Abstract
As the Wnt/β-catenin signaling pathway is linked to melanoma pathogenesis and to patient survival, we conducted a kinome siRNA screen in melanoma cells to expand our understanding of kinases that regulate this pathway, and to illuminate potential therapeutic directions. We found that BRAF signaling, which is constitutively activated in many melanomas by the BRAFV600E mutation, negatively regulates Wnt/β-catenin signaling in human melanoma cells. As inhibitors of BRAFV600E show promise in ongoing clinical trials we investigated whether altering Wnt/β-catenin signaling might enhance the efficacy of the BRAFV600E inhibitor, PLX4720. Surprisingly, endogenous β-catenin is required for PLX4720 to induce apoptosis in melanoma cells, while activation of Wnt/β-catenin signaling strongly synergizes with PLX4720 to decrease tumor growth in vivo and to increase apoptosis in vitro. This synergistic enhancement of apoptosis correlates with a reduction in the abundance of a β-catenin antagonist, AXIN1. In support of the hypothesis that AXIN1 is a mediator rather than a marker of apoptosis, melanoma cell lines that are resistant to apoptosis after treatment with a BRAFV600E inhibitor become susceptible, and undergo apoptosis, when AXIN1 is reduced by siRNA. These findings point to a role for Wnt/β-catenin signaling and AXIN1 in regulating the efficacy of inhibitors of BRAFV600E, and may stimulate consideration of potential combination therapies and biomarkers for use in conjunction with targeted BRAF therapy.
Introduction
The majority of both benign nevi and cutaneous melanomas harbor activating mutations in the BRAF oncogene, with BRAFV600E representing the most common of these mutations (1). The recent development of small molecule compounds designed to specifically target BRAFV600E, including PLX4720 (2), PLX4032 (vemurafenib) (3, 4), and GSK2118436 (5) has led to subsequent clinical trials that demonstrated an unprecedented tumor response rate in almost all patients with BRAFV600E tumors (5–7). However, despite the initial tumor response, only half of patients with BRAFV600E tumors meet established criteria for a confirmed objective clinical response. Furthermore, half of the patients exhibiting an initial response to BRAFV600E inhibitors develop resistant tumors and progressive disease within six months. These results highlight the need to identify regulatory interactions between BRAF signaling and other cellular pathways that may provide avenues for enhancing the long-term clinical effects of targeted BRAF inhibitors in melanoma treatment.
Activation of Wnt/β-catenin signaling promotes the nuclear functions of β-catenin (CTNNB1), resulting in the regulation of cell proliferation, differentiation, and behavior (8). The exact role of Wnt/β-catenin signaling in melanoma progression remains controversial. While transgenic mouse models expressing a melanocyte-specific, constitutively-active mutant β-catenin did not display any spontaneous melanomas, co-expression of a constitutively-active mutant Nras resulted in mice that exhibited enhanced immortalization of melanocytes and increased melanoma tumor promotion (9). By contrast, the decreased survival observed in patients exhibiting lower abundance of nuclear β-catenin in their tumors suggests that the loss of Wnt/β-catenin signaling plays an important role during melanoma evolution (10–14). Although benign nevi and a substantial number of melanoma tumors exhibit elevated nuclear β-catenin (10, 11, 13, 14), activating mutations in the Wnt/β-catenin pathway are rare in melanoma (5–17). Thus the mechanisms underlying elevated β-catenin in melanoma are unresolved as well as the functional significance of β-catenin in this context.
The extracellular signal-regulated kinases (ERKs), which are activated by multiple signals, represent another signaling pathway linked to melanoma (15). ERK signaling works via RAS small G proteins to activate RAF kinases, which phosphorylate and activate the kinases MEK1/2, which subsequently phosphorylate and activate the kinases ERK1/2. ERK1/2 phosphorylate and regulate numerous substrates leading to a variety of cell type and context-dependent responses (16). With regard to melanoma, constitutive activation of ERK1/2 by activating mutations in NRAS or BRAF is observed in the majority of melanomas and plays an integral role in the regulation of proliferation, invasiveness, and survival (17).
Several instances of crosstalk between Wnt/β-catenin and MAPK signaling have been reported with the majority revealing that Wnt/β-catenin regulates MAPK signaling (18). Conversely, others have reported that EGF-induced ERK activation in glioblastoma cell lines leads to phosphorylation of casein kinase-II (CSNK2) and to disruption of the interaction between β-catenin and α-catenin (19). Disruption of this complex then enhances β-catenin target gene trans-activation and subsequent tumor cell invasion.
Our current study reveals an unexpected cross-talk between BRAF and Wnt/β-catenin signaling in regulating apoptosis and the abundance of the scaffolding protein AXIN1 in melanoma. Specifically, we first demonstrate that activation of BRAF signaling by the BRAFV600E mutation negatively regulates Wnt/β-catenin signaling. Further supporting cross-talk between BRAF and Wnt/β-catenin signaling, we then show that endogenous β-catenin is required for the BRAFV600E inhibitor PLX4720 to induce apoptosis in melanoma. Moreover, activation of Wnt/β-catenin signaling enhances the ability of PLX4720 to reduce melanoma tumor growth in vivo and strongly synergizes with PLX4720 to reduce melanoma cell growth and to increase apoptosis in vitro. Mechanistically, we show that inhibition of BRAFV600E enhances Wnt-mediated reduction in the abundance of AXIN1, leading to elevation of Wnt/β-catenin signaling and to increases in β-catenin-mediated apoptosis of melanoma cells. Furthermore, knockdown of AXIN1 by siRNA sensitizes melanoma cell lines otherwise resistant to apoptosis following BRAFV600E inhibition. These results have implications for improving the efficacy of inhibitors of BRAFV600E in treating melanoma, as well as revealing functional cross-talk between Wnt/β-catenin and BRAF signaling in melanoma.
Results
BRAFV600E is a negative regulator of Wnt/β-catenin signaling in melanoma cells
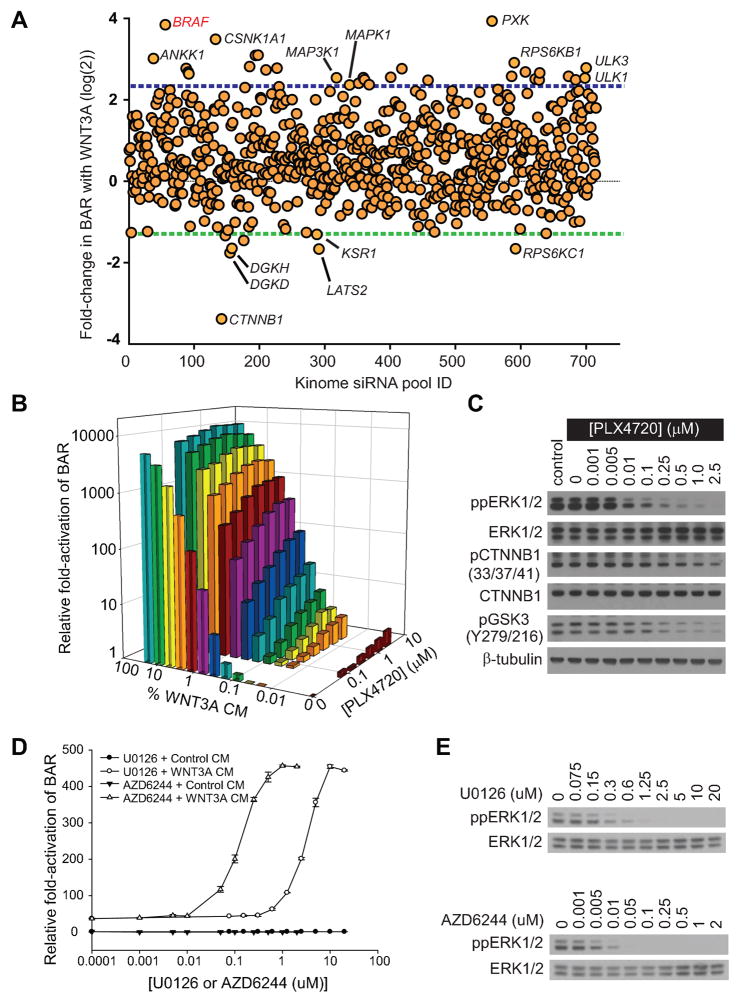
To identify new regulators of Wnt/β-catenin signaling in melanoma we employed A375 human melanoma cells (which harbor the BRAFV600E mutation) stably expressing the β-catenin-activated reporter (BAR) (20). These cells were then used in a high-throughput siRNA screen targeting 716 genes encoding known or predicted kinases. This screen revealed that BRAF siRNAs strongly synergize with WNT3A to activate the BAR reporter (Figure 1A and Supplemental Figure S1A-B and Supplemental Databases 1–2). This result was validated with four independent siRNAs targeting BRAF as well as with a published siRNA that specifically targets activated BRAFV600E (21) (Figure S2A-C and Supplemental Table S1). These data support the unexpected hypothesis that activated BRAFV600E negatively regulates Wnt/β-catenin signaling in melanoma.
Figure 1. BRAF signaling negatively regulates Wnt/β-catenin signaling in melanoma cells.
A) Scatter plot of a kinome-based siRNA screen in human A375 melanoma cells stably expressing the β-catenin-activated reporter (BAR) driving firefly luciferase, with each dot representing a known or predicted kinase. Blue- and green-dotted lines represent two mean absolute deviations above and below the mean, respectively. The full gene list is presented in Supplementary database S1–S2. B) An isobologram analysis shows a dose-dependent enhancement of Wnt/β-catenin signaling with the targeted BRAF inhibitor PLX4720 and WNT3A CM on BAR activity. C) Immunoblot analysis of the dose-dependent inhibition of dual-phosphorylated ERK1/2 (ppERK1/2), phosphorylated Ser33/Ser37/Thr41 β-catenin (pCTNNB1), and phosphorylated Tyr216 GSK3 following PLX4720 treatment. D) Two distinct MEK inhibitors, U0126 and AZD6244, both enhanced Wnt/β-catenin signaling in a dose-dependent manner. Symbols and error bars represent the mean and standard deviation, respectively, of three biologic replicates E) Immunoblot analysis of the dose-dependent inhibition of ppERK1/2 by MEK1/2 inhibitors U0126 and AZD6244. In (B–E), A375 cells were treated for 24 hours with the indicated conditions prior to harvesting and data are representative of at least three independent experiments.
