Abstract
Over the past decade, there has been much interest in the regulation of telomerase, the enzyme responsible for maintaining the integrity of chromosomal ends, and its crucial role in cellular immortalization, tumorigenesis, and the progression of cancer. Telomerase activity is characterized by the expression of the telomerase reverse transcriptase (TERT) gene, suggesting that TERT serves as the major limiting agent for telomerase activity. Recent discoveries have led to characterization of various interactants that aid in the regulation of human TERT (hTERT), including numerous transcription factors; further supporting the pivotal role that transcription plays in both the expression and repression of telomerase. Several studies have suggested that epigenetic modulation of the hTERT core promoter region may provide an additional level of regulation. Although these studies have provided essential information on the regulation of hTERT, there has been ambiguity of the role of methylation within the core promoter region and the subsequent binding of various activating and repressive agents. As a result, we found it necessary to consolidate and summarize these recent developments and elucidate these discrepancies. In this review, we focus on the co-regulation of hTERT via transcriptional regulation, the presence or absence of various activators and repressors, as well as the epigenetic pathways of DNA methylation and histone modifications.
Keywords: Human telomerase reverse transcriptase (hTERT), Telomerase, Cancer, Gene regulation, Telomeres
1. Introduction
Telomerase is a cellular ribonucleoprotein with reverse transcriptase activity (Liu, et al., 2004) that facilitates the capping of the ends of eukaryotic chromosomes via telomeres (Cukusić et al., 2008). The main function of these telomeres is to maintain chromosomal integrity and genome stability (Blackburn, 2001) via tandem hexameric repeats of a 5′-TTAGGG-3′ sequence ending in a 3′ single-stranded overhang or the G-strand overhang (Moyzis et al., 1988; Wellinger et al., 1997; Makarov et al., 1997; as reviewed by De Boeck et al., 2009). Telomerase remains inactive in most somatic cells but can be readily detectable in germ cells and other self-renewing tissues (Liu et al., 2004). In normal somatic cells, the erosion of telomeres is caused by incomplete replication at telomeric ends, leading to a loss of approximately 50 to 200 bp of telomeric DNA at each cell division until replicative senescence is reached (Chiu et al., 1997; Harley et al., 1990; Qi et al., 2011) (Fig. 1). However, in the presence of telomerase, telomere length is maintained via the addition of hexameric repeats that cap the ends of linear chromosomes and allows their evasion of replicative senescence (Liu et al., 2004). The dysregulation of telomerase leads to its activation in approximately 90% of human cancers, irrespective of tumor type (as reviewed by Kyo et al., 2008). This suggests that normal human somatic cells utilize regulatory factors to suppress the activity of telomerase; however, upon cellular immortalization, this regulation is either deviant or missing (Collins et al., 2002; Qi et al., 2011).
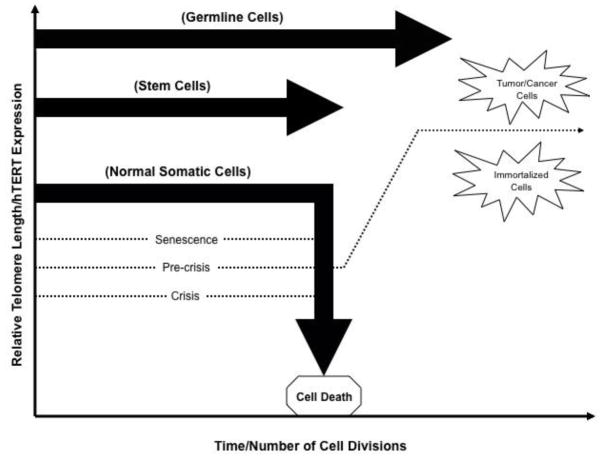
Figure 1. hTERT expression and telomere length during differentiation, immortalization, and tumorigenesis.
Diagram depicting the relative changes in telomere length and hTERT expression during differentiation, immortalization, and tumorigenesis. As indicated, germline cells exhibit relatively high levels of hTERT as well as lengthened telomeres, independently of time and the number of cellular divisions. In stem cells, the telomere lengths and expression of hTERT is slightly lower than that of germline cells, however, they do follow the same trend of independence relative to time and/or the number of cellular divisions. During the division of somatic cells, telomere length and hTERT expression generally decrease in both a time-dependent and division-dependent manner. Telomere lengths are shortened with each cellular division until senescence occurs followed by a crisis period then apoptosis/cellular death. Activation of telomerase at the pre-crisis stage allows for somatic cells to escape crisis and become immortalized. This occurrence is characteristic in approximately 90% of human cancers (as reviewed by Kyo et al., 2008).
Recent studies have identified several components of telomerase: the telomerase RNA (TER) molecules whose secondary structure is well defined; telomerase reverse transcriptase (TERT), the catalytic subunit; Es1p and Es3p which are categorized as additional protein subunits, more specifically the two units of the Ku heterodimer; as well as a vast majority of assembly and maturation proteins that contribute to the telomerase enzymatic complex (Liu et al., 2004). Dyskerin (DKC1) (Cohen et al., 2007) and telomerase protein component 1 (TEP1) (Saito et al., 1997) have also been identified as components of telomerase. However, it has been shown that only hTER and hTERT are necessary for the reestablishment of telomerase activity (Ishikawa, 1997; Weinrich et al., 1997; Beattie et al., 1998; Liu et al., 2004). On the other hand, recent evidence suggests that only hTERT is needed to restore telomerase activity in telomerase-negative normal cells, such as epithelial cells and human fibroblasts (Artandi et al., 2002; Gonzalez-Suarez et al., 2002; Gonzalez-Suarez et al., 2001; Stewart et al., 2002; Qi et al., 2011), and the ectopic expression of hTERT along with activated oncogenes results in tumorigenesis (Hahn et al., 1999; Qi et al., 2011). Although hTERT is tightly regulated during cellular differentiation and is expressed at very low levels in normal somatic cells (Masutomi et al., 2003), hTER is widely expressed in the majority of cell types including those that are telomerase-negative (Meyerson et al., 1997; Nakamura et al., 1997; Liu et al. 2004). This suggests the presence of additional mechanisms that aid in the protection of chromosomal ends. A study done by Buckhovich and Greider (1996) showed that lymphocytes, in particular, exhibit telomerase activity in response to stimulation (Chebel et al., 2009). Regulation of hTERT and subsequent telomerase expression in these specific cells is thought to occur via the inhibition of rapamycin during the G1 phase of the cell cycle (Chebel et al., 2009).
Studies have shown that telomeric ends form higher-order structures called T-loops that potentially protect these termini from the DNA repair system as well as regulate the access of telomerase to the telomere (Griffith et al., 1999; as reviewed by De Boeck et al., 2009). The TTAGGG duplex repeats found at the ends of mammalian telomeres are bound by two related proteins: TRF1 and TRF2, the TTAGGG repeat binding proteins (Chong et al., 1999; Bilaud et al., 1997; Broccoli et al., 1997; Griffith et al., 1999). TRF2 plays an essential role in both telomere end protection (van Steensel et al., 1998; Karlseder et al., 1999; Karlseder et al., 2002; as reviewed by De Boeck et al., 2009) and T-loop formation (Griffith et al., 1999). Subsequent studies have shown that the TRF2 complex promotes T-loop formation via its interaction with telomere overhang, notably the 3′ guanine-rich termini (Griffith et al., 1999; Stansel et al., 2001; Khan et al., 2007; as reviewed by De Boeck et al., 2009). Thus, this system could potentially play an essential role in the protection of telomeres via the formation of higher-order structures, concealing overhang and preventing the degradation of telomeres at DNA damage checkpoints (De Boeck et al., 2009). Further elucidation of these mechanisms is crucial in understanding the regulation of hTERT both in vivo and in vitro and the development of proactive anticancer therapies.
2. The hTERT promoter region
Transcription, alternate mRNA splicing, phosphorylation, and the maturation and modification of hTERT and hTER have all been shown to play vital roles in the regulation of telomerase activity (Cong et al., 2002; Cukusić et al., 2008). However, it is thought that the hTERT promoter is the most important regulatory element of telomerase expression (Cukusić et al., 2008). Cong et al. (1999) and Bryce et al. (2000) showed that the hTERT gene is located on the short arm of human chromosome 5 (5p15.33), more than 2 Mb away from the telomere (Leem et al., 2002). A recent study has also discovered an approximate 7 Mb telomerase repressor region located on the short arm of chromosome 3 (3p21.3) (Abe et al., 2010). The hTERT gene consists of 15 introns and 16 exons and is over 40 kb in length (Cukusić et al., 2008). Although rich in CpG dinucleotides and Sp1 sites, the hTERT promoter lacks both TATA and CAAT boxes (Horikawa et al., 1999; Takakura et al., 1999). The hTERT core promoter spans 330 bp upstream of the translational start site and 37 bp of exon 2 (Cong et al., 1999; as reviewed by Cukusić et al., 2008). Moreover, this regulatory region as well as upstream sequences interacts with both positive and negative regulators of hTERT via an abundance of transcriptional binding sites (Cukusić et al., 2008) (Table 1), further suggesting the compelling role of transcription in the regulation of hTERT.
Table 1.
