Abstract
Using molecular dynamics simulations and steady-state fluorescence spectroscopy, we have identified a conformational change in the active site of a thermophilic flavoenzyme, NADH oxidase from Thermus thermophilus HB8 (NOX). The enzyme’s far-UV circular dichroism spectrum, intrinsic tryptophan fluorescence, and apparent molecular weight measured by dynamic light scattering varied little between 25 and 75 °C. However, the fluorescence of the tightly bound FAD cofactor increased ~4-fold over this temperature range. This effect appears not to be due to aggregation, unfolding, cofactor dissociation, or changes in quaternary structure. We therefore attribute the change in flavin fluorescence to a temperature-dependent conformational change involving the NOX active site. Molecular dynamics simulations and the effects of mutating aromatic residues near the flavin suggest that the change in fluorescence results from a decrease in quenching by electron transfer from tyrosine 137 to the flavin.
Keywords: Thermophiles; thermophilic enzymes; Thermus thermophilus; flavoproteins; NADH oxidase; fluorescence; fluorescence quenching; electron transfer; temperature effects, molecular dynamics
Introduction
Thermophilic proteins typically reach their optimal activity at higher temperatures than homologous proteins from mesophiles,1 although the peak activities of the two proteins often are similar.2–4 The “corresponding-states” hypothesis5–9 explains these observations by positing that all enzymes require a certain level of flexibility in order to function, and that thermophilic enzymes reach this state only at the high temperatures where their host organisms thrive. Mesophilic enzymes are assumed to exist in an active state at the lower temperatures where their host organisms are found. Numerous studies comparing homologous mesophilic and thermophilic enzymes have been interpreted as supporting this hypothesis.3,4,7,10–13 However, others have found that the flexibility of thermophilic proteins at room temperature is similar to14 or even greater than15 that of homologous mesophilic proteins at the same temperature. Some thermophilic enzymes may simply have catalytic rate constants with smaller activation energies than their mesophilic homologues, and others may undergo conformational changes that favor catalysis at high temperatures. The hypothesis of temperature-dependent conformational changes is the focus of this paper.
In a previous study, we used NADH oxidase from Thermus thermophilus HB8 (NOX) and nitroreductase from Escherichia coli (NTR) as a model pair of homologous thermophilic and mesophilic enzymes for molecular dynamics studies.16 NOX is a dimeric flavoenzyme that catalyzes reduction of flavins and other compounds by NADH or NADPH, probably by a classical ping-pong mechanism in which a bound FMN or FAD coenzyme is first reduced by the pyridine nucleotide and then reoxidized by the oxidizing substrate.17 The effects of pH18 and kosmotropic and chaotropic anions19 and cations20 on NOX enzymatic activity and fluorescence have been characterized. Urea at moderate concentrations (~1 M) has been reported to increase the enzymatic activity and decrease the intrinsic tryptophan fluorescence,21 possibly by effects on the motion of tryptophan 47, which is in the active site.22 This body of work makes NOX an attractive system for investigating the roles of flexibility and conformational changes in a thermophilic protein.
In our previous work, we characterized the temperature dependence of several properties of NOX and NTR by MD simulations.16 We found that NOX, the thermophilic protein, was more resistant to temperature-induced conformational changes than NTR by every metric examined. However, the conformational properties of NOX did exhibit a temperature dependence, particularly in the active site (see Fig. 4 of reference 16). Here, we focus on the active site of NOX in more detail and we explore the effects of temperature by MD simulations, steady-state fluorescence spectroscopy and site-directed mutagenesis.
Materials and Methods
Molecular dynamics simulations
The MD simulations of NOX used here are described in Merkley et al.16 They were performed with in lucem molecular mechanics (ilmm),23 using the Levitt et al. potential function and flexible three-center water model24,25 and following the standard energy-minimization and solvation protocol.26 The crystal structure described by Hecht et al.27 (Protein Data Bank identification code 1NOX) was taken as the initial structure of the protein.
Mutagenesis, Expression and purification of NOX, and Measurements of Enzyme Activity
FAD and NADH were obtained from Sigma. Other chemicals were reagent grade. The expression plasmid pTNADOX28 was a gift from Prof. Mathias Sprinzl, University of Bayreuth, Germany. We sequenced the plasmid at the University of Washington Biochemistry DNA Sequencing Facility and confirmed the published sequence. The W47F, W52F, W131F and Y137F mutants of NOX were constructed using the QuikChange II Site-Directed Mutagenesis kit (Stratagene) with the following mutagenic primers and their reverse complements: 5’-GCGCCCTCGGCCTTTAACCTCCAGCCCT-3’ for W47F, 5’-TGGAACCTCCAGCCCTTTCGGATCGTGGTGGTGC-3’ for W52F, 5’-CGCGAAAGGCCTTTGCCTCCGGGCAGAGCTAC-3’ for W131F, and 5’-TCCGGGCAGAGCTTCATCCTCTTGGGC-3’ for Y137F. Clones were screened by DNA sequencing.
NOX was expressed in BL21*(DE3) Escherichia coli cells (Invitrogen). Cultures (1 L) were grown at 37 °C in LB medium with 100 µg/mL ampicillin to OD600 of 0.6–1.2, induced with 1 mM isopropyl β-D-1-thiogalactopyranoside, grown for another 3–5 hours at 37 °C, and harvested by centrifugation. Cells from 3–4 L were used for each preparation. Cell pellets were stored at −80 °C until lysis.
NOX was purified by a modification of the procedure of Park et al.28 Cells were resuspended in 100 mM Tris HCl (pH 7.4), 300 mM NaCl, 5 mM EDTA, 5 % v/v glycerol plus phenylmethanesulfonylfluoride and protease inhibitor cocktail (Sigma) and lysed by sonication on ice. The lysate was cleared by centrifugation, and FAD (0.5 mg/mL) was added to the soluble fraction (21 and E. Sedlák, personal communication), which was then incubated for 1 hr in a water bath at 72° C for wild-type NOX. The heat-treatment step was optimized for each NOX mutant by heating for various times at different temperatures and measuring the specific activity after Cibacron Blue chromatography. The sample was centrifuged (30 minutes, 23,500g) and the supernatant filtered through a 0.45-µm syringe-driven filter (Millipore). The sample was then applied to CL-6B Blue Sepharose (“Cibacron Blue,” GE Healthcare) column (3.2 × 1 cm) equilibrated in 50 mM Tris HCl (pH 7.2) containing 500 mM NaCl. The Cibacron Blue column was washed in the same buffer until the eluate absorbance at 280 nm was less than ~0.02 (~300 mL), and then NOX was eluted with ~70 mL of 10 mM NADH in the same buffer. Because of the UV absorbance of NADH, fractions were assayed by a modified Bradford assay (Coomassie Plus™ Protein Assay, Pierce). Protein-containing fractions were pooled and concentrated with an Amicon® Ultra-15 centrifugal filter device with a nominal molecular size cutoff of 10 kDa (Millipore). The concentrated Cibacron Blue eluate was applied to a Superdex 75 preparative grade size-exclusion column equilibrated in 50 mM sodium phosphate (pH 7.2) containing 500 mM NaCl. NOX-containing fractions were pooled, concentrated, and dialyzed for ~24 hours in 4 L of 50 mM sodium phosphate (pH 7.2) with three changes of buffer. Protein concentration was determined as described above, with bovine serum albumin (Pierce) as the standard.
