Abstract
The loss of telomere function can result in the fusion of telomeres with other telomeric loci, or non-telomeric double-stranded DNA breaks. Sequence analysis of fusion events between short dysfunctional telomeres in human cells has revealed that fusion is characterized by a distinct molecular signature consisting of extensive deletions and micro-homology at the fusion points. This signature is consistent with alternative error-prone end-joining processes. We have examined the role that Mre11 may play in the fusion of short telomeres in human cells; to do this, we have analysed telomere fusion events in cells derived from ataxia-telangiectasia-like disorder (ATLD) patients that exhibit hypomorphic mutations in MRE11. The telomere dynamics of ATLD fibroblasts were indistinguishable from wild-type fibroblasts and they were proficient in the fusion of short telomeres. However, we observed a high frequency of insertion of DNA sequences at the fusion points that created localized sequence duplications. These data indicate that Mre11 plays a role in the fusion of short dysfunctional telomeres in human cells and are consistent with the hypothesis that as part of the MRN complex it serves to stabilize the joining complex, thereby controlling the fidelity of the fusion reaction.
INTRODUCTION
Mammalian telomeres are comprised of TTAGGG repeats terminating in a single-stranded G-rich overhang at the terminus. The shelterin protein complex, which includes six core proteins, protects telomeres from aberrant non-homologous end joining and fusion (1). Telomeres rendered dysfunctional by the removal of TRF2 or by replicative erosion of telomeres are recognized as double-strand breaks (DSBs) activating both ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinase-dependent DNA damage response pathways (2–4). The Mre11 protein constitutes the core of the mammalian Mre11/Rad50/Nbs1 complex (MRN) (5,6) and is required for ATM signalling and the coordination of the DNA damage response to DSBs and telomere dysfunction (7). Mre11 exhibits structure-specific 3′–5′ exonuclease and endonuclease activities and controls the initiation of 5′–3′ resection (8–10). The DNA-binding activity of Mre11 brings DSBs together in order to facilitate repair (8,11,12). Mre11 is thus a key player in the major double DNA break repair pathways: homologous recombination and non-homologous end joining. Mre11 activity has also been implicated in end processing for micro-homology-mediated end joining (MMEJ). MMEJ is an error-prone Ku-independent non-homologous end-joining pathway that can lead to large-scale deletion events, presumably as a consequence of extensive nucleolytic resection of the broken ends (13–16). These alternative end-joining (A-EJ) processes are also associated with chromosome translocations and rearrangements in the absence of p53 (17) as well in class switch recombination (CSR) (18,19). The absence of Mre11 results in aberrant switch recombination junctions following CSR with increased micro-homology and additional sequences inserted at the junction point (20).
Telomere fusion is observed following the experimental abrogation of the shelterin component TRF2; in this situation telomere fusion is immediate and dependent on the key components of NHEJ include ligase IV and DNA-PK (21,22). Gradual replicative telomere erosion, as well telomeric deletion, results in short dysfunctional telomeres that can be subjected to fusion with other telomeres or non-telomeric double-stranded DNA breaks. This has been observed in experimental situations following the abrogation of the p53-dependent checkpoint responses where complete telomere loss leads to extensive telomere dysfunction and fusion (23,24). Telomere fusion has also been observed sporadically in normal cells (24) and during neoplastic progression (25). Direct sequence analysis of fusions derived from normal cells, cells undergoing crisis and cells derived from normal and neoplastic tissues has revealed a distinct mutational profile that accompanies telomere fusion (24–26). This consisted of significant micro-homology, similar to that observed at CSR junctions (19) and a G:C bias at the fusion point. Strikingly, all fusion events involved the deletion of one or both of the participating telomeres. The deletion events extended into the sub-telomeric DNA up to the limit of the assays (6 kb), the profile of sub-telomeric deletion indicated that deletion may extend beyond the limit of these assays (24,26). The mutational profile that accompanies the fusion of short dysfunctional telomeres in human cells is consistent with Ku-independent error-prone A-EJ processes. This is also supported by observations in Arabidopsis, mice and yeast that show short telomeres can undergo fusion in the absence of functional NHEJ (27–29). Given the role that Mre11 plays in end joining and its possible involvement with A-EJ processes, here we have investigated its role in mediating the fusion of short dysfunctional telomeres in human cells. To do this we have analysed telomere fusion events in cells derived from ataxia telangiectasia-like disorder (ATLD) patients that exhibit hypomorphic mutations in MRE11. The mutation in ATLD3/4 is characterized by full-length mutant hMre11 expressed from paternal allele with a miss-sense mutation at 350A/G, resulting in a 117 N/S amino acid change, and nonsense-mediated mRNA decay operating on the transcripts from the maternal allele (30,31). ATLD cells display impaired activation of the S phase DNA damage checkpoint, exhibit hypersensitivity to ionizing radiation and show an increased level of chromosome translocations in the peripheral blood (30–32). Here, we describe an analysis of telomere dynamics, replicative lifespan and fusion events in ATLD3/4 cells.
