Cell wall and cellulose structure is imperative for proper cell elongation and, consequently, the architecture of plants, but components regulating cellulose structure are still elusive. This article shows that the secreted CTL1/POM1 and its close homolog CTL2 interact with glucan-based polymers and influence cellulose crystallinity and cell expansion.
Abstract
Plant cells are encased by a cellulose-containing wall that is essential for plant morphogenesis. Cellulose consists of β-1,4-linked glucan chains assembled into paracrystalline microfibrils that are synthesized by plasma membrane–located cellulose synthase (CESA) complexes. Associations with hemicelluloses are important for microfibril spacing and for maintaining cell wall tensile strength. Several components associated with cellulose synthesis have been identified; however, the biological functions for many of them remain elusive. We show that the chitinase-like (CTL) proteins, CTL1/POM1 and CTL2, are functionally equivalent, affect cellulose biosynthesis, and are likely to play a key role in establishing interactions between cellulose microfibrils and hemicelluloses. CTL1/POM1 coincided with CESAs in the endomembrane system and was secreted to the apoplast. The movement of CESAs was compromised in ctl1/pom1 mutant seedlings, and the cellulose content and xyloglucan structures were altered. X-ray analysis revealed reduced crystalline cellulose content in ctl1 ctl2 double mutants, suggesting that the CTLs cooperatively affect assembly of the glucan chains, which may affect interactions between hemicelluloses and cellulose. Consistent with this hypothesis, both CTLs bound glucan-based polymers in vitro. We propose that the apoplastic CTLs regulate cellulose assembly and interaction with hemicelluloses via binding to emerging cellulose microfibrils.
INTRODUCTION
Plant cells are surrounded by a cell wall, which consists of a complex polysaccharide matrix. This structure is essential for plant morphogenesis, cell expansion and differentiation, intercellular communication, water movement, and responses to certain external stimuli (Vorwerk et al., 2004; Baskin, 2005; Somerville, 2006; Tsukaya and Beemster, 2006). Two main types of cell walls can be distinguished: the primary and the secondary cell wall. In both of these walls, mechanical strength is provided by cellulose microfibrils, consisting of hydrogen-bonded linear β-1,4-glucan chains, synthesized by cellulose synthase (CESA) complexes at the plasma membrane (Kimura et al., 1999). Mutant analyses and immunoprecipitation suggest that two triplexes of CESA proteins (CESA1, 3, and the 6-like CESAs as well as CESA4, 7, and 8) are necessary for cellulose synthesis during primary and secondary wall formation, respectively, in Arabidopsis thaliana (Taylor et al., 2000, 2003; Desprez et al., 2007; Persson et al., 2007b). The CESA complexes (CSCs) are presumed to be assembled in Golgi bodies and transported to the plasma membrane where they are guided by cortical microtubules during cellulose synthesis (Haigler and Brown, 1986; Paredez et al., 2006). Once they are inserted in the plasma membrane, the CSCs move at constant rates, which may be reduced in mutants impaired in cellulose synthesis (Paredez et al., 2006, 2008). A population of small post-Golgi CESA compartments, called microtubule-associated cellulose synthase compartments (MASCs; Crowell et al., 2009)/small CESA compartments (SmaCCs; Gutierrez et al., 2009), may regulate the insertion and internalization of CESAs at the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009).
After the unbranched β-1,4-glucan chains are extruded, they form microfibrils through inter- and intramolecular hydrogen bonds and Van der Waals forces. These interactions are not constant along the fibrils, leading to crystalline fibrils interspersed by amorphous zones, in which hemicelluloses, mainly xyloglucans (XGs) in dicot plants, become entrapped (Pauly et al., 1999a; Cosgrove, 2005). XGs can influence the structure of cellulose crystals (Whitney et al., 1995), and low levels of cellulose crystallinity lead to increased XG binding capacity (Chambat et al., 2005). In addition, XGs can be associated with cellulose microfibrils via hydrogen bonds (Hayashi, 1989; Pauly et al., 1999a).
The cellulose-hemicellulose network is crucial for maintenance of wall mechanical strength and for cell expansion (Fry, 1995; Cosgrove, 1997, 2000; Nishitani, 1998). Currently, three classes of proteins are known to affect cellulose-XG associations. Endoglucanases (Kaku et al., 2002) and XG endotransglycosylases (Fry, 1995) play key roles in wall expansion by modification of the primary XG structure, which affects interactions between XGs and cellulose fibrils. The third group of proteins, the expansins, can induce wall expansion by loosening hydrogen bonds at the XG-cellulose interface (Yuan et al., 2001; Marga et al., 2005). The cellulose crystalline/amorphous ratio also influences in muro interaction between XGs and cellulose (Hanus and Mazeau, 2006). Only one protein has been reported to alter that ratio; KORRIGAN (KOR), a membrane-bound β-1,4-glucanase, which increases the amount of noncrystalline cellulose (Nicol et al., 1998; Takahashi et al., 2009). The kor1/lion’s tail1 mutant was initially isolated in a screen for mutants with anisotropic growth defects (Nicol et al., 1998). Many other cellulose-deficient mutants with elongation defects, such as cobra (Hauser et al., 1995; Schindelman et al., 2001; Roudier et al., 2005), impaired in a glycosyl-phosphatidylinositol–anchored protein; kobito/elongation defective1, affecting a protein of unknown function (Pagant et al., 2002; Lertpiriyapong and Sung, 2003); and chitinase-like1 (ctl1)/pom-pom1 (pom1)/ectopic deposition of lignin in pith1 (elp1), which corresponds to a putative chitinase-like protein (Hauser et al., 1995; Zhong et al., 2000; Zhong et al., 2002), have been identified. Although it appears unlikely that all of these components are parts of the core cellulose synthesis machinery, it is evident that they execute functions that are important for cellulose synthesis.
The role of CTL1, and its close homolog CTL2, in cell wall biosynthesis is especially intriguing since associations between CTLs and primary or secondary cell wall synthesis have been reported in different plant species (Zhang et al., 2004). However, the biological function of the CTLs remains elusive. Mutations in CTL1/POM1 cause abnormal root swelling, ethylene overproduction, reduced tolerance to abiotic stresses, and ectopic deposition of lignin in Arabidopsis (Hauser et al., 1995; Schneider et al., 1997; Reed et al., 1998; Cary et al., 2001; Zhong et al., 2002; Rogers et al., 2005; Kwon et al., 2007; Hermans et al., 2010). In addition, cell walls of ctl1/pom1 mutants have an infrared spectrum that closely resembles cellulose-deficient mutants (Mouille et al., 2003). We show that mutations in CTL1/POM1 result in reduced velocities of plasma membrane–localized CSCs and that the CTL1/POM1 protein resides in the endomembrane system, is secreted to the apoplast, and colocalizes with a subpopulation of tethered MASCs/SmaCCs (Crowell et al., 2009; Gutierrez et al., 2009). Biochemical and nanostructural analyses revealed that the CTLs can bind glucans and that mutations in the CTLs change the crystalline cellulose content in the cell wall. These data suggest that the CTLs act as protein or carbohydrate scaffolds, which promote the interaction between glucan fibrils and therefore influence the cellulose-hemicellulose network.
RESULTS
CTL1 Affects Primary Wall Cellulose Deposition
CTL1 is coexpressed with the primary cell wall CESAs (see Supplemental Figure 1A online; Persson et al., 2005). We confirmed this by transforming plants with a β-glucuronidase (GUS) reporter gene under the control of a 1.4-kb CTL1 promoter. Strong GUS activity was observed in seedlings and mature roots, rosette leaves, various floral tissues, and siliques (see Supplemental Figures 1B to 1G online; Hossain et al., 2010). Rapidly elongating cells in etiolated seedlings also exhibited strong GUS activity (see Supplemental Figures 1K and 1O online). Consistent with the coexpression analysis, the CTL1 promoter is active in the same organs and developmental stages as the primary wall CESAs (see Supplemental Figures 1G to 1R online).
The coexpression of CTL1 and the primary wall CESAs, and the phenotypic characteristics of ctl1 mutants (Hauser et al., 1995; Zhong et al., 2002), suggest a role for CTL1 in primary wall cellulose synthesis. To corroborate this, we tested the sensitivity of a homozygous T-DNA insertion mutant, ctl1-1, to low levels of the cellulose inhibitor and herbicide isoxaben (Scheible et al., 2001). On normal growth medium, ctl1 seedlings exhibited reduced root growth (see Supplemental Figure 2C online; Hauser et al., 1995; Zhong et al., 2002). However, when grown on plates containing moderate levels of isoxaben (1.5 nM), which cause only minor growth phenotypes for wild-type seedlings, ctl1 mutant displayed severe root growth retardation, epidermal cell swelling, and a strong increase in lignin deposition (see Supplemental Figures 2A to 2F and 3B online). The ectopic lignification was similar to that of isoxaben-treated prc1-1 seedlings (see Supplemental Figures 2A to 2F online), a CESA6 mutant affecting primary wall cellulose synthesis (Fagard et al., 2000). The increased sensitivity of ctl1 to isoxaben supports a function for CTL1 in primary wall cellulose synthesis.
Due to functional redundancies of the CESA6-related position in the CSC, prc1-1 only displays modest phenotypes (Persson et al., 2007a). If CTL1 generally affects primary wall cellulose deposition, we would therefore expect the ctl1-1 mutant to have an additive phenotype when crossed to prc1-1. To test this, we made reciprocal crosses between prc1-1 and ctl1-1 and analyzed the homozygous progeny. The full-grown homozygous ctl1-1 prc1-1 double mutant plants were severely dwarfed and had reduced seed production, consistent with an additive behavior (see Supplemental Figure 2H online). In addition, seedlings of ctl1-1 prc1-1 double mutants developed dramatically short and swollen roots (see Supplemental Figure 2G online; Hauser et al., 1995). These data support a general role for CTL1 in primary wall cellulose deposition.
Similar phenotypes to those described here for ctl1-1 were previously reported for other ctl1 independent alleles, such as pom1 (Hauser et al., 1995) and elp1 (Zhong et al., 2002), which corroborates that the ctl1-1 T-DNA insertion is the cause of the phenotypes. To confirm this, ctl1-1 was complemented with a CTL1 genomic fragment (including the CTL1 open reading frame and a region 1.5 kb upstream of the predicted start codon). The transformants were indistinguishable from wild-type plants (see Supplemental Figure 3A online), confirming that the T-DNA insertion in CTL1 caused the phenotypic changes observed in ctl1-1.
Mutations in CTL1 Reduce the Velocity of Primary Wall CSCs
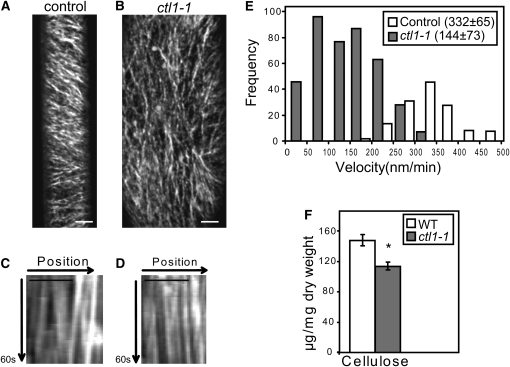
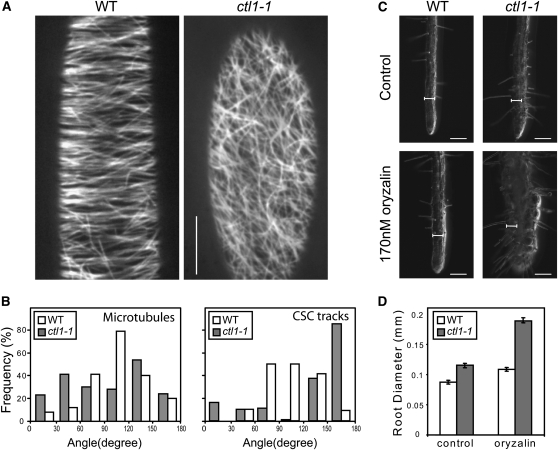
To investigate the role of CTL1 in primary wall cellulose synthesis, a yellow fluorescent protein (YFP)-tagged CESA6 (YFP:A6; Paredez et al., 2006) was crossed into ctl1-1. YFP:A6 labels small punctuate particles organized in linear arrays at the plasma membrane in elongating hypocotyl cells (Paredez et al., 2006). In these cells, we observed clear differences in CESA particle dynamics between the control and the ctl1-1 mutant in time-averaged projections (Figures 1A to 1D). Whereas the control line held CSCs that moved with constant velocities in parallel arrays at the plasma membrane, the ctl1-1 Y-A6 lines held disorganized arrays of slow, or even stalled, CSCs (cf. kymographs in Figures 1C and 1D; see Supplemental Movie 1 online). CSCs are guided at the plasma membrane by cortical microtubules (Paredez et al., 2006). Hence, the disorganized CSCs tracks may be due to defects in microtubule organization. To investigate this, we generated ctl1-1 lines that express the microtubule marker green fluorescent protein:microtubule-associated protein4 (GFP:MAP4; Marc et al., 1998). We observed a largely transverse to oblique microtubule array in elongating cells in wild-type hypocotyls, consistent with previous observations (Le et al., 2005; Paredez et al., 2006). However, corresponding cells in the ctl1-1 mutant displayed a less well organized microtubule array (Figures 2A and 2B). We reasoned that the less well ordered microtubule array in ctl1-1 should make the ctl1-1 mutant more sensitive to the microtubule inhibitor oryzalin. Indeed, ctl1-1 exhibited severe epidermal root cell swelling when grown in the presence of low levels of oryzalin (170 nM), while no apparent cell disturbance was observed in wild-type roots grown under the same conditions (Figures 2C and 2D). Interestingly, similar microtubule defects and increased sensitivity to oryzalin have been reported for other cellulose-deficient mutants, such as kor1 (Paredez et al., 2008). It is plausible that the mainly longitudinal CSC tracks observed in ctl1-1 might contribute to the epidermal cell swelling observed in ctl1-1. Nevertheless, the CSCs were clearly retarded in the ctl1-1 mutant compared with both elongating and mature wild-type cells, indicative of lower cellulose production. To estimate the overall retardation of the CSCs in the ctl1-1 background, we quantified the velocities of individual complexes by kymographic analyses (Figure 1E; Paredez et al., 2006). In control cells, the 332 ± 65 nm/min (n = 146) average velocity of individual complexes was similar to that previously reported (Paredez et al., 2006; Gu et al., 2010). However, CSCs typically migrated at 144 ± 73 nm/min (n = 404) in the ctl1-1 mutant (Figure 1E). These data show that CTL1 influences the velocity of the CSCs at the plasma membrane and suggest that the reduced CESA velocity in ctl1-1 is the cause for the reduced cellulose content in ctl1-1 seedlings (Figure 1F).
