Abstract
Sea surface temperatures (SST) are rising because of global climate change. As a result, pathogenic Vibrio species that infect humans and marine organisms during warmer summer months are of growing concern. Coral reefs, in particular, are already experiencing unprecedented degradation worldwide due in part to infectious disease outbreaks and bleaching episodes that are exacerbated by increasing SST. For example, Vibrio coralliilyticus, a globally distributed bacterium associated with multiple coral diseases, infects corals at temperatures above 27 °C. The mechanisms underlying this temperature-dependent pathogenicity, however, are unknown. In this study, we identify potential virulence mechanisms using whole genome sequencing of V. coralliilyticus ATCC (American Type Culture Collection) BAA-450. Furthermore, we demonstrate direct temperature regulation of numerous virulence factors using proteomic analysis and bioassays. Virulence factors involved in motility, host degradation, secretion, antimicrobial resistance and transcriptional regulation are upregulated at the higher virulent temperature of 27 °C, concurrent with phenotypic changes in motility, antibiotic resistance, hemolysis, cytotoxicity and bioluminescence. These results provide evidence that temperature regulates multiple virulence mechanisms in V. coralliilyticus, independent of abundance. The ecological and biological significance of this temperature-dependent virulence response is reinforced by climate change models that predict tropical SST to consistently exceed 27 °C during the spring, summer and fall seasons. We propose V. coralliilyticus as a model Gram-negative bacterium to study temperature-dependent pathogenicity in Vibrio-related diseases.
Keywords: Vibrio pathogens, coral disease, genome and proteome, quorum sensing, global climate change, temperature
Introduction
The correlation between temperature and disease is of an escalating concern because of observed and predicted changes attributed to global climate change (Hoegh-Guldberg and Bruno, 2010). Record breaking temperatures are occurring more frequently with the ten warmest years in recorded history experienced over the last 13 years (NOAA, 2011), and the average global temperature is predicted to increase from 1.8 to 4.0 °C in the 21st Century (IPCC, 2007). Concurrently, increased incidence and/or severity of diseases have been observed in human (Patz et al., 2005) and marine (Harvell et al., 2009) ecosystems. In coral reefs, an estimated one third of coral species are at a risk of extinction largely because of global warming and disease (Carpenter et al., 2008). Mass mortality of Caribbean coral ecosystems occurred in 1998 (Aronson et al., 2000) and 2005 (Eakin et al., 2010), two of the hottest years recorded, with record breaking sea surface temperatures (SST) (NOAA, 2011). Localized temperature-related bleaching episodes have also increased in frequency (Whiteman, 2010) and are predicted to occur biannually within 20 years (Donner et al., 2007).
Pathogens of the genus Vibrio are associated with temperature-related diseases exhibiting peak infection rates in humans (Igbinosa and Okoh, 2008) and corals (Vezzulli et al., 2010) following the warmer summer months. High summer temperatures correlate with increased V. cholerae outbreaks (Fernandez et al., 2009; Hashizume et al., 2011), as well as infections caused by V. parahaemolyticus and V. vulnificus (Iwamoto et al., 2010). Increased SST undoubtedly causes an increase in abundance of vibrios (Vezzulli et al., 2010); however, temperature also has a more direct role in Vibrio pathogenicity (Oh et al., 2009), although little is known regarding specific mechanisms involved in temperature-related infections.
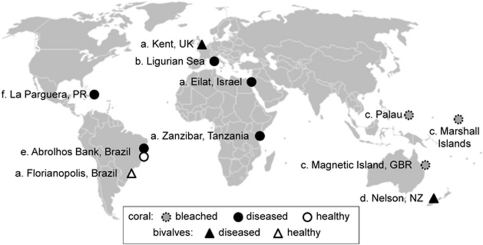
V. coralliilyticus is of interest because of its global distribution, broad host range and temperature-dependent pathogenicity in corals. V. coralliilyticus has been isolated from marine organisms in the Atlantic (Ben-Haim et al., 2003a; Alves et al., 2010; Vizcaino et al., 2010), Indian (Ben-Haim et al., 2003a) and Pacific Oceans (Sussman et al., 2008; Kesarcodi-Watson et al., 2009), as well as the Mediterranean (Vezzulli et al., 2010) and Red Seas (Ben-Haim et al., 2003a) (Figure 1). It causes fatal infections in a wide range of organisms, including unicellular algae (Ben-Haim et al., 2003b; de Oliveira Santos et al., 2011), corals (Ben-Haim et al., 2003b), oysters (Jeffries, 1982), shrimp (Austin et al., 2005; de Oliveira Santos et al., 2011), rainbow trout (Austin et al., 2005) and flies (Alves et al., 2010; de Oliveira Santos et al., 2011) during experimental infection assays. Although it is uncertain whether V. coralliilyticus is a primary or opportunistic coral pathogen, evidence strongly suggests that this endemic member of global coral holobionts (Pollock et al., 2010) has a role in coral disease (Rosenberg and Kushmaro, 2011). Infection experiments establish the ability of V. coralliilyticus to cause bacterial bleaching (Ben-Haim et al., 2003b), white syndrome (Sussman et al., 2008) and mortality in corals (Alves et al., 2010; Vezzulli et al., 2010), in addition to being associated with the microbial consortium of black band disease (Arotsker et al., 2009). V. coralliilyticus type strain ATCC (American Type Culture Collection, Manassas, VA, USA) BAA-450 (Vc450), isolated from bleached corals near Zanzibar, displays a tightly regulated temperature-dependent virulence; it is capable of invading and lysing coral tissue of Pocillopora damicornis at temperatures >27 °C, it attacks the symbiotic algae of this coral at temperatures between 24 °C and 26.5 °C and is avirulent at temperatures ⩽24 °C (Ben-Haim et al., 2003b). Further, Vc450 was recently shown to provoke a physiological response in P. damicornis during a temperature-induced infection experiment (Vidal-Dupiol et al., 2011). V. coralliilyticus P1 (VcP1), isolated from diseased corals in the Great Barrier Reef, infects corals at 28–31 °C (Sussman et al., 2008). It has been speculated that a zinc-metalloprotease may be driving these infections (Ben-Haim et al., 2003a; Sussman et al., 2009); however, recent infection experiments using a zinc-metalloprotease mutant of VcP1 (vcpA) revealed no significant differences in pathogenicity (de Oliveira Santos et al., 2011).
Figure 1.
Global distribution of V. coralliilyticus strains. The V. coralliilyticus strains represented here are (a) type strains (Ben-Haim et al., 2003a), as well as strains identified using (b) DnaJ PCR (Vezzulli et al., 2010), (c, d) 16S rRNA sequencing (Sussman et al., 2008; Kesarcodi-Watson et al., 2009), (e) multi-locus sequencing (Alves et al., 2010) and (f) multiple molecular analyzes, that is, 16S rRNA sequencing, recA PCR and repetitive extragenic palindromic - polymerase chain reaction (REP-PCR) (Vizcaino et al., 2010).