We then asked whether the enhancement of Wnt/β-catenin signaling observed with BRAF siRNAs could be phenocopied with PLX4720, a small molecule designed to selectively inhibit the constitutively-active BRAFV600E mutant kinase (2). Supportingly, PLX4720 enhanced Wnt/β-catenin signaling in a dose-dependent manner (Figure 1B and Supplemental Figure S3A–B) at doses similar to its dose-dependent inhibition of dual-phosphorylated ERK1/2 (ppERK1/2) (Figure 1C). Combination indices for WNT3A and PLX4720 were much less than 1 (Supplemental Figure S3C), supporting a synergistic interaction between these two drugs with respect to Wnt/β-catenin activation. In further support of a synergistic interaction, the addition of PLX4720 led to a calculated WNT3A dose-reduction index of 6.0 at a BAR response corresponding to the EC50 for WNT3A alone.
Consistent with its acting as an enhancer of Wnt/β-catenin signaling, PLX4720 treatment decreased phosphorylation of β-catenin at sites normally phosphorylated by glycogen synthase kinase-3 (GSK3) to target β-catenin for proteasomal degradation (Figure 1C). In support of this observation, the activating auto-phosphorylation of GSK3 at Tyr216 was lost upon treatment of cells with PLX4720 (Figure 1C). While these effects on phosphorylation of β-catenin and GSK3 did not result in increased abundance of cytosolic or nuclear β-catenin (Supplemental Figure S4), recent findings have established that activation of β-catenin function in melanoma correlates with the same changes in phosphorylation shown here (Figure 1C) rather than with changes in the abundance of β-catenin (22). As BRAF signals through the downstream kinase MEK, we next investigated the effects on β-catenin signaling of two independent small molecule MEK inhibitors, U0126 and AZD6244 (23). We found that both drugs synergistically enhanced Wnt/β-catenin activation as measured by the BAR assay (Figure 1D), and did so at doses similar to their dose-dependent inhibition of ppERK1/2 (Figure 1E). These results solidify the role of BRAF signaling via MEK as a regulator of Wnt/β-catenin signaling in melanoma.
WNT3A enhances the ability of an inhibitor of BRAFV600E to reduce tumor size
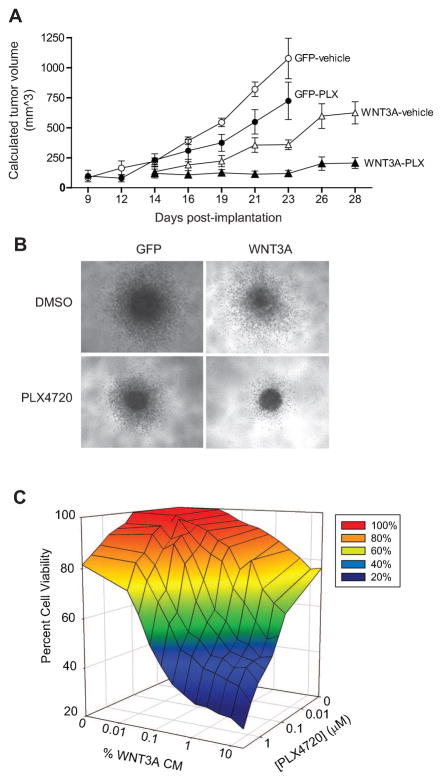
As targeted inhibition of BRAFV600E or activation of Wnt/β-catenin signaling reduces melanoma tumor size (2, 11, 24), and as we show above that the inhibition of BRAFV600E enhances Wnt/β-catenin signaling, we next asked whether concurrent inhibition of BRAFV600E and activation of Wnt/β-catenin signaling would cooperate to reduce tumor size. Immunosuppressed mice harboring subcutaneous xenografts generated from human A375:GFP cells (controls) or A375:WNT3A cells (expressing WNT3A-iresGFP) were treated by oral gavage with either vehicle or PLX4720. Inhibition of ppERK1/2 in vivo following PLX4720 treatment was confirmed using biochemical analysis of fine-needle aspirates sampled from tumors during treatment (Supplemental Figure S5A–B). Results of the xenograft study revealed that treatment of A375:GFP tumors with PLX4720 decreased tumor growth compared to drug vehicle (Figure 2A). The growth of A375:WNT3A tumors was slower compared to both A375:GFP tumors treated with vehicle and A375:GFP tumors treated with PLX4720. Remarkably, the effects of PLX4720 on A375:WNT3A tumor growth was even more pronounced than the effects on A375:GFP tumors, with near complete suppression of A375:WNT3A tumor growth over four weeks. Growth curves were significantly different upon one-way ANOVA with a post-test for linear trend (p=0.024). Direct comparisons of tumor volume between groups at day 23 (Supplemental Figure S5C) revealed a highly significant difference upon one-way ANOVA with post-test for linear trend (p<0.0001). These results paralleled the significant differences seen in mitotic index (p<0.0001) upon histological analysis of the xenografts (Supplemental Figure S5D). We conclude that WNT3A greatly enhances the ability of PLX4720 to reduce melanoma tumor size in this xenograft assay.
Figure 2. Wnt/β-catenin activation cooperates with targeted inhibition of mutant BRAF to inhibit tumor growth in vivo and in vitro.
A) WNT3A enhances the ability of the BRAF inhibitor, PLX4720, to reduce tumor size in vivo. Human A375 melanoma cells expressing either GFP or WNT3A(iresGFP) were grown as xenografts in NSG mice treated with either vehicle or 50 mg/kg PLX4720 after tumors had reached an initial size of 100mm3. For each treatment arm, the means and SEM are shown for five individual mice. B) WNT3A enhances the ability of the BRAF inhibitor PLX4720 to reduce spheroid size and in vitro. Human A375 melanoma cells expressing either GFP or WNT3A(iresGFP) were grown as spheroids in a three-dimensional collagen matrix, then treated with either DMSO or 2μM PLX4720 for 72 hours prior to imaging. Representative spheroids of greater than forty spheroids per treatment are shown in these light micrographs. C) WNT3A synergizes with PLX4720 to inhibit the viability of A375 melanoma cells. A375 melanoma cells were treated with the indicated combinations of WNT3A CM and PLX4720 concentrations for 48 hours. In (A–C), data are representative of at least three independent experiments with each data point assayed in triplicate.
To confirm and extend these results we turned to a three-dimensional spheroid assay of tumor cell growth and invasion within a collagen matrix. Treatment of both A375:GFP- and A375:WNT3A-derived spheroids with PLX4720 decreased spheroid size (Figure 2B), paralleling the decreased tumor sizes observed in xenograft studies (Figure 2A). Treatment of spheroids expressing WNT3A with PLX4720 led to a dramatic decrease in the number of invasive cells at 72 hours compared to either A375:WNT3A-derived spheroids treated with DMSO or to A375:GFP-derived spheroids treated with PLX4720 (Figure 2B).
We next tested for synergistic inhibition of melanoma cell growth by WNT3A and PLX4720 in two-dimensional cell culture. Cell viability was measured in A375 melanoma cells treated with combinations of WNT3A and PLX4720 at various concentrations (Figure 2C and Supplemental Figure S6A–B). The reduced number of viable cells following combined treatment with WNT3A and PLX4720 resulted in combination indices much less than 1 (Supplemental Figure S6C). At 50% growth inhibition, the drug reduction indices were 8.1 for PLX4720 and 117.4 for WNT3A CM, further supporting a synergistic effect of these two drugs. Together, these studies using three different assays demonstrate that the simultaneous activation of Wnt/β-catenin signaling and the targeted inhibition of BRAFV600E by PLX4720 functionally cooperate to decrease melanoma cell growth both in vivo and in vitro.
