Abstract
Cardiac hypertrophy is controlled by a complex signal transduction and gene regulatory network, containing multiple layers of crosstalk and feedback. While numerous individual components of this network have been identified, understanding how these elements are coordinated to regulate heart growth remains a challenge. Past approaches to measure cardiac myocyte hypertrophy have been manual and often qualitative, hindering the ability to systematically characterize the network's higher-order control structure and identify therapeutic targets. Here, we develop and validate an automated image analysis approach for objectively quantifying multiple hypertrophic phenotypes from immunofluorescence images. This approach incorporates cardiac myocyte-specific optimizations and provides quantitative measures of myocyte size, elongation, circularity, sarcomeric organization, and cell-cell contact. As a proof-of-concept, we examined the hypertrophic response to α-adrenergic, β-adrenergic, tumor necrosis factor (TNFα), insulin-like growth factor-1 (IGF-1), and fetal bovine serum pathways. While all five hypertrophic pathways increased myocyte size, other hypertrophic metrics were differentially regulated, forming a distinct phenotype signature for each pathway. Sarcomeric organization was uniquely enhanced by α-adrenergic signaling. TNFα and α-adrenergic pathways markedly decreased cell circularity due to increased myocyte protrusion. Surprisingly, adrenergic and IGF-1 pathways differentially regulated myocyte-myocyte contact, potentially forming a feed-forward loop that regulates hypertrophy. Automated image analysis unlocks a range of new quantitative phenotypic data, aiding dissection of the complex hypertrophic signaling network and enabling myocyte-based high-content drug screening.
Keywords: hypertrophy, image processing, neonatal cardiac myocyte, signal transduction
Introduction
Cardiac hypertrophy is the growth of individual cardiac myocytes in response to stress. Physiologic stresses like exercise and pregnancy cause an adaptive, reversible hypertrophy while pathologic stresses such as myocardial infarction lead to maladaptive hypertrophy and heart failure [1]. But major challenges remain in identifying, understanding, and ultimately controlling the molecular circuits that regulate heart growth [2]. Rather than acting through a common mechanism, these pathways form a dense web of interactions that has eluded therapeutic approaches to date [3]. The complexity of cardiac signaling networks indicates that integrative systems approaches will be critical for understanding and treating heart failure [4, 5].
Cultured neonatal rat ventricular myocytes have been widely used to study the signaling pathways that initiate cardiac hypertrophy [6]. While neonatal myocytes cannot replicate the native 3D environment and later stages of heart failure, most hypertrophic pathways and genes studied in vivo were first implicated in cultured cells [7]. Previous approaches to imaging hypertrophy in cultured myocytes have been low-throughput and subjective, which has generally limited individual studies to a single node or pathway. Systems-wide analysis of hypertrophy networks will require new quantitative, scalable phenotypic screening approaches [2]. One such approach is high-content cell imaging [8], which has emerged in the pharmaceutical industry for improved target validation compared with traditional biochemical screens [9]. Similarly, systems analyses of the myocyte hypertrophy network may benefit from automated imaging approaches that are amenable to testing larger numbers of pharmacologic or genetic perturbations.
Here, we develop a method for automated image analysis that enables high-content imaging of cardiac myocyte hypertrophy. In addition to myocyte size, several additional hypertrophic phenotypes are quantified including myocyte elongation, sarcomeric organization, and cell-cell contact. As a proof of principle, we measured phenotypic responses to five main classes of hypertrophic pathways: α-adrenergic, β-adrenergic, cytokine (tumor necrosis factor, TNFα), growth factor (insulin-like growth factor-1, IGF-1), and serum (fetal bovine serum, FBS) targeted pathways. Our image analysis approach reveals that while these pathways share a common response in myocyte size, their regulation of other hypertrophic metrics is remarkably distinct.
Methods
Cardiac myocyte hypertrophy experiments
Neonatal ventricular myocytes were isolated from 1-2 day old Sprague-Dawley rats using the Neomyts isolation kit (Cellutron, Baltimore MD). All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health and approved by the University of Virginia Institutional Animal Care and Use Committee. Myocytes were initially cultured in plating media (Dulbecco Modified Eagle Media, 17% M199, 10% Horse Serum, 5% Fetal Bovine Serum, 100U/mL penicillin, and 50 mg/mL streptomycin) on 35-mm glass-bottom dishes (MatTek, Ashland, MA) at a density of 750,000 cells/dish. Two days after isolation, myocytes were switched to serum-free maintenance media (Dulbecco Modified Eagle Media, 19% M199, 1% ITSS, 100U/mL penicillin, and 50 mg/mL streptomycin) for 24 hours. Then myocytes were rinsed and cultured in serum-free maintenance media containing one of five hypertrophic stimuli (1 μM isoproterenol, 100 μM phenylephrine, 10 ng/mL TNF-α, 10 ng/mL IGF-1, or 15% FBS) or negative control (serum-free media alone) for 48 hours, with solutions changed at 24 hours.
After 48 hours of hypertrophic agonist, myocytes were fixed with 4% paraformaldehyde for 20 minutes and then permeabilized with 0.1% Triton-X for 2 minutes. Using a 1% bovine serum albumin (BSA) in PBS solution, myocytes were blocked for 45 minutes. Then mouse anti-α-actinin primary antibody (Sigma-Aldrich, St. Louis MO) at a concentration of 1:200 was applied to the myocytes for 1 hour. Using a 2% donkey serum in PBS solution, myocytes were blocked for 30 minutes. Then Alexa Fluor-680-conjugated donkey anti-mouse secondary antibody (Invitrogen, San Diego CA) at a concentration of 1:200 was applied to the myocytes for 1 hour. Finally, myocytes were double-stained with DAPI and SlowFade with DAPI (Invitrogen, San Diego CA).