In this study, we identify potential virulence factors in Vc450 using whole genome sequencing and compare our results with that of the recently published VcP1 draft genome (de Oliveira Santos et al., 2011). In addition, we use two-dimensional liquid chromatography coupled with tandem mass spectrometry and bioassays to investigate the influence of nonpathogenic (24 °C) and pathogenic (27 °C) temperatures on the expression of virulence factors in Vc450. We demonstrate that Vc450 maintains a broad array of virulence mechanisms, similar yet distinct from VcP1, and provide evidence that increased temperature results in a significant increase in the number and expression level of numerous virulence factors, including flagellar-mediated motility, secretion systems, host degradation and antimicrobial resistance, as well as transcriptional regulators including quorum sensing (QS).
Materials and methods
Vc450 genome sequencing
The Vc450 genome was sequenced, assembled and finished at the Joint Genome Institute (Los Alamos, NM, USA). Draft sequences were obtained from paired-end Sanger sequencing on 8 kb plasmid libraries (5 times coverage) and 454 sequences (20 times coverage), providing 6.5 times total coverage. Details regarding sequencing and library construction can be found at http://www.jgi.doe.gov/. Gene finding and annotation were achieved using the RAST server (Aziz et al., 2008), and all genome comparisons were performed using SEED Viewer 2.0 (Overbeek et al., 2005). The Vc450 whole genome sequence data have been submitted to the GenBank database under the accession no. ACZN00000000. See Supplementary Materials and methods for further details.
Growth curves and protein quantification
Vc450 was grown on glycerol artificial sea water (GASW) agar (Smith and Hayasaka, 1982), with inocula taken from frozen glycerol stocks and tested for purity prior to use. Individual colonies were grown at either 24 °C or 27 °C overnight, and 4 ml of each Vc450 inoculum was transferred to 96 ml of GASW media. After 24 h, the optical density (OD610) was measured using a spectrophotometer (Beckman Coulter DU 800 Spectrophotometer, Fullerton, CA, USA), and 1 ml of 2.4 OD610 Vc450 inoculum was added to 99 ml GASW media. The OD610 and the number of colony forming units were determined for Vc450 cultures grown at 24 °C and 27 °C at 0, 2, 4, 6, 8, 12 and 24 h after inoculation. Total protein was quantified from aliquots collected concurrently with OD and CFU samples. Samples were centrifuged and cell pellets washed with 0.09% NaCl. Pellets were resuspended in 200 μl of 0.045% NaCl, 1 NaOH and boiled for 10 min. Extracted protein (100 μl) was added to 1 ml Coomassie Blue (Sigma, St Louis, MO, USA) and measured by a spectrophotometer at a 595-nm wavelength. The protein quantification was calculated using the regression equation of a bovine serum albumin standard curve.
Protein extraction
Vc450 cultures were grown as described above for 12 h to early stationary phase. The liquid culture was centrifuged and the resulting Vc450 pellets were resuspended in lysis buffer (40 m Tris pH 8.0, 10 m sodium fluoride and 1 × Complete Protease Inhibitor Cocktail Tablet stock (Roche Diagnostics, Pleasanton, CA, USA)). The resuspended pellet was vortexed with 0.1 mm silica beads (90 s) in a Mini-Bead Beater (BioSpec Products Inc., Bartlesville, OK, USA) three times, and the lysate was recovered and centrifuged to remove remaining beads. The protein concentration was determined with Commassie Plus—The Better Bradford Assay (Thermo Scientific, Rockford IL, USA). Isolated proteins were reduced using 10 m dithiothreitol and 1.6 mg l−1 RapiGest (Waters, Milford, MA, USA), and alkylated by adding 50 m iodoacetamide. An additional incubation was performed at room temperature for 30 min after adding 50 m dithiothreitol. The proteins were desalted and washed three times with Tris buffer (25 m Tris with 1 mg l−1 RapiGest) and concentrated using Ultrafree centrifuge tubes (membrane cutoff at >10 kDa; Millipore, Billerica, MA, USA). The concentrated proteins were resuspended in Tris buffer and digested overnight at 37 °C with a 1:50 ratio of trypsin and 1 mg l−1 RapiGest. To stop digestion, 3 HCl was added to each sample and incubated at 37 °C for 60 min. The supernatant was transferred to a fresh vial after centrifugation, and the protein was dried under vacuum and stored at −20 °C.
Two-dimensional liquid chromatography coupled with tandem mass spectrometry
In two independent experiments, Vc450 peptides were fractionated by strong cation exchange chromatography in a 2.1-mm Polysulfoethyl A ion-exchange column (PolyLC, Columbia, MD, USA). The peptides were separated at a flow rate of 200 μl min−1 using a 100-min gradient. Each fraction was further analyzed by LC-MS/MS using a reverse-phase C18 1 mm column (Waters) or a C18 75 μm column (Microtech Scientific, Anaheim, CA, USA) on an LTQ (linear trap quadruple) linear ion trap mass spectrometer (Thermo Fischer Scientific, San Jose, CA, USA). Mass spectra from both experiments were matched to predicted tryptic peptides from the Vc450 genome using Turbo SEQUEST (Eng et al., 1994). SEQUEST search result files (.srf) of the combined dataset (Supplementary Figure 1) were loaded into Scaffold (Proteome Software Inc., Portland, OR, USA; version Scaffold_2_05_01) for validation of peptide and protein identifications. Only proteins identified by Turbo SEQUEST and validated by Scaffold (Supplementary Figure 1) were included in the spectral counting analysis performed in Scaffold. We applied the G-test of independence, a likelihood ratio test for discreet data, to quantify the relative expression of proteins (that is, the number of spectral counts per protein) between Vc450 grown at 24 °C and 27 °C. Spectral counts were normalized according to the total number of spectral counts for both data sets, as suggested previously (Old et al., 2005; Hendrickson et al., 2006). See Supplementary Materials and methods for further details.
Electron microscopy
A Vc450 cell suspension (∼1010 cells ml−1) was prepared in 1 ml of fixative (3% glutaraldehyde in 0.1 sodium cacodylate, pH 7.2). After 24 h of incubation at 4 °C, the cells were washed twice in 0.9% saline and added to 200 μl of 1% phosphotungstic acid (pH 6.8). Fifteen μl of cells were applied to the surface of a 300-mesh, carbon-coated, formvar-coated copper grid. Excess stain was removed, and the grids were air-dried. A JEOL 1011 transmission electron microscope (JEOL USA Inc., Peabody, MA, USA) operating at an accelerating voltage of 80 kV was used to examine the Vc450 cells.
