Abstract
Substrate specificity of enzymes is frequently narrow and constrained by multiple interactions, limiting the use of natural enzymes in biocatalytic applications. Aldolases have important synthetic applications, but the usefulness of these enzymes is hampered by their narrow reactivity profile with unnatural substrates. To explore the determinants of substrate selectivity and alter the specificity of E. coli 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase, we employed structure-based mutagenesis coupled with library screening of mutant enzymes localized to the bacterial periplasm. We identified two active site mutations (T161S/S184L) that work additively to enhance the substrate specificity of this aldolase to include catalysis of retro-aldol cleavage of (4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (S-KHPB). These mutations improve the value of kcat/KMS-KHPB by >450-fold, resulting in a catalytic efficiency that is comparable to that of the wild-type enzyme with the natural substrate while retaining high stereoselectivity. Moreover, the value of kcatS-KHPB for this mutant enzyme, a parameter critical for biocatalytic applications, is 3-fold higher than the maximum value achieved by the natural aldolase with any substrate. This mutant also possesses high catalytic efficiency for the retro-aldol cleavage of the natural substrate, KDPG, and a >50-fold improved activity for cleavage of 2-keto-4-hydroxy-octonoate (KHO), a non-functionalized hydrophobic analog. These data suggest a substrate binding mode that illuminates the origin of facial selectivity in aldol addition reactions catalyzed by KDPG and 2-keto-3-deoxy-6-phosphogalactonate (KDPGal) aldolases. Furthermore, targeting mutations to the active site provides marked improvement in substrate selectivity, demonstrating that structure-guided active site mutagenesis combined with selection techniques can efficiently identify proteins with characteristics that compare favorably to naturally occurring enzymes.
Keywords: rational redesign, directed evolution, protein engineering, biocatalysis, stereoselectivity
Aldolases are a class of structurally and mechanistically diverse enzymes that catalyze reversible aldol addition reactions, and have demonstrated utility as biocatalysts.(1-3) This class of enzymes has long been used for the synthesis of carbohydrates because of their rapid catalysis, high stereospecificity, and regioselectivity. Four classes of aldolases have been described based on the identity of the preferred nucleophilic substrate (Table S1). The aldolases most widely used for biocatalysis prefer dihydroxyacetone phosphate (DHAP) as the nucleophilic substrate (DHAP aldolases). Although enzymes have been isolated that preferentially generate each of the four diasteromeric products at the bond forming carbon centers, these aldolases have limited synthetic usefulness due to difficulties associated with the absolute requirement for DHAP as the nucleophilic substrate, a species that is both difficult to synthesize and unstable, making it a poor choice for large-scale synthetic applications.(4, 5) For this reason, a number of researchers have worked on engineering DHAP aldolases or identifying alternative aldolases that do not require phosphorylated electrophiles.(6-9)
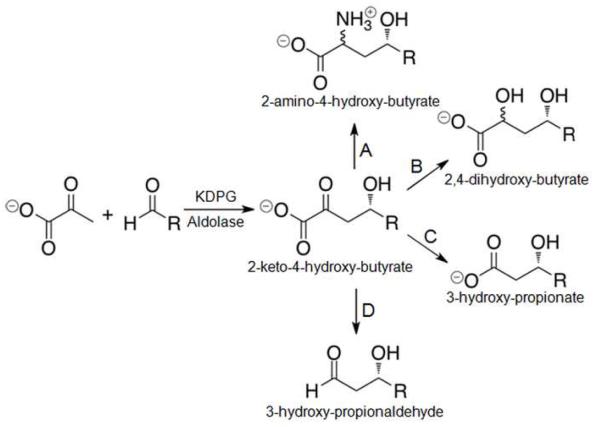
The pyruvate aldolases use pyruvate as the native nucleophilic substrate; an advantage for synthetic applications since it is both inexpensive and stable under most conditions. Additionally, the 2-keto-4-hydroxy-butyrate products that result from pyruvate aldolase-catalyzed aldol addition include all four commonly found oxidation states of carbon, giving the synthetic chemist differential handles for selective transformation in the subsequent steps. Figure 1 illustrates the range of transformations available from the product of pyruvate aldol-condensation. We have focused our efforts on optimizing the pyruvate aldolase, 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase, for use as a biosynthetic catalyst. This enzyme is attractive as a biocatalyst because it possesses a broad pH optimum, high tolerance of organic co-solvents, high stereoselectivity, and is easily over-expressed and purified.(10)
Figure 1.
Pyruvate aldolase-catalyzed stereospecific aldol reaction and synthetic elaborations of the 2-keto-4-hydroxy-butyrate product.(48) (A) Reductive amination. (B) Reduction. (C) Oxidative decarboxylation. (D) Non-oxidative decarboxylation.
The majority of aldolases studied to date catalyze transformations of carbohydrates, primarily catalyzing the retro-aldol cleavage of carbohydrates to three-carbon fragments.(4) BphI and HpaI aldolases are class II (metal-dependent) aldolases that catalyze reactions with unfunctionalized aliphatic aldehydes.(11, 12) Similarly, KDPG aldolase, the final enzyme in the Entner-Doudoroff pathway, is a class I aldolase that catalyzes the reversible aldol cleavage of the phosphorylated sugar acid, KDPG, to pyruvate and glyceraldehyde-3-phosphate (Scheme 1). In the synthetic direction, efficient catalysis is generally restricted to the reaction of pyruvate with a phosphorylated polyhydroxy aldehyde,(10) limiting the synthetic utility of the natural enzyme. A number of researchers have attempted to expand the substrate profile of KDPG(13-15) and the structurally and mechanistically related KDPGal(16) aldolases by directed evolution with modest success. Pyruvate aldolases that catalyze aldol reactions with unfunctionalized aliphatic and aromatic aldehydes remain a desirable target for biocatalytic applications.
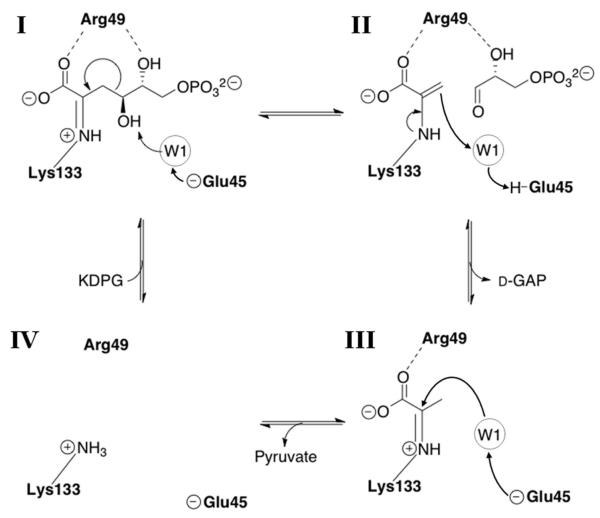
Scheme 1.
Catalytic mechanism of E. coli KDPG aldolase.(29, 30) The substrate is bound as a Schiff base adduct with Lys133 (I) and water W1 arises from the keto-group that is displaced by Schiff base formation.(30) Glu45 catalyzes proton removal from the C4 hydroxyl of KDPG. This proton transfer is mediated by W1. The C3-C4 bond is broken with the Schiff base stabilizing the formation of a carbanion at C4 to form D-glyceraldehyde-3-phosphate and a pyruvyl-enamine (II). The pyruvyl-enamine is protonated by the acidic form of Glu-45 via W1 to form a Schiff base (III). Finally, the pyruvyl-Schiff base is hydrolyzed resulting in the free enzyme (IV).
Here we use structure-guided mutagenesis to prepare KDPG aldolase mutants that increase the catalytic efficiency with hydrophobic aldehydes up to 450-fold while maintaining exquisite stereoselectivity. The high catalytic efficiency of these engineered enzymes with aliphatic aldehydes may aid future efforts in the chemo-enzymatic synthesis of antimicrobial compounds such as syringomycin E(17) and lyngbyaballin A(18)(Figure S1). Furthermore, the ability to catalyze the aldol addition of pyruvate and pyridine carboxaldehydes is useful for the synthesis of analogs of the antifungal agents, nikkomycins(19-21)(Figure S1). These studies also demonstrate that active site mutations can lead to large enhancements in both the breadth of substrate specificity and catalytic efficiency, and should therefore be targeted in protein engineering experiments.
EXPERIMENTAL PROCEDURES
Construction of periplasmic expression plasmid
The pBAD/giii-C (Invitrogen) and pUC-ECEDA(22) plasmids were digested with NcoI and XhoI (NEB) in NEB Buffer 4 and bovine serum albumin. The linearized pBAD vector and the E. coli eda gene were purified using a 1% agarose gel. The two DNA fragments were combined at equimolar concentrations and ligated using T4 DNA ligase (NEB) to prepare pBAD-giii-ECA. The resulting DNA was transformed into calcium competent SmartCells (Genelantis) and single colonies were picked. After propagation of each colony and isolation of the plasmid DNA, DNA sequencing data were used to confirm that the construct contained the full-length eda gene.
