Background: Cardiovascular implants can become infected with Staphylococcus aureus.
Results: Receptor proteins on S. aureus form a multivalent cluster bond with fibronectin, a human protein that coats implants.
Conclusion: A more resilient bond is associated with infections observed in vivo.
Significance: Normal microbial flora could be screened prior to surgery to determine risk in patients receiving cardiovascular implants.
Keywords: Atomic Force Microscopy, Bacteria, Biofilm, Cell Surface Protein, Kinetics, MSCRAMM, Cluster Bond, Fibronectin, Force Spectroscopy, Infection
Abstract
Staphylococcus aureus is part of the indigenous microbiota of humans. Sometimes, S. aureus bacteria enter the bloodstream, where they form infections on implanted cardiovascular devices. A critical, first step in such infections is a bond that forms between fibronectin-binding protein (FnBP) on S. aureus and host proteins, such as fibronectin (Fn), that coat the surface of implants in vivo. In this study, native FnBPs on living S. aureus were shown to form a mechanically strong conformational structure with Fn by atomic force microscopy. The tensile acuity of this bond was probed for 46 bloodstream isolates, each from a patient with a cardiovascular implant. By analyzing the force spectra with the worm-like chain model, we determined that the binding events were consistent with a multivalent, cluster bond consisting of ∼10 or ∼80 proteins in parallel. The dissociation rate constant (koff, s−1) of each multibond complex was determined by measuring strength as a function of the loading rate, normalized by the number of bonds. The bond lifetime (1/koff) was two times longer for bloodstream isolates from patients with an infected device (1.79 or 69.47 s for the 10- or 80-bond clusters, respectively; n = 26 isolates) relative to those from patients with an uninfected device (0.96 or 34.02 s; n = 20 isolates). This distinction could not be explained by different amounts of FnBP, as confirmed by Western blots. Rather, amino acid polymorphisms within the Fn-binding repeats of FnBPA explain, at least partially, the statistically (p < 0.05) longer bond lifetime for isolates associated with an infected cardiovascular device.
Introduction
Indwelling medical devices have had a significant positive impact on human health, and their use is increasing worldwide. Ironically, these life-saving devices can become the source of infection if bacteria colonize them. The average rate of infection of surgical implants is considerable, 2–40% (1). Furthermore, the incidence of infection of some types of medical devices is increasing at a rate faster than the actual implantation of the devices themselves (2). Mortality attributable to device-related infections is highest among patients with cardiovascular implants, particularly prosthetic heart valves (1, 3).
Bacterial adhesion to surfaces is the critical initial step in the infection of implanted medical devices. Staphylococcus aureus is currently the leading cause of infections of implanted cardiovascular devices (3, 4). S. aureus express surface protein receptors called microbial surface components recognizing adhesive matrix molecules (MSCRAMMs)3 (5) that allow them to adhere to host ligands like fibronectin (Fn). Fn is an extracellular matrix protein that is widely distributed in the tissues of vertebrates and is a potential ligand for a variety of bacterial cells (6). Fn is also known to coat the surface of implanted materials in contact with the human bloodstream (7, 8), particularly those that remain in the body for extended periods of time such as cardiac prostheses (1, 9). The bacterial surface proteins that bind to human Fn are referred to as fibronectin-binding proteins A and B (FnBPA and FnBPB). These FnBPs have been shown to play an important role in S. aureus infection of medical implants in vivo (10, 11).
In this study, we used atomic force microscopy (AFM) to investigate the binding reaction between an Fn-coated substrate and putative FnBPs on living S. aureus in buffer solution. Experiments were conducted on individual cells from each of 46 different S. aureus isolates obtained from bacteremic patients with cardiac devices. These isolates were grouped according to clinical evidence of S. aureus infection. One group of patients had infected cardiac devices (CDI; n = 26), and the other group of patients had uninfected cardiac devices (CDU; n = 20).
The AFM measurements presented herein complement other recent investigations that utilized techniques such as x-ray crystallography, NMR, and calorimetry (12–14) to study binding between fragments of Fn and FnBPs. Our force data on actual clinical isolates of S. aureus suggest that molecules of FnBPA and FnBPB, expressed in their native state on living bacteria, form a molecular cluster bond with Fn, and the tensile resilience of this bond determines, at least in part, which patients develop infections on implanted cardiovascular devices.
The dissociation rate constant (koff, s−1) of the Fn–FnBP bond was experimentally determined by measuring the rupture force at different loading rates. The binding mechanism could be modeled with the Bell equation of dissociation modified for multivalent interactions (15–17). The effective dissociation rate constant for the CDI isolates was significantly smaller than that of the CDU group (p < 0.05). This difference was not due to the increased amount of FnBP in CDI isolates, as determined by real-time PCR and Western blots. However, amino acid polymorphisms in FnBPA (E652D, H782Q, and K786N) from CDI isolates appear to explain, at least partially, the increased bond lifetime (1/koff). This was confirmed by conducting AFM analyses with a synthetic peptide containing two of the three polymorphisms.
EXPERIMENTAL PROCEDURES
Bacteria Isolates and Growth Conditions
Bacterial isolates came from the S. aureus bacteremia registry at Duke University Medical Center (Durham, NC). CDI (n = 26) isolates came from patients with an infected permanent pacemaker, implantable cardioverter defibrillator, or prosthetic cardiac valve. Infection was confirmed microbiologically with a positive culture from a prosthetic device, generator pocket, or electrode lead. Clinical confirmation of infection was assessed by echocardiogram (e.g. vegetations on valve or lead) or diagnosis of infective endocarditis (18). Cardiac device uninfected (CDU; n = 20) isolates were obtained from the bloodstream of patients in whom there was no evidence of device infection at the time of initial blood culture, in whom the cardiac device was not removed, and in whom there was no evidence of recurrent infection 12 weeks after the onset of bacteremia (or no evidence of device infection at autopsy).
Lower et al. (19) have shown that CDU is a valid control group as it has statistically similar characteristics as the CDI group. For example, demographic attributes of human patients (age, race, sex, time of implantation, type of device) are similar for both CDI and CDU (supplemental Table S1). Isolates from both groups share similar ancestral lineage as clonal complex 5, clonal complex 15, and clonal complex 30 account for ∼50% of all isolates from both CDI and CDU (supplemental Table S2). Finally, the CDI and CDU groups have similar genotypic characteristics in terms of 10 MSCRAMM adhesins including, bbp, clfA, clfB, cna, ebpS, efb, icaA, fnbA, fnbB, and map/eap (supplemental Table S2).