Myocytes were imaged using an Olympus IX81 inverted microscope with 20X PlanApo 0.85 NA oil objective, Orca-AG CCD camera (Hamamatsu, Bridgewater, NJ), and IPLab software (Scanalytics, Fairfax, VA). The α-actinin channel was acquired using an 480/40 excitation filter with 200 ms exposure time and recorded at 535/50-nm. The DAPI channel was acquired using a 350/50-nm excitation filter with 20 ms exposure time and recorded at 460/50-nm (Chroma filters; Optical Insights, Santa Fe, NM).
Automated image analysis
The algorithm for automated myocyte segmentation is composed of 6 main phases, which are outlined in Figure 1. In the first phase (Figure 1-1), the algorithm loads images of nuclei (stained using DAPI) and α-actinin, a myocyte-specific cytoskeletal protein. A median filter with an appropriately sized window (4 pixels in the experiments presented here) is applied to the α-actinin image to improve cell segmentation performance, and then all images are background-subtracted using a manual threshold that is constant across all image sets. In the second phase (Figure 1-2), nuclei are identified and segmented using an Otsu threshold of the DAPI image. The Otsu threshold is the optimum threshold in an intensity histogram that minimizes the variance between background and foreground pixels [10]. Next, the minor population of cardiac fibroblasts and other nonmyocytes is removed by filtering out nuclei that do not contain nuclear α-actinin signal (Figure 1-3). Some neonatal cardiac myocytes are binucleated, which causes difficulty for our nucleus-based cell segmentation. Therefore, in the fourth phase adjacent nuclei in the same cell are merged (Figure 1-4). Myocyte nuclei are dilated by a suitable margin width (3 pixels in this case), re-identified, and then eroded by the same margin width to restore their original size.
Figure 1. Approach for automated image segmentation of cardiac myocytes.
(1) Acquire images, primarily containing neonatal ventricular myocytes (MYO) but some fibroblasts (FB). Myocyte-specific α-actinin is labeled by immunofluorescence, while DNA is labeled with DAPI. (2) Smooth and threshold the α-actinin channel. Segment nuclei in the DAPI channel by Otsu thresholding. (3) Remove fibroblast/nonmyocyte nuclei. (4) Merge adjacent nuclei to form binucleates. (5) Segment myocyte boundaries by propagating from the nucleus outwards. (6) Eliminate myocytes touching the image border. The end result is a quantified nucleus and cell boundary for each myocyte.
In the fifth phase (Figure 1-5), cardiac myocyte cell boundaries are segmented and identified. Cell-background discrimination is based on an Otsu threshold of the α-actinin image. Then cell-cell boundary segmentation is performed using a nuclear propagation approach, in which the nucleus of each cell is used as the initial seed and then the algorithm propagates the cell boundary outward [11]. The final boundary between adjacent cells is determined using an equation that combines information about local minima (valleys) in the α-actinin image and the distance of the boundary from the nucleus (see Supplementary Methods). In the final phase (Figure 1-6), myocytes touching the edge of the image are removed. This is performed by creating a 1-pixel wide image border and then identifying myocytes that overlap with the border. Nuclei of myocytes touching the image edge are also removed at this stage. Once myocyte segmentation is complete, these objects can be used for subsequent shape and intensity-based measurements.
The automated cell segmentation algorithm was implemented using the open-source MATLAB-based CellProfiler software package [12]. Detailed steps of the algorithm are provided in Supplementary Table S1. The algorithm and example raw image data from this manuscript are freely available for download at http://bme.virginia.edu/saucerman/.
Manual segmentation
To assess the accuracy of the automated myocyte segmentation, manual segmentation was performed by two separate researchers blinded to the results of other segmentation results. As in the automated myocyte segmentation, pre-processing and automated nucleus segmentation was provided so that the only free variable was the cell segmentation itself. The performance of the segmentation was evaluated using the precision, recall and F-score of pixel overlap [13]. Subsequent comparisons of phenotypic metrics were performed automatically based on the manual segmentations.
Quantifying Hypertrophic Phenotypes
Cell area and cell perimeter were selected as metrics of cell size, while elongation and circularity were selected as metrics of cell shape. Elongation is the ratio of the major axis length to the minor axis length of a best-fit ellipse. Thus, a circle is defined to have an elongation of 1. Circularity is calculated as 4π*Area/Perimeter2. A circle has a circularity of 1 and all other shapes have circularities < 1. Circularity is used to characterize cell roundness with smaller circularity values indicating either elongation or increased cell protrusions. Sarcomeric organization was quantified using the pixel Uniformity within individual cells [14]. Cell-cell contact was quantified by the percentage of each myocyte's border that is shared with neighboring cells. Due to non-normal distributions of myocyte phenotypes, statistical significance was assessed by Kruskal-Wallis non-parametric analysis of variance followed by Dunn's multiple comparisons post-test in GraphPad Prism. P < 0.05 was considered significant.