Saturation mutagenesis libraries
Libraries containing all possible mutations at positions T161, G162, G163, S184 were made using whole plasmid amplification of the pBAD-giii-ECA construct and the follow primers (Integrated DNA Technologies) and their reverse complements: S184X 5′-GCTGTGCATCGGTGGTNNKTGGCTGGTTCCGGCAGATGC - 3′; T161X 5′- CCAGGTCCGTTTCTGCCCGNNKGGTGGTATTTCTCCGGCTAAC - 3′; G162X 5′ - GGTCCGTTTCTGCCCGACGNNKGGTATTTCTCCGGCTAACTACC - 3′; G163X 5′ - GGTCCGTTTCTGCCCGACGGGTNNKATTTCTCCGGCTAACTACC - 3′. The sequence of the position of interest was randomized by allowing the first two nucleotides in the codon to vary (N = A, T, G, C) and the third position was restricted to G or T (K = G or T).(23) Whole plasmid PCR amplification for each library catalyzed by PFU Turbo (Stratagene) was done using the primer pair for a single randomized codon under the following thermocycling conditions: 1 cycle of 95 °C for 30 s; 18 cycles of 95 °C for 30 s, 58 °C for 1 min, 68 °C for 11 min; and 1 cycle of 68 °C for 5 min. Following PCR amplification the template DNA was exhaustively digested overnight with DpnI and the resulting DNA was purified using the Wizard SV Gel and PCR Purification System (Promega). The DNA was transformed into SmartCells and plated on LB/ampicillin plates yielding hundreds of colonies. All of the colonies were scraped off and pooled together, and the plasmid DNA was purified. Each pool represents libraries of T161, G162, G163 or S184 mutants. DNA sequencing chromatograms of these libraries in aggregate indicated that the targeted sites contained a random assortment of nucleotides according to an NNK distribution. As a second test, the plasmid DNA from three individual colonies per library was isolated and sequenced. All 12 plasmids contained the full-length eda gene with different mutations at the targeted sites.
Selection conditions
The PB25 strain is derived from E. coli K12 JM101 and has disruptions in the pyruvate kinase genes, pykA and pykF; genotype [supE thi (lac-proAB) pykA::kanr pykF::catr].(24) PB25 cells transformed with pBAD-giii-ECA plasmids were plated on selective M9 minimal media that contained the following: 0.2% ribose, 1 μg/mL thiamine, 40 μg/mL proline, 50 μg/mL carbenecillin. The plates were augmented with additional nutrients that facilitate growth but do not negate the pyruvate auxotroph phenotype of PB25 cells, including: 1 μM FeCl3, 50 μM ZnSO4, 0.1 μM CaCl2, 40 μg/mL each of adenosine, guanosine, thymidine, cytosine and uracil, and 1 μg/mL each of D-biotin, cyanocobalamin, folic acid, niacinamide, D-pantothenate, pyridoxal hydrochloride and riboflavin. The library size was estimated by counting the number of recovered colonies on LB/carbenecillin plates. The plates were also supplemented with 0.1% l-arabinose (to induce expression of the aldolase gene) and 2.5 mM KHO or S-KHPB as a source for pyruvate. All colonies plated on selection media were grown at 34 °C.
As controls, plasmids containing the wild-type aldolase gene were transformed into PB25 cells and plated on selective media alongside the libraries. No significant background growth was seen on these plates until 72 and 120 hr of incubation for S-KHPB and KHO respectively. All colonies that grew in the first 72 or 120 hours on selection plates were picked, restreaked on selective media and then grown in 5 mL LB/carbenecillin liquid cultures to recover the plasmid DNA. The DNA was isolated using the Wizard SV Gel and PCR Purification System. The plasmids were retransformed and plated on selective media to verify that the beneficial activity was associated with the plasmid. The entire eda gene was sequenced at the University of Michigan DNA sequencing core.
Construction of mutants
For ease of expression and purification, the T161S, S184L, and S184F mutants were made in the pET-ECEDA plasmid. The QuikChange (Stratagene) method with the following primers and their reverse complements was used to construct the vectors: T161S 5′ - CCAGGTCCGTTTCTGCCCGTCGGGTGGTATTTCTCCGGCTAAC - 3′; S184F 5′- GCTGTGCATCGGTGGTTTCTGGCTGGTTCCGGCAG - 3′; and S184L 5′- GCTGTGCATCGGTGGTCTCTGGCTGGTTCCGGCAG - 3′. The pET-ECEDA plasmid(25) encodes the E. coli eda gene behind a T7 polymerase promoter and contains an N-terminal 6 × His-tag. After whole plasmid PCR amplification with PFU Turbo (Stratagene) and exhaustive digestion with DpnI (NEB), the products were transformed into SmartCells. Following overnight growth on Luria-Bertani media agar plates containing 100 μg/mL kanamycin, single colonies were cultured and the plasmid DNA was purified and sequenced. The T161S/S184L and T161S/S184F double mutants were constructed from the S184L and S184F plasmids with the primers for the T161S mutation using the whole plasmid PCR amplification method. The sequences of the double mutant plasmids were verified by DNA sequencing.
Protein expression and purification
The expression and purification of all KDPG aldolase mutants were done according to the protocol previously described(26) using metal affinity chromatography. Briefly, the pET-derived plasmids encoding each mutant eda gene were transformed into BL21(DE3) cells, grown on LB/kanamycin (100 μg/mL) plates and then cultured in Terrific Broth(27) containing 100 μg/mL kanamycin at 37 °C. At OD600 nm = 0.6-1.0, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and the cultures were incubated for an additional 4-8 h. The cells were pelleted, resuspended in HEPES (25 mM, pH 7.5) and lysed using a Sonic Dismembrator 550 (Fisher Sci). The crude cell extracts were clarified by centrifugation and the supernatant was loaded onto a Ni2+-charged Chelating Sepharose Fast Flow (GE Healthcare) column for fractionation using a stepwise gradient of 100 mM HEPES, pH 7.5, 100 mM NaCl containing: 5 mM imidazole (1.5 column volumes); 155 mM imidazole (2 column volumes); and 500 mM imidazole (2 column volumes). Fractions containing the his-tagged KDPG aldolase eluted during the 500 mM imidazole wash. These fractions were combined and concentrated using Amicon Ultra (Millipore) spin concentrators and extensively washed with 100 mM HEPES, pH 7.5, 100 mM NaCl to remove the imidazole. Enzymes stored at 4 °C in 100 mM HEPES, pH 7.5, 100 mM NaCl and 10% glycerol, at concentrations of 1-5 mM, are stable for up to 1 year.
The protein concentrations were determined by absorbance at OD280 nm where the extinction coefficient was calculated to be ε280 nm = 15510 M−1 cm−1 for both wild-type and mutant KDPG aldolases. The protein concentrations and purity were confirmed by 12% polyacrylamide gel electrophoresis followed by Coomassie staining and comparison of band intensity to a known standard.
Kinetic assay for retro-aldol cleavage
The mutants were assayed for retro-aldol cleavage of KDPG and related analogs (KDPGal, KHO, S-KHPB and R-KHPB) using a lactate dehydrogenase (LDH) coupled assay where the initial velocity for the production of pyruvate was measured by the decrease in NADH absorbance at 340 nm (ε = 6220 M−1cm−1). All assays were done using a CARY 100 Bio UV/Vis spectrometer fitted with a Peltier temperature controller in quartz microcuvettes (70 μL) at 25 °C. The reactions contained the following final concentrations: 100 mM HEPES pH 7.5, 250 μM NADH, 0.023 U/μL LDH (Sigma), 50 μM to 50 mM KDPG (or analogs) depending on the KM value and 50 nM to 10 μM enzyme, depending on the kcat value. Data were collected in 3 replicates. The Michaelis-Menten equation was fit to the data using the curve-fitting program GraphPad PRISM 4 and standard errors were determined.
Specificity assay with various electrophiles
Stopped assays were done to measure the efficiency of aldol synthesis by reaction of various aldehydes with pyruvate catalyzed by the T161S/S184L and wild-type KDPG aldolases. Each reaction contained 200 mM HEPES pH 7.5, 4 mM pyruvate and 50 mM aldehyde (D-GAP, D,L-glyceraldehydes, 2-, 3-, or 4-pyridine carboxaldehyde, valeraldehyde, benzaldehyde, 2-furaldehyde, or D- or L-erythrose). The stock solutions for benzaldehyde and 2-furaldehyde contained 20% DMSO to improve solubility in water and 10% DMSO was maintained in the reaction vessel for these two substrates. An aliquot was removed prior to adding the enzyme to serve as a zero point for the assay. The reaction was initiated by addition of enzyme (1 μM enzyme for D-GAP; 10 μM for glyceraldehyde and 2-pyridine carboxaldehyde; and 50 μM for the rest of the aldehydes) and the linear increase in product was monitored for a maximum of 3 hours. At 30 min intervals an aliquot of the reaction was diluted 20-fold into 100 mM HEPES pH 7.5 and 300 μM NADH. The change in absorbance at 340 nm after adding 0.8 U of LDH was measured to assay the remaining pyruvate concentration. The time-dependent decrease in pyruvate was used to calculate the initial velocity for the reaction of pyruvate with each electrophile. Two independent replicates were done and the initial velocity data were averaged.
