Abstract
Abasic (AP) sites are formed spontaneously and are inevitably intermediates during base excision repair of DNA base damages. AP sites are both mutagenic and cytotoxic and key enzymes for their removal are AP endonucleases. However, AP endonuclease independent repair initiated by DNA glycosylases performing β,δ-elimination cleavage of the AP sites has been described in mammalian cells. Here, we describe another AP endonuclease independent repair pathway for removal of AP sites in Schizosaccharomyces pombe that is initiated by a bifunctional DNA glycosylase, Nth1 and followed by cleavage of the baseless sugar residue by tyrosyl phosphodiesterase Tdp1. We propose that repair is completed by the action of a polynucleotide kinase, a DNA polymerase and finally a DNA ligase to seal the gap. A fission yeast double mutant of the major AP endonuclease Apn2 and Tdp1 shows synergistic increase in MMS sensitivity, substantiating that Apn2 and Tdp1 process the same substrate. These results add new knowledge to the complex cellular response to AP sites, which could be exploited in chemotherapy where synthetic lethality is a key strategy of treatment.
INTRODUCTION
One of the most frequent lesions in DNA is an abasic (AP) site, generated spontaneously by hydrolysis of the N-glycosylic bond between the base and the deoxyribose or as an intermediate in base excision repair (BER) by monofunctional DNA glycosylases removing damaged bases (1,2). More than 10 000 AP sites are estimated to arise spontaneously per mammalian cell per day (3). AP sites are cytotoxic to the cell by blocking DNA replication and transcription (4), and also mutagenic as bypass of AP sites by translesion polymerases can result in base substitutions and frameshift mutations (5). To maintain genomic integrity, the AP sites need to be repaired, and the main system for their removal is the BER pathway.
The major enzymes for incision of AP sites are AP endonucleases initiating BER by hydrolyzing the DNA at the 5′ side of the AP site leaving a single-stranded (ss) nick with 3′-hydroxyl (3′-OH) and 5′-deoxyribose phosphate (5′-dRP) ends (6). Further repair may follow by two different pathways. In short patch BER, the 5′-dRP terminus is processed to a 5′-phosphate (5′-P) by the 5′ AP lyase activity of DNA polymerase β(POLβ) (7). In long patch BER, flap endonuclease (FEN1) removes 2–8 nt from the 5′-dRP terminus generated by AP endonuclease incision, leaving a gap in the DNA (8,9). Bifunctional AP lyases/DNA glycosylases may also initiate short patch BER of AP sites. AP lyases cleave the DNA strand at the 3′ side of the AP site by β-elimination generating a 5′-P and a 3′-α,β-unsaturated aldehyde termini (3′-deoxyribose phosphate; 3′-dRP) (10). The 3′-dRP residue is further processed by an AP endonuclease leaving a 3′-OH terminus. Some AP lyases like Escherichia coli formamidopyrimidine DNA glycosylase (Fpg) and mammalian endonuclease VIII like DNA glycosylases 1 and 2 (NEIL1/2) also possess a δ-elimination reaction resulting in 5′-P and 3′-phosphate (3′-P) termini (11–13). The blocking 3′-P is removed by the phosphatase activity of polynucleotide kinase (PNKP) (14,15). In this situation, the repair is independent of AP endonucleases (16). Repair is completed by extension of the 3′-OH terminus by a DNA polymerase and gap sealing by a DNA ligase (17,18).
The same BER enzymes are also involved in the repair of DNA single-strand breaks. When resulting from free radical reaction of deoxyribose residues, such breaks invariably possess blocked termini that must be restored to the conventional 3′-OH and 5′-P moieties in order for gap filling and subsequent ligation to occur. For 5′ blocked termini POLβ (AP lyase) and FEN1, in addition to aprataxin, are the major cleansing enzymes. For ‘dirty’ 3′-ends, the key enzymes are AP endonuclease 1 (APE1) and PNKP together with tyrosyl phosphodiesterase 1, TDP1. The primary substrate for TDP1 is the product of abortive topoisomerase 1 (TOP1) reaction, namely, TOP1 covalently linked to the 3′-terminus of DNA (19–21). Tdp1 was first discovered in Saccharomyces cerevisiae (22) and was shown to hydrolyze the 3′-phosphotyrosyl bond between Top1 and DNA (23–25). The phosphodiesterase activity of TDP1 has also been implicated in the repair of other 3′-end alterations (26), including 3′-phosphoglycolates (27). The TDP1 reaction on these substrates leaves a 3′-P similar to the δ-elimination reaction product generated by Fpg/Nei like DNA glycosylases.
In Schizosaccharomyces pombe, Endonuclease III (Nth1) is known to be the major AP site incision enzyme, initiating the cleavage of AP sites by β-elimination (28,29). The 3′-dRP generated is further processed by the phosphodiesterase activity of the AP endonuclease Apn2 (28–31). In contrast to AP endonucleases in other eukaryotes and bacteria, the predominant function of the major AP endonuclease in S. pombe, Apn2 (30,31), is believed to be in the removal of 3′ blocks induced by Nth1 rather than incision of AP sites. Further, Nth1 is the only identified DNA glycosylase with AP lyase activity in S. pombe. No enzyme possessing β,δ-elimination activity like E. coli Fpg and Nei and human NEIL enzymes has been identified by sequence homology searches of the S. pombe genome (32). However, observation of a faint band migrating like a δ-elimination cleavage product (3′-P) has been reported in activity assays with an AP substrate and S. pombe wild-type extract (28,33), indicating the presence of an enzyme with δ-lyase activity in S. pombe.
In this study, we describe an AP endonuclease independent pathway for repair of AP sites in S. pombe. Nth1 initiates the repair by β-elimination of the AP site, whereas Tdp1 cleaves the 3′-dRP to generate a 3′-P that can be further processed by phosphatases. This newly identified branch of the BER pathway with Tdp1 working downstream of Nth1, seems to function as an important backup repair pathway in the absence of Apn2 in the repair of AP sites in S. pombe.
