Abstract
Background Disadvantaged socio-economic position (SEP) in childhood is associated with increased adult mortality and morbidity. We aimed to establish whether childhood SEP was associated with differential methylation of adult DNA.
Methods Forty adult males from the 1958 British Birth Cohort Study were selected from SEP extremes in both early childhood and mid-adulthood. We performed genome-wide methylation analysis on blood DNA taken at 45 years using MeDIP (methylated DNA immunoprecipitation). We mapped in triplicate the methylation state of promoters of approximately 20 000 genes and 400 microRNAs. Probe methylation scores were averaged across triplicates and differential methylation between groups of individuals was determined. Differentially methylated promoter sites of selected genes were validated using pyrosequencing of bisulfite-converted DNA.
Results Variably methylated probes (9112 from n = 223 359 on the microarray) corresponded to 6176 gene promoters with at least one variable probe. Unsupervised hierarchical clustering of probes obtained from the 500 most variable promoters revealed a cluster enriched with high SEP individuals confirming that SEP differences contribute to overall epigenetic variation. Methylation levels for 1252 gene promoters were associated with childhood SEP vs 545 promoters for adulthood SEP. Functionally, associations with childhood SEP appear in promoters of genes enriched in key cell signalling pathways. The differentially methylated promoters associated with SEP cluster in megabase-sized regions of the genome.
Conclusions Adult blood DNA methylation profiles show more associations with childhood SEP than adult SEP. Organization of these associations across the genome suggests a well-defined epigenetic pattern linked to early socio-economic environment.
Keywords: 1958 British birth cohort, adult DNA methylation profile, childhood socio-economic position
Introduction
Socio-economic circumstances are long-established determinants of health where better health is associated with socio-economic advantage.1–3 Some adult chronic diseases are influenced by circumstances in early life suggesting that the early environment has a long-lasting effect.4 Plausible explanations for these early-life influences have been proposed, involving both social and biological processes.5–9 However, the underlying biological mechanisms have yet to be established. Low early-life social class has been recently linked to enduring differences in adult gene expression patterns relating to glucocorticoid and inflammatory responses.10 A working hypothesis is that socio-economic circumstances leave their mark on the epigenome leading to stable changes in expression of genes critical for human health, such as those involved in cardiovascular, immune, stress response and behavioural pathologies.11
It is now well established that DNA sequence is complemented by epigenetic information including DNA methylation and histone modifications to determine gene expression.12,13 In several well-studied examples, methylation of regulatory regions results in gene silencing, whereas loss of methylation is associated with gene activity.12 Several mechanisms were proposed to explain how methylation of CpG sites in promoters results in gene silencing. These involve recruitment of methylated DNA-binding proteins which in turn recruit chromatin modifying enzymes precipitating an inactive chromatin configuration14 as well as direct interference in binding of transcription factors to genes.15
There is a growing body of evidence suggesting that the enzymatic machinery responsible for creating and maintaining DNA methylation patterns is responsive to environmental exposures during both intra-uterine development and after birth in animals16–22 and in humans.23–28 For example, early nurturing experiences influence epigenetic programming of the glucocorticoid receptor (GR) gene promoter in the hippocampus of rats21 and humans.27 Importantly, these types of observations are not restricted to neurons. The GR gene showed hypermethylation in human cord-blood DNA from newborns of depressed/anxious mothers.26 Increases in global DNA methylation levels in adult blood have been linked to maternal smoking during pregnancy.23 Several imprinted genes, including IGF2, showed altered (both hypo- and hyper-) methylation levels in blood DNA of 60-year-old individuals who had been prenatally exposed to famine.25,29 Moreover, there is recent evidence that many genomic sites show blood DNA methylation patterns that are stable within the individual over later stages of life but that nevertheless vary between individuals.30
It is plausible, therefore, that DNA methylation is involved in human developmental plasticity5 and may be studied using peripheral blood DNA. Our aim was to conduct a preliminary test of this hypothesis by creating and analysing genome-wide promoter methylation profiles of 40 adult males from high and low socio-economic position (SEP) in childhood and in adulthood. Specifically, we aimed to establish whether childhood SEP was associated with differential DNA methylation. Males in our study were all participants in the 1958 British birth cohort, an ongoing longitudinal study in which health differences have been demonstrated through to mid-adulthood in relation to both child and adult SEP.31,32
Methods
Study population
Participants were originally enrolled in the perinatal mortality survey (PMS) of all born in UK during 1 week in March 1958 with follow-up in childhood at 7, 11 and 16 years and in adulthood at 23, 33, 42 and 45 years. A total of 17 415 individuals were enrolled into the PMS from those eligible (N = 17 638) immigrants with the same birth dates were recruited up to 16 years (n = 920), thus the total study population is 18 558. At 45 years, 11 971 participants still in contact with the study, and those who at 42 years had not required a proxy interview, were invited to a clinical examination undertaken in their home by a trained nurse; 4665 males and 4712 females were seen from September 2002 to March 2004. From 4177 males providing consent to blood collection and DNA analysis, an eligible sample for epigenetic analysis was identified excluding: those reporting at 42 or 33 years that they had cancer (n = 43); elevated (8 mg/l) (n = 125) or missing data (n = 436) for C-reactive protein at 45 years; immigrants (n = 134) and others (n = 39) who lacked perinatal data; and non-White (n = 37) or missing data (n = 1) on ethnicity. This eligible sample (N = 3362) was classified according to SEP in early childhood and mid-adulthood as follows.
A childhood SEP score was derived [range 1 (least) to 12 (most) disadvantaged] from cross-classification of:
father's occupation in 1958. Six registrar general's (RG) occupational groups were used: professional (I), managerial/technical (II), other non-manual (IIInm), skilled manual (IIIm), partly skilled (IV) and unskilled manual (V), including those with no male head of household;
lacking or sharing access to household amenities (any of: hot water, bathroom and inside lavatory) or household overcrowding (more than one person/room) at age 7 years vs others.