WNT3A enhances the ability of an inhibitor of BRAFV600E to increase apoptosis
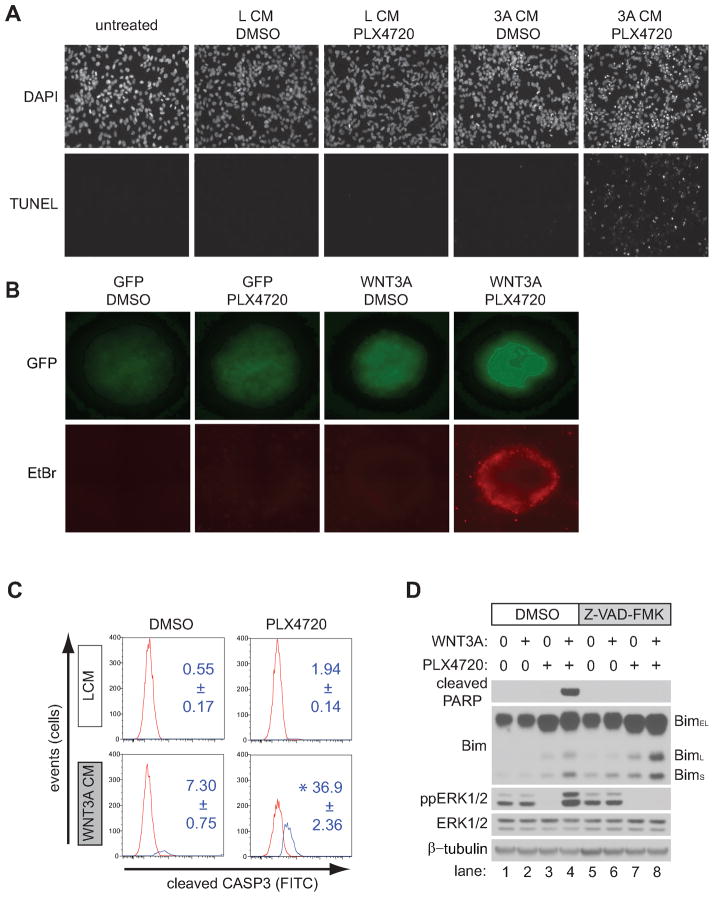
The data above establish that inhibition of BRAFV600E reduces tumor size, and that this effect is enhanced by activating Wnt/β-catenin signaling. We next asked whether this reduction in tumor size was the consequence of cell death. TUNEL staining of melanoma cells treated for 24 hours with WNT3A conditioned media (CM) and PLX4720 indicated the presence of apoptotic cell death (Figure 3A), a finding consistent with the detection of dead cells only in A375 spheroids concurrently expressing WNT3A and treated with PLX4720 (Figure 3B). These findings were confirmed by flow cytometry using an antibody that detects the cleaved (active) form of caspase-3 (Figure 3C). Consistent with the TUNEL and the spheroid assays, no apoptosis was seen in the presence of DMSO vehicle alone, and minimal increases in cleaved caspase-3 were seen with either PLX4720 or WNT3A CM alone. However, in the presence of both WNT3A CM and PLX4720, cleaved caspase-3 increased ~5–20-fold (Figure 3C). In support of a caspase-mediated apoptotic pathway, combined treatment with WNT3A and PLX4720 led to synergistic cleavage of the caspase-3 substrate, PARP1 (Figure 3D lane 4 versus lane 2 and 3). PARP1 cleavage was completely abolished by addition of the pan-caspase inhibitor, Z-VAD-FMK (Figure 3D, lane 8 versus lane 4). To establish that the effects of PLX4720 on apoptosis were specifically due to inhibition of BRAFV600E, we showed that knockdown of BRAF by siRNA mimics the ability of PLX4720 to enhance the cleavage of caspase-3 in the presence of WNT3A CM (Supplemental Figure S7). Together, these data demonstrate that simultaneous activation of Wnt/β-catenin signaling and inhibition of BRAFV600E functionally cooperate to induce caspase-mediated apoptosis of melanoma cells.
Figure 3. Wnt/β-catenin activation synergistically enhances apoptosis with BRAF inhibition.
A) TUNEL staining was used to visualize apoptosis in A375 cells following treatment with the indicated conditions, including WNT3A CM (3A CM) or L CM. B) Spheroids generated from A375 cells expressing either GFP or WNT3A(iresGFP) were grown in a three-dimensional collagen matrix and treated with the indicated conditions. Simultaneously, GFP was used to image all cells while EtBr staining was used to identify dead cells. Representative spheroids of greater than forty spheroids for each condition are shown in these panels. C) A flow cytometry-based assay for cleaved caspase-3 was used to detect apoptotic cells following treatment with the indicated conditions in A375 cells. Red and blue peaks on the representative histograms indicate the distribution of cells that were negative and positive for caspase-3 staining respectively. Numbers indicate the average percentage and standard deviation of caspase-3 positive cells from three biological replicates. D) Immunoblot analysis of the proapoptotic protein Bim in A375 cells treated with the indicated conditions. BimEL, BimL, and BimS represent the three major isoforms of Bim. In (A–D), cells were treated for 24 hours with the indicated conditions and data are representative of at least three independent experiments. Where indicated, cells were treated with 2μM PLX4720 and 100μM Z-VAD-FMK.
In order to understand how activation of Wnt/β-catenin signaling cooperates with inhibition of BRAFV600E to induce apoptosis in melanoma cells, we next explored the Bcl-2 homology domain 3 only (BH3-only) protein Bim (BCL2L11). Bim is an important regulator of melanoma apoptosis that can bind and inhibit all pro-survival Bcl-2 family members (25–32). The three major isoforms of Bim, BimEL, BimL, and BimS are generated by alternative splicing and vary in their pro-apoptotic activity, with BimS being the most potent followed by BimL (25). A recent study in melanoma cells expressing BRAFV600E showed that PLX4720 treatment increased the expression of all Bim isoforms and that the increased expression of BimS was the primary driver of apoptosis (26). We found that PLX4720 treatment of A375 melanoma cells enhanced the expression of all Bim isoforms (Figure 3D, lane 3 versus lane 1). In support of Wnt/β-catenin enhancing PLX4720-mediated apoptosis, co-treatment of cells with WNT3A led to a reproducible increase in BimL and BimS (Figure 3D, lane 4 versus 3). This increase in BimL and BimS is not blocked by Z-VAD-FMK (Figure 3D, lane 8 versus lane 4), consistent with its role as an upstream activator of caspase-3 during apoptosis. Taken together these data reveal that WNT3A increases the effectiveness of a BRAFV600E inhibitor to promote apoptosis in melanoma cells, through a mechanism that may involve Bim.
Endogenous β-catenin is required for PLX4720 to induce apoptosis
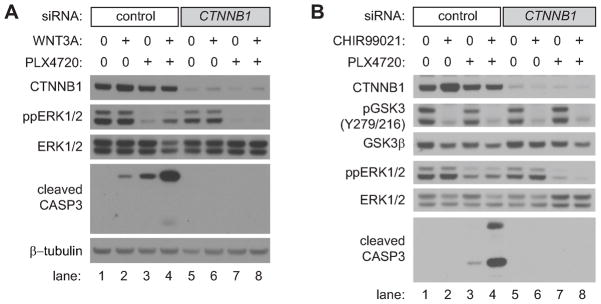
We then investigated whether the BRAFV600E inhibitor PLX4720 requires a functional Wnt/β-catenin pathway for its ability to induce apoptosis. Strikingly, β-catenin (CTNNB1) siRNA completely prevents apoptosis of A375 cells treated with PLX4720 (Figure 4A, lane 3 versus lane 7). This dependence on endogenous β-catenin for PLX4720 to elevate apoptosis was not overcome by exogenous WNT3A (Figure 4A, lane 4 versus lane 8). We then activated β-catenin signaling downstream of the Wnt-receptor complex by treating cells with the small molecule GSK3 inhibitor CHIR99021. Like WNT3A, CHIR99021 enhanced apoptosis in combination with PLX4720, and this apoptosis was completely inhibited upon siRNA knockdown of β-catenin (Figure 4B, lane 4 versus lane 8). These data support the unexpected conclusion that apoptosis mediated by targeted BRAF inhibition is dependent upon β-catenin, the primary downstream effector of Wnt/β-catenin signaling.
Figure 4. Apoptosis mediated by Wnt/β-catenin signaling and BRAF inhibition requires β-catenin.
A) An immunoblot analysis of cleaved caspase-3 from A375 cells pretreated with control or β-catenin (CTNNB1) siRNA followed by treatment with the indicated conditions. Cells were transfected with siRNAs, incubated for 48 hours, and then treated with the indicated conditions for 48 hours. Where indicated, cells were treated with 2μM PLX4720. B) Immunoblot analysis of cleaved caspase-3 from A375 cells pretreated with control or β-catenin (CTNNB1) siRNA followed by treatment with the indicated conditions. Cells were transfected with siRNAs, incubated for 48 hours, and then treated with the indicated conditions for 36 hours. Where indicated, cells were treated with 2μM PLX4720 and 5μM CHIR99021. Inhibition of GSK3 was confirmed by loss of the activating auto-phosphorylation at Tyr216. In (A–B), data are representative of at least three independent experiments.
PLX4720-mediated enhancement of Wnt/β-catenin signaling predicts apoptosis among melanoma cell lines
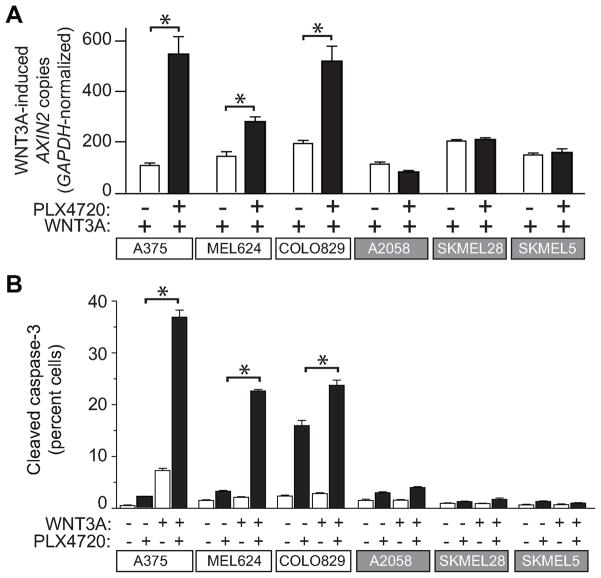
A considerable number of patients with tumors harboring activating BRAF mutations do not exhibit an objective clinical response to targeted BRAF inhibitors (7), suggesting the involvement of as yet unidentified proteins or signaling pathways that determine cellular susceptibility to therapy. We therefore asked whether any new insights into the heterogeneity of the response to targeted BRAF inhibitors could be gleaned by examining the interaction between Wnt/β-catenin and BRAF signaling in multiple melanoma cell lines that harbor the BRAFV600E mutation. In A375, MEL624 and COLO829 cells, treatment with WNT3A increased the abundance of AXIN2, a known target gene of Wnt/β-catenin signaling (11, 33), and co-treatment with PLX4720 led to further increases in levels of AXIN2 (Figure 5A). In contrast, treatment with PLX4720 did not elevate the WNT3A-mediated increases in AXIN2 transcripts in SKMEL5, SKMEL28, and A2058 cells (Figure 5A), despite the fact that these cells also harbor the BRAFV600E mutation (Supplemental Table S2). Interestingly, cell lines that display synergistic activation of Wnt/β-catenin signaling with WNT3A and PLX4720 also exhibit increased susceptibility to apoptosis as measured by cleaved caspase-3 (Figure 5B). These data are consistent with a model in which Wnt/β-catenin signaling is a major determinant of the apoptotic response to targeted BRAF inhibition (Figure 4A and 4B). Of note, the discrepancy in response among these cell lines cannot be accounted for by the allelic status of the BRAFV600E mutation (Supplemental Table S2). These data might be relevant to the observed variations in clinical response to targeted inhibitors of BRAF among tumors carrying the BRAFV600E mutation (5–7).
