Abstract
Many cell lines commonly used for biological studies have been found to harbor exogenous agents such as the human tumor viruses Epstein-Barr virus (EBV) and human papillomavirus. Nevertheless, broad-based, unbiased approaches to globally assess the presence of ectopic organisms within cell model systems have not previously been available. We reasoned that high-throughput sequencing should provide unparalleled insights into the microbiomes of tissue culture cell systems. Here we have used our RNA-seq analysis pipeline, PARSES (Pipeline for Analysis of RNA-Seq Exogenous Sequences), to investigate the presence of ectopic organisms within two EBV-positive B-cell lines commonly used by EBV researchers. Sequencing data sets from both the Akata and JY B-cell lines were found to contain reads for EBV, and the JY data set was found to also contain reads from the murine leukemia virus (MuLV). Further investigation revealed that MuLV transcription in JY cells is highly active. We also identified a number of MuLV alternative splicing events, and we uncovered evidence of APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G)-dependent DNA editing. Finally, reverse transcription-PCR analysis showed the presence of MuLV in three other human B-cell lines (DG75, Ramos, and P3HR1 Cl.13) commonly used by investigators in the Epstein-Barr virus field. We believe that a thorough examination of tissue culture microbiomes using RNA-seq/PARSES-like approaches is critical for the appropriate utilization of these systems in biological studies.
INTRODUCTION
The human body is a persistent host to a spectrum of not only bacterial organisms but also viruses such as herpesviruses. In some circumstances, especially in immunocompromised individuals, the balance of these organisms within the human ecosystem is altered, leading to pernicious diseases such as cancer. As a result, the human microbiome and the role of colonizing microbes in human health are becoming areas of significant interest (13).
Tissue culture cell lines have been important systems in the analysis of the fundamental inner workings of normal and diseased cellular regulatory processes. In many cases, however, commonly used cell lines play host to additional organisms such as viruses (e.g., Epstein-Barr virus [EBV] and human papillomavirus) or intracellular bacteria (e.g., mycobacteria). Oftentimes, the investigator is fully aware of the presence of ectopic organisms within a cell line and may in fact be studying biological aspects of the organism. Alternatively, the investigator may consider the presence of the ectopic organism to be irrelevant to the study being performed.
The presence of ectopic organisms within a cell line is frequently determined with the guidance of known associations between certain organisms and specific diseases/tissues. At other times, the presence of an ectopic organism is discovered by chance through visualization techniques such as electron microscopy. While exogenous organisms have been found in several cell model systems using these methods, the lack of a unified and unbiased approach for discovering passenger organisms leaves great uncertainty regarding their presence within any given cell system of interest.
Since all organisms carry genetic material, examination of the genetic composition of a biological sample should represent a comprehensive and relatively unbiased approach to uniformly assess the presence of ectopic organisms across sample types. Massive parallelization of sequencing provides the sequencing depth necessary to achieve a high level of sensitivity for interrogating the genetic composition of a specimen. We have previously developed a computational pipeline, PARSES (Pipeline for Analysis of RNA-Seq Exogenous Sequences), for the analysis of exogenous agents within human biological systems using RNA-seq data (1). Here we have applied PARSES to RNA-seq data from two EBV-positive human B-cell lines (i) to test the ability of the pipeline to successfully identify the presence of EBV and (ii) to assess the presence of other ectopic agents previously not known to be harbored in these cell lines. The results of this analysis demonstrate the ability of this approach to readily detect known and previously unknown associations between cell systems and ectopic agents harbored within these systems.
MATERIALS AND METHODS
Cell cultures.
All cells were grown in RPMI 1640 (catalog no. SH30027; Thermo Scientific) plus 10% fetal bovine serum (FBS; catalog no. 16000-044; Invitrogen-Gibco) with 0.5% penicillin-streptomycin (catalog no. 15140-122; Invitrogen-Gibco). Cells were grown at 37°C in a humidified, 5% CO2 incubator.
Akata cells were obtained from Kenzo Takada, who first described this cell line (20). Upon receipt, cells were cloned by limiting dilution over a period of approximately 3 months to obtain highly anti-immunoglobulin-responsive subclones. An individual freeze was thawed and cultured for 2 weeks prior to generating RNA for sequencing. JY cells were obtained from Jack Strominger (14, 21). The history of JY culture is more difficult to track since JY cells were generated more than 30 years ago and were obtained by us more than 20 years ago. Nevertheless, they are believed to have undergone multiple rounds of freeze-thaw cycles before we generated RNA in 2011.
