Abstract
The folding of nascent secretory and membrane proteins is monitored by the endoplasmic reticulum (ER) quality control system. Misfolded proteins are retained in the ER and can be removed by ER-associated degradation. As a model for the ER quality control of multispanning membrane proteins in yeast, we have been studying mutant forms of Ste6p. Here, we identify mislocalized mutant forms of Ste6p that induce the formation of, and localize to, prominent structures that are absent in normal cells. We have named these structures ER-associated compartments (ERACs), based on their juxtaposition to and connection with the ER, as observed by fluorescence and electron microscopy. ERACs comprise a network of tubulo-vesicular structures that seem to represent proliferated ER membranes. Resident ER lumenal and membrane proteins are present in ERACs in addition to their normal ER localization, suggesting there is no barrier for their entry into ERACs. However, the forms of Ste6p in ERACs are excluded from the ER and do not enter the secretory pathway; instead, they are ultimately targeted for ER-associated degradation. The presence of ERACs does not adversely affect secretory protein traffic through the ER and does not lead to induction of the unfolded protein response. We propose that ERACs may be holding sites to which misfolded membrane proteins are specifically diverted so as not to interfere with normal cellular functions. We discuss the likelihood that related ER membrane proliferations that form in response to certain other mutant or unassembled membrane proteins may be substantially similar to ERACs.
INTRODUCTION
Proteins that traffic via the exocytic pathway are translocated into the membrane or lumen of the ER and are then transported to the Golgi complex where sorting to a variety of cellular locations occurs. In addition to specific sequences that promote exit from the ER and delivery to the Golgi (reviewed in Barlowe, 2003), it is clear that for successful vesicular transport, proteins need to be properly folded and in some cases oligomerized. The surveillance system that detects improper folding has been referred to as “ER quality control” and is present in all eukaryotes, including yeast (reviewed in Ellgaard and Helenius, 2003; Kostova and Wolf, 2003). Misfolded proteins are retained in the ER and in certain cases sequestered; however, the mechanisms involved are only partially understood. Proteins that cannot fold properly are generally thought to be retrotranslocated from the ER and subjected to ER-associated degradation (ERAD) by the ubiquitin-proteasome system (reviewed in Hampton, 2002; Jarosch et al., 2003).
Although some ER quality control substrates are simply retained in the ER, others induce and localize to proliferated extensions of the ER. Examples of these ER proliferations include structures containing aggregates of misfolded lumenal proteins, called Russell bodies (Valetti et al., 1991; Umebayashi et al., 1997); proliferations of the ER-Golgi intermediate compartment (ERGIC) or vesicular-tubular clusters (VTCs) (Raposo et al., 1995); expanded ER exit sites, such as BiP bodies (Nishikawa et al., 1994) and related structures (Hobman et al., 1992, 1998; Kamhi-Nesher et al., 2001; Ferreira et al., 2002); and stacked cisternae, such as karmellae (Wright et al., 1988) or other multilayered structures (Elgersma et al., 1997; Zimmer et al., 1997; Becker et al., 1999). In some cases, mutant proteins that localize to these membrane proliferations may eventually fold properly and regain access to the normal secretory pathway. However, in most cases the fate of these proteins is unclear, and the nature of the membrane proliferations is poorly understood.
We are studying Ste6p as a model substrate for the quality control of multispanning membrane proteins in yeast. Ste6p, the a-factor pheromone transporter and a member of the ATP binding cassette (ABC) superfamily, is comprised of two homologous halves, each with six transmembrane spans and a cytosolic nucleotide binding domain. Ste6p traffics via the secretory pathway to the plasma membrane, undergoes rapid endocytosis and is delivered to the vacuole where it is degraded (Loayza and Michaelis, 1998; Shaw et al., 2001; Kelm et al., 2004). We have previously shown that mutant forms of Ste6p are subject to ER quality control, resulting in their retention in the ER and degradation by the ubiquitin-proteasome machinery with widely varying turnover rates (Loayza et al., 1998). Because the bulk of Ste6p is cytosolic, it is likely that machinery on the cytosolic face of the ER play an important role in its quality control, although recognition of ER lumenal and intramembrane regions could also be involved.
In this study, we have extended our previous mutant screen and identified additional mislocalized Ste6p mutant proteins. Interestingly, we find that several mutant proteins induce the formation of, and localize to, striking cellular structures designated ER-associated compartments (ERACs). Although the mutant Ste6p proteins in ERACs are excluded from the normal ER, resident ER proteins are present both in ERACs and the ER. By electron microscopy, ERACs seem to be comprised of a network of tubulo-vesicular structures that are directly connected to the ER. However, despite this physical connection, the mutant forms of Ste6p in ERACs do not enter the normal secretory pathway and instead are degraded by ERAD. The presence of ERACs does not adversely affect secretory protein traffic through the ER and does not lead to induction of the unfolded protein response. We propose that ERACs represent an important quality control compartment or subdomain of the ER to which mutant proteins are diverted, functioning as a “holding site” to protect the cells from the harmful accumulation of these proteins. Similarities and differences between ERACs and ER membrane proliferations induced by other mutant proteins are discussed.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
The yeast strains used in this study are listed in Table 1. Unless stated otherwise, all strains are isogenic to SM1058 (formerly designated EG123; Michaelis and Herskowitz, 1988). Plate and liquid complete, drop-out, or minimal media were prepared as described previously (Michaelis and Herskowitz, 1988; Kaiser et al., 1994). Yeast strains and cultures were grown at 30°C except where indicated.
Table 1.
Yeast strains used in this study
| Strain | Relevant genotypea | Reference/Source |
|---|---|---|
| SM1058 | MATatrp1 leu2 ura3 his4 can1 | Michaelis and Herskowitz, 1988 |
| SM1068 | MAT α lys1 | Michaelis and Herskowitz, 1988 |
| SM1646 | MATatrp1 leu2 ura3 his4 can1 ste6-Δ2::URA3 | Berkower and Michaelis, 1991 |
| SM1934 | MATaade2 leu2,3-112 his3 trp1 ura3 pep4::TRP1 | R. Schekman |
| SM2187 | MATaend4tsura3 his4 leu2 bar1-1 | Raths et al., 1993 |
| SM2544 | MATatrp1 leu2 ura3 his4 can1 ste6-Δ4 | Loayza et al., 1998 |
| SM2721 | MATatrp1 leu2 ura3 his4 can1 ste6-Δ5 | Berkower et al., 1996 |
| SM3205 | SM2721 [2μ LEU2 ste6-G414R::HAc] | Transformant of SM2721 with pSM1129 |
| SM3206 | SM2721 [2μ LEU2 ste6-G1092V::HAc] | Transformant of SM2721 with pSM1130 |
| SM3207 | SM2721 [2μ LEU2 ste6-L1239X::HAe] | Transformant of SM2721 with pSM1131 |
| SM3208 | SM2721 [2μ LEU2 ste6-G132R::HAc] | Transformant of SM2721 with pSM1132 |
| SM3209 | SM2721 [2μ LEU2 ste6-T1101R::HAc] | Transformant of SM2721 with pSM1133 |
| SM3210 | SM2721 [2μ LEU2 ste6-G38D::HAc] | Transformant of SM2721 with pSM1134 |
| SM3220 | SM2721 [2μ LEU2 STE6::HAc] | Transformant of SM2721 with pSM500 |
| SM3245 | SM1058 [2μ URA3 CFTR::HA] | Transformant of SM1058 with pSM1152 |
| SM3302 | SM2721 [2μ URA3 MDR1::HA] | Transformant of SM2721 with pSM812 |
| SM3317 | SM2544 [2μ URA3 ste6-Q1249X::HAe] | Transformant of SM2544 with pSM1082 |
| SM3897 | SM2721 [2μ URA3 STE6::GFPc] | Transformant of SM2721 with pSM1493 |
| SM3898 | SM2721 [2μ URA3 ste6-G38D::GFPc] | Transformant of SM2721 with pSM1508 |
| SM3863 | SM2187 [2μ URA3 STE6::GFPc] | Transformant of SM2187 with pSM1493 |
| SM3954 | SM2187 [2μ URA3 ste6-G38D::GFPc] | Transformant of SM2187 with pSM1508 |
| SM4031 | SM1934 [2μ URA3 STE6::GFPc] | Transformant of SM1934 with SM1493 |
| SM3966 | SM2721 [2μ LEU2 STE6::HAe] [2μ URA3 STE14] | Transformant of SM2721 with pSM694 and pSM1317 |
| SM3967 | SM2721 [2μ LEU2 ste6-L1239X::HAe] [2μ URA3 STE14] | Transformant of SM2721 with pSM1131 and pSM1317 |
| SM4031 | SM1934 [2μ URA3 STE6::GFPc] | Transformant of SM1934 with pSM1493 |
| SM4207 | SM2721 [CEN URA3 SEC63::GFP] [2μ LEU2 STE6::HAe] | Transformant of SM2721 with pSM1462 and pSM694 |
| SM4208 | SM2721 [CEN URA3 SEC63::GFP] [2μ LEU2 ste6-L1239X::HAe] | Transformant of SM2721 with pSM1462 and pSM1131 |
| SM4209 | SM2721 [CEN URA3 SEC63::GFP] [2μ LEU2 ste6-G38D::HAc] | Transformant of SM2721 with pSM1462 and pSM1134 |
| SM4213 | SM2721 [2μ URA3 ste6-G38D::GFPc] [2μ LEU2 STE14] | Transformant of SM2721 with pSM1508 and pSM1356 |
| SM4255 | MATaura3-52 leu2-3,112 ste6-Δ5 | This study |
| SM4256 | SM2721 [2μ LEU2 STE6::HAe] [2μ URA3 ste6-G38D::GFPc] | Transformant of SM2721 with pSM694 and pSM1508 |
| SM4460 | MATahis3 leu2 met15 ura3 | Invitrogen, Carlsbad, CA |
| SM4817 | MATahis3 leu2 met15 ura3 pep4::kanMX | Invitrogen, Carlsbad, CA |
| SM4821 | MATahis3 leu2 met15 ura3 ubc7::kanMX | Invitrogen, Carlsbad, CA |
| SM4922 | SM4460 [2μ LEU2 STE6::HAe] | Transformant of SM4460 with pSM694 |
| SM4923 | SM4460 [2μ LEU2 ste6-L1239X::HAe] | Transformant of SM4460 with pSM1131 |
| SM4924 | SM4460 [2μ LEU2 ste6-G38D::HAe] | Transformant of SM4460 with pSM1134 |
| SM4925 | SM4817 [2μ LEU2 STE6::HAe] | Transformant of SM4817 with pSM694 |
| SM4926 | SM4817 [2μ LEU2 ste6-L1239X::HAe] | Transformant of SM4817 with pSM1131 |
| SM4927 | SM4817 [2μ LEU2 ste6-G38D::HAe] | Transformant of SM4817 with pSM1134 |
| SM4928 | SM4821 [2μ LEU2 STE6::HAe] | Transformant of SM4821 with pSM694 |
| SM4929 | SM4821 [2μ LEU2 ste6-L1239X::HAe] | Transformant of SM4821 with pSM1131 |
| SM4930 | SM4821 [2μ LEU2 ste6-G38D::HAe] | Transformant of SM4821 with pSM1134 |
| SM4933 | SM4255 [CEN URA3 Pgal] [CEN LEU2 4 X UPRE-lacZ] | Transformant of SM4255 with pSM640 and pSM1408 |
| SM4934 | SM4255 [CEN URA3 Pgal STE6::HAe::GFPc] [CEN LEU2 4 X UPRE-lacZ] | Transformant of SM4255 with pSM1503 and pSM1408 |
| SM4935 | SM4255 [CEN URA3 Pgal ste6-G38D::GFPc] [CEN LEU2 4 X UPRE-lacZ] | Transformant of SM4255 with pSM1512 and pSM1408 |
HAc or GFPc (c for C-term) and HAe (e for ecto) refer to the presence of the epitope at the C terminus and in the first extracellular loop of Ste6p, respectively.