Results
Automated cell segmentation accurately identifies myocyte morphology
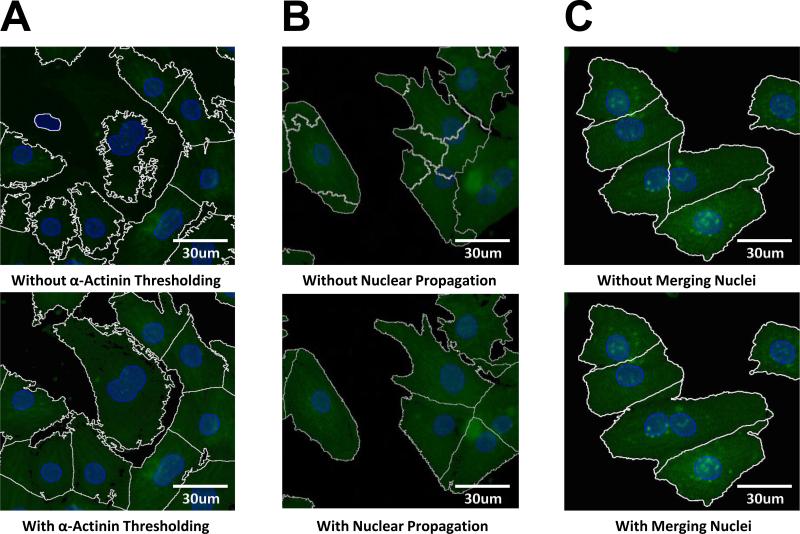
Several features of the automated cell segmentation algorithm (described above in Methods) were particularly important for accurate identification of cardiac myocytes. As shown in Figure 2A, thresholding the α-actinin channel before cell segmentation enabled more precise identification of cell-background borders. This is a challenging issue in α-actinin-labeled myocytes due to the complex periodic structure of the myofilaments. We evaluated several algorithms for automated segmentation of myocyte boundaries including watershed, active contours [15], and Otsu thresholds [10]. We found that a nucleus-propagation segmentation approach [11] was most robust for α-actinin-labeled cardiac myocytes (Figure 2B). However, the nucleus-propagation segmentation requires a single nucleus per cell, and neonatal myocytes are occasionally bi-nucleated. This could cause a single bi-nucleated cell to be incorrectly identified as two separate cells. We found that merging adjacent nuclei enabled correct segmentation of binucleated myocytes (Figure 2C).
Figure 2. Improvements to myocyte segmentation due to thresholding, merging adjacent nuclei, and nucleus-propagating segmentation steps.
Myocyte segmentation is shown both with and without each of three algorithm steps: A) α-actinin thresholding, B) nucleus-propagation segmentation and C) merging adjacent nuclei of binucleated myocytes. Each step is particularly beneficial to accurate segmentation of cardiac myocytes.
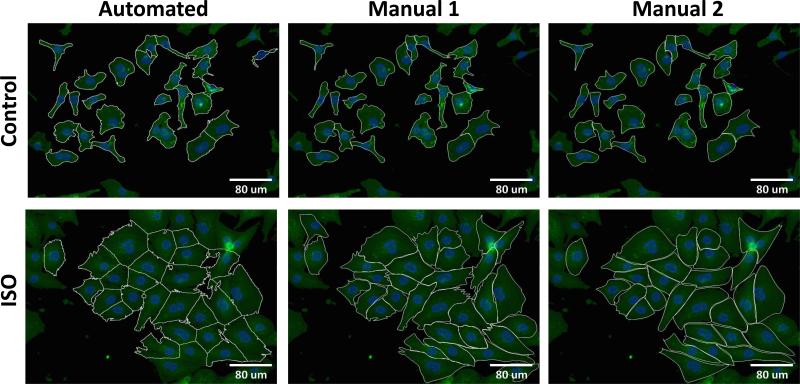
To assess the accuracy of automated segmentation, two researchers manually segmented 100 myocytes from a range of conditions (see Methods). As shown in Figure 3, under control conditions or regions of lower cell density the segmentation is straightforward. Under conditions of hypertrophy or higher regional cell density, automated segmentation is more challenging but the algorithm performs reasonably well. The most rigorous method to evaluate image segmentation is to evaluate the pixel overlap between individual automated and manual segmented cells [13]. As shown in Table 1, average precision (positive predictive value) is 89% and average recall (sensitivity) is 85%, producing an overall segmentation performance F-score of ~85%, which meets or exceeds the performance of previous cell segmentation studies [13, 16].
Figure 3. Visual comparison of automated and manual myocyte segmentation.
Representative images from control conditions and isoproterenol (ISO) treatment are shown. Myocytes were segmented either using the automated segmentation algorithm or manually by 2 separate researchers (Manual 1 and Manual 2).
Table 1.
Comparison of automated and manual cell segmentation by F-score analysis.