RNA-seq.
RNA samples were poly(A) selected, and libraries were prepared using the Illumina TruSeq RNA sample preparation protocol (catalog no. RS-930-2001). One hundred-base paired-end sequencing was performed using an Illumina HiSeq instrument.
PARSES.
PARSES (Pipeline for Analysis of RNA-Seq Exogenous Sequences) is an automated pipeline that seamlessly runs through all required stages of data processing. Using PARSES, RNA-seq data generated from either JY or Akata cells were first analyzed using the genome aligner Novoalign (version 2.07.07; Novocraft) to identify and isolate all sequences that mapped to the human genome. Junction-spanning reads were subsequently identified using the junction mapper TopHat (version 1.3.0) (22). The remaining no-aligned sequences were then analyzed by BLAST using the NCBI nonredundant nucleotide (NT) database. Results from this BLAST search were then input to the taxonomic classifier MEGAN 4 (MEtaGenome ANalyzer) (7) to produce taxonomic data for exogenous agents within the specimen being analyzed. PARSES was run in parallel on two 2- by 2.66-GHz 12-core Intel Xeon Mac Pro computers with 64 gigabytes of memory each.
PARSES was run using 1 gigabyte of RNA-seq data (approximately 3 million to 4 million reads) from JY and Akata cells. The sampling of a portion of the reads was done to reduce the run time and random access memory requirements incurred during the running of the BLAST component of PARSES.
Alignments to MuLV genomes.
For individual alignments to murine leukemia virus (MuLV) genomes, RNA-seq data generated from JY cells were aligned using the genome aligner Novoalign (version 2.07.07). Alignments were run using indices containing the hg19 version of the human genome plus one of the following MuLV genome sequences: a sequence with NCBI accession number AF221065, DQ399707, HQ246218, or U13766 or the JY version of MuLV determined here. Indices were built using a k-mer of 14 and a step size of 1. Novoalign was run using default parameters, which exclude reporting of reads mapping to repeats. Candidate MuLV splice junctions were identified through mapping of the JY RNA-seq data to the MuLV genome using the junction mapper TopHat.
Gene expression calculations and genome browser display.
Expression analysis was performed using our previously reported analysis software, SAMMate (24). Data were displayed using the Integrated Genome Viewer (IGV) (16).
Quantitative RT-PCR.
Total RNA was reverse transcribed with a SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions by using random hexamer primers (50 ng per 1 μg total RNA). For the incubation steps, a Mastercycler ep system (Eppendorf, Hamburg, Germany) was used. The resulting cDNA was subjected to quantitative (real-time) PCR using specific primers (Integrated DNA Technologies) (see Table S1 in the supplemental material). A master mix was prepared for each PCR run and included Platinum SYBR green SuperMix uracil DNA glycosylase (UDG; Invitrogen, Carlsbad, CA), 50 nM fluorescein-National Institute of Standards and Technology (NIST) traceable dye, 250 nM forward and reverse primers, and nuclease-free water. Amplification consisted of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s. Melt curve analysis was performed at the end of every quantitative reverse transcription-PCR (qRT-PCR) run. Samples were tested in triplicate. Real-time PCR was performed on a Bio-Rad (Hercules, CA) MyiQ iCycler apparatus, and data analysis was performed using Bio-Rad IQ5 (version 2.0) software. No-template controls and no-reverse-transcription controls were included with each PCR run. Relative detection levels were calculated using the ΔΔCT method (where CT represents threshold cycle), with the gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as the reference gene. Additionally, all PCR products were analyzed by gel electrophoresis on a 2.0% agarose gel with 5% ethidium bromide stain. Product size was estimated by comparing it to the TrackIt 100-bp DNA ladder (Invitrogen, Carlsbad, CA).
Splice junction validation in JY cells.
