Background: AGP31 is a multidomain plant cell wall hydroxyproline-rich glycoprotein. The position of Hyp and the distribution of carbohydrates is unknown.
Results: Most Hyp of the Pro-rich domain are isolated within repeated motifs and carry glycans of various sizes.
Conclusion: AGP31 glycoforms are very heterogeneous.
Significance: This might be the first evidence for new Hyp-O-glycans.
Keywords: Arabidopsis, Cell Wall, Galactose, Glycoprotein, Glycoprotein Structure, Lectin, Mass Spectrometry (MS), Post-translational Modification, Hydroxyproline-rich Glycoprotein, Pro-rich Protein
Abstract
Proteins are important actors in plant cell walls because they contribute to their architecture and their dynamics. Among them, hydroxyproline (Hyp)-rich glycoproteins constitute a complex family of O-glycoproteins with various structures and functions. In this study, we characterized an atypical Hyp-rich glycoprotein, AGP31 (arabinogalactan protein 31), which displays a multidomain organization unique in Arabidopsis thaliana, consisting of a short arabinogalactan protein (AGP) motif, a His stretch, a Pro-rich domain, and a C-terminal PAC (PRP-AGP containing Cys) domain. The use of various mass spectrometry strategies was innovative and powerful: it permitted us to locate Hyp residues, to demonstrate the presence of carbohydrates, and to refine their distribution over the Pro-rich domain. Most Hyp were isolated within repeated motifs such as KAOV, KSOV, K(PO/OP)T, K(PO/OP)V, T(PO/OP)V, and Y(PO/OP)T. A few extensin-like motifs with contiguous Hyp (SOOA and SOOT) were also found. The Pro-rich domain was shown to carry Gal residues on isolated Hyp but also Ara residues. The existence of new type Hyp-O-Gal/Ara-rich motifs not recognized by the β-glucosyl Yariv reagent but interacting with the peanut agglutinin lectin was proposed. In addition, the N-terminal short AGP motif was assumed to be substituted by arabinogalactans. Altogether, AGP31 was found to be highly heterogeneous in cell walls because arabinogalactans could be absent, Hyp-O-Gal/Ara-rich motifs of different sizes were observed, and truncated forms missing the C-terminal PAC domain were found, suggesting degradation in muro and/or partial glycosylation prior to secretion.
Introduction
Hydroxyproline (Hyp)-rich2 glycoproteins (HRGPs) are a superfamily of plant cell wall proteins (CWPs) identified several decades ago (1). Their study remains challenging because HRGPs constitute a very complex protein group with various structures and functions whose molecular bases are not fully elucidated (2–6). HRGPs fall into three families: (i) hyperglycosylated arabinogalactan proteins (AGPs) having diverse functions (5); (ii) moderately glycosylated extensins (EXTs) that can form cell wall scaffolds essential for cytokinesis (7); and (iii) Pro-rich proteins (PRPs) that may be nonglycosylated, weakly glycosylated, or highly glycosylated and possibly involved in various aspects of development (4, 8). Hybrid HRGPs are composed of different types of HRGP domains, and chimeric HRGPs contain at least one HRGP domain within a non-HRGP protein. A recent bioinformatics analysis achieved on the Arabidopsis thaliana genome identified 166 HRGPs classified in 85 AGPs, 59 EXTs, 18 PRPs, and 4 AGP/EXT hybrid proteins, highlighting the question why plants need so many HRGPs (9).
HRGPs are characterized by the presence of Hyp-rich repetitive motifs, called glycomodules, containing from 3 to 16 amino acids. Glycomodule sequences determine Pro hydroxylation and the subsequent Hyp O-glycosylation. These two consecutive sequence-driven post-translational modifications define the so-called Hyp-O-glycosylation code (3). AGPs have repetitive variants of (Xaa1–3-Hyp)n motifs (where Xaa is commonly Ser, Ala, or Thr) with O-linked arabinogalactan (AG) polysaccharides. EXTs display clusters of Ser-(Hyp)2,4 containing short O-Hyp arabinose (Ara)-oligosaccharides. Finally, PRPs are probably the most heterogeneous group, regarding their repetitive sequences and their carbohydrate patterns. They are characterized by the repeating occurrence of (Pro)2 or (Pro)3 motifs within a variety of larger repeated units. It was shown that PRPs contain approximately equimolar quantities of Pro and Hyp (8, 10–13). Many PRPs contain the repeated pentapeptide Pro-Hyp-Xaa-Yaa-Lys, where Xaa and Yaa can be Val, Tyr, His, or Glu (4, 14). PRPs were initially assumed to be non- or weakly glycosylated (11, 15). However, the characterization of a Gal-rich glycoprotein (GaRSGP) from Nicotiana alata styles revealed a new class of PRPs with repetitive motifs never described before and a carbohydrate content of 75%, unusual for PRPs (8). Recently, an A. thaliana homologue of GaRSGP has been described (12). This glycoprotein mostly contains Gal residues and was called AGP31 because of its positive interaction with the β-glucosyl Yariv reagent. This protein, encoded by At1g28290, was shown to be a multidomain protein and was classified as a chimeric AGP (9).
Here, we provide a detailed characterization of AGP31 extracted from cell walls of A. thaliana etiolated hypocotyls. Combining two isolation processes and three detection methods, we demonstrated that the native AGP31 displays a huge heterogeneity with various O-glycans and truncated forms. After enrichment, MS analyses were permitted to determine the location of Hyp residues within the Pro-rich domain, to demonstrate the presence of carbohydrates, and to refine their distribution.
EXPERIMENTAL PROCEDURES
Plant Material and Cell Wall Isolation
A. thaliana seeds (ecotype Columbia 0) were grown in the dark as described (16). Etiolated hypocotyls were collected at 11 days and used to isolate cell walls as reported (17).
AGP31 Isolation
CWPs were extracted from the cell wall fraction by successive steps of washing with a 0.2 m CaCl2 solution followed by a 2 m LiCl solution as described (16). Approximately 2 mg of CWPs were usually extracted from 1 g of dry cell walls. Two strategies were employed to isolate AGP31 from a total CWP extract: (i) a combination of cation exchange chromatography (CEC) and nickel affinity chromatography (NAC) and (ii) an affinity chromatography using a peanut agglutinin lectin from Arachis hypogaea (PNA)-agarose resin. PNA is a lectin specific for GalNAc, α-Gal, and β-Gal (18). For the first isolation strategy, 6 mg of CWPs were separated by CEC as previously described (16). AGP31 was identified by peptide mass fingerprinting (PMF) from MALDI-TOF MS analysis of CEC elution fraction aliquots separated by SDS-PAGE. The elution fractions containing AGP31 were pooled, desalted, lyophilized, and subsequently loaded onto a nickel-nitrilotriacetic acid column supplied in the nickel-nitrilotriacetic acid Fast Start Kit (Qiagen), following the manufacturer's recommendations. For the second isolation strategy, 2 mg of CWPs were incubated with 1 ml of PNA-agarose resin (Sigma). Chromatography was performed as previously reported (19). Protein content was estimated using the Bradford method (20).