An adulthood SEP score was derived [range 1 (least) to 12 (most) disadvantaged] from cross-classification of:
the participant's current or most recent occupation at 42 years (or 33 years if data were unavailable at 42 years), categorized using the six RG groups mentioned above for father's occupation;
housing tenure non-owner/buyer or financial difficulties at 45 years (ascertained from two questions: How often do you not have enough money to afford clothing or food for you or your family, and How much difficulty do you have meeting the payment of bills? Men who sometimes/often or always had difficulties were identified) vs others.
For both childhood and adult SEP, scores were assigned whereby individuals with professional occupations and without housing/financial adversity had a score of 1 (i.e. least disadvantaged); professional occupation with housing/financial adversity was scored 2 and so forth to a score of 12 (most disadvantaged) for unskilled manual background and housing/financial adversity. Ranking individuals from the top and bottom quintiles of the distribution of SEP in both childhood and adulthood, we selected groups from those with sufficient blood DNA for methylation analysis. Given our primary focus on child vs adult SEP, we selected four groups to provide a balanced design and options for analyses: low SEP in both childhood and adulthood (LL n = 10); childhood but not adulthood (LH n = 11); adulthood but not childhood (HL n = 11), and neither childhood nor adulthood (HH n = 8) (Figure S1, available as Supplementary Data at IJE online). Using the same SEP classification, we chose an additional 40 males to replicate selected findings.
DNA methylation analysis
DNA was extracted from whole blood collected in EDTA at 45 years using an in-house, manual guanidine hydrochloride and ethanol precipitation method. We used the well-established method of methylated DNA immunoprecipitation (MeDIP), which has been evaluated against other established technologies.33–39 Our MeDIP analysis was adapted from the method used by Keshet et al.40 We mapped the methylation state of the promoters of nearly 20 000 genes and 400 microRNAs printed on custom, high density oligonucleotide microarrays covering approximately 1000 bp upstream to 250 bp downstream at 100-bp spacing from the transcription start sites described in Ensembl (version 44). Three microarrays were generated per individual. Applying principal component analysis to the 500 most variable microarray probes (across all microarrays) we found the expected clustering of triplicates (Figure S2, available as Supplementary Data at IJE online). This reproducibility was confirmed with hierarchical clustering to the same set of probes and with only a few exceptions, triplicates were found in the same clusters (Figure S3, available as Supplementary Data at IJE online). Finally, we used the eigenR2 algorithm designed to estimate the proportion of variance explained by variables in microarray experiments (http://www.genomine.org/eigenr2/) and found that >70% of the variance in the 500 most variable probes is associated with differences between individuals. When microarray results were randomized (into sets of three) 100 times, differences between individuals explained 28–38% of the variance. Thus, using three different approaches, we have shown adequate reproducibility of the microarrays.
Microarray analysis
The principal statistic used to judge the statistical strength of our results is the ‘false discovery rate’ (FDR),41 which was designed to test the chances of an overall false discovery among a series of related results. The FDR is particularly useful for an exploratory analysis concerned with making general inferences from among a set of ‘discoveries’, rather than guarding against one or more individual false positives. Here, we use an FDR of 20% because our objective in this preliminary study is to generate hypotheses rather than to definitively characterize the epigenetic signatures of SEP.
Figure S4 (available as Supplementary Data at IJE online) summarizes the microarray analysis, with stages as follows. After microarray scanning, probe intensities were extracted from scan images using Agilent's Feature Extraction 9.5.3 Image Analysis Software. The extracted intensities were then analysed using the R software environment for statistical computing.42 Log-ratios of the bound (Cy5) and input (Cy3) microarray channel intensities were computed for each microarray and then microarrays were normalized to one another using quantile-normalization43 under the assumption that all samples have identical overall methylation levels. The resulting values for each probe are called ‘methylation scores’.
A probe was identified as ‘differentially methylated’ between two individuals if the methylation scores for all three triplicates for one participant were at least 0.5 greater or at least 0.5 less than the triplicate scores for the other participant. The ‘variability of a probe’ was quantified as the number of individual pairs for which it was variably methylated. A probe was identified as ‘variably methylated’ in the population if it was identified as differentially methylated between at least 100 pairs of participants (i.e. it had a variability of 100). Having differences between 100 pairs corresponds to splitting at least 20 participants into two groups. A ‘promoter’ was considered ‘variably methylated’ if it contained at least one variably methylated probe.
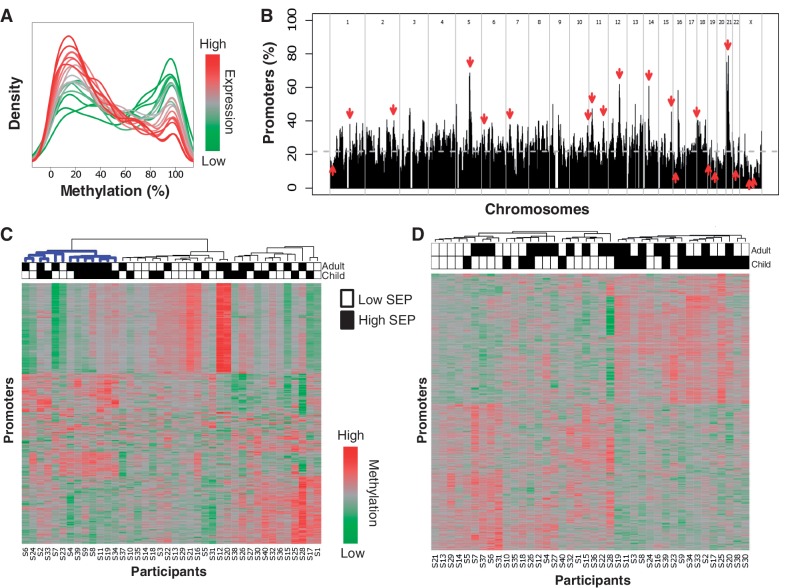
The heatmaps show probe methylation scores averaged across triplicate microarrays. Clustering was performed using Ward's hierarchical clustering algorithm with Pearson correlation distance as the distance metric. In Figure 1C, the heatmap consists of the 500 probes quantified as most variable with the condition that each probe represents a distinct promoter (i.e. for promoters with multiple highly variable probes, only the most variable was included). In Figure 1D, the heatmap consists of the 500 probes most associated with childhood SEP, once again such that each probe represents a distinct promoter. Hence, the clustering of male participants in Figure 1C is unsupervised and represents a class discovery approach, whereas the clustering in Figure 1D is supervised illustrating the results of class distinction.