MATERIALS AND METHODS
Strains and media
The S. pombe strains used in this study are derivatives of the wild-type strains FY526 h+ and FY527 h− and are outlined in Table 1. The strains were grown in complete yeast extract medium supplemented with 200 mg/l adenine, uracil, leucine, histidine, arginine and lysine (YES). When selection was required, pombe minimal glutamate (PMG) medium containing appropriate supplements, was used. To generate a mutant containing the kanMX cassette, the transformed cells were incubated in minimal sporulation liquid (MSL) medium (34) to give the marker time to be expressed, before the cells were challenged with geneticin. Escherichia coli BL21 Codon Plus (DE3) RIL cells (Stratagene) were used for expression and purification of recombinant Tdp1. The E. coli cells were grown in Luria–Bertani (LB) medium with antibiotic selection.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain name | Genotype | References |
|---|---|---|
| FY526 wt | h+ade6-M216 ura4-D18 leu1-32 his3-D1 | S. Forsburg (55) |
| FY527 wt | h−ade6-M216 ura4-D18 leu1-32 his3-D1 | S. Forsburg (55) |
| RHP302 | h−apn2::kanMX ade6-M216 ura4-D18 leu1-32 his3-D1 | FY527 |
| RHP357 | h+nth1::ura4+ade6-M216 ura4-D18 leu1-32 his3-D1 | FY526 |
| RHP378 | h+tdp1::kanMX nth1::ura4+ade6-M216 ura4-D18 leu1-32 his3-D1 | RHP357 |
| RHP379 | h?tdp1::kanMX ade6-M216 ura4-D18 leu1-32 his3-D1 | RHP378 × FY527 |
| RHP381 | h?tdp1::kanMX apn2::kanMX nth1::ura4+ade6-M216 ura4-D18 leu1-32 his3-D1 | RHP378 × RHP302 |
| RHP382 | h?tdp1::kanMX apn2::kanMX ade6-M216 ura4-D18 leu1-32 his3-D1 | RHP378 × RHP302 |
| RHP383 | h?nth1::ura4+apn2::kanMX ade6-M216 ura4-D18 leu1-32 his3-D1 | RHP357 × RHP302 |
h?, mating type not determined; wt, wild-type.
Schizosaccharomyces pombe cell-free extracts
Schizosaccharomyces pombe cells were grown in YES at 30°C to an OD600 of 1.5–2.0. The cells were harvested by centrifugation, washed with water, resuspended in extract buffer (25 mM HEPES pH 7.4, 0.1 mM EDTA, 100 mM KCl, 2 mM DTT and protease inhibitor cocktail) (Merck, Calbiochem) and frozen at −80°C. The cell-extract buffer suspension was thawed, glass beads added and the cells disrupted by the use of a Beadbeater for 1 min× 7, with 1 min incubation on ice in between. Cell debris was removed from the lysate by centrifugation at 12 000 rpm for 30 min at 4°C. The protein concentration was determined by the Bradford method using bovine serum albumin as reference.
Purification and identification of endogenous Tdp1
Cell-free extract from 60 l of S. pombe RHP357 (nth1−) cells was run through seven different columns in the following order; SP Sepharose Fast Flow 20 ml, Resource Q 6 ml, Heparin 2 ml, Resource S 1 ml, Hydroxyapatite 20 ml, MonoS SMART and Superdex75 SMART. From all of the columns except hydroxyapatite, proteins were eluted with a linear gradient of 0.05–1 M NaCl in HEPES buffer [25 mM HEPES pH 7.4, 50 mM NaCl, 10 mM β-mercaptoethanol (ME)]. For the hydroxyapatite column, proteins were eluted with a linear gradient of 0–400 mM KHPO4 pH 7.4 in HEPES buffer. After each column, the fractions were analyzed for 3′-deoxyribose (3′-dR) removing activity. Active fractions were pooled, dialyzed with HEPES buffer and loaded on the next column. After the final column, the proteins in the fractions flanking the activity peak were separated by 10% SDS–PAGE. The gel was stained with Imperial Protein Stain (Thermo Fischer), and visible bands in the most active fraction were cut from the gel, digested and analyzed by mass spectrometry (MS).
Disruption of nth1 and apn2
An nth1::ura4+ mutant (RHP357) and an apn2::kanMX mutant (RHP302) were generated by gene disruption of the nth1 or apn2 wild-type allele in FY526 (wt) or FY527 (wt) respectively as described (28,35). An nth1−apn2− double mutant (RHP383) was generated by random spore analysis by crossing RHP357 and RHP302. Genotypes of the mutants were confirmed by colony PCR. Four to six different isolates were screened for methyl methanesulfonate (MMS) sensitivity and for some mutants also camptothecin (CPT) sensitivity. A representative isolate was used in further crosses or survival assays.
Cloning and disruption of tdp1
The tdp1 gene (without start codon) was amplified by PCR of S. pombe genomic DNA using the primers 5′-cgatggatccgtctactcttgagcccgaaa and 5′-cgtactcgagttaccaattgggaggccaaac and cloned into the BamHI and XhoI sites of pET28b (Novagen). The resulting expression construct (pET28b-Tdp1) has a 6 × His tag in frame with the N terminus of Tdp1. Correct sequence of pET28b-Tdp1 was verified by DNA sequencing.