SDS-PAGE and Lectin Blot Analyses
AGP31-enriched fractions issued from the two isolation processes (CEC/NAC and PNA) were desalted using Econo-Pac 10 DG columns (Bio-Rad) and lyophilized prior to SDS-PAGE. Dried samples were dissolved in 150 μl of water. Fifty μl of each sample were loaded on a 10 × 12 × 0.15-cm SDS-polyacrylamide gel with a concentration of 12.5%/0.33% of acrylamide/bisacrylamide. Separation was performed according to Laemmli (21). Staining of gels with colloidal blue was carried out using PageBlueTM protein staining solution (Fermentas, Saint-Rémy-lès-Chevreuse, France) or the β-glucosyl Yariv reagent (22). For lectin blots, proteins separated by SDS-PAGE were transferred onto a nitrocellulose membrane (GE Healthcare) using a semi-dry transfer cell (Trans-Blot SD; Bio-Rad). Transfer was carried out at 4 °C in transfer buffer (48 mm Tris-HCl, 39 mm glycine, 0.0375% SDS, 20% methanol) for 1 h at 20 V. AGP31 bound to the membrane was revealed by a lectin blot method, using the digoxigenin (DIG)-labeled PNA provided in the DIG glycan differentiation kit, following the manufacturer's recommendations (Roche Applied Science).
Monosaccharide Composition Analyses
AGP31-enriched fractions issued from the two isolation processes (CEC/NAC and PNA) were desalted using Econo-Pac 10 DG columns (Bio-Rad) and lyophilized prior to acid hydrolysis carried out for 1 h at 110 °C using 2 n trifluoroacetic acid. Trimethylsilylated derivatives were prepared and analyzed by GC-MS as described (23).
Anhydrous Hydrogen Fluoride Deglycosylation
A sample of total CWPs extracted from 11-day-old etiolated hypocotyls was HF-deglycosylated for 1 h at 4 °C as described (24, 25). The HF was blown off under nitrogen gas, and the deglycosylated proteins were then separated by SDS-PAGE.
MS Analyses
Sample preparation for all MS analyses was performed as previously described (26). MALDI-TOF MS analyses were performed using a Voyager-DE STR mass spectrometer (Applied Biosystems/MDS, Sciex). The spectra were acquired in reflectron mode as previously reported (26, 27). MALDI-TOF/TOF MS/MS analyses were performed using a MALDI-TOF/TOF Voyager 4700 (Applied Biosystems/MDS, Sciex). MS/MS data were recorded using the following parameters: accelerating voltage of 8 and 15 kV (sources 1 and 2, respectively) and grid voltage of 86%. The mass selection of the precursor ion was achieved using a mass window of ±5 Da, and collision was performed in CID off mode. The data were acquired with 3750 shoots/spectrum.
For LC-MS/MS analyses, chip-based nano-LC separation was performed with an Agilent 1260 nano-pump system consisting of binary capillary flow and nano-flow pumps, a vacuum degasser, and an autosampler (Agilent Technologies Inc., Palo Alto, CA). Chromatographic separation of tryptic peptides was performed on a C18 chip (75-mm diameter, 43-mm length, 40-nl preconcentration trap, and 5-μm particle size). The chip column was equilibrated at room temperature with 97% eluent A (0.1% formic acid in water) and 3% eluent B (0.1% formic acid in 90% acetonitrile and 10% water) at a flow rate of 600 nl/min. The chip preconcentration trap was equilibrated with 0.1% formic acid in 97% water and 3% acetonitrile at a flow rate of 4 μl/min. After injection of the sample, the separation was carried out using a linear gradient from 3–55% mobile phase B for 20 min, 55–95% mobile phase B for 1 min, and 95% mobile phase B for 2.5 min. The LC system was interfaced to a Bruker Daltonics (Bruker Daltonics GmbH, Bremen, Germany) Amazon ion trap mass spectrometer via a ChipCube interface (Agilent Technologies Inc.). The spraying voltage was set to 1780 V. MS scans were performed in the “enhanced resolution” mode, and MS/MS scans were performed in “ultrascan” mode covering the 100–3000 m/z range for the analyses of all glycopeptides. For electron transfer dissociation (ETD) analyses, a negative chemical ionization source generated fluoranthene radical anions, which were transported to the ion trap and used as the electron transfer agents. To produce the radical anions, the reactant temperature was set at 60 °C, whereas the ionization energy and emission current were set at 70 eV and 4 μA, respectively. The reaction time for radical anions was set to 100 ms.
PMF was done by comparison to the non-redundant database of A. thaliana at NCBI using ProteinProspector MS-FIT. The criteria used for the database search were: a monoisotopic mass accuracy of 20 ppm, one missed cleavage, and hydroxylation of Pro residues. MS/MS results were analyzed using Protein Prospector MS-Product with Pro hydroxylation and glycosylation added as possible modifications.
RESULTS
Different Strategies to Isolate AGP31: Evidence for Huge Heterogeneity
In our previous cell wall proteomics analyses of A. thaliana etiolated hypocotyls, we identified AGP31 as an abundant protein (16, 19). This protein comprises several domains, from the N terminus to the C terminus (Fig. 1A): a predicted signal peptide, a short AGP motif, a His stretch, a Pro-rich domain, and a C-terminal Cys-rich domain also called the PAC (PRP-AGP containing Cys) domain (28).
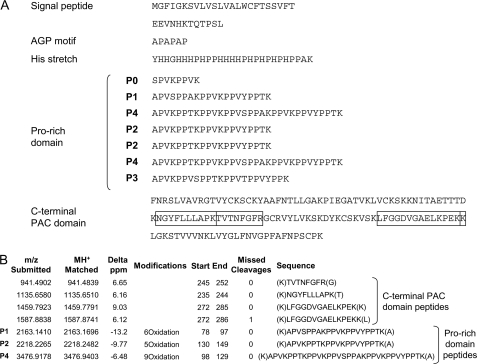
FIGURE 1.