Figure 1.
Whole-blood promoter methylation in a variable SEP population. (A) Distributions of promoter methylation levels by published expression level. Genes are divided into 20 levels by whole-blood expression percentiles (0–5, 5–10, … , 95–100) based on publicly available expression data.50 Shown are the distributions of methylation levels for each expression percentile. The distributions show that genes with low or no expression (represented in green) tend to have highly methylated promoters, whereas genes with high expression (represented in red) tend to have little or no promoter methylation. (B) Distribution of variably methylated promoters. Distribution of variably methylated promoters across the genome shown as percentage of promoters in each region. Downward pointing arrows indicate regions enriched with variably methylated promoters, and upward pointing arrows indicate regions with little variability (FDR < 0.05). The dashed grey line shows the expected percentage of variably methylated promoters. (C) Most variably methylated promoters. Heatmap showing methylation scores of the 500 most variably methylated promoters (rows) across all 40 individuals (columns). Each promoter is represented by its most variable probe. Blackened squares above the columns denote high SEP in childhood or adulthood. The cluster highlighted in blue contains a significant number of high childhood and adulthood SEP individuals (hypergeometric; P ≤ 0.009). Each probe represents a distinct promoter (i.e. for promoters with multiple highly variable probes, only the most variable was included). (D) Differentially methylated promoters associated with childhood SEP. The heatmap shows the methylation scores of the 500 probes most significantly associated with childhood SEP with the following qualifications: each probe satisfied the individual requirements to be called differentially methylated, each belonged to a promoter called differentially methylated, and no pair of probes belonged to the same promoter
Differential methylation between groups of participants was determined in several stages to ensure both statistical and biological relevance. In the first stage, linear models implemented in the ‘limma’ package44 of Bioconductor45 were used to compute a modified t-statistic at the individual probe level. An individual probe was called ‘differentially methylated’ if its t statistic gave a P ≤ 0.05 (uncorrected for multiple testing) and the associated difference of means between the groups was at least 0.25. Given that the DNA samples were sonicated prior to hybridization, we assumed that probes within 500 bp should approximately agree. Therefore, at each probe we centred a 1000-bp window and estimated the enrichment for high or low t-statistics of probes within that window. These enrichments were assessed using the Wilcoxon rank-sum test comparing t-statistics of the probes within the window against those of all the probes on the microarray. The statistical enrichment of differential methylation within each promoter was equated with the statistic of its strongest probe (at the window level). These promoter significance levels were then adjusted to obtain FDRs for each promoter. A promoter was then called ‘differentially methylated’ if it satisfied each of the following:
its FDR was at most 0.2; and
the 1000-bp window, centred at its most significant probe, contained at least one probe called differentially methylated.
The first requirement ensured that several probes in the promoter agreed on the methylation difference, and the second requirement ensured that the difference was not simply weakly distributed across the entire promoter and consequently difficult to validate.
All functional analysis was based on gene sets obtained from Gene Ontology (GO),46 Kyoto Encyclopedia of Genes and Genomes (KEGG),47 The Molecular Signatures Database (mSigDB)48 and Online Mendelian Inheritance in Man (OMIM).49 Enrichment for differential methylation was determined by applying the hypergeometric test to the overlap between defined gene sets and genes with differentially methylated promoters. FDRs were obtained by adjusting these significance levels over all gene sets and pathways considered.
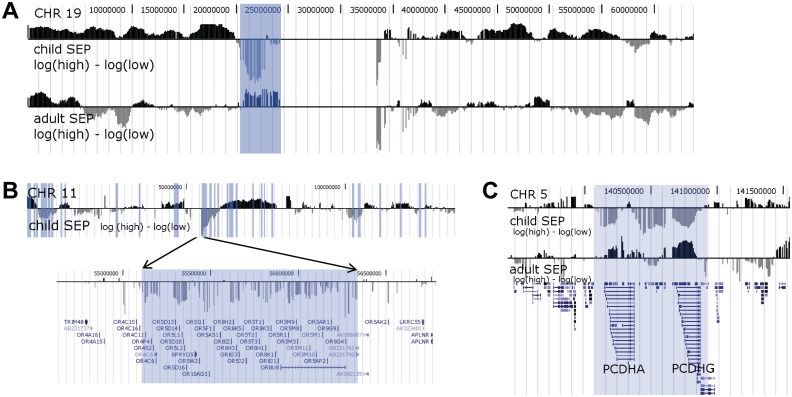
The mega-base methylation patterns of Figure 3 were obtained by computing the mean methylation score difference between groups for each probe, generating a University of California, Santa Cruz genome browser (UCSC) wiggle track file from these differences and then uploading it for display on the UCSC genome browser (http://genome.ucsc.edu/).
Figure 3.
