Abstract
Investigating processes and timing of differentiation of organisms is critical in the understanding of the evolutionary mechanisms involved in microevolution, speciation, and macroevolution that generated the extant biodiversity. From this perspective, the Telestes genus is of special interest: the Telestes species have a wide distribution range across Europe (from the Danubian district to Mediterranean districts) and have not been prone to translocation. Molecular data (mtDNA: 1,232 bp including the entire Cyt b gene; nuclear genome: 11 microsatellites) were gathered from 34 populations of the Telestes genus, almost encompassing the entire geographic range. Using several phylogenetic and molecular dating methods interpreted in conjunction with paleoclimatic and geomorphologic evidence, we investigated the processes and timing of differentiation of the Telestes lineages. The observed genetic structure and diversity were largely congruent between mtDNA and microsatellites. The Messinian Salinity Crisis (Late Miocene) seems to have played a major role in the speciation processes of the genus. Focusing on T. souffia, a species occurring in the Danube and Rhone drainages, we were able to point out several specific events from the Pleistocene to the Holocene that have likely driven the differentiation and the historical demography of this taxon. This study provides support for an evolutionary history of dispersal and vicariance with unprecedented resolution for any freshwater fish in this region.
Introduction
Three analytical levels can be defined when dealing with the processes of evolutionary differentiation [1]: microevolution (within and among populations), speciation, and supra-specific differentiation (macroevolution). One of the central questions of the study of genetic differentiation is to understand the putative barriers (preventing gene flow between populations) involved at these three levels [2]. The nature of the geomorphologic events that have fostered speciation and shape population structure of freshwater fishes is still strongly debated [3], [4]. A number of causes have been suggested, including (i) watersheds crossing by upstream river capture, (ii) downstream river connections (due to modification of the sea level), (iii) transient tributary connections (e.g. due to floods), (iv) periglacial lakes as vectors for colonisation processes, and (v) subsidence lakes.
For European freshwater fishes, these geomorphologic events are usually related to (i) the Messinian Salinity Crisis (MSC [5]), which started 5.96 Myr by the end of the Late Miocene [6] and was followed by the Lago Mare stage of the Mediterranean [5] between 5.50 and 5.33 Myr [6], or (ii) the Quaternary (from 1.8 Myr to present) glaciation cycles. The MSC and its final Lago Mare stage are considered as major events in the speciation and dispersal of freshwater fish taxa in the peri-Mediterranean area [7]–[9]. While the climatic oscillations of the Pleistocene are often invoked as major events in the formation of the geographic and genetic structure of current species via past range fragmentation, population contraction and re-colonisation (e.g. [10]–[12]).
The processes of colonisation of freshwater European ichthyofauna have received particular attention during the last decade, especially in Cyprinids. Many studies based on mitochondrial DNA (mtDNA) explored the major diversifications of this family in Europe and proposed an array of evolutionary scenarios for species or groups of species (reviewed in [13]). Other studies, which focused on finer scale analyses and were generally based on microsatellites, provided elements of the more recent evolutionary history of European ichthyofauna (e.g. [14]). The relative impact and importance of the MSC and the Pleistocene glaciations in the differentiation processes of European freshwater fishes is still debated (e.g. [15], [16]). More importantly, studies investigating the influence of Pleistocene glaciations are generally limited to challenging the impact of the Last Glacial Maximum (LGM) ∼20 Kyr ago. The detailed dynamics of demographic contraction, fragmentation and re-colonisation processes of European freshwater species during the Pleistocene remains poorly understood (but see [17], [18]). As a matter of fact, the Pleistocene has been shown to have been a critical period for the establishment of the present day genetic diversity of terrestrial fauna (e.g. [19], [20]). However, the Pleistocene (1.8 Myr) consisted of a series of well characterized glacials and interglacials [21] that have not systematically erased the prints and distribution patterns inherited from previous cycles but may, alternatively, have added new layers of complexity. Therefore, deciphering the relative importance of the successive historical events that may have shaped current species distribution and their genetic structure is challenging as it requires extensive sampling effort and enough data informativeness.
From this perspective, the Telestes genus is of special interest. A taxonomic revision and a thorough look at the differentiation pattern based on partial cytochrome b sequence, allozymes and morphology were recently undertaken for the Telestes genus [22], providing a solid foundation to investigate processes of differentiation in this genus. This study also suggested that the Telestes genus speciated mostly via allopatric isolation and that introgressive hybridization only had a marginal role [22]. Furthermore, the Telestes genus constitutes an attractive freshwater fish model considering its peculiar ecological niche. For instance, T. souffia generally occurs in ∼5 m wide rivers from ∼10 km downstream of the source [23], [24]. The mean discharge has no significant influence on its distribution and the relevant summer temperature varies between 11°C and 26.5°C [23], [24]. The species is rare or absent in large rivers [23] and dispersal is believed to be limited [25]. Telestes souffia is therefore considered as adapted to relatively small running rivers and cold-water. It is also noteworthy that species of the Telestes genus are rather small bodied (max. 160 mm). These species are thus not valuable for fishing and are therefore less prone to be translocated, an activity that is well known in economically valuable freshwater fish species [26]–[28] to alter original population structure [29].
Using 970 specimens, almost encompassing the entire geographic range of Telestes, we gathered molecular data from a 1232 bp long mitochondrial sequence (including the complete cytochrome b gene and 92 bp of flanking tRNA genes) and 11 nuclear microsatellite loci to investigate the major mechanisms and events responsible for the genetic structure and diversity pattern in Telestes. We explored whether (i) the MSC/Lago Mare event, (ii) the Pleistocene glaciation cycles, and/or (iii) the post-LGM/Holocene period could account for the current species distribution pattern and their genetic structure. Furthermore, we stressed the importance of watersheds crossing by upstream river capture periglacial lakes and downstream river connections in the Telestes genus evolution.
Materials and Methods
Ethics Statement
This study was conducted according to relevant national and international guidelines regarding the care and welfare of fishes. Field studies did not involve fish that were endangered or protected (The IUCN Red List of Threatened Species v. 2011.1; www.iucnredlist.org). The fishes collected for this study were killed rapidly (using clove oil as anesthetic) or were returned to the wild. The DDTs (Directions Départementales des Territoires) from the Alpes-de-Haute-Provence, Hautes-Alpes, Vaucluse, Ardèche, Haute-Savoie, Doubs, Ain and Saone districts issued the permits for France, where fishing was done in collaboration with Officers of the ONEMA (Office National de l'Eau et des Milieux Aquatiques, France). Permits for the localities in Romania, Italy, Greece and Montenegro were supplied respectively by the Institute of Biology of Bucarest (representative Petru Bănărescu), the Universita di Napoli ‘Federico II’ (representative Pier Giorgio Bianco), the Aristotle University, Thessaloniki (representative Panos Stavros Economidis) and the University of Montenegro, Podgorica (representative Drago Marić). The permit for the Slovenian sampling was supplied by the Angler Society of Idrija, the Angler Society of Sevnica and the Fisheries Research Institute of Ljubljana as part of the research project 9E0073 funded entirely by the Tour du Valat Foundation (representative Alain Crivelli) and the WWF International.
Sampling and DNA protocols
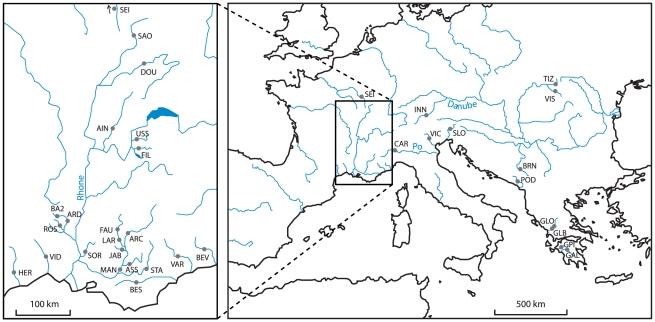
A total of 970 individuals from 34 locations were collected for this study (Fig. 1), covering almost the entire geographical range of the Telestes genus. The 34 locations encompassed six European biogeographical districts (as defined by [8], [30]): the Western-Greece district (WG), the Albanian district (AB), the Padano-Venetian district (PV), the Tuscano-Latium district (TL), the Danubian district (DB) and the Southern France district (SF). A more detailed sampling was performed in the Rhone drainage (in SF district) in order to conduct a finer scale analysis of the pattern of genetic variation in this area. Further details on samples are reported in Figure 1 and Table 1.
Figure 1. Location of the sampled Telestes populations with emphasize on the Rhone and French Mediterranean basins.
For samples ID, see Table 1.
Table 1. Sampled Telestes taxa, population locations and sample sizes.
| Taxa | Sample ID | Sampling year | River | River basin (sub-basin) | Districta | mtDNA | Microsatellites |
| Telestes alfiensis | GAL | 2003 | Alfios | Alfios | WG | 4 | 4 |
| GPI | 2003 | Pinios | Pinios | WG | 1 | 1 | |
| Telestes pleurobipunctatus | GLO | 2003 | Louros Ms | Louros | WG | 9 | 9 |
| GLB | 2003 | Louros Ps | Louros | WG | 23 | 22 | |
| Telestes montenigrinus | POD | 2001 | Zeta | Moraca | AB | 30 | 31 |
| Telestes muticellus | CAR | 1997 | Po | Po | PV | 29 | 25 |
| VIC | 1997 | Bacchiglione | Bacchiglione | PV | 27 | 24 | |
| BEV | 1996 | Bevera | Roya | TL | 29 | 29 | |
| Teleste souffia agassii 1 | BRN | 2001 | Lim | Danube | DB | 31 | 28 |
| SLO | 2000 | Soca | Soca | PV | 38 | 37 | |
| Teleste souffia agassii 2 | TIZ | 1999 | Tisza | Danube | DB | 21 | 15 |
| VIS | 1999 | Viseu | Danube | DB | 24 | 15 | |
| Telestes souffia agassii 3 | INN | 2002 | Inn | Danube | DB | 28 | 30 |
| Telestes souffia souffia | AIN | 1996 | Ain | Rhone | SF | 29 | 28 |
| ARC | 1996 | Durance | Rhone (Durance) | SF | 30 | 29 | |
| ARD | 1996 | Ardeche | Rhone (Ardeche) | SF | 20 | 20 | |
| ASS | 1996 | Asse | Rhone (Durance) | SF | 39 | 28 | |
| BA2 | 1996 | Ardeche | Rhone (Ardeche) | SF | 30 | 29 | |
| BES | 1996 | Argens | Argens | SF | 23 | 23 | |
| DOU | 1996 | Doubs | Rhone | SF | 33 | 23 | |
| FAU | 1996 | Buech | Rhone (Durance) | SF | 29 | 28 | |
| FIL | 1996 | Filliere | Rhone | SF | 31 | 33 | |
| HER | 1996 | Herault | Herault | SF | 33 | 33 | |
| JAB | 1996 | Jabron | Rhone (Durance) | SF | 22 | 21 | |
| LAR | 1996 | Buech | Rhone (Durance) | SF | 43 | 42 | |
| MAN | 1996 | Durance | Rhone (Durance) | SF | 22 | 20 | |
| ROS | 2008 | Beaume | Rhone (Ardeche) | SF | 39 | 39 | |
| SAO | 1996 | Saone | Rhone | SF | 30 | 24 | |
| SEI | 1996 | Ource | Seine | DB | 26 | 23 | |
| SOR | 1996 | Sorgue | Rhone | SF | 30 | 29 | |
| STA | 1996 | Verdon | Rhone (Durance) | SF | 35 | 28 | |
| USS | 1996 | Usses | Rhone | SF | 34 | 24 | |
| VAR | 1996 | Var | Var | SF | 67 | 65 | |
| VID | 1996 | Vidourle | Vidourle | SF | 31 | 30 |
see main text for abbreviations.