Identification of AGP31 by PMF. A, amino acid sequence of AGP31 displaying the different domains of the protein, i. e. signal peptide, AGP motif, His stretch, Pro-rich domain, and C-terminal PAC domain. The protein sequence should be read from left to right and from top to bottom. The sequence of the Pro-rich domain is arranged to emphasize the tryptic peptides (P0–P4) identified in this study. The four tryptic peptides of the C-terminal domain used for protein identification are indicated in boxes. Note that P1, P2, and P4 also contributed to protein identification as shown in B. B, ProteinProspector search results allowed the identification of AGP31 by PMF using MALDI-TOF MS. Among the seven matching tryptic peptides, four belong to the C-terminal domain and three to the Pro-rich domain (noted P1, P2, and P4). These three peptides are predicted to contain, respectively, six, five, and nine oxidations assumed to correspond to Pro hydroxylations. B shows the submitted and matched m/z of the peptides, the differences in mass between the two (Delta ppm), the proposed modifications, the position of the peptides on the protein (Start and End), the number of missed cleavages, and the peptide sequences.
Following a fractionation step using CEC, AGP31 was eluted at the end of the salt gradient, at ∼0.5 m NaCl, as determined by MALDI-TOF MS analyses of elution fraction aliquots separated by SDS-PAGE (Fig. 1B). The fractions containing AGP31 were collected for subsequent NAC. The combination of these two chromatographic steps was very efficient for isolating AGP31, which migrated as a smear from 50 to 90 kDa on a SDS-polyacrylamide gel (Fig. 2, lane 1a). This is much higher than the predicted molecular mass of AGP31, i.e. 38 kDa. In addition, an SDS-PAGE analysis of a similar fraction containing three times more proteins than previously (i.e. 36 μg) showed an additional smear from 30 to 40 kDa (Fig. 2, lane 1b). MALDI-TOF MS analyses of samples taken all along both smears (30–40 kDa and above 50 kDa) identified AGP31 as unique protein. Up to seven tryptic peptides were used for AGP31 identification by PMF (corresponding to a maximum coverage of 33.3%), among which four belonged to the C-terminal PAC domain and three to the Pro-rich domain (named P1, P2, and P4) (Figs. 1 and 3A). Truncated forms of AGP31 missing the C-terminal PAC domain were found in the 30–40-kDa smear.
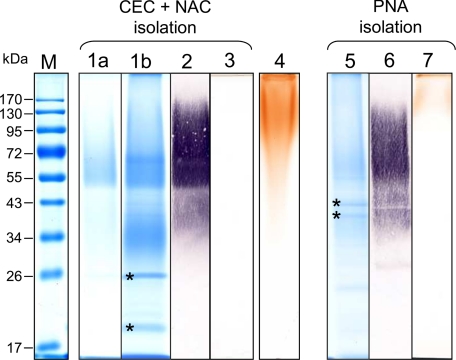
FIGURE 2.
Detection of AGP31 in enriched fractions after separation by SDS-PAGE. Two strategies were employed to enrich native AGP31 from a CWP extract of 11-day-old etiolated hypocotyls of A. thaliana: (i) a combination of CEC followed by NAC (lanes 1–3) and (ii) a PNA-lectin affinity chromatography (lanes 5–7). AGP31-enriched fractions were separated by SDS-PAGE. Approximately 13 μg of proteins were used for each analysis, except for lanes 1b, 2, and 3, where 36 μg of proteins were loaded. Minor contaminant proteins labeled with asterisks were identified in the elution fractions using both strategies (lanes 1b and 5). Molecular mass markers (M) are shown in the left lane. Lanes 1a, 1b, and 5, colloidal blue staining of AGP31-enriched fractions. Note that AGP31 migrated as a smear above 50 kDa but was also present as truncated forms in an additional smear from 30 to 40 kDa (lanes 1b and 5). Lanes 2 and 6, PNA-lectin blot analysis of AGP31-enriched fractions. After separation by SDS-PAGE, the proteins were electrotransferred onto a nitrocellulose membrane and treated as indicated under “Experimental Procedures.” PNA labeled a smear from 34 to 170 kDa. Lanes 3 and 7, β-glucosyl Yariv staining of AGP31-enriched fractions. Note that a signal was observed above 170 kDa in the PNA-fraction. Lane 4, β-glucosyl Yariv staining of the flow-through fraction from the CEC step.
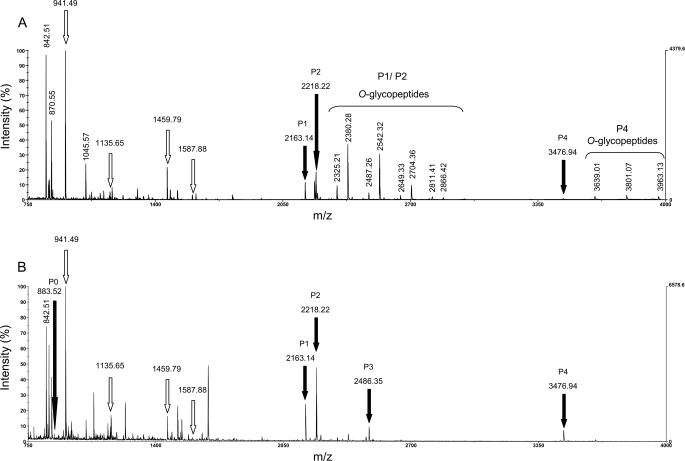
FIGURE 3.
Comparison of MALDI-TOF MS spectra of glycosylated and HF-deglycosylated forms of AGP31. A, identification of AGP31 by PMF using MALDI-TOF MS. After separation by SDS-PAGE of an AGP31-enriched fraction obtained combining CEC and NAC, bands containing glycosylated AGP31 were excised from the 50–60-kDa mass range and digested with trypsin. Unmodified tryptic peptides from the C-terminal domain (see Fig. 1B) are shown with white arrows, whereas tryptic peptides of the Pro-rich domain (P1, P2, and P4) are labeled with black arrows. O-Glycopeptides associated with these peptides are shown with a brace, and the masses of most intense ones are indicated. Note that other intense ions (842.51, 870.55, and 1045.57) correspond to trypsin or matrix peptides. B, identification of AGP31 after HF deglycosylation. After separation by SDS-PAGE of a total CWP extract, the bands containing deglycosylated AGP31 were excised, digested with trypsin, and analyzed by MALDI-TOF MS. AGP31 glycopeptides (shown in A) disappeared in favor of the corresponding deglycosylated peptides (P0–P4, black arrows). As expected, unmodified peptides from the C-terminal domain (white arrows) were not modified by the HF treatment.