Motility assay
A single colony each of Vc450 grown at 24 °C and 27 °C was inoculated into 3 ml GASW media and incubated at the corresponding temperature overnight at 180 r.p.m. Cell densities were adjusted to OD595 1.0, and 1 μl of the adjusted culture was stabbed into the center of a 0.35% GASW agar plates. The plates were then incubated at the respective temperatures for 24 h before the diameter of the growth zone was measured. Three independent cultures were performed in triplicate (N=9) for each temperature.
Chinese hamster ovary (CHO) cell assay
Vc450 was grown in casamino-yeast extract, Proteose peptone no. 3 and brain heart infusion medium with 2% salt at 24 °C, 27 °C and 30 °C, with agitation. Culture aliquots were taken after 6–8 h and after 24 h of growth. Cell supernatants were obtained by centrifugation of cells and cell lysates were prepared by incubating the cell pellet in tris-buffered saline containing 2 mg ml−1 polymixin B. CHO cells were grown in Eagle's minimum essential medium supplemented with 10% heat-inactivated fetal calf serum, 10% tryptose phosphate broth, penicillin (100 IU ml−1), 0.01% streptomycin and 0.14% sodium bicarbonate. The ability of the culture supernatant fluids and cell lysates to alter the morphology of CHO cells or to lyse them was determined using the same medium without the tryptose phosphate broth, but supplemented with 1% heat-inactivated fetal calf serum.
Hemolysis assay
Vc450 was grown on Trypticase Soy Agar amended with 5% Sheep Blood (Becton Dickinson, Sparks, MD, USA) at 24 °C and 27 °C in three independent cultures at each temperature. V. vulnificus and Escherichia coli were used as positive and negative controls, respectively.
Assays for autoinducer (AI)-1 and AI-2 signaling molecules
Vc450 was cultured as described above and grown in triplicate at 21 °C, 24 °C, 27 °C, 30 °C, 33 °C or 37 °C, and 1.5 ml was collected from each sample after 3, 12 and 24 h. The cell cultures were centrifuged, and the supernatant was filtered (0.2 μm) and stored at 4 °C. The V. harveyi reporter strains, BB886 (ATCC BAA-1118, luxPQ∷tn5kAN) and BB170 (ATCC BAA-1117, luxN∷tn5 kAN) were used to determine the presence of AI-1 and AI-2 signaling molecules, respectively. The bioluminescence assays were performed as described previously (Bassler et al., 1994). Luminescence measurements were taken using a luminometer (BMG Novostar, Ortenberg, Germany), normalized to background controls (that is, reporter strains with sterile media added), and presented as the fold change compared with endogenous levels of luminescence expressed by the reporter strains. See Supplementary Materials and methods for further details.
Results and Discussion
V. coralliilyticus genome
To investigate the presence of potential virulence factors, we performed whole genome sequencing of Vc450 and identified an asymmetrical, two-chromosome structure consistent among all Vibrio genomes examined (Okada et al., 2005; Chun et al., 2009). The larger (C1—3 416 103 bp) and smaller (C2—1 865 911 bp) chromosomes follow a gene distribution pattern typical for vibrios with C1 predominantly carrying genes for viability and growth, and C2 mostly bearing genes for adaptation to environmental change (Makino et al., 2003). A total of 5078 protein-coding sequences were identified (GenBank ACZN00000000): 3047 from C1 and 1656 from C2, with 81.1% and 74.7% showing sequence homology to proteins with known or putative functions, respectively. In addition, Vc450 contains a megaplasmid of ∼398 614 bp, encoding 369 coding sequences with 55% annotated as hypothetical proteins, indicating that the majority of the genes code for potentially novel proteins. The Vc450 genome lacks the IntI4 integrase associated with the superintegron cassette present in most vibrios; however, elements of the superintegron (for example, the RelEB toxin-antitoxin replicon stability system) are located on the megaplasmid, indicating acquisition and/or relocation of cassettes of the superintegron to a mobile conjugative replicon.
A gross comparison of the Vc450 and VcP1 genomes (Table 1) yields a conserved genome structure, consisting of two chromosomes, consistent for Vibrio species, a large plasmid and a conserved gene content (4478 shared genes). However, closer scrutiny reveals notable differences between the two strains. Although both genomes contain large plasmids, the Vc450 plasmid is considerably larger (∼399 kbp) than the one observed in VcP1, estimated at 252 kbp by comparative genomics (Supplementary Figure 2) and pulsed field gel electrophoresis (Supplementary Figure 3). Additionally, 12% of each genome is unique, representing 600 and 629 coding sequences in Vc450 and VcP1, respectively. This includes important virulence factors, such as a repeats-in-toxin (RTX) toxin, type 3 secretion genes and pilus proteins that are unique to Vc450. VcP1, in contrast, contains 12 unique prophage, transposon and integron regions, as well as a unique flagellar operon. This is consistent with a recent report that closely related Vibrio strains harbor unique integrons as a result of lateral gene transfer (Koenig et al., 2011). Further, genomic comparison at the nucleotide level shows a substantial level of divergence, ANIb (average nucleotide identity via BLAST)=96.6, between the two strains (Goris et al. 2007).
Table 1. Characteristics of the Vc450 and VcP1 genome.
| Characteristics | Vc450 |
VcP1 | |||
|---|---|---|---|---|---|
| C1 | C2 | pBAA-450 | Total | Total | |
| Size (bp) | 3 416 103 | 1 865 911 | 398 614 | 5 680 628 | 5 513 256 |
| Contigs | 17 | 2 | 1 | 20 | 230 |
| G+C content | 45.60% | 45.30% | 48.50% | 46.50% | 46% |
| CDS | 3053 | 1656 | 369 | 5078 | 5107 |
| Hypothetical | 607 | 419 | 173 | 1199 | 1245 |
| Identified | 2440 | 1237 | 196 | 3873 | 3862 |
| Total RNAs | 117 | 5 | 0 | 122 | 58 |
| tRNAs | 87 | 5 | 0 | 92 | 53 |
| rRNAs | 30 | 0 | 0 | 30 | 5 |
V. corallilyticus proteome
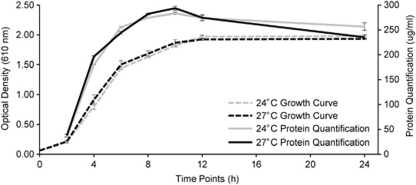
Vibrio pathogenicity is multifactorial, requiring the expression of numerous virulence factors and other essential genes for infectivity. Differential expression of these proteins regulates the multiple stages of microbial disease, including transmission, adhesion, penetration, survival and host injury. To assess gene expression in Vc450, we compared proteins expressed at its avirulent (24 °C) and virulent (27 °C) temperatures, using two-dimensional liquid chromatography coupled to tandem mass spectrometry. Growth curve analyses and protein assays did not reveal a significant difference in Vc450 growth at the temperatures utilized in this study (Figure 2), demonstrating that the differential regulation of virulence factors was not caused by changes in growth or abundance. Spectral counting provided quantification of the relative abundance of individual proteins between the two temperatures, revealing significant changes in gene expression (Supplementary Table 1). Our results revealed significant upregulation of 136 virulence-associated genes encoded in the genome of Vc450 grown at 27 °C, including factors involved in motility, host degradation, QS, antimicrobial resistance, secretion and transcriptional regulation (Table 2). In contrast, ribosomal proteins and general stress proteins (that is, heat shock and cold shock proteins) were downregulated at the higher temperature (Supplementary Table 1), providing additional evidence that the upregulation of these putative virulence factors is not a growth-dependent response.