Figure 5. Regulation of Wnt/β-catenin signaling by BRAF predicts apoptotic response to combined Wnt/β-catenin activation and BRAF inhibition.
A) In six melanoma lines harboring BRAFV600E mutations, synergistic enhancement of Wnt/β-catenin signaling by BRAFV600E inhibition was examined by quantitative PCR measurements of the endogenous target gene AXIN2. Data are expressed as copies of AXIN2 per 106 copies of GAPDH. B) Flow cytometry detection of cleaved caspase-3 was used to measure apoptosis in several melanoma cell lines harboring BRAFV600E mutations. In (A–B), cells were treated with the indicated conditions for 24 hours and 2μM PLX4720 was used where indicated. In (A–B) Columns and error bars represent the mean and standard deviation, respectively, of three biologic replicates. Asterixes (*) represent p<0.001 by one-way ANOVA with Tukey’s post-test. Data are representative of at least three independent experiments.
Reduction of AXIN1 predicts apoptosis with Wnt/β-catenin activation and BRAF inhibition
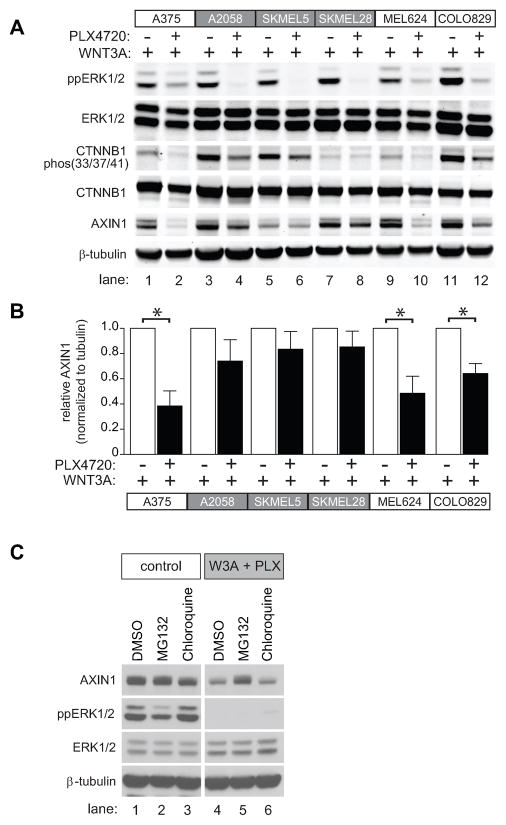
The correlation between Wnt/β-catenin signaling and apoptotic response (Figure 5A and 5B) led us to further investigate the underlying mechanisms. Interestingly, the three melanoma cell lines that displayed the greatest elevation in apoptosis in response to co-treatment with WNT3A plus PLX4720 (A375, MEL624, and COLO829), also displayed the greatest reduction in the abundance of the β-catenin antagonist AXIN1 when compared to WNT3A treatment alone (Figure 6A). This reduction was highly significant when the AXIN1 signal on immunoblots was quantified and normalized to β-tubulin from three separate experiments (Figure 6B). By contrast, in the melanoma cell lines that are resistant to apoptosis after treatment with WNT3A plus PLX4720 (SKMEL5, SKMEL28 and A2058) the relative abundance of AXIN1within each cell line did not significantly decrease when comparing treatment with WNT3A alone to WNT3A plus PLX4720 (Figure 6A and 6B). Together, these data demonstrate a direct correlation between melanoma cell apoptosis and the loss of AXIN1 in the presence of WNT3A and PLX4720. Of note, it is not the baseline levels of AXIN1, but rather the magnitude of reduction in AXIN1 abundance within each cell line, that is predictive of apoptosis with WNT3A and PLX4720. For example, MEL624 cells have relatively high baseline levels of AXIN1 (Figure 6A, lanes 9 and 10), and display a robust loss of AXIN1, and apoptosis, when treated with WNT3A and PLX4720. By contrast, SKMEL5 cells have very low baseline levels of AXIN1, and neither these levels of AXIN1, nor apoptosis, change markedly upon treatment with WNT3A and PLX4720 (Figure 6A, lanes 5 and 6).
Figure 6. AXIN1 abundance is positively regulated by BRAF signaling and reduction following BRAF inhibition predicts apoptotic response.
A) Immunoblot analysis of the same six melanoma cell lines analyzed in Figure 5 following treatment with the indicated conditions. B) The immunobimmlot of AXIN1 from (A) combined with immunoblots from two additional independent replicates were quantified by pixel intensity. In (A–B), A375 cells were treated with the indicated conditions for 24 hours. C) Reduced AXIN1 abundance following combined treatment with WNT3A and PLX4720 was rescued by the proteasome inhibitor MG132. A375 cells were treated with control conditions or WNT3A and PLX4720 in combination with DMSO or the proteasome inhibitor, MG132 or the lysosome inhibitor, chloroquine for 8 hours. In (A–C) 2μM PLX4720, 10μM MG132, and 10μM chloroquine were used where indicated. In (B) p-values were determined by two-tailed Student’s t-test and (*) represents p<0.005. In (A–C) data are representative of at least three independent experiments.
We next investigated how inhibition of BRAFV600E reduces the abundance of AXIN1. We first investigated whether the combination of WNT3A plus PLX4720 regulates the abundance of AXIN1 transcripts. Analysis of treated A375 melanoma cells by qRT-PCR revealed no changes in the abundance of AXIN1 transcripts after treatment of cells with WNT3A or PLX4720, either alone or in combination (Supplemental Figure S8). We next investigated whether WNT3A plus PLX4720 promotes proteasomal or lysosomal degradation of AXIN1. Treatment of A375 cells with MG132, but not chloroquine, rescued the decrease in AXIN1 seen upon treatment with WNT3A and PLX4720 (Figure 6C, lane 5 versus lane 4 and lane 6 versus lane 4). Together, these data reveal that BRAFV600E inhibition decreases the abundance of AXIN1 in the presence of WNT3A through a proteasome-dependent mechanism.
Loss of AXIN1 precedes apoptosis and can confer susceptibility to apoptosis with BRAF inhibition
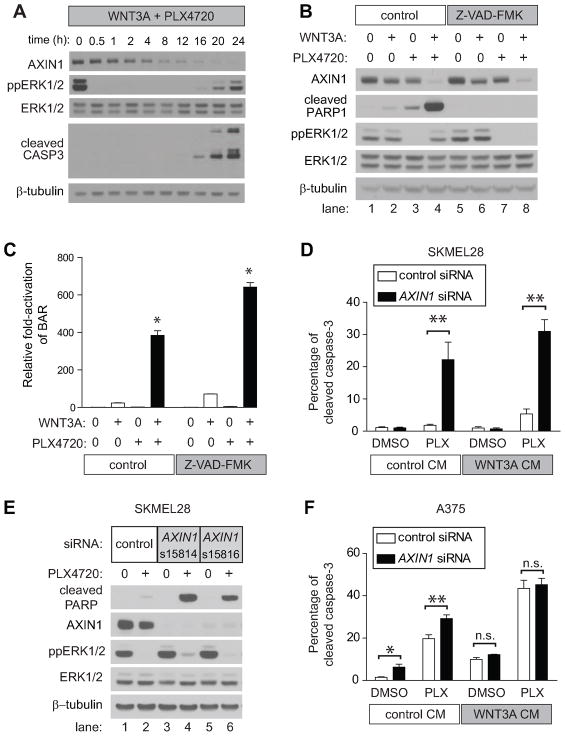
To determine whether decreases in AXIN1 abundance sensitize melanoma cells to PLX4720-mediated apoptosis, we first investigated the temporal coordination of ppERK1/2 relative to changes in AXIN1 abundance and to the onset of apoptosis. We performed a time course in A375 cells treated with WNT3A and PLX4720 followed by immunoblotting for AXIN1. A rapid decrease in AXIN1 abundance occurred within 1–2 hours of initiating treatment, with almost no detectable AXIN1 remaining after 16–20 hours of treatment (Figure 7A). This decrease in AXIN1 abundance followed the rapid inhibition of ppERK1/2, which occurred within 30 minutes of treatment. Apoptosis as measured by cleaved caspase-3 was first detected at 12–16 hours, and increased for the duration of the experiment (Figure 7A). These data suggest that loss of AXIN1 precedes caspase-mediated apoptosis. Furthermore, while the pan-caspase inhibitor Z-VAD-FMK was able to inhibit apoptosis in these cells (Figure 7B) it did not affect the loss of AXIN1, indicating that the decrease in AXIN1 is not a downstream consequence of caspase-3 activation. In agreement, inhibition of apoptosis by Z-VAD-FMK did not affect PLX4720-mediated enhancement of Wnt/β-catenin signaling as measured by BAR (Figure 7C). These data suggest that the enhanced Wnt/β-catenin signaling observed in cell lines that exhibit enhanced apoptosis with co-treatment of WNT3A and PLX4720 (Figure 5A and 5B) is upstream of caspase-3 activation. We conclude that decreases in AXIN1 abundance precede, and are independent of, the onset of apoptosis.