Characterization of pathways and constituent and other factors in combined regulation of hTERT transcription.
| Factor (hTERT binding sites) | Primary role | References |
|---|---|---|
|
Pathway – Cell Cycle
| ||
| CDK2 | Activator | (McArthur et al. 2002) |
| CDK4 | Activator | (Leng et al. 2002) |
| Cyclin D | Activator | (Leng et al. 2002) |
| Cyclin E | Activator | (McArthur et al. 2002) |
| p21 | Repressor | (Henderson et al. 2000; Lai et al. 2007; Harper et al. 1993) |
| p53 | Repressor | (Lai et al. 2007; Shats et al. 2004; Chun and Jin 2003) |
| p27 | Mostly repressor | (Ray et al. 2009) |
| Rb | Mostly repressor | (Wang et al. 2005) |
| E2F1 (2) | Mostly repressor | (Stanelle et al. 2002) |
| p14 (p19) | Repressor | (Moore et al. 2003; Guo et al. 2008; Wang et al. 2006) |
| MDM2 | Activator | (Moore et al. 2003; Wang et al. 2006) |
| p16 | Repressor | (Henderson et al. 2000) |
|
| ||
|
Pathway – TGF-β
| ||
| TGF-β | Repressor | (Wu et al. 2009) |
| TGF-βR1 | Repressor | (Eickelberg et al. 2002) |
| TGF-βR2 | Repressor | (Eickelberg et al. 2002) |
| SMAD3 | Repressor | (Chen et al. 2002) |
| SMAD4 | Repressor | (Wu et al. 2003; Ren et al. 2009) |
| E2F4 | Repressor | (Chen et al. 2002) |
| E2F5 | Repressor | (Chen et al. 2002) |
|
| ||
| Pathway – PI3K/Akt | ||
| PI3K | Activator | (Zhu et al. 2008; Bai et al. 2009 |
| Akt | Activator | (Bai et al. 2009; Chou et al. 2009) |
| PTEN | Repressor | (Feng et al. 2007; Zhou et al. 2006; Phuong et al. 2011) |
| mTOR | Activator | (Zhu et al. 2008) |
| IKK | Activator | (Bai et al. 2009) |
| hTERT | Activator | (Lai et al. 2007; Perrault et al. 2005; Takano et al. 2008) |
|
| ||
|
Pathway – NF-κB (canonical)
| ||
| NF-κB (p50) (1) | Activator | (Bai et al. 2009) |
| NF-κB (p65) (1) | Activator | (Bai et al. 2009) |
| IKK | Activator | (Bai et al. 2009; Kamata et al. 2002) |
| IκB | Repressor | (Bai et al. 2009) |
|
| ||
|
Pathway – NF-κB (non-canonical)
| ||
| NIK | Activator | (Bai et al. 2009; Varfolomeev et al. 2007) |
| IKK | Activator | (Bai et al. 2009) |
| NF-κB (p100) (1) | Activator | (Bai et al. 2009; Varfolomeev et al. 2007) |
| NF-κB (RelB) (1) | Activator | (Bai et al. 2009; Varfolomeev et al. 2007) |
|
| ||
|
Pathway – MAPK
| ||
| RAS | Activator | (Zhu et al. 2008) |
| MEK | Activator | (Zhu et al. 2008) |
| ERK | Activator | (Wang et al. 2008) |
|
| ||
|
Pathway – ErbB
| ||
| EGF | Activator | (Wang et al. 2008) |
| EGFR | Activator | (Wang et al. 2008) |
| HER2/neu | Activator | (Hsu et al. 2008) |
| PI3K | Activator | (Zhu et al. 2008) |
|
| ||
|
Pathway – TGF-β and Cell Cycle (a link between activation and repression pathways)
| ||
| p107 | Repressor | (Chen et al. 2002; Beijersbergen et al. 1994) |
|
| ||
|
Pathway – TGF-β and PI3K/Akt (a link between activation and repression pathways)
| ||
| Jab1 | Activator | (Wan et al. 2002; Tomoda et al. 2002; Oh et al. 2006) |
|
| ||
|
Pathway – Cell Cycle, TGF-β, PI3K/Akt and MAPK
| ||
| c-MYC (2) | Activator | (Chen et al. 2002; Casillas et al. 2003) |
| MAD1 (2) | Repressor | (Zhu et al. 2008; Casillas et al. 2003) |
|
| ||
|
Other
| ||
| AP2 (17) | Activator | (Kyo et al. 2008) |
| AP4 (9) | Activator | (Jung et al. 2008; Jung and Hermeking 2009) |
| BRCA1 | Repressor | (Li et al. 2002) |
| CCAC (1) | Activator | (Wick et al. 1999) |
| c-Ets-2 (2) | Activator | (Kyo et al. 2008) |
| c-IAPI | Activator | (Zhu et al. 2008) |
| c-Myb (2) | Activator | (Quintana et al. 2011; Drabsch et al. 2010) |
| CREB/ATF (1) | Activator | (Cong et al. 1999) |
| CTCF (2) | Mostly repressor | (Renaud et al. 2007) |
| AP1 | Mixed | (Cherlet and Murphy 2007) |
| ERα (2) | Activator | (Tee et al. 2004) |
| ERβ (2) | Mostly activator | (Tee et al. 2004) |
| IK2 (6) | Activator | (Cong et al. 1999) |
| INHBC | Activator | (Perrault et al. 2005) |
| MAD3 (2) | Repressor | (Jiang et al. 2008) |
| MAZ (3) | Activator | (Song et al. 1998; Song et al. 2003) |
| Menin (2) | Repressor | (Imachi et al. 2010; Suphapeetiporn et al. 2002) |
| MNT (2) | Repressor | (Hooker and Hurlin 2006) |
| MyoD (3) | Mixed | (Jin et al. 2011) |
| MZF-2 (4) | Repressor | (Fujimoto et al. 2000; Ogawa et al. 2003) |
| NF1 (9) | Activator | (Chikhirzhina et al. 2008) |
| NFAT (5) | Activator | (Chebel et al., 2009) |
| NF-E2 (1) | Activator | (Goerttler et al. 2005) |
| PITX1 | Repressor | (Qi et al. 2011) |
| Sp1 (14) | Mostly activator | (Parisi et al. 2007) |
| USF1/USF2 (2) | Mixed | (Goueli and Janknecht 2003) |
| WT1 (1) | Mostly repressor | (Sitaram et al. 2010) |
Recent discovery has shown that the G-rich sequences located within the hTERT promoter have a strong potential to form G-quadruplexes (Simonsson, 2001; Davis, 2004; Burge, 2006; Patel, 2007), which are helical structures composed of four-strands and formed via the stacking of GGGG tetrads (Lim et al., 2010; Gellert, 1962). Lim et al. (2010) discovered the coexistence of two major intramolecular G-quadruplex conformations within the G-rich region of the hTERT promoter, GTERT-060: Form 1, an intramolecular (3 + 1) G-quadruplex and Form 2, a propeller-type parallel-stranded G-quadruplex. Quadruplex formation within the telomeric G-rich overhang has also been shown to prevent its elongation by telomerase (Zahler et al., 1991), negatively impacting c-Myc expression and in turn the expression of hTERT (Grand et al., 2002), and altering the splicing pattern of hTERT RNA (Gomez et al., 2004) (Lim et al., 2010). In addition, a study done by Gros et al. (2008) revealed a quadruplex ligand that binds to hTER at its quadruplex-forming motif and can alter telomerase activity (Lim et al., 2010). These studies present a novel mechanism by which telomerase activity can be down-regulated or even inhibited in cancer models via G-quadruplex ligands that target the guanine-rich region at telomeric ends (Gros et al., 2008).
3. The hTERT promoter and associated transcription factors
After the successful cloning of the hTERT 5′-promoter region (Takakura et al., 1999; Horikawa et al., 1999; Cong et al., 1999; as reviewed by Kyo et al., 2008), studies were able to show an up-regulation of transcriptional activity in cancer cells and a lack of detectable activity in most normal cells via transient expression assays of the hTERT gene (Takakura et al., 1999; as reviewed by Kyo et al., 2008). This 5′ regulatory region contains numerous binding sites for a variety of transcription factors, including both activators and repressors of hTERT (Renaud et al., 2006) (Table 1). Renaud et al. (2006) found that there are 5′ exonic regions that may inhibit the transcriptional activity of hTERT and serve to regulate its expression. This finding is promising because the CCCTC-binding factor (CTCF) binds to these regions in telomerase-negative cells but not in telomerase-positive cells (Renaud et al., 2005), suggesting that CTCF may bind downstream of the transcriptional start site (Klenova et al., 1993; Lutz et al., 2000) and act as a repressor of hTERT in normal cells (Renaud et al., 2006).
A recent study showed that the proximal E-box, located +22 to +27 upstream of the transcriptional start site, is a repressive cis-element of the hTERT promoter in certain cell lines (Horikawa et al., 2002). The repressive nature of the E-box is thought to be associated with an interaction with the Mad1-Max complex in normal human somatic cells (Oh et al., 2000). Furthermore, several investigators have shown that c-Myc acts as a key regulator of hTERT transcription during carcinogenesis via its binding to the E-box and subsequent activation of transcription (Cong et al., 1999; Greenbery et al., 1999; Wang et al., 1998; Wu et al., 1999; as reviewed by Kyo et al., 2008). However, it should be noted that other studies have found that c-Myc does not necessarily affect hTERT expression in some cancer cells (Kirkpatrick et al., 2003; Günes et al., 2000). GC-boxes (GGGCGG) are another characteristic sequence of the hTERT promoter (as reviewed by Kyo et al., 2008). Xu et al. (2001) proved by electrophoretic mobility shift assay (EMSA) that Sp1, an activator of hTERT (Kyo et al., 2000), binds to these CG-boxes. Likewise, Filippova et al. (1996) showed that CTCF acts to inhibit hTERT expression when bound downstream of the transcriptional start site and preferentially to the CG boxes (Renaud et al., 2006). In this review, we will attempt to elucidate and summarize these findings and present the roles that transcription plays in the regulation of hTERT.