Plasmid Constructions
Plasmids used in this study are listed in Table 2. Yeast transformations were performed by the method of Elble (1992). Plasmid manipulations were performed in the Escherichia coli strains DH5α (Hanahan, 1983) and MH1 (Hall et al., 1984) by using standard media and techniques. The multicopy vectors (2μ URA3 and 2μ LEU2) used as the backbone for many of the plasmids constructed here have been described previously (Christianson et al., 1992; Chen et al., 1997).
Table 2.
Plasmids used in this study
| Plasmid | Relevant genotypea | Reference/Source |
|---|---|---|
| pSM500 | 2μ LEU2 STE6::HAc | Paddon et al., 1996 |
| pSM640 | CEN URA3 Pgal | P. Hieter |
| pSM694 | 2μ LEU2 STE6::HAe | This laboratory |
| pSM812 | 2μ URA3 MDR1::HA | This study |
| pSM1082 | 2μ URA3 ste6-Q1249X::HAe | Loayza et al., 1998 |
| pSM1129 | 2μ LEU2 ste6-G414R::HAc | This study |
| pSM1130 | 2μ LEU2 ste6-G1092V::HAc | This study |
| pSM1131 | 2μ LEU2 ste6-L1239X::HAe | This study |
| pSM1132 | 2μ LEU2 ste6-G132R::HAc | This study |
| pSM1133 | 2μ LEU2 ste6-T1101R::HAc | This study |
| pSM1134 | 2μ LEU2 ste6-G38D::HAc | This study |
| pSM1135 | 2μ LEU2 ste6-G397D::HAc | This study |
| pSM1152 | 2μ URA3 CFTR::HA | Zhang et al., 2001a |
| pSM1317 | 2μ URA3 STE14 | Romano et al., 1998 |
| pSM1356 | 2μ LEU2 STE14 | This laboratory |
| pSM1408 | CEN LEU2 4 X UPRE-lacZ | Sidrauski et al., 1996 |
| pSM1462 | CEN URA3 SEC63::GFP | Prinz et al., 2000 |
| pSM1493 | 2μ URA3 STE6::GFPc | This study |
| pSM1503 | CEN URA3 PgalSTE6::HAe::GFPc | This study |
| pSM1508 | 2μ URA3 ste6-G38D::GFPc | This study |
| pSM1512 | CEN URA3 Pgalste6-G38D::GFPc | This study |
HAc or GFPc (c for C-term) and HAe (e for ecto) refer to the presence of the epitope at the C terminus and in the first extracellular loop of Ste6p, respectively.
Plasmid pSM500 (2μ LEU2 STE6::HAc) contains the 120-base pair triply-iterated hemagglutinin epitope (triple-HA) at the C terminus of Ste6p (HAc), immediately preceding the termination codon (Paddon et al., 1996), and was used in the creation of novel ste6 mutants by chemical mutagenesis (see below). To avoid extraneous mutations, sequenced portions of ste6 containing the mutations were subcloned into wild-type STE6 vectors. Plasmids pSM1130 (ste6-G1092V::HAc) and pSM1133 (ste6-T1101R::HAc) were constructed by ligating 1.8-kb NcoI-HindIII fragments into pSM500 digested with the same enzymes, whereas for pSM1134 (ste6-G38D::HAc), a 2.5-kb SphI-PmlI fragment was subcloned into pSM500. For pSM1131 (ste6-L1239X::HAe), a 1.5-kb Bsu36I-NotI fragment was subcloned into pSM694 (2μ LEU2 STE6:: HAe). This plasmid contains the triple-HA epitope in the first extracellular loop (HAe) of Ste6p, between amino acids 68 and 69, as the C-terminal HA tag could not be used due to the premature truncation. Plasmids pSM1129 (ste6-G414R::HAc) and pSM1135 (ste6-G397D::HAc) were constructed by recombinational cloning (Oldenburg et al., 1997), in which 1.3-kb EcoRI-PflMI fragments and PmlI-linearized pSM500 were cotransformed into SM2721 (ste6Δ), and LEU+ transformants were selected and screened for those with the original mutant phenotype. Recombinational cloning was also used to construct pSM1132 (ste6-G132R::HAc) by cotransforming a 1.3-kb BglI-EcoRI fragment and StuI-linearized pSM500.
Plasmid pSM812 (2μ URA3 MDR1::HA) was constructed from pMDR2000XSG (Gottesman, National Institutes of Health; Kioka et al., 1989), which contains the human MDR1 coding sequence. HhaI sites at base -10 and before the termination codon of MDR1 were introduced by site-directed mutagenesis. The 4-kb HhaI fragment, consisting of the MDR1 coding sequence, was cloned into pSM688 (CEN LEU2 STE6) by using linkers, replacing the STE6 ORF and retaining a unique BamHI site immediately before the stop codon into which the triple-HA epitope was inserted. The entire construct, consisting of the STE6 promoter, the MDR1 coding sequence, the triple-HA epitope, and the STE6 3′ untranslated region, was then subcloned as a 6-kb SalI-NotI fragment into pSM217 (2μ URA3).
A green fluorescent protein (GFP)-tagged version of STE6 was constructed by recombinational cloning, by using a PCR product containing the coding sequence of GFP amplified from pQBI25 (Quantum Biotechnologies, Inc, Montreal, QC, Canada) and BamHI-digested pSM835 (2μ URA3 STE6::HAc). The resulting plasmid, pSM1493 (2μ URA3 STE6::GFPc), contains GFP fused immediately before the stop codon with BamHI sites flanking the GFP tag. Ste6p-GFP complements a ste6Δ mutation, showing that the GFP tag does not interfere with the normal function or trafficking of Ste6p; furthermore, the fluorescence localization pattern of wild-type Ste6p-GFP is identical to the indirect immunofluorescence patterns of its HA-tagged counterpart, and Ste6p-GFP is turned over at the same rate as Ste6p-HA (Mason, 2002). The plasmid pSM1508 (2μ URA3 ste6-G38D::GFPc) was constructed by recombinational cloning, with an EcoRI-BglI restriction fragment (containing the G38D mutation) from pSM1204 (2μ URA3 ste6-G38D::HAc) and AatII-linearized pSM1493.
To place STE6 under control of the GAL1 promoter, a HindIII fragment from pSM650 (CEN LEU2 STE6::HAe) encoding the complete STE6 ORF and ∼1 kb of 3′ untranslated region was cloned into the HindIII site of pSM640, creating pSM765. The GFP coding sequence was then fused to the 3′ end of the STE6 coding sequence by recombinational cloning between a SnaBI-HindIII fragment from pSM1493 and Bsu36I-digested pSM765, generating pSM1503. To introduce the G38D mutation into STE6, pSM1503 was digested with AatII and recombined with a SwaI-EcoRI fragment from pSM1134, generating pSM1512.
Mutant Isolation and Screen
To find new loss-of-function mutants of Ste6p, we mutagenized pSM500 (2μ LEU2 STE6::HAc) in vitro with hydroxylamine as described previously (Kaiser et al., 1994; Loayza et al., 1998). The mutagenized plasmid population was transformed into the ste6Δ strain SM1646, and transformants (∼2500) were screened for their mating capacity with the mating tester strain SM1068 by a colony replica mating assay. Twenty-two transformants were identified whose mating efficiency was <2% of wild-type at 37°C; all but two were also defective at 30°C (see below). Plasmids were isolated from the 22 transformants, and the ste6 mutation was mapped by gap repair and sequenced. Sixteen of the ste6 mutants contained premature translation termination codons and were not studied further, except for L1239X, which encodes the longest nonsense fragment (truncated 52 amino acids from the C terminus of Ste6p). Mutants T1101R and G414R, although defective for mating at 37°C, were capable of significant mating at 30°C (32% and 41% mating efficiency, respectively, compared with wild-type Ste6p). Interestingly, most of the nonsense mutants were still capable of a low level of mating (∼0.01% mating, as detected by papillation on mating plates), which is likely due to a small amount of readthrough of nonsense codons (although we did not detect any full-length Ste6p by metabolic labeling; our unpublished data), as has been observed previously for other ste6 nonsense mutants (Fearon et al., 1994). The remaining six missense mutants, together with L1239X, were analyzed in detail by microscopy and pulse-chase labeling, as described below. Several of these missense mutations overlap with those found in a separate study (Proff and Kolling, 2001).
Fluorescence and Electron Microscopy
For indirect immunofluorescence, cells were prepared and visualized as described previously (Loayza and Michaelis, 1998; Nijbroek and Michaelis, 1998). Antibodies were used at the following dilutions: anti-hemagglutinin (HA) (1:10,000), Ste14p (1:2,000), Kar2p (1:5,000), and Pma1p (1:200). The mouse anti-HA monoclonal antibody 12CA5 was purchased from Babco (Richmond, CA). The rabbit anti-Ste14p antibody has been described previously (Romano et al., 1998). The rabbit polyclonal anti-Kar2p and Pma1p antibodies were gifts from M. Rose (Princeton University, Princeton, NJ) and C. Slayman (Yale University, New Haven, CT), respectively. Fluorescent-conjugated secondary antibodies were purchased from Roche Diagnostics (Indianapolis, IN) (rhodamine-conjugated goat anti-mouse and fluorescein isothiocyanate [FITC]-conjugated goat anti-rabbit), and Jackson ImmunoResearch Laboratories (West Grove, PA) (Cy3-conjugated goat anti-mouse).