Figure 1.
Mutations in CTL1 Affect Cellulose Deposition.
(A) and (B) Time average of 60 confocal laser scanning microscopy (CLSM) image frames acquired over 5 min for line YFP:A6 (prc1-1 YFP:CESA6) (A) and ctl1-1 YFP:A6 (B).
(C) and (D) Kymographs from 60-s movies of YFP:A6 (C) and ctl1-1 YFP:A6 (D). Typical motility patterns of CSCs are shown in Supplemental Movie 1 online. Bars = 5 μm in (A) to (D).
(E) Histogram of particle velocities. Mean (±sd) velocity for each line is given in parentheses. n = 404 particles in 10 cells from nine seedlings analyzed for YFP:A6 in ctl1-1 background. n = 146 particles in four cells from three seedlings quantified for YFP:A6.
(F) Cellulose content in 6-d-old etiolated seedlings of the wild type (WT; Col-0) and ctl1-1 mutants. Data represent the average (±se) of n = 3 biological replicates, each with three technical repetitions. Asterisk indicates significant changes in relative peak intensities (Student’s t test; P < 0.05).
Figure 2.
Microtubule Defects in the ctl1-1 Mutant.
(A) Microtubule organization in wild-type (WT) and ctl1-1 etiolated hypocotyls visualized by confocal microscopy using the microtubule marker GFP:MAP4.
(B) Quantification of the orientation of microtubules and CSC tracks at the cell cortex and plasma membrane, respectively, of wild-type (white) and ctl1-1 (gray) etiolated hypocotyls. For microtubules, n = 200 in five cells from five seedlings; for CSCs, n = 160 in four cells from four seedlings. The plane perpendicular to the elongating direction is considered as reference (0° to 180°).
(C) and (D) ctl1-1 displays increased sensitivity to oryzalin.
(C) Root phenotypes of 5-d-old wild type and ctl1-1 grown on control- and 170 nM oryzalin-containing media.
(D) Measurement of root swelling in oryzalin-treated seedlings. Data represent the average (±se) of n = 50 seedlings.
Bars = 10 μm in (A), 100 μm in (C), and 66.5 μm for the internal scale.
CTL1 Colocalizes with Unknown Golgi-Derived Endomembranes That Move in an Actin-Dependent Manner
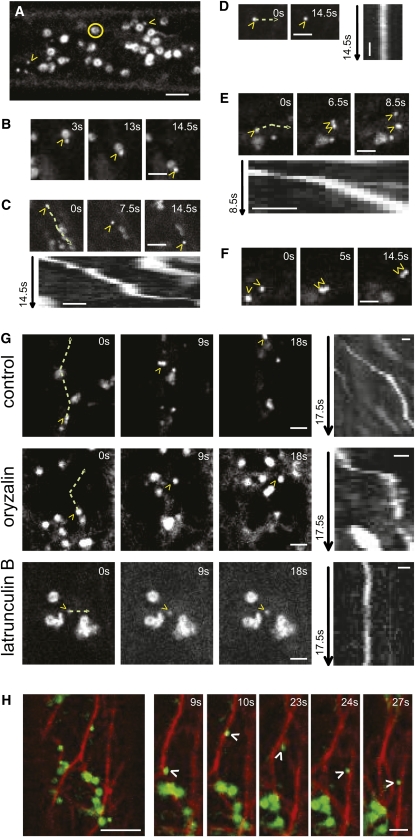
To investigate the subcellular localization of CTL1, we fused a GFP tag in frame to the C terminus of a genomic CTL1 fragment under the control of a 35S promoter. This construct rescued the ctl1 phenotypes, including the epidermal cell swelling in hypocotyls in etiolated ctl1 seedlings (see Supplemental Figures 3A to 3D online). Using spinning-disc confocal microscopy, the CTL1 localization was analyzed in elongating epidermal cells of 4-d-old etiolated hypocotyls. CTL1:GFP mainly labeled doughnut-shaped structures, presumably Golgi bodies (Paredez et al., 2006; Figures 3A to 3F; see Supplemental Movie 2 online). Moreover, a second population of particles was observed and named CTL1v (CTL1 vesicles; Figure 3; see Supplemental Movie 2 online). We confirmed that these particles were not due to overexpression of CTL1:GFP since CTL1vs were also present in lines expressing a CTL1:mCherryRFP controlled by the CTL1 promoter (see Supplemental Figure 3E online). However, the intensity of the RFP signal in the latter lines was very low; therefore, we used the 35S-driven construct for further analyses. CTL1v displayed erratic movement and behavior (Figure 3; see Supplemental Movie 2 online). Some CTLvs appeared to associate and dissociate with Golgi bodies (Figure 3B; see Supplemental Movie 2A online), others moved in linear patterns interrupted by short periods of fixed positions (Figure 3C; see Supplemental Movie 2B online), and yet others appeared fixed over longer time periods (Figure 3D; see Supplemental Movie 2C online). In addition, occasional bifurcation of CTL1v signals was observed suggesting a split of CTL1v into two vesicles (Figure 3E; see Supplemental Movie 2D online). In other instances, two CTL1v in close vicinity appeared to fuse (Figure 3F; see Supplemental Movie 2E online).
Figure 3.
CTL1 Localizes to Endomembranes That Move along Actin Filaments.
CLSM images of 4-d-old etiolated CTL1:GFP hypocotyl epidermal cells.
(A) CTL1 localizes to Golgi bodies (yellow circles) and unknown vesicles (yellow arrowheads; CTL1v).
(B) to (F) CTL1vs show different migratory patterns; the vesicles associate and dissociate with Golgi bodies (B), the vesicles’ linear movements are interrupted by short periods of fixed positions (C), sometimes the vesicles are immobile (D), they bifurcate (E), or two independent vesicles move together in close vicinity of each other (F). Typical motility patterns of CTL1vs are shown in Supplemental Movie 2 online.
(C) to (E) Kymographs from CTL1v over the indicated time in seconds (green line = migration path).
(G) and (H) CTL1v movement depends on an intact actin cytoskeleton.
(G) CLSM images of a cell from a seedling expressing CTL1:GFP after 16-h treatment with 20 μM oryzalin or 1 mM LatB for 3 h, which are microtubule and actin disruptors, respectively. CTLvs are indicated by yellow arrowheads. Control treatment is included for comparison. Kymographs from CTL1v over the indicated time in seconds (green line = migration path). Typical motility patterns of CTL1 are shown in Supplemental Movie 3 online.
(H) Seedlings expressing CTL1:GFP and mCherryRFP:FABD were imaged under standard (water) conditions. CTL1v moving over actin filaments is indicated by a white arrowhead. Typical movement of CTL1v along the actin cytoskeleton is shown in Supplemental Movie 4 online.
Bars = 5 μm in (A), (G), and (H), 2 μm in (B) to (F), and 1 μm in kymographs in (C) to (E).
The actin and microtubule cytoskeletons are important for delivery of the primary and secondary wall CESAs to the plasma membrane (Wightman and Turner, 2008; Crowell et al., 2009; Gutierrez et al., 2009). To elucidate the role of the cytoskeleton in the spatial control of CTL1 trafficking, CTL1:GFP seedlings were treated with oryzalin or the actin disruptor Latrunculin B (LatB). Incubation with 20 μM oryzalin for 16 h disrupted the microtubule array but did not alter the movement of the presumed CTL1 Golgi bodies, nor the CTL1v (Figures 3G; see Supplemental Figure 4 and Supplemental Movie 3 online). However, incubation with 1 μM LatB for 3 h impaired the distribution of the presumed CTL1 Golgi bodies similar to what has been observed for Golgi-localized CESAs (Figure 3G; see Supplemental Figure 4 and Supplemental Movie 3 online; Nebenführ et al., 1999; Crowell et al., 2009; Gutierrez et al., 2009). Moreover, the CTL1v displayed Brownian motion in actin-deficient cells (see Supplemental Movie 3 online), suggesting that both the tentative Golgi-localized CTL1 and the CTL1v need the actin cytoskeleton for motility. The movement of the CTL1v along the actin cytoskeleton was further corroborated by imaging dual-labeled CTL1:GFP and mCherryRFP:FABD (for F-Actin Binding Domain; Ketelaar et al., 2004) seedlings (Figure 3H; see Supplemental Movie 4 online)
CTL1 Is Secreted to the Apoplast
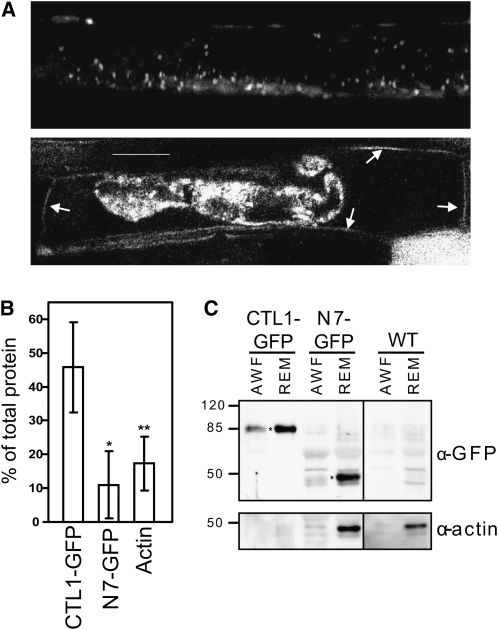
CTL1 harbors a putative N-terminal signal peptide, which suggests that CTL1 enters the secretory pathway (Passarinho and de Vries, 2002; Kwon et al., 2007). Consistent with this hypothesis, we observed a fluorescent signal in the periphery of the cell in both CTL1:GFP- and CTL1:mCherryRFP-expressing seedlings (Figures 3A and 4A; see Supplemental Figure 3E online). To resolve whether this signal resides at the plasma membrane or in the apoplast, we plasmolyzed the cells using 0.5 M Suc for 45 min. This treatment resulted in rapid shrinkage of the cytoplasm and revealed that the majority of fluorescence was retained in the constrained cytoplasm (Figure 4A), which is consistent with the intracellular labeling of the presumed Golgi bodies and CTL1v (Figures 3A to 3F). In addition, fluorescence was clearly visible at the cell wall (Figure 4A; Hermans et al., 2011). To confirm CTL1 secretion, we performed immunoblotting after harvesting apoplastic washing fluid (AWF) from CTL1:GFP seedlings. We could inmunodetect CTL1:GFP in the AWF and the remaining material (Figures 4B and 4C). As was expected, none of the controls (i.e., actin and N7-GFP that do not locate to the apoplastic space) were significantly detected in the AWF (Figures 4B and 4C). These results confirm that CTL1 is indeed secreted to the apoplast and the cell wall matrix.
