Abstract
Several genetic variants associated with platelet count and mean platelet volume (MPV) were recently reported in people of European ancestry. In this meta-analysis of 7 genome-wide association studies (GWAS) enrolling African Americans, our aim was to identify novel genetic variants associated with platelet count and MPV. For all cohorts, GWAS analysis was performed using additive models after adjusting for age, sex, and population stratification. For both platelet phenotypes, meta-analyses were conducted using inverse-variance weighted fixed-effect models. Platelet aggregation assays in whole blood were performed in the participants of the GeneSTAR cohort. Genetic variants in ten independent regions were associated with platelet count (N = 16,388) with p<5×10−8 of which 5 have not been associated with platelet count in previous GWAS. The novel genetic variants associated with platelet count were in the following regions (the most significant SNP, closest gene, and p-value): 6p22 (rs12526480, LRRC16A, p = 9.1×10−9), 7q11 (rs13236689, CD36, p = 2.8×10−9), 10q21 (rs7896518, JMJD1C, p = 2.3×10−12), 11q13 (rs477895, BAD, p = 4.9×10−8), and 20q13 (rs151361, SLMO2, p = 9.4×10−9). Three of these loci (10q21, 11q13, and 20q13) were replicated in European Americans (N = 14,909) and one (11q13) in Hispanic Americans (N = 3,462). For MPV (N = 4,531), genetic variants in 3 regions were significant at p<5×10−8, two of which were also associated with platelet count. Previously reported regions that were also significant in this study were 6p21, 6q23, 7q22, 12q24, and 19p13 for platelet count and 7q22, 17q11, and 19p13 for MPV. The most significant SNP in 1 region was also associated with ADP-induced maximal platelet aggregation in whole blood (12q24). Thus through a meta-analysis of GWAS enrolling African Americans, we have identified 5 novel regions associated with platelet count of which 3 were replicated in other ethnic groups. In addition, we also found one region associated with platelet aggregation that may play a potential role in atherothrombosis.
Author Summary
The majority of the variation in platelet count and mean platelet volume between individuals is heritable. We performed genome-wide association studies in more than 16,000 African American participants from seven population-based cohorts to identify genetic variants that correlate with variation in platelet count and mean platelet volume. We observed statistically significant evidence (p-value<5×10−8) that 10 genomic regions were associated with platelet count and 3 were associated with mean platelet volume. Of the regions that were significantly associated, we found 5 novel regions that were not reported previously in other populations. Three of these 5 regions were also associated with platelet count in European Americans and Hispanic Americans. All these regions contain genes that are either known to have or potentially may have a role in determining platelet count and/or mean platelet volume. We further found that one of these regions was also associated with agonist-induced platelet aggregation. Further studies will determine the exact role played by these genomic regions in platelet biology. The knowledge generated by this and other studies will not only help us better understand platelet biology but can also lead us to the discovery of new anti-platelet drugs.
Introduction
While platelets play a fundamental role in hemostasis, they are also important in the development of atherosclerosis and arterial thrombosis [1]. An elevated platelet count has been associated with adverse clinical outcomes after thrombolysis or coronary intervention in patients presenting with acute myocardial infarction and moderate reductions in platelet count by thrombopoietin inhibition were associated with reduced thrombogenesis in a primate model [2]–[4]. The heritability of variation in platelet count is substantial with estimates ranging from 54% to more than 80% [5]–[8]. In the GeneSTAR study, a cohort included in the current meta-analysis, the heritability of platelet count is 67% [9].
Like platelet count, an elevated mean platelet volume (MPV) is also associated with adverse cardiovascular events and its reported heritability is as high as 73% [8], [10]–[12]. The heritability of MPV in the GeneSTAR cohort was 71% [9]. Recent genome-wide association studies (GWAS) and meta-analyses have identified genetic variants associated with these two platelet traits in Caucasians and a Japanese population [13]–[15]. A recent meta-analysis in the CARe Project, involving genotyping of about 50,000 single nucleotide polymorphisms (SNPs) in 2,100 candidate genes, also reported two genetic variants associated with platelet count in African Americans [16]. The genetic variants reported to date explain only a small fraction of the heritability in platelet count and MPV, providing an opportunity for new studies to discover additional genetic variants of importance [15]. Moreover, African Americans have higher platelet counts than Caucasians and additional genetic variants may contribute to this difference [17]. Because of the different allele frequencies and linkage disequilibrium patterns in populations of European and African ancestry, we anticipated that we might discover new genetic loci associated with platelet count and MPV in an African American population compared to Caucasians [18].
We performed a meta-analysis of 7 GWAS studies that included African-American subjects in the Continental Origins and Genetic Epidemiology Network (COGENT) in order to identify novel genetic variants associated with platelet count and MPV.
Results
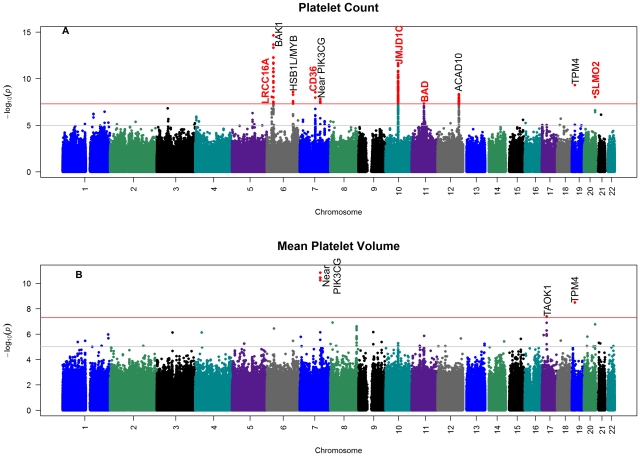
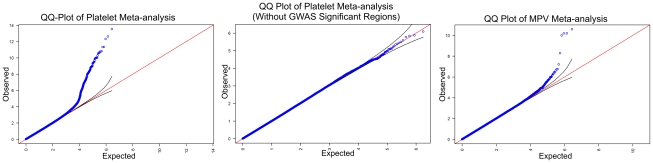
We performed a GWAS analysis of platelet count in an African American discovery sample of 16,388 individuals from 7 population-based cohorts (Table 1). The MPV meta-analysis included all subjects from three cohorts and a subset of subjects from two other cohorts (n = 4,531). Following stringent genotyping and imputation quality control procedures (as outlined in the Methods section), over 2.2 million SNPs were available for analysis in each cohort (Table 1). The results of association studies and the genomic-control corrected QQ plot for the combined African-African GWAS analysis for platelet count and MPV are shown in Figure 1 and Figure 2 and study specific QQ plots and genomic inflation factors are reported in Figures S3 and S4 and Table S1. The Jackson Heart Study (JHS) cohort contains a few hundred related individuals. This resulted in a high genomic inflation factor for platelet count and a few other traits, as previously described in Lettre et al [19]. Within the CARe Consortium, Lettre et al have done several analyses involving simulated phenotypes as well as empirical data (lipids, BMI) and have shown that for JHS, genomic control-correction is an appropriate way to control for the small sub-group of related individuals. A list of all genome-wide significant SNPs with regional plots for platelet count and MPV can be found in Tables S2 and S3 and Figure S1. Cohort-specific QQ-plots and association results for index SNPs associated with platelet count or MPV are summarized in Figure S2 and Table S4. Of the 10 loci on 7 chromosomes that reached GWAS threshold (p<5×10−8) in the platelet count meta-analysis, five have not been reported in previous platelet count GWAS studies in any population and 8 loci have not been reported previously in African Americans (Figure 1). The MPV meta-analysis identified three loci, each one on different chromosomes; two of these loci were also associated with platelet count at GWAS threshold in the current study (Figure 2). One MPV-associated locus has been reported in African Americans before, and two of these three loci have been associated with MPV in Caucasians in prior studies [15], [16]. A sex-specific meta-analysis did not reveal any heterogeneity for the allelic effect between the two sexes and did not uncover any additional loci. Thus, the sex-specific results are not reported here.
