co-workers (Litwin and Byrne 1998) demonstrated that the outer membrane protein, HupA is important for hemin uptake. Expression of the hupA gene that encodes the receptor protein is regulated at the transcriptional level by the iron-binding regulatory protein, Fur and a LysR homologue, HupR (Litwin and Quackenbush 2001). It was recently reported that expression of the hupA gene in addition to be iron-regulated, is also up-regulated at higher temperatures (Oh et al. 2009). Webster et al. (Webster and Litwin 2000) demonstrated that another outer membrane protein, VuuA, is responsible for the uptake of its main siderophore, vulnibactin and is also regulated by iron levels with the aid of Fur. Mutations in both receptors have been associated to decreased virulence in animal models, which further confirmed the importance of iron uptake in the pathogenesis of V. vulnificus infections. In this work, we report the existence in V. vulnificus of an additional iron regulated TonB-dependent hemin receptor, HvtA, which is a homologue of the V. cholera hemin receptor HutR and describe some of its properties.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Strains and plasmids used in this study are listed in Table 1. Bacteria were routinely grown in Trypticase soy broth supplemented with 1% NaCl (TSBS) or on TSAS agar (V. vulnificus), or on LB broth (E. coli) with appropriate antibiotics: chloramphenicol (2 and 10 µg/ml) for V. vulnificus and kanamycin (50 µg/ml), and chloramphenicol (30 µg/ml) for E. coli. M9 minimal medium (Crosa 1980) was used for iron-limiting conditions supplemented with 0.2% casaminoacids and 5% NaCl with the iron chelator ethylenediamine-di-(O-hydroxyphenylacetic) acid (EDDA) at indicated concentrations. Ferric ammonium citrate was added to the medium to obtain iron-rich growth conditions. Thiosulfate-citrate-bile-salts-sucrose agar (TCBS) (Preiser Scientific, Louisville, KY) was used for selection of V. vulnificus in conjugation and complementation experiments.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Phenotype | Source of reference |
|---|---|---|
| V. Vulnificus strains | ||
| CMCP6 | Wild type clinical isolate | Kim and Rhee (2003) |
| AA-16 | ΔvenB, ΔtonB1, ΔtonB2, ΔtonB3 | Alice et al. (2008) |
| AA-14 | ΔvenB | Alice et al. (2008) |
| VSSD5 | ΔvenB, ΔhvtR | This study |
| VSSD25 | ΔvenB, ΔhupA | This study |
| VSSD40 | ΔvenB, ΔhvtR, ΔhupA | This study |
| VSSD91 | VSSD40 harboring pMMB208-hvtR | This study |
| VSSD74 | ΔhvtR, ΔhupA | This study |
| VSSD61 | ΔvenB, ΔtonB1 | This study |
| VSSD62 | ΔvenB, ΔtonB2 | This study |
| VSSD53 | ΔvenB, ΔttpC2 | This study |
| VSSD57 | ΔvenB, ΔtonB1, ΔtonB2 | This study |
| VSSD64 | ΔvenB, ΔtonB1, ΔttpC2 | This study |
| VSSD92 | VSSD74 harboring pMMB208 | This study |
| VSSD93 | VSSD74 harboring pMMB208-hvtR | This study |
| E. Coli strains | ||
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ 80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S17-1λpir | Λpir lysogen; thi pro hsdR hsdM+ recA RP4-2 Tc:Mu-Km:Tn7(Tpr Smr) | Simon et al. (1983) |
| Plasmids | ||
| PCR II Blunt | Blunt end cloning vector: Kmr | Invitrogen |
| pPCR2.1 | TA cloning vector; Ampr, Kmr | Invitrogen |
| pDM4 | Suicide vector with oriR6K; CmrsacB | Milton et al. (1996) |
| pRK2013 | Helper plasmid; Kmr | Figurski and Helinski (1979) |
| pMMB208 | Broad-host-range expression vector; Cmr Ptac | Morales et al. (1991) |
| phupA | pDM4-ΔhupA | Alice et al. (2008) |
DNA manipulation and sequence analysis
Plasmid DNA was extracted using the Qiagen miniprep kit (Qiagen, Valencia, CA). Genomic DNA was isolated from the V. vulnificus CMCP6 strain using the DNeasy® Blood and Tissue kit (Qiagen, Valencia, CA). PCR reactions were carried out using a Mycycler ®Thermal Cycler as specified by the manufacturer (Bio-Rad Laboratories, Hercules, CA). Touchdown PCRs were performed using the Vent Polymerase (New England Biolabs Inc., Ipswich, MA), using the following conditions: 95°C for 4 min, 30 cycles of 95°C for 30 s, 63°C for 1 min (temperature of this step was decreased by 0.3°C at each cycle), and 72°C for 1 min, and an extension step of 72°C for 10 min. DNA sequencing reactions were carried out by the Oregon Health and Science University Vollum DNA Sequencing Core Facility.