Megabase co-clustering of differential methylation. (A) Co-clustering of differentially methylated promoters across megabases of DNA. Positive values (black bars) indicate increased methylation in high childhood SEP compared with low childhood SEP and negative values (grey bars) indicate the opposite. With the exception of one region next to the chromosome centromere (highlighted), chromosome 19 promoters are generally more methylated in high childhood SEP than in low childhood SEP. This is in contrast to adulthood SEP where the methylation differences are not significant. (B) Co-clustering of differentially methylated promoters with common function across megabases of DNA. Unlike chromosome 19, chromosome 11 has more directional variation with respect to childhood SEP. We highlight a region of 1 million bases that is consistently more methylated in low childhood SEP and contains about 40 genes, all olfactory receptor genes. Shaded vertical bars identify 500-kb regions of significant difference between childhood SEP groups (FDR ≤ 0.025). (C) Co-clustering of differential methylation with the protocadherins. Shaded is a nearly 1-Mb region containing mainly protocadherins whose promoters are consistently more methylated in low childhood SEP than in high. Most of this region was found to be consistently hypermethylated in breast tumours.53 We observe a similar but much weaker and inverted methylation difference between low and high adulthood SEP
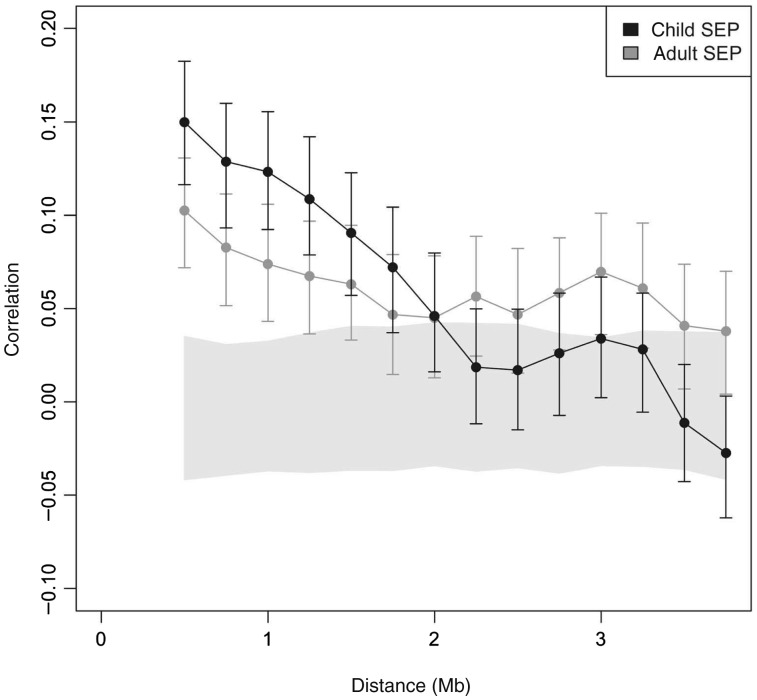
Differential methylation across 500-kb genomic regions with respect to childhood and adulthood SEP was computed by summarizing probe t-statistics across the entire region. T-statistics obtained for each probe using ‘limma’ (see above) and then a summary statistic (z-score) was computed for each 500-kb region by applying the Wilcoxon rank-sum test to the statistics assigned to its promoters in comparison to the statistics assigned to the set of all promoters across the genome. Figure 4 shows the correlations of the region z-scores at given distances apart. Error bars indicate the 95% confidence intervals (CIs) for the correlations obtained from 1000 bootstraps composed of random selections of region pairs with replacement. The grey region contains the 95% CI for correlations of independent regions. The interval was computed from 500 random permutations of the region coordinates, in order to simulate independence. The 500-kb region size was chosen to maximize the number of regions with a sufficient number of genes for the statistics to be meaningful. Regions with fewer than five genes were ignored.
Figure 4.
Methylation dependencies across megabases. Shown are correlations of methylation differences from 500-kb regions at various distances apart. The solid grey region contains the 95% CI, and error bars contain the 95% CI for correlation values
Details of DNA preparation, MeDIP (including DNA amplification), microarray design and hybridization, validation methods and further information on analysis of data are given in the Supplementary Methods, available as Supplementary Data at IJE online.
Comparison of SEP groups (physical and behavioural characteristics)
We compared variables denoting frequencies (e.g. percentage of smokers) for LH + LL vs HH + HL using Pearson's chi-square to test the null hypothesis that frequencies are uniform across these two main SEP groups. For variables with mean and standard deviation (SD) statistics, one-way ANOVA was used to test the null hypothesis that variation within SEP groups was the same as between SEP groups. Where variables were not normally distributed, we use quantiles and the non-parametric Kruskal–Wallis equality-of-populations rank test to test the same hypothesis.
Results
Characteristics of study participants in the SEP groups are shown in Table 1 (further details in Table S1, available as Supplementary Data at IJE online). As expected from the literature, there are differences between SEP groups, with poorer physical, cognitive and emotional status among males from low SEP in early life and mid-adulthood, although not reaching conventional P-values in this small sample.
Table 1.
Characteristics of the participants
| Characteristic | Age (years) | LL n = 10a | LH n = 11a | HL n = 11a | HH n = 8a | P | P (child SEP) |
|---|---|---|---|---|---|---|---|
| Childhood SEP score, mean (SD) | 0 + 7 | 10.0 (1.31) | 9.7 (1.42) | 3.0 (1.10) | 2.0 (1.41) | 10e-18 | 2e-19 |
| Adult SEP score, mean (SD) | 42 + 45 | 9.3 (1.34) | 2.8 (1.25) | 9.1 (2.07) | 2.5 (1.31) | 6e-14 | 0.72 |
| Birthweight, g, mean (SD) | 0 | 3490 (741) | 3335 (477) | 3490 (600) | 3771 (470) | 0.47 | 0.28 |
| Height, cm, mean (SD) | 7 | 119.7 (6.44) | 123.5 (6.25) | 122.9 (7.95) | 125.4 (6.08) | 0.41 | 0.34 |
| Alcohol drinks daily, n (%) | 42 | 1 (11%) | 2 (18%) | 3 (27%) | 3 (38%) | 0.59 | 0.22 |
| Height, cm, mean (SD) | 42 | 172.7 (3.36) | 178 (5.18) | 176.6 (1.01) | 182.4 (9.76) | 0.088 | 0.2 |
| Smokers, n (%) | 42 | 4 (44%) | 4 (36%) | 2 (18%) | 1 (12%) | 0.38 | 0.093 |
| BMI, kg/m2, mean (SD) | 45 | 26 (4.4) | 29 (4.7) | 28 (3.7) | 25 (2.1) | 0.083 | 0.54 |
| Diastolic blood pressure, mmHg, mean (SD) | 45 | 89 (12) | 85 (9.5) | 83 (14) | 75 (7.7) | 0.085 | 0.056 |
| Fibrinogen, median (Q1–Q3) | 45 | 3 (2.9–3.3) | 3 (2.5–3.9) | 2.9 (2.7–3.6) | 2.6 (2.4–3.5) | 0.68 | 0.43 |
| HbA1c, median (Q1–Q3) | 45 | 5.2 (4.8–5.7) | 5.5 (4.8–5.9) | 5.2 (5.1–5.7) | 5.2 (5–5.5) | 0.87 | 0.79 |
| HDL cholesterol, median (Q1–Q3) | 45 | 1.5 (1.4–1.7) | 1.1 (1.1–1.5) | 1.3 (1.1–1.6) | 1.5 (1.2–1.7) | 0.066 | 0.84 |
| Systolic blood pressure, mmHg, mean (SD) | 45 | 143 (19) | 136 (17) | 133 (21) | 120 (9) | 0.056 | 0.036 |
The column labelled ‘P (child SEP)’ denotes the P-value for tests (‘Methods’ section) between the childhood SEP groups, LL + LH and HL + HH The column labelled ‘P’ denotes the P-value resulting from tests across the four SEP groups: LL, LH, HL and HH.
aFor a few statistics, the number of participants per group is slightly less because statistics were not available for a few participants.