NOX samples purified in this way gave a band corresponding to an apparent molecular weight of 25,000 on sodium dodecylsulfate polyacrylamide gel electrophoresis, and were judged to be at least 90% pure by this procedure. The molecular weight of one sample was determined by mass spectrometry and found to be 22,739 ± 7, compared to the theoretical mass of 22,730. The molecular weight calculated from dynamic light-scattering measurements was 43,000, in agreement with the expected value for the dimeric enzyme.
Some NOX samples were purified further by cation-exchange chromatography (CEX) on a Source 15S 10/10 column (8 mL bed volume). NOX was eluted from the CEX column with a linear gradient from 0 to 0.5 M NaCl. Since NOX can be deflavinated by CEX at low pH,17 peaks containing NOX were concentrated, treated with FAD, and desalted (10 mL polyacrylamide D-Salt columns, Pierce) before being analyzed for activity and fluorescence.
Enzyme activity was measured in 50 mM sodium phosphate (pH 7.2) with enzyme concentrations in the range of 1–10 nM NOX dimer and substrate concentrations of 200 µM NADH and 120 µM FAD. The reaction was followed by measuring the loss of NADH absorbance at 340 nm, using a Shimadzu UV-1601 spectrophotometer. At room temperature (23 °C), the enzyme purified from wild-type cells had a specific activity of 36 ± 4 µmol NADH oxidized/min/mg protein, which is somewhat greater than the value of 11 µmol/min/mg reported by Žoldák et al.21
Circular dichroism, dynamic light scattering, and fluorescence spectroscopy
Circular dichroism (CD) was measured with an Aviv 62A DS spectropolarimeter equipped with a thermoelectric temperature-control module and a 2-mm quartz cell. The NOX dimer concentration was 5 µM. For thermal denaturation experiments, the CD signal at 222 nm was recorded from 20 to 98 °C at 2° intervals, with 30 s averaging time at each temperature. Dynamic light scattering (DLS) measurements were performed with 19.5 µM NOX dimer in a DynaPro DLS spectrometer equipped with temperature control.
Fluorescence was measured with a Perkin-Elmer LS-50B fluorescence spectrometer with a thermostatted cell holder. Temperature was measured with a copper-constantan thermocouple inserted into the cuvette. The usual protein concentration for fluorescence measurements was 0.5 µM. For protein tryptophan fluorescence, the excitation wavelength (λex) was 290 nm and the detection wavelength was 345 nm. For flavin fluorescence, λex was either 290 or 455 nm and the emission wavelength was either 525 or 523 nm. Excitation and emission band passes for measurements of quantum yields typically were both 2.5 nm. Fluorescence spectra were corrected for the wavelength dependence of the spectrometer optics by comparison with corrected spectra of standard fluorophores.29
Fluorescence quantum yields (Q) were measured by comparison with standards, using the equation
| (1) |
where the subscript ref indicates a compound with known Q, here either tryptophan (λex = 280 nm, Qref = 0.13)30 or tyrosine (λex=275 nm, Qref = 0.14);30 F is the integrated fluorescence emission intensity (305–544 nm for Trp, 283–420 nm for Tyr, 300–470.5 nm for NOX Trp residues, and 470 to 600 nm for flavin); A is the absorbance at the excitation wavelength; and n is the refractive index of the solvent.31 The ratio approaches unity for dilute protein solutions and was neglected.