Whole DNA was extracted from 25 mg of muscle or 0.25 cm2 of caudal fin using the Gentra® Puregen™ Tissue Kit (QIAGEN) following the manufacturer's instructions. A 1275 bp mitochondrial DNA (mtDNA) coding region was amplified by PCR using the Taq CORE Kit (MP Biomedicals) with primers L14350C (5′-ACCACCGTTGTAGTTCAACTAC-3′) and H15620U (5′-AGGGGTGGGAGTTAAAATCTC-3′). PCRs were conducted following standard conditions in a total volume of 40 µL with 1 U of Taq polymerase and 2 µL of 1∶10 diluted DNA extract (PCR cycling protocol: initial denaturation at 95°C for 5 min; denaturation at 95°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 1 min 30 s, repeated for 35 cycles; final extension at 72°C for 10 min). The amplicons were sequenced by GATC Biotech (Konstanz, Germany) using internal primers Cb-L500 (5′-CAATGAATCTGAGGCGGTTT-3′) and Cb-H650 (5′-GAGAAGTATGGGTGGAAAGA-3′), resulting in a 1232 bp long fragment including the entire cytochrome b (Cyt b) gene and 92 bp of flanking tRNA genes. Chromatograms were checked for quality and sequences were aligned using SeqScape 2.5 (Applied Biosystems). Sequences were deposited in Genbank (accession numbers JQ651395 to JQ652369).
Eleven microsatellite loci were successfully genotyped in 889 individuals. Ten out of the 11 primer pairs used in this work (Lsou5, Lsou8, Lsou10, Lsou34, BL1-2b, BL1-61, BL1-84, BL1-98, BL1-153 and BL1-T2) were described previously [31], [32] and one novel microsatellite locus was genotyped (BL1-36; core repeated motif: (AC)3; Genbank ID JQ652370) using primers BL1-36F (6FAM-5′-GATGACTGTGCGATGAATGC-3′) and BL1-36R (5′-TGTGTGTGCAGTTTGTGTGG-3′). The eleven loci were amplified through four multiplex PCRs. Amplicons were analyzed and allele sizes were scored using protocols and conditions described by [32]. All individuals with missing data (unscored loci) were discarded from the dataset and were not used for statistical analyses. To ensure reliability, ∼10% of the 889 individuals were genotyped twice, in all cases yielding identical results.
Population genetics analyses
In order to explore the diversity of the mitochondrial gene pool, Arlequin 3.11 [33] was used to compute the number of distinct haplotypes (k), the gene diversity (H) [34], as well as three estimators of the parameter θ = 2Nμ: θπ [35], θk [36] and θS [37]. To explore the diversity of microsatellite data, Genepop 4.0 [38] was used to: (i) test for Hardy-Weinberg (HW) equilibrium, (ii) estimate the heterozygosity for all loci and populations, and (iii) test linkage disequilibrium (LD) among loci within populations. Levels of significance for HW and LD tests were adjusted using the false discovery rate (FDR) procedure [39]. HW deficiencies were further tested using Micro-Checker 2.2.3 [40] in order to determine the causes of departures from HW equilibrium: genuine HW disequilibrium, null alleles, scoring errors (often resulting from stuttering) or allele dropout. For all loci, the observed (Ho) and expected (He) heterozygosities were estimated for each population sample using Arlequin 3.11. For all population samples and groups of populations, the mean number of alleles per locus (An), the average Ho and average He over loci were estimated with Arlequin 3.11. As the observed number of alleles in a sample is highly dependent on sample size, we used the rarefaction procedure [41] implemented in Adze 1.0 [42] to estimate the allelic richness (Ar) [43] and the private allelic richness (Ap) [41]. When several closely related populations are sampled, few alleles tend to be private to individual populations [41]. As proposed by [42], Ap was therefore estimated for groups of populations within the Rhone and Danube drainages.
Phylogenetic analyses
We collated 1232 bp long mtDNA sequences obtained from 970 individuals of the Telestes genus and from five other Leuciscinae species that were used as outgroups (Fig. 2). We selected one terminal for each haplotype and used Modeltest 3.7 [44] to obtain the substitution models that best fit each of the three codon positions of the Cyt b fragment and the tRNA collated flanking regions using the Akaike Information Criterion [45]. These four models (Supporting Information S1) were subsequently used for partitioned Bayesian analysis performed with MrBayes 3.1 [46] on the University of Canterbury Supercomputer (www.ucsc.canterbury.ac.nz). Bayesian analysis consisted of two independent runs of 2.0×107 generations with random starting trees and 10 Markov chains (one cold) sampled every 1000 generations. Adequate burn-in was determined by examining convergence onto stationarity on bivariate plots of the split frequencies, cumulative split frequency for all splits for the two runs of the simulation and symmetric tree-difference score [47] within and between runs using Awty [48]. We considered relationships with posterior probabilities ≥0.95 to be strongly supported.
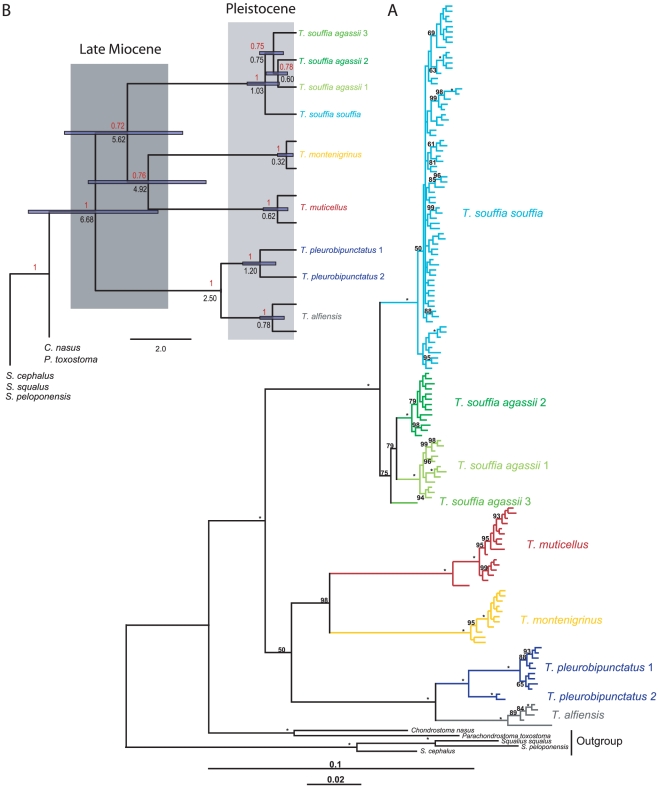
Figure 2. Phylogenetic analyses of mtDNA sequences.
A. Phylogenetic relationships among haplotypes of Telestes hypothesized from Bayesian analysis using 1232 bp of mtDNA. Posterior probability*100 are indicated, with “*” corresponding to pp = 0.1 while “-” are used to indicate pp<0.5. B. Bayesian time calibrated maximum clade credibility tree using relaxed clock with the same dataset but selected terminals representing major lineages. The calibration point used was the divergence between T. pleurobipunctatus and T. alfiensis. Posterior probabilities are indicated in red on the upper left of the nodes while the modes of the posterior distributions of the age of the nodes are indicated in Myr on the lower left in black. 95% CI are indicated with blue bars centered on the nodes.
To obtain an estimate of the molecular rates of evolution in the different Telestes lineages, as well as a time frame for the diversification, we used a relaxed Bayesian molecular clock with uncorrelated lognormal rates (Beast 1.4.8 [49]). We calibrated the tree reconstruction using an important and well-dated geological event: the formation of the Strait of Korinthos in the late Pliocene (2.5 Myr), which separates the Peloponnesus from mainland Greece [50]. Telestes pleurobipunctatus from the Arachthos River and T. alfiensis from the Alfios River are cyprinid taxa that were separated by the formation of the Strait of Korinthos [51]. The prior distribution of the age of the node was set as a normal distribution 2.5±0.1 Myr to account for uncertainty in the gene tree divergence. Unresolved relationships among haplotypes and intraspecific lineages impact the time estimates and rates of evolution [52]. Thus, we selected one representative of each major lineage (n = 17) according to the previous Bayesian analysis. According to the estimated models that best fit the dataset (Supporting Information S1), we used a GTR+I+G model partitioned for each codon position with unlinked parameters, and six Gamma shape categories. The tree prior used the Yule Process, with a UPGMA starting tree and operators optimized for each analysis by a preliminary run of 106 generations sampled every 1000 generations followed by two independent runs of the same length and sampling rate. Adequate burn-in was determined by examining a plot of the likelihood scores of the heated chain for convergence onto stationarity.
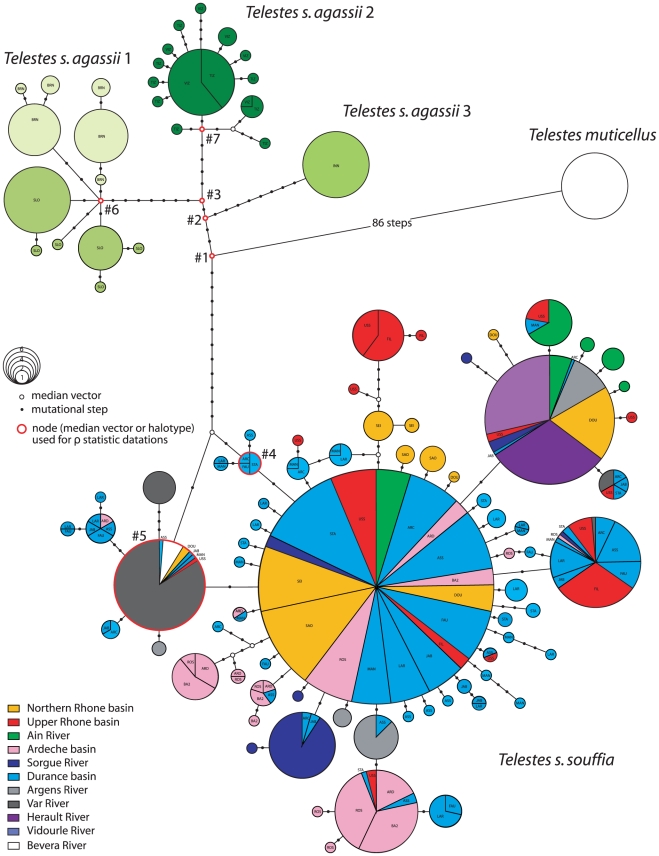
Network 4.05 (Fluxus Technology Ldt, www.fluxus-engineering.com) was used to compute a Median-Joining (MJ) network [53] of haplotypes from the T. souffia complex (T. muticellus haplotype from BEV sample was used to root the network). A statistical parsimony (SP) network of haplotypes [54] was also computed using Tcs 1.21 [55]. MJ and SP network reconstructions resulted in identical topologies, except that Tcs failed to resolve the topology of some parts of the phylogeny that were resolved by the MJ network. Furthermore, several TMRCA were estimated from the Median-Joining network through Network 4.05 using the ρ statistic methodology [56], [57] and the mean calibrated evolution rate estimated as described above. We therefore only show the MJ network reconstruction (Fig. 3).