Two minor contaminant proteins, a thaumatin encoded by At2g28790 and a glycine-rich protein encoded by At2g05580, were also found in this fraction, in bands of 26 and 20 kDa, respectively (labeled with asterisks on Fig. 2, lane 1b). These co-purified proteins may be AGP31 interactants or have similar behavior toward both chromatographic resins used. In particular, At2g28790 possesses a His stretch that may be responsible for its retention on the nickel resin. Altogether, an AGP31-enriched fraction containing ∼40 μg of proteins was routinely obtained from 6 mg of CWPs after CEC and NAC.
A PNA-lectin blot analysis of this enriched fraction showed a signal from 34 to 170 kDa, particularly strong above 50 kDa (Fig. 2, lane 2). This result corroborated the fact that AGP31 contains Gal residues, as previously reported (12) and showed that AGP31 displays a heterogeneous pattern of glycosylation. Monosaccharide composition analysis revealed that AGP31-enriched fraction mostly contains Gal (53.2%) but also Ara (39.5%) in a higher proportion than previously reported (12). Lesser amounts of Xyl (2.2%), Fuc (1.9%), Glc (1.8%), Man (1.3%), and GlcUA (0.3%) were also detected.
Finally, no signal was obtained with the β-glucosyl Yariv reagent, specific for AGPs, suggesting that AGP31 isolated by CEC and NAC does not display any AG (Fig. 2, lane 3). Interestingly, the analysis of the flow-through fraction of the CEC showed a positive response with the β-glucosyl Yariv reagent, demonstrating that AGPs did not bind to that resin, probably because their carbohydrate content hides their charge (Fig. 2, lane 4).
An alternative isolation strategy was carried out using a PNA-lectin agarose matrix, capitalizing on the remarkable affinity of AGP31 toward this lectin (19). Approximately 40 μg of proteins were routinely obtained in the PNA elution fraction from 2 mg of CWPs. AGP31 was identified by PMF using MALDI-TOF MS, with the same tryptic peptides as reported above (Fig. 1B). It migrates as two smears from 30 to 40 kDa and above 50 kDa, respectively, where AGP31 was the only protein identified (Fig. 2, lane 5). As expected, it was recognized by PNA, and the lectin blot signal was similar to that obtained using the CEC/NAC isolation strategy, confirming the large heterogeneity of AGP31 (Fig. 2, lane 6). The presence of residual Gal from the elution buffer (containing 0.5 m Gal) in the desalted AGP31-enriched fraction did not permit to determine a reliable monosaccharide composition of this fraction. Note that the At5g11420 and At5g25460 proteins containing a DUF642 domain of unknown function were found in bands of 43 and 40 kDa, respectively (labeled with asterisks on Fig. 2, lane 5). Finally, the β-glucosyl Yariv reagent gave a signal at the top of the SDS-polyacrylamide gel for the PNA elution fraction (Fig. 2, lane 7). This signal was assumed to be due to AGP31. Note that no other protein has been identified by PMF above 50 kDa in this fraction and that the flow-through fraction of the PNA chromatography showed a positive response with the β-glucosyl Yariv reagent, showing that other AGPs were not retained (data not shown).
Pro-rich Domain of AGP31: Domain Rich in O-Glycosylated Hyp
As mentioned above, among the seven tryptic peptides used for AGP31 identification, three belonged to the Pro-rich domain (named P1, P2, and P4) (Figs. 1 and 3A). P1, P2, and P4 were predicted to contain six, five, and nine hydroxyl groups, respectively. We assumed that these modifications correspond to hydroxylation of Pro residues, giving rise to Hyp. Additional peaks were observed on MS spectra of samples taken from 50 to 60 kDa and from 30 to 40 kDa (Fig. 2, lanes 1a, 1b, and 5). The masses of these peaks matched those of P1, P2, and P4 with regular mass increments of 162 and/or 132 Da, corresponding to the masses of a hexose and a pentose, respectively (Figs. 3A and 4). Because these peptides were predicted to contain Hyp residues, we assumed that they carried Hyp-O-glycosylations. It should be noted that mass increments of 132 Da corresponding to pentoses were only observed on P1 and P4.
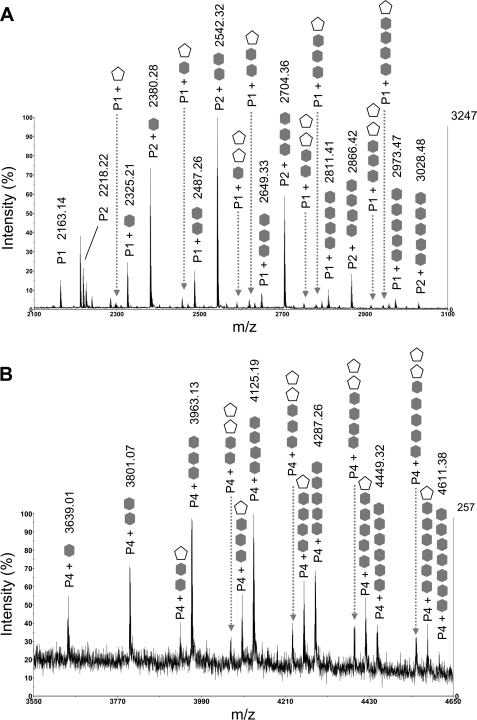
FIGURE 4.
Identification of O-glycopeptides of the AGP31 Pro-rich domain using MALDI-TOF MS. After separation by SDS-PAGE of an AGP31-enriched fraction obtained combining CEC and NAC, bands containing AGP31 were excised from the 50–60-kDa mass range and digested with trypsin. MALDI-TOF MS spectrum analysis showed the presence of tryptic peptides of the Pro-rich domain (P1, P2, and P4, see Figs. 1 and 3A) and associated glycopeptides, regularly spaced by 162 or 132 Da, corresponding to the mass of a hexose (gray hexagon) or a pentose (white pentagon), respectively. The numbers of hexoses and pentoses are indicated. For clarity, monoisotopic masses are indicated only for the most intense O-glycopeptides. A, MALDI-TOF MS spectrum zoom (m/z 2100–3100) on P1 and P2 O-glycopeptides. B, MALDI-TOF MS spectrum zoom (m/z 3550–4650) on P4 O-glycopeptides.
Samples taken along the SDS-polyacrylamide gel lanes, in the 50–60- and 30–40-kDa ranges, displayed various patterns of glycosylation (Fig. 2, lanes 1a, 1b, and 5). First, for a given sample, different glycoforms of each tryptic peptide (P1, P2, and P4) were observed, showing that O-glycosylation of the Pro-rich domain is heterogeneous (Fig. 4). Second, O-glycopeptides are usually larger in the upper part of these mass ranges than in their lower part, consistent with the fact that the smears are due to different degrees of Pro-rich domain O-glycosylation (supplemental Figs. S1, A and B). Note that P1, P2, and P4 were also found as nonglycosylated forms in bands taken at 30 and 50 kDa. Altogether, one to five hexoses were detected by MALDI-TOF MS on P1 and P2, and one to seven were detected on P4 (Table 1). One or two pentoses were detected on glycoforms of P1 and P4 carrying various number of hexoses (Fig. 4). It should be noted that no glycopeptides were observed on MS spectra of samples taken above 60 kDa where AGP31 was also found, even when increasing the m/z limit, suggesting an O-glycosylation pattern not observable by MALDI-TOF MS.