Total RNA from JY cells was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Five hundred nanograms of RNA was used in 20-μl reaction volumes according to the manufacturer's instructions. For the incubation steps, a Mastercycler ep system (Eppendorf, Hamburg, Germany) was used. The resulting cDNA was subjected to quantitative (real-time) PCR using sequence-specific forward and reverse primers (Integrated DNA Technologies) (see Table S1 in the supplemental material). For real-time PCR, 1 μl of the resulting cDNA was used in a 20-μl reaction volume that included 10 μl of SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA) and 500 nM (each) the forward and reverse primers. Amplification was carried out using the following conditions: 95°C for 1 min, followed by 40 cycles of 95°C for 5 s and 60°C for 5 s. Melt curve analysis was performed at the end of every qRT-PCR run. Samples were tested in triplicate. No-template controls and no-reverse-transcription controls were included in each PCR run. PCRs were performed on a Bio-Rad CFX96 real-time system, and data analysis was performed using CFX Manager (version 2.0) software. For cloning, 60 μl of the PCR mixture was run on a 2% agarose gel, and product size was estimated by comparing it to the TrackIt 100-bp DNA ladder (Invitrogen, Carlsbad, CA). Bands of interest were extracted from the gel in a 25-μl elution volume using a MinElute gel extraction kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. 3′ A overhangs were added to the PCR products by incubation with 1 μl DNA-free sensitive (DFS)-Taq DNA polymerase (Boca Scientific, Boca Raton, FL), 3 μl 10× reaction buffer, and 1 μl 10 mM deoxynucleoside triphosphate mix (Invitrogen, Carlsbad, CA). The incubation steps included 95°C for 5 min, followed by 72°C for 10 min, and were carried out on a Mastercycler ep system (Eppendorf, Hamburg, Germany). The reaction mix was purified using a MinElute PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations with an elution volume of 10 μl. A PCR Cloning Plus kit (Qiagen, Valencia, CA) was used for cloning of the PCR products. Colonies were selected by blue/white screening, and plasmid DNA was isolated from overnight cultures using a QIAprep spin miniprep kit (Qiagen, Valencia, CA) and analyzed by restriction digest. Plasmids containing the desired PCR product were subjected to Sanger sequencing (Genewiz, South Plainfield, NJ). Sequences were aligned to predicted splice junctions for validation using the BLASTAlign and DNASTAR Lasergene SeqBuilder programs.
Nucleotide sequence accession number.
Sequence data used here have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession number SRA 047981.2).
RESULTS
Detection of MuLV in JY cells using PARSES.
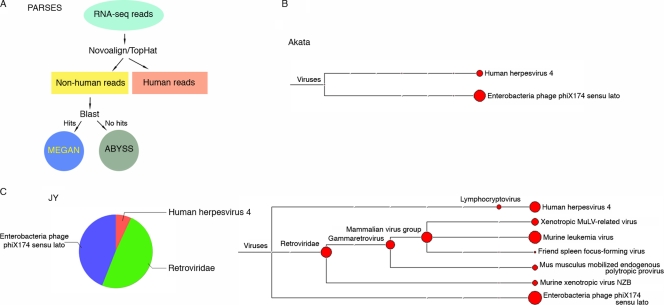
The central component of PARSES (Fig. 1A) (1) is the BLAST analysis of reads against a broad nucleotide database to identify reads that are derived from a nonhost organism. Because this component of the pipeline is computationally intensive, we reduce its burden by incorporating a preprocessing step to quickly identify and select out all reads that map to the human genome. This is accomplished through alignment to the human genome using the genome aligner Novoalign (Novocraft) and the junction mapper TopHat (22). Following alignments, all human and low-quality reads are set aside and the remaining reads are analyzed against the NT database with BLAST. After BLAST analysis, the hits are visualized using a taxonomic classifier, MEGAN 4 (7).
Fig 1.
PARSES analysis of RNA-seq data from the EBV-positive cell lines Akata and JY. (A) Schematic diagram showing essential components of PARSES pipeline. One gigabyte of RNA-seq read data from Akata (B) and JY (C) cells was analyzed using PARSES. The resulting virome data were displayed using MEGAN 4. Both data sets show the presence of EBV as well as ϕX174 (used as a spike-in sequencing control).
We have applied PARSES to RNA-seq data from two B-cell lines, Akata and JY, which are commonly used as models for EBV studies. Analysis of the virome component of the BLAST output showed the presence of the bacteria phage ϕX174 in both samples (Fig. 1B and C). This result is expected since the sequencing runs were spiked with 2% ϕX174 DNA. Epstein-Barr virus (shown in Fig. 1 as human herpesvirus 4) reads were readily detected in data sets from both cell lines (Fig. 1B and C) but not data sets from EBV-negative cell lines (CNE1 and 293 cells; data not shown), demonstrating the specificity of this approach.