To generate a tdp1 disruption construct, 930 bases of the tdp1 gene were removed by cleaving the pET28b-Tdp1 construct with BsrG1 and SpeI, and replacing it with the kanMX cassette PCR amplified from the pCore vector using primers 5′-cgtatgtacaccagtcgggaaacctgtcgt and 5′-cgcaactagtcatcgatgaattcgagctcgtt. The resulting construct contained 330 and 350 bp (up and downstream, respectively) of the tdp1 gene that flanks the kanMX cassette. For targeted gene disruption, the tdp1::kanMX fragment was PCR amplified by using the primers 5′-cgatggatccgtctactcttgagcccgaaa and 5′-cgtactcgagttaccaattgggaggccaaac. The ends were partly degraded with Bal31 nuclease and this tdp1::kanMX fragment was used for transformation of S. pombe RHP357 (nth1−) cells using the S.c. Easy Comp Transformation kit (Invitrogen). The transformed cells were incubated in MSL medium at 30°C for 16 h and plated on YES with 100 mg/l G418. The resulting tdp1−nth1− double mutant strain was designated RHP378. A tdp1− single mutant (RHP379), tdp1−apn2− double mutant (RHP382) and tdp1−apn2−nth1− triple mutant (RHP381) were constructed by random spore analysis after mating of RHP378 and FY527 to yield a tdp1− single mutant (RHP379) and mating of RHP378 and RHP302 to yield tdp1−apn2− (RHP382) and tdp1−apn2−nth1− (RHP381). Genotypes of the mutants were verified and a representative isolate selected as described earlier.
Tdp1 expression and purification
Escherichia coli BL21 Codon Plus (DE3) RIL cells transformed with pET28b-Tdp1 were grown in LB medium with 50 µg/ml kanamycin at 37°C to an OD600 of ~0.8. Protein expression was induced with 1 mM IPTG and cells grown for 2 h. The cells were harvested by centrifugation, washed with water and frozen at −80°C. The cell pellet was thawed and resuspended in sonication buffer (50 mM Na2HPO4/NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM ME), and the cells were broken by sonication. Cell debris was removed from the lysate by centrifugation at 13 000 rpm for 20 min at 4°C. The protein extract was loaded on to a Ni-NTA Agarose column (Qiagen) equilibrated with sonication buffer. The column was washed with sonication buffer with 50 mM imidazole, and the Tdp1 protein was eluted by increasing the imidazole concentration to 300 mM. Partially purified protein was visualized by separation on a 10% SDS–PAGE and Coomasie blue staining. Fractions containing Tdp1 were pooled, dialyzed with HEPES buffer and further purified on a Resource S 1 ml and a Superdex75 SMART column eluting the Tdp1 protein by a linear increase in the NaCl concentration from 0.05 to 1 M in the HEPES buffer. Fraction 14 from the Superdex75 column was used in the activity assays.
DNA cleavage activity assays
The sequence of the phosphotyrosine substrate was 5′-ctacgtcagatctgaggatg-pTyr, while for the uracil, 8oxoG and 5ohC substrates the sequence was 5′-gcatgcctgcacggXcatggccagatccccgggtaccgag, where X is the damaged base. The oligonucleotide substrates were end-labeled using T4 polynucleotide kinase (New England Biolabs) and [γ32P] ATP (3000 Ci/mmol, Amersham). Double-stranded substrates (ds; U:C, 8oxoG:C and 5ohC:G) were generated by annealing the labeled oligonucleotide to a complementary strand. To generate intact AP substrate or a nicked AP substrate with a 3′-dRP terminus, the uracil substrate was pretreated with uracil DNA glycosylase (Udg, NEB) or Udg and Nth (NEB), respectively, for 15 min at 37°C. Ten microliters of reaction mixtures contained reaction buffer (70 mM MOPS pH 7.5, 1 mM EDTA, 1 mM DTT and 5% glycerol), 10 fmol DNA substrate and protein concentrations as indicated. The reactions were incubated at 37°C for 30 min and stopped by addition of 10 µl formamide loading dye (80% formamide, 10 mM EDTA, 0.1% xylene cyanol and bromphenol blue). To hydrolyse uncleaved AP sites (in Udg glycosylase assay), the samples were incubated with 100 mM NaOH for 10 min at 70°C and subsequently neutralized with 100 mM HCl. The reactions were stopped by adding formamide loading dye. The oligonucleotides were denaturated at 90°C for 3 min, and the reaction products were separated on a 20% sequencing gel at 40 W for 1.5 h in 1× taurine buffer. The radiolabeled fragments were visualized by using a PhosphorImager (Typhoon 9410 Variable Mode Imager), and ImageQuant TL was used for quantification.
Schizosaccharomyces pombe survival assay
Schizosaccharomyces pombe cells were grown in YES medium to OD600 ~ 0.8, harvested by centrifugation and resuspended in water. The cells were 10-fold serial diluted in water and spotted on YES plates containing concentrations of MMS or CPT as indicated. The cells were incubated at 30°C for 3 days.
RESULTS
Identification of the δ-elimination enzyme in S. pombe
Certain bifunctional DNA glycosylases/AP lyases such as Fpg and Nei in E. coli and NEIL1 and 2 in mammalian cells, perform δ-elimination cleavage of AP sites. Despite neither encoding Fpg nor Nei homologs, δ-elimination cleavage (3′-P) is evident in protein extracts from S. pombe wild-type cells (Figure 1A, left panel) (28,33). In an attempt to identify the responsible enzyme, cell-free extract from S. pombe RHP357 (nth1−) cells was isolated. The nth1− strain was used to avoid interference from Nth1 which also acts on AP sites. When compared to total protein extract made from wild-type cells, the 3′-P cleavage product of a ds AP substrate was not observed in nth1− extracts (Figure 1A, left panel), suggesting that the enzyme responsible is either dependent on Nth1 to be active or that the enzyme is only processing the 3′-dRP terminus generated by Nth1 cleavage. When the AP substrate was pretreated with Nth, the 3′-P product was recovered (Figure 1A, right panel). This Nth-nicked substrate was used in the subsequent purification process.