Construction of chromosomal mutants
A hvtA (VV21549) deletion mutant in V. vulnificus was generated by allelic exchange using the pDM4 suicide plasmid (Milton et al. 1996). Upstream and downstream regions (700–800 bp) flanking the gene were amplified by specific primers, VV21549-1fwd and VV21549-3rev and VV21549-4fwd and VV21549-2rev, respectively (Table 2), and combined using Splicing by Overlapping Extension (SOE) PCR by using primers VV21549-1fwd and VV21549-2rev. The generated PCR product was cloned into the blunt PCR2.1 vector (Invitrogen, Carlsbad, CA), digested with ApaI and SpeI, and subcloned into the suicide vector pDM4 also linearized with the same restriction enzymes. The resulting pDM4 derivative was conjugated into V. vulnificus according to the procedure previously reported (Alice et al. 2008). For complementation, the hvtA gene was amplified by PCR with primers HvtA250compfwd and HvtA250comprev (Table 2), containing BamHI and EcoRI restriction sites, respectively. The PCR product was cloned into the pPCR2.1 vector (Invitrogen, Carlsbad, CA), and subsequently subcloned into the pMMB208 vector (Morales et al. 1991) linearized with BamHI/EcoRI restriction enzymes. The generated construct was transferred to the V. vulnificus VSSD40 strain (ΔvenBΔhupAΔhvtA) via triparental mating using the helper plasmid pRK2013 (Figurski and Helinski 1979). The cloned gene in pMMB208, which is under the control of the Ptac promoter, was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for bioassay experiments in the solid medium.
Table 2.
Primers used in this study
| Primers name | Nucleotide sequence (5′–3′) |
|---|---|
| Construction of mutants | |
| VV21549-1fwd | GTTCCAGTCACGCTGGCGTAC |
| VV21549-2rev | CTGCAGCGCTTGCAGATCCGC |
| VV21549-3rev | GAATTGGTATTTCACGTTGGCCGCTGAGACAGGCGTTAGTTTCAT |
| VV21549-4fwd | ATGAAACTAACGCCTGTCTCAGCGGCCAACGTGAAATACCAATTC |
| 1549fwd | CGAATTTCTCGACCAGCTTGTG |
| 1549rev | GGCTTTGAGATCCATATTAGGA |
| Complementation | |
| HvtA250compfwd | GGATCCCCTATCTCAACCTTCAATCGG (underline, BamHI site) |
| HvtA250comprev | GAATTCCATCAAAGTTATAGTGATGGCC (underline, EcoRI site) |
| qRT-PCR | |
| HupAForqRTPCR | TGATTCAGCATTCACAGGTCG |
| HupARevqRTPCR | GTTAGTGTAACCATGTCCCGG |
| HvtRForqRTPCR | TGATTCGTTTGAGGTAGGGC |
| HvtRRevqRTPCR | CCGTTGAGGTTTTGGTATTGC |
Hemin and hemoglobin utilization assays
Growth stimulation assay was performed to detect the utilization of hemin and hemoglobin by the V. vulnificus strains. Overnight cultures (50 µl) of the bacterial strains were mixed with 20 ml of CM9 media containing agar supplemented with 20 µM EDDA and poured onto petri dishes. After solidification, different iron sources, Ferric Ammonium Citrate (FAC) (Sigma Aldrich, MO), Hemin (Sigma Aldrich, MO) and Hemoglobin (Sigma Aldrich, MO) were spotted on the plate and incubated at 37°C, and appearance of the growth halo was monitored after 12–16 h. For growth analysis, hemin was used as the sole iron source to test its utilization by the V. vulnificus strains. Briefly, overnight cultures of the bacterial strains were inoculated (inoculation ratio 1:200) in iron-depleted CM9 minimal media containing 5 µM EDDA supplemented with 10 µMhemin (in 10 mM NaOH). Absorbance of the cultures was measured at A600 at regular time intervals.
RNA isolation and transcript analysis
Total RNA was extracted from the wild type V. vulnificus CMCP6 strain grown to log phase in CM9 minimal media (supplemented with either 10 µg/ml FAC, or 5 µM EDDA and 10 µM hemin) to log phase using the RNeasy® Mini Kit (Qiagen, Valencia, CA). Following extraction, the total RNA was subjected to DNase treatment with the TURBO DNA-free® kit (Ambion, Austin, TX) to remove any residual DNA. The reverse transcription reaction was performed using the SuperScript® II Reverse Transcriptase with random primers (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Quantitative PCRs were performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) to quantify the expressions of hupA and hvtA genes with primers HupAqRTPCRfor/Hup AqRTPCRrev and HvtAqRTPCRfor/HvtAqRTPCRrev (Table 2), respectively.
