Abstract
Environmental ‘light’ has a vital role in regulating plant growth and development. Transcriptomic profiling has been widely used to examine how light regulates mRNA levels on a genome-wide scale, but the global role of translational regulation in the response to light is unknown. Through a transcriptomic comparison of steady-state and polysome-bound mRNAs, we reveal a clear impact of translational control on thousands of genes, in addition to transcriptomic changes, during photomorphogenesis. Genes encoding ribosomal protein are preferentially regulated at the translational level, which possibly contributes to the enhanced translation efficiency. We also reveal that mRNAs regulated at the translational level share characteristics of longer half-lives and shorter cDNA length, and that transcripts with a cis-element, TAGGGTTT, in their 5′ untranslated region have higher translatability. We report a previously neglected aspect of gene expression regulation during Arabidopsis photomorphogenesis. The identities and molecular signatures associated with mRNAs regulated at the translational level also offer new directions for mechanistic studies of light-triggered translational enhancement in Arabidopsis.
Keywords: photomorphogenesis, polysome, translation
Introduction
The growth and development of a living organism is ultimately determined by the expression and interaction of genes. The expression of a protein-coding gene is a complex process stretching from the activation of transcription to the production of a protein, which functions in a defined time and place. This process requires precise and sophisticated regulatory mechanisms to ensure that gene expression is coordinated properly on a genome-wide level. In the post-genomic era, the development of high-throughput experimental platforms for transcriptome profiling has quickly expanded our knowledge of genome-wide gene expression at the steady-state mRNA level. However, researchers have observed a moderate or even poor correlation between mRNAs and their protein products in budding yeast and mammalian cells (Beyer et al, 2004; Tian et al, 2004). This finding indicates the existence of additional regulation at the levels of translation and post-translational degradation.
Because of their immobile nature, plants possess versatile strategies to interpret and respond to environmental signals. Light is one of the most influential environmental stimuli regulating numerous growth and developmental processes during a plant’s life cycle (for review, see Kami et al, 2010). Upon protruding from the soil, young seedlings proceed with photomorphogenesis, a developmental process transforming the seedlings into a vegetative state and required for photosynthetic activity. A successful photomorphogenic process determines whether a plant can survive in its growth habitat. At the molecular level, selective protein degradation regulated by COP9 signalosome and 26S proteasome regulates the accumulation of proteins for light perception and signaling in plants (for review, see Schwechheimer and Deng, 2000 and Henriques et al, 2009). Positive and negative transcriptional regulators orchestrate a sophisticated transcriptomic adjustment for photomorphogenic development (for review, see Casal and Yanovsky, 2005). However, whether the transcriptome observed could faithfully reflect the translated protein species is unclear. Previous studies indicated that mutation of the translation initiation factor 3 subunit H1 (eIF3h) or ectopic expression of eIF3e resulted in a partial constitutive photomorphogenic phenotype in dark-grown seedlings (Kim et al, 2004; Yahalom et al, 2008). eIF3h is needed for the ribosome to resume scanning after translating the upstream open reading frame(s) in 5′ untranslated regions (5′ UTRs) of some transcripts (Kim et al, 2007; Roy et al, 2010). Interestingly, the eIF3 complex is also structurally and evolutionarily similar to the lid complex of the 26S proteasome and the COP9 signalosome (Karniol and Chamovitz, 2000). These results suggest that translational control has an important regulatory role during photomorphogenesis.
Considerable efforts have also been invested in studies of genome-wide translational control in plants, with special focus on plants responding to abiotic stresses, including dehydration, elevated temperature, high salinity, oxygen deprivation, sucrose starvation and heavy metal (Kawaguchi et al, 2004; Branco-Price et al, 2005, 2008; Nicolai et al, 2006; Matsuura et al, 2010; Sormani et al, 2011). In these studies, plants under abiotic stresses show a global repression of translation. The expression of epitope-tagged ribosomal proteins under the control of cell type-specific promoters also helped with the precise identification of mRNAs associated with ribosomes (translatome) for specific cell types (Mustroph et al, 2009; Jiao and Meyerowitz, 2010).
For light-regulated responses in plants, translational control has been reported in regulating the expression of photosynthetic genes (Helliwell et al, 1997; Petracek et al, 1997; Dickey et al, 1998; Sherameti et al, 2002; Kim et al, 2003; Mussgnug et al, 2005; McKim and Durnford, 2006). A previous study elegantly compared the transcripts, protein level and enzymatic activity for 35 genes from central metabolic pathways in Arabidopsis rosette leaves experiencing diurnal changes (Piques et al, 2009). However, a global view of the translatome in early stages of photomorphogenesis is currently absent.
In this study, in addition to a survey of the steady-state transcriptome, we surveyed transcripts associated with the polysome to infer the translatome in photomorphogenic Arabidopsis seedlings. Our results indicate that 4 h of light treatment induces a massive increase in translation for one third of the transcripts present at this stage, an impact exceeding the previous estimation of transcriptome adjustment. This finding offers new directions for future mechanistic studies of light-regulated gene expression in Arabidopsis. Light-triggered translational control is evident for transcripts with TAGGGTTT or AAAACCCT in their 5′ UTRs but not for transcripts of high abundance. Therefore, this translational control may be highly selective. Our data confirm that plants adopt an evolutionarily conserved mechanism for translational control by favoring shorter and stable transcripts. Nonetheless, our data also imply that both common and unique translational controls co-exist to fine-tune the translation of diverse transcripts.
Results
Light enhances the global translation in Arabidopsis seedlings
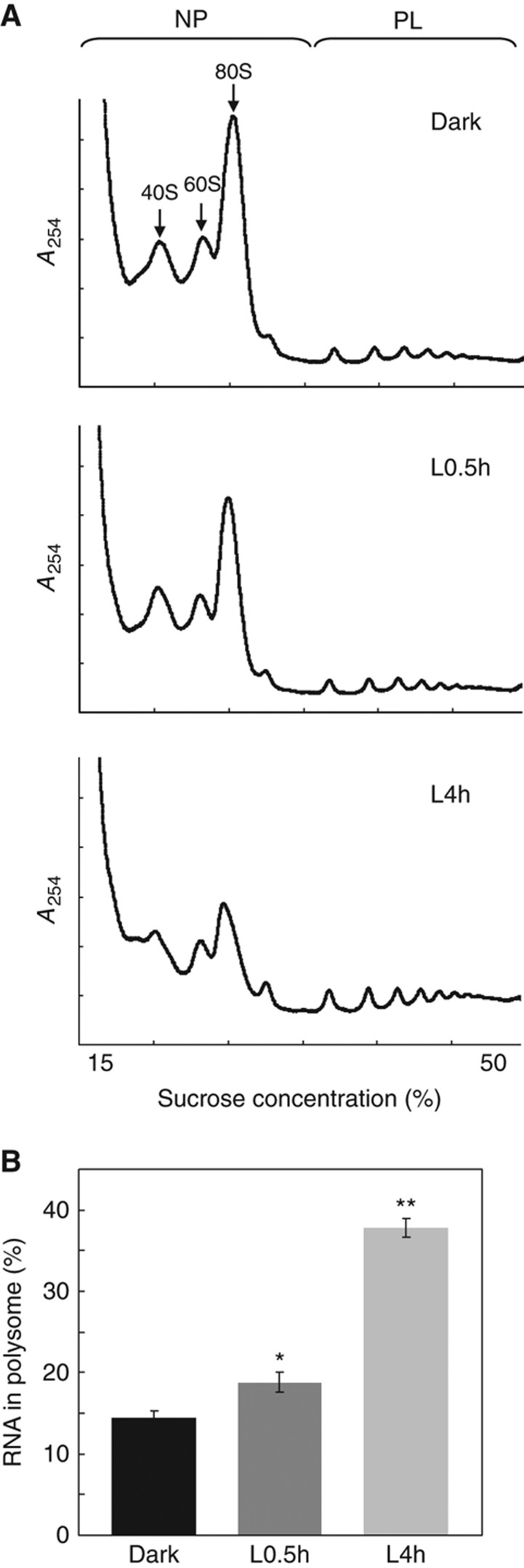
To investigate the effect of light on the global translation status during early photomorphogenesis, Arabidopsis 4-day-old etiolated seedlings were exposed to white light and harvested at 0 min, 10 min, 0.5 h, 1 h and 4 h. The translational status of the samples was examined by polysome profiling analyses, which allow for the differentiation of translating complexes with different numbers of ribosomes (Melamed and Arava, 2007). We designated fractions with no >2 ribosomes as the non-polysomal (NP) fraction (Figure 1A). mRNAs associated with NP fraction are likely in an inactive or less active translation state. On the other hand, the polysomal (PL) fraction includes mRNAs in a relatively more active translation status by associating with at least three ribosomes (Figure 1A).
Figure 1.
Light enhances the global translational efficiency in Arabidopsis. (A) Polysome profiles of Arabidopsis seedlings grown in the dark or treated with white light for 0.5 h (L0.5h) or 4 h (L4h). Ribosome subunits (40S and 60S), mono-ribosome (80S), NP and PL fractions are marked. (B) Bar graph shows the ribosome loading efficiency in seedlings grown in the Dark, L0.5h and L4h. Values are mean percentages±s.e. from three biological replicates. *P<0.05 and **P<0.0001, Student's t-test. A254, absorbance at 254 nm.
Etiolated seedlings (0 min) and seedlings with 10 min of light treatment did not differ in polysome profiles (Supplementary Figure S1). However, the extended light treatment caused a significant shift from NP to PL in polysome profiles (Figure 1A; Supplementary Figure S1). Therefore, translation activity in Arabidopsis seedlings increases with their transition from dark to light. From the results in Supplementary Figure S1, we chose three representative time points, 0 min (Dark), 0.5 h light (L0.5h) and 4 h light (L4h), for further studies of the light-triggered translational activation during photomorphogenesis.