| Automated vs. Manual 1 | Automated vs. Manual 2 | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-factor | Precision | Recall | F-factor | |
| Avg. | 0.88 | 0.84 | 0.85 | 0.89 | 0.84 | 0.86 |
| S.E. | 0.014 | 0.016 | 0.014 | 0.014 | 0.014 | 0.013 |
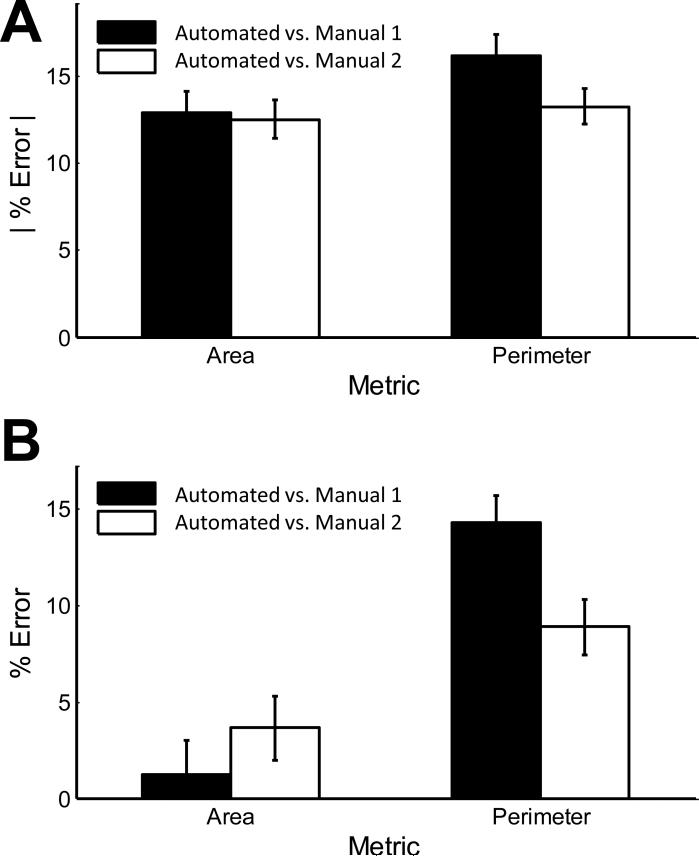
Automated segmentation can also be validated by the phenotypic metrics derived from the segmentation. As shown in Figure 4, the mean absolute error in cell area and cell perimeter is ~13%. While average error in cell area falls to ~1% due to cancelling over- and underestimations, average error in perimeter remains ~10%. We hypothesize that the consistently higher perimeters are caused by the pixel-by-pixel resolution of the algorithm, while manual segmenters have lower visuomotor resolution. This hypothesis is supported by inspection of magnified views provided in Supplementary Figure 1. Measures of elongation and circularity are shown in Supplementary Figure 2. The calculated error for circularity is high compared to the other metrics since it is a function of area and perimeter.
Figure 4. Quantitative comparison of automated and manual myocyte segmentation.
Automated and manual segmentation for 100 myocytes from a combination control, isoproterenol, and serum-treated conditions were compared. A) Absolute percent error in myocyte area and perimeter compared with manual segmentation 1 or 2. B) Percent error in myocyte area and perimeter compared with manual segmentation 1 or 2. Error bars are +/- SE.
Key advantages to automated myocyte segmentation are reproducibility and speed. As shown in Figure 3, there is some degree of inter-observer variability in manual myocyte segmentations. There is also potential for bias if the observer segmenting myocytes is not blinded to the condition. In contrast, the automated algorithm objectively performs segmentations and is completely reproducible. Another consideration is speed. Manual segmentation took on average 1.2 minutes/cell. Overall in this study 6895 cells were analyzed which would take ~138 hours to perform manually. In contrast the algorithm processes these cells in ~ 2 hours without user intervention.
Differential effects of hypertrophic pathways on myocyte size and shape
We next applied the automated segmentation algorithm to examine the hypertrophic response to receptor stimulation. β-adrenergic, α-adrenergic, tumor necrosis factor (TNFα), and insulin-like growth factor (IGF-1) signaling pathways have all been implicated in myocyte hypertrophy [2, 3]. These pathways were chosen as they represent distinct arms of the overall hypertrophic signaling network and are differentially associated with physiological and pathological hypertrophy. We hypothesized that while all of these pathways increase cell area, their diverse signaling mechanisms would result in divergent effects on other phenotypes relevant to cardiac hypertrophy. Myocytes were treated with a receptor agonist for one of these pathways for 48 h: 1 μM isoproterenol (ISO), 100 μM phenylephrine (PE), 10 ng/mL TNFα, 10 ng/mL IGF-1, or 15% FBS. Hypertrophic responses were quantified by myocyte area and perimeter (metrics of cell size) as well as elongation and circularity (metrics of cell shape) (see Figure 5).
Figure 5. Cell shape phenotypes quantified by automated segmentation.
Representative images are shown with myocytes that exhibit either small or large cell area, perimeter, elongation, or circularity. Elongation is the ratio of major-axis length to minor-axis length of a best-fit ellipse. Circularity is 4π*Area/Perimeter2.
As expected, each of the four hypertrophic agonists increased both cell area and cell perimeter (Figure 6). Note that the error bars in Figure 6 denote the 25-75th percentile range to illustrate the substantial heterogeneity at the single cell level. Excluding serum which contains a wide range of different growth factors, PE elicited the largest response in cell area and perimeter. But these agonists also affected cell shape, with decreasing circularity (or roundedness) in response to all five conditions. In contrast to the other morphological measures, cell elongation decreased significantly only in TNFα-treated myocytes. There was a trend towards increased elongation of PE-treated myocytes. A representative image for each condition is shown in Supplementary Figure 3.
Figure 6. Differential effects of diverse hypertrophic agonists on cell shape phenotypes.
Myocytes were serum-free (SF) or treated with isoproterenol (ISO), phenylephrine (PE), tumor necrosis factor-alpha (TNFα), insulin-like growth factor (IGF-1) or serum for 24 h. For each condition, cell area, perimeter, elongation and circularity were quantified. Error bars represent the 25-75th percentile range while the dot represents the median. *P<0.05 compared to control (SF). Number of myocytes analyzed for each condition was SF: 617, ISO, 639, PE: 534, TNFα: 883, IGF-1: 984, serum: 228.
Quantifying differential regulation of sarcomeric organization
In addition to myocyte size and shape, change to sarcomeric organization (increased or decreased regularity of sarcomere striations) is another phenotypic metric commonly associated with cardiac hypertrophy [17, 18]. However, sarcomeric organization is typically evaluated only qualitatively from representative images. We sought to develop a robust quantitative measure of sarcomeric organization using image texture analysis. Several measures of α-actinin image texture were tested, including Uniformity, Correlation, Contrast, and Fourier transforms [14]. When compared to manual scoring of 100 myocytes by two different researchers on a 1-4 scale, Uniformity had a Spearman rank correlation coefficient of -0.46, while Contrast and Correlation had coefficients of 0.41 and -0.26 respectively (see Supplementary Figure 4).