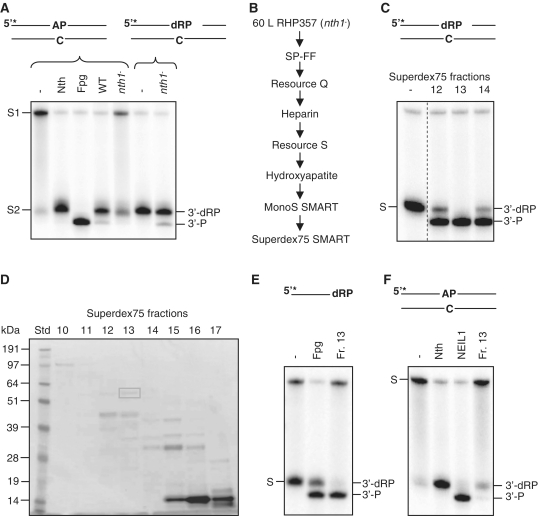
Figure 1.
Identification and characterization of endogenous Tdp1. (A) Cleavage of intact and nicked AP substrates in S. pombe whole cell extracts. Two micrograms of total protein extracts from FY526 (wt) and RHP357 (nth1−) cells were incubated with 10 fmol 5′-[32P]-labeled duplex DNA containing an AP site (opposite C) in reaction buffer for 30 min at 37°C (left panel). Two micrograms of the RHP357 (nth1−) extract was used in an equivalent reaction using the same substrate pretreated with Nth (right panel). The cleavage products were separated by 20% denaturing PAGE and visualized by phosphorimaging. The DNA substrates with an intact (S1) or Nth-nicked (S2) AP site and the cleavage products (3′-dRP and 3′-P) are indicated. Escherichia coli Nth and Fpg were used as positive controls for migration of the 3′-dRP and 3′-P products, respectively. (B) Schematic illustration of the columns used in the purification/identification process. (C) δ-elimination activity in Superdex75 SMART fractions. One microliter of fraction 12–14 from Superdex75 was incubated with Nth-nicked ds AP substrate and analyzed for cleavage activity as described in Figure 1A. The substrate (S; 3′-dRP) and the cleavage product (3′-P) are indicated. All samples were run on the same gel, but the control lane was cut from another part of the gel as indicated by the dotted line. (D) SDS–PAGE of Superdex75 SMART fractions. Proteins in fractions 10–17 (22 µl) from Superdex75 were separated by 10% SDS–PAGE, and bands in fraction 13 were excised from the gel and analyzed by MS. (E) δ-cleavage activity of Nth-nicked ss AP substrate. One microliter of fraction 13 from Superdex75 was incubated with an Nth-nicked ss AP substrate and analyzed for cleavage as described in Figure 1A. The substrate (S; 3′-dRP) and the cleavage product (3′-P) are indicated. Escherichia coli Fpg was used as a positive control for 3′-P terminus. (F) Assay for AP lyase activity. One microliter of fraction 13 from Superdex75 was incubated with an intact ds AP substrate and analyzed for cleavage as described in Figure 1A. The substrate (S) and the cleavage products (3′-dRP and 3′-P) are indicated. Escherichia coli Nth and purified human NEIL1 were used as positive controls for 3′-dRP and 3′-P, respectively.
To identify the enzyme responsible for the δ-elimination, the protein was purified by following the 3′-P cleavage product through a combination of seven different column matrices that separate proteins according to their size, charge and specific chemical affinities (Figure 1B). Proteins in the peak fraction (fraction 13, Figure 1C) from the last column (Superdex75 SMART) were separated by SDS–PAGE (Figure 1D) and visible bands excised from the gel. Analysis by mass spectroscopy of the marked band (~60 kDa) revealed the identity of only two proteins; tyrosyl DNA phosphodiesterase 1 (Tdp1) and ATP-dependent RNA helicase (Ded1). Of these, Tdp1 caught our interest. Schizosaccharomyces pombe Tdp1 has a molecular mass of 61.5 kDa and is known to process ‘dirty’ 3′-ends leaving a terminal phosphate like δ-lyases, making it a likely candidate. Activity assays with the partly purified Tdp1 (fraction 13) showed δ-cleavage activity on 3′-dRP termini in a ds context (Figure 1C) and also on a ss 3′-dRP substrate (Figure 1E). A very weak AP lyase activity toward an intact ds AP substrate is present in this partly purified fraction (Figure 1F).
No δ-cleavage of AP sites in protein extracts from tdp1− cells
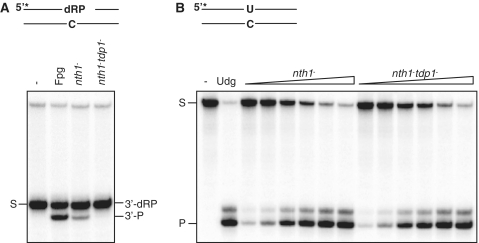
To verify that Tdp1 was responsible for the observed 3′-P cleavage product of the nicked AP substrate, a tdp1−nth1− double mutant (RHP378) was generated. Total protein extract from RHP378 (tdp1−nth1−) and also RHP357 (nth1−) cells were prepared and used in activity assay with an Nth-nicked ds AP substrate. The result showed that the 3′-P cleavage product was absent in the double mutant (Figure 2A), confirming that Tdp1 possesses the major δ-elimination activity in S. pombe. To exclude that the missing 3′-P cleavage product was due to an inactive protein extract, the extracts from both nth1− and tdp1−nth1− cells were analyzed for an independent DNA repair activity and a uracil DNA glycosylase (Udg) assay was performed. Udg is a monofunctional DNA glycosylase removing uracils from DNA. The two extracts were near to identical for Udg activity, confirming that both extracts were functional (Figure 2B).
Figure 2.