Table 1. Characteristics of COGENT African-American meta-analysis cohorts.** .
| ARIC | CARDIA | GeneSTAR | HANDLS | Health ABC | JHS | WHI | |
| Sample size | 2664 | 943 | 934 | 862 | 898 | 1992 | 8095 |
| Study design * | unrelated | unrelated | family | unrelated | unrelated | unrelated | unrelated |
| Age, years | 53.4 (5.8) | 24.4 (3.8) | 45.2 (12.6) | 48.2 (9.0) | 73.4 (2.8) | 50.0 (12.1) | 61.6 (7.0) |
| Female (%) | 63.2 | 58.7 | 61.6 | 56.0 | 58.8 | 61.2 | 100 |
| SNPs (N) | 2,799,937 | 2,813,829 | 3,181,434 | 3,021,329 | 3,129,972 | 2,819,255 | 2,486,528 |
| Platelet count (109/L) | 256.4 (65.7) | 279.7 (71.4) | 265.3 (67.0) | 268.7 (76.8) | 237.6 (71.6) | 256.6 (64.5) | 250.3 (60.1) |
| MPV *** | 9.3 (0.9) fL | NA | 7.9 (0.9) fL | 9.2 (1.0) fL | 10.9 (1.6) fL | 9.2 (0.9) fL | NA |
*: All studies are population-based, in addition, JHS has a small group of related individuals.
**: Mean (SD), and units (untransformed) that were used in the regression models for each trait.
***: For MPV, complete cohorts of GeneSTAR, HANDLS, and JHS were included, while only a subset of ARIC (n = 644) and Health ABC (n = 182) were included.
NA = not available; MPV = mean platelet volume.
Figure 1. Manhattan plot of the genome-wide association results for meta-analysis.
(a) platelet count; (b) mean platelet volume. SNPs are plotted on the x-axis according to their position on each chromosome against the negative log10 of p-values on y-axis. Names of the genes that contain the significant SNPs or are located close to the significant SNPs are indicated on the plot adjacent to the significant SNPs. Names of genes in the novel regions are in red.
Figure 2. Quantile–quantile (QQ) plots.
(a) platelet count meta-analysis with all SNPs included; (b) platelet count meta-analysis after removing 1 million base pairs around the top SNPs from the 10 loci; (c) mean platelet volume meta-analysis. Blue dots are SNPs plotted on the x-axis of expected p-value under the null hypothesis against the observed p-value in the study (p-values are plotted here as negative logarithm 10). The red diagonal line represents the line of unity, the region where expected and observed p-values are the same under the null hypothesis. Black lines above and below the red diagonal line bound 95% confidence intervals. Under the null hypothesis, SNPs should follow the line of unity closely except those SNPs for which the null hypothesis is rejected. SNPs in the QQ plot for the platelet count meta-analysis do not follow the line of unity closely and this appears to be due to large number of significant SNPs in the associated loci. When the chromosomal regions containing these loci are removed, the appearance of QQ plot improves considerably.
Identification of novel loci associated with platelet count and replication in European Americans and Hispanic Americans
The first of the novel loci from platelet count meta-analysis is located on chromosome 6p22. The best SNP (rs12526480; p = 9.1×10−9) in this region is located in the intron of the leucine-rich repeat containing 16A gene (LRRC16A). The minor allele (G) of rs12526480 was associated with decreased platelet count. Ten additional SNPs in the region had p<10−6 (Table 2 and Table S2). The LRRC16A gene encodes a protein called ‘capping protein ARP2/3 and myosin-I linker’ (CARMIL), which plays an important role in cell-shape change and motility. Genetic variants in LRRC16A have been previously reported to be associated with serum uric acid levels [20], nephrolithiasis [21] and markers of iron status [22] but there have been no reports of any association with either platelet count or other platelet phenotypes. In the three European American cohorts, rs12526480 was statistically significant in one cohort (p = 0.01) and near nominal significance in the combined meta-analysis (p = 0.06) with an effect size and direction similar to that observed in African Americans. In Hispanic Americans, rs12526480 was not significantly associated with platelet count (Table 3). Given the proximity of the LRRC16A gene to the hemochromatosis (HFE) gene and the well-known reciprocal relationship between platelet count and iron stores, we additionally assessed the association between rs12526480 and red cell phenotypes in the COGENT African Americans. There was no evidence of association between LRRC16A genotype and hemoglobin, hematocrit, red cell count or mean corpuscular volume in the 16,388 African Americans, nor was there any evidence of association between rs12526480 genotype and serum ferritin in 672 African Americans from CARDIA or 2,126 from JHS. Nor did adjustment for red cell phenotype or iron status alter the relationship between platelet count and rs12526480 genotype. Finally, we had uric acid levels available in 943 African Americans from CARDIA; again there was no association with LRRC16A genotype (Table S5).
Table 2. Novel and validated loci based on genome-wide association with platelet count and mean platelet volume in COGENT (novel loci are in bold).
| Locus | Significant SNPs (N)* | Top SNP in region | Position | Candidate gene | Maj/Min Allele | MAF | Effect size (SE) | p-value | Het-P (I2) |
| PLATELET COUNT | |||||||||
| 6p22 | 1 | rs12526480 | 25641513 | LRRC16A | G/T | 30.5% | −4.39 (0.76) | 9.15×10−9 | 0.62 (0) |
| 6p21 | 20 | rs210134 | 33648187 | BAK1 | A/G | 28.6% | −6.16 (0.78) | 2.32×10−15 | 0.18 (10.6) |
| 6q23 | 4 | rs9494145 | 135474245 | HBS1L, MYB | C/T | 7.3% | 8.19 (1.38) | 2.79×10−9 | 0.99 (0) |
| 7q11 | 2 | rs13236689 | 80073950 | CD36 | G/T | 43.6% | 4.18 (0.70) | 2.84×10−9 | 0.73 (0) |
| 7q22 | 4 | rs342293 | 106159455 | PIK3CG | G/C | 38.6% | −4.05 (0.72) | 1.58×10−8 | 0.18 (9) |
| 10q21 | 71 | rs7896518 | 64774506 | JMJD1C | G/A | 32.4% | 5.18 (0.74) | 2.26×10−12 | 0.14 (16) |
| 11q13 | 1 | rs477895 | 63805488 | BAD | C/T | 45.3% | −4.19 (0.77) | 4.91×10−8 | 0.17 (11) |
| 12q24 | 26 | rs6490294 | 110674821 | ACAD10 | C/A | 33.7% | −4.38 (0.75) | 4.78×10−9 | 0.71 (0) |
| 19p13 | 1 | rs8109288# | 16046559 | TPM4 | A/G | 9.7% | −8.72 (1.40) | 5.02×10−10 | 0.35 (0) |
| 20q13 | 1 | rs151361 | 57047397 | SLMO2, TUBB1 | G/A | 25.7% | 4.49 (0.78) | 9.44×10−9 | 0.04 (40) |
| MEAN PLATELET VOLUME | |||||||||
| 7q22 | 4 | rs342296 | 106160139 | PIK3CG | A/G | 37.2% | 0.16 (0.02) | 1.44×10−11 | 0.25 (0) |
| 17q11 | 1 | rs11653144 | 24699352 | TAOK1 | C/T | 44.2% | −0.13 (0.02) | 4.17×10−8 | 0.48 (0) |
| 19p13 | 1 | rs8109288 | 16046559 | TPM4 | A/G | 8.4% | 0.26 (0.04) | 3.30×10−9 | 0.84 (0) |
*: SNPs in locus reaching GWAS significant threshold (5×10−8).