Outer membrane protein expression
Outer membrane protein fractions from V. vulnificus cultures were isolated as reported previously with some modifications (Crosa and Hodges 1981). Briefly, overnight cultures of the bacterial strains were inoculated in iron-depleted minimal CM9 media supplemented with 5 µM EDDA and 10 µM hemin and grown at 37°C to early log phase. The cells were then harvested, resuspended in 500 µl of Tris-HCl (pH 7.5) buffer containing protease inhibitors and sonicated. Cell debris was removed by centrifugation at 8,000 rpm for 15 min and the supernatant was further centrifuged for 1 h at 4°C at 15,000 rpm to yield the total membrane proteins as a pellet. The pellet was then treated with 1.5% sodium lauroyl sarcosinate for 2 h at 4°C and centrifuged for 1 h at 15,000 rpm at 4°C. The pellets containing the outer membrane proteins were resuspended in 30 µl distilled water. Samples were then mixed with 2X-SDS loading buffer and analyzed by SDS-PAGE using Criterion® XT Precast Gel (10% Bis–Tris) (Bio-Rad Laboratories, Hercules, CA).
Results
Identification of a second receptor for hemin uptake in V. vulnificus
To study the hemin uptake system in V. vulnificus, a deletion of the vulnibactin biosynthesis gene venB (Litwin et al. 1996) was generated in all of the strains examined. This mutation allowed the elimination of the background growth mediated by the native siderophore vulnibactin in iron-restricted media. When hemin was used as an iron source, no growth stimulation was observed in the triple tonB mutant reporter strain of V. vulnificus, AA-16 (Table 3). In contrast, the hupA mutant strain (VSSD25) showed a smaller halo when compared to a positive control strain (AA-14). These results suggested that there must be another hemin receptor present in the organism. A BLAST search of the CMCP6 V. vulnificus genome database with the HupA amino acid sequence (accession no. AAO07241) yielded an open reading frame (ORF), VV21549 that corresponded to a still uncharacterized putative hemin receptor. This ORF was designated hvtA and the potential encoded protein HvtA. The HvtA protein showed considerable homology to the original hemin receptor proteins HutA (45% similarity and 26% identity) and HutR of V. cholerae (68% similarity and 51% identity) (Mey and Payne 2001) along with other hemin receptors from other vibrio species. Additionally, the protein sequence was also found to contain YRAP/NMDL motifs that are 75%/50% identical to the FRAP/NPNL motifs, respectively, and a conserved histidine residue located between the two motifs that are specific for outer membrane proteins to serve as hemin receptors (Bracken et al. 1999).
Table 3.
Bioassay to test hemin and hemoglobin utilization in V. vulnificus
| Strains | Iron sources | ||
|---|---|---|---|
| FAC | Hemin | Hemoglobin | |
| ΔvenB, ΔtonB1, ΔtonB2, ΔtonB3 | ++ | − | − |
| ΔvenB | ++ | ++ | ++ |
| ΔvenB, ΔhvtA | ++ | ++ | ++ |
| ΔvenB, ΔhupA | ++ | + | − |
| ΔvenB, ΔhvtA, ΔhupA | ++ | − | − |
| ΔvenB, ΔhvtA, ΔhupA, pMMB208-hvtA | ++ | + | + |
Strong growth after 18 h
weak growth after 18 h
no growth after 18 h. Indicated strains were mixed with CM9 media containing agar supplemented with 20 µM EDDA (and 1 mM IPTG for the strain containing the pMMB208 plasmid) and poured into petri dishes. After solidification, different iron sources (FAC, 1 mg/ml; hemin, 10 µM; and hemoglobin, 50 µM) were spotted on the plates and growth halos around the spots of the reporter strains were monitored after 18 h. FAC was used as a positive control
A double mutant strain was hence generated in a venB− background and tested for hemin and hemoglobin utilization through bioassay. Both single hupA and hvtA mutants showed hemin uptake that did not occur when combined to yield a double mutant. With hemoglobin, the hupA mutant did not grow whereas the hvtA mutant did indicating that while both HupA and HvtA serve as hemin receptors, HupA appears to be the sole receptor responsible for the transport of hemoglobin in V. vulnificus. The lack of growth of the double mutant strain with hemin was restored with the complementation of the hvtA gene in trans. It was also interesting to note that in the latter experiment the over-expression of the hvtA gene cloned into the pMB208 vector resulted in a weak uptake of hemoglobin in the complemented double mutant strain, hinting that HvtA may have a very low affinity for hemoglobin at high expression levels.