MATERIALS AND METHODS
Cell culture
ATLD1, 2, 3 and 4 cells (31) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin, 100 mg/ml streptomycin (Invitrogen) and 2 mM L-glutamine (Invitrogen). Primary cultures were infected at PD 7 (ATLD2 and 3) and at PD11 (ATLD4) with amphotropic retroviral vectors expressing HPV 16 E6E7 oncoproteins and selected with G418 (0.4 mg/ml) as described previously (33). Cells also were infected with retroviral empty vector pBabeNeo and used as a negative control in our study.
DNA extraction and single telomere length analysis
Genomic DNA from cells was extracted using standard proteinase K/phenol/chloroform protocols (34). For telomere length analysis at 17p and XpYp, including allele-specific XpYp analysis, we used the modified STELA protocol (24,34).
Telomere fusion assay
For analysis of telomere fusions we used a protocol and oligonucleotides described previously (24,26). Polymerase chain reaction (PCR) reactions with 17p6, XpYpM and 21q1group primers were performed to detect fusion products between 17p, XpYp and 21q group chromosomes with between 10 and 40 ng of DNA per reaction. All subsequent Southern blot hybridizations were carried out with random-primed [α33P]-labelled XpYp, 17p and 21q telomere-adjacent probes (24,26).
RESULTS
Telomere dynamics in ATLD cells
To study the role of Mre11 in telomere fusion we used primary fibroblasts cultures derived from ATLD patients. ATLD1, 2 3 and 4 cells express wild-type p53 and pRb (31). In order to inactivate the function of these major tumour suppressor genes, bypass replicative senescence and facilitate continued telomere erosion, the ATLD cells were transfected with human papilloma virus (HPV) 16 E6E7 oncoproteins. The abrogation of p53 and pRb extended the replicative life span by up to 19–24 PDs for ATLD4 E6E7 and ATLD3 E6E7, respectively. The empty vector controls underwent replicative senescence at PD15 in ATLD3 and at PD16 in ATLD4 (Supplementary Figure S1). The bulk population of cells reached crisis at PD27 (ATLD3) and PD30 (ATLD4), while the four clonal populations of cells surpassed them, approaching crisis at a maximum of PD 39 (ATLD 3) and PD 35 (ATLD 4, Supplementary Figure S1). ATLD 1 and 2 cells proliferated poorly in culture achieving just 0.8 PD and 7.4 PD, respectively, over a 90-day period; HPV E6E7 transfection of these cultures did not provide any evidence of life-span extension (data not shown). The lack of proliferation precluded an examination of telomere dynamics and fusion in ATLD 1 and 2 cells.