Cryogenically preserved isolates were cultured to exponential phase (A600 = 0.51 ± 0.02) at 37 °C in tryptic soy broth so that they expressed MSCRAMMs including FnBP (7, 20, 21). Cell suspensions (∼1 ml) were harvested via centrifugation (5000 × g for 3 min), washed three times in saline solution (0.1 m NaCl), and deposited onto clean glass (22) or Fn-coated slides (BD Biosciences). The slides were gently rinsed with phosphate-buffered saline (PBS; pH 7.2) after 5 min. Control experiments were performed with S. aureus DU5883, an fnbA fnbB double mutant that has lost the ability to attach to Fn, as well as strains that overexpress fnbA (DU5883 pFNBA4) and fnbB (DU5883 pFNBB4) (7).
Quantitative Real-time PCR Analysis of fnbA
Expression of fnbA, the gene for FnBPA, was determined as described by Lower et al. (19). Briefly, total RNA was isolated from midexponential phase cultures (26 CDI isolates, 20 CDU isolates) using the RNeasy kit (Qiagen). Approximately 2 μg of RNA was converted to cDNA using the iScript one-step RT-PCR kit (Bio-Rad). Real-time PCR (RT-PCR) reactions contained 2 μl of cDNA, 1 pmol of forward and reverse primers, 8.5 μl of nuclease-free deionized water, and 12.5 μl of IQ SYBR Green supermix (Bio-Rad). RT-PCR was performed using an iCycler detection system (Bio-Rad). The normalized amount of transcript for each gene was expressed as the n-fold difference relative to the control gene (2ΔCT, where ΔCT represents the difference in threshold cycle between the target gene and the 16 S rRNA gene). Samples were performed in triplicate.
Western Ligand Blots
Surface expression of FnBP in S. aureus was determined as described by Bisognano et al. (23). Briefly, exponential cultures of isolates (10 CDI isolates, 10 CDU isolates, and their duplicates) were washed twice in PBS, pelleted by centrifugation (10,000 × g, 10 min), and lysed in 1.5 ml of PBS with 1 mm Ca2+ and 0.5 mm Mg2+ containing 20 μg/ml lysostaphin, 20 μg/ml DNase, and a protease inhibitor mixture (Pierce). Protein concentration was determined by the BCA method (Pierce). The equivalent of 20 μg of protein per isolate was separated by SDS-PAGE (12% separating gel) and electroblotted onto polyvinylidene difluoride membranes (Invitrogen).
Membranes were blocked with StartingBlock T20 (TBS) blocking buffer (Invitrogen). Blots were incubated with 30 μg/ml Fn for 1 h followed by an anti-N-terminal Fn antibody (1:5,000; Chemicon) for 1 h. The secondary antibody was anti-mouse IgG coupled to peroxidase (Pierce). Detection was performed with an ECL kit (Pierce). FnBPA and FnBPB were identified by their apparent molecular weights as compared with those of reference strains DU5883 pFNBA4 and DU5883 pFNBB4, respectively (7). DU5883 (7) was used as a negative control. Due to the similar electrophoretic mobilities on SDS-PAGE gels, FnBPA and FnBPB were not discriminated from each other. Therefore, total FnBP concentrations were estimated with densitometry by ImageJ (version 1.45s). Band density values were normalized to the native band expressed on the reference strain DU5883 pFNBA4.
AFM Measurements on Live Bacteria
We used a Veeco/Digital Instruments Bioscope AFM and Nanoscope IV controller with an attached inverted microscope (Axiovert 200M, Zeiss) as described in Refs. 24 and 25. A single approach-retraction cycle took 0.5–22 s. Calibrated Si3N4 cantilevers were coated with a solution of 100 μg/ml Fn (Sigma-Aldrich) according to Refs. 24 and 25. Some cantilevers were coated with bovine serum albumin (BSA) to characterize nonspecific interactions according to Ref. 26 (see supplemental Experimental Procedures).
Force measurements were conducted in PBS solution at ambient temperature. An AFM tip was positioned over an S. aureus bacterium by using the inverted microscope attached to the AFM. An Fn-coated probe (tip radius 20–60 nm, spring constant 0.1 ± 0.06 nN nm−1) (27) was brought into contact with a bacterium and pushed against the cell wall so that each bacterium experienced the same contact force. The probe was then retracted away from the bacterium until complete separation. This process resulted in an approach force curve as well as a retraction force curve.
To ensure specificity and maintain a high level of stringency, retraction force spectra with two or more binding events were considered in this study. Only the final rupture peak (28–30) was included in the analysis of the Bell parameters (described below). Under this restriction, the percentage of spectra used was <10% for all S. aureus isolates.
Synthetic Peptides
Two 20-mer synthetic peptides (United Biosystems) were used in AFM experiments according to Ref. 19. Each peptide was linked to an AFM tip through a covalent bond between gold on the tip and the thiol group of a cysteine added at the C terminus of each peptide (31, 32). AFM measurements were performed in PBS with both commercially available Fn-coated slides (BD Biosciences) and homemade slides coated with 0.1% Fn-solution.
RESULTS AND DISCUSSION
S. aureus Infections on Cardiovascular Implants
Due to the increasing rate of cardiovascular device infections over the past decade, the American Heart Association and the European Society of Cardiology have published a list of recommendations regarding the prevention and management of infections caused by bacteria, particularly staphylococci such as S. aureus (33, 34). The source of such infections is a biofilm, or a sessile community of bacteria living on the surface of an implanted device (35). Biofilm-based infections are a multistep process that begins with the attachment of bacteria to the surface of the indwelling device.
Although some cardiovascular implants become infected, not all patients with implants develop an S. aureus infection even if bacteria are present in the bloodstream. We have collected bloodstream isolates of S. aureus from bacteremic patients who had permanent pacemakers, implantable cardioverter defibrillators, or prosthetic cardiac valves (n = 46). Although all of these patients had S. aureus in the bloodstream, only ∼60% developed an infected device. We hypothesized that these CDI isolates were able to form a stronger, more resilient bond with the surface of a cardiovascular implant, which allowed this group of isolates to form a biofilm-based infection.
