Abstract
Two mechanisms that play important roles in cell fate decisions are control of a “core transcriptional network” and repression of alternative transcriptional programs by antagonizing transcription factors. Whether these two mechanisms operate together is not known. Here we report that GATA-1, SCL, and Klf1 form an erythroid core transcriptional network by co-occupying >300 genes. Importantly, we find that PU.1, a negative regulator of terminal erythroid differentiation, is a highly integrated component of this network. GATA-1, SCL, and Klf1 act to promote, whereas PU.1 represses expression of many of the core network genes. PU.1 also represses the genes encoding GATA-1, SCL, Klf1, and important GATA-1 cofactors. Conversely, in addition to repressing PU.1 expression, GATA-1 also binds to and represses >100 PU.1 myelo-lymphoid gene targets in erythroid progenitors. Mathematical modeling further supports that this dual mechanism of repressing both the opposing upstream activator and its downstream targets provides a synergistic, robust mechanism for lineage specification. Taken together, these results amalgamate two key developmental principles, namely, regulation of a core transcriptional network and repression of an alternative transcriptional program, thereby enhancing our understanding of the mechanisms that establish cellular identity.
Keywords: ChIP sequencing, erythropoiesis, cross antagonism
Although cells have hundreds of transcriptional regulators, the function of only a few key factors has been proposed to be critical for establishing and/or maintaining cellular identity (1). Studies in just a few cell types, particularly embryonic stem (ES) cells, support this concept (2, 3). In ES cells, the “core pluripotency factors” Oct4, Nanog, and Sox2 co-occupy ∼300 genes that are enriched for developmental regulators and genes involved in self-renewal (2). The gene network formed by these three core ES cell factors exhibits several types of regulatory circuitry including a multi-input motif and feed-forward loops (2, 3). However, whether such a “core transcriptional network” exists in hematopoietic cells is not known.
GATA-1, SCL, and Klf1 are three essential erythroid promoting transcription factors that play critical roles in establishing erythroid identity through the up-regulation of erythroid-specific genes (4, 5). GATA-1 regulates expression of some erythroid-specific genes, such as globin genes, in association with SCL (6–8) and Klf1 (9–11). Recent studies of transcription factor occupancy in erythroid progenitors by chromatin immunoprecipitation and high-throughput sequencing (ChIP-Seq) revealed that GATA-1 bound regions are enriched for SCL binding elements (12, 13), SCL bound regions are enriched for potential GATA-1 binding sites (14), and Klf1 occupied regions are enriched for putative GATA-1 and SCL binding motifs (15). Although there is evidence that these three factors cooperatively regulate certain erythroid-specific genes, whether they form a network with features similar to the ES cell core transcriptional network is not known.
Whereas GATA-1, SCL, and Klf1 are essential for erythroid development, the myelo-lymphoid promoting transcription factor, PU.1, is a negative regulator of terminal erythroid differentiation (16–19). Surprisingly, PU.1 was found to occupy more genes in erythroid progenitors than the three erythroid-promoting factors (20). However, the extent of overlap between the genes bound by PU.1 and the three erythroid factors is not known.
In this study, we provide genomic evidence for the existence of a core erythroid network of >300 genes that are co-occupied and regulated by GATA-1, SCL, and Klf1. This network has characteristic features of core transcriptional networks, including a multi-input motif and feed-forward loops. Furthermore, we also find that PU.1 binds to and represses most of the genes in this network, indicating that PU.1 is a highly integrated negative regulator of the core erythroid network. Conversely, we also find that GATA-1 binds to and represses >100 PU.1 myelo-lymphoid gene targets in erythroid progenitors. Finally, mathematical modeling reveals that the dual mechanism used by both GATA-1 and PU.1 to repress an alternative lineage-specific transcriptional program provides a robust mechanism for lineage specification.
Results
GATA-1 Preferentially Binds Distal to Genes.
To begin to investigate the possible existence of a “core erythroid network,” we carried out ChIP-Seq experiments on endogenous GATA-1 in normal, murine ES cell-derived erythroid progenitors (ES-EP) (21) in both proliferating and differentiating conditions. The following results indicate that our ChIP-Seq data are of high quality. Several well-established GATA-1 binding sites are found in the dataset, including binding sites in the β-globin locus control region (LCR) (Fig. S1C). GATA-1 bound regions in both proliferating and differentiating conditions are highly enriched with a characteristic GATA-1 binding motif (Fig. S1D), similar to that observed in recent studies (12, 13). Using quantitative chromatin immunoprecipitation (qChIP) for GATA-1, we validated 100% (18/18) of the sites bound in both proliferating and differentiating conditions (Fig. S2).