The FAD fluorescence of NOX was concentration-dependent, leveling off at high protein concentrations instead of continuing to increase linearly (data not shown). To avoid complications from this effect, we measured the fluorescence yield for each sample at multiple protein concentrations and fit the data to a rectangular hyperbolic function of A. We then evaluated the analytical derivative at A = 0.001 (16 nM for wild-type NOX dimer), where the dependence on concentration was essentially linear. All quantum yield measurements were made at room temperature (22–23 °C).
Results
Molecular dynamics simulations
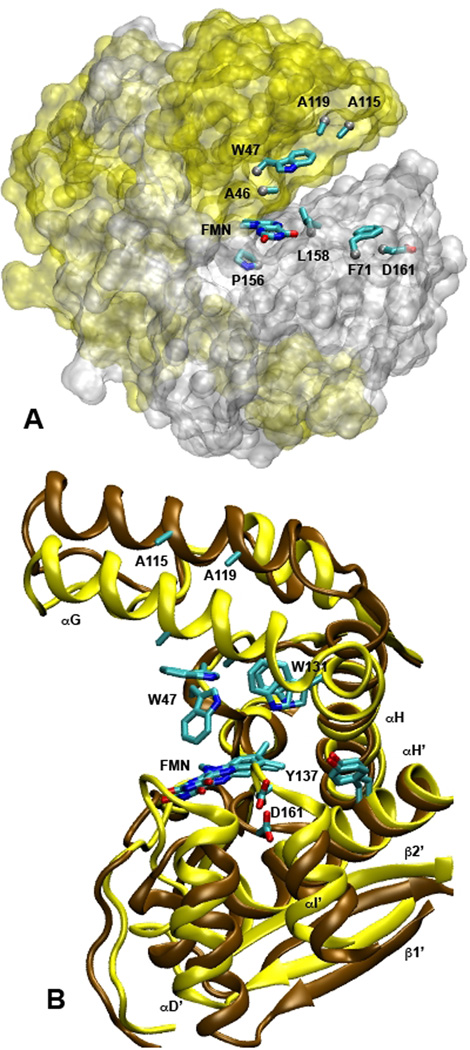
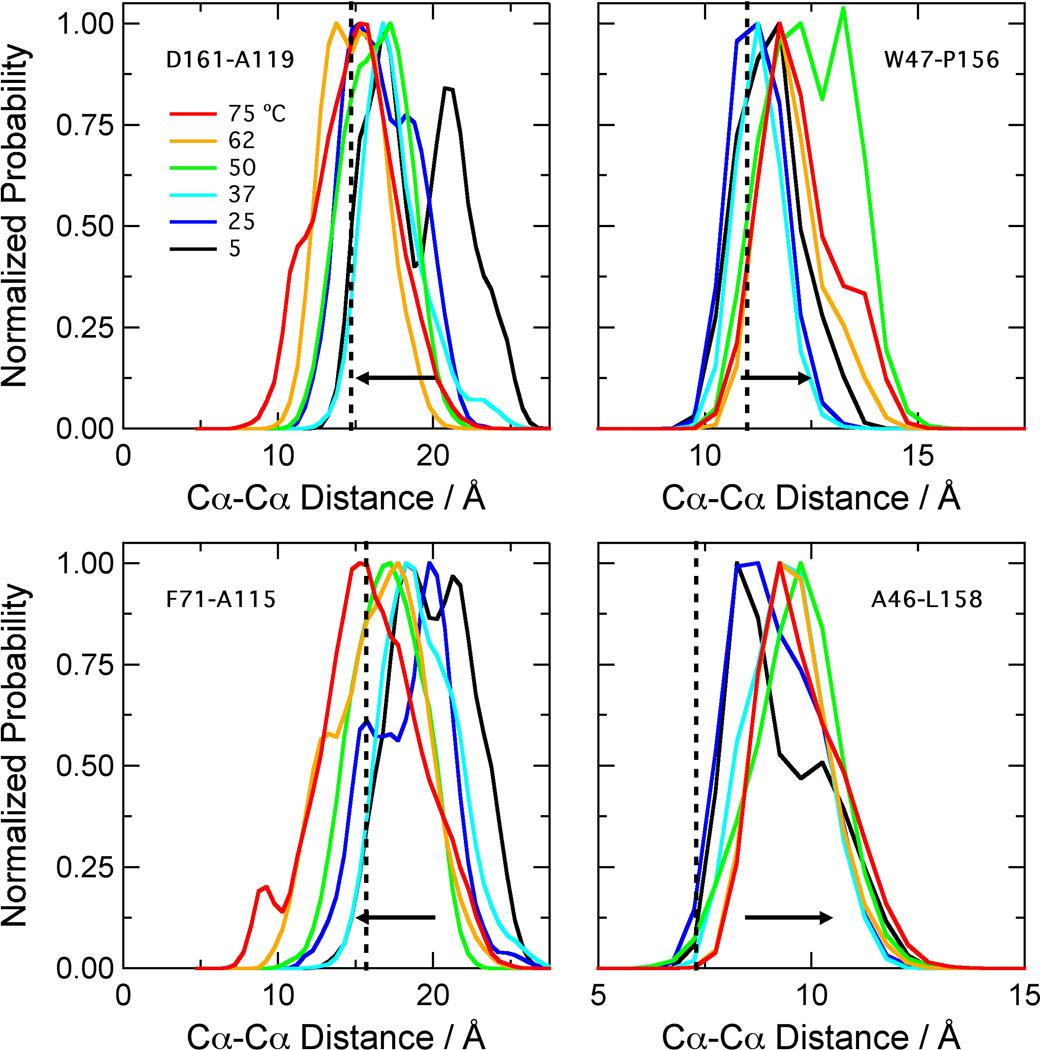
Our previous work16 showed that the simulations of NOX were stable over ~20 ns. The protein remained native-like at temperatures up to at least 75 °C, which is approximately the optimal growth temperature for T. thermophilus32 and the temperature of peak activity for the enzyme.17 To characterize the temperature dependence of the structure in more detail, we examined four inter-Cα distances that span the active site (Figure 1A). The gap between the Cα atoms of W47 and P156 reports on the size of the binding pocket for flavin and substrates as measured through the center of the FMN isoalloxazine ring, while the A46-L158 distance describes the part of the binding pocket that accommodates the hydrophobic dimethylbenzene portion of the ring. The D161-A119 and F71-A115 distances report on the entryway to the active site as measured from the C terminus of helix I’ to the C-terminus of helix G and from the helix D’/helix E’ loop to the center of helix G, respectively. The distance distributions (Figure 2) show that the binding site for flavin and substrates expands at higher temperatures, while the entryway contracts. The W47-P156 distance increases by 2 to 3 Å between 5 and 75 °C, and the A46-L158 distance increases by about 1 Å, while D161-A119 and F71-A115 both decrease by 2 to 3 Å. Some of these changes are illustrated in Figure 1B, which shows representative structures near the ends of simulations at 25 and 75 °C.
Figure 1.
Structures of the active site of NOX. (A) The protein surface at 25 °C as defined by a 1.4-Å probe (transparent representation with subunit A in yellow and B in silver), with the isoalloxazine ring of bound FMN and the side chains of the eight residues used for the distance calculations in Figure 2 superimposed (licorice representations colored by atom type). The phosphoribityl moiety of FMN is omitted for clarity. (B) Representative snapshots of the active site from a different viewpoint at 21.9 ns during MD simulations a t 2 5 ° C (cartoon representation of the protein backbone in brown) and 75 °C (yellow). The FMN and six of the residues considered in Figures 2 and 3 are superimposed (licorice). Helices αG and αH of subunit A are labeled, along with helices αD’, αH’ and αI’ and strands β1’ and β2’ of subunit B. Both figures were made with VMD43 using the crystal structure described by Hecht et al.27 (Protein Data Bank identification code 1nox).
Figure 2.
Distributions of backbone distances across the FMN binding site of NOX. The histograms were calculated with 0.5-A bins from data saved at 1-ps intervals from three ~21-ns simulations at each temperature. Because the protein is dimeric, this gave ~126,000 data points for each histogram. The distances in the crystal structure (1nox.pdb)27 are D161-A119, 14.7 Å; F71-A115, 15.7 Å; W47-P156, 11.0 Å; A46-L158, 7.3 Å (vertical dotted lines). The W47-P156 and A46-L158 distances increase with temperature; the D161-A119 and F71-A115 distances decrease (horizontal arrows).