In this study, we used AFM to examine the fundamental binding mechanism between a protein-coated substrate (i.e. proxy for implant) and receptor proteins expressed on the outer surface of each of the 46 different bloodstream isolates, which originated from two clinically distinct groups: (i) the CDI group mentioned above (n = 26), and (ii) bacteremic patients whose cardiovascular devices were not infected by the S. aureus present in their blood (CDU; n = 20). By focusing on clinical isolates of S. aureus, as opposed to type- or laboratory-derived strains, we are able to accentuate the relevancy to public health. Furthermore, by conducting AFM measurements on living bacteria, as opposed to purified receptor proteins, we preserve the native structure, orientation, architecture, number, etc. of proteins on the exterior cell wall of S. aureus.
Bond between Human Fibronectin and S. aureus Fibronectin-binding Protein
The binding mechanism that we chose to focus on is the reaction between human Fn and Fn-binding proteins produced by S. aureus. We chose to focus on Fn because it is a host protein that commonly coats the surface of cardiovascular implants and because Fn production is associated with endocardial trauma (1, 20, 35).
S. aureus produce two well characterized fibronectin-binding proteins (FnBPA and FnBPB). These cell wall-associated MSCRAMMs (5) enable S. aureus to attach to prostheses coated with ligands such as Fn. In fact, the binding reaction between host Fn and S. aureus FnBP is often cited as a critical initial step in prosthetic device infections (10, 11, 36).
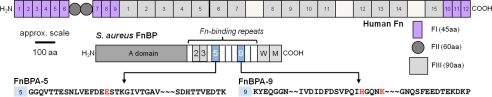
Fn and FnBPs are multivalent binding proteins that react with one another at a number of sites along their lengths (Fig. 1). The N-terminal type I modules (FI) of Fn are often identified as the binding site for FnBPs on S. aureus (37). A basic binding reaction was recently presented as a tandem β-sheet type of interaction between FI modules at the N terminus of Fn and a series of 11 ∼40-residue binding sites (FnBPA1 thru FnBPA11) located on FnBPA (12, 14). This prior work was based on elegant NMR and x-ray crystallography measurements on fragments of Fn and/or FnBP. More recent work with fragments and synthetic peptides of Fn and FnBP has revealed that each of the 11 Fn-binding repeats in FnBPA from S. aureus fall into one of two groups, low and high affinity sites (13, 38). Similar interactions were found between Fn and FnBPB also expressed on S. aureus (13, 38). It is worth noting that other binding sites have been identified along the lengths of Fn and FnBPs. For example, there is evidence that S. aureus binds to the C-terminal type I and III modules on Fn (39) through FnBPA (40).
FIGURE 1.
Structure of human Fn (at top) and S. aureus FnBP. FnBPA (shown here) has 11 Fn-binding repeats, each ∼40 residues (14). FnBPB is missing either the second or the third repeat (13). W, wall-spanning region; M, membrane-spanning region. The bottom panel shows the amino acid (AA) sequence of FnBPA-5 and -9 from the wild-type strain of S. aureus (NCTC 8325). Red letters highlight the location of three polymorphisms (E652D, H782Q, and K786N) that were statistically more common in FnBPA from S. aureus isolated from patients with infected cardiovascular devices (19).
Quantity of Fn-binding Protein Expressed by Clinical Isolates
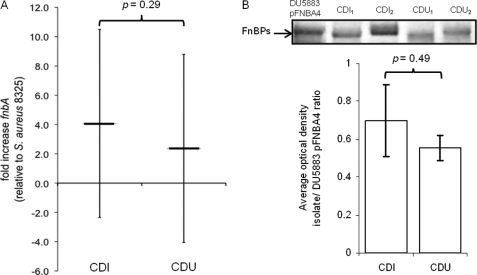
Because we were working with clinical isolates, we confirmed the presence and determined the amount of Fn-binding proteins on the various S. aureus specimens. PCR, as described in Ref. 19, was used to confirm that all isolates from the two groups had fnbA, the gene for FnBPA; all but four isolates from the CDI group also had fnbB, the gene for FnBPB (supplemental Table S2). The quantity of Fn-binding proteins was estimated with RT-PCR of fnbA expression and Western blot analysis. RT-PCR showed no statistical difference in the amount of fnbA expressed by isolates from the CDI versus CDU groups (p value = 0.29; Fig. 2A). Western blots confirm similar levels of FnBPs on the cell wall regardless of the clinical source of S. aureus (p value = 0.49; Fig. 2B). Taken together, these results indicate that all clinical isolates used in this study produce comparable levels of FnBPs on their external surface.
FIGURE 2.
A, quantitative real-time PCR of fnbA expression in S. aureus isolates from the CDI (n = 26) and CDU (n = 20) groups. Error bars indicate average (± 1 S.D.) -fold difference in fnbA expression level for each group relative to reference strain 8325-4. B, Western blot showing protein bands for FnBPs (top) from representative isolates of S. aureus from the CDI and CDU groups. Quantitative densitometry (bottom) of FnBP expression in CDI (n = 10) and CDU (n = 10) isolates normalized to FnBPA produced by S. aureus DU5883 pFNBA4, a strain that overexpresses fnbA.
Force Spectra of Fn–FnBP Bond
Fn-coated AFM tips were created by incubating a probe in a solution of 100 μg/ml Fn. This concentration of Fn was selected because it is close to that found in the human bloodstream (300–400 μg/ml (41, 42)), and other adhesion assays (e.g. optical microscopy counts, microtiter wells, and radiometric assays) have used a similar amount (10–50 μg/ml (36, 43)). Furthermore, the resulting Fn-coated tips produced only rare binding events on clean glass (≪30%), suggesting a surface coverage conducive to single molecule work (26).