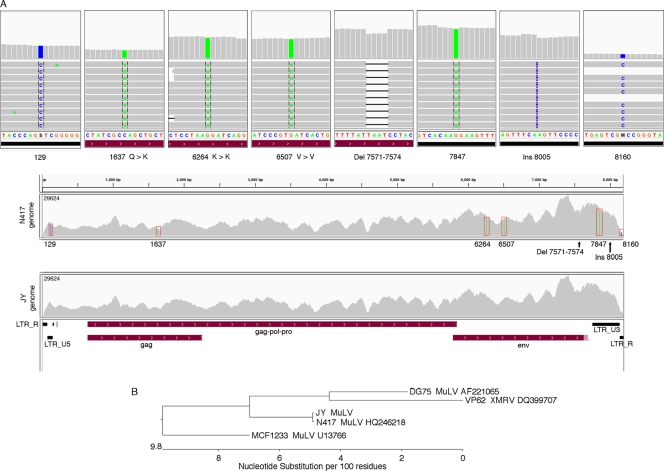
In the JY but not the Akata data set, we also detected a relative abundance of reads from the Retroviridae family (Fig. 1B and C). Since MuLV was the most highly represented member of the Retroviridae identified in this sample and since the others are MuLV related (Fig. 1C), we further explored the presence of MuLV in the JY RNA-seq data. Novoalign indices were built for the genomes of three MuLV strains (NCBI accession numbers AF221065 [15], HQ246218, and U13766 [17]) and one strain of the MuLV relative xenotropic murine leukemia virus-related virus (XMRV) (NCBI accession number DQ399707 [23]). The JY RNA-seq data were then aligned to each of these genomes using Novoalign. Although a large number of reads mapped to each of these genomes, the N417 strain (NCBI accession number HQ246218) showed the greatest homology to the JY reads. Alignment to the N417 genome showed a very high coverage across the genome, with as many as 29,000 reads covering some regions of the 3′ end (Fig. 2A). With this level of coverage, several differences with the N417 genome were easily determined. Four single-nucleotide differences between JY reads and the N417 genome were identified, and two ambiguities in the N417 sequence were resolved (Fig. 2A). In addition, an insertion and a partially penetrant deletion were observed (Fig. 2A; see Fig. S1 in the supplemental material). The sequence disparities in the N417 genome sequence were then corrected and the JY RNA-seq data were then aligned to the corrected N417 genome (referred to as the JY genome). No abundantly represented sequence differences were detected (Fig. 2A, bottom), except for the partially penetrant deletion (deletion of positions 7571 to 7574 [Del 7571 to 7574]), where an insertion was then observed for a significant proportion of the reads. We conclude from this analysis that the Retroviridae genome in JY cells is highly related to the N417 strain of MuLV (Fig. 2B; see Fig. S2 in the supplemental material).
Fig 2.
JY MuLV genome sequence analysis. (A) Alignment of JY RNA-seq reads to N417 and JY MuLV genomes. All mismatches, a small deletion, and a small insertion (Ins) observed from the alignment to the N417 genome are shown in the top eight panels. Coverage data for alignments to the N417 and JY genomes are shown in the lower two panels. The light red shaded region at the right side of the env feature represents the longer conserved env extension in the N417 genome which is partially penetrant in the JY data. (B) JY MuLV is highly related to the N417 strain of MuLV. Alignments were performed using the MegAlign program (DNAstar Inc., Madison, WI). LTR, long terminal repeat.
MuLV is highly expressed in JY cells.