Tdp1 possesses 3′-α,β-unsaturated aldehyde activity leaving a 3′-P terminus. (A) Assay for processing 3′-dRP termini. Ten micrograms total protein extracts from nth1− (RHP357) and tdp1−nth1− (RHP378) cells were analyzed for cleavage of an Nth-nicked ds AP substrate as described in Figure 1A. The substrate (S; 3′-dRP) and the cleavage product (3′-P) are indicated. Escherichia coli Fpg was used as a positive control for the 3′-P cleavage product. (B) Udg activity in the nth1− and tdp1−nth1− extracts. The nth1− and tdp1−nth1− extracts (0.03, 0.06, 0.12, 0.25, 0.5 and 1.0 µg; as in A) were incubated with 10 fmol duplex DNA containing an uracil (opposite C) in reaction buffer for 30 min at 37°C, following incubation with 100 mM NaOH for 10 min at 70°C. The cleavage products were separated on a sequencing gel and visualized by phosphorimaging. The substrate (S) and the cleavage product (P) are indicated. Escherichia coli Udg was used as a positive control.
Recombinant Tdp1 processes 3′-α,β-unsaturated aldehydes
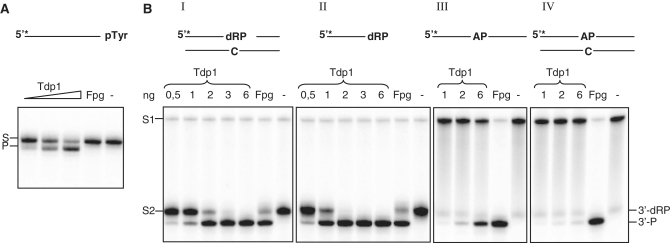
To further investigate the role of Tdp1 in processing 3′-dRP termini, recombinant S. pombe Tdp1 was purified from E. coli. The purified Tdp1 showed a single band of ~60 kDa when analyzed by SDS–PAGE (Supplementary Figure S1). To exclude interference from copurifying E. coli enzymes, cleavage of 8oxoG (Fpg) and 5ohC (Nei) substrates were examined. No activity toward these substrates was observed (Supplementary Figure S2A and B) with 50-fold more Tdp1 than used to obtain full cleavage with the nicked AP DNA. The main substrate for Tdp1 is a phosphotyrosyl moiety at the 3′-end of DNA, and recombinant Tdp1 cleaved such a substrate as expected (Figure 3A). In addition, Tdp1 showed δ-elimination activity toward both ds and ss AP substrate pre-treated with Nth (Figure 3B, panels I and II). It has been reported that human TDP1 has AP lyase activity toward both ds and ss AP substrates with the ss AP substrate as the preferred one (36). Activity assay with an intact ss or ds AP substrate revealed that S. pombe Tdp1 did not efficiently incise these substrates (Figure 3B, panels III and IV). Tdp1 cleaved only ~3 and 7% of the intact ds and ss AP substrates, respectively, when using the same amount of Tdp1 (2 ng) that gave full cleavage of Nth-nicked AP substrates.
Figure 3.
Recombinant Tdp1 possesses cleavage activity toward different substrates. (A) Cleavage assay of DNA containing a 3′-phosphotyrosine. 1, 2 and 6 ng recombinant Tdp1 was analyzed for cleavage of an ss oligonucleotide containing a phosphotyrosine at the 3′-end as described in Figure 1A. The substrate (S) and the cleavage product (P) are indicated. (B) Cleavage of ds AP and ss AP substrates with an intact or Nth-nicked AP site using increasing amounts of recombinant Tdp1 as indicated. Reaction products were analyzed as described in Figure 1A. The substrates with an intact (S1) or Nth1-nicked (S2) AP site and the cleavage products (3′-dRP and 3′-P) are indicated. Escherichia coli Fpg was used as a positive control for the 3′-P cleavage product. All panels are from the same gel but have been rearranged for clarity of presentation.
During the β-elimination reaction, a covalent imino enzyme–DNA complex is formed which can be trapped with sodium borohydride. Trapping assay with ss AP substrate did not reveal any trapped product (data not shown), confirming that Tdp1 does not possess robust AP lyase activity.
Tdp1 works in an alternative BER branch in S. pombe
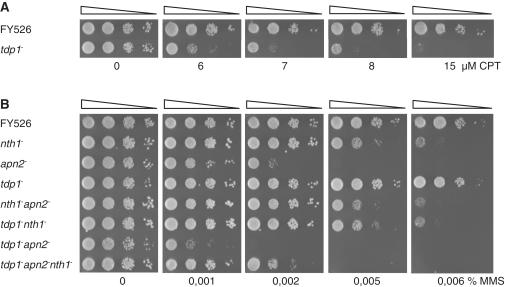
Processing of the 3′-dRP terminus generated by Nth1 cleavage of an AP site, suggests a role of Tdp1 in the BER pathway acting downstream of Nth1. To investigate this, a tdp1− single mutant (RHP379) was constructed. In addition, double and triple mutants of tdp1− and BER mutant cells were generated; RHP378 (tdp1−nth1−), RHP381 (tdp1−apn2−nth1−) and RHP382 (tdp1−apn2−). The anticancer drug camptothecin (CPT) acts by stabilizing Top1–DNA adducts, resulting in strand breaks in the DNA (37). Tdp1 is shown to be the key enzyme for removal of such adducts (24,38). When challenged with CPT, fission yeast cells lacking Tdp1 became sensitive as previously reported (Figure 4A) (39). Next, the tdp1− mutant was exposed to the alkylating agent MMS that via the Mag1 DNA glycosylase generates a heavy burden of AP sites. The tdp1− single mutant displayed no sensitivity to MMS and the tdp1−nth1− double mutant was as sensitive as the nth1− single mutant (Figure 4B). This is in accordance with Nth1 and Tdp1 acting in the same pathway. Further, lack of sensitization of the nth1− mutant by deletion of tdp1, confirmed our biochemical data that showed lack of a robust AP lyase activity of S. pombe Tdp1. In contrast, the tdp1−apn2− double mutant showed a synergistic increase in MMS sensitivity compared to the respective single mutants (Figure 4B), suggesting that Tdp1 and Apn2 act on the same substrate. In absence of both Tdp1 and Apn2, the 3′-blocking lesion (3′-dRP) produced by Nth1 is hardly processed and contributes to the cytotoxicity seen in the tdp1−apn2−mutant after MMS exposure. The importance of these two genes was also evident under normal conditions as the tdp1−apn2− double mutant was more slowly growing (Figure 4B, left panel). Deletion of nth1 in a tdp1−apn2− background partially relieved the MMS sensitivity of the tdp1−apn2− double mutant (Figure 4B). This is similar to what happens to the apn2− single mutant when nth1 is deleted [Figure 4B, and (28)]. This is probably due to accumulation of 3′-blocking lesions generated by Nth1 that cannot be further processed in the absence of Apn2 and Tdp1. 3′-dRP termini are more toxic to the cell than intact AP sites, which can be bypassed by translesions polymerases. The observed slow growing phenotype of the tdp1−apn2− double mutant in unstressed cells appears to be relieved in the tdp1−apn2−nth1− triple mutant (Figure 4B, left panel), demonstrating that Nth1 also acts on endogenously generated DNA damage.