3.1 Activators of hTERT
3.1.1 Transcription factors
There a several transcription factors that act a regulators of hTERT (Table 1). Sp1 is a zinc finger transcription factor (Suske, 1999) that has been shown to bind to the five GC-boxes on the TATA-less hTERT promoter (Cukusić et al., 2008) and activate hTERT (Kyo et al., 2000). Kyo et al. (2000) also localized these Sp1 binding sites between two E-boxes 110 bp upstream of the transcriptional start site. Luciferase reporter assays performed by Cong and Bacchetti (2000) showed that any mutations in the Sp1 binding sites drastically reduced hTERT promoter activity, further suggesting the regulatory role of Sp1 in the transcription of hTERT. However, it should be noted that because Sp1 is often universally expressed in normal cells, it is evident that Sp1 alone does not lead to the activation of hTERT (Horikawa et al., 2002). Sp1 has also been suggested to cooperate with the oncogene, c-Myc, to activate the transcription of the hTERT gene (Kyo et al., 2000). Liu et al. (2009) suggested a common mechanism in proliferative cancer cells by which Sp1 stimulates telomerase expression via the transcriptional function of the MBD1-containing chromatin-associated factor 1 (MCAF1). In the study, MCAF1 was shown to have a direct relationship with Sp1, and the depletion of either Sp1 or MCAF1 down-regulated the TERT and TER genes in cultured cells, resulting in decreased activity of telomerase (Liu et al., 2009). The high-mobility group A2 (HMGA2) gene, most frequently amplified in human cancers, has also been shown to regulate the expression of hTERT by interacting with Sp1 and impeding the recruitment of histone deacetylase 2 (HDAC2) to the hTERT proximal promoter (Li et al., 2011). In turn, this impediment stimulates hTERT expression and telomerase activity via the enhanced acetylation of histone H3-K9 (Li et al., 2011). Moreover, the study showed that HMGA2 partially replaced hTERT during the tumorigenic transformation of normal human fibroblasts (Li et al., 2011). A study done by Chebel et al. (2009) suggested a synergistic relationship between the nuclear factor of activated T cells (NFAT) and Sp1 in the transcriptional regulation of hTERT. The research showed that simultaneous mutations of both the −40 NFAT-responsive element and one or more Sp1-binding sites led to a greater decrease in hTERT promoter activity when compared to single mutations, suggesting NFAT as activator of hTERT apart and in the presence of Sp1, mainly through a consensus binding site localized within the core promoter (Chebel et al., 2009). Moreover, Chebel et al. (2010) proposed the activation of hTERT by NFAT at two levels: directly, at the level of transcription (Chebel et al., 2009) and indirectly via the activation of c-Myc (Buchholz et al., 2006) (Fig. 2). Sequential induction of Nuclear Factor-kappa B (NF-κB) and c-Myc has been suggested to aid human epidermal growth factor receptor 2 (HER2) in the activation of hTERT following irradiation (Papanikolaou et al., 2011). It was determined that the knockdown of HER2 led to the down-regulation of NF-κB and c-Myc-mediated hTERT and telomerase activity in human breast cancer cells (Papanikolaou et al., 2011). Deng et al. (2007) identified the activating enhancer binding protein-2β (AP-2β) as a unique transcriptional activator of hTERT in human lung cancer cells. Furthermore, it was shown that the inhibition of AP-2β down regulated telomerase activity, accelerated telomere degradation, and inhibited tumorigenesis, suggesting this pathway as a potentially novel therapeutic target in certain cancer lines (Deng et al., 2007).
Figure 2. Partial hTERT transcription regulation interactome.
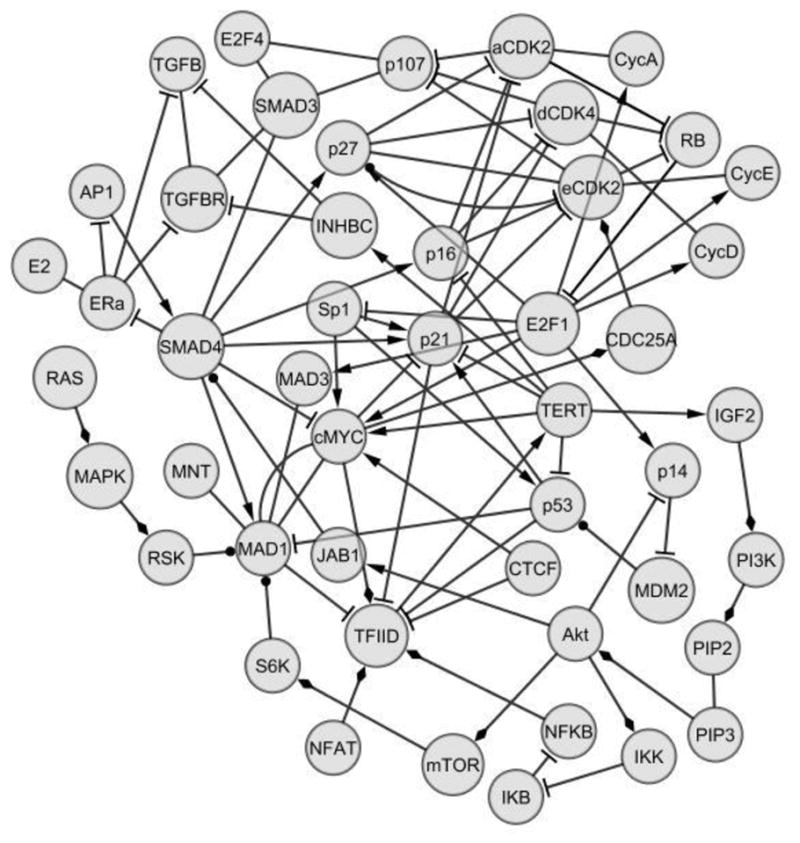
The principal protein-protein interactions that regulate hTERT involve cross-regulation between hTERT and the cell cycle, carried out largely by cyclin/cdk complexes and transcription factors that bind to the hTERT promoter. The cyclic nature of the cell cycle is reflected in the dual role of E2F-1 as both repressor and activator. Major regulatory pathways exert either repressive, as in the case of the TGF-b pathway, or activating influence on both the cell cycle and hTERT expression. The PI3K/Akt, NF-kB and MAP kinase pathways all activate. In addition, estrogen (E2) bound to estrogen receptor ERa blocks the repressive activity of the TGF-b pathway, as does cyclin/cdk phosphorylation of p107. In a positive feedback loop, expression of hTERT activates the PI3K/Akt pathway, which activates the cell cycle by MAD1 and p53 degradation, activates the NF-kB pathway and potentially blocks the TGF-b pathway.
| Arrow shape | Meaning |
|---|---|
| Delta | Increase, transcribe, synergy or assemble |
| Diamond | Activate |
| T | Decrease or block |
| Ball | Degrade |
Signaling and regulatory pathways with no component factors binding to the 5′-regulatory region have potentially decisive roles in the transcriptional regulation of hTERT through their regulation of critical hTERT transcription factors such as c-Myc and Mad1. In particular, the TGF-β pathway has been shown to down-regulate hTERT expression by increasing Mad1 and suppressing c-Myc (Wu et al., 2009; Chen et al., 2002; Hein et al., 2011). Several investigators demonstrated that the P13K/Akt and MAP kinase pathways up-regulate hTERT transcription via the phosphorylation of Mad1, leading to its ubiquitin-mediated proteolysis (Zhu et al., 2008; Chou et al., 2009). The role of the P13K/Akt pathway may, in fact, be central to the process of hTERT dysregulation and cellular immortalization. The pathway has been shown to potentially inhibit the TGF-β pathway by Jab1 activation and SMAD4 degradation, activate the NF-κB pathway, disrupt cell cycle control and advance cell cycle progression by either the degradation or down-regulation of p53, p21, and p27 (Ogawara et al., 2002; Bai et al., 2009; Li and Sarkar, 2002; Tomoda et al., 2002; Oh et al., 2006; Hsu et al., 2007; Johnson et al., 2010; Wan et al., 2002). A growing body of evidence now supports the existence of a positive feedback loop whereby the expression of hTERT activates the P13K/Akt pathway, which in turn initiates multiple mechanisms that increase hTERT expression and/or inactivate hTERT expression restraints (Lai et al., 2007; Perrault et al., 2005; Takano et al., 2008; Smith et al., 2003; Li et al., 2011; Farwell et al., 2000; Sasaki et al., 2009)(Table 1).
3.1.2 Oncogenes
Although c-Myc is an oncogene that is generally involved in the management of cellular proliferation, differentiation, and apoptosis in normal human cells, it is also often associated with the development of human cancers when mutated or over expressed and in instances of chromosome translocation and gene amplification (Cukusić et al., 2008). c-Myc protein levels are known to be elevated in undifferentiated and most neoplastic and transformed cells (Lie et al., 2001) but are minimal in differentiated somatic cells (Günes et al., 2000). Xu et al. (2001) demonstrated that c-Myc binds to the E-boxes (5′-CACGTG-3′) at the hTERT proximal promoter in exponentially proliferating HL60 cells, suggesting that the E-box occupancy by c-Myc is responsible for the activation of hTERT in this particular cell line. It has been shown that the product of c-Myc often complexes with Max protein to form a heterodimer (c-Myc/Max) that activates gene transcription (Blackwood and Eisenman, 1991; Liu et al., 2004). Kyo et al. (2000) demonstrated that the Myc/Max heterodimer binds to the E-box in the hTERT promoter and is essential in transactivation. In addition, Kyo et al. (2000) suggested a coactive function of c-Myc and Sp1 as important regulators of hTERT expression via the positive correlation between the transcriptional activity of hTERT and the expression of c-Myc and Sp1. However, because a large majority of these studies used overexpressed c-Myc when measuring hTERT promoter activity, it holds unclear as to the regulatory role that c-Myc plays in the transcription of hTERT when bound endogenously at its promoter (as reviewed by Kyo et al., 2008). Recent findings have challenged the exclusive role of Myc/Max in E-box dependent binding and activation of hTERT via the recognition of other E-box-binding proteins that regulate hTERT transcription, notably upstream stimulatory factor (USF) 1 and 2 (Goueli et al., 2003). The study suggested that the binding of USF1/2 precludes the binding of Myc/Max to the E-box in the hTERT promoter and indicated that USF1/2 binds more avidly to the hTERT E-boxes than does Myc/Max (Goueli et al., 2003). This regulation of hTERT at the E-boxes will be further discussed in combination with the repression of hTERT via the binding of Mad1.