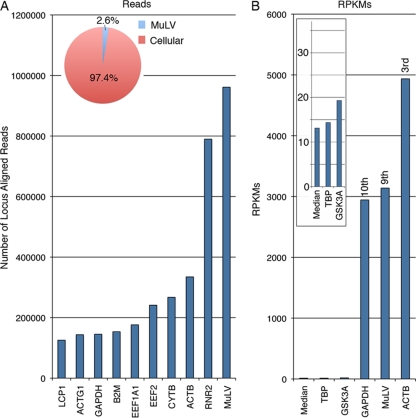
The depth of coverage of the MuLV genome using JY RNA-seq data was high, indicating robust expression of the MuLV genome in these cells. Analysis of MuLV and cellular read numbers revealed that MuLV accounts for 2.6% of all mapped reads derived from polyadenylated RNAs (Fig. 3A). Further, more reads were derived from MuLV than any annotated cellular gene (Fig. 3A). Since the transcribed region of the MuLV genome is larger than most spliced cellular transcripts, we calculated gene expression based on a normalization approach that takes into account the respective transcript length, the RPKM (number of reads per kilobase of exon model per million mapped reads) method. Using this approach, MuLV was found to be the 9th most highly expressed locus expressed in the cell, with an expression level that is slightly higher than that of the housekeeping gene GAPDH (Fig. 3B).
Fig 3.
MuLV transcript quantification. (A) Total read counts at most highly represented genomic loci. Read counts for MuLV include all reads mapping to the viral genome. Read counts for cellular genes include all reads mapping to the respective gene exons. (B) RPKMs are shown for 3 of the top 10 expressed loci (rightward three bars) and two genes with expression near the median of all expressed genes (leftward three bars and inset). Median expression was calculated on the basis of all genes with an expression level of greater than 1 RPKM.
Alternative splicing of MuLV transcripts.
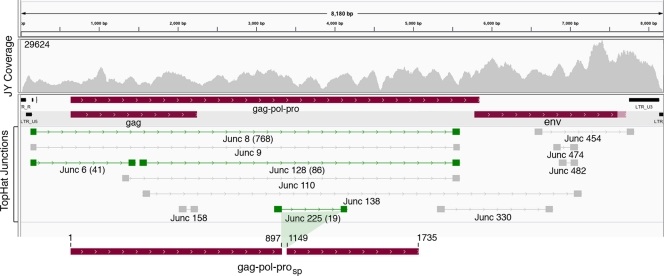
We reasoned that the sequencing depth observed for the MuLV genome in JY cells would allow us to identify abundant and moderately abundant MuLV splicing events. Junction candidates were identified using the junction mapper TopHat. Of the hundreds of TopHat junction predictions, we focused on rightward predicted junctions covered by more than 15 junction-spanning reads (Fig. 3). Of these, we chose 4 for validation. All 4 were found to represent bona fide junctions, as demonstrated by RT-PCR, cloning, and Sanger sequencing of the PCR products (green junctions in Fig. 4; see Fig. S3 in the supplemental material).
Fig 4.
Most abundant MuLV splicing events detected in JY cells. TopHat-predicted rightward junctions with greater than 15 reads per junction are displayed. Junctions in green shading were validated by RT-PCR, followed by cloning and Sanger sequencing. Junction 8 and junction 6 plus junction 128 represent possible splicing events leading to expression of the env gene. Junction 225 represents an in-frame splicing event predicted to give rise to a Gag-Pol-Pro polypeptide lacking functional regions of the reverse transcriptase. The number of reads spanning validated junctions is shown in parentheses.
The most abundant junction, junction 8, which had 768 junction-spanning reads, likely represents a splicing event that leads to the expression of the env open reading frame. The junction 8 splice donor location matches the splice donor annotated in the N417 genome (NCBI accession number HQ246218). In contrast, the junction 8 splice acceptor locus is clearly dislocated from the annotated N417 splice acceptor (NCBI accession number HQ246218) and may represent a previous annotation error. The splice acceptor site of junction 128 (a previously described alternative splice junction [2]) coincides with the splice acceptor site of junction 8 (Fig. 4; see Fig. S3 in the supplemental material). The tandem configuration of junctions 6 and 128 (Fig. 4) raises the possibility that these splicing events occur within single transcripts to give rise to an alternative isoform encoding the env reading frame. Lastly, junction 225 represents an in-frame splicing event that is predicted to give rise to a gag-pol-pro reading frame lacking critical regions of the reverse transcriptase catalytic domain (Fig. 4; see Fig. S4 in the supplemental material).
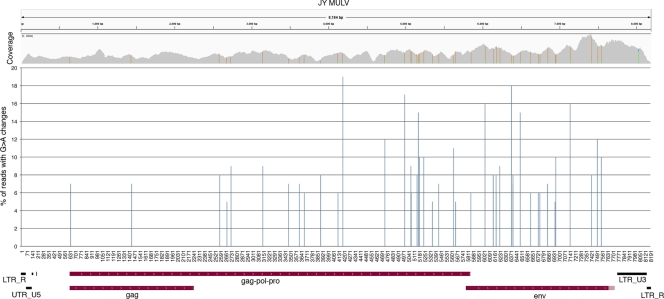
Identification of partially penetrant G-to-A changes in JY sequence reads.