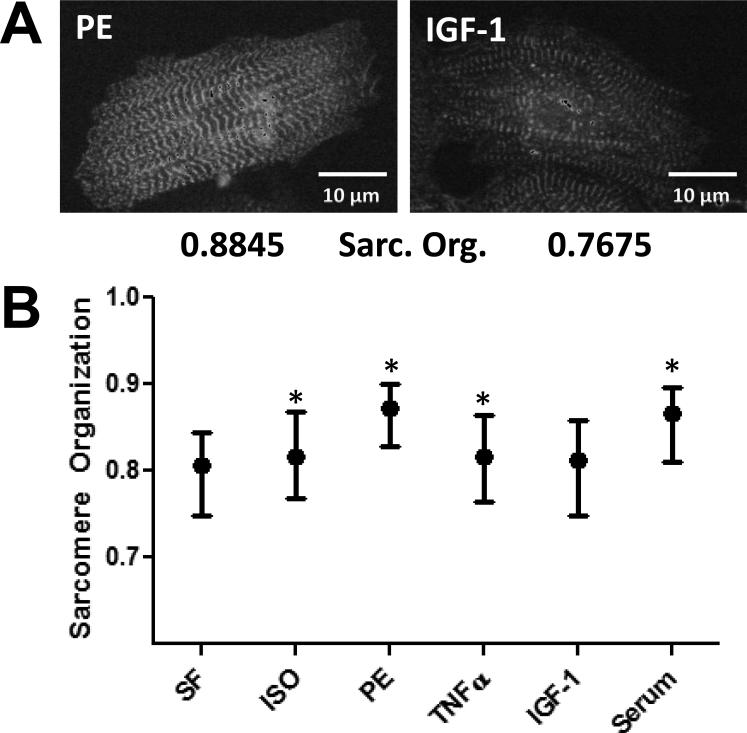
Uniformity is a measure of local homogeneity and has values ranging from 0 to 1, with a value of 1 representing completely uniform α-actinin labeling (see Supplementary Methods). Myocytes with highly organized sarcomeres exhibit sharp transitions in α-actinin intensity between z-lines, resulting in low Uniformity. Here, we calculated 1-Uniformity in order to transform this measure into a metric that positively correlates with sarcomeric organization (Figure 7A). Among the five hypertrophic stimuli, PE and serum stimulation resulted in a dramatic increase in sarcomeric organization, while IGF-1 did not affect sarcomeric organization (Figure 7B). Representative images are shown in Supplementary Figure 5. These results indicate that 1-Uniformity is a quantitative measure of sarcomeric organization and that hypertrophic pathways differentially regulate sarcomeric organization.
Figure 7. Automated measurement of sarcomeric organization.
A) Representative image of myocytes with low and high sarcomeric organization, along with corresponding quantification of intracellular contrast, a texture-based measure of sarcomeric organization. B) Effects of hypertrophic agonists ISO, PE, TNFα, IGF-1 and serum on intracellular contrast. Error bars represent the 25-75th percentile range while the dot represents the median. *P<0.05 compared to control (SF).
Differential effects of hypertrophic pathways on cell-cell contact
While analyzing the hypertrophic response, we noticed that some conditions appeared to cause clustering of myocytes, resulting in increased cell-cell contact. To quantify cell-cell contact, we measured the fraction of each myocyte's boundary that is shared with neighboring myocytes. For example a myocyte with no neighbors has 0% cell-cell contact, while a completely surrounded cell has 100% cell-cell contact (Figure 8A). The degree of cell-cell contact is expected to depend on average cell density and to correlate with increasing cell area. Indeed, serum, ISO, and PE significantly increased cell-cell contact compared to serum-free conditions. But changes to cell-cell contact were not explained by increased cell area alone. Inspection of ISO-treated cells in Figure 3 suggests nonrandom distribution of myocyte coverage. Further, TNFα and IGF-1 increased cell area but either did not change or trended towards decreased cell-cell contact. While myocytes do not regulate cell-cell contact in the normal adult heart, this appears to indicate either paracrine or contact-dependent cell-cell communication that is differentially regulated by hypertrophic pathways.
Figure 8. Automated measurement of cell-cell contact.
A) Representative image of myocytes along with the percent of each cell's perimeter that is shared with its neighbors. B) Differential effects of hypertrophic agonists ISO, PE, TNFα, IGF-1 and serum on cell-cell contact. Error bars represent the 25-75th percentile range while the dot represents the median. *P<0.05 compared to control (SF).