3.1.3 Hormones
Several studies have shown the up regulation of telomerase activity in breast and prostate cancer cells by estrogen (Kyo et al., 1999; Misiti et al., 2000; Nanni et al., 2002; Gao et al., 2003). The hTERT promoter contains two estrogen responsive elements; one at −2754 bp and the other at −950 bp upstream of the ATG start site (Cukusić et al., 2008). Some groups have found that estrogen receptor-α (ERα) binds to the estrogen-response element (ERE) in the hTERT promoter region and activates hTERT transcription, thus supporting the idea that estrogen up regulates telomrase activity (Kyo et al., 1999; Misiti et al., 2000; as reviewed by Kyo et al., 2008). Others have shown that ERα, when bound to the upstream hTERT regulatory region and activated by bound estrogen, can potentially block the repressive TGF-β pathway via interactions with the TGF-β receptor or with AP1 (Cherlet and Murphy, 2007; Welboren et al., 2009; Tee et al., 2004) (Fig. 2). Moreover, Kimura et al. (2004) demonstrated the activation of hTERT via phosphorylation, mediated by Akt signaling, in which hTERT is phosphorylated in an Akt-dependent manner. Tamoxifen, a selective modulator of ER, has also been shown to regulate hTERT expression in a cell-specific manner (Wang et al., 2002) by inhibiting the growth of breast cancer cells and down regulating the expression of hTERT mRNA in the presence of estrogen (as reviewed by Kyo et al., 2008). However, tamoxifen was shown to activate hTERT and stimulate the growth of endometrial cancer cells in both the absence and presence of estrogen (E2) (as reviewed by Kyo et al., 2008), solidifying its cell-specific manner. Previous studies conflicted on whether the induction of hTERT by estrogen was c-Myc dependent (Misiti et al., 2000; Kyo et al., 1999), but later studies by Kirkpatrick et al. (2003) and Wisman et al. (2003) agreed on the regulatory effects of estrogen on telomerase activity in certain tissues, markedly those targeted by estrogen. These studies did not, however, conclude whether the activation of hTERT is direct or a result of the up regulation of cMyc by estrogen (Cukusić et al., 2008). Interestingly, research of obese women has shown that adiposity increases the circulating levels of estrogen in adipose tissue via the conversion of androgens to estrogens by aromatase (Potischman et al., 1996), resulting in the stimulation of mammary epithelial cell mitosis and tumorigenesis (Han et al., 2005). Recent studies have drawn a link between leptin, an adipose derived hormone, obesity, and breast cancer risk (Rose et al., 2007; Rahmati-Yamchi et al., 2011). A correlation was observed between high serum levels of leptin and hTERT mRNA levels (Rahmati-Yamchi et al., 2011). In vitro studies have shown that in MCF-7 breast cancer and HepG2 liver carcinoma cell lines, hTERT is up regulated by leptin (Ren et al., 2010; Stefanou et al., 2010). Androgen has also been shown to be an activator of hTERT in androgen-sensitive prostate cancer cells (Guo et al., 2003; as reviewed by Kyo et al., 2008).
3.2 Repressors of hTERT
3.2.1 Transcription factors
CCCTC-binding factor (CTCF) is a ubiquitously expressed 11-zinc finger protein that binds to the hTERT proximal exonic region (Renaud et al., 2005) and is thought to be the only factor that can negatively regulate the transcription of hTERT irrespective of cell type (Renaud et al., 2005). Studies have shown that CTCF plays a pivotal role in the transcriptional activation of the APPB promoter (Vostrov et al., 1997), the imprinting control of the H19 region (Kanduri et al., 2000), and the silencing of cMyc (Filippova et al., 1996) However, a more recent study demonstrated that CTCF-binding at the cMyc locus does not repress the transcriptional activity of cMyc, but instead is required for its protection from DNA methylation (Gombert and Krumm, 2009) (Fig. 2). In one model, CTCF was revealed to be bound to hTERT in cells that did not express hTERT but not in telomerase-positive ones (Renaud et al., 2005), strongly suggesting that the binding of CTCF may serve as an important repressive mechanism of hTERT in normal cells. CTCF is known to bind directly to SIN3A, which recruits HDACs and thus prevents transcription (Lutz et al., 2000). However, Renaud et al., (2010) revealed evidence that BORIS (Brother of the Regulator of Imprinted Sites) may exert an opposing effect by opening chromatin around the transcriptional start site of hTERT, allowing for the expression of hTERT. In hTERT-negative BJ fibroblasts and HLF/hTERT cells, CTCF was demonstrated to bind to the first exon of the hTERT gene; however, CTCF did not exhibit binding in HeLa and SW480 tumor cell lines (Renaud et al., 2006). This study showed methylation patterns in the CpG islands and core promoter of hTERT to be the mechanism by which CTCF is prevented from binding in telomerase-positive cells (Renaud et al., 2006), implying an epigenetic role in the expression or repression of hTERT. Lee et al. (2003) demonstrated that IRF1 is a mediator for interferon-γ-induced inhibition of both hTERT expression and telomerase activity in cervical cancer cells. IRF1 increased by two-fold in the presence of over expressed WT1D, indicating its possible inhibitory effect on hTERT promoter activity (Sitaram et al., 2010). Jun and Fos family proteins form a heterodimeric complex to create transcriptional activator protein 1 (AP-1) which has been shown to lead to the transcriptional suppression of hTERT in cancer cells (Takakura et al., 2005). Chromatin immunoprecipitation (ChIP) analysis revealed the binding of JunD and c-Jun to two putative AP-1 binding sites between −2000 and −378 of the hTERT promoter region, suggesting that AP-1 directly associates with the hTERT promoter (Takakura et al., 2005). Ap1 has also been reported to enhance the repressive activity of the TGF-β pathway (Cherlet and Murphy, 2007). Interferon-β (IFN-β) signaling was shown by Lee et al. (2010) to repress telomerase activity and hTERT transcription in ovarian cancer. In addition to down regulating hTERT mRNA expression, IFN-β also induces p21 expression, independently of p53 (Lee et al., 2010). Shats et al. (2004) reported that p21 and E2F mediate the repression of hTERT via p53. Makorin-1 (MKRN1) is an E3 ubiquitin ligase that targets hTERT for proteasome processing (Kim et al., 2005). A study done by Salvatico et al. (2010) proposed an inverse relationship between the MKRN1 protein and telomerase activity, following terminal differentiation in HL-60 cells. This study is consistent with the idea that during differentiation or cell cycle arrest when telomerase action at chromosomal ends is no longer needed, MKRN1 represents an elimination pathway to rapidly reduce telomerase activity (Salvatico et al., 2010). Recently, PITX1 was identified as a repressor of hTERT promoter activity that binds directly to the promoter region (Qi et al., 2011). Several studies have reported reduced PITX1 expression in various types of human cancer, including gastric, bladder, and colon cancers, (Chen et al., 2007; Chen et al., 2008), strongly supporting the idea that PITX1 also plays a crucial role in the neoplastic development (Qui et al., 2011).
Research has shown that Mad1 can act as a transcription factor that binds with the E-box of the hTERT promoter and regulates its activity (Casillas et al., 2003). Mad1 also has the potential to interact and form a dimer with c-Myc at the specific E-box site sequence 5′-CACGTG-3′ and suppress the transcription of target genes (James et al., 2002). Furthermore, Mad1 has been shown to suppress hTERT transcription via competition with c-Myc for the CACGTG binding sites of the hTERT promoter (Zou et al., 2005). The research done by Gunes et al. (2000) demonstrated that the Myc-Mad1 dimer binds more easily to the E-box sites than does c-Myc or Mad1 alone. This additive relationship was shown to suppress hTERT expression in bladder cancer cells and illustrates a partial mechanism by which hTERT is regulated during carcinogenesis (Zou et al., 2005). A study by Won et al. (2002) found that Sp1 and Sp3 interact with histone deacetylase (HDAC) in human fibroblasts and play an important role in the repression of the hTERT promoter endogenously. This suggests that both Sp1 and Sp3 interact directly with the hTERT promoter by recruiting HDAC and repressing the transcription of hTERT in normal somatic cells (Won et al., 2002).
3.2.2 Tumor suppressors
p53 is likely the most characterized of the tumor suppressors. The gene acts as a tumor suppressor via its ability to induce cell cycle arrest and apoptosis in response to various stress stimuli (Asker et al., 1999; Cukusić et al., 2008). This suppressor gene is mutated in 50% of human tumors (Ko and Prives, 1996). Mechanisms of cell cycle inhibition and cancer prevention by p53 include the transcriptional activation of p21, inhibition of basal transcription factor TFIID, and the suppression of cyclin/cdk complex activity (Shats el al., 2004; Lai et al., 2007; Henderson et al., 2000) (Fig. 2).. Kanaya et al. (2000) showed that the hTERT gene has two p53 binding motifs upstream of the 5′ core promoter region, and an over expression of p53 and its subsequent binding with the help of transcription factor Sp1 at these two motifs leads to the repression of the hTERT promoter (Lai et al., 2007). Lai et al. (2007) also demonstrated that a knockdown of hTERT in HEK 293 cells resulted in elevated p53 transcription as well as a decrease in cellular proliferation, further supporting the antagonistic role of p53 in hTERT expression (Fig. 2). Shats et al. (2004) indicated that the DNA-binding domain of p53 is required for the repression of hTERT, but no direct binding of p53 to the hTERT core promoter was observed. In the absence of p53, an atypical E2F-binding site in the 5′ untranslated region of the hTERT promoter upregulated hTERT expression; however, hTERT expression was down regulated in the presence of p53 (Cukusić et al., 2008; Shats et al., 2004). Results from Shats et al. (2004) showed a repression of telomerase activity and hTERT expression in OVCAR and SKOV3 cell lines by p21, suggesting IFN-β/p21 signaling could possibly down regulate both hTERT expression and telomerase activity via a p53-independent pathway.