Figure 4.
Genetic interactions between tdp1− and BER mutants. (A) CPT sensitivity in the fission yeast tdp1− strain. Exponentially growing wild-type (FY526) and tdp1− (RHP379) cells were serial diluted in water and spotted onto YES plates containing CPT as indicated. The plates were incubated for 3 days at 30°C and the survival assessed. (B) MMS survival analysis of the tdp1− and BER single and double mutants. Exponentially growing wild-type (FY526), nth1− (RHP357), apn2− (RHP302), tdp1− (RHP379), nth1−apn2− (RHP383), tdp1−nth1− (RHP378), tdp1−apn2− (RHP382) and tdp1−apn2−nth1− (RHP381) cells were serial diluted in water and spotted onto YES plates containing MMS as indicated. The plates were incubated for 3 days at 30°C and the survival assessed.
In summary, our results show that Tdp1 plays a more general role in DNA repair than only removal of Top1-mediated DNA damage. Tdp1 can be placed in a new branch of the BER pathway (Figure 5) working downstream of Nth1 by processing the 3′-α,β-unsaturated aldehyde left after Nth1 cleavage. A 3′-P terminus is generated by the Tdp1 reaction and S. pombe Pnk1 has been reported to have both 5′-kinase and 3′-phosphatase activity (40). Pnk1 is thus a likely candidate for processing of the 3′-P terminus to generate a 3′-OH. Repair can now be completed by a DNA polymerase filling in a new base and a DNA ligase sealing the nick. The tdp1− mutant was not hypersensitive to MMS as observed for the apn2− mutant, indicating that the Apn2-dependent pathway is the preferred one, with Tdp1 working as an important backup of Apn2 in the repair of AP sites.
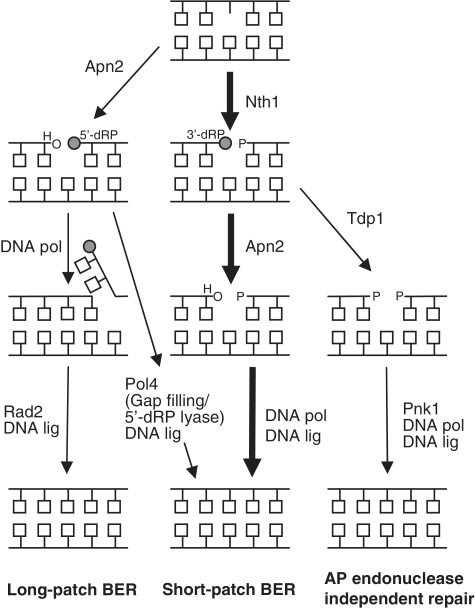
Figure 5.
BER of AP sites in S. pombe. AP sites in S. pombe are mainly repaired by the short-patch BER pathway (thick arrows), initiated by incision of the AP site by Nth1, leaving 5′-P and 3′-dRP ends. The lethal 3′-block is further processed to a 3′-OH by the 3′-phosphodiester activity of Apn2. A DNA polymerase and a DNA ligase finally complete the repair by filling the gap and sealing the nick. Alternatively, Apn2 initiates repair by incision of the AP site leaving a 5′-dRP end, which can be further processed by long-patch (strand displacement synthesis and subsequent cleavage of the 5′-flap by Rad2) or short-patch BER (gap insertion and 5′-dRP lyase by SpPol4). Apn2 participates in both long-patch and short-patch BER. In this study, we propose an AP endonuclease independent branch of BER in S. pombe with Tdp1 working as a backup of Apn2 in the repair of 3′-dRP termini left after Nth1 cleavage. Pnk1 is suggested to work downstream of Tdp1 by processing the 3′-P generated by Tdp1, leaving a 3′-OH. Finally, repair is completed by a DNA polymerase and a DNA ligase.
DISCUSSION
Abasic sites are one of the most frequent endogenous lesions in cellular DNA and their repair is critical for genomic stability and cellular survival. In S. pombe, Nth1 is the major AP site incision enzyme cleaving the AP sites by β-elimination. The S. pombe genome does not encode any bifunctional DNA glycosylases possessing both β- and δ-elimination similarly as E. coli Fpg/Nei and human NEIL enzymes. However, a cleavage product of an AP DNA substrate migrating similar as the δ-lyase product (3′-P) has been reported in protein extracts from fission yeast (28,33). In this work, we have revealed that S. pombe Tdp1 removes 3′-α,β-unsaturated aldehydes and we address the biological significance of Tdp1 in repair of AP sites.