We performed a more quantitative calculation to obtain the ribosome loading efficiency, designated as PL%. PL% is the percentage of total RNA (including mRNAs and rRNAs) in the PL fraction and is used to infer the proportion of rRNA/mRNA engaged in the active translation status. In etiolated seedlings (Dark), only 15% of total RNAs were committed to active translation, whereas in L0.5h and L4h seedlings, PL% increased to 18 and 35%, respectively (Figure 1B). These results suggest that the global translation was enhanced more than two-fold during the first 4 h of photomorphogenesis in Arabidopsis.
Translational control contributes to early Arabidopsis photomorphogenesis
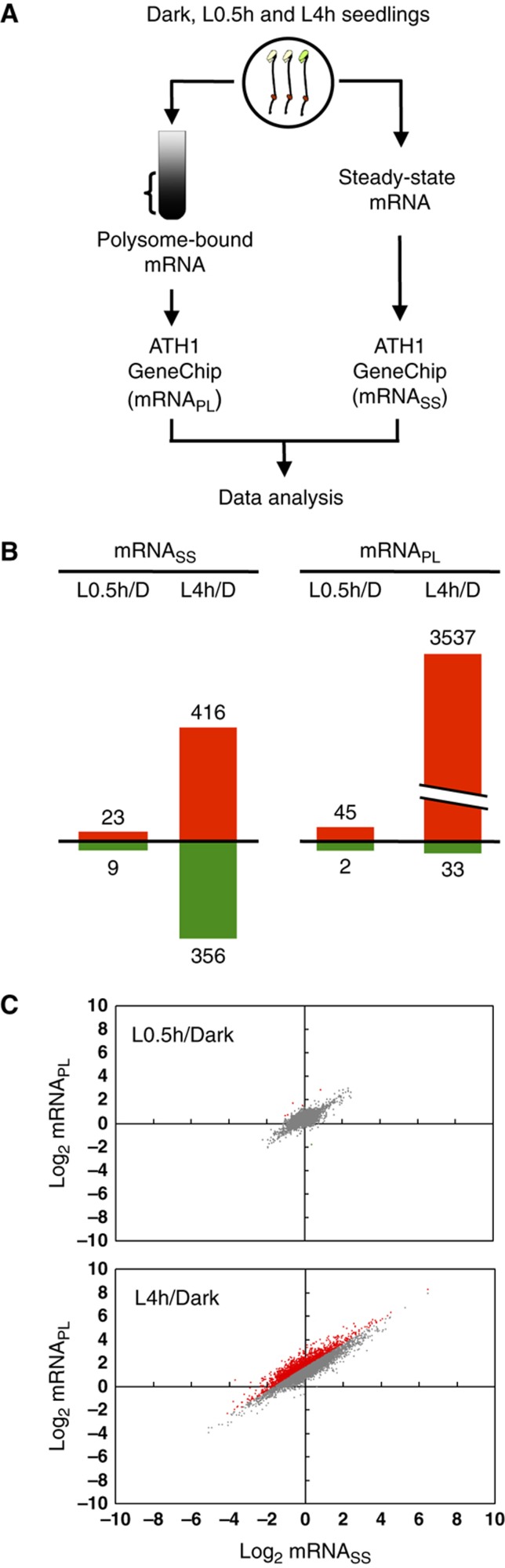
The increase in global translation could result from the net increase in mRNAs or the increased translation of pre-existing mRNAs. To assess whether one or both of these two causes contribute to the translation enhanced by light treatment, we performed transcriptomic analyses of steady-state mRNAs (mRNASS) and polysome-bound mRNAs (mRNAPL) by hybridization to the Arabidopsis whole-genome array ATH1 (Figure 2A). Three biological replicates were performed to reveal changes in mRNASS and mRNAPL in etiolated (Dark), L0.5h or L4h seedlings. Supplementary Figure S2 shows a flowchart of the microarray data analyses. With an arbitrary three-fold cutoff, 3537 mRNAs were upregulated at the mRNAPL level at L4h, whereas only 416 mRNAs were induced at the mRNASS level (Figure 2B; data in Supplementary Table S1). This finding suggests that the light-mediated translational control targets more mRNA species than ones with increased steady-state abundance.
Figure 2.
A comparison of light-induced changes in steady-state and polysome-bound mRNAs. (A) An illustration of the experimental design. Total and polysome-bound RNAs were isolated in parallel for hybridization to Affymetrix ATH1 GeneChips for transcriptomic profiling analyses. (B) The number of genes with ⩾three-fold changes at mRNASS or mRNAPL levels. Red and green colors represent genes upregulated and downregulated, respectively, by light treatment. (C) A genome-wide comparison of the gene expression at mRNASS and mRNAPL ratios between Dark- and L0.5h- or between Dark- and L4h-treated samples. The log2 values of the fold changes are plotted for the defined expressed genes (n=11 598). Red dots represent genes with ⩾three-fold differences between mRNASS and mRNAPL ratios.
We have also plotted the changes in mRNASS against mRNAPL populations as log2-transformed values between Dark and L0.5h or between Dark and L4h samples (Figure 2C). With 0.5 h of light treatment, the gene expression changes in both mRNASS and mRNAPL levels were modest, represented by the cluster of gene spots near (0.0). In contrast, 4 h of light treatment resulted in a range of 100-fold (∼27) upregulation or downregulation at the mRNASS level, which is comparable to previous reports (Tepperman et al, 2001, 2004; Chang et al, 2008). Similar fold changes were observed at the mRNAPL level (Figure 2C), which indicates a large increase in mRNAs in the PL fraction after 4 h of light. The result is consistent with the increased translation calculated (Figure 1B). Interestingly, 30% of expressed genes showed ⩾three-fold differences between mRNAPL and mRNASS ratios in pair-wise comparison of L4h and Dark (marked in red in Figure 2C). The above results highlight the previously neglected role of translational control in regulating gene expression during the early photomorphogenic stage.
Light triggers an increase in ribosome occupancy and ribosome density
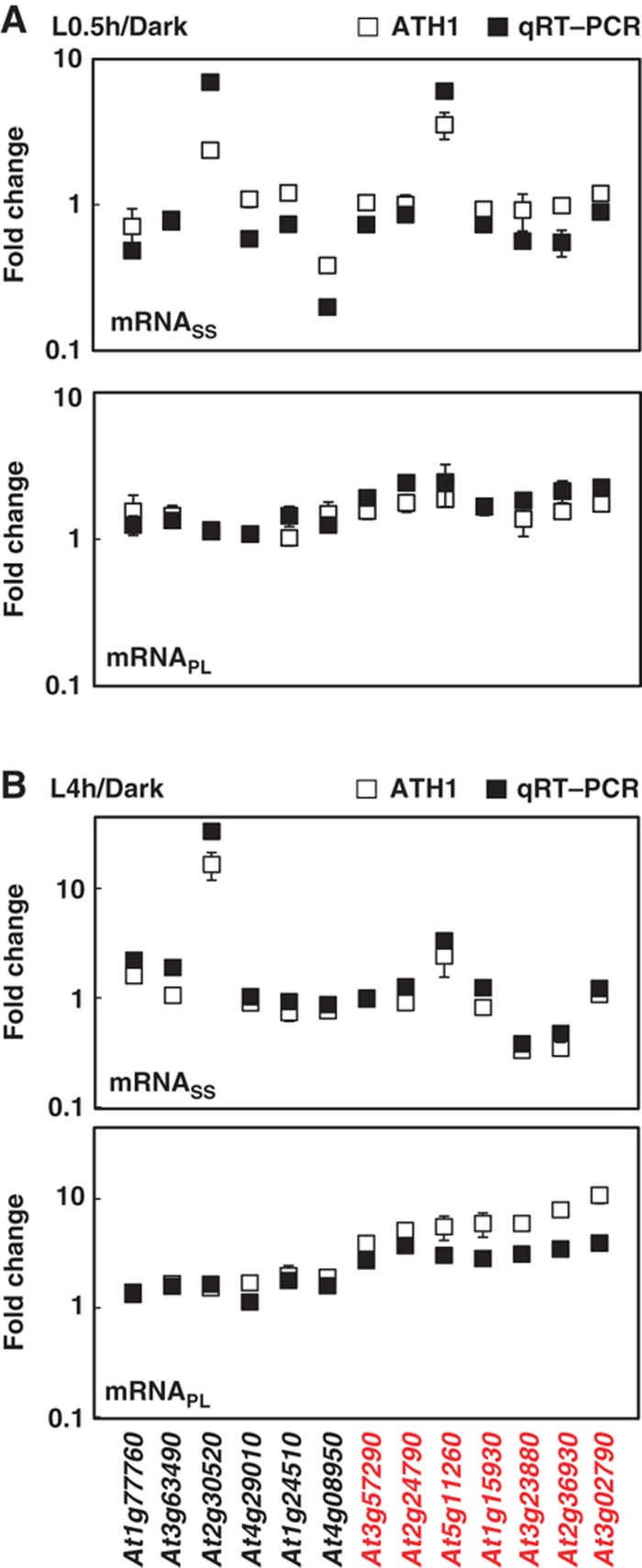
To validate the microarray data, we performed quantitative RT–PCR (qRT–PCR) to detect the level of selected transcripts in total RNA, NP and PL populations. We examined 13 genes covering various expression behaviors on light treatment with no change, upregulation or downregulation at the mRNASS level (Figure 3). In all, 7 of these 13 genes showed ⩾three-fold differences between mRNAPL and mRNASS ratios and were thus considered to be regulated at the translational level (highlighted in red in Figure 3; expression data listed in Source data for Figure 3). At the mRNASS level, qRT–PCR results were in good agreement with the microarray results for all 13 genes (Figure 3A and B). qRT–PCR also confirmed the microarray results for six genes that did not show light-induced increase in the PL fraction. For the seven genes with increased association with the PL fraction by microarray analyses, qRT–PCR also showed increased representation of these mRNAs in the PL fraction, although with slightly smaller fold changes for a few mRNAs at L4h (Figure 3B). The qRT–PCR data confirmed no biases imposed by the normalization methodology adopted. As well, the high percentage of mRNAs regulated at the translational level was indeed prominent during this physiological transition.