MAF = minor allele frequency; Chr = chromosome; SE = standard error; effect size = age-sex adjusted change in platelet count (109 L) or MPV (fL) per copy of minor allele; Maj = major; Min = minor; Het-P = Cochrane Q p-value to assess heterogeneity.
excluding cohorts that were included in the previous study16 that reported this SNP, the p-value was 8.6×10−7.
Table 3. Replication of the association of the best SNPs from each novel region with platelet count in three European American cohorts and a Hispanic American cohort.
| SNP | Candidate gene | Minor allele | Meta-analysis | ARIC-EA(N = 9274) | WHI-EA (N = 4243) | GeneSTAR-EA (N = 1392) | EA Meta-analysis | WHI-HA (N = 3462) | ||||||
| Effect size | p-value | Effect size | p-value | Effect size | p-value | Effect size | p-value | Effect size | p-value | Effect size | p-value | |||
| rs12526480 | LRRC16A | G | −4.39 | 9.15×10−9 | −2.29 | 0.01 | 0.79 | 0.54 | −2.57 | 0.32 | −1.34 | 0.06 | 0.28 | 0.65 |
| rs13236689 | CD36 | G | 4.18 | 2.84×10−9 | 0.56 | 0.53 | 2.4 | 0.05 | 1.67 | 0.51 | 1.24 | 0.07 | ||
| rs7896518 | JMJD1C | G | 5.18 | 2.26×10−12 | 5.07 | 9.65×10−9 | 4.02 | 0.10 | 4.95 | 2.61×10−9 | ||||
| rs477895 | BAD | C | −4.19 | 4.91×10−8 | −3.58 | 4.48×10−3 | −2.9 | 0.088 | −0.36 | 0.92 | −3.04 | 1.71×10−3 | −3.52 | 0.04 |
| rs8109288 | TPM4 | A | −8.72 | 5.02×10−10 | −18.98 | 1.49×10−7 | −11.26 | 0.03 | −8.31 | 0.33 | −15.58 | 2.60×10−8 | −12.38 | 0.02 |
| rs151361 | SLMO2 | G | 4.49 | 9.44×10−9 | 2.71 | 0.01 | 2.68 | 0.066 | 2.55 | 0.40 | 2.69 | 1.06×10−3 | 0.99 | 0.55 |
EA = European Americans; HA = Hispanic Americans.
The second locus is on chromosome 7q11 where two SNPs in intronic regions of the CD36 gene (rs13236689; p = 2.8×10−9 and rs17154155; p = 1.1×10−8) reached GWAS significance threshold, while 8 additional SNPs had p<10−6. rs13236689 and rs17154155 are in close linkage disequilibrium (r2 = 0.90 in the HapMap Yoruban population). After conditioning on rs13236689 in the association analysis, rs17154155 did not remain statistically significant (p = 0.39). Of the three European American cohorts, rs13236689 was statistically significant in the WHI cohort (p = 0.05) but not in the meta-analysis of all three studies (p = 0.07, Table 3). The CD36 gene encodes a thrombospondin receptor (platelet glycoprotein IV) which is present on the surface of platelets and several other cells [23]. rs17154155 has been reported to be associated with platelet function as well as with platelet expression of CD36 [24], [25].
In the third locus on chromosome 10q21, 71 SNPs reached GWAS threshold and 57 additional SNPs had p<10−6. Two non-synonymous common variants of unknown functional significance, rs 10761725 (resulting in serine to threonine substitution) and rs1935 (resulting in glutamate to aspartate substitution), in this region also crossed the GWAS threshold. All 128 SNPs in this region appear to be in strong linkage disequilibrium based on Yoruban HapMap data. The most significant SNP in this region, rs7896518 (p = 2.3×10−12), is located in an intron of the jumonji domain containing 1C (JMJD1C) gene. SNPs in this region have been reported to be associated with MPV (rs2393967) and with native platelet aggregation in platelet-rich plasma (rs10761741 in Caucasians and rs2893923 in African Americans) but not with platelet count [15], [26]. For rs7896518, data were available from 2 European American cohorts and meta-analysis found a significant association reaching GWAS threshold (p = 2.61×10−9) with similar direction of effect size (Table 3).
The fourth novel locus was located on chromosome 11q13. The most significant SNP (rs477895; p = 4.9×10−8) was in an intron of the BCL2-associated agonist of cell death (BAD) gene, while 23 other SNPs had p<10−6. For rs477895, all replication cohorts had effect sizes in a direction similar to African Americans and one European American and the Hispanic cohorts reached statistical significance (p = 4.48×10−3 and p = 0.04 respectively). Meta-analysis of the three European American cohorts also found significant association of rs477895 with platelet count (P = 1.71×10−3, Table 3). The protein encoded by the BAD gene inhibits the activity of the BCL-xL and BCL-2 proteins and thus has a pro-apoptotic effect [27]. Phospholipase C β3 protein encoded by another gene at this locus, PLCB3, is also known to be present in platelets and its deficiency results in impaired platelet function in mice [28]. This locus also contains SLC22A11 and SLC22A12, two genes that encode solute carrier proteins and previous GWAS have found association of genetic variants in these genes with serum uric acid levels [20]. Of the two genes, the transcript of SLC22A11 is present in significant amount in platelets as is the transcript for BAD [29]. Interestingly, a SNP about 20 kbp upstream of SLC22A11, rs4930420, almost reached GWAS threshold (p = 9.16×10−8, r2 with rs477895 = 0.21) and four additional SNPs in complete LD with rs4930420 (r2 = 1) had p-values<10−6. By examining the actual linkage disequilibrium patterns in this region in COGENT, and by performing conditional regression analysis in more than 8,400 African Americans from the WHI cohort simultaneously adjusting for BAD rs477895 and SLC22A11 rs4930420, we demonstrate that there are likely at least 2 independent platelet count association signals in this region and that the BAD and PLCB3 polymorphisms appear to represent the same association signal (Table S6).