RNA editing is a mechanism through which individual bases of select transcripts are modified posttranscriptionally to give rise to transcript variants with sometimes distinct function (3). Due to the typically incomplete nature of RNA-editing events, such events can be identified as partially penetrant nucleotide differences from the host genome. In our investigation of potential RNA-editing events in JY MuLV, we identified all partially penetrant nucleotide differences that were represented in more than 5% of all reads at each position (Fig. 5). Strikingly, of the 45 loci satisfying these criteria, all but 1 corresponded to G-to-A changes. The prevalence of G-to-A changes was unexpected, since A to I is the most common form of RNA editing (3, 6). On the other hand, these events could potentially represent C-to-U editing of transcripts oriented in the leftward direction; however, this form of editing is considered much more rare (6). Interestingly, DNA editing rather than RNA editing appears to be common in retroviruses and retrotransposons and the G-to-A change is the most frequent DNA-editing change observed (4, 26). The mechanism driving this change is the cellular retroviral defense protein APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G), which facilitates deamination of cytosines to uracils in the cytoplasm soon after the reverse transcription reaction (5, 10, 11). While most of these modified reverse-strand DNAs are degraded, some of them escape the degradation process and are passed on as viral mutants containing genomic G-to-A changes. We believe this to be the most plausible explanation for the G-to-A changes observed in the JY MuLV RNA-seq data.
Fig 5.
Partially penetrant G-to-A changes present in greater than 5% of reads. Nucleotide position is plotted on the x axis. UTR, untranslated region.
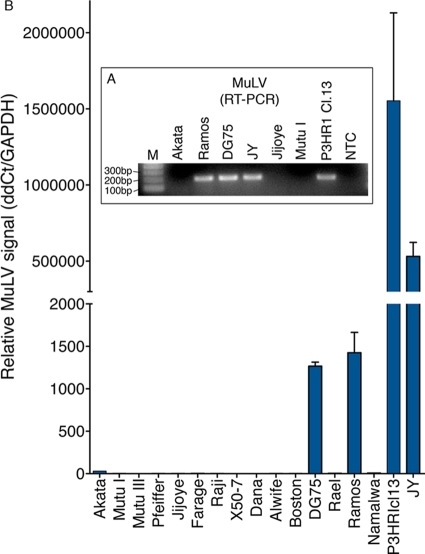
Detection of MuLV in a panel of human B-cell tissue culture lines.
Previous studies have similarly discovered MuLV-related virus in human B-cell lines such as DG75, Ramos, and P3HR1, although through different methods (8, 9, 15, 19, 25). Here we have found that JY MuLV displays significant metabolic activity which may potentially impact the interpretation of certain types of biological studies. We therefore investigated the presence of MuLV-related viruses in a panel of B-cell lines by RT-PCR. While MuLV was not detected in most cell lines, we detected MuLV in the three cell lines that were previously reported to have MuLV or MuLV-like virus (DG75, Ramos, and P3HR1 Cl.13) (Fig. 6). Notably, however, Ramos and P3HR1 cell lines obtained independently from the American Type Culture Collection (ATCC) were found to be MuLV negative (see Fig. S5 in the supplemental material). This observation is consistent with the hypothesis that MuLV infection likely occurred after the initial establishment of these cell lines. Based on our analysis and results from other studies, we propose that human B-cell lines should be tested for MuLV and its presence should be taken into consideration when performing experiments that may be impacted by this additional infectious agent.
Fig 6.
Detection of MuLV in other laboratory cell lines. (A) Nonquantitative RT-PCR detects MuLV-related virus in several laboratory cell lines. Lane M, molecular size marker; lane NTC, no-template control. (B) Quantitative RT-PCR analysis of a larger panel of B-cell lines shows the presence of MuLV-related virus in DG75, Ramos, P3HR1 Cl.13, and JY cells.