Cells were examined at 100× magnification on poly-lysine-coated slides using an Axioskop microscope equipped with fluorescence and Nomarski optics (Carl Zeiss, Thornwood, NY). Images were captured with a Cooke charge-coupled device camera and IP Lab Spectrum Software (Biovision Technologies, Exton, PA). To visualize FITC fluorescence, excitation and emission filters of 480 nm (40-nm bandwith) and 535 nm (50-nm bandwith), respectively, were used; for rhodamine and Cy3, excitation and emission filters of 545 nm (30-nm bandwith) and 610 nm (75-nm bandwith), respectively, were used. No FITC signal was observed in the rhodamine/Cy3 channel, and vice versa.
To visualize green fluorescent protein (GFP)-tagged constructs by fluorescence microscopy, live log phase cells were immobilized on poly-lysine-coated slides and examined directly as described above, with excitation and emission filters of 470 nm (40-nm bandwith) and 525 nm (50-nm bandwith), respectively. For covisualization of proteins by direct GFP fluorescence and indirect immunofluorescence, cells were prepared as described above for immunofluorescence by using secondary rhodamine- or Cy3-conjugated antibodies, and visualized with the appropriate filters for rhodamine/Cy3 and GFP. No bleed-through was observed between the rhodamine/Cy3 and GFP channels.
For transmission electron microscopy (EM) (Figure 5), cells were prepared as described previously (Koning et al., 1996; Wright, 2000). Briefly, cells were chemically fixed with glutaraldehyde and potassium permanganate, stained with uranyl acetate, infiltrated with Spurr's resin, subjected to thin sectioning, and stained with Reynold's lead citrate. For additional transmission EM (Figure 6), cells were processed as described previously (Rieder et al., 1996) using a modified KFeCN-OsO4-thiocarbohydrazide (OTO) fixation. Sections were prepared on a Leica UCT ultramicrotome and examined on a Philips EM 420 transmission electron microscope.
Figure 5.
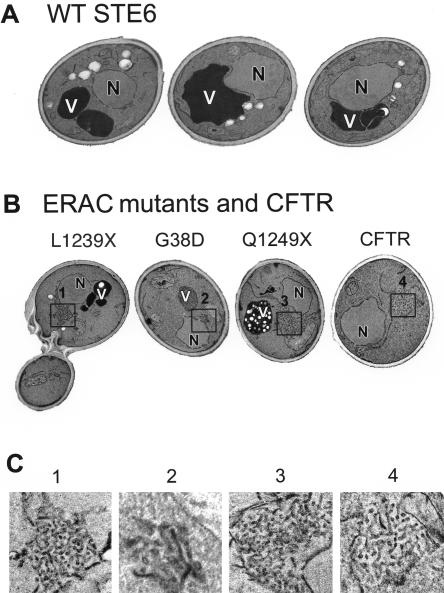
ERACs are distinctive membranous structures as visualized by transmission electron microscopy. Yeast strains were examined by transmission EM by using permanganate fixation to enhance visualization of membranes. (A) Micrographs of three cells expressing wild-type Ste6p. (B) Expression of the Ste6p mutants L1239X, G38D, and Q1249X, or CFTR induces formation of tubulo-vesicular structures that are absent in the wild-type strain. (C) Higher magnifications of the tubulo-vesicular areas boxed in B are shown. Cells were prepared for EM as described in MATERIALS AND METHODS. Strains are SM3220 (WT), SM3207 (L1239X), SM3210 (G38D), SM3317 (Q1249X), and SM3245 (CFTR). N, nucleus; V, vacuole.
Figure 6.
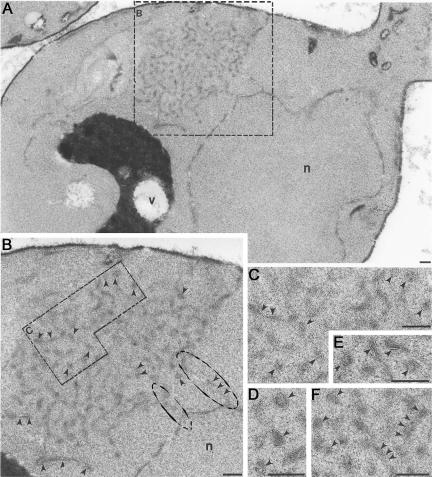
(A and B) ERACs exhibit direct connections to the ER, as detected by OTO fixation. (A) An ERAC is visible adjacent to the perinuclear ER (boxed) by transmission EM in a yeast cell overexpressing Ste6p-G38D. (B) Magnification of area boxed in A, showing direct connections between the ERAC and the ER (circled). (C-F) ERACs are membranous structures. At increased magnification, the membranous structures that comprise ERACs look like railroad tracks (indicated by arrowheads), characteristic of bilayers. C is an enlargement from B, whereas D-F are from other cells overexpressing Ste6p-G38D. Cells (strain SM3210) were prepared as described in MATERIALS AND METHODS. Bar, 0.1 μm.
For immunoelectron microscopy (Figures 7 and 8), cells were prepared as described previously (Rieder et al., 1996). Samples were labeled with mouse anti-HA (to detect Ste6p) and/or rabbit anti-Kar2p antibodies, followed by donkey anti-mouse (5 or 10 nm) and anti-rabbit (10 nm) silver conjugates.
Figure 7.
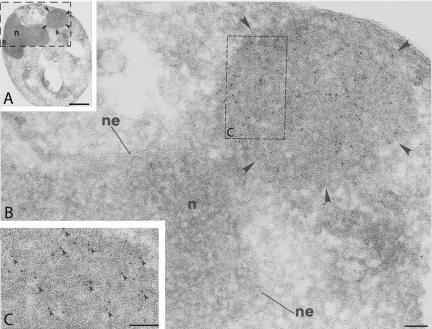
Ste6p-G38D-HA is concentrated in ERACs, as detected by indirect immunogold labeling of ultrathin cryosections. (A) Electron micrograph of a cell expressing Ste6p-G38DHA, with an ERAC marked by arrowheads (n, nucleus). (B) Higher magnification of the area boxed in A. Ste6p-G38D-HA is labeled with 10-nm gold particles and clusters in an ERAC (marked by arrowheads) adjacent to the nuclear envelope (ne)/ER region of the cell. (C) Further magnification of a portion of the ERAC boxed in B, with some of the gold particles labeling Ste6p-G38D-HA indicated by arrowheads. Cells (strain SM3210) were prepared as described in MATERIALS AND METHODS. Ste6p-G38D-HA was detected with a mouse anti-HA antibody followed by a donkey antimouse secondary antibody conjugated to 10-nm gold particles. Bar, 1 μM (A); 0.1 μm (B and C).
Figure 8.
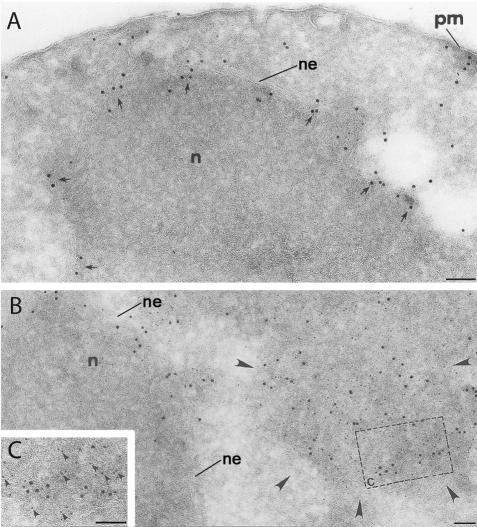
Ste6p-G38D-HA colocalizes with Kar2p in ERACs, but not in the ER, in double indirect immunogold labeling. (A) Immunogold labeling of Kar2p in a wild-type cell. Kar2p localizes to the nuclear envelope (ne)/perinuclear ER (arrows), and in some places extends out toward the plasma membrane (pm). (B) In a cell expressing Ste6p-G38D-HA, Kar2p (large gold) maintains its normal ER localization, but it is also found to cluster in an ERAC (marked by arrowheads) with Ste6p-G38D-HA (small gold), whereas Ste6p-G38D-HA is only observed in the ERAC. (C) A higher magnification of the colocalization is shown. Kar2p was labeled with 10-nm (large) gold particles, and Ste6p-G38D-HA with 5-nm (small) gold particles. Arrowheads indicate the small gold (i.e., Ste6p-G38D-HA) in proximity of the large gold (i.e., Kar2p). Cells (strain SM3210) were prepared as in Figure 7. Bar, 0.1 μm.
Metabolic Labeling and Immunoprecipitation
Cells were pulse labeled and immunoprecipitated as described previously (Loayza and Michaelis, 1998; Nijbroek and Michaelis, 1998). Briefly, log-phase cells were pulse labeled for 10 min with 30 μCi of35S-Express protein labeling mix (PerkinElmer Life Sciences, Boston, MA) per 1 OD600 unit of cells, and the label was chased with excess cold cysteine/methionine, removing 2.5 OD600 units of cells at the desired times. Wild-type and mutant forms of Ste6p, as well as CFTR and MDR1, were immunoprecipitated with anti-HA antibodies and resolved on 8% SDS-PAGE gels. Carboxypeptidase Y (CPY) was immunoprecipitated with rabbit polyclonal anti-CPY antibodies (gift of S. Emr, University of California, San Diego) and resolved on 10% SDS-PAGE gels. Proteins were visualized and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, or Bio-Rad, Hercules, CA) and Image Quant (Molecular Dynamics) or Quantity One software (Bio-Rad), and half-lives were calculated using Cricket Graph III (Islandia, NY) or KaleidaGraph (Synergy Software, Reading, PA).
UPRE-lacZ Assays
Cells were assayed for β-galactosidase expression as described previously (Guarente, 1983). β-Gal units are expressed in Miller units as 1000 × (A420)/[(tmin)(Vml) × (A600)].