Hypertrophic pathways have distinct phenotypic signatures
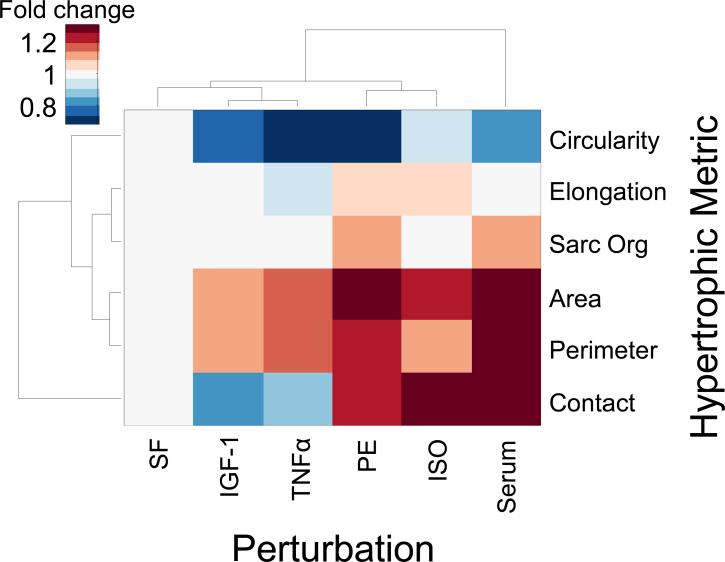
By integrating the above data into a single framework, we can begin to identify more complex relationships between hypertrophic pathways and phenotypic metrics. Highly correlated phenotypic metrics may be regulated by shared mechanisms in the relevant signaling pathways (“guilt by association”), whereas differentially regulated phenotypic metrics may indicate unique signaling mechanisms. Figure 9 illustrates the median effect of each hypertrophic agonist on the 6 phenotypic metrics. Cluster analysis was performed using a Euclidean distance metric to identify correlations in both phenotypic metrics and perturbations. The same qualitative dendrogram was obtained when clustering using a Manhattan distance metric (see Supplementary Methods). Strikingly, each particular phenotypic metric and each hypertrophic agonist had a qualitatively unique response pattern, or phenotypic signature. IGF-1 and TNFα were the most closely correlated hypertrophic agonists, but with qualitatively different effects on sarcomeric organization. The most closely correlated phenotypic metrics are cell area and cell perimeter, which is reasonable as they are both measures of cell size.
Figure 9. Hypertrophic agonists cause distinct forms of myocyte hypertrophy.
Effects of ISO, PE, TNFα, IGF-1 and serum are plotted against median changes in myocyte phenotype metrics quantified by automated segmentation. Phenotype metrics are cell area, perimeter, elongation, circularity, sarcomeric organization, and cell-cell contact. Hypertrophic agonists and phenotype metrics were grouped by hierarchical clustering. While all agonists increased cell area, they had qualitatively different effects on elongation, sarcomeric organization, and cell-cell contact.
While all hypertrophic agonists increased cell area, increased cell perimeter and decreased cell circularity, they did so do different degrees. For example ISO strongly regulated area but had a weaker effect on perimeter and circularity. Other hypertrophic metrics were regulated by agonists in opposite directions. Myocyte elongation was increased by PE yet decreased by TNFα. This indicates that unlike cell size, myocyte elongation is enhanced by mechanisms distinct to α-adrenergic signaling. Cell-cell contact was enhanced by ISO and PE yet decreased by IGF-1, indicating that cell-cell contact may be regulated by mechanisms that are shared by α- and β-adrenergic pathways but distinct from IGF and TNFα pathways. Sarcomeric organization (1-Uniformity) was strongly enhanced by PE yet unaffected by TNFα and IGF-1. This indicates that sarcomeric organization may be regulated by mechanisms specific to α-adrenergic signaling. Thus while cell size appears to be coordinately regulated by shared pathway mechanisms, cell shape and sarcomeric organization are more specifically regulated by distinct pathways. These complex relationships indicate that quantifying cell shape and texture metrics adds substantial new information above cell size alone.
Discussion
New unbiased phenotypic screening approaches are needed to more systematically characterize the hypertrophic signaling network and identify novel therapeutic targets [2]. Previous work has demonstrated high-content imaging of cancer cell lines and other cell types [8, 12], which enables systematic perturbation studies such as genome-wide RNA interference [19, 20]. Here, we developed an automated image analysis approach optimized for high-content imaging of cardiac myocyte hypertrophy. A related assay for screening miRNAs for effects on myocyte area was recently published [21]. Novel myocyte-specific features of our segmentation approach include smoothing and thresholds for segmenting despite irregular α-actinin patterns and merging of adjacent nuclei for binucleated myocytes. In addition to measuring cell area, this algorithm allowed measurement and quantitative validation of a number of new phenotypic metrics related to hypertrophy including perimeter, circularity, elongation, sarcomeric organization, and cell contact. As a proof-of-concept, the image analysis approach was tested by comparing the phenotypic response to five distinct classes of hypertrophic pathways: α-adrenergic, β-adrenergic, cytokine (TNFα), growth factor (IGF-1), and serum pathways. Despite a common qualitative increase in cell area under all conditions, these pathways exhibited divergent regulation of cell shape, sarcomeric organization, and cell-cell contact. Simultaneous quantification of multiple phenotypes in individual cardiac myocytes can aid identification of both common elements among hypertrophic pathways and phenotype-specific regulatory mechanisms.
Regulation of cardiac myocyte shape
Increases in myocyte area have been well documented for α-adrenergic [22], β-adrenergic [23], TNFα [24] and IGF-1 [25] receptor pathways. But while all five hypertrophic pathways stimulated increases in cell area and perimeter, we found that cell shape metrics of myocyte elongation and circularity were differentially regulated. Changes to elongation were modest, but TNFα decreased myocyte elongation while ISO and PE tended to cause myocyte elongation. Differential regulation of myocyte elongation has been of substantial interest due to its in vivo relevance to concentric and eccentric cardiac hypertrophy [1, 2].
Myocyte circularity decreased in response to all agonists in a manner that was inversely proportional to cell perimeter. ISO had a strong effect on area but a weaker effect on circularity, while TNFα had a larger effect on circularity than area. Circularity was previously used to quantify myocyte shape changes in response to substrate stiffness [26], and here we found that hypertrophic pathways can distinctly regulate elongation and circularity. Decreased circularity without a corresponding increase in elongation occurred when myocyte protrusions were not aligned in a particular direction, such as for TNFα, IGF-1 and serum.