HbA1c, a glycated or glycoslated haemoglobin; HDL, high density lipoprotein.
Variation in DNA methylation between all individuals
Whole genome promoter methylation profiles were correlated with published expression data for whole blood cells,50 and show an inverse correlation, as expected12 (Figure 1A). To determine the magnitude and breadth of inter-individual differences across the panel of methylation profiles, we identified all promoters on the microarray that showed differential methylation: 9112 variable probes were identified from the total of 223 359 on the microarray. These probes correspond to 6176 gene promoters each with at least one variable probe (Supplementary Spreadsheet 1, available as Supplementary Data at IJE online). Pathway analysis of this set of all variant promoters via Gene Ontology51 shows enrichment for genes involved in DNA packaging (FDR < 0.2). Figure 1B depicts the distribution of these promoters across the entire genome and highlights several regions with unexpectedly high or low numbers of variable promoters (FDR < 0.025). One example is the region highlighted on chromosome 5 (chr5:99024242–109024242), in which >60% of the genes were variably methylated.
To see if any aspect of SEP is captured by overall methylation variability, unsupervised hierarchical clustering was applied to probes obtained from the 500 most variable promoters. Figure 1C reveals a large cluster (n = 13; highlighted in blue) enriched with high SEP individuals, particularly the HH subgroup (P ≤ 0.009; hypergeometric). This unbiased, class discovery approach confirms that SEP differences contribute to the epigenetic variation between these 40 males.
Variation in DNA methylation between SEP groups
Methylation differences between SEP groups were obtained using linear models at the probe level and then a sliding window to identify short genomic regions enriched with similar differences. We identified 1252 gene promoters whose methylation levels are associated with childhood SEP, 666 more methylated in high childhood SEP and 586 promoters more methylated in low childhood SEP (Supplementary Spreadsheet 2, available as Supplementary Data at IJE online). A heatmap of the probes in the 500 gene promoters that best differentiate between low and high childhood SEP is shown in Figure 1D. We similarly identified 545 promoters containing methylation levels associated with adulthood SEP: 336 more methylated in high adult SEP; 209 more methylated in low adult SEP (Supplementary Spreadsheet 2, available as Supplementary Data at IJE online). Childhood and adulthood SEP-associated promoter sets overlap at only 63 promoters. Table 2 indicates functional signalling categories and pathways enriched with genes whose promoter methylation is associated with childhood SEP.
Table 2.
Functional signalling categories and pathways enriched (FDR ≤ 0.2) with genes whose promoter methylation is associated with childhood SEP
| More methylated in high childhood SEP | More methylated in low childhood SEP | |
|---|---|---|
| Extra-cellular signalling | NTRK2 (BDNF receptor) | Sensory perception of smell and taste, hormone-mediated signalling |
| Intra-cellular signalling | MAPK signalling | |
| DNA signalling | DNA packaging, targets of several transcription factors, MIR-510 targets | DNA methylation machinery (MBD4,HEMK2), DICER1 |
| Metabolism | Linoleic acid metabolism |
B- to T-cell ratios in blood are known to fluctuate so certain methylation differences between individuals could be caused by different cell ratios, particularly in promoters of genes with cell-type specific methylation. To rule out this possibility, we used recently published MeDIP datasets to compare methylation differences between B and T cells to methylation differences between SEP groups. More specifically, for each probe in our childhood SEP analysis, we identified the nearest probe in the T/B cell analysis (the two studies use different microarray formats). If B- to T-cell ratios explained the methylation differences in the childhood SEP analysis, then we would expect probes showing extreme methylation differences between B and T cells to correspond to similar methylation differences between the childhood SEP groups. Our results did not support this possible explanation: 215 of 318 probes have modified t-statistics >2 in one analysis and <−2 in the other analysis. Similar results were obtained for adulthood SEP.
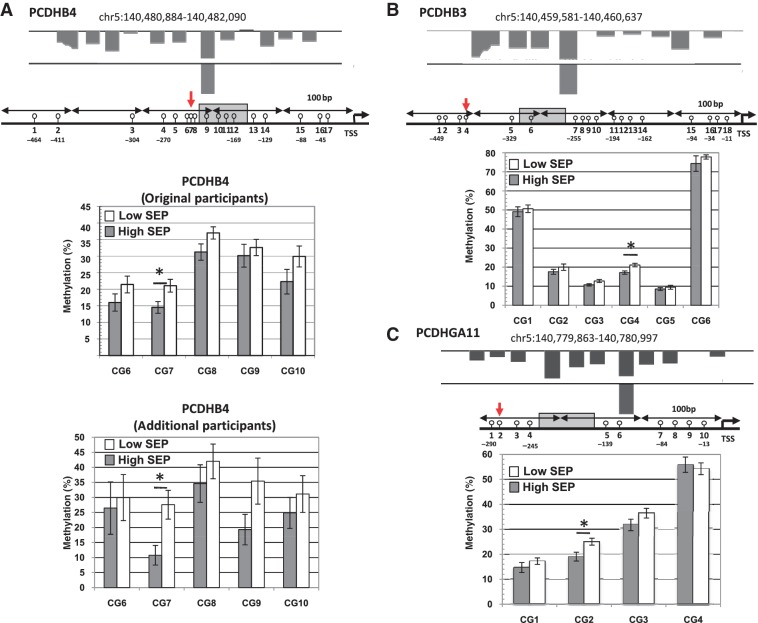
Among the differentially methylated promoters was the cluster of protocadherin genes on chromosome 5, with higher methylation associated with low childhood SEP (Figure 2). The protocadherin superfamily of genes encodes proteins involved in cell–cell adhesion/communication and synaptogenesis: three protocadherin gene promoters were selected for validation by pyrosequencing of bisulfite-treated DNA. Figure 2 shows consistently higher methylation for low child SEP across 14 of 15 CpG sites examined, thus confirming the differential methylation associations with childhood SEP observed using MeDIP. For PCDHB4 only, we used pyrosequencing of bisulfite-treated DNA in the additional 40 males: methylation associations with childhood SEP were replicated in this additional group of males (Figure 2). We genotyped the validated regions of PCDHB4 to verify that DNA methylation differences and not simply sequence polymorphism were responsible for the observed childhood SEP differences (Figure S5, available as Supplementary Data at IJE online). One sequence difference was uncovered, a T/G polymorphism at position −397 relative to the transcription start site of PCDHB4 that did not affect any CpG. Validation of additional genes is described in Supplementary information (Figure S7, available as Supplementary Data at IJE online).