TABLE 1.
MALDI-TOF MS analysis of the tryptic peptides of the AGP31 Pro-rich domain (P0–P4)
For each peptide, the m/z ratio is indicated, as well as the number of Hyp, hexose, and pentose residues determined by MALDI-TOF MS analysis of AGP31-enriched fraction issued from CEC/NAC and PNA isolation strategies. Samples were excised from SDS-polyacrylamide gels in the 30–40- and 50–60-kDa mass ranges (Fig. 2, lanes 1a, 1b, and 5).
| Peptide | m/z | Number of Hyp | Number of hexoses | Number of pentoses |
|---|---|---|---|---|
| P0 | 883.52 | 2 | NDa | ND |
| P1 | 2163.17 | 6 | 1–5 | 1–2 |
| P2 | 2218.25 | 5 | 1–5 | ND |
| P3 | 2486.35 | 6 | ND | ND |
| P4 | 3476.94 | 9 | 1–7 | 1–2 |
a ND, not detected.
An HF treatment was performed on a total CWP extract to remove glycans. After separation on SDS-polyacrylamide gel, bands containing deglycosylated AGP31 were excised, digested with trypsin, and analyzed by MALDI-TOF MS. The spectra obtained revealed that the peaks corresponding to AGP31 glycopeptides from untreated sample (Fig. 3A) disappeared in favor of those corresponding to unmodified peptides, confirming that the mass increments of 162 and 132 Da were due to glycosylations (Fig. 3B). In addition to the increase of intensity of peaks corresponding to P1, P2, and P4, this treatment revealed additional peaks corresponding to peptides of the Pro-rich domain, named P0 and P3 (Fig. 1A), which were not observed in untreated sample. The intensity of P0 was probably too low for detection in MALDI-TOF MS analysis of glycosylated AGP31. The m/z of P3 (2486.35) was close to that of P1 with two hexoses (2487.26), making difficult the identification of this peptide and the corresponding O-glycoforms in MS analyses of untreated sample. These additional MS data provided a full coverage of the Pro-rich domain. The information concerning the peptides of the Pro-rich region, namely P0–P4, is summarized in Table 1. Considering the whole sequence of the Pro-rich domain, a total of 42 Hyp and 29 Pro residues were found.
In Depth MS/MS Analyses of Pro-rich Domain: Location of Hyp and Hexose Residues
Our MALDI-TOF MS data provided a precise determination of Hyp number within the Pro-rich domain. However, the question of the arrangement of the Hyp and Pro residues (respectively 42 and 29), as well as that of the distribution of carbohydrates, was not yet addressed.
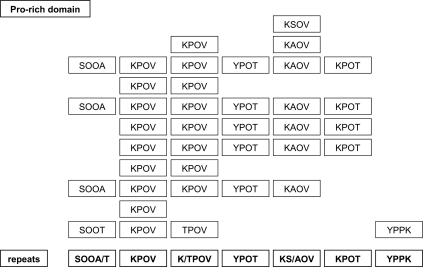
MALDI-TOF/TOF MS/MS experiments were carried out on deglycosylated AGP31 tryptic peptides obtained following gel excision of the HF sample (Fig. 3B). Fragmentation was successfully performed for P1, P2, P3, and P4 (supplemental Fig. S2). Note that it was not possible to perform fragmentation of P0 because of insufficient ion intensity. Analysis of b and y fragment ion masses allowed determination of the number of hydroxylations, i.e. the number of Hyp residues on each fragment. These results permitted precise location of some Hyp. However, it was not possible to determine the exact position of Hyp in some Pro-Pro motifs because fragmentation between two Pro (or two Hyp or Pro-Hyp) does not occur. Then we provided new features about repetitive motifs of the AGP31 Pro-rich domain (Table 2): (i) both Pro were hydroxylated in SPPA and SPPT motifs; (ii) Pro was invariably hydroxylated in KAPV and KSPV motifs; (iii) only one Pro was hydroxylated in KPPT, KPPV, TPPV, and YPPT motifs; and (iv) Pro was not hydroxylated in the YPPK motif. Note that the KPPV motif is the most widespread in the Pro-rich domain.
TABLE 2.
Amino acid sequences of the tryptic peptides of the AGP31 Pro-rich domain (P1–P4)
The locations of Pro (P) and Hyp (O) within the sequences were deduced from MALDI-TOF/TOF MS/MS results (supplemental Figs. S2). Fragmentation experiments were carried out on deglycosylated AGP31 tryptic peptides (P1–P4) obtained following gel excision of the HF sample (Fig. 3B). Pro and Hyp residues whose position was determined experimentally are shown in bold type. When it was not possible to determine the exact position of Hyp in some Pro-Pro motifs, both possible sequences (i.e. Pro-Hyp or Hyp-Pro) are indicated (PO/OP)
| Sequences of the Pro-rich domain tryptic peptides | |
|---|---|
| P1 | AOVSOOAK(PO/OP)VK(PO/OP)VY(PO/OP)TK |
| P2 | AOVK(PO/OP)TK(PO/OP)VK(PO/OP)VY(PO/OP)TK |
| P3 | AOVK(PO/OP)VSOOTK(PO/OP)VT(PO/OP)VYPPK |
| P4 | AOVK(PO/OP)TK(PO/OP)VK(PO/OP)VSOOAK(PO/OP)VK(PO/OP)VY(PO/OP)TK |
Additional LC-MS/MS experiments were performed on O-glycopeptides using ETD, a fragmentation mode that keeps post-translational modifications. They were successful on P2 carrying two, three, four, or five hexoses, which generated the most intense parent ions (Fig. 4A). Fragmentation patterns displaying c and z fragments were obtained, permitting determination of which Hyp residues carried hexoses (supplemental Fig. S3). It was shown from the fragmentation pattern of P2 carrying five hexoses that each of the five Hyp of this peptide was occupied by a hexose, revealing a uniform distribution of hexoses on isolated Hyp (supplemental Fig. S3D). Interestingly, none of the Hyp was shown to carry more than one hexose, demonstrating that the P2 species carrying five hexoses was homogeneous. The fragmentation pattern of P2 carrying two, three, or four hexoses indicated heterogeneous distribution of hexoses onto Hyp (supplemental Fig. S3, A–C). For instance, the fifth Hyp of P2 containing four hexoses was found under glycosylated and nonglycosylated forms (supplemental Fig. S3C). Here again, fragments with several hexoses on a Hyp were never observed.