Several recent reports describe genome-wide occupancy of GATA-1 in several murine and human erythroid cell lines (12, 13, 22, 23). Similar to these other reports, we find that GATA-1 most often binds distal to gene promoters; 80–90% of GATA-1 occupied sites are in intragenic or intergenic regions, and <20% of bound regions are within 2 kb of transcription start sites (TSS) (Fig. S1B). However, in contrast to the results in one of the studies with murine erythroleukemia (MEL) cells (22), we find that the number of sites occupied by GATA-1 increases during differentiation of erythroid progenitors, from 6,600 sites in proliferating progenitors to 10,600 sites in differentiating cells. In addition, we find that the magnitude of many GATA-1 ChIP-Seq peaks increases during differentiation at sites occupied in both proliferating and differentiating cells (Fig. S1 A and C). These results provide unique genome-wide occupancy maps of GATA-1 in normal erythroid progenitors, in both proliferating and differentiating conditions.
Associating GATA-1 Occupancy with Gene Regulation.
We next sought to understand the relationship between GATA-1 bound sites and genes. Because there is no well-established method for assigning transcription factor bound sites to genes (24, 25), we tested four different criteria and found that assigning genes to GATA-1 peaks that lie within the region spanning −20 kb of TSS to +10 kb of transcription end site (TES) leads to a greater median gene expression change than assigning peaks to the closest gene (P value = 3.5 × 10−2) (Fig. S3). The two other criteria, using smaller distances between the peak and TSS, did not improve upon this result (Fig. S3). Using the −20 kb of TSS to +10 kb of TES assignment, we associated GATA-1 binding with 5,293 genes (combined total in proliferating and differentiating conditions), including many well-characterized GATA-1 target genes such as GATA-1, GATA-2, and FOG1 (SI Materials and Methods).
GATA-1, SCL, and Klf1 Form a Core Erythroid Network.
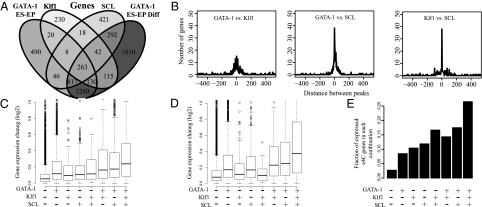
To explore the existence of a core erythroid network, we generated gene assignments for SCL and Klf1 using recently published genome-wide binding data for these two factors in fetal-liver erythroid progenitors (FL-EP) (14, 15), using the same criterion (−20 kb TSS to +10 kb TES). We found that each factor binds to a large number of genes independent of the other two factors. GATA-1 binds 3,560 (490 + 1,810 + 1,260) genes independently, and SCL and Klf1 bind independently to 421 and 230 genes, respectively (Fig. 1A). Importantly, however, the analysis revealed that 313 (263 + 8 + 42) genes were co-occupied by the three factors (P value <2.2 × 10−16) (Fig. 1B), which is strikingly similar to the number of genes that are co-occupied by the three core ES cell factors (353) (2). Furthermore, there is a strong bias for the three erythroid factors to bind in close proximity (within 200 bp) to one another near these 313 genes (Fig. 1B). This bias is significantly different from the pairwise peak distances computed for the genes bound by only two of the three factors (Fisher's exact test, P value <2.2 × 10−16). This finding suggests that the three erythroid factors form a dense overlapping regulon and, more specifically, a multi-input motif, similar to the core networks described in ES, pancreatic, and liver cells (2, 3). We also find that the genes co-occupied by GATA-1, SCL, and Klf1 exhibit the largest changes in gene expression during the differentiation of both ES-EP and MEL cells (Fig. 1 C and D). The expression change (a median of 1.2-fold) is significantly greater than the changes observed in genes either not occupied by any of the three factors (P value <3 × 10−56) or occupied by other combinations of these factors (P values <3 × 10−10 in comparisons to genes bound independently by each factor and between 0.08 and 2 × 10−6 in comparisons to genes occupied by two of the three factors). Furthermore, we find that genes co-occupied by the three factors are preferentially enriched for loci that have been reported to associate and form transcription factories with either the α- or the β-globin loci in erythroid cells (26) (Fig. 1E). In combination, these results suggest that genes bound by the three transcription factors may be enriched for genes involved in erythroid differentiation.