Synthesis of substrates
The syntheses of 2-keto-3-deoxy-6-phosphogluconate (KDPG), 2-keto-4-hydroxyoctonoate (KHO) and (4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (S-KHPB) were previously reported.(22, 26) 2-keto-3-deoxy-6-phosphogalactonate (KDPGal) and (4R)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (R-KHPB) with high enantiomeric purity were synthesized enzymatically using KDPGal aldolase as previously described(28) and were gifts from Matthew J. Walters (Duke University, NC).
Construction of substrate binding models for KDPG aldolases
Substrates were modeled into the published structures of E. coli KDPG aldolase (PDB: 1EUA(29)) using Accelrys Viewer Pro 4.2 software. The coordinates for the backbones of KDPGal-Schiff base (taken from the structure of KDPGal aldolase, PDB: 2V82(28)) were superimposed on the KDPG aldolase structure. The stereochemistry of the C4 hydroxyl was inverted and the C-C torsions were adjusted to avoid van der Waals contacts with the protein side chains. The torsion angles for the modeled KDPG substrate are: −178° (C1-C2); −93° (C2-C3); 171° (C3-C4); 109° (C4-C5); 102° (C5-C6), and 93° (C6-O6). The position of water W1 is inferred from the position of the carbinolamine oxygen, as described in Fullerton et al.(30) Water W1 was moved (up to 2 Å) to a position that approximates ideal geometries for proton transfer steps. No protein side chains positions were modified in this model.
The model of 2-pyridine carboxaldehyde bound to KDPG aldolase was generated using the drawing tools in Viewer Pro 4.2. The molecule was superimposed on the KDPG aldolase structure and adjusted to avoid van der Waals contacts and to maximize the orientation and geometry required for the aldol addition reaction. The torsion angle between Cα-C1 is 50°. The pyruvyl-enamine was modeled based on the structure of the pyruvyl-Schiff base(30) where the K133 Cδ-Cε and Cε-Nζ dihedral angles were modified by 10° and −26° respectively to avoid van der Waals clashes between the pyruvyl-C3 carbon and the aldehydic Cα carbon. Consequently, the pyruvyl-C3 is ~1.2 Å further away from the aldehyde binding pocket than the equivalent carbon of the pyruvyl-carbinolamine observed in the 1EUA crystal structure. Moving these atoms does not create any steric clashes or significantly alter the position of the pyruvyl-C1 carboxylate. The double mutant model was made by deleting the methyl side chain of T161 and replacing S184 with leucine using the drawing tools in Viewer Pro 4.2 and adjusting the torsions to minimize steric contacts.
RESULTS
Identification of KDPG aldolase mutants with altered substrate selectivity
We previously reported an active site mutation in KDPG aldolase that markedly improves the catalytic efficiency of aldol addition between pyruvate and 2-pyridine carboxaldehyde. In this mutant, serine-184, located in the phosphate binding site of KDPG aldolase, is substituted with leucine which enhances the reactivity of this enzyme with hydrophobic electrophiles, such as 2-pyridine carboxaldehyde, by up to 40-fold.(26) These mutagenesis data combined with structural studies identified the aldehyde binding pocket, allowing the use of structure-based approaches to redesign the substrate specificity of KDPG aldolase.(26, 30) Based on these studies, we mutagenized key amino acids that line the aldehyde binding loop to identify alterations that enhance reactivity with hydrophobic substrates.
Examination of every possible variant at every position for even a small enzyme like KDPG aldolase (213 AA) would require an astronomically large library (~20200 molecules). Limiting the variability to amino acids that are within 5 Å of the active site still results in an intractable library size of ~2030. Reasoning that alterations in residues that directly contact the substrate are more likely to alter substrate selectivity than distal amino acids, we limited the complexity of our libraries by varying only active site residues. Based upon structural insights about the aldehydic substrate binding site,(26, 30) we targeted four positions for mutagenesis: G162, G163 and S184, residues located on two loops that form the phosphate binding site, and T161, which bridges the pyruvate and aldehydic binding pockets and contributes to the stereospecificity of the enzyme(28) (Figure 2). The in vivo activity of the mutants encoded in these libraries was examined using a previously developed E. coli selection system based on a pyruvate auxotrophic strain (PB25).(22) This strain is unable to grow on minimal ribose plates without supplemental pyruvate.(24) Augmentation of selection plates with a compound that is cleaved by a mutant KDPG aldolase to generate pyruvate facilitates cell survival. Previous data suggested that this selection method mainly identifies enhancements in substrate binding due, presumably, to the low in vivo concentration of the substrates.(15) To circumvent this issue, KDPG aldolase was fused to the signal sequence from the filamentous phage fd pIII gene which localizes the protein to the periplasm(31) where the substrate concentration reflects the concentration in the medium. Using this method E. coli KDPG aldolase variants were selected for improved catalysis of two hydrophobic analogs of KDPG: (1) 2-keto-4-hydroxy-octonoate (KHO), a hydrophobic analog that lacks the C5 hydroxyl and C6 phosphate of KDPG; and (2) (4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (S-KHPB), an analog containing a pyridyl group that is an important precursor for nikkomycin synthesis (Figure 3).
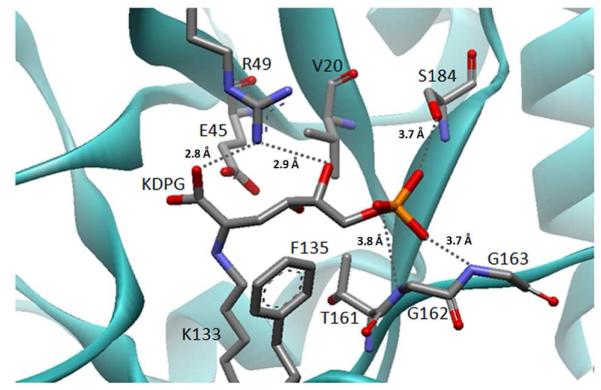
Figure 2.
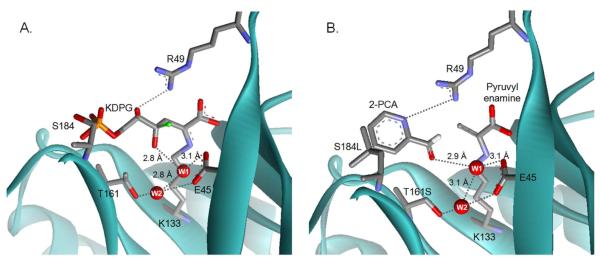
Model of KDPG bound as a Schiff base to E. coli KDPG aldolase. The model was built using PDB: 1EUA(29) as described in the Materials and Methods and the substrate conformation resembles the structure of KDPGal bound to E. coli KDPGal aldolase.(28) Proposed hydrogen bonds are indicated by grey dashed lines and distances. The catalytic waters have been omitted for clarity.
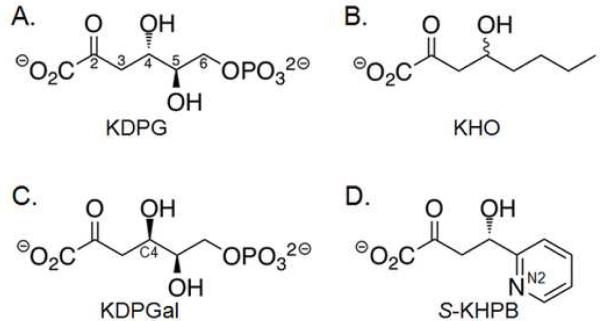
Figure 3.
Substrates for KDPG aldolase used in this study. (A) 2-keto-3-deoxy-6-phosphogluconate (KDPG) is the natural gluco sugar substrate. (B) 2-keto-4-hydroxy-octonoate (KHO) is a hydrophobic analog that lacks the C5 hydroxyl and C6 phosphate. (C) 2-keto-3-deoxy-6-phosphogalactonate (KDPGal) differs from KDPG by only the stereochemistry at C4. (D) (4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (S-KHPB) is a reactive analog.
To identify mutations that enhance reactivity with hydrophobic substrates, we prepared four libraries with the sequence of the codons at positions 161, 162, 163, and 184 individually randomized. By employing NNK degeneracy (N: all nucleotides, K: only guanosine and thymidine) in the primers, these libraries contain a total library size of 32 variants that encode all 20 amino acids and a stop codon. For such small libraries, screening 100 variants provides a 95% confidence level that every possible amino acid substitution is examined.(32) The mutant plasmid libraries were transformed into PB25 cells, plated on KHO or S-KHPB selection media and incubated for 72 hours. More than 100 clones were plated from each library, providing a high degree of confidence that all possible substitutions were examined. A small number of colonies (0 - 3) were isolated from each selection plate (Table S2) and the KDPG gene encoded on each plasmid was sequenced. Three single amino acid mutants were obtained from plasmids selected by growth on KHO plates: T161S, G162V and S184F. Only one mutation conferred growth on the S-KHPB selection plates, namely T161S. Unexpectedly, in this screen we did not identify the S184L mutation that we had previously demonstrated enhanced reactivity of this enzyme with 2-pyridine carboxyaldehyde.(26) This result suggests that another variable, such as the stability or expression level of the S184L compared to the S184F mutant, limited the ability to confer growth under the selection conditions.