Figure 2.
Mapping of the state of methylation of three protocadherin genes by pyrosequencing. Effect size (P-value) for three protocadherin promoters were as follows: [effect = log(mean probe intensity in high child SEP/mean probe intensity in low child SEP)] PCDHB4 effect = −0.47, P-value < 0.00114, q-value < 0.015; PCDHB3 effect = −0.34, P-value < 0.013, q-value < 0.023; PCDHGA11 effect = −0.53, P-value < 0.0051, q-value < 0.034. In the figure, the upper panel is a UCSC browser genomic display showing mean DNA methylation differences between high and low childhood SEP and the significant differential probe as detected by microarray analysis (A, B and C). Grey bars correspond to probes more methylated in the low childhood SEP group. The chromosomal location of each analysed region is indicated (hg18: human genome 18 assembly). CpG sites are annotated relative to the transcription start site of each gene. Each circle represents one CpG site. The grey box indicates the probe significantly different as shown by the microarrays data. The bar graphs show the average methylation levels of the CpG sites within the high and low childhood SEP groups as determined by pyrosequencing. Error bars show the SEM (*P < 0.05). (A) Methylation profile of the PCDHB4 promoter in childhood high and low SEP groups. In the PCDHB4 promoter, five CpG sites were analyzed by pyrosequencing (high SEP n = 17, low SEP n = 16). On average, all of the CpG sites have higher methylation in the low SEP group (P = 0.0043, Stouffer) with CG7 showing a significant difference (P = 0.018, t-test). The lower bar graph shows pyrosequencing average methylation levels for the same five CpGs in additional 30 participants (high SEP n = 13, low SEP n = 17). All five CpG sites are more methylated in the low SEP group with CG7 significantly different (P = 0.011, t-test). (B) Methylation profile of the PCDHB3 promoter in childhood high and low SEP groups. In the promoter of the PCDHB3 gene, six CpG sites were analysed by pyrosequencing (high n = 14, low n = 19). Though all CpG sites have higher methylation in the low SEP group, only CG4 is significantly different (P = 0.0049, t-test). (C) Methylation profile of the PCDHGA11 promoter in childhood high and low SEP groups. In the promoter of the PCDHGA11 gene, four CpG sites were analysed by pyrosequencing (high n = 16, low n = 20). CG2 shows significantly higher methylation in the low childhood SEP group (P = 0.010, t-test)
Potential functional relevance of childhood SEP-associated methylation
Having demonstrated differential promoter methylation associated with childhood SEP at 1252 gene promoters, raises questions about the functional relevance of these genes given that DNA methylation can influence gene expression. Whilst any comparison with disease outcome is highly tenuous, given the rudimentary understanding of genetic influences on complex diseases, such as those associated with SEP listed in Table S2 (available as Supplementary Data at IJE online), our analyses identified that the differentially methylated genes are overrepresented in particular pathways and functional categories. Cell signalling was prominent including pathways and categories involved in extra-cellular, intra-cellular, DNA and metabolic signalling (Table 2 and Supplementary Spreadsheet 3, available as Supplementary Data at IJE online). Childhood SEP-associated methylation is most overrepresented in the MAPK signalling pathway (Figure S6, available as Supplementary Data at IJE online). In general, high childhood SEP individuals have increased promoter methylation relative to those with low childhood SEP. Figure S6 (available as Supplementary Data at IJE online) also illustrates differential methylation in the chemokine signalling pathway that promotes signalling cascades to generate cellular responses, many of which involve the MAPK signalling pathway. Despite this functional relationship, promoters of genes in the chemokine signalling pathway are generally more methylated in low SEP individuals, the opposite of the pattern seen in the MAPK signalling pathway. Thus, functionally related pathways appear to be affected in distinct directions.
SEP-associated methylation clusters by genomic location
Aberrant DNA methylation and gene expression clusters across large genomic regions in various cancers.52,53 Our data reveal similar megabase-sized regions enriched for differentially methylated promoters between high- and low-SEP individuals. For example, Figure 3A shows methylation differences between SEP groups in gene promoters across chromosome 19, with higher methylation on average in the high compared with the low childhood SEP group. Exceptions include a 5-Mb region (highlighted in Figure 3A) that is consistently less methylated in high childhood SEP individuals and contains mainly zinc finger genes. Interestingly, this region is consistently more methylated in association with high adult SEP, suggesting that this region is epigenetically variable. A second example is a chromosome 11 region, where most genes are olfactory receptors, whose promoters are consistently more methylated in the low childhood SEP group (Figure 3B). A third example is a chromosome 5 region containing protocadherins alpha, beta and gamma, genes mainly involved in cell adhesion, with more methylation in low childhood SEP than in high (Figure 3C). Interestingly, this region overlaps almost exactly with two regions found to be consistently hypermethylated in breast tumours,53 and is differentially methylated in rat hippocampus that were exposed to differential maternal care early in life.54
We determined the range of co-clustering of SEP differentially methylated promoters. We covered the entire genome with 500-kb windows at 250-kb spacing. We then computed a summary statistic for each window, indicating its enrichment for promoters differentially methylated in the same direction. Figure 3B and 3C highlight such regions with enrichment (FDR < 0.05). Nearly 800 such 500-kb regions are enriched across the genome. We computed the correlations between differential probe statistics within 500-kb regions at set distances apart. Figure 4 shows that, on average, methylation differences in 500-kb regions tend to be unexpectedly similar at distances up to about two million bases apart. Childhood SEP has higher correlation for windows up to 2 Mb apart (P ≤ 0.008), whereas adulthood SEP has higher correlation for windows 2–4 Mb apart (P ≤ 0.008). Childhood SEP correlation drops into the 95% CI by 2 Mb, whereas adulthood SEP correlation remains outside up to 4 Mb. This clustering by genomic location in association with SEP is a notable finding of our study.