Figure 2.
The growth rate and total protein production of Vc450 is similar whether grown at 24 °C or 27 °C. Vc450 was grown in GASW media at 24 °C and 27 °C. The growth curves were prepared using optical density at time points 0, 2, 4, 6, 8, 10, 12 and 24 h for both 24 °C (  ) and 27 °C (- - - ). Protein production was measured using the Bradford assay for both 24 °C (
) and 27 °C (- - - ). Protein production was measured using the Bradford assay for both 24 °C (  ) and 27 °C (—) from time points 2–24 h. Error bars represent the standard deviation of three replicate samples.
) and 27 °C (—) from time points 2–24 h. Error bars represent the standard deviation of three replicate samples.
Table 2. Number of Vc450 genes and proteins identified within a given virulence category.
| Virulence categories | Genome | 24 °C proteomea | 27 °C proteomea |
|---|---|---|---|
| Chemotaxis/motility | |||
| Chemotaxis proteinsb | 57 | 20 (2) | 38 (31) |
| Flagellar proteinsb | 82 | 13 | 23 (16) |
| Host degradation | |||
| Toxinsb | 4 | 1 | 3 (3) |
| Hemolysin/cytolysinb | 14 | 3 | 4 (2) |
| Proteases | 45 | 18 (7) | 21 (7) |
| Resistance | |||
| Multidrug efflux pumps | 32 | 3 (2) | 4 (2) |
| Multidrug resistance | 11 | 4 (2) | 3 |
| Specific antibiotic | 14 | 1 | 2 |
| Secretion | |||
| T1b | 203 | 50 (11) | 68 (35) |
| T2 | 15 | 7 (1) | 9 (6) |
| Tfpb | 50 | 10 | 13 (10) |
| T3 | 21 | 1 | 1 |
| T4 | 14 | 0 | 0 |
| T6b | 42 | 10 (1) | 16 (12) |
| Regulation | |||
| Sigma factorsb | 28 | 7 (1) | 10(5) |
| H-NSb | 1 | 1 | 1 (1) |
| Quorum sensing signaling/receptors | 8 | 6 (1) | 5 (2) |
| Quorum sensing response regulatorsb | 12 | 4 (1) | 7 (4) |
The number of proteins significantly upregulated at the temperature indicated is given in parenthesis.
Virulence categories significantly upregulated as a whole at 27 °C using a G-test statistic (P>0.05).
In the initial stage of a Vibrio infection, chemotaxis and motility are essential for Vibrio species to locate and initiate infection in their host. V. vulnificus (Lee et al., 2004), V. anguillarum (Ormonde et al., 2000) and V. fischeri (Millikan and Ruby, 2004), all display attenuated infection in motility-deficient mutants. Similarly, nonmotile Vc450 mutants are unable to infect the coral, Pocillopora damicornis (Meron et al., 2009). Vc450 and VcP1 contain two adjacent regions involved in lateral flagella gene system (VIC_004722–VIC_004762); however, no gene products were present in the proteome and no lateral flagella were observed by electron microscopy. In contrast, the upregulation of polar flagellar proteins (Table 2) and increased motility observed at 27 °C (Supplementary Figure 4) demonstrate that temperature influences Vc450 motility via its single polar flagellum. Furthermore, there is an increase in methyl-accepting chemotaxis proteins that relay environmental signaling cues to flagellar motor controls, with 80% of these significantly upregulated (Table 2). Increased diversity of the methyl-accepting chemotaxis proteins allows a more sensitive response to changing conditions in the environment (Tran et al., 2008), whereas increased expression provides the ability to amplify a response signal (Parkinson, 2004). These results support the hypothesis that a temperature-dependent increase in chemotaxis and motility of Vc450 contributes to its increased virulence at higher temperatures.
Following transmission, secretion systems are utilized by vibrios to transport macromolecules necessary for the remaining stages of infection. The Vc450 genome encodes five of the six described bacterial secretion systems (T1-T6SS): T1-4SS and T6SS (two clusters). In addition, we identified type IV pilus (Tfp) subsystems, including three tight adherence (Tad) locus colonization islands, mannose-sensitive hemagglutinin (MSHA) genes and Pil components. VcP1 contains the same five secretion systems and three Tfp subsystems; however, there are notable differences. Whereas T2SS (that is, general secretion genes) and T6SS genes display 98–100% homology between Vc450 and VcP1, the T3SS genes (VIC_001039–VIC_001055) and one of the Tfp Tad clusters (VIC_001023–VIC_001030) located on the Vc450 megaplasmid display <50% sequence similarity with the corresponding VcP1 genes. Similarly, Vc450 and VcP1 T4SS conjugation genes of the megaplasmid exhibit only 67% sequence similarity.
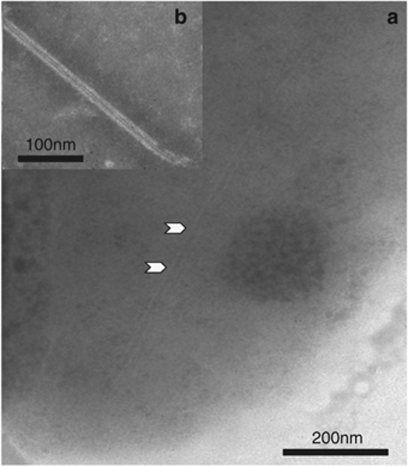
Expression of T1SS, T2SS, Tfp (MSHA and Tad) and T6SS genes are upregulated by Vc450 at 27 °C (Supplementary Table 1). Both pathogenic and nonpathogenic vibrios secrete factors necessary for host colonization, including RTX toxins, proteases and hemolysins via T1SS and T2SS, whereas the Tfp subsystems are important for biofilm formation, colonization and phage transductions by pathogens, including V. cholerae (Wooldridge, 2009). Of the two clusters of T6SS genes, 12 out of 19 genes from one cluster (VIC_003912–VIC_003930) are upregulated at 27 °C, whereas only 3 of the 23 genes from the second cluster (VIC_003136–VIC_003158), including the hcp effector gene (ZP_05886652.1), are upregulated at the higher temperature. Interestingly, VipA (annotated as ImpB) is expressed from both clusters (ZP_05886644.1 and ZP_05887414.1). It is unclear whether both copies need to be expressed to produce the T6SS tubules observed in Vc450 (Figure 3), which resemble those observed in other vibrios (Bonemann et al., 2009). The upregulation observed in such a broad array of secretion systems indicates an increased capacity for a variety of functions that facilitate establishment of Vc450 at higher temperatures.