The biochemical properties of the selected mutant KDPG aldolases were analyzed. All three mutants express to a high level in E. coli. The T161S and the S184F mutants were both easily purified but the G162V KDPG aldolase mutant precipitated during purification and was not analyzed further. To measure changes in substrate selectivity for the T161S and S184F mutants, the kinetic parameters for catalysis of retro-aldol cleavage of KDPG, KHO, and S- KHPB were determined (Table 1). The specificity constant (kcat/KM) is the most important parameter for consideration of substrate selectivity and catalytic efficiency while the turnover number may be most informative for identifying efficient biocatalysts. The steady state kinetic parameters for retro-aldol cleavage of KHO and S-KHPB catalyzed by wild-type KDPG aldolase (Table 1) indicate that the catalytic efficiency (kcat/KM) is decreased by 104-105 relative to KDPG due to both decreases in the value of kcat and increases in KM. Although KM frequently includes multiple terms and may not simply reflect binding affinity, the high value of this parameter coupled with the low turnover number suggests that wild-type KDPG aldolase binds these alternate substrates with low affinity.
Table 1.
Kinetic parameters for cleavage of KDPG, KHO, and S-KHPB catalyzed by E. coli KDPG aldolase mutants.a
| KDPG | KHOb | S-KHPB | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variants |
kcat s−1 |
KM mM |
kcat/KM M−1s−1 |
kcat s−1 |
KM mM |
kcat/KM M−1s−1 |
kcat s−1 |
KM mM |
kcat/KM M−1s−1 |
| Wild-type c | 83 | 0.10 | 830,000 | 2.0d | 146d | 14d | 11 | 32 | 330 |
| (2) | (0.01) | (20,000) | (0.6) | (50) | (1) | (1) | (2) | (9) | |
| T161S | 27 | 0.05 | 550,000 | 1.7 | 15 | 113 | 175 | 43 | 4,100 |
| (1) | (0.01) | (65,000) | (0.2) | (4) | (20) | (40) | (12) | (800) | |
| S184F | 40 | 1.7 | 24,000 | 1.5 | 25 | 60 | 19 | 5.7 | 3,300 |
| (2) | (0.2) | (2000) | (0.1) | (4) | (4) | (1) | (0.8) | (60) | |
| S184Lc | 74 | 0.35 | 210,000 | 2.9 | 26 | 110 | 40 | 3.0 | 13,000 |
| (7) | (0.10) | (10,000) | (0.2) | (4) | (10) | (1) | (0.2) | (600) | |
| T161S/S184F | 31 | 0.69 | 45,000 | e--- | --- | --- | 212 | 3.5 | 61,000 |
| (2) | (0.06) | (2,500) | (12) | (0.5) | (5000) | ||||
| T161S/S184L | 41 | 0.24 | 170,000 | 3.1 | 4.3 | 720 | 240 | 1.6 | 150,000 |
| (2) | (0.04) | (27,000) | (0.2) | (0.7) | (90) | (11) | (0.2) | (16,000) | |
Initial velocity for pyruvate formation was measured using a lactate dehydrogenase coupled assay as described in Materials and Methods. Steady state kinetic parameters were determined by fit of the Michaelis-Menten equation to the initial velocity as a function of the substrate concentration. These assays were done at 25 °C. Errors are shown in parenthesis.
KHO is synthesized in racemic form and the indicated concentration includes both C4 stereoisomers. Since KDPG aldolase preferentially catalyses the cleavage of (S)-KHO, the measured KM is likely to be higher than the actual value for (S)-KHO. Consequently the kcat/KM is an underestimate of the actual value.
These data were previously published.(26)
The initial velocity has a nearly linear dependence on the concentration of KHO up to the maximal measureable concentration of 50 mM.
Hypens indicate that data was not collected.
The T161S mutation improves the value of kcat/KMKHO for catalyzing cleavage of KHO by 8-fold, largely due to a 10-fold decrease in the value of KMKHO (Table 1). The T161S mutation causes an even larger enhancement in catalysis of retro-aldol cleavage of S-KHPB; the value of kcat/KMS-KHPB increases by 12-fold compared to the value for wild-type KDPG aldolase. Remarkably, this enhancement is caused by a 16-fold increase in the value of the turnover number. However, the T161S mutation does not increase the reactivity of KDPG aldolase with all substrates. This mutant has little effect on the catalytic efficiency with the natural substrate, KDPG, leading to comparable decreases in the values of kcatKDPG and KMKDPG (3- and 2-fold, respectively). Despite the large increases in the catalytic efficiency of KHO and S-KHPB cleavage, the T161S mutant retains selectivity for KDPG, with a cleavage efficiency (kcat/KMKDPG ) that is >100-fold larger than either of the other substrates.
The S184F mutation also significantly alters the substrate specificity of this aldolase, increasing the values of kcat/KMKHO and kcat/KMS-KHPB by 4-fold and 10-fold, respectively, mainly through lowering the value of KM. Yet, these enhancements in catalytic efficiency are 2 - 4-fold smaller than those observed for the S184L mutant previously measured(26) (Table 1). However, while the S184L mutation has a small effect (4-fold decrease) on the reactivity with KDPG, the S184F variant decreases the value of kcat/KMKDPG by ~35-fold, mainly by increasing the value of KMKDPG. Therefore the S184F substitution significantly increases the selectivity for cleavage of KHO and S-KHPB relative to KDPG compared to the S184L variant. These data are consistent with the proposal that the side chain at position 184 forms a portion of the aldehyde binding pocket(26) (Figure 2). Substitution of Ser184 with Phe or Leu presumably increases the hydrophobicity of the binding pocket, thereby improving the catalytic efficiency with hydrophobic aldehydes. However, the larger size of the Phe side chain compared to Leu or Ser interferes with the apparent binding affinity of KDPG.
Alterations in substrate selectivity are additive in double mutants
Since the T161S mutation primarily enhances the kcat parameter while the S184L/F mutations lower the KM parameter, the effects of these mutations could be additive, i.e. the first mutation makes beneficial contacts with the substrate while the second improves the cleavage efficiency of the bound substrate. To examine whether these mutations have an additive effect on catalysis, we prepared and purified two double mutants, T161S/S184L and T161S/S184F KDPG aldolase. Both proteins were readily expressed and stable.
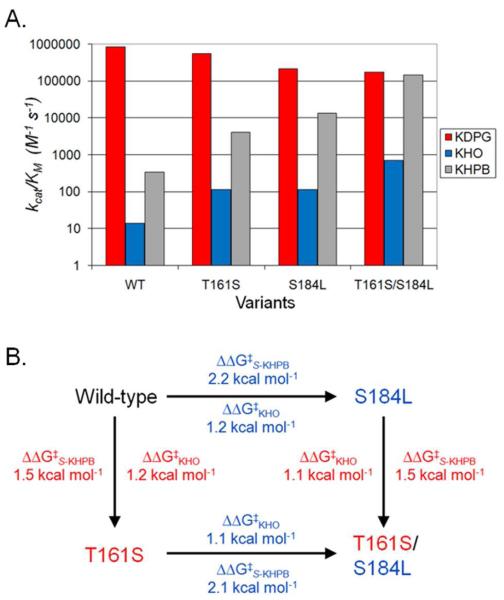
Combining mutations at positions 161 and 184 enhances the specificity of the aldolase for cleavage of KHO and S-KHPB relative to KDPG beyond that of either mutation alone. The T161S/S184L variant shows the most pronounced alterations in substrate selectivity. The value of kcat/KMKHO for cleavage of KHO is improved by >50-fold compared to that of the wild-type KDPG aldolase. Furthermore, these substitutions enhance the catalytic efficiency of S-KHPB cleavage by >450-fold, resulting in a value of kcat/KMKHPB that approaches (18%) the activity of the wild-type enzyme with KDPG. Since most biocatalytic processes are run under conditions where substrate is abundant, improving the turnover number is arguably the most important aspect of protein engineering, at least with respect to synthetic applications. Remarkably, the T161S/S184L mutant enhances the value of kcatS-KHPB for cleavage of S-KHPB by 22-fold, such that the turnover number (240 s−1) is nearly 3 times greater than that of wild-type KDPG aldolase with its natural substrate (83 s−1). The T161S/S184L mutant also retains significant activity with KDPG; the value of kcat/KMKDPG decreases by less than 5-fold. Therefore the T161S/S184L mutations significantly broaden the substrate selectivity of this aldolase, with comparable activities for KDPG and S-KHPB cleavage and enhanced activity with KHO (Figure 4A).
Figure 4.