Figure 4.
CTL1-GFP Is Secreted to the Apoplast.
(A) CLSM image of CTL1-GFP root cell (top part) and root cell plasmolyzed using 0.5 M Suc for 45 min (bottom part). After plasmolysis, GFP signal observed in the cell wall is indicated by arrows.
(B) and (C) CTL1-GFP localized to the AWF and in the remaining material (REM). A GFP-tagged nuclear protein, N7-GFP, and actin were included as controls.
(B) Percentage of protein detected in the AWF, as a percentage of total protein AWF+REM. Data represent the average (±sd) of at least n = 2 (n = 4 for CTL1-GFP) independent immunoblots. Asterisks indicate significant changes in percentage of colocalization compared with control treatment (Student’s t test; *P < 0.05; **P < 0.01).
(C) Immunoblot summary of AWFs and REMs. Top panel, immunodetection of proteins in AWF and REM. Asterisks mark the specific bands of CTL1-GFP and N7-GFP. Bottom panel, immunodetection of actin in AWF and REM. The amount of protein loaded in CTL1-GFP AWF and REM was too little to detect actin. The same experiment was repeated three times with similar results. WT, wild type. Bar = 20 μm.
The CTL1 Vesicles Represent an Unknown Secretory Compartment
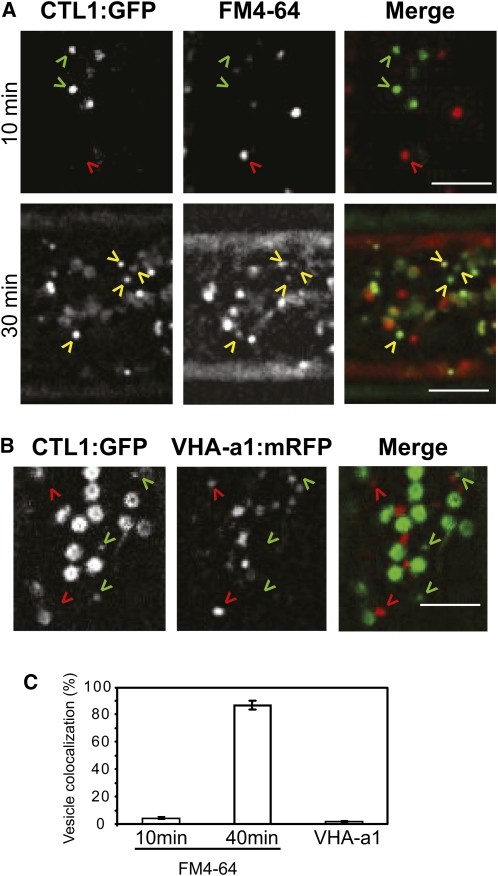
The dynamic regulation of the plasma membrane, and the cell wall content, is dependent on the organization of membrane trafficking. To investigate whether the CTL1vs are associated with endo- or exocytosis, we used the lipophilic tracer dye FM4-64. FM4-64 follows the endocytic pathway from the plasma membrane, but longer treatment redistributes the dye to other internal membranes, including Golgi bodies and the prevacuolar compartment (Bolte et al., 2004; Dettmer et al., 2006). Internalized FM4-64 did not label CTL1v in etiolated hypocotyls during the first 10 min of treatment (Figures 5A and 5C; see Supplemental Movie 5 online; Gutierrez et al., 2009). However, after longer incubations (30 min), the FM4-64 dye and CTL1v coincided (Figures 5A and 5C; see Supplemental Movie 5 online), suggesting that CTL1v are exocytosed rather than endocytosed. Interestingly, similar results have been described for tethered SmaCCs (Gutierrez et al., 2009).
Figure 5.
CTL1 Localizes to Nonendocytotic Vesicles.
(A) Four-day-old etiolated CTL1:GFP seedlings were incubated in 20 μM FM4-64 for 10 or 30 min. After 10 min, FM4-64 stains endosomes (red arrowheads) that do not overlap with CTL1v (green arrowheads). FM4-64 labels CTL1v (yellow arrowheads) after 30 min.
(B) Four-day-old etiolated seedlings expressing CTL1:GFP and VHA-a1:mRFP. CTL1v (green arrowheads) do not colocalize with VHA-a1 (red arrowheads) vesicles.
(C) Percentage of CTL1v that colocalize with FM4-64–stained vesicles and with VHA-a1 marker. Data represent the average (±se) of n = 3 cells from three different seedlings. In each cell, 40 CTL1vs were analyzed. Typical behavior of dual-labeled cells is shown in Supplemental Movie 5 online. Bars = 5 μm.
Viotti et al. (2010) have recently shown that both secretory and endocytic cargo pass through the trans-Golgi network/early endosome (TGN/EE). The VHA-a1 subunit of the V-ATPase is a marker of one of the TGN/EE pathways (Dettmer et al., 2006). To investigate whether the CTL1vs are associated with this TGN/EE route, we crossed the CTL1:GFP line with plants expressing a VHA-a1:RFP construct. Interestingly, CTL1v showed no colocalization with VHA-a1 (Figures 5B and 5C; see Supplemental Movie 5 online), indicating that CTL1vs do not pass through the TGN/EE pathway as defined by VHA-a1.
CTL1 Colocalizes with CESAs in Golgi Bodies and Tethered MASCs/SmaCCs
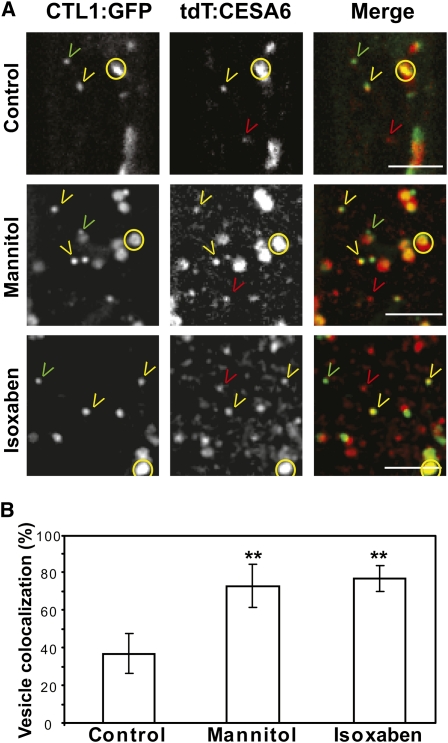
A subfraction of CESAs is associated with particles that move similarly to the CTL1v (i.e., that show alterations in motility by actin-depolymerizing agents) and that are labeled by FM4-64 after long incubations (Crowell et al., 2009; Gutierrez et al., 2009). We therefore investigated if the CTL1vs are associated with these CESA-containing vesicles in dual-labeled CTL1:GFP and tdTomato:CESA6 (tdT:A6) seedlings. The fluorescence of CTL1:GFP and tdT:A6 coincided in doughnut-shaped structures (Figure 6A; see Supplemental Movie 6 online), confirming that CTL1 is present in the Golgi. Interestingly, around 40% of CTL1vs colocalized with a population of MASCs/SmaCCs (Figure 6B; see Supplemental Movie 6 online; Crowell et al., 2009; Gutierrez et al., 2009). The MASCs/SmaCCs have been reported to accumulate at, and tether to, the cell cortex. Upon osmotic stress and treatment with cellulose synthesis inhibitors, such as isoxaben and CGA 325’615, MASCs/SmaCCs display sustained episodes of positional stability. This behavior is concomitant with the disappearance of CSCs from the plasma membrane (Paredez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009). To characterize further CTL1v behavior in response to stresses that cause alterations in MASC/SmaCC behavior, dual-labeled CTL1:GFP tdT:A6 seedlings were treated with mannitol and isoxaben. Both the osmotic and cellulose synthesis inhibitor treatments increased the colocalization of CTL1:GFP and tdT:A6 in the MASCs/SmaCCs to 75% of the total CTL1v (Figures 6A and 6B; see Supplemental Movie 6 online). Based on these observations, we conclude that CTL1v to a large extent colocalizes with Golgi-localized CESAs and tethered MASCs/SmaCCs. Most of the MASCs/SmaCCs move via microtubules (Crowell et al., 2009; Gutierrez et al., 2009), but disruption of the microtubule array by oryzalin treatment did not alter CTL1v movement (Figure 3G; see Supplemental Movie 3 online). To solve this apparent contradiction, we incubated CTL1:GFP tdT:A6 dual-labeled seedlings with oryzalin. As expected, the majority of MASCs/SmaCCs were immobile and none of them contained CTL1:GFP signal. In addition, another population of tdT:A6-containing vesicles maintained their motility and the majority of them colocalized with CTL1v (see Supplemental Figure 4 and Supplemental Movie 7 online). These data support previous results suggesting that a subpopulation of CesA3-containing vesicles move via actin filaments (Crowell et al., 2009). Moreover, we show that CTL1vs colocalize with CSC small actin-based vesicles.
Figure 6.
CTL1 Colocalizes with Tethered MASCs/SmaCCs in Secretory Vesicles.
Seedlings expressing CTL1:GFP and tdT:CESA6 were incubated in water (control), in 200 mM mannitol for 4 h, or in 100 nM isoxaben for 3 h and were imaged at the cell cortex of epidermal cells from etiolated hypocotyls.
(A) Golgi bodies are labeled with GFP and tdT (yellow circles). Other vesicles (SmaCCs/MASCs) are labeled with GFP (green arrowheads), with tdT only (red arrowheads), or with both fluorophores (yellow arrowheads).
(B) Percentage of CTL1v that colocalize with MASCs/SmaCCs in response to different treatments. Data represent the average (±se) of n = 3 cells from three different seedlings. In each cell, 25 to 40 CTL1vs were analyzed. Asterisks indicate significant changes in percentage of colocalization compared with control treatment (Student’s t test; P < 0.01). Typical behavior of dual-labeled cells is shown in Supplemental Movies 6 and 7 online.
Bars = 5 μm in (A) and (B).
CTL1 and Its Close Homolog CTL2 Are Functionally Equivalent and Are Not Essential for Cellulose Biosynthesis
CTL1 has only one close homolog, CTL2, which is coexpressed with the secondary wall CESAs (Persson et al., 2005). The public transcriptome data indicate that CTL2 is mainly expressed in stems and siliques (http://genecat.mpg.de; Mutwil et al., 2008). These data show that CTLs are likely to have a pairwise relationship with the primary and secondary wall CESAs, respectively. A similar relationship is also evident in other plant species, both in monocots and dicots, where different CTL homologs are coexpressed with the primary wall and secondary wall CESAs, respectively (see Supplemental Table 1 online). This indicates an important evolutionarily conserved role for the CTLs during primary and secondary wall cellulose production.
To remove completely any residual CTL activity, we generated double mutants between ctl1-1 and ctl2-1 (see Supplemental Figure 5 online). The ctl1-1 ctl2-1 (ctl1 ctl2) double mutant displayed subtle additive growth phenotypes compared with the single mutant parent lines, such as slight growth retardation of seedling hypocotyls and of root growth (see Supplemental Figures 5B and 5C online; Hossain et al., 2010). In addition, the double mutant held reduced levels of cellulose and displayed a concomitant increase in uronic acids compared with the single mutants (see Supplemental Figure 5D online). These data suggest that CTL activity is not essential for cellulose production, since complete block of primary wall cellulose synthesis results in collapsed pollen grains (Persson et al., 2007a), which was not observed in the ctl1 ctl2 double mutants.
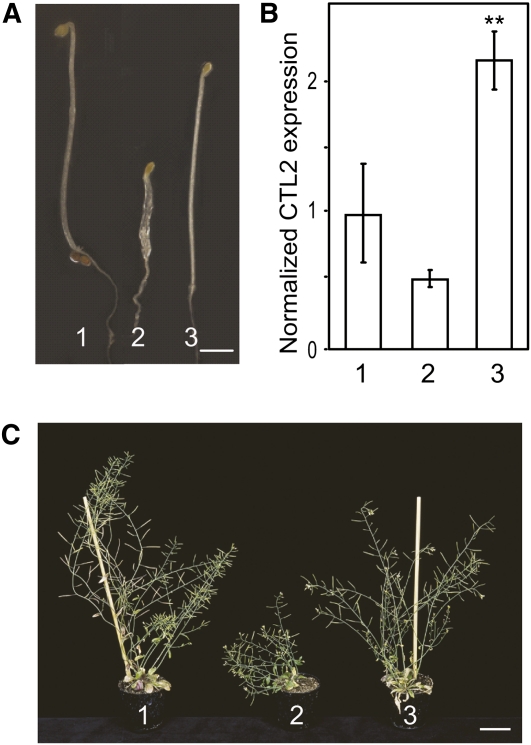
Given that CTL1 and CTL2 are coexpressed with primary and secondary wall CESAs, respectively, it appears likely that the respective proteins perform related functions in different tissue and cell types. To test this hypothesis, ctl1-1 was transformed with a CTL2 genomic clone under the control of the 1.4-kb CTL1 promoter. The construct completely restored the ctl1-1 phenotypes (Figure 7), which demonstrates that CTL1 and CTL2 are functionally equivalent.