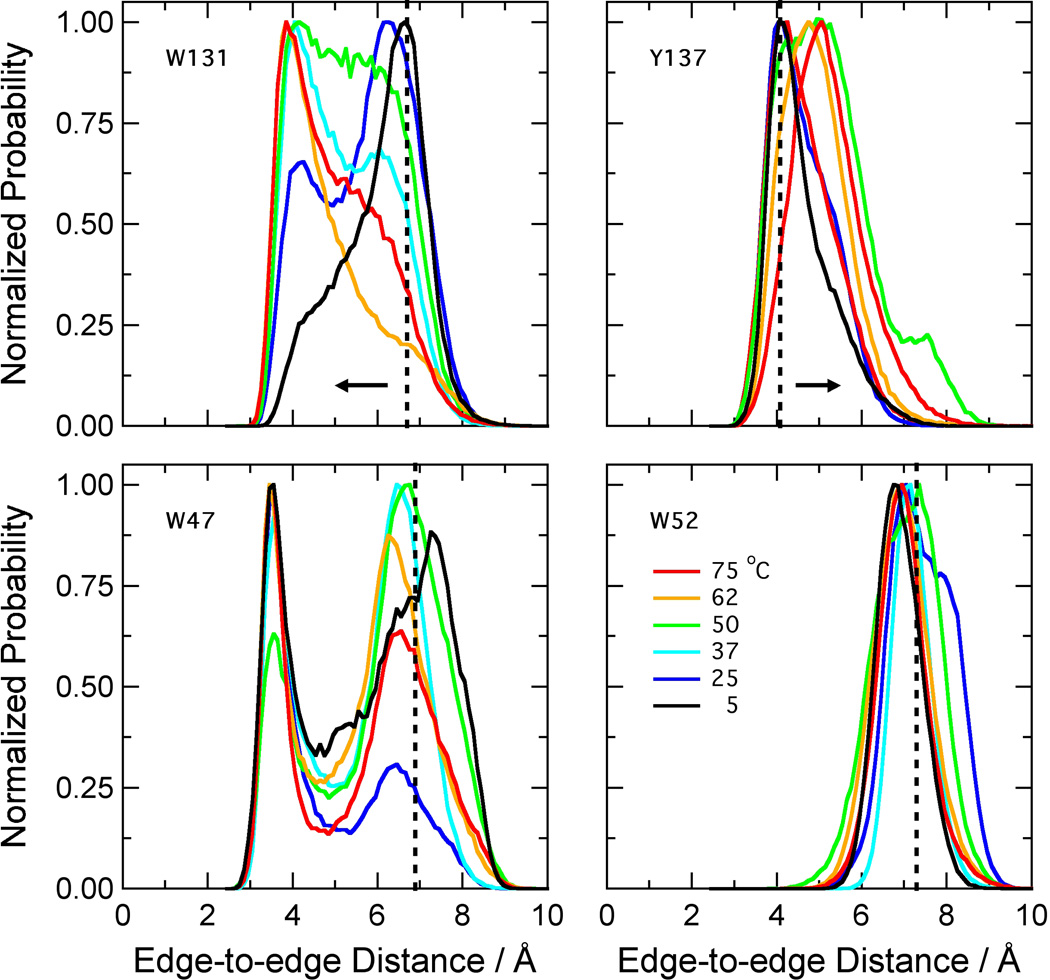
In concert with expansion of the active site, the MD simulations showed systematic changes in the edge-to-edge distances from the bound FMN to two nearby aromatic residues, W131 and Y137 (Figures 1B and 3). The distance to W131 decreased with increasing temperature, while that to Y137 increased. The distance to W52 remained essentially constant. The distance to W47 did not change systematically, although both this distance and the distance to W131 were distinctly bimodal in all the simulations (Figure 3). W47 has been proposed to occupy “open” and “closed” conformations, acting as a gate that either allows or blocks access of substrate molecules to the flavin.22 The distributions of dihedral angles χ1 and χ2 for the W47 side chain are shown in Supplemental Information Figures S1 and S2. The major component of these distributions (designated I in Figures S1 and S2) included both open conformations with a gap or a solvent molecule between W47 and the flavin ring, and closed conformations with the two rings in contact. The mean χ1χ2 coordinates of this component changed only slightly with temperature. Three minor components consisting of open conformations (II-IV) were seen at low or intermediate temperatures.
Figure 3.
Distributions of distances from the isoalloxazine ring of FMN to the nearest non-hydrogen atom of the side-chain of W47, W52, W131 and Y137. The histograms were calculated as in Figure 2. The distances in the crystal structure (1nox.pdb)27 are W47, 6.9 Å; W52, 7.2 Å; W131, 6.7 Å; Y137, 4.5 Å (vertical dotted lines). The distance to FMN increases with temperature for Y137, decreases with temperature for W131 (horizontal arrows), and has no clear trend for W47 or W52.
Spectroscopic measurements
Fluorescence of protein-bound FAD or FMN can be quenched by electron transfer to the flavin from a neighboring Trp or Tyr residue,33–38 and the rates of such electron-transfer reactions depend strongly on the edge-to-edge distance between the electron donor and acceptor.38–40 We therefore looked for temperature-dependent changes in the flavin fluorescence yield as a possible way of corroborating the conformational changes seen in the MD simulations.
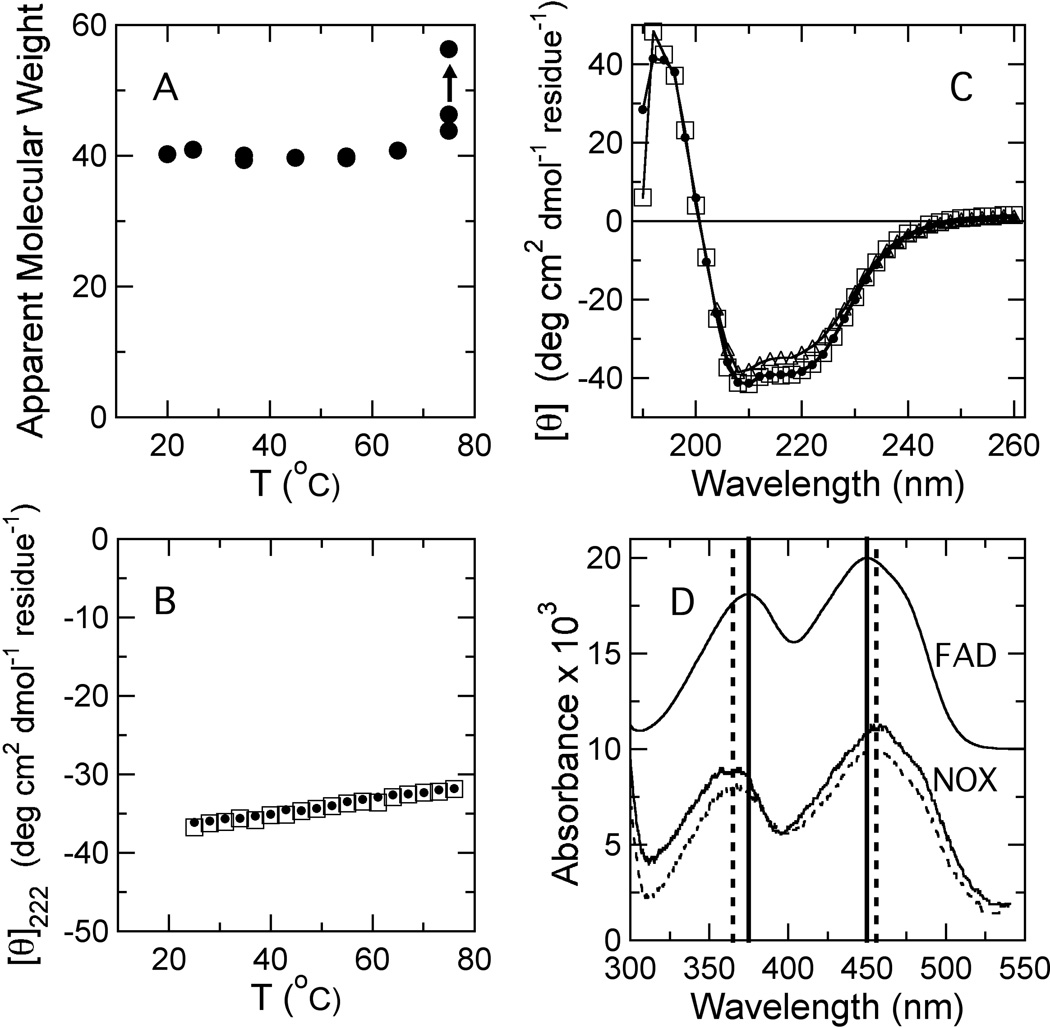
In accord with previous reports,17,21 purified NOX is extremely stable to thermal denaturation. The molecular weight obtained by DLS was independent of temperature up to 75 °C, indicating that the protein does not aggregate below this point (Figure 4A), and the midpoint temperature for denaturation (Tm) measured by CD was greater than 97 °C (Figures 4A and B). No precipitation was observed if the protein was heated to 75 °C, and heating and cooling scans of the ellipticity at 222 nm were superimposable (Figure 4B). The far-UV CD spectra before and after heating to 75 °C also were essentially identical, and spectra measured at 75 °C showed only a slight decrease in helicity relative to room temperature (Figure 4C).