(A) Comparison of kcat/KM values for E. coli KDPG aldolase wild-type and mutant enzymes with KDPG, KHO, and S-KHPB. The catalytic efficiencies are shown for KDPG cleavage (red), KHO cleavage (blue), and S-KHPB (gray). (B) Additive effect of individual mutations in the T161S/S184L mutant on catalytic efficiency of KHO and S-KHPB. ΔΔG‡ = −2.303RT log ([kcat/KMVariant1]/[kcat/KMVariant2]), where T = 298.15 K and R = 1.987 cal K−1 mol .
The T161S/S184F double mutant shows a similar trend to that observed for T161S/S184L aldolase, but the enhancement of S-KHPB cleavage efficiency is smaller (~180-fold), while the reduction in KDPG cleavage efficiency is more substantial (18-fold). As a result, the T161S/S184F mutant has a larger change in selectivity, altering the substrate selectivity ratio1 between KDPG and S-KHPB by 3400-fold (Table 2). These data are similar to the effects observed in the S184F single mutant, and are consistent with the phenylalanine substitution at position 184 partially occluding the KDPG binding site.
Table 2.
Changes in substrate selectivity of E. coli KDPG aldolase mutants.a
| Mutants | Substrate selectivity compared to KDPG cleavage |
|
|---|---|---|
| KHO | S-KHPB | |
| Wild-type | 1 | 1 |
| T161S/S184L | 250 | 2200 |
| T161S/S184F | --- | 3400 |
Selectivity ratio = [(kcat/KMKDPG)/(kcat/KMX)]Wild-type/[(kcat/KMKDPG)/(kcat/KMX)]Mutant
To further probe the substrate specificity of the T161S/S184L mutant, we measured the catalytic rate constant in the synthetic direction for the reaction of pyruvate and various electrophilic substrates at saturating concentrations of pyruvate (4 mM) and high concentrations of the aldehyde, mimicking plausible conditions for a synthetic application (Table 3). The most striking result is the switch in the identity of the highest activity aldehydic substrate from D-glyceraldehyde-3-phosphate for wild-type KDPG aldolase to 2-pyridine carboxaldehyde (2-PCA) for the T161S/S184L mutant, equivalent to a 24-fold change in selectivity2 for reaction with these two substrates. In general, under these assay conditions, reactivity with sugar electrophiles decreases (2- to 10-fold) in the mutant, while reactivity with more hydrophobic aldehydes is unchanged or increases, leading to a net increase in selectivity for hydrophobic aldehydes of 4 to 24-fold (Table 3). However, the selectivity for 2-PCA is the highest, suggesting that the selection identified a mutant that both broadens the specificity of the aldolase and forms a specific interaction with 2-PCA to enhance reactivity with this substrate.
Table 3.
Catalysis of aldol reaction between pyruvate and various aldehydes by E. coli KDPG aldolase and the T161S/S184L mutant.a
| Substrates | Enzymes |
Change in Selectivityb | |
|---|---|---|---|
| Wild-type | T161S/S184L | ||
| D-Glyceraldehyde-3-phosphate | 100 | 15% | -- |
| DL-Glyceraldehyde | 4% | 1% | 1.7 |
| D-Erythrose | 0.3% | 0.03% | 0.67 |
| L-Erythrose | 0.06% | 0.03% | 3.3 |
| 2-Pyridine carboxaldehyde | 25% | 89% | 24 |
| 3-Pyridine carboxaldehyde | 0.5% | 1% | 13 |
| 4-Pyridine carboxaldehyde | 3% | 2.5% | 5.6 |
| Valeraldehyde | 0.005% | 0.008% | 11 |
| 2-Furaldehyde | 0.005% | 0.003% | 4.0 |
| Benzaldehyde | 0% | 0% | -- |
This assay was done with 4 mM pyruvate and 50 mM aldehyde substrate. All of the data in this table were normalized to the activity of wild-type KDPG aldolase with its natural electrophilic substrate, D-glyceraldehyde-3-phosphate.
[(ActivityAldehyde)/(ActivityD-GAP)]Mutant/[(ActivityAldehyde)/ActivityD-GAP)]Wild-type
Aldolase mutants retain stereoselectivity
One important aspect of developing aldol catalysts for biocatalytic uses is to maintain the stereospecificity of the native reaction. To examine this property we compared the efficiency of mutant aldolases for aldol cleavage of substrates with epimeric C4 stereocenters: KDPG vs. KDPGal and S-KHPB vs. R-KHPB (Table 4). The T161S mutation increases the reactivity for cleaving KDPGal, reducing the observed stereoselectivity ratio3 from the wild-type value of 14,000 to 710. In contrast, the S184L substitution causes a larger decrease in the reactivity with KDPGal than KDPG, thus increasing the stereoselectivity ratio by >5-fold compared to wild-type KDPG aldolase. A priori, it was not clear how each of these effects would contribute to the stereoselectivity of the double mutants. However, the T161S/S184L double mutation also decreases the rate of KDPGal cleavage to a greater extent than KDPG (Table 4), leading to a stereoselectivity ratio that is almost 2-fold higher than that of the wild-type enzyme. Similar behavior is observed for the T161S/S184F mutant (data not shown). These data further suggest that the effects of the two mutations on substrate selectivity are roughly additive. Additionally, the two double mutants enhance the stereoselectivity ratio4 for cleavage of R-KHPB up to 150-fold relative to the wild-type aldolase (Table 4). Although the mutations slightly enhance (up to 3-fold) the reactivity with R-KHPB, the substantially larger increases in catalysis of cleavage of S-KHPB leads to increased enantioselectivity. The high activity and stereoselectivity of these mutants make them particularly useful for the stereospecific synthesis of pharmaceuticals and other high-value specialty and commodity chemicals.
Table 4.
Kinetic parameters for cleavage of KDPGal and (R)-KHPB catalyzed by E. coli KDPG aldolase mutantsa and the enantiomeric selectivity ratio
| Enzyme | kcat s−1 |
KM mM |
kcat/KMM−1s−1 | Stereoselectivity ratiob |
|---|---|---|---|---|
| KDPGal | ||||
| Wild-type c | 0.0063 ± 0.0001 | 0.10 ± 0.01 | 61 ± 3 | 14,000 |
| T161S | 0.23 ± 0.02 | 0.3 ± 0.05 | 770 ± 90 | 710 |
| S184L c | 0.0026 ± 0.0001 | 0.95 ± 0.17 | 2.7 ± 0.4 | 78,000 |
| T161S/S184L | 0.0060 ± 0.0008 | 1.0 ± 0.1 | 6.5 ± 0.7 | 26,000 |
|
| ||||
| R-KHPB d | ||||
| Wild-type | ---- | > 50 | 0.01 ± 0.003 | 33,000 |
| T161S/S184F | ---- | > 50 | 0.017 ± 0.008 | 3,600,000 |
| T161S/S184L | ---- | > 50 | 0.03 ± 0.004 | 5,000,000 |
The kinetic parameters were measured as described in Table 1.
The stereoselectivity ratio is defined as [(kcat/KMKDPG)/(kcat/KMKDPGal)] or [(kcat/KMS-KHPB)/(kcat/KMR-KHPB)] respectively. Values for kcat/KMKDPG and kcat/KMS-KHPB are listed in Table 1.
These data were previously published.(26)
The initial velocity is linearly dependent on the concentration of R-KHPB up to 50 mM, allowing measurement of only the value of kcat/KM.
DISCUSSION
Based on structure-guided mutagenesis, we previously prepared an active site mutant in KDPG aldolase (S184L) that markedly improves the catalytic efficiency of S-KHPB retro-aldol cleavage and enhances the reactivity with non-phosphorylated electrophiles.(26) Alteration of the serine residue located in the phosphate binding site of KDPG aldolase presumably enhances contacts between the hydrophobic tail of the substrate and the aldehydic substrate binding pocket to increase catalytic efficiency. Here we use targeted mutagenesis coupled with selection methods to identify two additional mutations that improve the catalytic efficiency of this enzyme for cleavage of S-KHPB. The S184F mutation most likely enhances catalytic efficiency for hydrophobic electrophiles in a similar fashion to S184L, by improving contacts with the substrate. Since the T161S mutation mainly increases the turnover number (kcat) rather than the apparent binding affinity (KM), this mutation has a different mode of action.