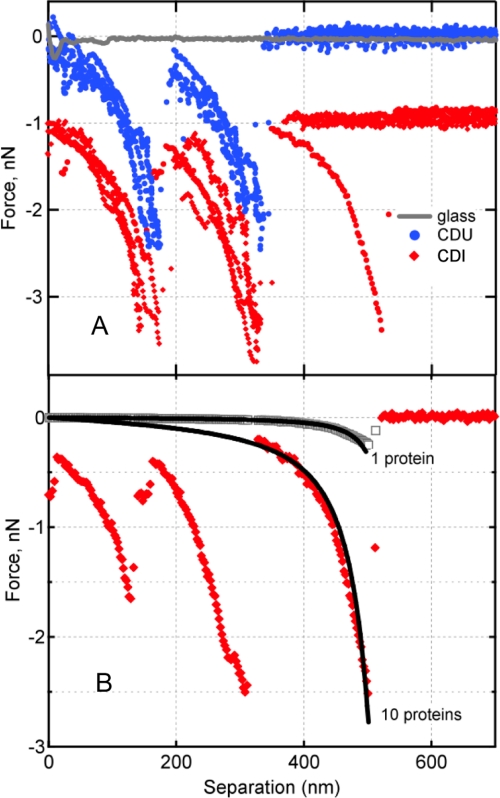
Although an Fn tip on glass produced very few binding events (Fig. 3A), something very different was observed when the Fn tip was used on S. aureus. As shown in Fig. 3A, for both CDI and CDU isolates, we observed distinct, nonlinear unbinding or rupture events of hundreds of piconewtons (10−12 N). Such nonlinear, force-distance profiles have been attributed to the forced extension or unfolding of linear polymers such as polypeptides (e.g. see Refs. 44–46), in this case, the unraveling of the Fn-FnBP complex.
FIGURE 3.
A, representative force spectra between Fn and putative FnBP expressed on the external envelop of S. aureus isolates from patients with an infected cardiac device (CDI, red diamonds) or the bloodstream of patients with an uninfected cardiac device (CDU, blue circles). Nonspecific adhesion (attractive interaction at separation ∼0 nm) (68) between Fn and a glass surface is shown in gray. By convention, negative values indicate attractive force in nanonewtons (10−9 N). The CDI traces are offset vertically to ease visual comparison. B, the worm-like chain model (Equation 1) was used to describe the force-extension profile for 1 protein or 10 parallel proteins (black curves). Following the protocol of Refs. 16 and 17, the 10-bond scenario was calculated as the summation of 10 individual polypeptide chains. Also shown are two experimentally measured protein extension traces preceding the specific bond rupture events. These show the rupture of one bond, observed rarely for Fn tip on clean glass (gray), versus 10 Fn–FnBP bonds, commonly observed for Fn tip on S. aureus (red).
Control experiments were performed to determine the specificity of these binding events. For example, an uncoated AFM tip did not elicit this response on S. aureus. However, this same type of binding event was observed when an Fn tip was used on strains of S. aureus that overexpress FnBPA or FnBPB (e.g. see Ref. 25). The frequency of these types of binding events was 0.43 ± 0.13 and 0.40 ± 0.07 for strains of S. aureus 8325-4 that overexpress FnBPA (DU5883 pFNBA4) and FnBPB (DU5883 pFNBB4), respectively. Binding events were significantly diminished (frequency = 0.12 ± 0.02) for a double mutant S. aureus strain (DU5883) that cannot produce either FnBPA or FnBPB. Taken together, this indicates that the AFM is able to probe a specific interaction between Fn and Fn-binding receptors on the surface of the S. aureus clinical isolates, and such interactions take the form of a unique, nonlinear force signature. Furthermore, the magnitude of these force signatures suggests that native FnBP on living cells forms a strong conformational structure when it binds to its ligand, as suggested by circular dichroism spectra, NMR, and x-ray crystallography of purified or recombinant FnBP (14, 37).
Number of Proteins That Participate in Bond
Force magnitudes near the nanonewton level, like those in Fig. 3A, are too large for single molecular pairs. Rather, they are indicative of multivalent bonds, which is not a surprising finding as one FnBP molecule can bind as many as nine molecules of Fn (47, 48). The mechanical signature observed in Fig. 3A can be used to estimate the number of proteins that participate in binding, as described by Refs. 16, 17, 49 and 50. This is accomplished by comparing the observed force spectra with a model (e.g. worm-like chain, WLC) describing the forced extension of a linear polymer such as a protein. The WLC model is
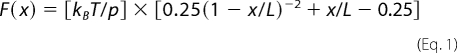 |
where F (newtons) is the force associated with the mechanical extension of a polypeptide to distance x (meters), kB is Boltzmann's constant (1.381 × 10−23 J K−1), T is temperature (kelvin), p is the persistence length (0.4 nm for a single protein molecule (45, 51)), and L is the contour length, or extended length of the protein when it breaks free of the AFM tip. Fig. 3B shows the force-extension relationship for one protein chain tethered between the AFM tip and a sample. The observed forces for S. aureus are much greater than the elastic force profile of a single protein. The WLC model can also be used to describe multiple, parallel protein tethers by simply summing individual WLC expressions for each protein molecule (e.g. 10 identical protein chains in parallel exert a force 10 times that of a single chain) (16, 22). Consistently, protein-stretching traces preceding the rupture event could be described by the WLC as multiple proteins in parallel (Fig. 3B). Analysis of all retraction curves by the WLC model yielded two modes: one corresponding to ∼10 and the other to ∼80 protein molecules linked in parallel between the AFM tip and the surface of an S. aureus bacterium. It is worth noting that simple geometry (i.e. the size of individual Fn molecules (52, 53), radiometric estimate of the number of FnBPs on S. aureus (20), and the radius of the AFM tip) predicts that the “reactive region” probed in these experiments contains 9–166 Fn molecules and 3–74 FnBPs (see supplemental Fig. S1).
It is no trivial feat to assign certain force signatures to specific domain(s) of Fn and/or FnBP. This is due to the multimodular nature of these proteins (Fig. 1) and the fact that both Fn and FnBP may unfold. Some information can be gleaned from the contour length of the final rupture event. Supplemental Fig. S2 shows the bond rupture lengths for each group of isolates. Six and seven maxima were observed for the CDU and CDI groups, respectively. Interestingly, of the 10–11 Fn-binding repeats in FnBP (Fig. 1), six have been identified as high affinity (13, 38). The maxima in supplemental Fig. S2 were fitted with Gaussian peaks and have an average Δlength of 77 ± 12.5 nm. This length scale corresponds to ∼200 residues, assuming 0.4 nm per amino acid (54). As noted above, the five FI modules that bind to FnBP consist of 225 residues. It is beyond the scope of this study to identify the exact regions of Fn and FnBP that are probed in our AFM experiments. Nonetheless, the “quantum” length scale shown in supplemental Fig. S2 confirms that we are observing the unfolding of distinct binding units along the lengths of each binding protein (i.e. a mechanical force signature).