RESULTS
Identification of Novel Mistrafficking ste6 Loss-of-Function Mutants
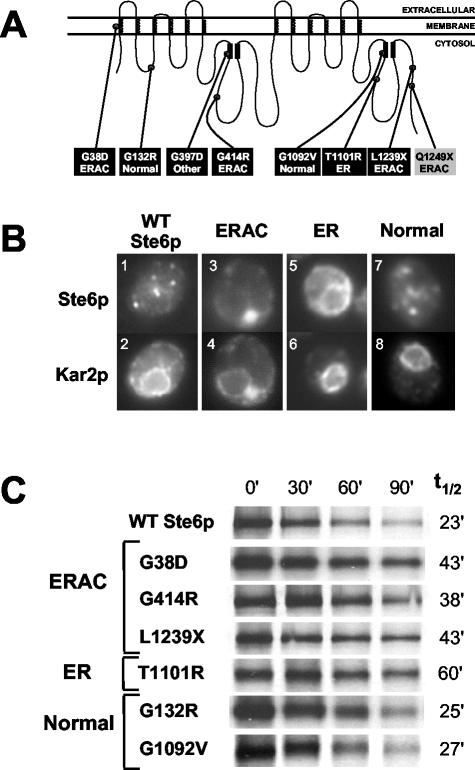
We previously identified three ste6 mutants that result in the retention of Ste6p in the ER, presumably due to misfolding (Loayza et al., 1998). These ste6 trafficking mutants [ste6-13 (A1201T, R1202I), ste6-90 (T1245M, H1246Y), and ste6-166 (Q1249X)] were identified by screening a collection of plasmid-borne ste6 loss-of-function mutants, either by immunofluorescence or by pulse-chase analysis. To identify additional mislocalized Ste6p mutants, we carried out a larger-scale mutagenesis of HA-tagged Ste6p here (described in MATERIALS AND METHODS). Of 22 loss-of-function mutants, we obtained six missense and 16 nonsense mutants. The locations of the missense mutations and the nonsense mutation encoding the longest truncation are shown in black boxes in Figure 1A. Whereas the loss-of-function mutations do not cluster in any particular region of STE6, it is of note that they are all on the cytosolic side of the protein, except for G38D, which lies in the first transmembrane span.
Figure 1.
(A) Location of mutations in Ste6p. Schematic showing the identity and positions within Ste6p of the six missense and one nonsense mutants identified in this study (black boxes). A previously identified nonsense mutant (ste6-166, Q1249X) also analyzed here is shown (gray box) (Loayza et al., 1998). The localization patterns of the mutant proteins as determined by immunofluorescence are indicated; “normal” refers to a wild-type localization, whereas “other” indicates an indeterminate localization. The coils in the schematic represent membrane spans of Ste6p and the black rectangles indicate the conserved Walker A and B motifs of the nucleotide binding-fold domains (NBDs). (B) ste6 mutants define three groups based on their localization properties. Examples of the coimmunofluorescence pattern of HA-tagged Ste6p (wild-type and mutants) and Kar2p (as a marker for the ER). Cells were costained with an anti-HA mouse antibody (top) and an anti-Kar2p rabbit antibody (bottom). The punctate localization of wild-type Ste6p (1) represents mainly endosomes, and possibly some Golgi (Kelm et al., 2004). The Ste6p mutant proteins G132R and G1092V also show this “normal” wild-type punctate localization pattern (7). The other Ste6p mutant proteins aberrantly localize to ERACs (G38D, G414R, and L1239X; 3) or the ER (T1101R; 5). Only one example from each group is shown: SM3220 (WT); SM3205 (G414R; ERAC); SM3209 (T1101R; ER); and SM3208 (G132R; normal). (C) Metabolic stability of wild-type Ste6p and Ste6p mutant proteins. Cells were pulse-labeled with35S-Met/Cys for 10 min and the label chased for the indicated times. Ste6p levels were analyzed by immunoprecipitation, SDS-PAGE, and PhosphorImager analysis as described in MATERIALS AND METHODS. Strains used are SM3220 (WT), SM3210 (G38D), SM3205 (G414R), SM3207 (L1239X), SM3209 (T1101R), SM3208 (G132R), and SM3206 (G1092V).
To ascertain whether the new missense and nonsense (L1239X) loss-of-function ste6 mutants exhibited a trafficking defect, we determined their Ste6p localization by immunofluorescence and classified them accordingly. Kar2p, a well-characterized ER lumenal chaperone protein, was used as a control for visualizing the ER. One of the mutant proteins (T1101R) was retained in the ER and displayed a perinuclear and peripheral localization coincident with Kar2p (Figure 1B, 5 and 6), similar to the mutant proteins we isolated in the original screen (Loayza et al., 1998). Interestingly, three of the mutant proteins (G38D, G414R, and L1239X) were excluded from the ER and instead localized to novel compartments adjacent to the ER (Figure 1B, 3 and 4). These structures are not normally present in cells and are only apparent when these particular mutant forms of Ste6p are expressed. We have designated these structures ERACs (ER-associated compartments) because of their close association with the ER. Coincidentally, the Ste6p-G38D mutation (and G38N) has been found in a separate screen by others for ste6 mating-defective mutants (Proff and Kolling, 2001), possibly because this codon represents a hotspot for chemical mutagenesis. In that study, the Ste6p-G38D mutant protein was described as mislocalizing to the ER, based on subcellular fractionation. However, our fractionation analysis (our unpublished data), and extensive characterization by microscopy (see below) indicated that Ste6p-G38D is in a distinct compartment from the ER.
Of the remaining mutants, two (G132R and G1092V) exhibited a localization pattern indistinguishable from wild-type Ste6p (Figure 1B, compare 7 and 8 with 1 and 2), indicative of normal trafficking. One of the mutant proteins (G397D) localized to an indeterminate region of the cell (our unpublished data) and was not characterized further. Together, of the seven mating-defective ste6 mutants analyzed here, it is notable that five encode mislocalized proteins.
We have previously shown that ER-retained mutant forms of Ste6p fall into two classes based on their metabolic stabilities, namely, hyperstable and highly unstable relative to wild-type Ste6p (Loayza et al., 1998). We therefore assessed the metabolic stability of the mutants by carrying out pulse-chase analysis (Figure 1C). The stability of the ERAC- and ER-retained mutants was similar to the previously described hyperstable mutants, and none exhibited the highly unstable phenotype of Ste6p-Q1249X (t1/2 ∼6 min) (Loayza et al., 1998).
ER Resident Proteins Are Also Present in ERACs
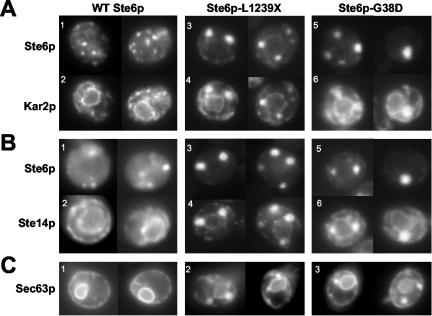
Of the Ste6p mutant proteins isolated, the ERAC-forming mutants were the most striking and novel, warranting further characterization. We chose two of the ERAC-forming mutants, Ste6p-L1239X and Ste6p-G38D, for detailed analysis by immunofluorescence and direct fluorescence microscopy. In cells expressing these mutant proteins, the resident ER chaperone Kar2p was found in ERACs as well as its normal perinuclear ER pattern (Figure 2A, compare 2 to 4 and 6; see also Figure 1B), suggesting that ERACs are directly connected to the ER and freely accessible by resident ER proteins. However, it is possible the Kar2p chaperone may be specifically recruited to ERACs because of the presence of mutant, and presumably misfolded, forms of Ste6p, although this is unlikely considering the topology of Ste6p (i.e., very little in the ER lumen). Therefore, we examined whether other ER resident proteins with nonchaperone functions are also present in ERACs. First, we analyzed the localization of the ER membrane protein Ste14p, the isoprenylcysteine methyl transferase that mediates carboxyl methylation of a-factor and Ras proteins in Saccharomyces cerevisiae (Sapperstein et al., 1994). Like Kar2p, Ste14p also localized to ERACs, in addition to its characteristic perinuclear staining pattern (Figure 2B, compare 2 to 4 and 6). A similar result was observed with the ER membrane protein Ste24p, the endoprotease that processes a-factor at the C- and N-termini (Schmidt et al., 1998) (our unpublished data). Finally, a GFP-tagged form of Sec63p, part of the ER translocon pore, was also found both in the ER and in ERACs when coexpressed with the Ste6p mutants (Figure 2C, 2 and 3), whereas only a typical ER pattern was observed when coexpressed with wild-type Ste6p (Figure 2C, 1). Together, these findings support the conclusion that ERACs are directly connected to the ER and derived from the ER membrane and that there is no barrier for entry into ERACs of both ER lumenal and membrane proteins.
Figure 2.
ER resident proteins localize to the ER and to ERACs, whereas mutant Ste6p is confined to ERACs and excluded from the ER. (A) The localization pattern of wild-type and mutant forms of Ste6p with Kar2p is shown. For HA-tagged wild-type Ste6p and Ste6p-L1239X, cells were costained with an anti-HA mouse antibody to detect Ste6p and an anti-Kar2p rabbit antibody. For GFP-tagged Ste6p-G38D, Ste6p was detected by direct fluorescence, whereas Kar2p was detected by immunofluorescence as described above. Strains used are SM3220 (1 and 2), SM3207 (3 and 4), and SM3898 (5 and 6). (B) The localization pattern of wild-type and mutant forms of Ste6p with the ER membrane protein Ste14p is shown. Cells were prepared as described above, except that an anti-Ste14p rabbit antibody was used. Strains used are SM3966 (1 and 2), SM3967 (3 and 4), and SM4213 (5 and 6). (C) The fluorescence pattern of Sec63p-GFP is shown in cells expressing wild-type, L1239X, or G38D Ste6p. Strains used are SM4207 (1), SM4208 (2), and SM4209 (3).