Figure 3.
Transmission electron photomicrograph of Vc450 T6SS tubular structure. Vc450 cell stained with 0.5% sodium phosphotungstic acid, pH 6.8. (a) A VipA/VipB-like T6SS tubular structure, similar to that described for V. cholerae, is evident in the cytoplasm (black arrowheads). (b) A Vc450 VipA/VipB-like T6SS tubular structure found outside of a cell.
Once associated with a host, pathogenic Vibrio species employ various mechanisms, including antibiotic resistance, to maintain competitiveness against other microorganisms and to ward off host defenses. Vc450 exhibits a temperature-dependent increase in resistance to the antimicrobial activity of coral-associated bacteria, as well as to therapeutic antibiotics (Vizcaino et al., 2010). We observed a larger number of multidrug-resistance efflux pump proteins expressed at 27 °C, with two of these (ZP_05885878.1 and ZP_05883974.1) displaying significant upregulation (Supplementary Table 1). Multidrug efflux pumps have a significant role in virulence of V. cholerae, and are required for resistance to the host innate immune system (Bina et al., 2008). This may be relevant to Vc450, as the innate immune system of cnidarians, including corals, shares some conserved defenses with that of higher vertebrates, including humans (Dunn, 2009). Evidence also suggests that multidrug-resistance efflux pumps provide an alternative function in bacterial pathogenicity, including transport of virulence factors (Piddock, 2006). The convergence of temperature-dependent virulence and increased antibiotic resistance in Vc450 highlights its unique attributes as a model organism in a warming environment compounded by multiple stressors.
Host degradation factors, such as proteases and toxins, also contribute to Vibrio pathogenicity (Thompson et al., 2004). In the Vc450 genome, there are 45 annotated proteases, 2 of which (VIC_003472 thermolysin/zinc-metalloprotease and VIC_002633 neutral protease precursor) have homologous regions with metalloproteases previously identified in V. coralliilyticus infection studies (Ben-Haim et al., 2003b; Sussman et al., 2009). Out of the 21 proteases identified in the Vc450 proteome, neither VIC_003472 nor VIC_002633 were detected. This could be the result of strict parameters employed in identifying proteins in the Vc450 proteome, or it could indicate that there is redundancy in the metalloprotease functionality as suggested previously (de Oliveira Santos et al., 2011).
Toxins, including hemolysins, have a significant role in Vibrio pathogenicity (Thompson et al., 2004; Igbinosa and Okoh, 2008), and their activity has been shown to be directly affected by temperature (for example, enterohaemorrhagic E. coli) (Li et al., 2008). Vc450 exhibits upregulation of two hemolysins (ZP_05886322.1 and ZP_05888459.1) at 27 °C, with corresponding enhanced hemolytic activity (Supplementary Figure 5). Vc450 also displays significant upregulation of the RTX toxin (ZP_05887531.1), which is not present in VcP1. Further screening of Vc450 supernatants and cell lysates with CHO cells revealed the production of multiple active proteins. Vc450 produced a cell elongation factor at all temperatures assayed early in its growth curve and into stationary phase. At 24 h of growth, Vc450 secreted a nonhemolytic cytotoxic substance at higher temperatures (27 °C and 30 °C) and a hemolysin, which had low cytotoxic activity against CHO cells, present at all temperatures tested. Although we could not predict the cell elongation factor of Vc450 from the proteome, we hypothesize that the RTX toxin is the most likely candidate as the cytotoxic substance, as it is a pore-forming toxin and the CHO cells were not completely lysed during the assay. In V. cholerae, RTX toxin acts as a virulence cofactor disrupting the cell wall integrity of the host cells (Olivier et al., 2007), whereas the V. vulnificus RTX toxin causes cell lysis through pore formation, resulting in the degradation of phagocytic host cells (Lo et al., 2011). An increased expression of RTX toxin in Vc450 at the virulent temperature of 27 °C may allow for increased survival of Vc450 owing to degradation of the host's innate immune system.
V. coralliilyticus pathogenicity islands
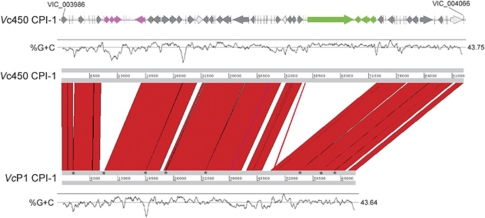
Pathogenic vibrios, like many bacteria, commonly acquire virulence factors via horizontal transfer of bacteriophages and pathogenicity islands (Chun et al., 2009). We identified two novel pathogenicity islands in the genome of Vc450. Coralliilyticus pathogenicity island-1 (CPI-1) is located on C1 at an integration site consistent with the dif-like region of V. cholerae and V. parahaemolyticus, the insertion site for CTX and f237 phages in these species, respectively. CPI-1 contains homologs of the VvhA cytolysin (VIC_004014) and the associated secretory protein VvhB (VIC_004013), which produce and secrete the primary toxin of V. vulnificus, respectively. CPI-1 additionally carries a putative bacteriocin (VIC_004006) and the RTX toxin mentioned above (VIC_004043). This integration locus also possesses two T3SS clusters, presumably for secretion of the different effector proteins within the island. The VcP1 genome carries a similar pathogenicity island to the CPI-1 (Figure 4), with most corresponding genes sharing >90% homology. However, the RTX toxin (ZP_05887531.1), one of 12 CPI-1 proteins upregulated at 27 °C in the Vc450 proteome, is unique to the Vc450 CPI-1. The upregulation of numerous proteins from the CPI-1 at 27 °C indicates that CPI-1 contributes to Vc450 pathogenicity at a higher temperature, whereas the low homology between the Vc450 and VcP1 RTX toxins indicates a potential difference in virulence between the two strains.
Figure 4.
Schematic representation of Vc450 CPI-1 and comparison with VcP1 using Artemis Comparison Tool (ACT, Wellcome Trust Sanger Institute, Hinxton, UK). Coding sequences of CPI-1 from Vc450 are shown on the top, with G+C content directly below. Two large regions are present in Vc450 and not in VcP1, one with mostly hypothetical proteins (fuschia) and the other containing an RTX homolog, transporter and associated genes (green). Additionally, there were three regions of gene-level divergence (light gray) at VIC_4016, VIC_4037 and within VIC_4065. CPI-1 of VcP1 is composed of nine contigs: AEQS01000075, −105, −183, −138, −135, −025, −161, −224 and −071, with contig gaps (asterisks) indicated on the bottom ACT scale.