Figure 7.
CTL1 and CTL2 Are Functionally Equivalent.
(A) and (C) pCTL1:CTL2 rescues the ctl1-1 mutant phenotype in 6-d-old etiolated seedlings (A) and 7-week-old plants (C). Bars = 1 mm in (A) and 5 cm in (C).
(B) Relative quantification of CTL2 levels in 6-d-old etiolated seedlings. Values are represented as n-fold CTL2 cDNA levels compared with the wild type. Data represent the average (±se) of n = 3 biological replicates, each containing three technical repetitions. Asterisks indicate significant changes in relative CTL2 expression compared with the wild type (Student’s t test; P < 0.01). 1, the wild type (Col-0); 2, ctl1-1; 3, ctl1-1 complemented with PCTL1:CTL2.
CTL1 and CTL2 Bind Glucan-Containing Polymers
The CTLs are named based on their amino acid sequence similarity to known chitinases or GH19 glycosyl hydrolases (Graham and Sticklen, 1994). However, CTL1 and CTL2 lack amino acids that are important for chitin binding and catalytic activity (Zhang et al., 2004).
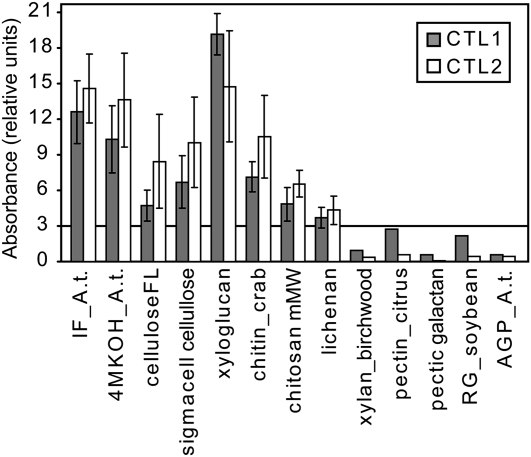
To investigate whether the CTLs can bind carbohydrate-based polymers, we expressed the proteins fused to C-terminal MYC- and His-tags in Pichia pastoris. The expression was confirmed by dot blotting using an anti-MYC antibody (Bauer et al., 2006) and by subsequent protein gel blots. The two affinity-purified recombinant proteins were assayed for binding activity with dark-grown Columbia-0 (Col-0) Arabidopsis seedling cell wall fractions (containing mainly XG as hemicellulose) and various oligosaccharides (see Supplemental Table 2 online). The binding efficiency was quantified with ELISA and anti-MYC antibodies. Both CTL1 and CTL2 bound to the etiolated seedling insoluble cell wall fraction, commercial XG, and fibrous cellulose (Figure 8). These results indicate that CTL1 and CTL2 can bind mainly glucan-based polysaccharides (Figure 8). To confirm these data, we performed immunoblots including all the substrates that gave positive results in the ELISA and polysaccharides representing the other cell wall components (i.e., pectins and xylan) (see Supplemental Figure 6 online). Incubation with the CTLs corroborated binding of the proteins to glucan-based substrates (see Supplemental Figures 6B and 6C online). To verify that the nonbound substrates (e.g., pectins and xylan) were properly linked to the matrix, we incubated the membranes with specific antibodies (i.e., JIM5) (antipectin; see Supplemental Figure 6D online), LM5 (antigalactan; see Supplemental Figure 6E online), and LM11 (antixylan; see Supplemental Figure 6F online). These immunoblots confirmed that the substrates were present on the membranes; hence, the CTLs preferentially bind to glucan-based polysaccharides.
Figure 8.
CTL1 and CTL2 Bind Glucan Polymers.
CTL1 and CTL2 binding activity toward substrates with relative absorbance >3 and three selected substrates with relative absorbance <3 (for complete list, see Supplemental Table 2 online). Relative absorbance units were calculated by the ratio of the (control-subtracted) absolute measured absorbance at 450 nm and normalized with respect to the concentration of protein used for each replicate (in mg/mL). IF_A.t., insoluble fraction of Arabidopsis cell wall; 4MKOH_A.t., 4 M KOH fraction of Arabidopsis cell wall; celluloseFL, cellulose, fibrous, long; RG, rhamnogalacturonan; AGP_A.t., arabinogalactanproteins from Arabidopsis.
Data represents the average (±se) of n = 3 independent ELISA replicates.
Even though important amino acids for catalytic activity appear to have been changed in the CTLs (Passarinho and de Vries, 2002; Hermans et al., 2010; Hossain et al., 2010), it is still possible that the proteins have retained catalytic activities. However, chitinase activity of recombinant CTL1 protein purified from Escherichia coli was tested in vitro with negative results (Zhong et al., 2002; Hermans et al., 2010). Nevertheless, other hydrolytic activities cannot be ruled out. We therefore incubated the purified recombinant CTLs with the substrates that they could bind in our ELISA and immunoblot-based studies (Figure 8; see Supplemental Figure 6 online) (i.e., the cell wall insoluble fractions, XG, cellulose, and other glucan-based polymers) and monitored hydrolytic products using capillary electrophoresis. However, we were unable to detect any peaks resulting from hydrolytic activity using these substrates. Hence, at least when expressed in yeast, the CTLs lack hydrolytic activity against glucan-containing substrates.
Mutations in the CTLs Affect Cellulose Fibril Assembly
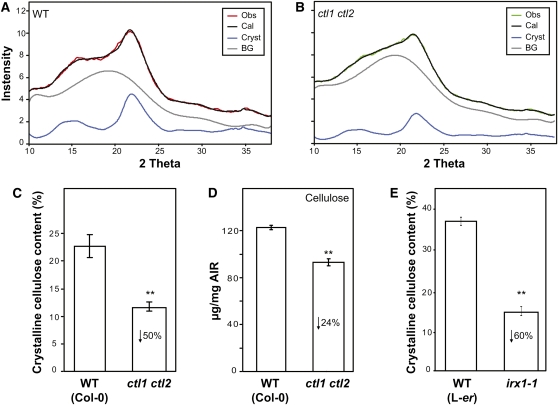
To determine whether the CTLs have an impact on the structure of cellulose and the total cellulose content, wide-angle x-ray diffraction was performed on the first centimeter from the lower part of 7-week-old stems of Col-0 plants and ctl1-1, ctl2-1, and ctl1 ctl2 mutants. X-ray diffraction has previously been used for Arabidopsis samples (Fujita et al., 2011). With x-ray diffraction, information on crystallinity, crystallite size, and orientation of cellulose can be gained with very little sample preparation (Jakob et al., 1995). We analyzed intact stems rather than fractionated cellulose as cellulose crystallinity (Hermans and Weidinger, 1949) and crystallite size (Haase et al., 1976) are sensitive to acid treatments. To analyze the scattering signal, the two-dimensional scattering images were integrated azimuthally and radially. The azimuthal profile (intensity versus azimuthal angle φ) of the (200) Bragg reflection of cellulose (indexing the reflection planes of cellulose according to Gardner and Blackwell [1974]) gives information on the orientation of the cellulose. The radial profile (intensity versus scattering angle 2θ) provides information on the structure of the cellulose. The position of the Bragg peaks originates from the structure of the cellulose crystalline unit cell, whereas the widths of the peaks are inversely proportional to crystallite size. The total scattering signal represents the sum of the scattering contributions of crystalline cellulose and of amorphous parts of the cell wall. The contribution to amorphous scattering may be caused by amorphous cellulose and also by noncellulosic amorphous cell wall components.
In the radial profile of the wild type (Figure 9A), two peaks were observed at scattering angles of 16° and 21.5°. The peak at 21.5° can be assigned to the (200) Bragg reflection of crystalline cellulose, whereas the peak at 16.5° represents a merged peak originating from the (110) and (1-10) Bragg reflections. The width of the Bragg peaks indicates small cellulose crystallites and may explain the rather low angular resolution of the radial profile. The radial profiles of the wild type and the single mutants ctl1 and ctl2 were very similar, whereas the radial profile of the double mutant ctl1 ctl2 was different (see Supplemental Figure 7A online). In the profile of the double mutant, the different Bragg peaks can hardly be distinguished. Nevertheless, by comparing the profiles of the double mutant with those of the single mutant and the wild type, two important conclusions can be made: (1) The larger scattering contribution in the profile of the double mutant, which leads to the masking of the Bragg peaks, occurred within a range of 2θ where no significant Bragg peak of cellulose exists. In this range of 2θ, the maximum scattering contribution of amorphous cellulose can be observed (Paakkari et al., 1989). (2) The slopes of the (200) Bragg peak at 21.5° do not differ between the wild type and the double mutant, which indicates that the mutation did not result in a change in crystallite size. Hence, it can be stated qualitatively from these observations that the double mutant showed an increased scattering contribution of amorphous cell wall components, while the crystallite size of the cellulose was not altered.
Figure 9.
ctl1 ctl2 Double Mutant Stems Show Differences in Crystalline Cellulose Content Compared with the Wild Type.
(A) and (B) Final best fit for wild-type (WT) (A) and ctl1 ctl2 double mutant (B) radial profiles, obtained by the DFA approach. Obs (observed), x-ray diffraction data (red for the wild type and green for ctl1 ctl2); Cal (calculated), simulated pattern (black); Cryst, crystalline contribution (blue); BG, background (gray).
(C) and (E) Percentage of crystalline cellulose content calculated for the ctl1 ctl2 double mutant, irx1 plants, and the corresponding wild-type controls (Col-0 and Landsberg erecta [L-er], respectively).
(D) Total cellulose contents (μg/mg alcohol-insoluble residue [AIR]) in the same samples of wild-type and ctl1 ctl2 stems used for x-ray measurement. In (C) to (E), data represent the average (±se) of n = 5 biological replicates. Asterisks indicate significant changes in relative peak intensities (Student’s t test; P < 0.01).
To quantify the crystallite size and the ratio of crystalline components to total cell wall components in the wild type and the mutants, Debye function analysis (DFA) was applied to the radial profiles. For the wild type and the double mutant, a crystallite size of 2.46 ± 0.11 nm and 2.46 ± 0.31 nm was obtained, respectively (see Supplemental Figure 7C online; crystallite size). The crystalline cellulose content was calculated to be 22.7% ± 3.8% for the wild type and 11.6% ± 1.1% for the double mutant ctl1 ctl2 (Figures 9A to 9C). These results are in line with the interpretation of the observed differences in the scattering profiles of the wild type and the double mutant. The total cellulose content was also reduced in cell walls of ctl1 ctl2 stems (Figure 9D), which could contribute to the reduced crystalline cellulose content. To test whether this was the case, we quantified the total amount of cellulose in the samples that were used for the x-ray analysis. The total cellulose content was reduced by ~30% and the crystalline cellulose content was reduced by ~50% in the ctl1 ctl2 double mutant compared with the wild type (Figures 9C and 9D). Hence, these data suggest that the reduction in crystalline cellulose content in the ctl1 ctl2 mutant can only partly be explained by the reduction in total cellulose content.
Contrary to the alterations in crystalline cellulose content, the analysis of the azimuthal profiles of all samples indicated no changes in the orientation of cellulose fibrils in the ctl1-1, ctl2-1, and ctl1 ctl2 mutants compared with the wild type (see Supplemental Figure 7B online).
To determine whether this alteration in cellulose fibril structure is specific for plants impaired in CTL activity, similar analyses were performed for the secondary CESA mutant, irx1, which presumably completely lacks secondary wall cellulose (Turner and Somerville, 1997). irx1 (Landsberg erecta background) showed a significant reduction in crystalline cellulose content compared with the wild type (Figure 9E; see Supplemental Figure 7D online). Indeed, the crystallinity was so low that little orientation could be observed in the azimuthal profile (see Supplemental Figure 7E online). Interestingly, a proportional decrease in total cellulose was previously reported for this mutant (~60%; Turner and Somerville, 1997), indicating that the lower crystalline cellulose content quantified in irx1 could be a direct consequence of its reduction in cellulose content.
The obtained data indicate that the cellulose structure in the ctl1 ctl2 mutant is distinguishable from that in other cellulose-deficient mutants and the wild type and support an influence of the CTLs on the assembly of cellulose microfibrils.