Hemin and hemoglobin uptake depends on either the TonB1 or TonB2-TtpC2 systems
To determine if hemin and hemoglobin transport was TtpC2-dependent, a combination of the TonB system mutants were generated in the venB− background and tested with bioassays (Table 4). Mutation in ttpC2 alone showed hemin and hemoglobin utilization as observed in ΔtonB1 or ΔtonB2 control strains, but no growth halos were detected with the double mutant ΔtonB1ΔttpC2 strain, where double mutant ΔtonB1 ΔtonB2 and the triple mutant ΔtonB1ΔtonBΔtonB3 were used as negative control strains. These data suggest that TtpC2 and TonB1play important roles in the transport of hemin and hemoglobin.
Table 4.
TonB-dependent hemin and hemoglobin utilization in V. vulnificus
| Strains | Iron sources | ||
|---|---|---|---|
| FAC | Hemin | Hemoglobin | |
| ΔvenB, ΔtonB1 | ++ | ++ | ++ |
| ΔvenB, ΔttpC2 | ++ | ++ | ++ |
| ΔvenB, ΔtonB2 | ++ | ++ | ++ |
| ΔvenB, ΔtonB1, ΔtonB2 | ++ | − | − |
| ΔvenB, ΔtonB1, ΔttpC2 | ++ | − | − |
Strong growth after 18 h
no growth after 18 h. Indicated strains were mixed with CM9 media containing agar supplemented with 40 µM EDDA. After solidification, different iron sources (FAC, 1 mg/ml; hemin, 10 µM; and hemoglobin, 50 µM) were spotted on the plates and growth halos around the spots of the reporter strains were monitored after 18 h. FAC was used as a positive control
Effect of the hvtA mutation on hemin utilization as sole iron source
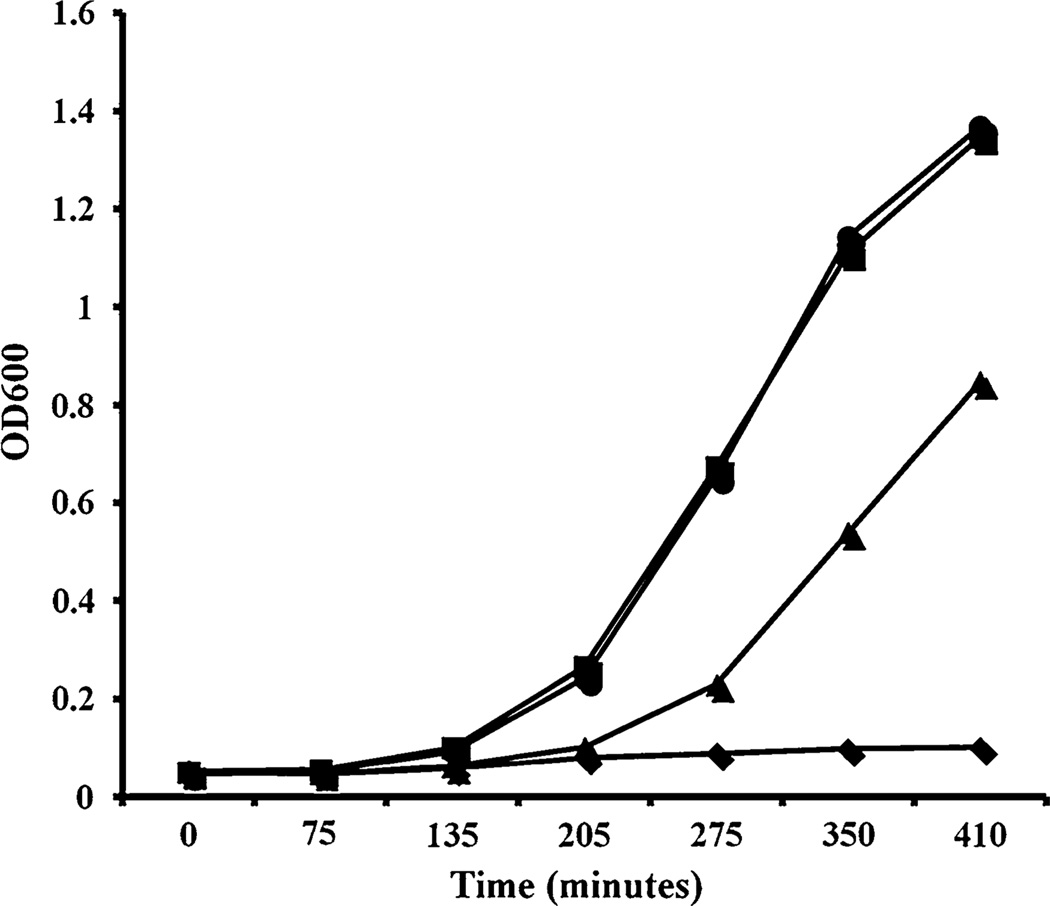
To further characterize HvtA, experiments were performed to compare the growth pattern of the wild-type strain with those of the mutants in the presence of hemin as sole iron source. Mutation in hvtA (VSSD5) did not alter the growth pattern using hemin as an iron source as compared to the wild-type strain (VSSD4). However, growth of the single hupA mutant strain (VSSD25) was observed although mitigated when hemin was provided as the sole iron source (Fig. 1). Further, the double hupAhvtA mutant strain (VSSD40) was not able to grow. These results underscore the fact that both HvtA and HupA by themselves are responsible for hemin uptake in V. vulnificus.