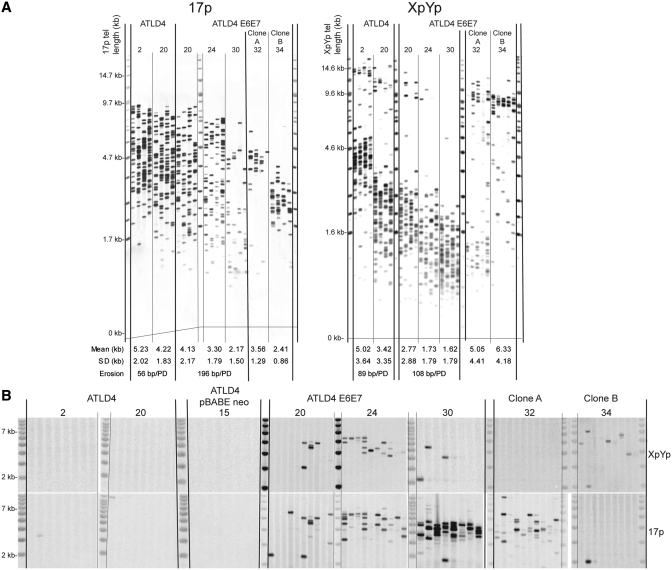
Telomere dynamics were monitored in ATLD 3 and 4 cells using single telomere length analysis (STELA), a high-resolution single-molecule approach to determine telomere length at specific chromosome ends (24,34,35). Analysis of the XpYp and 17p telomere revealed that the telomere dynamics of ATLD 3 and 4 fibroblasts were indistinguishable from those observed in normal primary diploid fibroblast cell strains. The mean rate of erosion at the 17p and XpYp telomere was 73 and 97 bp/PD for ATLD 3 and 4, respectively (Figure 1A and Supplementary Figure S2A) within the range of erosion rates in normal fibroblast cultures (63±39 bp/PD, mean ± SD) documented previously using STELA (34). The expression of HPV E6E7 facilitated ongoing cell division and continued telomere erosion beyond the point of replicative senescence. Consistent with the telomere dynamics observed in cell populations entering crisis (24), the ATLD E6E7 expressing cells displayed an increased rate of telomere erosion (Figure 1A and Supplementary Figure S2A).
Figure 1.
Telomere dynamics and fusion in ATLD4 cells. (A) XpYp and 17p STELA, each sample is analysed with four separate reactions; the population doubling (PD) point of the culture is indicated above each gel. Mean telomere length, standard deviation and rate of telomere erosion as a function of PD is indicated below the gel. (B) Telomere fusion analysis, single molecule fusion events are detected by southern hybridization with the XpYp and 17p telomere-adjacent probes as indicated on the right. The PD points of the cultures are indicated above.
The extent of telomere erosion indicated that we would be able to detect fusion of critically eroded telomeres in ATLD cells. To characterize fusion events we used a highly specific and sensitive single-molecule telomere fusion assay allowing the identification and sequence characterization of telomere fusion events between the XpYp, 17p and a family of telomeres related to the 21q telomere which includes the following chromosomes: 21q, 1q, 2q, 5q, 6q, 6p, 8p, 10q, 13q, 17q, 19p, 19q and 22q (24,26). We did not detect fusion in ATLD 3 or 4 cells in the early stages of the replicative life span of the culture. However, with ongoing cell division beyond the point of replicative senescence there was an increasing frequency of fusions as both the ATLD 3 and 4 cells expressing HPV E6E7 approached crisis (Figure 1B and Supplementary Figure S2B). In ATLD3 cells the highest frequency of fusions was observed at crisis with a maximum frequency of 3 × 10−4 which was similar to the frequencies of fusion observed in MRC5 fibroblasts undergoing crisis (26). The frequency of fusion in ATLD4 E6E7 was higher, with a maximum frequency of 2.3 × 10−3; this was over 5× higher than that observed in MRC5 cells (26). Fusion was also observed in the clonal E6E7 populations and the pattern of fusion events appeared to match the telomere dynamics, with the appearance of fusion events coinciding with the apparent loss of telomeric alleles (Figure 1B and Supplementary Figure S2).
Taken together, these data indicate that ATLD cells exhibit normal rates of telomere erosion and that telomere erosion to critical lengths results in telomere–telomere fusion.
Telomere fusion in ATLD cells
In order to further examine telomere fusion in ATLD cells we purified and characterized the DNA sequence of a total of 102 independent telomere fusion events in both ATLD3/4 E6E7 cells. This allowed us to examine the mutational profile that accompanies telomere fusion in the ATLD fibroblast cultures and compare this to similar data sets derived from normal human fibroblast cultures (24,26).