Figure 7. AXIN1 depletion sensitizes melanoma cells to apoptosis mediated by BRAF inhibition.
A) Immunoblot analysis of a time-course of A375 cells treated with WNT3A and PLX4720. B) The decrease in AXIN1 abundance following WNT3A and PLX4720 treatment is not rescued by Z-VAD-FMK. A375 cells were treated for 24 hours with the indicated conditions. C) PLX4720 enhancement of Wnt/β-catenin signaling is not dependent on caspase activation. A375 cells containing the BAR reporter were treated for 24 hours with the indicated conditions. The (*) represents p<0.001 when compared to all other conditions by one-way ANOVA with Tukey’s post-test. D) Flow cytometry detection of cleaved caspase-3 in SKMEL28 cells transfected with control or AXIN1 siRNA and treated with the indicated conditions for 24 hours. E) Knockdown of AXIN1 by siRNA sensitizes SKMEL28 cells to PLX4720-induced apoptosis. Immunoblots show lysates from SKMEL28 cells transfected with either control or two non-overlapping independent siRNAs targeting AXIN1 and treated with the indicated conditions for 24 hours. F) Flow cytometry detection of cleaved caspase-3 in A375 cells transfected with control or AXIN1 siRNA and treated with the indicated conditions for 24 hours. In (D) and (F), p-values were determined by two-way ANOVA with Bonferroni post-test; (*) represents p<0.05 and (**) represents p<0.001. In (A–F) 2μM PLX4720, 10μM MG132, and 100μM Z-VAD-FMK were used where indicated, and data are representative of at least three independent experiments.
Given that decreases in AXIN1 abundance (Figure 6A, 6B and 7A) precede apoptosis and seem to predict both susceptibility to Wnt and PLX4720-driven apoptosis (Figure 5B) as well as enhancement of Wnt/β-catenin signaling by BRAF inhibition (Figure 5A), we hypothesized that reducing the abundance of AXIN1 in the three melanoma cell lines that are more resistant to apoptosis (SKMEL28, A2058 and SKMEL5) would render them newly susceptible to apoptosis in the presence of PLX4720. Indeed, siRNA-mediated knockdown of AXIN1 (Supplemental Figure S9) led to increased apoptosis with PLX4720 as measured by cleaved caspase-3 in all three cell lines (Table 1 and Figure 7D). This result is consistent with our hypothesis that the reduction of AXIN1 abundance seen with β-catenin activation facilitates PLX4720-mediated apoptosis. The ability of AXIN1 knockdown to confer susceptibility to apoptosis with PLX4720 was confirmed using two independent and validated siRNAs (Figure 7E). In the A375 cell line that responds with robust apoptosis upon WNT3A and PLX4720 treatment, knockdown of AXIN1 by siRNA enhanced apoptosis with PLX4720 while apoptosis with either WNT3A or the combination of WNT3A and PLX4720 is not further enhanced by siRNA-mediated knockdown of AXIN1 (Figure 7F). These results strongly argue that the decrease in AXIN1 abundance observed with the combination of BRAF inhibition and Wnt/β-catenin activation plays an important and previously unsuspected role in the regulation of apoptosis in melanoma cells.
Table 1. AXIN1 siRNAs confer sensitivity to PLX4720-mediated apoptosis in previously unresponsive cell lines.
A2058, SMKEL5, and SKMEL28 cell were transfected with control siRNA or siRNA targeting AXIN1. 48 hours following transfection, cells were treated with the indicated conditions for 24 hours and analyzed for apoptosis by flow cytometry for cleaved caspase-3. L CM and W3A CM indicate control and WNT3A conditioned media, respectively. 2μM PLX4720 was used where indicated. Values represent the average number of cleaved caspase-3-positive cells ± standard deviation in three or more biologic replicates. Statistically significant differences between control and AXIN1 siRNA for different conditions within each cell line are indicated by an asterix (*) denoting a p<0.001 by two-way ANOVA with Bonferonni’s post-test comparison.
| Control siRNA | AXIN1 siRNA | |||||||
|---|---|---|---|---|---|---|---|---|
| Melanoma Cell Line | L CM DMSO |
W3A CM DMSO |
L CM PLX4720 |
W3A CM PLX4720 |
L CM DMSO |
W3A CM DMSO |
L CM PLX4720 |
W3A CM PLX4720 |
| A2058 | 1.18 ± 0.30 | 1.37 ± 0.08 | 2.20 ± 0.643 | 3.16 ± 0.40 | 3.78 ± 0.51 | 5.23 ± 0.94 | *21.27 ± 2.11 | *21.77 ± 4.36 |
| SKMEL28 | 1.13 ± 0.25 | 1.00 ± 0.40 | 1.81 ± 0.35 | 5.34 ± 1.55 | 1.03 ± 0.24 | 0.71 ± 0.32 | *22.20 ± 5.43 | *30.97 ± 3.65 |
| SKMEL5 | 1.01 ± 0.14 | 0.96 ± 0.15 | 2.20 ± 0.17 | 2.72 ± 0.18 | 2.25 ± 0.52 | 2.45 ± 0.12 | *10.80 ± 1.06 | *12.10 ± 1.15 |
Discussion
The development and initial clinical success of targeted BRAF inhibitors such as vemurafenib and GSK2118436 represents a milestone in cancer treatment that will likely pave the way for other mutation-specific cancer therapies. While the response rates in early clinical trials with these two drugs are extremely promising, there are still obstacles to achieving long-term disease control with this approach. For example, the variability of responses to targeted BRAF inhibitors among patients with BRAFV600E tumors remains unexplained (5–7). Our finding that AXIN1 abundance can predict the apoptotic response to inhibition of BRAFV600E begins to shed light on the mechanisms underlying these variable responses, and points to potential approaches for enhancing the effectiveness of vemurafenib therapy.
Another ongoing clinical problem is the eventual development of resistant tumors and the progression of the disease even in patients who respond well to initial therapy (7). This raises the question of whether targeting multiple signaling pathways may lead to a durable clinical result. While combination targeting of BRAF signaling has been suggested with other pathways implicated in melanoma, such as the PI3K pathway (34, 35), our data support the evaluation of inhibition of BRAF signaling concurrent with activation of Wnt/β-catenin signaling.
The recent characterization of a large panel of melanoma cells treated with PLX4032 (36) supports the need for evaluating the efficacy of combination therapies for melanoma. Consistent with our observation that β-catenin is required for PLX4720 to promote apoptosis, the transcriptional profiling of melanoma lines revealed that cell lines that are more resistant to growth inhibition by PLX4032 exhibit the loss of genes related to active Wnt/β-catenin signaling (36). These observations may facilitate the identification of patients who will benefit from concurrent activation of Wnt/β-catenin signaling and inhibition of BRAFV600E.
While the notion of activating Wnt/β-catenin signaling in any cancer patient may seem initially contra-indicated in light of its frequent role as an oncogenic pathway in colorectal carcinoma (37), it is likely that activating β-catenin has context-dependent effects biochemically, leading to distinct cellular responses. For example, context-dependent differences in the role of β-catenin in preventing versus promoting programmed cell death have been reported (38–41). Given that β-catenin signaling can elicit context-dependent effects, and given the lack of consensus on the effects of activating β-catenin signaling from in vitro cell models of melanoma (42–46), it is difficult to accurately predict the consequences of systemic activation of this pathway in melanoma patients, pointing to the need for additional research.
The observed effects of BRAF inhibition on AXIN1 abundance and GSK3 phosphorylation and activation (Figure 1C) intuitively predict that the abundance of β-catenin would likely change in response to BRAF inhibition. However, the ability of PLX4720 to enhance Wnt/β-catenin signaling, as monitored by both the luciferase reporter assay (BAR) and by monitoring the increased expression of the endogenous target gene AXIN2, does not require additional β-catenin accumulation (47–49). While this seems perplexing based on general models of Wnt signaling, such general models do not always fit with observed data. Indeed, the recent report that loss of phosphorylation of β-catenin at Thr41 is sufficient to enhance Wnt/β-catenin signaling in melanoma cells independent of detected increases in nuclear β-catenin is entirely consistent with our current observations (22). While decreased phosphorylation of β-catenin with PLX4720 is seen across all cell lines tested (Figure 6A), enhanced expression of AXIN2 with PLX4720 was observed in only half of these lines (Figure 5A) suggesting that decreased β-catenin phosphorylation alone is not sufficient to enhance Wnt/β-catenin signaling with targeted BRAF inhibition.
These studies uncover a novel reciprocal relationship between Wnt/β-catenin and BRAF signaling in melanoma, highlighting the potential impact of both pathways on therapeutic efforts to target BRAF in metastatic melanoma with drugs such as vemurafenib. The discovery that regulation of AXIN1 provides the basis for mediating functional cross-talk between these two pathways provides not only a new model for studying Wnt/β-catenin and BRAF signaling in melanoma, but it also provides a novel foundation for identifying tumor-specific determinants of the response to vemurafenib and other targeted BRAF inhibitors. The identification of these tumor-specific determinants may facilitate the development of new therapies and prognostic biomarkers that can further extend the promising clinical results recently seen with pioneering mutation-specific drugs such as vemurafenib, with the ultimate goal of developing therapies that provide sustained long-term clinical responses for patients with metastatic melanoma.