The Vc450 genome contains a second novel pathogenicity island, coralliilyticus pathogenicity island-2, which is not found in VcP1. Coralliilyticus pathogenicity island-2 is similar to vibrio seventh pandemic island-II of V. cholerae and located on C1, inserted between a tRNA (Uracil54-C5-)-methyltransferase (VIC_000153) and an adenosine triphosphatases (VIC_000179) of the AAA+ class (Supplementary Figure 6). The tandem arrangement of these two genes is highly conserved among Vibrio, Allivibrio and Photobacterium species, with only four species (Vc450, V. furnissii CIP102972, V. alginolyticus 12G01 and V. cholerae RC385) known to contain a genomic island at this site.
Temperature-dependent regulation
The altered regulation of virulence factors and phenotypic changes documented in this study indicate that temperature affects Vc450 virulence mechanisms independent of growth or abundance. We hypothesize that global transcriptional regulators, which by definition are capable of affecting the expression of numerous genes from multiple pathways, are driving the temperature modulation observed. The Vc450 genome contains bacterial thermosensors and regulators known to influence downstream virulence signaling in other vibrios, and these transcriptional regulators are differentially expressed between 24 °C and 27 °C (Table 2). For example, nucleoid-associated protein (H-NS) binds DNA at lower temperatures blocking the transcription of multiple genes, whereas higher temperatures cause loosening in the DNA structure allowing transcription to occur. H-NS suppresses virulence-associated genes, such as RTX and CTX, in V. vulnificus (Liu et al., 2009) and V. cholerae (Stonehouse et al., 2011), respectively. The concurrent upregulation of H-NS (ZP_05887985.1) and RTX toxin (ZP_05887531.1) on CPI-1 in the Vc450 proteome at 27 °C could be the result of a conserved Vc450 H-NS unable to regulate a more recently acquired RTX toxin. Alternatively, decoupling of the H-NS protein from the RTX toxin DNA could allow greater detection of H-NS protein concurrent with the resulting upregulation of RTX toxin.
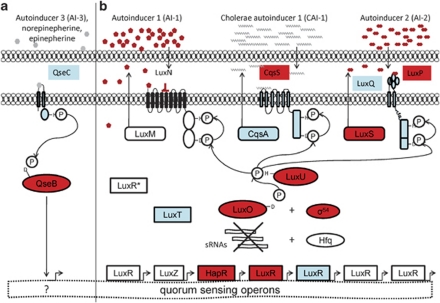
QS is another global mechanism by which temperature can directly regulate virulence in Vc450. In V.cholerae, V. harveyi and V. parahaemolyticus QS is achieved through AI stimulation of histidine kinase receptor pathways (Ng and Bassler, 2009). Activation of these pathways result in the transcription of small RNAs, which subsequently degrade the messenger RNA of virulence factors (Ng and Bassler, 2009). Temperature has recently been shown to affect QS mechanisms (Tait et al., 2010). For example, V. mediterranei produces four N-Acyl homoserine lactone at 18 °C compared with only two at 25 °C and 30 °C (Tait et al., 2010), and there is evidence that temperature can affect the level of AHL production positively (Hasegawa et al., 2005; Latour et al., 2007) and negatively (Tait et al., 2010). The Vc450 genome possesses three (AI-1/LuxMN, AI-2/LuxSPQ and CAI-1/CqsAS) two-component histidine kinase QS pathways (Figure 5) and three small RNAs (Supplementary Figure 7), characteristic of those identified in the regulation of other Vibrio species QS (Lenz et al., 2004). In addition, the AI-3/QseBC QS pathway, originally described in enterohemorrhagic E. coli is present (Figure 5), providing a potential mechanism for direct interactions with a host (Hughes and Sperandio, 2008). Although this system is not well characterized in vibrios, genomic comparisons reveal the presence of qseBC genes in numerous Vibrio species (Supplementary Figure 8). The four QS systems present in Vc450 are also present in VcP1 and share >98% homology, indicating that these systems are common between two geographically distinct strains of V. coralliilyticus.
Figure 5.
Proposed Vc450 QS systems. This figure illustrates the potential QS mechanisms utilized by V. coralliilyticus at high density. (a) The QseBC system is a two-component system, in which the QseC histidine kinase receptor becomes phosphorylated when bound to cognate ligands and subsequently activates QseB through phosphorelay. The activated QseB molecule binds DNA, acting as a direct transcriptional regulator. (b) The three two-component histidine kinase receptor systems previously described in Vibrio species, each produce and detect a specified class of AI. Ligand binding, at levels above the density threshold, blocks the kinase activity of the membrane-bound receptors, reversing the phosphorelay. This results in the dephosphorylation of the response regulator, LuxO, via LuxU. In its unactivated state, LuxO is unable to transcriptionally activate the sRNAs that degrade the messenger RNA (mRNA) of LuxR-type genes. Thus, LuxR-type mRNA is stabilized and proteins are produced. The LuxR-type proteins in turn act as transcriptional regulators of virulence-associated genes. All of the proteins shown are present in the Vc450 genome. The shaded (blue and red) proteins are present in the Vc450 proteome, with (red) proteins representing those significantly affected by temperature.
The AI2/LuxSPQ, CAI-1/CqsAS and the AI-3/QseBC QS systems are detected in the Vc450 proteome (Figure 5), whereas AI signaling from AI-1/LuxMN and AI-2/LuxSPQ QS systems were established using bioluminescence reporter assays (Supplementary Figure 9). Collectively, these results indicate that all four QS pathways found in the Vc450 genome are active (Figure 5). Furthermore, the Vc450 proteome exhibits upregulation at 27 °C of numerous QS proteins (Supplementary Table 1), including the following: histidine kinase receptors (ZP_05879449.1, ZP_05886587.1), response regulators (ZP_05888198.1, ZP_05888199.1) and transcriptional regulators (ZP_05887548.1, ZP_05884374.1). Bioluminescence reporter assays also reveal significant temperature effects on AI-1 and AI-2 (Supplementary Figure 9) signaling, indicating that temperature has a direct effect on Vc450 QS.