ERAC Formation Does Not Perturb Normal Secretory Traffic from the ER
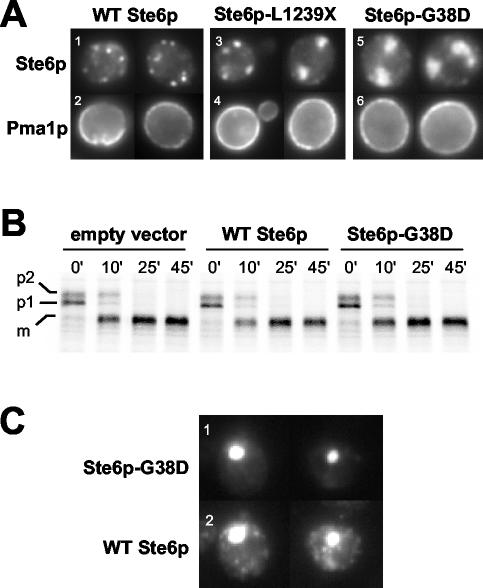
To determine whether membrane and secretory proteins destined for post-ER locations are detained in ERACs, we examined the trafficking of Pma1p (the plasma membrane ATPase) and CPY (the vacuolar carboxypeptidase Y) in strains expressing the ERAC-inducing Ste6p-L1239X and Ste6p-G38D mutant proteins. By coimmunofluorescence, no Pma1p was seen to localize to ERACs with either Ste6p mutant protein; instead, we only observed the characteristic plasma membrane staining pattern for Pma1p (Figure 3A, bottom). Thus, the steady-state localization of Pma1p is unaffected by the presence of ERACs.
Figure 3.
(A) Localization of the plasma membrane protein Pma1p is not affected in cells expressing ERAC-forming Ste6p mutant proteins. The coimmunofluorescence localization pattern of Pma1p with wild-type and mutant forms of Ste6p is shown. Cells were costained with an anti-HA mouse antibody (top) and an anti-Pma1p rabbit antibody (bottom). Strains used are SM3220 (WT), SM3207 (L1239X), and SM3210 (G38D). (B) CPY trafficking is not perturbed when ERACs are present. The trafficking of CPY was followed by pulse-chase analysis in a strain containing an empty vector and strains expressing either wild-type or G38D Ste6p-GFP under the control of a galactose-inducible promoter. Ste6p expression was induced by overnight incubation with 4% galactose, after which ERACs could be observed in ≥50% of cells in the population by fluorescence microscopy. Cells were pulse labeled with35S-Met/Cys for 10 min and the label chased for the indicated times (minutes). CPY was immunoprecipitated, subjected to SDS-PAGE, and analyzed as described in MATERIALS AND METHODS. Strains used are SM4933 (empty vector), SM4934 (WT Ste6p), and SM4935 (Ste6p-G38D). (C) Wild-type Ste6p localizes to ERACs and to endosomes when coexpressed with Ste6p-G38D. To detect wild-type Ste6p-HA, cells were stained with an anti-HA mouse primary antibody and a Cy3-conjugated anti-mouse secondary antibody. Ste6p-G38D-GFP was visualized by direct fluorescence. There is no bleed-through by the GFP signal from Ste6p-G38D into the Cy3 filter used to detect wild-type Ste6p-HA (our unpublished data). The strain used was SM4256.
To determine whether the kinetics of trafficking through the secretory pathway are affected by ERACs, we followed the maturation of CPY by performing metabolic labeling and pulse-chase analysis. To ensure that the majority of cells contained ERACs, Ste6p-G38D-GFP was expressed under the control of a galactose-inducible promoter in a CEN plasmid. An overnight induction with galactose resulted in ERACs being observed in ≥50% of cells by fluorescence microscopy (our unpublished data). The kinetics of appearance of mature (m) CPY in strains expressing Ste6p-G38D were similar to that seen in a strain expressing wild-type Ste6p (Figure 3B). Thus, the presence of ERACs did not perturb CPY trafficking. Interference with CPY trafficking at the ER (p1 precursor) or Golgi (p2 precursor) would have been observed if the formation of the ERACs significantly affected the normal ER-to-Golgi trafficking of CPY. For example, in a parallel experiment with a sec18-1 mutant (defective in ER-to-Golgi trafficking), the p1 precursor of CPY accumulated at the nonpermissive temperature, indicative of a defect in CPY trafficking (our unpublished data).
Mistrafficked mutant forms of Pma1p, including Pma1p-D378N (Harris et al., 1994) and Pma1p-G381A (Ferreira et al., 2002), have been shown to cause wild-type Pma1p to mislocalize as well, reflecting the ability of these proteins to homo-oligomerize. We asked whether mutant forms of Ste6p that induced ERAC formation had any effect on the trafficking of wild-type Ste6p. For colocalization studies, we coexpressed HA-tagged wild-type Ste6p and GFP-tagged Ste6p-G38D. Using direct fluorescence microscopy to visualize Ste6p-G38D-GFP, we observed structures indicative of ERACs (Figure 3C, 1). When wild-type Ste6p-HA was visualized in the same cells by indirect immunofluorescence with anti-HA antibodies, the wild-type Ste6p-HA protein was generally found solely in the normal punctate endosomal location. However, in ∼20% of cells examined, wild-type Ste6p-HA was also retained in ERACs, colocalized with Ste6p-G38D-GFP (Figure 3C, 2). We have previously observed interactions between full-length and partial Ste6p molecules (Berkower et al., 1996); thus, as for Pma1p, it is reasonable that the mislocalization of wild-type Ste6p is a consequence of its association with the mutant form.
Distinct Fates of Human ABC Proteins Expressed in Yeast
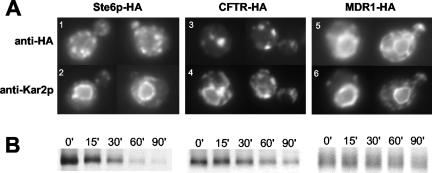
We, and others, have used yeast for high-level expression of heterologous mammalian ABC transporters (Raymond et al., 1992; Kiser et al., 2001; Zhang et al., 2001a,b). To compare the fate of two heterologously expressed ABC transporters in yeast, we examined HA-tagged forms of human CFTR and MDR1 for their localization and metabolic stability (Figure 4). These proteins, which normally reside at the plasma membrane in mammalian cells (Ambudkar et al., 1999; Gelman and Kopito, 2002), are both mislocalized in yeast. CFTR was present in several large structures adjacent to the ER that also contained Kar2p, indistinguishable from ERACs induced by the Ste6p mutant proteins G38D, G414R, and L1239X (Figure 4A, 3 and 4; see also Figure 5B). In fact, ERAC-like structures have previously been noted by us and others when expressing CFTR in yeast (Kiser et al., 2001; Zhang et al., 2001b; Fu and Sztul, 2003; Sullivan et al., 2003). MDR1, however, exhibited solely the characteristic perinuclear staining pattern of the ER and no ERACs (Figure 4A, 5 and 6), similar to the ER-retained Ste6p mutant T1101R. Both CFTR and MDR1 were expressed at levels comparable with wild-type Ste6p and were relatively stable as evidenced by pulse-chase analysis (Figure 4B).
Figure 4.
Human CFTR expressed in yeast induces the formation of and localizes to ERACs, whereas human MDR1 does not. (A) The coimmunofluorescence localization pattern of HA-tagged Ste6p, human CFTR, and human MDR1 with Kar2p is shown. Cells were costained with an anti-HA mouse antibody (top) and an anti-Kar2p rabbit antibody (bottom). (B) The metabolic stability of HA-tagged Ste6p, human CFTR, and human MDR1 was examined by metabolic pulse-chase labeling, immunoprecipitation with anti-HA antibodies, and SDS-PAGE as described in MATERIALS AND METHODS. Strains used are SM3220 (STE6), SM3245 (CFTR), and SM3302 (MDR1).
It is striking that CFTR expressed in yeast induces ERAC formation and localizes exclusively to these structures, whereas MDR1 localizes to the ER and does not cause ERACs to form. These distinct fates of mammalian ABC proteins are similar to the retention of different Ste6p mutant proteins in ERACs or the ER, suggesting specific modes of recognition and retention machinery for discrete ER quality control substrates.
ERACs Are Collections of Tubulo-vesicular Structures
To characterize ERACs further, we examined ERAC-forming mutants by transmission EM (Figure 5). In cells expressing either Ste6p-L1239X or Ste6p-G38D, extensive networks of tubulo-vesicular structures were observed in ∼55% of the cells (Figure 5, B and C, 1 and 2). These structures were absent in the wild-type strain (Figure 5A); we saw only one ERAC-containing cell among several hundred wild-type cells examined by transmission EM. Interestingly, the hyperunstable mutant Ste6p-Q1249X (Loayza et al., 1998) also formed tubulo-vesicular networks corresponding to ERACs (Figure 5, B and C, 3) in ∼35% of the cells, suggesting that ERAC formation is not simply due to the persistent overaccumulation of mutant Ste6p molecules. Finally, ERACs were observed in ∼55% of the cells when human CFTR was expressed in yeast (Figure 5, B and C, 4).
We performed additional conventional and immunoelectron microscopy analyses of ERACs in cells expressing HA-tagged Ste6p-G38D. Cells fixed with OTO showed an accumulation of membranous structures in these cells (based on the “railroad-track” appearance of the proliferated structures) (Figure 6). Direct connections between the ERACs and the ER were also readily apparent (Figure 6B, circled). Importantly, HA-tagged Ste6p-G38D localized to ERACs by indirect immunogold labeling of ultrathin cryosections (as detected by gold-labeled anti-HA antibodies) but was excluded from the ER (Figure 7), as we had observed by immunofluorescence microscopy. To confirm the restricted localization of Ste6p-G38D, we performed double indirect immunogold labeling to localize Kar2p and Ste6p-G38D in the same cells. In cells not expressing Ste6p-G38D, the lumenal ER marker Kar2p localized strictly to the perinuclear and peripheral ER (Figure 8A), whereas in cells expressing Ste6p-G38D, Kar2p was found to cluster in ERACs in addition to its normal ER localization (Figure 8, B and C, large gold particles). However, Ste6p-G38D was only observed in ERACs and not in the ER (Figure 8, B and C, small gold particles), consistent with the immunofluorescence microscopy results.