Figure 3. Median-Joining network of the T. souffia mitochondrial haplotypes.
Population samples ID detailed in haplotypes charts when necessary.
Genetic relationships between populations based on microsatellite allele frequencies were also investigated using the Cavalli-Sforza chord distance (Dc) [58] calculated with the maximum likelihood algorithm implemented in Phylip 3.69 [59]. Node support was tested using 1000 bootstrapped datasets.
Population structure analyses
A Bayesian-based approach was used to search for the occurrence of independent genetic groups (K) in the Telestes microsatellite dataset (implemented in Structure 2.3.3 [60]–[63]). The burn-in length was set to 50,000 followed by 200,000 iterations within a Markov Chain Monte Carlo (MCMC). The “admixture model” was used with priors on population sampling location. Using priors on population sampling location allows genetic structure to be detected at lower levels of divergence and/or with a limited number of loci (i.e. <15 loci). Moreover, the model is not biased towards detecting non existing genetic structure [63]. The “I-model” (independent allele frequencies) was used when investigating different species, and the “C-model” (correlated allele frequencies) was used when investigating the structure of the T. s. souffia lineage. Ten repeats were conducted for each K value, with K = 1–15. Each individual was assigned to the inferred clusters according to the results from the simulation procedures. The way for determining the most meaningful value of K is greatly debated (see [64]). Actually, multiple biologically meaningful K values are likely for one data set depending on the biological questions asked [65]. Additionally, forcing Structure to place individuals into too few clusters often results in an inferred genetic structure that is not consistent with the evolutionary history of the populations [66]. Using CorrSieve 1.6–2 [64] we combined three approaches to determine K: (i) choosing the K value that maximizes the posterior probability of the data Ln P(D) [60], (ii) the ΔK test [67], and (iii) the ΔFst test [64]. Structure does not export Fst values between clusters when the I-model is used. Only the first two approaches were thus used for the analyses dealing with the different Telestes species. Finally, T. alfiensis individuals were discarded from the Structure analyses since assignment tests are biased for populations of limited sample size [68].
In order to graphically summarize the allelic frequencies variation for the different populations, Factorial Correspondence Analyses (FCA), which are well suited for qualitative variables (here, genotypes) [69], were conducted using Genetix 4.05 [70].
Population differentiation was estimated by pairwise F-statistics (Fst) [71] calculated using Arlequin 3.11. In order to test isolation by distance (IBD) within the Rhone basin we used Mantel test [72] with 1000 permutations using Arlequin 3.11. As recommended for analyzing differentiation in elongated habitats [73], a one-dimensional model was used and we analyzed the linear relationship between Fst/(1−Fst) and (unmodified) geographical distance. A Mantel test was also conducted to explore the correlation between the microsatellite Fst/(1−Fst) and the mtDNA Fst/(1−Fst). Since null alleles were detected at two loci for a few populations (see below), FreeNA [74] was also used to calculate Fst including a correction for null alleles. Samples VAR, BES, HER, VID and SEI were discarded from the IBD analyses, since these are populations that are currently disconnected from the Rhone river system.
Demographic analyses
Bayesian skyline plots (BSP) [75] permit the estimation of the effective population size (Ne) through time. We used the BSPs implemented in Beast 1.4.8 [49] for each major mitochondrial lineage in the Danube and Rhone basins. We first undertook preliminary runs for each lineage. Given previous estimations of the best-fitting model we applied a GTR+I+G model, estimated based frequencies, four Gamma categories, the previously estimated rate of molecular evolution (see before) for the species group in a strict clock model, 10 grouped coalescent intervals (m), and priors for the phylogenetic model and population sizes were uniformly distributed. These analyses were sampled every 1000th iteration for 10 million generations. In a second step we undertook 40 million generation runs with bounded prior distributions and optimized tuning. Plots for each analysis were visualized using Tracer 1.4 [76].
Using mtDNA sequences, the demographic history of populations and major lineages was also investigated by computing Tajima's D (DT) [77] and Fu's Fs neutrality tests [78] with Arlequin 3.11. Using microsatellites, we tested for potential recent reduction in Ne through the Wilcoxon test implemented in Bottleneck 1.2 [79]: a transient excess of heterozygotes signals a recent (0.2Ne-4Ne generations) bottleneck event [80], [81]. Several alleles in our dataset testified that most loci depart from the stepwise mutation model (SMM) [82]. The infinite allele model (IAM) [83] was therefore used for Bottleneck analyses, as well as a two-phase mutation model (TPM) [84] assuming 70% of single-step mutation and variance among multiple steps of 30% (as recommended by [81]).
Results
Checking the microsatellites: low polymorphic loci, HWD, null alleles and LD
All the microsatellite loci were polymorphic in at least one population, and only two loci (BL1-36 and Lsou10) were found to be monomorphic in several populations. BL1-36 presented only two alleles (namely 195 and 197). Allele 195 was found in T. alfiensis, T. pleurobipunctatus and T. muticellus populations, while allele 197 was found in T. montenigrinus and T. souffia populations. Heterozygote individuals were only found in BEV (T. muticellus) and INN (T. souffia) populations. Lsou10 was polymorphic in T. muticellus (BEV, CAR and VIC), T. montenigrinus (POD) and T. s. agassii 1 (BRN and SLO) only.
After applying the FDR controlling procedure, significant departure from HW equilibrium was observed at several loci in 12 populations (including both cases of heterozygote excess and deficiency; Supporting Information S2). Significant excess of heterozygotes was observed in three populations: ARC (one locus), GLB (one locus) and INN (six loci; see below, results of Bottleneck analyses). MicroChecker analyses were conducted on populations with significant heterozygote deficiency. Neither allele dropout nor scoring errors were detected. Null alleles were detected in three loci: BL1-T2, BL1-98 and BL1-153. BL1-T2, which was originally developed from a T. souffia genomic library [32], presents null alleles for all three T. muticellus populations. BL1-98 and BL1-153 present null alleles in all eight populations with heterozygote deficiency. Null alleles are believed to have a negligible impact on assignment tests [85]. However, null alleles may bias Fst [74] and genetic diversity summary statistics [86]. In this study, Fst were estimated for T. s. souffia populations (IBD analyses), for which only three populations (FIL, VAR and ARD) present null alleles at one locus each. We therefore assumed a limited impact on Fst and genetic diversity summary statistics estimations. With regard to Fst, this limited impact was confirmed by the FreeNA estimation of Fst including a correction for null alleles: the corrected Fst were highly correlated to uncorrected Fst (Supporting Information S3).
A total of 1185 pairwise comparisons were submitted to LD analyses. Interestingly, LD was found between BL1-2b and BL1-T2 in ten populations representing four Telestes species or sub-species (Supporting Information S4). This pair of loci previously showed significant LD in four other European cyprinid species [87]. Since the karyology and chromosome topology are well conserved across cyprinid species (e.g. [88]), the prevalence of LD between BL1-2b and BL1-T2 may indicate a loose vicinity on the chromosome. Alternatively, BL1-2b and BL1-T2 loci may be linked to genes under selection in cyprinids. However, after FDR correction, only 19 pairs of loci exhibited LD, i.e. 1.6% of the total pairwise comparisons (Supporting Information S4), which indicates that LD is far from dominating our data set. We therefore assumed a negligible impact of LD on our analyses (see [89]).
Population-level analyses and genetic diversity
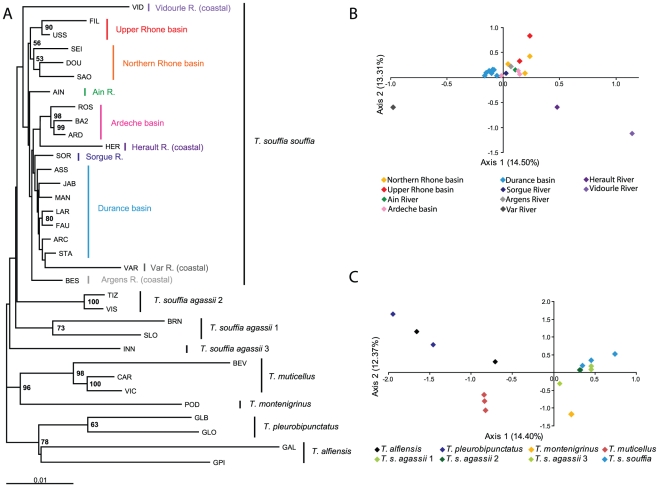
All but three populations (INN, HER and VID) were polymorphic for mtDNA data (Supporting Information S5). From θπ and θS indices, BEV had an extremely high genetic diversity but harbored only two haplotypes. However, these two haplotypes were nested within two distinct species in phylogenetic reconstructions: one haplotype within T. muticellus (not shared with, but one mutational step from haplotypes sampled in CAR and VIC), and one haplotype within T. s. souffia (shared with six populations of the Rhone drainage and very frequent in the Var River) (Fig. 3). This confirms hybridization between T. muticellus and T. s. souffia in the Bevera River [90]. However, Structure failed to identify individuals with a possible nuclear genomic component from T. s. souffia in our BEV sample (Fig. 4A).
Figure 4. Phylogenetic and multivariate analyses of microsatellites data.
Unrooted Maximum Likelihood tree based on microsatellites Dc distances (A) and FCA scatter plots for T. s. souffia population samples (B) and population samples representative of Telestes genus (C).
θk is known to be very sensitive to Ne fluctuations, more than H, θπ or θS [91]. For instance, although BEV had the highest θπ and θS, this sample exhibited a very low θk (0.28) when compared to most of the other Telestes population samples (up to 8.48 in T. muticellus and 12.46 in T. souffia). This may indicate a hybridization event associated with a sharp bottleneck event, which is also suggested by the very low Ar value.
Within the Danube drainage, it is noteworthy that although mtDNA diversity and Ar were similar to those encountered in the Rhone drainage (except for INN), the values of Ap were about two-fold higher (even for INN). In the Rhone basin, θk values tended to be higher in populations from the Durance basin (Southern Rhone drainage) than in most of the other more northern Rhodanian populations, whether considering individual populations or groups of populations. This pattern was similar when Ar (estimated from microsatellites) was considered, except for the Ardeche basin, Sorgue River and Ain River that were very similar to the Durance basin. On the contrary, Ap tended to be lower in the Durance basin (0.60) than in the northern areas of the Rhone drainage (except the Upper Rhone), with a peak of Ap in the Ardeche basin (0.95). Contrasted diversity patterns were observed within the French Mediterranean coastal rivers (BES, VAR, HER and VID). Their genetic diversity indices ranged from comparable to or slightly higher than some Rhodanian populations (in BES) to very low genetic diversity (in VID).