The fifth novel locus was on chromosome 20q13 where one SNP in the SLMO2 gene exceeded GWAS significance threshold (rs151361; p = 9.4×10−9) while 2 other SNPs had p<10−6. One of these two SNPs was located in the first intron of TUBB1 gene (rs6070696; p = 2.5×10−7) and was 16.3 kbp downstream of the lead SNP (YRI HapMap r2 = 0.6). The TUBB1 gene encodes a beta1 tubulin, which plays an important role in megakaryopoiesis [30]. All replication cohorts had effect sizes in the direction similar to African Americans for rs151361 but only one European American study reached statistical significance (p = 0.01). The meta-analysis of the three European American replication cohorts also found a statistically significant association between rs151361 and platelet count (p = 1.1×10−3, Table 3).
Validation of previously reported loci for platelet count
In addition to identifying novel loci, we also replicated 5 previously reported loci at GWAS significance threshold and 3 other loci that were highly significant in our study but not at GWAS significance level (Table S7). The strongest signal in our platelet count meta-analysis was from chromosome 6p21 (SNP with the lowest p-value = rs210134; p = 2.3×10−15) located in the BAK1 gene, a locus that has been reported previously in Caucasians, Japanese, and African American populations [13]–[16]. We also found strong associations between platelet count and loci on chromosomes 6q23 (rs9494145; p = 2.8×10−9), 7q22 (rs342293; p = 1.6×10−8), and 12q24 (rs6490294; p = 4.8×10−9), all of which have been previously reported for Caucasians but not for African Americans [15]. Finally, we confirmed the association of a genetic variant rs8109288 (p = 5.0×10−10) in the tropomyosin 4 (TPM4) gene at chromosome 19p13 that has been previously reported for African Americans in a candidate gene study [16]. In our replication cohorts, rs8109288 was associated with platelet count in meta-analysis of European American cohorts and in Hispanic Americans (p = 2.6×10−8 and 0.02 respectively). We were also able to confirm the association of all previously reported SNPs (or a nearby SNP in the same LD block) with platelet count at a p<0.05 (Table S5).
Identification of loci for MPV
Of the three loci we identified at GWAS significance level for MPV, 2 have been previously reported to be associated with MPV in Caucasians, and one has been reported previously in African Americans. The association which has been previously reported in African Americans was of the A-allele of rs8109288 in TPM4 with increased MPV (p = 3.3×10−9); the same SNP was also associated with platelet count in this study. TPM4, a protein with a major role in stabilizing the cellular cytoskeleton, is present in platelets [31]. In the 7q22 region, we found that the SNP with the lowest p-value for MPV (rs342296; p = 1.4×10−11) was different from the SNP most associated with platelet count (rs342293; p-value = 5.84×10−11) although the two SNPs were only 684 bp apart and are in the same LD block (r2 = 0.92 based on HapMap II YRI) [15]. We also replicated a locus associated with MPV on 17q11 (rs11653144; p = 4.2×10−8) at GWAS significance threshold [15]. Of the 10 additional previously reported loci for MPV, we found statistically significant associations with 7 of them although these associations did not reach GWAS significance threshold (Table S8). For the loci that we were unable to replicate, we found other nearby SNPs with p<0.05. The direction of effect for all SNPs was not similar to the previously reported study of individuals of European ancestry suggesting that the alleles at the causal loci may be different between the two populations.
Platelet aggregation studies
Three regions (7q11, 7q22, 10q21) containing four SNPs (rs13236689, rs342296, rs342293, rs7896518) have already been shown to be associated with platelet aggregation [24]–[26], [32]. Therefore, the SNPs with the lowest p-values in each of the remaining 8 regions (Table 4) identified for either platelet count or MPV were examined for their association with platelet aggregation in 832 African-American individuals from the GeneSTAR study. Of the 8 SNPs, 3 were associated with a significant change in agonist-induced platelet aggregation but only one exceeded the Bonferroni-corrected significance threshold of 0.005 (Table 4). The minor allele (C) of rs6490294 in the ACAD10 gene (12q24) was associated with increased ADP-induced platelet aggregation (p = 0.002). Variants in this region have been previously reported to be associated with coronary artery disease [15]. The minor allele (A) of the 2nd SNP, rs8109288, in the TPM4 gene, was associated with decreased arachidonic-induced platelet aggregation (p = 0.03) and a trend towards decreased aggregation with ADP (p = 0.09). The minor allele (G) of the 3rd SNP, rs151361, in the SLMO2 gene, was associated with increased ADP-induced platelet aggregation (p = 0.008). The last 2 SNPs were nominally significant but did not exceed the Bonferroni-corrected significance threshold.
Table 4. Association of the top SNP from each locus with agonist-induced platelet aggregation in whole blood.
| SNP | Candidate gene (s) | Allele (MAF) | Arachidonic acid | ADP | Collagen | |||
| P-value | ES | P-value | ES | P-value | ES | |||
| rs12526480 | LRRC16A | G (0.30) | 0.38 | 0.320 | 0.13 | 0.453 | 0.51 | 0.252 |
| rs210134 | BAK1 | A (0.29) | 0.42 | −0.272 | 0.94 | −0.024 | 0.46 | −0.304 |
| rs9494145 | HBS1L, MYB | C (0.07) | 0.85 | 0.117 | 0.41 | 0.412 | 0.87 | −0.121 |
| rs477895 | BAD | C (0.45) | 0.33 | −0.333 | 0.25 | −0.338 | 0.93 | 0.034 |
| rs6490294 | ACAD10 | C (0.34) | 0.14 | 0.460 | 1.77×10−3 | 0.827 | 0.07 | 0.580 |
| rs11653144 | TAOK1 | C (0.44) | 0.57 | −0.181 | 0.15 | −0.414 | 0.12 | −0.659 |
| rs8109288 | TPM4 | A (0.10) | 0.03 | −1.171 | 0.09 | −0.698 | 0.35 | −0.533 |
| rs151361 | SLMO2, TUBB1 | G (0.26) | 0.54 | 0.213 | 7.95×10−3 | 0.698 | 0.11 | 0.578 |
Arachidonic acid 0.5 mmol/L, ADP 10 µmol/L, and collagen 5 µg/mL; MAF = minor allele frequency; ES = effect size.
Maximal aggregation was measured in ohms with impedance aggregometry 5 minutes after introducing agonist.
Discussion
We report the first meta-analysis of GWA studies of platelet count and MPV in a large number of African American participants from 7 population-based cohorts. We have identified 5 novel loci associated with platelet count of which three were replicated in the European American cohorts and one in the Hispanic cohort. None of these new African-American platelet loci have been reported previously in any racial group. In addition, we have confirmed that several loci previously reported in Europeans or Japanese are also associated with these platelet phenotypes in African Americans. We have further shown that 3 of the 8 loci (with one exceeding Bonferroni-corrected threshold), for which there have been no previously known association with platelet aggregation, are also associated with differences in platelet function using a subset of our African American sample.