Ste6p Mutant Proteins in ERACs Do Not Enter the Secretory Pathway
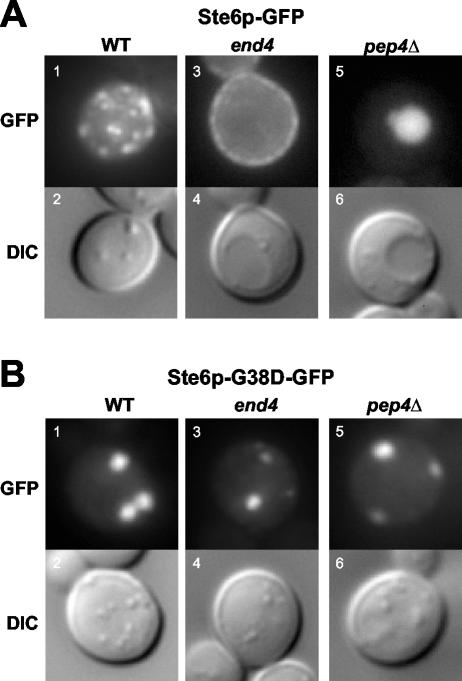
Wild-type Ste6p traffics through the ER and Golgi to the plasma membrane, is rapidly internalized to endosomes, and ultimately degraded in the vacuole. It is conceivable that the Ste6p mutant proteins that localize to ERACs still traffic like wild-type Ste6p but are simply delayed in their export from the ER, only temporarily accumulating in hypertrophied ER exit sites as has been demonstrated for other misfolded plasma membrane proteins (Ferreira et al., 2002). To examine this issue, we used the end4 mutation to prevent Ste6p internalization and vacuolar degradation, thereby trapping any Ste6p in the secretory pathway at the plasma membrane.
As expected, in contrast to the punctate (endosomal) pattern of Ste6p-GFP in wild-type cells, Ste6p-GFP accumulated at the plasma membrane in an endocytosis-defective end4 mutant at the nonpermissive temperature (Figure 9A, 1 and 3). If any Ste6p-G38D-GFP escaped from ERACs into the secretory pathway, then it too would be expected to accumulate at the plasma membrane in the end4 mutant. However, Ste6p-G38D-GFP was only observed in ERACs, whether in the wild-type or end4 strains (Figure 9B, 1 and 3). In addition, Ste6p-G38D-GFP was not found in the vacuole in a pep4Δ strain, indicating that it also does not transit from ERACs to the vacuole via a novel route (Figure 9B, 5). The lack of plasma membrane or vacuolar fluorescence for Ste6p-G38D-GFP in the end4 and pep4Δ mutants demonstrates that this mutant protein, once retained in ERACs, does not reenter the secretory pathway.
Figure 9.
Ste6p-G38D does not exit from ERACs into the secretory pathway. The fluorescence pattern and corresponding Nomarski (differential interference contrast) images are shown for cells expressing Ste6p-GFP or Ste6p-G38D-GFP in wild-type, end4, and pep4Δ strains. (A) Ste6p-GFP localizes in a punctate (endosomal) pattern in a wild-type strain (SM3897, 1 and 2), is trapped at the plasma membrane in an end4 strain (SM3863, 3 and 4), and fills the vacuole in a pep4Δ strain (SM4031, 5 and 6). (B) Ste6p-G38D-GFP localizes exclusively to ERACs in all three strains: wild-type (SM3898, 1 and 2), end4 (SM3954, 3 and 4), and pep4Δ (SM1934/pSM1508, 5 and 6).
ERACs Are Holding Sites for Ste6p Mutant Proteins before ERAD
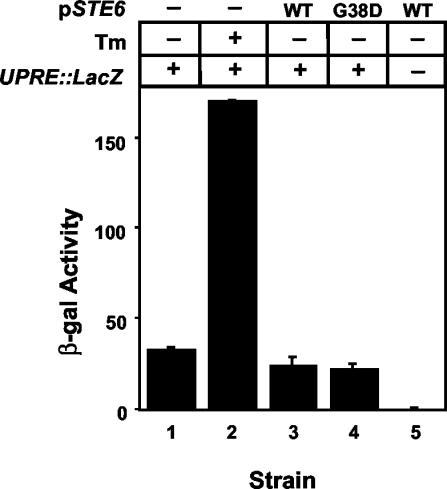
The formation of ERACs in response to Ste6p mutant proteins suggested that ERACs are specialized subdomains of the ER that form to allow the cell to cope with the presence of mutant proteins, i.e., a form of ER quality control. The accumulation of misfolded proteins in the ER generally leads to induction of the unfolded protein response (UPR) (Casagrande et al., 2000; Friedlander et al., 2000; Ng et al., 2000; Travers et al., 2000); thus, we were interested in knowing whether ERAC-forming Ste6p mutant proteins also induce the UPR. We transformed a UPRE-lacZ reporter construct into strains expressing GFP-tagged wild-type Ste6p or Ste6p-G38D under the control of a galactose-inducible promoter in a CEN plasmid. By fluorescence microscopy, ERACs were visible in ≥50% of cells expressing Ste6p-G38D after an overnight induction with galactose (our unpublished data). The amount of lacZ expression remained at the basal level regardless of whether wild-type Ste6p or Ste6p-G38D was expressed, whereas the UPR inducer tunicamycin strongly induced lacZ expression (Figure 10). Similar results were observed when Ste6p-G38D was expressed on a multicopy 2μ plasmid under its native promoter (our unpublished data). Thus, ERAC formation does not induce the UPR.
Figure 10.
The UPR is not induced by ERAC-forming Ste6p mutant proteins. Strains transformed with a UPRE-lacZ reporter construct and with either empty PGAL vector (-), PGALSTE6 (WT), or PGALste6-G38D (G38D) were induced overnight in 4% galactose and assayed for β-Gal activity. As a control, cells transformed with the empty PGAL vector were treated with the UPR inducer tunicamycin (Tm) for 6 h. Strains used were SM4933 (1 and 2), SM4934 (3), SM4935 (4), and SM4255/pSM1503 (5).
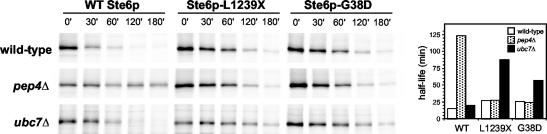
The mutant forms of Ste6p that induce ERACs are relatively stable but are still degraded, albeit relatively slowly (Figure 1C). Because the Ste6p mutant proteins in ERACs do not traffic through the secretory pathway and therefore cannot reach the vacuole, we hypothesized that they are instead degraded by the proteasome via ERAD. To test this possibility, we compared the turnover of wild-type Ste6p with Ste6p-L1239X and Ste6p-G38D in pep4Δ and ubc7Δ strains to inhibit vacuolar and proteasomal degradation, respectively. Pep4p is the master vacuolar protease, and Ubc7p is an E2 ubiquitin-conjugating enzyme that is required for ubiquitination of lumenal and membrane-associated ER proteins before ERAD (Jungmann et al., 1993). Wild-type Ste6p is degraded in the vacuole, and as expected it was strongly stabilized in the absence of Pep4p and unaffected by the absence of Ubc7p (Figure 11). By contrast, both Ste6p-L1239X and Ste6p-G38D were stabilized in the ubc7Δ strain and unaffected in the pep4Δ strain (Figure 11). This result is consistent with what we have observed with Ste6p-Q1249X, namely, that it is degraded by the ubiquitin-proteasome machinery (Loayza et al., 1998). Significantly, the lack of stabilization in the pep4Δ strain also confirms that these Ste6p mutant proteins are not escaping from ERACs into the secretory pathway and trafficking like wild-type Ste6p. Thus, ERACs represent holding sites into which the Ste6p mutant proteins are diverted away from normal secretory traffic before ultimately being degraded by ERAD.
Figure 11.
Ste6p-G38D and Ste6p-L1239X undergo ubiquitin-mediated ERAD and not Pep4p-dependent vacuolar degradation. Isogenic wild-type, ubc7Δ, and pep4Δ strains overexpressing HA-tagged wild-type, L1239X, or G38D Ste6p were metabolically labeled and analyzed for Ste6p levels by immunoprecipitation and SDS-PAGE as described in MATERIALS AND METHODS. The bar graph shows the half-lives of Ste6p in the indicated strain backgrounds, calculated as described in MATERIALS AND METHODS. Strains used are wild-type: SM4922, SM4923, SM4924; pep4Δ: SM4925, SM4926, SM4927; and ubc7Δ: SM4928, SM4929, and SM4930.
DISCUSSION
ERACs: A Quality Control Subcompartment of the ER
An Overview of ERACs. Our studies of mislocalized Ste6p mutant proteins have revealed that a striking subcompartment of the ER is specifically induced in response to certain mutant proteins. We refer to this subcompartment as an ERAC (ER-associated compartment); generally, we observe one to four ERACs per cell. Fluorescence and electron microscopy show that ERACs are extended proliferations of the ER, because direct connections can be visualized by EM between ERACs and the ER. Interestingly, Ste6p mutant proteins that induce ERACs are excluded from the ER per se and instead are confined to ERACs, whereas resident ER lumenal and membrane proteins are present both in ERACs and the ER. The mutant Ste6p proteins in ERACs that we examined here are not released into the secretory pathway and instead are ultimately degraded by ERAD. Thus, ERACs seem to be a quality control subcompartment of the ER, at the interface between recognition of ER quality control substrates and their sequestration and degradation.
It should be noted that ERACs are not simply an artifact of persistent accumulation of mutant Ste6p proteins, because ERACs were also observed in response to Ste6p-Q1249X. This hyperunstable mutant (t1/2 ∼6 min) has a very low steady-state level, making its visualization by fluorescence microscopy very difficult. In fact, we had originally classified Ste6p-Q1249X as being ER-retained (Loayza et al., 1998) based on our immunofluorescence microscopy of the small number of cells that exhibited visible staining; however, the EM analysis performed here (Figure 5) revealed that it in fact induces formation of ERACs. Mutations in the ERAD pathway that stabilize Ste6p-Q1249X greatly enhance its visualization by fluorescence microscopy in ERACs (our unpublished data), consistent with the functional connection between ERACs and ERAD.
Comparison of Previously Reported ER Proliferations to ERACs. ER membrane proliferations that form in response to mutant or heterologously expressed proteins have been reported in a number of studies, both in yeast and in mammalian cells. At first glance, these ER membrane proliferations all seem to be different, further reinforced by the lack of any unified nomenclature. However, upon closer examination many of the previously reported structures seem to be similar to the ERACs we have described here. For example, overexpression of yeast Sec12p, a protein involved in COPII coat formation, induces BiP bodies, punctate regions of the ER that accumulate Kar2p (Nishikawa et al., 1994). Similarly, the glucose and galactose transporters Hxt1p and Gal2p, respectively, accumulate in ER-associated punctate structures in the absence of Gsf2p, a putative folding and/or vesicle packaging factor specific for these transporters (Sherwood and Carlson, 1999). Certain mutant forms of the yeast plasma-membrane ATPase Pma1p, as well as overexpressed Pma2p, induce and localize to tubulo-vesicular extensions of the ER that bear a striking resemblance to ERACs (Supply et al., 1993; Harris et al., 1994; Ferreira et al., 2002). In mammalian cells, proliferations of the ER called Russell bodies have been described that contain aggregates of misfolded immunoglobulins (Valetti et al., 1991); similar structures were observed in yeast expressing a mutant form of RNAP-I (an aspartic protease from Rhizopus niveus) (Umebayashi et al., 1997). ERAC-like structures were also observed by Hobman et al. (1992, 1998) when expressing the rubella virus E1 glycoprotein subunit in the absence of the E2 subunit, and by Raposo et al. (1995) examining cells lacking the TAP transporter.