Determining Whether Rupture Force Varies with Loading Rate
In 1978, Bell (55) published a seminal study that predicted that the unbinding force of a ligand-receptor bond should depend logarithmically on the loading rate of the bond. In this model, an applied force f distorts the energy landscape of a ligand-receptor complex, resulting in a lowering of the activation barrier as follows (56, 57).
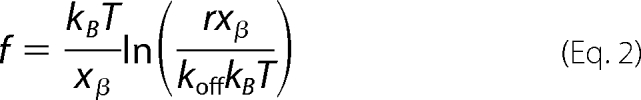 |
where f is the most probable rupture force (supplemental Fig. S3), r is the instantaneous loading rate (N s−1) determined as the product of the slope of the final, nonlinear rupture event (pN/nm) and the velocity of the tip (nm/s) (26, 58), koff (s−1) is the dissociation rate constant in the absence of the applied force, xβ (m) is the potential barrier position, and kBT is 4.1 pN·nm at room temperature. After plotting f versus ln r, the unstressed off-rate (koff) and the separation distance along the reaction coordinate (xβ) are calculated from the abscissa intercept (at f = 0) and the slope of the fitted line, respectively (28, 30).
Although Equation 2 is ideal for single ligand-receptor pairs, it has been applied to multivalent interactions and bond clusters (e.g. see Refs. 59–62). Sulchek et al. (16, 17) have shown that Equation 2 can provide an accurate measure of the kinetic off-rate of multivalent interactions by normalizing the loading rate by 1/Nb, where Nb is the number of bonds. As noted above, our clinical isolates formed bond clusters consisting of ∼10 or ∼80 proteins in parallel.
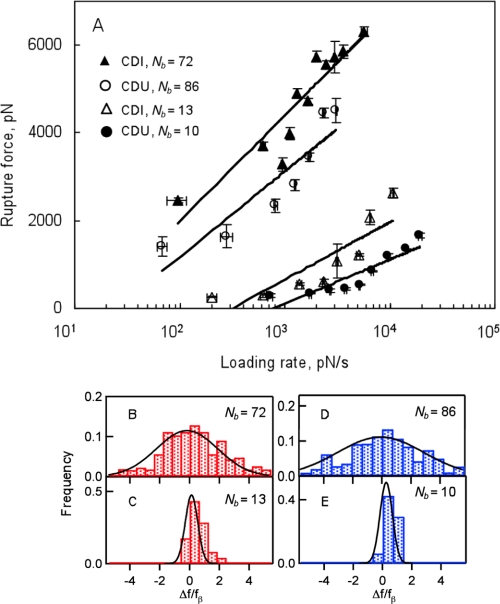
We used this assumption to produce dynamic force spectra for each clinical group (CDI and CDU) and ∼10- or ∼80-protein ruptures (Fig. 4A). The effective kinetic off-rates from these multivalent Fn-FnBP interactions were determined by fitting the spectra to Equation 2 (Table 1). As expected, the off-rates dropped with an increase in the number of bonds. These data clearly indicate that one of the main benefits of multivalent interactions is a reduction in koff and corresponding increase in bond lifetime (1/koff). Additionally, Bustanji et al. (63) determined a bond lifetime of 0.21 s, comparable with the values shown in Table 1, for a single molecular bond between Fn on an AFM tip and an Fn receptor on Staphylococcus epidermidis.
FIGURE 4.
A, dynamic force spectra for the rupture of multivalent Fn–FnBP bonds for S. aureus from the CDI (triangles; n = 26 different isolates) and CDU (circles; n = 20 different isolates) groups. Error bars represent S.D. of the data. Loading rates for the multibond rupture events were normalized by the number of bonds (Nb) according to Refs. 16 and 17. Most probable rupture forces were determined as shown in supplemental Fig. S3. B–E, histograms of normalized residuals from A for different number of bonds according to Ref. 15. CDI are in red for Nb = 72 (B) and Nb = 13 (C); CDU are in blue for Nb = 86 (D) and Nb = 10 (E). The residuals were normalized by the corresponding value of the force scale fβ. Solid lines are Gaussian fits.
TABLE 1.
Dissociation rate constants (koff) and bond lifetimes (1/koff; τ) for ∼10- and ∼80-bond clusters of Fn in complex with Fn receptors on living S. aureus
These values were determined by probing the binding reaction on 46 different S. aureus isolates. These isolates came from two different groups of human patients, all of whom had cardiovascular implants (e.g. prosthetic heart valve, permanent pacemaker, or defibrillator). One group of patients had S. aureus infection of the implanted cardiac device (CDI), whereas the other group (CDU) did not have an infected device, although S. aureus were present in their blood. p values were calculated with the Student's t test.
| Dissociation rate constant (koff) and bond lifetime (1/koff) | Clinical isolates of S. aureus |
p value | ||
|---|---|---|---|---|
| Cardiac device infected (n = 26) | Cardiac device uninfected (n = 20) | |||
| ∼10-Bond cluster | koff (s−1) | 0.56 | 1.04 | <0.05 |
| τ (s) | 1.79 | 0.96 | ||
| ∼80-Bond cluster | koff (s−1) | 0.014 | 0.029 | <0.05 |
| τ (s) | 69.47 | 34.02 | ||
The effective bond width or “reactive compliance” (xβ), determined by fitting Equation 2 to the lines in Fig. 4A, varied from 0.07 to 0.13 Å for the ∼10-bond clusters, and from 0.04 to 0.05 Å for the ∼80-bond clusters. These values are smaller than “typical” values for xβ, ∼1–10 Å (64). This discrepancy is explained quite simply by the fact that multivalent binding decreases xβ (65, 66).