Figure 3.
Confirmation of increased ribosome occupancy by qRT–PCR. Changes in steady-state and polysome-bound mRNA abundance between Dark and L0.5h (A) or between Dark and L4h (B) samples (filled square) with standard deviations were calculated from three technical repeats of one representative biological repeat. Results for an independent biological repeat were shown in Supplementary Figure S3. Transcriptome data obtained from ATH1 hybridization were also plotted (open square), with error bars calculated from three biological repeats. Red color represents genes under translational control. At1g77760, nitrate reductase 1 (NIA1); At3g63490, ribosomal protein L1p/L10e family; At2g30520, root phototropism 2 (RPT2); At4g29010, abnormal inflorescence meristem (AIM1); At1g24510, TCP-1/cpn60 chaperonin family protein; At4g08950, phosphate-responsive 1 family protein; At3g57290, eukaryotic translation initiation factor 3E (eIF3e); At2g24790, constans-like 3 (COL3); At5g11260, elongated hypocotyl 5 (HY5); At1g15930, ribosomal protein S12e (RPS12e); At3g23880, F-box family protein; At2g36930, zinc finger (C2H2 type) family protein; At3g02790, zinc finger (C2H2 type) family protein. Source data is available for this figure in the Supplementary Information.
Notably, translational control could affect mRNAs with variable fold changes at the mRNASS level. For example, high induction of At2g30520 (RPT2) with light treatment does not guarantee its high translation capacity at L0.5h (Figure 3A). However, At3g23880 was downregulated at the mRNASS level but had higher association with the PL fraction at L4h (Figure 3B). Also, light-triggered translational activation can result from the increase in ribosome occupancy of mRNAs with similar steady-state abundance before and after light treatment; examples are At3g57290 (eIF3e), At2g24790 (COL3), At1g15930 (RPS12e) and At3g02790.
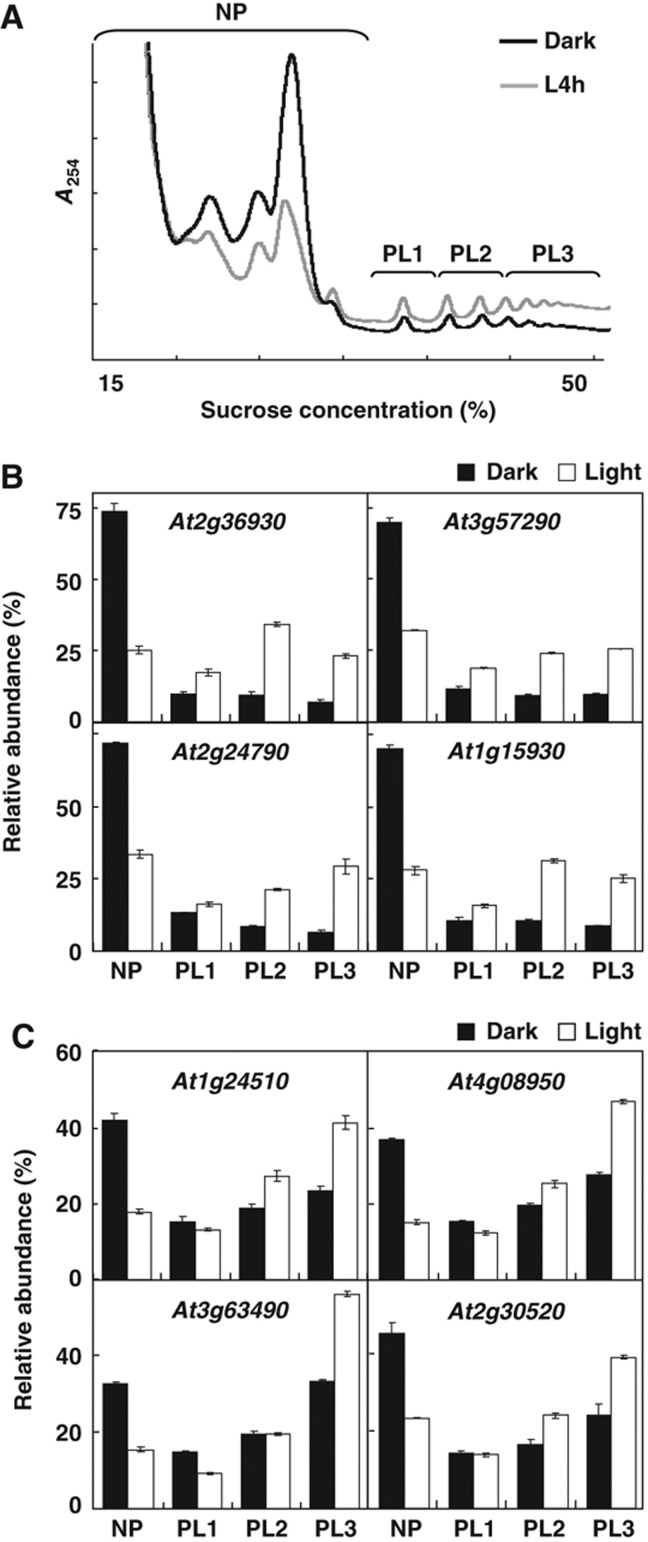
Ribosome occupancy refers to the association of mRNAs with polysomes (Arava et al, 2003; Lackner et al, 2007; Piques et al, 2009). In addition to ribosome occupancy, the changes in ribosome density of a given mRNA could also enhance its translation rate. Indeed, previous studies have shown that, through the control of ribosome density, yeast under amino-acid starvation or Arabidopsis under daily light/dark changes could alter their gene expression patterns (Ingolia et al, 2009; Piques et al, 2009). To further explore the effect of ribosome density on the translational control during photomorphogenesis, we examined the ribosome numbers per transcript in detail for selected transcripts. According to the number of ribosomes associated with mRNAs, the PL fraction was further divided into three subfractions, PL1, PL2 and PL3 (Figure 4A). In all, 8 of the 13 mRNAs in Figure 3 were also examined for their corresponding ribosome density in PL1, PL2 and PL3 fractions by qRT–PCR. Among them, At2g36930, eIF3e, COL3 and RPS12e, representing mRNAs with a significant increase in ribosome occupancy (Figure 3), showed mRNAs evenly distributed among the three PL subfractions (Figure 4B). In contrast, the other four mRNAs with relatively minor increase in ribosome occupancy (Figure 3) showed a primary association with the PL3 fraction with 4 h light (Figure 4C). Whether this represents an increased translation rate or the result of ribosome pausing could not be differentiated with the present study. Nevertheless, this result implies that the translational control of these mRNAs could be achieved by shifting mRNAs to a higher order of ribosome fractions, rather than by an overall increase in ribosome occupancy.
Figure 4.
Light triggers an increase in ribosome density. (A) An illustration showing NP, and PL subfractions, PL1, PL2 and PL3, corresponding to the polysome profiles of the Dark and L4h seedlings. (B, C) qRT–PCR analysis of relative mRNA abundance (%) of selected genes in each fraction. The filled and open bars represent expression data from Dark and L4h samples, respectively. Error bars represent the standard deviation calculated from three technical repeats of one representative biological repeat. Results for two independent biological repeats were shown in Supplementary Figure S4. Source data is available for this figure in the Supplementary Information.
These data suggest that the light-enhanced translation could be achieved by adjusting both the ribosome occupancy and ribosome density, similarly to a previous report based on 35 genes in Arabidopsis rosette leaves (Piques et al, 2009). Our current transcriptome analyses mostly revealed mRNA species with a marked increase in ribosome occupancy. More detailed polysome fractionation is needed to better reveal mRNAs with altered ribosome density in photomorphogenic Arabidopsis.