Conclusions
V. coralliilyticus is considered an endemic member of coral reef ecosystems, consisting of geographically distinct strains that exhibit genetic variations (Pollock et al., 2010). Vc450, isolated from the Indian Ocean, and VcP1, isolated from the GBR, represent two geographically distinct strains that cause different coral infections (that is, bleaching and white syndrome, respectively), indicating that each of them harbor unique virulence mechanisms in addition to shared virulence characteristics. The complexity of virulence-associated factors expressed by Vc450, similar to the complexity described for VcP1 (de Oliveira Santos et al., 2011), suggests that V. coralliilyticus infections depend on the coordinated expression of multiple factors. Although the two strains share conserved virulence-associated genetic components, such as flagellar, secretion and QS systems, we describe here a number of genomic differences between Vc450 and VcP1, namely RTX toxin, proteases and pathogenicity islands, that most likely account for their unique physiological characteristics. Further, the ANIb of 96.6 between the two genomes argues that although Vc450 and VcP1 are strains of the same species, they have diverged significantly in a vertical fashion, in addition to the lateral differences described above, and may represent distinct ecotypes or subspecies. Much remains unknown, however, regarding the ecological and physiological differences among strains of V. coralliilyticus.
In a warming ocean, the confluence of genetic mobility, temperature-dependent virulence and increased antimicrobial resistance makes V. coralliilyticus a formidable global pathogen with broad host specificity. Vc450 exhibits resistance to many common antibiotics (that is, tetracycline, erythromycin and quinolones (Vizcaino et al., 2010)) and to date, phage therapy is the only proposed strategy for mitigation of V. coralliilyticus infections (Efrony et al., 2009). Elucidating temperature-dependent virulence mechanisms of V. coralliilyticus may assist in the design of antivirulence therapies (Cegelski et al., 2008) for this organism, as well as for other vibrios, which exhibit temperature-related disease outbreaks, including V. cholerae. With the world's oceans changing rapidly (Hoegh-Guldberg and Bruno, 2010), we hypothesize that V. coralliilyticus will become a sustained threat to coral reefs and propose that V. coralliilyticus establishes a model to further elucidate temperature-dependent virulence mechanisms.
Acknowledgments
This work was supported by NSF Biodiversity Surveys and Inventories (DEB 0516347, DEB 0964997) to PJM, a NSF Foundation Graduate Research Fellowship to NEK, the NOAA OHHI Distinguished Scholars program to RCC, and NOAA (SO660009) and NIH (1R01A139129-01) to RRC. Sequencing support was received from the Office of the Chief Scientist (USA), University of Maryland Vibrio Genome Sequencing Project and the Los Alamos National Laboratory. The Fellowship for Interpretation of Genomes (FIG, Argonne National Laboratory) and the National Institute of Allergy and Infectious Diseases (NIH) were instrumental in supporting the RAST and the SEED data analysis environments. We thank Veronika Vonstein and Ross Overbeek for their assistance with the RAST system, Lisa Kilpatrick (NIST) and Kevin Schey/Jennifer Bethard (MUSC Mass Spectrometry Facility) for the use of their facilities to perform the two-dimensional liquid chromatography coupled with tandem mass spectrometry experiments, and Jana Lee (Proteome Software) for assistance in using the Scaffold software.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alves N, Neto OSM, Silva BSO, de Moura RL, Francini-Filho RB, Castro CB, et al. Diversity and pathogenic potential of vibrios isolated from Abrolhos Bank corals. Environ Microbiol Rep. 2010;2:90–95. doi: 10.1111/j.1758-2229.2009.00101.x. [DOI] [PubMed] [Google Scholar]
- Aronson RB, Precht WF, Macintyre IG, Murdoch TJ. Coral bleach-out in Belize. Nature. 2000;405:36. doi: 10.1038/35011132. [DOI] [PubMed] [Google Scholar]
- Arotsker L, Siboni N, Ben-Dov E, Kramarsky-Winter E, Loya Y, Kushmaro A. Vibrio sp. as a potentially important member of the black band disease (BBD) consortium in Favia sp. corals. FEMS Microbiol Ecol. 2009;70:515–524. doi: 10.1111/j.1574-6941.2009.00770.x. [DOI] [PubMed] [Google Scholar]
- Austin B, Austin D, Sutherland R, Thompson FL, Swings J. Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ Microbiol. 2005;7:1488–1495. doi: 10.1111/j.1462-2920.2005.00847.x. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, et al. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Internatl J Syst Evol Microbiol. 2003a;53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003b;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Provenzano D, Nguyen N, Bina JE. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun. 2008;76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter KE, Abrar M, Aeby G, Aronson R, Banks S, Bruckner AW, et al. One-third of reef-building corals face extinction risk from climate change and local impacts. Science. 2008;321:560–563. doi: 10.1126/science.1159196. [DOI] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–28. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Santos E, Alves N, Jr, Dias GM, Mazotto AM, Vermelho A, Vora GJ, et al. Genomic and proteomic analyses of the coral pathogen Vibrio coralliilyticus reveal a diverse virulence repertoire. ISME J. 2011;5:1471–1483. doi: 10.1038/ismej.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner SD, Knutson TR, Oppenheimer M. Model-based assessment of the role of human-induced climate change in the 2005 Caribbean coral bleaching event. Proc Natl Acad Sci USA. 2007;104:5483–5488. doi: 10.1073/pnas.0610122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SR. Immunorecognition and immunoreceptors in the Cnidaria. Invertebr Surv J. 2009;6:7–14. [Google Scholar]
- Eakin CM, Morgan JA, Heron SF, Smith TB, Liu G, Alvarez-Filip L, et al. Caribbean corals in crisis: record thermal stress, bleaching, and mortality in 2005. PLoS One. 2010;5:e13969. doi: 10.1371/journal.pone.0013969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efrony R, Atad I, Rosenberg E. Phage therapy of coral white plague disease: properties of phage BA3. Curr Microbiol. 2009;58:139–145. doi: 10.1007/s00284-008-9290-x. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fernandez MAL, Bauernfeind A, Jimenez JD, Gil CL, Omeiri NE, Guibert DH. Influence of temperature and rainfall on the evolution of cholera epidemics in Lusaka, Zambia, 2003–2006: analysis of a time series. Trans R Soc Trop Med Hyg. 2009;103:137–143. doi: 10.1016/j.trstmh.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Harvell CD, Altizer S, Cattadori IM, Harrington L, Weil E. Climate change and wildlife diseases: when does the host matter the most. Ecology. 2009;90:912–920. doi: 10.1890/08-0616.1. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Chatterjee A, Cui Y, Chatterjee AK. Elevated temperature enhances virulence of Erwinia carotovora subsp. carotovora strain EC153 to plants and stimulates production of the quorum sensing signal, N-acyl homoserine lactone, and extracellular proteins. Appl Environ Microbiol. 2005;71:4655–4663. doi: 10.1128/AEM.71.8.4655-4663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume M, Faruque ASG, Terao T, Yunus M, Streatfield K, Yamamoto T, et al. The Indian Ocean dipole and cholera incidence in Bangladesh: a time series. Environ Health Perspect. 2011;119:239–244. doi: 10.1289/ehp.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Bruno JF. The impact of climate change on the world's marine ecosystems. Science. 2010;328:1523–1528. doi: 10.1126/science.1189930. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa EO, Okoh AI. Emerging Vibrio species: an unending threat to public health in developing countries. Res Microbiol. 2008;159:495–506. doi: 10.1016/j.resmic.2008.07.001. [DOI] [PubMed] [Google Scholar]