Regulation of sarcomeric organization
Enhanced sarcomeric organization is often considered a hallmark of the hypertrophic response [17]. But some forms of hypertrophy cause sarcomeric disarray, decreasing contractile efficiency in hypertrophic cardiomyopathy [18, 27]. While sarcomeric organization in cultured myocytes has previously been measured only qualitatively [17, 28], we developed a novel quantitative measure of sarcomeric organization based on image texture of α-actinin immunofluorescence. Sarcomeric organization was strongly enhanced by α-adrenergic signaling or serum and unaffected by IGF-1 signaling. These results are consistent with data that Gq-coupled pathways enhance sarcomeric organization via Rho and myosin light chain phosphorylation, which are regulated independently from cell size [28].
Regulation of cell-cell contact
Previous studies have shown that myocyte plating density can modulate hypertrophic signaling pathways due to enhanced paracrine or contact-dependent cell-cell communication. In particular β-adrenergic stimulation of hypertrophy [23] and p38 regulation of ANF expression [29] have been reported to require higher cell densities. Serendipitously, we observed that some hypertrophic agonists appeared to induce myocyte clustering, resulting in enhanced cell-cell contact. Therefore we developed a quantitative measure of cell-cell contact, finding that serum, β-adrenergic, and α-adrenergic signaling strongly enhanced cell-cell contact. In contrast, IGF-1 decreased cell-cell contact despite increasing cell area. Increased cell-cell contact was not strongly correlated with increased cell area, indicating distinct regulation. These results suggest a potential positive feed-forward loop, where α- and β-adrenergic signaling enhance cell area directly by intracellular signaling and indirectly by enhancing cell-cell contact. While increased myocyte-myocyte contact is not clearly relevant to adult hypertrophy in vivo, cardiac myocyte migration and aggregation is an important aspect of developmental heart growth [30] and the spontaneous assembly of engineered heart tissues [31].
In summary, we developed an automated image analysis approach for simultaneously quantifying multiple hypertrophic metrics from immunofluorescence images of cardiac myocytes. Automated image analysis is a critical step for the development of phenotypic screens that allow systematic and quantitative analysis of crosstalk in the cardiac hypertrophy signaling network. These approaches may also be potentially extended to human stem-cell derived cardiac myocytes [32] for high-throughput drug screening applications [9].
Supplementary Material
Supplementary Figure Legends
Supplementary Figure 1. Example magnified views of automated and manual segmentations. This magnified view demonstrates that the manual segmentations (performed at lower magnification) oversimplify cell borders, which explains why the automated segmentations consistently have higher perimeters.
Supplementary Figure 2. Quantitative comparison of automated and manual myocyte segmentation for area, perimeter, elongation and circularity. Automated and manual segmentation for 100 myocytes from a combination control, isoproterenol, and serum-treated conditions were compared. A) Absolute percent error in each phenotypic metric compared with manual segmentation. B) Percent error in in each phenotypic metric compared with manual segmentation. Error bars are +/- SE.
Supplementary Figure 3. Representative images of myocytes treated with hypertrophic agonists. Below each cell are the values of the phenotypic metrics (area, perimeter, circularity and elongation) for that particular cell, which overall agree well with the median for that condition.
Supplementary Figure 4. Comparison of manual scoring of sarcomeric organization with automated measures of image texture. Two researchers manually scored for the overall level of sarcomeric organization in each cell (100 myocytes) on a 1-4 scale, and the average result was taken. Manual scores are compared with 3 automated measures of image texture within in each cell: Uniformity, Contrast, and Correlation.
Supplementary Figure 5. Representative images of sarcomeric organization from myocytes treated with hypertrophic agonists. Above each cell is listed the quantitative value of sarcomeric organization (1-Uniformity) for that cell. Representative cells were chosen by identifying cells close to the median of that population.
Highlights.
Automated image analysis method for cardiac myocyte hypertrophy
New quantitative measure of sarcomeric organization
TNFα and α-adrenergic signaling have opposite effects on myocyte elongation.
Sarcomeric organization is uniquely enhanced by α-adrenergic signaling.
α- and β-adrenergic pathways enhance myocyte-myocyte contact.