Figure 4.
Effects of temperature on the structure and spectroscopic properties of wild-type NOX. (A) Apparent molecular weight of NOX determined by dynamic light scattering at a protein concentration of 19.5 µM in 50 mM sodium phosphate buffer (pH 7.2). The data point indicated by an arrow was measured after prolonged incubation at 75 °C. (B) Heating (filled circles) and cooling (open squares) scans of the ellipticity of NOX at 222 nm between 25 and 75 °C. Note that heating and cooling scans are essentially superimposed. (C) Far-UV circular dichroism spectra of NOX measured at 25 °C before and after heating to 76 °C (filled circles and open squares, respectively), and measured at 75 °C (open triangles). (D) Absorption spectra of NOX at 24 and 65 °C (solid and dashed curves, respectively) and of free FAD at 24 °C (shifted upward for clarity). The solid vertical lines indicate the absorption peaks for free FAD (375 and 450 nm); the dashed lines, the corresponding peaks for NOX (365 and 456 nm).
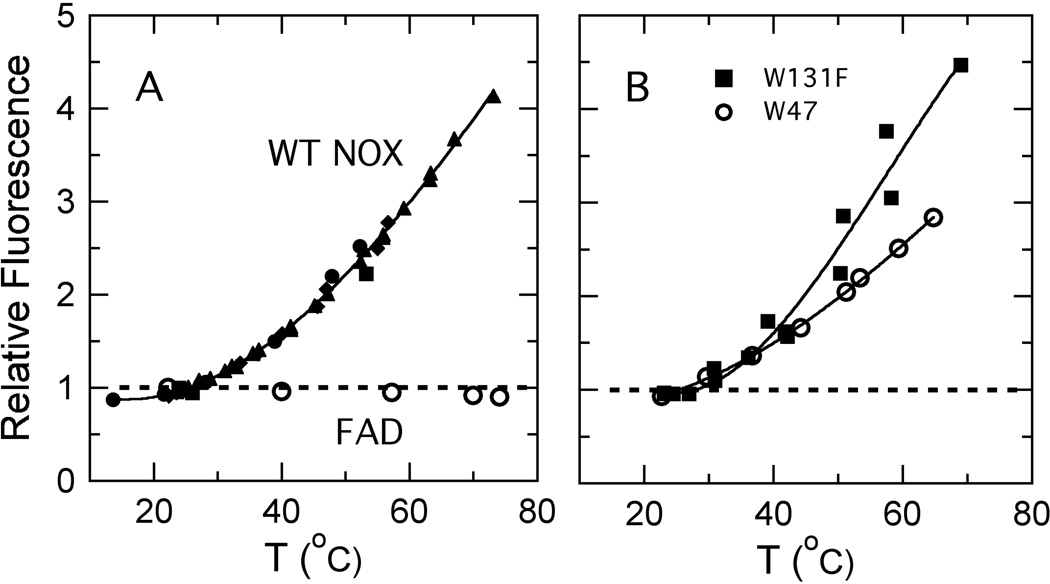
The tryptophan fluorescence of wild-type NOX varies by less than 15% between 20 and 75 °C (not shown). The FAD fluorescence, however, increases approximately 4-fold over this temperature range (Figure 5A, filled symbols). The fluorescence of free FAD, in contrast, is essentially independent of temperature between 20 and 75 °C (Figure 5A, open circles). The change in fluorescence of the protein-bound flavin is reversible if the sample is allowed to cool slowly at room temperature (data not shown). The flavin absorption peaks at 375 and 450 nm, which shift to 365 and 456 nm when FAD binds to NOX, do not shift significantly when NOX-bound FAD is heated to 65 °C, indicating that the flavin probably remains bound to the protein (Figure 4D). There is a small (~10%) decrease in the strength of the absorption band at 270 nm with increasing temperature, but no wavelength shift (not shown). Since the solubility of molecular oxygen, a known quencher of fluorescence, decreases with temperature, we measured the temperature-dependent fluorescence of a degassed NOX sample and found it to be similar to that of samples in equilibrium with air (not shown).
Figure 5.
(A) Temperature dependence of the amplitude of fluorescence of FAD bound to wild-type NOX (filled symbols) and free FAD (open circles), both expressed relative to the amplitudes at 25 °C. Filled circles, squares, triangles and diamonds represent four independent preparations of NOX. NOX fluorescence was excited at 455 nm and detected at 523 nm; fluorescence of free FAD was excited at 290 and detected at 525 nm. Measurements were made in 50 mM sodium phosphate buffer (pH 7.2) at a protein or FAD concentration of 0.5 µM. (B) Temperature dependence of the amplitude of fluorescence of FAD bound to the W47F and W131F mutants (diamonds and squares, respectfully), both expressed relative to the amplitudes at 25 °C.
NOX mutants
With the goal of assigning the temperature dependence of the flavin fluorescence to changes in quenching by a particular Trp or Tyr residue, we constructed the W47F, W52F, W131F, and Y137F mutants. These were the only Trp and Tyr residues that came within 5 Å of the cofactor during the simulations, and as noted above, the distances from W131 and Y137 to the bound FMN changed systematically with temperature. If one of them quenches the flavin fluorescence strongly at low temperatures, replacing it by Phe would be expected to increase the fluorescence yield and change the dependence of the fluorescence on temperature. All four mutants were enzymatically active (Table I), well folded as judged by CD spectroscopy, and thermostable as judged by CD thermal denaturation experiments (data not shown). The W47F and W131F mutants both showed an increase in flavin fluorescence with temperature similar to that of the wild-type enzyme (Figure 5B), and neither of these mutations increased the fluorescence yield at low temperatures (Table 1), indicating that quenching by W47 or W131 probably is not responsible for the temperature dependence of the fluorescence in the wild-type protein. The W52F and Y137F mutants had diminished flavin binding affinity, as judged by decreased flavin absorbance at 455 nm and increased Trp fluorescence. (Resonance energy transfer to the flavin probably quenches Trp fluorescence in the wild-type protein.) This binding defect precluded reliable measurements of the flavin fluorescence yields for these two mutants and raised the concern that structural changes caused by the mutations could complicate interpretation of the results. The fluorescence of both mutants tended to increase with temperature, but the signal intensity was weak and the changes were poorly reproducible (not shown). Y137 nevertheless seems the most likely candidate to account for the temperature dependence of the fluorescence in wild-type NOX because, in the MD simulations, it is the only one of the four residues that moved away from the flavin with increasing temperature, the direction required in order to slow electron transfer (Figure 3).