Inactivation of the retinoblastoma protein Rb is common in many types of cancers (Murphree and Benedict, 1984). pRb binds to and inhibits transcription factors of the E2F family (Cukusić et al., 2008). Rb creates a complex and acts as a growth suppressor when bound to E2F, causing cell cycle arrest and recruiting HDAC to further suppress DNA synthesis (Cukusić et al., 2008). Zheng and Lee (2002) showed that Rb is active when in a hypophosphorylated state and inactive when phosphorylated, supporting several studies that indicated a down-regulation of telomerase activity in human carcinoma cell lines via an over expression of hypophosphorylated Rb (Xu et al., 1997; Nguyen and Crowe, 1999). Meeran et al. (2011) revealed that the acetylated green tea polyphenol, pEGCG, inhibits hTERT expression partially through the binding of E2F-1 and Mad1 to the hTERT promoter in both ER (+) and ER (−) breast cancer cells.
Wilms’ tumor 1 (WT1) is a tumor suppressor gene, potentially oncogenic, that acts as an important regulator of cellular processes, particularly cell growth and development, as well as a strong transcriptional regulator of hTERT (Sitaram et al., 2010). WT1 has been shown to repress the transcription of hTERT in specific cell types, notably the virally transformed human embryonic kidney 293 cells identified by Oh et al. (1999). There is strong indication that there are multiple pathways for hTERT regulation by WT1 in clear cell renal cell carcinoma (ccRCC), though the repression of the hTERT promoter generated by WT1 is complex (Sitaram et al., 2010). WT1 has been shown to bind to the promoters of hTERT, c-Myc, and SMAD3, and suppress c-Myc at both a transcriptional and protein level; suppress hTERT expression; and up-regulate SMAD3 and Jun (Sitaram et al., 2010). Furthermore, direct repression of hTERT by SMAD3 via TGF-β has also been reported (Lacerte et al., 2008).
4. Epigenetic mechanisms regulating hTERT
The cluster of CpG sites found within the hTERT promoter has prompted many researchers to believe that methylation plays a key role in the regulation of hTERT (Kyo et al., 2008). However, contradictory results have been reported. Some groups showed that methylation of the hTERT promoter resulted in the silencing of the gene (Shin et al., 2003; Lopatina et al., 2003; Liu e al., 2004), while others have drawn no significant correlation between the methylation of the hTERT promoter and subsequent expression of the gene (Devereux et al., 1999; Dessain et al., 2000). Meeran et al. (2008) and Renaud et al. (2007) have shown that in most cancer cells, the regulatory region of hTERT is hypermethylated, which is associated with increased expression, whereas demethylation of this region inhibits hTERT transcription. This pattern is contrary to what commonly occurs during the regulation of genes via methylation, in which methylation of the cytosines in a promoter region generally inhibits gene transcription (Kikuno et al., 2008; Majid et al., 2008; Meeran et al., 2010). Hypomethylation of the hTERT promoter can be seen in certain hTERT negative cells, markedly those that have not been differentiated or transformed, suggesting a mechanism independent of promoter methylation by which these cells tightly repress the transcription of hTERT (Dessain et al., 2000; Liu et al., 2004; Lopatina et al., 2003; Shin et al., 2003). Zinn et al. (2007) indentified little to no methylation of alleles around their transcriptional start sites in telomerase-positive cancer cells, despite an abundance of methylation in more upstream regions. This suggests that the absence of methylation at and around the transcriptional start site allows for the transcription and expression of hTERT, which shows some consistency with the usual dynamics of gene expression (as reviewed by Kyo et al., 2008).
Research has shown that total methylation of hTERT inhibits transcription and that a region of the hTERT minimal promoter must be hypomethylated in order for transcription to proceed (Renaud et al., 2007). CTCF, a repressor of hTERT activity, has shown to be regulated through the methylation of its recognition sequence (Renaud et al., 2007). In this study, ChIP assays revealed that CTCF binds to the first exon of hTERT when its CpG island is not methylated and no longer binds when its regulation sequence is methylated (Renaud et al., 2007). Therefore, it can be postulated that hypermethylation of the CTCF recognition sequence suppresses the factor’s repressor activity, and that the main purpose of methylating the hTERT CpG island is likely to prevent the binding of CTCF and allow for the transcription of hTERT (Renaud et al., 2007). Choi et al. (2010) found that trichostatin A (TSA) induced the demethylation of site-specific CpGs on the hTERT promoter via the down regulation of DNA methyltransferase 1 (DNMT1). Within this demethylated region is a binding site for CTCF, located between the 31st and 33rd CpGs; ChIP analysis revealed that the demethylation of these particular CpG sites by TSA promoted CTCF binding to the hTERT promoter and led to the repression of hTERT transcription. Previous studies have shown that certain dietary compounds, notably genistein and EGCG, down-regulate DNMTs which is directly associated with the repression of hTERT via hTERT promoter demethylation in breast cancer cells (Berletch et al., 2008; Li et al., 2009; Meeran et al., 2011). It has also been suggested that EGCG-induced down-regulation of DNMTs is not only involved in the regulation of hTERT in the process of anti-carcinogenesis, but also in enhancing the binding of methylation-sensitive transcription factors such as E2F-1 to the hTERT promoter region (Crowe et al., 2001; Meeran et al., 2011). Furthermore, Meeran et al. (2010) found that sulforaphane (SFN), an isothiocyanate found in cruciferous vegestables, aids in the down-regulation of DNMT1 and DNMT3a, resulting in site-specific CpG demethylation in the hTERT promoter and thereby facilitating the binding of CTCF and subsequent repression of hTERT. A study by Iliopoulos et al. (2009) revealed that hTERT expression levels were inversely related with DNA methylation levels in hepatocellular carcinoma (HCC) and normal tissues. Moreover, hTERT expression was determined to be regulated by DNA methylation and histone H3-K9 modifications, affecting the ability of cMyc to bind to E-box 1 on the hTERT promoter and further supporting the proposal that hTERT is regulated both epigenetically via mechanisms of DNA methylation and histone modifications and by cMyc in certain cancers (Iliopoulos et al., 2009).
Modification of histones by acetylation/deacetylation is known to regulate the structure of chromatin and thereby affect gene transcription (Stein et al., 2000; as reviewed by Kyo et al., 2008). Histone acetylases (HATs) and histone deacetylases (HDACs) generally cause the acetylation and deacetylation of chromatin, respectively, and thus play pivotal roles in the regulation of hTERT expression (Choi et al., 2009; Meeran et al., 2010). Meeran et al. (2010) revealed that SFN-induced hyperacetylation of the hTERT promoter facilitated the binding of MAD1, CTCF, and several other repressor proteins to the hTERT regulatory region. One study indicated that HDAC inhibitors activate the hTERT promoter in normal cells in a Sp1-dependent manner (as reviewed by Kyo et al., 2008). A role for histone methylation in the regulation of hTERT has also been demonstrated. A recent study reported that highly trimethylated H3-K4 is associated with active transcription of the hTERT gene in telomerase-proficient tumor cells (Atkinson et al., 2005). Furthermore, Liu et al. (2007) found that SET-and MYND-domain-containing protein-3 (SMYD3) are highly important in the regulation of the hTERT promoter. SMYD3-mediated trimethylation of histone H3-K4 may lead to the further recruitment of HAT via its function as a recruitment element for the consequent binding of transcription factors to the hTERT promoter (as reviewed by Kyo et al., 2008). A recent study by Mao et al. (2011) showed that Sirt1, a NAD-dependent class III histone deacetylase, interacts with the C-terminus of cMyc, causing its deacetylation both in vitro and in vivo. Furthermore, this deacetylation of cMyc by Sirt1 was shown to promote its association with Max, an essential partner for cMyc activation, and facilitate cMyc transactivation activity on the promoter of hTERT, suggesting a potential role of Sirt1 in tumorigenesis, notably in K562 human leukemia cells (Mao et al., 2011). Epigenetic control and modulation of the hTERT promoter via methylation and acetylation qualifies as a promising method in understanding the regulation of hTERT, though further research is needed to elucidate these mechanisms.
5. Conclusion
Recent studies have presented a broad range of mechanisms by which hTERT is controlled and regulated in both cancer and normal cell lines. Transcriptional regulation via the recruitment of activators or repressors is most commonly associated with the expression or silencing, respectively, of the hTERT gene. Epigenetics has also been shown to play a pivotal role in the regulation of hTERT through various methylation and acetylation mechanisms. However, the abundance of regulatory models reveals the need for further research in determining how hTERT expression and telomerase activity are regulated in vivo and in vitro.
Understanding the mechanisms of hTERT regulation is imperative for many reasons. Telomerase plays in important role in the maintenance, protection, and stabilization of linear chromosomes (Cukusić et al., 2008). Unfortunately, this diversity can lead to numerous opportunities for cancers to activate hTERT during tumorigenesis and escape cell senescence (Cukusić et al., 2008). Breakthroughs in the discovering the roles of various regulatory elements of hTERT have provided new approaches in the treatment of certain cancers and a better understanding of tumorigenesis. Various dietary compounds, such as genistein, EGCG, and sulforaphane show promising avenues by which hTERT is down-regulated and cellular apoptosis induced in human breast cancer cells (Meeran et al., 2010; Meeran et al., 2011). Denotation of the genomic sequence of hTERT and elucidation of its gene organization (Cong et al., 1999; Horikawa et al., 1999; Takakura et al., 1999; Wick et al., 2003) has led to the characterization of hTERT as the limited factor for telomerase activity. Recent developments have paved the way for various cancer-specific diagnostic and therapeutic approaches as well. Telomerase-specific replicative adenovirus (Telomelysis, OBP-301) has been developed as an oncolytic virus that replicates specifically in cancer cells and causes cell death (as reviewed by Kyo et al., 2008). The hypothesis establishing hTERT regulation as a key mechanism in cell immortalization, neoplasms, and the progression of cancers seems solid, however, considerably more research is needed to evolve and elucidate more concrete regulatory mechanisms.
Highlights.