A common feature of almost all these structures is the presence of Kar2p/BiP; the exceptions (Valetti et al., 1991; Sullivan et al., 2003) may simply result from the high density of the ERAC-localized mutant protein making immunodetection of resident ER chaperones difficult. Some studies of mammalian cells suggest that these structures may be similar to VTCs/ERGIC, based on the colocalization of ERGIC-53 (Raposo et al., 1995), whereas others suggest that they are distinct (Hobman et al., 1998; Kamhi-Nesher et al., 2001) because β-COP and ERGIC-53 are excluded. The difficulty in clearly visualizing and describing VTCs may contribute to the conflicting reports.
Multiple Fates of Proteins Diverted to ERACs. An important difference between our findings with Ste6p mutant proteins localized to ERACs and other studies of misfolded proteins localized to ERAC-like structures is the fate of these proteins. For Ste6p mutant proteins in ERACs, it is clear that they are targeted for degradation by ERAD and do not enter the secretory pathway. However, certain other mutant proteins that localize to ERAC-like structures eventually exit into the normal secretory system. For example, Pma1p-G381A resides only transiently in the ERAC-like structures it induces, because it is eventually packaged into COPII vesicles and delivered to the plasma membrane, albeit still in a misfolded state (Ferreira et al., 2002). The authors propose that the ER membrane proliferations induced by Pma1p-G381A are expanded ER exit sites, further supported by the localization of Lst1p, a COPII coat component, to these structures. In addition, Kahmi-Nesher et al. (2001) describe a “quality-control” subcompartment of the ER in mammalian cells in which ERAD substrates accumulate when the proteasome is inhibited. This compartment contains ER-resident proteins and seems to be functionally connected to the ER, because proteins retained here are also secreted from the cell. Finally, BiP bodies in yeast (Nishikawa et al., 1994), and the ER membrane proliferations observed by Hobman et al. (1998) have also been proposed to represent expanded ER exit sites based on the localization of marker proteins.
A Working Model for ERACs. We propose that ERACs are a quality-control subcompartment of the ER that can form in response to misfolded proteins, acting as a holding site to allow the proteins additional time to fold. An unknown surveillance mechanism presumably exists to decide whether to release proteins from ERACs into the secretory pathway (e.g., Pma1p-G381A) or to target them for destruction by ERAD (e.g., our Ste6p mutant proteins). It will be revealing to examine whether Pma1p-G381A and our ERAC-forming Ste6p mutant proteins colocalize, suggesting that sorting occurs within an ERAC. Alternatively, proteins with different fates may be sorted to distinct ERACs, each destined for a particular fate (i.e., ERAD or exit into secretory system). It is interesting to note that ERACs can occupy a significant portion of cells as evident by immunofluorescence and EM, yet they do not seem to affect the general health of the cells, and their presence does not affect normal secretory traffic through the ER. Thus, ERACs seem to provide the cell with a controlled mechanism for dealing with misfolded ER membrane proteins that might otherwise have seriously negative consequences for the cell.
Other ER Membrane Proliferations and Quality Control Structures
Stacked cisternae of the ER, called karmellae, represent a separate group of ER membrane proliferations that are morphologically distinct from ERACs. These structures have been best described in yeast, resulting from overexpression of HMG-CoA reductase (Wright et al., 1988). Other proteins that induce karmellae-like structures in yeast when overexpressed include cytochrome P450 proteins (Zimmer et al., 1997), Pex15p (Elgersma et al., 1997), and the canine 180-kDa ribosome receptor (Becker et al., 1999). Although these structures share some similarities in their appearance by microscopy, it is unclear whether they are functionally related and whether they are ER quality control compartments per se.
It is also notable that under certain conditions, some ER quality control substrates can be sequestered outside of the ER, in the form of cytosolic aggregates that are not membrane-enclosed. In yeast, overexpression of catalase A causes its accumulation in electron-dense cytosolic aggregates called inclusion bodies (Binder et al., 1991). In mammalian cell lines, overexpression of CFTR seems to overwhelm the proteasome, leading to the accumulation of stable, high-molecular-weight, detergent-insoluble, multiubiquitinated forms of CFTR at distinct pericentriolar structures in the cytosol surrounded by a cage of intermediate filament protein, called aggresomes (Johnston et al., 1998). By contrast, we find that CFTR overexpressed in yeast induces and localizes to ERACs and not aggresomes, as described above.
ERACs May Provide Clues about ER Quality Control and ERAD Machinery
How Are ERACs Formed? The expression of ERAC-inducing Ste6p mutant proteins reveals an important response mechanism in yeast for dealing with the presence of aberrant ER membrane-spanning proteins. It will be of great interest to further investigate how ERACs are formed; how aberrant proteins are directed to ERACs; and how these proteins are retained in ERACs and kept out of the ER per se, despite direct continuity between ERACs and the ER that allow resident ER proteins to freely enter ERACs. A recent report shows that CFTR expressed in yeast is sequestered into ERAC-like subdomains of the ER in a sec18ts mutant, and therefore sequestration does not depend on ER-to-Golgi transport; however, in a sec23ts mutant, CFTR remains in the ER, implying a role for the Sar1p/COPII machinery in sequestration of CFTR (Fu and Sztul, 2003). We similarly observe that ERAC formation by Ste6p-G38D occurs in a sec18ts mutant and therefore does not depend on ER-to-Golgi transport; however, a sec23ts mutant has no effect on Ste6p-G38D ERAC formation, suggesting that the COPII machinery does not play a role in sorting it into ERACs (our unpublished data). Thus, the COPII machinery may actively sort some, but not all, ER-retained quality control substrates into ERACs, away from normal secretory traffic. It may be possible to fractionate and purify ERACs from yeast, allowing further characterization of these structures as was done by Hobman et al. (1998). The isolation of ERAC-specific protein components will provide further insight into how the ER quality control machinery sorts mutant and wild-type proteins to ensure normal ER-to-Golgi trafficking.
What Signals within a Protein Direct It into ERACs? It is puzzling why some Ste6p mutant proteins are retained in the ER but do not induce ERAC formation, whereas others do. Notably, in this study we also directly compared two similar wild-type human ABC proteins expressed in yeast, CFTR and MDR1: CFTR localizes to ERACs (Figure 4; see also Kiser et al., 2001; Zhang et al., 2001b; Fu and Sztul, 2003; Sullivan et al., 2003), whereas MDR1 is retained in the ER. Thus, two structurally similar heterologous wild-type proteins are recognized and handled very differently by the yeast quality control machinery. Furthermore, we have observed that both metabolically stable and unstable Ste6p mutants induce and localize to ERACs (Loayza et al., 1998; our unpublished data). Regardless of the stability of a particular Ste6p mutant, it seems to be ultimately targeted for ERAD by the ubiquitin-proteasome machinery. Why then should mutant forms of Ste6p that localize to ERACs have varying stabilities? One possibility is that subsets of ERACs associate with proteasome components. How the decision is made to divert some but not all Ste6p mutant proteins into these subsets of ERACs is unclear. Alternatively, some Ste6p mutant proteins may be more easily extracted from the ER membrane and/or degraded by the ubiquitin-proteasome system than others, because of the varied physical properties of the mutant protein itself or because of interaction(s) with regulatory machinery.
Why Is the UPR Not Induced by Mutant Forms of Ste6p? Importantly, ERAC formation does not coincide with induction of the UPR. Accumulation of certain ERAD substrates in the ER (e.g., upon chemical treatment, or by expression of CPY* or unassembled MHC class I heavy chains) has been shown to induce the UPR in yeast, demonstrating a functional connection between these processes (Casagrande et al., 2000; Friedlander et al., 2000; Ng et al., 2000; Travers et al., 2000). However, even though the mutant forms of Ste6p that induce and localize to ERACs are degraded by ERAD, the UPR is not induced. Similarly, Pma1p-G381A does not induce the UPR (Ferreira et al., 2002), nor does CFTR expressed in yeast (Zhang et al., 2001b), although both induce and localize to ERAC-like structures. The UPR is induced when misfolded proteins bind the chaperone Kar2p/BiP in the ER lumen, titrating the chaperone away from Ire1p which can then dimerize and become activated (Hampton, 2000). In the case of Ste6p, Pma1p, and CFTR, the bulk of these multispanning transmembrane proteins is cytosolic and therefore Kar2p likely does not bind to the mutant proteins. Indeed, ERAD of a Ste6p mutant (Ste6p-Q1249X) is unaffected in a kar2 mutant, whereas its degradation is inhibited when the cytosolic Hsp70-like chaperones (ssa1-4) are mutated (our unpublished data), consistent with the UPR not being induced because of the lack of involvement of Kar2p. It will be important to determine what genes, if any, are induced by the presence of ERACs and what signaling mechanism leads to their induction.
Summary
In conclusion, we have shown that prominent ER membrane proliferations called ERACs are induced in response to the expression of various Ste6p mutant proteins. Whereas ERACs are directly connected to the ER, the mutant proteins that are sequestered in ERACs are excluded from the normal ER. Proteins diverted into ERACs may eventually exit into the secretory pathway, whereas others are degraded by ERAD without ever entering the secretory pathway. ERACs represent an important quality control subcompartment of the ER, and the Ste6p mutant proteins reported here should prove to be valuable tools for dissecting the specialized machinery involved in their formation and their role in ER quality control of multispanning membrane proteins.
Acknowledgments
We thank Mark Rose, Carolyn Slayman, and Scott Emr for providing antibodies; Randy Schekman and Howard Riezman for strains; and Peter Walter, Michael Gottesman, Phil Hieter, and Pam Silver for plasmids. We also thank Gwen Phan for technical assistance; Diego Loayza, Danny Taglicht, and Julia Romano for plasmid constructions; and Doug Murphy for the use of a microscope. This work was supported by a predoctoral minority fellowship from the National Institute of General Medical Sciences to G.L.L. and a National Institutes of Health grant (GM-51508) to S.M.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0546. Article and publication date are available at www. molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0546.