Fig. 1.
Growth of V. vulnificus strains (circles ΔvenB, squares ΔvenBΔhvtR, triangles ΔvenBΔhupA, diamonds ΔvenBΔhvt RΔhupA) in media containing hemin as sole iron sources. Overnight cultures of strains were inoculated in CM9 minimal media containing 10 µM of hemin and optical densities were measured at indicated time points
Transcription of HvtA is regulated by iron
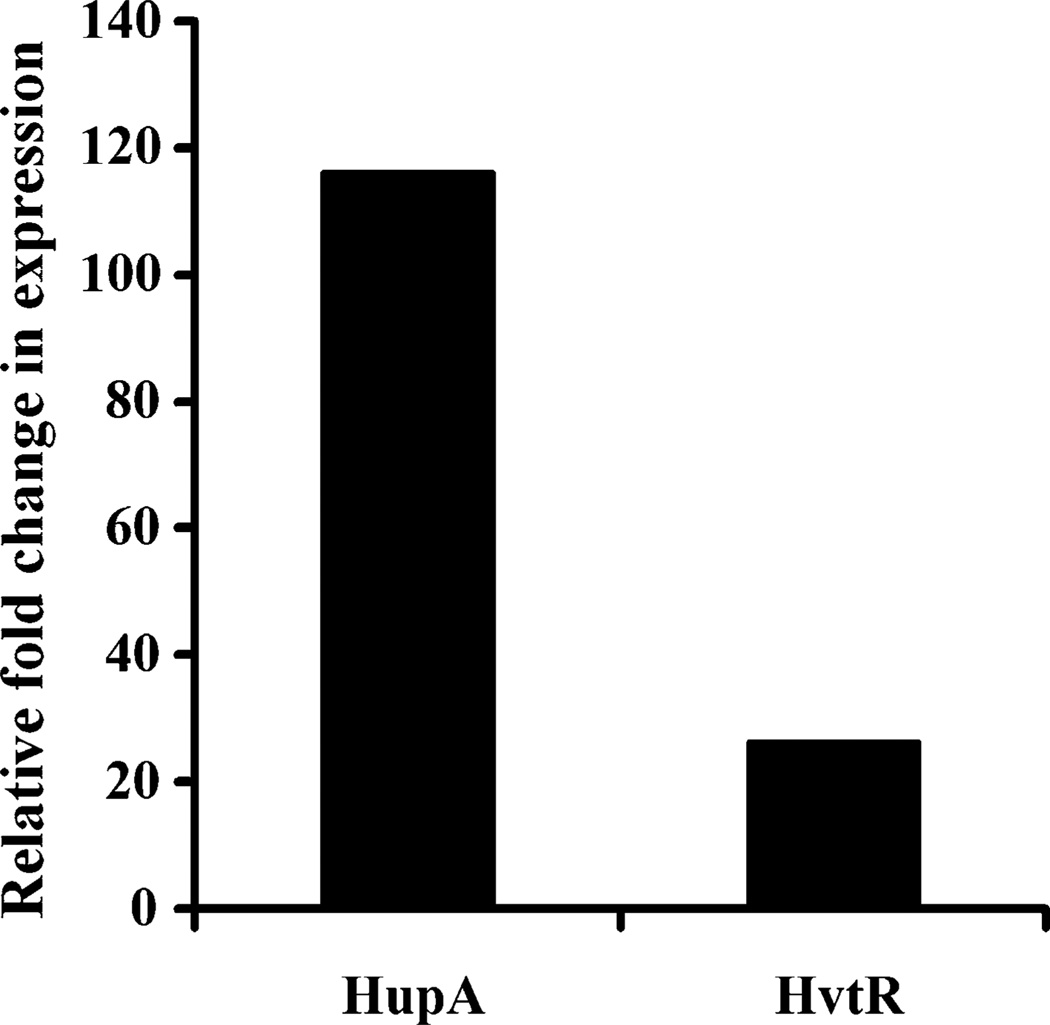
To test if hvtA transcription was regulated by iron, wild-type V. vulnificus CMCP6 strain was grown at 37°C in both iron-rich (10 µg/ml FAC) and iron-depleted media containing hemin as iron source. The levels of hupA and hutR mRNA in the total RNA extracted from the cultures were determined using qRT-PCR. Figure 2 shows a 26-fold increment in the hvtA mRNA levels under iron-limiting condition, whereas the hupA transcript was up-regulated to a even greater extent (116-fold).