TTAGGG repeat content, micro-homology and sub-telomeric deletion
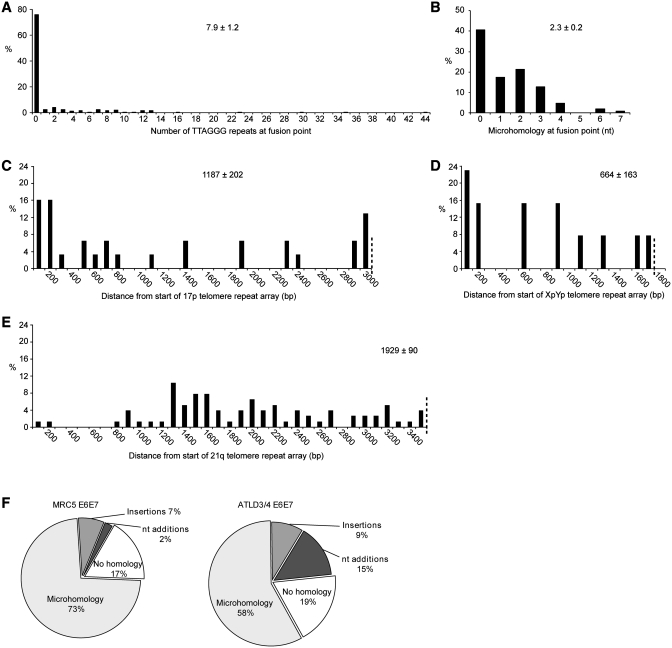
Of the fusion events characterized from ATLD3/4 E6E7 cells 54% did not contain telomere repeats (TTAGGG)n immediately adjacent to the fusion point (Figure 2A). Among those that did contain TTAGGG repeats, the length ranged from 0.17 to 43.17 repeats with a mean 7.9±1.2 (SE), this was not significantly different from that observed using the same assays in MRC5 with wild-type Mre11 (P = 0.37) (26). Again, consistent with our previous data in Mre11 wild-type cells (P = 0.25, Χ2), the ATLD3/4 cells exhibited micro-homology at the fusion point in 58% of the total number of fusion events (Figure 2B) which was less than that observed in Mre11 wild-type cells (73%, Figure 2F). The mean length of micro-homology was 2.3 nucleotides, ranging from 1 to 7 nucleotides, this distribution was not significantly different from Mre11 wild-type cells (P = 0.25, Χ2). All fusions in ATLD3/4 E6E7 cells involved the deletion of one or both telomeres with deletion extending into the telomere-adjacent DNA up to the limits of the assays (3.4 kb from the telomere repeat array), again the extent of sub-telomeric deletion was not significantly different from Mre11 wild-type cells (P = 0.54 for XpYp and P = 0.08 for 17p) (Figure 2C–E).
Figure 2.
Illustrating the mutational profile that accompanies telomere fusion in ATLD 3/4 cells. (A) The frequency of TTAGGG repeats at the fusion point. (B) Micro-homology at the fusion point. (C–E) Representing the distance of sub-telomeric deletion from the start of the telomere repeat arrays of 17p, XpYp and 21q. The extent of sub-telomeric deletion that can be detected with the assays is indicated by a dotted line across the x-axis. The means ± standard error are indicated on each panel. (F) Pie chart displaying the proportions of telomere fusion events displaying sequence homology, nucleotide insertions and identifiable insertions at the fusion point in MRC5 and ATLD 3/4 cells.
Thus, compared to that observed in Mre11 wild-type cells, telomere fusion in ATLD cells appears to involve telomeres within the same length range and there was no detectable difference in the profile of telomere fusion in terms of micro-homology and sub-telomeric deletion.
Insertion
We have previously documented that telomere fusion can result in the insertion of sequences from identifiable non-telomeric loci between the two telomeres involved in the fusion events; these constitute 7–9% of fusion events in Mre11 wild-type and ATLD cells, respectively (Figure 2F) (24,26). However, in ATLD cells we observed an additional class of event that resulted in the insertion of short tracts of apparently random nucleotides. We detected these insertions in 15 out of 102 fusion events sequenced (15%) in ATLD cells, a significant increase on that observed in mre11 wild-type cells where just 3 out of 139 events sequenced where of this type (2%, P < 0.001, Figure 2F).