Discussion
Our study provides evidence of extensive, clustered and genome-wide variation in promoter DNA methylation in middle-aged males, some of which associates with SEP. Clustering of all participants by methylation levels of the 500 most variably methylated promoters revealed a cluster of high SEP in early and adult life, thus showing that SEP, or something correlated with it, contributes to general DNA methylation variation. More surprising was the number of promoters (1252 promoters) associated with childhood SEP, given that blood was taken at age 45 years. Importantly, there was little overlap in the differentially methylated promoters associated with child and adult SEP.
Several methodological considerations arise in our study. First, there is currently no ‘gold standard’ for measuring the methylome, yet MeDIP is a well-established genome-wide method that has been evaluated,33–39 our analyses included triplicate arrays and methylation differences were confirmed in a few selected genes using other gene-specific methods. Secondly, with DNA available only in adulthood, we cannot establish the life stage when SEP associations with methylation were manifest. For example, our findings for childhood SEP may reflect prenatal environment or even earlier parental circumstances as summarized by commonly used occupation and wealth (housing)-based indicators.55,56 Thirdly, with access only to whole-blood DNA, we cannot know the extent to which our results relate to other tissues (e.g. in the hippocampus27) or to gene expression. Use of whole blood also raises the possibility that SEP-linked differences in B- to T-cell ratios might account for some of our observations. We have partly addressed this issue by noting that B-cell and T-cell expression and methylation profiles50,57 do not differ for many of the genes whose promoters contain SEP-associated methylation levels. Fourthly, our study design does not allow us to extrapolate from methylation differences to disease risk, with the small sample ascertained according to exposure (SEP) and not health outcomes. Notably, the study benefits from prospective data ascertainment for SEP from a population-based birth cohort. But such cohort studies following individuals over many decades of life are potentially vulnerable to bias associated with loss to follow-up, as reported previously for this cohort.58 Ranking male participants, we achieved a balanced design for child and adult SEP, except for a minor imbalance within the child SEP groups where the proportions with high adult SEP varied (i.e. 42 and 52%, respectively for high and low child SEP). A major impact of this slight imbalance on our results is unlikely in this study, which describes a preliminary, rather than definitive, epigenetic signature associated with childhood SEP.
Our finding of a methylation signature of early-life SEP in adult blood DNA taken at 45 years is consistent with epigenetic mechanisms contributing to the association between early-life SEP and adult health. Our observation of an early-life methylation signature evident decades later is in line with other evidence for epigenetic associations with prenatal famine in 60-year-old adults.25,29 Using a broad composite SEP measure, we found a distinct contribution of child from adult SEP, with little overlap of differentially methylated promoters, justifying further studies focussing on specific early-life exposures.
Another striking feature of our study is the fact that SEP-associated methylation levels are not uniformly distributed across the genome but instead form clusters in large genomic regions, as seen in various cancers,52,53 with childhood SEP showing the largest and most pronounced clusters. This finding suggests that the methylation differences between SEP groups are the result of a systematic epigenomic organization rather than simply the methylation differences one might expect to see between groups of randomly selected individuals. Genes showing SEP-associated promoter differential methylation are enriched for basic cell functions including several different modes of signalling, which in turn suggest plausible routes by which early environment might become biologically embedded and influence future disease. Our preliminary study therefore suggests that further investigations are warranted that explore epigenomic responses to social environment in humans, highlighting the importance of early-life experience.
Supplementary Data
Supplementary Data are available at IJE online.
Funding
Grants from the Canadian Institute of Health Research; the National Cancer Institute of Canada; the Canadian Institute for Advanced Research; the Sackler program in Psychobiology and Epigenetics at McGill University (to M.S.); fellowship from the Canadian Institute of Health Research and the Fragile X Research Foundation of Canada (to N.B.); the UK Medical Research Council (MRC) (grant G0000934 to the clinical examination and DNA banking of the 1958 cohort). Work undertaken at Great Ormond Street Hospital/University College London, Institute of Child Health is in part supported by funding from the Department of Health's National Institute of Health Research (‘Biomedical Research Centres' funding), and MRC provides funds for the MRC Centre of Epidemiology for Child Health. Laboratory work in the Department of Social Medicine, University of Bristol was supported in part by a grant to MP from the Welcome Trust.
KEY MESSAGES.
Genome-wide, DNA methylation analysis of adult blood DNA shows substantial differences in gene promoter methylation status between 40 males from the UK 1958 birth cohort selected for the extremes of SEP in childhood and/or adulthood.
Participants with high SEP as both children and adults have a distinct DNA methylation profile within the overall variation in gene promoter methylation in this group of 40 males, showing SEP differences contribute to overall epigenetic variation in these individuals.
Despite the analysis being on adult DNA, the greatest number of promoter methylation differences were associated with differences in childhood SEP rather than adult SEP.
Genomically, the SEP promoter methylation associations cluster in megabase-sized regions.
Supplementary Material
Acknowledgements
This study was designed by C.P., C.H. and M.Sz. Participants were selected from socio-economic extremes by C.P. and C.H. Development of DNA bank and access to participant blood samples was by C.P., M.P. and W.M. DNA banking and processing was by W.M. Methylation analysis was completed by N.B. with the assistance of A.R. Bioinformatic analysis was performed by M.Su. under the supervision of M.H. The entire process was overseen by M.Sz. N.B., M.H., C.H., M.P., C.P., M.Su. and M.Sz. all contributed to writing the article.