DISCUSSION
Forty-four out of 45 partially penetrant base changes identified in the JY RNA-seq data are G-to-A changes. Notably, three out of four of the fully penetrant base changes identified are also G to A. These, too, are likely candidate mutations resulting from the action of APOBEC3G. The fully penetrant nature of these changes, however, indicates that they occurred upstream from the partially penetrant G-to-A changes. The fully penetrant changes were likely incorporated in either the producer cells that gave rise to infection of JY cells or even further up in the infection cascade. The finding of partially penetrant G-to-A changes indicates the presence of a mixed population of MuLV genomes in JY cells. The partially penetrant deletion (Del 7571 to 7574) further supports the idea that JY cells harbor a mixed population of MuLV genomes. Such a mixed population could have resulted from infection of the JY cell line with multiple different MuLV particles carrying variant genomes. On the other hand, it is possible that these changes occurred subsequent to the initial infection event(s) through the exchange of viral particles produced in JY cells to adjacent cells or through intracellular retrotransposition. As determined from our sequencing data, APOBEC3G transcripts are expressed at 50 RPKMs in JY cells, which is almost four times higher than the median level of expressed genes (Fig. 3). This raises the possibility that MuLV continues to evolve through an APOBEC3G-dependent mechanism during culture.
In this study, we have demonstrated the utility of our computational pipeline, PARSES, to rapidly identify ectopic organisms that potentially coexist, unbeknownst to the researcher, within common cell model systems. In the example presented here, we readily detected not only the presence of EBV, as expected, but also the presence of MuLV in the EBV-positive lymphoblastoid cell line JY. RT-PCR detected MuLV-like genomes not only in JY cells but also in three other B-cell lines commonly used by EBV and other researchers. Although to our knowledge no study has previously reported the detection of MuLV within the JY cell line, MuLV or MuLV-like virus has previously been detected in some lineages of all three of these other B-cell lines (P3HR1 Cl.13, Ramos, and DG75) (Fig. 6) (8, 9, 15).
In the case of DG75, whereas some laboratory sources of these cells were found to be MuLV positive, DG75 cells from another lab were found to be MuLV negative (15). Further, we were able to purchase the Ramos cell line and the P3HR1 Cl.13 progenitor cell line, P3HR1, from ATCC, and these lots were found to be MuLV negative (see Fig. S5 in the supplemental material). Unfortunately, since ATCC does not carry JY cells, we were unable to perform a similar analysis with this cell line. Nevertheless, the batch specificity of MuLV presence in the DG75, Ramos, and P3HR1/P3HR1 Cl.13 cell lines implicates laboratory contamination as the most likely mechanism of infection. This is consistent with previous findings pertaining to the MuLV-related virus XMRV within prostate cancer cell lines (12). In these cases, it appears that the XMRV-positive prostate cancer cell lines were passaged in mice, where they acquired XMRV (12). Rather than XMRV being implicated in prostate cancer development, these findings indicate that the presence of XMRV in these cell lines is a laboratory artifact.
We contend that it is important for researchers using human B-cell lines to be aware of the presence of MuLV (or possibly other ectopic organisms). First, at least in JY cells, the transcriptional activity of the MuLV genome is high (Fig. 3) and MuLV RNAs and proteins are likely to impact overall cellular metabolism as well as specific regulatory circuits. As a result, the presence of MuLV in EBV-infected cells, for example, may influence the results of EBV-related studies in unforeseen ways that may cause invalid or skewed conclusions. Second, it is readily apparent that MuLV, a mouse leukemia virus, can infect human B cells. Knowledge of the presence of MuLV or other organisms harbored within cell model systems is important for establishing appropriate biohazard safety precautions.
Although the host status of many cell model systems is known, these associations have oftentimes been guided by prior knowledge, which confines the discovery to the organism being investigated. The RNA-seq approach outlined here is much less confined by prior knowledge and is primarily limited only by the need for genetic information for the respective ectopic organism. Notably, however, PARSES has an optional component involving the de novo assembly of reads (using the assembler ABySS [18]) that do not match during the BLAST step (Fig. 1A). This process results in larger sequence contigs that are translated in all six frames and analyzed with BLAST against a protein database to identify homologies to known related species. As a result, PARSES can also reveal the presence of previously undiscovered organisms or organisms for which there is no known genetic information. In our current study using the Akata and JY cell lines, this arm of the pipeline did not detect any novel agents (data not shown). Nevertheless, this component may prove highly informative in other studies.