Tdp1 was first isolated in S. cerevisiae by its ability to hydrolyze the phosphodiester bond between the Top1 tyrosine residue and the DNA 3′-phosphate originating from abortive Top1–DNA complexes. Later on, it has been shown that human and yeast Tdp1 also process a variety of other 3′-alterations such as phosphoglycolates (27), tetrahydrofuran (AP site mimic) (26) and mononucleosides (26), emphasizing the general lack of specificity for the 3′-leaving group. To this list, we add 3′-dRP residues that we show are processed both by endogenous and recombinant S. pombe Tdp1. Recombinant Tdp1 removed 3′-dR residues from ss and ds substrates equally efficiently and at a comparable rate to tyrosyl residues, the recognized Tdp1 substrate, indicating that this is a biologically relevant substrate. Further, the increased alkylation sensitivity of the tdp1−apn2− double mutant supports the idea that Tdp1 plays a significant role in processing of AP sites. Though, detailed kinetic experiments are needed to carefully determine the substrate preferences for Tdp1. Recently, Lebedeva et al. (36) showed removal of 3′-dRPs also by recombinant human TDP1, however, the in vivo significance of this observation was not addressed. Lebedeva et al. (36) also reports AP lyase activity for human TDP1. In contrast, S. pombe Tdp1 showed negligible AP lyase activity toward the ss and ds AP substrates. Deletion of tdp1 in the nth1− mutant did not sensitize the nth1− cells when challenged with MMS, further supporting that S. pombe Tdp1 is not an AP lyase. Also S. cerevisiae Tdp1 displayed no cleavage of intact AP sites (41).
The product of Tdp1 activity is DNA with a 3′-P terminus, which needs to be further processed by a 3′-phosphatase to create termini compatible for repair synthesis. In humans, polynucleotide kinase catalyzes removal of 3′-P from DNA ends (15,16,42) and repair of TOP1–DNA complexes can be reconstituted with TDP1, PNKP and DNA ligase (20). Furthermore, TDP1 has been shown to exist in complex with PNKP (20), suggesting that TDP1 and PNKP function in the same pathway in human cells. Also in S. cerevisiae, a link between PNKP and Tdp1 exists as the PNKP homolog Tpp1 is epistatic to Tdp1 (43). The genome of S. pombe encodes a polynucleotide kinase, Pnk1 showing phosphatase activity (40,44), and a pnk1− mutant is CPT sensitive (40), suggesting a role in repair of Top1–DNA damage. We propose that Pnk1 in S. pombe is responsible for removal of the 3′-P left after Tdp1 cleavage of 3′dRP termini as also suggested for repair of oxidative stress-induced DNA damage (39). Further, in human cell-free extracts TDP1 has been reported to interact with the BER proteins DNA ligase III and XRCC1 (20,45), and XRCC1 was shown to stimulate both 5′-kinase and 3′-phosphatase activity of PNKP (46). Overall, these observations suggest a role for TDP1 in an APE1-independent BER pathway in mammals similar to that for the NEIL proteins (16,47). It is also tempting to speculate whether Fpg/NEIL enzymes could act in parallel with Tdp1 in removing 3′-tyrosyl residues from DNA termini. At least for Fpg, this appears not to be the case, as in our experiments Fpg could process 3′-dRPs but not 3′-tyrosines (Figure 3A and B).
TDP1 has an essential role in humans as mutation in the TDP1 gene results in the hereditary disease SCAN1 (spinocerebellar ataxia with axonal neuropathy-1), a degenerative neurological syndrome specifically affecting neurons (48). It is believed that TOP1 single-strand breaks accumulate in the absence of TDP1 and that neurons are particularly vulnerable due to high levels of oxidative stress, low levels of antioxidant enzymes and high transcriptional activity (20,24). Mammalian cells lacking TDP1 are consequently hypersensitive to the TOP1–inhibitor CPT (24). Further, it was shown that these cells have reduced repair capacity for oxidative DNA single-strand breaks (20), suggesting that the repair function of TDP1 is not restricted only to TOP1–DNA adducts. The S. cerevisiae tdp1 mutant has been analyzed for sensitivity toward CPT and also agents inducing oxidative stress. However, unless combined with other DNA repair defect mutants, little or no increase in sensitivity is seen, probably reflecting redundancy for the repair of these lesions in budding yeast [CPT: Rad1and Rad9; (49) and Rad9; (25)]. Similarly, in our study, MMS sensitivity for the tdp1− mutant was dependent on Apn2. We propose that Apn2 is the preferred enzyme removing the 3′-dRP generated by Nth1, and that in the absence of Apn2, Tdp1 activity is important. Similarly, in S. cerevisiae, deletion of apn1 and apn2 in the tdp1 mutant sensitizes this strain to bleomycin, possibly indicating overlapping substrate specificities for these three enzymes (49). To our knowledge, MMS was not tested for these mutants. The β-lyase function of Ntg1 and Ntg2 is also important in S. cerevisiae for repair of MMS-induced damage (50), and also budding yeast lack DNA glycosylases performing δ-elimination. Further studies are needed to evaluate a role for Tdp1 in processing alkylation-induced DNA damage in other species.
Cells have evolved a complex network of DNA repair pathways for removal of DNA damage to ensure maintenance of genomic integrity. Understanding the roles of different human or mammalian DNA repair proteins in such a genetic network is a formidable challenge. This is partly because genetic studies in mammalian systems have been limited due to the lack of readily available tools including defined mutant cell lines, regulatory expression systems and appropriate selectable markers. To circumvent these difficulties, model genetic systems in lower eukaryotes have become an attractive choice. The mechanisms of the DNA repair have been largely conserved from bacteria to mammals both when it comes to structure and function, making simpler organisms suitable models. Conclusions derived from genetic and biochemical analysis in yeast can often directly be extrapolated to human systems and indeed, results from such studies have been instrumental for elucidating corresponding pathways in mammalian cells.