Materials and Methods
Reagents
Detailed information on the β-catenin activated reporter (BAR) has been previously described (20). Briefly, the β-catenin activated reporter (pBAR) is a lentiviral plasmid that contains 12 TCF/LEF binding sites (5′-AGATCAAAGG-3′) each separated by distinct 5 base-pair linkers upstream of a minimal promoter and the firefly luciferase open reading frame. The reporter also contains a separate PGK promoter that constitutively drives the expression of a puromycin resistance gene for mammalian cell selection. Transient transfection of siRNA was performed with RNAiMAX, as directed by the manufacturer (13778-075, Invitrogen). siRNA sequences used are listed in Supplemental Table S1. Protease (#11873580001) and phosphatase (#04906845001) inhibitor tablets were purchased from Roche (Indianapolis, IN). Con A Sepharose was purchased from GE Healthcare (Uppsala, Sweden #17-0440-03). U0126 was purchased from LC labs (Woburn, MA cat# U-6770). AZD6244 was purchased from Selleck Chemicals (Houston, TX cat# S1008). PLX4720 was purchased from Symansis (Australia cat# SY- PLX4720). CHIR99021 was purchased from Axon MedChem (Gronigen, Netherlands catalog #Axon1386). Z-VAD-FMK was purchased from R&D systems (Minneapolis, MN cat# FMKSP01). Anti-ERK (p42/44) (#9102), anti-phopho-ERK (p42/44) (#9101S), anti-phospho-β-catenin S33/37/T41 (#9561S), anti-BRAF (#9434), anti-cleaved CASP3 (#9661S), anti-cleaved PARP1 (#9541), anti-Bim (#2933), and anti-cleaved CASP3 Alexafluor 488 conjugate (#9669) antibodies were purchased from Cell Signaling (Cell Signaling, Beverly MA). Anti-β-tubulin (T7816) and anti-β-catenin (C2206) antibodies were purchased from Sigma Aldrich (Sigma Aldrich St. Louis, MO). Anti-phospho-GSK3 Y279/216 (05-413) was purchased from Upstate Biotechnology (Waltham, Massachusetts). The anti-AXIN1 (AF3287) antibody was purchased from R&D Systems (Minneapolis, MN). In Situ Cell Death Detection kit (cat# 12 156 792 910) was purchased from Roche (Indianapolis, IN).
Cell Lines
The human melanoma cell lines A375, A2058 and MEL624 were a generous gift from Cassian Yee (Fred Hutchinson Cancer Research Institute; Seattle, WA). The human melanoma cell lines COLO-829, SKMEL28, SKMEL5 were purchased from ATCC (Manassas, VA). Human Epidermal Melanocytes, adult, lightly pigmented donor, (HEMa-LP) (C0245C) were purchased from Invitrogen (Carlsbad, CA). Stable BAR cell lines were generated as previously described(20). BAR luciferase cell lines were also infected with a lentivirus carrying Renilla luciferase driven by a constitutive EF1alpha promoter.
Cell Culture
The human melanoma lines A375, A2058 were cultured in DMEM supplemented with 5% FBS and 1% antibiotic. The human melanoma lines SK-MEL-5 and SK-MEL-28 were grown in EMEM supplemented with 10% FBS and 1% antibiotic. The human melanoma lines COLO-829 and MEL624 were grown in RPMI supplemented with 10% FBS and 1% antibiotic HEMa-LP cells were cultured in medium 254 supplemented with 1% HMGS and 1% antibiotic (Invitrogen Carlsbad, CA). Synthetic siRNAs were transfected into cultured cells at a final concentration of 20nM using RNAiMAX (Invitrogen; Grand Island, NY).
High Throughput Screening
Screening was performed at the Quellos high throughput screening facility at the University of Washington’s Institute for Stem Cells and Regenerative Medicine (Seattle, WA). A library of siRNAs targeting primarily the human kinome was screened in A375 melanoma cells stably expressing the β-catenin activated reporter (BAR). The kinome siRNA library was purchased from Sigma Aldrich (Sigma Aldrich St. Louis, MO) and resuspended in RNase free water. The library consists of a pool of three independent non-overlapping siRNAs for each mRNA target. siRNA pools were screened in quadruplicate at 9.5nM, 1.9nM, 0.38nM, and 0.08nM final concentration. Cell viability was assessed by adding resazurine (Sigma Aldrich St. Louis, MO) at a final concentration of 1.25ug/ml (PBS vehicle) and measuring fluorescence intensity (Ex=530nM Em=580nM) on an Envision multilabel plate reader (Perkin Elmer Waltham, MA). Luciferase activity was assessed by adding 5uL/well SteadyGlo (Promega Madison, WI) and measuring total luminescence on an Envision multilabel plate reader (Perkin Elmer Waltham, MA) The screen workflow was as follows:
On day 1, 1.5 uL of the appropriate concentration of siRNA was added to 28.5uL of Optimem (Invitrogen, Carlsbad, CA) containing 3.125uL/mL RNAiMAX (Invitrogen, Carlsbad, CA). 5uL of this mix was transferred to a 384 well plate containing 15uL of growth media (DMEM/5%FBS/1%PenStrep). 20uL of cells at 75 cells/uL was added to each well for a final cell number of 1500 cells/well. On day 3, 10uL of WNT3A conditioned media diluted 1:12.8 with growth media was added for a final dilution of 1:64. On Day 4, 10uL of 6X resazurine was added to each well, incubated at 37°C for three hours, and fluorescence intensity was measured. Immediately following, 5uL of SteadyGlo was added, incubated at room temperature for 10 minutes and total luminescence was measured. Data are represented as a ratio of BAR reporter activity (Luminescence) to cell viability (Resazurine fluorescence intensity).
Low throughput BAR reporter assays
Cells were plated in 96-well plates. 24 hours following plating, cells were treated with the indicated conditions and luciferase activity was measured 24 hours later with the a dual luciferase reporter assay kit (Promega; Madison, WI) and an Envision multi-label plate reader (Perkin Elmer, Waltham, MA) per manufactures suggestions. For BAR assays involving siRNAs, siRNAs were transfected 48 hours prior to treatment.
Low throughput siRNA transfections
Cells were reverse transfected with 20nM siRNA (final concentration) in 6-well plates using RNAiMAX reagent per manufactures suggestions (Invitrogen, Carlsbad, CA). Cells were incubated for 48 hours following transfection and then treated with the indicated conditions for the indicated amount of time.
Cytosolic and Nuclear β-catenin Fractionation
Cells were plated in 100mm dishes. 24 hours following plating, cells were treated with the indicated conditions for 24 hours. Cells were gently rinsed with PBS and harvested by scraping in 500uL of hypotonic lysis buffer (50mM HEPES pH 8.0, 1mM EDTA, 1mM DTT) containing protease and phosphatase inhibitors. Cells were swelled on ice for 30 minutes and then passed through a 27 gauge needle ten times and checked for complete lysis with a microscope. Lysates were centrifuged at 10,000xg for 20 minutes and supernatant was collected as the cytosolic fraction. Pelleted membranes were washed 5 times with hypotonic lysis buffer and then solubilized with solubilization buffer (50mM Tris pH 8.0, 150mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) containing protease and phosphatase inhibitors. After a 30 minute incubation on ice, lysates were centrifuged at 16,000xg for 20 minutes. The protein concentration of the cleared supernatant was determined by BCA analysis and an equal amount of protein and volume was then incubated with pre-washed Con A sepharose beads overnight at 4degree C. Supernatant was collected as the nuclear fraction.
RNA purification and qRT-PCR analysis
RNA was purified using the RNeasy kit following the manufacturer’s protocol (Qiagen; Maryland, MD). cDNA was synthesized using RevertAid™ M-MuLV Reverse Transcriptase (Fermentas; Ontario, CAN). Light Cycler FastStart DNA Master SYBR Green1 (Roche; Mannheim, Germany) was used for real-time PCR as previously described (50). Quantitative PCR results presented in the manuscript are averages of a minimum of three biologic replicates.
Isobologram Analysis of Cell Viability
A375 melanoma cells were seeded in 96-well plates at a concentration of 8,000 cells per well in 100μl of growth media. 24 hours after plating, cells were treated with all combinations of 2-fold dilutions of WNT3A CM ranging from 20% to 0% and 2-fold dilutions of PLX4720 ranging from 5μM-0μM for 48 hours. 10uL of CellTiter-Glo (Promega Madison, WI) was added to each well and total luminescence was measured on an Envision multilabel plate reader (Perkin Elmer Waltham, MA). Each condition within an experiment was assayed in triplicate wells and three independent experiments were performed.
Flow cytometry for Active Caspase-3
Cells were seeded in a 6-well dish at a density to achieve 90–100% confluence at harvest. 24 hours after seeding, cells were treated with the indicated conditions for the indicated amount of time. At the time of collection, supernatants were collected and pooled with trypsinized cells. Cells were fixed with 4% paraformaldehyde and permeabilized according to vendor’s protocol for Cleaved Caspase-3 (Asp175) Antibody (AlexaFluor 488 Conjugate) (catalog # 9669) (Cell Signaling, Beverly MA). The antibody was used at a final dilution of 1:100. Flow was performed on a BD FACSCanto II, and data analysed with FlowJo 8.8.6 (Tree Star) software. Experiments were performed with biological triplicates and data are representative of at least three independent experiments.
For experiments involving siRNAs, cells were reverse transfected with 20nM siRNA in 6-well dishes in triplicate with RNAiMax according to manufacturer’s protocol. 48 hours following transfection, cells were treated with the indicated conditions for 24 hours and then harvested for analysis. Cells were harvested, stained, and analyzed as described above.
TUNEL
Glass coverslips were coated with poly-L-lysine in a 24-well dish, rinsed with PBS, and dried. Cells were seeded at a density to achieve 90–100% at harvest. 24 hours after seeding, cells were treated with the indicated conditions and incubated for 24 hours. TUNEL staining was performed according to vendor’s protocol (cat# 12 156 792 910) (Roche Indianapolis, IN). Briefly, media was gently aspirated and cells were fixed in 4% paraformaldehyde for 1 hour at room temperature. Cells were gently rinsed twice with PBS and permeabilized with 0.1% Triton X-100in 0.1% sodium citrate for 2 minutes on ice. Cells were rinsed twice with PBS, and 40uL of TUNEL reaction mixture was added directly on top of the slide and incubated for 1 hour at 37°C in a humidified incubator. Slips were rinsed 3 times and mounted on superfrost plus glass slides (cat# 48311-703 VWR West Chester, PA) with Prolong Gold anti-fade mounting media containing DAPI (cat# P36931 Invitrogen; Grand Island, NY). Images were obtained on a Nikon TiE inverted widefield high-resolution microscope (Nikon Melville, NY).