Mutations in CTLs Affect the Extractability of Hemicelluloses
The results obtained from the ELISA assay suggest that CTL1 and CTL2 can bind glucan-based polysaccharides (Figure 8) (i.e., cellulose and XG). To evaluate if alterations in the XG polymers also occurred in the ctl1 mutants, cell walls of 6-d-old etiolated wild-type and ctl1-1 seedlings were treated with a xyloglucan endoglucanase (XEG; Pauly et al., 1999b). This enzyme cleaves the XG backbone behind nonsubstituted Glc residues with respect to the nonreducing end and releases mainly heptasaccharide XXXG to decasaccharide XLFG fragments (Fry et al., 1993). The treatment resulted in a significant relative increase of the peak at 1247 mass-to-charge ratio (m/z) in ctl1-1 compared with wild-type cell wall material (see Supplemental Figures 8A to 8C online). This peak corresponds to the sodium adduct of isomeric galactosylated XG fragments containing three unsubstituted xylose branches and one galactosyl-substituted xylose branch (such as XXLG/XLXG; Lerouxel et al., 2002). The increase in the 1247 peak simultaneously resulted in a relative decrease in the peaks 1393 and 1555 m/z (see Supplemental Figures 8A to 8C online). These peaks correspond to the fucosylated units XXFG and XLFG, respectively. In addition, we observed a small but significant reduction in the 1085 peak, characteristic of XXXG units, in the ctl1-1 mutant (see Supplemental Figure 8C online). It is therefore apparent that either the ratio of galactosylated and fucosylated XG side chains is changed, or the accessibility of the enzymes to the XG polymer is altered, in ctl1-1. Interestingly, a similar XEG digestion pattern was observed in wild-type plants grown on isoxaben (see Supplemental Figures 8D to 8F online), which supports the hypothesis that the accessibility of XEG to the XG is altered when cellulose synthesis is impaired.
Complex signaling and feedback events occur between the cell wall and the cytosol that lead to redistribution of carbon between various wall polymers (Haigler et al., 2001; see Supplemental Figure 6D online). Therefore, we quantified how the ctl mutations affected cell wall polysaccharides by analyzing fractionated cell walls of 7-week-old plant stems. Comparative analyses of alditol acetates showed that ctl mutants, especially the double mutant ctl1 ctl2, had higher xylose, galactose, and arabinose levels and lower glucose levels after trifluoroacetic acid (TFA) treatment of the residue in the insoluble fraction (Table 1). Most of these sugars are typical components of hemicelluloses. Since TFA treatment dissolves only the noncrystalline polysaccharides (Selvendran and O’Neill, 1987; Kakegawa et al., 1998), and based on the cellulose-hemicellulose interaction model suggested by Pauly et al. (1999a), the insoluble fraction might be enriched in hemicelluloses entrapped in the amorphous cellulose (Hayashi et al., 1994; Cosgrove, 2005). This result suggests an altered extractability of hemicelluloses in the ctl mutants or an enrichment of these polysaccharides in the noncrystalline cellulose.
Table 1.
Glycosyl Residue Composition of Fractionated Cell Walls from 7-Week Old Wild-Type (Col-0), ctl1-1, ctl2-1, and ctl1-1 ctl2-1 Stems
| Sugars (μg/mg) |
||||||||||||||||
| Fraction | Rhamnose | Fuc | Ara | Xyl | Man | Gal | Glc | Uronic Acids | ||||||||
| CDTA | Av | se | Av | se | Av | se | Av | se | Av | se | Av | se | Av | se | Av | se |
| Wild type | 18.10 | 0.32 | 2.42 | 0.24 | 20.46 | 0.22 | 4.12 | 0.22 | 3.80 | 0.28 | 25.29 | 0.64 | 25.81 | 0.60 | 113.15 | 39.57 |
| ctl1-1 | 20.13 | 0.44 | 1.77 | 0.10 | 17.78 | 0.18 | 5.40 | 0.17 | 4.99 | 0.07 | 23.70 | 0.19 | 26.23 | 0.87 | 129.80 | 30.17 |
| ctl2-1 | 17.65 | 0.57 | 1.83 | 0.06 | 18.91 | 0.20 | 11.09 | 0.25 | 3.35 | 0.09 | 24.07 | 0.41 | 23.10 | 0.52 | 128.28 | 20.74 |
| ctl1-1ctl2-1 | 15.36 | 0.26 | 1.86 | 0.10 | 17.18 | 0.15 | 14.04 | 0.10 | 6.49 | 0.15 | 23.58 | 0.25 | 21.49 | 0.42 | 98.30 | 29.98 |
| Na2CO3 | ||||||||||||||||
| Wild type | 26.15 | 0.92 | 1.81 | 0.12 | 15.60 | 0.59 | 14.93 | 0.82 | 2.21 | 0.24 | 17.01 | 0.36 | 22.30 | 1.44 | 84.58 | 13.70 |
| ctl1-1 | 20.33 | 1.00 | 1.24 | 0.08 | 13.00 | 0.59 | 17.64 | 0.51 | 1.56 | 0.13 | 19.97 | 0.35 | 26.26 | 1.51 | 102.30 | 7.81 |
| ctl2-1 | 23.64 | 0.48 | 1.94 | 0.11 | 14.82 | 0.12 | 16.27 | 0.61 | 1.91 | 0.11 | 18.54 | 0.32 | 22.87 | 0.54 | 130.21 | 28.58 |
| ctl1-1ctl2-1 | 22.21 | 0.79 | 1.93 | 0.17 | 15.18 | 0.42 | 18.59 | 1.65 | 1.84 | 0.33 | 17.89 | 0.10 | 22.36 | 1.56 | 173.03 | 14.04 |
| 1 M KOH | ||||||||||||||||
| Wild type | 3.89 | 0.21 | – | – | 2.86 | 0.06 | 81.68 | 0.73 | 0.53 | 0.06 | 2.30 | 0.12 | 8.74 | 0.53 | 21.03 | 5.07 |
| ctl1-1 | 3.33 | 0.06 | – | – | 2.73 | 0.02 | 78.60 | 0.70 | 0.50 | 0.05 | 2.96 | 0.25 | 11.87 | 0.39 | 32.29 | 3.12 |
| ctl2-1 | 3.00 | 0.16 | – | – | 2.37 | 0.04 | 84.65 | 0.42 | 0.31 | 0.01 | 2.10 | 0.05 | 7.57 | 0.53 | 33.41 | 3.03 |
| ctl1-1ctl2-1 | 2.69 | 0.03 | – | – | 2.27 | 0.05 | 82.94 | 0.56 | 0.46 | 0.04 | 2.28 | 0.10 | 9.35 | 0.51 | 28.39 | 1.33 |
| 4 M KOH | ||||||||||||||||
| Wild type | 5.10 | 0.42 | – | – | 3.14 | 0.37 | 63.77 | 0.65 | 4.12 | 0.35 | 4.17 | 0.30 | 19.71 | 1.07 | 3.41 | 0.22 |
| ctl1-1 | 3.63 | 0.06 | – | – | 2.70 | 0.04 | 65.03 | 0.24 | 4.32 | 0.10 | 4.53 | 0.06 | 19.79 | 0.32 | 4.10 | 0.53 |
| ctl2-1 | 3.63 | 0.21 | – | – | 2.34 | 0.03 | 64.39 | 0.73 | 4.46 | 0.03 | 4.57 | 0.07 | 20.24 | 0.31 | 4.93 | 0.86 |
| ctl1-1ctl2-1 | 4.55 | 0.45 | – | – | 2.84 | 0.17 | 60.27 | 0.27 | 6.53 | 0.20 | 5.20 | 0.03 | 20.61 | 0.39 | 6.30 | 1.13 |
| Insolubles | ||||||||||||||||
| Wild type | 6.75 | 1.03 | – | – | 2.54 | 0.09 | 11.15 | 1.16 | 9.70 | 0.16 | 3.74 | 0.22 | 66.12 | 1.18 | 22.91 | 2.33 |
| ctl1-1 | 9.54 | 0.63 | – | – | 4.02 | 0.12 | 15.48 | 0.74 | 10.03 | 0.27 | 5.83 | 0.37 | 55.09 | 1.86 | 24.84 | 3.07 |
| ctl2-1 | 8.73 | 0.10 | – | – | 3.28 | 0.16 | 16.00 | 0.52 | 9.92 | 0.52 | 4.79 | 0.50 | 57.27 | 0.62 | 20.55 | 3.59 |
| ctl1-1ctl2-1 | 12.96 | 0.11 | – | – | 5.15 | 0.11 | 18.48 | 0.27 | 11.35 | 0.17 | 6.72 | 0.24 | 45.35 | 0.64 | 17.22 | 0.71 |
| Totala | ||||||||||||||||
| Wild type | 5.00 | 0.37 | 1.01 | 0.09 | 7.21 | 0.20 | 59.52 | 1.43 | 11.26 | 0.36 | 16.37 | 0.68 | 46.71 | 1.31 | 61.69 | 1.8654 |
| ctl1-1 | 5.89 | 0.11 | 1.27 | 0.02 | 7.89 | 0.16 | 53.98 | 1.32 | 10.08 | 0.18 | 18.65 | 0.46 | 47.00 | 0.97 | 78.79 | 1.6735 |
| ctl2-1 | 5.47 | 0.41 | 1.18 | 0.05 | 6.88 | 0.09 | 57.26 | 2.07 | 11.13 | 0.42 | 16.34 | 0.60 | 52.05 | 3.15 | 66.26 | 2.1743 |
| ctl1-1ctl2-1 | 6.66 | 0.30 | 1.25 | 0.18 | 7.95 | 0.15 | 60.48 | 1.42 | 12.03 | 0.19 | 16.89 | 0.59 | 43.19 | 1.83 | 92.69 | 2.7299 |
Average (Av) of at least three biological replicates per genotype. The se is included. Numbers in bold indicate significant differences between mutant and wild-type control (t test; P < 0.01). –, Not detectable.
Total cell wall material, without fractionation.
DISCUSSION
Plant cell walls are highly organized networks of extended polysaccharides, proteins, and aromatic compounds (Somerville et al., 2004). Cellulose microfibrils, cross-linked by hemicelluloses, represent the main scaffolding elements in this matrix, creating a strong but flexible polysaccharide framework (Cosgrove, 2005). However, many questions regarding cellulose synthesis and its interactions with other cell wall glycans remain unresolved. In this work, we describe the role of CTL1 and CTL2 in cellulose biosynthesis. We propose that the CTLs influence the cellulose assembly and that they are involved in cross-linking of glucan-based polysaccharides (i.e., cellulose and hemicelluloses).
Several lines of evidence, including coexpression, phenotypic resemblance, and reduced cellulose content, have suggested a functional link between the cell wall CESAs and the CTLs (Hauser et al., 1995; Zhong et al., 2002; Mouille et al., 2003; Zhang et al., 2004). Using live-cell imaging, we found that mutations in CTL1 reduced the velocities of the primary wall CSCs (Figure 1; see Supplemental Movie 1 online), indicative of reduced cellulose deposition in the mutant cell walls (Figure 1D). However, if the CTL1 is necessary for cellulose production in the primary cell wall, we would anticipate that complete removal of its activity would lead to pollen lethality (Persson et al., 2007b). CTL1 has only one close homolog in Arabidopsis, CTL2, and it is therefore plausible that the latter could execute CTL-related functions in the absence of CTL1. Indeed, the complementation of the ctl1 mutant by CTL2 under the CTL1 promoter showed that CTL1 and CTL2 are functionally equivalent (Figure 7). Thus, it is likely that CTL2, at least partially, could take over the role of CTL1 in the ctl1 plants, although CTL2 expression levels do not increase in ctl1 mutants compared with the wild type (Figure 7B). Generation of ctl1 ctl2 double mutants should in theory abolish CTL activity in the plant and therefore also remove CTL function during cellulose production. However, although the double mutant displayed additive phenotypes, for example, reduced crystalline cellulose content in stems (Figure 9, Table 1; see Supplemental Figure 5 online), it was viable and fertile. These data suggest that the function of CTL1 and CTL2 is not essential for cellulose synthesis per se, but rather that they may work in conjunction with the primary and secondary CSC, respectively, to contribute to the cellulose structure.
CTL1 localized to Golgi-derived bodies (Figure 2A; see Supplemental Movie 3 online) and in a heterogeneous population of vesicles, CTL1v, that move along the actin cytoskeleton (Figures 2B to 2H; see Supplemental Movies 2 to 4 online). The behavior of these vesicles resembled a population of small CESA-containing vesicles (Crowell et al., 2009; Gutierrez et al., 2009). Indeed, visualization of the CTL1 together with a fluorescently labeled CESA6 showed an interesting colocalization pattern of the unknown vesicles, a pattern that was enhanced upon stress treatment (Figure 6; see Supplemental Movie 6 online). While we could not determine the exact identity of these vesicles, we can discard the endocytic nature of CTL1v based on the FM4-64 staining behavior of CTL1v (Figure 5A; see Supplemental Movie 5 online). In addition, CTL1:GFP was also visible and immunodetected in the cell wall, confirming secretion of the protein to the apoplast and cell wall (Figure 4).