Interestingly, the 5 novel platelet count loci are intragenic and 4 of these genes are known to have some role in platelet formation or biology. Platelets are small anucleate blood cells that are released from the cytoplasm of much larger bone marrow precursor cells known as megakaryocytes. One of the novel findings is the association of LRRC16A gene with platelet count. The protein encoded by the LRRC16A gene, capping protein ARP2/3 and myosin-I linker (CARMIL), plays an important role in actin-based cellular processes. Actin filaments are essential for end-amplification of pro-platelet processes during megakaryocyte maturation [33]. CARMIL exposes the barbed ends of actin filaments by binding to and then dislodging the capping protein from the actin filament [34]. Capping proteins are up-regulated during megakaryocyte maturation and LRRC16A is differentially expressed in megakaryocytes compared to other blood cells [35], [36]. The capping protein binding region of the CARMIL protein resides in the later part of the protein (940–1121 amino acid residues), which is a highly conserved region from protozoa to vertebrates. The majority of the residues in this region are critical for the anti-capping protein activity of CARMIL [37]. The rs12526480 genetic variant identified in our study is located in the latter part of the gene and may be in LD with a functional mutation in this conserved region. Any mutation that decreases the ability of CARMIL to dislodge capping protein from the barbed ends of the actin filament may result in abnormal megakaryocyte maturation and decreased platelet formation which is consistent with the direction of effect we observed in our study.
Another novel finding not reported in earlier GWA studies is the association of platelet count with CD36, a gene that encodes a receptor present on the surface of platelets, megakaryocytes, and several other cells. CD36 has a wide variety of ligands including thrombospondin [23]. Both CD36 and thrombospondin genes are up-regulated during megakaryocyte maturation and binding of thrombospondin-I to CD36 inhibits megakaryopoiesis, thus potentially providing a feedback mechanism for control of megakaryopoiesis [34], [36], [38]. The exact mechanism through which activation of CD36 inhibits megakaryopoiesis is unclear but may involve activation of extrinsic apoptotic mechanisms [39].
The most significant SNP associated with platelet count (rs210134 in BAK1) in our study is in complete LD with the most significant BAK1 SNP reported to be associated with platelet count in individuals of European ancestry (rs210135, r2 = 1 with rs210134 in HapMap II YRI, p = 2.18×10−14 in the current study). While the magnitude of effect is similar, the direction of effect is opposite suggesting that the allele at the causal locus is different in the two ethnic groups. A candidate gene study in African Americans has reported another SNP (rs449242, r2 = 0.81 with rs210134 in HapMap II YRI) in BAK1 and the direction of effect is similar to our study (Table S5) [16]. In addition to confirming the association of genetic variants in the pro-apoptotic BAK1 gene with low platelet count, we have identified and replicated a variant in another pro-apoptotic gene, BAD, that is associated with low platelet count. The protein encoded by BAD acts as a sensor for apoptotic signals upstream of BAK and activates BAK through indirect mechanisms [27]. The identification of these two genes in the intrinsic apoptotic pathway highlights the importance of the apoptotic process in modulating platelet lifespan in the circulation, which is one of the mechanisms that regulate platelet count [40]. Interestingly, this region also contains genetic variants associated with serum uric acid levels [20], however, the mechanism through which uric acid levels may be associated with platelet count remains unclear.
Genetic variants in the JMJD1C gene have been previously reported to be associated with MPV in Caucasians but not with platelet count. Conversely, we found several SNPs in this region that reached GWAS significance threshold for association with platelet count but none with MPV and we replicated the lead SNP in European Americans at GWAS threshold. In a GWAS study of platelet aggregation in Caucasians, the minor allele (T) of rs10761741 was associated with an increase in epinephrine-induced platelet aggregation in Caucasians [26]. JMJD1C gene is a histone demethylase and appears to be involved in steriodogenesis [41]. In addition to its association with platelet aggregation and MPV, previous GWAS have found genetic variants in this gene to be associated with serum levels of alkaline phosphatase and lipoprotein particle size and content [42]–[44].
In addition to confirming the finding of association of A-allele of rs8109288 in TPM4 gene with lower platelet count [16] and replicating this finding in European Americans, we also confirmed the association of the A-allele of this SNP with increased MPV and found a nominally significant association with decreased platelet aggregation. TPM4 gene expression is higher in megakaryocytes than other blood cells or other hematopoietic cells [35], [45]. Tropomyosin proteins play a central role in actin-based cytoskeletal changes and there appears to be biological plausibility for an effect of genetic variants on megakaryocyte maturation and platelet aggregation [46].
The final novel locus in the SLMO2 gene was also replicated in European Americans but SLMO2 gene has no known role in megakaryocyte biology. However, the variant is located within 13 kb of the TUBB1 gene, which is essential in the formation of normal mature platelets. The TUBB1 gene encodes beta1-tubulin that is exclusively expressed in platelets and megakaryocytes and forms a component of microtubules [30]. Loss of function mutations in TUBB1 gene have been reported in the literature and result in thrombocytopenia, large platelets, and increased risk of intracranial hemorrhage in men [47], [48]. The G-allele of the rs1513691 variant is associated with increased platelet count, decreased MPV, and increased aggregation, which may point towards a gain in function mutation in this region.
All previously reported loci that were also significantly associated with platelet count or MPV at GWAS threshold in our study have known biological roles in platelet biology. Two of these regions, 6q23 and 12q24, have pleiotropic effects with the 6q23 region associated with several hematological traits [13], [15], [49] and the 12q24 region associated with celiac disease and coronary artery disease [15]. More importantly, we also found that the 12q24 locus was associated with platelet aggregation after Bonferroni adjustment for multiple comparisons and thus may provide a mechanistic explanation of its role in development of coronary artery disease. The GG genotype of the most significant SNP in the 7q22 region, rs342293, is known to be associated with higher PIK3CG mRNA levels in platelets [32]. SNPs at this locus are also associated with platelet aggregation, pulse pressure, and carotid artery plaque [26], [50], [51]. TAOK1 is an important regulator of the mitotic progression and may also play a role in the apoptosis of cells [52], [53].
Our study included over 16,000 participants with platelet count and over 4500 participants with MPV measured and we were able to identify loci that explain between 0.16–0.33% of the variance in platelet count and loci that explain 1–1.5% of the variance of MPV (Table S9). Overall, the loci we identified explain up to 7% of the variance in platelet count and up to 6% of the variance in MPV, assuming that the each of these loci is independent. However, for both platelet count and MPV, the estimated heritability is >50%. Therefore, for each of these traits, the majority of heritability remains unexplained. One of the limitations of GWA studies is the limited power to detect effects caused by genetic variants with frequency <5%. We hypothesize that a significant proportion of the heritability of platelet count and MPV may be explained by variants with frequency <5%. Alternatively, there may be a large number of additional common variants that affect these traits, but have more modest effects.
In conclusion, we have conducted a meta-analysis of GWAS studies of platelet count and MPV in a large African American population and identified novel genetic variants in regions with genes that are likely to have a role in platelet formation. Furthermore, we have replicated 3 of the 5 novel loci in European Americans and one in Hispanic Americans. The novel regions identified may provide a focus for further research in improving our understanding of the biology of megakaryocyte maturation and platelet survival. In addition, we examined the effect of the genetic variants associated with platelet count and MPV on platelet function, and found 3 of these genetic variants to be associated with agonist-induced platelet aggregation of which one crossed Bonferroni-corrected significance threshold. Whether these newly identified genetic variants contribute to the risk of coronary artery disease or myocardial infarction, or to disorders associated with hyper- or hypo-aggregation of platelets, merits further investigation.