Spheroid Assay
A375 cells were used for the spheroid assays. Spheroids were formed and implanted in collagen as previously described (51). Spheroids were treated with indicated conditions 30 minutes after collagen polymerization. Images were obtained on a Nikon TiE inverted widefield high-resolution microscope (Nikon Melville, NY). For comparison of growth effects such as shown in Figure 2, spheroids were imaged at 72 hours after spheroid implantation. For live-dead imaging assays such as shown in Figure 3, imaging was performed at 24 hours after spheroid implantation.
Xenograft assays
NSG (NOD/SCID/IL2r-gamma (null)) mice were injected with 5×105 A375 cells stably expressing GFP or 5×105 A375 cells stably expressing WNT3A-IRES-GFP. Tumors were allowed to establish to approximately 100 mm3, after which mice where tumor size-matched and allocated to five per treatment group (vehicle or PLX4720). WNT3A-IRES-GFP tumors grew slower and therefore the first day of treatment was day 14 while GFP expressing tumors were first treated on day 9. Treatment was by oral gavage once daily with 5% DMSO in 1% carboxymethyl cellulose or 50mg/kg PLX4720 in 1% carboxymethyl cellulose (604mM PLX4720 in DMSO was diluted 1:20 in 1% carboxymethylcellulose). Tumor size was determined by caliper measurements of tumor length and width every 3 to 4 days. Tumor volume was then calculated using the following formula: volume = (width) 2 × length/2. Tumors were harvested 2 hours after the last dose and fixed in neutral-buffered formalin overnight at room temperature.
Mitotic index
Hematoxylin- and eosin-stained tumor sections were scored for mitotic activity by a board-certified pathologist who was blinded to the treatment conditions. For each treatment condition, five tumors were evaluated and a range of 26–60 high-powered fields (hpf’s) per individual tumor were scored (average of 44 hpf’s per tumor). Areas with fixation artifact were excluded a priori from the final analysis, accounting for differences in the number of hpf’s per individual tumor. Analysis was performed using a one-way ANOVA followed by a post-test for linear trend.
Statistical analysis
Standard statistical analysis was performed using GraphPad Prizm (GraphPad Inc., LaJolla CA) version 5.01. Dose-effect analyses, including combination indices, dose reduction indices and median-effect analysis were performed using the method of Chou and Talay (52) via the CalcuSyn software suite (Biosoft, Cambridge UK), version 2.1.
Supplementary Material
Supplemental Database S1. A375 melanoma cells expressing the BAR reporter were screened using siRNAs for kinases that regulate Wnt/β-catenin signaling.
Supplemental Database S2. Screen data from Supplemental Database S1 without resazurine normalization. This is the raw BAR reporter firefly luciferase counts from A375 cells treated with 1.8nM siRNA.
Supplemental Figure S1. BRAF and other members of the MAPK family are identified as regulators of Wnt/β-catenin signaling in melanoma cells.
Supplemental Figure S2. BRAF is a negative regulator of Wnt/β-catenin signaling and inhibition of BRAF decreases the phosphorylation of CTNNB1 at S33/S37/T41 in melanoma cells.
Supplemental Figure S3. Isobologram analysis of WNT3A and PLX4720 shows a synergistic activation of Wnt/β-catenin signaling in melanoma cells.
Supplemental Figure S4. BRAF inhibition reduces the abundance of phosphorylated Ser33/Ser37/Thr41 β-catenin with no effect on total β-catenin abundance.
Supplemental Figure S5. Activation of Wnt/β-catenin signaling cooperates with BRAF inhibition to inhibit proliferation of melanoma cells in vivo.
Supplemental Figure S6. Isobologram analysis of WNT3A and PLX4720 shows a synergistic inhibition of melanoma cell viability.
Supplemental Figure S7. Activation of Wnt/β-catenin signaling combined with siRNA-mediated knockdown of BRAF promotes apoptosis of melanoma cells.
Supplemental Figure S8. BRAF inhibition does not regulate AXIN1 mRNA abundance.
Supplemental Figure S9. siRNA targeting AXIN1 effectively reduces AXIN1 protein abundance.
Supplemental Table S1. siRNA sequences used for the described studies.
Supplemental Table S2. Allelic status of BRAFV600E in different melanoma cell lines.
Acknowledgments
The authors thank Mr. Jeffrey D. Lebowski for administrative assistance with preparation of the manuscript.
Funding: T.L.B. is funded in part through a training grant from NIH/NIAMS (T32AR056969). A.J.C. is funded by the NIH/NCI (K08CA128565). R.M.K. is supported by an administrative supplemental grant through the American Recovery and Relief Act and the NIH/NCI. R.T.M. is an Investigator of the Howard Hughes Medical Institute. We are indebted to these funding agencies for their continued support of our work. The contents of this manuscript are the sole responsibility of the authors and do not necessarily represent the official views of the NIAMS, NCI, NIH or the Howard Hughes Medical Institute.
Footnotes
“This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Conflicts of interest: None declared
Author contributions: T.L.B, A.J.C, and R.T.M conceived the project. T.L.B, A.J.C, and R.T.M designed the experiments and analyzed the data. T.L.B and R.M.K performed most of the experiments. R.A.T. performed the in vivo studies. O.M.L and R.D.S performed in vitro spheroid assays. R.G.J and N.C.R helped with the siRNA screening. D.W.D analyzed the xenograft histology. T.L.B., A.J.C, and R.T.M wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949. doi: 10.1038/nature00766. published online EpubJun 27. nature00766 [pii]) [DOI] [PubMed] [Google Scholar]
- 2.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, Sproesser K, Li L, Smalley KS, Fong D, Zhu YL, Marimuthu A, Nguyen H, Lam B, Liu J, Cheung I, Rice J, Suzuki Y, Luu C, Settachatgul C, Shellooe R, Cantwell J, Kim SH, Schlessinger J, Zhang KY, West BL, Powell B, Habets G, Zhang C, Ibrahim PN, Hirth P, Artis DR, Herlyn M, Bollag G. Discovery of a selective inhibitor ofoncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041. doi: 10.1073/pnas.0711741105. published online EpubFeb 26. 0711741105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596. doi: 10.1038/nature09454. published online EpubSep 30. nature09454 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT, Li L, Brafford PA, van den Eijnden M, Halloran MB, Sproesser K, Haass NK, Smalley KS, Tsai J, Bollag G, Herlyn M. PLX4032, a potent inhibitor of the B-Raf V600Eoncogene, selectively inhibits V600E-positive melanomas. Pigment Cell Melanoma Res. 2010;23:820. doi: 10.1111/j.1755-148X.2010.00763.x. published online EpubDec (PCR763 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kefford RF, Arkenau H, Brown MP, Millward M, Infante JR, Long GV, Ouellet D, Curtis M, Lebowitz PF, Falchook GS. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010;28:abstract 8503. published online Epub2010 ( [Google Scholar]
- 6.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AMM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. New England Journal of Medicine. 2011 doi: 10.1056/NEJMoa1103782. online advanced publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty K, Puzanov I, Kim K, Ribas A, McArthur GA, Sosman JA, O’Dwyer P, Lee R, Grippo JF, Nolop K, Chapman P. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129:1614. doi: 10.1038/jid.2008.445. published online EpubJul. jid2008445 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, Denat L, Goodall J, Luciani F, Viros A, Demirkan N, Bastian BC, Goding CR, Larue L. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 2007;21:2923. doi: 10.1101/gad.450107. published online EpubNov 15. 21/22/2923 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progressionof melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11:8606. doi: 10.1158/1078-0432.CCR-05-0011. published online EpubDec 15. 11/24/8606 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193. doi: 10.1073/pnas.0811902106. published online EpubJan 27, 0811902106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould Rothberg BE, Berger AJ, Molinaro AM, Subtil A, Krauthammer MO, Camp RL, Bradley WR, Ariyan S, Kluger HM, Rimm DL. Melanoma prognostic model using tissue microarrays and genetic algorithms. J Clin Oncol. 2009;27:5772. doi: 10.1200/JCO.2009.22.8239. published online EpubDec 1. JCO.2009.22.8239 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kageshita T, Hamby CV, Ishihara T, Matsumoto K, Saida T, Ono T. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Br J Dermatol. 2001;145:210. doi: 10.1046/j.1365-2133.2001.04336.x. published online EpubAug (bjd4336 [pii]) [DOI] [PubMed] [Google Scholar]
- 14.Maelandsmo GM, Holm R, Nesland JM, Fodstad O, Florenes VA. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin Cancer Res. 2003;9:3383. published online EpubAug 15 ( [PubMed] [Google Scholar]
- 15.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. MAP kinases. Chem Rev. 2001;101:2449. doi: 10.1021/cr000241p. published online EpubAug (cr000241p [pii]) [DOI] [PubMed] [Google Scholar]
- 16.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21. doi: 10.1080/02699050500284218. published online EpubMar. N13017H076M340R1 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Smalley KS. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? Int J Cancer. 2003;104:527. doi: 10.1002/ijc.10978. published online EpubMay 1. [DOI] [PubMed] [Google Scholar]