DISCUSSION
AGP31 is a remarkable cell wall protein with a multidomain organization unique in A. thaliana. Although it was already reported that AGP31 was a glycoprotein, no data about the distribution of Hyp and O-glycans on the protein were available so far (12). The use of diverse MS technologies was innovative and provided an in depth characterization of the AGP31 Pro-rich domain. Combining various experimental approaches, we demonstrated that native AGP31 displays a huge heterogeneity in cell walls, with various O-glycans and truncated forms of its protein core.
Our results provided for the first time a full MS coverage of the Pro-rich region of a native HRGP, whereas most available data were obtained by Edman sequencing of small peptides (reviewed in Refs. 2, 4, and 29) and on recombinant HRGPs (30). Combining literature data with our fragmentation results on tryptic peptides from the Pro-rich domain, we could locate all of the Hyp residues in the Pro-rich domain of AGP31. Indeed, the so-called Hyp-O-glycosylation code based on protein sequence data assumes that Lys, Phe, and Tyr prevent hydroxylation of the following Pro (3). In addition, TPK motifs were found in the maize THRGP (31). Then we propose that KPOT, KPOV, TPOV, and YPOT motifs are present in AGP31 (Fig. 5). The sequences of other repetitive motifs determined experimentally, i.e. SOOA, SOOT, KSOV, and KAOV, are consistent with the Hyp-O-glycosylation code. It should be noted that SOOA/T motifs are similar to those found in EXTs (3, 24). The only exception to the Hyp-O-glycosylation code is the YPPK motif in which none of the Pro is hydroxylated (Fig. 5). The amino acids preceding Pro are assumed to provide a specific local conformation permitting or preventing the action of prolyl 4-hydroxylases involved in Pro hydroxylation. Enzymatic characterization of A. thaliana prolyl 4-hydroxylases provided insight into their substrate specificity (32, 33). Recently, the crystal structure of an algal prolyl 4-hydroxylase complexed with a Ser-Pro5 substrate highlighted the molecular bases of Pro hydroxylation (34). The mechanisms of Pro hydroxylation within Pro-rich domains of PRPs will require further investigation to check whether it follows the same rules as in EXTs and AGPs. As suggested, this code may also depend on the organ and/or plant (35, 36).
FIGURE 5.
Location of Hyp residues within the Pro-rich domain. Hyp residues were located combining MALDI-TOF/TOF MS/MS fragmentation results on deglycosylated tryptic peptides of the AGP31 Pro-rich domain (see supplemental Fig. S2, A–D) and literature data (3, 29). The sequence should be read from left to right and from top to bottom. The Hyp residues of the AGP31 Pro-rich domain are represented by O. The repeated motifs (repeats) are indicated in bold type at the bottom of the figure.
Our experimental data gave new insight into the O-glycosylation of the Pro-rich domain of AGP31. MALDI-TOF MS analyses showed the presence of hexoses and pentoses, assumed to be Gal and Ara according to the PNA-lectin blot detection and the monosaccharide composition of AGP31. The ETD fragmentation technology was employed to describe the distribution of carbohydrates onto the Pro-rich domain, providing the first successful study of a plant protein O-glycosylation using this method. ETD MS/MS experiments performed onto P2 O-glycopeptides containing hexoses showed that Gal are uniformly distributed on isolated Hyp within the KAOV, KPOT, KPOV, and YPOT motifs. Because such experiments were not possible on P1 and P4 O-glycopeptides containing pentoses, we could not locate Ara on these peptides. However, we suggest that Ara or short Ara-oligosaccharides may be carried by contiguous Hyp in SOOA/T motifs, like in EXTs (3, 24). In addition, we cannot exclude that Ser residues within these EXT-like motifs could also be O-galactosylated (3, 24).
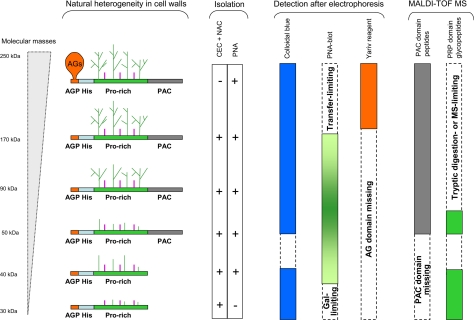
An important feature highlighted in this study is the huge heterogeneity of AGP31. After separation by SDS-PAGE and various detection methods, AGP31, whose predicted molecular mass is 38 kDa, was found as a smear from 30 to ∼250 kDa. Combining all of our results, we could propose structural models schematized on Fig. 6. The Pro-rich domain on which carbohydrates were detected by MALDI-TOF MS is probably also substituted by larger O-glycans that escaped MS analyses because of the large size of the corresponding tryptic glycopeptides. According to the monosaccharide composition of AGP31-enriched fraction obtained using CEC and NAC (53.2% Gal and 39.5% Ara), we propose to call them Hyp-O-Gal/Ara-rich motifs. Note that our monosaccharide analysis showed a higher proportion of Ara than previously reported, probably because our expression and purification procedures were different (12). These Pro-rich domain O-glycans were specifically recognized by PNA as a smear from 34 to 170 kDa, but not by the β-glucosyl Yariv reagent, suggesting a structure different from that of type II arabino-3,6-galactans predominantly found in AGPs (37). The higher molecular mass glycoforms of AGP31 (Fig. 6, smear above 170 kDa) are also assumed to carry AG motifs on the short N-terminal AGP sequence (APAPAP) as suggested by the positive signal with the β-glucosyl Yariv reagent. The absence of a signal above 170 kDa by PNA-lectin blot analysis was probably due to electrophoretic transfer limitations. Finally, truncated forms of AGP31 missing the C-terminal PAC domain also exist (Fig. 6, smear of 30–40 kDa after SDS-PAGE). These latter forms were found to be weakly O-glycosylated on their Pro-rich domain.
FIGURE 6.
AGP31 structural models. The different domains of the mature AGP31 are represented from left to right: N-terminal AGP motif (AGP), His stretch (His), Pro-rich domain (Pro-rich), and C-terminal PAC domain (PAC). The various glycoforms and truncated forms of AGP31 are schematized, as well as the results obtained for isolating, detecting, and analyzing each of them using the different approaches carried out in this study. The limits encountered at each step are indicated. AG motifs assumed to substitute the short N-terminal AGP motif are indicated. The size-heterogeneous Hyp-O-Gal/Ara-rich motifs expected on the Pro-rich domain are represented with green lines, and putative short Ara-oligosaccharides are represented with purple lines.