Phylogeography and genetic structures
A total of 140 distinct mitochondrial haplotypes were found in the entire Telestes dataset, with 96 in the T. souffia complex and 71 in T. s. souffia alone. Bayesian phylogenetic tree reconstruction (Fig. 2A) provided a well resolved topology with most of the posterior probabilities (pp) of the nodes between the different taxonomic entities in the Telestes genus being >0.95. Three major clades were recovered: (i) T. souffia complex comprising four highly divergent lineages, (ii) an Italian/Balkan clade with T. muticellus+T. montenigrinus, and (iii) a Greek clade with T. pleurobipunctatus+T. alfiensis. Nevertheless, a few ambiguities remain: the basal split among these three Telestes clades and the relationships among the four lineages within the T. souffia complex remain poorly supported. Moreover, T. pleurobipunctatus, T. muticellus and T. alfiensis are also each subdivided into highly divergent lineages that do not spatially overlap (except for T. muticellus). The same topology (except the basal split among Telestes clades) was recovered in the Bayesian relaxed molecular clock time calibration (Fig. 2B). Most nodes are strongly supported (pp≥0.95), except the dichotomies between the T. souffia complex and T. muticellus+T. montenigrinus (pp = 0.72), between T. muticellus and T. montenigrinus (pp = 0.76), between T. s. agassii 3 and T. s. agassii 1+T. s. agassii 2 (pp = 0.75) and between T. s. agassii 1 and T. s. agassii 2 (pp = 0.78).
The topology of the ML tree based on microsatellites (Fig. 5A) was congruent with the mitochondrial gene trees. The three main mitochondrial clades were also recovered: T. muticellus+T. montenigrinus (BP = 96), T. pleurobipunctatus+T. alfiensis (BP = 78) and the Telestes souffia complex (although BP<50) with INN (T. s. agassii 3) displaying basal position. Within T. pleurobipunctatus, the GLB and GLO populations formed differentiated clusters (BP = 63). Moreover, the 21 populations of T. s. souffia form a clade (although BP<50). As for the FCA conducted on populations (Fig. 5C), it clustered all three T. muticellus populations together on the one hand, and all but two (INN, T. s. agassii 3; and VAR, T. s. souffia) T. souffia populations together on the other hand.
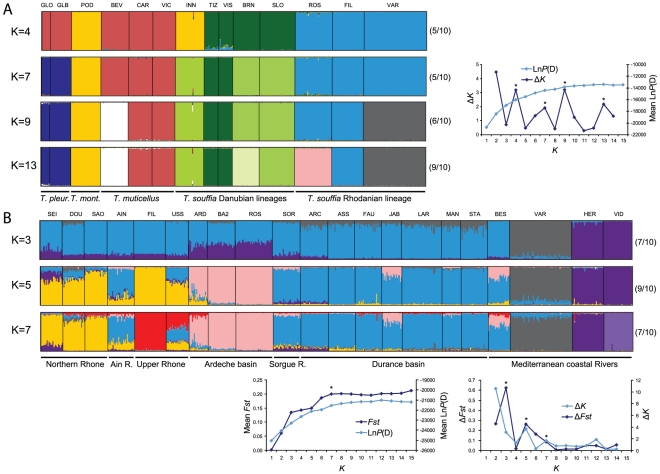
Figure 5. Bayesian clustering analyses of microsatellites using Structure.
Estimated population structure for population samples representative of Telestes genus (A) and T. s. souffia (B). “*” indicates K values retained from ΔK [67] and ΔFst [64] tests and when maximizing the posterior probability value [60]. Between parentheses, the number of convergent runs associated to the structure displayed.
Four values of K were found to fit the data from ΔK analyses of Structure runs conducted on 14 populations representative of the Telestes genus (Fig. 4A). However, as expected when relatively long divergence times separate the investigated evolutionary lineages [66], results are hardly interpretable with K<13: in fact, clustering is phylogenetically questionable (T. s. agassii 3 and T. montenigrinus grouped together for K = 4) and no more than half of the runs were convergent. On the contrary, for K = 13, 9/10 runs converged and all population samples were discriminated, except CAR+VIC (T. muticellus) and GLO+GLB (T. pleurobipunctatus).
Our dataset allowed us to focus further on the genetic pattern and structure of the T. souffia complex. From the Bayesian phylogenetic tree reconstructions (Fig. 2), the T. souffia complex is structured into four population segments that display non-overlapping lineages. Telestes s. souffia is distributed in the eastern half of France (in the Rhone basin and some Mediterranean coastal rivers). The three other clusters are distributed in distinct patches in the headwaters of the Danube River catchment with INN population harboring an exclusive lineage assigned to T. s. agassii 3; VIZ and TIZ in Romania assigned to T. s. agassii 2 and BRN and SLO from the Lim and Soca rivers assigned to T. s. agassii 1. From the MJ network, these Danubian clades are separated by 12 to 16 mutational steps, while they are separated from T. s. souffia haplotypes by 19 to 21 mutational steps (Fig. 3). The same genetic structure is observed from microsatellites using ML tree reconstruction (Fig. 5A) and using Structure (for K = 7; Fig. 4A) in the T. souffia complex. Furthermore, from Structure analyses, no detectable gene flow could be observed between the three Danubian headwaters (i.e. between VIS+TIZ, BRN and INN).
The most readily sampled group is T. s. souffia with 23 populations and 681 individuals. All these populations display some degree of haplotype sharing and the T. s. souffia haplotype network (Fig. 3) has a noticeable star-like shape with a major haplotype having a high frequency (∼30%, n = 192). However, most populations have private haplotypes. Moreover, some populations harbor particular features on the network and peripheral haplotypes are also strongly represented: the south-eastern population VAR display one derived haplotype (3 steps from closest haplotype) that is not found elsewhere; the northern populations USS and FIL also have two haplotypes clearly distinct from the rest of the network (4 steps from closest haplotype); and populations from the Ardeche basin share very few haplotypes with the rest of the populations. On the contrary, the south Rhone populations (Durance basin) are much diversified and their haplotypes are found all around the network.
Bayesian clustering using microsatellites revealed distinct subpopulations. Three values of K (3, 5 and 7) were retained from the analysis of the Structure runs (Fig. 4B): K = 3 and K = 5 correspond to peaks of both ΔK and ΔFst, while K = 7 corresponds to a peak for ΔK and to the plateau reached by mean Fst and mean LnP(D) values. For K = 3, the Mediterranean coastal river Var on the one hand and the rivers Herault and Vidourle on the other hand are discriminated from the rest of the Rhone basin. These three coastal rivers are characterized by very low or null mitochondrial diversity and low microsatellite diversity (see Supporting Information S5). Moreover, the FCA analysis (Fig. 5B) showed that VAR, HER and VID are extremely differentiated from the other populations. Structure therefore tended to discriminate first populations that experienced deep genetic drift during their evolutionary history. For K = 5, the two additional clusters discriminated Ardeche populations (ARD, BA2 and ROS) and populations from the North of the Rhone basin (DOU, SAO, USS and FIL) and from the South Rhodanian populations (SOR and Durance basin). Interestingly, both AIN (Northern Rhone basin) and BES (Mediterranean coastal river) are hardly differentiable from the South Rhodanian populations, and SEI (from the Seine basin) is similar to the North Rhodanian populations. For K = 7, VID was discriminated from HER and a new cluster characterized the Upper Rhone (USS and FIL). Nevertheless, the genetic structure was less clear and gene flow between the different parts of the Rhone drainage could be inferred from the results: (i) AIN sample, from the Northern part of the Rhone drainage, was hardly differentiable from the South Rhodanian populations for all K values, and (ii) some traces of gene flow from the southern and northern parts of the Rhone drainage were detectable in USS and (to a lesser extent) in ARD.
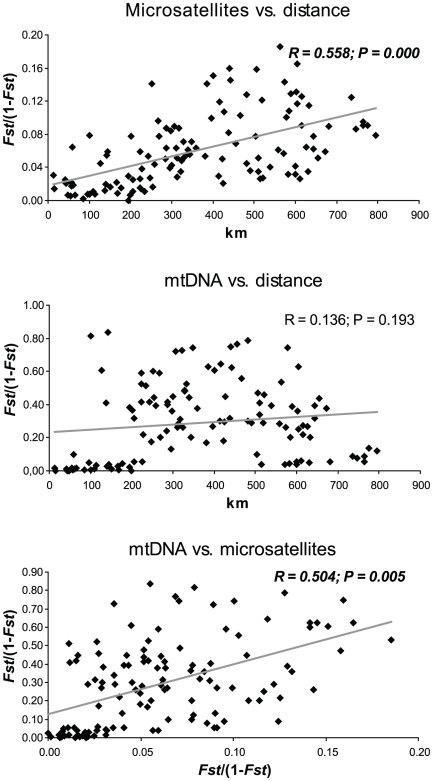
IBD was observed in the Rhone drainage using linear distances vs. Fst/(1−Fst) values estimated from microsatellites (R = 0.558; P = 0.000) (Fig. 6). Although such a correlation was not observed using Fst values obtained from mtDNA data (R = 0.136; P = 0.193), a Mantel test found the correlation between microsatellites Fst/(1−Fst) and mtDNA Fst/(1−Fst) to be significant (R = 0.504; P = 0.005). This minor discrepancy is probably due to the four-fold lower effective size of the mtDNA that makes this marker more prone to local genetic drift, which may have distorted the imprint of IBD. When using Fst corrected for null alleles (microsatellites data), Mantel tests gave similar results: R = 0.561 and P = 0.000 for linear distances vs. Fst/(1−Fst); R = 0.508 and P = 0.005 for microsatellites Fst/(1−Fst) vs. mtDNA Fst/(1−Fst).
Figure 6. Pairwise relationships between mtDNA, microsatellites and geographical distances in the Rhone drainage.
Pairwise relationship between Fst/(1−Fst) (genetic distance) vs. geographical distances (km) using microsatellites (A) and mtDNA (B) or between microsatellites vs. mtDNA genetic distances in the Rhone drainage. The correlation coefficient (R) and the P-value (P) obtained from Mantel test are reported for each bivariate analysis.
Molecular dates and differentiation events
The estimated calibrated rate of evolution of the Telestes species is 1.055E10−2 subs/site/Myr (95%CI 0.738–1.402E10−2). Our evolutionary rate is in agreement with previous estimates for Leuciscins Cyt b gene [51], [92], but is two-fold higher than recently estimated evolutionary rates [15], [16]. To obtain an estimation of the ages of the major cladogenic events, three different methods were used that gave remarkably similar results (Table 2).