The spread in the rupture force was compared with model predictions that show deviations in the normalized rupture force scale as the number of bonds (15, 16). Fig. 4, B--E, show the Gaussian distributions with the width Nb × σ, where σ is the residual S.D. from the ∼10-bond case. This analysis provides an additional validation for the multivalent binding model used to estimate koff in our experiments.
A bacterium in a human cardiovascular system is subjected to a range of loading rates in vivo. A mechanical heart valve, for example, experiences loading rates of 700–3000 mm Hg/s (67), which is equivalent to ∼0.09–0.40 pN nm−2 s−1. In our experiments, this would be similar to loading rates of ∼100–4000 pN s−1 (Fig. 4A) for an AFM tip that has a radius of 20–60 nm. This suggests that the koff values determined herein are relevant in vivo.
Reason for Different koff in CDI Group
All bacteria used in this study were very similar (e.g. see supplemental Table S2). All isolates were the same species expressing the same Fn-binding proteins. However, Table 1 shows that the bond lifetime (1/koff) for the CDI group was ∼2 times longer than the CDU isolates. This difference cannot be explained by the quantity of Fn-binding proteins as all groups produced statistically the same amount (Fig. 2).
This difference could be due to the “quality” of the binding proteins, specifically the primary sequence of FnBPA or FnBPB. In a recent publication, we found three amino acid polymorphisms in FnBPA produced by the CDI isolates: E652D, H782Q, and K786N (19). All three of these polymorphisms (Fig. 1) occur in Fn-binding repeats designated as high affinity (13, 38).
Therefore, we determined the effective koff for S. aureus that had multiple (2–3) of these polymorphisms in FnBPA versus those that did not (supplemental Fig. S4). This was accomplished by simply grouping force data based on the sequence of FnBPA for a particular isolate as opposed to the clinical classification of that isolate (i.e. CDI or CDU). When grouped in this fashion, we found that the relative bond lifetime was ∼10 times longer for isolates that had at least two of the key FnBPA polymorphisms shown in Fig. 1.
Binding of Peptides Mimicking FnBR9
A final check was performed to determine whether the polymorphisms in FnBPA could be responsible, at least partly, for the longer bond lifetime. AFM was used to probe the rupture force between an Fn-coated tip and each of two peptides that were identical in sequence to a portion of Fn-binding repeat 9, a repeat that contains two of the three key polymorphisms (H782Q and K786N; Fig. 1). The peptide sequences were: VPQIHGQNKGNQSFEEDTEC and VPQIQGQNNGNQSFEEDTEC. As determined from supplemental Fig. S5, the bond lifetime (1/koff) of the double mutant was indeed 2.5 times longer than the “wild-type” peptide lacking these two polymorphisms.
Conclusion
S. aureus are able to form infectious biofilms on implanted cardiovascular devices. This type of infection begins with the formation of a bond between receptors on the cell wall of the bacterium and human proteins, such as Fn, that coat the implant. The strength of this bond is dependent upon its loading rate. AFM was used to directly probe the fundamental binding mechanism between an Fn-coated substrate (i.e. proxy for cardiovascular implant) and Fn-binding receptors on the surface of living S. aureus at loading rates that mimicked physiological conditions.
The interaction between Fn and putative FnBPs does not appear to form a single ligand-receptor pair, but rather, a multivalent cluster bond consisting of ∼10 or ∼80 proteins bound in parallel. The dissociation rate constant (koff) and corresponding bond lifetime (1/koff) were determined by applying a multivalent version of the Bell equation (Equation 2) to a plot of rupture force versus the log loading rate. A significantly longer bond lifetime (1/koff) was observed for a group of S. aureus isolates from patients with confirmed clinical infection of implanted cardiovascular devices. This longer bond lifetime could not be explained by increased amounts of FnBP, but rather, the presence of three polymorphisms in high affinity, Fn-binding regions of FnBPA explained, at least partially, the reason cardiovascular implants in the CDI group became infected with S. aureus.
By working with isolates that were actually associated with disease, we have shown a direct link between a fundamental measure of bond strength and the clinical presentation of disease in humans (i.e. biofilm-based infection of implanted medical devices). Additional work is, therefore, warranted on laboratory strains of S. aureus that are engineered to produce different constructs of FnBPs and/or other MSCRAMMs, as well as different ligands. On a more practical level, the risk of biofilm-based infection could be determined by screening the natural flora of a patient prior to surgical implantation of a device. Such as assay could be tailored even further by coating an AFM tip with blood plasma collected from the same patient.
Supplementary Material
Acknowledgments
We thank T. J. Foster for providing reference strains of S. aureus for control experiments and the two anonymous reviewers for critical reading of the manuscript. S. K. L. thanks J. Tak for support.
This work was supported in part by National Institutes of Health Grant R21HL086593 (to S. K. L.). This work was also supported by National Science Foundation Grant EAR0745808 (to S. K. L.).

This article contains supplemental Experimental Procedures, Tables S1 and S2, and Figs. S1–S5.
- MSCRAMMs
- microbial surface components recognizing adhesive matrix molecules
- Fn
- fibronectin
- FnBP
- fibronectin-binding protein
- AFM
- atomic force microscopy
- CDI
- cardiac device infected
- CDU
- cardiac device uninfected
- WLC
- worm-like chain
- N
- newtons.