References
- Ambudkar, S.V., Dey, S., Hrycyna, C.A., Ramachandra, M., Pastan, I., and Gottesman, M.M. (1999). Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39, 361-398. [DOI] [PubMed] [Google Scholar]
- Barlowe, C. (2003). Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13, 295-300. [DOI] [PubMed] [Google Scholar]
- Becker, F., Block-Alper, L., Nakamura, G., Harada, J., Wittrup, K.D., and Meyer, D.I. (1999). Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J. Cell Biol. 146, 273-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower, C., and Michaelis, S. (1991). Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 10, 3777-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower, C., Taglicht, D., and Michaelis, S. (1996). Functional and physical interactions between partial molecules of STE6, a yeast ATP-binding cassette protein. J. Biol. Chem. 271, 22983-22989. [DOI] [PubMed] [Google Scholar]
- Binder, M., Schanz, M., and Hartig, A. (1991). Vector-mediated overexpression of catalase A in the yeast Saccharomyces cerevisiae induces inclusion body formation. Eur. J. Cell Biol. 54, 305-312. [PubMed] [Google Scholar]
- Casagrande, R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zuniga, M., Brown, P.O., and Ploegh, H. (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5, 729-735. [DOI] [PubMed] [Google Scholar]
- Chen, P., Sapperstein, S.K., Choi, J.D., and Michaelis, S. (1997). Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 136, 251-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119-122. [DOI] [PubMed] [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13, 18-20. [PubMed] [Google Scholar]
- Elgersma, Y., Kwast, L., van den Berg, M., Snyder, W.B., Distel, B., Subramani, S., and Tabak, H.F. (1997). Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 16, 7326-7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- Fearon, K., McClendon, V., Bonetti, B., and Bedwell, D.M. (1994). Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J. Biol. Chem. 269, 17802-17808. [PubMed] [Google Scholar]
- Ferreira, T., Mason, A.B., Pypaert, M., Allen, K.E., and Slayman, C.W. (2002). Quality control in the yeast secretory pathway: a misfolded PMA1 H+-ATPase reveals two checkpoints. J. Biol. Chem. 277, 21027-21040. [DOI] [PubMed] [Google Scholar]
- Friedlander, R., Jarosch, E., Urban, J., Volkwein, C., and Sommer, T. (2000). A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2, 379-384. [DOI] [PubMed] [Google Scholar]
- Fu, L., and Sztul, E. (2003). Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 160, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman, M.S., and Kopito, R.R. (2002). Rescuing protein conformation: prospects for pharmacological therapy in cystic fibrosis. J. Clin. Investig. 110, 1591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L. (1983). Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101, 181-191. [DOI] [PubMed] [Google Scholar]
- Hall, M.N., Hereford, L., and Herskowitz, I. (1984). Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell 36, 1057-1065. [DOI] [PubMed] [Google Scholar]
- Hampton, R.Y. (2000). ER stress response: getting the UPR hand on misfolded proteins. Curr. Biol. 10, R518-R521. [DOI] [PubMed] [Google Scholar]
- Hampton, R.Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476-482. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557-580. [DOI] [PubMed] [Google Scholar]
- Harris, S.L., Na, S., Zhu, X., Seto-Young, D., Perlin, D.S., Teem, J.H., and Haber, J.E. (1994). Dominant lethal mutations in the plasma membrane H(+)ATPase gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91, 10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman, T.C., Woodward, L., and Farquhar, M.G. (1992). The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J. Cell Biol. 118, 795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman, T.C., Zhao, B., Chan, H., and Farquhar, M.G. (1998). Immunoisolation and characterization of a subdomain of the endoplasmic reticulum that concentrates proteins involved in COPII vesicle biogenesis. Mol. Biol. Cell 9, 1265-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, E., Lenk, U., and Sommer, T. (2003). Endoplasmic reticulum-associated protein degradation. Int. Rev. Cytol. 223, 39-81. [DOI] [PubMed] [Google Scholar]
- Johnston, J.A., Ward, C.L., and Kopito, R.R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann, J., Reins, H.A., Schobert, C., and Jentsch, S. (1993). Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature 361, 369-371. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics: A Cold Spring Harbor Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Kamhi-Nesher, S., Shenkman, M., Tolchinsky, S., Fromm, S.V., Ehrlich, R., and Lederkremer, G.Z. (2001). A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioka, N., Tsubota, J., Kakehi, Y., Komano, T., Gottesman, M.M., Pastan, I., and Ueda, K. (1989). P-glycoprotein gene (MDR1) cDNA from human adrenal: normal P-glycoprotein carries Gly185 with an altered pattern of multidrug resistance. Biochem. Biophys. Res. Commun. 162, 224-231. [DOI] [PubMed] [Google Scholar]
- Kiser, G.L., Gentzsch, M., Kloser, A.K., Balzi, E., Wolf, D.H., Goffeau, A., and Riordan, J.R. (2001). Expression and degradation of the cystic fibrosis trans-membrane conductance regulator in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 390, 195-205. [DOI] [PubMed] [Google Scholar]
- Koning, A.J., Roberts, C.J., and Wright, R.L. (1996). Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol. Biol. Cell 7, 769-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova, Z., and Wolf, D.H. (2003). For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22, 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza, D., and Michaelis, S. (1998). Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza, D., Tam, A., Schmidt, W.K., and Michaelis, S. (1998). Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 2767-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, D.L. (2002). Functional Analysis of MRP Transporters in Saccharomyces cerevisiae. Ph.D. Thesis, The Johns Hopkins University School of Medicine, Baltimore, MD.
- Michaelis, S., and Herskowitz, I. (1988). The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell Biol. 8, 1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D.T., Spear, E.D., and Walter, P. (2000). The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150, 77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijbroek, G.L., and Michaelis, S. (1998). Functional assays for analysis of yeast ste6 mutants. Methods Enzymol. 292, 193-212. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S., Hirata, A., and Nakano, A. (1994). Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol. Biol. Cell 5, 1129-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K.R., Vo, K.T., Michaelis, S., and Paddon, C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon, C., Loayza, D., Vangelista, L., Solari, R., and Michaelis, S. (1996). Analysis of the localization of STE6/CFTR chimeras in a Saccharomyces cerevisiae model for the cystic fibrosis defect CFTR delta F508. Mol. Microbiol. 19, 1007-1017. [DOI] [PubMed] [Google Scholar]
- Prinz, W.A., Grzyb, L., Veenhuis, M., Kahana, J.A., Silver, P.A., and Rapoport, T.A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proff, C., and Kolling, R. (2001). Functional asymmetry of the two nucleotide binding domains in the ABC transporter Ste6. Mol. Gen. Genet. 264, 883-893. [DOI] [PubMed] [Google Scholar]
- Raposo, G., van Santen, H.M., Leijendekker, R., Geuze, H.J., and Ploegh, H.L. (1995). Misfolded major histocompatibility complex class I molecules accumulate in an expanded ER-Golgi intermediate compartment. J. Cell Biol. 131, 1403-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths, S., Rohrer, J., Crausaz, F., and Riezman, H. (1993). end3 and end4, two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120, 55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, M., Gros, P., Whiteway, M., and Thomas, D.Y. (1992). Functional complementation of yeast ste6 by a mammalian multidrug resistance mdr gene. Science 256, 232-234. [DOI] [PubMed] [Google Scholar]
- Rieder, S.E., Banta, L.M., Kohrer, K., McCaffery, J.M., and Emr, S.D. (1996). Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell 7, 985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, J.D., Schmidt, W.K., and Michaelis, S. (1998). The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell 9, 2231-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein, S., Berkower, C., and Michaelis, S. (1994). Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol. Cell Biol. 14, 1438-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, W.K., Tam, A., Fujimura-Kamada, K., and Michaelis, S. (1998). Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 95, 11175-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J.D., Cummings, K.B., Huyer, G., Michaelis, S., and Wendland, B. (2001). Yeast as a model system for studying endocytosis. Exp. Cell Res. 271, 1-9. [DOI] [PubMed] [Google Scholar]
- Sherwood, P.W., and Carlson, M. (1999). Efficient export of the glucose transporter Hxt1p from the endoplasmic reticulum requires Gsf2p. Proc. Natl. Acad. Sci. USA 96, 7415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski, C., Cox, J.S., and Walter, P. (1996). tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405-413. [DOI] [PubMed] [Google Scholar]
- Sullivan, M.L., Youker, R.T., Watkins, S.C., and Brodsky, J.L. (2003). Localization of the BiP molecular chaperone with respect to endoplasmic reticulum foci containing the cystic fibrosis transmembrane conductance regulator in yeast. J. Histochem. Cytochem. 51, 545-548. [DOI] [PubMed] [Google Scholar]
- Supply, P., Wach, A., Thines-Sempoux, D., and Goffeau, A. (1993). Proliferation of intracellular structures upon overexpression of the PMA2 ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 268, 19744-19752. [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., Hirata, A., Fukuda, R., Horiuchi, H., Ohta, A., and Takagi, M. (1997). Accumulation of misfolded protein aggregates leads to the formation of Russell body-like dilated endoplasmic reticulum in yeast. Yeast 13, 1009-1020. [DOI] [PubMed] [Google Scholar]
- Valetti, C., Grossi, C.E., Milstein, C., and Sitia, R. (1991). Russell bodies: a general response of secretory cells to synthesis of a mutant immunoglobulin which can neither exit from, nor be degraded in, the endoplasmic reticulum. J. Cell Biol. 115, 983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R. (2000). Transmission electron microscopy of yeast. Microsc. Res. Tech. 51, 496-510. [DOI] [PubMed] [Google Scholar]
- Wright, R., Basson, M., D'Ari, L., and Rine, J. (1988). Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107, 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Michaelis, S., and Brodsky, J.L. (2001a). CFTR expression and ER-associated degrdation in yeast. In: Cystic Fibrosis Methods and Protocols, vol. 70, ed. W.R. Skach, Totowa, NJ: Humana Press, 257-265. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Nijbroek, G., Sullivan, M.L., McCracken, A.A., Watkins, S.C., Michaelis, S., and Brodsky, J.L. (2001b). Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, T., Vogel, F., Ohta, A., Takagi, M., and Schunck, W.H. (1997). Protein quality - a determinant of the intracellular fate of membrane-bound cytochromes P450 in yeast. DNA Cell Biol. 16, 501-514. [DOI] [PubMed] [Google Scholar]