Table 2. Molecular datations from mtDNA sequences.
| Coalescence time (TMRCA) | Expansion time events (BSP) | ||||||||
| Lineage | BEAST | 95% CI | (#)a | ρ | 95% CI | BSP | 95% CI | Event 1 | Event 2 |
| Telestes | 6.68 Myr | [4.60–8.92] | - | ||||||
| (T. muticellus+T. montenigrinus) vs. T. souffia | 5.62 Myr | [3.77–7.71] | - | ||||||
| T. muticellus vs. T. montenigrinus | 4.92 Myr | [3.00–6.92] | - | ||||||
| T. souffia (Danube and Rhone drainages) | 1.03 Myr | [0.56–1.63] | (#1) | 1.01 Myr | [0.57–1.46] | ||||
| T. souffia (Danube drainage only) | 0.75 Myr | [0.42–1.21] | (#2) | 0.64 Myr | [0.37–0.91] | 0.56 Myr | [0.32–0.83] | ||
| T. s. agassii 1 vs. T. s. agassii 2 | 0.60 Myr | [0.28–1.00] | (#3) | 0.59 Myr | [0.35–0.83] | - | |||
| T. s. souffia | - | (#4) | 159.2 Kyr | [3.2–315.2] | 161.3 Kyr | [70.1–274.0] | ∼75 Kyr | ∼10 Kyr | |
| T. s. souffia | (#5) | 140.7 Kyr | [0.0–286.3] | ||||||
| T. s. agassii 1 | - | (#6) | 125.6 Kyr | [33.5–217.7] | 158.9 Kyr | [58.7–278.1] | ∼60 Kyr | none | |
| T. s. agassii 2 | - | (#7) | 110.7 Kyr | [0.0–243.6] | 88.3 Kyr | [27.7–185.0] | none | ∼10 Kyr | |
Nodes of the MJ network used to estimate TMRCAs from ρ statistic (see Figure 3).
From time calibrated phylogenetic reconstruction, diversification in Telestes that corresponds with the separation between the four Telestes species groups (T. souffia, T. muticellus, T. montenigrinus and T. pleurobipunctatus+T. alfiensis) ranged from 6.7 [8.9-4.6] Myr to 4.9 [6.9-3.0] Myr ago, which corresponds to Late Miocene and matches the age of the MSC (6.0-5.3 Myr) (Table 2; Fig. 2B). Most of the divergences occurred during the Pleistocene, from 1.2 (within T. pleurobipunctatus) to ∼0.1 Myr ago (within T. souffia) (Table 2; Fig. 2B).
When focusing on the T. souffia complex, we obtained an estimation of the divergence between T. s. souffia and the other T. souffia lineages of ∼1 Myr ago, while the sampled Danubian lineages split between 0.7 and 0.6 Myr ago. These differentiation events fall within the Early-Middle Pleistocene transition (∼0.5–1.2 Myr), the period of some of the most severe glacial stages of the Pleistocene [93]. The TMRCA of the T. souffia lineages were younger with means comprised between 160 Kyr (T. s. souffia) and 90 Kyr (T. s. agassii 2), which correspond to the Late Pleistocene (Table 2).
Demographic inferences
For demographic inferences, we focused on Rhone and Danube lineages and populations. BSPs of the T. souffia complex lineages revealed different demographic histories (Table 2; Supporting Information S6). In T. s. souffia, two phases of population expansion are likely: Ne seems to have increased steadily from ∼75 Kyr and a sudden burst was detected in the last 10 Kyr. On the contrary, T. s. agassii 2 and T. s. agassii 1 seem to have increased only once: while T. s. agassii 2 had a very stable demographic trend with a very recent increase during the last 10 Kyr, T. s. agassii 1 BSP indicated a population growth starting at ∼60 Kyr.
Tajima and Fu's neutrality tests suggested population expansion (both DT and Fs tests significantly negative) in T. s. souffia and T. s. agassii 2 populations only (Supporting Information S5). These results are in agreement with the star-like phylogeny of the MJ networks (Fig. 3) for these two lineages, which testified to demographic expansions. Interestingly, in the Rhone drainage, only the populations from the Durance basin (except FAU) showed signs of population expansion (from DT and Fs tests).
No signal of recent population contraction was detected from Bottleneck analyses, except in two populations: INN and BES. This indicates that anthropogenic factors have had minor impacts on the genetic diversity of the Telestes populations that we sampled, and that most of the observed patterns are due to the (natural) evolutionary history of the species and populations.
Discussion
Several studies demonstrated extensive introgressions in Leuciscinae (e.g. [94], [95]) and cases of punctual hybridization involving Telestes species have been reported (e.g.: Squalius peloponensis X Telestes alfiensis [22], [96]; Parachondrostoma toxostoma X T. s. souffia; Chondrostoma soetta X T. muticellus [87]; T. muticellus X T. s. souffia [90]). In the present study we did detect evidence of hybridization from mtDNA but in only one river (BEV), between T. muticellus and T. s. souffia. However, we did not recover any corresponding evidence from nuclear markers (at locus BL1-36, one individual with allele 197, but no significant LD between loci or significant HW disequilibrium). This asymmetric pattern suggests a past hybridization event with the subsequent sorting of the nuclear genome from T. s. souffia while both mitochondrial genomes subsisted. Our sampling scheme aimed at better characterizing the introgression between T. muticellus and T. s. souffia previously detected in the Var basin [90], [97]. Although 67 individuals were analyzed, we were neither able to detect any T. muticellus mtDNA haplotypes in VAR, nor any evidence from microsatellites. Furthermore, any sign of hybridization between T. s. agassii 1 and T. muticellus was found in the Soca River (SLO), which represents a contact zone between these two species according to [97]. Therefore, our study strongly suggests that hybridization has only played a minor role in the evolutionary history of Telestes. This statement is not trivial considering the importance of this process in Cyprinid speciation [98].
Tempo and mode of differentiation in Telestes
The patterns of genetic structure and diversity obtained from mtDNA and microsatellites are largely congruent. We are therefore confident that our mtDNA-based time estimates represent the Telestes genome evolution well.
The Late Miocene
The mtDNA-based estimations of divergence time between the four Telestes species groups (T. souffia, T. muticellus, T. montenigrinus and T. pleurobipunctatus+T. alfiensis) ranged from 6.7 [8.9-4.6] Myr to 4.9 [6.9-3.0] Myr. These dates are consistent with those found from 16S rDNA sequences (split between T. souffia and T. muticellus) [90], from partial Cyt b sequences (split between T. souffia and its sister species T. beoticus and T. pleurobipunctatus) [51] and from the mtDNA Control Region (divergence of T. muticellus, T. souffia and T. turskyi) [97]. These dates are ∼5 Myr and are very close to the end of the MSC (6.0 - 5.3 Myr). The importance of this geomorphologic event in the dispersal of the Telestes genus is also suggested by the distribution of the different species in independent ichthyological districts of the Mediterranean area: Rhone and Danube Rivers representing the SF and DB districts, the Po River for the PV district, the Moraca River for the AB district and the Louros and Alfios Rivers for the WG district. The Lago Mare phase that occurred at the end of the MSC may have favored the dispersal of Telestes across these Mediterranean districts. The Lago Mare phase then ended abruptly during the Zanclean ∼5.33 Myr [99] when the waters of the Atlantic entered the Mediterranean by the Strait of Gibraltar and re-established open marine conditions in the Mediterranean. The Zanclean therefore initiated a phase of vicariance for the Telestes populations that had scattered across the Mediterranean rivers.
Furthermore, during the Lago Mare, both the Mediterranean and the Parathetys were reduced to a network of lakes, among which was the Pannonian Lake (covering the current Hungary). And yet, the freshwater Parathetys drained into the freshwater Mediterranean, connecting the Adriatic/Ionian area with the forthcoming Danube drainage [5]. The split between T. souffia (Danubian) and T. montenigrinus+T. muticellus (both Adriatic) ∼5.6 Myr suggests that Telestes colonized the Danube drainage as early as the Lago Mare stage.
The Pleistocene
Divergences among T. souffia sub-species range between 0.6 and 1.0 Myr. The estimation of the divergence time between the Rhodanian T. s. souffia and its Danubian sister sub-species is 1.0 [0.6–1.6] Myr from time calibrated phylogeny and 1.0 [0.6–1.5] Myr from the ρ statistic method. These dates are consistent with the mtDNA Control Region based estimate of [97]: 1.0 [0.7–1.3] Myr. Many changes in upstream river capture involving the Danube, Rhine and Rhone drainages are recorded for the Pliocene and the Early-Middle Pleistocene transition (e.g. [100]). For example, in the Late Pliocene, waters forming the current Upper Rhine were carried out eastward and fed the Danube drainage [101], [102], at the start of the Middle Pleistocene, the waters of the current Alpine Rhine were diverted northward and fed the Rhine drainage [100]. Telestes souffia might have benefited a series of similar upstream river capture events to colonize the Rhone drainage from the Danube, possibly via the Rhine drainage. Moreover, the formation of periglacial lakes at the edge of glaciers are known to have transiently connected the Rhine and Rhone drainages during the LGM or the Younger Dryas promoting the dispersal of Cottus gobio lineages from the Rhine drainage to the Danube drainage [14], [103]. Similar periglacial lakes may also have favored the dispersal of T. souffia in the Rhone drainage during the Middle Pleistocene.