The side chains of S184 and T161 are 7 Å apart, and mutations at these positions largely affect different steady state kinetic parameters. Therefore, these mutations might work independently and provide complementary benefits in substrate binding and catalysis. In accord with this hypothesis, the contributions from each mutation are largely additive in the double mutants, as illustrated in Figure 4B for T161S/S184L. The T161S mutation decreases the activation energy for S-KHPB catalysis (ΔΔG‡S-KHPB) by 1.5 kcal/mol.5 The analogous mutation in the S184L background (T161S/S184L mutant) yields a ΔΔG‡S-KHPB of 1.4 kcal/mol compared to the reaction catalyzed by the S184L variant. Likewise, the S184L mutation in the wild-type background decreases the activation energy by 2.2 kcal/mol, while the same mutation in the T161S mutant results in a 2.1 kcal/mol change in ΔΔG‡S-KHPB. Catalysis of KHO cleavage reflects a similar trend for ΔΔG‡KHO (Figure 4B). On the other hand, the effect on stereoselectivity does not show a consistent quantitative pattern. Although the T161S mutation reduces the stereoselectivity of the enzyme while the S184L mutation increases stereoselectivity, the magnitude of the effect is dependent on the context. This result suggests stereoselectivity in this enzyme is determined by complex protein-substrate recognition, rather than by interactions at a single key location. In contrast, the stereoselectivity of BphI aldolase (a class II metal dependent enzyme) is controlled mainly by a single residue.(11)
Origin of stereoselectivity in KDPG aldolase
Since KDPG aldolase and a structural homologue, KDPGal aldolase, catalyze aldol addition of substrates differing only in facial selectivity of addition, a comparison of these enzymes might provide insight into the structural basis of stereochemical control. Previous studies of KDPG(29) and KDPGal aldolases(28) identified numerous subtle differences in the active site of these two proteins and demonstrated that the mutations in the residue at 161 (equivalent to V154 in KDPGal aldolase) alter the overall stereoselectivity of the two enzymes.(28) In particular, the T161A mutant in KDPG aldolase relaxes the stereoselectivity of the mutant enzyme more than 3000-fold by both enhancing KDPGal cleavage and decreasing KDPG cleavage rates. Furthermore, mutation of T161 to valine essentially abolished catalytic activity for both epimeric sugars. In contrast, the T161S mutant identified in this work using selection methods retains significant enantioselectivity (Table 4) since the efficiency of cleaving KDPGal increases modestly (13-fold) without adversely affecting the reaction with KDPG. Taken together, these data suggest that a hydroxyl group at position 161 plays an important role in determining stereoselectivity.
To clarify the role of T161 in stereoselectivity we constructed models of enzyme-substrate complexes. Based upon the crystal structure of KDPGal bound to E. coli KDPGal aldolase (PDB:2V82(28)) we modeled the gluco-sugar, KDPG, bound as a Schiff base in the active site of E. coli KDPG aldolase (PDB: 1EUA(29)) (Figure 2). We further postulated that the two substrates bind in similar orientations and no major conformational shifts occur during the catalytic cycle so that the positions of the side chains are unchanged. (This is a reasonable assumption since all crystal structures of KDPG and KDPGal aldolases with or without bound substrates have similar overall folds with only minor differences in the orientation of catalytic residues.(28-30)) In this model, the C4 (epimeric carbon) hydroxyl of KDPG points down into the cavity towards the methyl group of T161 (Figure 5A). This conformation is stabilized by interaction of the C4 hydroxyl (O4) directly with water W1 and indirectly with water W2 via W1. (W2 is unique to KDPG aldolases and is not observed in KDPGal aldolases. In the crystal structure,(29) W2 forms hydrogen bonds with T161 and E45, and is positioned to form a hydrogen bond with W1.) In this model of the gluco-sugar, W1 is positioned to mediate proton transfer from O4 to E45 (distance of 2.8 Å and a proton abstraction angle of 125°) to initiate retro-aldol cleavage (Scheme 1). By contrast, in a model similarly constructed of the galacto-sugar bound to KDPG aldolase, the C4 hydroxyl points towards R49 where unfavorable van der Waals contacts with R49 and the side chain of V20 are likely (Figure 5A and Figure S2). Furthermore, in this model W1 is poorly positioned to facilitate proton transfer (3.8 Å distance and a proton abstraction angle of 76°). These models provide a rationale for the enhanced affinity and reactivity of KDPG aldolase with the gluco- vs. galacto- sugar as well as the effects of mutations at T161. We posit that the T161S mutant preserves the hydrogen bond network between E45, W1, and W2 and therefore maintains high efficiency for catalyzing KDPG cleavage (Table 1). However, the threonine to serine mutation widens the tunnel to the active site, allowing for increased substrate promiscuity.
Figure 5.
Models of substrates bound to E. coli KDPG aldolase. (A) A model of KDPG bound to the active site of KDPG aldolase. A proposed hydrogen bond network of waters that stabilize the complex is shown in gray dashed lines with key distances indicated. Water W1 is proposed to preferentially enhance the affinity of the gluco-sugar and mediate catalytically important proton transfer steps. Water W2 is proposed to help position W1. The C4 hydroxyl from a model of a galacto-sugar bound to KDPG aldolase is superimposed as a green ball. This model suggests that the galacto-epimer is a less efficient substrate for KDPG aldolase because O4 bumps into R49, the angle for deprotonation of O4 by W1 is poor, and the distance between these atoms is long (3.8 Å). (B) A model of 2-PCA bound to the T161S/S184L mutant. The model suggests that hydrophobic contacts are made between the aromatic heterocycle of 2-PCA and leucine at position 184 and that the T161S mutation removes a steric clash with the aldehydic oxygen of 2-PCA. Also the side chain of R49 is positioned to form a hydrogen bond with N2 of 2-PCA, similar to the hydrogen bond between KDPG O4 and R49.
A role for catalytic waters has been proposed for many class I aldolases,(30, 33-37) while a role for water in controlling stereochemistry has been suggested in the tagatose 1,6-bisphosphate and fructose-1,6-bisphosphate aldolases.(35) Likewise, in KDPGal aldolase, subtle structural differences in the active site position W1 differently than in KDPG aldolase, making occupancy of the upper site by O4 preferred, and presumably giving rise to the preference for reaction with substrates containing inverted C4 stereochemistry (see Figure S3 for an explanation of the KDPGal stereoselectivity). The careful positioning of water to mediate stereospecific proton transfers may be a common strategy in class I aldolase enzymes.
Improving activity with 2-pyridine carboxaldehyde
Remarkably, the T161S/S184L mutant has a maximal turnover number for retro-aldol cleavage of S-KHPB that is 3 times higher than that of the wild-type enzyme with any substrate (Table 1) and 2-PCA is the best electrophilic substrate for the mutant enzyme (Table 3). The enhanced reactivity of 2-PCA with the T161S/S184L KDPG aldolase mutant can be rationalized by modeling the structure of 2-PCA at the active site of pyruvate-bound wild-type and mutant E. coli KDPG aldolase(29) assuming that the protein structure was unchanged (except for minor rotations around the Schiff base lysine (K133) dihedral angles Cδ-Cε and Cε-Nζ). The model of 2-PCA bound to the wild-type enzyme suggests that steric interactions between the methyl group of T161 and the substrate aldehyde do not allow the formation of optimal reaction distances between the nucleophilic carbon C3 in pyruvate and the electrophilic carbon in 2-PCA (C3-pyruvate to Cα of 2-PCA distance is 4.3 Å). The hydrogen bond between W1 and the carbonyl oxygen of 2-PCA (distance is 3.5 Å) is also too long for efficient catalysis. In the model of PCA bound to the T161S/S184L mutant (Figure 5B) N2 of the pyridyl moiety of 2-PCA forms a hydrogen bond with the side chain of R49 and the aldehydic oxygen is positioned in the hole left by removal of the methyl group of T161. This repositioning of 2-PCA decreases the distance between the reacting carbon atoms (C3 to Cα distance is 3.3 Å,) and shortens the hydrogen bond with W1 to 2.9 Å, presumably enhancing the catalytic activity. Furthermore, the S184L substitution is positioned to make hydrophobic contacts with the aromatic ring of 2-PCA, as previously suggested from structure-reactivity studies of mutations at this position.(26) This proposed binding configuration provides a reasonable explanation for the enhanced reactivity of the mutant enzymes with S-KHPB.
The models of the KDPG aldolase - substrate complexes also suggest that the side chain of R49 forms a hydrogen bond with the C5 hydroxyl of KDPG (Figure 2 and 5A) or N2 of the aromatic ring of 2-PCA. An analogous hydrogen bond between the C5 hydroxyl of KDPGal and R17 has been observed in the crystal structure of KDPGal aldolase complexed with substrate.(28) The low reactivity of KDPG aldolase with substrates incapable of forming a hydrogen bond with R49, including 3-PCA, 4-PCA, valeraldehyde, and benzaldehyde (Table 3) suggest that this interaction is important for activity. Consistent with this, deletion of the arginine-49 side chain in the R49A mutant of KDPG aldolase decreases the efficiency of retro-aldol cleavage of KDPG enormously (2.3 × 104-fold; kcat/KMKDPG = 36 ± 5 M−1s−1) which comes as a result of a 140-fold decrease in kcat (kcatKDPG = 0.6 ± 0.04 s−1) and a 170-fold increase in KM (KMKDPG = 17 ± 3 mM) (unpublished data). These observations suggest that R49 makes contacts with the substrate or an important negatively charged intermediate, such as the enamine/carbanion, that enhance ligand affinity and catalytic reactivity.