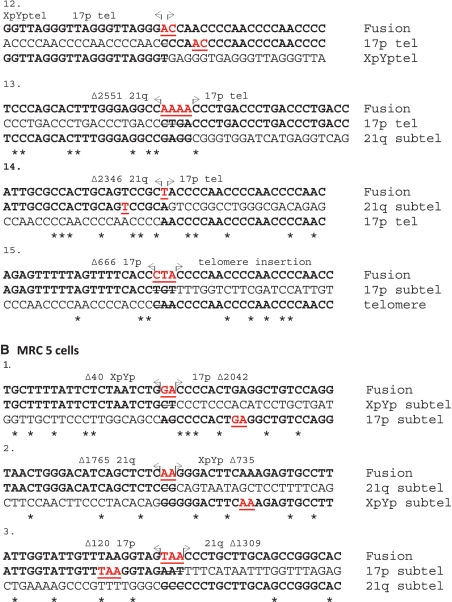
We examined the sequence of the nucleotide insertions up to 20 nt either side of the breakpoint in the participating telomeres. This revealed that in 13 of the 15 fusion events that contained nucleotide insertions (86%), the sequences across the junction, including both the inserted DNA and nucleotides flanking the break point, were duplicated in the adjacent DNA of one of the participating telomeres (Figure 3A). This was also apparent in the three fusion events of this type observed in Mre11 wild-type cells, although the insertion size was smaller (Figure 3B).
Figure 3.
Displaying the DNA sequence at the fusion point in a subset of fusions that display nucleotide insertions in ATLD cells (A) and MRC5 cells (B). The fusion sequence is displayed on the top lines, with the fusion partners displayed below. Arrows indicate the fusion point, the DNA between the arrows represent the DNA insertions, the participating chromosomes and the deletion sizes are stated above, asterisks below indicate homology between the fusion partners, sequence highlighted in red and underlined indicate duplicated sequences across the fusion point. No duplications could be detected in fusions 13 and 15.
Our data allow us to conclude that Mre11 is involved in the fusion of short dysfunctional telomeres and that telomere fusion in ATLD 3/4 cells is more error prone, resulting in insertion and duplication of sequences around the fusion point.
DISCUSSION
We took advantage of the Mre11 mutations in ATLD cells to study of the role of this protein in the fusion of short dysfunctional telomeres. The mutations in ATLD are hypomorphic and the documented DNA repair defects in these cells are subtle (31). This may also be reflected in the telomere length analysis of ATLD 3/4 cells described in this study in which we observed no overt differences in telomere dynamics. This included the rates of telomere erosion as a function of population doubling that were entirely normal. Moreover the telomeres at the point of senescence in ATLD cells were also within the normal length range detected in Mre11 wild-type cells undergoing p53-dependent replicative senescence (34,35). Furthermore, the expression of HPV E6E7 in ATLD cells extended the replicative lifespan of these cells by up to 24 PDs, again this is within the range observed in Mre11 wild-type cells (24). These data indicate that ATLD 3/4 cells undergo an otherwise normal p53-dependent replicative life span; this is consistent with the observation that ATLD cells exhibit a normal p53 response to ionizing radiation (31). In contrast, ATLD 1 and 2 cells exhibited a severely truncated replicative life span; this may reflect the more severe phenotype of these cells specifically the defective checkpoint responses to DNA damage (36,37).
The extended replicative life span following the expression of HPV E6E7 facilitated continued telomere erosion to a point at which complete telomere loss could be detected. This was accompanied by an increase in the frequency of telomere fusion molecules to rates slightly higher than that observed in Mre11 wild-type cells (26). Our data on telomere fusion indicate that the expression of HPV 16 E6E7 does not impact on telomere fusion (24,25,26). These data show that the mutational spectrum (deletion size, number of TTAGGG repeats and micro-homology) of fusion observed in HPV E6E7 fibroblast cultures undergoing ‘crisis’ is indistinguishable from sporadic fusion events in normal cell cultures in vitro or during tumour progression in vivo. Thus, we consider that the fusion of short telomeres following extended proliferation as a consequence of HPV E6E7 expression represents a physiologically relevant model. The extent of sub-telomeric deletion observed in ATLD cells was indistinguishable from wild-type cells; however, because ATLD 3/4 contains hypomorphic Mre11 alleles these data do not allow us to draw any conclusions about the involvement, or not, of Mre11 nuclease activity in the DNA resection that may accompany the fusion of short dysfunctional telomeres. The nuclease activity of Mre11 is required double-strand DNA break repair (38) and for telomere fusion in context of TRF2 knockdown in mouse models (39).