Fig. 1.
GATA-1, SCL, and Klf1 co-occupy many genes and bind in close proximity to each other. (A) Using occupancy maps derived from ChIP-Seq analysis of GATA-1 in proliferating and differentiating ES-EP along with SCL (14) and Klf1 (15) occupancy maps in FL-EP, we determined what genes were occupied by each factor. Occupancy was defined as a factor binding within −20 kb of TSS through +10 kb of TES of a given gene. A four-set Venn diagram is displayed, comparing the number of genes occupied by each factor. (B) The distance between bound sites of each factor was calculated for genes that are occupied by SCL, Klf1, and GATA-1 (in proliferating and/or differentiating cells). (C and D) Boxplots show the relative gene expression changes that occur during the differentiation of ES-EP (C) (20) or MEL cells (D) (20) for genes occupied by the indicated combination of factors. (E) The fraction of genes with the indicated occupancy of the three erythroid factors that were found to be expressed and form transcription factories with either α- or β-globin loci in erythroid cells as identified in a recent study (26).
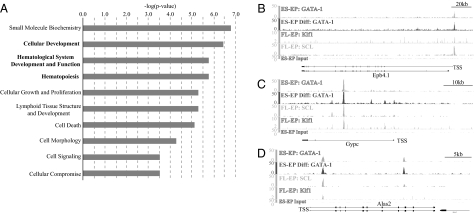
To test this possibility, we performed pathway analysis (Ingenuity Systems) to assess the biological functions of the genes bound by GATA-1, SCL, and Klf1. The analysis showed that these genes are indeed enriched for genes involved in development (P value = 3.24 × 10−7) and hematopoiesis (P value = 1.52 × 10−6) (Fig. 2A). Moreover, comparison of the gene list with the Gene Ontology (GO) term Erythrocyte Differentiation (GO term 0030218) showed that the genes occupied by the three factors are enriched for erythroid-specific genes (binomial P value = 1.0 × 10−5). Consistent with these findings, we also find that these triply occupied genes are overrepresented (binomial P value <2.2 × 10−16) in the group of genes that were previously found to be highly expressed in the erythroid lineage (27). Three such genes are band 4.1 (Epb4.1), glycophorin C (Gypc), and aminolevulinic acid synthase 2 (Alas2) (Fig. 2 B–D). These results indicate that the genes bound by GATA-1, SCL, and Klf1 constitute an erythroid core network.
Fig. 2.
Genes bound by GATA-1, SCL, and Klf1 are enriched for erythroid genes. (A) Ingenuity pathway analysis (IPA) was performed on genes co-occupied by all three erythroid factors. The 10 most significantly enriched molecular and cellular functions/physiological system development and functions within this set of genes are displayed. (B) Occupancy maps are shown for GATA-1, SCL, and Klf1 in the vicinity of the genes that encode for band 4.1 (Epb4.1), glycophorin C (Gypc), and aminolevulinic acid synthase 2 (Alas2), which are three erythroid-specific genes.
As in the genes encoding the core ES cell factors, expression of the GATA-1 and SCL genes is subject to autoregulation (28, 29). Although the GATA-1 and SCL proteins exhibit very similar binding patterns near the GATA-1, SCL, and Klf1 genes, only GATA-1 up-regulates expression of all three of these erythroid promoting factors, indicating that GATA-1 is upstream of SCL and Klf1 (Fig. S4). This relationship is consistent with GATA-1 forming a coherent type I feed-forward loop with both SCL and Klf1 (Fig. S4F), similar to that reported in the other core transcriptional networks (2, 3). These findings may also help to explain the observation that overexpression of GATA-1 is sufficient to reprogram nonerythroid cells (30–32), whereas, as far as we are aware, this property has not been attributed to SCL or Klf1.
PU.1 Is a Highly Integrated, Negative Regulator of the Core Erythroid Network.