Categorization of mRNA species regulated at the steady-state RNA and/or translationally active levels
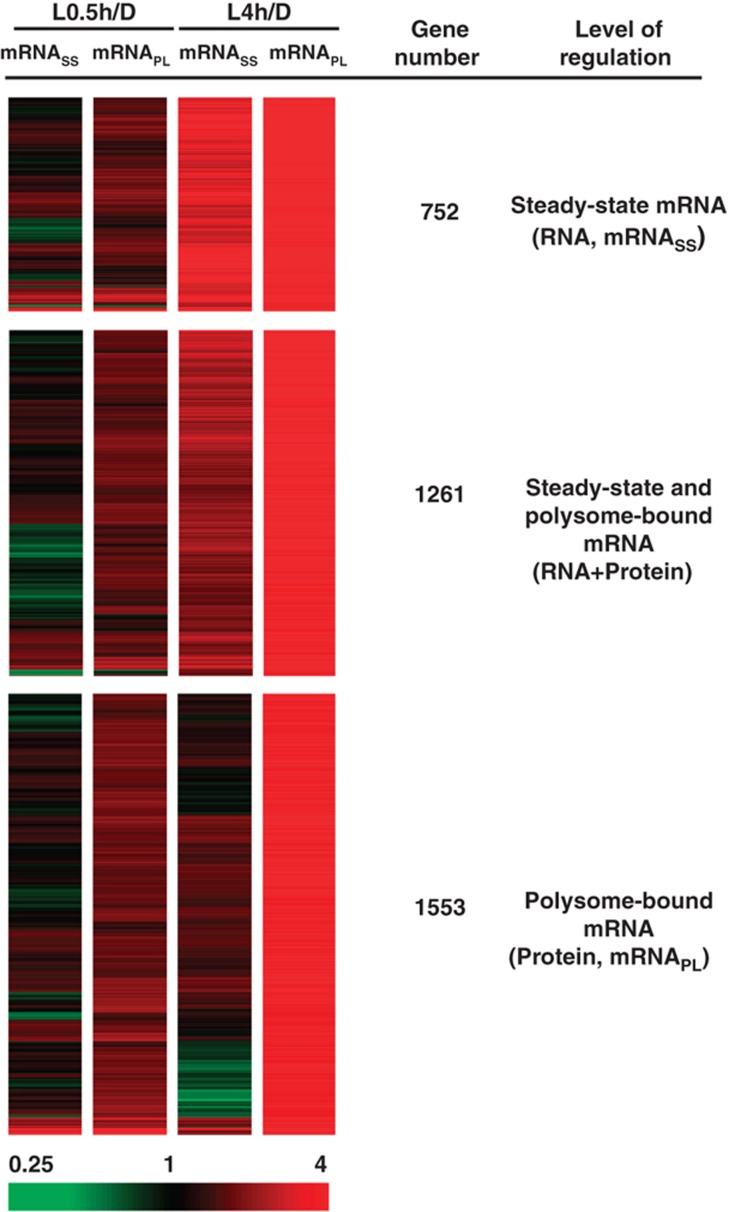
Our transcriptomic analysis revealed 3566 genes upregulated at the mRNASS and/or mRNAPL levels with light treatment (Supplementary Figure S2). As a first step to investigate the biological impact resulting from gene expression regulated at various levels, we performed k-means cluster analysis to categorize these genes and revealed four expression groups with distinct expression patterns (Supplementary Figure S5; Supplementary Table S2). mRNAs in cluster 1 (n=752) had a concordant induction at mRNASS and mRNAPL levels in response to light. The translational increase of mRNAs in cluster 1 may simply reflect their increase at the mRNASS level and thus were designated the ‘RNA’ group (Figure 5; Supplementary Figure S5). mRNAs in the RNA group are primarily regulated at the mRNASS level. mRNAs in cluster 2 (n=1261) showed a moderate but clear induction at the mRNASS level and a more prominent increase at the mRNAPL level and were designated the ‘RNA+Protein’ group (Figure 5; Supplementary Figure S5). mRNAs in the RNA+Protein group have increased transcript levels and are preferentially associated with polysomes. mRNAs in both clusters 3 (n=1076) and 4 (n=477) showed enrichment at the mRNAPL level, but their mRNASS levels were maintained (cluster 3) or even slightly repressed (cluster 4) (Supplementary Figure S5; Supplementary Table S2). Therefore, the main regulation of mRNAs in clusters 3 and 4 likely comes from their enhanced association with the PL fraction, regardless of their steady-state mRNA abundance, for efficient translation after light treatment, and thus were designated the ‘Protein’ group (n=1553) (Figure 5).
Figure 5.
Categorization of light-responsive genes regulated at mRNA or protein levels. The 3566 light-upregulated genes (with ⩾ three-fold changes at mRNASS or mRNAPL levels) were categorized into three groups, preferentially regulated at the steady-state mRNA level (RNA), the polysome-bound mRNA level (Protein) or both (RNA+Protein). Extreme red and green colors indicate four-fold upregulation and downregulation, respectively.
Translational control is independent of mRNA abundance but favors mRNAs with longer half-lives
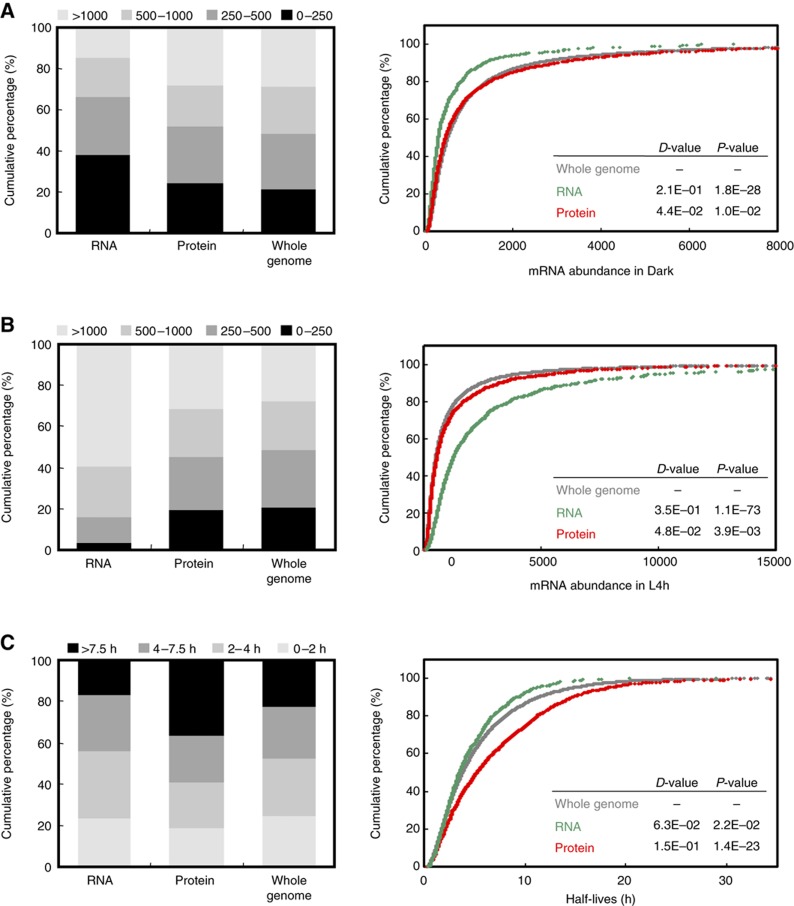
The mRNA abundance for different genes could span several orders of magnitude. We next examined whether abundant mRNAs are more inclined to engage in translation. We categorized mRNAs in the ‘Whole-genome’, RNA and Protein groups into four expression levels by their abundance according to ATH1 hybridization signal intensity. Proportions of mRNAs at each level of expression were similar between the Protein and Whole-genome groups in both Dark and L4h samples (left panels in Figure 6A and B, expression data shown in Supplementary Table S3). However, mRNAs in the RNA group showed a significant increase in abundance in the L4h sample. The proportion of mRNAs with expression levels >1000 was increased from 15 to 59%, whereas that of mRNAs with levels <250 was decreased from 38 to 3% after 4 h of light (left panels in Figure 6A and B).
Figure 6.
Light activates translation of transcripts with longer half-lives. Genes in each defined regulatory group were partitioned according to their mRNA abundances in Dark (A), L4h (B) samples and mRNA half-lives (C) and expressed as relative percentages (left panel). The bin size was arbitrarily determined so that the percentage of each category for the whole genome data set was about 25%. The cumulative curves (right panel) show the distribution of mRNA abundances and half-lives for genes in each group. The D-values of the K–S test represent the difference in distribution between the ‘Whole genome’ (gray) and ‘RNA’ (green) or ‘Whole genome’ and ‘Protein’ (red) groups. P-values are also calculated to determine statistically significant differences.
The Kolmogorov–Smirnov (K–S) test (Davis, 1986) was used to statistically examine differences in levels of expression for mRNAs in the Whole-genome, RNA or Protein groups as defined in Figure 5. Cumulative mRNA abundance curves were plotted separately for genes in RNA and Protein groups and in all expressed genes (Whole genome). Results showed negligible differences between the cumulative curves for mRNAs under translational control (Protein group) and Whole-genome mRNAs expressed in the Dark or L4h conditions (right panels in Figure 6A and B). These data indicated no correlation of transcripts with high abundance and translational regulation. K–S test also confirmed the clear distribution difference between mRNAs in the RNA and the Whole-genome groups in the dark (D=0.21, P=1.8E−28) and in L4h (D=0.35, P=1.1E−73) (right panels of Figure 6A and B). The analyses showed genes primarily regulated at the mRNASS level with more fluctuating transcript abundance in response to environmental treatments.
The steady-state mRNA abundance is determined by both the rates of transcription and mRNA decay. Information on genome-wide transcriptional rate for Arabidopsis genes is currently unavailable. Despite different growth and developmental conditions, the mRNA decay rates of 13 012 genes in cultured Arabidopsis cells were reported previously (Narsai et al, 2007). Information for mRNA half-lives is available for 90% of mRNAs in the Protein group and 80% of mRNAs in the RNA group (Supplementary Table S3). Of mRNAs in the Protein group, 37% have longer half-lives (>7.5 h), as compared with 22% in the Whole-genome group (left panel in Figure 6C). K–S test confirmed significantly longer half-lives for mRNAs in the Protein than in Whole-genome group (D=0.15, P=1.4E−23; right panel in Figure 6C). However, the distribution of mRNAs half-lives was comparable between the mRNAs in the RNA and Whole-genome groups (Figure 6C). These data imply that light may trigger the association of polysomes with mRNA species with higher stability.