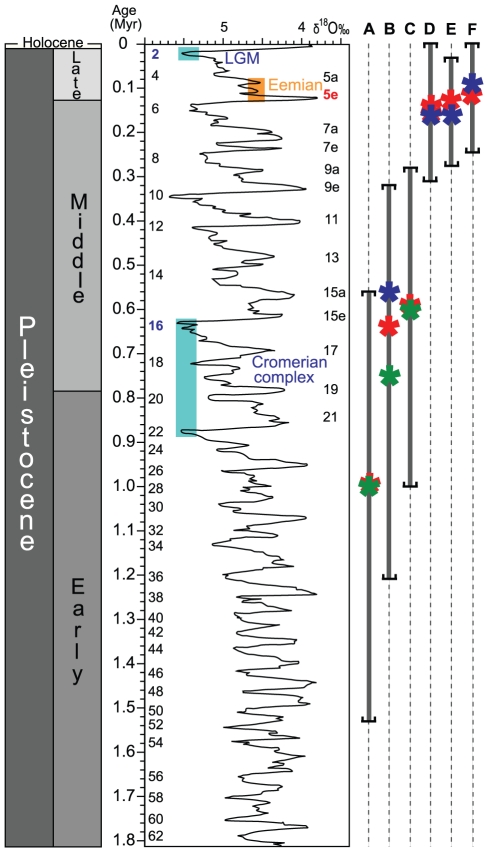
The crown ages of the different T. souffia lineages in the Danube are comprised between 0.7 and 0.6 Myr. T. souffia does not currently occur in the main course of the Danube [7]. Our data suggest this was the case even before anthropogenic modifications. In fact, the different populations in the Danube are highly differentiated and have no sign of admixture based on microsatellites or mtDNA (T. s. agassii 1, T. s. agassii 2 and T. s. agassii 3 are reciprocally monophyletic). The estimated TMRCAs fall within the transition between Early and Middle Pleistocene (0.5–1.2 Myr [93]), when the most extensive glaciations in the Quaternary occurred. During this period, the Northern Hemisphere experienced two of the most severe and longest glacial periods [93]: the Marine oxygen-Isotopic Stage 22 (MIS22; c. 870-880 Kyr) and MIS16 (c. 650-620 Kyr) (see Fig. 7). During glacial periods, European meandering rivers (such as the Danube) tended to adopt braided courses due to the increase of periglacial-derived gravel and sand discharge and a decrease in water flow [104]. The Early-Middle Pleistocene transition therefore may correspond to the spread of suitable habitats for T. souffia in the main course of the Danube. Such conditions have likely allowed the small river and cold adapted T. souffia to colonize downstream Danubian sub-drainages such as the Sava (including the Lim River) and Tisza river systems. Subsequent milder climate conditions, notably the interglacial MIS15 (c. 620-560 Kyr; see Fig. 7), have probably restricted T. souffia suitable habitats to the Danube peripheral tributaries, preventing further dispersals and gene flows. The glacial periods in the Middle and Late Pleistocene were less severe in amplitude and duration and, given the observed genetic structure, have unlikely permitted subsequent spread of favorable habitats and dispersal. Although times of divergences were found older (i.e. Pliocene) than in T. souffia, a similar pattern was also found in the cold-adapted and rheophilic Barbatula barbatula, for which populations were maintained in the upstream portion of the rivers [105]. As a corollary, the thermophilic Rhodeus amarus, a species common in lotic and lentic habitats [106], has been documented to harbor rather homogenous populations along the Danube basin. Our results therefore suggest that the Pleistocene glaciation cycles acted very differently on the dispersal and vicariance of European freshwater fish, as a function of their ecology. Accumulating detailed specific patterns may allow the inference of general evolutionary histories related to groups of species with similar ecological features.
Figure 7. Time frame of the Quaternary period and the TMRCA obtained for T. souffia lineages.
The mean and 95% CI maximum interval of the TMRCA obtained for T. souffia lineages using mtDNA sequences and MIS are indicated. A. T. souffia (Rhone and Danube); B. T. souffia (Danube only); C. T. s. agassii 1 vs. T. s. agassii 2; D. T. s. souffia; E. T. s. agassii 1; F. T. s. agassii 2. Green “*”, means of TMRCA obtained from BEAST; red “*”, means of TMRCA obtained from the ρ statistic methodology; blue “*”, means of TMRCA obtained from BSP methodology.
The TMCRAs for T. s. souffia, T. s. agassii 1 and T. s. agassii 2 populations are all very similar (Table 2; Fig. 7), with means ranging from ∼160 Kyr to ∼90 Kyr. They straddle the Eemian interglacial period (127-117 Kyr [107]) that is increasingly considered as critical in the establishment of the genetic pattern of some extant or recently extinct species [20]. The Eemian started with a very warm stage (MIS5e, when mean surface temperatures were at least 2°C warmer than present [108]). Subsequent events of demographic expansion were detected during the cooler Eemian stages that followed MIS5e for T. s. souffia and T. s. agassii 1 (at ∼75 and ∼60 Kyr respectively; Table 2, Supporting Information S6). This suggests that, both in Rhone River and Danube River, the climatic and ecological conditions during the MIS5e have been quite unfavorable for the cold-adapted T. souffia, while cooler climates promoted demographic expansion.
The T. s. agassii 1 population from the Soca River constitutes the only known occurrence of this lineage outside the Danube drainage. Upstream river capture seems the most likely cause of this current pattern. However, the age of this event remains ambiguous. The mean TMRCA of the T. s. agassii 1 (involving both Danubian and Adriatic populations) is ∼120-160 Kyr old, but Danubian and Adriatic populations share a population expansion ∼60 Kyr ago. Despite lack of precision, molecular dating suggests that the upstream river capture responsible for the occurrence of T. s. agassii 1 in an Adriatic river likely dates back to the Late Pleistocene. Upstream river catchment has been previously suggested for T. muticellus [109]. As supported by nuclear data, such a geomorphologic event favored exchanges between the Po river system and some Ligurian coastal rivers. As well, it could explain the co-occurrence of a divergent mtDNA lineage in the Po River [110].
The Holocene and anthropogenic factors
Demographic expansion was detected in T. s. agassii 1 and T. s. souffia ∼10 Kyr (Table 2, Supporting Information S6). The LGM (MIS2) spanned 26.5-19.0 Kyr [111]. After a transient stage of global warming, climate reverted to glacial conditions during the Younger Dryas (12.8-11.5 Kyr [112]). The age for the latest expansion events within T. souffia lineages therefore suggest that the end of the Younger Dryas (rather than the end of the LGM) was critical (as suggested for Cottus gobio [103]) in promoting demographic expansion.
For T. s. souffia (Rhone drainage), the processes involved in this expansion could be investigated in more detail given the completeness of the sampling. When all populations are considered, a clear signal of demographic expansion is detected using mtDNA (from BSP analysis and neutrality tests). However, only samples from the Durance River (in the southern Rhone basin) exhibit signs of population expansion when considered individually. This result suggests a glacial refugium during the LGM/Younger Dryas in the South of the Rhone drainage. This southern Rhone refugium hypothesis is further supported by higher mtDNA diversity in Durance (with respect to θk) than in the other parts of the range. Considering microsatellites, the pattern of Ap also supports a range expansion from the Southern Rhone: higher Ap values in populations from the Northern Durance River (Ardeche basin, Ain River, Upper Rhone and Northern Rhone) fit the so called allele surfing model [113]. This model of range expansion assumes that, during the colonisation stages, the allele frequencies within the wave of advance could be sharply modified and that new alleles have a higher probability of being retained and to propagate. For T. s. souffia, we can hypothesize a demographic and range expansion that would have started in the Southern Rhone drainage at the beginning of the Holocene.
Along with this major glacial refugium in the Southern Rhone basin for T. s. souffia, several elements suggest that two potential microrefugia also co-occurred during the LGM/Younger Dryas. As a matter of fact, the Ardeche basin and the Upper Rhone are currently connected to the rest of the Rhone basin but hold the genetic signature of past isolation: high microsatellite Ar values and multiple divergent and private mtDNA haplotypes. As previously recognized for some other taxa [114], [115], microrefugia could have actually played a critical role during the post-glacial recolonisation processes.
Interestingly, the Bayesian clustering analysis (Fig. 4B) does not distinguish SOR from the Durance basin and suggests that AIN and USS constituted examples of mixed populations between northern and southern clusters. These results indicate that the Rhone seems to have acted as a corridor, probably until very recently, among T. s. souffia populations that are isolated today. In fact, before the Rhone River was channelized and fragmented during the 19th and the 20th centuries, T. s. souffia occurred in the main course of the Rhone River [116].
The differentiation pattern and lack of genetic diversity observed in the isolated populations of the French Mediterranean coastal rivers (VAR, HER, VID) could be explained by recent downstream connections during phases of sea level drops linked to the glacial periods. A subsequent rise in sea level at the beginning of the Holocene would have led to isolation of the populations and genetic differentiation. MtDNA haplotype sharing, the Bayesian clustering analysis (K = 3 and K = 5) and (to a lesser extent) the FCA, suggest that HER and VID stemmed from the same source population. However, this scenario is unlikely for the Var River population. In fact, the topology of the seafloor in this region does not support the possibility of a past connection between the Var and the Rhone rivers, even when the sea was at its lowest level. Alternatively, the topography of the Var drainage and of the Verdon drainage (Durance basin) supports possible episodic upstream connections either linked to the Alpine orogenesis or to periglacial lakes. Moreover, these three coastal river populations clearly experienced sharp genetic drift, suggesting that the connection was very limited in time and that a founder effect characterized their settlement processes.
However, these scenarios were not supported for the Argens coastal river (BES). The fact that BES is both indistinguishable from the Durance basin (based on Bayesian clustering approach) and has experienced a recent bottleneck (Fig. 4B, Supporting Information S5) supports a very recent colonisation involving quite few individuals. In fact, Argens basin was connected to Durance basin in the mid-20th century by a channel devoted to irrigation and human water consumption, the Canal de Provence [117], [118], which could have led to the colonisation of the Argens River by T. s. souffia. A similar scenario involving human influence could also be invoked for the pattern observed in SEI, which is indistinguishable from the other Northern Rhone drainage populations, DOU and SAO (based on Bayesian clustering approach applied to microsatellites). In the mid-19th century, the Seine, Rhone, Rhine and Loire basins were connected through the construction of a network of navigation channels. These channels have been previously proven to have allowed some cyprinid species to cross basins. They were notably instrumental in the invasion of French hydrographic systems by Chondrostoma nasus [119], [120].
A case of parapatric differentiation?
Although they are found in the same river system, the two populations of T. pleurobipunctatus from the Louros River exhibit an important genetic differentiation based on microsatellites and harbor very distinct mitochondrial lineages. These results confirmed previous work that used allozymic data [22] and indicate the existence of reproductive isolation between the two lineages. In fact, these lineages are found in two distinct habitats of the Louros river system: GLO was sampled in the mainstream river whereas GLB was sampled in flood plain ponds that skirt the mainstream river. Our results therefore support a parapatric differentiation stemming from ecological specialization of the two lineages. This evolutionary process has started in the Pleistocene, likely from 1.2 Myr as suggested by the age of the two T. pleurobipunctatus mtDNA lineages (Fig. 2B, Table 2). Additionally, the case of T. pleurobipunctatus in the Louros River illustrates that, although allopatric differentiation (vicariance) seems to have been the major process, parapatric differentiation may have played a non-negligible role in the Telestes genus evolution.
Conclusions
We were able to establish several clear links between major historical events and the genetic differentiation in Telestes. These events range from geomorphologic, climatic to anthropogenic factors and date back from Late Miocene to Holocene. Most of them constituted a series of alternations between dispersal and vicariance that critically impacted the evolution of the Telestes genus. Additionally, the dynamics of recolonisation of a species from glacial refugial zones in a hydrographic system are still poorly understood (but see [121]). The detailed analysis of the Rhone drainage suggested that a combination between range expansion and IBD can explain current patterns of differentiation at finer geographical scales. Our study also suggests that several climatic oscillations (MIS16/MIS15, Eemian, LGM/Younger Dryas and Holocene) have had a critical impact on the demography and the range of several Telestes lineages. Moreover, in the current context of global climate change, our data related to the Eemian may suggest a potential risk of population decline for T. souffia in the near future (the mean global temperature during MIS5e having been at least 2°C higher than the current mean temperature). These results could constitute important foundation information for improving the conservation strategies for European freshwater fishes.
Furthermore, the existence of general patterns of colonisation for the European freshwater fishes (as inferred for terrestrial fauna, e.g. [12]) were recently questioned (e.g. [13], [122]). We believe that the approach developed in this study, establishing fine relationships between time, patterns and processes, should assist in determining the existence of such general patterns.
Supporting Information
Models of mtDNA sequence evolution.
(XLS)
Results from HW equilibrium tests conducted on microsatellites.