ELISA and immunoblot assays of heterologously expressed CTL1 and CTL2 revealed that the proteins bind commercial glucan-based substrates and the cellulose- and XG-enriched fractions (insoluble and 4 M KOH fractions, respectively) of Arabidopsis etiolated cell walls (Figures 8; see Supplemental Figure 6 online). However, we could not detect any in vitro hydrolytic activity for the CTLs. One explanation could be that the CTLs modify glucan-based polysaccharide precursors. If this is the case, the CTLs could cut certain branches that are necessary for the production of the oligomers. This hypothesis is supported by the large amount of CTL1 in the endomembrane system (Figures 3A and 4A) and may explain why we could not detect any hydrolytic activity of cell wall–related substrates. Given that CTLs bind to glucan-based polymers, the CTLs could modify XGs during trafficking to the cell wall. Such activity could alter the XG structure and, perhaps, their ability to interact with cellulose in the apoplast. We did observe structural changes of the XG in the ctl1 mutant after cell wall digestion with XEG (see Supplemental Figures 8A to 8C online). However, the alterations shown in the relative peak area followed the same XEG hydrolytic pattern as those observed in seedlings grown on the cellulose inhibitor isoxaben (see Supplemental Figures 8C and 8F online). Therefore, this indicates that the alteration of the XEG digestion patterns in the ctl1 mutant probably is due to modifications of the cellulose. In addition, CTL2 appears to be present during secondary wall synthesis. Since the major hemicellulose in the secondary wall is xylan, which has a xylose-based backbone and is not recognized by CTLs in the in vitro assay (Figure 8; see Supplemental Figure 6 online), we again find it unlikely that the CTLs act on the XGs and favor a functionality associated with cellulose. Such a scenario is further corroborated by the tight coexpression of CESAs and CTLs in Poaceae species (see Supplemental Table 1 online), which largely lack XGs (Sarkar et al., 2009).
A more plausible scenario is that the CTLs bind to emerging cellulose microfibrils in the apoplast. The CTLs could then affect either the crystallinity of the cellulose microfibrils or the association of hemicelluloses with amorphous regions. The latter could in turn also affect structural features of the cellulose. Consequently, the hemicellulose-cellulose network domains might change, and this may alter the ability of, for example, the XEG to digest XGs. In this case, the high amount of CTL1 observed in cytoplasmic compartments may signify a high rate of synthesis and recycling of the proteins, similar to what is observed for the primary wall CESAs. CTL activity would then be likely to affect the crystalline cellulose content and the cellulose-hemicellulose interaction. In fact, we did observe a significant reduction in the crystalline cellulose content in the ctl double mutants (Figure 9). Previous in vitro results have shown that a decrease in cellulose crystallinity causes an increase in XG binding capacity to the cellulose (Chambat et al., 2005). Consistent with this and with the cellulose-hemicellulose interaction model suggested by Pauly et al. (1999a), we observed a greater degree of hemicellulose-related sugars in the insoluble cell wall fraction in the ctl1 ctl2 mutant (Table 1). Whereas we did not observe any changes in the x-ray pattern of the ctl1 or ctl2 single mutant cell walls compared with the wild type (see Supplemental Figure 7A online), it is important to note that the method most likely is unable to detect small changes in crystalline cellulose content. Thus, although each ctl mutation may contribute to the reduction in fibril assembly, only impairment of both proteins may result in a strong enough effect to be detected by x-ray diffraction. Consistent with this, the biochemical analyses showed that the composition of ctl1 and ctl2 cell walls appears to be intermediate to the wild type and the double mutant (see Supplemental Figure 7 online; Table 1).
Biophysical models indicate that the microfibril polymerization and crystallization might drive the movement of the CSCs in the plasma membrane (Diotallevi and Mulder, 2007). Thus, impairment of cellulose fibril assembly could alter the speed of CSCs, which may explain the reduced CESA6 velocities observed in ctl1-1 (Figure 1). Similar retardation in the CSC movement in the plasma membrane was observed in a kor1 mutant (Paredez et al., 2008). Interestingly, KOR1 also plays a role in cellulose crystallinity, presumably by increasing the amount of amorphous cellulose (Takahashi et al., 2009). Both kor1 and ctl1 show dramatically reduced cellulose synthesis activity compared with the wild type as quantified by the relative incorporation of 14C-Gluc into the cellulosic fractions (Mouille et al., 2003). Perhaps CTL1 and KOR1 work in conjunction during cellulose synthesis to proofread the assembly of the cellulose microfibrils. In addition, mutations in both KOR1 and in CTL1 caused microtubule disorganization, presumably through a feedback mechanism between the CSC and the microtubules (Paredez et al., 2008).
In conclusion, we propose that the CTLs can bind to glucan chains, presumably to emerging cellulose microfibrils, and that these interactions affect the cellulose microfibril polymerization, which in turn modulates interactions between the cellulose and hemicelluloses.
METHODS
Plant Material and Genetic Analysis
Arabidopsis thaliana plants were grown as described by Persson et al. (2005). For confocal microscopy, plants were grown as described by Gutierrez et al. (2009). Insertion lines for ctl1-1 (At1g05850; SALK_093049) and ctl2-1 (At3g16920; SALK_055713) were obtained from the ABRC (https://abrc.osu.edu/resources; Alonso et al., 2003). Primers used for PCR to obtain homozygous insertion lines are listed in Supplemental Table 3 online. The irx1 mutant (Turner and Somerville, 1997) and prc1-1 mutant (Fagard et al., 2000) were previously described. The pom1-9 mutant line was obtained in the lab of M.-T.H. pCESAs:GUS (Persson et al., 2007a), pCESA6:YFP:CESA6 (Paredez et al., 2006), GFP:MAP4 (Marc et al., 1998), N7-GFP, and VHA-a1:mRFP lines (Dettmer et al., 2006) were previously described.
The PCESA6:tdTomato:CESA6 (Sampathkumar et al., 2012) line was kindly provided by R. Gutierrez (Carnegie Institution for Science, Stanford, CA).
Constructs
A genomic DNA fragment extending 1.4 kb upstream of the ATG starting codon for CTL1 was amplified (see Supplemental Table 3 online) and inserted using HindIII and NcoI in front of the GUS gene of the pCAMBIA1305:GUS-Plus vector (http://www.cambia.org/daisy/cambia/585.html), generating pCTL1:GUS. The construct was transformed into wild-type plants by Agrobacterium tumefaciens–mediated transformation. Transgenic plants were selected on hygromycin and used for GUS reporter analyses.
For the CTL1/POM1:GFP construct, a genomic fragment of the accession Col was amplified with primers harboring a NcoI site (see Supplemental Table 3 online) and inserted in front of the GFP gene with the NcoI site of the pGreenII0029:35S:GFP_RL vector (Hellens et al., 2000). The construct was transformed into pom1-9, which has a nonsense mutation leading to a stop at amino acid 69. Kanamycin was used for selection and segregation analysis for single insertions lines that were further characterized for complementation and microscopy analyses.
An mCherry clone was amplified from the PGWB2CHE vector as template (see Supplemental Table 3 online), which was a kind gift from Thierry Desprez (Institut National de la Recherche Agronomique, Centre de Versailles, Grignon, France). The amplified fragment was digested using BamHI and SpeI and ligated into the pCAMBIA1390 vector (Hajdukiewicz et al., 1994; http://www.cambia.org/daisy/cambia/585.html), resulting in the vector pCAMBIA1390:mCherry. A CTL1 genomic clone including the complete coding region and 1.4 kb upstream of the ATG start codon was amplified from genomic wild-type DNA with primers harboring a SalI and a BamHI restriction site, respectively (see Supplemental Table 3 online). After digestion with those enzymes, the fragment was ligated either into pCAMBIA1390, generating pCAMBIA1390:CTL1, or into pCAMBIA1390:mCherry, generating pCAMBIA1390:CTL1:mCherry. The constructs were transformed into ctl1-1 plants and selected on hygromycin.
For the mCherry:FABD construct, mCherry was amplified from mCherry:TUA5 (Gutierrez et al., 2009) using primers containing KpnI and AscI restriction sites (see Supplemental Table 3 online). The PCR product was digested, purified, and cloned into pMDC43 (Curtis and Grossniklaus, 2003) previously digested with the same enzymes. FABD was amplified from pCBGFP:ABD (Ketelaar et al., 2004) using primers containing Gateway recombination sequences (see Supplemental Table 3 online) and recombined into pDONR221 (http://products.invitrogen.com/ivgn/product/12536017). The mCherry:FABD plasmid was generated by a Gateway LR reaction. The construct was transformed into wild-type plants by Agrobacterium-mediated transformation.
For the pCTL1:CTL2 construct, a genomic CTL2 clone was generated from wild-type DNA (see Supplemental Table 3 online) digested with BspHI and MluI and ligated into the pCTL1:GUS vector previously digested with NcoI (BspHI compatible) and MluI. The construct was transformed into ctl1-1 plants by Agrobacterium-mediated transformation.
For the constructs used for heterologous expression in yeast, the CTL1 and CTL2 cDNAs were cloned from mRNAs extracted from Arabidopsis leaf and stem material using a one-step RT-PCR kit (see Supplemental Table 3 online). The products were digested with PmlI and XbaI and cloned into pPICZαC (Invitrogen).
Drug Treatments
Seedlings were immersed in 2 mL of water containing different drugs or sugars in 12-well cell culture plates in darkness and were subsequently imaged as described by Gutierrez et al. (2009). For testing the stress response to isoxaben (Dr. Ehrenstorfer GmbH) and oryzalin (Dr. Ehrenstorfer GmbH), the seeds were directly germinated on plates containing 2 nM isoxaben or 170 nM oryzalin, respectively.
Microscopy
Light microscopy was performed using a Leica stereomicroscope (Leica MZ12.5 and Leica DFC420 digital camera). Seedlings expressing GFP:MAP4, Fimbrin:GFP, mCherry:TUA5, CTL1:GFP, CTL1:mCherry, and dual-labeled lines of CTL1:GFP and tdTomato:CESA6, VHA-a1:mRFP, or mCherry:FABD were grown and mounted as described by Gutierrez et al. (2009) and imaged on a confocal microscope equipped with a Yokogawa spinning disc head fitted to a Nikon Ti-E inverted microscope and mounted on the microscope stage as described by Gutierrez et al. (2009)
Environmental scanning electron microscopy was performed as described by Persson et al. (2007a).
All images were processed using ImageJ software (W.S. Rasband, National Institutes of Health). Background correction was performed using the “subtract background” tool (rolling ball radius 30 to 50 pixels), and StackReg was used to correct for focus drifts. YFP:CESA6 particle dynamics and velocities were measured as by Paredez et al. (2006).
Expression of CTL1 and CTL2
The expression of CTL1 in different organs and tissues was studied using the GUS reporter gene of PCTL1:GUS transgenic plants in GUS staining solution and observed as described by Persson et al. (2007b).
Quantitative RT-PCR were performed using primers in Supplemental Table 3 online. The expression of the GAPDH gene was used as a cDNA synthesis control (GAPDH_600b and GAPDH_3end) and as amplification control (GAPDH_3end). Gene expression analysis was determined as described by Czechowski et al. (2005)
CTL1 Apoplastic Quantification
The secretion of CTL1 to the apoplast was confirmed adapting the method already published (Rico and Preston, 2008). To harvest the AWF, 16-d-old seedlings cultivated on Murashige and Skoog 2.5 were infiltrated with distilled water. After carefully drying the surface, the seedlings were centrifuged for 5 min, 4°C, 800g. For the remaining plant material fraction (REM), proteins were extracted with buffer (25 mM potassium phosphate buffer, pH 7.5, 1 mM EDTA, 1 mM DTT, 5 mM phenylmethylsulfonyl fluoride, and 1× plant proteinase inhibitor mix) from the infiltrated seedlings. Protein concentrations were quantified according to the manufacturer’s protocol of the Qubit protein assay system (Invitrogen). Equal amounts of AWF and REM proteins were resolved on homemade 10% SDS-polyacrylamide gels and transferred to 0.45 μm polyvinylidene fluoride membranes in a tank blotting apparatus (Bio-Rad) using a 50 mM Tris, 50 mM boric acid buffer, pH 8.3, at 30 V, 4°C, overnight. Membranes were blocked in 5% nonfat milk in TBS-T (20 mM Tris-HCl, pH 7.6, and 137 mM NaCl) for 60 min. The GFP-labeled proteins were detected with 1:2000 mouse α-GFP primary antibody (Roche) in TBS-T for 60 min at room temperature. After three washes with TBS-T for 10 min each, the membranes were incubated with 1:10,000 goat α-mouse IgG horseradish peroxidase–conjugated secondary antibody (Pierce) in TBS-T for 60 min. After repetition of the washing steps, Roti-Lumin (Carl Roth) was used for chemiluminescent detection in a ChemiDoc XRS molecular imager (Bio-Rad). Membranes were stripped using Roti-Free ready-to-use stripping buffer (Carl Roth) and used to detect actin with 1:1000 mouse α-actin primary antibody (Affinity Bioreagents) following the same procedure as above. Immunoblot signals were quantified with the Quantity One software (Bio-Rad).