Translational control favors shorter mRNAs
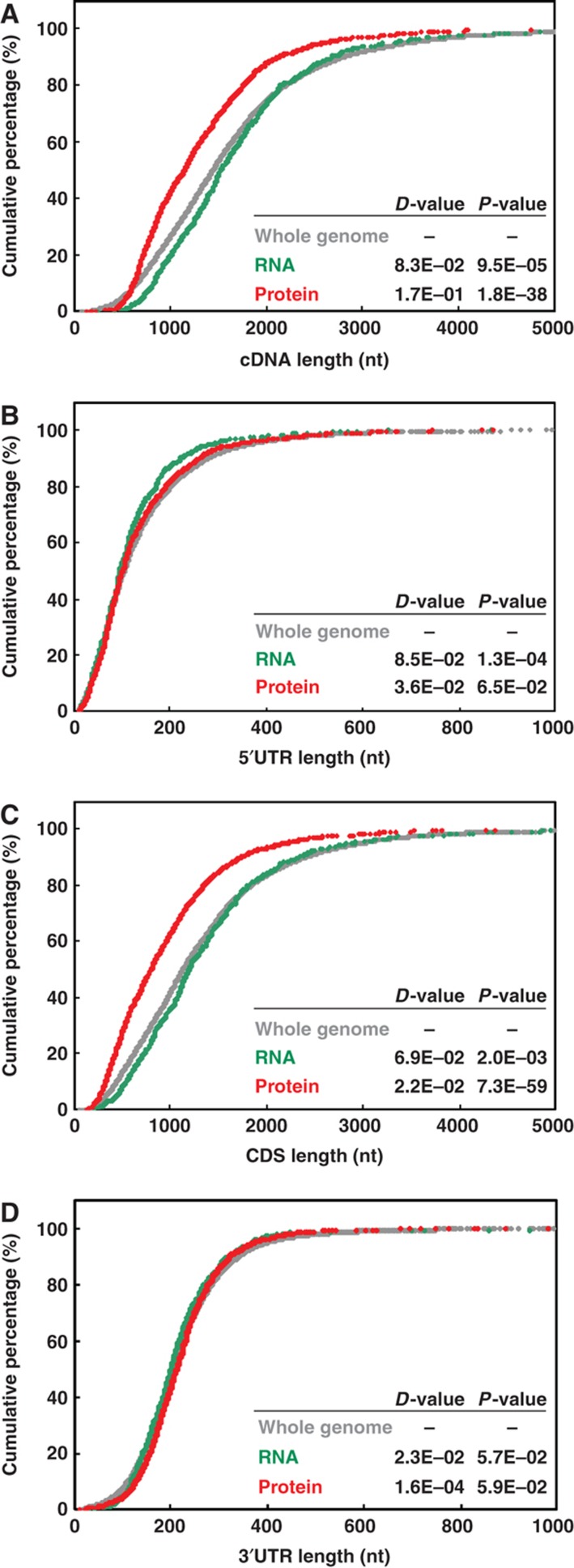
Previous studies of both yeast and human suggested a negative correlation of transcript length and tendency to associate with polysomes (Arava et al, 2003; Lackner et al, 2007; Vogel et al, 2010). We next examined whether translational control in Arabidopsis has similar characteristics through evolution. Results shown in Figure 7A indicate that genes in the Protein group produced shorter transcripts (cDNAs) (D=0.17, P=1.8E−38) and genes in RNA group slightly longer cDNAs (detailed cDNA length for each gene is in Supplementary Table S3). Because a cDNA consists of a 5′ UTR, coding sequence and a 3′ UTR, we further examined whether any or all of the regions contribute to the difference observed in cDNA length in mRNAs regulated at the translational level. K–S tests showed that, except for a slightly shorter 5′ UTR in the RNA group, genes in Whole-genome, RNA or Protein groups share similar 5′ and 3′ UTR lengths (Figure 7B and D). In contrast, genes in the Protein group had significantly shorter CDS length as compared with the other groups (D=0.22, P=7.3E−59; Figure 7C). Therefore, as observed in other model species, selective translation in light-treated Arabidopsis seedlings favors shorter transcripts. Effective translation of shorter cDNAs appears to be a conserved phenomenon across a wide spectrum of eukaryotic species.
Figure 7.
Light activates translation of transcripts with shorter CDS length. The cumulative curves were plotted for genes in ‘Whole genome’, ‘RNA’ or ‘Protein’ groups according to mRNA features: (A) cDNA length, (B) 5′ UTR length, (C) CDS length and (D) 3′ UTR length. The statistical difference was measured by the K–S test as described in Figure 6.
Cis-elements enriched in the 5′ UTR of the preferentially translated mRNAs
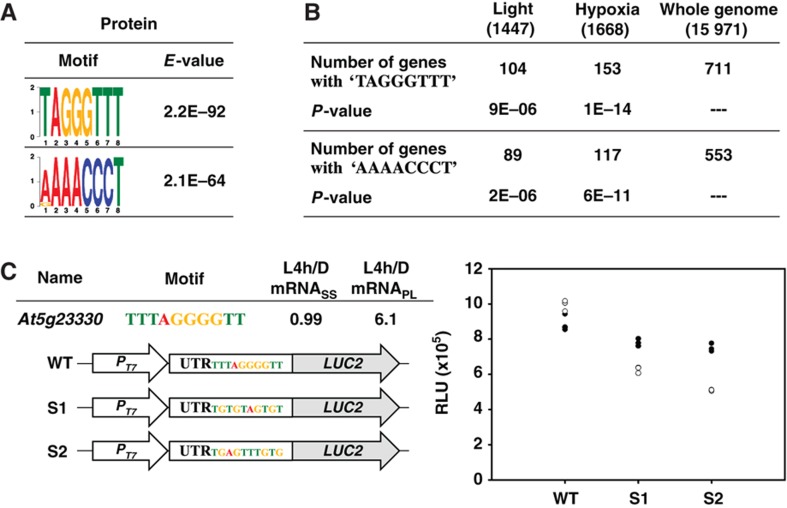
Translation discriminates against mRNAs with a 5′ terminal oligopyrimidine tract in their 5′ UTR regions in both plants and animals (Shama and Meyuhas, 1996). We next sought to investigate whether the 5′ UTRs of transcripts have cis-elements that can benefit the translation. We searched for 6- to 8-mer sequences preferentially present in the 5′ UTR regions of mRNAs in the Protein group. Input 5′ UTR sequences in each group were shuffled randomly and used as a reference data set. Only cis-elements with significantly lower E-values (E-value <1E−53) in the 5′ UTR than in the shuffled data set were selected. Supplementary Figure S6 shows cis-elements overrepresented in both RNA and Protein groups. These cis-elements may simply represent common features in 5′ UTRs of Arabidopsis mRNAs. The analysis also revealed two distinct cis-elements, TAGGGTTT and AAAACCCT, overrepresented only in the Protein group (Figure 8A). Alignment of these cis-elements and their flanking sequences are shown in Supplementary Figure S7.
Figure 8.
Cis-elements contributing to selective translation. (A) Distinct motifs overrepresented in the ‘Protein’ group are shown as the sequence LOGO. (B) Occurrences of ‘TAGGGTTT’ and ‘AAAACCCT’ elements in the 5′ UTR of the translationally regulated mRNAs under light or hypoxia treatments. P-values were determined by two-tailed Fisher's exact test. (C) mRNASS and mRNAPL ratios of At5g23330 obtained from ATH1 hybridization in this study are shown. Sequences and constructs illustrated were used for evaluating the translatability of LUC2 transcript harboring ‘TAGGGGTT’ element (WT) or sequences of scrambled cis-elements (S1 and S2) in its 5′ UTR region. T7 promoter (PT7) was used for in-vitro transcription. The LUC2 activity was expressed as relative luminescence unit (RLU) in an in-vitro transcription and translation assay as described in Materials and methods. Three technical repeats for each of the two biological repeats are plotted (marked as filled and open circles). Source data is available for this figure in the Supplementary Information.
Whether mRNAs harboring these two cis-elements tend to be translationally regulated in response to different environmental stimuli is unknown. Therefore, we compared the frequency of the occurrences for these two cis-elements in 15 971 Arabidopsis genes with available 5′ UTR sequence information and mRNAs regulated at the translational level in light-treated Arabidopsis (this study) and in Arabidopsis undergoing hypoxia stress (Branco-Price et al, 2008). Two-tailed Fisher's exact test (Agresti, 1992) was used to evaluate whether the frequency of occurrence in the genes regulated at the translational level significantly differed from that in the whole genome. The number of transcripts with TAGGGTTT in their 5′ UTRs was significantly higher in both the light-regulated group (P=9E−06) and hypoxia group (P=1E−14) (Figure 8B). Similarly, AAAACCCT was overrepresented in translationally regulated genes identified by both treatments (Figure 8B). Therefore, instead of being specific to the light signal, these two cis-elements may exert functions in translational control when Arabidopsis is orchestrating its proteome in response to external environmental stimuli.
We next examined whether the conserved motif indeed functions to selectively increase the translation efficiency of its downstream CDS. We used the 5′ UTR of At5g23330, a gene regulated primarily at the translational level (Figure 8C) and containing a TTTAGGGGTT motif to assess whether this cis-element has a positive regulatory role in translation control. Constructs harboring the 5′ UTR of At5g23330 with wild-type (WT) or mutated cis-elements (S1 and S2) were fused with coding regions of a reporter gene, LUC2, as illustrated in Figure 8C for an in-vitro translation assay. With an equal amount of transcript inputs, WT:LUC2 transcripts were more efficiently translated than were S1:LUC2 or S2:LUC2 transcripts in an in-vitro translation system (Figure 8C). Taken together, we identified a cis-element TAGGGTTT overrepresented in transcripts regulated at the translational level (Figure 8A). When present in the 5′ UTR of a reporter transcript, the cis-element could serve as a general enhancer in an in-vitro translation assay.
Transcriptional and translational regulations have complementary and distinct impacts on biochemical pathways and biological processes
The photomorphogenesis process is achieved via the seamless integration and precise commitment of biochemical pathways and biological processes. To address whether translational control regulates specific aspects of cellular functions, genes showing regulation at the RNA, RNA+Protein and Protein levels were examined for overrepresentation of specific biochemical pathways and gene ontology assignments (genes in each overrepresentative pathway and ontology group are in Supplementary Table S4).