- 18.Bikkavilli RK, Malbon CC. Mitogen-activated protein kinases and Wnt/beta-catenin signaling: Molecular conversations among signaling pathways. Commun Integr Biol. 2009;2:46. doi: 10.4161/cib.2.1.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H, Wang J, Nika H, Hawke D, Keezer S, Ge Q, Fang B, Fang X, Fang D, Litchfield DW, Aldape K, Lu Z. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36:547. doi: 10.1016/j.molcel.2009.09.034. published online EpubNov 25. S1097-2765(09)00694-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- 21.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198. published online EpubSep 1 ( [PubMed] [Google Scholar]
- 22.Kuphal S, Bosserhoff AK. Phosphorylation of beta-catenin results in lack of beta-catenin signaling in melanoma. Int J Oncol. 2011;39:235. doi: 10.3892/ijo.2011.1028. published online EpubJul. [DOI] [PubMed] [Google Scholar]
- 23.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, Chresta C, McCormack R, Byrne N, Cockerill M, Graham A, Beran G, Cassidy A, Haggerty C, Brown H, Ellison G, Dering J, Taylor BS, Stark M, Bonazzi V, Ravishankar S, Packer L, Xing F, Solit DB, Finn RS, Rosen N, Hayward NK, French T, Smith PD. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264. doi: 10.1158/0008-5472.CAN-09-1577. published online EpubMar 15. 0008-5472.CAN-09-1577 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeflich S, Herter KP, Tien J, Wong L, Berry L, Chan J, O’Brien C, Modrusan Z, Seshagiri S, Lackner M, Stern H, Choo E, Murray L, Friedman LS, Belvin M. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69:3042. doi: 10.1158/0008-5472.CAN-08-3563. published online EpubApr 1. 0008-5472.CAN-08-3563 [pii] [DOI] [PubMed] [Google Scholar]
- 25.O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384. doi: 10.1093/emboj/17.2.384. published online EpubJan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang CC, Lai F, Tay KH, Croft A, Rizos H, Becker TM, Yang F, Liu H, Thorne RF, Hersey P, Zhang XD. Apoptosis of human melanoma cells induced by inhibition of B-RAFV600E involves preferential splicing of bimS. Cell Death Dis. 2010;1:e69. doi: 10.1038/cddis.2010.48. cddis201048 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008;27:3301. doi: 10.1038/sj.onc.1211003. published online EpubMay 22. 1211003 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, McMahon M. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res. 2008;21:534. doi: 10.1111/j.1755-148X.2008.00491.x. published online EpubOct. PCR491 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393. doi: 10.1016/j.molcel.2004.12.030. published online EpubFeb 4. S1097276505010403 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118:3651. doi: 10.1172/JCI35437. published online EpubNov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response tooncogene-targeted therapeutics. FEBS J. 2009;276:6050. doi: 10.1111/j.1742-4658.2009.07329.x. published online EpubNov. EJB7329 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Sheridan C, Brumatti G, Martin SJ. Oncogenic B-RafV600E inhibits apoptosis and promotes ERK-dependent inactivationof Bad and Bim. J Biol Chem. 2008;283:22128. doi: 10.1074/jbc.M800271200. published online EpubAug 8. M800271200 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172. doi: 10.1128/MCB.22.4.1172-1183.2002. published online EpubFeb ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008;68:3429. doi: 10.1158/0008-5472.CAN-07-5867. published online EpubMay 1. 68/9/3429 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D’Andrea K, Pushparajan A, Hayden JE, Brown KD, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683. doi: 10.1016/j.ccr.2010.11.023. published online EpubDec 14. S1535-6108(10)00484-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tap WD, Gong KW, Dering J, Tseng Y, Ginther C, Pauletti G, Glaspy JA, Essner R, Bollag G, Hirth P, Zhang C, Slamon DJ. Pharmacodynamic characterization of the efficacy signals due to selectiveBRAF inhibition with PLX4032 in malignant melanoma. Neoplasia. 2010;12:637. doi: 10.1593/neo.10414. published online EpubAug ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucero OM, Dawson DW, Moon RT, Chien AJ. A re-evaluation of the “oncogenic” nature of Wnt/beta-catenin signaling in melanoma and othercancers. Curr Oncol Rep. 2010;12:314. doi: 10.1007/s11912-010-0114-3. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorsky RI, Moon RT, Raible DW. Environmental signals and cell fate specification in premigratory neural crest. Bioessays. 2000;22:708. doi: 10.1002/1521-1878(200008)22:8<708::AID-BIES4>3.0.CO;2-N. published online EpubAug ( [DOI] [PubMed] [Google Scholar]
- 39.Ellies DL, Church V, Francis-West P, Lumsden A. The WNT antagonist cSFRP2 modulates programmed cell death in the developing hindbrain. Development. 2000;127:5285. doi: 10.1242/dev.127.24.5285. published online EpubDec ( [DOI] [PubMed] [Google Scholar]
- 40.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087. doi: 10.1242/dev.129.9.2087. published online EpubMay ( [DOI] [PubMed] [Google Scholar]
- 41.Yeo W, Gautier J. Early neural cell death: dying to become neurons. Dev Biol. 2004;274:233. doi: 10.1016/j.ydbio.2004.07.026. published online EpubOct 15. S0012-1606(04)00502-0 [pii]) [DOI] [PubMed] [Google Scholar]
- 42.Mikheev AM, Mikheeva SA, Rostomily R, Zarbl H. Dickkopf-1 activates cell death in MDA-MB435 melanoma cells. Biochem Biophys Res Commun. 2007;352:675. doi: 10.1016/j.bbrc.2006.11.079. published online EpubJan 19. S0006-291X(06)02550-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinnberg T, Menzel M, Ewerth D, Sauer B, Schwarz M, Schaller M, Garbe C, Schittek B. beta-Catenin Signaling Increases during Melanoma Progression andPromotes Tumor Cell Survival and Chemoresistance. PLoS One. 2011;6:e23429. doi: 10.1371/journal.pone.0023429. PONE-D-11-04261 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinnberg T, Menzel M, Kaesler S, Biedermann T, Sauer B, Nahnsen S, Schwarz M, Garbe C, Schittek B. Suppression of casein kinase 1alpha in melanoma cells induces a switch in beta-catenin signaling to promote metastasis. Cancer Res. 2010;70:6999. doi: 10.1158/0008-472.CAN-10-0645. published online EpubSep 1. 0008-5472.CAN-10-0645 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, He X, Fisher DE. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol. 2002;158:1079. doi: 10.1083/jcb.200202049. published online EpubSep 16. jcb.200202049 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You L, He B, Xu Z, Uematsu K, Mazieres J, Fujii N, Mikami I, Reguart N, McIntosh JK, Kashani-Sabet M, McCormick F, Jablons DM. An anti-Wnt-2 monoclonal antibody induces apoptosis in malignant melanoma cells and inhibits tumor growth. Cancer Res. 2004;64:5385. doi: 10.1158/0008-5472.CAN-04-1227. published online EpubAug 1. 64/15/5385 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Guger KA, Gumbiner BM. A mode of regulation of beta-catenin signaling activity in Xenopus embryos independent of its levels. Dev Biol. 2000;223:441. doi: 10.1006/dbio.2000.9770. published online EpubJul 15. S0012-1606(00)99770-7 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Nelson RW, Gumbiner BM. A cell-free assay system for beta-catenin signaling that recapitulates direct inductive events in the early xenopus laevis embryo. J Cell Biol. 1999;147:367. doi: 10.1083/jcb.147.2.367. published online EpubOct 18 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staal FJ, Noort MMv, Strous GJ, Clevers HC. Wnt signals are transmitted through N-terminally dephosphorylated beta-catenin. EMBO Rep. 2002;3:63. doi: 10.1093/embo-reports/kvf002. published online EpubJan. kvf002 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, Angers S, Moon RT. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043. doi: 10.1126/science/1141515. published online EpubMay 18 ( [DOI] [PubMed] [Google Scholar]
- 51.Smalley KS, Lioni M, Herlyn M. Life isn’t flat: taking cancer biology to the next dimension. In Vitro Cell Dev Biol Anim. 2006;42:242. doi: 10.1290/0604027.1. published online EpubSep-Oct. 0604027 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621. doi: 10.1124/pr.58.3.10. published online EpubSep. 58/3/621 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Database S1. A375 melanoma cells expressing the BAR reporter were screened using siRNAs for kinases that regulate Wnt/β-catenin signaling.
Supplemental Database S2. Screen data from Supplemental Database S1 without resazurine normalization. This is the raw BAR reporter firefly luciferase counts from A375 cells treated with 1.8nM siRNA.
Supplemental Figure S1. BRAF and other members of the MAPK family are identified as regulators of Wnt/β-catenin signaling in melanoma cells.
Supplemental Figure S2. BRAF is a negative regulator of Wnt/β-catenin signaling and inhibition of BRAF decreases the phosphorylation of CTNNB1 at S33/S37/T41 in melanoma cells.
Supplemental Figure S3. Isobologram analysis of WNT3A and PLX4720 shows a synergistic activation of Wnt/β-catenin signaling in melanoma cells.
Supplemental Figure S4. BRAF inhibition reduces the abundance of phosphorylated Ser33/Ser37/Thr41 β-catenin with no effect on total β-catenin abundance.
Supplemental Figure S5. Activation of Wnt/β-catenin signaling cooperates with BRAF inhibition to inhibit proliferation of melanoma cells in vivo.
Supplemental Figure S6. Isobologram analysis of WNT3A and PLX4720 shows a synergistic inhibition of melanoma cell viability.
Supplemental Figure S7. Activation of Wnt/β-catenin signaling combined with siRNA-mediated knockdown of BRAF promotes apoptosis of melanoma cells.
Supplemental Figure S8. BRAF inhibition does not regulate AXIN1 mRNA abundance.
Supplemental Figure S9. siRNA targeting AXIN1 effectively reduces AXIN1 protein abundance.
Supplemental Table S1. siRNA sequences used for the described studies.
Supplemental Table S2. Allelic status of BRAFV600E in different melanoma cell lines.