- IPCC . Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press: Cambridge, UK; 2007. [Google Scholar]
- Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev. 2010;23:399–411. doi: 10.1128/CMR.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries VE. Three Vibrio strains pathogenic to larvae of Crassostrea gigas and Ostrea edulis. Aquaculture. 1982;29:201–226. [Google Scholar]
- Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L. Two pathogens of Greenshell (TM) mussel larvae, Perna canaliculus: Vibrio splendidus and a V.coralliilyticus/neptunius-like isolate. J Fish Dis. 2009;32:499–507. doi: 10.1111/j.1365-2761.2009.01006.x. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Bourne DG, Curtis B, Dlutek M, Stokes HW, Doolittle WF, et al. Coral-mucus-associated Vibrio integrons in the Great Barrier Reef: genomic hotspots for environmental adaptation. ISME J. 2011;5:962–972. doi: 10.1038/ismej.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour X, Diallo S, Chevalier S, Morin D, Smadja B, Burini J, et al. Thermoregulation of N-acyl homoserine lactone-based quorum sensing in the soft rot bacterium Pectobacterium atrosepticum. Appl Environ Microbiol. 2007;73:4078–4081. doi: 10.1128/AEM.02681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Rho JB, Park KS, Kim CB, Han Y, Choi SH, et al. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect Immun. 2004;72:4905–4910. doi: 10.1128/IAI.72.8.4905-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Li H, Granat A, Stewart V, Gillespie JR. RpoS, H-NS, and DsrA inluence EHEC hemolysin operon(ehxCABD) transcription in Escherichia coli O157:H7 strainEDL933. FEMS Microbiol Lett. 2008;285:257–262. doi: 10.1111/j.1574-6968.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- Liu MC, Naka H, Crosa JH. HlyU acts as an H-NS antirepressor in the regulation of the RTX toxin gene essential for the virulence of the human pathogen Vibrio vulnificus CMCP6. Mol Microbiol. 2009;72:491–505. doi: 10.1111/j.1365-2958.2009.06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H, Lin J, Chen Y, Chen C, Shao C, Lai Y, et al. RTX toxin enhances the survival of Vibrio vulnificus during infection by protecting the organism from phagocytosis. J Infect Dis. 2011;203:1866–1874. doi: 10.1093/infdis/jir070. [DOI] [PubMed] [Google Scholar]
- Makino K, Oshima K, Kurokawa K, Katsushi Y, Honda T, Shinagawa H, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from V. cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- Meron D, Efrony R, Johnson WR, Schaefer AL, Morris PJ, Rosenberg E, et al. Role of flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2009;75:5704–5707. doi: 10.1128/AEM.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan DS, Ruby E. Vibrio fischeri Flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA 2011National Climatic Data Center, State of the climate: global analysis annual 2010published online January 2011, retrieved on 20 January 2011 from http://www.ncdc.noaa.gov/sotc/gloabl/ .
- Oh MH, Lee SM, Lee DH, Choi SH. Regulation of the Vibrio vulnificushupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect Immun. 2009;77:1208–1215. doi: 10.1128/IAI.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Iida T, Kita-Tsukamotot K, Honda T. Vibrios commonly possess two chromosomes. J Bacteriol. 2005;187:752–757. doi: 10.1128/JB.187.2.752-757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, et al. Comparison of lable-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Olivier V, Haines K, Tan Y, Satchell KJF. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor 01 strains. Infect Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormonde P, Horstedt P, O'toole R, Milton D. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J Bacteriol. 2000;182:2326–2328. doi: 10.1128/jb.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signal amplification in bacterial chemotaxis through receptor teamwork. ASM News. 2004;70:575–582. [Google Scholar]
- Patz JA, Campbell-Lendrum D, Holloway T, Foley JA. Impact of regional climate change on human health. Nature. 2005;438:310–317. doi: 10.1038/nature04188. [DOI] [PubMed] [Google Scholar]
- Piddock LJV. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Pollock JF, Wilson B, Johnson WR, Morris PJ, Willis BL, Bourne DG. Phylogeny of the coral pathogen Vibrio coralliilyticus. Environ Microbiol Rep. 2010;2:172–178. doi: 10.1111/j.1758-2229.2009.00131.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Kushmaro A.2011Microbial diseases of corals: pathology and physiologyIn: Dubinsky Z, Stambler N (eds).Coral Reefs: An Ecosystem In Transition Springer: New York; 451–464. [Google Scholar]
- Smith G, Hayasaka SS. Nitrogenase activity associated with Halodule wrightii roots. Appl Environ Microbiol. 1982;43:1244–1248. doi: 10.1128/aem.43.6.1244-1248.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK. H-NS binding and repression of the ctx promoter in Vibrio cholerae. J Bacteriol. 2011;193:979–988. doi: 10.1128/JB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Mieog JC, Doyle J, Victor S, Willis B, Bourne DG. Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS Biol. 2009;4:e4511. doi: 10.1371/journal.pone.0004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Willis B, Victor S, Bourne D. Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. PLoS One. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait K, Hutchinson Z, Thompson FL, Munn CB. Quorum sensing signal production and inhibition bt coral-associated vibrios. Environ Microbiol Rep. 2010;2:145–150. doi: 10.1111/j.1758-2229.2009.00122.x. [DOI] [PubMed] [Google Scholar]
- Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Krushkal J, Antommattei FM, Lovley DR, Weis RM. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BCM Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L, Previati M, Pruzzo C, Marchese A, Bourne DG, Cerrano C, et al. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbiol. 2010;12:2007–2019. doi: 10.1111/j.1462-2920.2010.02209.x. [DOI] [PubMed] [Google Scholar]
- Vidal-Dupiol J, Ladriere O, Meistertzheim A, Foure L, Adjeroud M, Mitta G. Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J Experimental Biol. 2011;214:1533–1545. doi: 10.1242/jeb.053165. [DOI] [PubMed] [Google Scholar]
- Vizcaino MI, Johnson WR, Kimes NE, Williams K, Torralba M, Nelson KE, et al. Antimicrobial resistance of the coral pathogen Vibrio coralliilyticus and Caribbean sister phylotypes isolated from a diseased octocoral. Microb Ecol. 2010;59:646–657. doi: 10.1007/s00248-010-9644-3. [DOI] [PubMed] [Google Scholar]
- Whiteman E. A fatal switch for corals. PLoS Biol. 2010;8:e1000346. doi: 10.1371/journal.pbio.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge K. Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. Caister Academic Press: Portland; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.