Results in Table I show that pathways or processes for the biosynthesis of pigments, such as porphyrins and xanthophyll, are largely regulated at the mRNASS level. Genes dedicated to photosynthesis are primarily regulated at the transcript level but are also augmented by regulation at the translational level. Translational control appears to mainly apply to genes involved in the biogenesis of ribosome and the translational machinery (Table I). Intuitively, transcripts of these genes receive higher priority in engaging translation before the increase in their transcript levels, if any. The effective translation of these transcripts will then fuel the translation capacity for the enhancement of global translation during photomorphogenesis, as observed in Figure 2C. Therefore, a highly coordinated regulation at both transcription and translation levels is needed to guarantee a successful transition from skotomorphogenesis to photomorphogenesis.
Table 1. List of biochemical pathways, gene ontology (GO) and light signaling genes regulated at various levels of light-mediated gene expression.
| Regulatory group | RNA | RNA+Protein | Protein |
|---|---|---|---|
| Only pathways and GOs with P<1.0E−05 are listed. | |||
| KEGG pathway | Photosynthesis | Photosynthesis | Ribosome |
| (P-value <1.0E−05) | Porphyrin and chlorophyll metabolism | Ribosome | |
| Gene ontology-biological process (P-value <1.0E−05) | Chloroplast organization | Photosynthesis | Chloroplast organization |
| Chlorophyll biosynthetic process | Photosynthetic electron transport in photosystem | Response to heat | |
| Photosynthesis | Protein folding | Response to water deprivation | |
| Photosynthetic electron transport in photosystem | Response to cadmium ion | Ribosome biogenesis | |
| Protein folding | Ribosome biogenesis | Translation | |
| PSII-associated LHC catabolic process | Transmembrane receptor | ||
| Response to | PTK signaling pathway | ||
| blue light | |||
| cadmium ion | |||
| cold | |||
| far-red light | |||
| heat | |||
| red light | |||
| UV-B | |||
| Xanthophyll biosynthetic process | |||
| Genes for perceiving and transducing light signals | ATAB2, COP1, CR88, ELF4, BBX22/ LZF1, PIF4, SPA1, SPA4, BBX24/STO | ADO2, ARR4, ATABC1, FUS5, GI, PHYB, PHYE, SBH1, SPA1 | APRR5, COL3, DET1, eIF3e, FAR1, HY2, HY5, LHY, NDPK2, PKS1, SHL1, SHW1, SRR1, TED3, ZTL |
Translational control of light-sensing and signaling molecules
The identification of key regulators in light-sensing/signaling pathways primarily depends on forward genetics and transcriptome profiling (Casal and Yanovsky, 2005; Chory, 2010). Among the 92 genes reported to be involved in light perception or signaling pathways (see Materials and methods), 33 showed differential regulation at the mRNASS or mRNAPL level in our study. Our steady-state transcriptomic study confirmed that some key regulators regulated at the transcript level include COP1, PIF4, BBX22/LZF1 and BBX24/STO. Nevertheless, our results indicated that translational control has an even stronger impact on genes known to have key roles in light perception and signaling (Table I). After transfer from dark to light for 4 h, transcription and translation act synergistically to regulate the expression of, for example, PHYB, PHYE, SPA1 and GI. Interestingly, translational control is responsible for regulating most genes contributing to light signaling, including COL3, DET1, FAR1, HY2, HY5, LHY, NDPK2, PKS1 and ZTL (Table I).
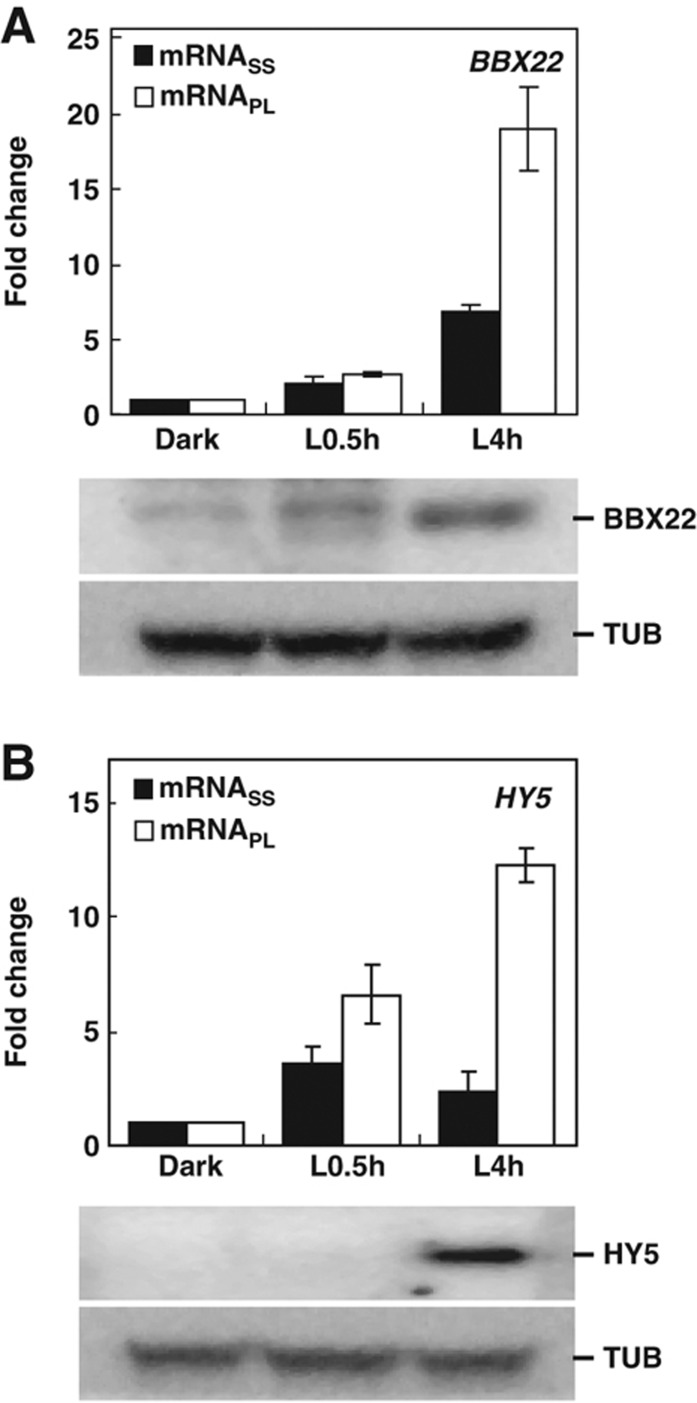
BBX22 and HY5, two positive regulators in light signaling pathways (Chattopadhyay et al, 1998; Chang et al, 2008), were assigned to groups regulated at the transcript and protein levels, respectively (Table I). Transcriptome analyses revealed BBX22 transcripts associated with the PL fraction mainly through its increase at the mRNASS level (Figure 9A, upper panel; Table I). Immunoblot analyses confirmed that the accumulation of BBX22 protein coincided with the association of BBX22 transcripts with the polysome fraction (Figure 9A, lower panel). However, HY5 exhibited a transient, light-triggered upregulation at the mRNASS level during the initial exposure to light (Figure 9B, upper panel; Chang et al, 2008); yet, HY5 protein remained undetectable (Figure 9B, lower panel). At L4h, although the HY5 mRNA level start to decline, it showed preferential association with the PL fraction, which resulted in the accumulation of HY5 protein (Figure 9B, lower panel). The HY5 protein accumulation kinetics were comparable between Arabidopsis seedlings treated with or without the proteasome inhibitors MG115/132 (Supplementary Figure S8). This finding further supports that translation control is primarily responsible for the HY5 protein detected.
Figure 9.
Experimental validation of light-responsive transcription factors regulated at transcriptional and translational levels. ATH1 expression data for BBX22 (A) and HY5 (B) at mRNASS and mRNAPL levels in Dark, L0.5h and L4h samples are plotted as fold change, with the Dark sample arbitrarily set to 1 (upper panel). Immunoblotting was used to detect BBX22 and HY5 proteins at different times. Endogenous α-tubulin (TUB) was used as a loading control (lower panel). Source data is available for this figure in the Supplementary Information.
These data validated that transcripts associated with the PL fraction are actively translated into protein products. This result also confirms that translational control is an important regulatory role for the key transcriptional regulator HY5 in the light signaling pathway (Table I).
Discussion
Translational control has an important role in regulating gene expression in photomorphogenic Arabidopsis
Although transcriptomic alteration is a hallmark in photomorphogenic Arabidopsis, modest efforts have been made to investigate other aspects of gene expression regulation. We provide the first genome-wide survey of translational control in gene expression regulation during photomorphogenesis in Arabidopsis. Translational control appears to incur greater gene expression regulation than with steady-state mRNA transcriptome regulation. After light treatment for 4 h, the translation capacity increased more than two-fold (Figure 1B) and resulted in a preferential translation for about one third of the total transcriptome at this stage (Figure 2B). The quick implementation of selective translation allows for timely production of proteins in translation and photosynthesis complexes (Table I). With this process, Arabidopsis can meet the demand of photosynthesis and the massive production of proteins for growth and development after exposure to environmental light signals.
By comparing the mRNASS and mRNAPL levels, we could classify many known light perception or signaling molecules into various regulatory groups (Table I). As expected, changes in transcript levels are responsible for the regulation of some key regulators in the light signaling pathway. Interestingly, we found even more ‘light’ genes regulated at the translational level. Our study revealed a new repertoire of genes that contribute to photomorphogenesis (Supplementary Table S2). The identities of these genes could not be revealed by traditional transcriptome study that only measures steady-state mRNAs. Exploring the functions of these genes would be useful for a more comprehensive understanding of light-mediated growth and development in Arabidopsis.