The heterogeneity of AGP31 in cell walls raises the question of its origin. O-Glycans might be processed by glycoside hydrolases. Galactosidases and arabinosidases were found in A. thaliana cell wall proteomes (38, 39), and several studies provided evidence for enzymatic digestion of AGP O-glycans (40–43). Their degradation has been assumed to produce signal molecules involved in plant development (6, 37). The turnover of AGP O-glycans was also described as part of a salvage pathway allowing the recycling of sugars for the synthesis of new polymers (44). This turnover of O-glycans could explain the observed low molecular mass glycoforms of AGP31. It should be noted that truncated forms of AGP31 missing the C-terminal PAC domain are (i) not glycosylated on the AGP motif and (ii) weakly O-glycosylated on their Pro-rich domain. We suggest that the PAC domain may be degraded by proteases present in A. thaliana cell wall proteomes (45) and that O-glycosylation protects AGP31 from proteolysis, as assumed for AGPs (4). Alternatively, the heterogeneity of the Pro-rich domain O-glycosylation may reflect a partial O-glycosylation of AGP31 along the secretory pathway. Then our ETD MS/MS data suggest that isolated Hyp of the Pro-rich domain may be first galactosylated before the subsequent elongation step. Galactosylation would require two different galactosyltransferases (GalTs) as recently reported for AGPs in A. thaliana and Nicotiana tabacum (46, 47). Two distinct GalT activities were identified from in vitro assays: a Hyp:GalT activity catalyzing the addition of Gal onto peptidyl-Hyp residues and a Gal:GalT activity extending the sugar chain. Twenty putative β-(1,3)-GalTs were predicted by bioinformatics in A. thaliana (48). It will be necessary to characterize the substrate specificity of each of them to elucidate the mechanisms of O-galactosylation of AGPs and PRPs.
Altogether, AGP31 is a glycoprotein far more complex than previously assumed. The next challenge will be to perform the structural characterization of the new Hyp-O-Gal/Ara-rich motifs of its Pro-rich domain. Such work is mostly performed using biochemical approaches like methylation analysis and NMR spectroscopy and requires large amounts of pure protein. In this way, several models were proposed for type II AGs isolated from recombinant AGPs (30). Interestingly, Tryfona et al. (49) carried out an alternative approach to elucidate the structure of native wheat flour AGP, combining AG enzymatic digestion and product analysis using polysaccharide analysis by carbohydrate gel electrophoresis and MS. Note that type III AGs have also been described in Art v 1 and Amb a allergens, which have Pro-rich domains (50, 51).
Finally, it will be important to elucidate the function of each of the AGP31 domains and the role of the various glycans. As a first clue, this work highlighted the remarkable affinity between AGP31 and PNA. Hyp-O-Gal/Ara-rich motifs of the AGP31 Pro-rich domain may interact with cell wall lectins in muro. Legume lectins, which are the closest homologues of PNA in A. thaliana, were found in A. thaliana cell wall proteomes (16, 39). This trail may be interesting to explore the structure/function relationships of AGP31 and other proteins with O-glycosylated Pro-rich domains.
Supplementary Material
Acknowledgments
We thank Dr. S. Fournier and Dr. A. Lemassu for advice on trimethylsilylated derivative analyses, Dr. G. Boudart for HF deglycosylation experiments, the late Prof. R. Pont-Lezica for critical reading of the manuscript, and Dr. H. Canut for stimulating discussions. We also thank Prof. M. Kieliszewski (University of Ohio) for welcoming Dr. G. Boudart into her laboratory.
This work was supported by the Université Paul Sabatier (Toulouse, France), the CNRS (France), and a grant from the Lebanon ecological association (to M. H.). This work has been done in the LRSV, part of the “Laboratoire d'Excellence” (LABEX) entitled TULIP (ANR-10-LABX-41).

This article contains supplemental Figs. S1–S3.
- Hyp
- hydroxyproline(s)
- AG
- arabinogalactan
- AGP
- arabinogalactan protein
- CEC
- cation exchange chromatography
- CWP
- cell wall protein
- ETD
- electron transfer dissociation
- EXT
- extensin
- GalT
- galactosyl transferase
- HRGP
- hydroxyproline-rich glycoprotein
- NAC
- nickel affinity chromatography
- PAC
- PRP-AGP containing Cys
- PMF
- peptide mass fingerprinting
- PNA
- peanut agglutinin lectin
- PRP
- Pro-rich protein
- Ara
- arabinose
- Gal
- galactose
- DIG
- digoxigenin.
REFERENCES
- 1. Lamport D., Northcote D. (1960) Hydroxyproline in primary cell walls of higher plants. Nature 188, 665–66613757790 [Google Scholar]
- 2. Kieliszewski M., Lamport D. (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172 [DOI] [PubMed] [Google Scholar]
- 3. Kieliszewski M. (2001) The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57, 319–323 [DOI] [PubMed] [Google Scholar]
- 4. Showalter A. (1993) Structure and function of plant cell wall proteins. Plant Cell 5, 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seifert G. J., Roberts K. (2007) The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161 [DOI] [PubMed] [Google Scholar]
- 6. Nothnagel E. (1997) Proteoglycans and related components in plant cells. Int. Rev. Cytol. 174, 195–291 [DOI] [PubMed] [Google Scholar]
- 7. Cannon M. C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B. L., Chen L., Lamport D. T., Chen Y., Kieliszewski M. J. (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. U.S.A. 105, 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sommer-Knudsen J., Clarke A. E., Bacic A. (1996) A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 9, 71–83 [DOI] [PubMed] [Google Scholar]
- 9. Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Averyhart-Fullard V., Datta K., Marcus A. (1988) A hydroxyproline-rich protein in the soybean cell wall. Proc. Natl. Acad. Sci. U.S.A. 85, 1082–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datta K., Schmidt A., Marcus A. (1989) Characterization of two soybean repetitive proline-rich proteins and a cognate cDNA from germinated axes. Plant Cell 1, 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu C., Mehdy M. (2007) A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiol. 145, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kleis-San Francisco S., Tierney M. (1990) Isolation and characterization of a proline-rich cell wall protein from soybean seedlings. Plant Physiol. 94, 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fowler T., Bernhardt C., Tierney M. (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol. 121, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kieliszewski M., de Zacks R., Leykam J. F., Lamport D. T. (1992) A repetitive proline-rich protein from the gymnosperm Douglas fir is a hydroxyproline-rich glycoprotein. Plant Physiol. 