Cell Wall Biochemical Analyses
Cell wall monosaccharides were assayed after hydrolysis with 2 M TFA as alditol acetate derivatives (Neumetzler, 2010) using a modified protocol from Albersheim et al. (1967) by gas chromatography performed on an Agilent 6890N GC system coupled with an Agilent 5973N mass selective detector. Myo-Inositol was added as an internal standard. Cellulose was determined on the fraction resistant to extraction with 2 M TFA by Seaman hydrolysis (Selvendran and O’Neill, 1987) using Glc equivalents as standard. The hexose content was determined with the anthrone assay (Dische, 1962). Uronic acids were colorimetrically quantified using the soluble 2 M TFA fraction using 2-hydroxydiphenyl as reagent (Vilím, 1985) using galacturonic acid as standard (Filisetti-Cozzi and Carpita, 1991). Statistical significance was determined using Student’s t test.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight
Crude cell wall material from 6-d-old etiolated seedlings of the mutant and wild type was treated with a xyloglucanendohydrolase (0.02 units; Pauly et al., 1999b) as described by Neumetzler (2010) via matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (Lerouxel et al., 2002). The analysis was performed with a Voyager DE-Pro matrix-assisted laser desorption ionization–time-of-flight instrument (Applied Biosystems) using the positive reflectron mode with an acceleration voltage of 20 kV and a delay time of 350 ns. For the downstream data analysis, the relative peak areas of the spectra from a control and a sample set were compared in a pairwise fashion. Alterations were proven for statistical significance using Student’s t test.
X-Ray Diffraction Analysis
Arabidopsis stems were washed in chloroform for ~5 s to remove cuticular waxes that would cause strong Bragg peaks superimposing the cellulose peaks. The stems were then dried at 60°C for 36 h. The first centimeter from the rosette was used for the analysis.
Wide-angle x-ray diffraction experiments were performed on a Nanostar instrument (Bruker AXS) using CuKα radiation with a wavelength λ = 0.154 nm. The samples (n = 5) were placed in vacuum at a sample detector distance of 5 cm, and two-dimensional scattering patterns were collected in with a position sensitive area detector (HiStar) and exposure times of 60 min.
The two-dimensional scattering images were integrated azimuthally to obtain radial intensity profiles for structural analysis of cellulose regarding crystallite size and crystalline cellulose content. Prior to the structural analysis, a texture correction was applied to the radial profiles to account for the cylindrical geometry of the cellulose microfibril. Structural analysis was performed by DFA using Debussy software (Cervellino et al., 2010). This software creates sampled interatomic distance databases based on the atomic positions in the cellulose crystal lattice as given by Nishiyama et al. (2003). With these databases, radial profiles can be simulated. By varying structural parameters, such as size distribution of the cellulose crystallites and contribution of diffuse background scattering, the measured radial profile is fitted. Radial integration within the 2θ range of the (200) Bragg peak of cellulose led to azimuthal intensity profiles that gave information on cellulose orientation.
Protein Expression
Pichia pastoris cells were grown in 50 mL BMGY (buffered minimal glycerol complex medium) in a 250-mL flask in an orbital shaker (200 rpm) at 29°C for 36 h, and cells were diluted with 500 mL of MMY (minimal methanol complex medium) and incubated for 3 d in a 2.8-liter flask with daily addition of 3 mL of methanol (Bauer et al., 2005). The suspension was centrifuged (10 min, 5000g), and the pH of the supernatant was adjusted to pH 7.5 to 7.8 and filtered (0.22 μm polyethersulfone). After addition of 5 mL of nickel-NTA agarose (Qiagen), the suspension was stirred overnight at 4°C on a magnetic stirrer. The resin was collected by centrifugation (5 min, 5000g) and transferred into a glass column and washed with 3 × 5 mL of buffer A (500 mM NaCl and 20 mM Tris-Cl, pH 7.9) containing 20 mM imidazole. The enzyme was eluted with 3 × 5 mL of 250 mM imidazole in buffer A, pH 7.9. The eluate was concentrated in centrifugal units (Amicon; 10 kD cutoff) and repeatedly washed with water (4000g, 4°C) to remove imidazole. Protein content was measured using a Nanodrop spectrophotometer (NanoDrop ND-800; Thermo Scientific), and A280 was calculated into concentration in milligrams/milliliter.
ELISA
Substrates (see Supplemental Table 2 online; each 50 μg/mL in 50 mM sodium carbonate, pH 9.6) were incubated (100 μL/per well) overnight with moderate agitation (100 rpm) in 96-well EIA/RIA flat-well high binding plates (Costar). For the blanks, only sodium carbonate solution was used. All substrates and the blanks were run in duplicate. Substrate and blank solutions were replaced with 300 μL 5% milk (fat-free, in TBS [10 mM Tris-Cl, 150 mM NaCl, pH 7.4, containing 0.1% Tween 20]) and incubated for 6 h with moderate agitation (100 rpm). The solutions were removed, and the wells were washed with TBS. For assays and enzyme blanks, 100 μL protein in 5% milk was used per well. For substrate blanks, only 100 μL of 5% milk were used. After an overnight incubation at 4°C with moderate agitation (100 rpm), wells were washed extensively with TBS and incubated overnight at 4°C with rabbit anti-c-myc antibody (Sigma-Aldrich) in 5% milk, extensively washed again with TBS, and incubated overnight at 4°C with ImmunoPure goat anti-rabbit secondary antibody fused to horseradish peroxidase (rabbit IgG [H+L]; Thermo Scientific) in 5% milk and washed extensively with TBS. Wells were incubated with 150 μL tetramethylbenzidine (Sigma-Aldrich), and the absorbance at 450 nm (after stopping the reaction by adding of 35 μL 2 n sulfuric acid after 30 min) was read using a plate reader (Paradigm Detection Platform; Beckman Coulter). All commercial substrates were from Sigma-Aldrich except rye arabinoxylan, galactan (lupin), XG (tamarind), rhamnogalacturonan I (from potato [Solanum tuberosum]), rhamnogalacturonan I (from soybean [Glycine max]), pectic galactan (lupin), and isoprimeverose, which were purchased from Megazyme.
Carbohydrate Microarrays
Cadoxen was prepared as described in Fry (1988). Substrate stock solutions were prepared at a concentration of 1 mg/mL in Dulbecco’s PBS buffer (pectin esterified from citrus, pectin from apple [Malus domestica], soybean rhamnogalacturonan, xylan from birchwood, XG, lichenan, chitosan medium molecular weight, chitin from crab shells, and carboxymethyl cellulose) or Cadoxen (cellulose fibrous long, Sigmacell cellulose type 20, avicel PH-101, 4 M KOH fraction plant cell wall, and insoluble fraction plant cell wall) and diluted to 0.5 mg/mL and 0.1 mg/mL with PBS. One microliter of each diluted solution was spotted in duplicate on a nitrocellulose membrane (Protran; Whatman). Membranes were incubated at room temperature in 5% milk for 1 h and then incubated with antibodies (LM5 [1:50], LM11 [1:20], or JIM5 [1:50]; all obtained from Paul Knox’s lab; http://www.plantprobes.net/; Jones et al., 1997; Clausen et al., 2003; McCartney et al., 2005) for 2 h at room temperature or with CTL1 (0.007 mg/mL) or CTL2 (0.011 mg/mL) in 5% milk at 4°C overnight. After extensive washing with TBS, membranes were incubated with anti-rat IgG peroxidase produced in rabbit (Sigma-Aldrich) for LM5 and JIM5 or peroxidase-conjugated AffiniPure goat anti-rat IgG (H+L) for LM11 or successive rabbit anti-c-myc (Sigma-Aldrich) and ImmunoPure goat anti-rabbit secondary antibody fused to horseradish peroxidase (rabbit IgG [H+L]; Thermo Scientific) for CTL1 and CTL2 at room temperature each for 2 h and then incubated with SuperSignal West Pico Chemiluminescence solutions (Pierce). Chemiluminescence was measured using an LAS-4000 device (Fujifilm)
Capillary Electrophoresis
Mixtures containing 50 μL acetate buffer (100 mM, pH 5.5), 30 μL substrate (1 mg/mL), and 20 μL protein solution (10 μg/mL) were incubated at 37°C overnight. For substrate blank, the protein solution was replaced by 20 μL of water. The mixture was centrifuged (10 min, 16,000g), and 50 μL of the supernatant was evaporated (speed-vac, 60°C). The residue was resuspended in 2 μL of 8-aminopyrene, 1,3,6-trisulfonic acid (100 mM in 15% acetic acid), and 5 μL of sodium cyanoborohydride (NaBH3CN, 1 M in dimethylsulfoxide) and kept at 65°C. After 1 h, 50 μL of water was added, and 20 μL of the mixture was diluted with 180 μL of water and analyzed using capillary electrophoresis (PA800; Beckman Coulter) equipped with a 30-cm eCap capillary (75 μm i.d., 375 μm o.d.; Beckman Coulter) and laser detection (488 nm excitation, 520 nm emission) at 25°C and an applied voltage of 25 kV using carbohydrate separation buffer (Beckman Coulter).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CTL1 (At1g05850), ctl1-1 (SALK_093049), CTL2 (At3g16920), ctl2-1 (SALK_055713), irx1 (At4g18780; Turner and Somerville, 1997), and prc1-1 (At5g64740; Fagard et al., 2000). All the fluorescence-labeled lines included were obtained from the corresponding references indicated.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Analyses of CTL1.
Supplemental Figure 2. Genetic Interactions between ctl1-1 and prc1-1.
Supplemental Figure 3. ctl1-1 Complementation.
Supplemental Figure 4. CTL1 Colocalizes with Oryzalin-Insensitive MASCs/SmaCCs in Secretory Vesicles.
Supplemental Figure 5. ctl1 ctl2 Double Mutant Displays Subtle Additive Phenotypes.
Supplemental Figure 6. Carbohydrate Microarray.
Supplemental Figure 7. X-Ray Diffraction Analysis of 7-Week-Old Stems of Wild-Type Plants and ctl and irx1 Mutants.
Supplemental Figure 8. Oligosaccharide Mass Profiling Analyses of ctl1-1 and Isoxaben-Grown Seedlings.
Supplemental Table 1. Identification of CTL-Like Homologs That Are Coexpressed with Primary and Secondary CESA Genes in Other Plant Species Using PlanNet and BLAST.
Supplemental Table 2. List of Substrates Tested in ELISA Binding Studies with CTL1 and CTL2 and Resulting Relative Absorbance Units.
Supplemental Table 3. Primer Sequences Used for Analyses.
Supplemental Movie 1. YFP:CESA6 Movement in the Plasma Membrane Is Altered in ctl1-1.
Supplemental Movie 2. Dynamics of CTL1 Vesicles at the Cell Cortex.
Supplemental Movie 3. Dynamics of CTL1 Vesicles Are Affected by LatB.
Supplemental Movie 4. CTL1 Vesicles Move through Actin Filaments.
Supplemental Movie 5. CTL1 Localize to Endosome Vesicles Not Transported via VHA-a1 Marked TGN Vesicles.
Supplemental Movie 6. CTL1 Vesicles Colocalize with Tethered MASCs/SmaCCs.
Supplemental Movie 7. CTL1 Colocalizes with Oryzalin-Insensitive MASCs/SmaCCs.