Table 1.
Fluorescence Quantum Yields and Specific Activities
| Construct | Quantum Yield | Specific Activitya | |
|---|---|---|---|
| FAD | Trp | ||
| WT | 0.06 | 0.08 | 36±4 |
| 0.05b | 0.08 | ||
| W47F | 0.05 | 0.05 | 48±3 |
| 0.05 | 0.05 | ||
| W131F | 0.02 | 0.11 | 20±3 |
| 0.04 | 0.10 | ||
| W52F | n.d.c | 0.12 | 42±10 |
| n.d.c | 0.14 | ||
| Y137F | n.d.c | 0.17 | 32±4 |
Specific activity (µmol NADH oxidized per minute per mg enzyme) was measured at 22–23 °C with 200 µM NADH and 120 µM FAD.
Duplicate entries represent experiments with independent preparations of NOX. The experimental errors of replicate measurements of the quantum yield with the same preparation were approximately ±0.002 for FAD and ±0.01 for Trp.
Flavin fluorescence for these constructs was too low to measure QFAD reliably.
The finding that the Y137F and W52F mutants retained high specific activities in spite of their decreased flavin binding bears some comment. The A455/A280 ratio and fluorescence properties were measured with NOX that had been purified by chromatography and dialysis without additional flavins, following addition of FAD to the cell lysate (see Methods); they therefore depended on flavin that remained tightly bound to the protein. The activity assays, in contrast, were conducted with 120 µM FAD. At this concentration, FAD evidently binds to Y137F and W52F NOX sufficiently to allow full enzymatic activity. (The Kd for FMN in wildtype NOX is reported to be 0.09 µM,27 and that for FAD likely is comparable.)
Discussion
Our previous MD simulations showed that NOX retains its overall structure remarkably well at elevated temperatures.16 However, the distributions of active-site distances shown in Figure 2 change with temperature. Most importantly, the distance between residues on opposite sides of the flavin’s isoalloxazine ring (W47-P156 and A46-L158) increases. This separation does not appear to reflect a global expansion of the protein, because residues on opposite sides of the entryway to the active site (D161-A119 and F71-A115) move closer together. It is unlikely to reflect gross disordering of the protein at high temperatures, because A46, W48, P156 and L158 are in internal regions with well-defined helical or β secondary structure, where the RMS fluctuations of the Cα carbons are relatively small (0.5 to 0.7 Å) at all the temperatures considered (see Figure 6a of reference 16). There also is little or no increase in the overall Cα RMSD up to 75 °C.16
The W47-P156 and A46-L158 distances are potentially important because they provide a metric of the space available for insertion of NADH and the oxidizing substrate. Co-crystals of the homologous protein NTR with bound nicotinic acid41 or a benzoate inhibitor42 indicate that the nicotinamide ring of NADH probably inserts between the re face of the flavin and residues A46 and W47, and suggest that this same slot accommodates the oxidizing substrate after NAD+ departs. The expansion observed in the MD simulations thus could facilitate entry and departure of both substrates. However, further experimental and computational studies will be needed to determine the effect of this structural change on the rate of the enzymatic reaction.
W47 has been proposed to act as a “conformational gate” that controls access to the active site.21,22 In simulations of NOX at 298 K, Hritz et al.22 found W47 most often in a “closed” conformation, raising the possibility that the enzyme might shift to an “open” conformation with a more accessible flavin at higher temperatures. Our simulations showed a W47 rotamer distribution with four components (I-IV) at 278 K, reducing to three components by 298 K and two (I and IV) by 323 K (Supplementary Figures S1 and S2). The main component (I) included both open and closed conformations, and there was no systematic change in these subpopulations with temperature (Figure 3). While we do not rule out a role for W47 as a conformational gate, the backbone motions that expand the active site with increasing temperature evidently are not coupled tightly to particular rotamers of W47.
The 4-fold increase in flavin fluorescence between 25 and 75 °C (Figure 5A) provides independent experimental evidence for a temperature-dependent conformational change in the active site. Several other possible interpretations of this effect can be excluded. The CD data (Figure 4B and C) indicate that it is not due to global unfolding of the protein; the DLS results (Figure 4A) indicate that it is not due to aggregation; and the reversibility of the effect argues against a covalent modification. The fluorescence increase also is unlikely to be due to dissociation of the flavin cofactor, because the 375- and 450-nm absorbance peaks of FAD, which shift when FAD binds to NOX, do not change significantly when NOX is heated to 75 °C (Figure 4D).
We also considered the possible presence of a contaminant that quenches NOX fluorescence at low temperatures and dissociates at higher temperatures. However, further purification of NOX by CEX chromatography under conditions that (reversibly) remove the bound flavin17 had no significant effect on the temperature dependence of the fluorescence, although the overall fluorescence was slightly increased (data not shown). In addition, NOX samples dialyzed for ~24 hrs at either 4 or 25 °C had identical specific activities and very similar fluorescence yields.
A temperature-dependent equilibrium between monomers and dimers could potentially cause a change in fluorescence. However, a variety of measurements made with NOX concentrations ranging from low nanomolar to high micromolar all support the view that the dimer is the predominant species in solution. These experiments include analytical ultracentrifugation,18 DLS measurements (Figure 4), steady-state kinetic measurements (a linear dependence of the initial rate on enzyme concentration over the range of concentrations where the kinetics could be measured), and crosslinking with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (data not shown). Because the active site is located at the dimer interface, dissociation almost certainly would disrupt catalytic activity. The physiological requirement for activity at high temperatures thus also argues that the dimer remains intact.
Our findings therefore suggest that the temperature-dependent increase in flavin fluorescence reflects a conformational change in the active site. The change appears to occur gradually and continuously, rather than by a cooperative or two-state process. The structural change may be akin to the difference between the native (N) states of proteins at moderate temperatures and the corresponding states (N′) seen at elevated temperatures in MD simulations of chymotrypsin inhibitor 2,44 ribonuclease A,45 and the engrailed homeodomain.46 These N′ states generally have the same topology and tertiary contacts as the N states, but are somewhat expanded. For thermophilic proteins, however, the high-temperature state is the more important state physiologically and might aptly be called N rather than N’; the low-temperature conformation is relatively inactive. Cast in these terms, the difference between thermophilic and mesophilic proteins becomes not so much a question of dynamics (the corresponding-states hypothesis), but one of active and inactive conformational states.