Fig. 2.
Regulation of hupA and hvtA genes by iron. Wild-type V. vulnificus strain was grown in iron-rich and iron-depleted media containing hemin and relative levels of hupA and hvtA mRNA were analyzed by qRT-PCR (See materials and methods). The levels of hupA and hvtA transcripts are presented relative to the amount of transcripts yielded in iron-rich condition
HvtA is an outer membrane protein
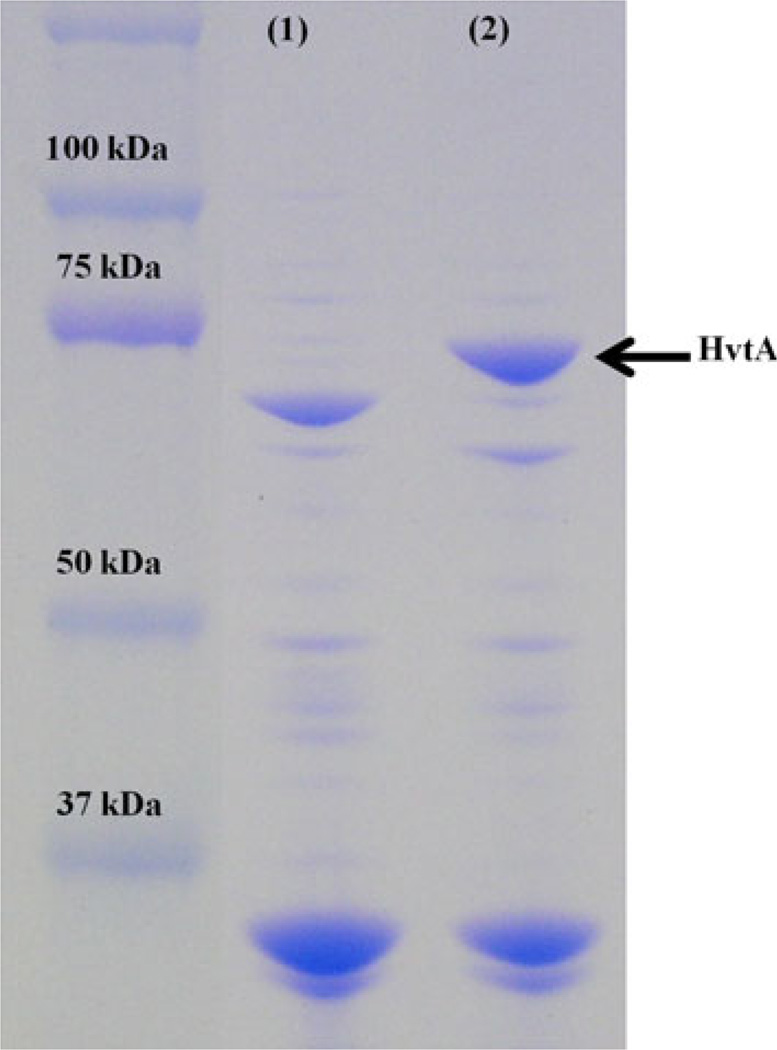
The presence of terminal phenylalanine and an arginine residue in the 11th position from the C-terminus of the HvtA protein sequence suggested the possibility of its localization in the outer membrane (Struyvé et al. 1991). Besides, the sequence was also predicted to be in the outer membrane using the PSORTb (Yu et al. 2010) and ProtCompB (http://linux1.softberry.com/berry.phtml?topic=protcompan &group=programs&subgroup) computational tools that are commonly used to predict subcellular localization of proteins in Gram-negative bacteria. Although strongly predicted to be an outer membrane protein, in practice it was fairly difficult to detect the protein in outer membrane fractions when the bacterial strain was grown under iron-limiting conditions probably due to its low level of expression. Thus, an over-expressing strain was used to induce HvtA expression and determine its localization. Since the predicted molecular weight of HvtA and HupA are very similar, a ΔhupAΔhvtA strain was used as the background strain to avoid confusion in the identification of the proteins. The hvtA gene was cloned into the pMMB208 vector and expressed in this double mutant. Figure 3 shows that a band of approximately 76.9 kDa, corresponding to the mature HvtA protein was detected in the outer membrane fraction in the over-expressed strain, which was not detected in the negative control (ΔhupAΔhvtA–pMMB208).