Supplementary Material
Acknowledgments
We thank Norma Funke, Natalia Khitrov, and Alexandra Dotson-Fagerström for assistance with the CTL expression in P. pastoris, maintenance of plants, and advice with the dot-blot binding experiments, respectively. We thank Carl Douglas, Christina Lang-Mladek, Wiebke Öhr, Agata Mansfeld, Marek Mutwil, Araceli Díaz, Anja Thalhammer, and Dirk Hincha for help at various steps of the project. We also thank Tijs Ketellar and Hannie S. van der Honing for the mCherry:FABD line. We thank Ingrid Zenke for her support during the x-ray measurements and Barbara Aichmayer, Michaela Eder, and Peter Fratzl for helpful discussions on the data. This work was supported in part by a grant from the U.S. Department of Energy (DOE-FG02-03ER20133), by the Carnegie Institution for Science and the Energy Biosciences Institute, by the Austrian Science Found (P1477-B12) to M.-T.H., by the Spanish Ministerio de Ciencia e Innovación (2008-0861) to C.S.-R., and by the Max-Planck Gesellschaft to S.P., A.S., F.S., M.R., and L.N.
AUTHOR CONTRIBUTIONS
C.S.-R. and S.P. performed research, analyzed data, and wrote the article. S.B., K.H., and F.S. performed research and analyzed data. A.B.I., V.V., C.K., A.S., E.A., and L.N. performed research. M.R., I.B., and C.S. analyzed data. M.-T.H. analyzed data and wrote the article.
References
- Albersheim P., Nevins D.J., English P.D., Karr A. (1967). A method for the analysis of sugars in plant cell-wall polysaccharides by gas-liquid chromatography. Acer pseudoplatanus tissue culture cells. Carbohydr. Res. 5: 340–345 [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baskin T.I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bauer S., Vasu P., Mort A.J., Somerville C.R. (2005). Cloning, expression, and characterization of an oligoxyloglucan reducing end-specific xyloglucanobiohydrolase from Aspergillus nidulans. Carbohydr. Res. 340: 2590–2597 [DOI] [PubMed] [Google Scholar]
- Bauer S., Vasu P., Persson S., Mort A.J., Somerville C.R. (2006). Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 103: 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Cary A., Uttamchandani S.J., Smets R., Van Onckelen H.A., Howell S.H. (2001). Arabidopsis mutants with increased organ regeneration in tissue culture are more competent to respond to hormonal signals. Planta 213: 700–707 [DOI] [PubMed] [Google Scholar]
- Cervellino A., Giannini C., Guagliardi A. (2010). DEBUSSY: A Debye user system for nanocrystalline materials. J. Appl. Crystallogr. 43: 1543–1547 [Google Scholar]
- Chambat G., Karmous M., Costes M., Picard M., Joseleau J.P. (2005). Variation of xyloglucan substitution pattern affects the sorption on celluloses with different degrees of crystallinity. Cellulose 12: 117–125 [Google Scholar]
- Clausen M.H., Willats W.G.T., Knox J.P. (2003). Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (1997). Assembly and enlargement of the primary cell wall in plants. Annu. Rev. Cell Dev. Biol. 13: 171–201 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2000). Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotallevi F., Mulder B. (2007). The cellulose synthase complex: A polymerization driven supramolecular motor. Biophys. J. 92: 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dische Z. (1962). General color reactions. In Methods in Carbohydrate Chemistry, R.L. Whistler and M.L. Wolfrom, eds (New York: Academic Press), pp. 478–481 [Google Scholar]
- Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filisetti-Cozzi T.M., Carpita N.C. (1991). Measurement of uronic acids without interference from neutral sugars. Anal. Biochem. 197: 157–162 [DOI] [PubMed] [Google Scholar]
- Fry S.C. (2000). The Growing Plant Cell Wall: Chemical and Metabolic Analysis. (Caldwell, NJ: The Blackburn Press), p. 68 [Google Scholar]
- Fry S.C. (1995). Polysaccharide-modifying enzymes in the plant cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 497–520 [Google Scholar]
- Fry S.C., York W.S., Albersheim P. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharide. Physiol. Plant. 89: 1–3 [Google Scholar]
- Fujita M., Himmelspach R., Hocart C.H., Williamson R.E., Mansfield S.D., Wasteneys G.O. (2011). Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J. 66: 915–928 [DOI] [PubMed] [Google Scholar]
- Gardner K.H., Blackwell J. (1974). The hydrogen bonding in native cellulose. Biochim. Biophys. Acta 343: 232–237 [DOI] [PubMed] [Google Scholar]
- Graham L.S., Sticklen M.B. (1994). Plant chitinases. Can. J. Bot. 72: 1057–1083 [Google Scholar]
- Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T.I., Persson S., Somerville C.R. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Haase J., Hosemann R., Renwanz B. (1976). Crystallite size, lattice-defects and long periods of acid hydrolyzed regenerated cellulose. Colloid Polym. Sci. 254: 199–204 [Google Scholar]
- Haigler C.H., Brown R.M. (1986). Transport of rosettes from the golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma 134: 111–120 [Google Scholar]
- Haigler C.H., Ivanova-Datcheva M., Hogan P.S., Salnikov V.V., Hwang S., Martin K., Delmer D.P. (2001). Carbon partitioning to cellulose synthesis. Plant Mol. Biol. 47: 29–51 [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hanus J., Mazeau K. (2006). The xyloglucan-cellulose assembly at the atomic scale. Biopolymers 82: 59–73 [DOI] [PubMed] [Google Scholar]
- Hauser M.T., Morikami A., Benfey P.N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (1989). Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 139–168 [Google Scholar]
- Hayashi T., Ogawa K., Mitsuishi Y. (1994). Characterization of the adsorption of xyloglucan to cellulose. Plant Cell Physiol. 35: 893–899 [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hermans C., Porco S., Verbruggen N., Bush D.R. (2010). Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 152: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C., Porco S., Vandenbussche F., Gille S., De Pessemier J., Van Der Straeten D., Verbruggen N., Bush D.R. (2011). Dissecting the role of CHITINASE-LIKE1 in nitrate-dependent changes in root architecture. Plant Physiol. 157: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P.H., Weidinger A. (1949). Change in crystallinity upon heterogeneous acid hydrolysis of cellulose fibers. J. Pol. Sci. IV: 317–322 [Google Scholar]
- Hossain M.A., Noh H.N., Kim K.I., Koh E.J., Wi S.G., Bae H.J., Lee H., Hong S.W. (2010). Mutation of the chitinase-like protein-encoding AtCTL2 gene enhances lignin accumulation in dark-grown Arabidopsis seedlings. J. Plant Physiol. 167: 650–658 [DOI] [PubMed] [Google Scholar]
- Jakob H.F., Fengel D., Tschegg S.E., Fratzl P. (1995). The elementary cellulose fibril in Picea abies: Comparison of transmission electron microscopy, small-angle X-ray scattering, and wide-angle X-ray scattering results. Macromolecules 28: 8782–8787 [Google Scholar]
- Jones L., Seymour G.B., Knox J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol. 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa K., Edashige Y., Ishii T. (1998). Xyloglucan from xylem-differentiating zones of Cryptomeria japonica. Phytochemistry 47: 767–771 [DOI] [PubMed] [Google Scholar]
- Kaku T., Tabuchi A., Wakabayashi K., Kamisaka S., Hoson T. (2002). Action of xyloglucan hydrolase within the native cell wall architecture and its effect on cell wall extensibility in azuki bean epicotyls. Plant Cell Physiol. 43: 21–26 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Kimura S., Laosinchai W., Itoh T., Cui X., Linder C.R., Brown R.M., Jr (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Kim S.H., Jung M.S., Kim M.S., Oh J.E., Ju H.W., Kim K.I., Vierling E., Lee H., Hong S.W. (2007). Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J. 49: 184–193 [DOI] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., Verbelen J.-P. (2005). Cell elongation and microtubule behavior in the Arabidopsis hypocotyl. Responses to ethylene and auxin. J. Plant Growth Regul. 24: 166–178 [Google Scholar]
- Lerouxel O., Choo T.S., Séveno M., Usadel B., Faye L., Lerouge P., Pauly M. (2002). Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol. 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertpiriyapong K., Sung Z.R. (2003). The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Mol. Biol. 53: 581–595 [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marga F., Grandbois M., Cosgrove D.J., Baskin T.I. (2005). Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J. 43: 181–190 [DOI] [PubMed] [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Mouille G., Robin S., Lecomte M., Pagant S., Höfte H. (2003). Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J. 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Mutwil M., Obro J., Willats W.G., Persson S. (2008). GeneCAT–novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36: W320–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Gallagher L.A., Dunahay T.G., Frohlick J.A., Mazurkiewicz A.M., Meehl J.B., Staehelin L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumetzler L. (2010). Identification and Characterization of Arabidopsis Mutants Associated with Xyloglucan Metabolism. (Berlin: Rhombos-Verlag) [Google Scholar]
- Nicol F., His I., Jauneau A., Vernhettes S., Canut H., Höfte H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. (1998). Construction and restructuring of the cellulose-xyloglucan framework in the apoplast as mediated by the xyloglucan-related protein family - A hypothetical scheme. J. Plant Res. 111: 159–166 [Google Scholar]
- Nishiyama Y., Sugiyama J., Chanzy H., Langan P. (2003). Crystal structure and hydrogen bonding system in cellulose I(alpha) from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 125: 14300–14306 [DOI] [PubMed] [Google Scholar]
- Paakkari T., Serimaa R., Fink H.P. (1989). Structure of amorphous cellulose. Acta Polymerica. 40: 731–734 [Google Scholar]
- Pagant S., Bichet A., Sugimoto K., Lerouxel O., Desprez T., McCann M., Lerouge P., Vernhettes S., Höfte H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho P.A., de Vries S.C. (2002). Arabidopsis chitinases: A genomic survey. The Arabidopsis Book 1: e0023, doi/10.1199/tab.0023 [Google Scholar]
- Pauly M., Albersheim P., Darvill A., York W.S. (1999a). Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M., Andersen L.N., Kauppinen S., Kofod L.V., York W.S., Albersheim P., Darvill A. (1999b). A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Persson S., Caffall K.H., Freshour G., Hilley M.T., Bauer S., Poindexter P., Hahn M.G., Mohnen D., Somerville C. (2007a). The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007b). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G.P., Somerville C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Elumalai R.P., Chory J. (1998). Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 148: 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico A., Preston G.M. (2008). Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant Microbe Interact. 21: 269–282 [DOI] [PubMed] [Google Scholar]
- Rogers L.A., Dubos C., Surman C., Willment J., Cullis I.F., Mansfield S.D., Campbell M.M. (2005). Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 168: 123–140 [DOI] [PubMed] [Google Scholar]
- Roudier F., Fernandez A.G., Fujita M., Himmelspach R., Borner G.H., Schindelman G., Song S., Baskin T.I., Dupree P., Wasteneys G.O., Benfey P.N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A., Lindeboom Jelmer, J., Debolt S., Gutierrez R., Ehrhardt D.W., Ketelaar T., Persson S. (2012). Live-cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23: 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P., Bosneaga E., Auer M. (2009). Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 60: 3615–3635 [DOI] [PubMed] [Google Scholar]
- Scheible W.R., Eshed R., Richmond T., Delmer D., Somerville C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G., Morikami A., Jung J., Baskin T.I., Carpita N.C., Derbyshire P., McCann M.C., Benfey P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Wells B., Dolan L., Roberts K. (1997). Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124: 1789–1798 [DOI] [PubMed] [Google Scholar]
- Selvendran R.R., O’Neill M.A. (1987). Isolation and analysis of cell walls from plant material. Methods Biochem. Anal. 32: 25–153 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Takahashi J., et al. (2009). KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol. 50: 1099–1115 [DOI] [PubMed] [Google Scholar]
- Taylor N.G., Howells R.M., Huttly A.K., Vickers K., Turner S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.G., Laurie S., Turner S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H., Beemster G.T. (2006). Genetics, cell cycle and cell expansion in organogenesis in plants. J. Plant Res. 119: 1–4 [DOI] [PubMed] [Google Scholar]
- Turner S.R., Somerville C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilím V. (1985). Colorimetric estimation of uronic acids using 2-hydroxydiphenyl as a reagent. Biomed. Biochim. Acta 44: 1717–1720 [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S., Somerville S., Somerville C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Whitney S.E.C., Brigham J.E., Darke A.H., Reid J.S.G., Gidley M.J. (1995). ln vitro assembly of cellulose/xyloglucan networks: Ultrastructural and molecular aspects. Plant J. 8: 491–504 [Google Scholar]
- Wightman R., Turner S.R. (2008). A novel mechanism important for the alignment of microtubules. Plant Signal. Behav. 3: 238–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Wu Y., Cosgrove D.J. (2001). A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 127: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Hrmova M., Wan C.H., Wu C., Balzen J., Cai W., Wang J., Densmore L.D., Fincher G.B., Zhang H., Haigler C.H. (2004). Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol. Biol. 54: 353–372 [DOI] [PubMed] [Google Scholar]
- Zhong R., Kays S.J., Schroeder B.P., Ye Z.H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ripperger A., Ye Z.H. (2000). Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 123: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.