Telomerase is regulated by the hTERT gene.
The hTERT gene is controlled by genetic and epigenetic factors.
>hTERT gene control is important in cancer and aging.
Acknowledgments
This work was supported in part from grants by the NCI (ROI CA129415) and the Norma Livingston Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe S, Tanaka H, et al. Localization of an hTERT repressor region on human chromosome 3p21.3 using chromosome engineering. Genome Integr. 2010;1(1):6. doi: 10.1186/2041-9414-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Alson S, et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci U S A. 2002;99(12):8191–6. doi: 10.1073/pnas.112515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SP, Hoare SF, et al. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65(17):7585–90. doi: 10.1158/0008-5472.CAN-05-1715. [DOI] [PubMed] [Google Scholar]
- Bai D, Ueno L, et al. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int J Cancer. 2009;125(12):2863–70. doi: 10.1002/ijc.24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, et al. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8(3):177–80. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Beijersbergen RL, Hijmans EM, et al. Interaction of c-Myc with the pRb-related protein p107 results in inhibition of c-Myc-mediated transactivation. EMBO J. 1994;13(17):4080–6. doi: 10.1002/j.1460-2075.1994.tb06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Liu C, et al. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103(2):509–19. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, et al. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17(2):236–9. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106(6):661–73. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Godley LA, et al. Telomerase activation in mouse mammary tumors: lack of detectable telomere shortening and evidence for regulation of telomerase RNA with cell proliferation. Mol Cell Biol. 1996;16(7):3765–72. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce LA, Morrison N, et al. Mapping of the gene for the human telomerase reverse transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in situ hybridization. Neoplasia. 2000;2(3):197–201. doi: 10.1038/sj.neo.7900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz M, Schatz A, et al. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25(15):3714–24. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich KJ, Greider CW. Telomerase regulation during entry into the cell cycle in normal human T cells. Mol Biol Cell. 1996;7(9):1443–54. doi: 10.1091/mbc.7.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, et al. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34(19):5402–15. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas MA, Brotherton SL, et al. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene. 2003;316:57–65. doi: 10.1016/s0378-1119(03)00739-x. [DOI] [PubMed] [Google Scholar]
- Casillas MA, Lopatina N, et al. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252(1–2):33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- Chebel A, Ffrench M. Transcriptional regulation of the human telomerase reverse transcriptase: New insights. Transcription. 2010;1(1):27–31. doi: 10.4161/trns.1.1.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebel A, Rouault JP, et al. Transcriptional activation of hTERT, the human telomerase reverse transcriptase, by nuclear factor of activated T cells. J Biol Chem. 2009;284(51):35725–34. doi: 10.1074/jbc.M109.009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CR, Kang Y, et al. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110(1):19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen Y, Knösel T, et al. Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer. 2007;55(3):287–94. doi: 10.1016/j.lungcan.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Chen YN, Chen H, et al. Expression of pituitary homeobox 1 gene in human gastric carcinogenesis and its clinicopathological significance. World J Gastroenterol. 2008;14(2):292–7. doi: 10.3748/wjg.14.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherlet T, Murphy LC. Estrogen receptors inhibit Smad3 transcriptional activity through Ap-1 transcription factors. Mol Cell Biochem. 2007;306(1–2):33–42. doi: 10.1007/s11010-007-9551-1. [DOI] [PubMed] [Google Scholar]
- Chikhirzhina GI, Al’-Shekhadat RI, et al. [Transcription factors of the nuclear factor 1 (NF1) family. Role in chromatin remodelation] Mol Biol (Mosk) 2008;42(3):388–404. [PubMed] [Google Scholar]
- Chiu CP, Harley CB. Replicative senescence and cell immortality: the role of telomeres and telomerase. Proc Soc Exp Biol Med. 1997;214(2):99–106. doi: 10.3181/00379727-214-44075. [DOI] [PubMed] [Google Scholar]
- Choi JH, Min NY, et al. TSA-induced DNMT1 down-regulation represses hTERT expression via recruiting CTCF into demethylated core promoter region of hTERT in HCT116. Biochem Biophys Res Commun. 2010;391(1):449–54. doi: 10.1016/j.bbrc.2009.11.078. [DOI] [PubMed] [Google Scholar]
- Choi KC, Jung MG, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69(2):583–92. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, et al. A human telomeric protein. Science. 1995;270(5242):1663–7. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- Chou CK, Lee DF, et al. The suppression of MAD1 by AKT-mediated phosphorylation activates MAD1 target genes transcription. Mol Carcinog. 2009;48(11):1048–58. doi: 10.1002/mc.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun AC, Jin DY. Transcriptional regulation of mitotic checkpoint gene MAD1 by p53. J Biol Chem. 2003;278(39):37439–50. doi: 10.1074/jbc.M307185200. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315(5820):1850–3. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21(4):564–79. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275(46):35665–8. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wen J, et al. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8(1):137–42. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wright WE, et al. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66(3):407–25. doi: 10.1128/MMBR.66.3.407-425.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe DL, Nguyen DC, et al. E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 2001;29(13):2789–94. doi: 10.1093/nar/29.13.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukusić A, Skrobot Vidacek N, et al. Telomerase regulation at the crossroads of cell fate. Cytogenet Genome Res. 2008;122(3–4):263–72. doi: 10.1159/000167812. [DOI] [PubMed] [Google Scholar]
- Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43(6):668–98. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- De Boeck G, Forsyth RG, et al. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J Pathol. 2009;217(3):327–44. doi: 10.1002/path.2500. [DOI] [PubMed] [Google Scholar]
- Deng WG, Jayachandran G, et al. Tumor-specific activation of human telomerase reverses transcriptase promoter activity by activating enhancer-binding protein-2beta in human lung cancer cells. J Biol Chem. 2007;282(36):26460–70. doi: 10.1074/jbc.M610579200. [DOI] [PubMed] [Google Scholar]
- Dessain SK, Yu H, et al. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60(3):537–41. [PubMed] [Google Scholar]
- Devereux TR, Horikawa I, et al. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59(24):6087–90. [PubMed] [Google Scholar]
- Drabsche Y, Ramsay RG, et al. MYB supresses differentiation and apoptosis of human breast cancer cells. Breast Cancer Research. 2010;12:R55. doi: 10.1186/bcr2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickelberg O, Centrella M, et al. Betaglycan inhibits TGF-beta signaling by preventing type I-type II receptor complex formation. Glycosaminoglycan modifications alter betaglycan function. J Biol Chem. 2002;277(1):823–9. doi: 10.1074/jbc.M105110200. [DOI] [PubMed] [Google Scholar]
- Farwell DG, Shera KA, et al. Genetic and epigenetic changes in human epithelial cells immortalized by telomerase. Am J Pathol. 2000;156(5):1537–47. doi: 10.1016/S0002-9440(10)65025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67(7):3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16(6):2802–13. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Kyo S, et al. Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (hTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nucleic Acids Res. 2000;28(13):2557–62. doi: 10.1093/nar/28.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Chen D, et al. Effect of estrogen on telomerase activity in human breast cancer cells. J Huazhong Univ Sci Technolog Med Sci. 2003;23(3):286–7. 293. doi: 10.1007/BF02829516. [DOI] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, et al. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–8. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerttler PS, Kreutz C, et al. Gene expression profiling in polycythaemia vera: overexpression of transcription factor NF-E2. Br J Haematol. 2005;129(1):138–50. doi: 10.1111/j.1365-2141.2005.05416.x. [DOI] [PubMed] [Google Scholar]
- Gombert WM, Krumm A. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One. 2009;4(7):e6109. doi: 10.1371/journal.pone.0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Lemarteleur T, et al. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32(1):371–9. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E, Flores JM, et al. Cooperation between p53 mutation and high telomerase transgenic expression in spontaneous cancer development. Mol Cell Biol. 2002;22(20):7291–301. doi: 10.1128/MCB.22.20.7291-7301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E, Samper E, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20(11):2619–30. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22(39):8042–7. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
- Grand CL, Han H, et al. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol Cancer Ther. 2002;1(8):565–73. [PubMed] [Google Scholar]
- Griffith JD, Comeau L, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Gros J, Guédin A, et al. G-Quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem. 2008;9(13):2075–9. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- Günes C, Lichtsteiner S, et al. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60(8):2116–21. [PubMed] [Google Scholar]
- Guo C, Armbruster BN, et al. In vivo regulation of hTERT expression and telomerase activity by androgen. J Urol. 2003;170(2 Pt 1):615–8. doi: 10.1097/01.ju.0000074653.22766.c8. [DOI] [PubMed] [Google Scholar]
- Guo Y, Pajovic S, et al. Expression of p14ARF, MDM2, and MDM4 in human retinoblastoma. Biochem Biophys Res Commun. 2008;375(1):1–5. doi: 10.1016/j.bbrc.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400(6743):464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Han DF, Zhou X, et al. Polymorphisms of estrogen-metabolizing genes and breast cancer risk: a multigenic study. Chin Med J (Engl) 2005;118(18):1507–16. [PubMed] [Google Scholar]