Translational control prefers stable or shorter mRNAs
Stable and abundant mRNAs are more efficiently translated in fission yeast (Lackner et al, 2007). However, budding yeast shows no association of ribosome density and mRNA half-lives or abundance (Arava et al, 2003). In our study, polysomes prefer binding to transcripts of high stability but not high abundance (Figure 6). The stability of mRNAs might be a cause, a consequence or both, of the selective translation. In mammals, mRNA targeted by miRNAs could be stabilized by ribosomes located around the target sites (Gu et al, 2009). If ribosomes could protect mRNAs from the degrading enzymes, the high stability of actively translated mRNAs is likely a consequence. Alternatively, high mRNA stability could render a transcript with high translation efficiency under specific environmental cues. Sequestration of transcripts to a ‘non-translating’ pool was proposed to explain the globally reduced translation in yeast under high salinity (Melamed et al, 2008). The stable existence of these transcripts in the ‘non-translating’ pool was suggested to allow for efficient translation when recovering from the stress. This may also explain why transcripts repressed at the translational level under hypoxia could quickly resume the translation efficiency after 1 h of reoxygenation (Branco-Price et al, 2008; Supplementary Table S5). As well, some mRNAs sequestered to cytosolic foci, such as stress granules (SGs) or processing bodies (PBs), showed repressed translation (for review, see Parker and Sheth, 2007 and Bailey-Serres et al, 2009). Previous studies showed that SGs or PBs were preferentially present in dark-grown than in light-grown plants (Pomeranz et al, 2010). Certain transcripts in dark-grown seedlings may be temporally associated with SGs or PBs and remain stable. Light treatment might trigger the disruption of SGs or PBs or the release of these transcripts for active translation. Transcriptome profiling of SGs and/or PBs in etiolated seedlings will help address this possibility.
Interestingly, 477 transcripts showed preferential association with polysome, but their transcript levels were slightly reduced (Supplementary Figure S5). It will be of special interest to study whether these transcripts are subjected to the regulation of co-translational mRNA decay, as was previously reported in yeast (Hu et al, 2009).
Light-mediated translational control also favors shorter cDNAs (Figure 7), similarly to the inverse correlation of ribosome density and ORF length observed in Arabidopsis under hypoxia stress (Branco-Price et al, 2005). The closed-loop model for mRNA translation may explain this phenomenon in part (Kahvejian et al, 2001). In this model, the interaction between proteins bound to the 5′-cap structure and 3′-poly(A) tail of a transcript could enhance the translation efficiency (for reviews, see Kawaguchi and Bailey-Serres, 2002 and Lackner and Bahler, 2008). This intramolecular interaction favors shorter transcripts. Alternatively, long transcripts have a higher likelihood of forming secondary structures that may inhibit translation initiation or even elongation.
Sequence features of translationally regulated mRNAs
Two cis-elements are overrepresented in the 5′ UTRs of transcripts regulated at the translational level (Figure 8A). A previous report of 12 749 Arabidopsis genes showed the cis-element AAACCCTA present in 242 promoters (−50 to −1 regions) and 316 5′ UTRs (+1 to +50 region), whereas the cis-element TAGGGTTT was found in 203 5′ UTRs (Molina and Grotewold, 2005). Deletion of AAACCCTA or the complementary TAGGGTTT elements in the promoter regions of eEF1A or the gene encoding 60S ribosomal protein L15 abolished their transcript accumulation (Tremousaygue et al, 1999; Tatematsu et al, 2005). Therefore, these elements were required for the promoter activities of these two genes.
Our study indicated that TAGGGTTT, when residing in the 5′ UTR of a transcript, could enhance the translatability of this transcript in vitro (Figure 8C). AAACCCTA and TAGGGTTT elements were found to be overrepresented within the 5′ UTR region of many genes encoding the components of protein synthesis (Tremousaygue et al, 1999; Tatematsu et al, 2005). These elements may contribute to the prompt translation of the ribosome apparatus to meet the demand of translation.
These findings suggest that the cis-element TAGGGTTT, depending on its location relative to a gene, could promote transcription as well as translation. How this element could function in distinct levels of gene expression regulation requires the identification and characterization of its trans protein factors.
Also, the sequence complementarities of these two elements suggested that they might function by forming a hairpin-like structure on an mRNA to control the translational process. However, on examining the occurrence of these elements on the 5′ UTRs of transcripts in the Protein group, we found only four genes containing both AAAACCCT and TAGGGTTT elements (data not shown). Therefore, the base-pairing structure formed by these two cis-elements could not explain the translational regulation of most transcripts identified. Also, these two elements may function independently to regulate the translation process.
Common and parallel mechanisms modulate translational control in Arabidopsis
Translations are enhanced by light treatment (this study) or are repressed by dehydration and hypoxia (Kawaguchi et al, 2004; Branco-Price et al, 2005). Interestingly, transcripts regulated at the translational level in response to environmental stimuli have shorter cDNA length (this study and Branco-Price et al, 2005). Furthermore, cis-elements were enriched in the 5′ UTRs of the transcripts differentially translated under light- or hypoxia-triggered translational control (Figure 8B). These results suggest the existence of a common regulatory mechanism to modulate immediate translational reprogramming for timely responsiveness to environmental changes.
Although significantly overrepresented, the two cis-elements identified are present only in a small fraction of transcripts undergoing translational control (Figure 8B). Because of only limited numbers of cis-elements identified and low numbers of genes with these cis-elements, additional regulatory mechanisms may enhance translation of transcripts with sequence or structural features such as shorter cDNA length and stability of transcripts. The current approach is unable to reveal conserved RNA sequence signatures based on the secondary or tertiary structures. Also, whether specific ‘cargos’ exist for batch translation of certain transcript species in response to environmental signals is unknown. Studies in these unexplored areas may provide new insights into the molecular mechanisms of translational control and further elucidate their impact on optimizing the growth fitness of organisms living in the ever-changing environment.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 was surface sterilized, sown on half-strength MS medium plates (0.8% agar, pH 5.7), and kept at 4°C in darkness for 4 days for stratification. For the dark treatment, the seeds were grown in darkness at 22°C for 4 days (Dark). For the light treatment, 4-day-old etiolated seedlings were illuminated with white light (70–80 μmol m−2 s−1) at 22°C for 0.5 h (L0.5 h) or 4 h (L4 h).
Isolation of total RNA and polysomal RNA
The aerial part of seedlings was harvested and ground by liquid nitrogen. Total RNA was isolated as described previously (Chang et al, 2008).
For the isolation of polysomal RNA, about 750 μl of frozen tissue powder was extracted with 375 μl of polysome extraction buffer (200 mM Tris–HCl, pH 8.5, 50 mM KCl, 25 mM MgCl2, 100 μg ml−1 heparin, 50 μg ml−1 cycloheximide, 400 U ml−1 RNasin (Promega, Madison, WI), 2% polyoxyethylene 10 tridecyl ether, 1% deoxycholic acid; recipe modified from Davies and Abe, 1995). The resuspended mixture was incubated on ice for 5 min, then spun at 15 000 g for 5 min at 4°C. In all, 400 μl supernatant was loaded on a 10-ml continuous sucrose gradient (15–50%) prepared by a gradient maker (ISCO, Lincoln, NE), and spun at 210 000 g for 3.5 h at 4°C. The distribution of the nucleic acids was examined by a UV254 absorbance profile (model #UV-6, ISCO). Total RNA from different polysomal fractions was purified with phenol/chloroform extraction, LiCl precipitation and dissolved in RNase-free H2O for further analyses.
For polysome-bound (PL) RNA, PL% was used as a calibration factor for ‘Per-Chip’ normalization as described previously (Kawaguchi et al, 2004; Branco-Price et al, 2005) except estimating PL% from the RNA quantity in NP and PL fractions. PL% is the percentage of the polysomal RNA relative to the total RNA as calculated by the following formula:
Affymetrix GeneChip hybridization and data analyses
Three biological replicates were performed for total or polysomal RNA isolated from each treatment indicated. Total RNAs were labeled and hybridized to Arabidopsis ATH1 Genome Array (Affymetrix Inc., San Jose, CA) as suggested by the manufacturer. Gene expression data for Affymetrix ATH1 were extracted with MAS5.0 (Affymetrix Inc.) and analyzed as described previously (Lin and Wu, 2004) with the following modifications. First, probes marked as ‘_s_’ or ‘_x_’, indicating they are less specific, were removed. Second, probe sets had to be marked as ‘present’ or ‘marginal’ in all 18 data sets. Third, the average intensity had to be ⩾50 in all three biological replicates from at least one of the time points examined. The 11 598 genes shown in Figure 2C passed these three criteria and were considered as ‘expressed genes’ at total or polysomal RNA levels.
For calculating the changes in both steady-state and polysome-bound RNA, the average intensity in all 18 chips was scaled to 500. All measurements with intensity <0.01 were raised to 0.01. For per-chip normalization, 50th percentile for the chips of steady-state mRNA measurement and the PL% for the chips of polysome-bound mRNAs were used, respectively. For per-gene normalization, ratios of light to dark samples belonging to the same biological experiment were calculated. The above normalizations were performed with the use of GeneSpring 7.3 (Agilent, Santa Clara, CA). Two-class Significance Analysis of Microarrays (SAM) was used for identifying genes with significantly different expressions (Tusher et al, 2001). Only genes that passed the criterion of a false discovery rate (FDR) listed in Supplementary Figure S2 were selected for further studies. Genes with expression ratios of ⩾three-folds or ⩽1/3-fold were considered upregulated or downregulated, respectively. A k-means cluster analysis (GeneSpring 7.3; Agilent) was performed to categorize the differentially expressed genes as shown in Supplementary Figure S5 and Figure 5. Parameters included the specification of 4 clusters and 200 iterations, and Pearson's correlation was used for similarity measurement. The data sets were deposited in NCBI′s Gene Expression Omnibus (Edgar et al, 2002) and accessible through the Gene Expression Omnibus series accession number GSE29657.
qRT–PCR analyses
The synthesis of cDNAs from total RNA and qRT–PCR were as described (Wu et al, 2008) with primers listed in Supplementary Table S6.