Acknowledgements
The authors thank Matthew Kraeutler, Renata Polanowska-Grabow, and Jason Yang for technical assistance and discussion. This work was supported by National Institutes of Health (grant HL094476), the American Heart Association (grant 0830470N), and the University of Virginia FEST program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: none declared
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 3.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–28. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 4.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–70. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JH, Saucerman JJ. Computational models reduce complexity and accelerate insight into cardiac signaling networks. Circ Res. 2011;108:85–97. doi: 10.1161/CIRCRESAHA.110.223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook SA, Clerk A, Sugden PH. Are transgenic mice the ‘alkahest’ to understanding myocardial hypertrophy and failure? J Mol Cell Cardiol. 2009;46:118–29. doi: 10.1016/j.yjmcc.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Molkentin JD, Robbins J. With great power comes great responsibility: using mouse genetics to study cardiac hypertrophy and failure. Journal of molecular and cellular cardiology. 2009;46:130–6. doi: 10.1016/j.yjmcc.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans JG, Matsudaira P. Linking microscopy and high content screening in large-scale biomedical research. Methods Mol Biol. 2007;356:33–8. doi: 10.1385/1-59745-217-3:33. [DOI] [PubMed] [Google Scholar]
- 9.Bickle M. The beautiful cell: high-content screening in drug discovery. Anal Bioanal Chem. 2010 doi: 10.1007/s00216-010-3788-3. [DOI] [PubMed] [Google Scholar]
- 10.Otsu N. Threshold Selection Method from Gray-Level Histograms. Ieee T Syst Man Cyb. 1979;9:62–6. [Google Scholar]
- 11.Jones TR, Carpenter A, Golland P. Voronoi-based segmentation of cells on image manifolds. Lect Notes Comput Sc. 2005;3765:535–43. [Google Scholar]
- 12.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du X, Dua S. Segmentation of fluorescence microscopy cell images using unsupervised mining. Open Med Inform J. 2010;4:41–9. doi: 10.2174/1874431101004020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haralick RM. Statistical and Structural Approaches to Texture. P Ieee. 1979;67:786–804. [Google Scholar]
- 15.Li B, Acton ST. Active contour external force using vector field convolution for image segmentation. Ieee T Image Process. 2007;16:2096–106. doi: 10.1109/tip.2007.899601. [DOI] [PubMed] [Google Scholar]
- 16.Ruusuvuori P, Aijo T, Chowdhury S, Garmendia-Torres C, Selinummi J, Birbaumer M, et al. Evaluation of methods for detection of fluorescence labeled subcellular objects in microscope images. BMC Bioinformatics. 2010;11:248. doi: 10.1186/1471-2105-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zechner D, Thuerauf DJ, Hanford DS, McDonough PM, Glembotski CC. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization, and cardiac-specific gene expression. J Cell Biol. 1997;139:115–27. doi: 10.1083/jcb.139.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, et al. A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:2800–11. doi: 10.1172/JCI2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods. 2004;1:127–32. doi: 10.1038/nmeth711. [DOI] [PubMed] [Google Scholar]
- 20.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 21.Jentzsch C, Leierseder S, Loyer X, Flohrschutz I, Sassi Y, Hartmann D, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Simpson P. Norepinephrine-stimulated hypertrophy of cultured rat myocardial cells is an alpha 1 adrenergic response. J Clin Invest. 1983;72:732–8. doi: 10.1172/JCI111023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishopric NH, Kedes L. Adrenergic regulation of the skeletal alpha-actin gene promoter during myocardial cell hypertrophy. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:2132–6. doi: 10.1073/pnas.88.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long CS, Henrich CJ, Simpson PC. A growth factor for cardiac myocytes is produced by cardiac nonmyocytes. Cell Regul. 1991;2:1081–95. doi: 10.1091/mbc.2.12.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito H, Hiroe M, Hirata Y, Tsujino M, Adachi S, Shichiri M, et al. Insulin-like growth factor-I induces hypertrophy with enhanced expression of muscle specific genes in cultured rat cardiomyocytes. Circulation. 1993;87:1715–21. doi: 10.1161/01.cir.87.5.1715. [DOI] [PubMed] [Google Scholar]
- 26.Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophysical journal. 2008;95:3479–87. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varnava AM, Elliott PM, Mahon N, Davies MJ, McKenna WJ. Relation between myocyte disarray and outcome in hypertrophic cardiomyopathy. Am J Cardiol. 2001;88:275–9. doi: 10.1016/s0002-9149(01)01640-x. [DOI] [PubMed] [Google Scholar]
- 28.Aoki H, Sadoshima J, Izumo S. Myosin light chain kinase mediates sarcomere organization during cardiac hypertrophy in vitro. Nat Med. 2000;6:183–8. doi: 10.1038/72287. [DOI] [PubMed] [Google Scholar]
- 29.Hines WA, Thorburn J, Thorburn A. Cell density and contraction regulate p38 MAP kinase-dependent responses in neonatal rat cardiac myocytes. Am J Physiol. 1999;277:H331–41. doi: 10.1152/ajpheart.1999.277.1.H331. [DOI] [PubMed] [Google Scholar]
- 30.Hakim ZS, DiMichele LA, Doherty JT, Homeister JW, Beggs HE, Reichardt LF, et al. Conditional deletion of focal adhesion kinase leads to defects in ventricular septation and outflow tract alignment. Mol Cell Biol. 2007;27:5352–64. doi: 10.1128/MCB.00068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eschenhagen T, Zimmermann WH. Engineering myocardial tissue. Circ Res. 2005;97:1220–31. doi: 10.1161/01.RES.0000196562.73231.7d. [DOI] [PubMed] [Google Scholar]
- 32.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure Legends
Supplementary Figure 1. Example magnified views of automated and manual segmentations. This magnified view demonstrates that the manual segmentations (performed at lower magnification) oversimplify cell borders, which explains why the automated segmentations consistently have higher perimeters.
Supplementary Figure 2. Quantitative comparison of automated and manual myocyte segmentation for area, perimeter, elongation and circularity. Automated and manual segmentation for 100 myocytes from a combination control, isoproterenol, and serum-treated conditions were compared. A) Absolute percent error in each phenotypic metric compared with manual segmentation. B) Percent error in in each phenotypic metric compared with manual segmentation. Error bars are +/- SE.
Supplementary Figure 3. Representative images of myocytes treated with hypertrophic agonists. Below each cell are the values of the phenotypic metrics (area, perimeter, circularity and elongation) for that particular cell, which overall agree well with the median for that condition.
Supplementary Figure 4. Comparison of manual scoring of sarcomeric organization with automated measures of image texture. Two researchers manually scored for the overall level of sarcomeric organization in each cell (100 myocytes) on a 1-4 scale, and the average result was taken. Manual scores are compared with 3 automated measures of image texture within in each cell: Uniformity, Contrast, and Correlation.
Supplementary Figure 5. Representative images of sarcomeric organization from myocytes treated with hypertrophic agonists. Above each cell is listed the quantitative value of sarcomeric organization (1-Uniformity) for that cell. Representative cells were chosen by identifying cells close to the median of that population.