Information-guided library screening
Previously, we concluded that the intracellular concentration of targeted aldol products was likely nM to μM, far lower than the KM value of the wild-type enzyme for these substrates.(15) Therefore random mutagenesis coupled to in vivo selection identified mutations that lowered the value of KM, but did not improve kcat. Albery and Knowles(38) suggest that the first mutations in the evolution of a new activity often result in “uniform binding” where a mutation has a similar stabilizing effect on both the ground and transition states of the rate-limiting step, leaving the value of kcat unchanged. Many types of interactions, both near and far from the active site, can produce uniform binding enhancements (electrostatic effects, hydrogen bonding, hydrophobic interactions, etc.).(38) Furthermore, these mutations may be dominant when the substrate concentration is significantly lower than the value of KM. To increase the likelihood of identifying mutations that improve the value of kcat, we expressed KDPG aldolase in the periplasm where the concentration of aldol substrate will more nearly reflect the millimolar concentration of substrate added to the media. This method was particularly successful for identification of the T161S mutation that increased the value of kcat for S-KHPB cleavage. For the KHO selections, the identified mutations had little or no effect on the value of kcat, likely resulting from the concentration of KHO (2.5 mM) in the medium being lower than the wild-type KM value. These data suggest that selection methods using periplasmic expression may be a useful strategy to increase the odds of finding mutants with higher turnover numbers, an important parameter for biocatalytic applications.
The successful engineering of KDPG aldolase substrate selectivity demonstrates that mutagenesis efforts focused on active site residues can have a profound impact on the specificity and reactivity of an enzyme. While whole gene directed evolution experiments are extremely useful for improving thermal and chemical stability of enzymes,(39) employing a similar strategy to alter the specificity of aldolase/transaldolase genes has generally led to only modest improvements (6 to 80-fold) in substrate selectivity and reactivity.(13, 15, 16, 40-43) Additionally, the optimized mutant enzymes often possess improved kcat/KM and KM values but lower catalytic turnover numbers. In contrast, efforts combining structural information and site-specific saturation mutagenesis have led to large alterations in the enzymatic properties of various aldolase/transaldolases with minimal mutagenesis.(8, 11, 44-47) In a recent example, Schneider and colleagues(46) used site-saturation mutagenesis to identify a single amino acid substitution in E. coli TalB transaldolase that improves the aldol cleavage reaction by 70-fold compared to wild-type. In another example, Royer et al(47) employed active site-directed mutagenesis to create two variants of Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase that improve the enantioselectivity of the enzyme, preferentially catalyzing the formation of either D-2-keto-3-deoxygluconate or D-2-keto-3-deoxygalactonate substrate. However, in both examples the enhanced properties are obtained at a significant cost to the overall catalytic efficiency and turnover number of the mutant when compared to the natural reaction catalyzed by each enzyme. These examples illustrate the power of information-guided evolution approaches to alter the catalytic characteristics of an enzyme with minimal screening effort. Our results add to a growing body of evidence arguing that the targeting of active sites is a highly efficient method for obtaining large enhancements in catalytic activity and substrate selectivity. Remarkably, in this case, the catalytic characteristics of the engineered enzyme match or exceed that of the wild-type enzyme reacting with the natural substrate in terms of efficiency, maximal turnover number and stereospecificity.
CONCLUSIONS
The development of novel biocatalysts requires balancing many parameters. The engineered enzymes should have high catalytic activity and fidelity for the chemical reaction (including stereochemistry), yet also possess broad substrate tolerance and high protein stability. Frequently, when the substrate tolerance of an enzyme is broadened, the reactivity, fidelity and stereoselectivity are compromised. Here we have used structure-guided active site mutagenesis to engineer an aldolase that shows significant tolerance for unnatural electrophilic substrates, yet retains near wild-type activity. Impressively, this enzyme also maintains exquisite stereoselectivity in the aldol product. This enzyme should prove useful for the synthesis of nikkomycins and other important natural products. Our study highlights the substantial benefit that minor remodeling of the active site architecture can have on substrate specificity, and suggests that structure guided mutagenesis coupled with biochemical analysis and selection techniques provide a useful avenue for effective protein engineering.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew J. Walters for providing KDPGal and R-KHPB and Lance W. Rider for measuring the kinetic parameters of the R49A KDPG aldolase mutant.
Funding
This work was supported by National Institutes of Health grant GM61596. M.C. received support from the Chemical Biology Interface training program GM08597.
ABBREVIATIONS
- DHAP
dihydroxyacetone phosphate
- DMSO
dimethyl sulfoxide
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)
- KDPG
2-keto-3-deoxy-6-phosphogluconate
- KDPGal
2-keto-3-deoxy-6-phosphogalactonate
- KHO
2-keto-4-hydroxy-octonoate
- LDH
lactate dehydrogenase
- NADH
β-nicotinamide adenine dinucleotide
- PCA
pyridine carboxaldehyde
- R-KHPB
(4R)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate
- S-KHPB
(4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate
Footnotes
Selectivity ratio = [(kcat/KMKDPG)/(kcat/KMKHX)]Wild-type/[(kcat/KMKDPG)/(kcat/KMKHX)]Mutant
Selectivity ratio = [(ActivityAldehyde)/(ActivityD-GAP)]Mutant/[(ActivityAldehyde)/ActivityD-GAP)]Wild-type
Stereoselectivity ratio = [(kcat/KMKDPG)/(kcat/KMKDPGal)]
Stereoselectivity ratio = [(kcat/KMS-KHPB)/(kcat/KMR-KHPB)]
ΔΔ‡ = −2.303RT log (kcat/KMVariant 1/kcat/KMVariant 2), Where T = 298.15 K and R = 1.987 cal K−1 mol−1
Current address MC: New England Biolabs, Ipswich, MA 01938
Supporting Information
Figure S1 shows pharmaceutical targets that could be partially synthesized by enzyme-catalyzed aldol condensations. Figure S2 shows the structure of E. coli KDPG aldolase to explain the low activity of the galacto-sugar and S3 shows the structure of E. coli KDPGal aldolase to explain the stereoselectivity of this enzyme. Table S1 lists synthetically useful aldolases, and S2 lists the number of mutants selected in this study. The supporting figures and tables referenced in this article are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Samland AK, Sprenger GA. Microbial aldolases as C-C bonding enzymes-unknown treasures and new developments. Appl Microbiol Biotechnol. 2006;71:253–264. doi: 10.1007/s00253-006-0422-6. [DOI] [PubMed] [Google Scholar]
- 2.Fessner WD, Walter C. Enzymatic C-C bond formation in asymmetric synthesis. Top Cur Chem. 1997;184:97–194. [Google Scholar]
- 3.Bolt A, Berry A, Nelson A. Directed evolution of aldolases for exploitation in synthetic organic chemistry. Arch Biochem Biophys. 2008;474:318–330. doi: 10.1016/j.abb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faber K. Biotransformations in Organic Chemistry. 5th ed Springer; Berlin: 2004. [Google Scholar]
- 5.Clapes P, Fessner WD, Sprenger GA, Samland AK. Recent progress in stereoselective synthesis with aldolases. Curr Opin Chem Biol. 2010;14:154–167. doi: 10.1016/j.cbpa.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Sugiyama M, Hong Z, Greenberg WA, Wong CH. In vivo selection for the directed evolution of L-rhamnulose aldolase from L-rhamnulose-1-phosphate aldolase (RhaD) Bioorg Med Chem. 2007;15:5905–5911. doi: 10.1016/j.bmc.2007.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schurmann M, Sprenger GA. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J Biol Chem. 2001;276:11055–11061. doi: 10.1074/jbc.M008061200. [DOI] [PubMed] [Google Scholar]
- 8.Schneider S, Gutierrez M, Sandalova T, Schneider G, Clapes P, Sprenger GA, Samland AK. Redesigning the active site of transaldolase TalB from Escherichia coli: new variants with improved affinity towards nonphosphorylated substrates. Chembiochem. 2010;11:681–690. doi: 10.1002/cbic.200900720. [DOI] [PubMed] [Google Scholar]
- 9.Garrabou X, Joglar J, Parella T, Crehuet R, Bujons J, Clapes P. Redesign of the phosphate binding site of L-rhamnulose-1-phosphate aldolase towards a dihydroxyacetone dependent aldolase. Adv Synth Catal. 2011;353:89–99. [Google Scholar]
- 10.Shelton MC, Cotterill IC, Novak ST, Poonawala RM, Sudarshan S, Toone EJ. 2-keto-3-deoxy-6-phosphogluconate aldolases as catalysts for stereocontrolled carbon-carbon bond formation. J Am Chem Soc. 1996;118:2117–2125. [Google Scholar]