Methods
Subjects
The 7 studies included in this meta-analysis belonged to COGENT and enrolled 16,388 African American participants. The supplementary text contains a detailed description of each participating COGENT study cohort (Text S1). All participants self-reported their racial category. Additional clinical information was collected by self-report and clinical examination. All participants provided written informed consent as approved by local Human Subjects Committees. Study participants who were pregnant or had a diagnosis of cancer or AIDS at the time of blood count were excluded. We also excluded subjects who were outliers in the analysis of genetic ancestry (as determined by cluster analysis performed using principal component analysis or multi-dimensional scaling) or who had an overall SNP missing rate >10%.
Platelet count and MPV measurements
Fasting blood samples for complete blood count (CBC) analysis were obtained by venipuncture and collected into tubes containing ethylenediaminetetraacetic acid. Platelet counts and MPV were performed at local laboratories using automated hematology cell counters and standardized quality assurance procedures. Methods used to measure the blood traits analyzed in this study have been described previously for ARIC, CARDIA, JHS, Health ABC, WHI, and GeneSTAR [54]–[58]. Platelet count was reported as 109 cells per liter, and was recorded in all 16,388 study participants. Information on MPV was available in a subset of 4,612 participants from five COGENT study cohorts (ARIC, GeneSTAR, Health ABC, HANDLS, and JHS) and was reported in femto liters (10−15 L). All the phenotypes were approximately normally distributed and we did not perform any data transformations.
Genotype data and quality control
Genotyping was performed within each COGENT cohort using methods described in Text S1. Affymetrix chips were used in the ARIC, CARDIA, JHS, and WHI studies and Illumina chips were used in GeneSTAR, HANDLS, and Health ABC. DNA samples with a genome-wide genotyping success rate <95%, duplicate discordance or sex mismatch between genetic estimates of gender and self-report, SNPs with genotyping failure rate >10%, monomorphic SNPs, SNPs with minor allele frequency (MAF) <1%, and SNPs that mapped to several genomic locations were removed from the analyses. Because African-American populations are recently admixed, we did not filter on Hardy-Weinberg equilibrium p-value. Instead, significantly associated SNPs were later examined for strong deviations from Hardy–Weinberg equilibrium and/or raw genotype data was examined for abnormal clustering. Participants and SNPs passing basic quality control were imputed to >2.2 million SNPs based on HapMap II haplotype data using a 1∶1 mixture of Europeans (CEU) and Africans (YRI) as the reference panel. Details of the genotype imputation procedure are described further under Supplemental Methods. Prior to meta-analyses, SNPs were excluded if imputation quality metrics (equivalent to the squared correlation between proximal imputed and genotyped SNPs) were less than 0.50.
Platelet aggregation assays
Differences in platelet count may affect platelet function and aggregation [59]. In addition, younger platelets have higher MPV than older platelets and are more reactive [60]. We hypothesized that the genetic variants that determine platelet count and MPV may also affect platelet aggregation. To examine this hypothesis, we used agonist-mediated platelet aggregation assays, which can provide information about the different aspects of platelet aggregation. For these assays, platelet aggregation agonists, such as collagen or ADP, are added to whole blood or platelet-rich plasma and platelet aggregation is measured after a specified amount of time (300 seconds). We performed platelet aggregation assays in the participants of the GeneSTAR cohort. Blood samples were obtained as described above, and platelet aggregation in whole blood was measured as reported previously [57]. Briefly, in vitro whole blood impedance in a Chrono-Log dual-channel lumiaggregometer (Havertown, Pa) was performed after samples were stimulated with arachidonic acid (0.5 mmol/L, intra-assay CV = 24%), collagen (5 µg/mL; intra-assay CV = 9%), or ADP (10 µmol/L; intra-assay CV = 46%). Maximal aggregation within 5 minutes of agonist stimulation was recorded in ohms.
Data analysis
For all cohorts, genome-wide association (GWAS) analysis was performed using linear regression adjusted for covariates, implemented in either PLINK v1.07, R v2.10, or MACH2QTL v1.08 [61], [62]. Allelic dosage at each SNP was used as the independent variable, adjusted for primary covariates of age, age-squared, sex, and clinic site (if applicable). The first 10 principal components were also incorporated as covariates in the regression models to adjust for population stratification (Text S1). For GeneSTAR, family structure was accounted for in the association tests using linear mixed effect (LME) models implemented in R [63]. Although the JHS has a small number of related individuals, extensive analyses have shown that results were concordant using linear regression or LME, after genomic control [19]. Therefore, results are presented for JHS using linear regression. For imputed genotypes, we used dosage information (i.e. a value between 0.0–2.0 calculated using the probability of each of the three possible genotypes) in the regression model implemented in PLINK or MACH2QTL (for cohorts with unrelated individuals) or the Maximum Likelihood Estimation (MLE) routines (for GeneSTAR).
For both platelet phenotypes, meta-analyses were conducted using inverse-variance weighted fixed-effect models to combine beta coefficients and standard errors from study level regression results for each SNP to derive a combined p-value and effect estimate [64]. Study level results were corrected for genomic inflation factors (λGC) by incorporating study specific λGC estimates into the scaling of the standard errors (SE) of the regression coefficients by multiplying the SE by the square-root of the genomic inflation factor. The inflation factors for all completed analyses are presented in Table S1. To maintain an overall type 1 error rate of 5%, a threshold of α = 5×10−8 was used to declare genome-wide statistical significance. Between-study heterogeneity of results was assessed by using Cochrane's Q statistic and the I2 inconsistency metric. Meta-analyses were implemented in the software METAL [64] and were performed independently by two analysts to confirm results. To examine whether there were any differences between males and females, sex-specific GWAS were conducted in each cohort. The results for each SNP were pooled and heterogeneity of allelic effects between females and males was examined using the meta-analysis methods as implemented in GWAMA software [65].
To assess whether the loci previously reported to be associated with the platelet phenotypes in Europeans, Japanese, and African Americans were replicated in the COGENT African-Americans, we examined the meta-analysis results for each index SNP in the regions previously reported, including consistency of direction of effect. If the reported index SNP was not significant at p<0.05 we examined adjacent SNPs and reported the closest SNP with p<0.05 along with its distance from the index SNP.
To examine the association of genotype on platelet aggregation in the GeneSTAR cohort, linear mixed models were used with additive models adjusting for age and sex, and taking into account familial correlation between the individuals.
Supporting Information
Negative log(10) statistical significance plots of the each local region with 500 kbp on either side of the top SNP significantly associated with platelet count.
(PDF)
Negative log(10) statistical significance plots of the each local region with 500 kbp on either side of the top SNP significantly associated with mean platelet volume.
(PDF)
QQ plots of individual studies for platelet count (PLT).
(PDF)
QQ plots of individual studies for mean platelet volume (MPV).
(PDF)
Genomic inflation factors for all GWAS analyses included in the meta-analysis.
(PDF)
List of all SNPs with p-values<10−6 in the regions that were significant at GWAS threshold in platelet count meta-analysis.
(PDF)
List of all SNPs that were significant at p-value<10−6 in regions with at least one SNP with GWAS threshold in mean platelet volume meta-analysis.
(PDF)
Individual study results for the top significant SNP from each region.
(PDF)
Association analysis of rs12526480 SNP with selected phenotypes.
(PDF)
Conditional analysis of the significant SNPs in at the 11q13 locus for platelet count.
(PDF)
Association of loci previously reported with platelet count in Caucasians, Japanese, or African American populations (from references 13, 15, and 16).