(XLS)
Pairwise relationship between uncorrected Fst and Fst corrected for null alleles (Fst c).
(PDF)
Results from LD tests conducted on microsatellites.
(XLS)
Genetic diversity and demographic parameters obtained from mtDNA and microsatellites data.
(XLS)
Bayesian Skyline Plots for T. s. souffia, T. s. agassii 1 and T. s. agassii 2.
(PDF)
Acknowledgments
Data used in this work were partly produced through molecular genetic analysis technical facilities of the SFR ‘Montpellier Environnement Biodiversité’ (France). Warm thanks go to Jean-François Martin (Montpellier SupAgro, France) for his logistical help with the genotyping work, to Vladimir Mencl and the University of Canterbury for help with using the grid, to Walter Salzburger (University of Basel, Switzerland) for providing material from the Inn River and to Alain Crivelli (Tour du Valat, France) for sharing samples from the Soca River. We express our gratitude to the late Pr. Petru Bănărescu (Institute of Biology, Bucarest), Pier Giorgio Bianco (Universita di Napoli ‘Federico II’, Napoli), Panos Stavros Economidis (Aristotle University, Thessaloniki), Drago Marić (University of Montenegro, Podgorica) and the French ONEMA for their help with the sample collection.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Cracraft J. Geographic differentiation, cladistics, and vicariance biogeography: Reconstructing the tempo and mode of evolution. Am Zool. 1982;22:411–424. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates; 2004. 545 [Google Scholar]
- 3.Bănărescu P. Zoogeography of fresh waters, 1. Wiesbaden: AULA-Verlag; 1990. 511 [Google Scholar]
- 4.Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: Fish DNA sheds light on time-dependency. Mol Biol Evol. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
- 5.Hsü K, Montabert L, Bernoulli D, Cita MB, Erickson A, et al. History of the Mediterranean salinity crisis. Nature. 1977;267:399–403. [Google Scholar]
- 6.Krijgsman W, Hilgen FJ, Raffi I, Sierro FJ, Wilson DS. Chronology, causes and progression of the Messinian salinity crisis. Nature. 1999;400:652–655. [Google Scholar]
- 7.Bănărescu P. Zoogeography and history of the freshwater fish fauna of Europe. In: Holcik J, editor. The freshwater fishes of Europe. Wiesbaden: AULA-Verlag; 1989. pp. 88–103. [Google Scholar]
- 8.Bianco PG. Potential role of the palaeohistory of the Mediterranean and Paratethis basins on the early dispersal of Euro-Mediterranean freshwater fishes. Ichthyol Explor Freshw. 1990;1:167–184. [Google Scholar]
- 9.Reyjol Y, Hugueny B, Pont D, Bianco PG, Beier U, et al. Patterns in species richness and endemism of European freshwater fish. Global Ecol Biogeogr. 2007;16:65–75. [Google Scholar]
- 10.Avise JC, Walker D, Johns GC. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc R Soc B. 1998;265:1707–1712. doi: 10.1098/rspb.1998.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernatchez L, Wilson CC. Comparative phylogeography of Nearctic and Palearctic fishes. Mol Ecol. 1998;7:431–452. [Google Scholar]
- 12.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 13.Costedoat C, Gilles A. Quaternary pattern of freshwater fishes in Europe: Comparative phylogeography and conservation perspective. Open Conserv Biol J. 2009;3:36–48. [Google Scholar]
- 14.Vonlanthen P, Excoffier L, Bittner D, Persat H, Neuenschwander S, et al. Genetic analysis of potential postglacial watershed crossings in Central Europe by the bullhead (Cottus gobio L.). Mol Ecol. 2007;16:4572–4584. doi: 10.1111/j.1365-294X.2007.03531.x. [DOI] [PubMed] [Google Scholar]
- 15.Levy A, Doadrio I, Almada VC. Historical biogeography of European leuciscins (Cyprinidae): Evaluating the Lago Mare dispersal hypothesis. J Biogeogr. 2009;39:55–65. [Google Scholar]
- 16.Perea S, Böhme M, Zupancic P, Freyhof J, Sanda R, et al. Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol Biol. 2010;10:265. doi: 10.1186/1471-2148-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkaert FAM, Hänfling B, Hellman B, Carvalho GR. Timing of the population dynamics of bullhead Cottus gobio (Teleostei: Cottidae) during the Pleistocene. J Evol Biol. 2002;15:930–944. [Google Scholar]
- 18.Cortey M, Vera M, Pla C, García-Marín JL. Northern and Southern expansions of Atlantic brown trout (Salmo trutta) populations during the Pleistocene. Biol J Linn Soc. 2009;97:904–917. [Google Scholar]
- 19.Tougard C, Renvoisé E, Petitjean A, Quéré JP. New insight into the colonization processes of common voles: Inferences from molecular and fossil evidence. PLoS ONE. 2008;3:e3532. doi: 10.1371/journal.pone.0003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofreiter M, Stewart J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Cur Biol. 2009;19:R584–R594. doi: 10.1016/j.cub.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Gibbard P, van Kolfschoten T. The Pleistocene and Holocene epochs. In: Gradstein FM, Ogg JG, Smith AG, editors. A geologic time scale. Cambridge: Cambridge University Press; 2004. pp. 441–452. [Google Scholar]
- 22.Gilles A, Costedoat C, Barascud B, Voisin A, Bănărescu P, et al. Speciation pattern of Telestes souffia complex (Teleostei, Cyprinidae) in Europe using morphological and molecular markers. Zool Scr. 2010;39:225–242. [Google Scholar]
- 23.Changeux T, Pont D. Current status of the riverine fishes of the French Mediterranean basin. Biol Conserv. 1995;72:137–158. [Google Scholar]
- 24.Schwarz M. Biologie, menaces et protection du blageon (Leuciscus souffia) en Suisse. Bern: OFEFP; 1998. 98 [Google Scholar]
- 25.Wocher H, Rosch R. Migrations of soufie (Leuciscus souffia agassizii, Val. 1844) in a natural river and a tributary determined by mark-recapture. Arch Hydrobiol. 2006;165:77–87. [Google Scholar]
- 26.Hoffmann RC. Environmental change and the culture of common carp in medieval Europe. Guelph Ichthyol Rev. 1995;3:57–85. [Google Scholar]
- 27.Sastre C. Les introductions d'espèces dans le milieu aquatique. Le contexte biogéographique: Aspects généraux et paléohistoriques. Bull Fr Pêche Piscic 344/ 1997;345:9–14. [Google Scholar]
- 28.Solar II. Use and exchange of salmonid genetic resources relevant for food and aquaculture. Rev Aquacult. 2009;1:174–196. [Google Scholar]
- 29.Lajbner Z, Linhart O, Kotlík P. Human-aided dispersal has altered but not erased the phylogeography of the tench. Evol Appl. 2011;4:545–561. doi: 10.1111/j.1752-4571.2010.00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ketmaier V, Bianco PG, Cobolli M, Krivokapic M, Caniglia R, et al. Molecular phylogeny of two lineages of Leuciscinae cyprinids (Telestes and Scardinius) from the peri-Mediterranean area based on cytochrome b data. Mol Phylogenet Evol. 2004;32:1061–1071. doi: 10.1016/j.ympev.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Muenzel FM, Sanetra M, Salzburger W, Meyer A. Microsatellites from the vairone Leuciscus souffia (Pisces: Cyprinidae) and their application to closely related species. Mol Ecol Notes. 2007;7:1048–1050. [Google Scholar]
- 32.Dubut V, Martin JF, Costedoat C, Chappaz R, Gilles A. Isolation and characterization of polymorphic microsatellite loci in the freshwater fishes Telestes souffia and Telestes muticellus (Teleostei: Cyprinidae). Mol Ecol Resour. 2009;9:1001–1005. doi: 10.1111/j.1755-0998.2009.02539.x. [DOI] [PubMed] [Google Scholar]
- 33.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 34.Nei M. Molecular evolutionary genetics. New-York: Columbia University Press; 1987. 512 [Google Scholar]
- 35.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewens WJ. The sampling theory of selectively neutral alleles. Theor Pop Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- 37.Watterson G. On the number of segregating sites in genetical models without recombination. Theor Pop Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 38.Rousset F. Genepop'007: A complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate –A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 40.van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- 41.Kalinowski ST. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv Genet. 2004;5:539–543. [Google Scholar]
- 42.Szpiech ZA, Jakobsson M, Rosenberg NA. Adze: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petit R, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv Biol. 1998;12:844–855. [Google Scholar]
- 44.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 45.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Cont. 1974;19:716–723. [Google Scholar]
- 46.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 47.Penny D, Hendy MD. The use of tree comparison metrics. Syst Zool. 1985;34:75–82. [Google Scholar]
- 48.Wilgenbusch JC, Warren DL, Swofford DL. Awty: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. Tallahassee: School of Computation Sciences, Florida State University; 2004. Available: http://ceb.csit.fsu.edu/awty. [Google Scholar]
- 49.Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dermitzakis DM. Paleogeography, geodynamic processes and event stratigraphy during the late Cenozoic of the Aegean area. Atti dei Convegni Lincei. 1990;85:263–288. [Google Scholar]
- 51.Zardoya R, Doadrio I. Molecular evidence of the evolutionary and biogeographical patterns of European cyprinids. J Mol Evol. 1999;49:227–237. doi: 10.1007/pl00006545. [DOI] [PubMed] [Google Scholar]
- 52.Won H, Renner SS. Dating dispersal and radiation in the gymnosperm Gnetum (Gnetales) –Clock calibration when outgroup relationships are uncertain. Syst Biol. 2006;55:610–622. doi: 10.1080/10635150600812619. [DOI] [PubMed] [Google Scholar]
- 53.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 54.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III: Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clement M, Posada D, Crandall KA. Tcs: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 56.Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: A reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 57.Saillard J, Forster P, Lynnerup N, Bandelt HJ, Nørby S. MtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis: Models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 59.Felsenstein J. Confidence limits on phylogenies-an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 60.Pritchard JK, Stephens M, Donnelly PJ. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campana MG, Hunt HV, Jones H, White J. CorrSieve: Software for summarizing and evaluating Structure output. Mol Ecol Resour. 2011;11:349–352. doi: 10.1111/j.1755-0998.2010.02917.x. [DOI] [PubMed] [Google Scholar]