References
- 1.Acheson D. Inequalities in Health: Report of an Independent Inquiry. London: HMSO; 1998. [Google Scholar]
- 2.Berkman LF, Kawachi IO. Social Epidemiology. New York: Oxford University Press; 2000. [Google Scholar]
- 3.WHO Commission on Social Determinants of Health, World Health Organization. Closing the Gap in a Generation: Health Equity through Action on the Social Determinants of Health: Commission on Social Determinants of Health Final Report: Executive Summary. Geneva, Switzerland: World Health Organization, Commission on Social Determinants of Health; 2008. [Google Scholar]
- 4.Galobardes B, Lynch JW, Davey Smith G. Childhood socioeconomic circumstances and cause-specific mortality in adulthood: systematic review and interpretation. Epidemiol Rev. 2004;26:7–21. doi: 10.1093/epirev/mxh008. [DOI] [PubMed] [Google Scholar]
- 5.Gluckman PD, Hanson MA, Bateson P, et al. Towards a new developmental synthesis: adaptive developmental plasticity and human disease. Lancet. 2009;373:1654–57. doi: 10.1016/S0140-6736(09)60234-8. [DOI] [PubMed] [Google Scholar]
- 6.Hertzman C, Power C. Health and human development: understandings from life-course research. Dev Neuropsychol. 2003;24:719–44. doi: 10.1080/87565641.2003.9651917. [DOI] [PubMed] [Google Scholar]
- 7.Kuh D, Ben-Shlomo Y. A Life Course Approach to Chronic Disease Epidemiology. 2nd edn. New York: Oxford University Press; 2004. [Google Scholar]
- 8.Graham H, Power C. Childhood disadvantage and health inequalities: a framework for policy based on lifecourse research. Child Care Health Dev. 2004;30:671–78. doi: 10.1111/j.1365-2214.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- 9.Hertzman C. The biological embedding of early experience and its effects on health in adulthood. Ann N Y Acad Sci. 1999;896:85–95. doi: 10.1111/j.1749-6632.1999.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller GE, Chen E, Fok AK, et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA. 2009;106:14716–21. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mutagen. 2008;49:46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- 12.Razin A. CpG methylation, chromatin structure and gene silencing - a three-way connection. EMBO J. 1998;17:4905–08. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg AP. Epigenomics reveals a functional genome anatomy and a new approach to common disease. Nat Biotechnol. 2010;28:1049–52. doi: 10.1038/nbt1010-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan X, Cross S, Bird A. Gene silencing by methyl-CpG-binding proteins. Novartis Found Symp. 1998;214:6–16. doi: 10.1002/9780470515501.ch2. [DOI] [PubMed] [Google Scholar]
- 15.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–82. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–72. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLennan NK, James SJ, Melnyk S, et al. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics. 2004;18:43–50. doi: 10.1152/physiolgenomics.00042.2004. [DOI] [PubMed] [Google Scholar]
- 20.Ke X, Lei Q, James SJ, et al. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics. 2006;25:16–28. doi: 10.1152/physiolgenomics.00093.2005. [DOI] [PubMed] [Google Scholar]
- 21.Weaver IC, Diorio J, Seckl JR, Szyf M, Meaney MJ. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann NY Acad Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- 22.Sinclair KD, Allegrucci C, Singh R, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104:19351–56. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terry MB, Ferris JS, Pilsner R, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borghol N, Lornage J, Blachere T, Sophie Garret A, Lefevre A. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics. 2006;87:417–26. doi: 10.1016/j.ygeno.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–49. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 27.McGowan PO, Sasaki A, D'Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–48. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterland RA, Kellermayer R, Laritsky E, et al. Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 2011;6:e1001252. doi: 10.1371/journal.pgen.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feinberg AP, Irizarry RA, Fradin D, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2:49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Power C, Matthews S. Origins of health inequalities in a national population sample. Lancet. 1997;350:1584–89. doi: 10.1016/S0140-6736(97)07474-6. [DOI] [PubMed] [Google Scholar]
- 32.Power C, Atherton K, Strachan DP, et al. Life-course influences on health in British adults: effects of socio-economic position in childhood and adulthood. Int J Epidemiol. 2007;36:532–39. doi: 10.1093/ije/dyl310. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–90. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia J, Pekowska A, Jaeger S, Benoukraf T, Ferrier P, Spicuglia S. Assessing the efficiency and significance of Methylated DNA Immunoprecipitation (MeDIP) assays in using in vitro methylated genomic DNA. BMC Res Notes. 2010;3:240. doi: 10.1186/1756-0500-3-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- 37.Rajendram R, Ferreira JC, Grafodatskaya D, et al. Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms. Epigenetics. 2011;6:410–15. doi: 10.4161/epi.6.4.14763. [DOI] [PubMed] [Google Scholar]
- 38.Robinson MD, Stirzaker C, Statham AL, et al. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res. 2010;20:1719–29. doi: 10.1101/gr.110601.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Zhang K, Dai W, et al. Systematic evaluation of genome-wide methylated DNA enrichment using a CpG island array. BMC Genomics. 2011;12:10. doi: 10.1186/1471-2164-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–53. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 41.Soric B. Statistical ‘discoveries' and effect-size estimation. J Amer Statist Assoc. 1989;84:608–10. [Google Scholar]
- 42.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 45.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.OMIM. Online Mendelian Inheritance in Man. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University and National Center for Biotechnology Information, National Library of Medicine; 2009. p. April 15, 2009. http://www.ncbi.nlm.nih.gov/omim/ (5 August 2009, date last accessed)
- 50.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–67. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–49. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 53.Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68:8616–25. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGowan PO, Suderman M, Sasaki A, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galobardes B, Shaw M, Lawlor DA, Lynch JW, Davey Smith G. Indicators of socioeconomic position (part 2) J Epidemiol Community Health. 2006;60:95–101. doi: 10.1136/jech.2004.028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galobardes B, Shaw M, Lawlor DA, Lynch JW, Davey Smith G. Indicators of socioeconomic position (part 1) J Epidemiol Community Health. 2006;60:7–12. doi: 10.1136/jech.2004.023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakyan VK, Down TA, Thorne NP, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–29. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atherton K, Fuller E, Shepherd P, Strachan DP, Power C. Loss and representativeness in a biomedical survey at age 45 years: 1958 British birth cohort. J Epidemiol Community Health. 2008;62:216–23. doi: 10.1136/jech.2006.058966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.