One of the main advantages of using yeast as an experimental system is the ease of generation and characterization of double, triple and multiple mutants. Such studies have revealed that there is significant overlap between the different DNA repair pathways as discussed earlier. Defects in two or more DNA repair pathways often result in dramatically increased sensitivity (synthetic lethality) toward DNA damaging agents as seen for the tdp1−apn2− double mutant. As cancer cells often are defective or dysregulated in one or more DNA repair pathways, there is a potential to exploit the ‘synthetic lethality’ phenotype in cancer therapy by the use of specific inhibitors (51). CPT is in use against certain cancers and TDP1 is counteracting the CPT effects. Inhibitors of TDP1 have already been identified, with the intention to enhance the activity of anticancer agents like CPT (52). In human cells, the BER enzyme APE1 is the main enzyme for cleavage of AP sites, and overexpression of APE1 is often observed in tumor cells (53). Subsequently, APE1 inhibitors could enhance the effect of chemotherapeutic agents in APE1 overexpressing tumors (54). By using a combination of both APE1 and TDP1 inhibitors based on targeting/inactivating the BER pathways, chemotherapy may be more successful. Corresponding studies based on mutant analysis as reported in this study needs to be executed in mammalian cell lines to confirm the existence of a similar AP endonuclease independent BER of AP sites involving TDP1 and PNKP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: The Research Council of Norway (171564/V40, 191577/V40).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Henning Cederkvist for MS analysis.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair-recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 4.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 5.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 6.Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 8.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katcher HL, Wallace SS. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983;22:4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- 11.Bailly V, Verly WG, O'Connor T, Laval J. Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine]DNA glycosylase. Biochem. J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor TR, Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc. Natl Acad. Sci. USA. 1989;86:5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 15.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le Beau MM, et al. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 16.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 18.Crespan E, Amoroso A, Maga G. DNA polymerases and mutagenesis in human cancers. Subcell. Biochem. 2010;50:165–188. doi: 10.1007/978-90-481-3471-7_9. [DOI] [PubMed] [Google Scholar]
- 19.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 21.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair. 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme-DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 26.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 28.Alseth I, Korvald H, Osman F, Seeberg E, Bjoras M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugimoto T, Igawa E, Tanihigashi H, Matsubara M, Ide H, Ikeda S. Roles of base excision repair enzymes Nth1p and Apn2p from Schizosaccharomyces pombe in processing alkylation and oxidative DNA damage. DNA Repair. 2005;4:1270–1280. doi: 10.1016/j.dnarep.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Ribar B, Izumi T, Mitra S. The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:115–126. doi: 10.1093/nar/gkh151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanihigashi H, Yamada A, Igawa E, Ikeda S. The role of Schizosaccharomyces pombe DNA repair enzymes Apn1p and Uve1p in the base excision repair of apurinic/apyrimidinic sites. Biochem. Biophys. Res. Commun. 2006;347:889–894. doi: 10.1016/j.bbrc.2006.06.191. [DOI] [PubMed] [Google Scholar]
- 32.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair. 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 33.Laerdahl JK, Korvald H, Nilsen L, hl-Michelsen K, Rognes T, Bjoras M, Alseth I. Schizosaccharomyces pombe encodes a mutated AP endonuclease 1. DNA Repair. 2011;10:296–305. doi: 10.1016/j.dnarep.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Egel R, Willer M, Kjaerulff S, Davey J, Nielsen O. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 1994;10:1347–1354. doi: 10.1002/yea.320101012. [DOI] [PubMed] [Google Scholar]
- 35.Osman F, Bjoras M, Alseth I, Morland I, McCready S, Seeberg E, Tsaneva I. A new Schizosaccharomyces pombe base excision repair mutant, nth1, reveals overlapping pathways for repair of DNA base damage. Mol. Microbiol. 2003;48:465–480. doi: 10.1046/j.1365-2958.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- 36.Lebedeva NA, Rechkunova NI, Lavrik OI. AP-site cleavage activity of tyrosyl-DNA phosphodiesterase 1. FEBS Lett. 2011;585:683–686. doi: 10.1016/j.febslet.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 38.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben HS, Arcangioli B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009;28:632–640. doi: 10.1038/emboj.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D. Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:4050–4055. doi: 10.1074/jbc.M109383200. [DOI] [PubMed] [Google Scholar]
- 41.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 42.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 43.Vance JR, Wilson TE. Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3′ phosphatases. Mol. Cell Biol. 2001;21:7191–7198. doi: 10.1128/MCB.21.21.7191-7198.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jilani A, Ramotar D. Purification and partial characterization of a DNA 3′-phosphatase from Schizosaccharomyces pombe. Biochemistry. 2002;41:7688–7694. doi: 10.1021/bi012213m. [DOI] [PubMed] [Google Scholar]
- 45.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 46.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 47.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair. 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair. 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011;117:6074–6082. doi: 10.1182/blood-2011-01-313734. [DOI] [PubMed] [Google Scholar]
- 52.Huang SY, Pommier Y, Marchand C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert. Opin. Ther. Pat. 2011;21:1285–1292. doi: 10.1517/13543776.2011.604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Mohammed MZ, Vyjayanti VN, Laughton CA, Dekker LV, Fischer PM, Wilson DM, III, Abbotts R, Shah S, Patel PM, Hickson ID, et al. Development and evaluation of human AP endonuclease inhibitors in melanoma and glioma cell lines. Br. J. Cancer. 2011;104:653–663. doi: 10.1038/sj.bjc.6606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang TD, Hodson JA, Forsburg SL. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 1999;112:559–567. doi: 10.1242/jcs.112.4.559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.