- 65.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalinowski ST. The computer program Structure does not reliably identify the main genetic clusters within species: Simulations and implications for human population structure. Heredity. 2010;106:625–632. doi: 10.1038/hdy.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 68.Fogelqvist J, Niittyvuopio A, Ågren J, Savolainen O, Lascoux M. Cryptic population genetic structure: The number of inferred clusters depends on sample size. Mol Ecol Resour. 2010;10:314–323. doi: 10.1111/j.1755-0998.2009.02756.x. [DOI] [PubMed] [Google Scholar]
- 69.Guinand B. Use of a multivariate model using allele frequency distribution to analyse patterns of genetic differentiation among populations. Biol J Linn Soc. 1996;58:173–195. [Google Scholar]
- 70.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Genetix 4.05: Logiciel sous Windows TM pour la génétique des populations. Montpellier: Laboratoire Génome Populations Interactions (UMR 5000, CNRS, Université de Montpellier II); 2004. Available: http://kimura.univ-montp2.fr/genetix/ [Google Scholar]
- 71.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 72.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 73.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 75.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 76.Rambaut A, Drummond A. Tracer, version 1.2. Oxford: Department of Zoology, University of Oxford; 2007. Available: http://evolve.zoo.ox.ac.uk/tracer. [Google Scholar]
- 77.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piry S, Luikart G, Cornuet JM. Bottleneck: A computer program for detecting recent reductions in effective population size from allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- 80.Nei M, Marumaya T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 81.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kimura M, Ohta T. Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci USA. 1978;75:2868–2872. doi: 10.1073/pnas.75.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, et al. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlsson J. Effects of microsatellite null alleles on assignment testing. J Hered. 2008;99:616–623. doi: 10.1093/jhered/esn048. [DOI] [PubMed] [Google Scholar]
- 86.Chapuis MP, Lecoq M, Michalakis Y, Loiseau A, Sword GA, et al. Do outbreaks affect genetic population structure? A worldwide survey in Locusta migratoria, a pest plagued by microsatellite null alleles. Mol Ecol. 2008;17:3640–3653. doi: 10.1111/j.1365-294X.2008.03869.x. [DOI] [PubMed] [Google Scholar]
- 87.Dubut V, Sinama M, Martin JF, Meglécz E, Fernandez J, et al. Cross-species amplification of 41 microsatellites in European cyprinids: A tool for evolutionary, population genetics and hybridization studies. BMC Res Notes. 2010;3:135. doi: 10.1186/1756-0500-3-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ráb P, Rábová M, Pereira CS, Collares-Pereira MJ, Pelikánová Š. Chromosome studies of European cyprinid fishes: Interspecific homology of leuciscine cytotaxonomic marker-the largest subtelocentric chromosome pair as revealed by cross-species painting. Chromosome Res. 2008;16:863–873. doi: 10.1007/s10577-008-1245-3. [DOI] [PubMed] [Google Scholar]
- 89.Prichard JK, Wen X, Falush D. Documentation for Structure software: Version 2.3. Chicago: University of Chicago; 2010. 39 available: http://pritch.bsd.uchicago.edu/structure.html. [Google Scholar]
- 90.Gilles A, Chappaz R, Cavalli L, Lortscher M, Faure E. Genetic differentiation and introgression between putative subspecies of Leuciscus soufia (Teleostei: Cyprinidae) of the region of the Mediterranean Alps. Can J Fish Aquat Sci. 1998;55:2341–2354. [Google Scholar]
- 91.Helgason A, Nicholson G, Stefánsson K, Donnelly P. A reassessment of genetic diversity in Icelanders: Strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann Hum Genet. 2003;67:281–297. doi: 10.1046/j.1469-1809.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- 92.Machordom A, Doadrio I. Evidence of a Cenozoic Betic-Kabilian connection based on freshwater fish phylogeography (Luciobarbus, Cyprinidae). Mol Phylogenet Evol. 2001;18:252–263. doi: 10.1006/mpev.2000.0876. [DOI] [PubMed] [Google Scholar]
- 93.Head JH, Gibbard PL. Early-Middle Pleistocene transitions: An overview and recommendation for the defining boundary. In: Head JH, Gibbard PL, editors. Early-Middle Pleistocene transitions: The land-ocean evidence. London: The Geological Society of London; 2005. pp. 1–18. [Google Scholar]
- 94.Brito RM, Briolay J, Galtier N, Bouvet Y, Coelho MM. Phylogenetic relationships within genus Leuciscus (Pisces, Cyprinidae) in Portuguese freshwaters, based on mitochondrial DNA cytochrome b sequences. Mol Phylogenet Evol. 1997;8:435–442. doi: 10.1006/mpev.1997.0429. [DOI] [PubMed] [Google Scholar]
- 95.Gilles A, Lecointre G, Faure E, Chappaz R, Brun G. Mitochondrial phylogeny of the European cyprinids: Implications for their systematics, reticulate evolution, and colonization time. Mol Phylogenet Evol. 1998;10:132–143. doi: 10.1006/mpev.1997.0480. [DOI] [PubMed] [Google Scholar]
- 96.Bianco PG. Leuciscus cephalus (Linnaeus) with record of fingerling adult males, Leuciscus pleurobipunctatus (Stephanidis) and their hybrids from Western Greece. J Fish Biol. 1988;32:1–16. [Google Scholar]
- 97.Salzburger W, Brandstatter A, Gilles A, Parson W, Hempel M, et al. Phylogeography of the vairone (Leuciscus souffia, Risso 1826) in Central Europe. Mol Ecol. 2003;12:2371–2386. doi: 10.1046/j.1365-294x.2003.01911.x. [DOI] [PubMed] [Google Scholar]
- 98.Dowling TE, Demarais BD. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature. 1993;362:444–446. [Google Scholar]
- 99.Garcia-Castellanos D, Estrada F, Jiménez-Munt I, Gorini C, Fernàndez M, et al. Catastrophic flood of the Mediterranean after the Messinian salinity crisis. Nature. 2009;462:778–781. doi: 10.1038/nature08555. [DOI] [PubMed] [Google Scholar]
- 100.Lóczy D. The Danube: Morphology, evolution and environmental issues. In: Gupta A, editor. Large rivers: Geomorphology and management. Chichester: Wiley; 2008. pp. 235–260. [Google Scholar]
- 101.Sommerwerk N, Baumgartner C, Bloesch J, Hein T, Ostojić A, et al. The Danube River Basin. In: Tockner K, Uehlinger U, Robsinson CT, editors. Rivers of Europe. London: Academic Press; 2009. pp. 59–112. [Google Scholar]
- 102.Uehlinger U, Wantzen KM, Leuven RSEW, Arndt H. The Rhine River Basin. In: Tockner K, Uehlinger U, Robsinson CT, editors. Rivers of Europe. London: Academic Press; 2009. pp. 199–245. [Google Scholar]
- 103.Neuenschwander S, Largiadèr CR, Ray N, Currat M, Vonlanthen P, et al. Colonization history of the Swiss Rhine basin by the bullhead (Cottus gobio): Inference under a Bayesian spatially explicit framework. Mol Ecol. 2008;17:757–772. doi: 10.1111/j.1365-294X.2007.03621.x. [DOI] [PubMed] [Google Scholar]
- 104.Gibbard P. The history of the great northwest European rivers during the past 3 million years. Phil Trans R Soc B. 1988;318:559–602. [Google Scholar]
- 105.Šedivá A, Janko K, Šlechtová V, Kotlík P, Simonović P, et al. Around or across the Carpathians: Colonization model of the Danube basin inferred from genetic diversification of stone loach (Barbatula barbatula) populations. Mol Ecol. 2008;17:1277–1292. doi: 10.1111/j.1365-294X.2007.03656.x. [DOI] [PubMed] [Google Scholar]
- 106.Bryja J, Smith C, Konečný A, Reichard M. Range-wide population genetic structure of the European bitterling (Rhodeus amarus) based on microsatellite and mitochondrial DNA analysis. Mol Ecol. 2010;19:4708–4722. doi: 10.1111/j.1365-294X.2010.04844.x. [DOI] [PubMed] [Google Scholar]
- 107.Sirocko F, Seelos K, Schaber K, Rein B, Dreher F, et al. A late Eemian aridity pulse in central Europe during the last glacial inception. Nature. 2005;436:833–836. doi: 10.1038/nature03905. [DOI] [PubMed] [Google Scholar]
- 108.Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A. Simulating Arctic climate warmth and icefield retreat in the last interglaciation. Science. 2006;311:1751–1753. doi: 10.1126/science.1120808. [DOI] [PubMed] [Google Scholar]
- 109.Marchetto F, Zaccara S, Muenzel FM, Salzburger W. Phylogeography of the Italian vairone (Telestes muticellus, Bonaparte 1837) inferred by microsatellite markers: Evolutionary history of a freshwater fish species with a restricted and fragmented distribution. BMC Evol Biol. 2010;10:111. doi: 10.1186/1471-2148-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaccara S, Stefani F, Delmastro Phylogeographical structure of vairone Telestes muticellus (Teleostei, Cyprinidae) within three European peri-Mediterranean districts. Zool Scr. 2007;36:443–453. [Google Scholar]
- 111.Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, et al. The Last Glacial Maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 112.Muscheler R, Kromer B, Björck S, Svensson A, Friedrich M, et al. Tree rings and ice cores reveal 14C calibration uncertainties during the Younger Dryas. Nat Geosci. 2008;1:263–267. [Google Scholar]
- 113.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 114.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 115.Rull V. Microrefugia. J Biogeogr. 2009;36:481–484. [Google Scholar]
- 116.Olivier JM, Carrel G, Lamouroux N, Dole-Olivier MJ, Malard F, et al. The Rhône River Basin. In: Tockner K, Uehlinger U, Robsinson CT, editors. Rivers of Europe. London: Academic Press; 2009. pp. 247–295. [Google Scholar]
- 117.Clébert JP, Rouyer JP. La Durance. Toulouse: Privat; 1991. 189 [Google Scholar]
- 118.Société du Canal de Provence (SCP) Carte des aménagements hydrauliques. Aix-en-Provence: SCP; 1995. 1 [Google Scholar]
- 119.Nelva A. Origine et biogéographie des deux Chondrostomes français: Chondrostoma nasus et C. toxostoma (Pisces, Cyprinidae). Cybium. 1988;12:287–299. [Google Scholar]
- 120.Nelva A. La pénétration du hotu, Chondrostoma nasus nasus (poisson cyprinidé), dans le réseau hydrographique français et ses conséquences. Bull Fr Pêche Piscic. 1997;344/345:253–259. [Google Scholar]
- 121.Stepien CA, Murphy DJ, Lohner RN, Sepulveda-Villet OJ, Haponski AE. Signatures of vicariance, postglacial dispersal and spawning philopatry: Population genetics of the walleye Sander vitreus. Mol Ecol. 2009;18:3411–3428. doi: 10.1111/j.1365-294X.2009.04291.x. [DOI] [PubMed] [Google Scholar]
- 122.Costedoat C, Chappaz R, Barascud B, Guillard O, Gilles A. Heterogeneous colonization pattern of European cyprinids, as highlighted by the dace complex (Teleostei: Cyprinidae). Mol Phylogenet Evol. 2006;41:127–148. doi: 10.1016/j.ympev.2006.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Models of mtDNA sequence evolution.
(XLS)
Results from HW equilibrium tests conducted on microsatellites.
(XLS)
Pairwise relationship between uncorrected Fst and Fst corrected for null alleles (Fst c).
(PDF)
Results from LD tests conducted on microsatellites.
(XLS)
Genetic diversity and demographic parameters obtained from mtDNA and microsatellites data.
(XLS)
Bayesian Skyline Plots for T. s. souffia, T. s. agassii 1 and T. s. agassii 2.
(PDF)