98, 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana. Confirmed actors and newcomers. BMC Plant Biol. 8, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feiz L., Irshad M., Pont-Lezica R. F., Canut H., Jamet E. (2006) Evaluation of cell wall preparations for proteomics. A new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arockia Jeyaprakash A., Jayashree G., Mahanta S. K., Swaminathan C. P., Sekar K., Surolia A., Vijayan M. (2005) Structural basis for the energetics of jacalin-sugar interactions. Promiscuity versus specificity. J. Mol. Biol. 347, 181–188 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y., Giboulot A., Zivy M., Valot B., Jamet E., Albenne C. (2011) Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 20. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 21. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22. Willats W. G., Knox J. P. (1996) A role for arabinogalactan-proteins in plant cell expansion. Evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925 [DOI] [PubMed] [Google Scholar]
- 23. Merkle R. K., Poppe I. (1994) Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 230, 1–15 [DOI] [PubMed] [Google Scholar]
- 24. Shpak E., Leykam J. F., Kieliszewski M. J. (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. U.S.A. 96, 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mort A. J., Lamport D. T. (1977) Anhydrous hydrogen fluoride deglycosylates glycoproteins. Anal. Biochem. 82, 289–309 [DOI] [PubMed] [Google Scholar]
- 26. Borderies G., Jamet E., Lafitte C., Rossignol M., Jauneau A., Boudart G., Monsarrat B., Esquerré-Tugayé M. T., Boudet A., Pont-Lezica R. (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures. A critical analysis. Electrophoresis 24, 3421–3432 [DOI] [PubMed] [Google Scholar]
- 27. Boudart G., Jamet E., Rossignol M., Lafitte C., Borderies G., Jauneau A., Esquerré-Tugayé M. T., Pont-Lezica R. (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes. Identification by mass spectrometry and bioinformatics. Proteomics 5, 212–221 [DOI] [PubMed] [Google Scholar]
- 28. Baldwin T., Domingo C., Schindler T., Seetharaman G., Stacey N., Roberts K. (2001) DcAGPI, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol. Biol. 45, 421–435 [DOI] [PubMed] [Google Scholar]
- 29. Sommer-Knudsen J., Bacic A., Clarke A. (1998) Hydroxyproline-rich plant glycoproteins. Phytochemistry 47, 483–497 [Google Scholar]
- 30. Tan L., Varnai P., Lamport D. T., Yuan C., Xu J., Qiu F., Kieliszewski M. J. (2010) Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kieliszewski M. J., Leykam J. F., Lamport D. T. (1990) Structure of the threonine-rich extensin from Zea mays. Plant Physiol. 92, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hieta R., Myllyharju J. (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor α-like peptides. J. Biol. Chem. 277, 23965–23971 [DOI] [PubMed] [Google Scholar]
- 33. Tiainen P., Myllyharju J., Koivunen P. (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280, 1142–1148 [DOI] [PubMed] [Google Scholar]
- 34. Koski M. K., Hieta R., Hirsilä M., Rönkä A., Myllyharju J., Wierenga R. K. (2009) The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J. Biol. Chem. 284, 25290–25301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278 [DOI] [PubMed] [Google Scholar]
- 36. Estevez J., Kieliszewski M., Khitrov N., Somerville C. (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol. 142, 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaspar Y., Johnson K., McKenna J., Bacic A., Schultz C. (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176 [PubMed] [Google Scholar]
- 38. Minic Z., Jouanin L. (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 44, 435–449 [DOI] [PubMed] [Google Scholar]
- 39. Jamet E., Canut H., Boudart G., Pont-Lezica R. (2006) Cell wall proteins. A new insight through proteomics. Trends Plant Sci. 11, 33–39 [DOI] [PubMed] [Google Scholar]
- 40. Kotake T., Tsuchiya K., Aohara T., Konishi T., Kaneko S., Igarashi K., Samejima M., Tsumuraya Y. (2006) An α-l-arabinofuranosidase/β-d-xylosidase from immature seeds of radish (Raphanus sativus L.). J. Exp. Bot. 57, 2353–2362 [DOI] [PubMed] [Google Scholar]
- 41. Kotake T., Dina S., Konishi T., Kaneko S., Igarashi K., Samejima M., Watanabe Y., Kimura K., Tsumuraya Y. (2005) Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1–3)- and β-(1–6)-galactosyl residues of arabinogalactan protein. Plant Physiol. 138, 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hata K., Tanaka M., Tsumuraya Y., Hashimoto Y. (1992) α-l-Arabinofuranosidase from radish (Raphanus sativus L.) seeds. Plant Physiol. 100, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eudes A., Mouille G., Thévenin J., Goyallon A., Minic Z., Jouanin L. (2008) Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 49, 1331–1341 [DOI] [PubMed] [Google Scholar]
- 44. Gibeaut D., Carpita N. (1991) Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol. 97, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jamet E., Albenne C., Boudart G., Irshad M., Canut H., Pont-Lezica R. (2008) Recent advances in plant cell wall proteomics. Proteomics 8, 893–908 [DOI] [PubMed] [Google Scholar]
- 46. Oka T., Saito F., Shimma Y., Yoko-o T., Nomura Y., Matsuoka K., Jigami Y. (2010) Characterization of endoplasmic reticulum-localized UDP-d-galactose. Hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol. 152, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang Y., Faik A., Kieliszewski M., Tan L., Xu W. L., Showalter A. (2010) Identification and characterization of in vitro galactosyltransferase activities involved in arabinogalactan-protein glycosylation in tobacco and Arabidopsis. Plant Physiol. 154, 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qu Y., Egelund J., Gilson P. R., Houghton F., Gleeson P. A., Schultz C. J., Bacic A. (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 68, 43–59 [DOI] [PubMed] [Google Scholar]
- 49. Tryfona T., Liang H. C., Kotake T., Kaneko S., Marsh J., Ichinose H., Lovegrove A., Tsumuraya Y., Shewry P. R., Stephens E., Dupree P. (2010) Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr. Res. 345, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 50. Léonard R., Wopfner N., Pabst M., Stadlmann J., Petersen B. O., Duus J. Ø., Himly M., Radauer C., Gadermaier G., Razzazi-Fazeli E., Ferreira F., Altmann F. (2010) A new allergen from ragweed (Ambrosia artemisiifolia) with homology to art v 1 from mugwort. J. Biol. Chem. 285, 27192–27200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leonard R., Petersen B. O., Himly M., Kaar W., Wopfner N., Kolarich D., van Ree R., Ebner C., Duus J. Ø., Ferreira F., Altmann F. (2005) Two novel types of O-glycans on the mugwort pollen allergen Art v 1 and their role in antibody binding. J. Biol. Chem. 280, 7932–7940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.