Fig. 3.
Cellular localization of the HvtA protein. Outer membrane fractions were isolated from strains ΔhvtAΔhupA, pMMB208 (lane 1) and ΔhvtAΔhupA, pMMB208-hvtA (lane 2) after growing them in CM9 minimal media containing 5 µM EDDA and 10 µM hemin with 1 mM IPTG to log phase. Samples were analyzed by SDS-PAGE using Criterion® XTP-recast gel (10% Bis–Tris) for 7 h at 50 V. Precision Plus Protein Standards (Bio-rad)
Discussion
Virulence in Vibrio vulnificus requires a TonB-dependent iron-acquisition system, which involves siderophore-mediated uptake of iron via specific outer membrane receptors. Additionally, it has also been demonstrated previously by various research groups that this organism utilizes hemoglobin and hemin-compounds as iron sources for its infection into the host via the HupA receptor protein (Helms et al. 1984; Oh et al. 2009). In this study, we report the identification of a second iron regulated hemin receptor, HvtA, in V. vulnificus.
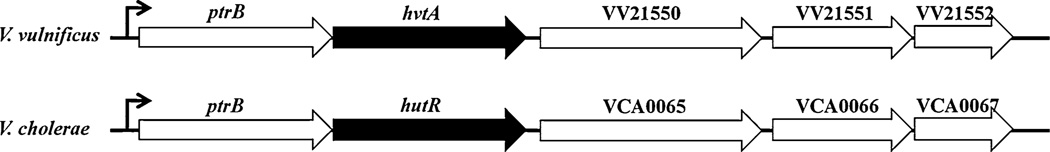
Existence of multiple hemin receptors is not uncommon in vibrio species. Payne and co-workers demonstrated the presence of more than one hemin substrate receptor in V. cholerae. Our detailed analysis of the hvtA gene showed that it is located in the second chromosome where it is predicted to be the second ORF in a putative five-member operon. This operon is significantly similar to the one in V. cholerae containing the hutR hemin receptor gene (Mey and Payne 2001). Additionally, the operon includes a type II protease gene, ptrB, upstream of the hvtA gene, which is highly homologous (63% similar and 46% identical) to the protease gene that is co-transcribed with the hutR gene in V. cholerae (Fig. 4). Identification of a Fur box upstream of the ptrB gene in the V. vulnificus chromosome suggested that the expression of hvtA may be iron-regulated, which is confirmed from the results of growth assays and transcriptional analysis. Whether the hvtA gene is also co-transcribed along with the other genes in the operon remains to be determined.
Fig. 4.
Schematic map and comparison of the hvtA in V. vulnificus (top) and hutR in V. cholerae (bottom). Black and white thick arrows show the ORFs along with their orientations and thin right-angle arrow points to the putative promoter of the two operons
However, by using growth assays we showed that HupA transports hemin more efficiently than HvtA, since deletion of hvtA did not impair the ability of V. vulnificus to utilize hemin to a great extent as compared to the hupA mutant strain. This observation may be explained due to the difference in their transcription levels in iron-limiting condition, which in turn results in different expression levels of the two proteins. This is also consistent with the visibility of the HupA protein in the outer membrane fractions, whereas HvtA was only visible when over-expressed from a plasmid in trans.
In this work we also demonstrated that hemin and hemoglobin uptake mechanisms in the wild type strain are either TonB1 or TonB2-TtpC2-dependent. In this vein, the HvtA protein sequence contains a conserved TonB box (DEVVVSA) found in the N-terminus from amino acid positions 28–34, which is similar to those identified in other hemin utilization proteins in various vibrio species including HutA and HutR in V. cholera (accession nos. AAF95978 and NP232966, respectively), HutA in V. parahaemolyticus (accession no. NP800392), and HuvA in V. anguillarum (accession no. CAC28362), suggesting that the HvtA protein is likely to interact directly with either of these TonB proteins.
Vibrio vulnificus is an opportunistic pathogen for both humans and marine organisms that are known to live in distinct and variant set of conditions. Thus, each receptor may have evolved and adapted differently to different hosts and environments and under other conditions it might be possible that the hvtA gene may be the dominant hemin uptake receptor.
Acknowledgment
This project was supported by a National Institutes of Health grant AI 65981 to J.H.C.