(PDF)
Association of loci previously reported with mean platelet volume in Caucasians (N = 13943)(from reference [15]).
(PDF)
Percentage variance in phenotype explained by each SNP for each study.
(PDF)
Supporting Methods.
(DOCX)
Acknowledgments
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the involved research institutions, study investigators, field staff, and study participants of Atherosclerosis Risk in Communities (ARIC), Coronary Artery Risk in Young Adults (CARDIA), Jackson Heart Study (JHS), and Broad Institute in creating the Candidate-gene Association Resource for biomedical research (CARe; http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx). The authors also wish to thank the investigators, staff, and participants of GeneSTAR, Health ABC, Healthy Aging in Neighborhoods of Diversity across the Life Span Study (HANDLS), and Women Health Initiative (WHI) for their important contributions. A listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf. The authors also wish to acknowledge the support of the GARNET Collaborative Research Group.
Footnotes
The authors have declared that no competing interests exist.
The ARIC, CARDIA, and JHS studies contributed data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226) to create genotype/phenotype data base for wide dissemination to the biomedical research community. The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The Coronary Artery Risk in Young Adults (CARDIA) was supported by grants to University of Alabama at Birmingham (N01-HC-48047, N01-HC-95095), University of Minnesota (N01-HC-48048), Northwestern University (N01-HC-48049), Kaiser Foundation Research Institute (N01-HC-48050), Tufts-New England Medical Center (N01-HC-45204), Wake Forest University (N01-HC-45205), Harbor-UCLA Research and Education Institute (N01-HC-05187), and University of California Irvine (N01-HC-45134, N01-HC-95100). The Jackson Heart Study (JHS) was supported by grants to Jackson State University (N01-HC-95170), University of Mississippi (N01-HC-95171), and Tougaloo College (N01-HC-95172). The Healthy Aging in Neighborhoods of Diversity across the Life Span Study (HANDLS) was supported by the Intramural Research Program of the NIH, National Institute on Aging and the National Center on Minority Health and Health Disparities (project # Z01-AG000513 and human subjects protocol # 2009-149). Data analyses for the HANDLS study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The Health ABC study was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The GeneSTAR study was supported by the National Heart, Lung, and Blood Institute (NHLBI) through the PROGENI (U01 HL72518) and STAMPEED (R01 HL087698-01) consortia. The Women's Health Initiative (WHI) program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services, through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. Additional support for this work was provided by the Fondation de l'Institut de Cardiologie de Montréal (to GL), and by NIH R01 HL71862-06 (to APR) NIH 1K23HL105897-01 (to RQ) and ARRA N000949304 (to APR). Funding support for WHI-GARNET was provided through the NHGRI Genomics and Randomized Trials Network (GARNET) (Grant Number U01 HG005152). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH Genes, Environment, and Health Initiative [GEI] (U01 HG004424). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davi G, Patrono C. Platelet activation and atherothrombosis. N Engl J Med. 2007;357:2482–2494. doi: 10.1056/NEJMra071014. [DOI] [PubMed] [Google Scholar]
- 2.Nikolsky E, Grines CL, Cox DA, Garcia E, Tcheng JE, et al. Impact of baseline platelet count in patients undergoing primary percutaneous coronary intervention in acute myocardial infarction (from the CADILLAC trial). Am J Cardiol. 2007;99:1055–1061. doi: 10.1016/j.amjcard.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 3.Turakhia MP, Murphy SA, Pinto TL, Antman EM, Giugliano RP, et al. Association of platelet count with residual thrombus in the myocardial infarct-related coronary artery among patients treated with fibrinolytic therapy for ST-segment elevation acute myocardial infarction. Am J Cardiol. 2004;94:1406–1410. doi: 10.1016/j.amjcard.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Tucker EI, Marzec UM, Berny MA, Hurst S, Bunting S, et al. Safety and antithrombotic efficacy of moderate platelet count reduction by thrombopoietin inhibition in primates. Sci Transl Med. 2010;2:37ra45. doi: 10.1126/scitranslmed.3000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biino G, Balduini CL, Casula L, Cavallo P, Vaccargiu S, et al. Analysis of 12,517 inhabitants of a Sardinian geographic isolate reveals that predispositions to thrombocytopenia and thrombocytosis are inherited traits. Haematologica. 2011;96:96–101. doi: 10.3324/haematol.2010.029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley MF, James JW, Brown DE, Whyte GS, Dean MG, et al. A novel approach to the assessment of variations in the human platelet count. Thromb Haemost. 2000;83:480–484. [PubMed] [Google Scholar]
- 7.Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 8.Traglia M, Sala C, Masciullo C, Cverhova V, Lori F, et al. Heritability and demographic analyses in the large isolated population of Val Borbera suggest advantages in mapping complex traits genes. PLoS One. 2009;4:e7554. doi: 10.1371/journal.pone.0007554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray PF, Mathias RA, Faraday N, Yanek LR, Fallin MD, et al. Heritability of platelet function in families with premature coronary artery disease. J Thromb Haemost. 2007;5:1617–1623. doi: 10.1111/j.1538-7836.2007.02618.x. [DOI] [PubMed] [Google Scholar]
- 10.Chu SG, Becker RC, Berger PB, Bhatt DL, Eikelboom JW, et al. Mean platelet volume as a predictor of cardiovascular risk: a systematic review and meta-analysis. J Thromb Haemost. 2010;8:148–156. doi: 10.1111/j.1538-7836.2009.03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncalves SC, Labinaz M, Le May M, Glover C, Froeschl M, et al. Usefulness of mean platelet volume as a biomarker for long-term outcomes after percutaneous coronary intervention. Am J Cardiol. 2011;107:204–209. doi: 10.1016/j.amjcard.2010.08.068. [DOI] [PubMed] [Google Scholar]
- 12.Klovaite J, Benn M, Yazdanyar S, Nordestgaard BG. High platelet volume and increased risk of myocardial infarction: 39 531 participants from the general population. J Thromb Haemost. 2011;9:49–56. doi: 10.1111/j.1538-7836.2010.04110.x. [DOI] [PubMed] [Google Scholar]
- 13.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 14.Meisinger C, Prokisch H, Gieger C, Soranzo N, Mehta D, et al. A genome-wide association study identifies three loci associated with mean platelet volume. Am J Hum Genet. 2009;84:66–71. doi: 10.1016/j.ajhg.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo KS, Wilson JG, Lange LA, Folsom AR, Galarneau G, et al. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segal JB, Moliterno AR. Platelet counts differ by sex, ethnicity, and age in the United States. Ann Epidemiol. 2006;16:123–130. doi: 10.1016/j.annepidem.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 18.Casto AM, Feldman MW. Genome-wide association study SNPs in the human genome diversity project populations: does selection affect unlinked SNPs with shared trait associations? PLoS Genet. 2011;7:e1001266. doi: 10.1371/journal.pgen.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tore S, Casula S, Casu G, Concas MP, Pistidda P, et al. Application of a new method for GWAS in a related case/control sample with known pedigree structure: identification of new loci for nephrolithiasis. PLoS Genet. 2011;7:e1001281. doi: 10.1371/journal.pgen.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, et al. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 2009;84:60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A, Murugesan G, Chen K, Zhang L, Wang Q, et al. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood. 2011;117:6355–6366. doi: 10.1182/blood-2011-02-338582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CI, Bray S, Garner SF, Stephens J, de Bono B, et al. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114:1405–1416. doi: 10.1182/blood-2009-02-202614. [DOI] [PubMed] [Google Scholar]