In addition to interacting with SCL and Klf1, GATA-1 directly interacts with the myelo-lymphoid promoting factor PU.1 (16, 33, 34). The interplay between PU.1 and GATA-1 has served as an important model for understanding the mechanisms underlying lineage specification (35). Although PU.1 is essential for myeloid and B-cell development (36, 37), it is also expressed in erythroid progenitors, where it plays an important role in regulating the terminal differentiation decision (18, 38). PU.1 blocks erythroid differentiation by inhibiting GATA-1 transcriptional activity (39, 40), as well as by directly regulating many genes in immature erythroid cells (20). Unexpectedly, ChIP-Seq studies of PU.1 in erythroid progenitors revealed that PU.1 occupies many more sites in these cells than any of the three erythroid-promoting factors (20). However, whether PU.1 affects erythroid differentiation by regulating the core erythroid network formed by the three erythroid factors is not known.
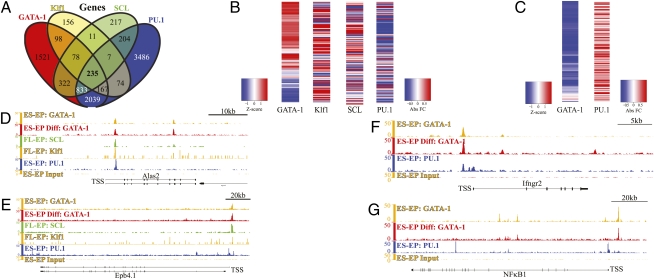
To test this possibility, we used our recently published ChIP-Seq data for PU.1 in ES-EP (20) to interrogate PU.1 occupancy near the 313 genes bound by the three erythroid factors. Strikingly, we find that 75% (235/313) of these genes are occupied by PU.1 (Fig. 3A). Using recently published data for GATA-1– (13, 23), SCL- (14), Klf1- (41), and PU.1-dependent (20) gene expression changes in erythroid progenitors, we investigated the response of the 235 genes to the four factors. Remarkably, we find that GATA-1, SCL, and Klf1 act to positively regulate expression of 68%, 67%, and 52% of these genes, respectively, whereas PU.1 represses expression of 72% of the genes (Fig. 3B). For example, Alas2 and Epb4.1 are two erythroid-specific genes that are activated by the three erythroid factors and repressed by PU.1 (Fig. 3 C and D). These results reveal a unique feature of a core transcriptional network, namely that it can be subject to negative regulation by a transcription factor that promotes other closely related lineages.
Fig. 3.
PU.1 is an integral repressor of a core erythroid network, and conversely, GATA-1 represses numerous PU.1 myelo-lymphoid gene targets in erythroid progenitors. (A) PU.1 ChIP-Seq analysis in ES-EP (20) was used to determine the genes occupied by PU.1 in erythroid progenitor cells. Using the same criterion for associating bound regions with genes as in Fig. 1A, a four-way Venn diagram displays the overlap of genes occupied by PU.1, GATA-1 (in proliferating and/or differentiating ES-EP cells), SCL, and Klf1. (B) GATA-1– (13, 23), Klf1- (41), SCL- (14), and PU.1-dependent (20) gene expression datasets were used to determine how each factor regulates the expression of genes co-occupied by all four factors in erythroid progenitor cells. GATA-1–dependent gene expression was generated by performing expression array analysis at multiple time points after the introduction of GATA-1 into GATA-1 null erythroblasts (13, 23). The heatmap displayed for GATA-1 represents the final time point of this induction series represented as a Z-score relative to all time points. Heatmaps for Klf1, SCL, and PU.1 represent the absolute fold change in gene expression in fetal-liver–derived erythroid progenitors from wild-type animals relative to those from mutant animals. (C and D) Occupancy maps from ChIP-Seq data for GATA-1, SCL, Klf1, and PU.1 in the vicinity of the genes encoding (C) aminolevulinic acid synthase 2 (Alas2) and (D) band 4.1 (Epb4.1). (E) Heatmaps displayed show GATA-1– (13, 23) and PU.1-dependent (20) gene regulation of 151 myelo-lymphoid genes bound by both proteins in erythroid progenitors. Myelo-lymphoid genes were identified as genes that are highly expressed in granulocytic, monocytic, T-cell, or B-cell lineages compared with other hematopoietic cells (27). The heatmap displayed for GATA-1 represents the 21-h time point of this induction series represented as a Z-score relative to all time points, whereas the PU.1 heatmap represents the absolute fold change in gene expression in fetal-liver–derived early erythroid progenitor cells from wild-type animals compared with that in mice that have ∼70% reduction in PU.1 levels. (F and G) Occupancy maps of GATA-1 and PU.1 in ES-EP cells are displayed in the vicinity of the myelo-lymphoid genes that encode for (F) Ifngr2 and (G) NfκB1.