The MD simulations suggest a mechanism whereby a temperature-dependent conformational change could lead to a change in cofactor fluorescence: the well-known quenching of flavin fluorescence by excited-state electron transfer from nearby tryptophan or tyrosine residues.33–38 A case in point is E. coli flavin reductase, in which Y35 is 4.5 Å from the bound flavin, approximately the same as the distance betweeen Y137 and FMN in the NOX crystal structure. Mutating Y35 to serine dramatically increases the fluorescence lifetime, despite the presence of three other tyrosines at distances of approximately 7 Å.37 Replacing Y177 of glutathione reductase by phenylalanine has a similar effect.33 Callis and Liu38 have reviewed such studies of “non-fluorescent” (highly quenched) flavoproteins, and concluded that strong quenching by electron transfer occurs only if the edge-to-edge distance of the tyrosine or tryptophan side chain to the flavin is less than 5–6 Å. Y137 is the only tyrosine within this distance in the NOX crystal structure and, in our MD simulations, the only nearby tyrosine or tryptophan residue whose distance from the flavin increases systematically with temperature (Figure 3). The distribution of its edge-to-edge distance peaks slightly below 4 Å at low temperatures, and shifts to longer distances by about 1 Å at higher temperatures.
The MD simulations thus suggest that movement of Y137 away from the flavin decreases quenching by electron transfer from the Tyr side chain to the flavin at high temperatures. We tested this hypothesis by mutagenesis experiments. The results ruled out changes in quenching by W47 or W131 as dominant factors, because replacing these residues by Phe had little effect on either the yield or the temperature dependence of the fluorescence (Table I and Figure 5). The experiments were inconclusive with regard to W52 and Y137, because weakened binding of FAD to the W52F and Y137F mutants prevented reliable measurements of the fluorescence yield. However, W52 is unlikely to be the dominant quencher because it is considerably farther from the flavin than the other residues and the MD simulations indicate that the W52-FMN distance changes little with temperature (Figure 3). It is possible that two or more residues contribute to the quenching, or that temperature-dependent structural changes affect the fluorescence through independent effects on the energy of electron transfer to the flavin or on the availability of a critical proton donor or acceptor. Time-resolved fluorescence measurements might help to resolve these uncertainties.
Although the mutagenesis experiments do not prove either that electron transfer from a Tyr or Trp residue underlies the temperature dependence of the fluorescence, or that Y137 is the putative electron donor, the combination of experimental and simulation data appear to us to provide compelling support for the conclusion that the active site of NOX experiences a temperature-dependent conformational change. This finding has important implications for the study of thermophilic enzymes. First, the low temperatures (often cryogenic temperatures) at which protein crystal structures usually are determined are far from the temperatures where thermophilic proteins are most active. Thus, the crystal structures of some thermophilic enzymes may not represent the active states of these enzymes. Second, conformational changes provide an alternative to the “corresponding-states” hypothesis for explaining the temperature dependence of some of these enzymes.
Supplementary Material
Acknowledgments
We thank Dr. David Beck for ilmm analysis software, Andrew McMillan, Prof. Rachel Klevit, Dr. Kathryn Scott, and Prof. Eric Sedlak for helpful discussions, Dr. Stewart Turley for help with the DLS measurements, and Bennett Addison for assistance in protein expression. The pTNADOX expression plasmid was a kind gift from Prof. Mathias Sprinzl. This work was supported by National Institutes of Health grant GM 50789 (to V.D.), the Molecular Biophysics Training Grant 5 T32 GM008268-19 (to E.D.M.), and National Science Foundation grant 0641640 (to W.W.P.). Some of the MD trajectories were produced using resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231; other trajectories were produced using computer resources provided by the NIH through the Multi-Tiered Proteomics Compute Cluster (NCRR 1S10RR023044-01).
Abbreviations
- CD
circular dichroism
- CEX
cation exchange chromatography
- DLS
dynamic light scattering
- λex
excitation wavelength
- NOX
Thermus thermophilus NADH oxidase
- MD
molecular dynamics
- NTR
Escherichia coli nitrate reductase
- Q
fluorescence quantum yield
- RMSF
root mean square fluctuation
References
- 1.Vieille C, Zeikus GJ. Hyperthermophilic Enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varley PG, Pain RH. Relation between stability, dynamics and enzyme activity in 3-phosphoglycerate kinase from yeast and Thermus thermophilus. J Mol Biol. 1991;220:531–538. doi: 10.1016/0022-2836(91)90028-5. [DOI] [PubMed] [Google Scholar]
- 3.Brown SH, Sjolholm C, Kelley RM. Purification and characterization of a highly stable glucose isomerase produced by the extremely thermophilic eubacterium, Thermotoga maritima. Biotechnol Bioeng. 1993;41:878–886. doi: 10.1002/bit.260410907. [DOI] [PubMed] [Google Scholar]
- 4.Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 5.Alexandrov VY. Cells, Molecules, and Temperature: Conformational Flexibility of Macromolecules and Ecological Adaptation. New York: Springer-Verlag; 1977. [Google Scholar]
- 6.Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem. 1991;202:715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- 7.Jaenicke R, Böhm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 8.Vihinen M. Relationship of protein flexibility to thermostability. Protein Eng. 1987;1:477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- 9.Somero GN. Temperature adaptation of enzymes: Biological optimization through structure-function compromises. Annual Review of Ecological Systems. 1978;9:1–29. [Google Scholar]
- 10.Tehei M, Franzetti B, Madern D, Ginzberg M, Ginzberg BZ, Giudici-Orticoni M-T, Bruschi M, Zaccai G. Adaptation to extreme environments: macromolecular dynamics in bacteria compared in vivo by neutron scattering. EMBO Reports. 2004;5:66–70. doi: 10.1038/sj.embor.7400049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrba A, Schweiger A, Schultes V, Jaenicke R, Zavodsky P. Extremely thermostable D-glyceraldehyde-3-phosphate dehydrogenase from the eubacterium Thermotoga maritima. Biochemistry (Moscow) 1990;29:7584–7592. doi: 10.1021/bi00485a007. [DOI] [PubMed] [Google Scholar]
- 12.Zavodsky P, Kardos J, Svingor A, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci U S A. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohen A, Klinman JP. Protein Flexibility correlates with degree of hydrogen tunneling in thermophilic and mesophilic alcohol dehydrogenases. J Am Chem Soc. 2000;122:10738–10739. [Google Scholar]