The JY and Akata cDNA libraries used for this study were generated from poly(A)-selected RNA to reduce the burden of sequencing large amounts of rRNA fragments. Poly(A) selection has been a common method to achieve a high percentage of non-rRNA reads, and most RNA-seq data in current public repositories are from poly(A)-selected libraries. Most human viruses express genes that are polyadenylated. Bacterial transcripts are also polyadenylated (although to a lesser extent), and we have readily detected robust amounts of bacterial transcripts in poly(A) RNA-seq libraries from a cohort of publically available clinical tumor samples using PARSES (M. J. Strong and E. K. Flemington, unpublished data). Importantly, however, some viral transcripts are not polyadenylated, and in some cases, nonpolyadenylated transcripts are the only viral RNAs expressed in a given cell or tissue sample. With the recent advances in alternative methods to deplete rRNAs, the ability to broadly detect cohabitating organisms will be enhanced even further.
Together, we believe that PARSES or PARSES-like RNA-seq approaches represent powerful, sensitive, and relatively unbiased means to determine the microbiome of not only clinical samples but also cell lines commonly used in biomedical research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01CA124311, R01CA130752, and R01CA138268 to E.K.F. and a grant from the Ladies Leukemia League to E.K.F.
Footnotes
Published ahead of print 11 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Coco JR, Flemington EK, Taylor CM. 2011. PARSES: a Pipeline for Analysis of RNA-Seq Exogenous Sequences, p 196–200 Abstr. ISCA 3rd Int. Conf. Bioinform. Comput. Biol., New Orleans, LA [Google Scholar]
- 2. Dejardin J, et al. 2000. A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing. J. Virol. 74:3709–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis 32:1569–1577 [DOI] [PubMed] [Google Scholar]
- 4. Esnault C, et al. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433 [DOI] [PubMed] [Google Scholar]
- 5. Harris RS, et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 6. Hogg M, Paro S, Keegan LP, O'Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 73:87–120 [DOI] [PubMed] [Google Scholar]
- 7. Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kotler M, Balabanova H, Ben-Moyal Z, Friedman A, Becker Y. 1977. Properties of the oncornavirus particles isolated from P3HR-1 and Raji human lymphoblastoid cell lines. Isr. J. Med. Sci. 13:740–746 [PubMed] [Google Scholar]
- 9. Lasky RD, Troy FA. 1984. Possible DNA-RNA tumor virus interaction in human lymphomas: expression of retroviral proteins in Ramos lymphoma lines is enhanced after conversion with Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 81:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 11. Mangeat B, et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 12. Paprotka T, et al. Recombinant origin of the retrovirus XMRV. Science 333:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pflughoeft KJ, Versalovic J. 25 January2011. Human microbiome in health and disease. Annu. Rev. Pathol. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 14. Ploegh HL, Cannon LE, Strominger JL. 1979. Cell-free translation of the mRNAs for the heavy and light chains of HLA-A and HLA-B antigens. Proc. Natl. Acad. Sci. U. S. A. 76:2273–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raisch KP, et al. 2003. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology 308:83–91 [DOI] [PubMed] [Google Scholar]
- 16. Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sijts EJ, et al. 1994. Cloning of the MCF1233 murine leukemia virus and identification of sequences involved in viral tropism, oncogenicity and T cell epitope formation. Virus Res. 34:339–349 [DOI] [PubMed] [Google Scholar]
- 18. Simpson JT, et al. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun R, et al. 1995. Transmissible retrovirus in Epstein-Barr virus-producer B95-8 cells. Virology 209:374–383 [DOI] [PubMed] [Google Scholar]
- 20. Takada K, Ono Y. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terhorst C, Parham P, Mann DL, Strominger JL. 1976. Structure of HLA antigens: amino-acid and carbohydrate compositions and NH2-terminal sequences of four antigen preparations. Proc. Natl. Acad. Sci. U. S. A. 73:910–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Urisman A, et al. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Xu G, et al. 2011. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol. Med. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yaniv A, Gotlieb-Stematsky T, Vonsover A, Perk K. 1980. Evidence for type-C retrovirus production by Burkitt's lymphoma-derived cell line. Int. J. Cancer 25:205–211 [DOI] [PubMed] [Google Scholar]
- 26. Zaranek AW, Levanon EY, Zecharia T, Clegg T, Church GM. A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing. PLoS Genet. 6:e1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.