REFERENCES
- 1. Darouiche R. O. (2004) Treatment of infections associated with surgical implants. N. Engl. J. Med. 350, 1422–1429 [DOI] [PubMed] [Google Scholar]
- 2. Voigt A., Shalaby A., Saba S. (2006) Rising rates of cardiac rhythm management device infections in the United States: 1996 through 2003. J. Am. Coll. Cardiol. 48, 590–591 [DOI] [PubMed] [Google Scholar]
- 3. Wang A., Athan E., Pappas P. A., Fowler V. G., Jr., Olaison L., Paré C., Almirante B., Muñoz P., Rizzi M., Naber C., Logar M., Tattevin P., Iarussi D. L., Selton-Suty C., Jones S. B., Casabé J., Morris A., Corey G. R., Cabell C. H. (2007) Contemporary clinical profile and outcome of prosthetic valve endocarditis. JAMA 297, 1354–1361 [DOI] [PubMed] [Google Scholar]
- 4. Margey R., McCann H., Blake G., Keelan E., Galvin J., Lynch M., Mahon N., Sugrue D., O'Neill J. (2010) Contemporary management of and outcomes from cardiac device-related infections. Europace 12, 64–70 [DOI] [PubMed] [Google Scholar]
- 5. Gordon R. J., Lowy F. D. (2008) Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson B., Nair S., Pallas J., Williams M. A. (2011) Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 35, 147–200 [DOI] [PubMed] [Google Scholar]
- 7. Greene C., McDevitt D., Francois P., Vaudaux P. E., Lew D. P., Foster T. J. (1995) Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 8. Foster T. J., Höök M. (1998) Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6, 484–488 [DOI] [PubMed] [Google Scholar]
- 9. Arrecubieta C., Asai T., Bayern M., Loughman A., Fitzgerald J. R., Shelton C. E., Baron H. M., Dang N. C., Deng M. C., Naka Y., Foster T. J., Lowy F. D. (2006) The role of Staphylococcus aureus adhesins in the pathogenesis of ventricular assist device-related infections. J. Infect. Dis 193, 1109–1119 [DOI] [PubMed] [Google Scholar]
- 10. Que Y. A., François P., Haefliger J. A., Entenza J. M., Vaudaux P., Moreillon P. (2001) Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 69, 6296–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Que Y. A., Haefliger J. A., Piroth L., François P., Widmer E., Entenza J. M., Sinha B., Herrmann M., Francioli P., Vaudaux P., Moreillon P. (2005) Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., Potts J. R. (2003) Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423, 177–181 [DOI] [PubMed] [Google Scholar]
- 13. Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., Höök M., Speziale P., Potts J. R. (2007) The tandem β-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 14. Bingham R. J., Rudiño-Piñera E., Meenan N. A., Schwarz-Linek U., Turkenburg J. P., Höök M., Garman E. F., Potts J. R. (2008) Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. U.S.A. 105, 12254–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams P. M. (2003) Analytical descriptions of dynamic force spectroscopy: behaviour of multiple connections. Anal. Chim. Acta 479, 107–115 [Google Scholar]
- 16. Sulchek T. A., Friddle R. W., Langry K., Lau E. Y., Albrecht H., Ratto T. V., DeNardo S. J., Colvin M. E., Noy A. (2005) Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc. Natl. Acad. Sci. 102, 16638–16643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sulchek T., Friddle R. W., Noy A. (2006) Strength of multiple parallel biological bonds. Biophys. J. 90, 4686–4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J. S., Sexton D. J., Mick N., Nettles R., Fowler V. G., Jr., Ryan T., Bashore T., Corey G. R. (2000) Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30, 633–638 [DOI] [PubMed] [Google Scholar]
- 19. Lower S. K., Lamlertthon S., Casillas-Ituarte N. N., Lins R. D., Yongsunthon R., Taylor E. S., DiBartola A. C., Edmonson C., McIntyre L. M., Reller L. B., Que Y. A., Ros R., Lower B. H., Fowler V. G., Jr. (2011) Polymorphisms in fibronectin-binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc. Natl. Acad. Sci. 108, 18372–18377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Proctor R. A., Mosher D. F., Olbrantz P. J. (1982) Fibronectin binding to Staphylococcus aureus. J. Biol. Chem. 257, 14788–14794 [PubMed] [Google Scholar]
- 21. Lowy F. D. (1998) Staphylococcus aureus infections. New Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 22. Lower S. K., Yongsunthon R., Casillas-Ituarte N. N., Taylor E. S., DiBartola A. C., Lower B. H., Beveridge T. J., Buck A. W., Fowler V. G., Jr. (2010) A tactile response in Staphylococcus aureus. Biophys. J. 99, 2803–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bisognano C., Vaudaux P. E., Lew D. P., Ng E. Y., Hooper D. C. (1997) Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41, 906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yongsunthon R., Fowler V. G., Jr., Lower B. H., Vellano F. P., 3rd, Alexander E., Reller L. B., Corey G. R., Lower S. K. (2007) Correlation between fundamental binding forces and clinical prognosis of Staphylococcus aureus infections of medical implants. Langmuir 23, 2289–2292 [DOI] [PubMed] [Google Scholar]
- 25. Buck A. W., Fowler V. G., Jr., Yongsunthon R., Liu J., DiBartola A. C., Que Y. A., Moreillon P., Lower S. K. (2010) Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir 26, 10764–10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanley W., McCarty O., Jadhav S., Tseng Y., Wirtz D., Konstantopoulos K. (2003) Single molecule characterization of P-selectin/ligand binding. J. Biol. Chem. 278, 10556–10561 [DOI] [PubMed] [Google Scholar]
- 27. Tortonese M., Kirk M. (1997) Characterization of application specific probes for SPMs. Proc. Soc. Photo. Instrument. Eng. 3009, 53–60 [Google Scholar]
- 28. Strunz T., Oroszlan K., Schäfer R., Güntherodt H. J. (1999) Dynamic force spectroscopy of single DNA molecules. Proc. Natl. Acad. Sci. 96, 11277–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baumgartner W., Hinterdorfer P., Ness W., Raab A., Vestweber D., Schindler H., Drenckhahn D. (2000) Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. 97, 4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwesinger F., Ros R., Strunz T., Anselmetti D., Güntherodt H. J., Honegger A., Jermutus L., Tiefenauer L., Pluckthun A. (2000) Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. 97, 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ros R., Schwesinger F., Anselmetti D., Kubon M., Schäfer R., Plückthun A., Tiefenauer L. (1998) Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. 95, 7402–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baddour L. M., Epstein A. E., Erickson C. C., Knight B. P., Levison M. E., Lockhart P. B., Masoudi F. A., Okum E. J., Wilson W. R., Beerman L. B., Bolger A. F., Estes N. A., 3rd, Gewitz M., Newburger J. W., Schron E. B., Taubert K. A. (2010) Update on cardiovascular implantable electronic device infections and their management: a scientific statement from the American Heart Association. Circulation 121, 458–477 [DOI] [PubMed] [Google Scholar]