Total RNA in the NP and PL fractions and PL subfraction were isolated as described above, except 20 μl of 1:5000 diluted spike-in DAP mRNA was added before RNA extraction (GeneChip Eukaryotic Poly-A RNA control kit, Ambion, Foster City, CA). For the detection of changes in ribosome occupancy, the expression of spike-in DAP was used as a control to normalize the efficiency of the RNA purification, cDNA synthesis and PCR process. The RNA quantities of NP and PL fractions were summed and set as 100% for the calculation of the relative percentage of each transcript in the PL fraction.
mRNA feature analyses
Data sets for the 5′ UTR, CDS, 3′ UTR, full-length cDNA sequences and the representative gene models for all Arabidopsis genes were retrieved from The Arabidopsis Information Resources (TAIR9; ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR9_genome_release). The representative gene model was used as a reference gene model for analyses. For whole-genome gene lists shown in Figures 6–8, 15 971 5′ UTR, 21 119 CDS, 22 176 cDNA and 16 996 3′ UTR were used. An Excel Macro for the calculation of K–S statistic was downloaded from the Arizona Laserchron Center at the University of Arizona (http://docs.google.com/View?id=dcbpr8b2_7c3s6pxft).
The prediction of the overrepresented cis-element(s) involved use of Multiple Em for Motif Elicitation (MEME; Bailey and Elkan, 1994). The minimum and maximum motif width was set to 6 and 8, respectively, and the E-value was used to evaluate the significance of occurrence for the putative cis-elements. For genes regulated at the translational level on 9 h hypoxia treatment, the expression data were retrieved from a previous report (Branco-Price et al, 2008), filtered and clustered according to the criteria used in this study to reveal 1746 genes regulated at the ‘Protein’ level (Supplementary Table S5). Sequence information for 5′ UTRs is available for 1447 and 1668 of the 1553 and 1746 genes responsive to light and hypoxia, respectively.
Gene ontology and pathway analyses
The Gene Ontology category of ‘biological process’ (ATH_GO_GOSLIM_20100427) was downloaded from TAIR. The enriched functional groups were revealed with use of the ‘elim’ method from the topGO package (v1.16.2) (Alexa et al, 2006) that is part of the Bioconductor project (v2.6) in R (v2.11.0). The ‘Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY’ database was downloaded from the KEGG website (ftp://ftp.genome.jp/pub/kegg/pathway/organisms/ath; version 9 October 2009) and integrated into GeneSpring 7.3 (Agilent) for discovering pathways enriched in selected regulatory groups. We retrieved 92 genes involved in the light signal pathway from TAIR (Keyword:photomorphogenesis) (GO:0009640) or from previous studies of the phytochrome signal mechanism (Wang and Deng, 2002).
Assays of cis-element
Full-length 5′ UTR (65 bp) of At5g23330 was amplified from Arabidopsis genomic DNA with use of primers T7-WT-F and WT-R in Supplementary Table S6 to create a fusion of T7 promoter and the 5′ UTR sequence. This amplicon was used to replace the SalI-NcoI fragment between the 35S promoter and the LUC2 gene in pJD301 to create the WT-LUC2 construct as illustrated in Figure 8C. S1-LUC2 and S2-LUC2 constructs with scrambled cis-elements were generated with the WT-LUC2 construct used as a template and primers S1-F, S2-F and Scrm-R (Supplementary Table S6) by use of a Phusion site-directed mutagenesis kit (ThermoFisher Scientific, Waltham, MA).
For in-vitro transcription and translation, the WT-LUC2, S1-LUC2 and S2-LUC2 plasmids were linearized by SacI for in-vitro transcription with the AmpliScribeTM T7 High Yield Transcription Kit (EPICENTRE, Madison, WI). In total, 10 ng of the transcripts derived from WT-LUC2, S1-LUC2 or S2-LUC2 was translated for 40 min by use of wheat germ extract (Promega). LUC2 activity was assayed with the Luciferase Assay System (Promega) and measured with an EG&G Berthold Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany).
Immunoblot analyses
BBX22-specific antibody (Chang et al, 2011) and HY5-specific antibody generated against a synthetic peptide (MQEQATSSLAASSLPSS) were used for detecting BBX22 and HY5 protein, respectively. Plant samples were collected from 4-day-old etiolated seedlings treated with white light for 0 min, 0.5 h and 4 h. The total protein extraction and detection procedure were as described (Chang et al, 2011).
Supplementary Material
Supplementary Figures S1-8, Supplementary Tables S4 and S6
Acknowledgments
We thank Wen-Dar Lin for technical assistance with bioinformatics analyses, and Tien-Hsien Chang and Hua Dai for helpful discussion on the polysome profiling experiments. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Lab (http://ipmb.sinica.edu.tw/affy/) supported by Academia Sinica. This research was supported by a grant to S-H Wu from Academia Sinica and fellowship to M-J Liu and S-H Wu from the Taiwan International Graduate Program.
Author contributions: M-JL designed and performed research, analyzed data and wrote the paper. Szu-Hsien Wu designed and performed research and analyzed data. H-MC designed research. Shu-Hsing Wu designed research and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agresti A (1992) A survey of exact inference for contingency tables. Stat Sci 7: 131–177 [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey-Serres J, Sorenson R, Juntawong P (2009) Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci 14: 443–453 [DOI] [PubMed] [Google Scholar]
- Beyer A, Hollunder J, Nasheuer HP, Wilhelm T (2004) Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics 3: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Yanovsky MJ (2005) Regulation of gene expression by light. Int J Dev Biol 49: 501–511 [DOI] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, Somerville S, Wu SH (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH (2011) COP1-Mediated Degradation of BBX22/LZF1 Optimizes Seedling Development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J (2010) Light signal transduction: an infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E, Abe S (1995) Methods for isolation and analysis of polyribosomes. Methods Cell Biol 50: 209–222 [DOI] [PubMed] [Google Scholar]
- Davis JC (1986) Statistics and Data Analysis in Geology. New York: John Wiley & Sons [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF (1998) Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA (2009) Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol 16: 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Webster CI, Gray JC (1997) Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J 12: 499–506 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J (2009) Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM (2010) Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Karniol B, Chamovitz DA (2000) The COP9 signalosome: from light signaling to general developmental regulation and back. Curr Opin Plant Biol 3: 387–393 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J (2002) Regulation of translational initiation in plants. Curr Opin Plant Biol 5: 460–465 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carre IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Bahler J (2008) Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell Mol Biol 271: 199–251 [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JF, Wu SH (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Matsuura H, Ishibashi Y, Shinmyo A, Kanaya S, Kato K (2010) Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol 51: 448–462 [DOI] [PubMed] [Google Scholar]
- McKim SM, Durnford DG (2006) Translational regulation of light-harvesting complex expression during photoacclimation to high-light in Chlamydomonas reinhardtii. Plant Physiol Biochem 44: 857–865 [DOI] [PubMed] [Google Scholar]
- Melamed D, Arava Y (2007) Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol 431: 177–201 [DOI] [PubMed] [Google Scholar]
- Melamed D, Pnueli L, Arava Y (2008) Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 14: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Grotewold E (2005) Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug JH, Wobbe L, Elles I, Claus C, Hamilton M, Fink A, Kahmann U, Kapazoglou A, Mullineaux CW, Hippler M, Nickelsen J, Nixon PJ, Kruse O (2005) NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 17: 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O′Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai M, Roncato MA, Canoy AS, Rouquie D, Sarda X, Freyssinet G, Robaglia C (2006) Large-scale analysis of mRNA translation states during sucrose starvation in arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol 141: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Hohne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC (2010) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Vaughn JN, Kim BH, Zhou F, Gilchrist MA, Von Arnim AG (2010) The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW (2000) The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin Cell Dev Biol 11: 495–503 [DOI] [PubMed] [Google Scholar]
- Shama S, Meyuhas O (1996) The translational cis-regulatory element of mammalian ribosomal protein mRNAs is recognized by the plant translational apparatus. Eur J Biochem 236: 383–388 [DOI] [PubMed] [Google Scholar]
- Sherameti I, Nakamura M, Yamamoto YY, Pfannschmidt T, Obokata J, Oelmuller R (2002) Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transport. Plant J 32: 631–639 [DOI] [PubMed] [Google Scholar]
- Sormani R, Delannoy E, Lageix S, Bitton F, Lanet E, Saez-Vasquez J, Deragon JM, Renou JP, Robaglia C (2011) Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol 52: 436–447 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE (2004) Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics 3: 960–969 [DOI] [PubMed] [Google Scholar]
- Tremousaygue D, Manevski A, Bardet C, Lescure N, Lescure B (1999) Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J 20: 553–561 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Phytochrome signaling mechanism. In The Arabidopsis Book, Somerville CR, Meyerowitz EM (eds), Rockville, MD: American Society of Plant Biologists [Google Scholar]
- Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148: 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom A, Kim TH, Roy B, Singer R, von Arnim AG, Chamovitz DA (2008) Arabidopsis eIF3e is regulated by the COP9 signalosome and has an impact on development and protein translation. Plant J 53: 300–311 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-8, Supplementary Tables S4 and S6