- 11.Baker P, Carere J, Seah SY. Probing the molecular basis of substrate specificity, stereospecificity, and catalysis in the class II pyruvate aldolase, BphI. Biochemistry. 2011;50:3559–3569. doi: 10.1021/bi101947g. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Baker P, Seah SY. Comparison of two metal-dependent pyruvate aldolases related by convergent evolution: substrate specificity, kinetic mechanism, and substrate channeling. Biochemistry. 2010;49:3774–3782. doi: 10.1021/bi100251u. [DOI] [PubMed] [Google Scholar]
- 13.Fong S, Machajewski TD, Mak CC, Wong CH. Directed evolution of D-2-keto-3-deoxy-6-phosphogluconate aldolase to new variants for the efficient synthesis of D- and L-sugars. Chem Biol. 2000;7:873–883. doi: 10.1016/s1074-5521(00)00035-1. [DOI] [PubMed] [Google Scholar]
- 14.Wymer N, Buchanan LV, Henderson D, Mehta N, Botting CH, Pocivavsek L, Fierke CA, Toone EJ, Naismith JH. Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli. Structure. 2001;9:1–9. doi: 10.1016/s0969-2126(00)00555-4. [DOI] [PubMed] [Google Scholar]
- 15.Cheriyan M, Walters MJ, Kang BD, Anzaldi LL, Toone EJ, Fierke CA. Directed evolution of a pyruvate aldolase to recognize a long chain acyl substrate. Bioorg Med Chem. 2011;19:6447–6453. doi: 10.1016/j.bmc.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran N, Frost JW. Directed evolution of 2-keto-3-deoxy-6-phosphogalactonate aldolase to replace 3-deoxy-D-arabino-heptulosonic acid 7-phosphate synthase. J Am Chem Soc. 2007;129:6130–6139. doi: 10.1021/ja067330p. [DOI] [PubMed] [Google Scholar]
- 17.Stock SD, Hama H, Radding JA, Young DA, Takemoto JY. Syringomycin E inhibition of Saccharomyces cerevisiae: Requirement for biosynthesis of sphingolipids with very-long-chain fatty acids and mannose- and phosphoinositol-containing head groups. Antimicro Agents & Chemo. 2000;44:1174–1180. doi: 10.1128/aac.44.5.1174-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luesch H, Yoshida WY, Moore RE, Paul VJ. Isolation and structure of the cytotoxin lyngbyabellin B and absolute configuration of lyngbyapeptin A from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1437–1439. doi: 10.1021/np000104n. [DOI] [PubMed] [Google Scholar]
- 19.Schlüter U. Ultrastructural evidence for inhibition of chitin synthesis by nikkomycin. Develop Gen Evol. 1982;91:205–207. doi: 10.1007/BF00848337. [DOI] [PubMed] [Google Scholar]
- 20.Henderson DP, Shelton MC, Cotterill IC, Toone EJ. Stereospecific preparation of the N-terminal amino acid moiety of nikkomycins Kx and Kz via a multiple enzyme synthesis. J Org Chem. 1997;62:7910–7911. doi: 10.1021/jo971549s. [DOI] [PubMed] [Google Scholar]
- 21.Ling H, Wang G, Tian Y, Liu G, Tan H. SanM catalyzes the formation of 4-pyridyl-2-oxo-4-hydroxyisovalerate in nikkomycin biosynthesis by interacting with SanN. Biochem Biophys Res Commun. 2007;361:196–201. doi: 10.1016/j.bbrc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths JS, Cheriyan M, Corbell JB, Pocivavsek L, Fierke CA, Toone EJ. A bacterial selection for the directed evolution of pyruvate aldolases. Bioorg Med Chem. 2004;12:4067–4074. doi: 10.1016/j.bmc.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 23.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Research. 2004;32 doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponce E, Flores N, Martinez A, Valle F, Bolivar F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J Bacteriol. 1995;177:5719–5722. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchanan LV, Mehta N, Pocivavsek L, Niranjanakumari S, Toone EJ, Naismith JH. Initiating a structural study of 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1999;55:1946–1948. doi: 10.1107/s0907444999011166. [DOI] [PubMed] [Google Scholar]
- 26.Cheriyan M, Toone EJ, Fierke CA. Mutagenesis of the phosphate binding pocket of KDPG aldolase enhances selectivity for hydrophobic substrates. Protein Science. 2007;16:2368–2377. doi: 10.1110/ps.073042907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. 3rd ed Vol. 3. Cold Spring Harbor Laboratory; Cold Spring Harbor: 2001. [Google Scholar]
- 28.Walters MJ, Srikannathasan V, McEwan AR, Naismith JH, Fierke CA, Toone EJ. Characterization and crystal structure of Escherichia coli KDPGal aldolase. Bioorg Med Chem. 2008;16:710–720. doi: 10.1016/j.bmc.2007.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allard J, Grochulski P, Sygusch J. Covalent intermediate trapped in 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase structure at 1.95-angstrom resolution. Proc Natl Acad Sci USA. 2001;98:3679–3684. doi: 10.1073/pnas.071380898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fullerton SWB, Griffiths JS, Merkel AB, Cheriyan M, Wymer NJ, Hutchins MJ, Fierke CA, Toone EJ, Naismith JH. Mechanism of the class I KDPG aldolase. Bioorg Med Chem. 2006;14:3002–3010. doi: 10.1016/j.bmc.2005.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapoza MP, Webster RE. The filamentous bacteriophage assembly proteins require the bacterial SecA protein for correct localization to the membrane. J Bacteriol. 1993;175:1856–1859. doi: 10.1128/jb.175.6.1856-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchison CA, Nordeen SK, Vogt K, Edgell MH. A complete library of point substitution mutations in the glucocorticoid response element of mouse mammary tumor virus. Proc Natl Acad Sci USA. 1986;83:710–714. doi: 10.1073/pnas.83.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heine A, Luz JG, Wong CH, Wilson IA. Analysis of the class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase at 0.99 Å resolution. J Mol Biol. 2004;343:1019–1034. doi: 10.1016/j.jmb.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 34.Jia J, Schorken U, Lindqvist Y, Sprenger GA, Schneider G. Crystal structure of the reduced Schiff-base intermediate complex of transaldolase B from Escherichia coli: mechanistic implications for class I aldolases. Protein Sci. 1997;6:119–124. doi: 10.1002/pro.5560060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LowKam C, Liotard B, Sygusch J. Structure of a class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity. J Biol Chem. 2010;285:21143–21152. doi: 10.1074/jbc.M109.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St-Jean M, Lafrance-Vanasse J, Liotard B, Sygusch J. High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit. J Biol Chem. 2005;280:27262–27270. doi: 10.1074/jbc.M502413200. [DOI] [PubMed] [Google Scholar]
- 37.Radaev S, Dastidar P, Patel M, Woodard RW, Gatti DL. Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase. J Biol Chem. 2000;275:9476–9484. doi: 10.1074/jbc.275.13.9476. [DOI] [PubMed] [Google Scholar]
- 38.Albery WJ, Knowles JR. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 39.O’Fagain C. Engineering protein stability. Methods Mol Biol. 2011;681:103–136. doi: 10.1007/978-1-60761-913-0_7. [DOI] [PubMed] [Google Scholar]
- 40.DeSantis G, Liu J, Clark DP, Heine A, Wilson IA, Wong CH. Structure-based mutagenesis approaches toward expanding the substrate specificity of D-2-deoxyribose-5-phosphate aldolase. Bioorg Med Chem. 2003;11:43–52. doi: 10.1016/s0968-0896(02)00429-7. [DOI] [PubMed] [Google Scholar]
- 41.Hsu CC, Hong Z, Wada M, Franke D, Wong CH. Directed evolution of D-sialic acid aldolase to L-3-deoxy-manno-2-octulosonic acid (L-KDO) aldolase. Proc Natl Acad Sci USA. 2005;102:9122–9126. doi: 10.1073/pnas.0504033102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jennewein S, Schurmann M, Wolberg M, Hilker I, Luiten R, Wubbolts M, Mink D. Directed evolution of an industrial biocatalyst: 2-deoxy-D-ribose 5-phosphate aldolase. Biotechnol J. 2006;1:537–548. doi: 10.1002/biot.200600020. [DOI] [PubMed] [Google Scholar]
- 43.Williams GJ, Domann S, Nelson A, Berry A. Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution. Proc Natl Acad Sci U S A. 2003;100:3143–3148. doi: 10.1073/pnas.0635924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams GJ, Woodhall T, Nelson A, Berry A. Structure-guided saturation mutagenesis of N-acetylneuraminic acid lyase for the synthesis of sialic acid mimetics. Protein Eng Des Sel. 2005;18:239–246. doi: 10.1093/protein/gzi027. [DOI] [PubMed] [Google Scholar]
- 45.Toscano MD, Muller MM, Hilvert D. Enhancing activity and controlling stereoselectivity in a designed PLP-dependent aldolase. Angew Chem Int Ed Engl. 2007;46:4468–4470. doi: 10.1002/anie.200700710. [DOI] [PubMed] [Google Scholar]
- 46.Schneider S, Sandalova T, Schneider G, Sprenger GA, Samland AK. Replacement of a phenylalanine by a tyrosine in the active site confers fructose-6-phosphate aldolase activity to the transaldolase of Escherichia coli and human origin. J Biol Chem. 2008;283:30064–30072. doi: 10.1074/jbc.M803184200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Royer SF, Haslett L, Crennell SJ, Hough DW, Danson MJ, Bull SD. Structurally informed site-directed mutagenesis of a stereochemically promiscuous aldolase to afford stereochemically complementary biocatalysts. J Am Chem Soc. 2010;132:11753–11758. doi: 10.1021/ja104412a. [DOI] [PubMed] [Google Scholar]
- 48.Cotterill IC, Shelton MC, Machemer DE, Henderson DP, Toone EJ. Effect of phosphorylation on the rate of unnatural electrophiles with KDPG aldolase. J Chem Soc Perkin Trans. 1998;1:1335–1341. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.