- 26.Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42:608–613. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2008;27(Suppl 1):S53–70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- 28.Lian L, Wang Y, Draznin J, Eslin D, Bennett JS, et al. The relative role of PLCbeta and PI3Kgamma in platelet activation. Blood. 2005;106:110–117. doi: 10.1182/blood-2004-05-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugert P, Kluter H. Profiling of gene transcripts in human platelets: an update of the platelet transcriptome. Platelets. 2006;17:503–504. doi: 10.1080/09537100600901491. [DOI] [PubMed] [Google Scholar]
- 30.Italiano JE, Bergmeier W, Tiwari S, Falet H, Hartwig JH, et al. Mechanisms and implications of platelet discoid shape. Blood. 2003;101:4789–4796. doi: 10.1182/blood-2002-11-3491. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill EE, Brock CJ, von Kriegsheim AF, Pearce AC, Dwek RA, et al. Towards complete analysis of the platelet proteome. Proteomics. 2002;2:288–305. doi: 10.1002/1615-9861(200203)2:3<288::aid-prot288>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 32.Soranzo N, Rendon A, Gieger C, Jones CI, Watkins NA, et al. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood. 2009;113:3831–3837. doi: 10.1182/blood-2008-10-184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Italiano JE, Hartwig JH. Megakaryocyte Development and Platelet Formation. In: Michelson AD, editor. Boston: Elsevier; 2007. pp. 27–34. [Google Scholar]
- 34.Yang C, Pring M, Wear MA, Huang M, Cooper JA, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raslova H, Kauffmann A, Sekkai D, Ripoche H, Larbret F, et al. Interrelation between polyploidization and megakaryocyte differentiation: a gene profiling approach. Blood. 2007;109:3225–3234. doi: 10.1182/blood-2006-07-037838. [DOI] [PubMed] [Google Scholar]
- 37.Uruno T, Remmert K, Hammer JA., 3rd CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281:10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- 38.Lim CK, Hwang WY, Aw SE, Sun L. Study of gene expression profile during cord blood-associated megakaryopoiesis. Eur J Haematol. 2008;81:196–208. doi: 10.1111/j.1600-0609.2008.01104.x. [DOI] [PubMed] [Google Scholar]
- 39.De Botton S, Sabri S, Daugas E, Zermati Y, Guidotti JE, et al. Platelet formation is the consequence of caspase activation within megakaryocytes. Blood. 2002;100:1310–1317. doi: 10.1182/blood-2002-03-0686. [DOI] [PubMed] [Google Scholar]
- 40.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 41.Kim SM, Kim JY, Choe NW, Cho IH, Kim JR, et al. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 2010;38:6389–6403. doi: 10.1093/nar/gkq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf SS, Patchev VK, Obendorf M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch Biochem Biophys. 2007;460:56–66. doi: 10.1016/j.abb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Yuan X, Waterworth D, Perry JR, Lim N, Song K, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komor M, Guller S, Baldus CD, de Vos S, Hoelzer D, et al. Transcriptional profiling of human hematopoiesis during in vitro lineage-specific differentiation. Stem Cells. 2005;23:1154–1169. doi: 10.1634/stemcells.2004-0171. [DOI] [PubMed] [Google Scholar]
- 46.Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 47.Kunishima S, Kobayashi R, Itoh TJ, Hamaguchi M, Saito H. Mutation of the beta1-tubulin gene associated with congenital macrothrombocytopenia affecting microtubule assembly. Blood. 2009;113:458–461. doi: 10.1182/blood-2008-06-162610. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira MA, Hottenga JJ, Warrington NM, Medland SE, Willemsen G, et al. Sequence variants in three loci influence monocyte counts and erythrocyte volume. American Journal of Human Genetics. 2009;85:745–749. doi: 10.1016/j.ajhg.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, et al. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet. 2011;43:940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 53.Wu MF, Wang SG. Human TAO kinase 1 induces apoptosis in SH-SY5Y cells. Cell Biol Int. 2008;32:151–156. doi: 10.1016/j.cellbi.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Folsom AR, Rosamond WD, Shahar E, Cooper LS, Aleksic N, et al. Prospective study of markers of hemostatic function with risk of ischemic stroke. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Circulation. 1999;100:736–742. doi: 10.1161/01.cir.100.7.736. [DOI] [PubMed] [Google Scholar]
- 55.Margolis KL, Manson JE, Greenland P, Rodabough RJ, Bray PF, et al. Leukocyte count as a predictor of cardiovascular events and mortality in postmenopausal women: the Women's Health Initiative Observational Study. Arch Intern Med. 2005;165:500–508. doi: 10.1001/archinte.165.5.500. [DOI] [PubMed] [Google Scholar]
- 56.Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qayyum R, Becker DM, Yanek LR, Moy TF, Becker LC, et al. Platelet inhibition by aspirin 81 and 325 mg/day in men versus women without clinically apparent cardiovascular disease. Am J Cardiol. 2008;101:1359–1363. doi: 10.1016/j.amjcard.2007.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimakawa T, Bild DE. Relationship between hemoglobin and cardiovascular risk factors in young adults. J Clin Epidemiol. 1993;46:1257–1266. doi: 10.1016/0895-4356(93)90090-n. [DOI] [PubMed] [Google Scholar]
- 59.Jennings LK, White MM. Platelet Aggregation. In: Michelson AD, editor. Platelets. 2nd ed. Boston: Elsevier; 2007. pp. 495–507. [Google Scholar]
- 60.Harker LA, Slichter SJ. The bleeding time as a screening test for evaluation of platelet function. N Engl J Med. 1972;287:155–159. doi: 10.1056/NEJM197207272870401. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34:846–853. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative log(10) statistical significance plots of the each local region with 500 kbp on either side of the top SNP significantly associated with platelet count.
(PDF)
Negative log(10) statistical significance plots of the each local region with 500 kbp on either side of the top SNP significantly associated with mean platelet volume.
(PDF)
QQ plots of individual studies for platelet count (PLT).
(PDF)
QQ plots of individual studies for mean platelet volume (MPV).
(PDF)
Genomic inflation factors for all GWAS analyses included in the meta-analysis.
(PDF)
List of all SNPs with p-values<10−6 in the regions that were significant at GWAS threshold in platelet count meta-analysis.
(PDF)
List of all SNPs that were significant at p-value<10−6 in regions with at least one SNP with GWAS threshold in mean platelet volume meta-analysis.
(PDF)
Individual study results for the top significant SNP from each region.
(PDF)
Association analysis of rs12526480 SNP with selected phenotypes.
(PDF)
Conditional analysis of the significant SNPs in at the 11q13 locus for platelet count.
(PDF)
Association of loci previously reported with platelet count in Caucasians, Japanese, or African American populations (from references 13, 15, and 16).
(PDF)
Association of loci previously reported with mean platelet volume in Caucasians (N = 13943)(from reference [15]).
(PDF)
Percentage variance in phenotype explained by each SNP for each study.
(PDF)
Supporting Methods.
(DOCX)