- 14.Hernandez G, Jenney FE, Adams MWW, LeMaster DM. Millisecond time scale conformational flexibility in a hyperthermophile protein at ambient temperature. Proc Natl Acad Sci U S A. 2000;97:3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinhold L, Clement D, Tehei M, Daniel R, Finney JL, Smith JC. Protein dynamics and stability: The distribution of atomic fluctuations in thermophilic and mesophilic dihydrofolate reductase derived using elastic incoherent neutron scattering. Biophys J. 2008;94:4812–4818. doi: 10.1529/biophysj.107.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkley ED, Parson WW, Daggett V. Temperature dependence of the flexibility of thermophilic and mesophilic members of the nitroreductase fold family. Prot Eng Des Sel. 2010;23:327–336. doi: 10.1093/protein/gzp090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park HJ, Reiser COA, Kondruweit S, Erdmann H, Schmid RD, Sprinzl M. Purification and characterization of a NADH oxidase from the thermophile Thermus thermophilus Hb8. Eur J Biochem. 1992;205:881–885. doi: 10.1111/j.1432-1033.1992.tb16853.x. [DOI] [PubMed] [Google Scholar]
- 18.Žoldák G, Musatov A, Stupak M, Sprinzo M, Sedlák E. pH-induced changes in activity and conformation of NADH oxidase from Thermus thermophilus. Gen Physiol Biophys. 2005;24:279–298. [PubMed] [Google Scholar]
- 19.Žoldák G, Sprinzl M, Sedlák E. Modulation of activity of NADH oxidase from Thermus thermophilus through change in flexibility in the enzyme active site induced by Hofmeister series anions. Eur J Biochem. 2004;271:48–57. doi: 10.1046/j.1432-1033.2003.03900.x. [DOI] [PubMed] [Google Scholar]
- 20.Toth K, Sedlák E, Sprinzl M, Žoldák G. Flexibility and enzyme activity of NADH oxidase from Thermus thermophilus in the presence of monovalent cations of Hofmeister series. Biochim Biophys Acta. 2008;1784:789–795. doi: 10.1016/j.bbapap.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Žoldák G, Šut'ák R, Antalik M, Sprinzl M, Sedlák E. Role of conformational flexibility for enzymatic activity in NADH oxidase from Thermus thermophilus. Eur J Biochem. 2003;270:4887–4897. doi: 10.1046/j.1432-1033.2003.03889.x. [DOI] [PubMed] [Google Scholar]
- 22.Hritz J, Žoldák G, Sedlák E. Cofactor assisted gating mechanism in the active site of NADH oxidase from Thermus thermophilus. Proteins-Structure Function And Bioinformatics. 2006;64:465–476. doi: 10.1002/prot.20990. [DOI] [PubMed] [Google Scholar]
- 23.Beck DAC, Alonso DOV, Daggett V. ilmm, in lucem molecular mechanics. Seattle: University of Washington; 2000–2008. [Google Scholar]
- 24.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput Phys Commun. 1995;91:215–231. [Google Scholar]
- 25.Levitt M, Hirshberg M, Sharon R, Laidig K, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. J Phys Chem B. 1997;101:5051–5056. [Google Scholar]
- 26.Beck DAC, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Hecht HJ, Erdmann H, Park HJ, Sprinzl M, Schmid RD. Crystal structure of NADH oxidase from Thermus thermophilus. Nat Struct Biol. 1995;2:1109–1114. doi: 10.1038/nsb1295-1109. [DOI] [PubMed] [Google Scholar]
- 28.Park HJ, Kreutzer R, Reiser COA, Sprinzl M. Molecular cloning and nucleotide sequence of the gene encoding a H2O2-forming NADH oxidase from the extreme thermophilic Thermus thermophilus Hb8 and its expression in Escherichia coli. Eur J Biochem. 1992;205:875–879. doi: 10.1111/j.1432-1033.1992.tb16852.x. [DOI] [PubMed] [Google Scholar]
- 29.Gardecki JA, Maroncelli M. Set of secondary emission standards for calibration of the spectral responsivity in emission spectroscopy. Applied Spectroscopy. 1998;52:1179–1189. [Google Scholar]
- 30.Chen RF. Fluorescence quantum yields of tryptophan and tyrosine. Analytical Letters. 1967;1:35–42. [Google Scholar]
- 31.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
- 32.Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov.: A Nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–112. [Google Scholar]
- 33.van den Berg PAW, van Hoek A, Walentas CD, Perham RN, Visser AJWG. Flavin fluorescence dynamics and photoinduced electron transfer in Escherichia coli glutathione reductase. Biophys J. 1998;74:2046–2058. doi: 10.1016/S0006-3495(98)77911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mataga N, Chosrowjan H, Shibata T, Tanaka F. Ultrafast fluorescence quenching dynamics of flavin chromophores in protein nanospace. Journal of physical Chemistry B. 1998;102:7081–7084. [Google Scholar]
- 35.Mataga N, Chosrowjan T, Tanaka F, Nishina Y, Shiga K. Dynamics and mechanisms of ultrafast fluorescence quenching reactions of flavin chromophores in protein nanospace. Journal of Physical Chemistry B. 2000;104:10667–10677. [Google Scholar]
- 36.Mataga N, Chosrowjan H, Tanaka F, Taniguchi S. Femtosecond fluorescence dynamics of flavoproteins: Comparative studies on flavodoxin, Its site-directed mutants, and riboflavin binding protein regarding ultrafast electron transfer in protein nanospaces. J Phys Chem. 2002;106:8917–8920. [Google Scholar]
- 37.Yang H, Luo G, Karnchanaphanurach P, Louie T-M, Rech I, Cova S, Xun L, Xie XS. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 38.Callis PR, Liu TQ. Short range photoinduced electron transfer in proteins: QM-MM simulations of tryptophan and flavin fluorescence quenching in proteins. Chemical Physics. 2006;326:230–239. [Google Scholar]
- 39.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. The Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 40.Page C, Moser CC, Chen XX, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 41.Lovering AL, Hyde EI, Searle PF, White SA. The structure of Escherichia coli nitroreductase complexed with nicotinic acid: Three crystal forms at 1.7 Å, 1.8 Å and 2.4 Åresolution. J Mol Biol. 2001;309:203–213. doi: 10.1006/jmbi.2001.4653. [DOI] [PubMed] [Google Scholar]
- 42.Haynes CA, Koder RL, Miller A-F, Rodgers DW. Structures of nitroreductase in three states. J Biol Chem. 2002;277:11513–11520. doi: 10.1074/jbc.M111334200. [DOI] [PubMed] [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38.. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]; Day R, Daggett V. Direct observation of microscopic reversibility in single-molecule protein unfolding. J Mol Biol. 2007;366:677–686. doi: 10.1016/j.jmb.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merkley ED, Bernard B, Daggett V. Conformational changes below the Tm: Molecular dynamics studies of the thermal pretransition of ribonuclease A. Biochemistry. 2008;47:880–892. doi: 10.1021/bi701565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCully ME, Beck DA, Daggett V. Microscopic reversibility of protein folding in molecular dynamics simulations of the engrailed homeodomain. Biochemistry. 2008;47:7079–7089. doi: 10.1021/bi800118b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.