PU.1 Directly Represses Expression of GATA-1, SCL, Klf1, and Important GATA-1 Cofactors.
As mentioned, two features of the core erythroid network formed by GATA-1, SCL, and Klf1 are regulation of SCL and Klf1 by GATA-1 (Fig. S4) and autoregulation of the GATA-1 and SCL genes (28, 29). Therefore, it was of interest to determine whether PU.1, in addition to repressing many downstream targets of the three erythroid factors, also negatively regulates expression of the factors themselves. Indeed, we find that PU.1 binds in close proximity to the GATA-1, SCL, and Klf1 genes (Fig. S5). PU.1 represses expression of the three genes, albeit to a greater degree for GATA-1 and Klf1 than SCL (Fig. S5). Moreover, we find that PU.1 binds near the TSS of genes encoding two critical GATA-1 cofactors, FOG1 (Zfpm1), and Gfi-1b, and represses their expression (Fig. S6). Thus, the negative effect of PU.1 on the erythroid core network extends to a group of factors that help drive the network.
GATA-1 Binds to and Represses Many PU.1 Myeloid–Lymphoid Gene Targets in Erythroid Progenitors.
We previously found that PU.1 occupies several myelo-lymphoid–specific genes in erythroid progenitors (20). Given the mutual antagonism between PU.1 and GATA-1 (16, 33, 34), one might predict that GATA-1 represses these gene targets in erythroid cells. Indeed, we find 151 myelo-lymphoid genes that are occupied by GATA-1 and PU.1 and that are positively regulated by PU.1 and repressed by GATA-1 (Fig. 3E). For example, we find that GATA-1 binds in close proximity to and represses the myelo-lymphoid genes NFκB1 and the IFN-γ receptor 2 (Ifngr2), which are essential for immune cell function (Fig. 3 F and G). Interestingly, a recent report demonstrated that during erythroid differentiation GATA-1 represses expression of PU.1 itself (42). Taken together with all of the aforementioned effects of PU.1 on the erythroid core network, these results suggest that an important aspect of lineage specification is negative cross-regulation directed at both the downstream gene targets of the factors and the genes encoding the factors themselves.
Mathematical Modeling of the GATA-1 and PU.1 Transcriptional Interaction.
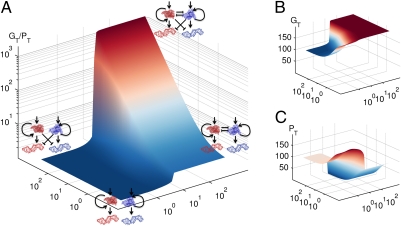
To better understand the consequences of this dual mechanism of repressing both the opposing upstream activator and its downstream targets, we developed a mathematical model (SI Materials and Methods) to describe the dynamics and steady-state expression profiles of GATA-1 and PU.1 target genes. Fig. 4A shows the equilibrium expression-level ratio [GATA-1 targets/PU.1 targets (GT/PT); Fig. 4 B and C], following a transient stimulus favoring GATA-1, over the plane representing the 2D continuum of possible antagonistic effects: (i) between the factors themselves and (ii) on the downstream targets of the opposing factor (Figs. S7–S11 and Tables S1–S2). Whereas mutual inhibition between GATA-1 and PU.1 alone increases the GT/PT ratio (right corner of Fig. 4A), the model behavior illustrates that mutual inhibition and repression of opposing downstream targets act synergistically (center top corner of Fig. 4A) to maximize the GT/PT ratio. These results suggest that the dual mechanism identified here provides, in comparison with either cross-inhibition or target inhibition alone, more robust suppression of an alternative gene expression program during lineage specification.
Fig. 4.