- Harley CB, Futcher AB, et al. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–60. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, et al. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hein N, Jiang K, et al. TGFβ1 enhances MAD1 expression and stimulates promoter-bound Pol II phosphorylation: basic functions of C/EBP, SP and SMAD3 transcription factors. BMC Mol Biol. 2011;12(9) doi: 10.1186/1471-2199-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson YC, Breau RL, et al. Telomerase activity in head and neck tumors after introduction of wild-type p53, p21, p16, and E2F-1 genes by means of recombinant adenovirus. Head Neck. 2000;22(4):347–54. doi: 10.1002/1097-0347(200007)22:4<347::aid-hed6>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Hooker CW, Hurlin PJ. Of Myc and Mnt. J Cell Sci. 2006;119(Pt 2):208–16. doi: 10.1242/jcs.02815. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Cable PL, et al. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59(4):826–30. [PubMed] [Google Scholar]
- Horikawa I, Cable PL, et al. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol Biol Cell. 2002;13(8):2585–97. doi: 10.1091/mbc.E01-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MC, Chai CY, et al. Jab1 is overexpressed in human breast cancer and is a downstream target for HER-2/neu. Mod Pathol. 2008;21(5):609–16. doi: 10.1038/modpathol.2008.23. [DOI] [PubMed] [Google Scholar]
- Hsu MC, Chang HC, et al. HER-2/neu transcriptionally activates Jab1 expression via the AKT/beta-catenin pathway in breast cancer cells. Endocr Relat Cancer. 2007;14(3):655–67. doi: 10.1677/ERC-07-0077. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Satra M, et al. Epigenetic regulation of hTERT promoter in hepatocellular carcinomas. Int J Oncol. 2009;34(2):391–9. [PubMed] [Google Scholar]
- Imachi H, Murao K, et al. Menin, a product of the MENI gene, binds to estrogen receptor to enhance its activity in breast cancer cells: possibility of a novel predictive factor for tamoxifen resistance. Breast Cancer Res Treat. 2010;122(2):395–407. doi: 10.1007/s10549-009-0581-0. [DOI] [PubMed] [Google Scholar]
- Ishikawa F. Regulation mechanisms of mammalian telomerase. A review. Biochemistry (Mosc) 1997;62(11):1332–7. [PubMed] [Google Scholar]
- James L, Eisenman RN. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci U S A. 2002;99(16):10429–34. doi: 10.1073/pnas.162369299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Hein N, et al. Regulation of the MAD1 promoter by G-CSF. Nucleic Acids Res. 2008;36(5):1517–31. doi: 10.1093/nar/gkn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Liu Y, et al. Involvement of MyoD and c-myb in regulation of basal and estrogen-induced transcription activity of the BRCA1 gene. Breast Cancer Res Treat. 2011;125(3):699–713. doi: 10.1007/s10549-010-0876-1. [DOI] [PubMed] [Google Scholar]
- Johnson TL, Lai MB, et al. Inhibition of Cell Proliferation and MAP Kinase and Akt Pathways in Oral Squamous cell Carcinoma by Genistein and Biochanin A. Evid Based Complement Alternat Med. 2010;7(3):351–8. doi: 10.1093/ecam/nen011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Hermeking H. The c-MYC-AP4-p21 cascade. Cell Cycle. 2009;8(7):982–9. doi: 10.4161/cc.8.7.7949. [DOI] [PubMed] [Google Scholar]
- Jung P, Menssen A, et al. AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci U S A. 2008;105(39):15046–51. doi: 10.1073/pnas.0801773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata H, Manabe T, et al. Hydrogen peroxide activates IkappaB kinases through phosphorylation of serine residues in the activation loops. FEBS Lett. 2002;519(1–3):231–7. doi: 10.1016/s0014-5793(02)02712-6. [DOI] [PubMed] [Google Scholar]
- Kanaya T, Kyo S, et al. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6(4):1239–47. [PubMed] [Google Scholar]
- Kanaya T, Kyo S, et al. hTERT is a critical determinant of telomerase activity in renal-cell carcinoma. Int J Cancer. 1998;78(5):539–43. doi: 10.1002/(sici)1097-0215(19981123)78:5<539::aid-ijc2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10(14):853–6. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, et al. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283(5406):1321–5. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, et al. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295(5564):2446–9. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- Khan SJ, Yanez G, et al. Interactions of TRF2 with model telomeric ends. Biochem Biophys Res Commun. 2007;363(1):44–50. doi: 10.1016/j.bbrc.2007.08.122. [DOI] [PubMed] [Google Scholar]
- Kikuno N, Shiina H, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123(3):552–60. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- Kim JH, Park SM, et al. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005;19(7):776–81. doi: 10.1101/gad.1289405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Ohmichi M, et al. Induction of hTERT expression and phosphorylation by estrogen via Akt cascade in human ovarian cancer cell lines. Oncogene. 2004;23(26):4505–15. doi: 10.1038/sj.onc.1207582. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick KL, Ogunkolade W, et al. hTERT expression in human breast cancer and non-cancerous breast tissue: correlation with tumour stage and c-Myc expression. Breast Cancer Res Treat. 2003;77(3):277–84. doi: 10.1023/a:1021849217054. [DOI] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13(12):7612–24. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10(9):1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kusumoto M, Ogawa T, et al. Adenovirus-mediated p53 gene transduction inhibits telomerase activity independent of its effects on cell cycle arrest and apoptosis in human pancreatic cancer cells. Clin Cancer Res. 1999;5(8):2140–7. [PubMed] [Google Scholar]
- Kyo S, Takakura M, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99(8):1528–38. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo S, Takakura M, et al. Estrogen activates telomerase. Cancer Res. 1999;59(23):5917–21. [PubMed] [Google Scholar]
- Kyo S, Takakura M, et al. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28(3):669–77. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerte A, Korah J, et al. Transforming growth factor-beta inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal. 2008;20(1):50–9. doi: 10.1016/j.cellsig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lai SR, Cunningham AP, et al. Evidence of extra-telomeric effects of hTERT and its regulation involving a feedback loop. Exp Cell Res. 2007;313(2):322–30. doi: 10.1016/j.yexcr.2006.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee SY, et al. p21 WAF1 is involved in interferon-β-induced attenuation of telomerase activity and human telomerase reverse transcriptase (hTERT) expression in ovarian cancer. Mol Cells. 2010;30(4):327–33. doi: 10.1007/s10059-010-0131-y. [DOI] [PubMed] [Google Scholar]
- Leng X, Noble M, et al. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol Cell Biol. 2002;22(7):2242–54. doi: 10.1128/MCB.22.7.2242-2254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AY, Lin HH, et al. High-Mobility Group A2 Protein Modulates hTERT Transcription To Promote Tumorigenesis. Mol Cell Biol. 2011;31(13):2605–17. doi: 10.1128/MCB.05447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lee TH, et al. A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer. J Biol Chem. 2002;277(23):20965–73. doi: 10.1074/jbc.M112231200. [DOI] [PubMed] [Google Scholar]
- Li J, Huang X, et al. Human telomerase reverse transcriptase regulates cyclin D1 and G1/S phase transition in laryngeal squamous carcinoma. Acta Otolaryngol. 2011;131(5):546–51. doi: 10.3109/00016489.2011.557393. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu L, et al. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125(2):286–96. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sarkar FH. Inhibition of nuclear factor kappaB activation in PC3 cells by genistein is mediated via Akt signaling pathway. Clin Cancer Res. 2002;8(7):2369–77. [PubMed] [Google Scholar]
- Lim KW, Lacroix L, et al. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc. 2010;132(35):12331–42. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- Liu C, Fang X, et al. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67(6):2626–31. doi: 10.1158/0008-5472.CAN-06-4126. [DOI] [PubMed] [Google Scholar]
- Liu L, Ishihara K, et al. MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem. 2009;284(8):5165–74. doi: 10.1074/jbc.M807098200. [DOI] [PubMed] [Google Scholar]
- Liu L, Lai S, et al. Genetic and epigenetic modulation of telomerase activity in development and disease. Gene. 2004;340(1):1–10. doi: 10.1016/j.gene.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Liu L, Saldanha SN, et al. Epigenetic regulation of human telomerase reverse transcriptase promoter activity during cellular differentiation. Genes Chromosomes Cancer. 2004;41(1):26–37. doi: 10.1002/gcc.20058. [DOI] [PubMed] [Google Scholar]
- Lopatina NG, Poole JC, et al. Control mechanisms in the regulation of telomerase reverse transcriptase expression in differentiating human teratocarcinoma cells. Biochem Biophys Res Commun. 2003;306(3):650–9. doi: 10.1016/s0006-291x(03)01033-7. [DOI] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28(8):1707–13. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Kikuno N, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68(8):2736–44. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, et al. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88(5):657–66. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Mao B, Zhao G, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43(11):1573–81. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Masutomi K, Yu EY, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114(2):241–53. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- McArthur GA, Foley KP, et al. MAD1 and p27(KIP1) cooperate to promote terminal differentiation of granulocytes and to inhibit Myc expression and cyclin E-CDK2 activity. Mol Cell Biol. 2002;22(9):3014–23. doi: 10.1128/MCB.22.9.3014-3023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeran SM, Ahmed A, et al. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics. 2010;1(3–4):101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeran SM, Katiyar S, et al. Berberine-induced apoptosis in human prostate cancer cells is initiated by reactive oxygen species generation. Toxicol Appl Pharmacol. 2008;229(1):33–43. doi: 10.1016/j.taap.2007.12.027. [DOI] [PubMed] [Google Scholar]
- Meeran SM, Patel SN, et al. A Novel Prodrug of Epigallocatechin-3-gallate: Differential Epigenetic hTERT Repression in Human Breast Cancer Cells. Cancer Prev Res (Phila) 2011 doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeran SM, Patel SN, et al. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5(7):e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–95. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Misiti S, Nanni S, et al. Induction of hTERT expression and telomerase activity by estrogens in human ovary epithelium cells. Mol Cell Biol. 2000;20(11):3764–71. doi: 10.1128/mcb.20.11.3764-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Venkatachalam S, et al. Cooperativity of p19ARF, Mdm2, and p53 in murine tumorigenesis. Oncogene. 2003;22(49):7831–7. doi: 10.1038/sj.onc.1206985. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–9. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nanni S, Narducci M, et al. Signaling through estrogen receptors modulates telomerase activity in human prostate cancer. J Clin Invest. 2002;110(2):219–27. doi: 10.1172/JCI15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DC, Crowe DL. Intact functional domains of the retinoblastoma gene product (pRb) are required for downregulation of telomerase activity. Biochim Biophys Acta. 1999;1445(2):207–15. doi: 10.1016/s0167-4781(99)00044-5. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Murayama A, et al. Regulation of myeloid zinc finger protein 2A transactivation activity through phosphorylation by mitogen-activated protein kinases. J Biol Chem. 2003;278(5):2921–7. doi: 10.1074/jbc.M207615200. [DOI] [PubMed] [Google Scholar]