References
- Alice AF, Naka H, Crosa JH. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun. 2008;76:4019–4037. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro C, Biosca EG, Fouz B, Alcaide E, Esteve C. Evidence that water transmits Vibrio vulnificus biotype 2 infections to eels. Appl Environ Microbiol. 1995;61:1133–1137. doi: 10.1128/aem.61.3.1133-1137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CS, Baer MT, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Hodges LL. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect Immun. 1981;31:223–227. doi: 10.1128/iai.31.1.223-227.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig PA, Bourdage KL, Starks AM. Molecular Pathogenesis of Vibrio vulnificus. J Microbiol. 2005;43:118–131. [PubMed] [Google Scholar]
- Helms SD, Oliver JD, Travis JC. Role of heme compounds and haptoglobin in Vibrio vulnificus pathogenicity. Infect Immun. 1984;45:345–349. doi: 10.1128/iai.45.2.345-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlady WG, Klontz KC. The epidemiology of Vibrio infections in Florida, 1981–1993. J Infect Dis. 1996;173:1176–1183. doi: 10.1093/infdis/173.5.1176. [DOI] [PubMed] [Google Scholar]
- Kim YR, Rhee JH. Flagellar basal body flg operon as a virulence determinant of Vibrio vulnificus. Biochem Biophys Res Commun. 2003;304:405–410. doi: 10.1016/s0006-291x(03)00613-2. [DOI] [PubMed] [Google Scholar]
- Kothary MH, Kreger AS. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J Gen Microbiol. 1987;133:1783–1791. doi: 10.1099/00221287-133-7-1783. [DOI] [PubMed] [Google Scholar]
- Lee JH, Rho JB, Park KJ, Kim CB, Han YS, Choi SH, Lee KH, Park SJ. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect Immun. 2004;72:4905–4910. doi: 10.1128/IAI.72.8.4905-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim MW, Kim BS, Kim SM, Lee BC, Kim TS, Choi SH. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J Microbiol. 2007;45:146–152. [PubMed] [Google Scholar]
- Litwin CM, Byrne BL. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect Immun. 1998;66:3134–3141. doi: 10.1128/iai.66.7.3134-3141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin CM, Quackenbush J. Characterization of a Vibrio vulnificus LysR homologue, HupR, which regulates expression of the haem uptake outer membrane protein, HupA. Microb Pathog. 2001;31:295–307. doi: 10.1006/mpat.2001.0472. [DOI] [PubMed] [Google Scholar]
- Litwin CM, Rayback TW, Skinner J. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun. 1996;64:2834–2838. doi: 10.1128/iai.64.7.2834-2838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Alice AF, Naka H, Crosa JH. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect Immun. 2007;75:3282–3289. doi: 10.1128/IAI.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Naka H, Crosa JH. HlyU acts as an H-NS anti-repressor in the regulation of the RTX toxin gene essential foRthe virulence of the human pathogen Vibrio vulnificus CMCP6. Mol Microbiol. 2009;72:491–505. doi: 10.1111/j.1365-2958.2009.06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol. 2001;42:835–849. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Morris JGJ. Vibrio vulnificus—a new monster of the deep? Ann Intern Med. 1988;109:261–263. doi: 10.7326/0003-4819-109-4-261. [DOI] [PubMed] [Google Scholar]
- Nishina Y, Miyoshi S, Nagase A, Shinoda S. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect Immun. 1992;60:2128–2132. doi: 10.1128/iai.60.5.2128-2132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Lee SM, Lee DH, Choi SH. Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect Immun. 2009;77:1208–1215. doi: 10.1128/IAI.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7:109–116. doi: 10.1007/BF00140480. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhle RA. A broad host range mobilization system for in vivo genetic-engineering-transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:787–796. [Google Scholar]
- Simpson LM, Oliver JD. Siderophore production by Vibrio vulnificus. Infect Immun. 1983;41:644. doi: 10.1128/iai.41.2.644-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, Siebeling RJ. Identification of genetic loci required for capsular expression in Vibrio vulnificus. Infect Immun. 2003;71:1091–1097. doi: 10.1128/IAI.71.3.1091-1097.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, Paranjpye RN. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2000;2:177–188. doi: 10.1016/s1286-4579(00)00270-7. [DOI] [PubMed] [Google Scholar]
- Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Webster AC, Litwin CM. Cloning and characterization of vuuA, a gene encoding the Vibrio vulnificus ferric vulnibactin receptor. Infect Immun. 2000;68:526–534. doi: 10.1128/iai.68.2.526-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AC, Simpson LM, Oliver JD. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AC, Powell JL, Tanner MK, Ensor LA, Karpas AB, Morris JG, Jr, Sztein MB. Differential expression of Vibrio vulnificus capsular polysaccharide. Infect Immun. 1999;67:2250–2257. doi: 10.1128/iai.67.5.2250-2257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]