- 34. Habib G., Hoen B., Tornos P., Thuny F., Prendergast B., Vilacosta I., Moreillon P., de Jesus Antunes M., Thilen U., Lekakis J., Lengyel M., Müller L., Naber C. K., Nihoyannopoulos P., Moritz A., Zamorano J. L. (2009) Guidelines on the prevention, diagnosis, and treatment of infective endocarditis (new version 2009): the Task Force on the Prevention, Diagnosis, and Treatment of Infective Endocarditis of the European Society of Cardiology (ESC); endorsed by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the International Society of Chemotherapy (ISC) for Infection and Cancer. Eur. Heart. J. 30, 2369–2413 [DOI] [PubMed] [Google Scholar]
- 35. Donlan R. M., Costerton J. W. (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peacock S. J., Foster T. J., Cameron B. J., Berendt A. R. (1999) Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145, 3477–3486 [DOI] [PubMed] [Google Scholar]
- 37. House-Pompeo K., Xu Y., Joh D., Speziale P., Höök M. (1996) Conformational changes in the fibronectin-binding MSCRAMMs are induced by ligand binding. J. Biol. Chem. 271, 1379–1384 [DOI] [PubMed] [Google Scholar]
- 38. Provenza G., Provenzano M., Visai L., Burke F. M., Geoghegan J. A., Stravalaci M., Gobbi M., Mazzini G., Arciola C. R., Foster T. J., Speziale P. (2010) Functional analysis of a murine monoclonal antibody against the repetitive region of the fibronectin-binding adhesins fibronectin-binding protein A and fibronectin-binding protein B from Staphylococcus aureus. FEBS J. 277, 4490–4505 [DOI] [PubMed] [Google Scholar]
- 39. Kuusela P., Vartio T., Vuento M., Myhre E. B. (1984) Binding sites for streptococci and staphylococci in fibronectin. Infect. Immun. 45, 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakata N., Jakab E., Wadström T. (1994) Human plasma fibronectin possesses second binding site(s) to Staphylococcus aureus on its C-terminal region. J. Biochem. 115, 843–848 [DOI] [PubMed] [Google Scholar]
- 41. Pankov R., Yamada K. M. (2002) Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 42. Mosher D. F. (2006) Plasma fibronectin concentration: a risk factor for arterial thrombosis? Arterioscler. Thromb. Vasc. Biol. 26, 1193–1195 [DOI] [PubMed] [Google Scholar]
- 43. Grundmeier M., Hussain M., Becker P., Heilmann C., Peters G., Sinha B. (2004) Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72, 7155–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 45. Müller D. J., Baumeister W., Engel A. (1999) Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl. Acad. Sci. 96, 13170–13174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lower B. H., Shi L., Yongsunthon R., Droubay T. C., McCready D. E., Lower S. K. (2007) Specific bonds between an iron oxide surface and outer membrane cytochromes MtrC and OmcA from Shewanella oneidensis MR-1. J. Bacteriol. 189, 4944–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fröman G., Switalski L. M., Speziale P., Höök M. (1987) Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 262, 6564–6571 [PubMed] [Google Scholar]
- 48. Schwarz-Linek U., Höök M., Potts J. R. (2004) The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 49. Kellermayer M. S., Smith S. B., Granzier H. L., Bustamante C. (1997) Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 276, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 50. Dietz H., Rief M. (2004) Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. 101, 16192–16197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lower B. H., Yongsunthon R., Vellano F. P., 3rd, Lower S. K. (2005) Simultaneous force and fluorescence measurements of a protein that forms a bond between a living bacterium and a solid surface. J. Bacteriol. 187, 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Engel J., Odermatt E., Engel A., Madri J. A., Furthmayr H., Rohde H., Timpl R. (1981) Shapes, domain organizations, and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J. Mol. Biol. 150, 97–120 [DOI] [PubMed] [Google Scholar]
- 53. Erickson H. P., Carrell N., McDonagh J. (1981) Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J. Cell Biol. 91, 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mueller H., Butt H. J., Bamberg E. (1999) Force measurements on myelin basic protein adsorbed to mica and lipid bilayer surfaces done with the atomic force microscope. Biophys. J. 76, 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bell G. I. (1978) Models for the specific adhesion of cells to cells. Science 200, 618–627 [DOI] [PubMed] [Google Scholar]
- 56. Evans E., Ritchie K. (1997) Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. (1999) Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 [DOI] [PubMed] [Google Scholar]
- 58. Zhang X., Wojcikiewicz E., Moy V. T. (2002) Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 83, 2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evans E., Leung A., Hammer D., Simon S. (2001) Chemically distinct transition states govern rapid dissociation of single L-selectin bonds under force. Proc. Natl. Acad. Sci. 98, 3784–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cheung L. S., Konstantopoulos K. (2011) An analytical model for determining two-dimensional receptor-ligand kinetics. Biophys. J. 100, 2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gomez-Casado A., Dam H. H., Yilmaz M. D., Florea D., Jonkheijm P., Huskens J. (2011) Probing multivalent interactions in a synthetic host-guest complex by dynamic force spectroscopy. J. Am. Chem. Soc. 133, 10849–10857 [DOI] [PubMed] [Google Scholar]
- 62. Teulon J. M., Delcuze Y., Odorico M., Chen S. W., Parot P., Pellequer J. L. (2011) Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 24, 490–502 [DOI] [PubMed] [Google Scholar]
- 63. Bustanji Y., Arciola C. R., Conti M., Mandello E., Montanaro L., Samorí B. (2003) Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. 100, 13292–13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Evans E. (1998) Energy landscapes of biomolecular adhesion and receptor anchoring at interfaces explored with dynamic force spectroscopy. Faraday Discuss. 111, 1–16 [DOI] [PubMed] [Google Scholar]
- 65. Tees D. F., Waugh R. E., Hammer D. A. (2001) A microcantilever device to assess the effect of force on the lifetime of selectin-carbohydrate bonds. Biophys. J. 80, 668–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hanley W. D., Wirtz D., Konstantopoulos K. (2004) Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J. Cell Sci. 117, 2503–2511 [DOI] [PubMed] [Google Scholar]
- 67. Chandran K. B., Dexter E. U., Aluri S., Richenbacher W. E. (1998) Negative pressure transients with mechanical heart valve closure: correlation between in vitro and in vivo results. Ann. Biomed. Eng. 26, 546–556 [DOI] [PubMed] [Google Scholar]
- 68. Baumgartner W., Hinterdorfer P., Schindler H. (2000) Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy 82, 85–95 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.