- Ogawara Y, Kishishita S, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277(24):21843–50. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- Oh S, Song YH, et al. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene. 2000;19(11):1485–90. doi: 10.1038/sj.onc.1203439. [DOI] [PubMed] [Google Scholar]
- Oh W, Lee EW, et al. Jab1 induces the cytoplasmic localization and degradation of p53 in coordination with Hdm2. J Biol Chem. 2006;281(25):17457–65. doi: 10.1074/jbc.M601857200. [DOI] [PubMed] [Google Scholar]
- Papanikolaou V, Athanassiou E, et al. hTERT regulation by NF-κB and c-myc in irradiated HER2-positive breast cancer cells. Int J Radiat Biol. 2011;87(6):609–21. doi: 10.3109/09553002.2011.572112. [DOI] [PubMed] [Google Scholar]
- Parisi F, Wirapati P, et al. Identifying synergistic regulation involving c-Myc and sp1 in human tissues. Nucleic Acids Res. 2007;35(4):1098–107. doi: 10.1093/nar/gkl1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, et al. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35(22):7429–55. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault SD, Hornsby PJ, et al. Global gene expression response to telomerase in bovine adrenocortical cells. Biochem Biophys Res Commun. 2005;335(3):925–36. doi: 10.1016/j.bbrc.2005.07.156. [DOI] [PubMed] [Google Scholar]
- Phuong NT, Kim SK, et al. Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Res Treat. 2011;130(1):73–83. doi: 10.1007/s10549-010-1304-2. [DOI] [PubMed] [Google Scholar]
- Potischman N, Swanson CA, et al. Reversal of relation between body mass and endogenous estrogen concentrations with menopausal status. J Natl Cancer Inst. 1996;88(11):756–8. doi: 10.1093/jnci/88.11.756. [DOI] [PubMed] [Google Scholar]
- Qi DL, Ohhira T, et al. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol. 2011;31(8):1624–36. doi: 10.1128/MCB.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana AM, Liu F, et al. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer. 2011;11(30) doi: 10.1186/1471-2407-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati-Yamchi M, Zarghami N, et al. Plasma Leptin, hTERT Gene Expression, and Anthropometric Measures in Obese and Non-Obese Women with Breast Cancer. Breast Cancer (Auckl) 2011;5:27–35. doi: 10.4137/BCBCR.S6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, James MK, et al. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol Cell Biol. 2009;29(4):986–99. doi: 10.1128/MCB.00898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao T, et al. Leptin upregulates telomerase activity and transcription of human telomerase reverse transcriptase in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2010;394(1):59–63. doi: 10.1016/j.bbrc.2010.02.093. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wu L, et al. Dual effects of TGF-beta on ERalpha-mediated estrogenic transcriptional activity in breast cancer. Mol Cancer. 2009;8(111) doi: 10.1186/1476-4598-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Loukinov D, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35(4):1245–56. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Loukinov D, et al. BORIS/CTCFL-mediated transcriptional regulation of the hTERT telomerase gene in testicular and ovarian tumor cells. Nucleic Acids Res. 2011;39(3):862–73. doi: 10.1093/nar/gkq827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Loukinov D, et al. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33(21):6850–60. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose DP, Haffner SM, et al. Adiposity, the metabolic syndrome, and breast cancer in African-American and white American women. Endocr Rev. 2007;28(7):763–77. doi: 10.1210/er.2006-0019. [DOI] [PubMed] [Google Scholar]
- Saito T, Matsuda Y, et al. Comparative gene mapping of the human and mouse TEP1 genes, which encode one protein component of telomerases. Genomics. 1997;46(1):46–50. doi: 10.1006/geno.1997.5005. [DOI] [PubMed] [Google Scholar]
- Salvatico J, Kim JH, et al. Differentiation linked regulation of telomerase activity by Makorin-1. Mol Cell Biochem. 2010;342(1–2):241–50. doi: 10.1007/s11010-010-0490-x. [DOI] [PubMed] [Google Scholar]
- Sasaki R, Narisawa-Saito M, et al. Oncogenic transformation of human ovarian surface epithelial cells with defined cellular oncogenes. Carcinogenesis. 2009;30(3):423–31. doi: 10.1093/carcin/bgp007. [DOI] [PubMed] [Google Scholar]
- Shats I, Milyavsky M, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J Biol Chem. 2004;279(49):50976–85. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- Shin KH, Kang MK, et al. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer. 2003;89(8):1473–8. doi: 10.1038/sj.bjc.6601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson T. G-quadruplex DNA structures--variations on a theme. Biol Chem. 2001;382(4):621–8. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- Sitaram RT, Degerman S, et al. Wilms’ tumour 1 can suppress hTERT gene expression and telomerase activity in clear cell renal cell carcinoma via multiple pathways. Br J Cancer. 2010;103(8):1255–62. doi: 10.1038/sj.bjc.6605878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LL, Coller HA, et al. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5(5):474–9. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- Song J, Murakami H, et al. Genomic organization and expression of a human gene for Myc-associated zinc finger protein (MAZ) J Biol Chem. 1998;273(32):20603–14. doi: 10.1074/jbc.273.32.20603. [DOI] [PubMed] [Google Scholar]
- Song J, Ugai H, et al. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements. Int J Mol Med. 2003;11(5):547–53. [PubMed] [Google Scholar]
- Stanelle J, Stiewe T, et al. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30(8):1859–67. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, et al. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20(19):5532–40. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanou N, Papanikolaou V, et al. Leptin as a critical regulator of hepatocellular carcinoma development through modulation of human telomerase reverse transcriptase. BMC Cancer. 2010;10(442) doi: 10.1186/1471-2407-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Montecino M, et al. Nuclear structure-gene expression interrelationships: implications for aberrant gene expression in cancer. Cancer Res. 2000;60(8):2067–76. [PubMed] [Google Scholar]
- Stewart SA, Hahn WC, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci U S A. 2002;99(20):12606–11. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suphapeetiporn K, Greally JM, et al. MEN1 tumor-suppressor protein localizes to telomeres during meiosis. Genes Chromosomes Cancer. 2002;35(1):81–5. doi: 10.1002/gcc.10113. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238(2):291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Takakura M, Kyo S, et al. Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol Cell Biol. 2005;25(18):8037–43. doi: 10.1128/MCB.25.18.8037-8043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura M, Kyo S, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59(3):551–7. [PubMed] [Google Scholar]
- Takano H, Murasawa S, et al. Functional and gene expression analysis of hTERT overexpressed endothelial cells. Biologics. 2008;2(3):547–54. doi: 10.2147/btt.s2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee MK, Rogatsky I, et al. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors alpha and beta. Mol Biol Cell. 2004;15(3):1262–72. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277(3):2302–10. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, et al. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–13. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131(4):669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272(52):33353–9. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- Wan M, Cao X, et al. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3(2):171–6. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Hou X, et al. Activation of p27Kip1 Expression by E2F1. A negative feedback mechanism. J Biol Chem. 2005;280(13):12339–43. doi: 10.1074/jbc.C400536200. [DOI] [PubMed] [Google Scholar]
- Wang J, Barnes RO, et al. Jab1 is a target of EGFR signaling in ERalpha-negative breast cancer. Breast Cancer Res. 2008;10(3):R51. doi: 10.1186/bcr2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Greiner TC, et al. Decreased Mdm2 expression inhibits tumor development induced by loss of ARF. Oncogene. 2006;25(26):3708–18. doi: 10.1038/sj.onc.1209411. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kyo S, et al. Tamoxifen regulates human telomerase reverse transcriptase (hTERT) gene expression differently in breast and endometrial cancer cells. Oncogene. 2002;21(22):3517–24. doi: 10.1038/sj.onc.1205463. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17(4):498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33(5):735–49. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- Wick M, Zubov D, et al. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232(1):97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- Wisman GB, Hollema H, et al. Telomerase in relation to expression of p53, c-Myc and estrogen receptor in ovarian tumours. Int J Oncol. 2003;23(5):1451–9. doi: 10.3892/ijo.23.5.1451. [DOI] [PubMed] [Google Scholar]
- Won J, Yim J, et al. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J Biol Chem. 2002;277(41):38230–8. doi: 10.1074/jbc.M206064200. [DOI] [PubMed] [Google Scholar]
- Wu L, Wu Y, et al. Smad4 as a transcription corepressor for estrogen receptor alpha. J Biol Chem. 2003;278(17):15192–200. doi: 10.1074/jbc.M212332200. [DOI] [PubMed] [Google Scholar]
- Wu S, Hultquist A, et al. TGF-beta enforces senescence in Myc-transformed hematopoietic tumor cells through induction of Mad1 and repression of Myc activity. Exp Cell Res. 2009;315(18):3099–111. doi: 10.1016/j.yexcr.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Xu D, Popov N, et al. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci U S A. 2001;98(7):3826–31. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Wang Q, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19(45):5123–33. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- Xu HJ, Zhou Y, et al. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene. 1997;15(21):2589–96. doi: 10.1038/sj.onc.1201446. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, et al. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350(6320):718–20. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zhou C, Bae-Jump VL, et al. The PTEN tumor suppressor inhibits telomerase activity in endometrial cancer cells by decreasing hTERT mRNA levels. Gynecol Oncol. 2006;101(2):305–10. doi: 10.1016/j.ygyno.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Zhu J, Blenis J, et al. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci U S A. 2008;105(18):6584–9. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn RL, Pruitt K, et al. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67(1):194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- Zou L, Zhang PH, et al. Transcript regulation of human telomerase reverse transcriptase by c-myc and mad1. Acta Biochim Biophys Sin (Shanghai) 2005;37(1):32–8. doi: 10.1093/abbs/37.1.32. [DOI] [PubMed] [Google Scholar]