The key observation in our data set was the presence of additional nucleotides at the telomere fusion points in ATLD cells. These nucleotide insertions were distinct from the insertion of clearly identifiable loci from elsewhere in the genome, as we have previously described in Mre11 wild-type cells. The insertion of additional nucleotides has also been described at CSR junctions in ATLD cells (20). Together, these data are consistent with the role of Mre11 in A-EJ processes and that these processes may underlie the fusion of short dysfunctional telomeres. An increase in small unidentified insertions at fused telomeres have also been described in Arabidopsis tert ku70 mre11 triple mutants; however, the origin of these sequences could not be determined (28). In ATLD cells, the nucleotide insertions resulted in the duplication of the adjacent DNA of one of the participating telomeres. This implies that during the processing of telomere–telomere fusion events there is DNA synthesis templated from the adjacent DNA of one of the partner chromosomes. The miss-sense mutations within Mre11 in ATLD 3/4 cells impair Nbs1 binding (6,31) and this may occur by distorting the surface of Mre11 and interfering with protein–protein interactions (12). It is apparent that Mre11 dimerization is stabilised by DNA binding (12). Thus, in the context of the ATLD 3/4 Mre11 mutations, the MRN complex may be less stable and less able to maintain the synapses of the two participating telomeres. We consider this may allow for aberrant DNA synthesis, by, for example, polymerases Polλ and Polμ that are implicated in NHEJ during DNA repair (40,41). This may serve to increase the localized homology and base pairing to stabilize the complex prior to ligation.
The fusion of short dysfunctional telomeres has been implicated in driving instability during tumour progression (25). The data presented here allow us to conclude that, like telomeres rendered dysfunctional by the experimental abrogation of TRF2 (39), the fusion of short dysfunctional telomeres can involve Mre11 function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Cancer Research UK (C1799/A6932; C1799/A5603; C17199/A13490). Funding for open access charge: Cancer Research UK (C17199/A13490).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at the sequencing facility of the Cardiff University Central Biotechnology Service.
REFERENCES
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 3.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 4.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 5.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J. Biol. Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 8.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 9.Paull TT, Gellert M. The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 10.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 18.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 20.Lahdesmaki A, Taylor AM, Chrzanowska KH, Pan-Hammarstrom Q. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 2004;279:16479–16487. doi: 10.1074/jbc.M312796200. [DOI] [PubMed] [Google Scholar]
- 21.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 22.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell. Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 23.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, Fegan C, Pepper C, Baird DM. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukaemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 26.Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterised by extensive deletion and microhomology and can result in complex rearrangements. Nucleic Acids Res. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maser RS, Wong KK, Sahin E, Xia H, Naylor M, Hedberg HM, Artandi SE, DePinho RA. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitts SA, Kullar HS, Stankovic T, Stewart GS, Last JI, Bedenham T, Armstrong SJ, Piane M, Chessa L, Taylor AM, et al. hMRE11: genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum. Mol. Genet. 2001;10:1155–1162. doi: 10.1093/hmg/10.11.1155. [DOI] [PubMed] [Google Scholar]
- 31.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 32.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair. 2004;3:1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Bond JA, Haughton MF, Rowson JM, Smith PJ, Gire V, Wynford-Thomas D, Wyllie FS. Control of replicative life span in human cells: barriers to clonal expansion intermediate between M1 senescence and M2 crisis. Mol. Cell. Biol. 1999;19:3103–3114. doi: 10.1128/mcb.19.4.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 35.Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- 36.Delia D, Piane M, Buscemi G, Savio C, Palmeri S, Lulli P, Carlessi L, Fontanella E, Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum. Mol. Genet. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 37.Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- 38.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.