Mathematical model demonstrating the synergistic effect of repressing both the upstream activator and its downstream targets during lineage specification. (A) We determined the equilibrium expression-level ratio of GATA-1 targets/PU.1 targets (GT/PT, on the z axis using logarithmic scale) following a transient stimulation of GATA-1 for different combinations of the parameter values that independently modulate the mutual inhibition between GATA-1 and PU.1 (right corner, x axis) and inhibition of the other's downstream targets (left corner, y axis) from the base network topology represented in the bottom center corner (all other parameter values are constant and tabulated in SI Materials and Methods). The top center corner represents simultaneous high-level mutual inhibition and repression of the downstream targets. (B and C) GATA-1 target (GT) and PU.1 target (PT) gene expression levels, respectively, used to compute the GT/PT ratio. The highest GT/PT ratio observed in the top center corner of A is due to sustained elevated GT concentration with decreased PT concentration relative to all other corners.
Discussion
Work in a limited number of cell types suggests that one important aspect of cellular identity is determined by the concerted actions of a few key transcriptional regulators controlling a subset of genes referred to as a core transcriptional network (1). In this study, we find that GATA-1, SCL, and Klf1, three essential erythroid transcription factors, form such a network in erythroid cells (Fig. 1). Several characteristics of this network are highly reminiscent of the ES cell core network (2), including the existence of a multi-input motif formed by the three factors (Fig. 1B), as well as coherent type I feed-forward loops (Fig. S4). Because both inputs are required for optimal expression of the output signal, coherent type I feed-forward loops may be important for combinatorial transcriptional regulation (43). Indeed, absence of SCL leads to a reduction in GATA-1 binding at some erythroid-specific genes that results in suboptimal expression of these genes (14). Interestingly, in all cases studied thus far, the critical factors co-occupy ∼300 genes (2, 3) (Fig. 1A). The fact that these factors often bind in close proximity to one another (2) (Fig. 1B) suggests that the cis-binding elements may have coevolved together. In addition to binding in close proximity, another feature shared by the core ES factors and the three erythroid factors is the ability to physically interact. Nanog and Oct4 interact with one another (44), whereas GATA-1 interacts with both SCL (6) and Klf1 (10). This observation raises an interesting question. Did the ability of these transcription factors to interact favor the evolution of cis-elements in close proximity or did the evolution of cis-binding sites facilitate the ability of these factors to interact? Phylogenetic analysis of the binding elements, along with studies of how the protein interaction interfaces evolved, could provide novel insights into the evolution of these core transcriptional networks.
Interestingly, we also find that PU.1, a negative regulator of terminal erythroid differentiation, is a highly integrated component of the erythroid core network (Fig. 3A). Furthermore, we show that, in addition to negatively regulating the expression of many erythroid core network genes, PU.1 also represses expression of GATA-1, SCL, and Klf1 themselves (Fig. S5), as well as some key GATA-1 cofactors (Fig. S6). In this way, PU.1 appears to antagonize both the erythroid-specific transcriptional and proteomic networks in erythroid progenitors. Importantly, we also find that GATA-1 represses many of the myelo-lymphoid downstream gene targets of PU.1 (Fig. 3C), as well as the PU.1 gene itself (42). Mathematical modeling reveals that this dual mechanism of repressing both the opposing upstream activator and its downstream targets provides for a robust method of silencing alternative gene expression programs (Fig. 4). This result suggests that such a mechanism may be used by antagonizing transcription factors in other lineages.
The work reported in this study unifies two key concepts that are important for establishing cellular identity, namely the core transcriptional network and the mutual antagonism between master transcriptional regulators. Our findings demonstrate that a “core transcriptional network” can be subject to negative regulation by a master regulatory transcription factor from a closely related lineage. In the future, it will be important to determine whether the types of positive and negative effects on a core transcriptional network found here in the erythroid lineage are also present in other developmental systems.
Materials and Methods
Cell Culture Conditions.
ES-EP were cultured as previously described (21). Briefly, cells were grown in StemPro34 medium (Invitrogen) supplemented with 2 units/mL Epogen (Amgen), 40 ng/mL hIGF-1 (Sigma), 1 μM dexamethasone (Sigma), 100 ng/mL murine SCF (R&D Systems/Invitrogen), and 0.1% β-mercaptoethanol (Gibco) at 37 °C in a humidified 10% CO2 atmosphere. The cell concentration was maintained between 2 × 106 and 6 × 106 cells/mL by daily medium changes. ES-EP cells were differentiated for 24 h by culturing in StemPro34 media supplemented with 10 units/mL Epogen, 10 μg/mL insulin (Sigma), 3 μM mifepristone (Sigma), and 0.1% β-mercaptoethanol.
qChIP and ChIP-Seq.
qChIP was performed as previously described (45). Briefly, cross-linked chromatin from 2.5 × 106 cells was immunoprecipated with Protein A agarose beads (Roche), using 2 μg of HA (Y-11; Santa Cruz) or GATA-1 (46) antisera. Chromatin bound to beads was eluted with 1% SDS and 0.1 M NaCHO3 and the eluate was incubated at 65 °C overnight. Protein and RNA were digested with Proteinase K (Invitrogen) and RNase A (Roche) following the manufacturers’ instructions and DNA was isolated using a PCR purification column (Qiagen). qPCR was performed using the primers indicated in Table S3. All qChIP experiments were performed using two independent chromatin preparations.
ChIP-Seq samples were prepared similarly, using chromatin from 5 × 107 cells and immunoprecipitation with 40 μg of GATA-1 antiserum. ChIP-Seq was performed in duplicate, using two independent chromatin preparations. DNA was isolated before immunoprecipitation and used as an input control sample. DNA was isolated and libraries were prepared for sequencing as described previously (47). Libraries were sequenced using an Illumina Analyzer GAII and processed with the Illumina ELAND pipeline. Uniquely mapped reads were aligned to the mouse genome (mm9). Further details on processing of ChIP-Seq samples can be found in ref. 20.
Data Analysis and Mathematical Modeling.
ChIP-Seq data analysis; assignment of GATA-1, SCL, Klf1, and PU.1 binding sites to genes; integrated analysis of gene expression data and e-4C interaction data; statistical analysis; and mathematical modeling are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Perkins for kindly providing access to Klf1-dependent gene expression data. We thank Drs. Ghia Euskirchen, Hannah Monahan, Debasish Raha, and Minyi Shi for their help with Illumina sequencing. In addition, we thank Drs. Matthew Scharff and Ulrich Steidl for helpful comments on the manuscript. Work presented in this report was supported by National Institutes of Health Grants HL078381/CA154239 (to A.I.S.) and DK68634 (to E.H.B.) and National Eye Institute Grant EY012200 (to D.Z.). S.N.W. was supported by National Institutes of Health/Medical Scientist Training Program Grant 5T32GM07288. A.I.S. also receives support from National Cancer Institute Cancer Center Grant 2P30CA13330. This work was also partially supported by startup funds from Albert Einstein College of Medicine of Yeshiva University (to D.Z.).
Footnotes
The authors declare no conflict of interest.
Database deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35385).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121019109/-/DCSupplemental.
References
- 1.Cole MF, Young RA. Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:183–193. doi: 10.1101/sqb.2008.73.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 5.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: Model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- 6.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anguita E, et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahlil R, Lécuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez P, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory RC, et al. Functional interaction of GATA1 with erythroid Krüppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996;87:1793–1801. [PubMed] [Google Scholar]
- 11.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu M, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kassouf MT, et al. Genome-wide identification of TAL1's functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tallack MR, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 18.Pop R, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010;8:e1000484. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes J, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Wontakal SN, et al. A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet. 2011;7:e1001392. doi: 10.1371/journal.pgen.1001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolznig H, et al. Expansion and differentiation of immature mouse and human hematopoietic progenitors. Methods Mol Med. 2005;105:323–344. doi: 10.1385/1-59259-826-9:323. [DOI] [PubMed] [Google Scholar]
- 22.Soler E, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerenyi MA, Orkin SH. Networking erythropoiesis. J Exp Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macquarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: Beyond direct target regulation. Trends Genet. 2011;27(4):141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 29.Ogilvy S, et al. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol Cell Biol. 2007;27:7206–7219. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 31.Visvader JE, Elefanty AG, Strasser A, Adams JM. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 1992;11:4557–4564. doi: 10.1002/j.1460-2075.1992.tb05557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwasaki H, et al. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 35.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 36.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 37.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 38.Back J, Dierich A, Bronn C, Kastner P, Chan S. PU.1 determines the self-renewal capacity of erythroid progenitor cells. Blood. 2004;103:3615–3623. doi: 10.1182/blood-2003-11-4089. [DOI] [PubMed] [Google Scholar]
- 39.Rekhtman N, et al. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23:7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodge D, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou ST, et al. Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood. 2009;114:983–994. doi: 10.1182/blood-2009-03-207944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 45.Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates cdk6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. J Biol Chem. 2010;285:3044–3052. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Im H, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.